- 1Institute of Molecular Biology, University of Oregon, Eugene, OR, USA
- 2Institute of Neuroscience, University of Oregon, Eugene, OR, USA
Alkaline phosphatases (Alps) are well-studied enzymes that remove phosphates from a variety of substrates. Alps function in diverse biological processes, including modulating host-bacterial interactions by dephosphorylating the Gram-negative bacterial cell wall component lipopolysaccharide (LPS). In animals, Alps are encoded by multiple genes characterized by either ubiquitous expression (named Alpls for their liver expression, but a key to proper bone mineralization), or their tissue-specific expression, for example in the intestine (Alpi). We previously characterized a zebrafish alpi gene (renamed here alpi.1) that is regulated by Myd88-dependent innate immune signaling and that is required to prevent a host’s excessive inflammatory reactions to its resident microbiota. Here we report the characterization of two new alp genes in zebrafish, alpi.2 and alp3. To understand their origins, we investigated the phylogenetic history of Alp genes in animals. We find that vertebrate Alp genes are organized in three clades with one of these clades missing from the mammals. We present evidence that these three clades originated during the two vertebrate genome duplications. We show that alpl is ubiquitously expressed in zebrafish, as it is in mammals, whereas the other three alps are specific to the intestine. Our phylogenetic analysis reveals that in contrast to Alpl, which has been stably maintained as a single gene throughout the vertebrates, the Alpis have been lost and duplicated multiple times independently in vertebrate lineages, likely reflecting the rapid and dynamic evolution of vertebrate gut morphologies, driven by changes in bacterial associations and diet.
Introduction
Alkaline phosphatases (Alps) are a superfamily of metalloenzymes that catalyze the hydrolytic removal of phosphate from a variety of molecules (Millán, 2006). Alps have been extensively studied biochemically, but the full spectrum of their biological functions is not known. In animals, Alps are encoded by multiple genes that can be classified by their expression patterns into two general groups, the tissue-non-specific Alps (known as Alpl, for their liver expression, also known as TNAP for tissue non-specific alkaline phosphatase), and the tissue-specific Alps [placental Alp (Alpp), intestinal Alp (Alpi), etc.]. The best studied biological function of Alps is the role of mammalian Alpl in osteogenesis by promoting bone mineralization, as demonstrated by the hypophosphatasia that results from ALPL deficiency in humans and mice (Whyte, 2010). More recently, the Alpis have been implicated in mediating host-bacterial interactions through their ability to dephosphorylate lipid A of the Gram-negative bacterial cell wall component lipopolysaccharide (LPS; Lalles, 2010).
In mammals, Alpi is expressed by intestinal epithelial cells (IECs) and is enriched in vesicles that are actively released from IEC microvillar tips into the intestinal lumen (McConnell et al., 2009; Shifrin et al., 2012). Thus Alpi is located at the interface between the intestinal tissue, the ingesta and the vast microbiota, which suggests its involvement in a variety of biological processes. Recent studies have discovered that Alpi regulates metabolism by controlling the uptake of nutrients such as lipids (Narisawa et al., 2003; Lynes et al., 2011) and calcium (Brun et al., 2012), affects gut physiology by maintaining protective surface microclimate pH in the duodenum (Akiba et al., 2007; Mizumori et al., 2009), and impacts innate immunity by modulating bacterial LPS-induced inflammation (Poelstra et al., 1997b; Bates et al., 2007; Campbell et al., 2010).
Lipopolysaccharide, also commonly referred to as endotoxin, is a component of Gram-negative bacterial cell walls and is abundantly present in the intestinal lumen of animals. LPS is a classic microbial associated molecular pattern (MAMP) and potent inducer of innate immune signaling in both vertebrates and invertebrates (Beutler and Rietschel, 2003). In mammals, LPS binds specifically to a complex consisting of Toll-like receptor 4 (TLR4) and MD-2 through two phosphate groups of its lipid A moiety (Kim et al., 2007), and induces innate immune responses by activating two distinct pathways, namely NF-kB (through MyD88-dependent and independent pathways) and IRF-3 (through TRIF/TRAM). Although the specifics of LPS binding do not seem to be conserved between mammals and teleosts (Sullivan et al., 2009), this MAMP elicits similar pro-inflammatory responses through a Myd88-dependent mechanism in zebrafish (Bates et al., 2007) as in mammals.
Alps have been shown to remove the lipid A phosphates of LPS at physiological pH levels (Poelstra et al., 1997a,b), thereby reducing its affinity for TLR4 and, correspondingly, its endotoxic properties. Our studies in zebrafish larvae (Bates et al., 2006, 2007) demonstrated the functional significance of Alpi’s LPS dephosphorylating activity in the intestine, and showed that this gene plays an integral role in modulating innate immune responses in the gut through a Myd88-dependent negative feedback loop. We found that LPS incubation as well as Gram-negative bacterium inoculation upregulated zebrafish Alpi, a process that required MyD88. We showed that Alpi functioned in the detoxification of LPS because treatment with the Alpi-specific inhibitor L-phenylalanine or alpi.1-specific morpholino rendered zebrafish more sensitive in an LPS killing assay, whereas fish were resistant to LPS pretreated with calf Alpi. Furthermore, zebrafish with reduced Alpi activity exhibited elevated levels of pro-inflammatory cytokines and intestinal neutrophil influx, both Myd88-dependent processes. However, when Alpi deficient zebrafish were derived germ-free, removing microbiota-associated LPS, no excess neutrophil influx was observed. Collectively these results show that zebrafish intestinal colonization by Gram-negative bacteria upregulates the host enzyme Alpi, which functions to reduce host inflammatory responses to resident microbiota.
The anti-inflammatory function of Alpi is supported by many other observations from mammalian systems. Cell culture studies showed that in IECs (i.e., HT-29, T84, and IEC-6) overexpressing Alpi, LPS-activated NF-κB nuclear translocation was significantly inhibited (Goldberg et al., 2008). At the whole animal level, administration of bovine Alpi proved to reduce local/systemic inflammation and improve tissue morphology in the mouse polymicrobial sepsis model induced by cecal ligation and puncture (Van Veen et al., 2005), in the rat liver ischemia–reperfusion model (Van Veen et al., 2006), in the murine chronic colitis model induced by dextran sulfate sodium (DSS; Tuin et al., 2009; Campbell et al., 2010; Ramasamy et al., 2011) or TNBS (Martinez-Moya et al., 2012), and in the neonatal necrotizing enterocolitis rat model (Rentea et al., 2012). In clinical trials in humans, exogenous Alpi exerted protective anti-inflammatory effects on patients after cardiopulmonary surgery (Kats et al., 2009), patients with moderate to severe ulcerative colitis (Lukas et al., 2010), and patients with severe sepsis or septic shock (Heemskerk et al., 2009; Pickkers et al., 2012). Collectively, these findings confirm the importance of Alpi as an innate immune regulator, locally and systemically. LPS-detoxification by Alpi is also confirmed in cells (Goldberg et al., 2008) and animals (Beumer et al., 2003). The anti-inflammatory role of Alps is not restricted to the intestinal type, since Alps from other sources (e.g., placental Alp) protected mice against Escherichia coli-induced sepsis (Verweij et al., 2004).
Alp’s role in lipid A dephosphorylation and modulation of LPS recognition appears to be an ancient function for this family of enzymes, as demonstrated by recent work in the Hawaiian bobtail squid Euprymna scolopes (Rader et al., 2012). The squid acquires its Gram-negative bacterial symbiont Vibrio fischeri from the environment at the juvenile stage and thereafter enters a lifelong partnership with the luminous marine microbe. Rader et al. characterized two E. scolopes Alps (EsAlps), which are closely related to other mollusk Alps. EsAlp is highly active at the lumina of crypt spaces where the bacteria reside. Interestingly, EsAlp activity remained at low levels before and during the lipid A-induced tissue regression at the initial animal-bacteria contact. This enzyme regulation at the early stage proved important for the formation of the symbiotic relationship as demonstrated by two observations: (i) that inhibition of Alp by levamisole compromised the normal colonization of the symbiont and (ii) that V. fischeri lipid A pretreated by Alp failed to cause early stage apoptosis that is necessary for persistent colonization of V. fischeri. After colonization, however, the squid continuously adjusts EsAlp activity in accordance with the diel rhythm of bacterial population density, i.e., high at dusk and low at dawn, a pattern the authors suggest is strategically governed to render the animal insensitive to lipid A signaling by Alp dephosphorylation of LPS, and therefore protect the animal from excessive inflammation and tissue damage. Taken together, the data from E. scolopes presented an elegant example of the conserved role of Alps in tuning host immune recognition of LPS.
All animals live in close associations with microbial communities. Most frequently, the vast majority of these microbes reside in the digestive tract, where they assist the host in the breakdown of ingested food. Gut microbiota are highly species-specific, based on host phylogeny, diet, and digestive tract morphology (Ley et al., 2008a,b). Further microbial community specialization occurs along the length of the gut. Considering the conserved role of Alps in host-bacteria interactions, we imagine that intestinal Alps have been under continuous selective pressure to accommodate changing host-microbe interactions. For example, evolution of host digestive tract physiologies, driven by dietary changes, could spur Alpi gene duplication and diversification to buffer host inflammatory responses during the acquisition and compartmentalization of novel bacterial communities that facilitate food digestion.
In this report, we characterized two new alp genes in zebrafish and investigated their evolutionary history through the lens of the animal Alp gene phylogenies. We find that unlike the Alpl clade, the other Alp genes, which are frequently intestinally expressed, have been dynamically lost and duplicated throughout animal lineages, consistent with dynamically changing host-microbe interactions. These results suggest that Alp gene evolution has played an important role in shaping innate immune response to the intestinal microbiota.
Results
The Four Zebrafish alp Genes are Distributed Among Three Vertebrate alp Gene Clades
We had previously characterized two zebrafish alp genes, which we called alp (accession number NM_201007.1) on chromosome 11 and alpi (accession number NM_001014353.1) on chromosome 22, and had shown that the former gene is ubiquitously expressed and the latter is intestinal-specific (Bates et al., 2007). BLAST searches against the subsequent refinement of the zebrafish genome sequence revealed two additional alp-related genes: zgc:110409 (accession number NM_001025188.1) on chromosome 22, and the most recently described alp-related gene (accession number XM_003201677.1) on unassembled Scaffold Zv9 NA903 that Ensembl annotated as “alpl.” All four genes likely encode enzymatically active Alps based on their conserved Alp motifs. Protein sequence alignment against human ALPL and ALPI showed that active site residues and proposed metal-binding residues are invariant in the proteins encoded by the two human genes and four zebrafish genes (Figure 1).
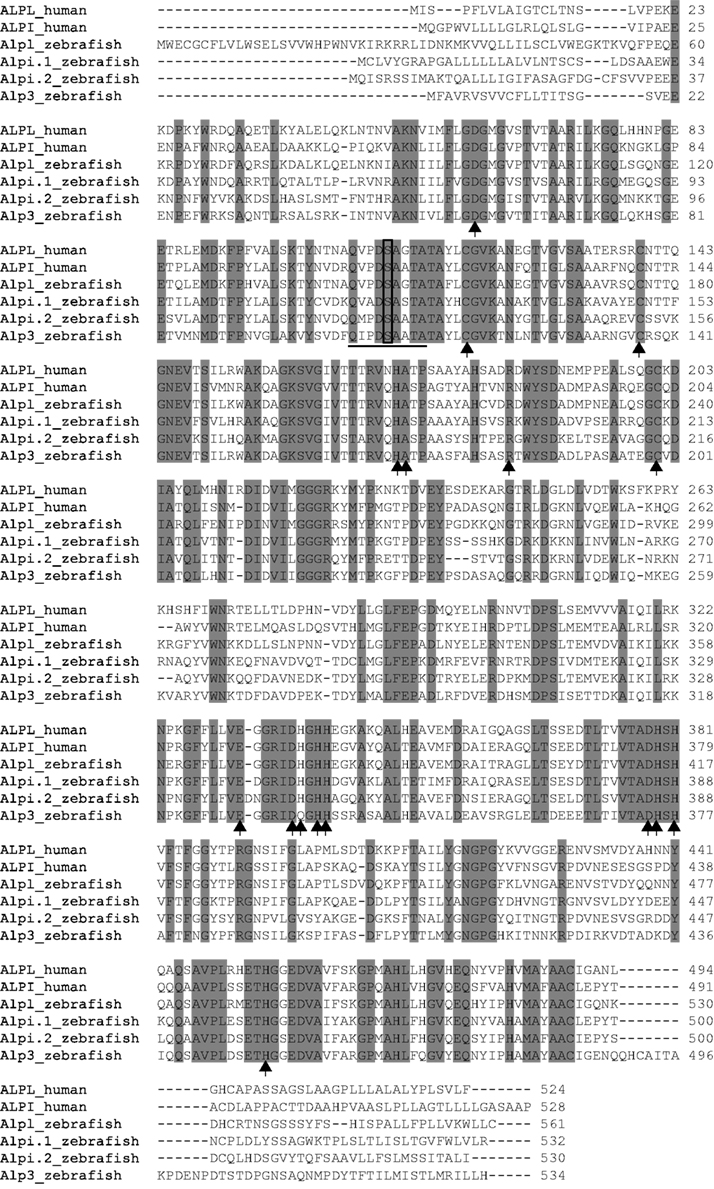
Figure 1. ClustalW Sequence alignment of human and zebrafish Alps. Identical amino acids are shaded gray, putative metal-binding sites are indicated by arrows, active sites are underlined, and the conserved serine required for enzyme activity is boxed. The proteins used in the alignment are human ALPL (ENSP00000363973) and ALPI (ENSP00000295463); zebrafish Alpl (ENSDARP00000117214), Alpi.1 (ENSDARP00000016216), Alpi.2 (ENSDARP00000070354), and Alp3 (ENSDARP00000019098).
To better understand the identity and origin of the four zebrafish alp-related genes, we investigated their evolutionary history. Phylogenetic analysis of vertebrate Alp protein sequences rooted on non-vertebrate chordate sequences revealed three distinct clades of Alp genes (Figure 2). The first of these clades, which we call Alp1, contains the human tissue non-specific gene ALPL and gene sequences from both lineages of bony vertebrates, the Sarcopterygii (lobe fin fish, including the basally diverging coelacanth and lungfish as well as tetrapods) and the Actinopterygii (ray fin fish, including the basally diverging gar as well as teleosts). The second clade, Alp2, contains the human intestinal-specific gene ALPI, the two human placental Alps, ALPP and ALPP2, as well as representative sequences from both lobefins (e.g., mammals) and ray fins (e.g., teleosts). The third clade, Alp3, contains genes exclusive to the fishes, including ray fins and the basally diverging lobe fin, the coelacanth.
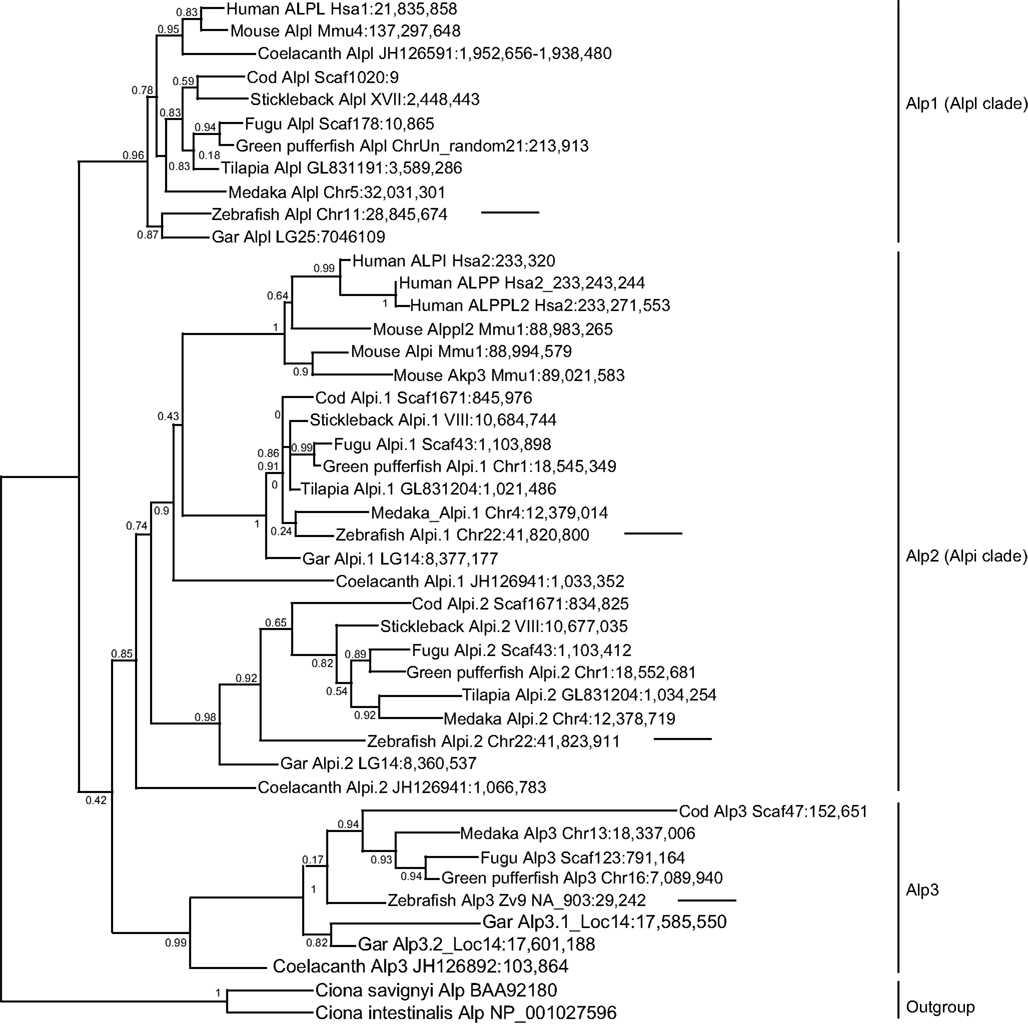
Figure 2. Diversification of vertebrate Alp protein sequences. Sequences are indicated on this maximum likelihood tree with the species name, gene name, and genomic location. For example, the taxon “Human ALPL Hsa1:21,835,858” indicates the start of the human ALPL gene encoding this protein on human chromosome 1 at nucleotide position 21,835,858 according to Ensembl Release 68 (July 2012) and Pre-Ensembl Release 66 (April 2012). The tree has three major clades [Alp1 (Alpl clade), Alp2 (Alpi clade), and Alp3]. Arrows point to the four zebrafish Alp genes.
Examination of the vertebrate Alp gene phylogeny suggests the following hypothesis for the origin of this gene family. A single Alp gene in a pre-vertebrate chordate duplicated to form four copies after the first and second vertebrate genome duplications (VGD1 and VGD2); we can call these genes Alp1, Alp2, Alp3, all of which persisted in some lineages, and Alp4, which was subsequently lost before the divergence of rayfins and lobefins. Rayfins and lobefins both retained Alp1, which became Alpl, and preserved Alp2, which experienced several lineage-specific tandem duplication events to become Alpi-like genes. Alp3 persisted in rayfins and a basally diverging lobefin, the coelacanth, but was lost from crown lobefins, the tetrapods.
Analysis of the four zebrafish alp genes within this phylogeny revealed that (i) the chromosome 11 “alp” is a genuine ortholog of the human tissue non-specific gene ALPL; (ii) the chromosome 22 “alpi” and the neighboring “zgc: 110409” are tandem duplicates derived from the ancestral alpi gene and represent coorthologs of human ALPI; and (iii) the new Scaffold Zv9 NA903 alp annotated in Ensembl as “alpl” belongs to the Alp3 clade maintained in teleosts but lost in tetrapods. Based on these findings, we developed a new nomenclature for the four genes, i.e., the current “alp” is renamed alpl, “alpi” is alpi.1, “zgc:110409” becomes alpi.2, and “alpl” is alp3. We cloned and sequenced the complete coding DNA sequences of the four zebrafish alp genes and submitted to GenBank: alpl (accession number JX847415), alpi.1 (accession number JX847416), alpi.2 (accession number JX847417), and alp3 (accession number JX847418).
The Vertebrate Alpl and Alpi Genes Arose during the Vertebrate Genome Duplications
At least two alternative models can explain the origin of multiple Alp paralogs in vertebrates. Under one hypothesis, the Alpi and Alpl genes arose from gene duplication in pre-vertebrate ancestors and were inherited by the vertebrates, but under an alternative hypothesis, vertebrate Alp paralogs arose as ohnologs (paralogs derived from genome duplication) in the two rounds of whole genome duplication VGD1 and VGD2 that occurred at the base of the vertebrate radiation (Dehal and Boore, 2005). These two hypotheses and more complicated alternatives can be winnowed down by examining phylogenetic trees that include both invertebrate and vertebrate taxa. Figure 3A shows that, while non-vertebrate chordates (sea squirts Ciona intestinalis and Ciona savignyi and amphioxus) have multiple Alp genes, these diverge basal to the vertebrates, as would be expected if the vertebrate Alp genes arose after the divergence of vertebrates from non-vertebrate chordates. Likewise, except for one fly (Drosophila melanogaster) sequence, Alp genes of insects diverge basal to the chordates, while the non-bilaterian Cnidarian sequences (the sea anemone Nematostella vectensis and the hydra Hydra magnipapillata) root the tree. The preponderance of evidence leads to the conclusion that the Alp paralogs arose in the vertebrate genome duplication events.
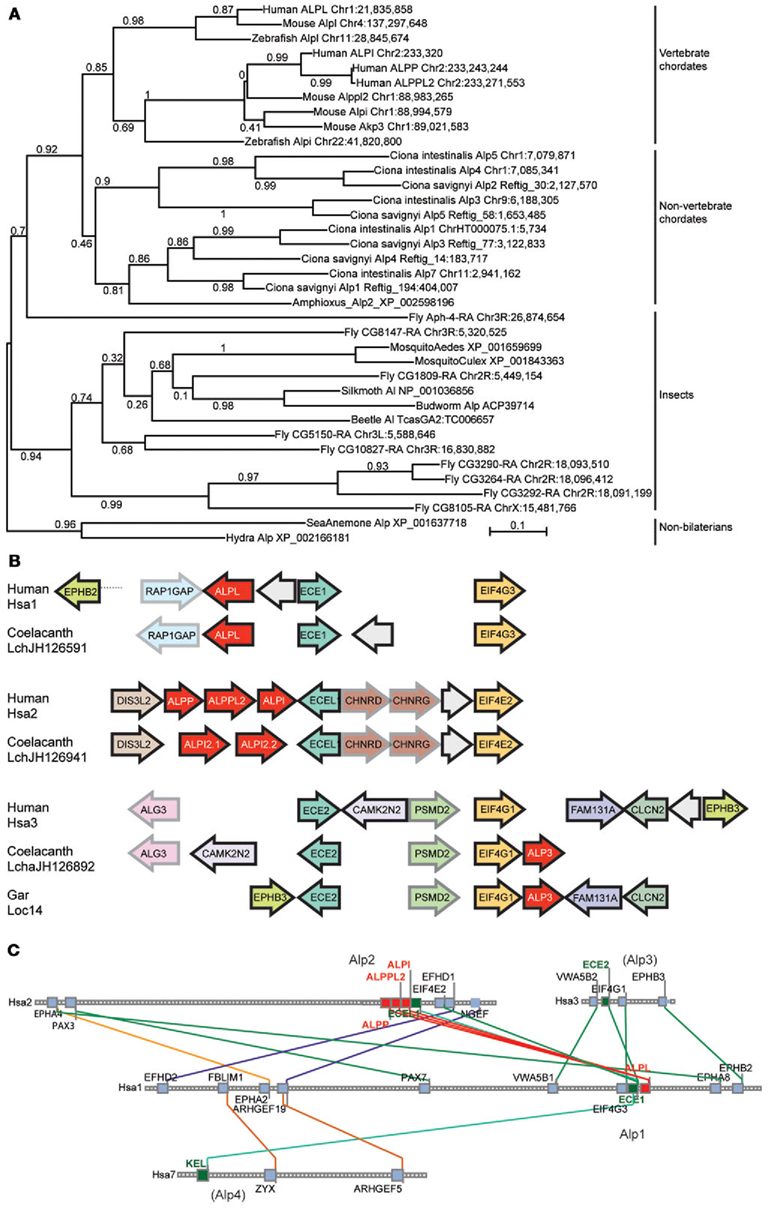
Figure 3. Invertebrate Alp protein sequences and conserved syntenies. (A) Alp genes amplified independently in vertebrate chordate and non-vertebrate chordate lineages, and in deuterostomes (including vertebrates) and protostomes (including insects). (B) Conserved syntenies among paralogons including human, coelacanth, and gar Alp genes. Alp genes in red, and paralogs of other gene families, such as Ece1, Ecel1, and Ece2 genes and Eif4 genes shown in unifying colors. Arrows represent direction of transcription. (C) Conserved syntenies in four paralogons in the human genome, only two of which have ALP genes. The figure shows four human chromosomes, Hsa1, Hsa2, Hsa3, and Hsa7, with lines between chromosomes connecting paralogs. Each square represents a gene; small gray squares from which no lines extend are genes without paralogs in the regions shown.
Analysis of conserved syntenies comparing the human genome to the genomes of coelacanth and gar supports this model for Alp gene history. Located near ALP3 in the coelacanth genome are the genes EIF4G1 and ECE2, which are paralogs of genes located near the human ALPL and ALPI genes, namely EIF4G3 – ECE1 and EIF4E2 – ECEL1, respectively (Figure 3B). At least five genes immediately adjacent to coelacanth ALP3 are adjacent in the human genome, although inversions have altered gene order. These three paralogons containing ALP – ECE – EIF4 genes likely resulted from two rounds of duplication, most parsimoniously explained as happening in the first and second vertebrate genome duplication events VGD1 and VGD2 (Dehal and Boore, 2005). The Synteny Database (Catchen et al., 2009) identifies four chromosome segments containing ECE paralogs, two with ALP gene neighbors and two without, including regions on Hsa3 and Hsa7 (Figure 3C). These data are consistent with the hypothesis that an ancestral chordate chromosome segment contained ALP, ECE, and EIF4 genes became four paralogons after VGD2, followed in the human lineage by the loss of two of the ALP paralogs from the chromosome segments that eventually became the relevant part of Hsa3 and Hsa7 and the diversification of the eventual human chromosome 1 (Hsa1) and Hsa2 genes as the ALPI-like (Alp2) and ALPL-like (Alp1) genes of today’s vertebrates.
Three of the Zebrafish alp Genes Show Enriched Expression in the Intestine
We further explored the four zebrafish alp genes by investigating their tissue expression patterns. We used semi-quantitative reverse transcription PCR to estimate the abundance of the transcripts in intestinal tissue (“I”) versus the rest of the body (referred to as carcass, “C”). Transcript levels of alpl were abundant in the carcass as well as the intestine (Figure 4A). In contrast, transcripts of the other three genes were enriched (alpi.1 and alpi.2) or exclusively expressed (alp3) in intestinal tissue (Figure 4A). We next performed in situ hybridization with gene-specific RNA probes to further examine the expression patterns of the four alp transcripts. Consistent with our previous analysis (Bates et al., 2007), we observed that alpl was diffusely expressed in many tissues (Figure 4B). Also, as we showed previously (Bates et al., 2007), alpi.1 was highly expressed in the intestine (Figure 4C). We also observed high levels of intestinal-specific expression of alpi.2 (Figures 4C,D), indicating that the tissue-specific expression of this alpi coortholog has been maintained. Finally, we observed intestinal-specific expression of the teleost-specific alp3 gene (Figures 4C,E), suggesting that intestinal-specific expression of alp genes is an ancestral trait possessed by the single Alp gene found in non-vertebrate chordates before the VGD1 and VGD2 events about 650 million years ago (Hedges et al., 2006; Braasch and Postlethwait, 2012).
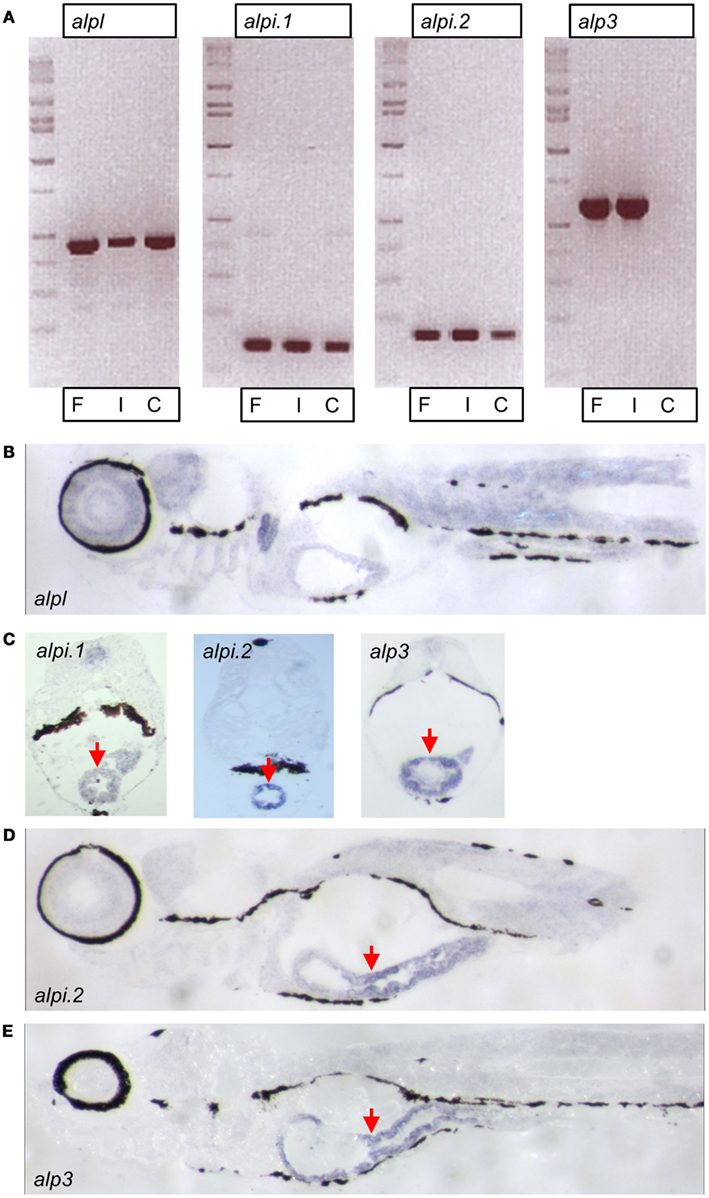
Figure 4. Expression of zebrafish alp genes. (A) Semi-quantitative reverse transcription PCR analysis of alp gene transcript levels at 7 days post fertilization (dpf) in whole fish (F), dissected intestines (I), and carcasses with intestine removed (C). In situ hybridization of 7 dpf larval sagittal (B,D,E) and transverse (C) sections with probes to alpl, alpi.1, alpi.2, and alp3 as indicated. The hybridization is visible in blue. Arrows point to the intestinal epithelium.
Mammalian Alpi Genes Have Undergone Rapid and Dynamic Evolution
Our phylogenetic analysis of the Alp gene family (Figure 1) suggested a dynamic evolutionary history of clades containing the intestinal-specific zebrafish alp genes. We examined this further through a phylogenetic analysis of mammalian Alp genes. All mammals examined in this study had a single Alp gene at the Alpl-like locus, which was surrounded by genes that were orthologs in all species with locally well-assembled genomes, showing that this region of the genome has been well-conserved among mammals. The genomic situation at the Alpi-like locus, however, differed greatly among taxa, with several species having multiple Alpi-related genes (Figure 5A).
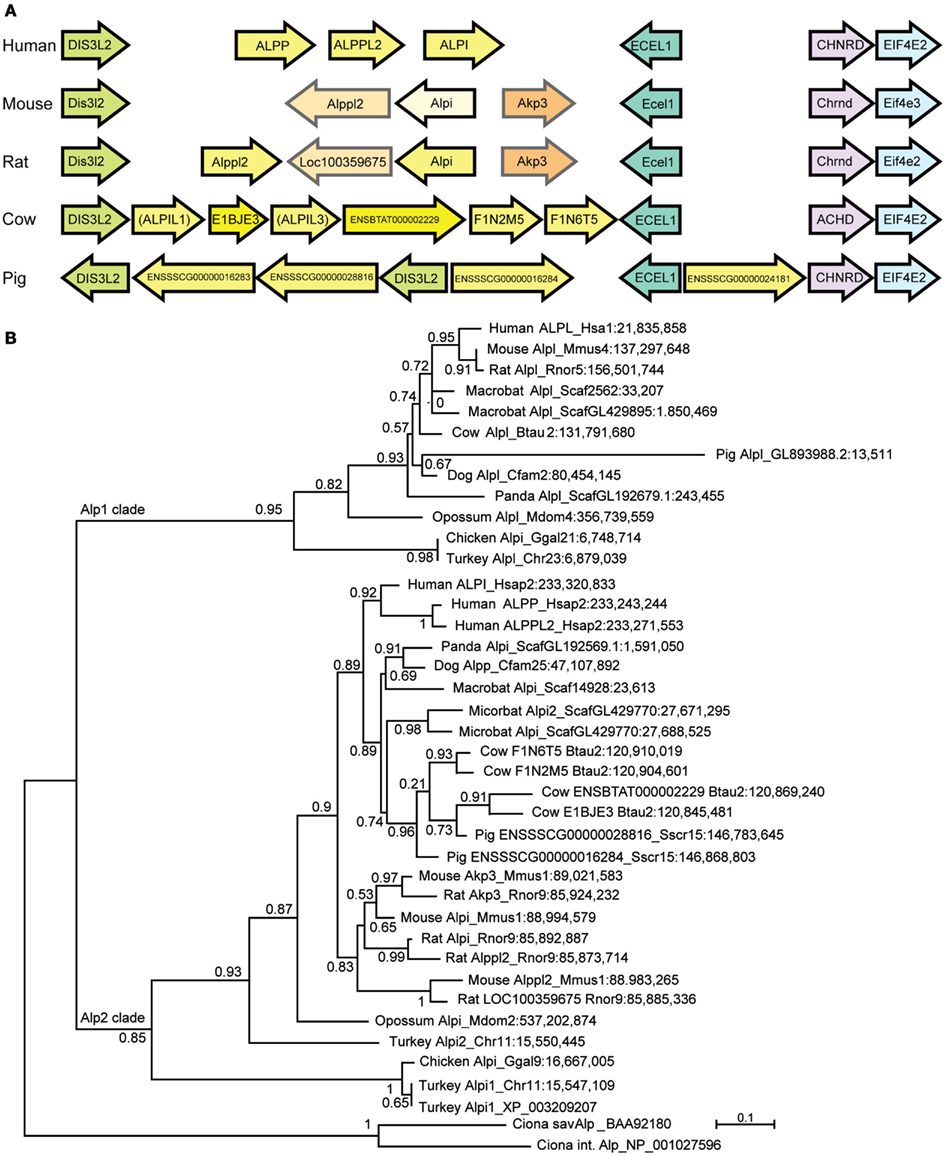
Figure 5. The diversification of mammalian Alp protein sequences. (A) The Alpi-related locus in five mammals showing Alp genes in yellow. Orthologs between mouse and rat shown in similar shades. (B) Maximum likelihood tree showing mammalian and bird Alp genes. In general, although the Alpi (Alp2) locus is orthologous and the flanking genes are orthologs among the various mammals, the tree shows that the Alpi-related genes themselves are usually not one-to-one orthologs.
Phylogenetic analysis rooted on bird and non-vertebrate chordate Alp sequences showed that many of these Alpi-like genes have arisen very recently (Figure 5B). For example, none of the three human genes (ALPI, ALPP, and ALPP2) is a unique ortholog of any non-primate Alpi-like gene despite names in common use.
The rodents mouse and rat also show shared and independent Alpi-like gene duplications (Figure 5B). The mouse Alpi-like locus contains three genes called Alpi, Alppl2, and Akp3. Although the human genome has genes called ALPI and ALPPL2, the mouse and human genes are not one-to-one orthologs according to phylogenetic analysis (see also Figure 3A). If this were the case, then the human and mouse ALPI/Alpi genes would group together in the tree and the ALPPLP/Appl2 genes would group together; instead, the tree clearly shows that the human and mouse lineages diverged long before ALPI diverged from ALPP and ALPPL2 and before Alpi diverged from Alppl2 and Akp3.
The phylogenetic tree suggests that mouse and rat genomes contain at least two pairs of orthologous Alpi-like genes called Akp3/Akp3 and Alppl2/LOC100359675, respectively (Figure 5B). In addition, the tree shows that rat has two genes derived from a recent duplication called Alpi and Alppl2. The tree shows with strong support that rat Alpi and Alppl2 arose in a tandem gene duplication event and that the rat Alpi sequence is more closely related to the rat Alppl2 sequence than it is to the mouse Alpi sequence, despite the names. In addition, although genes flanking these mammal’s Alpi-like locus are orthologs (Figure 5A), gene orientations are consistent with non-orthology or a gene-specific inversion event for the rodent genes called Alpi.
Similar findings come from a detailed phylogenetic analysis of the primate Alp genes (Figure 6). Although each primate had a single Alpl gene, the Alpi-like clade displayed several cases of independent tandem duplication events. The three Alpi-related genes of mouse formed a strong outgroup for the primate Alpi genes, suggesting that these three murine genes duplicated independently from the primate genes, confirming that none is a unique ortholog of any of the human paralogs. The single Alpi genes in the two Strepsirrhini species – lemur and bushbaby – grouped together at the base of the primate Alpi clade, which would be expected if ancestral primates had a single Alpi gene (Figure 6). A tandem duplication after the divergence of Strepsirrhini and Haplorrhini produced an Alpi clade and an Alpp clade; subsequently, after the divergence of the human and chimpanzee lineage about 6 million years ago, the human lineage experienced a tandem duplication in the ALPP clade and the chimpanzee lineage had independent duplications in the ALPP lineage, evidence of a dynamic diversification of ALP-related sequences in our recent history.
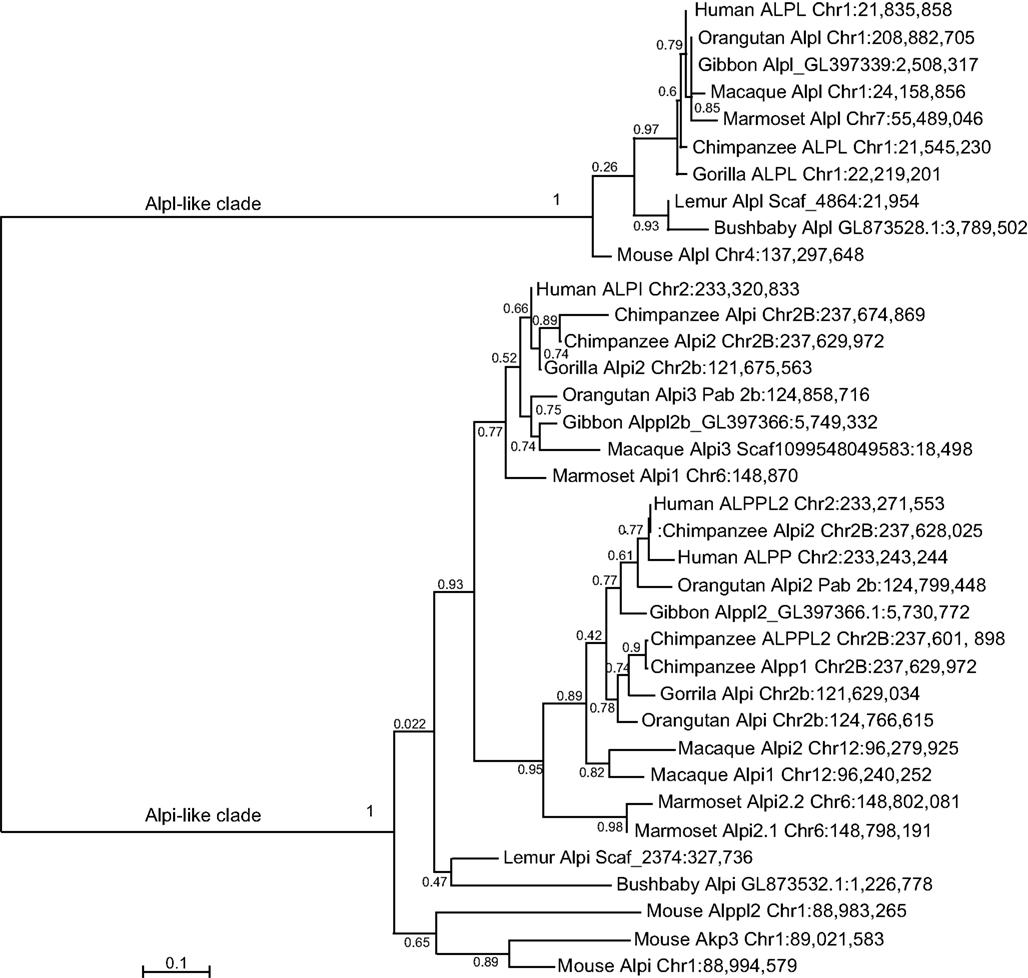
Figure 6. Maximum likelihood tree of primate Alp sequences. After the divergence of the Strepsirrhini (lemur and bushbaby) lineage from the Haplorrhini lineages, a tandem gene duplication produced an Alpi-like gene and an Alpp-like gene with further duplications within some of the Haplorrhini sub-lineages.
Variable Copy Numbers of Alpi Genes Among Mammals
We speculate that the rapid gains and losses of Alpi genes manifest in vertebrate lineages are driven by dynamic changes in host-microbe associations. In particular, if Alps play a conserved function in detoxifying LPS, then requirements for this gene could change rapidly with changing proportions of Gram-negative bacterial associates, possibly driven by adaptations to different diets that require different microbial physiologies for their metabolism. To explore this hypothesis, we determined the representation of Gram-negative phyla present in a published dataset of fecal samples from 60 mammalian species (Ley et al., 2008a; Figure 7). These samples contained an enormous range of proportional representation of Gram-negative phyla from 90% (rock hyrax) to 0% (cow). Even within one host species (humans), proportions ranged from 50 to 9%, emphasizing the variable nature of gut-associated microbial communities. We found no significant correlation between the proportional representation of Gram-negative bacteria and gut physiologies. We note, however, that fecal sampling is unlikely to capture the microbial composition of specialized gut compartments. For example, whereas the cow fecal sample from this study contained no Gram-negative phyla, surveys of cow rumen content typically find a high proportion of Gram-negative species (Jami and Mizrahi, 2012).
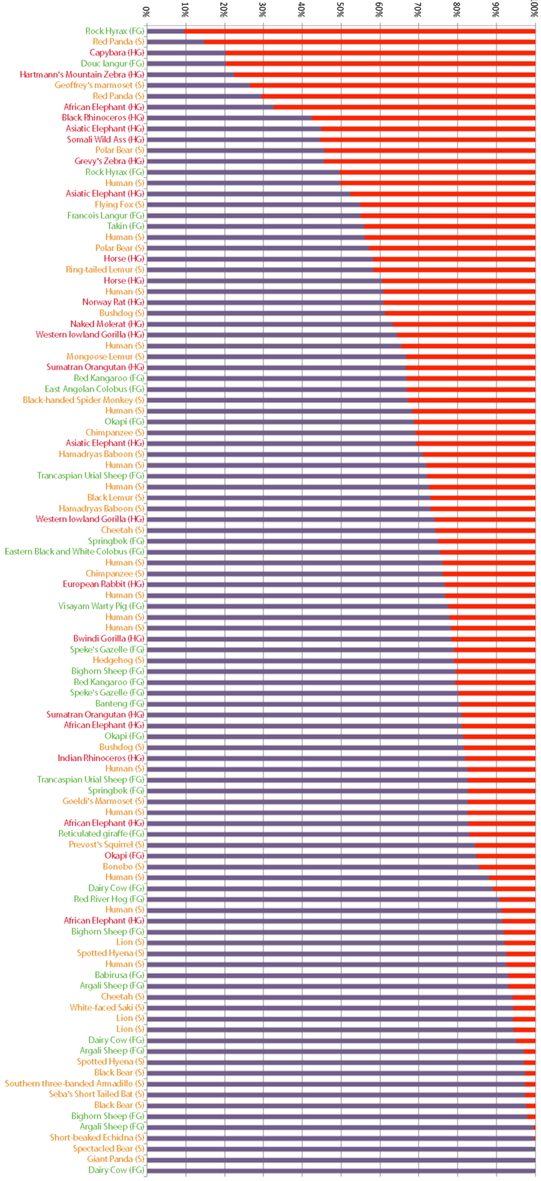
Figure 7. Proportional representation of Gram-negative phyla in fecal samples from different mammals. Bacterial composition of mammalian fecal samples, as previously reported (Ley et al., 2008a), are represented as the proportion of phyla composed of Gram-negative (red) and Gram-positive (purple) bacteria. Host animals’ gut morphologies are indicated and color coded (FG, foregut fermenter in green; S, simple in yellow; HG, hindgut fermenter in red).
The limited number and sometimes low quality of whole genome sequences from mammalian species precluded us from performing a systematic correlation between Alpi gene copy number and gut morphology, but we noted some interesting trends. Although many of the mammals have multiple Alpi-like genes, three members of the order Carnivora that have simple gut morphologies – the giant panda, domestic dog, and domestic cat – all have only one Alpi gene (despite the fact that the panda is an herbivore). In contrast, Artiodactyls, which have more complex digestive systems, showed more complex Alpi-like genomics. The cow has six Alpi-related genes (called here Alpi-like 1–6), only four of which (Alpil2;E1BJE3, Alpil4;ENSBTAT000002229, Alpil5;F1N2M5, and Alpil6;F1N6T5) are annotated in Ensemble. These four annotated genes are arranged in neighboring pairs oriented in the same direction (Figure 5A), with Alpil5 and Alpil6 falling as sisters in the tree and as neighbors in the genome and Alpil4 and Alpil5 also diverging as sisters and neighbors, suggesting tandem duplication events (Figure 5B). The pig has about three Alpi-like genes, although only two of them are well-assembled in Ensemble. One of the pig genes (Alpil2; ENSSSCG00000028816) appears to be coorthologous to at least two of the cow genes (Alpi2 and 4), suggesting that tandem gene duplications occurred in the Artiodactyl lineage before the divergence of swine and bovine lineages. The tree suggests the loss of ancestral Artiodactyl genes in the pig lineage and the tree and gene orientations indicate that independent duplications occurred at least in the cow lineage as well as local inversions during lineage divergence. An open and interesting question is whether different bovine Alpi-like genes are expressed in specific portions of the cow’s complex digestive system and are adapted to the different microbial contents of each compartment.
Discussion
Our investigation into the phylogenetic relationship between Alp genes from zebrafish and other vertebrates reveals a remarkable evolutionary history for this gene family. As summarized in Figure 8, our analysis supports the model that the vertebrate Alp genes arose from a single Alp gene in ancestral chordates that duplicated during two rounds of genome duplication events (VGD1 and VGD2) that preceded the diversification of extant vertebrates after the divergence of vertebrates from non-vertebrate chordates and initially yielded four genes. One of these genes became the modern Alpl. This gene was faithfully maintained as a single copy throughout the vertebrates, with the coorthologous gene that would have been generated during a third round of genome duplication, the teleost whole genome duplication (TGD), having been lost. Indeed none of the three predicted coorthologous Alp genes are found within the teleost genomes we examined, but this could be due to chance; because about 75% of zebrafish coorthologues have been lost, the likelihood of all three Alp coorthologues being lost by chance is 42%, clearly a frequent occurrence for any three genes taken at random. The predicted fourth Alp generated during the two vertebrate genome duplications was lost before the divergence of lobefin and rayfin bony vertebrates. Alp3 persisted in the rayfin lineage where it underwent tandem duplication in the gar and can be found in the basally diverging lobefin, the coelacanth, but was lost from crown group lobefins, the tetrapods. The evolutionary history of Alp2 (Alpi-like) is the most dynamic. Examination of phylogenies of fishes, mammals, and primates reveals that this gene has undergone multiple independent losses and tandem duplications in its history. The role of gene conversion in homogenizing sequences after tandem duplications is as yet unstudied.
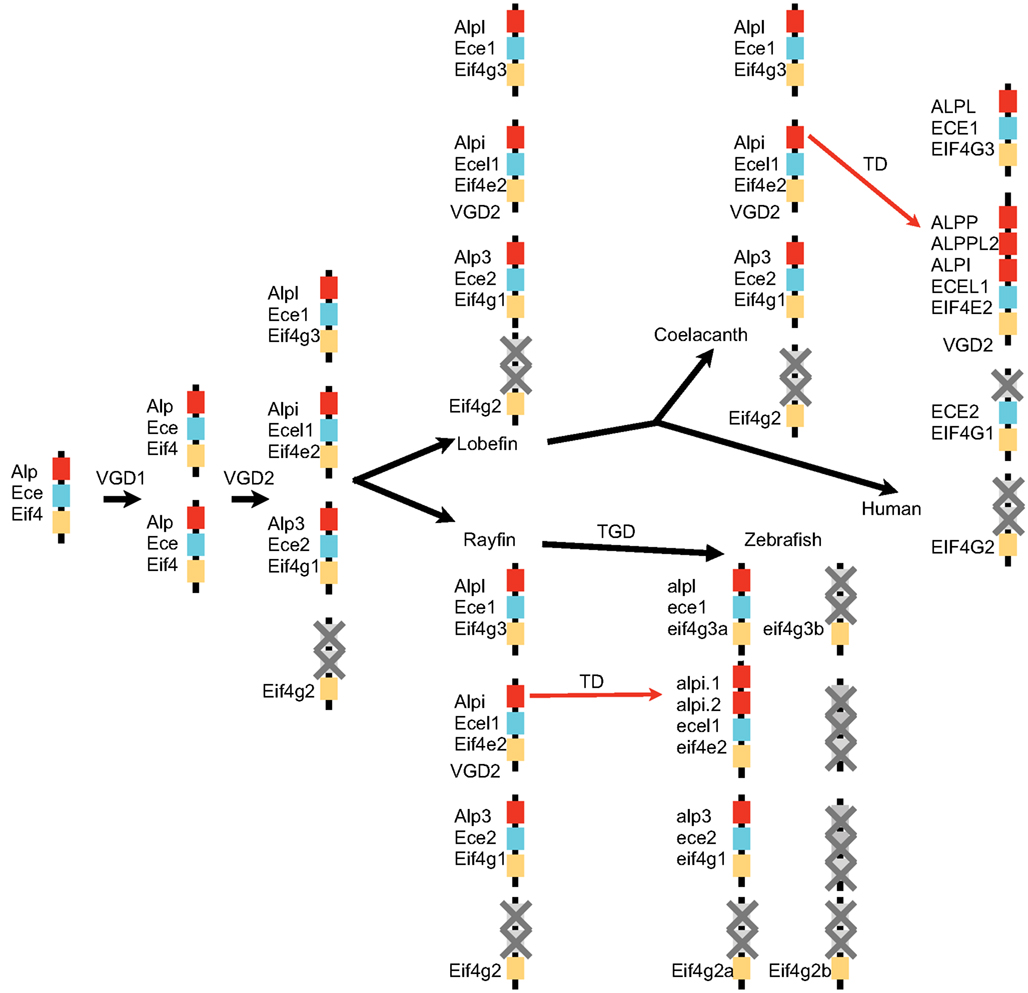
Figure 8. A model for the history of Alp genes in vertebrates. A chromosome segment in a non-vertebrate chordate contained Ece, Alp, and Eif4 genes and duplicated twice in the vertebrate genome duplication events (VGD1 and VGD2) to make four paralogons, one of which lost its Alp gene. After the divergence of rayfin and lobefin bony fish lineages, the teleost genome duplication event (TGD) did not result in further extant Alp genes, but tandem duplications (TD) in the Alpi locus provided a number of separate Alp genes.
We speculate that the basis for the dramatically different evolutionary histories of the tissue non-specific Alpl and intestinal Alpi genes, which encode enzymes with well-conserved catalytic activities, lies in their different patterns of tissue expression. The ubiquitously expressed Alpl plays an important function in bone mineralization in mammals (Golub and Boesze-Battaglia, 2007). In humans, over 261 different mutations in ALPL have been linked to hypophosphatasia and skeletal abnormalities, of which 75% are missense mutations1. Several of these mutations result in dominant inheritance of the disease (Mornet et al., 2011), suggesting that modest changes in Alp function at sites of bone mineralization can lead to deleterious phenotypes, possibly restricting the evolution of copy number variation of this gene once it became dedicated to this function.
The frequent gains and losses of the intestinally expressed Alp genes across vertebrate lineages indicate much greater plasticity in the requirements for the enzyme encoded at this locus. The rapid evolution of gene copy number in the Alpi gene clade is especially striking among the mammals, which have undergone dramatic changes in their gut morphologies that accommodate different microbial fermentation strategies during adaptations to different diets, with innovations such as foregut fermentation arising independently multiple times in the mammalian tree (Stevens and Hume, 2004). Possibly the loss of Alp3 prior to the divergence of the tetrapods put extra pressure on Alpi to accommodate changing requirements for Alp activity in the intestine. We hypothesize that requirements for Alpi function in detoxification of luminal LPS from Gram-negative gut bacteria changed dramatically with alterations in associated microbial communities, driven by dietary changes. Additionally, dietary changes may have altered the selective pressures on Alpi’s lipid absorption function. Alpi gene expansion may have been a way to tune the levels and spatial distribution of this enzyme within the gut and to allow the evolution of different gut compartments adapted to different bacterial communities or other gut functions. For example, one of the three mouse Alpi-like genes, Akp3, is restricted in its expression to the proximal duodenum (Narisawa et al., 2007). It would be interesting to determine whether the six Alpi genes in the cow are restricted to different regions of the elaborate gastrointestinal tract anatomy of this foregut fermenter. The gut is also a major site of infections, and the capacity of intestinal Alp to detoxify LPS is likely to be important for modulating immune responses to pathogens as well as resident beneficial bacteria. Epidemics of infections with Gram-negative enteric pathogens may have been additional driving forces that shaped the rapid evolution of the intestinal Alp genes.
In summary, the stark contrast between the evolutionary history of the vertebrate Alpl and Alpi clades suggests that these genes diversified in the functions they perform in organisms. We propose that the highly dynamic pattern of gene evolution of the Alpi clade is indicative of a gene family that serves important functions in mediating host-microbe interactions, which can change dramatically over short periods of time and impose strong selective pressures on animals.
Materials and Methods
Use of Vertebrate Animals
All zebrafish experiments were performed using protocols approved by the University of Oregon Institutional Care and Use Committee, and following standard protocols (Westerfield, 2007).
Sanger Sequencing of Zebrafish Alp Gene Coding Sequences
Fresh RNA was extracted from 7 days post fertilization (dpf) zebrafish larvae (illustra RNAspin mini kit, GE Healthcare). Total cDNA was synthesized using RNA as template (SuperScript III reverse transcription kit, Life technologies). The coding sequences of Alp genes were amplified from cDNA in PCR (Phusion DNA polymerase, Thermo Scientific). Primers used in PCR included alplF: 5′-ATGTGGGAATGTGGATGCTTTCTTG-3′, alplR: 5′-TCAGCAAAGCAGCCATTTGACC-3′; alpi.1F: 5′-ATGTGTTTGGTTTACGGTCGGGC-3′, alpi.1R: 5′-TCATCTCAAAACAAGCCAAAACACG-3′; alpi.2F: 5′-ATGGCCAAAACACAAGCCCTG-3′, alpi.2R: 5′-CTAAATAAGAGCAGTAATGGAGGACATCAG-3′; alp3F: 5′-ATGTTTGCTGTCCGTGTGTCC-3′, alp3R: 5′-TCAGTGCAGTAAAATCCTCATCAGTG-3′. PCR products were evaluated by DNA gel electrophoresis for purity and then extracted from gel. The purified PCR products were cloned into the pCR® – Blunt II TOPO® vector (ZeroBlunt® TOPO® PCR Cloning kit, Life Technologies). Clones carrying Alp gene coding sequences were sequenced (Sequetech). The complete coding sequences were assembled using the plasmid editor ApE2 and submitted to GenBank.
PCR Detection of Zebrafish Alp Gene Transcription
Total cDNA was synthesized from fresh RNA at 7 dpf from whole fish, dissected intestines, or carcasses with guts removed. Gene-specific primers were used in PCR to test the presence of gene transcripts (alplF: 5′-TATTTCTTGGAGATGGGATGGGTG-3′, alplR: 5′-TTCAAAGAGTTGTCTGGCGATGTC-3′; alpi.1F: 5′-GCACCGCGCCAAAGCACAAG-3′, alpi.1R: 5′-CGGGCTTCGGAGGGCACATC-3′; alpi.2F: 5′-TGCGCTTTACGGAAACGGTCCA-3′, alpi.2R: 5′ – TGCGCCATCGGGCCTTTAGC-3′; alp3F: 5′-ATGTTTGCTGTCCGTGTGTCC-3′, alp3R: 5′-ACGA GAAACCGCCTCATCCAG-3′). Equal amounts of cDNA (200 ng/μl, NanoDrop 1000) were added as template in PCR.
In situ Hybridization
Zebrafish cDNA was obtained as describe above and used in PCR to amplify a unique fragment of alpl (alplF: TTCCAGAGCAAGAGAAGCGG; alplR: GTCTTAGAGAGGGCGACGTG), alpi.1 (alpi.1F: CGACCGGGCGATTCAGAGAG; alpi.1R: TGGTGTACGGCTCAAGGCAC), alpi.2 (alpi.2F: TCACTAACGGGACTCGACCT; alpi.2R: AGGCCATAGCGTGAGGAATG), and alp3 (alp3F: CAGGGTCATCACTCCAGTCG; alp3R: TCTGGACGCTTGTTGTTGGT). The purified PCR product was cloned into the pCR® – Blunt II TOPO® vector (ZeroBlunt® TOPO® PCR Cloning kit, Life Technologies) and validated by sequencing (Sequetech). The verified plasmid was used in PCR to add T7 RNA polymerase binding sites to the gene-specific fragment (alplF: TTCCAGAGCAAGAGAAGCGGC, alplT7-R: GTAATACGACTCACTATAGGGGTCTTAGAGAGGG; ali.1p-F: CGACCGGGCGATTCAGAGAG, alpi.1T7-R: GTAATACGACTCACTATAGGGTGGTGTACGGCTCAA; alpi.2F: TCACTAACGGGACTCGACCTGATGT, alpi.2T7-R: GTAATACGACTCACTATAGGGAGGCCATAGCGTGAG; alp3F: CAGGGTCATCACTCCAGTCGGGC, alp3T7-R: GTAATACGACTCACTATAGGGTCTGGACGCTTGTTG). The purified PCR product was used as template to synthesize the DIG labeled RNA probe (DIG RNA Labeling Mix (T7), Roche Applied Science).
Seven dpf zebrafish larvae were fixed in 4% PFA overnight at 4°C and then washed in 1×PBST. Rinsed larvae were embedded in 1.5% agar and cryo-cut into 16 μm thick sections. Sections were defrosted and air died overnight at room temperature. Diluted RNA probes were added on slides and incubated overnight at 70°C. After hybridization, slides were rinsed with the wash solution (1×SSC, 50% formamide, 0.1% Tween-20). Slides were then treated with the block solution (MABT, 2% blocking reagent, 20% heat inactivated sheep serum) for 3 h. Following that, first antibody solution (AP conjugated anti-DIG) was applied to slides and incubated overnight at 4°C. Slides were then washed in AP staining buffer (5 M NaCl, 1 M MgCl2, 1 M Tris pH 9.5, 20% Tween-20) and incubated with NBT/BCIP color regents in the dark at 37°C. The color developing process was stopped by washing slides in PBST. Slides were then rinsed in ddH2O, dehydrated in ethanol and eventually mounted in Permount. Images were taken under a Nikon TE2000 inverted microscope with a CoolSNAP camera (Photometrics).
Genomic Analyses
ALP-related sequences were obtained from sequenced genomes present in Ensembl3 and Pre-Ensembl4 by BLASTP searches using the human ALPI (ENSG00000163295) and ALPL (ENSG00000162551) sequences as query. Protein sequences for non-annotated genes in Ensembl were assembled using GeneMark5 (Lomsadze et al., 2005). Protein sequences were retrieved and subjected to phylogenetic analysis using MUSCLE for multiple alignment (Edgar, 2004), Gblocks to remove poorly aligned positions and divergent regions (Castresana, 2000), the PhyML maximum likelihood method for tree building (Guindon and Gascuel, 2003), and TreeDyn6 for tree rendering (Chevenet et al., 2006; Dereeper et al., 2008). Conserved synteny analyses were performed using the Synteny Database (Catchen et al., 2009). The sequences used in phylogeny study were provided in supplements.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ruth Ley for sharing microbiota data, Rose Sockol and the staff of the University of Oregon Zebrafish Facility for excellent fish husbandry, and Poh Kheng Loi and the staff of the University of Oregon histology facility for histology services. This research was supported by NIH grants 1R01DK075667 (to Karen Guillemin), 1R01AG031922 (to John H. Postlethwait), and 8R01OD011116 (to John H. Postlethwait). NIH grant HD22486 provided support for the Oregon Zebrafish Facility.
Author Note
New sequences submitted to GenBank: zebrafish alpl complete CDS (accession number JX847415), zebrafish alpi.1 complete CDS (accession number JX847416), zebrafish alpi.2 complete CDS (accession number JX847417), and zebrafish alp3 complete CDS (accession number JX847418).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Molecular_Innate_Immunity/10.3389/fimmu.2012.00314/abstract
Data sheet S1. Alp sequences used to generate Figure 2.
Data sheet S2. Alp sequences used to generate Figure 3.
Data sheet S3. Alp sequences used to generate Figure 5.
Data sheet S4. Alp sequences used to generate Figure 6.
Footnotes
References
Akiba, Y., Mizumori, M., Guth, P. H., Engel, E., and Kaunitz, J. D. (2007). Duodenal brush border intestinal alkaline phosphatase activity affects bicarbonate secretion in rats. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G1223–G1233.
Bates, J. M., Akerlund, J., Mittge, E., and Guillemin, K. (2007). Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2, 371–382.
Bates, J. M., Mittge, E., Kuhlman, J., Baden, K. N., Cheesman, S. E., and Guillemin, K. (2006). Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 297, 374–386.
Beumer, C., Wulferink, M., Raaben, W., Fiechter, D., Brands, R., and Seinen, W. (2003). Calf intestinal alkaline phosphatase, a novel therapeutic drug for lipopolysaccharide (LPS)-mediated diseases, attenuates LPS toxicity in mice and piglets. J. Pharmacol. Exp. Ther. 307, 737–744.
Beutler, B., and Rietschel, E. T. (2003). Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 3, 169–176.
Braasch, I., and Postlethwait, J. H. (2012). “Polyploidy in fish and the teleost genome duplication,” in Polyploidy and Genome Evolution, eds P. S. Soltis and D. E. Soltis (New York: Springer) (in press).
Brun, L. R., Brance, M. L., and Rigalli, A. (2012). Luminal calcium concentration controls intestinal calcium absorption by modification of intestinal alkaline phosphatase activity. Br. J. Nutr. 108, 229–233.
Campbell, E. L., MacManus, C. F., Kominsky, D. J., Keely, S., Glover, L. E., Bowers, B. E., et al. (2010). Resolvin E1-induced intestinal alkaline phosphatase promotes resolution of inflammation through LPS detoxification. Proc. Natl. Acad. Sci. U.S.A. 107, 14298–14303.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552.
Catchen, J. M., Conery, J. S., and Postlethwait, J. H. (2009). Automated identification of conserved synteny after whole-genome duplication. Genome Res. 19, 1497–1505.
Chevenet, F., Brun, C., Banuls, A. L., Jacq, B., and Christen, R. (2006). TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7, 439.
Dehal, P., and Boore, J. L. (2005). Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3, e314.
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469.
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113.
Goldberg, R. F., Austen, W. G. Jr., Zhang, X., Munene, G., Mostafa, G., Biswas, S., et al. (2008). Intestinal alkaline phosphatase is a gut mucosal defense factor maintained by enteral nutrition. Proc. Natl. Acad. Sci. U.S.A. 105, 3551–3556.
Golub, E. E., and Boesze-Battaglia, K. (2007). The role of alkaline phosphatase in mineralization. Curr. Opin. Orthop. 18, 444–448.
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704.
Hedges, S. B., Dudley, J., and Kumar, S. (2006). TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972.
Heemskerk, S., Masereeuw, R., Moesker, O., Bouw, M. P., Van Der Hoeven, J. G., Peters, W. H., et al. (2009). Alkaline phosphatase treatment improves renal function in severe sepsis or septic shock patients. Crit. Care Med. 37, 417–423, e411.
Jami, E., and Mizrahi, I. (2012). Composition and similarity of bovine rumen microbiota across individual animals. PLoS ONE 7, e33306.
Kats, S., Brands, R., Seinen, W., De Jager, W., Bekker, M. W., Hamad, M. A., et al. (2009). Anti-inflammatory effects of alkaline phosphatase in coronary artery bypass surgery with cardiopulmonary bypass. Recent Pat. Inflamm. Allergy Drug Discov. 3, 214–220.
Kim, H. M., Park, B. S., Kim, J. I., Kim, S. E., Lee, J., Oh, S. C., et al. (2007). Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell 130, 906–917.
Lalles, J. P. (2010). Intestinal alkaline phosphatase: multiple biological roles in maintenance of intestinal homeostasis and modulation by diet. Nutr. Rev. 68, 323–332.
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008a). Evolution of mammals and their gut microbes. Science 320, 1647–1651.
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R., and Gordon, J. I. (2008b). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788.
Lomsadze, A., Ter-Hovhannisyan, V., Chernoff, Y. O., and Borodovsky, M. (2005). Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33, 6494–6506.
Lukas, M., Drastich, P., Konecny, M., Gionchetti, P., Urban, O., Cantoni, F., et al. (2010). Exogenous alkaline phosphatase for the treatment of patients with moderate to severe ulcerative colitis. Inflamm. Bowel Dis. 16, 1180–1186.
Lynes, M., Narisawa, S., Millan, J. L., and Widmaier, E. P. (2011). Interactions between CD36 and global intestinal alkaline phosphatase in mouse small intestine and effects of high-fat diet. Am. J. Physiol. Regul. Integr. Comp. Physiol. 301, R1738–R1747.
Martinez-Moya, P., Ortega-Gonzalez, M., Gonzalez, R., Anzola, A., Ocon, B., Hernandez-Chirlaque, C., et al. (2012). Exogenous alkaline phosphatase treatment complements endogenous enzyme protection in colonic inflammation and reduces bacterial translocation in rats. Pharmacol. Res. 66, 144–153.
McConnell, R. E., Higginbotham, J. N., Shifrin, D. A. Jr., Tabb, D. L., Coffey, R. J., and Tyska, M. J. (2009). The enterocyte microvillus is a vesicle-generating organelle. J. Cell Biol. 185, 1285–1298.
Millán, J. L. (2006). Mammalian Alkaline Phosphatases: From Biology to Applications in Medicine and Biotechnology. Weinheim: Wiley-VCH.
Mizumori, M., Ham, M., Guth, P. H., Engel, E., Kaunitz, J. D., and Akiba, Y. (2009). Intestinal alkaline phosphatase regulates protective surface microclimate pH in rat duodenum. J. Physiol. (Lond.) 587, 3651–3663.
Mornet, E., Yvard, A., Taillandier, A., Fauvert, D., and Simon-Bouy, B. (2011). A molecular-based estimation of the prevalence of hypophosphatasia in the European population. Ann. Hum. Genet. 75, 439–445.
Narisawa, S., Hoylaerts, M. F., Doctor, K. S., Fukuda, M. N., Alpers, D. H., and Millan, J. L. (2007). A novel phosphatase upregulated in Akp3 knockout mice. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G1068–G1077.
Narisawa, S., Huang, L., Iwasaki, A., Hasegawa, H., Alpers, D. H., and Millan, J. L. (2003). Accelerated fat absorption in intestinal alkaline phosphatase knockout mice. Mol. Cell. Biol. 23, 7525–7530.
Pickkers, P., Heemskerk, S., Schouten, J., Laterre, P. F., Vincent, J. L., Beishuizen, A., et al. (2012). Alkaline phosphatase for treatment of sepsis-induced acute kidney injury: a prospective randomized double-blind placebo-controlled trial. Crit. Care 16, R14.
Poelstra, K., Bakker, W. W., Klok, P. A., Hardonk, M. J., and Meijer, D. K. (1997a). A physiologic function for alkaline phosphatase: endotoxin detoxification. Lab. Invest. 76, 319–327.
Poelstra, K., Bakker, W. W., Klok, P. A., Kamps, J. A., Hardonk, M. J., and Meijer, D. K. (1997b). Dephosphorylation of endotoxin by alkaline phosphatase in vivo. Am. J. Pathol. 151, 1163–1169.
Rader, B. A., Kremer, N., Apicella, M. A., Goldman, W. E., and McFall-Ngai, M. J. (2012). Modulation of symbiont lipid A signaling by host alkaline phosphatases in the squid-vibrio symbiosis. MBio 3.
Ramasamy, S., Nguyen, D. D., Eston, M. A., Alam, S. N., Moss, A. K., Ebrahimi, F., et al. (2011). Intestinal alkaline phosphatase has beneficial effects in mouse models of chronic colitis. Inflamm. Bowel Dis. 17, 532–542.
Rentea, R. M., Liedel, J. L., Welak, S. R., Cassidy, L. D., Mayer, A. N., Pritchard, K. A. Jr., et al. (2012). Intestinal alkaline phosphatase administration in newborns is protective of gut barrier function in a neonatal necrotizing enterocolitis rat model. J. Pediatr. Surg. 47, 1135–1142.
Shifrin, D. A. Jr., McConnell, R. E., Nambiar, R., Higginbotham, J. N., Coffey, R. J., and Tyska, M. J. (2012). Enterocyte microvillus-derived vesicles detoxify bacterial products and regulate epithelial-microbial interactions. Curr. Biol. 22, 627–631.
Stevens, C. E., and Hume, I. D. (2004). Comparative Physiology of the Vertebrate Digestive System. Cambridge, New York: Cambridge University Press.
Sullivan, C., Charette, J., Catchen, J., Lage, C. R., Giasson, G., Postlethwait, J. H., et al. (2009). The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J. Immunol. 183, 5896–5908.
Tuin, A., Poelstra, K., De Jager-Krikken, A., Bok, L., Raaben, W., Velders, M. P., et al. (2009). Role of alkaline phosphatase in colitis in man and rats. Gut 58, 379–387.
Van Veen, S. Q., Dinant, S., Van Vliet, A. K., and Van Gulik, T. M. (2006). Alkaline phosphatase reduces hepatic and pulmonary injury in liver ischaemia – reperfusion combined with partial resection. Br. J. Surg. 93, 448–456.
Van Veen, S. Q., Van Vliet, A. K., Wulferink, M., Brands, R., Boermeester, M. A., and Van Gulik, T. M. (2005). Bovine intestinal alkaline phosphatase attenuates the inflammatory response in secondary peritonitis in mice. Infect. Immun. 73, 4309–4314.
Verweij, W. R., Bentala, H., Huizinga-Van Der Vlag, A., Van Loenen-Weemaes, A.-M., Kooi, K., Meijer, D. K., et al. (2004). Protection against an Escherichia coli-induced sepsis by alkaline phosphatase in mice. Shock 22, 174–179.
Keywords: zebrafish, intestinal alkaline phosphatase, vertebrate, evolution, microbiota
Citation: Yang Y, Wandler AM, Postlethwait JH and Guillemin K (2012) Dynamic evolution of the LPS-detoxifying enzyme intestinal alkaline phosphatase in zebrafish and other vertebrates. Front. Immun. 3:314. doi: 10.3389/fimmu.2012.00314
Received: 31 August 2012; Accepted: 21 September 2012;
Published online: 12 October 2012.
Edited by:
Larry J. Dishaw, University of South Florida, USAReviewed by:
Tatsuya Ota, The Graduate University for Advanced Studies, JapanJeffrey A. Yoder, North Carolina State University, USA
Copyright: © 2012 Yang, Wandler, Postlethwait and Guillemin. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: John H. Postlethwait, Institute of Neuroscience, University of Oregon, Eugene, OR 97403, USA. e-mail: jpostle@uoneuro.uoregon.edu; Karen Guillemin, Institute of Molecular Biology, University of Oregon, Eugene, OR 97403, USA. e-mail: guillemin@molbio.uoregon.edu
†Current address: Anica M. Wandler, Department of Pediatrics, University of California San Francisco, San Francisco, CA, USA.
