- 1Academic Unit of Cancer Sciences, Faculty of Medicine and Institute for Life Sciences, University of Southampton and NIHR Cancer Research UK Experimental Cancer Medicine Centre, Southampton, UK
- 2Wessex Investigational Sciences Hub (WISH) Laboratory, Department of Allergy, Asthma and Clinical Immunology, University Hospital Southampton NHS Foundation Trust, Southampton, UK
Innate T cells are a heterogeneous group of αβ and γδ T cells that respond rapidly (<2 h) upon activation. These innate T cells also share a non MHC class I or II restriction requirement for antigen recognition. Three major populations within the innate T cell group are recognized, namely, invariant NKT cells, mucosal associated invariant T cells, and gamma delta T cells. These cells recognize foreign/self-lipid presented by non-classical MHC molecules, such as CD1d, MR1, and CD1a. They are activated during the early stages of bacterial infection and act as a bridge between the innate and adaptive immune systems. In this review, we focus on the functional properties of these three innate T cell populations and how they are purposed for antimicrobial defense. Furthermore, we address the mechanisms through which their effector functions are targeted for bacterial control and compare this in human and murine systems. Lastly, we speculate on future roles of these cell types in therapeutic settings such as vaccination.
A successful immune response to foreign pathogen requires a rapid activation of innate immunity, which directs the subsequent development of a productive adaptive immune response. Innate T cells represent a group of T lymphocytes that are able to act during the lag time while effective adaptive immune responses develop (1). Similar to conventional T cells, innate T cells undergo T cell receptor (TCR) rearrangement and thymic selection. Unlike their conventional counterparts, innate T cells rapidly recognize foreign pathogen signals and manifest immediate effector functions after activation. This allows innate T cells to perform effector immune responses much earlier than conventional T cells, and act as an additional “bridge” between innate and adaptive immune responses (2).
Classically, T cells are subdivided into two major populations based upon their TCR expression, namely alpha beta (αβ) T cells and gamma delta (γδ) T cells. Conventional αβ T cells recognize a broad range of peptide antigens typically presented by Major Histocompatibility Complex (MHC I and II) complexes, enabled through their highly diverse TCR arrangement. In contrast, the αβ innate T cells that have been identified display a restricted T cell repertoire characterized by the expression of an invariant or semi-invariant TCR α chain. In humans, two well-defined αβ innate T cell populations have been identified in recent years, namely, mucosal-associated invariant T (MAIT) cells and invariant natural killer T (iNKT) cells. These two T cell populations together with γδ T cells form the three major types of innate T cell (1). All three innate T cell populations express a C-type lectin molecule CD161. CD161 was initially identified on CD4, CD8, and γδ subsets in the 1990s (3, 4). CD161 is variably expressed across human T cells, and three populations can be identified, expressing negative, intermediate, and high levels of CD161 (2). The expression of CD161 in human T cells populations is summarized in Figure 1. The level of CD161 expression is distinctive between conventional T cells and innate T cells, with MAIT cells displaying the highest levels (5).
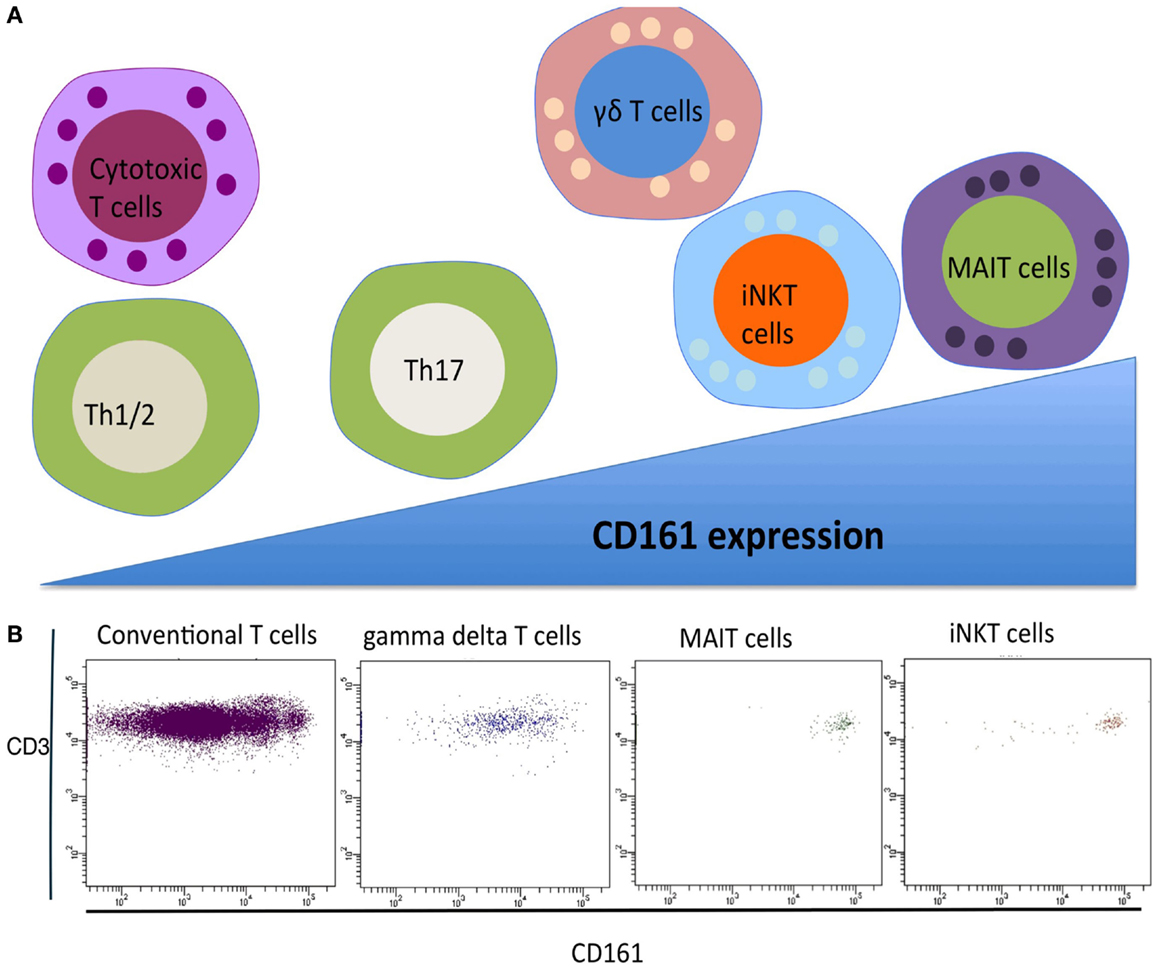
Figure 1. (A) CD161 expression of T cell subsets. Gamma delta T cells, iNKT cells, and MAIT cells express CD161 at a higher level compared to conventional T helper and T cytotoxic T cells. (B) FACS analysis of CD161 expressing in different human T cell subsets.
While innate T cells are distinctive as a subpopulation of T cells, they have other distinct features, which do not overlap. They share the absence of MHC I/II peptide restriction but differ in their respective antigen presentation modalities. iNKT and MAIT cells respond to the MHC-like molecules, CD1d and MR1, respectively, while CD1c can present antigens to γδ T cells. The nature of the antigens recognized by innate T cells is also diverse and broadly non-overlapping involving metabolites, bacterial products, and lipids. iNKT cells have been principally shown to respond to glycolipids, γδ T cells are potently activated by (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate (HMBPP), and MAIT cells can be activated by riboflavin metabolites – reduced 6-hydroxy methyl-8-d-ribitylumazine (rRL-6-CH2OH), as well as folic acid metabolite, 6-formyl pterin (6FP). Finally, the sites of development, residence, and frequency within the T cell poll are distinctive and summarized in Table 1.
iNKT Cells
Invariant NKT cells are one of the most well-studied innate T cell populations (6–8). These cells are defined by their semi variant TCR, CD1d antigen restriction, and glycolipid recognition. Numerous studies have been undertaken with these cells following the discovery of their specific ligand, alpha-galactosylceramide, in 1997 (7, 9–11). Over the subsequent years, a range of endogenous and exogenous lipid antigens has been identified, which may change the effector responses of this innate T cell population (12–16). This cell population is also notable for its expression of previously considered NK cell specific markers such as CD161, which has subsequently been recognized on other innate T cell populations (17).
Invariant NKT cells develop in the thymus and are present at a very low number in most tissues. They are selected by CD1d, which is expressed on double-positive (CD4, CD8) thymocytes, through the recognition of endogenous lipids. In the thymus, iNKT cells acquire a memory/effector phenotype prior to exiting to the circulation. Recent studies have suggested that post-thymic education is required for iNKT cells to become fully mature and achieve functional competency (18, 19). In human peripheral blood, approximately 0.01–1% of the T lymphocytes are iNKT cells, characterized by their hallmark TCR-invariant chain Vα24-Jα18 and variant Vβ11; and Vα14-Jα18 in mice with a limited number of β chains, including Vβ8.2, Vβ7, and Vβ2 (20). Despite their presence in relatively low numbers in humans, iNKT cells can be very effective in early host defense mechanisms and are involved in a variety of disease settings (6, 21–23). A key feature of iNKT cells is their rapid release of a wide array of cytokines and chemokine following ligand activation (17). This plays an important role in their early effector and regulatory properties. Our understanding of the importance of iNKT cells is largely based on disease studies undertaken in iNKT deficient mice (24). Previous studies have shown that iNKT cells play an important role in the detection of various pathogens, including Pseudomonas aeruginosa, Streptococcus pneumoniae, Salmonella typhimurium, Mycobacterium tuberculosis, Listeria monocytogenes, and Borrelia burgdorferi (1, 25, 26). In addition to bacterial infections, iNKT cells have also been found to play an important role in viral infections, including influenza, cytomegalovirus, and coxsackie B3 viral diseases (25, 27). Finally, they also play an important role in tumor immunity (28) and autoimmune disease (22). In human studies, a link between defects in iNKT cells may lead to susceptibility to certain infectious diseases, such as tuberculosis (29, 30), EBV (31–33), allergy (34), atherosclerosis (35), and immunodeficiency. (10, 36–38).
Role during Bacterial Infections
The presence of functional iNKT cells during bacterial sepsis has been shown in a number of different murine settings. In S. pneumoniae infection, a much higher level of bacteria were identified in the Jα18 knockout mice compared to the iNKT competent wild type mice, resulting in significant survival rate differences between the two strains (39). In the iNKT knockout mice, a defect was found in neutrophil recruitment to the lung together with a reduced production of neutrophil chemo-attractants, including TNF-alpha and MIP-2. A reconstitution of iNKT cells from wild-type mice to iNKT deficient mice was able to restore the production of TNF-alpha and MIP-2, leading to improved neutrophil and bacterial clearance (40). In a further bacterial infection caused by Chlamydia pneumoniae, accumulations of iNKT cells within the lung were visible within hours of acute infection, demonstrating IFN-gamma production at the site of infection (41). An extension of conventional bacterial challenge studies has recently been undertaken by Wong et al., which suggested that iNKT cells might play a role in control of bacterial infections associated with stoke. Compared to their WT little mate, iNKT deficient mice were found to be more susceptible to bacterial infection post transient midcerebral artery occlusion. This was related to the ability of iNKT cells to act as a suppressor for neurotransmitter release post-stroke, which is lost in iNKT deficient mice, making them more susceptible to the bacterial infection (42). In humans, several studies have established the link between iNKT cells and M. tuberculosis infection with both the function and number of iNKT cells reduced in these patients (43). Two distinct pathways have been proposed for iNKT cell activation during infection. They can either be directly activated through TCR-CD1d-glycoplid recognition or indirectly through their response to innate cytokines that are released from other innate cells.
Indirect Activation of iNKT Cells by Gram-Negative Bacteria
Early secretion of IFN-gamma can be induced by iNKT cells following an encounter with both Gram-negative and Gram-positive bacteria. Innate receptors that recognize bacterial signals have a crucial role in triggering the antigen presenting cells, which subsequently direct the activation of iNKT cells (9, 44–49). The activated antigen presenting cells stimulate the iNKT cells by signaling through toll like receptors (i.e., TLR4, TLR7, and TLR9) leading to the production of IL-12, also other inflammatory cytokines. Studies by De Libero and Paget have suggested that TLR signaling through APCs are not only important for cytokine production but also the accumulation of self-lipid antigen for CD1d presentation (47, 50). A study by Darmoise et al. showed that the TLR signaling triggered the accumulation of self-lipid including iGb3 in the lysosome, leading to an enhanced iNKT cell activation (51).
Direct Activation of iNKT Cells by Gram-Negative Bacteria
Another mechanism that allows iNKT cells to respond to bacterial infection occurs through the direct recognition of the glycosphingolipid in the cell wall of Gram-negative bacteria. One such example is Sphingomonas/Novosphingobium spp., where the glycosphingolipids present in the bacteria cell wall are alphagalacturonylceramides and alphaglucuronylceramides (52). These glycosphingolipids contain one sugar ring and have been showed to activate iNKT cells in vitro, while multi-sugar ring glycosphingolipids have not been able to activate iNKT cells in co-culture. Murine studies suggested that CD1dKO mice were able to clear infections with Sphingomonas/Novosphingobium as well as some other LPS-negative bacteria, but at a much slower rate compared to the wild type mice (45, 53, 54). This would suggest the iNKT cells are one of the major innate cell types involved in bacterial clearance and playing a major role in the early response.
γδ T Cells
γδ T cells are another group of innate T cells that have been found to play an important role during bacterial infections. Unlike conventional αβ T cells, γδ T cells do not usually express a CD4 or CD8 lineage marker and they do not require conventional antigen presentation via MHC class molecules (55). Different subtypes of γδ T cells have been described often identified by the different arrangement of their TCRs in early development. The differences in TCR arrangement directly influence their eventual principle tissue of residence. In human, the majority of the γδ T cells present in the peripheral blood express the TCR Vγ9Vδ2, whereas Vδ1 and Vδ3 TCR are primarily expressed at the mucosal surfaces. In mice, Vγ1 and Vγ4 are present in the lymphoid tissues; Vγ5 is found to be present in the skin; Vγ6 in the reproductive tract; and Vγ1, Vγ4, and Vγ6 present in the lung (56). A number of mechanisms have been described linking γδ cells and bacterial infections. Similar to iNKT cells, γδ are able to sense danger signals in both a TCR dependent and TCR independent way. γδ T cells can be activated by Class I like molecules such as T10/T22 (in mice) and members of CD1 family; they can also be activated by MHC-unrelated molecules such as viral glycoproteins and F1-ATPase complex in human (57–59). In addition to TCR recognition, γδ T cells also express pattern recognition receptor and receptor typically associated with NK cells.
γδ T cells may expand in the patient’s peripheral blood during bacterial infections with studies identifying up to 12% in listeriosis, 14% in tuberculosis, and 29% in brucellosis (60). Human γδ T cells respond to bacterial infections by recognizing (E)-4-hydroxy-3-methyl-but-2enyl pyrophosphate (HMBPP) derived from various bacteria. γδ T cells were shown to be particularly important in response to intracellular bacterial pathogens including M. tuberculosis and Legionella micdadei. In the case of L. micdadei, Vγ9Vδ2 T cells were found to be depleted from the circulation upon bacterial infection, followed by a sharp increase, then a slow decline over a 6-month-period (61). This dynamic change may indicate Vγ9Vδ2 might be important in contributing to the pathophysiological changes of Pontiac fever-like disease. A similar kinetic pattern is seen with M. tuberculosis infection, following the Vγ9Vδ2 T cell response to the metabolite IPP (62).
Early studies found that Vγ9Vδ2 T cells were the most important group that led to the eradication of bacteria (63, 64). Seminal studies identified the antigens involved in the recognition were intermediates in isoprenoid biosynthesis, namely (E)-4-hydroxy-3-methyl-but-2enyl pyrophsphate (HMBPP) (65). The level of HMBPP directly influences the magnitude of Vγ9Vδ2 T cell activation and proliferation (66). Recently, a major breakthrough in discovering the mechanisms for activating the Vγ9Vδ2 T cells was made by Bonneville and Scotet’s group. They identified that a member of butyrophilin molecule family CD277 played a crucial role during γδ T cells activation (67–71).
γδ T cells have also been found to be able to promote self-activation through cell to cell interaction (72). However, it was demonstrated that the self-activation mechanism is not as effective as formal presentation through antigen presenting cells (73–75). One important aspect of γδ T cells is that they can trigger the maturation of dendritic cells. Devilder et al. showed that Vγ9Vδ2 T cells can stimulate the maturation signal on mycobacterial infected DCs, through a Fas–Fas ligand interaction (76) and/or TCR-CD1 contact (77). Other than dendritic cells, γδ T cells have also been found be important in macrophage recruitment. During infection with listeriosis, γδ T cells were found to be a key player in controlling the production of key macrophage chemo attractants (78). Skeen et al. also showed that macrophages failed to undergo maturation in the absence of γδ T cells (79). Direct engagement of γδ T cells may facilitate pathogen clearance through their production of bacteriostatic and lytic molecules, such as granulysin and defensins. During Staphylococcus aureus respiratory infection, γδ T cells sense the dysregulation of the mevalonate pathway within the infected cells. This leads to the activation and expansion of γδ T cells, in particular, Vγ9Vδ2 T cells. The active γδ T cells then produce cytokines such as IL-17, which leads to airway protection. γδ T cells also play a role during M. tuberculosis infection, producing a variety of cytokines including IFN- γ, TNF-α, and IL-17. IFN-γ and TNF-α play are essential in host protection against M. tuberculosis enabling granuloma formation and disease containment.
MAIT Cells
Mucosal-associated invariant T cells are the newest members of the innate T cell family. They were first described by Tilloy et al. (80) and represent the most abundant innate T cells in humans. They express a canonical Va7.2-Ja33 chain in humans and the orthologous Va19-Ja33 in mice. The development of MAIT cells is parallel to the development of iNKT cells and both express the transcription factor ZBTB16 (81). In adults, they display an effector phenotype, whereas MAIT cells possess a naïve phenotype in cord blood. In both cord and adult blood, MAIT cells express CD161, IL-18Ra, CCR6, and about 50% of the MAIT cells express the T cells co-receptor CD8 (82–84). Recent studies also show that MAIT cells express the ABC binding cassette (ABC) B1 drug resistance transporter (85, 86). MAIT cells have a further unique antigen recognition system recognizing a MHC Class I related molecule (MR1), which is able to present bacterial derived ligand. Study by Kjer-Nielsen et al. showed that 6-formyl pterin (6-FP), a metabolite on the folic acid pathway, could stabilize the MR1 molecule but failed to activate the cells. Full activation of primary MAIT cells was achieved with ligand reduced 6-hydroxymethyl-8-d-ribityllumazine (rRL-6-CH2OH), a riboflavin metabolite. Related products 7-hydroxy-6-methyl-8-d-ribityllumazine (RL-6-Me-7-OH) and 6,7-dimethyl-8-d-ribityllumazine (RL-6.7-diMe) have also shown similar agonistic activity for MAIT cells, leading to the rapid production of cytokines (87). In recent years, studies on MAIT cells have associated their number and function with diverse of disease settings, including bacterial infections and autoimmune disorder.
The first hint that MAIT cells have anti-bacterial activities was described in 2010, where studies by Gold et al. and Le Bourhis et al. showed that MAIT cells could recognize a range of bacteria species through MR1 (88, 89). In the study by Gold et al., MAIT cells could respond to M. tuberculosis even in unexposed individuals. They further showed that MAIT cells responded to Salmonella enteria, Escheichia coli, and S. aureus infected APC (90). Le Bourhis et al. showed that MAIT cells could MAIT cells are able to respond to a wide array of bacteria including Gram-positive Bacteria S. aureus, Staphylococcus epidermidis, Lactobacillus acidophilus, and Gram-negative Bacteria E.coli, Klebsiella pneumonia, Pseudomonas aeruginosa, and Mycobacterium abscessus. Importantly, some bacterial species were shown not to activate MAIT cells, namely Enterococcus faecalis and Streptococcus pyogenes, suggesting a novel specificity. The importance of their role in bacterial defense was suggested by a study by Georgel et al. demonstrating that during the Klebsiella pneumonia infection, MR1 deficient mice succumbed to disseminated infection whereas the WT mice achieved full bacteria clearance within 2 days (91). Similarly, in the Va19 transgenic mice, enhanced control of the E.coli and M. abscessus infection were observed. Further studies performed by Chua et al. and Meierovics et al. also showed that MAIT cells were needed in the early control of Mycobacterium bovis, BCG, and Francisella tularensis infection (92, 93). In humans, how MAIT cells play a role in infectious disease is less well understood. A number of studies have associated the frequency of MAIT cells in different infectious diseases (94). MAIT cell numbers were found to be lower in peripheral blood of patients with M. tuberculosis infection (95). Also, in a study of critically ill septic and non-septic patients, the patients with severe bacterial infections, but not viral infections, had a much lower MAIT count compared to healthy controls (96).
One of the most well-studied examples of MAIT cells in bacterial infection is during Salmonella infection (97, 98). Upon activation, MAIT cells produce IFN-gamma, TNF α, and IL-17. These cytokines have been shown to be critical in controlling Salmonella infections, with IL-17 preventing the dissemination of infection (99). MAIT cells may also play a role during Salmonella infection through their early cytotoxic activity (100), although further studies are needed as MAIT cells were not able to directly kill Salmonella infected cell lines (94, 101).
Over the last 5–10 years, there has been advancement in the understanding and description of unconventional T cells. These studies demonstrate that unconventional T cells do indeed play an important role during bacteria infection and contribute the ability of host organism to clear and control certain bacterial infections (Figure 2). These cells are able to efficiently traffic to the sites of inflammation, and initiate rapid responses by means of cytokine production and cytotoxic activities. Further studies will elucidate the molecular details of this cellular control suggesting novel approaches to how we may harness these cells through therapeutic vaccination and pharmaceutical manipulations.
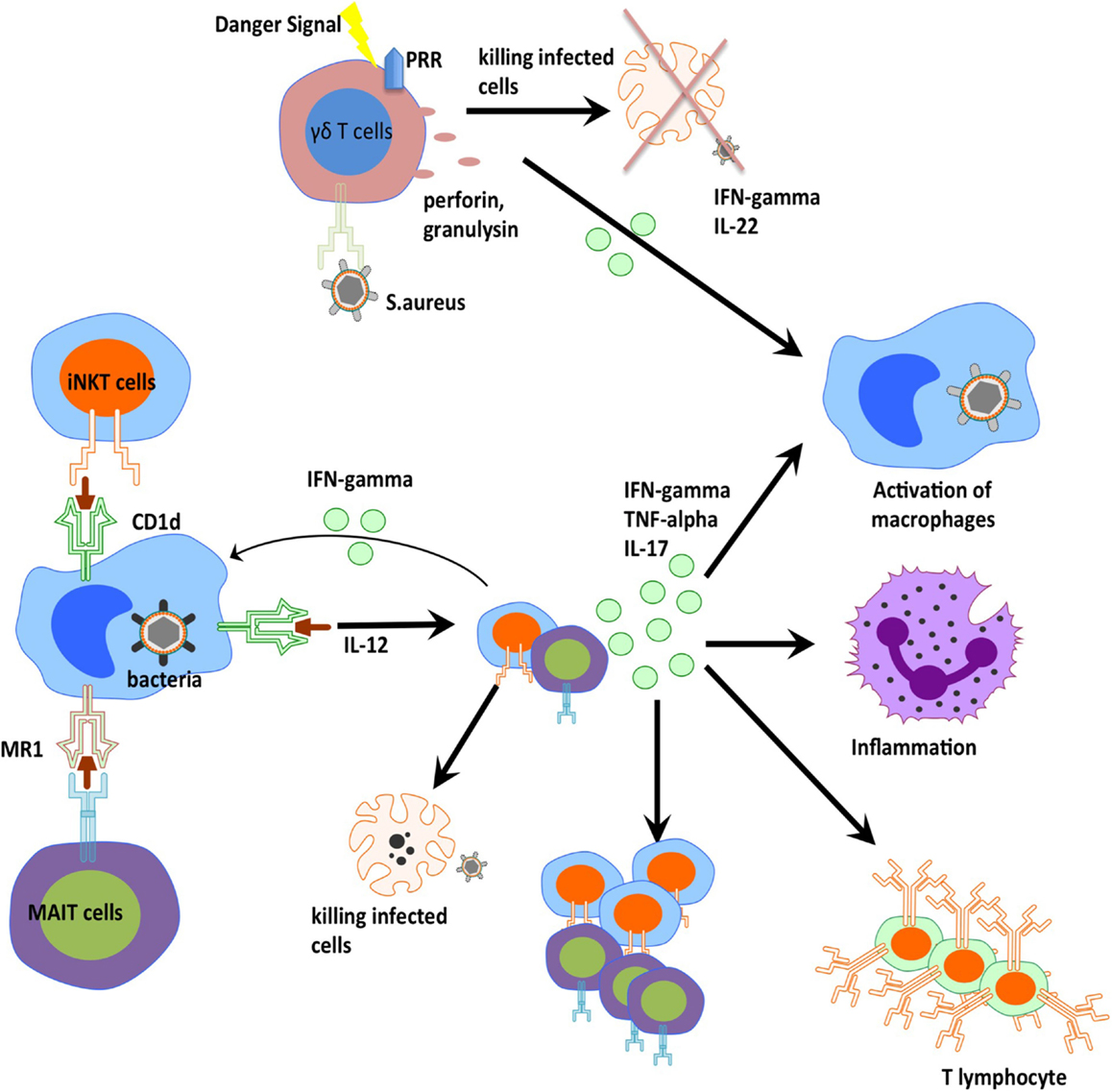
Figure 2. Orchestration of innate T cells in anti-bacterial immunity. iNKT cells, MAIT cells, and gamma delta T cells play an important role in anti-bacterial defense through cytokine production, perforin release, and cross-talk to other innate and adaptive cells.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
YG and APW were supported by Cancer Research UK and the Experimental Cancer Medicine Centre, Southampton.
References
1. Ivanov S, Paget C, Trottein F. Role of non-conventional T lymphocytes in respiratory infections: the case of the pneumococcus. PLoS Pathog (2014) 10(10):e1004300. doi:10.1371/journal.ppat.1004300
2. Fergusson JR, Smith KE, Fleming VM, Rajoriya N, Newell EW, Simmons R, et al. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep (2014) 9(3):1075–88. doi:10.1016/j.celrep.2014.09.045
3. Lanier LL, Chang C, Phillips JH. Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol (1994) 153(6):2417–28.
4. Battistini L, Borsellino G, Sawicki G, Poccia F, Salvetti M, Ristori G, et al. Phenotypic and cytokine analysis of human peripheral blood gamma delta T cells expressing NK cell receptors. J Immunol (1997) 159(8):3723–30.
5. Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol (2009) 7(3):e54. doi:10.1371/journal.pbio.1000054
6. Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol (2007) 25:297–336. doi:10.1146/annurev.immunol.25.022106.141711
7. Cohen NR, Garg S, Brenner MB. Antigen presentation by CD1 lipids, T cells, and NKT cells in microbial immunity. Adv Immunol (2009) 102:1–94. doi:10.1016/S0065-2776(09)01201-2
8. Kinjo Y, Kitano N, Kronenberg M. The role of invariant natural killer T cells in microbial immunity. J Infect Chemother (2013) 19(4):560–70. doi:10.1007/s10156-013-0638-1
9. Brigl M, Tatituri RV, Watts GF, Bhowruth V, Leadbetter EA, Barton N, et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med (2011) 208(6):1163–77. doi:10.1084/jem.20102555
10. Carvalho KI, Melo KM, Bruno FR, Snyder-Cappione JE, Nixon DF, Costa-Carvalho BT, et al. Skewed distribution of circulating activated natural killer T (NKT) cells in patients with common variable immunodeficiency disorders (CVID). PLoS One (2010) 5(9):e12652. doi:10.1371/journal.pone.0012652
11. Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol (2011) 12(10):966–74. doi:10.1038/ni.2096
12. Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruìz Y, Bachi A, Kronenberg M, et al. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol (2005) 175(2):977–84. doi:10.4049/jimmunol.175.2.977
13. Horikoshi M, Goto D, Segawa S, Yoshiga Y, Iwanami K, Inoue A, et al. Activation of Invariant NKT cells with glycolipid ligand alpha-galactosylceramide ameliorates glucose-6-phosphate isomerase peptide-induced arthritis. PLoS One (2012) 7(12):e51215. doi:10.1371/journal.pone.0051215
14. Joyce S, Girardi E, Zajonc DM. NKT cell ligand recognition logic: molecular basis for a synaptic duet and transmission of inflammatory effectors. J Immunol (2011) 187(3):1081–9. doi:10.4049/jimmunol.1001910
15. Park JJ, Lee JH, Ghosh SC, Bricard G, Venkataswamy MM, Porcelli SA, et al. Synthesis of all stereoisomers of KRN7000, the CD1d-binding NKT cell ligand. Bioorg Med Chem Lett (2008) 18(14):3906–9. doi:10.1016/j.bmcl.2008.06.036
16. Voyle RB, Beermann F, Lees RK, Schümann J, Zimmer J, Held W, et al. Ligand-dependent inhibition of CD1d-restricted NKT cell development in mice transgenic for the activating receptor Ly49D. J Exp Med (2003) 197(7):919–25. doi:10.1084/jem.20021615
17. Carreno LJ, Kharkwal SS, Porcelli SA. Optimizing NKT cell ligands as vaccine adjuvants. Immunotherapy (2014) 6(3):309–20. doi:10.2217/imt.13.175
18. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science (2012) 336(6080):489–93. doi:10.1126/science.1219328
19. Wingender G, Stepniak D, Krebs P, Lin L, McBride S, Wei B, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology (2012) 143(2):418–28. doi:10.1053/j.gastro.2012.04.017
20. Godfrey DI, Rossjohn J. New ways to turn on NKT cells. J Exp Med (2011) 208(6):1121–5. doi:10.1084/jem.20110983
21. Aggarwal A, Sharma A, Bhatnagar A. Bi(o)communications among peripheral blood fractions: a focus on NK and NKT cell biology in rheumatoid arthritis. Autoimmunity (2013) 46(4):238–50. doi:10.3109/08916934.2012.755959
22. Fletcher MT, Baxter AG. Clinical application of NKT cell biology in type I (autoimmune) diabetes mellitus. Immunol Cell Biol (2009) 87(4):315–23. doi:10.1038/icb.2009.5
23. Berzins SP, Smyth MJ, Baxter AG. Presumed guilty: natural killer T cell defects and human disease. Nat Rev Immunol (2011) 11(2):131–42. doi:10.1038/nri2904
24. Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol (2013) 13(2):101–17. doi:10.1038/nri3369
25. Paget C, Ivanov S, Fontaine J, Blanc F, Pichavant M, Renneson J, et al. Potential role of invariant NKT cells in the control of pulmonary inflammation and CD8+ T cell response during acute influenza A virus H3N2 pneumonia. J Immunol (2011) 186(10):5590–602. doi:10.4049/jimmunol.1002348
26. van der Sluijs KF, van der Poll T, Lutter R, Juffermans NP, Schultz MJ. Bench-to-bedside review: bacterial pneumonia with influenza – pathogenesis and clinical implications. Crit Care (2010) 14(2):219. doi:10.1186/cc8893
27. Tessmer MS, Fatima A, Paget C, Trottein F, Brossay L. NKT cell immune responses to viral infection. Expert Opin Ther Targets (2009) 13(2):153–62. doi:10.1517/14712590802653601
28. Berzofsky JA, Terabe M. The contrasting roles of NKT cells in tumor immunity. Curr Mol Med (2009) 9(6):667–72. doi:10.2174/156652409788970706
29. Sada-Ovalle I, Chiba A, Gonzales A, Brenner MB, Behar SM. Innate invariant NKT cells recognize Mycobacterium tuberculosis-infected macrophages, produce interferon-gamma, and kill intracellular bacteria. PLoS Pathog (2008) 4(12):e1000239. doi:10.1371/journal.ppat.1000239
30. Chackerian A, Alt J, Perera V, Behar SM. Activation of NKT cells protects mice from tuberculosis. Infect Immun (2002) 70(11):6302–9. doi:10.1128/IAI.70.11.6302-6309.2002
31. He Y, Xiao R, Ji X, Li L, Chen L, Xiong J, et al. EBV promotes human CD8 NKT cell development. PLoS Pathog (2010) 6(5):e1000915. doi:10.1371/journal.ppat.1000915
32. Yuling H, Ruijing X, Li L, Xiang J, Rui Z, Yujuan W, et al. EBV-induced human CD8+ NKT cells suppress tumorigenesis by EBV-associated malignancies. Cancer Res (2009) 69(20):7935–44. doi:10.1158/0008-5472.CAN-09-0828
33. Xiao W, Li L, Zhou R, Xiao R, Wang Y, Ji X, et al. EBV-induced human CD8(+) NKT cells synergise CD4(+) NKT cells suppressing EBV-associated tumours upon induction of Th1-bias. Cell Mol Immunol (2009) 6(5):367–79. doi:10.1038/cmi.2009.48
34. Eguchi T, Kumagai K, Kobayashi H, Shigematsu H, Kitaura K, Suzuki S, et al. Accumulation of invariant NKT cells into inflamed skin in a novel murine model of nickel allergy. Cell Immunol (2013) 284(1–2):163–71. doi:10.1016/j.cellimm.2013.07.010
35. Tupin E, Nicoletti A, Elhage R, Rudling M, Ljunggren HG, Hansson GK, et al. CD1d-dependent activation of NKT cells aggravates atherosclerosis. J Exp Med (2004) 199(3):417–22. doi:10.1084/jem.20030997
36. Fernandez CS, Chan AC, Kyparissoudis K, De Rose R, Godfrey DI, Kent SJ. Peripheral NKT cells in simian immunodeficiency virus-infected macaques. J Virol (2009) 83(4):1617–24. doi:10.1128/JVI.02138-08
37. Fujita K, Sandford AP, Kobayashi M, Hanafusa T, Herndon DN, Suzuki F. Role of natural killer T (NKT) cells lacking interleukin (IL)-4 producing abilities on the CC-chemokine ligand 2-associated herpes simplex virus type 1 infection in human severe combined immunodeficiency (SCID) mouse chimeras. Burns (2005) 31(2):145–52. doi:10.1016/j.burns.2004.09.006
38. Gao Y, Workman S, Gadola S, Elliott T, Grimbacher B, Williams AP. Common variable immunodeficiency is associated with a functional deficiency of invariant natural killer T cells. J Allergy Clin Immunol (2014) 133(5):1420–1428, 1428 e1421. doi:10.1016/j.jaci.2013.10.059
39. Kawakami K, Yamamoto N, Kinjo Y, Miyagi K, Nakasone C, Uezu K, et al. Critical role of Valpha14+ natural killer T cells in the innate phase of host protection against Streptococcus pneumoniae infection. Eur J Immunol (2003) 33(12):3322–30. doi:10.1002/eji.200324254
40. Stanic AK, De Silva AD, Park JJ, Sriram V, Ichikawa S, Hirabyashi Y, et al. Defective presentation of the CD1d1-restricted natural Va14Ja18 NKT lymphocyte antigen caused by beta-D-glucosylceramide synthase deficiency. Proc Natl Acad Sci U S A (2003) 100(4):1849–54. doi:10.1073/pnas.0430327100
41. Joyee AG, Qiu H, Wang S, Fan Y, Bilenki L, Yang X. Distinct NKT cell subsets are induced by different chlamydia species leading to differential adaptive immunity and host resistance to the infections. J Immunol (2007) 178(2):1048–58. doi:10.4049/jimmunol.178.2.1048
42. Wong CH, Jenne CN, Lee WY, Leger C, Kubes P. Functional innervation of hepatic iNKT cells is immunosuppressive following stroke. Science (2011) 334(6052):101–5. doi:10.1126/science.1210301
43. Kee SJ, Kwon YS, Park YW, Cho YN, Lee SJ, Kim TJ, et al. Dysfunction of natural killer T cells in patients with active Mycobacterium tuberculosis infection. Infect Immun (2012) 80(6):2100–8. doi:10.1128/IAI.06018-11
44. Paget C, Ivanov S, Fontaine J, Renneson J, Blanc F, Pichavant M, et al. Interleukin-22 is produced by invariant natural killer T lymphocytes during influenza A virus infection: potential role in protection against lung epithelial damages. J Biol Chem (2012) 287(12):8816–29. doi:10.1074/jbc.M111.304758
45. Mattner J, Debord KL, Ismail N, Goff RD, Cantu C III, Zhou D, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature (2005) 434(7032):525–9. doi:10.1038/nature03408
46. Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol (2003) 4(12):1230–7. doi:10.1038/ni1002
47. Paget C, Mallevaey T, Speak AO, Torres D, Fontaine J, Sheehan KC, et al. Activation of invariant NKT cells by toll-like receptor 9-stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity (2007) 27(4):597–609. doi:10.1016/j.immuni.2007.08.017
48. Salio M, Speak AO, Shepherd D, Polzella P, Illarionov PA, Veerapen N, et al. Modulation of human natural killer T cell ligands on TLR-mediated antigen-presenting cell activation. Proc Natl Acad Sci U S A (2007) 104(51):20490–5. doi:10.1073/pnas.0710145104
49. Paget C, Bialecki E, Fontaine J, Vendeville C, Mallevaey T, Faveeuw C, et al. Role of invariant NK T lymphocytes in immune responses to CpG oligodeoxynucleotides. J Immunol (2009) 182(4):1846–53. doi:10.4049/jimmunol.0802492
50. De Libero G, Moran AP, Gober HJ, Rossy E, Shamshiev A, Chelnokova O, et al. Bacterial infections promote T cell recognition of self-glycolipids. Immunity (2005) 22(6):763–72. doi:10.1016/j.immuni.2005.04.013
51. Darmoise A, Teneberg S, Bouzonville L, Brady RO, Beck M, Kaufmann SH, et al. Lysosomal alpha-galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity (2010) 33(2):216–28. doi:10.1016/j.immuni.2010.08.003
52. Van Kaer L, Parekh VV, Wu L. Invariant natural killer T cells as sensors and managers of inflammation. Trends Immunol (2013) 34(2):50–8. doi:10.1016/j.it.2012.08.009
53. Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature (2005) 434(7032):520–5. doi:10.1038/nature03407
54. Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol (2005) 35(6):1692–701. doi:10.1002/eji.200526157
55. Morita CT, Lee HK, Leslie DS, Tanaka Y, Bukowski JF, Marker-Hermann E. Recognition of nonpeptide prenyl pyrophosphate antigens by human gammadelta T cells. Microbes Infect (1999) 1(3):175–86. doi:10.1016/S1286-4579(99)80032-X
56. Bonneville M, O’Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol (2010) 10(7):467–78. doi:10.1038/nri2781
57. Kalyan S, Kabelitz D. Defining the nature of human gammadelta T cells: a biographical sketch of the highly empathetic. Cell Mol Immunol (2013) 10(1):21–9. doi:10.1038/cmi.2012.44
58. Russano AM, Bassotti G, Agea E, Bistoni O, Mazzocchi A, Morelli A, et al. CD1-restricted recognition of exogenous and self-lipid antigens by duodenal gammadelta+ T lymphocytes. J Immunol (2007) 178(6):3620–6. doi:10.4049/jimmunol.178.6.3620
59. Uldrich AP, Le Nours J, Pellicci DG, Gherardin NA, McPherson KG, Lim RT, et al. CD1d-lipid antigen recognition by the gammadelta TCR. Nat Immunol (2013) 14(11):1137–45. doi:10.1038/ni.2713
60. Wu YL, Ding YP, Tanaka Y, Shen LW, Wei CH, Minato N, et al. Gammadelta T cells and their potential for immunotherapy. Int J Biol Sci (2014) 10(2):119–35. doi:10.7150/ijbs.7823
61. Kroca M, Johansson A, Sjostedt A, Tarnvik A. V gamma 9V delta 2 T cells in human legionellosis. Clin Diagn Lab Immunol (2001) 8(5):949–54. doi:10.1128/CDLI.8.5.949-954.2001
62. Zheng J, Liu Y, Lau YL, Tu W. Gammadelta-T cells: an unpolished sword in human anti-infection immunity. Cell Mol Immunol (2013) 10(1):50–7. doi:10.1038/cmi.2012.43
63. Pechhold K, Wesch D, Schondelmaier S, Kabelitz D. Primary activation of V gamma 9-expressing gamma delta T cells by Mycobacterium tuberculosis. Requirement for Th1-type CD4 T cell help and inhibition by IL-10. J Immunol (1994) 152(10):4984–92.
64. Behr C, Poupot R, Peyrat MA, Poquet Y, Constant P, Dubois P, et al. Plasmodium falciparum stimuli for human gammadelta T cells are related to phosphorylated antigens of mycobacteria. Infect Immun (1996) 64(8):2892–6.
65. Arigoni D, Sagner S, Latzel C, Eisenreich W, Bacher A, Zenk MH. Terpenoid biosynthesis from 1-deoxy-D-xylulose in higher plants by intramolecular skeletal rearrangement. Proc Natl Acad Sci U S A (1997) 94(20):10600–5. doi:10.1073/pnas.94.20.10600
66. Espinosa E, Belmant C, Pont F, Luciani B, Poupot R, Romagné F, et al. Chemical synthesis and biological activity of bromohydrin pyrophosphate, a potent stimulator of human gamma delta T cells. J Biol Chem (2001) 276(21):18337–44. doi:10.1074/jbc.M100495200
67. Harly C, Guillaume Y, Nedellec S, Peigné CM, Mönkkönen H, Mönkkönen J, et al. Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human gammadelta T-cell subset. Blood (2012) 120(11):2269–79. doi:10.1182/blood-2012-05-430470
68. Luoma AM, Castro CD, Mayassi T, Bembinster LA, Bai L, Picard D, et al. Crystal structure of Vdelta1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human gammadelta T cells. Immunity (2013) 39(6):1032–42. doi:10.1016/j.immuni.2013.11.001
69. Silva A, Mount A, Krstevska K, Pejoski D, Hardy MP, Owczarek C, et al. The combination of ISCOMATRIX adjuvant and TLR agonists induces regression of established solid tumors in vivo. J Immunol (2015) 194(5):2199–207. doi:10.4049/jimmunol.1402228
70. Sandstrom A, Peigné CM, Léger A, Crooks JE, Konczak F, Gesnel MC, et al. The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vgamma9Vdelta2 T cells. Immunity (2014) 40(4):490–500. doi:10.1016/j.immuni.2014.03.003
71. Vavassori S, Kumar A, Wan GS, Ramanjaneyulu GS, Cavallari M, El Daker S, et al. Butyrophilin 3A1 binds phosphorylated antigens and stimulates human gammadelta T cells. Nat Immunol (2013) 14(9):908–16. doi:10.1038/ni.2665
72. Miyagawa F, Tanaka Y, Yamashita S, Minato N. Essential requirement of antigen presentation by monocyte lineage cells for the activation of primary human gamma delta T cells by aminobisphosphonate antigen. J Immunol (2001) 166(9):5508–14. doi:10.4049/jimmunol.166.9.5508
73. Brandes M, Willimann K, Moser B. Professional antigen-presentation function by human gammadelta T Cells. Science (2005) 309(5732):264–8. doi:10.1126/science.1110267
74. Shin S, El-Diwany R, Schaffert S, Adams EJ, Garcia KC, Pereira P, et al. Antigen recognition determinants of gammadelta T cell receptors. Science (2005) 308(5719):252–5. doi:10.1126/science.1106480
75. Sarikonda G, Wang H, Puan KJ, Liu XH, Lee HK, Song Y, et al. Photoaffinity antigens for human gammadelta T cells. J Immunol (2008) 181(11):7738–50. doi:10.4049/jimmunol.181.11.7738
76. Collins C, Wolfe J, Roessner K, Shi C, Sigal LH, Budd RC. Lyme arthritis synovial gammadelta T cells instruct dendritic cells via fas ligand. J Immunol (2005) 175(9):5656–65. doi:10.4049/jimmunol.175.9.5656
77. Leslie DS, Vincent MS, Spada FM, Das H, Sugita M, Morita CT, et al. CD1-mediated gamma/delta T cell maturation of dendritic cells. J Exp Med (2002) 196(12):1575–84. doi:10.1084/jem.20021515
78. DiTirro J, Rhoades ER, Roberts AD, Burke JM, Mukasa A, Cooper AM, et al. Disruption of the cellular inflammatory response to Listeria monocytogenes infection in mice with disruptions in targeted genes. Infect Immun (1998) 66(5):2284–9.
79. Skeen MJ, Freeman MM, Ziegler HK. Changes in peritoneal myeloid populations and their proinflammatory cytokine expression during infection with Listeria monocytogenes are altered in the absence of gamma/delta T cells. J Leukoc Biol (2004) 76(1):104–15. doi:10.1189/jlb.1103574
80. Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med (1999) 189(12):1907–21.
81. Seach N, Guerri L, Le Bourhis L, Mburu Y, Cui Y, Bessoles S, et al. Double-positive thymocytes select mucosal-associated invariant T cells. J Immunol (2013) 191(12):6002–9. doi:10.4049/jimmunol.1301212
82. Billerbeck E, Kang YH, Walker L, Lockstone H, Grafmueller S, Fleming V, et al. Analysis of CD161 expression on human CD8+ T cells defines a distinct functional subset with tissue-homing properties. Proc Natl Acad Sci U S A (2010) 107(7):3006–11. doi:10.1073/pnas.0914839107
83. Takahashi T, Dejbakhsh-Jones S, Strober S. Expression of CD161 (NKR-P1A) defines subsets of human CD4 and CD8 T cells with different functional activities. J Immunol (2006) 176(1):211–6. doi:10.4049/jimmunol.176.1.211
84. Maggi L, Santarlasci V, Capone M, Peired A, Frosali F, Crome SQ, et al. CD161 is a marker of all human IL-17-producing T-cell subsets and is induced by RORC. Eur J Immunol (2010) 40(8):2174–81. doi:10.1002/eji.200940257
85. Turtle CJ, Swanson HM, Fujii N, Estey EH, Riddell SR. A distinct subset of self-renewing human memory CD8+ T cells survives cytotoxic chemotherapy. Immunity (2009) 31(5):834–44. doi:10.1016/j.immuni.2009.09.015
86. Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood (2011) 117(4):1250–9. doi:10.1182/blood-2010-08-303339
87. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature (2012) 491(7426):717–23. doi:10.1038/nature11605
88. Gold MC, Eid T, Smyk-Pearson S, Eberling Y, Swarbrick GM, Langley SM, et al. Human thymic MR1-restricted MAIT cells are innate pathogen-reactive effectors that adapt following thymic egress. Mucosal Immunol (2013) 6(1):35–44. doi:10.1038/mi.2012.45
89. Le Bourhis L, Martin E, Péguillet I, Guihot A, Froux N, Coré M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol (2010) 11(8):701–8. doi:10.1038/ni.1890
90. Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol (2010) 8(6):e1000407. doi:10.1371/journal.pbio.1000407
91. Georgel P, Radosavljevic M, Macquin C, Bahram S. The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Mol Immunol (2011) 48(5):769–75. doi:10.1016/j.molimm.2010.12.002
92. Chua WJ, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun (2012) 80(9):3256–67. doi:10.1128/IAI.00279-12
93. Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci U S A (2013) 110(33):E3119–28. doi:10.1073/pnas.1302799110
94. Ussher JE, Klenerman P, Willberg CB. Mucosal-associated invariant T-cells: new players in anti-bacterial immunity. Front Immunol (2014) 5:450. doi:10.3389/fimmu.2014.00450
95. Wong EB, Akilimali NA, Govender P, Sullivan ZA, Cosgrove C, Pillay M, et al. Low levels of peripheral CD161++CD8+ mucosal associated invariant T (MAIT) cells are found in HIV and HIV/TB co-infection. PLoS One (2013) 8(12):e83474. doi:10.1371/journal.pone.0083474
96. Grimaldi D, Le Bourhis L, Sauneuf B, Dechartres A, Rousseau C, Ouaaz F, et al. Specific MAIT cell behaviour among innate-like T lymphocytes in critically ill patients with severe infections. Intensive Care Med (2014) 40(2):192–201. doi:10.1007/s00134-013-3163-x
97. Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med (2013) 210(11):2305–20. doi:10.1084/jem.20130958
98. Reantragoon R, Kjer-Nielsen L, Patel O, Chen Z, Illing PT, Bhati M, et al. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med (2012) 209(4):761–74. doi:10.1084/jem.20112095
99. Nauciel C, Espinasse-Maes F. Role of gamma interferon and tumor necrosis factor alpha in resistance to Salmonella typhimurium infection. Infect Immun (1992) 60(2):450–4.
100. Lo WF, Ong H, Metcalf ES, Soloski MJ. T cell responses to Gram-negative intracellular bacterial pathogens: a role for CD8+ T cells in immunity to Salmonella infection and the involvement of MHC class Ib molecules. J Immunol (1999) 162(9):5398–406.
Keywords: iNKT cells, MAIT cells, gamma delta T cells, innate T cells, bacterial infections
Citation: Gao Y and Williams AP (2015) Role of innate T cells in anti-bacterial immunity. Front. Immunol. 6:302. doi: 10.3389/fimmu.2015.00302
Received: 13 April 2015; Accepted: 27 May 2015;
Published: 11 June 2015
Edited by:
S. M. Mansour Haeryfar, Western University, CanadaReviewed by:
Paul Klenerman, University of Oxford, UKDieter Kabelitz, Christian-Albrechts University Kiel, Germany
Copyright: © 2015 Gao and Williams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yifang Gao, Cancer Sciences Unit, Southampton General Hospital, Somers Cancer Research Building, Mail Point 824, Hampshire, Southampton, UK, yg13g11@soton.ac.uk
 Yifang Gao
Yifang Gao Anthony P. Williams1,2
Anthony P. Williams1,2
