- Division of Gastroenterology, Infectious Diseases and Rheumatology, Medical Department 1, Charité – Universitätsmedizin Berlin, Berlin, Germany
Crohn’s disease (CD) is a chronic, relapsing inflammatory disorder that can occur anywhere along the gastrointestinal tract. The precise etiology of CD is still unclear but it is widely accepted that a complex series of interactions between susceptibility genes, the immune system and environmental factors are implicated in the onset and perpetuation of the disease. Increasing evidence from experimental and clinical studies implies the intestinal microbiota in disease pathogenesis, thereby supporting the hypothesis that chronic intestinal inflammation arises from an abnormal immune response against the microorganisms of the intestinal flora in genetically susceptible individuals. Given that CD patients display changes in their gut microbiota composition, collectively termed “dysbiosis,” the question raises whether the altered microbiota composition is a cause of disease or rather a consequence of the inflammatory state of the intestinal environment. This review will focus on the crosstalk between the gut microbiota and the innate immune system during intestinal inflammation, thereby unraveling the role of the microbiota in CD pathogenesis.
Introduction
Crohn’s disease (CD) is a chronic, relapsing inflammatory disorder affecting the gastrointestinal tract and together with ulcerative colitis (UC) commonly included in the collective term inflammatory bowel diseases (IBD). Inflammation associated with CD is characterized by a discontinuous, transmural pattern, and can affect any part of the gastrointestinal tract (1). Although the precise etiology of CD remains unclear, several factors are believed to play a role in its development and progression. Given the results from genome-wide association studies (GWAS), it is undisputed that genetic susceptibility plays an important role in disease development (2, 3). Among immune-related conditions, CD is special in that the genetic contribution to disease is high with concordance rates up to 50% among monozygotic twins (4, 5). However, in countries that have adopted a “modernized” lifestyle the incidence rates of IBD have steeply increased in the last few decades. These epidemiological observations clarify that the host genotype alone accounts for a significant but limited proportion of CD risk and rather underline the multifactorial nature of the disease (6–10). The role of the intestinal microbiota in disease pathogenesis has become more and more appreciated. Evidence from genetic, immunological, and microbial studies implicates that chronic intestinal inflammation results from a dysregulated immune response toward components of the commensal intestinal microbiota in genetically susceptible individuals (11). This perception is supported by the identification of several risk loci associated with CD, including genes involved in intracellular processing and killing of bacteria (e.g., NOD2, ATG16L1, IRGM) (12, 13).
Starting with the intestinal lumen, the first part of this review will focus on the intestinal microbiota composition and its changes during inflammation. In the following sections, we will then take a closer look at deeper layers of the intestinal tissue to dissect the interplay of the intestinal microbiota with cells of the host immune system.
Human Intestinal Microbiota
Intestinal Microbiota – Composition
The assembly of the human gut microbiota starts during birth with Bifidobacterium and Lactobacillus derived from the vaginal canal and breast milk displaying the microorganisms that initially colonize the human intestinal tract (14). During adolescence, phylogenetic diversity of the microbial community increases leading to a complex, diverse, and dynamic microbiota. The adult human gastrointestinal tract contains an abundant microflora, including bacteria, archaea, eukarya, and viruses (15). Recent development of culture-independent molecular profiling methods has greatly advanced our understanding of the microbiome (16). Nowadays, the composition of microbial communities is typically evaluated by targeting the bacterial 16S ribosomal RNA gene as a phylogenetic marker (15). These culture-independent molecular techniques indicate that the human microbiota contains about 200 strains of bacteria comprising over 100 different bacterial species, dominated by just a few phyla (16). Even though these techniques revealed a high level of variability between individuals at the bacterial species level, Firmicutes and Bacteroidetes represent the predominant phyla across all vertebrates, representing over 90% of all intestinal bacteria. It is assumed that the individual composition of the microbiota is relatively stable over time, a term called “resilience.” Factors that promote microbial diversity in healthy adults include diet, environment, gender, and genetics among others (17, 18). Studies focusing on gut microbiota profiles of adult humans with varying degrees of genetic relatedness effectively demonstrated the impact of genetic and environmental factors on gut microbiota development. The intestinal microbiota composition of monozygotic twins indicated a high degree of similarity, but was yet distinct. Individuals who were living in the same environment and shared similar eating habits showed the least similarity, while siblings displayed an increased similarity in their species profile (1, 19). Recent studies in nucleotide-binding oligomerization domain 2 (Nod2)-deficient mice and humans carrying NOD2 variants point to an essential role of Nod2 for the development and composition of the host microbiota. Nod2-deficient mice displayed an increased load of commensal microbiota and an altered microbiota composition. Additional studies in weaning mice illustrated that NOD2 may affect the microbial community early in life. Furthermore, the substantial changes in the microbiota composition caused by Nod2 deficiency was paralleled by an increased colitis severity following chemically induced injury. Subsequent co-housing and cross-feeding experiments revealed the transmissibility of the observed genotype-dependent disease risk (20). Taken together, these studies clearly indicated that the host genotype has a lasting effect on the intestinal microbiota composition and vice versa the intestinal microbiota is capable of determining the host phenotype.
Intestinal Microbiota – Shaping the Gastrointestinal Immune System
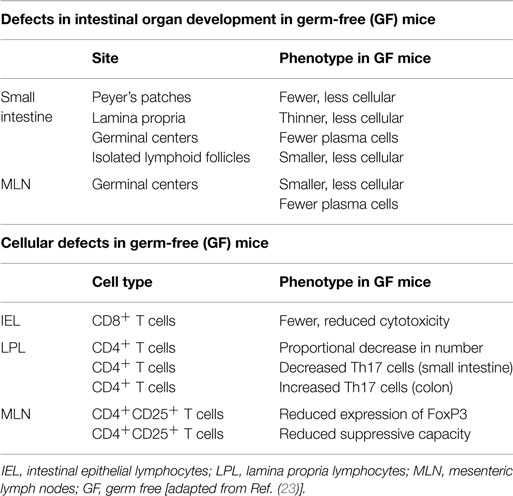
In health, the relationship between the host and the intestinal microbiota provides mutual benefits. On the one hand, the microbiota benefits from the nutrient-rich environment of the gut paving the way for the establishment of a relatively stable ecosystem. In turn, the intestinal microbiota enriches the host with vital functions that the host itself cannot perform. The intestinal microbiota is substantial for mucosal barrier function, affects the development of the mucosal immune system, and is essential for a number of physiological metabolic processes as described further on (21). The profound effects of the commensal microbiota on intestinal and immune cell development have best been highlighted by the engraving phenotype of germ-free (GF) mice. One of the first deficiencies observed in GF mice was a profound reduction of secretory immunoglobulin A (IgA) levels in the intestine. Subsequent mono-association of these mice with various bacteria was shown to lead to an increased IgA expression (22). In addition to numerous defects in antibody production, GF mice display various morphological tissue defects in their intestines [Table 1; (23)]. These developmental impairments are attenuated following the introduction of gut bacteria, once more illustrating the indispensable connection between the ultrastructural development of the intestine and the commensal microbiota. Next to defects in intestinal organ development, investigations on GF mice also revealed cellular defects in intestinal epithelial and lamina propria lymphocytes as well as in mesenteric lymph nodes (Table 1). Normal functioning of intestinal epithelial cells, including the expression of microbial recognition receptors, defensins, and antimicrobial peptides (AMPs), was shown to be impaired in GF animals compared to their conventionally raised counterparts (23).
Detailed investigations on certain members of the intestinal microbiota served to unravel mechanisms by which commensal bacteria induce immune tolerance. For example, investigations on segmented filamentous bacteria (SFB) revealed that these Gram-positive bacteria are sufficient to promote T helper 17 cells (Th17) development in the small intestinal lamina propria (24). In addition, colonization of GF mice with SFB resulted in an increased production of serum amyloid A in the terminal ileum, which in turn enhanced IL-6 and IL-23 production by lamina propria dendritic cells (DCs), thereby stimulating a Th17 inducing environment (24). Th17 effector cytokines enhance epithelial cell tight junctions, induce mucin production, and have been associated with induction of AMPs. Colonization of GF mice with SFB resulted in the induction of multiple AMP genes, for example, RegIIIγ (24). Even though Th17 cells are crucial for protecting the host against pathogenic infection, it should be noted that these cells also display an inflammatory potential as observed in different murine models of autoimmune diseases (25, 26). Recently, certain strains within Clostridia clusters XIVa, IV, and XVIII were shown to induce regulatory T cell (Treg) responses in the colon (27). Another prominent human commensal, Bacteroides fragilis, was found to direct the development of FoxP3+ Tregs via the immunomodulatory molecule polysaccharide A. Monocolonization of GF mice with B. fragilis was accompanied by an increased suppressive Treg capacity and the induction of an anti-inflammatory cytokine profile emerging from FoxP3+ T cells in the gut (28).
Intestinal Microbiota – Metabolic Functions
Next to their ability to promote immune system development and maturation as outlined above, the intestinal microbiota enriches the host with metabolic functions by synthesizing vitamins and degrading complex indigestible dietary carbohydrates and proteins. Fermentation of dietary fiber leads to the production of short chain fatty acids (SCFAs), primarily acetate, propionate, and butyrate (21, 29). The functions of SCFAs in promoting colonic health range from displaying a unique energy source for colonocytes to mediating anti-inflammatory and anti-tumorigenic effects (30). Therefore, it is easily comprehensible that conditions coming along with reduced SCFA-levels, including diversion colitis, fiber-free diet, or GF conditions, present with metabolic starvation and consecutive colonic atrophy (31). Among SCFAs, butyrate has received most attention for its effects on colonic health. Butyrate displays an anti-inflammatory effect by decreasing the expression of pro-inflammatory cytokines via inhibition of NF-κB activation and was shown to exert the ability to influence gene expression in the colon through histone deacetylase inhibition (32, 33). Moreover, butyrate elicits biological effects by binding to G protein-coupled receptors, namely GPR109A and GPR43. Butyrate serves as an endogenous agonist for GPR109A, expressed in the colonic epithelium, adipose tissue, and on immune cells, while GRP43 is activated by all three SCFAs (34). The activation of GPR109A results in a decrease of intracellular cAMP-levels accompanied by controlling electrolyte and water absorption, thereby potentially affecting the incidence of diarrhea (35). Activation of GPR109A by butyrate was found to impose anti-inflammatory properties in colonic macrophages and DCs and enabled them to induce the differentiation of Tregs and IL-10 producing T cells. Furthermore, GPR109A signaling was shown to be decisive for butyrate-mediated induction of IL-18 in the colonic epithelium (36). Comparative analysis in GF mice and their conventionally colonized counterparts revealed markedly reduced SCFA concentrations in the intestines of GF mice. Application of SCFAs via drinking water restored colonic Treg homeostasis and function in GF mice, suggesting that their lack of SCFAs may account at least partially for their immune defects, especially their reduced colonic Treg numbers (37). In a murine model of colitis, dextran sulfate sodium (DSS)-induced inflammation in GF mice was ameliorated through additional application of acetate. Conventionally colonized mice lacking Gpr43 showed a markedly increased inflammatory response following DSS application as compared to wild-type (wt) mice. Subsequent acetate application via drinking water resulted in a decrease of colonic inflammation but only in wt mice. Additional acetate application in Gpr43−/− mice did not lead to alterations in the severity of inflammation indicating that the protective effect of acetate seen in wt mice occurred through binding to GPR43 (38). Most recently, Mackay and colleagues provided evidence that diet deficient or low in fiber content exacerbates colitis induced by DSS, whereas a high-fiber diet was shown to exert a protective effect. Moreover, it was demonstrated that fiber mediates its protective properties via activating the NLRP3 inflammasome (39).
Given the aforementioned tremendous metabolic functions of the intestinal microbiota, the question raises whether inflammation in peripheral tissues is also influenced by intestinal microbiota-derived metabolites. Recently, Marsland and colleagues found evidence that dietary fermentable fiber and SCFAs are capable of shaping the immunological environment not only in the murine intestine but also in the lung (40). Oral administration of a high-fiber diet was accompanied by increased circulating SCFA levels and concomitant protection against allergic inflammation in the lung following exposure to house mite extract (HM) through intranasal administration. By contrast, feeding mice with a low-fiber diet resulted in decreased SCFA levels and an increase of allergic airway disease in response to HM application. Accordingly, histological sections revealed enhanced eosinophilic and lymphocytic infiltrates in the airways of mice fed a low-fiber diet as well as enhanced concentrations of IL-4, IL-5, IL-13, and IL-17A in lung tissue homogenates. The detected differing outcome in response to HM exposure underlines the impact of SCFAs on shaping the immunological environment in the lung, thereby impinging on the severity of allergic inflammation.
Intestinal Microbiota in Disease – Changes in the Gut Microbial Ecosystem
Changes in the microbiota composition display a hallmark of CD, commonly described as dysbiosis. Individuals with IBD have been characterized by marked qualitative and quantitative changes in their microbiota composition (41). Several culture-dependent and -independent analyses focusing on the microbiota profile of CD patients revealed less complex microbiota profiles and higher numbers of mucosa-associated bacteria compared to healthy individuals (41–45). Metagenomic-based studies have reported a reduction in members of the phyla Bacteroidetes and Firmicutes in patients suffering from CD or UC (41, 46, 47). Among members of the Firmicutes phyla, significant reductions in the butyrate-producing bacterium Faecalibacterium prausnitzii (F. prausnitzii) have been observed repeatedly in CD patients (7, 48). This bacterium displays anti-inflammatory properties, which have been intensively studied both in vitro and in vivo. Stimulation of peripheral blood mononuclear cells with F. prausnitzii induced very low levels of the pro-inflammatory cytokines IFNγ and IL-12 paralleled by high levels of IL-10 (48). These in vitro effects were confirmed in vivo by using a 2,4,6-trinitrobenzenesulphonic acid (TNBS)-induced model of colitis. Oral administration of F. prausnitzii markedly reduced the severity of TNBS colitis and tended to rectify the dysbiosis associated with colitis in this model. Follow-up studies after ileal resection provided evidence that a low proportion of F. prausnitzii on resected ileal mucosa of CD patients was associated with an increased risk of endoscopic recurrence after 6 months.
The changes in abundance and biodiversity of intestinal microbiota during inflammation are further characterized by an increase of members associated with the Proteobacteria and Actinobacteria phyla. CD patients were shown to harbor increased loads of Enterobacteriaceae, in particular Escherichia coli belonging to the taxonomic lineages B2 and D (49). Adherent-invasive E. coli (AIEC) pathovar has been commonly identified in the intestinal mucosa of patients with CD, particularly associated with ileal mucosal lesions (50, 51). Isolates of these certain strains of E. coli were shown to not only adhere to epithelial cells but also to invade and replicate intracellularly. Moreover, AIEC is able to survive and replicate within macrophages without triggering host cell death accompanied by the release of large amounts of tumor necrosis factor (TNF)-α (52). Compared to healthy controls, mucosal biopsy specimens from CD patients displayed a 10-fold higher presence of bacteria that penetrate the mucus layer. These results from fluorescent in situ hybridization analyses indicate that in CD the microbiota might have closer contact with the mucosa, a hypothesis that will be discussed in a later section. However, this finding could be explained by the increased numbers of mucolytic bacteria, such as Ruminococcus gnavus and Ruminococcus torques, observed in macroscopically and histologically unaffected intestinal epithelium of CD patients (53). In 2011, the so far largest cohort study provided a detailed description of the microbiome in CD patients (54). A lower occurrence of F. prausnitzii, Bifidobacterium adolescentis, Dialister invisus, and an uncharacterized species belonging to Clostridium cluster XIVa as well as a higher number of R. gnavus characterized the dysbiosis signature of CD patients (54). From a metabolic point of view, this comes along with an increased mucolytic and reduced butyrate-producing capacity. Moreover, this study not only confirmed previous findings but also propounded that these changes were markedly characteristic for the disease as this profile was not found in unaffected relatives of analyzed patients.
Intestinal Microbiota in the Pathogenesis of Crohn’s Disease
Experimental and Clinical Evidence
Over the last decades, a consistent body of evidence has accumulated supporting the role of the intestinal microbiota in precipitating IBD. Studies in IL-2- and IL-10-deficient mice were one of the first highlighting the role of the intestinal microbiota in the induction and perpetuation of chronic inflammation. Spontaneous colitis observed in IL-2 deficient mice when raised under specific-pathogen-free (SPF) conditions was unverifiable in animals with the same genetic background bred under GF conditions. Similarly, IL-10-deficient animals developed an attenuated inflammation with regard to disease severity and expansion when kept in a facility with a defined microbial environment (55, 56). Subsequently, Tbet-deficient mice were shown to develop chronic colitis with a histopathological similarity to human UC. The study revealed also a correlation of the presence of Proteus mirabilis and Klebsiella pneumoniae and colitis in these animals. Furthermore, healthy wt mice developed chronic colitis upon co-housing with T-bet−/−/Rag2−/− mice (TRUC) or following gavage of feces from TRUC mice (57, 58). These data indicate that loss of T-bet influences bacterial populations to become colitogenic and that colitis is transferable to genetically intact hosts. More recently, studies in TNFdelta ARE mice provided clear experimental evidence for the causal role of bacterial dysbiosis in the development of chronic small intestinal inflammation. Deletion in the TNF adenosine–uracil (AU)-rich elements (ARE) leads to TNF-driven spontaneous small intestinal inflammation, characterized by both transmural manifestation and a predominant ileal involvement, thereby mimicking key features of human CD pathology (59). Haller and colleagues assessed the impact of intestinal bacteria in this model of ileitis by using different hygienic conditions (GF, SPF, and conventional housing) as well as antibiotics. CD-like ileitis development was completely absent in GF mice. Antibiotic treatment resulted in an amelioration of disease severity and a recurrence of inflammation was observed following the relapse of microbiota composition. Moreover, transfer of dysbiotic cecal microbial communities from SPF–TNFdelta ARE mice into GF recipients resulted in the development of ileitis, mimicking inflammation severity of corresponding donors (60). These aforementioned studies just provide examples for the experimental evidence that has accumulated over the last years.
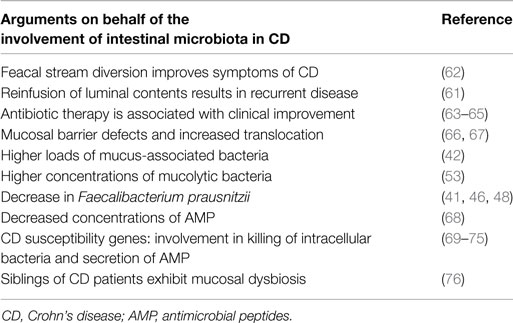
Event though CD can manifest anywhere along the human alimentary tract, it is mainly observed in areas containing the highest concentrations of bacteria (i.e., terminal ileum and colon). One of the first clinical references for the involvement of the intestinal microbiota in disease pathogenesis came from experiments showing that diversion of the fecal stream from an inflamed segment of the small intestine improved symptoms of CD patients. Furthermore, restoration of fecal stream and postoperative exposure of the neoterminal ileum to luminal contents induced inflammation, indicating that the microbiota acts as a trigger in postoperative recurrence of CD (61, 62). Numerous arguments in favor of the involvement of the human microbiota in disease pathogenesis have emerged over the last years (Table 2).
Although there has accumulated a large body of experimental and clinical evidence supporting the role of dysbiosis in the pathogenesis of CD, it is still up for discussion whether these alterations in the gut microbiota composition represent a cause or a consequence of chronic intestinal inflammation. Taking into account that the development of inflammation was shown to be dependent on the presence of a gut microbiota in different murine models and given the insufficient data to recommend probiotics for use in CD implies a pathogenetic role (55, 56, 77, 78). Investigations on fecal microbiota composition in first-degree relatives of patients with IBD have coined the term “predysbiosis.” As already outlined above, several studies indicated a significant impact of the host genotype on the composition of the intestinal microbiota. Relatives of CD patients are at much higher risk of developing CD as compared with the general population. The risk of falling ill is highest in first-degree relatives, especially siblings, and also extends to more distant relatives (54, 79). The relative risk of developing CD was shown to be over 30-fold higher in siblings of patients with CD compared to that of the general population (76). Moreover, an over 50-fold increase in the incidence of IBD was reported within multiply affected families (80, 81). Unaffected relatives of CD patients were found to have a different composition of their intestinal microbiota compared to healthy controls without familial predisposition (54). This subclinical dysbiosis observed in asymptomatic relatives was different from the dysbiosis detected in CD patients and was characterized by lower numbers of both Collinsella aerofaciens and an unspecified member of the E. coli – Shigella group as well as higher numbers of the mucolytic bacterium R. torques compared with healthy subjects. R. torques, a non-butyrate-producing member of the Clostridium cluster XIVa is capable of degrading gastrointestinal mucin. In contrast to the dysbiosis detected in CD patients, the subclinical dysbiosis in their relatives was not characterized by a diminished butyrate-producing capacity but enhanced mucin degradation might be assumed in these individuals. Given the differences between the observed dysbiosis in CD patients and the subclinical dysbiosis in unaffected relatives, the results from this cohort study suggest that a “predysbiosis” may precede the clinical manifestation of CD. Moreover, the increased numbers in mucolytic bacteria let one suppose that an enhanced mucin degradation capacity of the intestinal microbiota represents an interim step, leading from normobiosis to the investigated dysbiosis among the patient cohort (54). In a recently published study, Hedin and colleagues focused on the mucosal microbiota in healthy siblings of CD patients and described a mucosal dysbiosis characterized by a reduced diversity of core microbiota and a lower abundance of F. prausnitzii in these at risk individuals. The lower abundance of F. prausnitzii is also one of the convincingly observed species-specific findings in CD patients’ dysbiosis, therefore also suggesting the causative role rather than being a consequence of chronic inflammation (76). Further arguments in favor for an etiological contribution of dysbiosis in CD are provided by the transmissibility of inflammation to genetically susceptible hosts as demonstrated in numerous in vivo models. However, similar patterns of microbial changes in diverse hosts as well as the fact that inflammation per se is capable of inducing dysbiosis pleads for the theory of dysbiosis as a consequence of the inflammatory milieu. The fact remains that the altered microbial composition entails metabolic consequences and subsequent changes in the intestinal milieu deriving from the variety of essential functions provided by the intestinal microbiota and the crosstalk between the microbiota and the host immune system.
Host–Microbiota Crosstalk – Innate Mechanisms for Maintaining Intestinal Homeostasis
The innate immune system consists of the intestinal epithelium and cells of the innate immune system, such as neutrophils, DCs, monocytes/macrophages, and innate lymphoid cells (ILCs). The luminal surface of the gastrointestinal tract with approximately 300–400 m2 represents the largest interface between the host and the environment. The diverse and abundant indigenous microflora exists in close proximity to the immune system of the intestinal mucosa and immune cells in the underlying tissue. Moreover, the gastrointestinal mucosa is continuously exposed to antigens derived from the food and has to deal with pathogenic microorganisms that can cause tissue damage. The intestinal immune system therefore possesses multiple layers of protection to respond against invading bacteria, thereby limiting their exposure to the systemic immune system while maintaining tolerance toward luminal bacterial antigens and food antigens. Given that a dysregulated immune response toward components of the intestinal microbiota is thought to be a fundamental pillar of chronic intestinal inflammation development, the complex interaction of the host with the plentiful intestinal microbiota has to be precisely regulated.
Barriers of Protection
The Intestinal Mucus Layer – A Protective Blanket
The intestinal epithelium lies at the interface between the intestinal lumen and the gastrointestinal-associated lymphoid tissue, thereby building the first barrier against excessive microbial translocation to the lamina propria (82). The intestinal epithelium itself is covered with a mucus layer constituting the first physical barrier to luminal antigens. Mucus is secreted by goblet cells and typically contains several major components, which in the intestine comprise of MUC2 and MUC5AC. These densely glycosylated proteins are resistant to digestive enzymes and give the mucus its gel-like properties (7). The type of mucus organization in the small and large intestine are clearly different. The small intestinal mucus fills the luminal space between the villi and is not attached to the epithelial surface. Antimicrobial products such as AMPs derived from epithelial cells and Paneth cells as well as IgA interfuse with the mucus secreted from the crypts, thereby generating an antibacterial gradient that acts to keep luminal bacteria away from epithelial surfaces. Of note, the small intestinal mucus layer is penetrable to bacteria but still provides a diffusion barrier (83). The mammalian large intestinal mucus is organized in a two-layer system. The loose outer layer of the mucus is composed of mucin, diluted antimicrobials, and is the normal habitat for commensal bacteria, whereas the inner layer is firmly attached to the epithelial cells, rich in antimicrobials, and displays a low bacterial density (84). The physiological relevance and protective function of the mucus layer has best been highlighted in studies using MUC2−/− mice. Muc2-deficiency was shown to lead to colonic inflammation accompanied by the presence of bacteria in direct contact with the intestinal epithelium (85). Moreover, these animals displayed a much higher susceptibility to infection by pathogens (86). From studies in GF mice, it emerged that intestinal microbiota have a role in shaping the colonic mucus barrier (87). Mice housed under GF conditions have been characterized by an extremely thin adherent colonic mucus layer. Exposure to bacterial products, such as lipopolysaccharide or peptidoglycan, leads to the re-storage of the mucus thickness to levels observed in conventionally housed mice. A recently published work reinforced the assumption that intestinal bacteria affect the host mucus barrier properties. In this study, two colonies of C57BL/6 mice were housed and bred in different rooms but both under SPF conditions. Analysis of the microbiota composition by 16S rRNA gene sequencing revealed significant differences at multiple taxonomic levels between the two separately housed colonies. Interestingly, the two colonies not only differed in their microbiota composition but also have a different mucus phenotype that was shown to be specific for each colony. Whereas the thickness of the mucus layer was similar in both groups, a major difference regarding permeability was detected. One colony displayed a mucus layer impenetrable to bacteria, the other colony had an inner mucus layer that was penetrable to both bacteria and beads. The causal role of the intestinal microbiota in the detected mucus phenotypes was underpinned by subsequent experiments showing that the different mucus properties were transmissible by transfer of cecal microbiota to GF mice (88). Although genetic risk loci in MUC1 and MUC19 have been identified in IBD, there is only limited and widely varying data regarding changes of the mucus layer during CD (3). One study investigated the thickness and continuity of the mucus barrier in rectal biopsies from CD patients but only provided a small number of cases (89). In this study, no significant differences between CD patient specimens and controls were found. Another study reported a depletion of the mucus layer at inflamed sites of the gut of CD patients compared to healthy controls as well as compared to non-inflamed areas in the same patient (90). Of note, there were no significant differences regarding the thickness of the mucus layer between non-inflamed areas of CD patients and healthy controls. These results are in direct contrast to previously published data measuring the thickness of the colonic mucus in surgically resected specimens from CD patients. Pullan and colleagues reported increased values compared to healthy controls (91). As briefly outlined above, the colonic mucus is organized in two layers. Given the contrary results, one has to consider that in the study showing a thicker mucus layer in CD patients a PAS/AB staining technique was used. During PAS/AB staining variable amounts of the loose outer mucus layer might remain thus contributing to the larger thickness observed in CD specimens. The different results might also reflect different stages of disease and moreover, inflammation in CD is characterized by a discontinuous appearance therefore a general mucus barrier dysfunction is unlikely to occur.
Antimicrobial Peptides and Antibodies
A substantial mechanism for controlling the contact between intestinal epithelial cells and luminal antigens is the secretion of AMPs. One of the intensive studied proteins among AMPs is the antibacterial lectin RegIIIγ produced by multiple epithelial lineages, including enterocytes and Paneth cells and capable of mediating direct killing of Gram-positive bacteria (92). Mouse RegIIIγ and its human counterpart HIP/PAP are primarily expressed in the small intestine. Inflammatory conditions were shown to increase the expression of RegIIIγ in the mouse intestine, likewise HIP/PAP expression is increased in the mucosa of IBD patients (93). Reconstitution of GF mice with an intestinal microbiota from conventionally raised mice resulted in a sharp increase in the abundance of RegIIIγ transcripts in Paneth cells (92). This study also explored the mRNA expression of RegIIIγ during weaning reflecting an early stage of intestinal development. RegIIIγ mRNA levels showed a sharp upward movement that was missing in GF controls. To further unravel the host–microbial interactions underlying the regulation of RegIIIγ, Cash and colleagues investigated the dynamic of this antibactericidal protein in mice lacking IgA. Bacteroides thetaiotaomicron and Listeria innocua – under state conditions compartmentalized in the intestinal lumen – showed an increased mucosal adherence in the absence of IgA and were found to substantially trigger a vast RegIIIγ mRNA increase in GF mice lacking IgA. These results indicate that an increased contact between microbiota and the intestinal epithelium drives RegIIIγ expression. The role of RegIIIγ as a key element of the intestinal mucosal defense has further been validated in murine RegIIIγ-deficiency studies that revealed an increased bacterial colonization of the intestinal epithelial surface (94).
Another major group of AMPs is represented by α-defensins, largely produced by small intestinal Paneth cells and β-defensins produced by most epithelial cells (95). Human defensin (HD)-5 and HD-6, both belonging to the group of α-defensins as well as the human β-defensin (HBD)-1, are expressed constitutively (16). HBD-2 and HBD-3 belong to the group of inducible defensins and are expressed in the large intestine. The induction was shown to be mediated by pro-inflammatory cytokines, such as IL-1β, involving NF-κB pathways (96, 97). Induction was also observed following the recognition of intestinal bacteria by pattern recognition receptors (PRRs) (98). Decreased defensin levels can result in a weakening of the intestinal epithelial barrier and might be involved in the pathophysiology of chronic inflammation. Ileal CD is associated with reduced expression of Paneth cell-derived α-defensins and colonic CD was reported to be associated with reduced expression of β-defensins by enterocytes (68). Next to their antimicrobial property, α-defensins possess an additional homeostatic role in regulating and shaping the composition of the small intestinal microbiota. Transgenic expression of DEFA5 (α-defensin) comes along with a significant decrease of members of the phylum Firmicutes, paralleled by an increase in the percentage of Bacteroidetes. Moreover, transgenic expression of DEFA5 also causes a loss of SFB and concomitantly Th17 in the lamina propria (99).
A third immune mechanism serving to control the microbiota and reduce the contact between intestinal bacteria and epithelial cells is the secretion of IgA by lamina propria plasma cells (100). DCs within the Peyer’s patches and subepithelial dome region continuously sample luminal bacteria. Bacteria- and antigen-loaded DCs interact with and prime T- and B-cells in the Peyer’s patches or find their way to the gut-draining mesenteric lymph nodes via the afferent lymphatics. The interaction of antigen-bearing DCs with naïve B cells induces the activation and differentiation of naïve B cells to IgA producing plasma cells. These plasma cells leave the Peyer’s patches or mesenteric lymph nodes, respectively, enter the systemic circulation via the efferent lymphatics and return to the intestinal lamina propria where they secrete IgA into the intestinal interstitium. The secreted IgA is specific for commensal microbiota, subsequently taken up by epithelial cells and delivered to their apical surface. When released into the intestinal lumen, IgA binds to commensal microbiota, thereby limiting their capability to penetrate the epithelial barrier (7, 101). Remarkably, live bacteria transported by DCs do not gain access to the systemic circulation at any time thus are not able to induce systemic immune responses. Recent studies provide evidence that goblet cells, supposedly purely secretory cells, also act as luminal sensors for the innate immune system. McDole and colleagues were able to show that in the absence of inflammation, small intestinal goblet cells act as passages delivering low molecular weight soluble antigens from luminal to CD103+ DCs in the underlying lamina propria (102). The interconnection between goblet cells and DCs with tolerogenic potential illustrate that goblet cells contribute to the communication between luminal antigens and the innate immune system.
Taken together, these aforementioned innate barriers of protection are crucial for intestinal homeostasis. Moreover, a finely balanced interplay between the intestinal microbiota and cells of the host immune system is indispensable for ensuring the functionality of these barrier mechanisms.
Epithelial Barrier and the Crosstalk Underneath
In contact with the inner mucus layer, there is the intestinal epithelium consisting of four main types of epithelial cells, namely enterocytes, enteroendocrine cells, goblet cells, and Paneth cells. The intestinal epithelium contributes to absorption, digestion as well as secretion, and functions as a mucosal barrier (84). Mucosal barrier integrity is maintained by tight junctions, desmosomes, and adherence junctions. Impairment of the epithelial barrier is accompanied by increased intestinal permeability and bacterial translocation leading to persistent immune activation, a condition that has been observed in both CD and UC patients (103). In health, only small amounts of luminal antigens are allowed to pass across the epithelium. Murine models displaying an impaired barrier function were shown to develop intestinal inflammation. Junctional adhesion molecule-A (JAM-A) is a key structure of tight junctions and mandatory for controlling cell migration into the underlying tissues. Studying of CD tissue specimens for JAM-A expression revealed a loss of epithelial JAM-A expression (104). In line with these findings are the decreased levels of further tight junction proteins, such as claudins, observed in CD (66).
Lying between the luminal microbiota on the one side and immune cells in the lamina propria on the other, the intestinal epithelium functions to communicate with both. Sensing of microbial antigens by epithelial cells as well as by innate immune cells, such as macrophages and DCs, is crucial for maintaining intestinal homeostasis and is mediated by PRRs (95). These receptors include the family of toll-like receptors (TLRs) as well as intracytoplasmatic receptors, such as NOD-like receptors (NLRs). Polymorphisms in TLRs and NLRs have been implicated in increased susceptibility to IBD (3, 69). Whereas TLRs are capable of detecting a variety of bacterial components, such as lipopolysaccharide, lipoproteins, and CpG DNA, NLRs recognize peptidoglycan molecules on the bacterial cell wall (105). Activation of PRRs results in down-stream signaling cascades that are widely mediated by Myd88. MyD88 represents a cytosolic adaptor protein, equipped with a toll/IL-1 receptor (TIR) domain, thereby able to directly bind TLRs (106). Once engaging TLRs, MyD88 mediates MAP kinase activation, recruitment of a variety of signaling molecules and drives nuclear translocation of NF-κB and subsequent production of pro-inflammatory cytokines. Given the abundance of commensal antigens in the intestinal lumen, it is crucial that activation of PRRs not exclusively drives inflammatory responses. Excessive stimulation can lead to detrimental inflammation. Otherwise, repetitive stimulation of TLRs due to commensal bacterial exposure was shown to result in a down-regulation of the NF-κB pathway and stimulation of AMP production (1). Moreover, triggering of PRRs also promotes antigen presenting cell maturation and is involved in proliferation of Tregs (107). Already about one decade ago, Rakoff-Nahoum and colleagues provided experimental evidence that the recognition of the commensal microbiota by TLRs is crucial for maintenance of intestinal epithelial homeostasis and required for regeneration after mucosal injury (108). TLR expression and activation has to be tightly regulated and this regulation is exerted by a number of mechanisms, such as adjustment of their expression, their localization, and their positioning within the tissue. Most surface TLRs are principally found on the basolateral side of IEC and are only up-regulated during inflammation (109).
Punctilious sampling and processing of luminal antigens is essential for avoiding the development of intestinal inflammation and establishing and sustaining a healthy relationship between the commensal microbiota and the host. DCs are an indispensable element in this crosstalk as they are continuously exposed to antigens derived from commensal microbiota, dietary products, and intestinal pathogens. The ductility of DCs is warranted by their ability to adapt to influences of the microenvironment. In this context, different DC populations with different functionality have been described. Gut-resident DCs display tolerogenic functions as they encounter and sense commensal bacteria and act, for example, by inducing a highly tolerogenic response through differentiation and expansion of Tregs (110, 111). The triangular crosstalk between epithelial cells, DCs, and the intestinal microbiota is orchestrated by PPR signaling. Using an in vitro model, intestinal epithelial cells were shown to release cytoprotective factors, such as thymic stromal lymphopoietin and transforming-growth factor-β upon activation of TLR-signaling, leading to the presence of a predominantly tolerogenic DC phenotype. These tolerogenic DCs were found to secrete IL-10, leading to an immune response dominated by Tregs (112).
Even though the intestinal epithelium with its numerous lines of protection provides a barrier separating luminal antigen from the underlying tissue, it is inevitable that antigens gain access to the lamina propria. As mentioned above, this might not only occur through injury or infection but also happens when commensals undergo epithelial cell transcytosis or translocation via M cells or DCs (113). In response to transforming-growth factor-β and IL-8, chemokines abundant in the extracellular matrix of the lamina propria, circulating blood monocytes gather to the non-inflamed lamina propria where they become resident intestinal macrophages. Located strategically in the subepithelial lamina propria, intestinal macrophages are part of the armament of innate defense mechanisms. The interaction between macrophages and microorganisms is mediated by PRRs, including TLRs and NLRs. In contrast to peripheral macrophages, intestinal macrophages were shown to perform their defense activities without releasing inflammatory cytokines while retaining their phagocytic and bacteriocidal activity. Consequently, intestinal macrophages do not provoke a pro-inflammatory cytokine milieu in the non-inflamed mucosa despite the close proximity to and interaction with immunostimulatory microorganisms. This so-called “inflammatory-anergy” was shown to be at least in part attributable to matrix-bound transforming-growth factor-β and Tregs causing a down-regulation of PRRs and related adapter proteins, resulting in NF-κB inactivation (113, 114). In the case of inflammation or intestinal infection, blood monocytes accumulate in the lamina propria and actively combat invading microorganisms through uptake and degradation, including the release of inflammatory mediators.
Over the last decade, an emerging family of innate immune cells, termed ILCs has been characterized. Since ILCs reside primarily at mucosal sites and are enriched at mammalian barrier surfaces, including the intestine, these cells are in close proximity to environmental antigens and commensal microbiota in particular. Commensal microbiota were found to provide signals for the development, differentiation, and function of ILCs and this crosstalk, the interplay between the microbiota and ILCs is indeed an area of intense research. ILCs have been shown to contribute to tissue repair and remodeling at barrier surfaces and were found to influence inflammatory conditions, thereby orchestrating host–commensal relationships (115). Being derived from an Id2-dependent lymphoid progenitor cell population, ILCs are of lymphoid origin but do not require antigen receptors generated by somatic recombination (116). Based on transcription factors required for their development, their cytokine production patterns and surface markers, the family of ILCs is categorized in three main branches. In brief, group 1 ILCs (ILC1) are Tbet- dependent and are composed of ILC1s and NK cells. ILC1s respond to IL-12 and IL-18 and produce IFN-γ. Group 2 or GATA3+ ILCs secrete IL-5, IL-9, IL-13, and amphiregulin in response to IL-33 and IL-25. Finally, group 3 ILCs (ILC3) express RORγt and include lymphoid tissue-inducer (LTi) cells and natural cytotoxicity receptor (NCR) expressing ILCs. Group 3 ILCs respond to IL-1β and IL-23 and are the main producers of IL-17 and IL-22 (117). Given the spatial proximity of ILCs and commensal microbiota in the intestine, multiple groups have investigated the possible role for commensal microbiota in the development of ILCs. Studies using GF mice indicated that NK cells and GATA3+ ILCs develop properly without receiving signals from commensal microbiota (118, 119). Regarding RORγt+ ILCs, there have been controversial reports on the requirement of commensal microbiota for their development. It seems that the development of at least subsets of RORγt+ ILCs occurs regardless of the presence of a commensal flora as evident by the presence of LTi cells as well as the generation of secondary lymphoid structures in the sterile environment before birth (115, 120, 121). However, the ripening of intestinal cryptopatches into isolated lymphoid follicles was found to be compromised in GF mice, suggesting at least a functional impairment in some populations of LTi-like RORγt+ ILCs (122). Several groups reported normal development of RORγt+ ILC subsets in both GF and antibiotic treated mice (115, 123, 124). By contrast, other studies proposed that commensal bacteria are capable of enhancing the development of NCR+ RORγt+ ILCs and IL-22 production. IL-22 produced by this population in turn contributed to epithelial homeostasis by regulating genes involved in tissue repair and antimicrobial defense mechanisms, such as RegIIIβ and RegIIIγ (125–127). Factors accounting for these differing results may relate to host genetics, a differential presence of other cell types and cytokines and not least to a varying exposure to non-live bacterial- or diet-derived signals. Although it seems that the development of most ILCs is not reliant on bacterial interaction, signals derived from the commensal microbiota directly affect the function of ILCs. The regulation of ILC function by commensal bacteria can happen indirectly or directly through engagement of PRRs on ILCs themselves. For instance, human NK cells were shown to express functional TLR2 and TLR9 (128, 129). Moreover, stimulation of human RORγt+ ILCs with TLR2 agonists resulted in IL-2 production which in turn enhanced IL-22 expression (130). In addition, there is accumulating evidence that NCRs of NK cells and NCR+ RORγt+ ILCs act as direct sensors of components of the commensal bacteria (131–133). Next to direct signaling through TLRs and NCRs, different groups provided evidence that the aryl hydrocarbon receptor (AhR), expressed by RORγt+ ILC3s and well known for its critical function in ILC development and IL-22 production has also a role in the commensals regulatory function. More specifically, the AhR can be stimulated by ligands derived from the tryptophan metabolism of commensal microbiota, thereby having an intermediate position between direct and indirect regulation (134). In addition to direct regulation, commensal bacteria also take advantage of an indirect route of influencing the function of ILCs via triggering cytokine signals from epithelial cells or other innate immune cells. As one example, commensal bacteria captured by DCs lead to the induction of IL-12 that acts on ILC1s by stimulating their IFN-γ production that in turn accelerates macrophage phagocytosis (116). Next to the effects of the commensal microbiota on ILCs, there is evidence that ILCs are capable of influence the microbial community. This reciprocal interaction is, for example, simply attributed to signals derived from ILCs that act on the intestinal permeability and thus affect bacterial translocation.
Due to displaying a major source of IL-17 and IL-22, ILC3s are supposed to be critical for the promotion of inflammation and tissue repair in the intestine as well as for dealing with extracellular bacteria (135). Murine models provided evidence that NCR+ ILC3s rapidly response to infection with the enteric pathogen Citrobacter rodentium by producing IL-22, which is essential for host protection. This ILC3 response is promoted by the occurrence of a positive feedback loop stimulating DCs to the production of both lymphotoxin-β and IL-23 (124, 136). IL-17 and IL-22 derived from ILC3s promotes neutrophil recruitment to the intestine and stimulates inter alia the production of RegIIIγ, RegIIIβ, as well as mucus production, thereby also pointing out the important role of ILC3s in tissue repair (135, 137). Moreover, ILC3s were shown to have the potential to regulate not only innate but also adaptive immune responses, a capacity that might be crucial especially with regard to chronic inflammatory disorders like IBD. Mortha and colleagues recently reported that ILC3s are the primary source of granulocyte-macrophage colony-stimulating factor (GM-CSF) in the intestine. GM-CSF production was depend on and regulated by the ability of macrophages to sense commensal microbiota and produce IL-1β. ILC3-derived GM-CSF influenced myeloid cell homeostasis and was essential for the generation of subsequent Treg responses toward food antigens and maintenance of oral tolerance (138). Furthermore, mouse and human ILC3s were also found to express MHCII, and directly interact with CD4+ T cells. Most interestingly, murine studies revealed that depletion of ILC3-intrinsic MHCII leads to the development of spontaneous CD4+ T cell-driven microbiota-dependent inflammation (139, 140).
Of note, this only provides a brief insight into the complex world of ILCs but states incontrovertibly that the emerging family of ILCs is essential in the context of the crosstalk between the microbiota and the innate immune system, thereby displaying a key determinant in regulating the host–commensal relationship.
Conclusion
The innate immune system provides multiple layers of protection to regulate and control interactions between the intestinal microbiota and the host. However, the functionality of these protective mechanisms depends on and is positively affected by finely balanced signals derived from the commensal microbiota, thereby ensuring their reliability and performance. In CD, these mechanisms of defense and tolerance are impaired at multiple levels. Intestinal dysbiosis and concomitant changes of the intestinal luminal milieu and environment weaken the intestinal epithelial barrier and entail an increased epithelial permeability. Epithelial barrier dysfunction is followed by an increase of translocation to the lamina propria, where defective handling of antigens might elicit a strong inflammatory response, maintained and enhanced by ineffective phagocytosis, and bacterial clearance as well as impairment of adaptive immune responses. Although it remains unclear, if dysbiosis precedes disease or results from active inflammation, the changes observed during active CD have a lasting and debilitating effect on the numerous host mechanisms that function to maintain homeostasis. Two recent randomized controlled trials provide additional insight by indicating that re-establishing of the intestinal microbiota composition by fecal microbiota transplantation (FMT) ameliorates active UC in a patient subgroup. The first trial conducted by Moayyedi et al. randomly assigned patients with active UC to receive either FMT via enema from healthy anonymous donors or placebo (water via enema) once weekly for a period of 6 weeks (141). The second study also investigated the therapeutic impact of FMT in patients with UC (142). Remarkably, in this study by Rossen et al., autologous fecal microbiota served as control and the administration route of FMT was via nasoduodenal tube. In brief summary and without discussing all study characteristics and statistical outcomes in detail, the decisive insight obtained from these studies is that responsiveness to FMT was accompanied with changes in the microbiota composition and especially with a significant increase of the patients microbial diversity. Moreover, patients with a recent diagnosis of UC seem to be more likely to respond to FMT. Given that seven of the nine responding patients in the study conducted by Moayyedi et al. received fecal material from the same single donor, this leads to the suggestion of a donor-dependent effect and underlines the relevance of the microbiota composition in intestinal homeostasis. Furthermore, it points out that intestinal dysbiosis – irrespective of being cause or consequence – comes along with changes in the intraluminal milieu and together with a genetic predisposition might be capable of triggering a vicious cycle resulting in an abnormal inflammatory response.
This increasing evidence on the interplay of the intestinal microbiome, barrier, and intraepithelial as well as mucosal immune cells displays a complex network that is tightly regulated. Nevertheless, the data discussed indicate several target structures that require in-depth exploration, such as targeting the luminal site via a defined change in the microbiota composition or via selected nutritional restriction that strengthen barrier function and exert anti-inflammatory effects on immune cells. FMT will have to be replaced by a defined administration of a bacterial mix. The definition of this “mix” or additional food compounds that influence the mucosal balance will be the center of research and might ultimately provide novel therapeutic targets not only for IBD.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe? World J Gastroenterol (2011) 17(5):557–66. doi:10.3748/wjg.v17.i5.557
2. Ellinghaus D, Bethune J, Petersen BS, Franke A. The genetics of Crohn’s disease and ulcerative colitis – status quo and beyond. Scand J Gastroenterol (2015) 50(1):13–23. doi:10.3109/00365521.2014.990507
3. Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet (2010) 42(12):1118–25. doi:10.1038/ng.717
4. Tysk C, Lindberg E, Jarnerot G, Floderus-Myrhed B. Ulcerative colitis and Crohn’s disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut (1988) 29(7):990–6. doi:10.1136/gut.29.7.990
5. Halfvarson J, Bodin L, Tysk C, Lindberg E, Jarnerot G. Inflammatory bowel disease in a Swedish twin cohort: a long-term follow-up of concordance and clinical characteristics. Gastroenterology (2003) 124(7):1767–73. doi:10.1016/S0016-5085(03)00385-8
6. Hedin CR, McCarthy NE, Louis P, Farquharson FM, McCartney S, Taylor K, et al. Altered intestinal microbiota and blood T cell phenotype are shared by patients with Crohn’s disease and their unaffected siblings. Gut (2014) 63(10):1578–86. doi:10.1136/gutjnl-2013-306226
7. Koboziev I, Reinoso Webb C, Furr KL, Grisham MB. Role of the enteric microbiota in intestinal homeostasis and inflammation. Free Radic Biol Med (2014) 68:122–33. doi:10.1016/j.freeradbiomed.2013.11.008
8. Sartor RB. Key questions to guide a better understanding of host-commensal microbiota interactions in intestinal inflammation. Mucosal Immunol (2011) 4(2):127–32. doi:10.1038/mi.2010.87
9. Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology (2008) 134(2):577–94. doi:10.1053/j.gastro.2007.11.059
10. Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol (2010) 28:573–621. doi:10.1146/annurev-immunol-030409-101225
11. Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature (2011) 474(7351):298–306. doi:10.1038/nature10208
12. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature (2011) 474(7351):307–17. doi:10.1038/nature10209
13. Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology (2013) 145(2):293–308. doi:10.1053/j.gastro.2013.05.050
14. Solis G, de Los Reyes-Gavilan CG, Fernandez N, Margolles A, Gueimonde M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe (2010) 16(3):307–10. doi:10.1016/j.anaerobe.2010.02.004
15. Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vazquez-Baeza Y, et al. Meta-analyses of studies of the human microbiota. Genome Res (2013) 23(10):1704–14. doi:10.1101/gr.151803.112
16. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science (2005) 308(5728):1635–8. doi:10.1126/science.1110591
17. Rajilic-Stojanovic M, Heilig HG, Tims S, Zoetendal EG, de Vos WM. Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol (2013) 15(4):1146–59. doi:10.1111/1462-2920.12023
18. Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol (2011) 12(5):R50. doi:10.1186/gb-2011-12-5-r50
19. Zoetendal EG, Akkermans ADL, Akkermans-van-Vliet WM, de Visser JAGM, de Voss WM. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb Ecol Health Dis (2001) 13:129–34. doi:10.1080/089106001750462669
20. Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut (2011) 60(10):1354–62. doi:10.1136/gut.2010.216259
21. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (2005) 307(5717):1915–20. doi:10.1126/science.1104816
22. Moreau MC, Ducluzeau R, Guy-Grand D, Muller MC. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun (1978) 21(2):532–9.
23. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol (2009) 9(5):313–23. doi:10.1038/nri2515
24. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell (2009) 139(3):485–98. doi:10.1016/j.cell.2009.09.033
25. Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol (2007) 19(6):377–82. doi:10.1016/j.smim.2007.10.009
26. Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction and effector functions of T(H)17 cells. Nature (2008) 453(7198):1051–7. doi:10.1038/nature07036
27. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature (2013) 500(7461):232–6. doi:10.1038/nature12331
28. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A (2010) 107(27):12204–9. doi:10.1073/pnas.0909122107
29. Thibault R, Blachier F, Darcy-Vrillon B, de Coppet P, Bourreille A, Segain JP. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: a transport deficiency. Inflamm Bowel Dis (2010) 16(4):684–95. doi:10.1002/ibd.21108
30. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther (2008) 27(2):104–19. doi:10.1111/j.1365-2036.2007.03562.x
31. Cherbuy C, Darcy-Vrillon B, Morel MT, Pegorier JP, Duee PH. Effect of germfree state on the capacities of isolated rat colonocytes to metabolize n-butyrate, glucose, and glutamine. Gastroenterology (1995) 109(6):1890–9. doi:10.1016/0016-5085(95)90756-4
32. Segain JP, Raingeard de la Bletiere D, Bourreille A, Leray V, Gervois N, Rosales C, et al. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut (2000) 47(3):397–403. doi:10.1136/gut.47.3.397
33. Thangaraju M, Karunakaran SK, Itagaki S, Gopal E, Elangovan S, Prasad PD, et al. Transport by SLC5A8 with subsequent inhibition of histone deacetylase 1 (HDAC1) and HDAC3 underlies the antitumor activity of 3-bromopyruvate. Cancer (2009) 115(20):4655–66. doi:10.1002/cncr.24532
34. Blad CC, Tang C, Offermanns S. G protein-coupled receptors for energy metabolites as new therapeutic targets. Nat Rev Drug Discov (2012) 11(8):603–19. doi:10.1038/nrd3777
35. Field M, Rao MC, Chang EB. Intestinal electrolyte transport and diarrheal disease (2). N Engl J Med (1989) 321(13):879–83. doi:10.1056/NEJM198909283211307
36. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity (2014) 40(1):128–39. doi:10.1016/j.immuni.2013.12.007
37. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science (2013) 341(6145):569–73. doi:10.1126/science.1241165
38. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature (2009) 461(7268):1282–6. doi:10.1038/nature08530
39. Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun (2015) 6:6734. doi:10.1038/ncomms7734
40. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med (2014) 20(2):159–66. doi:10.1038/nm.3444
41. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A (2007) 104(34):13780–5. doi:10.1073/pnas.0706625104
42. Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology (2002) 122(1):44–54. doi:10.1053/gast.2002.30294
43. Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut (2006) 55(2):205–11. doi:10.1136/gut.2005.073817
44. Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology (2011) 140(6):1720–8. doi:10.1053/j.gastro.2011.01.054
45. Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M, et al. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis (2010) 16(12):2034–42. doi:10.1002/ibd.21319
46. Mondot S, Kang S, Furet JP, Aguirre de Carcer D, McSweeney C, Morrison M, et al. Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflamm Bowel Dis (2011) 17(1):185–92. doi:10.1002/ibd.21436
47. Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis (2006) 12(12):1136–45. doi:10.1097/01.mib.0000235828.09305.0c
48. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A (2008) 105(43):16731–6. doi:10.1073/pnas.0804812105
49. Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut (2007) 56(5):669–75. doi:10.1136/gut.2006.099796
50. Rolhion N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis (2007) 13(10):1277–83. doi:10.1002/ibd.20176
51. Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology (1998) 115(6):1405–13. doi:10.1016/S0016-5085(98)70019-8
52. Barnich N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli and Crohn’s disease. Curr Opin Gastroenterol (2007) 23(1):16–20. doi:10.1097/MOG.0b013e3280105a38
53. Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol (2010) 105(11):2420–8. doi:10.1038/ajg.2010.281
54. Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut (2011) 60(5):631–7. doi:10.1136/gut.2010.223263
55. Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell (1993) 75(2):263–74. doi:10.1016/0092-8674(93)80068-P
56. Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell (1993) 75(2):253–61. doi:10.1016/0092-8674(93)80067-O
57. Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe (2010) 8(3):292–300. doi:10.1016/j.chom.2010.08.004
58. Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell (2007) 131(1):33–45. doi:10.1016/j.cell.2007.08.017
59. Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity (1999) 10(3):387–98. doi:10.1016/S1074-7613(00)80038-2
60. Schaubeck M, Clavel T, Calasan J, Lagkouvardos I, Haange SB, Jehmlich N, et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut (2015). doi:10.1136/gutjnl-2015-309333
61. D’Haens GR, Geboes K, Peeters M, Baert F, Penninckx F, Rutgeerts P. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology (1998) 114(2):262–7. doi:10.1016/S0016-5085(98)70476-7
62. Rutgeerts P, Goboes K, Peeters M, Hiele M, Penninckx F, Aerts R, et al. Effect of faecal stream diversion on recurrence of Crohn’s disease in the neoterminal ileum. Lancet (1991) 338(8770):771–4. doi:10.1016/0140-6736(91)90663-A
63. Selby W, Pavli P, Crotty B, Florin T, Radford-Smith G, Gibson P, et al. Two-year combination antibiotic therapy with clarithromycin, rifabutin, and clofazimine for Crohn’s disease. Gastroenterology (2007) 132(7):2313–9. doi:10.1053/j.gastro.2007.03.031
64. Feller M, Huwiler K, Schoepfer A, Shang A, Furrer H, Egger M. Long-term antibiotic treatment for Crohn’s disease: systematic review and meta-analysis of placebo-controlled trials. Clin Infect Dis (2010) 50(4):473–80. doi:10.1086/649923
65. Thia KT, Mahadevan U, Feagan BG, Wong C, Cockeram A, Bitton A, et al. Ciprofloxacin or metronidazole for the treatment of perianal fistulas in patients with Crohn’s disease: a randomized, double-blind, placebo-controlled pilot study. Inflamm Bowel Dis (2009) 15(1):17–24. doi:10.1002/ibd.20608
66. Zeissig S, Burgel N, Gunzel D, Richter J, Mankertz J, Wahnschaffe U, et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut (2007) 56(1):61–72. doi:10.1136/gut.2006.094375
67. Hollander D, Vadheim CM, Brettholz E, Petersen GM, Delahunty T, Rotter JI. Increased intestinal permeability in patients with Crohn’s disease and their relatives. A possible etiologic factor. Ann Intern Med (1986) 105(6):883–5. doi:10.7326/0003-4819-105-6-883
68. Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, et al. Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc Natl Acad Sci U S A (2005) 102(50):18129–34. doi:10.1073/pnas.0505256102
69. Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature (2001) 411(6837):599–603. doi:10.1038/35079107
70. Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet (2007) 39(2):207–11. doi:10.1038/ng1954
71. Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet (2007) 39(5):596–604. doi:10.1038/ng2032
72. Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature (2001) 411(6837):603–6. doi:10.1038/35079114
73. Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature (2008) 456(7219):259–63. doi:10.1038/nature07416
74. Wehkamp J, Harder J, Weichenthal M, Schwab M, Schaffeler E, Schlee M, et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut (2004) 53(11):1658–64. doi:10.1136/gut.2003.032805
75. Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell (2008) 134(5):743–56. doi:10.1016/j.cell.2008.07.021
76. Hedin C, van der Gast CJ, Rogers GB, Cuthbertson L, McCartney S, Stagg AJ, et al. Siblings of patients with Crohn’s disease exhibit a biologically relevant dysbiosis in mucosal microbial metacommunities. Gut (2015). doi:10.1136/gutjnl-2014-308896
77. Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun (1998) 66(11):5224–31.
78. Ghouri YA, Richards DM, Rahimi EF, Krill JT, Jelinek KA, DuPont AW. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin Exp Gastroenterol (2014) 7:473–87. doi:10.2147/CEG.S27530
79. Russell RK, Satsangi J. IBD: a family affair. Best Pract Res Clin Gastroenterol (2004) 18(3):525–39. doi:10.1016/j.bpg.2003.12.006
80. Joossens M, Van Steen K, Branche J, Sendid B, Rutgeerts P, Vasseur F, et al. Familial aggregation and antimicrobial response dose-dependently affect the risk for Crohn’s disease. Inflamm Bowel Dis (2010) 16(1):58–67. doi:10.1002/ibd.20985
81. Van Kruiningen HJ, Joossens M, Vermeire S, Joossens S, Debeugny S, Gower-Rousseau C, et al. Environmental factors in familial Crohn’s disease in Belgium. Inflamm Bowel Dis (2005) 11(4):360–5. doi:10.1097/01.MIB.0000158536.31557.90
82. Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology (2011) 140(6):1729–37. doi:10.1053/j.gastro.2011.02.012
83. Johansson ME, Hansson GC. Microbiology. Keeping bacteria at a distance. Science (2011) 334(6053):182–3. doi:10.1126/science.1213909
84. Wallace KL, Zheng LB, Kanazawa Y, Shih DQ. Immunopathology of inflammatory bowel disease. World J Gastroenterol (2014) 20(1):6–21. doi:10.3748/wjg.v20.i1.6
85. Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology (2006) 131(1):117–29. doi:10.1053/j.gastro.2006.04.020
86. Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog (2010) 6(5):e1000902. doi:10.1371/journal.ppat.1000902
87. Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver physiol (2011) 300(2):G327–33. doi:10.1152/ajpgi.00422.2010
88. Jakobsson HE, Rodriguez-Pineiro AM, Schutte A, Ermund A, Boysen P, Bemark M, et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep (2015) 16(2):164–77. doi:10.15252/embr.201439263
89. Strugala V, Dettmar PW, Pearson JP. Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn’s disease. Int J Clin Pract (2008) 62(5):762–9. doi:10.1111/j.1742-1241.2007.01665.x
90. Fyderek K, Strus M, Kowalska-Duplaga K, Gosiewski T, Wedrychowicz A, Jedynak-Wasowicz U, et al. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J Gastroenterol (2009) 15(42):5287–94. doi:10.3748/wjg.15.5287
91. Pullan RD, Thomas GA, Rhodes M, Newcombe RG, Williams GT, Allen A, et al. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut (1994) 35(3):353–9. doi:10.1136/gut.35.3.353
92. Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science (2006) 313(5790):1126–30. doi:10.1126/science.1127119
93. Ogawa H, Fukushima K, Naito H, Funayama Y, Unno M, Takahashi K, et al. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm Bowel Dis (2003) 9(3):162–70. doi:10.1097/00054725-200305000-00003
94. Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science (2011) 334(6053):255–8. doi:10.1126/science.1209791
95. Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev (2014) 13(1):3–10. doi:10.1016/j.autrev.2013.06.004
96. Wehkamp J, Harder J, Wehkamp K, Wehkamp-von Meissner B, Schlee M, Enders C, et al. NF-kappaB- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium. Infect Immun (2004) 72(10):5750–8. doi:10.1128/IAI.72.10.5750-5758.2004
97. O’Neil DA, Porter EM, Elewaut D, Anderson GM, Eckmann L, Ganz T, et al. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol (1999) 163(12):6718–24.
98. Abreu MT, Fukata M, Arditi M. TLR signaling in the gut in health and disease. J Immunol (2005) 174(8):4453–60. doi:10.4049/jimmunol.174.8.4453
99. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol (2010) 11(1):76–83. doi:10.1038/ni.1825
100. Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science (2000) 288(5474):2222–6. doi:10.1126/science.288.5474.2222
101. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (2012) 336(6086):1268–73. doi:10.1126/science.1223490
102. McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature (2012) 483(7389):345–9. doi:10.1038/nature10863
103. Salim SY, Soderholm JD. Importance of disrupted intestinal barrier in inflammatory bowel diseases. Inflamm Bowel Dis (2011) 17(1):362–81. doi:10.1002/ibd.21403
104. Vetrano S, Rescigno M, Cera MR, Correale C, Rumio C, Doni A, et al. Unique role of junctional adhesion molecule-a in maintaining mucosal homeostasis in inflammatory bowel disease. Gastroenterology (2008) 135(1):173–84. doi:10.1053/j.gastro.2008.04.002
105. Cario E. Bacterial interactions with cells of the intestinal mucosa: toll-like receptors and NOD2. Gut (2005) 54(8):1182–93. doi:10.1136/gut.2004.062794
106. Weng M, Walker WA. The role of gut microbiota in programming the immune phenotype. J Dev Orig Health Dis (2013) 4(3):203–14. doi:10.1017/S2040174412000712
107. Bhaskaran N, Cohen S, Zhang Y, Weinberg A, Pandiyan P. TLR-2 signaling promotes IL-17A production in CD4+CD25+Foxp3+ regulatory cells during oropharyngeal candidiasis. Pathogens (2015) 4(1):90–110. doi:10.3390/pathogens4010090
108. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell (2004) 118(2):229–41. doi:10.1016/j.cell.2004.07.002
109. Dupaul-Chicoine J, Dagenais M, Saleh M. Crosstalk between the intestinal microbiota and the innate immune system in intestinal homeostasis and inflammatory bowel disease. Inflamm Bowel Dis (2013) 19(10):2227–37. doi:10.1097/MIB.0b013e31828dcac7
110. Rutella S, Locatelli F. Intestinal dendritic cells in the pathogenesis of inflammatory bowel disease. World J Gastroenterol (2011) 17(33):3761–75. doi:10.3748/wjg.v17.i33.3761
111. Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid- dependent mechanism. J Exp Med (2007) 204(8):1757–64. doi:10.1084/jem.20070590
112. Zeuthen LH, Fink LN, Frokiaer H. Epithelial cells prime the immune response to an array of gut-derived commensals towards a tolerogenic phenotype through distinct actions of thymic stromal lymphopoietin and transforming growth factor-beta. Immunology (2008) 123(2):197–208. doi:10.1111/j.1365-2567.2007.02687.x
113. Smith PD, Smythies LE, Shen R, Greenwell-Wild T, Gliozzi M, Wahl SM. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol (2011) 4(1):31–42. doi:10.1038/mi.2010.66
114. Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest (2005) 115(1):66–75. doi:10.1172/JCI200519229
115. Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity (2012) 37(4):601–10. doi:10.1016/j.immuni.2012.10.003
116. Moro K, Koyasu S. Innate lymphoid cells, possible interaction with microbiota. Semin Immunopathol (2015) 37(1):27–37. doi:10.1007/s00281-014-0470-4
117. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells – a proposal for uniform nomenclature. Nat Rev Immunol (2013) 13(2):145–9. doi:10.1038/nri3365
118. Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity (2012) 37(1):171–86. doi:10.1016/j.immuni.2012.05.020
119. Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol (2011) 12(11):1045–54. doi:10.1031/ni.2131
120. Mebius RE, Rennert P, Weissman IL. Developing lymph nodes collect CD4+CD3- LTbeta+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity (1997) 7(4):493–504.
121. van de Pavert SA, Mebius RE. New insights into the development of lymphoid tissues. Nat Rev Immunol (2010) 10(9):664–74. doi:10.1038/nri2832
122. Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature (2008) 456(7221):507–10. doi:10.1038/nature07450
123. Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, Langa F, et al. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science (2010) 330(6004):665–9. doi:10.1126/science.1194597
124. Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science (2012) 336(6086):1321–5. doi:10.1126/science.1222551
125. Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol (2009) 10(1):83–91. doi:10.1038/ni.1684
126. Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity (2008) 29(6):958–70. doi:10.1016/j.immuni.2008.11.001
127. Philip NH, Artis D. New friendships and old feuds: relationships between innate lymphoid cells and microbial communities. Immunol Cell Biol (2013) 91(3):225–31. doi:10.1038/icb.2013.2
128. Martinez J, Huang X, Yang Y. Direct TLR2 signaling is critical for NK cell activation and function in response to vaccinia viral infection. PLoS Pathog (2010) 6(3):e1000811. doi:10.1371/journal.ppat.1000811
129. Sivori S, Falco M, Della Chiesa M, Carlomagno S, Vitale M, Moretta L, et al. CpG and double-stranded RNA trigger human NK cells by toll-like receptors: induction of cytokine release and cytotoxicity against tumors and dendritic cells. Proc Natl Acad Sci U S A (2004) 101(27):10116–21. doi:10.1073/pnas.0403744101
130. Crellin NK, Trifari S, Kaplan CD, Satoh-Takayama N, Di Santo JP, Spits H. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by toll-like receptor 2. Immunity (2010) 33(5):752–64. doi:10.1016/j.immuni.2010.10.012
131. Chaushu S, Wilensky A, Gur C, Shapira L, Elboim M, Halftek G, et al. Direct recognition of Fusobacterium nucleatum by the NK cell natural cytotoxicity receptor NKp46 aggravates periodontal disease. PLoS Pathog (2012) 8(3):e1002601. doi:10.1371/journal.ppat.1002601
132. Esin S, Batoni G, Counoupas C, Stringaro A, Brancatisano FL, Colone M, et al. Direct binding of human NK cell natural cytotoxicity receptor NKp44 to the surfaces of mycobacteria and other bacteria. Infect Immun (2008) 76(4):1719–27. doi:10.1128/IAI.00870-07
133. Vankayalapati R, Wizel B, Weis SE, Safi H, Lakey DL, Mandelboim O, et al. The NKp46 receptor contributes to NK cell lysis of mononuclear phagocytes infected with an intracellular bacterium. J Immunol (2002) 168(7):3451–7. doi:10.4049/jimmunol.168.7.3451
134. Stockinger B, Hirota K, Duarte J, Veldhoen M. External influences on the immune system via activation of the aryl hydrocarbon receptor. Semin Immunol (2011) 23(2):99–105. doi:10.1016/j.smim.2011.01.008
135. Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol (2011) 12(5):383–90. doi:10.1038/ni.2025
136. Ota N, Wong K, Valdez PA, Zheng Y, Crellin NK, Diehl L, et al. IL-22 bridges the lymphotoxin pathway with the maintenance of colonic lymphoid structures during infection with Citrobacter rodentium. Nat Immunol (2011) 12(10):941–8. doi:10.1038/ni.2089
137. Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med (2015) 21(7):698–708. doi:10.1038/nm.3892
138. Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science (2014) 343(6178):1249288. doi:10.1126/science.1249288
139. Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature (2013) 498(7452):113–7. doi:10.1038/nature12240
140. Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity (2014) 40(4):594–607. doi:10.1016/j.immuni.2014.03.005
141. Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology (2015) 149(1):102–9e6. doi:10.1053/j.gastro.2015.04.001
Keywords: Crohn’s disease, innate immune system, inflammation, microbiota, dysbiosis
Citation: Haag L-M and Siegmund B (2015) Intestinal microbiota and the innate immune system – a crosstalk in Crohn’s disease pathogenesis. Front. Immunol. 6:489. doi: 10.3389/fimmu.2015.00489
Received: 01 June 2015; Accepted: 07 September 2015;
Published: 22 September 2015
Edited by:
Amélia M. Sarmento, Universidade Fernando Pessoa, PortugalReviewed by:
Geanncarlo Lugo-Villarino, Centre National de la Recherche Scientifique, FranceKlaudia Farkas, University of Szeged, Hungary
Copyright: © 2015 Haag and Siegmund. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Britta Siegmund, Medical Department 1 (Gastroenterology/Infectious Diseases/Rheumatology), Charité – Universitätsmedizin Berlin, Campus Benjamin Franklin, Hindenburgdamm 30, Berlin DE-12200, Germany, britta.siegmund@charite.de
 Lea-Maxie Haag
Lea-Maxie Haag Britta Siegmund
Britta Siegmund