- 1Primate Genetics Laboratory, German Primate Center, Leibniz Institute for Primate Research, Göttingen, Germany
- 2Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, GA, USA
Natural killer lymphocytes are essentially involved as the first line of defense against agents such as viruses and malignant cells. The activity of these cells is regulated via interaction of specific and diverse killer cell immunoglobulin-like receptors (KIR) with the highly polymorphic cognate MHC class I proteins on target cells. Genetic variability of both KIR and MHC-I ligands has been shown to be associated with resistance to many diseases, including infection with the immunodeficiency virus. Disease course and progression to AIDS after infection with human immunodeficiency virus-1 (HIV-1) is essentially influenced by the presence of the stimulatory KIR3DS1 receptor in combination with HLA-Bw4. Knowledge of such genetic interactions that contribute to not only disease resistance but also susceptibility are just as important. Such combined genetic factors were recently reported in the rhesus macaque AIDS model. Here, we review the rhesus macaque MHC class I and KIR gene systems and the role of their polymorphisms in the SIV infection model.
Introduction
Natural killer (NK) cells are lymphoid cells that are part of the innate immune system. They are known to play important roles that include the elimination of infected cells (1), immune surveillance of cancer cells (2), the regulation of adaptive immune responses (3), and human reproduction (4). The activation of NK cells is regulated by input from both inhibitory and stimulatory receptors with many of those showing clonal expression patterns (5). NK cell receptors belong to two divergent protein families, the killer cell lectin-like receptors (KLR) and the killer cell immunoglobulin-like receptors (KIR), which are characterized by their extracellular C-type lectin-like and immunoglobulin-like domains, respectively. While keeping their KLR system relatively conserved (6), platyrrhine (New World monkeys) and catarrhine primates (Old Word monkeys, apes, human) have substantially diversified their KIR system (7–9). Important ligands for many KLRs and KIRs are members of the family of major histocompatibility complex (MHC) class I proteins. Both the KIRs and their natural cognate ligands display enormous genetic variability (10–12) and combinations of respective allotypes show differential binding strengths and can, therefore, substantially impact NK cell function (13–18). Consequently, certain polymorphisms of MHC class I ligands and KIR are associated with various diseases, including infections with the immunodeficiency viruses [for reviews, see Ref. (19, 20)]. Progression to AIDS is significantly delayed in HIV-1 infected individuals carrying the HLA-Bw4 serological epitope, in particular those having isoleucine at amino acid position 80 (Bw4-80I), and certain allotypes of the inhibitory KIR3DL1 or the activating KIR3DS1 receptors (20–22). These genetic and epidemiologic data were supported by in vitro functional data showing enhanced cytolytic activity of KIR3DS1+ NK cells toward HIV-infected Bw4-80I+ target cells and elevated control of HIV-1 replication compared to KIR3DS1− NK cells (23). The expansion of KIR3DL1+ and KIR3DS1+ NK cells that were dependent on the presence of the cognate Bw4 epitope were also monitored in patients during acute HIV-1 infection (24). Evidence for successful interaction between KIR3DS1 and HLA-Bw4-80I has only recently been documented (25) despite numerous attempts in the past. Interestingly, this interaction is peptide dependent and includes peptides from HIV-1 proteins pol and nef (25).
MHC Class I Gene Polymorphisms and the Outcome of Experimental SIV Infection in Rhesus Macaques
The first human AIDS-like symptoms were observed in a colony of macaques (26). Subsequently, it was demonstrated that AIDS could be induced by experimental inoculation of naïve macaques with tissues from the diseased animals (27, 28). Since then infection of macaques with the simian immunodeficiency virus (SIV) has become an established and excellent animal model for studying HIV infection and AIDS. Similar to human HIV infection, macaques are not natural hosts of SIV and have therefore not adapted to it. Of the currently known 23 species of macaques, the rhesus macaques (Macaca mulatta), long-tailed (or cynomolgus) macaques (Macaca fascicularis), and pig-tailed macaques (Macaca nemestrina) are the predominant species utilized for studies following experimental infection with a number of SIV isolates (29, 30). Notably, rhesus macaques of Chinese and Indian geographic origin differ genetically (31) and these differences are expected to underlie the known phenotypic alterations of Chinese and Indian SIV-infected rhesus macaques [reviewed in Ref. (32)]. Particularly influencing disease course and progression to AIDS are polymorphisms of MHC class I genes due to their crucial role in regulating cytotoxic T cell and NK cell responses. Indeed, there are striking differences in the frequencies of MHC class I alleles between rhesus macaques of different geographic origin (33–35).
The class I genes of the rhesus macaque MHC (Mamu) belong to the MHC-A, MHC-B, MHC-E, MHC-G, and MHC-F types. The characterization of both the MHC-A and MHC-B genes shows that there is a substantial degree of genetic variability with duplications/deletions of genes and presence of multiple alleles (36–45). Seven Mamu-A genes are known. While Mamu-A1 is present on almost all haplotypes and is the most polymorphic gene in terms of number of alleles (41) (see also IPD database1), the other six genes (Mamu-A2–Mamu-A7) show considerable variability with respect to presence/absence on Mamu haplotypes but display only low allelic polymorphism. Thus, in contrast to humans, chimpanzees, and gorillas, the number of MHC class I genes on haplotypes is not fixed and contributes significantly to the genetic variability of the rhesus macaque MHC (37). Up to six Mamu-A genes per haplotype have been observed (43). Assignment of new sequences to one of the seven types of Mamu-A genes is rather straightforward and is usually accomplished by phylogenetic tree analysis (39). Similar to Mamu-A, the Mamu-B genes also show different region configurations and some degree of allelic polymorphism (38, 42–45). So far, only a single MHC haplotype has been completely sequenced and contains 19 Mamu-B genes of which at least five appear non-functional (36). Studies of segregating sequences in offspring has identified up to 12 transcribed Mamu-B genes per haplotype (38, 45). The transcription strength of both Mamu-A and Mamu-B genes differs, leading to the assignment of genes either as a “major” or “minor” (37, 39). Notably, Mamu-A1 is a major and the other Mamu-A genes are minors. The majority of haplotypes contains only 1–3 Mamu-B majors, whereas the number of “minor” Mamu-B genes is variable and ranges from 1 to 10 (43). Thus, although the rhesus macaque genome contains a substantial number of MHC-A and MHC-B type of genes, there appear to be only up to 4 or 5 genes with substantial mRNA production and, most probably, high protein expression on the cell surface.
Mamu-E and Mamu-F are single copy genes and clear orthologs of the human HLA-E and HLA-F, respectively, with Mamu-E being more polymorphic than HLA-E (see IPD database2). All Mamu-G genes are pseudogenes and the so-called Mamu-AG genes, which are a result of a combination of characteristics of both the Mamu-A and Mamu-G genes, have overtaken their individual function (46, 47). Four potentially functional Mamu-AG genes have been identified in the sequenced Mamu haplotype (36).
Early studies identified the MHC class I allotype Mamu-A1*001 (previous designations Mamu-A1 or Mamu-A*01) to induce and serve as targets of SIV gag-specific CD8 T cell responses (48–50). Mamu-A1*001-positive rhesus macaques experimentally infected with either SIVmac239 or SIVmac251 exhibit a delayed progression to AIDS, longer survival times, and lower viral loads at set point as compared to Mamu-A1*001-negative animals (51–54) (Table 1). Similarly, Mamu-A1*002 and A1*011 were found significantly enriched in a cohort of elite controller rhesus macaques infected with SIVmac239 (55). By contrast, Mamu-A1*004 (56) and A1*004-positive haplotypes (57) were identified as negatively influencing SIV infection and subsequent progression to disease [odds ratio 11.0 (56)]. Mamu-B*008, B*017, and B*029 have also been identified as beneficial (54, 55, 58) (Table 1). Of note, B*017 and B*029 are frequently observed together in individual rhesus macaques (43, 54, 56, 57, 59) and both genes have been observed as well together with Mamu-A1*001 (54). Thus, the individual contribution of B*017 and B*029 to SIV resistance is difficult to distinguish. Actually, rhesus macaques with identical Mamu-B*017 haplotypes (which probably carry B*029 as well) display substantial variations in chronic-phasic viremia, making it unlikely to predict disease outcome based on the presence of Mamu-B*017 (60). This indicates that a set of combinations of individual Mamu-A1 alleles and Mamu-B region configurations instead of single gene analyses might be more powerful in SIV disease association studies. However, previous association studies had to rely on complex and labor-intensive (Sanger) sequence-based typing of MHC class I genes. As a result of this limitation, only a select number of genes were typed in the cohorts of infected and control animals. This limitation has recently been overcome by the advent of massive parallel sequencing, which enables comprehensive MHC class I genotyping (42, 56, 61, 62). This technological progress avoids the need to restrict genotyping to a few loci/alleles and, therefore, allows the inclusion of new and previously ignored rhesus macaque MHC class I alleles in studies of association analysis.
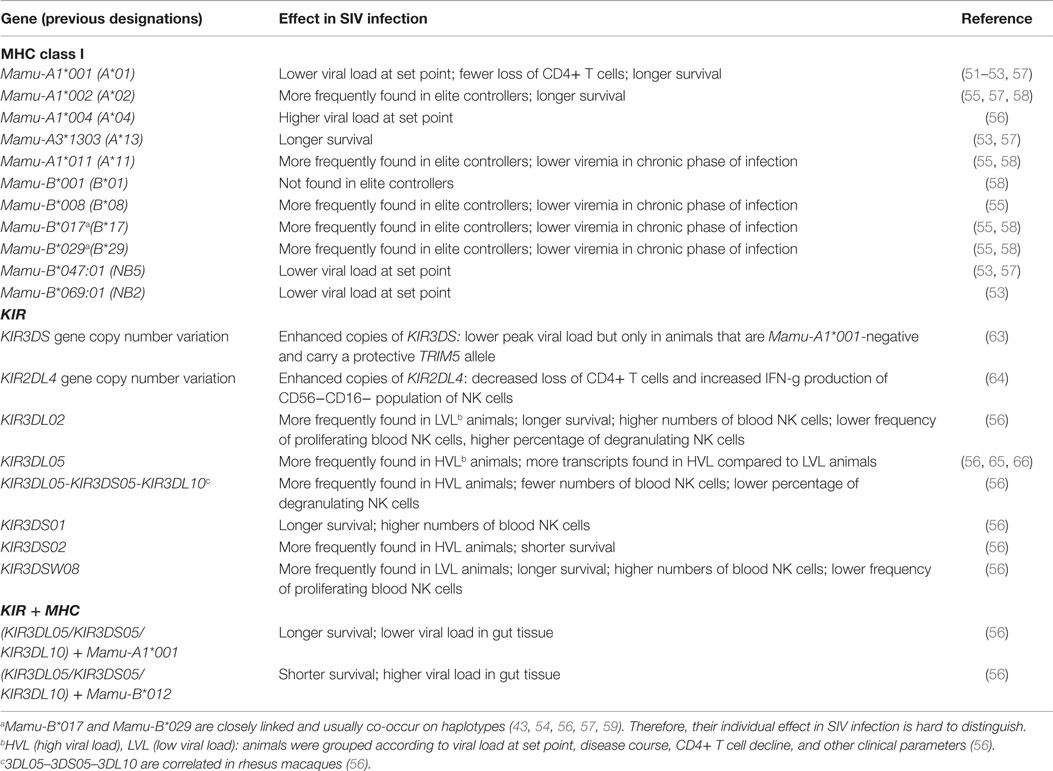
Table 1. Associations of MHC class I and KIR genes with experimental SIV infection (SIVmac239, SIVmac251) in rhesus macaques.
KIR Gene Polymorphisms and the Outcome of Experimental SIV Infection in Rhesus Macaques
The rhesus macaque KIR region has been completely sequenced using two overlapping BAC clones (67). This sequenced KIR haplotype contains five KIR genes: 1D, 2DL4, 3DL01, 3DL10, and 3DL20. The open reading frame of the KIR1D gene mRNA contains exons for the D1 domain, a truncated D2 with premature termination, stem, trans-membrane, and cytoplasmic parts, respectively. However, evidence to date is lacking as to whether this transcribed gene is expressed at the protein level as well. The rhesus macaque KIR2DL4 gene is orthologous to KIR2DL4 of hominoids (humans, great, and small apes) and belongs to the KIR lineage I. The two rhesus macaque KIR3D genes are not particularly related to any of the human or the great ape KIR3D genes but are also assigned to the KIR lineage II. Phylogenetic tree analysis indicates that there is a mixed relationship between the rhesus macaque KIR3DL20 with KIR genes of human lineage I (2DL4, 2DL5) and lineage V (3DL3) genes. Exon 3 (D0 domain) of KIR3DL20 shows closest similarity to human KIR2DL5, while the exons 4 and 5 (D1, D2 domain) of KIR3DL20 are more closely related to human KIR3DL3. The KIR3DL20 exons encoding the stem, trans-membrane, and cytoplasmic regions are more closely related to macaque KIR genes (67). Support for a shared phylogenetic origin of the human KIR3DL3 and rhesus macaque KIR3DL20 is based on the presence of an identical position at the centromeric end in the KIR region of both species (67).
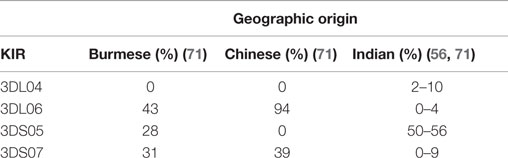
Interestingly, the sequenced haplotype does not appear to encode for any known activating KIR. Further analyses of additional rhesus macaques indicated the presence of activating KIR genes, documented the fact that the lineage II KIR genes have substantially expanded, and demonstrated differential KIR gene content between individuals (56, 62, 65, 67–72). Besides KIR1D, KIR2DL4, and KIR3DL20, 10 inhibitory KIR genes (3DL01, 3DL02, 3DLW03, 3DL04, 3DL05, 3DL06, 3DL07, 3DL08, 3DL10, and 3DL11) and 9 activating KIR genes (3DS01, 3DS02, 3DS03, 3DS04, 3DS05, 3DS06, 3DS07, 3DS08, and 3DS09) were identified. In addition to differential gene content, there is also significant allelic polymorphism of the various KIR genes (62, 65, 70, 72). The frequencies of the individual rhesus macaque KIR genes within and between populations differ considerably. For example, KIR3DL01 is present in most animals [84–98% (56, 62, 71)], whereas KIR3DL04, KIR3DL06, KIR3DS01, KIR3DS04, KIR3DS06, and KIR3DS07 are rather rare or even absent in the studied populations (56, 62, 71). As already pointed out, rhesus macaques from different geographic origins differ genetically (31). In accord with this, there are striking differences in the presence of KIR3DL04, 3DL06, 3DS05, and 3DS07 genes between Burmese, Chinese, and Indian rhesus macaques (Table 2). Additional differences in the frequencies of other KIR genes inherited by macaques of Burmese, Chinese, and Indian origin have been described (71). Such data are indeed useful and important information when analyzing cohorts of animals with known (but also unknown or mixed) geographic origin for studies of genetic association with SIV disease outcome.
Specific interactions between MHC class I proteins and a couple of KIR proteins have been described. Thus, the rhesus macaque KIR3D molecules bind to Mamu-A and Mamu-B proteins (15–17). The specificity of the interactions between the lineage II KIR proteins and the corresponding MHC-A and MHC-B ligands is conserved in evolution and has been traced to the divergence of the lineages leading to macaques and humans, i.e., about 31.5 million years ago (73). Similar to human KIR, the binding of rhesus macaque KIR proteins to their cognate ligand is influenced by the peptide content of the ligand (15). Of interest in the context of KIR and MHC class I polymorphism and SIV infection, Colantonio et al. (15) showed that allotypes of KIR3DL05 differ in their specificity for SIV peptide-loaded Mamu-A1*002 tetramers. Such differences likely contribute to variations in disease susceptibility and the further elucidation of ligand/peptide specificity of the rhesus macaque KIR/MHC ligand system is central to an understanding of the mechanisms underlying the epidemiological disease association data.
The first studies to show an association between KIR genes and SIV-induced disease progression identified KIR3DL05 to be more frequent in animals that rapidly progressed to AIDS (65, 66). However, KIR3DL05 was later found to be part of a group of closely linked KIR genes (KIR3DL05–KIR3DS05–KIR3DL10) that were identified in a correlation analysis (56), which makes it difficult to clearly identify one member of this cluster as the responsible gene. Nevertheless, at least one KIR gene of this cluster is thought to be responsible for the observed association with various parameters such as high viral load, decreased number of blood NK cells and the lower frequencies of degranulating NK cells (Table 1).
Follow-up studies identified the protective role of KIR3DL02 and KIR3DSW08 in SIV infection (56). Thus, animals carrying either one of these two genes (a) slowly progressed to disease or became elite controllers (odds ratio 0.09 and OR = 0.1, respectively), (b) showed longer survival times (hazard ratio 0.24 and 0.29, respectively), (c) exhibited higher number of blood NK cells, and (d) lower numbers of proliferating NK cells (Table 1). Animals having the KIR3DL02 gene were additionally characterized by a higher conservation of the lytic potential of NK cells. A further beneficial gene might be ascribed to the KIR3DS01 gene, but the frequency of this gene in the studied cohort was too low to provide robust association data. Future studies are needed that include sufficient number of KIR3DS01-positive animals to clarify and establish this association.
Besides gene association studies, several studies have examined copy number variation (CNV) of KIR genes to investigate their association with SIV infection outcome (63, 64). It was shown that enhanced copies of KIR2DL4 is associated with decreased loss of CD4+ T cells and an increase in IFN-gamma production by the CD56−CD16− population of NK cells during the acute phase of SIV infection (64). However, these studies did not investigate whether the CD56−CD16− population indeed expressed the KIR2DL4 protein. It is also conceivable that the observed CNV of KIR2DL4 is associated with certain KIR haplotypes and genes other than KIR2DL4 could potentially contribute to the associated phenotype. Consistent with this assumption, Hermes et al. (74) found that the CD56−CD16− NK cell subset expressed readily detectable levels of the KIR3D proteins. A further study by Hellmann et al. (63) indicated that higher copy numbers of activating KIR genes is associated with lower peak viral load upon SIV infection but only in animals that are Mamu-A1*001-negative and carry a protective TRIM5 allele.
Combinations of KIR and MHC Class I Gene Polymorphisms and the Outcome of Experimental SIV Infection in Rhesus Macaques
There had been no information about epistasis of KIR and MHC class I genes in rhesus macaques compared to respective information in the human KIR/HLA system and HIV-1 infection until a recent study by Albrecht et al. (56) who used 52 SIV-infected rhesus macaques of Indian origin. While not all KIR genes could be included in the epistasis analysis due to limited numbers of animals carrying these genes such as KIR3DL02 or KIR3DSW08, the most consistent results were obtained for the linked KIR3DL05–KIR3DS05–KIR3DL10 genes. The odds ratio of rapid progression to AIDS and presence of KIR3DS05 or KIR3DL10 [odds ratio 21.6 (56)] is nullified in animals carrying in addition Mamu-A1*001 [OR = 0.035 for KIR3DS05; OR0.036 for KIR3DL10 (56)]. It appears rather unlikely that the effect is attributable to only Mamu-A1*001, as the risk estimate (i.e., rapid progression to AIDS) for Mamu-A1*001 in animals negative for either KIR3DS05 or KIR3DL10 was OR = 1.5 and a considerable number of Mamu-A1*001-positive animals rapidly progressed to disease. Therefore, the authors speculated that the known protective effect of Mamu-A1*001 is more a consequence from epistasis with KIR genes than from Mamu-A1*001 alone (56). In addition, there was a potential epistasis of both KIR3DS05 and KIR3DL10 with Mamu-B*012 [OR = 25.0, p = 0.036 (56)]. Support for the epistases on SIV outcome was obtained by studying also viral copy numbers in gut tissue and the survival time: the simultaneous presence of either KIR3DS05 and Mamu-B*012 or KIR3DL10 and Mamu-B*012 is associated with significantly higher viral copy numbers in gut tissue and shorter survival time (hazard ratio 11.1).
Epistasis of KIR and MHC class I genes and outcome of immunodeficiency virus infection were rather hard to interpret, because interactions between the KIR and corresponding MHC molecules could not be demonstrated (KIR3DS1 and HLA-Bw4-80I) (75). In addition, functional interactions were not performed (KIR3DS05 or KIR3DL10 with Mamu-B*012) (56). However, O’Connor et al. (25) recently demonstrated binding of HLA-B*57 (Bw4-80I positive) tetramers loaded with HIV-1 peptides Pol839–847 and Nef82–90 to KIR3DS1. This finding is consistent with the idea of activating KIR’s having much lower affinity to self-peptides than pathogen-derived peptides presented on cognate MHC class I proteins. It is generally thought that presentation of peptides influences the relative affinity to both inhibitory and stimulatory KIR’s and, thereby, on the activation of NK cells (76). Higher affinity of inhibitory KIR’s and lower affinity of stimulatory KIR’s to self-peptides would secure tolerance of the NK cell, while presentation of viral peptides would then lead to loss of inhibitory KIR binding (“missing self”) and involvement of activating KIR’s (76–80). It has been speculated that viruses try to evade this NK cell activation by selection of escape mutations enabling stronger binding to inhibitory receptors (76–80). However, it is rather difficult to attribute such escape mutations to only the KIR system and NK cell recognition and not to T cell receptor and CD8+ T cell recognition as both lymphocyte populations use MHC class I proteins as ligands. Nevertheless, from a virus point of view, mutations of MHC class I-presented peptides could kill two birds with one stone and potentially lead to immune escape of both NK and CD8+ T cell responses.
Conclusion
Products of a number of host genes such as MHC class I and KIR play crucial roles in the defense of viruses as they are directly involved in the recognition of infected cells and in the restriction of viruses. The enormous genetic variability of both KIR and MHC class I genes in rhesus macaques impacts the relative susceptibility to SIV infection. The genetic complexity of such host genes, however, formed a major obstacle in the past for molecular typing and comprehensive genetic association analyses. With the advent of next-generation sequencing methods, such limitations are now overcome. Both detrimental and beneficial KIR and MHC class I genes and alleles as well as combinations of both have already been identified. Further studies will show whether the identified associations can be confirmed in other cohorts and whether further risk or beneficial genes/alleles can be identified. In addition, the KIR/MHC class I ligand system needs further detailed characterization at the protein level, which should allow for a better understanding of the observed genetic associations. Such knowledge would also further strengthen the rhesus macaque SIV infection model of AIDS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
1. Jost S, Altfeld M. Control of human viral infections by natural killer cells. Annu Rev Immunol (2013) 31:163–94. doi: 10.1146/annurev-immunol-032712-100001
2. Zamai L, Ponti C, Mirandola P, Gobbi G, Papa S, Galeotti L, et al. NK cells and cancer. J Immunol (2007) 178:4011–6. doi:10.4049/jimmunol.178.7.4011
3. Rydyznski C, Daniels KA, Karmele EP, Brooks TR, Mahl SE, Moran MT, et al. Generation of cellular immune memory and B-cell immunity is impaired by natural killer cells. Nat Commun (2015) 6:6375. doi:10.1038/ncomms7375
4. Moffett A, Colucci F. Uterine NK cells: active regulators at the maternal-fetal interface. J Clin Invest (2014) 124:1872–9. doi:10.1172/JCI68107
5. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol (2008) 9:495–502. doi:10.1038/ni1581
6. Averdam A, Kuhl H, Sontag M, Becker T, Hughes AL, Reinhardt R, et al. Genomics and diversity of the common marmoset monkey NK complex. J Immunol (2007) 178:7151–61. doi:10.4049/jimmunol.178.11.7151
7. Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos Trans R Soc Lond B Biol Sci (2012) 367:800–11. doi:10.1098/rstb.2011.0266
8. Walter L. Major histocompatibility complex class-I-interacting natural killer cell receptors of non-human primates. J Innate Immun (2011) 3:236–41. doi:10.1159/000323932
9. Cadavid LF, Lun C-M. Lineage-specific diversification of killer cell Ig-like receptors in the owl monkey, a new world primate. Immunogenetics (2009) 61:27–41. doi:10.1007/s00251-008-0342-y
10. Kelley J, Walter L, Trowsdale J. Comparative genomics of major histocompatibility complexes. Immunogenetics (2005) 56:683–95. doi:10.1007/s00251-004-0717-7
11. Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet (2005) 1:129–39. doi:10.1371/journal.pgen.0010027
12. Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol (2013) 13:133–44. doi:10.1038/nri3370
13. Yawata M, Yawata N, Draghi M, Little A-M, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med (2006) 203:633–45. doi:10.1084/jem.20051884
14. Moesta AK, Parham P. Diverse functionality among human NK cell receptors for the C1 epitope of HLA-C: KIR2DS2, KIR2DL2, and KIR2DL3. Front Immunol (2012) 3:336. doi:10.3389/fimmu.2012.00336
15. Colantonio AD, Bimber BN, Neidermyer WJ, Reeves RK, Alter G, Altfeld M, et al. KIR polymorphisms modulate peptide-dependent binding to an MHC class I ligand with a Bw6 motif. PLoS Pathog (2011) 7:e1001316. doi:10.1371/journal.ppat.1001316
16. Rosner C, Kruse PH, Hermes M, Otto N, Walter L. Rhesus macaque inhibitory and activating KIR3D interact with Mamu-A-encoded ligands. J Immunol (2011) 186:2156–63. doi:10.4049/jimmunol.1002634
17. Schafer JL, Colantonio AD, Neidermyer WJ, Dudley DM, Connole M, O’Connor DH, et al. KIR3DL01 recognition of Bw4 ligands in the rhesus macaque: maintenance of Bw4 specificity since the divergence of apes and old world monkeys. J Immunol (2014) 192:1907–17. doi:10.4049/jimmunol.1302883
18. Maloveste SM, Chen D, Gostick E, Vivian JP, Plishka RJ, Iyengar R, et al. Degenerate recognition of MHC class I molecules with Bw4 and Bw6 motifs by a killer cell Ig-like receptor 3DL expressed by macaque NK cells. J Immunol (2012) 189:4338–48. doi:10.4049/jimmunol.1201360
19. Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol (2005) 5:201–14. doi:10.1038/nri1570
20. Martin MP, Carrington M. Immunogenetics of HIV disease. Immunol Rev (2013) 254:245–64. doi:10.1111/imr.12071
21. Martin MP, Gao X, Lee J-H, Nelson GW, Detels R, Goedert JJ, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet (2002) 31:429–34. doi:10.1038/ng934
22. Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet (2007) 39:733–40. doi:10.1038/ng2035
23. Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med (2007) 204:3027–36. doi:10.1084/jem.20070695
24. Alter G, Rihn S, Walter K, Nolting A, Martin M, Rosenberg ES, et al. HLA class I subtype-dependent expansion of KIR3DS1 + and KIR3DL1 + NK cells during acute human immunodeficiency virus type 1 infection. J Virol (2009) 83:6798–805. doi:10.1128/JVI.00256-09
25. O’Connor GM, Vivian JP, Gostick E, Pymm P, Lafont BAP, Price DA, et al. Peptide-dependent recognition of HLA-B*57:01 by KIR3DS1. J Virol (2015) 89:5213–21. doi:10.1128/JVI.03586-14
26. Letvin NL, Eaton KA, Aldrich WR, Sehgal PK, Blake BJ, Schlossmann SF, et al. Acquired immundeficiency syndrome in a colony of macaque monkeys. Proc Natl Acad Sci U S A (1983) 80:2718–22. doi:10.1073/pnas.80.9.2718
27. London WT, Sever JL, Madden DL, Henrickson RV, Gravell M, Maul DH, et al. Experimental transmission of simian acquired immunodeficiency syndrome (SAIDS) and Kaposi-like skin lesions. Lancet (1983) 322:869–73. doi:10.1016/S0140-6736(83)90867-X
28. Letvin NL, Aldrich WR, King NW, Blake BJ, Daniel MD, Hunt RD. Experimental transmission of macaque AIDS by means of inoculation of macaque lymphoma tissue. Lancet (1983) 322:599–602. doi:10.1016/S0140-6736(83)90682-7
29. Gardner MB, Luciw PA. Macaque models of human infectious disease. ILAR J (2008) 49:220–55. doi:10.1093/ilar.49.2.220
30. Misra A, Thippeshappa R, Kimata JT. Macaques as model hosts for studies of HIV-1 infection. Front Microbiol (2013) 4:176. doi:10.3389/fmicb.2013.00176
31. Hernandez RD, Hubisz MJ, Wheeler DA, Smith DG, Ferguson B, Rogers J, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science (2007) 316:240–3. doi:10.1126/science.1140462
32. Zhou Y, Bao R, Haigwood NL, Persidsky Y, Ho W-Z. SIV infection of rhesus macaques of Chinese origin: a suitable model for HIV infection in humans. Retrovirology (2013) 10:89. doi:10.1186/1742-4690-10-89
33. Naruse TK, Chen Z, Yanagida R, Yamashita T, Saito Y, Mori K, et al. Diversity of MHC class I genes in Burmese-origin rhesus macaques. Immunogenetics (2010) 62:601–11. doi:10.1007/s00251-010-0462-z
34. Liu Y, Li A, Wang X, Sui L, Li M, Zhao Y, et al. Mamu-B genes and their allelic repertoires in different populations of Chinese-origin rhesus macaques. Immunogenetics (2013) 65:273–80. doi:10.1007/s00251-012-0673-6
35. Li A, Wang X, Liu Y, Zhao Y, Liu B, Sui L, et al. Preliminary observations of MHC class I A region polymorphism in three populations of Chinese-origin rhesus macaques. Immunogenetics (2012) 64:887–94. doi:10.1007/s00251-012-0645-x
36. Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res (2004) 14:1501–15. doi:10.1101/gr.2134504
37. Otting N, Heijmans CMC, Noort RC, de Groot NG, Doxiadis GGM, van Rood JJ, et al. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci U S A (2005) 102:1626–31. doi:10.1073/pnas.0409084102
38. Otting N, Heijmans CMC, van der Wiel M, de Groot NG, Doxiadis GGM, Bontrop RE. A snapshot of the Mamu-B genes and their allelic repertoire in rhesus macaques of Chinese origin. Immunogenetics (2008) 60:507–14. doi:10.1007/s00251-008-0311-5
39. Otting N, de Vos-Rouweler AJM, Heijmans CMC, de Groot NG, Doxiadis GGM, Bontrop RE. MHC class I A region diversity and polymorphism in macaque species. Immunogenetics (2007) 59:367–75. doi:10.1007/s00251-007-0201-2
40. Bonhomme M, Doxiadis GG, Heijmans CM, Vervoort V, Otting N, Bontrop RE, et al. Genomic plasticity of the immune-related Mhc class I B region in macaque species. BMC Genomics (2008) 9:514. doi:10.1186/1471-2164-9-514
41. Doxiadis GGM, de Groot N, Otting N, Blokhuis JH, Bontrop RE. Genomic plasticity of the MHC class I A region in rhesus macaques: extensive haplotype diversity at the population level as revealed by microsatellites. Immunogenetics (2011) 63:73–83. doi:10.1007/s00251-010-0486-4
42. Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, et al. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med (2009) 15(11):1322–6. doi:10.1038/nm.2038
43. Karl JA, Bohn PS, Wiseman RW, Nimityongskul FA, Lank SM, Starrett GJ, et al. Major histocompatibility complex class I haplotype diversity in Chinese rhesus macaques. G3 (Bethesda) (2013) 3:1195–201. doi:10.1534/g3.113.006254
44. Karl JA, Wiseman RW, Campbell KJ, Blasky AJ, Hughes AL, Ferguson B, et al. Identification of MHC class I sequences in Chinese-origin rhesus macaques. Immunogenetics (2008) 60:37–46. doi:10.1007/s00251-007-0267-x
45. Wiseman RW, Karl JA, Bohn PS, Nimityongskul FA, Starrett GJ, O’Connor DH. Haplessly hoping: macaque major histocompatibility complex made easy. ILAR J (2013) 54:196–210. doi:10.1093/ilar/ilt036
46. Boyson JE, Iwanaga KK, Golos TG, Watkins DI. Identification of a novel MHC class I gene, Mamu-AG, expressed in the placenta of a primate with an inactivated G locus. J Immunol (1997) 159:3311–21.
47. Slukvin II, Lunn DP, Watkins DI, Golos TG. Placental expression of the non-classical MHC class I molecule Mamu-AG at implantation in the rhesus monkey. Proc Natl Acad Sci U S A (2000) 97:9104–9. doi:10.1073/pnas.97.16.9104
48. Miller MD, Yamamoto H, Hughes AL, Watkins DI, Letvin NL. Definition of an epitope and MHC class I molecule recognized by gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J Immunol (1991) 147:320–9.
49. Kuroda MJ, Schmitz JE, Barouch DH, Craiu A, Allen TM, Sette A, et al. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med (1998) 187:1373–81. doi:10.1084/jem.187.9.1373
50. Allen TM, Sidney J, del Guercio MF, Glickman RL, Lensmeyer GL, Wiebe DA, et al. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol (1998) 160:6062–71.
51. Pal R, Venzon D, Letvin NL, Santra S, Montefiori DC, Miller NR, et al. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J Virol (2002) 76:292–302. doi:10.1128/JVI.76.1.292-302.2002
52. Mothé BR, Weinfurter J, Wang C, Rehrauer W, Wilson N, Allen TM, et al. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol (2003) 77:2736–40. doi:10.1128/JVI.77.4.2736-2740.2003
53. Mühl T, Krawczak M, Haaft Ten P, Hunsmann G, Sauermann U. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J Immunol (2002) 169:3438–46. doi:10.4049/jimmunol.169.6.3438
54. O’Connor DH, Mothé BR, Weinfurter JT, Fuenger S, Rehrauer WM, Jing P, et al. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J Virol (2003) 77:9029–40. doi:10.1128/JVI.77.16.9029-9040.2003
55. Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol (2007) 81:8827–32. doi:10.1128/JVI.00895-07
56. Albrecht C, Malzahn D, Brameier M, Hermes M, Ansari AA, Walter L. Progression to AIDS in SIV-infected rhesus macaques is associated with distinct KIR and MHC class I polymorphisms and NK cell dysfunction. Front Immunol (2014) 5:600. doi:10.3389/fimmu.2014.00600
57. Sauermann U, Siddiqui R, Suh Y-S, Platzer M, Leuchte N, Meyer H, et al. Mhc class I haplotypes associated with survival time in simian immunodeficiency virus (SIV)-infected rhesus macaques. Genes Immun (2008) 9:69–80. doi:10.1038/sj.gene.6364448
58. Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, et al. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol (2006) 80:5074–7. doi:10.1128/JVI.80.10.5074-5077.2006
59. Doxiadis GGM, de Groot N, Otting N, de Vos-Rouweler AJM, Bolijn MJ, Heijmans CMC, et al. Haplotype diversity generated by ancient recombination-like events in the MHC of Indian rhesus macaques. Immunogenetics (2013) 65:569–84. doi:10.1007/s00251-013-0707-8
60. Wojcechowskyj JA, Yant LJ, Wiseman RW, O’Connor SL, O’Connor DH. Control of simian immunodeficiency virus SIVmac239 is not predicted by inheritance of Mamu-B*17-containing haplotypes. J Virol (2006) 81:406–10. doi:10.1128/JVI.01636-06
61. O’Leary CE, Wiseman RW, Karl JA, Bimber BN, Lank SM, Tuscher JJ, et al. Identification of novel MHC class I sequences in pig-tailed macaques by amplicon pyrosequencing and full-length cDNA cloning and sequencing. Immunogenetics (2009) 61:689–701. doi:10.1007/s00251-009-0397-4
62. Moreland AJ, Guethlein LA, Reeves RK, Broman KW, Johnson RP, Parham P, et al. Characterization of killer immunoglobulin-like receptor genetics and comprehensive genotyping by pyrosequencing in rhesus macaques. BMC Genomics (2011) 12:295. doi:10.1186/1471-2164-12-295
63. Hellmann I, Lim S-Y, Gelman RS, Letvin NL. Association of activating KIR copy number variation of NK cells with containment of SIV replication in rhesus monkeys. PLoS Pathog (2011) 7:e1002436. doi:10.1371/journal.ppat.1002436
64. Hellmann I, Letvin NL, Schmitz JE. KIR2DL4 copy number variation is associated with CD4 + T-cell depletion and function of cytokine-producing NK cell subsets in SIV-infected Mamu-A*01-negative rhesus macaques. J Virol (2013) 87:5305–10. doi:10.1128/JVI.02949-12
65. Bostik P, Kobkitjaroen J, Tang W, Villinger F, Pereira LE, Little DM, et al. Decreased NK cell frequency and function is associated with increased risk of KIR3DL allele polymorphism in simian immunodeficiency virus-infected rhesus macaques with high viral loads. J Immunol (2009) 182:3638–49. doi:10.4049/jimmunol.0803580
66. Chaichompoo P, Bostik P, Stephenson S, Udompunturuk S, Kobkitjaroen J, Pattanapanyasat K, et al. Multiple KIR gene polymorphisms are associated with plasma viral loads in SIV-infected rhesus macaques. Cell Immunol (2010) 263:176–87. doi:10.1016/j.cellimm.2010.03.014
67. Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, et al. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res (2005) 15:25–35. doi:10.1101/gr.2381205
68. Grendell RL, Hughes AL, Golos TG. Cloning of rhesus monkey killer-cell Ig-like receptors (KIRs) from early pregnancy decidua. Tissue Antigens (2001) 58:329–34. doi:10.1034/j.1399-0039.2001.580507.x
69. Hershberger KL, Shyam R, Miura A, Letvin NL. Diversity of the killer cell Ig-like receptors of rhesus monkeys. J Immunol (2001) 166:4380–90. doi:10.4049/jimmunol.166.7.4380
70. Blokhuis JH, van der Wiel MK, Doxiadis GGM, Bontrop RE. The mosaic of KIR haplotypes in rhesus macaques. Immunogenetics (2010) 62:295–306. doi:10.1007/s00251-010-0434-3
71. Blokhuis JH, van der Wiel MK, Doxiadis GGM, Bontrop RE. The extreme plasticity of killer cell Ig-like receptor (KIR) haplotypes differentiates rhesus macaques from humans. Eur J Immunol (2011) 41:2719–28. doi:10.1002/eji.201141621
72. Kruse PH, Rosner C, Walter L. Characterization of rhesus macaque KIR genotypes and haplotypes. Immunogenetics (2010) 62:281–93. doi:10.1007/s00251-010-0433-4
73. Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MAM, et al. A molecular phylogeny of living primates. PLoS Genet (2011) 7:e1001342. doi:10.1371/journal.pgen.1001342
74. Hermes M, Albrecht C, Schrod A, Brameier M, Walter L. Expression patterns of killer cell immunoglobulin-like receptors (KIR) of NK-cell and T-cell subsets in old world monkeys. PLoS One (2013) 8:e64936. doi:10.1371/journal.pone.0064936
75. O’Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol (2007) 178:235–41. doi:10.4049/jimmunol.178.1.235
76. Das J, Khakoo SI. NK cells: tuned by peptide? Immunol Rev (2015) 267:214–27. doi:10.1111/imr.12315
77. Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, Cazaly A, et al. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci U S A (2010) 107:10160–5. doi:10.1073/pnas.0913745107
78. Borhis G, Ahmed PS, Mbiribindi B, Naiyer MM, Davis DM, Purbhoo MA, et al. A peptide antagonist disrupts NK cell inhibitory synapse formation. J Immunol (2013) 190:2924–30. doi:10.4049/jimmunol.1201032
79. Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature (2011) 476:96–100. doi:10.1038/nature10237
Keywords: KIR, MHC class I, SIV infection, genetic association, rhesus macaque
Citation: Walter L and Ansari AA (2015) MHC and KIR polymorphisms in rhesus macaque SIV infection. Front. Immunol. 6:540. doi: 10.3389/fimmu.2015.00540
Received: 28 August 2015; Accepted: 08 October 2015;
Published: 21 October 2015
Edited by:
Can Kesmir, Utrecht University, NetherlandsReviewed by:
Carsten Watzl, Leibniz Research Centre for Working Environment and Human Factors – IfADo, GermanyNatasja G. De Groot, Biomedical Primate Research Centre, Netherlands
Copyright: © 2015 Walter and Ansari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lutz Walter, walter@dpz.eu
 Lutz Walter
Lutz Walter Aftab A. Ansari
Aftab A. Ansari