- 1Fraunhofer Institute for Molecular Biology and Applied Ecology IME, Aachen, Germany
- 2Institute for Phytopathology and Applied Zoology, Justus-Liebig University Giessen, Giessen, Germany
- 3RWTH Aachen University, Institute for Molecular Biotechnology, Aachen, Germany
- 4Indiana Biosciences Research Institute (IBRI), Indianapolis, IN, United States
The blood-stage malaria vaccine candidate Plasmodium falciparum apical membrane antigen 1 (PfAMA1) can induce strong parasite growth-inhibitory antibody responses in animals but has not achieved the anticipated efficacy in clinical trials. Possible explanations in humans are the insufficient potency of the elicited antibody responses, as well as the high degree of sequence polymorphisms found in the field. Several strategies have been developed to improve the cross-strain coverage of PfAMA1-based vaccines, whereas innovative concepts to increase the potency of PfAMA1-specific IgG responses have received little attention even though this may be an essential requirement for protective efficacy. A previous study has demonstrated that immunization with a complex of PyAMA1 and PyRON2, a ligand with an essential functional role in erythrocyte invasion, leads to protection from lethal Plasmodium yoelli challenge in an animal model and suggested to extend this strategy toward improved strain coverage by using multiple PfAMA1 alleles in combination with PfRon2L. As an alternative approach along this line, we decided to use PfRon2L in combination with three PfAMA1 diversity covering variants (DiCo) to investigate the potential of this complex to induce more potent parasite growth inhibitory immune response in combination with better cross-strain-specific efficacy. Within the limits of the study design, the ability of the PfAMA1 DiCo-Mix to induce cross-strain-specific antibodies was not affected in all immunization groups, but the DiCo–PfRon2L complexes did not improve the potency of PfAMA1-specific IgG responses.
Introduction
Malaria remains a major global health problem affecting >200 million people and killing more than 500,000 per year (1). An effective malaria vaccine is regarded as an essential component of any eradication strategy. Plasmodium falciparum apical membrane antigen 1 (PfAMA1), a Plasmodium protein functionally involved in human erythrocyte invasion, is one of the leading blood-stage vaccine candidates. Many studies indicate that PfAMA1-specific antibodies contribute to naturally acquired semi-immunity, so the capacity of this protein to induce parasite growth-inhibitory responses has been investigated in animals (2–6) and humans (7–10).
Both in vitro and in vivo studies show that antibody responses induced by single PfAMA1 alleles achieve significantly lower efficacy against heterologous strains (11). Several epidemiological studies in different countries have revealed large numbers of different PfAMA1 haplotypes even in defined endemic areas (12, 13). This high degree of polymorphism in the field is likely to be a parasite strategy to evade the immune system (11), thus presenting a serious challenge for the development of effective PfAMA1-based vaccine candidates. The problem has been tackled by different groups using either mixtures of up to seven PfAMA1 alleles (14–18) or by the design of three so-called diversity covering (DiCo) variants (19). These artificial sequences were generated based on the analysis of over 300 different PfAMA1 sequences from field isolates, and cover 97% of the observed amino acid variability affecting around 10% of the amino acid residues. Additionally, all potential N-glycosylation sites were removed using preferentially natural occurring mutations. Both strategies are successful in eliciting antibodies with a broader range of specificity by focusing the immune response toward conserved regions of the molecule. The outcome of several studies performed with PfAMA1-based vaccines in animals and humans suggest that besides cross-strain efficacy, also the potency of the immune IgG needs to be improved to induce sufficient protection. Since variations in dose, adjuvant, formulation [protein in adjuvant, DNA, viral vectored as well as combinations thereof (20–22)] have shown only moderate improvements, it is believed that alternative strategies are required to improve the potency of PfAMA1-specific antibodies.
Plasmodium falciparum apical membrane antigen 1 plays an important role in the erythrocyte invasion machinery and an essential step during invasion is the formation of a moving junction between the merozoite and the erythrocyte membrane. During this process, the connection between the two cells is maintained by the interaction between PfAMA1 located on the surface of the parasite and PfRon2, another P. falciparum protein, which is translocated to the erythrocyte membrane early in the process (23–25). Previous studies have shown that antibodies (26–28), peptides (29, 30), and drugs (31, 32) that interfere with the AMA1–Ron2 interaction in different plasmodium species inhibit the growth of the parasite. Additionally, structural analysis of the PfAMA1–PfRon2 complex has revealed extensive conformational changes in the PfAMA1 variable loops surrounding the PfRon2-binding pocket compared to PfAMA1 alone (25, 33–35) making those regions particularly interesting as targets for potent parasite growth-inhibitory antibodies. A complex of AMA1 and Ron2L (a synthetic peptide, representing the PfAMA1-binding domain of PfRon2) as the immunogen achieved higher efficacy than AMA1 alone in an in vitro parasite growth inhibition assay (GIA) using P. falciparum and also protected mice against a lethal challenge with P. yoelii (35). Even though this strategy improves the potency of AMA1-based vaccines, it does not address the need for cross-strain protection, leading the authors of the abovementioned study to suggest the use of multiple PfAMA1–PfRon2 complexes representing different PfAMA1 alleles (35). As an alternative, we chose to investigate a scenario involving the minimum number of different recombinant molecules by using the three DiCo PfAMA1 variants (19) in a complex with PfRon2L.
Materials and Methods
Bacteria, Plants, and Parasites
Agrobacterium tumefaciens strain GV3101:pMP90RK (GmR, KmR, RifR) (36) was used for the production of recombinant proteins in Nicotiana benthamiana plants by agroinfiltration. Parasite strains P. falciparum 3D7, FCR3, and HB3 (MR4, Manassas, VA, USA) were used for the GIAs.
Construct Cloning and Transient Expression in N. benthamiana
The DiCo1-3 sequences (Figure S1 in Supplementary Material) were amplified from their source constructs (37) and introduced into the plant expression vector pTRAkc-ERH (linearized with NcoI/NotI) in-frame with an upstream signal peptide sequence and a downstream His6 tag and SEKDEL signal for retention in the endoplasmic reticulum (38). Additionally, six different alleles of PfAMA1 (PfAMA1-3D7, PfAMA1-FCR3, PfAMA1-HB3, PfAMA1-Dd2, PfAMA1-7G8, and PfAMA1-RO33) were obtained as synthetic genes codon optimized for N. benthamiana from Geneart (Thermo Fisher Scientific, Waltham, MA, USA) and introduced into the plant expression vector pTRAkc-ERH using the cloning strategy mentioned above. All cloning steps were confirmed by DNA sequencing. The transformation and cultivation of A. tumefaciens as well as transient expression in N. benthamiana plants was carried out as previously described (38).
Purification of Recombinant Proteins
The three DiCo variants and six PfAMA1 alleles (PfAMA1-3D7, PfAMA1-FCR3, PfAMA1-HB3, PfAMA1-Dd2, PfAMA1-7G8, and PfAMA1-RO33) were purified by immobilized metal affinity chromatography (IMAC) followed by size exclusion chromatography (SEC) on a Superdex 75 column (GE Healthcare Life Sciences, Little Chalfont, UK) as previously described (39).
Analysis of DiCo–PfRon2L Complex Formation
The concentration of PfRon2L (DITQQAKDIGAGPVASCFTTRMSPPQQICLNSVVNTALS), purchased in the oxidized form from Pepscan (Lelystad, The Netherlands) required for equilibrium saturation of the three purified DiCo molecules was determined by surface plasmon resonance (SPR)-based competition analysis using a Biacore T200 instrument (Biacore, Uppsala, Sweden). We mixed 796 nM of each purified DiCo variant with serial 1:3 dilutions of PfRon2L starting at 12.5 µM and descending to a minimum of 5.7 nM. The residual binding of the remaining free DiCo molecules was quantified using an S-series streptavidin chip coated with biotinylated PfRon2L.
Formulation of Antigens
Immunization with DiCo-Mix (group D), DiCo-Mix, and PfRon2L at different injection sites (group D + R) or the Dico-Mix–PfRon2L complex (group C) was achieved using different vaccine dose formulations. The purified DiCo variants and the PfRon2L peptide were lyophilized (PBS, pH 7.4) in single-dose scale glass vials and stored at −20°C. For immunization, the lyophilized proteins were reconstituted in sterile water. To facilitate complex formation, the DiCo–PfRon2L mixture was incubated for 30 min at room temperature prior to final formulation with the adjuvant.
SDS-PAGE and Immunoblot Analysis
Proteins were separated on 4–12% (w/v) NuPage polyacrylamide gradient gels (Thermo Fisher Scientific, Waltham, MA, USA) and either stained with Coomassie Brilliant Blue or transferred onto a nitrocellulose membrane (Whatmann, Dassel, Germany) for immunoblot analysis as previously described (39).
Rabbit Immunization and IgG Purification
Rabbits were housed, immunized, and sampled by Biogenes GmbH (Berlin, Germany) according to national animal welfare regulations. Four rabbits were immunized with either DiCo-Mix (D, 50 µg), or DiCo-Mix (50 µg) and PfRon2L (50 µg) at different injection sites (D + R), or the Dico-Mix–PfRon2L complex (C, prepared by mixing 50 µg of DiCo-Mix with 50 µg PfRon2L), each formulated with the Biogenes proprietary adjuvant, on days 0, 28, and 56. Serum samples were collected on day 70. IgG purification and quantification was carried out as previously described (39).
Analysis of Immune Sera
Antibody titers against the different PfAMA1 variants were determined by direct-coating enzyme-linked immunosorbent assay (ELISA) as previously described (39), using DiCo-Mix, single DiCo variants, and six different PfAMA1 alleles (PfAMA1-3D7, PfAMA1-FCR3, PfAMA1-HB3, PfAMA1-Dd2, PfAMA1-7G8, and PfAMA1-RO33). To measure the avidity of the immune sera for the DiCo-Mix, we used the NaSCN displacement ELISA, an adapted protocol called Avidity ELISA (40). Based on previous titer determinations, all serum samples were diluted to OD405 nm = 0.6–0.8. The avidity index is the molar NaSCN concentration at which 50% of the bound serum antibodies can be eluted.
Calibration-Free Concentration Analysis (CFCA) of Purified Rabbit Immune IgG
Antigen-specific antibody concentrations were measured in the purified rabbit antibody preparations by CFCA (41) using a Biacore T200 instrument. The antigens (DiCo-Mix, PfAMA1-3D7, PfAMA1-FCR3, and PfAMA1-HB3) were separately covalently coupled to CM5 S-Series sensor chips using standard EDC–NHS chemistry as previously described (39).
SPR-Based Competition Analysis
To confirm the CFCA results, a competition assay was carried out using the Biacore T200 instrument and the DiCo-Mix surface. To determine the quantity of DiCo and allele-specific antibodies, the purified IgG preparations were mixed either with running buffer, or a molar excess of DiCo-Mix, single DiCo (1-3), PfAMA1-3D7, PfAMA1-FCR3, or PfAMA1-HB3, as well as a mixture of three alleles (PfAMA1-3D7, PfAMA1-FCR3, PfAMA1-HB3) and a mixture of six alleles (PfAMA1-3D7, PfAMA1-FCR3, PfAMA1-HB3, PfAMA1-Dd2, PfAMA1-7G8, and PfAMA1-RO33). The 90-s injections were conducted under mass transport limitation, and the DiCo-Mix surface was regenerated between injections using 20-s pulses with 30 mM HCl. The binding signal resulting from competition mixtures was normalized against the end-point values for the corresponding buffer controls, which were set to 100%.
Parasite Culture and GIA
P. falciparum strains 3D7A, HB3, and FCR3 were cultured under routine culture conditions and synchronized as described before (42). The ability of purified polyclonal rabbit IgGs to inhibit the growth of P. falciparum strains 3D7A, FCR3, and HB3 was determined by conducting GIAs as previously described (43, 44). Highly synchronous parasites were treated with eight serial dilutions of the rabbit IgG (1:2, starting at a final concentration of 6 mg/ml) at schizont stage. The parasites cultures were harvested at 42–44 h of coculture. As controls, BG98 (positive control, kindly provided by Ed Remarque, BPRC, Rijswijk, Netherlands) (45) and purified IgG from non-immunized rabbits (negative control) were used at a concentration of 6 mg/ml. Parasite growth was estimated using the pLDH-assay (42).
Statistical Analysis
Titers, avidities, allele-specific antibody concentrations, competition data, and GIA IC50 values derived from the three different immunization groups (D, D + R, and C) were compared by one-way analysis of variance (ANOVA) using Origin data analysis software (OriginLab, Northampton, MA, USA). GIAs were analyzed using GraphPad Prism software package v7.02. For the determination of IC50-values, the growth curves were fitted using a 4-parameter logistic curve fit and the IC50-value estimated using the Hill equation. The level of statistical significance for all analyses was set at 0.05.
Results
Transient Expression and Purification of PfAMA1 Variants
After generating the expression constructs and the corresponding recombinant A. tumefaciens cultures, small-scale transient expression was carried out in N. benthamiana allowing the provision of recombinant proteins within a few days (38). All three DiCo proteins (DiCo1-3), as well as the six alleles (PfAMA1-3D7, PfAMA1-FCR3, PfAMA1-HB3, PfAMA1-Dd2, PfAMA1-7G8, and PfAMA1-RO33) accumulated to high levels and were successfully purified from leaf tissue by IMAC and SEC.
Analysis of DiCo–PfRon2L Complex Formation
The full equilibrium saturation of each purified DiCo variant as well as the DiCo-Mix was achieved at 12.5 µM (50 µg/ml) PfRon2L as illustrated by the complete reduction of the binding signal in SPR measurements (Figure 1). Although almost full saturation was achieved at a concentration >1,000 nM for DiCo1, DiCo2, and DiCo-Mix, a higher concentration was required for the equilibrium saturation of DiCo3 (>10,000 nM), probably reflecting the lower PfRon2L-binding affinity of this variant. The DiCo concentration (796 nM or 50 µg/ml) used for equilibrium saturation analysis was identical to the conditions used for the constitution of the immunization complex (50 µg DiCo-Mix plus 50 µg PfRon2L).
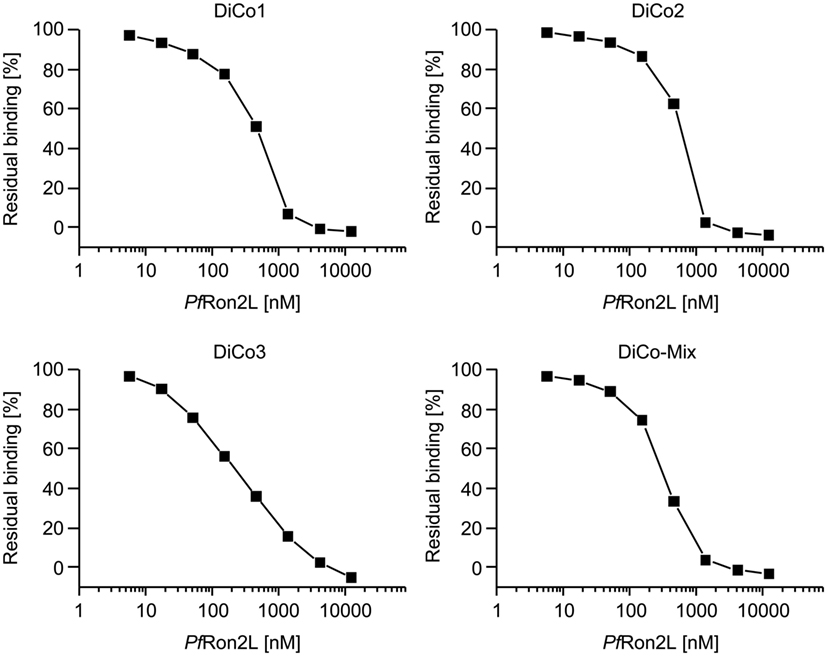
Figure 1. Analysis of DiCo–PfRon2L complex formation. To determine the equilibrium saturation of the DiCo–PfRon2L complex, surface plasmon resonance-based competition experiments were conducted using a biotinylated PfRon2L peptide immobilized on a Series S sensor chip streptavidin. Equivalent concentrations of purified DiCo1, DiCo2, DiCo3 as well as a balanced DiCo-Mix were mixed 1:1 with buffer (reference) or decreasing concentrations of the PfRon2L peptide. The final concentration in the competition assays was 50 µg/ml for the DiCo proteins with a molecular weight of approximately 62.5 kDa (796 nM) and 50 µg/ml (12,500 nM)—0.022 µg/ml (5.7 nM) for the PfRon2L peptide. The competition samples were incubated for 4 h at room temperature and the residual binding to the immobilized PfRon2L was analyzed. The buffer control (reference) was set to 100%, and residual binding was expressed as a percentage compared to the control.
Rabbit Immunizations and Characterization of Immune Sera
Serum samples collected after the immunization of rabbit groups were analyzed by ELISA to determine specific IgG titers for DiCo-Mix, the three individual DiCo variants, and the different PfAMA1 alleles (Figure 2). No significant differences in IgG titer were observed among the three groups (D, D + R, and C). Geometric mean titers against DiCo-Mix and the individual DiCo variants were approximately 2.5 × 105 (Figure 2). Immune sera were also compared by avidity ELISA, revealing no significant differences among the immunization groups (Figure 3).
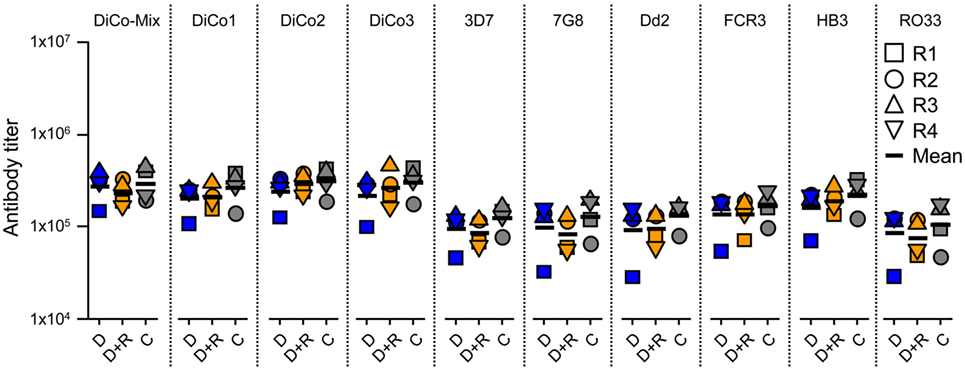
Figure 2. Determination of antibody titers in the serum samples. Four rabbits (R1–R4) in each group were immunized using a one prime (day 0) and two boost (day 28 and day 56) immunization schedule and the serum samples were collected on day 70. The antibody titers were assessed by direct-coating enzyme-linked immunosorbent assay using the DiCo-Mix and single DiCo variants as well as six different Plasmodium falciparum apical membrane antigen 1 alleles as coating antigens (indicated above each lane). D: group of rabbits that received 50 µg DiCo-Mix; D + R: group of rabbits that received 50 µg DiCo-Mix and 50 µg of PfRon2L peptide at different injection sites; C: group of rabbits that were immunized with the DiCo–PfRon2L complex formed by mixing 50 µg of DiCo-Mix with 50 µg of PfRon2L. To facilitate complex formation, the DiCo–PfRon2L mixture was incubated for 30 min at room temperature before formulation with the adjuvant and immunization. The end-point titers were defined as the highest dilution that gave double the value of the background (pre-immune serum).
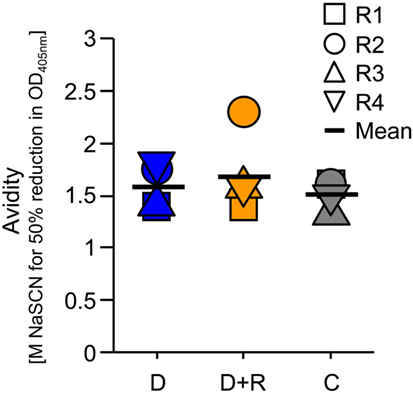
Figure 3. Determination of antibody avidity in the serum samples. The antibody avidity for the DiCo-Mix was assessed using the NaSCN-displacement enzyme-linked immunosorbent assay protocol and is defined as the NaSCN concentration (molar) required to reduce the OD405 nm by 50% compared to the reference sample incubated without NaSCN. D: group of rabbits that received 50 µg DiCo-Mix; D + R: group of rabbits that received 50 µg DiCo-Mix and 50 µg of PfRon2L peptide at different injection sites; C: group of rabbits that were immunized with the DiCo–PfRon2L complex formed by mixing 50 µg of DiCo-Mix and 50 µg of PfRon2L. To facilitate the complex formation, the DiCo–PfRon2L mixture was incubated for 30 min at room temperature before formulation with the adjuvant and immunization.
Quantification and Analysis of Purified Immune IgG
Analytical SEC was used to determine the total quantity of IgG in the rabbit immune IgG purified by Protein A affinity chromatography. Sensor chips functionalized with DiCo-Mix, PfAMA1-3D7, PfAMA1-FCR3, or PfAMA1-HB3 were used to determine the concentrations of antigen-specific antibodies in all preparations by CFCA. Table 1 shows the concentration of total IgG (milligrams per milliliter) and the quantity of antigen-specific antibodies indicated both by the concentration (milligrams per milliliter) and the proportion relative to total IgG (%). The quantity of allele-specific IgG is also shown relative to the quantity of DiCo-Mix-specific IgG (%). As already observed for the avidity index (Figure 3), there were no significant differences in immunogenicity among the three immunization groups (Figure 4A). Using the combination of PfAMA1 and PfRon2L either as complex (C) or at separate injections sites (D + R) also had no quantitative effect on the strain specificity of the induced immune responses, which ranged from 60 to 70% (PfAMA1-3D7 and PfAMA1-HB3) up to 80% (PfAMA1-FCR3) allele-specific IgG relative to the DiCo-Mix-specific immune IgG (Figure 4B). This result was also confirmed by SPR-based competition assays (Figure 4C). In the SPR-based competition assay using the DiCo-Mix surface, we also tested mixtures of three and six alleles to investigate the reactivity profile of the DiCo-Mix-specific antibody preparations. As also shown in Figure 4C, the use of DiCo-Mix as a competitor led to the complete abolition of binding, whereas three alleles (PfAMA1-3D7, PfAMA1-FCR3, PfAMA1-HB3) neutralized 80% of the binding signal and six alleles (PfAMA1-3D7, PfAMA1-FCR3, PfAMA1-HB3, PfAMA1-Dd2, PfAMA1-7G8, and PfAMA1–RO33) neutralized 90% of the binding signal.
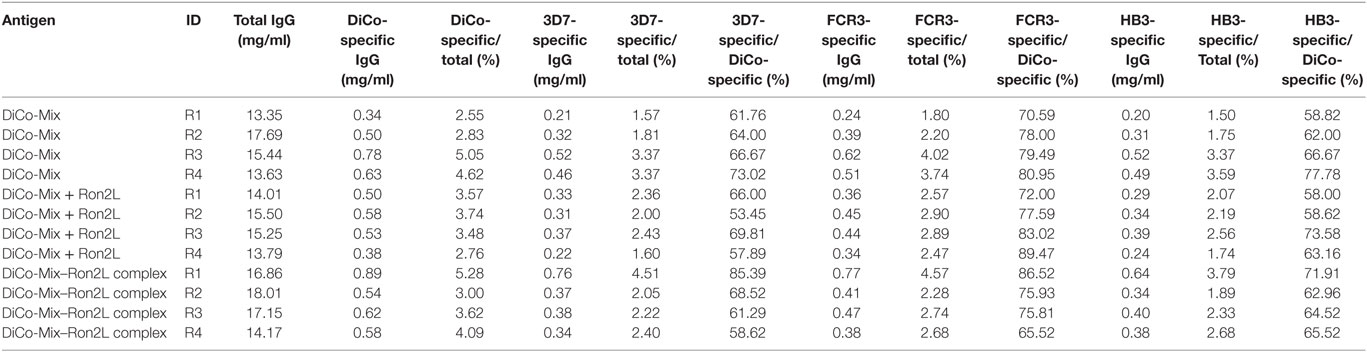
Table 1. Summary of total and antigen-specific antibody concentrations in the purified immune IgG preparations.
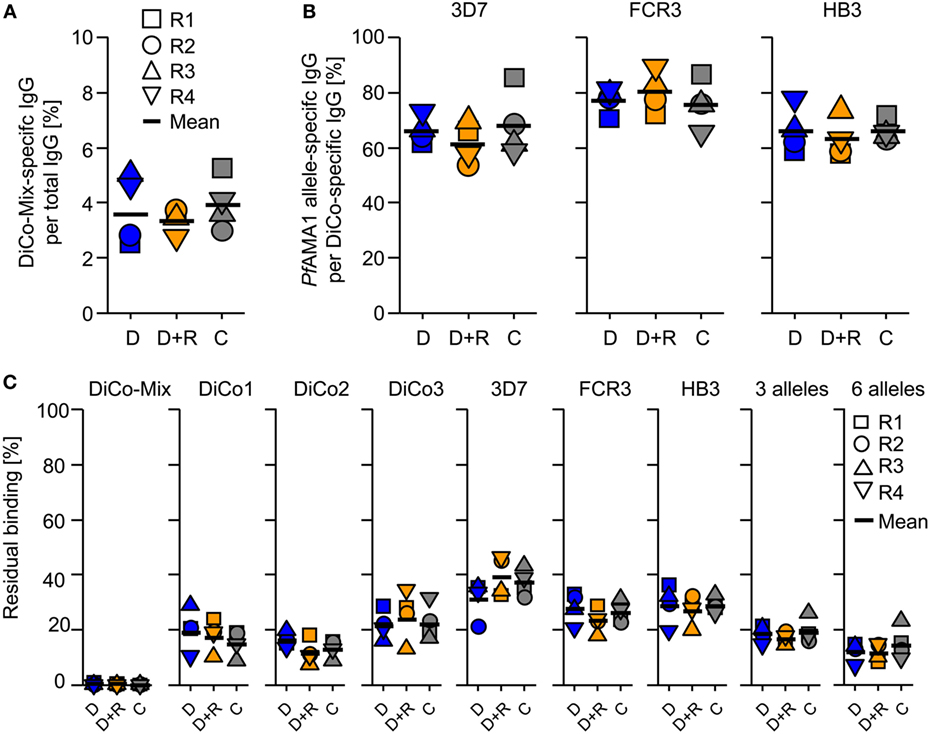
Figure 4. Quantification and analysis of purified immune IgG. The total IgG concentration in the purified IgG preparation was determined by analytical size exclusion chromatography. The antigen-specific IgG concentration for the immunization antigen (DiCo-Mix) as well as for three Plasmodium falciparum apical membrane antigen 1 (PfAMA1) alleles (3D7, FCR3, and HB3) was quantified by surface plasmon resonance (SPR) spectroscopy (Biacore T200) using the calibration-free concentration analysis (CFCA) module. The DiCo-specific IgG concentration is expressed as a percentage of total IgG (A) and as an indication for balanced PfAMA1 allele coverage as a percentage PfAMA1 allele-specific response per DiCo-Mix-induced IgG response (B). (C) SPR-based competition experiments were performed to confirm the CFCA results and to further characterize the purified IgG preparation after immunization with the different DiCo formulations (D: group of rabbits that received 50 µg DiCo-Mix; D + R: group of rabbits that received 50 µg DiCo-Mix and 50 µg of PfRon2L peptide at different injection sites; C: group of rabbits that were immunized with the DiCo–PfRon2L complex formed by mixing 50 µg of DiCo-Mix and 50 µg of PfRon2L). The DiCo-Mix was immobilized by EDC–NHS chemistry to a CM5 sensor chip. Purified IgG preparations were diluted 1:100 and mixed 1:1 with 30 µg/ml of each competitor antigen (indicated above each lane). For the DiCo-Mix and the three-allele mixture (3D7, FCR3, and HB3), the concentration of each antigen was 10 µg/ml, whereas in the six-allele mixture (3D7, FCR3, HB3, Dd2, 7G8, and RO33), the concentration of each individual PfAMA1 allele was 5 µg/ml. Competition samples were incubated for 1 h at room temperature and residual binding to the DiCo-mix surface was determined. The reference (IgG sample mixed with buffer) was set to 100% and residual binding is expressed as a percentage compared to the reference.
Growth Inhibition Assays
Growth inhibition assays were used to compare the ability of the immune IgG preparations derived from the three different rabbit groups (D, D + R, and C) to inhibit parasite growth. The data were used to calculate IC50 values for three different P. falciparum strains (3D7A, FCR3, and HB3) that have previously been used to characterize PfAMA1 strain-specific antibodies. Figure 5 shows the IC50 values as normalized to the DiCo-specific IgG (Figure 5A) as well as to allele-specific IgG (Figure 5B). The results clearly show that for both, DiCo-specific IgG, as well as strain-specific IgG, there is no significant difference between the different groups regarding the IC50 values observed for each of the three different strains. It is also obvious that the mean IC50 values (between 120 and 180 µg/ml) observed for the DiCo-specific IgG are in the expected range and do not differ significantly for the three strains, which proves the induction of a cross-strain-specific immune response. The differences between DiCo and single allele-specific IC50 values are proportional to the fraction of allele-specific IgG within the DiCo-specific antibody response (Figure 4B).
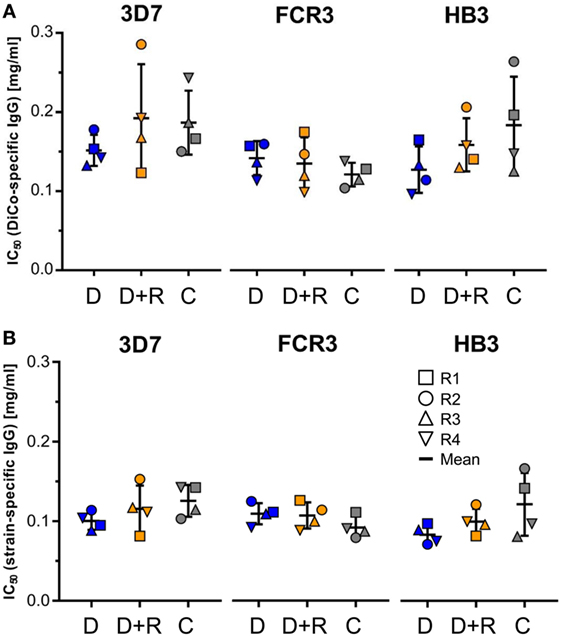
Figure 5. Growth inhibition assay. The parasite growth-inhibitory activity of the immune IgG preparations from the three different rabbit groups (see below) were assessed using three P. falciparum strains (3D7A, FCR3, and HB3). A serial dilution of purified IgG preparations starting at 6 mg/ml total IgG were used to determine IC50 values for the three P. falciparum strains. Based on the antigen-specific antibody concentrations (Table 1), IC50 values are expressed in micrograms per milliliter DiCo-specific IgG (A) and micrograms per milliliter allele-specific IgG (B). D: group of rabbits that were vaccinated using 50 µg DiCo-Mix; D + R: group of rabbits that were vaccinated using 50 µg DiCo-Mix and 50 µg of PfRon2L peptide at different injection sites; C: group of rabbits that were immunized with the DiCo–PfRon2L complex formed by mixing 50 µg of DiCo-Mix and 50 µg of PfRon2L. To facilitate the complex formation, the DiCo–PfRon2L mixture was incubated for 30 min at room temperature before formulation with the adjuvant and immunization.
Discussion
A preparation consisting of recombinant PfAMA1 and its peptide ligand PfRon2L was recently shown to achieve greater efficacy than PfAMA1 alone in an in vitro parasite GIA using P. falciparum and to protect mice from a lethal challenge with P. yoelii (35). We, therefore, tested the same strategy to determine whether it could enhance the potency of antibody responses when combining recombinant DiCo variants of PfAMA1 with PfRon2L. The available epidemiological (46), preclinical (14), and clinical data (47, 48) clearly show that both greater potency (by increasing specificity or immunogenicity) and improved cross-strain coverage are required to develop an efficient PfAMA1-based blood-stage vaccine (19, 49). We immunized three groups of four rabbits with a balanced mixture of the three DiCo variants (D), or DiCo-Mix and PfRon2L at different injection sites (D + R), or the DiCo-Mix in complex with PfRon2L (C), and conducted a detailed quantitative and qualitative analysis of the resulting immune IgG, using different ELISA formats, SPR-based binding assays and parasite GIAs.
In agreement with our earlier DiCo-based studies (37, 44) and those reported by other groups (19, 45), we observed balanced IC50 values (~150 μg/ml) against different strains. This is in contrast to using single alleles for immunization, where the IC50 values in GIA are significantly higher (greater than twofold) for heterologous compared to homologous (vaccine-like) strains (18).
On one hand, comparison among the three immunization groups (D, D + R, and C) showed that co-immunization with PfRon2L [either at a different injection site (D + R) or in a complex (C)] does not reduce the ability of the DiCo-Mix to induce cross-strain parasite inhibitory responses. On the other hand, there was no significant difference in IC50 values between the DiCo–PfRon2L complex group and the other two groups, as reported for homologous GIAs using P. falciparum and a lethal challenge using P. yoelii following the immunization of mice with single-allele AMA1–Ron2L complexes (35).
Even though these results appear contradictory, there may be a common explanation. The authors of the abovementioned study provided evidence that the improved efficacy of the AMA1–Ron2L complex relies on antibody responses, and that antibodies against certain variable loops surrounding the Ron2-binding pocket play an important role in this scenario (35). Parasite growth-inhibitory antibodies interfering with the AMA1–Ron2L interaction by recognizing these variable (or even hypervariable) loops are most probably strain specific and, therefore, less favorable when aiming for cross-strain efficacy. This is further illustrated by the monoclonal antibody 1F9 (50), a murine antibody raised by immunization with PfAMA1-3D7, which interferes with the PfAMA1–PfRon2 interaction by binding to a reduction-sensitive epitope including the most polymorphic residue of the antigen. Point mutations at this residue, such as those found in alleles PfAMA1-HB3 and PfAMA1-W2mef, prevent 1F9 binding to the corresponding alleles and eliminate growth inhibitory activity against these strains (28). Immunization with multiple PfAMA1 alleles improved cross-strain efficacy by increasing the proportion of conserved face-specific antibodies that recognize epitopes shared by the majority or even all known P. falciparum strains (14, 15, 17, 18). Taken together, these experiments suggest that at least four (18) or five (17) different alleles must be combined to induce cross-strain-specific responses covering diverse naturally occurring strains.
Each additional allele provided as a recombinant protein in the context of a vaccine formulation adds to the costs and complexity of process development, manufacturing, and regulatory approval, so, three DiCo variants have been designed to cover the allelic diversity of PfAMA1 comprehensively using the smallest number of recombinant proteins (19). As discussed above for the conventional multi-allele approach (14, 15, 17, 18), the DiCo approach successfully increases the induction of conserved region-specific antibodies by dilution of the strain-specific variable epitopes (51). Alternative approaches that drive the immune response toward conserved PfAMA1 epitopes include the immunodampening of the hypervariable loop Id (52) as well as glycan masking of the variable regions (Boes et al., in preparation). If the improved potency of the AMA1–Ron2L complex results from the induction of antibodies targeting variable loops near the Ron2-binding pocket that undergo conformational changes when Ron2 binds (35), then a strategy favoring cross-strain specific epitopes by overrepresentation of the conserved regions may reduce the induction of such antibodies below effective concentrations, which is probably why the IC50-values we observed could not be improved by the combination of DiCo-Mix with PfRon2L. Alternatively, the artificial mixed allele design approach of the PfAMA1 DiCo variants could affect the conformational changes normally induced by PfRon2-binding in the variable loop region of natural PfAMA1 alleles and thus fail to induce efficacious antibodies. Although these explanations are speculative and require conformational studies, they highlights the complexity associated with PfAMA1 as a vaccine target.
In our setting, co-formulation with the PfRon2L peptide did not improve the in vitro efficacy of the DiCo-Mix, a vaccine that aims for cross-strain coverage by eliciting higher levels of constant region-specific antibodies. However, our results provide additional insight into DiCo-specific antibody responses. The competition experiment revealed that single PfAMA1 alleles neutralize between 65% (PfAMA1-3D7) and 75% (PfAMA1-FCR3) of DiCo-specific immune IgG. Whereas conserved region-specific immune IgG will be neutralized by all alleles, antibodies against polymorphic, strain-specific regions will only be neutralized by alleles that present the corresponding epitopes. The observed trend toward different degrees of competition of the three alleles (Pf3D7, PfFCR3, and PfHB3), although not statistically significant, most probably reflects the ability of the DiCo-Mix to induce corresponding strain-specific IgG in addition to commonly neutralizing cross-strain-specific antibodies, given that constant region-specific antibodies will be neutralized equally by all PfAMA1 alleles and DiCo variants. The trend indicates that DiCo-specific immune IgG may contain a lower proportion of strain-specific IgG directed toward PfAMA1-3D7 than PfAMA1-FCR3 or PfAMA1-HB3. The comparison of GIA IC50 values (~125 μg/ml) for PfAMA1-3D7-specific antibodies in immune IgG preparations derived from the three immunization groups (D, D + R, and C) and the corresponding IC50 values for PfAMA1-3D7-specific antibodies (~40 μg/ml) generated by single-allele immunization with PfAMA1-3D7 (38) suggests that DiCo-derived PfAMA1-3D7-specific antibodies have a lower in vitro efficacy. This contrasts with the results of a PfAMA1 multi-allele study in which identical IC50 values were observed for affinity-purified PfAMA1-3D7-specific antibodies derived from rabbits immunized with either single-allele PfAMA1-3D7 or with a mixture (Quadvax) of the four different PfAMA1 alleles 3D7, FVO, HB3, and W2mef (18). Even though Quadvax seems to induce predominantly conserved region-specific antibodies, it is possible that the presence of PfAMA1-3D7 within the vaccine formulation leads to the induction of potent, allele-specific growth inhibitory antibodies, which account for this difference. Looking at an alignment of sequences featuring the three DiCo variants, as well as the three alleles PfAMA1-3D7, PfAMA1-FCR3, and PfAMA1-HB3 (Figure S1 in Supplementary Material), we find a glutamic acid residue (E197) in the hypervariable loop Id (33), which is not present in any of the three DiCo variants or strains PfAMA1-3D7, PfAMA1-FCR3, and PfAMA1-HB3. E197, the most polymorphic residue in PfAMA1, is a key residue within an epitope targeted by the Pf3D7 growth-inhibitory monoclonal antibody 1F9 (28). Replacing this residue with random amino acids or those found in other allelic variants of PfAMA1 abolishes 1F9 binding and consequently growth-inhibitory activity. Another hint that antibodies directed to the Id loop may play an essential role was shown by replacing all polymorphic residues within the Id loop with alanine, aiming to reduce Id-specific reactivity and generate improved cross-strain-specific responses targeting the conserved region (52). The improved cross-strain efficacy was achieved at the cost of reduced overall growth-inhibitory activity for the homologous strain, which is analogous to our observations. All these results and conclusions together suggest that it would be more promising to work on the induction of highly potent or maximally cross-strain-specific antibodies by investing resources into concepts that target these goals separately, and, if reasonable, combine them once both approaches show promising improvements beyond the current state of the art.
Ethics Statement
Rabbits were housed, immunized, and sampled by Biogenes GmbH (Berlin, Germany) according to national animal welfare regulations. The animal facilities and protocols were reviewed and approved by: Landesamt für Landwirtschaft, Lebensmittelsicherheit, und Fischerei MecklenburgVorpommern (LALLF M-V) (Approval No: 7221.3-2-030-13).
Author Contributions
AB and HS conceived the study, performed the experiments, analyzed the data, and wrote the manuscript. RF performed and analyzed the GIA experiments and contributed to data analysis and writing the manuscript. AR, SS, and RFi conceived the overall study design and contributed to writing the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Richard M. Twyman for manuscript editing. We are grateful for the BG98 standard, which was kindly provided by BPRC (Rijswijk, the Netherlands). Plasmodium falciparum 3D7A, MRA-151, deposited by D. Walliker; Plasmodium falciparum HB3, MRA-155, deposited by T. E. Wellems; and Plasmodium falciparum FCR-3/Gambia Subline F-86, MRA-731, deposited by W. Trager, were obtained through the MR4 as part of the BEI Resources Repository, NIAID, NIH. We also would like to thank Anh-Tuan Pham for technical help performing the culture of Plasmodium falciparum parasites and the growth inhibitions assays. This work was supported by the “Fraunhofer-Zukunftsstiftung.”
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.00743/full#supplementary-material.
References
2. Anders RFR, Crewther PEP, Edwards SS, Margetts MM, Matthew MLM, Pollock BB, et al. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine (1997) 16:240–7. doi:10.1016/S0264-410X(97)88331-4
3. Collins WE, Pye D, Crewther PE, Vandenberg KL, Galland GG, Sulzer AJ, et al. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am J Trop Med Hyg (1994) 51:711–9. doi:10.4269/ajtmh.1994.51.711
4. Deans JA, Knight AM, Jean WC, Waters AP, Cohen S, Mitchell GH. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol (1988) 10:535–52. doi:10.1111/j.1365-3024.1988.tb00241.x
5. Narum DL, Ogun SA, Thomas AW, Holder AA. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect Immun (2000) 68:2899–906. doi:10.1128/IAI.68.5.2899-2906.2000
6. Remarque EJE, Faber BWB, Kocken CHMC, Thomas AWA. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol (2008) 24:11–11. doi:10.1016/j.pt.2007.12.002
7. Thera MA, Doumbo OK, Coulibaly D, Diallo DA, Kone AK, Guindo AB, et al. Safety and immunogenicity of an AMA-1 malaria vaccine in Malian adults: results of a phase 1 randomized controlled trial. PLoS One (2008) 3:e1465. doi:10.1371/journal.pone.0001465
8. Pierce MA, Ellis RD, Martin LB, Malkin E, Tierney E, Miura K, et al. Phase 1 safety and immunogenicity trial of the Plasmodium falciparum blood-stage malaria vaccine AMA1-C1/ISA 720 in Australian adults. Vaccine (2010) 28:2236–42. doi:10.1016/j.vaccine.2009.12.049
9. Ellis RD, Wu Y, Martin LB, Shaffer D, Miura K, Aebig J, et al. Phase 1 study in malaria naïve adults of BSAM2/Alhydrogel®+CPG 7909, a blood stage vaccine against P. falciparum malaria. PLoS One (2012) 7:e46094. doi:10.1371/journal.pone.0046094
10. Dicko A, Sagara I, Ellis RD, Miura K, Guindo O, Kamate B, et al. Phase 1 study of a combination AMA1 blood stage malaria vaccine in Malian children. PLoS One (2008) 3:e1563. doi:10.1371/journal.pone.0001563
11. Healer J, Murphy V, Hodder AN, Masciantonio R, Gemmill AW, Anders RF, et al. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol Microbiol (2004) 52:159–68. doi:10.1111/j.1365-2958.2003.03974.x
12. Polley SD, Chokejindachai W, Conway DJ. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics (2003) 165(2):555–61.
13. Polley SD, Conway DJ. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics (2001) 158(4):1505–12.
14. Kusi KA, Faber BW, Riasat V, Thomas AW, Kocken CHM, Remarque EJ. Generation of humoral immune responses to multi-allele PfAMA1 vaccines; effect of adjuvant and number of component alleles on the breadth of response. PLoS One (2010) 5:e15391. doi:10.1371/journal.pone.0015391
15. Kusi KA, Faber BW, Thomas AW, Remarque EJ. Humoral immune response to mixed PfAMA1 alleles; multivalent PfAMA1 vaccines induce broad specificity. PLoS One (2009) 4:e8110. doi:10.1371/journal.pone.0008110
16. Kusi KA, Faber BW, van der Eijk M, Thomas AW, Kocken CHM, Remarque EJ. Immunization with different PfAMA1 alleles in sequence induces clonal imprint humoral responses that are similar to responses induced by the same alleles as a vaccine cocktail in rabbits. Malar J (2011) 10:40. doi:10.1186/1475-2875-10-40
17. Miura K, Herrera R, Diouf A, Zhou H, Mu J, Hu Z, et al. Overcoming allelic specificity by immunization with five allelic forms of Plasmodium falciparum apical membrane antigen 1. Infect Immun (2013) 81:1491–501. doi:10.1128/IAI.01414-12
18. Dutta S, Dlugosz LS, Drew DR, Ge X, Ge X, Ababacar D, et al. Overcoming antigenic diversity by enhancing the immunogenicity of conserved epitopes on the malaria vaccine candidate apical membrane antigen-1. PLoS Pathog (2013) 9:e1003840. doi:10.1371/journal.ppat.1003840
19. Remarque EJE, Faber BWB, Kocken CHMC, Thomas AWA. A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infect Immun (2008) 76:2660–70. doi:10.1128/IAI.00170-08
20. Sedegah M, Hollingdale MR, Farooq F, Ganeshan H, Belmonte M, Kim Y, et al. Sterile immunity to malaria after DNA prime/adenovirus boost immunization is associated with effector memory CD8+ T cells targeting AMA1 class I epitopes. PLoS One (2014) 9:e106241. doi:10.1371/journal.pone.0106241
21. Hodgson SH, Choudhary P, Elias SC, Milne KH, Rampling TW, Biswas S, et al. Combining viral vectored and protein-in-adjuvant vaccines against the blood-stage malaria antigen AMA1: report on a phase 1a clinical trial. Mol Ther (2014) 22:2142–54. doi:10.1038/mt.2014.157
22. Biswas S, Dicks MDJ, Long CA, Remarque EJ, Siani L, Colloca S, et al. Transgene optimization, immunogenicity and in vitro efficacy of viral vectored vaccines expressing two alleles of Plasmodium falciparum AMA1. PLoS One (2011) 6:e20977. doi:10.1371/journal.pone.0020977
23. Lamarque M, Besteiro S, Papoin J, Roques M, Vulliez-Le Normand B, Morlon-Guyot J, et al. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog (2011) 7:e1001276. doi:10.1371/journal.ppat.1001276
24. Srinivasan P, Beatty WL, Diouf A, Herrera R, Ambroggio X, Moch JK, et al. Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proc Natl Acad Sci U S A (2011) 108:13275–80. doi:10.1073/pnas.1110303108
25. Vulliez-Le Normand B, Tonkin ML, Lamarque MH, Langer S, Hoos S, Roques M, et al. Structural and functional insights into the malaria parasite moving junction complex. PLoS Pathog (2012) 8:e1002755. doi:10.1371/journal.ppat.1002755
26. Vulliez-Le Normand B, Faber BW, Saul FA, van der Eijk M, Thomas AW, Singh B, et al. Crystal structure of Plasmodium knowlesi apical membrane antigen 1 and its complex with an invasion-inhibitory monoclonal antibody. PLoS One (2015) 10:e0123567. doi:10.1371/journal.pone.0123567
27. Maskus DJ, Królik M, Bethke S, Spiegel H, Kapelski S, Seidel M, et al. Characterization of a novel inhibitory human monoclonal antibody directed against Plasmodium falciparum apical membrane antigen 1. Sci Rep (2016) 6:39462. doi:10.1038/srep39462
28. Coley AM, Parisi K, Masciantonio R, Hoeck J, Casey JL, Murphy VJ, et al. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect Immun (2006) 74:2628–36. doi:10.1128/IAI.74.5.2628-2636.2006
29. Wang G, Drinkwater N, Drew DR, MacRaild CA, Chalmers DK, Mohanty B, et al. Structure-activity studies of β-Hairpin peptide inhibitors of the Plasmodium falciparum AMA1-RON2 interaction. J Mol Biol (2016) 428:3986–98. doi:10.1016/j.jmb.2016.07.001
30. Wang G, MacRaild CA, Mohanty B, Mobli M, Cowieson NP, Anders RF, et al. Molecular insights into the interaction between Plasmodium falciparum apical membrane antigen 1 and an invasion-inhibitory peptide. PLoS One (2014) 9:e109674. doi:10.1371/journal.pone.0109674
31. Srinivasan P, Yasgar A, Luci DK, Beatty WL, Hu X, Andersen J, et al. Disrupting malaria parasite AMA1-RON2 interaction with a small molecule prevents erythrocyte invasion. Nat Commun (2013) 4:2261. doi:10.1038/ncomms3261
32. Pihan E, Delgadillo RF, Tonkin ML, Pugnière M, Lebrun M, Boulanger MJ, et al. Computational and biophysical approaches to protein-protein interaction inhibition of Plasmodium falciparum AMA1/RON2 complex. J Comput Aided Mol Des (2015) 29:525–39. doi:10.1007/s10822-015-9842-7
33. Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, et al. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci U S A (2005) 102:12736–41. doi:10.1073/pnas.0501808102
34. Lim SS, Yang W, Krishnarjuna B, Kannan Sivaraman K, Chandrashekaran IR, Kass I, et al. Structure and dynamics of apical membrane antigen 1 from Plasmodium falciparum FVO. Biochemistry (2014) 53:7310–20. doi:10.1021/bi5012089
35. Srinivasan P, Ekanem E, Diouf A, Tonkin ML, Miura K, Boulanger MJ, et al. Immunization with a functional protein complex required for erythrocyte invasion protects against lethal malaria. Proc Natl Acad Sci U S A (2014) 111:10311–6. doi:10.1073/pnas.1409928111
36. Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet (1986) 204:383–96. doi:10.1007/BF00331014
37. Spiegel H, Boes A, Kastilan R, Kapelski S, Edgue G, Beiss V, et al. The stage-specific in vitro efficacy of a malaria antigen cocktail provides valuable insights into the development of effective multi-stage vaccines. Biotechnol J (2015) 10(10):1651–9. doi:10.1002/biot.201500055
38. Boes A, Reimann A, Twyman RM, Fischer R, Schillberg S, Spiegel H. A plant-based transient expression system for the rapid production of malaria vaccine candidates. Methods Mol Biol (2016) 1404:597–619. doi:10.1007/978-1-4939-3389-1_39
39. Boes A, Spiegel H, Edgue G, Kapelski S, Scheuermayer M, Fendel R, et al. Detailed functional characterization of glycosylated and nonglycosylated variants of malaria vaccine candidate PfAMA1 produced in Nicotiana benthamiana and analysis of growth inhibitory responses in rabbits. Plant Biotechnol J (2014) 13(2):222–34. doi:10.1111/pbi.12255
40. Remarque EJ, Roestenberg M, Younis S, Walraven V, van der Werff N, Faber BW, et al. Humoral immune responses to a single allele PfAMA1 vaccine in healthy malaria-naïve adults. PLoS One (2012) 7:e38898. doi:10.1371/journal.pone.0038898
41. Pol E, Karlsson R, Roos H, Jansson A, Xu B, Larsson A, et al. Biosensor-based characterization of serum antibodies during development of an anti-IgE immunotherapeutic against allergy and asthma. J Mol Recognit (2007) 20:22–31. doi:10.1002/jmr.804
42. Maskus DJ, Bethke S, Seidel M, Kapelski S, Addai-Mensah O, Boes A, et al. Isolation, production and characterization of fully human monoclonal antibodies directed to Plasmodium falciparum MSP10. Malar J (2015) 14:276. doi:10.1186/s12936-015-0797-X
43. Boes A, Spiegel H, Voepel N, Edgue G, Beiss V, Kapelski S, et al. Analysis of a multi-component multi-stage malaria vaccine candidate-tackling the cocktail challenge. PLoS One (2015) 10:e0131456. doi:10.1371/journal.pone.0131456
44. Boes A, Spiegel H, Kastilan R, Bethke S, Voepel N, Chudobová I, et al. Analysis of the dose-dependent stage-specific in vitro efficacy of a multi-stage malaria vaccine candidate cocktail. Malar J (2016) 15:279. doi:10.1186/s12936-016-1328-0
45. Faber BW, Younis S, Remarque EJ, Garcia RR, Riasat V, Walraven V, et al. Diversity covering AMA1-MSP119 fusion proteins as malaria vaccines. Infect Immun (2013) 81(5):1479–90. doi:10.1128/IAI.01267-12
46. Osier FHA, Weedall GD, Verra F, Murungi L, Tetteh KKA, Bull P, et al. Allelic diversity and naturally acquired allele-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 in Kenya. Infect Immun (2010) 78:4625–33. doi:10.1128/IAI.00576-10
47. Ouattara A, Mu J, Takala-Harrison S, Saye R, Sagara I, Dicko A, et al. Lack of allele-specific efficacy of a bivalent AMA1 malaria vaccine. Malar J (2010) 9:175. doi:10.1186/1475-2875-9-175
48. Payne RO, Milne KH, Elias SC, Edwards NJ, Douglas AD, Brown RE, et al. Demonstration of the blood-stage Plasmodium falciparum controlled human malaria infection model to assess efficacy of the P. falciparum apical membrane antigen 1 vaccine, FMP2.1/AS01. J Infect Dis (2016) 213:1743–51. doi:10.1093/infdis/jiw039
49. Barry AE, Arnott A. Strategies for designing and monitoring malaria vaccines targeting diverse antigens. Front Immunol (2014) 5:359. doi:10.3389/fimmu.2014.00359
50. Coley AM, Campanale NV, Casey JL, Hodder AN, Crewther PE, Anders RF, et al. Rapid and precise epitope mapping of monoclonal antibodies against Plasmodium falciparum AMA1 by combined phage display of fragments and random peptides. Protein Eng (2001) 14(9):691–8.
51. Ouattara A, Barry AE, Dutta S, Remarque EJ, Beeson JG, Plowe CV. Designing malaria vaccines to circumvent antigen variability. Vaccine (2015) 33:7506–12. doi:10.1016/j.vaccine.2015.09.110
Keywords: agroinfiltration, growth inhibition assay, plant molecular farming, Plasmodium falciparum, strain-transcending immune responses, surface plasmon resonance spectroscopy, calibration-free concentration analysis
Citation: Spiegel H, Boes A, Fendel R, Reimann A, Schillberg S and Fischer R (2017) Immunization with the Malaria Diversity-Covering Blood-Stage Vaccine Candidate Plasmodium falciparum Apical Membrane Antigen 1 DiCo in Complex with Its Natural Ligand PfRon2 Does Not Improve the In Vitro Efficacy. Front. Immunol. 8:743. doi: 10.3389/fimmu.2017.00743
Received: 24 March 2017; Accepted: 12 June 2017;
Published: 27 June 2017
Edited by:
Xu Huji, Second Military Medical University, ChinaReviewed by:
Raffael Nachbagauer, Icahn School of Medicine at Mount Sinai, United StatesUrszula Krzych, Walter Reed Army Institute of Research, United States
Copyright: © 2017 Spiegel, Boes, Fendel, Reimann, Schillberg and Fischer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Holger Spiegel, holger.spiegel@ime.fraunhofer.de
†These authors have contributed equally to this work.
 Holger Spiegel
Holger Spiegel Alexander Boes
Alexander Boes Rolf Fendel
Rolf Fendel Andreas Reimann
Andreas Reimann Stefan Schillberg
Stefan Schillberg Rainer Fischer
Rainer Fischer