- 1Division of Vaccine Discovery, La Jolla Institute for Allergy and Immunology, La Jolla, CA, United States
- 2Laboratorio Nacional de Virología, Centro Nacional de Diagnóstico y Referencia, Ministerio de Salud, Managua, Nicaragua
- 3Division of Clinical Immunology and Allergy, School of Medicine, University of São Paulo, São Paulo, Brazil
- 4Johns Hopkins University Bloomberg School of Public Health, Baltimore, MD, United States
- 5Vaccine Testing Center, Department of Medicine, Larner College of Medicine, University of Vermont, Burlington, VT, United States
- 6Institute for Immunology and Infectious Diseases, Murdoch University, Perth, WA, Australia
- 7Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, United States
- 8Genetech Research Institute, Colombo, Sri Lanka
- 9Chantal BIYA International Reference Centre for Research on the Prevention and Management of HIV/AIDS CIRCB, Yaoundé, Cameroon
- 10Department of Microbiology and Immunology, University of North Carolina School of Medicine, Chapel Hill, NC, United States
- 11Emerging Infectious Disease Programme, Duke-NUS Medical School, Singapore, Singapore
- 12Immunology Programme, Department of Microbiology and Immunology, Life Sciences Institute, National University of Singapore, Singapore, Singapore
- 13Institute of Infection and Global Health, University of Liverpool, Liverpool, United Kingdom
- 14National Institute for Health Research, Health Protection Research Unit in Emerging and Zoonotic Infections, University of Liverpool, Liverpool, United Kingdom
- 15Department of Microbiology and Cell Biology, Indian Institute of Science, Bangalore, India
- 16Neurovirology, National Institute of Mental Health and Neurosciences (NIMHANS), Bangalore, India
- 17Division of Infectious Diseases and Vaccinology, School of Public Health, University of California, Berkeley, Berkeley, CA, United States
Background: Dengue is a major public health problem worldwide. Assessment of adaptive immunity is important to understanding immunopathology and to define correlates of protection against dengue virus (DENV). To enable global assessment of CD4+ T cell responses, we mapped HLA-DRB1-restricted DENV-specific CD4+ T cell epitopes in individuals previously exposed to DENV in the general population of the dengue-endemic region of Managua, Nicaragua.
Methods: HLA class II epitopes in the population of Managua were identified by an in vitro IFNγ ELISPOT assay. CD4+ T cells purified by magnetic bead negative selection were stimulated with HLA-matched epitope pools in the presence of autologous antigen-presenting cells, followed by pool deconvolution to identify specific epitopes. The epitopes identified in this study were combined with those previously identified in the DENV endemic region of Sri Lanka, to generate a “megapool” (MP) consisting of 180 peptides specifically designed to achieve balanced HLA and DENV serotype coverage. The DENV CD4MP180 was validated by intracellular cytokine staining assays.
Results: We detected responses directed against a total of 431 epitopes, representing all 4 DENV serotypes, restricted by 15 different HLA-DRB1 alleles. The responses were associated with a similar pattern of protein immunodominance, overall higher magnitude of responses, as compared to what was observed previously in the Sri Lanka region. Based on these epitope mapping studies, we designed a DENV CD4 MP180 with higher and more consistent coverage, which allowed the detection of CD4+ T cell DENV responses ex vivo in various cohorts of DENV exposed donors worldwide, including donors from Nicaragua, Brazil, Singapore, Sri Lanka, and U.S. domestic flavivirus-naïve subjects immunized with Tetravalent Dengue Live-Attenuated Vaccine (TV005). This broad reactivity reflects that the 21 HLA-DRB1 alleles analyzed in this and previous studies account for more than 80% of alleles present with a phenotypic frequency ≥5% worldwide, corresponding to 92% phenotypic coverage of the general population (i.e., 92% of individuals express at least one of these alleles).
Conclusion: The DENV CD4 MP180 can be utilized to measure ex vivo responses to DENV irrespective of geographical location.
Introduction
Infection with dengue virus (DENV) is an ever-increasing public health issue of global concern. DENV is endemic in more than 120 countries, with more than 40% of the world population at risk of DENV transmission (1, 2). Overall, it has been estimated that there are up to 100 million cases of dengue infection per year (3). Infections with any of the four different DENV serotypes can either be asymptomatic, as it is in more than 75% of the cases, or lead to clinical manifestations ranging from mild febrile illness (dengue fever, DF) to severe disease (dengue hemorrhagic fever, DHF) and shock syndrome with plasma leakage (dengue shock syndrome) (2, 4).
The role of T cells in severe dengue disease has been a topic of spirited debate and remains controversial. A role for T cells in the immunopathogenesis of the virus has been proposed based on the higher TNFα vs IFNγ production seen in DHF, and the suboptimal degranulation and cytokine production with heterologous peptides stimulation (5–7). Conversely, several studies have proposed a protective role for DENV-specific CD4+ and CD8+ T cells with increased frequency in subclinical infection relative to severe disease, effective antiviral activity of CD8+ T cells during secondary DENV infection and HLA-restricted protective responses associated with specific phenotypes and polyfunctionality (8–12). The general consensus is that in addition to antibodies, CD8+ and CD4+ T cell responses should be taken into consideration in accounting for a complete DENV-specific adaptive immunity.
Several issues need to be addressed to enable the global assessment of T cell responses. First, accurate assessment of responses requires the identification of a sufficient number of T cell epitopes. We and others (12–16) have studied responses observed in the general population in endemic areas to define the epitope repertoire associated with at least partial natural immunity. Second, geographic variations in HLA frequencies underscore the necessity of defining sets of epitopes that can elicit responses restricted by the most representative and common HLA alleles worldwide. Thus, it is important that epitope identification studies are conducted in different geographical locations, to enable identification of epitopes restricted by a diverse set of HLA molecules.
We previously determined the repertoire of DENV-specific CD8+ T cell responses in the endemic region of Colombo (Sri Lanka) and reported that different HLA alleles are associated with differential magnitudes of response (13). When the repertoire of CD8+ T cell responses was assessed in a different cohort of donors derived from the endemic region of Managua, Nicaragua, we observed a very similar HLA-associated hierarchy of responses (11).
Similarly, when the repertoire of DENV-specific CD4+ T cell responses in the Colombo region of Sri Lanka was determined, different HLA-DRB1 alleles were associated with different magnitudes of responses (12). In this study, we examined the DENV-specific CD4+ T cell responses in the Managua region and asked whether HLA-specific CD4+ T cell responses in Nicaragua and Sri Lanka were also similarly correlated.
In addition to a repertoire of epitopes spanning the most common HLA types, it is important to be able to assess responses directly ex vivo, so that the phenotypes are not altered. It is also critical to evaluate T cell responses in small sample volumes, as often only small volumes are available in large scale vaccine trials or study of severe DENV disease, especially in pediatric patients. To meet these challenges, we developed the megapool (MP) approach, which is based on a large numbers of peptides pooled and formulated via sequential lyophilization (17). DENV MPs for both CD8+ and CD4+ T cell epitopes have been previously reported (12, 14). The study of T cell responses in different settings and geographical locations is important in the definition of MPs that are truly global. Accordingly, we leveraged the combined information derived from our previous studies and this investigation of the CD4+ T cell repertoire in the Managua region to specifically design a comprehensive and widely applicable DENV CD4 MP that can detect T cell immune responses ex vivo independently of the HLA class II haplotypes represented in a given population.
Materials and Methods
Human Blood Samples
Blood bank samples from endemic areas were obtained from healthy adult blood donors from the Nicaraguan National Blood Center, Managua and from National Blood Center, Ministry of Health, Colombo, Sri Lanka, in an anonymous fashion, as previously described (11–14). We also studied a cohort of volunteers from Burlington, VT, Baltimore, MD, and Washington, DC, who were flavivirus naïve at the time of vaccination with a live-attenuated tetravalent DENV vaccine formulation (DLAV, TV005), as previously described (18, 19). Negative controls were healthy volunteers from San Diego, CA, who were tested and found to be seronegative for all DENV serotypes and yellow fever virus, and therefore defined in this study as “flavivirus naïve.” To verify the reactivity of the DENV epitopes, additional independent cohorts of DENV cases were selected in several endemic regions from Brazil, Singapore, and India. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation and frozen in fetal bovine serum supplemented with 10% DMSO (13, 14). Seropositivity to DENV was determined by testing for total anti-DENV antibodies by inhibition ELISA or antigen capture ELISA and modified microFRNT assay for neutralization titer (conducted at the National Virology Laboratory of the Nicaraguan Ministry of Health and the University of North Carolina, Chapel Hill, respectively), as previously described (20–22). All samples were collected and used following guidelines from the Institutional Review Boards (IRBs) of the La Jolla Institute for Allergy and Immunology (LIAI), the University of California, Berkeley, the Nicaraguan Ministry of Health, and the Medical Faculty, University of Colombo (serving as the National Institutes of Health-approved IRB for Genetech Research Institute) and clinical trials registered at ClinicalTrials.gov under registration numbers NCT01506570 and NCT01436422.
Brazil
Longitudinal samples were collected during the 2014 and 2015 outbreaks at the Hospital das Clinicas Universidade de São Paulo with acute and convalescent dengue specimens stored in the sample repository. The infecting DENV serotype was determined in all patients by RT-PCR. Samples obtained at least 1 month after the onset of acute dengue symptoms were made available for this study. All participants signed the IRB-approved informed consent document (IRB document 0652/09, CAPPesq, Hospital das Clínicas, Universidade de São Paulo).
Singapore
A cohort of DF cases was established at Tan Tock Seng Hospital enrolling patients with suspected DENV. DENV diagnosis was confirmed by the detection of DENV RNA by reverse transcription polymerase chain reaction, or of NS1 antigen by enzyme-linked immunosorbent assay (Bio-Rad), as previously described (9). The 16 samples used in this study were collected in the late convalescent phase 60–160 days after fever onset and all participants gave informed consent. Collection of PBMCs was approved by the Singapore National Healthcare Group ethical review board (DSRB 2008/00293).
India
Healthy adults were recruited into the study after advertisement by the Indian Institute of Science (IISc) and the National Institute of Mental Health and Neurosciences (NIMHANS), both in Bengaluru, Karnataka State, India, as part of a study on Japanese encephalitis (JE) and JE vaccine. The study was conducted according to the principles of the Declaration of Helsinki. All participants gave written informed consent and the protocol was approved by the IISc Institutional Human Ethics Committee (ref 5/2011). Previous exposure to DENV was determined by neutralization titers against all four serotypes, as previously described (23).
HLA Typing and Phenotype Frequency Calculations
Donors were HLA typed by an ASHI-accredited laboratory at Murdoch University (Western Australia). HLA typing was performed for Class I (HLA A; B; C) and Class II (DQA1; DQB1, DRB1 3, 4, 5; DPB1), as previously described (12, 19, 24). Average phenotype frequencies for individual DRB1 alleles in the worldwide population are based on data available at DbMHC [NCBI (25)] and allelefrequencies.net (26), and were calculated as previously described (27–29). Briefly, for each HLA allele, an average gene frequency (gf) across Europe, North Africa, North-East Asia, South Pacific (Australia and Oceania), Hispanic North and South America, American Indian, South-East Asia, South-West Asia, and Sub-Saharan Africa populations was calculated, and an average phenotypic frequency (pf) was then derived utilizing the binomial distribution formula: pf = 1 − (1 − Σgf)2.
MHC Class II-Binding Predictions and Peptide Selection
A set of DENV peptides predicted to bind various HLA-DRB1 allelic variants (DRB1*01:01, 01:02, 03:01, 04:03, 04:07, 07:01, 08:02, 09:01, 11:01, 11:04, 13:01, 14:02, 14:06, 15:01) was chosen according to the criteria described in the Section “Results” to assess DENV-specific HLA-restricted CD4+ T cell responses. Fifteen-mer peptides from all four serotypes were selected based on their predicted binding affinity to the selected MHC class II molecules (12, 30, 31). This resulted in the synthesis of more than 1,900 predicted HLA-DRB1-binding peptides (Mimotopes, VIC, Australia). Of a total of 1,931 peptides tested, 398 (21%) were predicted to bind two or more DRB1 alleles, and only 138 (7%) were predicted to bind to three or more. For screening studies, the class II peptides were combined into pools of approximately 20 individual peptides, according to their predicted HLA restriction, resulting in approximately 7 pools per HLA allele. Table S1 in Supplementary Material lists the number of peptides synthetized for each allele and serotype. For MP generation, each individual peptide was resuspended in 40 mg/ml DMSO. All resuspended peptides were then pooled and re-lyophilized. The resulting lyocake was resuspended in DMSO corresponding to a concentration of 1 mg/ml of each individual peptide and then diluted 1:4 in distilled water. Similarly, a set of 122 previously defined CMV- and EBV-specific peptides was resuspended as a MP (1 mg/ml) and used as a control pool in the intracellular cytokine staining (ICS) experiments (17).
In Vitro Expansion of DENV-Specific T Cells
CD4+ T cells isolated from thawed PBMCs by magnetic bead negative selection and autologous antigen-presenting cells (APCs) were cultured in RPMI 1640 (Omega Scientific) supplemented with 5% human serum (Cellgro) at a 2:1 ratio and a density of 2 × 106 and 1 × 106 cells/ml, respectively, in 24-well plates (BD Biosciences). Cells were stimulated with DENV-specific peptide pools (averaging 20 peptides per pool) and incubated at 37°C in 5% CO2. IL-2 (10 U/ml; eBioscience) was added 4, 7, and 11 days after initial antigenic stimulation. Cells were harvested and screened for individual DENV-specific peptide reactivity on day 14.
IFNγ ELISPOT Assay
Peripheral blood mononuclear cells (5 × 104) were incubated in triplicates with 0.1 ml complete RPMI 1640 in the presence of HLA-matched peptide pools and individual peptides (2 µg/ml) after 14 days of in vitro expansion. After 20 h of incubation at 37°C, cells were incubated with biotinylated IFNγ mAb (mAb 7-B6-1 Mabtech, Stockholm, Sweden) for 2 h and developed as previously described (10, 13).
Flow Cytometry
For ICS, PBMCs were cultured in the presence DENV CD4 MPs and CMV/EBV MP (1–2 µg/ml) for 2 h prior to the addition of GolgiPlug containing brefeldin A (BD Biosciences, San Diego, CA, USA) and further incubation for 4 h. At the end of the stimulation, cells were stained for surface markers, then permeabilized, and finally stained for intracellular markers, and then analyzed as previously described (13). Gating strategy is summarized in Figure S1 in Supplementary Material.
Results
Analysis of HLA Frequencies in Nicaragua and Selection of Target Alleles
Previous studies in the DENV endemic region of Colombo, Sri Lanka, analyzed a total of 16 HLA-DRB1 alleles present at frequencies of ≥5% in that population (12). Here, we expanded our analysis of HLA-DRB1-restricted responses to the Managua region in Nicaragua, which is also endemic for DENV infections. As a first step, we performed HLA typing on PBMCs from 334 healthy blood bank donors. The frequency of different HLA-DRB1 alleles was analyzed to identify alleles frequent in Nicaragua and not covered by the previous Sri Lanka study.
Figure 1 shows the phenotypic frequency of alleles found at a frequency of 5% or greater in the Nicaraguan cohort (Figure 1, gray bars) and in the previously assessed Sri Lanka cohort (Figure 1, white bars) (12). The average allele frequency in the global worldwide population, calculated as described in the Section “Materials and Methods,” is also shown for comparison (Figure 1, black bars). To expand coverage of the Nicaraguan cohort, five HLA-DRB1 alleles that were not previously studied, but present at frequency ≥5% in Nicaragua (Figure 1, red arrows), were targeted for the epitope identification studies described below.
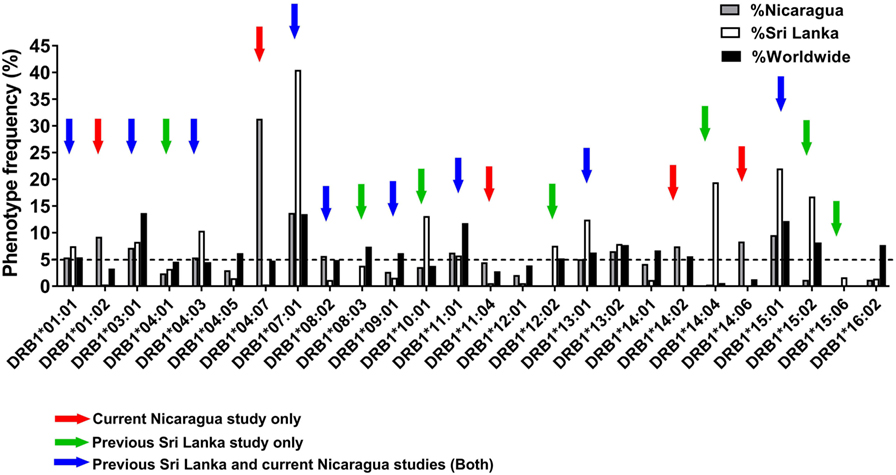
Figure 1. Phenotypic frequency of different HLA-DRB1 allelic variants in Nicaragua, Sri Lanka, and worldwide populations. The HLA-DRB1 phenotypic frequency from 334 Nicaraguan donors (gray bars) were compared to frequencies in Sri Lanka donors [(12); white bars] and worldwide (calculated as described in Section “Materials and Methods”; black bars). Red arrows indicate DRB1 alleles studied for epitope identification in this study only (not previously studied in Sri Lanka). Green arrows indicate DRB1 alleles not considered for epitope identification in this study, but previously addressed by the Sri Lankan studies. Blue arrows indicate DRB1 alleles studied for epitope identification in this study, and also previously addressed in the Sri Lankan studies.
To examine potential differences or similarities in HLA-DRB1-associated responses in Nicaragua and Sri Lanka, we selected nine HLA-DRB1 alleles previously studied in Sri Lanka which were also present at a frequency ≥5% in Nicaragua (blue arrows in Figure 1). An additional seven HLA-DRB1 alleles for which the epitope repertoire was previously studied in Sri Lanka (green arrows in Figure 1) were not further analyzed in this study. Overall, the 21 HLA-DRB1 alleles targeted for epitope identification studies by this and previous Sri Lankan study (marked by the colored arrows in Figure 1) cover 81.7% of alleles with phenotype frequency ≥5% worldwide. Combined, these alleles afford for 92% phenotypic coverage worldwide (i.e., 92% of individuals express at least one of these alleles).
Overall CD4 Reactivity in the Nicaragua Cohort for the HLA-DRB1 Alleles Studied
Previous experiments analyzed CD4+ T cell responses restricted by various HLA-DRB1 alleles (12) in a cohort of Sri Lankan donors. Potential epitopes from each of the four DENV serotypes predicted to bind at least 1 of 15 HLA-DRB1 alleles were selected, as previously described (12). An overall summary of the number of predicted binders for each DENV serotype and HLA allele is presented in Table S1 in Supplementary Material. For each HLA-DRB1 allelic variant considered, approximately 130 predicted peptides were tested in HLA-DRB1 matched donors, also as previously described (10).
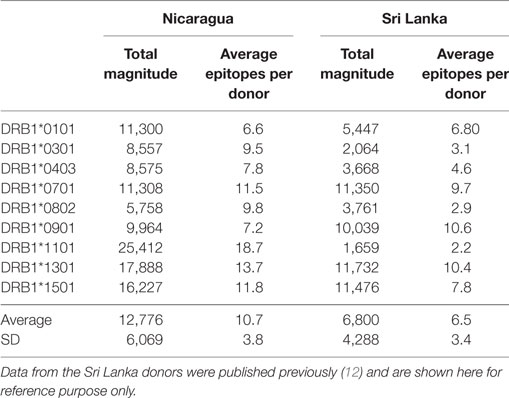
CD4+ T cells purified through negative selection using magnetic beads were cultured with autologous APCs and stimulated with HLA-matched DENV-specific pools. On day 14, cells were harvested and screened for reactivity against individual DENV-specific peptides in an IFNγ ELISPOT assay. A total of 4–10 donors for each of the HLA-DRB1 allelic variants were tested. A complete listing of all peptides tested, including response frequency and HLA restriction has been submitted to the Immune Epitope Database (IEDB)1,2 and the Immport database3 (Submission ID: SDY1109). The overall reactivity as a function of the different HLA alleles and different proteins is shown in Table 1. The average magnitude of responses for each allele (total magnitude of responses detected for that allele corrected for the number of donors tested) was 13,764 ± 6,451 (IFNγ SFC/106 PBMC) and the average number of epitopes/donor recognized in the context of each allele was 11 ± 4 epitopes/donor.
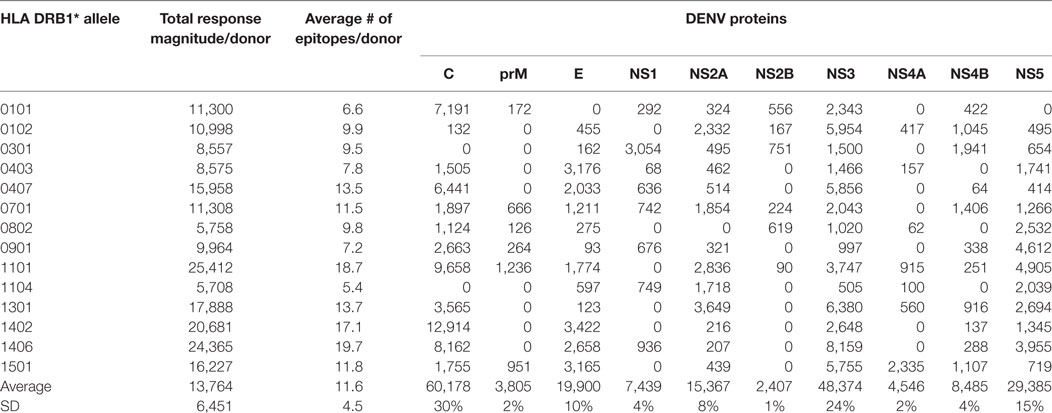
Table 1. CD4+ T cell dengue virus (DENV) immunoreactivity in peripheral blood mononuclear cells from the Nicaraguan cohort studied.
Similar Patterns of Protein Immunodominance and Higher Magnitude of DENV-Specific Responses in the Nicaraguan Cohort
Next, we considered the pattern of epitope immunoreactivity in the Nicaraguan cohort as a function of location within the DENV genome, and in comparison to the pattern previously observed in the Sri Lankan cohort (12). Pie charts in Figure 2 show data from the Nicaragua (Figure 2A) and Sri Lankan (Figure 2B) cohorts. The pattern of immunodominance in the two cohorts is remarkably similar, with the C, NS5, and NS3 antigens being most dominant in both, and accounting for 30 vs 31%, 24 vs 21%, and 15 vs 13% of the total response in the Nicaraguan and Sri Lankan cohorts, respectively.
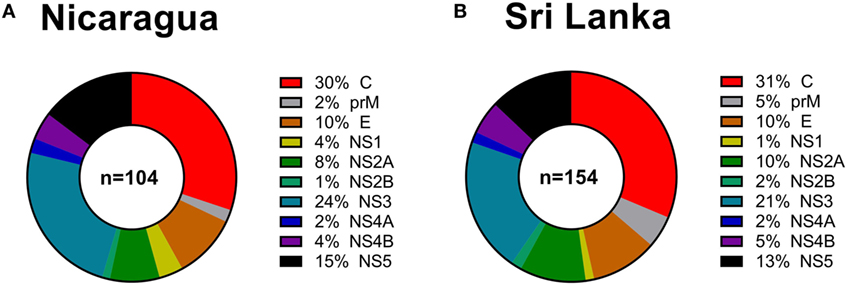
Figure 2. Patterns of protein immunodominance of CD4+ T cells responses in Nicaragua and Sri Lanka. Pie charts indicate the percentage of total magnitude of responses per protein observed in 104 Nicaragua (A) and 154 Sri Lanka (B) donors tested using identical methodology. Data from the Sri Lanka donors were published previously (12) and are shown here for reference purposes only.
We further compared the magnitude of CD4+ T cell responses in the Nicaraguan cohort to that previously observed in Sri Lankan donors (Table 2). When the 9 alleles tested in both cohorts are considered, the average magnitude of Nicaraguan responses was 12,776 ± 6,069 (IFNγ SFC/106 PBMC), while in the Sri Lankan study the average magnitude was 6,800 ± 4,288 (IFNγ SFC/106 PBMC) (Table 2). The response observed in the Nicaraguan cohort was almost twofold higher than in the Sri Lankan cohort (p = 0.0315 in a two-tailed Mann–Whitney test). A similar trend was observed when comparing the number of epitopes recognized for each allele in the two cohorts (10 ± 4 vs 6 ± 3; p = ns).
Differences in Patterns of HLA-Linked Immunodominance of CD4 Reactivity in Sri Lanka and Nicaragua Cohorts
CD8+ T cell responses measured in Sri Lankan and Nicaraguan healthy blood donor cohorts are associated with a very similar HLA-associated hierarchy of responses (11). The total response magnitude per donor for each HLA-DRB1 alleles analyzed in this study was then compared to the total magnitude of the same HLA-DRB1 previously tested in a Sri Lankan cohort using the (12). No correlation of HLA-DRB1 restriction and magnitude of responses was apparent (Figure 3A; R2 = 0.07291, p = 0.4823). Removing the “outlier” HLA-DRB1*11:01 allele with higher total responses magnitude per donor in the Nicaraguan cohort compared to the Sri Lankan cohort led to a significant, albeit weak correlation (Figure 3A; R2 = 0.5537, p = 0.0343). Even in this case, the correlation is much less than that previously observed in CD8+ T cell responses (R2 = 0.98) (11). The mechanism for this poor (or lack of) correlation was further investigated in the case of the HLA-DRB1*11:01 “outlier.”
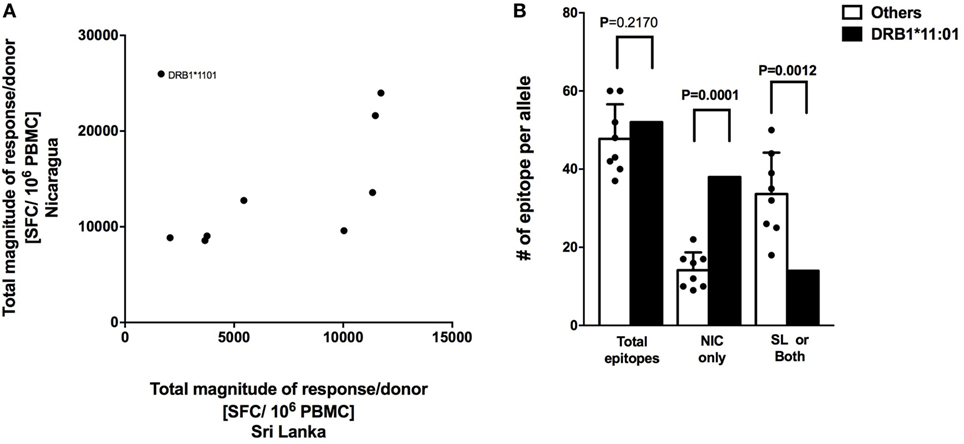
Figure 3. Correlation of HLA-restricted CD4+ T cells responses in Nicaragua and Sri Lanka cohorts. (A) Correlation of total responses magnitude per donor as a function of restricting HLA molecule, between Nicaraguan and Sri Lankan cohorts (R2 = 0.07291, p = 0.4823; if HLA-DRB1*11.01 is removed, R2 = 0.5537, p = 0.0343). (B) Comparison of the number of epitopes detected in the case of the HLA-DRB1*11:01 allele and in other eight HLA-DRB1 alleles (*01:01, *03:01, *04:03, *07:01, *08:02, *09:01, *13:01, and *15:01) considered in the study using one-sample t test. Bars refer to total epitopes, epitopes detected only in Nicaragua (NIC), and epitopes detected in Sri Lanka (SL) only or both cohorts.
We specifically tested whether the higher HLA-DRB1*11:01 CD4+ T cell responses in the Nicaraguan cohort were due to recognition of the same epitopes with higher magnitude of responses or, alternatively, whether the greater response to HLA-DRB1*11:01 was due to recognition of a new and/or broader set of epitopes. When looking at the total number of epitopes identified in the 2 cohorts, 52 total HLA-DRB1*11:01 epitopes and on average 48 (range 39–60) total epitopes restricted to the other 8 HLA-DRB1 alleles were identified. Therefore, in terms of total number of epitopes, DRB1*11:01 does not seem to recognize more epitopes respect to the other alleles in this study. We then looked and the number of epitopes recognized only in Nicaragua, or only in Sri Lanka or both populations. Out of 52 total HLA-DRB1*11:01 epitopes, 38 (73.1%) were recognized uniquely by CD4+ T cells from Nicaragua subjects, and only 14 (27.9%) were recognized in both populations or only the Sri Lankan cohort (Figure 3B).
In conclusion, when including all alleles studied, we did not observe a clear HLA-DRB1 response hierarchy of CD4+ T cell responses, which is in contrast to what we have previously detected at the level of CD8+ T cell responses (11). Our results also suggest that the HLA-DRB1*11:01 allele was able to recognize a broader array of epitopes presented only in the Nicaraguan cohort, in contrast to what was observed for the other alleles, where epitope-specific responses were detected in both cohorts (11).
Epitope Selection for a Global DENV CD4 MP
Pools of large numbers of epitopes, generated by sequential lyophilization (17), have been utilized as a tool to globally characterize both CD4+ and CD8+ T cell responses. These MPs have been validated in a number of contexts, including allergies (32), latent tuberculosis infection (33), tetanus/pertussis vaccination (34, 35), and DENV immunity (12, 14). We previously constructed CD4 DENV MP based on epitopes recognized by CD4+ T cells from Sri Lankan subjects (12). Here, we reevaluated the DENV CD4 MP by studying a broader set of epitopes than previously examined, including epitopes restricted by additional HLA alleles deriving from Nicaraguan subjects.
The previously published MP (DENV CD4 MP363) was composed of 363 epitopes defined in the Sri Lankan cohort (12). Here, we took a more structured approach to derive a new DENV CD4 MP (Figure 4A). First, we selected the highest response magnitude epitopes, considering responses in both the Sri Lankan and Nicaraguan cohorts, but also a previously described cohort of individuals immunized with a Tetravalent Dengue Live-Attenuated Vaccine (TV005) (19), until coverage of at least 50% of the total response was reached for each of the 21 HLA-DRB1 alleles identified above. If needed, additional epitopes were selected to ensure that each DENV serotype was represented by at least four epitopes for each HLA-DRB1 allele (some peptides are conserved in multiple serotypes, and thus fewer epitopes were required to meet this criteria). Additional epitopes were also selected to ensure that each DENV protein was represented by at least five epitopes. This process resulted in selection of a total of 209 epitopes. In instances where sequences overlapped by at least 13 residues, longer peptides encompassing both sequences were synthetized. This resulted in a final selection of 180 epitopes, which allowed coverage of 62% of the total responses of all cohorts combined.
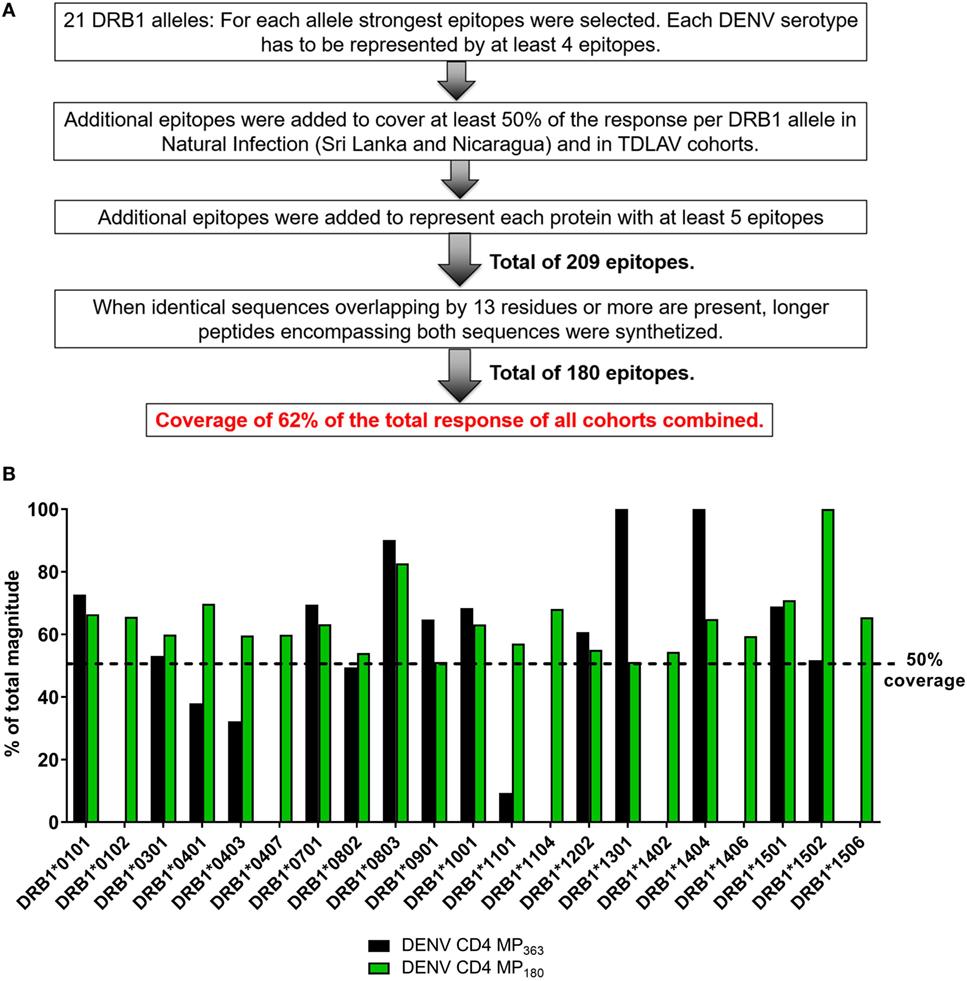
Figure 4. Characteristics of dengue virus (DENV) CD4 MP180. (A) Strategy used to select the epitopes composing the DENV CD4 MP180. (B) Comparison of HLA coverage in terms of magnitude of responses for epitopes belonging to the two DENV CD4 MPs.
The HLA coverage and magnitude of responses of the new optimized DENV CD4 MP180 was compared to the previous DENV CD4 MP363 (Figure 4B). Across the different HLA alleles, the DENV CD4 MP180 provided an average coverage of 62.4% (±13.6) of the total response, which is higher and more consistent than the previous DENV CD4 MP363 (42.0 ± 35.7%). DENV serotype coverage and protein representation had very similar patterns of coverage, with both DENV CD4 MPs providing fairly balanced coverage across the four serotypes (Figure S2A in Supplementary Material) and very similar patterns of protein representation (Figure S2B in Supplementary Material).
The DENV CD4 MP180 Affords Higher and More Consistent Coverage in Different Donor Cohorts
To assess the capacity of the DENV CD4 MP180 to detect T cell responses ex vivo from DENV exposed individuals, the DENV CD4 MPs were next tested in ICS assays utilizing PBMCs from Nicaraguan and Sri Lankan donor cohorts, as well as a cohort of donors vaccinated with TV005 (19) (Figure 5). As expected, the DENV CD4 MP180 performed better in the Nicaraguan cohort. Specifically, significant reactivity was detected in 6 out of 20 donors for the DENV CD4 MP363 and 11 out of 20 donors for the DENV CD4 MP180. The overall magnitude of response between the two MPs is significant by a two-tailed Wilcoxon test (Figure 5A). We did not expect to see a significant difference in reactivity in the Sri Lankan cohort, as Sri Lankan epitopes were contained in both DENV CD4 MPs. Indeed, in the Sri Lankan cohort, significant reactivity was detected in 8 out of 20 donors for both DENV CD4 MPs, with no significant difference in the magnitude of responses (Figure 5B). Similarly, TV005-induced responses were detected in seven out of eight donors for both DENV CD4 MPs, with no significant difference in response magnitude (Figure 5C).
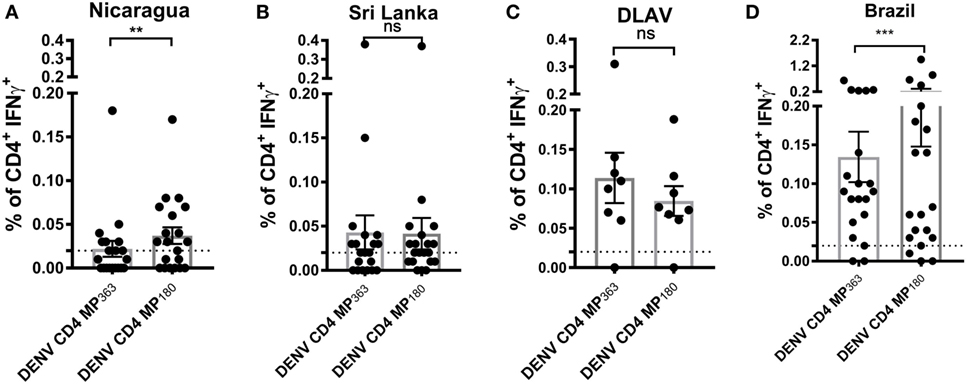
Figure 5. Comparison of CD4+ T cell reactivity to dengue virus (DENV) CD4 MP363 and DENV CD4 MP180. Percentage of CD4+ T cells producing IFNγ upon stimulation with the two DENV CD4 MPs in Nicaraguan (n = 20) (A), Sri Lankan (n = 20) (B) DLAV (n = 8) (C), and Brazilian (n = 20) (D) donor cohorts. Data are expressed as mean ± SEM. Statistical analyses were performed using two-tailed Wilcoxon test. *p < 0.05; **p < 0.01; ns, not significant. The dashed lines at 0.02% represent cutoff of positivity.
Next, we were interested to compare the performance of both DENV CD4 MP’s in a cohort that was not originally used to perform epitope identification. Accordingly, both pools were tested in a Brazilian cohort. Interestingly, also in a cohort that was not used to define the epitopes the MP180 pool works significantly better when compared to the previously defined CD4 MP, confirming the better performance of DENV CD4 MP180 when compared to DENV CD4 MP363 (Figure 5D).
Finally, we asked whether the DENV CD4 MP180 was capable of globally capturing DENV reactivity when multiple cohorts from different geographical locations were considered. We specifically considered locations that are endemic for DENV infections and that had not provided samples for our epitope identification studies. Accordingly, we analyzed responses in PBMCs derived from Brazilian, Singaporean, and Indian cohorts. As a negative control, we assayed healthy volunteers from San Diego who had been tested and found to be seronegative for all DENV serotypes and yellow fever virus (“flavivirus naïve” controls). As a positive control, we included a previously described CMV/EBV MP (17) (Figure 6).
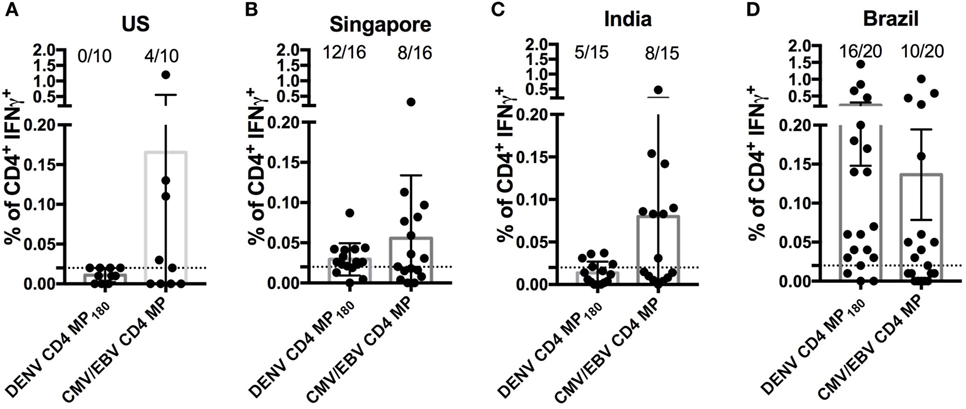
Figure 6. Assessment of dengue virus (DENV) CD4 MP180 reactivity in diverse geographical locations. Percentage of CD4+ T cells producing IFNγ upon stimulation with DENV CD4 MP180 in flavivirus naïve (n = 10) (A) and DENV exposed Singaporean (n = 16) (B), Indian (n = 9) (C), and Brazilian (n = 20) (D) donor cohorts. CMV/EBV CD4 MP stimulation was used as a control. To allow for comparison of the responses frequencies against the CMV/EBV and the dengue-specific response in all new cohorts the data showing response frequencies against the DENV CD4 MP180 in the Brazilian cohort from Figure 5D has been re-plotted in Figure 6D. Data are expressed as mean ± SEM, and the dashed lines at 0.02% represent the cutoff of positivity.
As expected, flavivirus naïve donors did not show reactivity to DENV CD4 MP180. Also, as expected, all the cohorts showed similar frequencies of reactivity, in the 45–55% range, against the CMV/EBV MP (Figure 6). DENV exposed cohorts were associated with a range of reactivity between 33 and 75% against DENV CD4 MP180. When frequencies and magnitude of response between cohorts were compared, magnitude but not frequency of response from Brazil was significantly higher compared to all other cohorts (Table S2 in Supplementary Material). Since the Brazilian cohort was not used to define the epitopes included in MP180, the most likely explanation is that these differences are due infection history and not MP composition. Nevertheless, the data should be interpreted with this caveat in mind.
Discussion
In this study, we analyzed the epitopes recognized by DENV-specific CD4+ T cells in the context of different HLA alleles in the general population of Nicaragua, where DENV is highly endemic. Comparison of the patterns of responses to those previously defined using identical methodology in the Colombo endemic area of Sri Lanka revealed a striking similarity in the overall pattern of recognition at the antigenic level (12). Using the knowledge derived from the combined analysis in the two locations, we derived a MP of epitopes (DENV CD4 MP180) and showed that it can be applied to the study of DENV responses in PBMC samples derived from disparate geographical locations.
Several lines of evidence indicate that T cells play an important role in the control of DENV infection, disease pathogenesis, and responses to vaccination (36, 37). This suggests that, in parallel with antibody response, T cell responses should be measured and analyzed to achieve a complete assessment of DENV-specific adaptive immunity. In addition, peripheral CD4+ T cells probably contain a fraction of circulating memory follicular T helper cells that are relevant for B cell differentiation and the production of neutralizing antibodies (38, 39). Since DENV infection and disease is a worldwide and ever-increasing problem, a global approach to assess T cell responses in different human populations is required. While several studies have analyzed CD8+ T cell responses against DENV (11, 13, 15, 40, 41), less information is available on their CD4+ T cell counterparts (12, 42). In this context, investigating DENV-specific HLA-restricted CD4+ T cell epitopes recognized in different DENV endemic populations and restricted by a broad variety of HLAs is necessary to evaluate potential genetic and geographical factors affecting DENV reactivity.
Our strategy expands previous studies on HLA-DRB1 locus-specific responses in the DENV endemic region of Colombo, Sri Lanka (12), to increase the HLA-DRB1 coverage from 16 to 21 of the most common alleles. We estimate that together these alleles provide phenotypic coverage of 92% of individuals in the general worldwide population, thus accomplishing our goal of enabling global coverage of the HLA-DRB1 locus. This statement should be interpreted with two caveats in mind: first, certain alleles not particularly common on a worldwide basis might be common in particular populations. In these cases, additional epitope identification studies may be required to fill gaps in coverage for a specific population. Second, our strategy does not include the DRB3/4/5, DP, and DQ loci. We have previously shown that epitope predictions performed on the main DRB1 molecules should be able to cover approximately 50% of the total CD4+ T cell response (43). Despite this evidence, future studies are planned to expand the coverage of DENV CD4+ T cell epitopes by considering those loci. Third, we note that our assignment of these responses to the HLA DRB1 locus is based on binding predictions and correspondence with donor HLA type. However, it is possible, given the well-known crossreactivity of HLA class II, that some of the response is in fact due to coexpressed DP or DQ specificities. However, we believe that this collection of epitopes will provide wide coverage based on the fact that different HLA class II molecules exhibit significant inter- and intra-loci repertoire overlap (28, 29, 44, 45) and that our epitope sets were tested in various geographical locations. An issue of relevance for this study is the accuracy and reliability of epitope prediction software, the IEDB runs periodic benchmarking of its tools, the latest published benchmarking for HLA class II (31) found AUC values in the 0.622–0.818 range for the DRB1 alleles, the HLA class II molecules of interest for this study. However, it is certainly possible that differences in predictive capacity or performance may influence the results and should therefore be interpreted with this caveat in mind.
Nicaraguan DENV-specific CD4+ T cell responses preferentially targeted C, NS3, and NS5. This pattern is in agreement with what we previously observed in Sri Lanka (12), as well as what was also detected by Rivino et al. in DENV infected donors from Singapore (46), suggesting a similar protein source for immunodominant epitopes in different endemic areas. This finding is not only significant for the study of DENV CD4+ T cell immunity but also to the design and evaluation of vaccines. Indeed, this pattern of immunodominance is also observed in individuals vaccinated with an experimental tetravalent live-attenuated vaccine formulation (TV005) and the attenuated DENV2 virus (Tonga 74) used as a challenge strain in a DENV human challenge model (19, 24). Accordingly, we conclude that the patterns of immunodominance induced by vaccination with attenuated DENV vaccines should be able to mimic natural immunity regardless of geographical location.
The patterns of HLA-linked immunodominance previously observed at the level of CD8+ T cell responses (11) in the Sri Lankan and Nicaraguan cohorts were virtually indistinguishable. Here, at the level of HLA class II, we did not detect a significant correlation between response magnitudes between the two cohorts. A caveat to this is that our results were based on a limited number of HLA allelic variants. In pursuing this line of inquiry, we uncovered the unique case of the HLA-DRB1*11:01 allele, which exhibited much higher reactivity in the Nicaragua cohort as compared to the Sri Lanka cohort. Furthermore, most of the differences were related to recognition of a new set of epitopes recognized specifically in the Nicaragua cohort. The mechanism of this is as yet unclear. Strong T cell responses/donor relative to other alleles were observed in the case of HLA-DRB1*11:01, *14:02, and *14:06 alleles in the Nicaragua population. HLA-DRB1*11:01 recognized a new and broader set of epitopes that is unique to the cohort from Nicaragua when compared to previously identified epitope sets in Sri Lanka. In the case of HLA-DRB1*14:02+ and *14:06+ donors with similar strong responses, this comparison was not possible as those alleles are not present in the Sri Lanka populations (Figure 1).
Thus, it is possible that that the differences observed for HLA-DRB1*11:01 between the two populations could be observed for other alleles with similar magnitude of T cell response/donor. It is also possible that these differences may relate to the potential contribution of other loci in linkage disequilibrium with the HLA-DRB1 locus, and sharing some degree of repertoire overlap. The future studies of DENV CD4+ T cell epitopes restricted by HLA-DRB3/4/5, DP, and DQ loci will address this hypothesis.
Our original goal was to identify reagents and assay strategies to allow assessment of DENV CD4+ T cell responses regardless of geographical location in the specific context of DENV disease or vaccination. This first required the identification of a repertoire of epitopes spanning the most common HLA types. In addition, it was important to adopt methodologies amenable to direct ex vivo analysis to avoid altering T cell phenotypes and to sample-preserving approaches given that volume is often limiting in vaccine studies and pediatric cohorts. To meet these challenges, our group has developed the MP approach, based on large numbers of peptides pooled and formulated to take into account sequential lyophilization (17). In this study, we refined the previously described DENV CD4 MP363 by incorporating information from the Nicaraguan study and using a rational, optimized, epitope selection strategy targeting balanced HLA, DENV serotype, and protein composition coverage in the process of epitope selection. We validated this optimized DENV MP by showing that it can be used to assess DENV-specific CD4+ T cell responses across donors independently from HLA type, geographical region, or infecting serotype. It is apparent that different cohorts vary somewhat in reactivity to the MP180 pool. This differential reactivity might be due to insufficient coverage or due to the different history of DENV infection and burden of the disease. The DENV cohorts from Singapore and India (Figures 6B,C) are associated with relatively lower levels of secondary DENV infection (47, 48), as compared with the Brazilian cohort where secondary infections were more prevalent, reflecting a higher burden of infection (49, 50). We have previously shown that multiple exposures to DENV increase the magnitude of CD4+ T cell DENV-specific responses relative to primary infection suggesting that infection history can influence magnitude of DENV-specific T cell responses (10). Most importantly, we showed that the assessment can be performed directly ex vivo using ICS assays. This has multiple advantages compared to other assays such as ELISPOT. Gating strategies allow the exclusion of CD8+ T cell responses that might respond to nested MHC class I epitopes within the tested 15mer peptide. Furthermore, it will allow parallel phenotypic assessments across different studies and capture of antigen-specific T cells for transcriptomic analysis on a large scale. Future studies will address the secretion of multiple cytokines and marker expression revealing potential additional dengue-specific CD4+ T cell responses but also specific polarizations of T cell responses between the different cohorts.
Ethics Statement
All samples were collected and used following guidelines from the Institutional Review Boards (IRBs) of the La Jolla Institute for Allergy and Immunology (LIAI), the University of California, Berkeley, the Nicaraguan Ministry of Health, and the Medical Faculty, University of Colombo (serving as the National Institutes of Health-approved IRB for Genetech Research Institute) and clinical trials registered at ClinicalTrials.gov under registration numbers NCT01506570 and NCT01436422. All Brazilian participants signed the IRB-approved informed consent document (IRB document 0652/09, CAPPesq, Hospital das Clínicas, Universidade de São Paulo). Collection of PBMCs from Singapore was approved by the Singapore National Healthcare Group ethical review board (DSRB 2008/00293). All Indian participants gave written, informed consent and the protocol was approved by the IISc Institutional Human Ethics Committee (ref 5/2011).
Author Contributions
AG, MA, BL, PHoR, CC, CS, AM, PC, MC, QL, FT, AD, and LR performed experiments, reviewed data, and planned the experimental strategy. JS and BP performed bioinformatics analyses. EP and SM performed and coordinated HLA typing and related analysis. AB, APD, SD, ADdS, GN, CN, AMdS, PM, TS, VR, VS, JC, EK, LT, and EH collected samples and provided clinical information. AG, AS, and DW conceived and directed the study, and wrote the manuscript. All the authors have critically read and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Nicaraguan Blood Center and National Blood Center, Ministry of Health, Colombo, Sri Lanka, for providing buffy coat samples used in this study. We further wish to acknowledge the help provided by Dr. Manjunatha Venkataswamy with this study.
Funding
This work was supported by National Institutes of Health contracts Nr. HHSN272200900042C and HHSN27220140045C to AS, 1P01AI106695 to EH, and the BMGF grant 457 OPP1104710 to APD. The Singapore work was funded by the Cooperative Basic Research Grant-New Investigator Grant (CBRG-NIG R-913-301-289-213) of the Singapore National Medical Research Council to LR. LT is supported by a Wellcome Trust clinical research career development fellowship, grant number 205228/Z/16/Z. TS is supported by the National Institute of Health Research Health Protection Unit (NIHR HPRU) in Emerging and Zoonotic Infections. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, the Department of Health, or Public Health England.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/article/10.3389/fimmu.2017.01309/full#supplementary-material.
Footnotes
References
1. Roth A, Mercier A, Lepers C, Hoy D, Duituturaga S, Benyon E, et al. Concurrent outbreaks of dengue, chikungunya and Zika virus infections – an unprecedented epidemic wave of mosquito-borne viruses in the Pacific 2012-2014. Euro Surveill (2014) 19(41). doi:10.2807/1560-7917.ES2014.19.41.20929
2. Cucunawangsih, Lugito NPH. Trends of dengue disease epidemiology. Virology (Auckl) (2017) 8:1178122X17695836. doi:10.1177/1178122X17695836
3. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature (2013) 496(7446):504–7. doi:10.1038/nature12060
4. Khetarpal N, Khanna I. Dengue fever: causes, complications, and vaccine strategies. J Immunol Res (2016) 2016:6803098. doi:10.1155/2016/6803098
5. Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, Jairungsri A, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol (2006) 176(6):3821–9. doi:10.4049/jimmunol.176.6.3821
6. Imrie A, Meeks J, Gurary A, Sukhbataar M, Kitsutani P, Effler P, et al. Differential functional avidity of dengue virus-specific T-cell clones for variant peptides representing heterologous and previously encountered serotypes. J Virol (2007) 81(18):10081–91. doi:10.1128/JVI.00330-07
7. Duangchinda T, Dejnirattisai W, Vasanawathana S, Limpitikul W, Tangthawornchaikul N, Malasit P, et al. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc Natl Acad Sci U S A (2010) 107(39):16922–7. doi:10.1073/pnas.1010867107
8. Hatch S, Endy TP, Thomas S, Mathew A, Potts J, Pazoles P, et al. Intracellular cytokine production by dengue virus-specific T cells correlates with subclinical secondary infection. J Infect Dis (2011) 203(9):1282–91. doi:10.1093/infdis/jir012
9. Rivino L, Kumaran EA, Thein TL, Too CT, Gan VC, Hanson BJ, et al. Virus-specific T lymphocytes home to the skin during natural dengue infection. Sci Transl Med (2015) 7(278):278ra235. doi:10.1126/scitranslmed.aaa0526
10. Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, de Silva AM, et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci U S A (2015) 112(31):E4256–63. doi:10.1073/pnas.1505956112
11. de Alwis R, Bangs DJ, Angelo MA, Cerpas C, Fernando A, Sidney J, et al. Immunodominant dengue virus-specific CD8+ T cell responses are associated with a memory PD-1+ phenotype. J Virol (2016) 90(9):4771–9. doi:10.1128/JVI.02892-15
12. Weiskopf D, Angelo MA, Grifoni A, O’Rourke PH, Sidney J, Paul S, et al. HLA-DRB1 alleles are associated with different magnitudes of dengue virus-specific CD4+ T-cell responses. J Infect Dis (2016) 214(7):1117–24. doi:10.1093/infdis/jiw309
13. Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci U S A (2013) 110(22):E2046–53. doi:10.1073/pnas.1305227110
14. Weiskopf D, Cerpas C, Angelo MA, Bangs DJ, Sidney J, Paul S, et al. Human CD8+ T cell responses against the four dengue virus serotypes are associated with distinct patterns of protein targets. J Infect Dis (2015) 212:1743–51. doi:10.1093/infdis/jiv289
15. Chandele A, Sewatanon J, Gunisetty S, Singla M, Onlamoon N, Akondy RS, et al. Characterization of human CD8 T cell responses in dengue virus-infected patients from India. J Virol (2016) 90(24):11259–78. doi:10.1128/JVI.01424-16
16. Rivino L, Lim MQ. CD4+ and CD8+ T-cell immunity to dengue – lessons for the study of Zika virus. Immunology (2017) 150(2):146–54. doi:10.1111/imm.12681
17. Carrasco Pro S, Sidney J, Paul S, Lindestam Arlehamn C, Weiskopf D, Peters B, et al. Automatic generation of validated specific epitope sets. J Immunol Res (2015) 2015:763461. doi:10.1155/2015/763461
18. Kirkpatrick BD, Whitehead SS, Pierce KK, Tibery CM, Grier PL, Hynes NA, et al. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci Transl Med (2016) 8(330):330ra336. doi:10.1126/scitranslmed.aaf1517
19. Angelo MA, Grifoni A, O’Rourke PH, Sidney J, Paul S, Peters B, et al. Human CD4+ T cell responses to an attenuated tetravalent dengue vaccine parallel those induced by natural infection in magnitude, HLA restriction, and antigen specificity. J Virol (2017) 91(5):e02147–16. doi:10.1128/JVI.02147-16
20. Fernandez RJ, Vazquez S. Serological diagnosis of dengue by an ELISA inhibition method (EIM). Mem Inst Oswaldo Cruz (1990) 85(3):347–51. doi:10.1590/S0074-02761990000300012
21. Harris E, Videa E, Perez L, Sandoval E, Tellez Y, Perez ML, et al. Clinical, epidemiologic, and virologic features of dengue in the 1998 epidemic in Nicaragua. Am J Trop Med Hyg (2000) 63(1–2):5–11. doi:10.4269/ajtmh.2000.63.5
22. Collins MH, McGowan E, Jadi R, Young E, Lopez CA, Baric RS, et al. Lack of durable cross-neutralizing antibodies against Zika virus from dengue virus infection. Emerg Infect Dis (2017) 23(5):773–81. doi:10.3201/eid2305.161630
23. Turtle L, Tatullo F, Bali T, Ravi V, Soni M, Chan S, et al. Cellular immune responses to live attenuated Japanese encephalitis (JE) vaccine SA14-14-2 in adults in a JE/dengue co-endemic area. PLoS Negl Trop Dis (2017) 11(1):e0005263. doi:10.1371/journal.pntd.0005263
24. Grifoni A, Angelo M, Sidney J, Paul S, Peters B, de Silva AD, et al. Patterns of cellular immunity associated with experimental infection with rDEN2Delta30 (Tonga/74) support its suitability as a human dengue virus challenge strain. J Virol (2017) 91(8):e02133–16. doi:10.1128/JVI.02133-16
25. Meyer D, Singe RM, Mack SJ, Lancaster A, Nelson MP, Erlich H, et al. Single locus polymorphism of classical HLA genes. In: Hansen J, editor. Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference. Seattle: IHWG Press (2007). p. 653–704.
26. Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens (2003) 61(5):403–7. doi:10.1034/j.1399-0039.2003.00062.x
27. Sidney J, Grey HM, Southwood S, Celis E, Wentworth PA, del Guercio MF, et al. Definition of an HLA-A3-like supermotif demonstrates the overlapping peptide-binding repertoires of common HLA molecules. Hum Immunol (1996) 45(2):79–93. doi:10.1016/0198-8859(95)00173-5
28. Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, et al. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J Immunol (2010) 184(5):2492–503. doi:10.4049/jimmunol.0903655
29. Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, et al. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J Immunol (2010) 185(7):4189–98. doi:10.4049/jimmunol.1001006
30. Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol (2008) 4(4):e1000048. doi:10.1371/journal.pcbi.1000048
31. Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics (2010) 11:568. doi:10.1186/1471-2105-11-568
32. Hinz D, Seumois G, Gholami AM, Greenbaum JA, Lane J, White B, et al. Lack of allergy to timothy grass pollen is not a passive phenomenon but associated with the allergen-specific modulation of immune reactivity. Clin Exp Allergy (2016) 46(5):705–19. doi:10.1111/cea.12692
33. Lindestam Arlehamn CS, McKinney DM, Carpenter C, Paul S, Rozot V, Makgotlho E, et al. A quantitative analysis of complexity of human pathogen-specific CD4 T cell responses in healthy M. tuberculosis infected South Africans. PLoS Pathog (2016) 12(7):e1005760. doi:10.1371/journal.ppat.1005760
34. Bancroft T, Dillon MB, da Silva Antunes R, Paul S, Peters B, Crotty S, et al. Th1 versus Th2 T cell polarization by whole-cell and acellular childhood pertussis vaccines persists upon re-immunization in adolescence and adulthood. Cell Immunol (2016) 30(4–305):35–43. doi:10.1016/j.cellimm.2016.05.002
35. da Silva Antunes R, Paul S, Sidney J, Weiskopf D, Dan JM, Phillips E, et al. Definition of human epitopes recognized in tetanus toxoid and development of an assay strategy to detect ex vivo tetanus CD4+ T cell responses. PLoS One (2017) 12(1):e0169086. doi:10.1371/journal.pone.0169086
36. Mathew A, Rothman AL. Understanding the contribution of cellular immunity to dengue disease pathogenesis. Immunol Rev (2008) 225:300–13. doi:10.1111/j.1600-065X.2008.00678.x
37. Rivino L. T cell immunity to dengue virus and implications for vaccine design. Expert Rev Vaccines (2016) 15(4):443–53. doi:10.1586/14760584.2016.1116948
38. Ma CS, Deenick EK. Human T follicular helper (Tfh) cells and disease. Immunol Cell Biol (2014) 92(1):64–71. doi:10.1038/icb.2013.55
39. Muema DM, Macharia GN, Olusola BA, Hassan AS, Fegan GW, Berkley JA, et al. Proportions of circulating follicular helper T cells are reduced and correlate with memory B cells in HIV-infected children. PLoS One (2017) 12(4):e0175570. doi:10.1371/journal.pone.0175570
40. Piazza P, Campbell D, Marques E, Hildebrand WH, Buchli R, Mailliard R, et al. Dengue virus-infected human dendritic cells reveal hierarchies of naturally expressed novel NS3 CD8 T cell epitopes. Clin Exp Immunol (2014) 177(3):696–702. doi:10.1111/cei.12373
41. de Matos AM, Carvalho KI, Rosa DS, Villas-Boas LS, da Silva WC, Rodrigues CL, et al. CD8+ T lymphocyte expansion, proliferation and activation in dengue fever. PLoS Negl Trop Dis (2015) 9(2):e0003520. doi:10.1371/journal.pntd.0003520
42. Goncalves AJ, Oliveira ER, Costa SM, Paes MV, Silva JF, Azevedo AS, et al. Cooperation between CD4+ T cells and humoral immunity is critical for protection against dengue using a DNA vaccine based on the NS1 antigen. PLoS Negl Trop Dis (2015) 9(12):e0004277. doi:10.1371/journal.pntd.0004277
43. Paul S, Lindestam Arlehamn CS, Scriba TJ, Dillon MB, Oseroff C, Hinz D, et al. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J Immunol Methods (2015) 422:28–34. doi:10.1016/j.jim.2015.03.022
44. Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol (1998) 160(7):3363–73.
45. Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics (2011) 63(6):325–35. doi:10.1007/s00251-011-0513-0
46. Rivino L, Kumaran EA, Jovanovic V, Nadua K, Teo EW, Pang SW, et al. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol (2013) 87(5):2693–706. doi:10.1128/JVI.02675-12
47. Low SL, Lam S, Wong WY, Teo D, Ng LC, Tan LK. Dengue seroprevalence of healthy adults in Singapore: serosurvey among blood donors, 2009. Am J Trop Med Hyg (2015) 93(1):40–5. doi:10.4269/ajtmh.14-0671
48. Singla M, Kar M, Sethi T, Kabra SK, Lodha R, Chandele A, et al. Immune response to dengue virus infection in pediatric patients in New Delhi, India – association of viremia, inflammatory mediators and monocytes with disease severity. PLoS Negl Trop Dis (2016) 10(3):e0004497. doi:10.1371/journal.pntd.0004497
49. Fares RC, Souza KP, Anez G, Rios M. Epidemiological scenario of dengue in Brazil. Biomed Res Int (2015) 2015:321873. doi:10.1155/2015/321873
50. Faria NR, Solorzano VE, Nogueira RM, Chouin-Carneiro T, Nunes PC, Simoes JB, et al. Dengue epidemics in two distinct periods reveal distinct epidemiological, laboratorial and clinical aspects in a same scenario: analysis of the 2010 and 2013 epidemics in Mato Grosso do Sul, Brazil. Trans R Soc Trop Med Hyg (2016) 110(4):228–36. doi:10.1093/trstmh/trw013
Keywords: dengue virus, CD4+ T cells, HLA, epitope, adaptive immunity
Citation: Grifoni A, Angelo MA, Lopez B, O’Rourke PH, Sidney J, Cerpas C, Balmaseda A, Silveira CGT, Maestri A, Costa PR, Durbin AP, Diehl SA, Phillips E, Mallal S, De Silva AD, Nchinda G, Nkenfou C, Collins MH, de Silva AM, Lim MQ, Macary PA, Tatullo F, Solomon T, Satchidanandam V, Desai A, Ravi V, Coloma J, Turtle L, Rivino L, Kallas EG, Peters B, Harris E, Sette A and Weiskopf D (2017) Global Assessment of Dengue Virus-Specific CD4+ T Cell Responses in Dengue-Endemic Areas. Front. Immunol. 8:1309. doi: 10.3389/fimmu.2017.01309
Received: 07 September 2017; Accepted: 28 September 2017;
Published: 13 October 2017
Edited by:
Loretta Tuosto, Sapienza Università di Roma, ItalyReviewed by:
Benny Chain, University College London, United KingdomArnaud Moris, Université Pierre et Marie Curie, France
Copyright: © 2017 Grifoni, Angelo, Lopez, O’Rourke, Sidney, Cerpas, Balmaseda, Silveira, Maestri, Costa, Durbin, Diehl, Phillips, Mallal, De Silva, Nchinda, Nkenfou, Collins, de Silva, Lim, Macary, Tatullo, Solomon, Satchidanandam, Desai, Ravi, Coloma, Turtle, Rivino, Kallas, Peters, Harris, Sette and Weiskopf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Weiskopf, daniela@lji.org
†Present address: Aruna D. De Silva, Faculty of Medicine, Department of Paraclinical Sciences, Kotelawala Defense University, Ratmalana, Sri Lanka
 Alba Grifoni1
Alba Grifoni1 Michael A. Angelo
Michael A. Angelo Benjamin Lopez
Benjamin Lopez Patrick H. O’Rourke
Patrick H. O’Rourke Alvino Maestri
Alvino Maestri Anna P. Durbin
Anna P. Durbin Sean A. Diehl
Sean A. Diehl Godwin Nchinda
Godwin Nchinda Matthew H. Collins
Matthew H. Collins Mei Qiu Lim
Mei Qiu Lim Filippo Tatullo
Filippo Tatullo Vijaya Satchidanandam
Vijaya Satchidanandam Vasanthapram Ravi
Vasanthapram Ravi Laura Rivino
Laura Rivino Eva Harris
Eva Harris Daniela Weiskopf
Daniela Weiskopf