- 1Institute for Virology and Immunobiology, University of Würzburg, Würzburg, Germany
- 2Institute of Pathophysiology and Immunology, Medical University of Graz, Graz, Austria
- 3Department of Dermatology, University Hospital Erlangen, Erlangen, Germany
- 4Department of Dermatology, Venereology and Allergology, Medical University of Innsbruck, Innsbruck, Austria
Dendritic cells (DCs) and macrophages (Mph) share many characteristics as components of the innate immune system. The criteria to classify the multitude of subsets within the mononuclear phagocyte system are currently phenotype, ontogeny, transcription patterns, epigenetic adaptations, and function. More recently, ontogenetic, transcriptional, and proteomic research approaches uncovered major developmental differences between Flt3L-dependent conventional DCs as compared with Mphs and monocyte-derived DCs (MoDCs), the latter mainly generated in vitro from murine bone marrow-derived DCs (BM-DCs) or human CD14+ peripheral blood monocytes. Conversely, in vitro GM-CSF-dependent monocyte-derived Mphs largely resemble MoDCs whereas tissue-resident Mphs show a common embryonic origin from yolk sac and fetal liver with Langerhans cells (LCs). The novel ontogenetic findings opened discussions on the terminology of DCs versus Mphs. Here, we bring forward arguments to facilitate definitions of BM-DCs, MoDCs, and LCs. We propose a group model of terminology for all DC subsets that attempts to encompass both ontogeny and function.
Introduction
Dendritic cells (DCs) are major players to direct adaptive immunity or tolerance. More recently, the origins and possible subdivisions into DC subsets and the DC commonalities with macrophages (Mphs) have been discussed by numerous papers and reviews (1–7).
In their peripheral tissue-resident state, DCs act as immune sensors that recognize pathogens and then convert into a mature or activated state enabling their migration to the draining lymph node to stimulate T cell immunity (8). By contrast, during homeostasis lymphatic organ-resident DCs and steady-state migratory DCs contribute to immune tolerance (9).
This functional capacity of antigen transport from the periphery to the lymph nodes has been one of their major cellular characteristics distinguishing them from Mphs. Marked differences between DCs and Mphs have recently also been observed by quantitative proteomic analyses that point out differential and specific epigenetic programming of each cell type (10) or, at a more functional level, by dissecting and defining differential cellular mechanisms in endocytic recycling pathways of MHC I molecules for cross-presentation (11).
Several data indicate that murine bone marrow-derived DCs (BM-DCs), human monocyte-derived DCs (MoDCs), and Langerhans cells (LCs) show considerable transcriptional overlap and a common ontogenetic origin with Mphs (12–15). This raised doubts whether the name “DC” is still correct. On the other hand, there are also data showing mouse and human LC hierarchical clustering with subsets of conventional DCs (cDCs) (16). Here, we recall several aspects of the biology of these cell types, and we suggest to retain their name, not least for historic reasons allowing a better online search on a cell type. For example, follicular DC and pDC neither share overlaps in transcriptional profiling with cDCs nor do they exert typical DC functions. Yet, there is no discussion that they are both called DCs. In fact, human and mouse pDC share less transcriptional overlap with cDCs as compared with LC and MoDCs (17, 18), while follicular DCs are derived from marginal reticular cells, a population of mesenchymal stroma cells lining the lymph node subcapsular sinus (19). Evidence suggests that a subset of pDCs may derive from common lymphoid progenitors (CLP) and not from a common myeloid progenitor (CMP) (20) or might even derive from an own lineage (21) Nevertheless, all these cells retain their name “DC” independent from either their function or their origin from CMP, CLP, CDP (common DC progenitor), or even from non-hematopoietic progenitors.
To restructure the DC nomenclature on the basis of ontogenetic data alone may be highly confusing since two names or designations for the same cell will be perpetuated. For example, LCs would appear in older literature as “epidermal DCs” and newly “epidermal Mphs.” Here, we review LCs as well as GM-CSF-dependent DC and Mph generation from BM or monocytes and their functions. We believe that that functional aspects can and should be integrated in the definition of DCs (Figure 1).
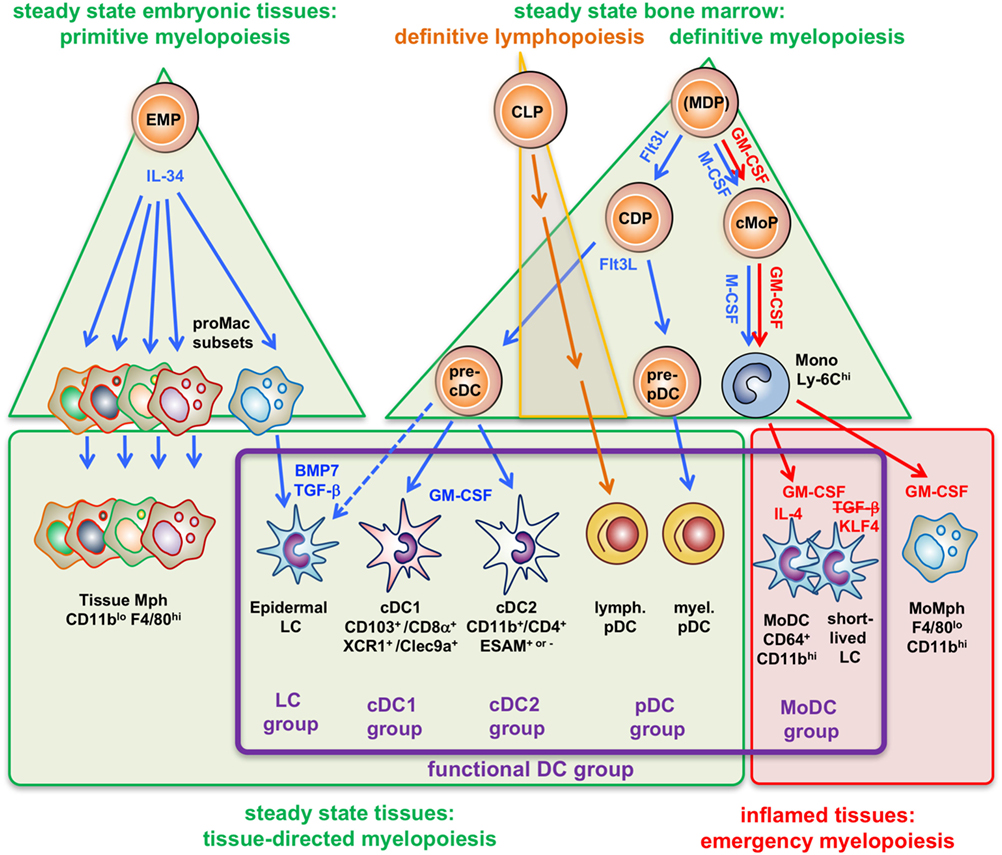
Figure 1. Four types of myelopoiesis and local cytokines control steady-state and inflammatory generation of grouped dendritic cell (DC) types. Early embryonic macrophage-erythrocyte precursors (EMPs) in the yolk sac and fetal liver responding to IL-34 develop into different preformed macrophage (Mph) progenitors (proMac) that generate most Mph populations and the epidermal DC subset, called Langerhans cell (LCs), which persist there throughout adulthood. As an example, differentiation of LCs requires additional cytokines such as TGF-β or BMP7 produced within the epidermis to reach the final stage of tissue-directed myelopoiesis. This primitive hematopoiesis is substituted by the definitive hematopoiesis in the BM in the adult. There, under steady-state conditions the growth factor Flt3L promotes the development of myeloid precursors giving rise to macrophages and DCs (MDPs) into common DC precursors (CDPs) that split into pre-pDCs (22) and pre-cDCs released into the blood. There is currently not yet full consensus about the potential of the cells designated as “MDPs” (2, 23, 24). Upon migration into the spleen (or other lymphatic tissues) or peripheral non-lymphatic tissues the CD103+ or CD8α+ cDC1 groups and CD11b+ or CD4+ cDC2 groups further acquire different phenotypes by tissue-derived factors, e.g., GM-CSF (in blue). By contrast, (MDPs) sensing M-CSF under steady-state conditions will develop into common monocyte precursors (cMoPs) that predominantly develop into classical Ly-6Chi monocytes found in the murine blood. Under inflammatory conditions, activated CD4+ T-helper cells produce large amounts of GM-CSF (red) at systemic levels, thereby initiating so-called emergency myelopoiesis (25) driving MDPs and cMoPs into cell cycle and releasing increased amounts of classical monocytes into the blood. After extravasation, these monocytes can differentiate into inflammatory types of MoMph or, in the additional presence of IL-4 (red), into cells of the monocyte-derived DC (MoDC) group. Thus, differential developmental pathways merge in the generation of functional DCs (functional DC group, violet frame), independent from their origin.
GM-CSF in DC Growth, Survival, and Functional Activation
GM-CSF supplemented BM-DC cultures for murine DC generation are under debate for their usefulness to study DC biology, mainly since Mphs and neutrophils are generated by the same cytokine in these cultures. Similarly, human MoDCs may more closely mimic Mphs rather than DCs. Factors promoting DC versus Mph development from monocytes and myeloid progenitor cells have been reported. Human monocytes differentiate toward Mphs upon exposure to IL-6, which upregulated the M-CSF receptor (M-CSFR) expression to enable consumption of their autocrine M-CSF (26). Human MoDCs can be selectively generated from monocytes in cultures by combined use of GM-CSF and IL-4 (27, 28) by inducing TNF-converting enzyme (TACE) that cleaves the M-CSFR thereby disabling autocrine M-CSF-dependent Mph generation (29). IL-4 imprinted differential epigenetic signatures for both DCs and Mphs influencing their further response to LPS (30, 31). Addition of TNF can further stabilize GM-CSF/IL-4 mediated DC skewing (32). MoDC development from monocytes is characterized by specific epigenetic programming such as histone H4K16 acetylation that was not observed in monocytes or Mphs (10). Thus, M-CSF/IL-6 promotes Mph growth while GM-CSF/IL-4 suppresses M-CSF signals and thereby support DC development.
Murine BM cells cultured with GM-CSF contain neutrophils for up to 5 days before they die and finally only loosely adherent MHC IIlow cells and strongly adherent MHC IIneg Mphs remain (33–35). The MHC IIlow cells are composed of immature DCs with the potential to become mature DCs and Mph progenitors developing into MHC IIneg Mphs (36, 37). Unlike for human MoDC cultures, the addition of IL-4 to murine BM-DCs does not prevent Mph growth but fulfills other functions reviewed elsewhere (38). Reversely, the generation of human BM-DCs with GM-CSF with or without IL-4 can be used to generate immunogenic or tolerogenic DCs similar as found in murine settings (39–42). Comparative analyses showed that murine BM-DCs and human MoDCs are highly similar and therefore can be considered as functional homologs (43, 44). BM-DC cultures contain proliferating cells (35). The proliferating cells mostly represent macrophage-DC progenitors (MDP) and common monocyte progenitors while differentiated Ly-6Chigh monocytes fail to proliferate in GM-CSF cultures and therefore do not contribute substantially to the BM-DC progeny (37) and as observed in mice under steady-state conditions (45). The lack of proliferation has been described for human CD14+ monocytes undergoing MoDC differentiation (46).
Murine BM-DCs are typically generated in vitro with GM-CSF. However, protocols are available that employ Flt3L instead of GM-CSF to generate bulk populations containing mixtures of CD103+ cDCs, CD11b+ cDCs, and pDCs from mouse bone marrow (47–49) or similarly but less well defined from human peripheral blood (50, 51). The generation of such DC subtypes in vitro is similar to what is observed in vivo (Figure 2). Two articles nicely dissected the precursors of human pDCs and CD1c+ cDCs as well as CD141+ cDC and claimed to provide a method to selectively generate all three cell types from CD34+ progenitors (52, 53). Already earlier, a protocol for the bulk generation of all three human cDC subsets had been reported also using CD34+ cells (54).
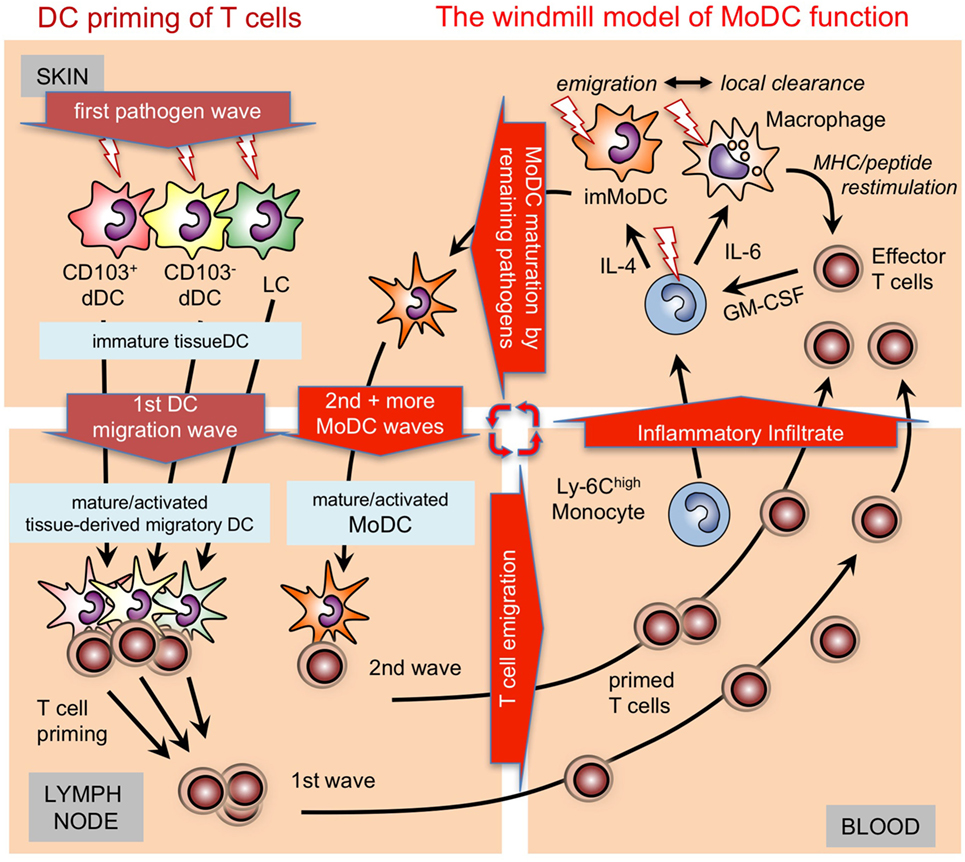
Figure 2. Time-dependent activation/maturation of tissue dendritic cells (DCs) and perpetuated generation of monocyte-derived DCs (MoDCs). A model showing cooperation of preexisting tissue DCs with newly generated MoDCs from infiltrating monocytes as shown before (55, 56) in a windmill-like schematic manner. Initial pathogens invading the skin as depicted here will first encounter epidermal Langerhans cell (LCs) and dermal DC subsets (dDCs). All these DC subsets are capable of capturing pathogens, undergoing maturation and can migrate CCR7 dependent into the draining lymph nodes to initiate T cell priming. The first wave of T cells will arrive together with monocytes and other cells of the inflammatory infiltrate in the infected skin. Local pathogen-specific MHC/peptide dependent reactivation of T cells, e.g., by resident or infiltrating macrophages will lead to their GM-CSF release and, together with cytokines in the environment, promote MoDC generation from monocytes. The resulting immature MoDCs follow the tissue DCs into the lymph node to perpetuate T cell priming in secondary and subsequent waves. Since the local reconstitution of emigrated tissue DCs is slow, MoDC generation by T cell-derived GM-CSF is continued as long as the infection persists as depicted graphically as a windmill model, i.e., as long as the “pathogen wind blows.”
A massive expansion of monocyte and dendritic cell progenitor (MDP), but very low effects on common DC progenitors (CDPs), have been found in GM-CSF supplemented BM-DC cultures (37), confirming major effects of GM-CSF on myelomonocytic cells rather than committed DC precursors (CDP) developing into Zbtb46 expressing cDCs. Although the transcription factor Zbtb46 had been considered to be specific for cDCs (57), recent data indicate that LCs co-express Zbtb46 in addition to the Mph-specific transcription factor KLF4 (58). Moreover, Ly-6ChiTREML4neg monocytes can differentiate into Zbtb46+ MoDCs in response to GM-CSF and IL-4. This occurred independent of Batf3 but dependent on Irf4 and although IL-4 induced both transcription factors in murine MoDCs (59). Thus, the so far DC subset-specific transcription factors may not be restricted to a DC subset defined by ontogeny but induced by environmental cytokine signals or factors inducing specific functional activation.
However, GM-CSF has a major impact on the steady-state cDC generation from preDCs in vivo since mice deficient for the GM-CSF receptor (csf2r) have severe deficits for both subsets of cDCs (60). This is partially in agreement with the finding that also the selective in vitro generation of murine CD103+Clec9A+XCR1+ cDCs from BM cells with Flt3L was enabled by addition of only very low doses of GM-CSF (61). Moreover, CD8+ T cell activation during lung infection was abrogated in csf2r−/− mice (60). In fact, some data indicate that Flt3L alone may not be sufficient to generate fully functional cDCs. Functional studies with human in vitro-generated cDC subsets on allogeneic T cell proliferation and cross-priming obtained only poor stimulatory effects (54). Murine immature Flt3L-generated DCs were unable to induce T cell anergy or convert regulatory T cells as compared with immature GM-CSF-generated DCs (49). Additional GM-CSF signals were required by Flt3L cultured cDCs from murine BM to acquire cross-presenting capacities, which was associated but not functionally linked with the further upregulation of CD103 and an increase the frequency of CD103+ cells in culture (62), or CD8α+ and CD8α− cDCs from spleen to upregulate costimulatory molecules and cytokines after activation by pathogens (63), a process presumably regulated by the STAT5 target gene cytokine inducible SH2-domain protein (CISH) (64). GM-CSF also contributes to pDC maturation/activation by inducing PU.1 dependent MHC II upregulation in pDCs (65). However, GM-CSF (with or without TNF) impaired Flt3L-induced pDC generation from murine myeloid progenitors in favor of myeloid DCs and Mphs (66).
Together, the functional data available indicate that in vitro GM-CSF cultures of BM cells or monocytes generate different myeloid cell types and among them a fraction clearly shows characteristics of DCs. Additional cytokine use or specific culturing/harvesting procedures further support the selective yield of DCs. Moreover, GM-CSF also controls some cDC and pDC functions.
Heterogeneity of BM-DC Cultures
BM harbors heterogeneous cellular sources of different developmental stages of cell types including myeloid cells responsive to GM-CSF. Accordingly, the in vitro exposure of BM cells to GM-CSF generates different waves of BM-DC development. This can be demonstrated by culturing specific early and late myeloid precursors with GM-CSF and measuring the time required to develop into CD11c+ DCs (37, 67). BM precursors for two different DC subsets and Mphs responding to GM-CSF differ in their endocytosis capacities and expression of the surface markers E-cadherin, scavenger receptor A (2F8, CD204), CD11b, and Gr-1. The sorted cells with an endocytosishighMHC IIlow profile gave rise either exclusively to one subset of MHC IIhigh DCs, while endocytosislowMHC IIlow cells resulted in two populations, one upregulating and one downregulating MHC II molecules after two further days of culture in GM-CSF, indicative for further development into another DC subset and Mphs, respectively (36). Stimulating sorted MHC IIlow cells with LPS further indicated that some MHC IIlow cells within this population turned into MHC IIneg and adherent Mphs, while other MHC IIlow cells (immature DCs) matured into MHC IIhigh DCs (36). Thus, these findings indicate that bulk cultures as well as MHC IIlow cells generated in response to GM-CSF are not uniform. Rather they include cells with differentiation potential for Mphs and at least two different subsets of immature DCs. Since such an MHC IIlow Mph/DC mix will blur the results of mRNA profiling for cellular subset identification careful dissection of all subsets is a prerequisite. Additional factors may bias results from murine BM-DCs, such as components in the culture medium or fetal calf serum which can influence the outgrowth of DCs (68). While maturation of isolated murine LCs was not influenced under serum-free conditions (69), serum-free BM cell cultures failed to generate fully functional murine BM-DCs (70). BM-DC heterogeneity has also been reported by others (37) and similarly human DC/LC cultures supplemented with GM-CSF are reportedly heterogenous (71). These phenotypically distinct in vitro-generated DC subsets now require more detailed -omics profiling but importantly, also distinguishing between immature/resting and mature/activated stages. DCs expressing different levels of MHC II- and costimulatory molecules will certainly differ in transcriptional patterns although belonging to the same ontogenetic DC subset (72). Such data allow then further comparisons with the same maturation stages of Flt3L in vitro-generated or ex vivo-isolated cDC and pDC subsets.
Since some DC subsets share their capacities for pathogen recognition (73, 74), they also may translate them into same polarized pathogen-specific T-helper cell (Th) response. The treatment of bulk BM-DC cultures with different maturation stimuli appeared valuable to determine distinct response patterns by mRNA analyses, which could be attributed to DC-mediated Th1 and Th2 polarization (75).
Monocytes as a Source for DCs
Under steady-state conditions, MoDCs are hardly found in mice and man (76). However, epithelia and mucosal tissues do contain detectable amounts of MoDCs presumably induced by commensals (77). Clearly, inflammatory and infectious conditions can recruit Ly-6Chigh monocytes into tissues, which then develop into DCs that initiate T cell priming in the draining lymph nodes (55, 78–81). Similar results were obtained for the appearance of MoDCs in humans from synovia of rheumatoid arthritis patients and ascites from cancer patients (44). Thus, fully functional MoDCs can be differentiated under inflammatory or infectious conditions from monocytes in mice and humans.
In the absence of or early after depletion of Batf3-dependent CD103+ DCs in C57BL/6 mice with Leishmania infection a Th1 response does not develop (82). However, during later stages 4 weeks after infection, dermal MoDCs were the only DC subset at the site of skin infection and in the draining lymph nodes to present Leishmania antigens and to produce IL-12 to maintain the Th1 response (56). Together, these data led us to establish a “windmill model” of MoDC function (Figure 2).
In vitro studies showed a strict dependency of murine BM-DCs and human MoDCs on GM-CSF. When such in vitro-generated cells were injected s.c. or i.d. they homed to the T cell areas of the draining lymph nodes (83), a typical DC function that requires CCR7 expression (84). In comparison, inert particles (85) were flushed into the lymph nodes by the lymph fluid; they only penetrated the subcapsular sinus but did not reach the T cell zone. From these studies, it had been concluded that GM-CSF is also critical for driving inflammatory MoDC generation in vivo.
Surprisingly, abrogation of GM-CSF or its receptor in mice did not affect MoDC generation and activation of CD8+ T cell responses. Conversely, deficiency of the M-CSFR (csf1) impaired inflammatory MoDC recruitment and CD80/CD86 surface expression (60). Therefore, the role of GM-CSF for the generation of inflammatory MoDC in vivo remains questionable. On the other hand, Mphs but not DCs arise from human monocytes in response to M-CSF or IL-34 in vitro (86), the latter representing a recently discovered new ligand for the M-CSFR. Thus, the precise roles of M-CSF and GM-CSF for MoDC generation in vitro and in vivo are not fully understood and additional factors from the local inflammatory environment may critically contribute to the monocyte-to-DC conversion.
In Vitro-Generated MoDCs as Models for In Vivo cDC Function?
Human and mouse MoDCs can be generated from blood or BM monocytic cells and precursors in large amounts. This has enabled early studies to investigate them by biochemical techniques. However, one should be careful to simply extrapolate these functional analyses generalized from MoDCs to cDCs. The different cDCs subsets appear to have specialized preferences for cross-presentation (CD8α+/CD103+ cDC1) or MHC II dependent presentation (CD4+/CD11b+ cDC2) (87). More recent studies further dissected cDC2 into ESAM+ cDC2 inducing Th17 and ESAM+ cDC2 specialized to promote Th2 polarization (5). By contrast, in vitro-generated MoDCs (BM-DCs) appear more versatile in their functional adaptation to generate specific Th1, Th2, and Th17 responses in vitro or after injection into mice depending on their stimulation with LPS or TNF (75), cholera toxin (88), or in cross-presenting antigens to CD8+ T cells (89) or presenting glycolipids on CD1d molecules for polarizing NKT cells into either IFN-γ or IL-4 producing subtypes (90, 91). However, indirect cross-priming and NKT cell priming by injected BM-DCs with endogenous spleen DC subsets has been observed (92), which may point to DC–DC cooperation as observed in lymph nodes after virus infection (93).
Several early biochemical findings with MoDCs/BM-DCs could not be confirmed using more recent mouse models for cDCs in vivo. This includes the role of the transcription factor CIITA for the regulation of MHC II genes and DC development (94) and the transcriptional regulation of cross-presentation (59, 95). Clearly, MoDC functions should not be merely extrapolated to cDC functions. Nevertheless, if sensibly used and in a critical manner, these cells certainly retain value for studying certain aspects of DC biology.
GM-CSF-Generated MoDC as Tumor Vaccines
GM-CSF/IL-4 generated MoDCs in vitro, or the direct use of GM-CSF as adjuvant to promote MoDC generation in situ, remain both promising concepts in antitumor vaccine trials (96–98), especially in potential future combinations with modern “immune checkpoint inhibitors” such as anti-CTLA-4 or anti-PD-1 (99, 100). MoDC treatment of melanoma appears highly successful in melanoma patients and showed the same 19% 12 year survival rate as compared with achieved by ipilimumab treatment (101). Under steady-state conditions monocytes rarely immigrate into peripheral tissues to develop into MoDCs due to the absence of inflammation and high GM-CSF concentrations in the local environment (76). Thus, efficient monocyte immigration and their conversion into MoDC allowing migration from peripheral tissues or injection sites into the lymph node may benefit from an inflammatory environment (Figure 2). Preinjection of the DC injection site with TNF or repetitive DC injections into the same site have been shown to dramatically improve DC homing in mice by upregulating CCR7 on DCs and also its ligand CCL21 in lymphatic endothelial cells (102). In a recent clinical study of DC vaccination in glioblastoma patients, the injection site was pretreated with tetanus/diphtheria toxoid before injection of MoDCs, which dramatically improved vaccine efficacy as observed by the patients’ survival rate (103). The benefits of low but not high doses of local GM-CSF as an adjuvant have also been elaborated (104). It would be interesting to investigate the functional role of GM-CSF produced at immunization sites by infiltrating T cells both to enhance cDC responsiveness and maturation (63) as well as in the local conversion of monocytes into MoDCs.
Epidermal LCs as DCs
Recently, it was found that LCs were derived from EMC precursors in the yolk sac by stimulation of the M-CSFR through IL-34 (105–107) together with various tissue Mph populations (108–110). This led to a frequently expressed notion that LCs represent tissue-resident Mphs. However, functionally LCs also show a strong overlap with DCs since they are migratory both in the steady state to induce tolerance and during inflammation. They capture selective pathogens and migrate to initiate T cell responses in the lymph nodes (6, 111, 112). Moreover, microglia and in part LC are the only yolk sac-derived cells, and all other early Mph populations are generated in the aorta-gonad-mesonephros and fetal liver (113), thus further challenging the pure Mph identity of microglia and LCs. Taken together, LCs were found to share both characteristics of Mphs and DCs (6, 58).
It remains to be solved, whether a further subset division into Mph-like LCs and DC-like LCs may exist, because under infectious or inflammatory conditions never all LCs emigrate from the epidermis, even under harsh conditions. Evidence for a “dual identity” of LCs was recently provided from isolated murine bulk LCs expressing both the Mph-specific transcription factor Mafb and the transcription factor Zbtb46 specific for cDCs (58). In the future, single cell mRNA sequencing may reveal whether both factors are expressed in the same cells or in different LC subsets.
Under inflammatory conditions, blood monocytes do replenish the local pool of epidermal and mucosal LCs to compensate for the loss of LCs that emigrated toward the lymph node (114–116) (Figure 2). In the case of the murine oral cavity, a steady-state population of cDC and monocyte-derived LCs have also been described (116). Murine monocyte-derived LCs enabled an Id2-independent short-term reconstitution of the epidermal LC pool (115). Similarly, human CD14+ monocytes can be induced to acquire LC phenotypic characteristics in vitro. LC phenotype induction required TGF-β1, GM-CSF, and IL-4 (117). Later, it was shown that a combination of TGF-β, GM-CSF, and Notch ligand (Delta-1 Jagged2) allows efficient generation of LC-like cells from monocytes (118, 119). Whether monocytes take over long-term reconstitution of epidermal LCs remains unclear (115, 120, 121) According to murine studies, monocytes may only transiently replenish LC-like cells exhibiting maintained expression of monocyte markers and reduced expression of LC markers. Conversely, another so far undefined BM-derived precursor may lead to a long lasting replenishment of LCs in an Id2-dependent manner (115). Human CD1c+ circulating peripheral blood DCs rapidly acquired LC characteristics in vitro, and these cells did not require Notch ligand for LC differentiation (122, 123). Whether these in vitro-generated candidate non-monocyte LC precursors replenish human LCs in vivo remains unknown (124, 125). It must be considered that human LCs lack Mph-associated markers and cross-species transcriptional analyses of skin DC subsets revealed that human LCs are more closely related to human and murine DCs rather than to murine LC, the latter exhibiting Mph markers (126). We recently observed that CD14+ human blood monocytes lose expression of the transcription factor KLF4 during LC commitment (in response to GM-CSF/TGF-β and Notch ligand), and loss of KLF4 is accompanied by loss of monocyte markers. Moreover, loss of KLF4 may represent a prerequisite for TGF-β1-mediated induction of RUNX3, a master transcription factor inducing LC lineage commitment (127). Given that KLF4 restores monocyte/Mph differentiation from fetal liver progenitor cells lacking PU.1 together with driving human monocyte differentiation, KLF4 can be considered a lineage identity factor for monocytes/Mphs (128). Consistently, abovementioned murine short-term LCs (Id2-independent) expressed KLF4, whereas long-term LCs (Id2-dependent) lacked KLF4 (115). Although the earliest LC precursors develop in parallel with embryonic Mphs and only differentiate into the LC phenotype within the epidermis, they specifically acquire typical DC transcription factors. Ahr and Runx3, which are not shared with any other Mph subset (15) have been identified in DCs before, e.g., Ahr in BM-DCs and splenic DCs (129–131) and Runx3 in splenic EsamhiCD11b+ cDCs (132). Also at a global transcriptional level human and murine LC resemble more closely cDCs than monocytes or monocyte-derived cells (16). Thus, it appears that a cell of mononuclear phagocyte ontogeny and identity can convert into epidermal DCs, called LCs (Figure 1).
Mo-derived (or CD1c+ blood DC-derived) LCs and DCs may fulfill an important task to replenish tissues or compartments after the resident cDCs have migrated out of the tissue as illustrated by the “windmill model” of MoDC function (Figure 2). Tissue-resident or blood precursors both contribute to reconstitute LC in the murine or human system (122, 123, 133). The local potential for replenishment may be limited especially under chronic inflammatory conditions. In such a situation GM-CSF-driven emergency myelopoiesis in the BM and monocyte conversion to MoDCs in inflamed or infected peripheral tissues is required for replenishment with DCs. Although not directly tested, both populations are assumed to fulfill the same functions in immune surveillance in spite of being derived from different lineage origins.
The huge body of data that has been acquired using these models and omics data has critically contributed to our current profound knowledge on DC biology. Clearly, there is a need for clarification and simplification of DC nomenclature. However, it is important to take into account all available knowledge—new and old—when attempting to design a new DC nomenclature to avoid uncertainty and irritation in the field. This applies even more so to LCs. It was LCs that, for the first time, allowed to perceive and observe the concept of DC maturation (134), a true hallmark of the DC nature. Thirty years later, DC maturation is understood in much more detail (135). Also, we have come to realize that LCs can also induce tolerance, depending on the circumstances and the quality of maturation (136–138). Successful approaches to employ their DC-like migratory and T cell priming potential for vaccine technology may underscore their DC-like functions (139, 140). Thus, the LC example emphasizes the importance to consider also functional lineage plasticity besides ontogenetic data.
Evidence for DCs Arising from Emergency Granulopoiesis
Neutrophilic granulocytes represent by far the most abundant leukocyte subtype in human bone marrow. They arise from hematopoietic stem cells via granulocyte/macrophage progenitor cells, a cell stage hierarchically upstream of macrophage/DC progenitors (MDP). Steady-state BM contains various differentiation stages of neutrophils. In response to acute inflammation or trauma, neutrophils are rapidly mobilized from bone marrow into peripheral blood to meet the bodies’ high demand on these cells to fight microbial infections. These emergency neutrophils mainly exhibit a band-shaped nucleus and differ from polymorphonuclear neutrophils (PMN) observed in the steady state. Such neo-recruited neutrophils in human peripheral blood were shown to possess in vitro DC differentiation potential in response to 5- to 9-day culture with GM-CSF, IL-4, and TNF, a process occurring without cell proliferation (141). While most generated cells lacked neutrophil markers they still expressed myeloperoxidase, a lysosomal protein found in granulopoietic cells and blood monocytes. Interestingly, so-called “neutrophil-DC hybrids” were generated from murine bone marrow in response to GM-CSF (142) and appeared in vivo in experimentally induced inflammatory lesions in mice (143). In line with this, neutrophils from G-CSF mobilized blood “trans”-differentiated into monocytic cells in vivo in mice or for human cells in vitro in response to GM-CSF, TNF, IL-1β (144) or GM-CSF, M-CSF, TNF, IL-4, IFN-γ (145). This neutrophil plasticity seems to be confined to immature neutrophils characterized by their band-like-shaped nuclei, since PMN from the blood of healthy individuals lacked monocyte or DC differentiation potential (141, 144). Inflammatory signals encountered within lesions may transcriptionally reprogram neutrophils into monocytes/Mphs given that the selective activation of p38MAPK (induced by inflammatory signals) was sufficient to induce monocyte differentiation from granulocytic cells (144). Therefore, left-shifted band-stage neutrophilic granulocytes seem to possess a potential to differentiate into cells of the mononuclear phagocyte system including DCs rapidly within inflammatory lesions [recently reviewed in Ref. (146)]. Although this granulopoietic pathway for DCs still remains poorly defined, to term the resulting monocytes and DCs still “neutrophils,” only based on their ontogeny, would be misleading.
Conclusion
Bone marrow-derived DC, MoDC, and LC share ontogenic and transcriptional similarities with the Mph lineage but also with cDCs. They possess strong phenotypes and functions as known for cDCs (2). Further dissection of the functional plasticity of monocytes in their acquisition of DC-like or Mph-like functions is required. A hematopoietic model including ontogenetic and functional DC/Mph differences is proposed here (Figure 1). Recent data focusing on steady-state distributions of DC subsets and DC in vivo function with respect to defined cytokine deficiencies (147) or the interplay between GM-CSF, M-CSF, and IL-3 (60) may point to alternative approaches toward a better understanding of GM-CSF-derived cells. In any case, the purity of DC populations defined on the basis of all published data as well as differences in activation/maturation stages have to be considered to obtain meaningful RNA sequencing data on DC subsets. Why GM-CSF-based protocols are so successful for generation of murine and human MoDCs in vitro, but GM-CSF-deficient mice (60) or GM-CSF injected mice (148) fail to show such a role requires further investigation. Together, based on these considerations, we propose that a nomenclature for DCs and Mphs may benefit from considering all available and therefore also functional characteristics of cells in addition to their developmental or hematopoietic origination.
Author Contributions
All authors listed have a made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This publication was funded by the German Research Foundation (DFG) and the University of Wuerzburg in the funding programme Open Access Publishing.
References
1. Merad M, Ginhoux F, Collin M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat Rev Immunol (2008) 8:935–47. doi:10.1038/nri2455
2. Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science (2010) 327:656–61. doi:10.1126/science.1178331
3. Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat Immunol (2012) 13:1145–54. doi:10.1038/ni.2467
4. Malissen B, Tamoutounour S, Henri S. The origins and functions of dendritic cells and macrophages in the skin. Nat Rev Immunol (2014) 14:417–28. doi:10.1038/nri3683
5. Murphy TL, Grajales-Reyes GE, Wu X, Tussiwand R, Briseno CG, Iwata A, et al. Transcriptional control of dendritic cell development. Annu Rev Immunol (2016) 34:93–119. doi:10.1146/annurev-immunol-032713-120204
6. Doebel T, Voisin B, Nagao K. Langerhans cells – the macrophage in dendritic cell clothing. Trends Immunol (2017) 17:30120–123. doi:10.1016/j.it.2017.06.008
7. See P, Dutertre CA, Chen J, Gunther P, Mcgovern N, Irac SE, et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science (2017) 356:eaag3009. doi:10.1126/science.aag3009
8. Steinman RM. Dendritic cells: understanding immunogenicity. Eur J Immunol (2007) 37(Suppl 1):S53–60. doi:10.1002/eji.200737400
9. Steinman RM, Nussenzweig MC. Avoiding horror autotoxicus: the importance of dendritic cells in peripheral T cell tolerance. Proc Natl Acad Sci U S A (2002) 99:351–8. doi:10.1073/pnas.231606698
10. Nicholas D, Tang H, Zhang Q, Rudra J, Xu F, Langridge W, et al. Quantitative proteomics reveals a role for epigenetic reprogramming during human monocyte differentiation. Mol Cell Proteomics (2015) 14:15–29. doi:10.1074/mcp.M113.035089
11. Blander JM. The comings and goings of MHC class I molecules herald a new dawn in cross-presentation. Immunol Rev (2016) 272:65–79. doi:10.1111/imr.12428
12. Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol (2012) 13:1118–28. doi:10.1038/ni.2419
13. Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol (2014) 14:571–8. doi:10.1038/nri3712
14. Guilliams M, Malissen B. A death notice for in-vitro-generated GM-CSF dendritic cells? Immunity (2015) 42:988–90. doi:10.1016/j.immuni.2015.05.020
15. Mass E, Ballesteros I, Farlik M, Halbritter F, Gunther P, Crozet L, et al. Specification of tissue-resident macrophages during organogenesis. Science (2016) 353:aaf4238. doi:10.1126/science.aaf4238
16. Carpentier S, Vu Manh TP, Chelbi R, Henri S, Malissen B, Haniffa M, et al. Comparative genomics analysis of mononuclear phagocyte subsets confirms homology between lymphoid tissue-resident and dermal XCR1(+) DCs in mouse and human and distinguishes them from Langerhans cells. J Immunol Methods (2016) 432:35–49. doi:10.1016/j.jim.2016.02.023
17. Crozat K, Guiton R, Guilliams M, Henri S, Baranek T, Schwartz-Cornil I, et al. Comparative genomics as a tool to reveal functional equivalences between human and mouse dendritic cell subsets. Immunol Rev (2010) 234:177–98. doi:10.1111/j.0105-2896.2009.00868.x
18. Collin M, Mcgovern N, Haniffa M. Human dendritic cell subsets. Immunology (2013) 140:22–30. doi:10.1111/imm.12117
19. Jarjour M, Jorquera A, Mondor I, Wienert S, Narang P, Coles MC, et al. Fate mapping reveals origin and dynamics of lymph node follicular dendritic cells. J Exp Med (2014) 211:1109–22. doi:10.1084/jem.20132409
20. Manz MG, Traver D, Miyamoto T, Weissman IL, Akashi K. Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood (2001) 97:3333–41. doi:10.1182/blood.V97.11.3333
21. Shortman K, Sathe P, Vremec D, Naik S, O’Keeffe M. Plasmacytoid dendritic cell development. Adv Immunol (2013) 120:105–26. doi:10.1016/B978-0-12-417028-5.00004-1
22. Scott CL, Soen B, Martens L, Skrypek N, Saelens W, Taminau J, et al. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J Exp Med (2016) 213:897–911. doi:10.1084/jem.20151715
23. Schraml BU, Van Blijswijk J, Zelenay S, Whitney PG, Filby A, Acton SE, et al. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell (2013) 154:843–58. doi:10.1016/j.cell.2013.07.014
24. Sathe P, Metcalf D, Vremec D, Naik SH, Langdon WY, Huntington ND, et al. Lymphoid tissue and plasmacytoid dendritic cells and macrophages do not share a common macrophage-dendritic cell-restricted progenitor. Immunity (2014) 41:104–15. doi:10.1016/j.immuni.2014.05.020
25. Boettcher S, Manz MG. Sensing and translation of pathogen signals into demand-adapted myelopoiesis. Curr Opin Hematol (2016) 23:5–10. doi:10.1097/MOH.0000000000000201
26. Berard F, Blanco P, Davoust J, Neidhart-Berard EM, Nouri-Shirazi M, Taquet N, et al. Cross-priming of naive CD8 T cells against melanoma antigens using dendritic cells loaded with killed allogeneic melanoma cells. J Exp Med (2000) 192:1535–44. doi:10.1084/jem.192.11.1535
27. Romani N, Gruner S, Brang D, Kampgen E, Lenz A, Trockenbacher B, et al. Proliferating dendritic cell progenitors in human blood. J Exp Med (1994) 180:83–93. doi:10.1084/jem.180.1.83
28. Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med (1994) 179:1109–18. doi:10.1084/jem.179.4.1109
29. Hiasa M, Abe M, Nakano A, Oda A, Amou H, Kido S, et al. GM-CSF and IL-4 induce dendritic cell differentiation and disrupt osteoclastogenesis through M-CSF receptor shedding by up-regulation of TNF-alpha converting enzyme (TACE). Blood (2009) 114:4517–26. doi:10.1182/blood-2009-04-215020
30. Jansen JH, Wientjens GJ, Fibbe WE, Willemze R, Kluin-Nelemans HC. Inhibition of human macrophage colony formation by interleukin 4. J Exp Med (1989) 170:577–82. doi:10.1084/jem.170.2.577
31. Vento-Tormo R, Company C, Rodriguez-Ubreva J, De La Rica L, Urquiza JM, Javierre BM, et al. IL-4 orchestrates STAT6-mediated DNA demethylation leading to dendritic cell differentiation. Genome Biol (2016) 17:4. doi:10.1186/s13059-015-0863-2
32. Chomarat P, Dantin C, Bennett L, Banchereau J, Palucka AK. TNF skews monocyte differentiation from macrophages to dendritic cells. J Immunol (2003) 171:2262–9. doi:10.4049/jimmunol.171.5.2262
33. Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med (1992) 176:1693–702. doi:10.1084/jem.176.6.1693
34. Scheicher C, Mehlig M, Zecher R, Reske K. Dendritic cells from mouse bone marrow: in vitro differentiation using low doses of recombinant granulocyte-macrophage colony-stimulating factor. J Immunol Methods (1992) 154:253–64. doi:10.1016/0022-1759(92)90199-4
35. Lutz MB, Kukutsch N, Ogilvie ALJ, Rößner S, Koch F, Romani N, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods (1999) 223:77–92. doi:10.1016/S0022-1759(98)00204-X
36. Menges M, Baumeister T, Rossner S, Stoitzner P, Romani N, Gessner A, et al. IL-4 supports the generation of a dendritic cell subset from murine bone marrow with altered endocytosis capacity. J Leukoc Biol (2005) 77:535–43. doi:10.1189/jlb.0804473
37. Helft J, Bottcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, et al. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c(+)MHCII(+) macrophages and dendritic cells. Immunity (2015) 42:1197–211. doi:10.1016/j.immuni.2015.05.018
38. Lutz MB. IL-3 in dendritic cell development and function: a comparison with GM-CSF and IL-4. Immunobiology (2004) 209:79–87. doi:10.1016/j.imbio.2004.03.001
39. Reid CD, Stackpoole A, Meager A, Tikerpae J. Interactions of tumor necrosis factor with granulocyte-macrophage colony-stimulating factor and other cytokines in the regulation of dendritic cell growth in vitro from early bipotent CD34+ progenitors in human bone marrow. J Immunol (1992) 149:2681–8.
40. Young JW, Szabolcs P, Moore MA. Identification of dendritic cell colony-forming units among normal human CD34+ bone marrow progenitors that are expanded by c-kit-ligand and yield pure dendritic cell colonies in the presence of granulocyte/macrophage colony-stimulating factor and tumor necrosis factor alpha. J Exp Med (1995) 182:1111–9.
41. Bai L, Feuerer M, Beckhove P, Umansky V, Schirrmacher V. Generation of dendritic cells from human bone marrow mononuclear cells: advantages for clinical application in comparison to peripheral blood monocyte derived cells. Int J Oncol (2002) 20:247–53. doi:10.3892/ijo.20.2.247
42. Berger TG, Schulze-Koops H, Schäfer M, Müller E, Lutz MB. Immature and maturation-resistant human dendritic cells generated from bone marrow require two stimulations to induce T cell anergy in vitro. PLoS One (2009) 4:e6645. doi:10.1371/journal.pone.0006645
43. Xu Y, Zhan Y, Lew AM, Naik SH, Kershaw MH. Differential development of murine dendritic cells by GM-CSF versus Flt3 ligand has implications for inflammation and trafficking. J Immunol (2007) 179:7577–84. doi:10.4049/jimmunol.179.11.7577
44. Segura E, Touzot M, Bohineust A, Cappuccio A, Chiocchia G, Hosmalin A, et al. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity (2013) 38:336–48. doi:10.1016/j.immuni.2012.10.018
45. Hettinger J, Richards DM, Hansson J, Barra MM, Joschko AC, Krijgsveld J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol (2013) 14:821–30. doi:10.1038/ni.2638
46. Bender A, Sapp M, Schuler G, Steinman RM, Bhardwaj N. Improved methods for the generation of dendritic cells from nonproliferating progenitors in human blood. J Immunol Methods (1996) 196:121–35. doi:10.1016/0022-1759(96)00079-8
47. Masurier C, Pioche-Durieu C, Colombo BM, Lacave R, Lemoine FM, Klatzmann D, et al. Immunophenotypical and functional heterogeneity of dendritic cells generated from murine bone marrow cultured with different cytokine combinations: implications for anti-tumoral cell therapy. Immunology (1999) 96:569–77. doi:10.1046/j.1365-2567.1999.00728.x
48. Naik SH, Proietto AI, Wilson NS, Dakic A, Schnorrer P, Fuchsberger M, et al. Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol (2005) 174:6592–7. doi:10.4049/jimmunol.174.11.6592
49. Pletinckx K, Lutz MB. Dendritic cells generated with Flt3L and exposed to apoptotic cells lack induction of T cell anergy and Foxp3(+) regulatory T cell conversion in vitro. Immunobiology (2014) 219:230–40. doi:10.1016/j.imbio.2013.10.006
50. Brossart P, Grunebach F, Stuhler G, Reichardt VL, Mohle R, Kanz L, et al. Generation of functional human dendritic cells from adherent peripheral blood monocytes by CD40 ligation in the absence of granulocyte-macrophage colony-stimulating factor. Blood (1998) 92:4238–47.
51. Hubert P, Greimers R, Franzen-Detrooz E, Doyen J, Delanaye P, Boniver J, et al. In vitro propagated dendritic cells from patients with human-papilloma virus-associated preneoplastic lesions of the uterine cervix: use of Flt3 ligand. Cancer Immunol Immunother (1998) 47:81–9. doi:10.1007/s002620050507
52. Breton G, Lee J, Zhou YJ, Schreiber JJ, Keler T, Puhr S, et al. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J Exp Med (2015) 212:401–13. doi:10.1084/jem.20141441
53. Lee J, Breton G, Oliveira TY, Zhou YJ, Aljoufi A, Puhr S, et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J Exp Med (2015) 212:385–99. doi:10.1084/jem.20141442
54. Proietto AI, Mittag D, Roberts AW, Sprigg N, Wu L. The equivalents of human blood and spleen dendritic cell subtypes can be generated in vitro from human CD34(+) stem cells in the presence of fms-like tyrosine kinase 3 ligand and thrombopoietin. Cell Mol Immunol (2012) 9:446–54. doi:10.1038/cmi.2012.48
55. Randolph GJ, Inaba K, Robbiani DF, Steinman RM, Muller WA. Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity (1999) 11:753–61. doi:10.1016/S1074-7613(00)80149-1
56. León B, López-Bravo M, Ardavín C. Monocyte-derived dendritic cells formed at the infection site control the induction of protective T helper 1 responses against Leishmania. Immunity (2007) 26:519–31. doi:10.1016/j.immuni.2007.01.017
57. Satpathy AT, Kc W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med (2012) 209:1135–52. doi:10.1084/jem.20120030
58. Wu X, Briseno CG, Durai V, Albring JC, Haldar M, Bagadia P, et al. Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J Exp Med (2016) 213:2553–65. doi:10.1084/jem.20160600
59. Briseno CG, Haldar M, Kretzer NM, Wu X, Theisen DJ, Kc W, et al. Distinct transcriptional programs control cross-priming in classical and monocyte-derived dendritic cells. Cell Rep (2016) 15:2462–74. doi:10.1016/j.celrep.2016.05.025
60. Greter M, Helft J, Chow A, Hashimoto D, Mortha A, Agudo-Cantero J, et al. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity (2012) 36:1031–46. doi:10.1016/j.immuni.2012.03.027
61. Mayer CT, Ghorbani P, Nandan A, Dudek M, Arnold-Schrauf C, Hesse C, et al. Selective and efficient generation of functional Batf3-dependent CD103+ dendritic cells from mouse bone marrow. Blood (2014) 124:3081–91. doi:10.1182/blood-2013-12-545772
62. Sathe P, Pooley J, Vremec D, Mintern J, Jin JO, Wu L, et al. The acquisition of antigen cross-presentation function by newly formed dendritic cells. J Immunol (2011) 186:5184–92. doi:10.4049/jimmunol.1002683
63. Min L, Mohammad Isa SAB, Shuai W, Piang CB, Nih FW, Kotaka M, et al. Cutting edge: granulocyte-macrophage colony-stimulating factor is the major CD8+ T cell-derived licensing factor for dendritic cell activation. J Immunol (2010) 184:4625–9. doi:10.4049/jimmunol.0903873
64. Miah MA, Yoon CH, Kim J, Jang J, Seong YR, Bae YS. CISH is induced during DC development and regulates DC-mediated CTL activation. Eur J Immunol (2012) 42:58–68. doi:10.1002/eji.201141846
65. Miura R, Kasakura K, Nakano N, Hara M, Maeda K, Okumura K, et al. Role of PU.1 in MHC class II expression via CIITA transcription in plasmacytoid dendritic cells. PLoS One (2016) 11:e0154094. doi:10.1371/journal.pone.0154094
66. Gilliet M, Boonstra A, Paturel C, Antonenko S, Xu XL, Trinchieri G, et al. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J Exp Med (2002) 195:953–8. doi:10.1084/jem.20020045
67. Nikolic T, De Bruijn MF, Lutz MB, Leenen PJ. Developmental stages of myeloid dendritic cells in mouse bone marrow. Int Immunol (2003) 15:515–24. doi:10.1093/intimm/dxg050
68. Lutz MB, Rößner S. Factors influencing the generation of murine dendritic cells from bone marrow: the special role of fetal calf serum. Immunobiology (2008) 212:855–62. doi:10.1016/j.imbio.2007.09.001
69. Kocikova A, Kolesaric A, Koszik F, Stingl G, Elbe-Burger A. Murine Langerhans cells cultured under serum-free conditions mature into potent stimulators of primary immune responses in vitro and in vivo. J Immunol (1998) 161:4033–41.
70. Rößner S, Zinser E, Menges M, Wiethe C, Littmann L, Hänig J, et al. Minor role of bystander tolerance to fetal calf serum in a peptide-specific dendritic cell vaccine model against autoimmunity: comparison with serum-free cultures. J Immunother (2008) 31:656–64. doi:10.1097/CJI.0b013e31818283ef
71. Strunk D, Rappersberger K, Egger C, Strobl H, Kromer E, Elbe A, et al. Generation of human dendritic cells/Langerhans cells from circulating CD34+ hematopoietic progenitor cells. Blood (1996) 87:1292–302.
72. Lutz MB, Inaba K, Schuler G, Romani N. Still alive and kicking: in-vitro-generated GM-CSF dendritic cells! Immunity (2016) 44:1–2. doi:10.1016/j.immuni.2015.12.013
73. Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, et al. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J Exp Med (2001) 194:863–9. doi:10.1084/jem.194.6.863
74. Shortman K, Heath WR. The CD8+ dendritic cell subset. Immunol Rev (2010) 234:18–31. doi:10.1111/j.0105-2896.2009.00870.x
75. Pletinckx K, Stijlemans B, Pavlovic V, Laube R, Brandl C, Kneitz S, et al. Similar inflammatory DC maturation signatures induced by TNF or Trypanosoma brucei antigens instruct default Th2-cell responses. Eur J Immunol (2011) 41:3479–94. doi:10.1002/eji.201141631
76. Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity (2013) 39:599–610. doi:10.1016/j.immuni.2013.08.007
77. Varol C, Landsman L, Fogg DK, Greenshtein L, Gildor B, Margalit R, et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med (2007) 204:171–80. doi:10.1084/jem.20061011
78. Randolph GJ, Beaulieu S, Lebecque S, Steinman RM, Muller WA. Differentiation of monocytes into dendritic cells in a model of transendothelial trafficking. Science (1998) 282:480–3. doi:10.1126/science.282.5388.480
79. Cheong C, Matos I, Choi J-H, Dandamudi DB, Shrestha E, Longhi MP, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell (2010) 143:416–29. doi:10.1016/j.cell.2010.09.039
80. Hammad H, Plantinga M, Deswarte K, Pouliot P, Willart MA, Kool M, et al. Inflammatory dendritic cells – not basophils – are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J Exp Med (2010) 207:2097–111. doi:10.1084/jem.20101563
81. Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity (2012) 37:1076–90. doi:10.1016/j.immuni.2012.08.026
82. Ashok D, Schuster S, Ronet C, Rosa M, Mack V, Lavanchy C, et al. Cross-presenting dendritic cells are required for control of Leishmania major infection. Eur J Immunol (2014) 44:1422–32. doi:10.1002/eji.201344242
83. Inaba K, Metlay JP, Crowley MT, Witmer-Pack M, Steinman RM. Dendritic cells as antigen presenting cells in vivo. Int Rev Immunol (1990) 6:197–206. doi:10.3109/08830189009056630
84. Ohl L, Mohaupt M, Czeloth N, Hintzen G, Kiafard Z, Zwirner J, et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity (2004) 21:279–88. doi:10.1016/j.immuni.2004.06.014
85. Baumjohann D, Hess A, Budinsky L, Brune K, Schuler G, Lutz MB. In vivo magnetic resonance imaging of dendritic cell migration into the draining lymph nodes of mice. Eur J Immunol (2006) 36:2544–55. doi:10.1002/eji.200535742
86. Foucher ED, Blanchard S, Preisser L, Garo E, Ifrah N, Guardiola P, et al. IL-34 induces the differentiation of human monocytes into immunosuppressive macrophages. Antagonistic effects of GM-CSF and IFNgamma. PLoS One (2013) 8:e56045. doi:10.1371/journal.pone.0056045
87. Pooley JL, Heath WR, Shortman K. Cutting edge: intravenous soluble antigen is presented to cd4 t cells by cd8(-) dendritic cells, but cross-presented to cd8 t cells by cd8(+) dendritic cells. J Immunol (2001) 166:5327–30. doi:10.4049/jimmunol.166.9.5327
88. Silva-Vilches C, Pletinckx K, Lohnert M, Pavlovic V, Ashour D, John V, et al. Low doses of cholera toxin and its mediator cAMP induce CTLA-2 secretion by dendritic cells to enhance regulatory T cell conversion. PLoS One (2017) 12:e0178114. doi:10.1371/journal.pone.0178114
89. Albert ML, Sauter B, Bhardwaj N. Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature (1998) 392:86–9. doi:10.1038/32183
90. Wiethe C, Schiemann M, Busch D, Haeberle L, Kopf M, Schuler G, et al. Interdependency of MHC class II/self-peptide and CD1d/self-glycolipid presentation by TNF-matured dendritic cells for protection from autoimmunity. J Immunol (2007) 178:4908–16. doi:10.4049/jimmunol.178.8.4908
91. Wiethe C, Debus A, Mohrs M, Steinkasserer A, Lutz MB, Gessner A. Dendritic cell differentiation state and their interaction with NKT cells determine Th1/Th2 differentiation in the murine model of Leishmania major infection. J Immunol (2008) 180:4371–81. doi:10.4049/jimmunol.180.7.4371
92. Petersen TR, Sika-Paotonu D, Knight DA, Simkins HM, Hermans IF. Exploiting the role of endogenous lymphoid-resident dendritic cells in the priming of NKT cells and CD8+ T cells to dendritic cell-based vaccines. PLoS One (2011) 6:e17657. doi:10.1371/journal.pone.0017657
93. Brewitz A, Eickhoff S, Dahling S, Quast T, Bedoui S, Kroczek RA, et al. CD8+ T cells orchestrate pDC-XCR1+ dendritic cell spatial and functional cooperativity to optimize priming. Immunity (2017) 46:205–19. doi:10.1016/j.immuni.2017.01.003
94. Anderson DA III, Grajales-Reyes GE, Satpathy AT, Hueichucura CEV, Murphy TL, Murphy KM. Revisiting the specificity of the MHC class II transactivator CIITA in classical murine dendritic cells in vivo. Eur J Immunol (2017) 47:1317–23. doi:10.1002/eji.201747050
95. Theisen D, Murphy K. The role of cDC1s in vivo: CD8 T cell priming through cross-presentation. F1000Res (2017) 6:98. doi:10.12688/f1000research.9997.1
96. Gupta R, Emens LA. GM-CSF-secreting vaccines for solid tumors: moving forward. Discov Med (2010) 10:52–60.
97. Bol KF, Schreibelt G, Gerritsen WR, De Vries IJ, Figdor CG. Dendritic cell-based immunotherapy: state of the art and beyond. Clin Cancer Res (2016) 22:1897–906. doi:10.1158/1078-0432.CCR-15-1399
98. Saxena M, Bhardwaj N. Turbocharging vaccines: emerging adjuvants for dendritic cell based therapeutic cancer vaccines. Curr Opin Immunol (2017) 47:35–43. doi:10.1016/j.coi.2017.06.003
99. Romero P, Banchereau J, Bhardwaj N, Cockett M, Disis ML, Dranoff G, et al. The human vaccines project: a roadmap for cancer vaccine development. Sci Transl Med (2016) 8:334s339. doi:10.1126/scitranslmed.aaf0685
100. Garg AD, Coulie PG, Van Den Eynde BJ, Agostinis P. Integrating next-generation dendritic cell vaccines into the current cancer immunotherapy landscape. Trends Immunol (2017) 38:577–93. doi:10.1016/j.it.2017.05.006
101. Gross S, Erdmann M, Haendle I, Voland S, Berger T, Schultz E, et al. Twelve-year survival and immune correlates in dendritic cell-vaccinated melanoma patients. JCI Insight (2017) 2:e91438. doi:10.1172/jci.insight.91438
102. Martin-Fontecha A, Sebastiani S, Hopken UE, Uguccioni M, Lipp M, Lanzavecchia A, et al. Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J Exp Med (2003) 198:615–21. doi:10.1084/jem.20030448
103. Mitchell DA, Batich KA, Gunn MD, Huang MN, Sanchez-Perez L, Nair SK, et al. Tetanus toxoid and CCL3 improve dendritic cell vaccines in mice and glioblastoma patients. Nature (2015) 519:366–9. doi:10.1038/nature14320
104. Parmiani G, Castelli C, Pilla L, Santinami M, Colombo MP, Rivoltini L. Opposite immune functions of GM-CSF administered as vaccine adjuvant in cancer patients. Ann Oncol (2007) 18:226–32. doi:10.1093/annonc/mdl158
105. Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, et al. Stroma-derived interleukin-34 controls the development and maintenance of Langerhans cells and the maintenance of microglia. Immunity (2012) 37:1050–60. doi:10.1016/j.immuni.2012.11.001
106. Hoeffel G, Wang Y, Greter M, See P, Teo P, Malleret B, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med (2012) 209:1167–81. doi:10.1084/jem.20120340
107. Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol (2012) 13:753–60. doi:10.1038/ni.2360
108. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science (2010) 330:841–5. doi:10.1126/science.1194637
109. Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science (2012) 336:86–90. doi:10.1126/science.1219179
110. Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity (2016) 44:439–49. doi:10.1016/j.immuni.2016.02.024
111. Lutz MB, Dohler A, Azukizawa H. Revisiting the tolerogenicity of epidermal Langerhans cells. Immunol Cell Biol (2010) 88:381–6. doi:10.1038/icb.2010.17
112. Igyarto BZ, Kaplan DH. Antigen presentation by Langerhans cells. Curr Opin Immunol (2013) 25:115–9. doi:10.1016/j.coi.2012.11.007
113. Sheng J, Ruedl C, Karjalainen K. Most tissue-resident macrophages except microglia are derived from fetal hematopoietic stem cells. Immunity (2015) 43:382–93. doi:10.1016/j.immuni.2015.07.016
114. Ginhoux F, Tacke F, Angeli V, Bogunovic M, Loubeau M, Dai XM, et al. Langerhans cells arise from monocytes in vivo. Nat Immunol (2006) 7:265–73. doi:10.1038/ni1307
115. Sere K, Baek JH, Ober-Blobaum J, Muller-Newen G, Tacke F, Yokota Y, et al. Two distinct types of Langerhans cells populate the skin during steady state and inflammation. Immunity (2012) 37:905–16. doi:10.1016/j.immuni.2012.07.019
116. Capucha T, Mizraji G, Segev H, Blecher-Gonen R, Winter D, Khalaileh A, et al. Distinct murine mucosal Langerhans cell subsets develop from pre-dendritic cells and monocytes. Immunity (2015) 43:369–81. doi:10.1016/j.immuni.2015.06.017
117. Geissmann F, Prost C, Monnet JP, Dy M, Brousse N, Hermine O. Transforming growth factor beta1, in the presence of granulocyte/macrophage colony-stimulating factor and interleukin 4, induces differentiation of human peripheral blood monocytes into dendritic Langerhans cells. J Exp Med (1998) 187:961–6. doi:10.1084/jem.187.6.961
118. Hoshino N, Katayama N, Shibasaki T, Ohishi K, Nishioka J, Masuya M, et al. A novel role for Notch ligand Delta-1 as a regulator of human Langerhans cell development from blood monocytes. J Leukoc Biol (2005) 78:921–9. doi:10.1189/jlb.1204746
119. Kriehuber E, Bauer W, Charbonnier AS, Winter D, Amatschek S, Tamandl D, et al. Balance between NF-kappaB and JNK/AP-1 activity controls dendritic cell life and death. Blood (2005) 106:175–83. doi:10.1182/blood-2004-08-3072
120. Heinz LX, Platzer B, Reisner PM, Jorgl A, Taschner S, Gobel F, et al. Differential involvement of PU.1 and Id2 downstream of TGF-beta1 during Langerhans-cell commitment. Blood (2006) 107:1445–53. doi:10.1182/blood-2005-04-1721
121. Sparber F, Scheffler JM, Amberg N, Tripp CH, Heib V, Hermann M, et al. The late endosomal adaptor molecule p14 (LAMTOR2) represents a novel regulator of Langerhans cell homeostasis. Blood (2014) 123:217–27. doi:10.1182/blood-2013-08-518555
122. Martinez-Cingolani C, Grandclaudon M, Jeanmougin M, Jouve M, Zollinger R, Soumelis V. Human blood BDCA-1 dendritic cells differentiate into Langerhans-like cells with thymic stromal lymphopoietin and TGF-beta. Blood (2014) 124:2411–20. doi:10.1182/blood-2014-04-568311
123. Milne P, Bigley V, Gunawan M, Haniffa M, Collin M. CD1c+ blood dendritic cells have Langerhans cell potential. Blood (2015) 125:470–3. doi:10.1182/blood-2014-08-593582
124. Ito T, Inaba M, Inaba K, Toki J, Sogo S, Iguchi T, et al. A CD1a+/CD11c+ subset of human blood dendritic cells is a direct precursor of Langerhans cells. J Immunol (1999) 163:1409–19.
125. Bigley V, Mcgovern N, Milne P, Dickinson R, Pagan S, Cookson S, et al. Langerin-expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells. J Leukoc Biol (2015) 97:627–34. doi:10.1189/jlb.1HI0714-351R
126. Artyomov MN, Munk A, Gorvel L, Korenfeld D, Cella M, Tung T, et al. Modular expression analysis reveals functional conservation between human Langerhans cells and mouse cross-priming dendritic cells. J Exp Med (2015) 212:743–57. doi:10.1084/jem.20131675
127. Jurkin J, Krump C, Koffel R, Fieber C, Schuster C, Brunner PM, et al. Human skin dendritic cell fate is differentially regulated by the monocyte identity factor Kruppel-like factor 4 during steady state and inflammation. J Allergy Clin Immunol (2017) 139:1873–84.e10. doi:10.1016/j.jaci.2016.09.018
128. Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J (2007) 26:4138–48. doi:10.1038/sj.emboj.7601824
129. Esser C, Rannug A, Stockinger B. The aryl hydrocarbon receptor in immunity. Trends Immunol (2009) 30:447–54. doi:10.1016/j.it.2009.06.005
130. Quintana FJ, Murugaiyan G, Farez MF, Mitsdoerffer M, Tukpah AM, Burns EJ, et al. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A (2010) 107:20768–73. doi:10.1073/pnas.1009201107
131. Julliard W, Fechner JH, Mezrich JD. The aryl hydrocarbon receptor meets immunology: friend or foe? A little of both. Front Immunol (2014) 5:458. doi:10.3389/fimmu.2014.00458
132. Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity (2014) 40:642–56. doi:10.1016/j.immuni.2014.04.016
133. Mielcarek M, Kirkorian AY, Hackman RC, Price J, Storer BE, Wood BL, et al. Langerhans cell homeostasis and turnover after nonmyeloablative and myeloablative allogeneic hematopoietic cell transplantation. Transplantation (2014) 98:563–8. doi:10.1097/TP.0000000000000097
134. Schuler G, Steinman RM. Murine epidermal Langerhans cells mature into potent immunostimulatory dendritic cells in vitro. J Exp Med (1985) 161:526–46. doi:10.1084/jem.161.3.526
135. Dalod M, Chelbi R, Malissen B, Lawrence T. Dendritic cell maturation: functional specialization through signaling specificity and transcriptional programming. EMBO J (2014) 33:1104–16. doi:10.1002/embj.201488027
136. Pletinckx K, Dohler A, Pavlovic V, Lutz MB. Role of dendritic cell maturity/costimulation for generation, homeostasis, and suppressive activity of regulatory T cells. Front Immunol (2011) 2:39. doi:10.3389/fimmu.2011.00039
137. Romani N, Brunner PM, Stingl G. Changing views of the role of Langerhans cells. J Invest Dermatol (2012) 132:872–81. doi:10.1038/jid.2011.437
138. Ardouin L, Luche H, Chelbi R, Carpentier S, Shawket A, Montanana Sanchis F, et al. Broad and largely concordant molecular changes characterize tolerogenic and immunogenic dendritic cell maturation in thymus and periphery. Immunity (2016) 45:305–18. doi:10.1016/j.immuni.2016.07.019
139. Toke ER, Lorincz O, Csiszovszki Z, Somogyi E, Felfoldi G, Molnar L, et al. Exploitation of Langerhans cells for in vivo DNA vaccine delivery into the lymph nodes. Gene Ther (2014) 21:566–74. doi:10.1038/gt.2014.29
140. Ali MA, Thrower SL, Hanna SJ, Coulman SA, Birchall JC, Wong FS, et al. Topical steroid therapy induces pro-tolerogenic changes in Langerhans cells in human skin. Immunology (2015) 146:411–22. doi:10.1111/imm.12518
141. Oehler L, Majdic O, Pickl WF, Stöckl J, Riedl E, Drach J, et al. Neutrophil granulocyte-committed cells can be driven to acquire dendritic cell characteristics. J Exp Med (1998) 187:1019–28. doi:10.1084/jem.187.7.1019
142. Matsushima H, Geng S, Lu R, Okamoto T, Yao Y, Mayuzumi N, et al. Neutrophil differentiation into a unique hybrid population exhibiting dual phenotype and functionality of neutrophils and dendritic cells. Blood (2013) 121:1677–89. doi:10.1182/blood-2012-07-445189
143. Geng S, Matsushima H, Okamoto T, Yao Y, Lu R, Page K, et al. Emergence, origin, and function of neutrophil-dendritic cell hybrids in experimentally induced inflammatory lesions in mice. Blood (2013) 121:1690–700. doi:10.1182/blood-2012-07-445197
144. Koffel R, Meshcheryakova A, Warszawska J, Hennig A, Wagner K, Jorgl A, et al. Monocytic cell differentiation from band-stage neutrophils under inflammatory conditions via MKK6 activation. Blood (2014) 124:2713–24. doi:10.1182/blood-2014-07-588178
145. Araki H, Katayama N, Yamashita Y, Mano H, Fujieda A, Usui E, et al. Reprogramming of human postmitotic neutrophils into macrophages by growth factors. Blood (2004) 103:2973–80. doi:10.1182/blood-2003-08-2742
146. Takashima A, Yao Y. Neutrophil plasticity: acquisition of phenotype and functionality of antigen-presenting cell. J Leukoc Biol (2015) 98:489–96. doi:10.1189/jlb.1MR1014-502R
147. Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KW, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol (2014) 15:1181–9. doi:10.1038/ni.3006
Keywords: dendritic cells, GM-CSF, monocytes, Langerhans cells, macrophages
Citation: Lutz MB, Strobl H, Schuler G and Romani N (2017) GM-CSF Monocyte-Derived Cells and Langerhans Cells As Part of the Dendritic Cell Family. Front. Immunol. 8:1388. doi: 10.3389/fimmu.2017.01388
Received: 10 August 2017; Accepted: 09 October 2017;
Published: 23 October 2017
Edited by:
Irina Caminschi, Burnet Institute, AustraliaReviewed by:
Susan Kovats, Oklahoma Medical Research Foundation, United StatesKen Shortman, Walter and Eliza Hall Institute of Medical Research, Australia
Copyright: © 2017 Lutz, Strobl, Schuler and Romani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manfred B. Lutz, m.lutz@vim.uni-wuerzburg.de
 Manfred B. Lutz
Manfred B. Lutz Herbert Strobl2
Herbert Strobl2 Nikolaus Romani
Nikolaus Romani