
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 04 December 2017
Sec. Vaccines and Molecular Therapeutics
Volume 8 - 2017 | https://doi.org/10.3389/fimmu.2017.01647
Salmonella infections remain a big problem worldwide, causing enteric fever by Salmonella Typhi (or Paratyphi) or self-limiting gastroenteritis by non-typhoidal Salmonella (NTS) in healthy individuals. NTS may become invasive and cause septicemia in elderly or immuno-compromised individuals, leading to high mortality and morbidity. No vaccines are currently available for preventing NTS infection in human. As these invasive NTS are restricted to several O-antigen serogroups including B1, D1, C1, and C2, O-antigen polysaccharide is believed to be a good target for vaccine development. In this study, a strategy of O-serotype conversion was investigated to develop live attenuated S. Typhimurium vaccines against the major serovars of NTS infections. The immunodominant O4 serotype of S. Typhimurium was converted into O9, O7, and O8 serotypes through unmarked chromosomal deletion–insertion mutations. O-serotype conversion was confirmed by LPS silver staining and western blotting. All O-serotype conversion mutations were successfully introduced into the live attenuated S. Typhimurium vaccine S738 (Δcrp Δcya) to evaluate their immunogenicity in mice model. The vaccine candidates induced high amounts of heterologous O-polysaccharide-specific functional IgG responses. Vaccinated mice survived a challenge of 100 times the 50% lethality dose (LD50) of wild-type S. Typhimurium. Protective efficacy against heterologous virulent Salmonella challenges was highly O-serotype related. Furthermore, broad-spectrum protection against S. Typhimurium, S. Enteritidis, and S. Choleraesuis was observed by co-vaccination of O9 and O7 O-serotype-converted vaccine candidates. This study highlights the strategy of expressing heterologous O-polysaccharides via genetic engineering in developing live attenuated S. Typhimurium vaccines against NTS infections.
Salmonellae are facultative intracellular pathogens that are capable of infecting a wide range of animals and are responsible for high mortality and morbidity worldwide (1, 2). More than 2,500 Salmonella serovars have so far been identified, while 99% of human and animal infections are caused solely by one subspecies, Salmonella enterica subsp. enterica (S. enterica) (3). With regard to human disease, S. enterica has traditionally been divided into a small number of human-restricted invasive typhoidal Salmonella and thousands of non-typhoidal Salmonella (NTS) (4). Human host-restricted S. enterica serovars Typhi and Paratyphi A are the leading causes of typhoid and paratyphoid enteric fevers, respectively, while NTS predominantly cause a self-limiting gastroenteritis in healthy individuals. Although NTS generally produce diarrhea, they can become invasive and cause septicemia, as well as focal infections such as meningitis, endocarditis, and osteomyelitis (5). Typically, these invasive NTS are restricted to several O-antigen serogroups, including B1, D1, C1, and C2. In Sub-Saharan Africa, invasive NTS (iNTS) have emerged as a prominent cause of bloodstream infections (6), with S. enterica serovars Typhimurium (serogroup B1) and Enteritidis (serogroups D1) being the most prevalent. Clinical diagnosis in these regions is difficult, as there are no signs or symptoms to distinguish NTS from a number of other common infections, such as endemic malaria (7). In developed countries, NTS infection is mainly foodborne and causes gastroenteritis, with bacteremia typically occurring as a rare complication associated with immunodeficiency. Serogroup C is becoming the most common serogroup in the USA and has been increasing in Europe over the last decade (8). Additionally, increasing frequencies of multi-drug resistance among invasive isolates threaten the effectiveness of amenable antibiotic treatments (9). To date, vaccines are regarded as the most economical and effective ways to prevent salmonellosis (10, 11).
The first clinical vaccine against Salmonella was an inactivated whole-cell vaccine (TAB vaccine), which was used extensively by the British and US military to prevent typhoid fever and associated deaths (11, 12). However, these vaccines are no longer used due to their high reactogenicity (13). As an improvement, the following three types of vaccines have been licensed: the live attenuated vaccine Ty21a, a purified unconjugated Vi polysaccharide, and a Vi polysaccharide conjugated to tetanus toxoid (14). Except for S. Typhi, no vaccines against other Salmonella serovars are currently licensed for use in humans. Vaccines against NTS are even further behind in the development pipeline. Studies of NTS vaccines are mainly focused on live attenuated Salmonella vaccine and O-antigen polysaccharide-based subunit vaccine.
Live attenuated vaccines have a number of potential advantages, including an excellent ability to elicit T-cell responses, a convenient oral vaccination route, and good capacities to induce mucosal immunity (15, 16). The major challenge in developing live attenuated vaccines lies in attaining an optimal level of attenuation without compromising immunogenicity (17). To date, the only live attenuated NTS vaccine that has completed a Phase 1 study is WT05, a S. Typhimurium vaccine containing attenuated aroC and ssaV (18), though other live NTS vaccine candidates are in preclinical development. These include S. Typhimurium and S. Enteritidis, lacking the guaBA and clpP genes, that were shown to protect mice against a lethal homologous challenge (19). Attenuation strategies target global regulators of gene expression, such as mutations in cya and crp, are also promising (20, 21). Another potential strategy is to introduce mutations in Salmonella that lead to regulated delayed attenuation in vivo via dependence on key nutrients that are not available in host tissues, thus leading to attenuation after invading and colonizing host (22, 23). However, vaccines based on these strategies have not yet reached clinical trials.
O-antigen polysaccharide, a portion of lipopolysaccharide, is responsible for Salmonella serovar specificity and is considered to be an excellent protective antigen (24, 25). Many efforts have been made to develop vaccines that contain repeating O-polysaccharide polymers conjugated to a range of protein carriers (10) including tetanus toxin (TT), diphtheria toxin (DT), and the non-toxic recombinant form of DT (CRM197) (10, 11). The conjugation of Salmonella O-polysaccharide to Salmonella proteins is likely to be a more effective alternative to exogenous carriers (26, 27), as they could induce immune responses against two Salmonella antigens instead of one. However, these attenuated NTS vaccines and glycoconjugate vaccines are largely restricted to a single serovar or group of serovars, mainly against serovars Typhimurium and Enteritidis and unable to or only partially provide cross-protection against heterologous Salmonella infections. Moreover, these serovar-specific vaccines may suffer the potential risk of changing the epidemiology of NTS. The clinical evidence has shown that S. Choleraesuis (group C) is becoming prevalent in the USA and Europe (8). However, little work has been done to develop vaccines against Salmonella serogroup C infections (28, 29).
O-antigens of Salmonella groups B and D share a common trisaccharide backbone of α-Man(1→4)-α-Rha-(1→3)-α-Gal-(1→2), which serologically contributes to epitopes 1 and 12 (30). In each case, a unique dideoxyhexose sugar contributes to their immunodominance in serogroup specificity, namely, O4 (group B, α-Abe(1→3)Man), O8 (group C2, α-Abe(1→3)Rha), and O9 (group D, α-Tyv(1→3)Man). The unique sugar components and linkages in the O-unit of group C1 contribute to factor O7 (31). The O-antigenic characteristics of S. Typhimurium, S. Enteritidis, S. Choleraesuis, and S. Newport are hereafter referred to as O4, O9, O7, and O8, rather than their full O-antigen formulae. Passive protection studies demonstrated that IgG or IgM directed against the immunodominant group-specific epitope O4 played more important for protection than antibodies specific to epitope O1 and O12 (32). Immunodominant O-antigens also play an important role in eliciting protective memory responses against Salmonella (33). In this study, we modified S. Typhimurium O-antigen structure, converting its native B1 group immunodominant O4 serotype to the D1 group O9, C1 group O7, and C2 group O8 via chromosomal genetic manipulation. We expect to combine these heterologous O-polysaccharides in live attenuated S. Typhimurium vaccines to protect against serogroups B, D, C1, and C2, thereby preventing the majority of NTS infection. Our work highlights the possibilities of achieving a broad protective coverage against S. Typhimurium, S. Enteritidis, S. Choleraesuis and S. Newport by live attenuated S. Typhimurium vaccines based on O-serotype conversion.
All bacteria and plasmids used in this study are listed in Table 1. The O-serotype-converted Salmonella mutants were all derived from wild-type S. Typhimurium S100. Escherichia coli and S. enterica were grown at 37°C in Luria-Bertani (LB) broth or in LB agar. sacB gene-based counter selection in allelic exchange experiments was performed on LB agar containing 10% sucrose with no sodium chloride added and grown at 30°C (34). Media were supplemented with 25 µg/ml chloramphenicol for selection. Diaminopimelic acid (50 µg/ml) was added for the growth of the Δasd strain (35). Electrocompetent E. coli or S. enterica cells were prepared as described previously (36). In vitro growth rates of Salmonella strains were determined by optical density measurements.
DNA manipulations were performed using standard methods (37) and were approved by Division of Environmental Health and Safety of Sichuan Agricultural University. No restriction endonuclease sites were introduced when amplifying DNA fragments from chromosomes or plasmids. DNA concentrations and purity were measured using a Nanodrop ND-2000 spectrophotometer (Thermo Fisher Scientific). DNA fragments were assembled using Gibson Assembly Master Mix according to the manufacturer’s instructions (New England Bio Labs). PCR was performed in 20 µl reaction volumes in a Thermo Scientific Arktik Thermal Cycler. Reaction mixtures contained one volume of 1 µM (each) forward and reverse primers and DNA template (~50 ng plasmid DNA or ~100 ng chromosomal DNA) with an equal volume of 2× premix PrimeSTAR Max DNA polymerase (TaKaRa). Thermal cycler conditions were 98°C for 2 min; 30 cycles of 98°C for 1 min, 55°C for 30 s, and 72°C for 15–30 s/kb; and a final extension at 72°C for 10 min.
All primers used in this study are listed in Table S1 in Supplementary Material. We used a sacB gene-based suicide vector to counter-select deletion and/or insertion mutations. For deletion mutation suicide vector construction, two homologous DNA fragments, upstream and downstream of the gene or operon to be deleted, were amplified using primer pairs designated D-N-1F/D-N-1R and D-N-2F/D-N-2R (N represents the name of the gene or operon to be deleted). After DNA purification, these two fragments were fused by PCR using primer pairs designated D-N-1F/D-N-2R and linked into the suicide vector pYA4278 (38). For insertion mutation suicide vector construction, the allelic gene or operon to be inserted in a directed site was amplified using primer pairs designated (G)In-I-F/(G)In-I-R (I represents the name of the gene or operon to be inserted). The backbone of the suicide vector, which contained the homologous upstream and downstream sequence of the site to be inserted, was amplified using primer pairs designated (G)Vec-D-N-F/(G)Vec-D-N-R. All purified DNA fragments were assembled in order using Gibson Assembly (39), resulting in targeted gene or operon inserted into the directed site in a suicide plasmid. All allelic gene exchanges or whole-operon replacement in S. Typhimurium were achieved in two steps (Figure 1): deletion of the original gene or operon followed by insertion of the target gene or operon. Chromosomal modifications were introduced by suicide plasmids using standard methods (40, 41). All of the constructed mutants, either intermediates or final constructs, were routinely sequenced.
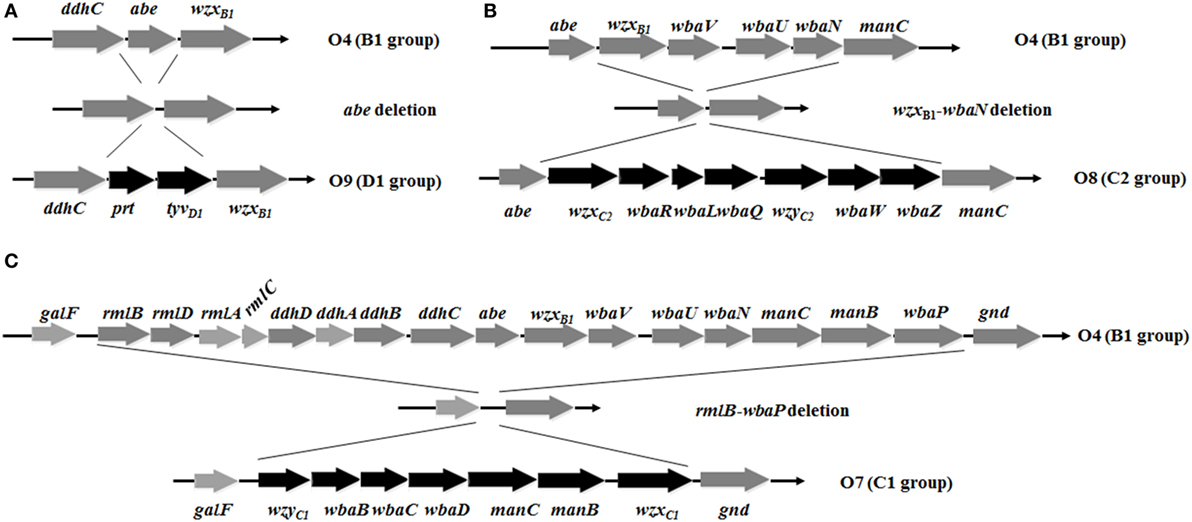
Figure 1. Deletion-insertion mutations resulting in O-serotype conversion in Salmonella Typhimurium. (A) The abe gene was deleted and inserted by prt-tyvD1 from S. Enteritidis, converting the O4 serotype to O9. (B) Genes wzxB1, wbaV, wbaU, and wbaN were deleted and inserted by wzxC2, wbaR, wbaL, wbaQ, wzyC2, wbaW, and wbaZ from S. Newport, converting the O4 serotype to O8. (C) The entire O-antigen gene cluster of S. Typhimurium was deleted and inserted by the C1 serogroup O-antigen gene cluster from S. Choleraesuis, converting the O4 serotype to O7. Genes inserted into the S. Typhimurium O-antigen gene cluster for O-serotype conversion are indicated by black arrows and native genes are depicted as gray arrows. Diagrams were drawn to scale.
LPS were prepared and visualized by the method of Hitchcock and Brown (42). LPS samples were separated via 12.5% SDS-PAGE gels and transferred to nitrocellulose membranes using a Trans-Blot SD semidry transfer system (Bio-Rad, Hercules, CA, USA). Membranes were first incubated with O-antigen signal-factor rabbit antisera (BD Biosciences) or vaccinated murine pooled sera (1:100 dilution) followed by secondary anti-rabbit or anti-mouse horseradish peroxidase-conjugated antibody (Sigma) at a 1:1,000 dilution. Patterns were detected by chemiluminescence using western ECL blotting substrates (Bio-Rad).
P22HT int was propagated on S. Typhimurium S100 carrying the chromosomal integrated suicide vector pSS241 (43), which confers chloramphenicol resistance. Strains to be tested were grown overnight in LB broth at 37°C. Cultures were diluted 1:100 into fresh LB broth and grown at 37°C to an OD600 of 0.6. Then, 10 µl of phage (1 × 108 PFU) was mixed with 1 ml of bacteria (~5 × 106 CFU) and incubated at 37°C for 30 min. After the incubation, the mixture was centrifuged and resuspended in 1 ml of PBS. A 100-µl aliquot was spread onto LB agar plate containing 25 µg/ml chloramphenicol. Colonies were counted after an overnight incubation at 37°C.
Motility assays were performed on LB plates containing 0.3% agar. The plates were allowed to dry at room temperature for approximately 2 h before the assays. Then, 6 µl of freshly grown bacteria (~5 × 106 CFU) was spotted onto the middle of the plates and incubated at 37°C for 6 h. The diameters of the colonies (in millimeters) were measured.
The MICs of deoxycholate (DOC) and polymyxin B were determined using 96-well microtiter plates. Two-fold serial dilutions of DOC (0.39–59 mg/ml) and polymyxin B (0.078–10 µg/ml) were made along the plates. Bacteria were grown to an OD600 of 0.6 and diluted to ~5.0 × 104 CFU/ml in LB broth. Then, 100 µl of diluted bacteria suspension was added to each well and followed by an overnight incubation at 37°C. The optical density of each well was determined using an iMark™ Microplate Reader (Bio-Rad, Hercules, CA, USA). The threshold of inhibition was 0.1 at OD600.
The human epithelial type 2 (Hep-2) cell line (ATCC strain CCL-6) was used to perform bacterial attachment and invasion assays as described previously (44). The bacteria were added to each well at a multiplicity of infection of 10:1. The percentage of attachment rate was calculated as follows: percentage of attachment = 100 × (number of cell-associated bacterial/initial number of bacterial added). The percentage of invasion rate was calculated as follows: percentage of invasion = 100 × (number of bacteria resistant to gentamicin/initial number of bacteria added).
All animal studies were conducted in compliance with the Animal Welfare Act and regulations stated in the Guide for the Care and Use of Laboratory Animals, which was approved by Sichuan Agricultural University Institutional Animal Care and Use Committee (Ya’an, China; Approval No. 2011028).
Six-week-old female BALB/c mice were purchased from Dashuo Biotechnology Co., Ltd. (Chengdu, China). To determine the 50% lethality dose (LD50), bacteria were grown statically overnight at 37°C. Overnight cultures were diluted 1:100 into fresh LB media, grown at 37°C until reaching an OD600 of 0.8–0.9. Cells were harvested by centrifugation at 3,452 × g at room temperature, washed once, and adjusted to the required inoculum density in buffered saline with gelatin (BSG). Groups of six mice each were infected orally with 20 µl of BSG containing various doses of S. Typhimurium S100 or its derivatives, ranging from 1 × 104 to 1 × 109 CFU. Animals were observed for 4 weeks after infection, and deaths were recorded daily. The LD50 for each strain was calculated using the method of Reed and Muench (45). To evaluate colonization, groups of three mice were orally inoculated with 20 µl of BSG containing 1 × 109 CFU bacteria. On days 4 and 8 post-inoculation, Peyer’s patches, spleen, and liver samples were collected. Samples were homogenized, and dilutions were plated onto MacConkey and LB agar to determine viable counts.
Groups of 12 mice each were inoculated orally with 20 µl of BSG containing approximately 1 × 109 CFU vaccine strains on day 0 and boosted on day 14 with the same dose. Blood samples were collected after 28 days. Mice were challenged orally on day 56 with 5 × 107 CFU of S. Typhimurium, S. Choleraesuis, or S. Enteritidis (~100 times LD50).
S. Typhimurium and S. Enteritidis LPS were purchased from Sigma (St. Louis, MO, USA). S. Choleraesuis and S. Newport LPS were purified as described previously (46). A quantitative enzyme-linked immunosorbent assay (ELISA) was performed to determine serum antibody concentrations with the following modifications. Microtiter plates were coated with Salmonella LPS. The capture antibody, unlabeled goat antimouse IgG (H + L) (BD Pharmingen, San Diego, CA, USA) at 1 µg/ml in PBS, was added to extra uncoated wells to generate the standard curve. The plates were incubated overnight at 4°C, followed by blocking with PBS containing 5% BSA for 1 h at room temperature. For the LPS-coated wells, 100 µl of diluted serum was added to individual wells in triplicate. For the capture antibody-coated wells, the purified mouse IgG standard (for the standard curve quantification, BD Pharmingen, San Diego, CA, USA) was added, followed by two-fold serial dilutions starting at 0.5 µg/ml. The plates were incubated for 1 h at 37°C and then treated with biotinylated goat anti-mouse IgG (Southern Biotechnology Associates, Birmingham, AL, USA). The wells were developed with a streptavidin-alkaline phosphatase conjugate (Southern Biotechnology Associates, Birmingham, AL, USA), followed by a p-nitrophenylphosphate substrate (Sigma-Aldrich, St. Louis, MO, USA). Color development was recorded at 405 nm using an iMark™ Microplate Reader (Bio-Rad, Hercules, CA, USA). The ELISA standard curve was drawn using Curve Expert software (Hyams DG, Starkville, MS, USA). Serum antibody concentrations were calculated based on absorbance values and the standard curve.
Sera used for complement deposition assays were pooled sera taken from mice after the second immunization and were heated at 56°C for 30 min to inactivate endogenous complement. Bacteria were grown to an OD600 of 0.8 and harvested by centrifugation at 6,000 rpm for 2 min. Bacterial pellets were washed, centrifuged, and resuspended to approximately 5 × 108 CFU/ml in PBS. Then, 20 µl of bacterial sample was incubated with 80 µl of complement-inactivated sera at 37°C for 30 min. Bacteria were then washed once with PBS, resuspended and incubated with 100 µl of fresh naive BALB/c mouse sera at 37°C for 30 min. After another wash with PBS, the samples were incubated with 100 µl of FITC-conjugated goat anti-mouse complement C3c (Abcam) at a dilution of 1:100 on ice for 30 min in the dark. After incubation, the bacteria were washed with PBS, resuspended in 1% formaldehyde, and latter analyzed with a flow cytometer (BD FACSVerse™). The negative control was wild-type S. Typhimurium incubated with non-vaccinated complement-inactivated mice sera, and the positive control was wild-type S. Typhimurium incubated with complement-inactivated rabbit anti-O4 Salmonella sera (BD Biosciences). All other processes were the same as the test groups.
An in vitro assay was performed to analyze the differential uptake of S. Typhimurium, S. Enteritidis, S. Choleraesuis, and S. Newport by the RAW264.7 macrophage cell line. Briefly, 1 × 105 RAW264.7 cells in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS (Newborn calf serum) and Pen/Strep were allowed to adhere to a 24-well plate for 24 h. Each well contained approximately 5 × 105 RAW264.7 cells. Approximately 30 min prior to infection, the old media were replaced with fresh DMEM containing only 10% FBS. In each well, 5 × 106 Salmonella opsonized with relative vaccinated mice pooled sera or naive serum (1 µl serum for a 100-µl volume of Salmonella in PBS) were added. Gentamicin was added to each well at a final concentration of 100 µg/ml at different time intervals of 0, 20, 40, and 60 min, and the plates were incubated for 1 h to kill non-phagocytized bacterial cells. After three washes with PBS, the macrophages were lysed with 1% Triton X-100 and appropriate dilutions were plated on LB agar plates. Colonies were enumerated the next day.
Data were analyzed using the GraphPad Prism 5 software (Graph Software, San Diego, CA, USA) by one-way or two-way ANOVA followed by Tukey’s multiple-comparison post-test. Kaplan–Meier survival curve comparisons were calculated by comparing two groups at each time point through the log-rank (Mantel–Cox) test. The data were expressed as the mean ± SEM. P < 0.05 was considered statistically significant.
To develop effective live attenuated vaccines against invasive NTS infections, we targeted the structurally hyper-variable O-antigens. The distinctive O-antigen gene clusters of groups B1, D1, C1, and C2 were compared (Figure S1 in Supplementary Material), together with their chemical structures, to identify the relevant sugar components and glycosidic linkages (Figure S2 in Supplementary Material). The genetic modifications we used to achieve the desired S. Typhimurium O-serotype conversions are shown in Figure 1. Specifically, (1) the abe gene in S. Typhimurium was replaced with prt-tyvD1 from S. Enteritidis to convert the O4 serotype to O9, resulting in S1031 (Δabe:prt-tyvD1) (O9) (Figure 1A); (2) the genes wzxB1-wbaN were replaced with wzxC2-wbaZ from S. Newport to convert O4 into O8, resulting in S1131 [Δ(wzxB1-wabN):(wzxC2-wbaZ)] (O8) (Figure 1B); and (3) the entire O-antigen gene cluster of group B1 was replaced with C1 from S. Choleraesuis to convert O4 into O7, resulting in S1124 [Δ(rmlB-wbaP):(wzyC1-wzxC1)] (O7) (Figure 1C). The LPS profiles of all these O-serotype-converted mutant strains were examined by silver staining and confirmed by western blotting (Figure 2). Notably, the LPS profiles of S1031 and S1131 differed from their parent S. Typhimurium, but exhibited similar patterns to S. Enteritidis (Figure 2A) and S. Newport (Figure 2C), respectively. However, the LPS profile of S1124 matched neither that of S. Typhimurium nor that of S. Choleraesuis (Figure 2B), but western blotting showed that S1124 generated LPS reactive with anti-O7 factor serum, indicating that O-antigen polysaccharide of S. Choleraesuis was successfully produced and ligated to S. Typhimurium core moiety in S1124 (Figure 2B). These O-serotype conversion mutations were later introduced into a live attenuated S. Typhimurium vaccine strain, S738 (Δcrp Δcya) (O4), resulting in S1075 (Δabe:prt-tyvD1 Δcrp Δcya) (O9), S1157 [Δ(rmlB-wbaP):(wzyC1-wzxC1) Δcrp Δcya] (O7), and S1116 [Δ(wzxB1-wabN):(wzxC2-wbaZ) Δcrp Δcya] (O8), respectively, for further evaluation of immunogenicity and protective efficacy.
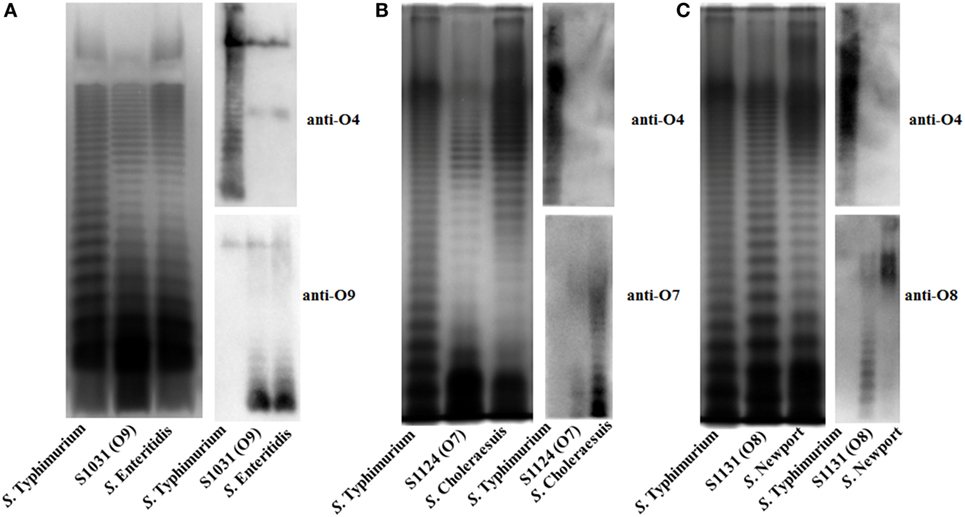
Figure 2. LPS profiles of O-serotype-converted mutants. (A) The LPS profiles of wild-type Salmonella Typhimurium, S. Enteritidis. and S1031 (O9) were compared. The O-serotype of S1031 was confirmed by incubation with Salmonella D1 serogroup-characterized O9 single-factor antisera, as indicated. (B) The LPS profiles of wild-type S. Typhimurium, S. Choleraesuis, and S1124 (O7) were compared. The O-serotype of S1124 was confirmed by incubation with Salmonella C1 serogroup-characterized O7 single-factor antisera, as indicated. (C) The LPS profiles of wild-type S. Typhimurium, S. Newport, and S1131 (O8) were compared. The O-serotype of S1131 was confirmed by incubation with Salmonella C2 serogroup-characterized O8 single-factor antisera, as indicated.
In vitro assays were done in triplicate. Phage P22 infections were performed to further examine the O-antigen structure of the mutants. The mutants were grown in LB broth and used as recipients for transduction assays. The number of transductions obtained from S1031 (O9) was similar to that from wild-type S100. However, we did not obtain any transductions from S1124 (O7) or S1131(O8) (Table 2).
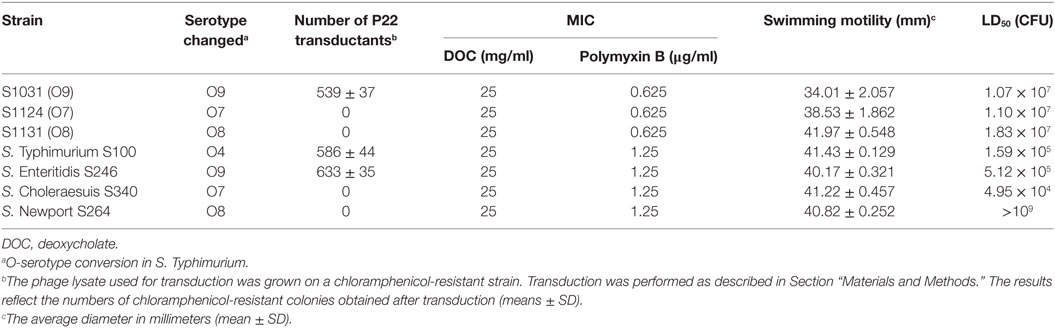
Table 2. Transduction efficiencies, minimum inhibitory concentration (MIC) of deoxycholate (DOC) and Polymyxin B, swimming motility, and virulence of wild-type Salmonella and its derivatives.
We next evaluated the impact of the O-antigen modifications on virulence and survival attributes. The mutants were evaluated for their sensitivity to the bile salt DOC and the cationic antimicrobial peptide polymyxin B. The DOC MICs did not differ among these strains, whereas the polymyxin B MICs for wild-type S100 were twofold higher than those for S1124 (O7), S1031 (O9), and S1131 (O8) (Table 2). We observed slightly slower growth rates for S1031 (O9), S1124 (O7), and S1131 (O8) compared to wild-type S100, but the differences were not significant (Figure S3 in Supplementary Material). All of the mutants retained wild-type or near wild-type motility (Table 2). To be effective, a live attenuated Salmonella vaccine needs interact with host epithelial cells. Thus, we examined the ability of our Δcya Δcrp derivatives to attach to and invade Hep-2 cells. No significant differences among strains were observed (Figure S4 in Supplementary Material).
Wild-type S. Typhimurium S100, S. Enteritidis S246, and S. Choleraesuis S340 displayed high virulence, with LD50 values of approximately 105 CFU (47), whereas the LD50 of wild-type S. Newport S264 was greater than 109 CFU, indicating a non-virulent phenotype of S264 in the murine model. The LD50 values of S1031 (O9), S1124 (O7), and S1131 (O8) were of a similar order of magnitude, approximately 107 (Table 2). The colonization of each Δcya Δcrp vaccine candidate in murine Peyer’s patches, spleens, and livers was determined on days 4 and 8 after oral inoculation. All of the candidates displayed good colonization in Peyer’s patches, livers, and spleens, and no significant differences were observed among these groups. No deaths occurred during this period (Figure S5 in Supplementary Material).
To assess the immunogenicity of these vaccine candidates, mice were inoculated orally with approximately 109 CFU of each strain on day 0 and boosted on day 14 with the same doses. Anti-S. Typhimurium, anti-S. Enteritidis, anti-S. Choleraesuis, and anti-S. Newport LPS serum antibodies were measured on day 28. The results are depicted in Figure 3. Mice vaccinated with S1075 (O9) mounted a significantly higher anti-S. Enteritidis LPS immune response than those vaccinated with S738 (O4). A similar result was observed in mice vaccinated with S1157 (O7) or S1116 (O8), which mounted significantly higher anti-S. Choleraesuis or anti-S. Newport LPS immune responses, respectively, than those vaccinated with S738 (O4). All vaccines induced a significantly higher IgG2a response than IgG1. The low level of IgG1/IgG2a ratio indicated that the cellular immunity was biased to Th1-type immune response (Figure S6 in Supplementary Material), consistent with our and other previous observations that Salmonella induced a predominant Th1-type response to either heterologous antigens or Salmonella own antigens (48, 49). Negative control groups (BSG) did not mount a detectable immune response. Apart from ELISA, we also performed western blotting to evaluate the sensitivity of polyclonal antibodies using pooled sera from vaccinated mice (Figure S7 in Supplementary Material). The sera from S738 (O4)- and S1075 (O9)-vaccinated mice were cross-reactive to LPS from S. Typhimurium and S. Enteritidis, while those from S1157 (O7)- and S1116 (O8)-vaccinated mice were specific to LPS from S. Choleraesuis and S. Newport, respectively. No positive bands were detected using pooled sera from the BSG control group.
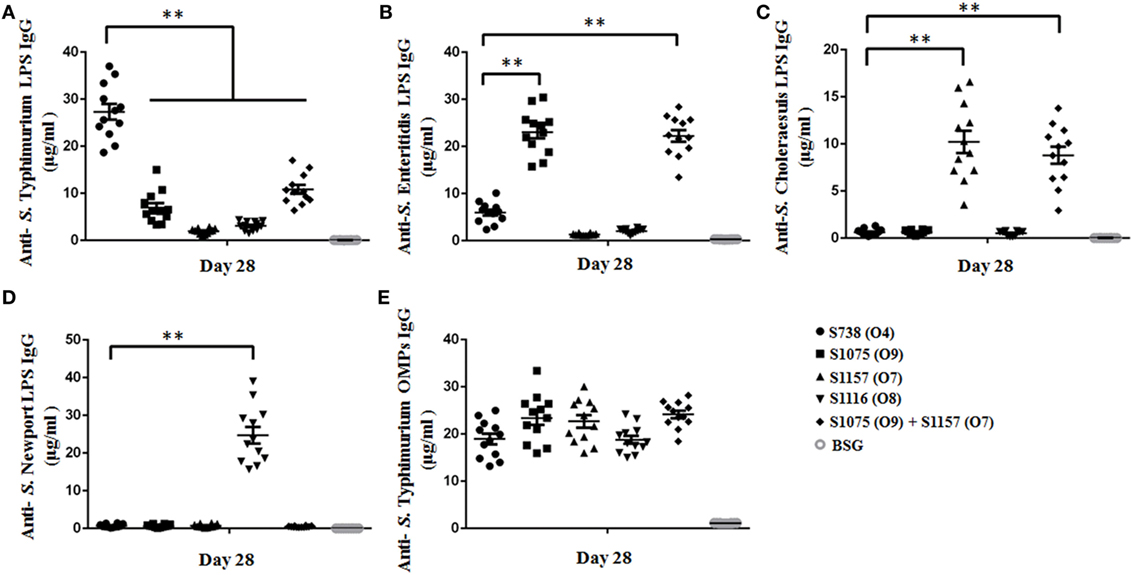
Figure 3. Sera IgG responses. Anti-Salmonella Typhimurium (A), anti-S. Enteritidis (B), anti-S. Choleraesuis (C), and anti-S. Newport (D) LPS IgG antibody concentrations in vaccinated mice sera were measured. (A) Anti-S. Typhimurium LPS IgG levels induced by the parent strain S738 (O4) were significantly higher than those induced by other vaccine candidates (**, P < 0.01). (B) Anti-S. Enteritidis LPS IgG levels induced by S1075 (O9) were significantly higher than those induced by the parent strain S738 (O4) (**, P < 0.01). (C) Anti-S. Choleraesuis LPS IgG levels induced by S1157 (O7) were significantly higher than those induced by the parent strain S738 (O4) (**, P < 0.01). (D) Anti-S. Newport LPS IgG levels induced by S1116 (O8) were significantly higher than those induced by the parent strain S738 (O4) (**, P < 0.01). (E) There were no significant differences in anti-S. Typhimurium OMP IgG levels among the different groups of vaccinated mice sera. Antibody concentrations were calculated using a standard curve. All concentrations of the measured samples were within the range of the standard curve. Error bars represent the SEs of the means.
C3 complement deposition is the key process for antibodies targeting surface antigens, leading to complement activation and subsequent serum bactericidal activity. Therefore, we determined the ability of serum antibodies from vaccinated mice to direct complement deposition on the surface of different wild-type Salmonella. Sera used in this assay were boosted pooled sera from mice vaccinated with S738 (O4), S1075 (O9), S1157 (O7), S1116 (O8), and equal-volume-mixed S1075 (O9) and S1157 (O7). The percentage of bacteria coated with C3 was determined by flow cytometry (Figure 4). Compared to negative controls, a high percentage of bacteria deposited with C3 complement on the surfaces of wild-type S. Typhimurium (Figure 4A), S. Enteritidis (Figures 4B,E), S. Choleraesuis (Figures 4C,F), and S. Newport (Figure 4D) were detected when incubated with mice sera induced by S738 (O4), S1075 (O9), S1157 (O7), and S1116 (O8), respectively. These results indicated that antibodies in mice sera induced by live vaccine candidates were able to trigger the classical pathway of complement activation. Furthermore, an in vitro assay was performed to analyze the differential uptake of S. Typhimurium, S. Enteritidis, S. Choleraesuis, and S. Newport by RAW264.7 macrophages. The aim of this assay was to evaluate the role of vaccine-induced antibody opsonization in the early stages of opsonophagocytosis. Inoculation with sera primed with a specific O-serotype-converted vaccine resulted in significantly increased uptake of the same O-serotype wild-type Salmonella by macrophages (Figure 5). The uptake of S. Enteritidis inoculated with sera from mice primed with S1075 (O9) was significantly higher than naive sera or other non-specific sera. Similar results were observed with S. Choleraesuis and S. Newport when opsonized with sera from mice primed with S1157 (O7) and S1116 (O8), respectively. In particular, the uptake of both S. Enteritidis and S. Choleraesuis by macrophages was significantly increased when opsonized with sera from mice co-vaccinated by S1075 (O9) and S1157 (O7).
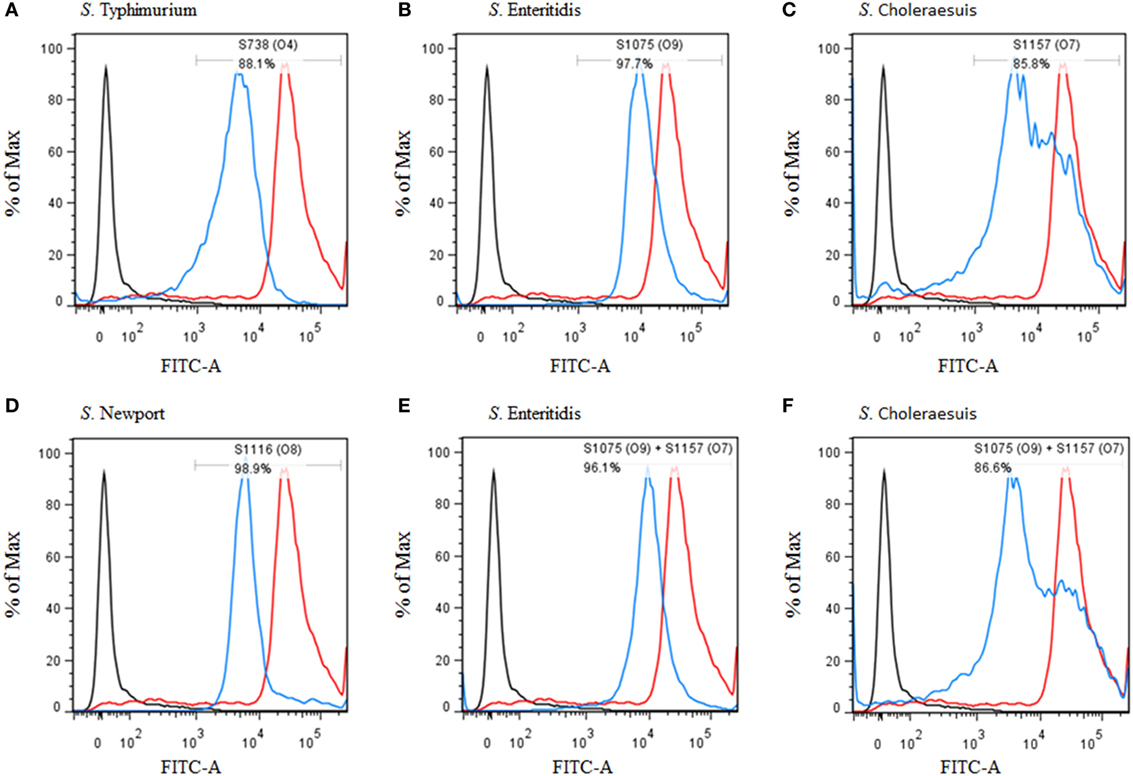
Figure 4. Flow cytometry histograms of C3 complement deposition. The percentage of FITC-positive bacteria was determined by flow cytometry. (A) Wild-type S. Typhimurium was incubated with sera from mice vaccinated with S738 (O4). Approximately 88.1% of bacteria were positive. (B) Wild-type S. Enteritidis was incubated with sera from mice vaccinated with S1075 (O9). Approximately 97.7% of bacteria were positive. (C) Wild-type S. Choleraesuis was incubated with sera from mice vaccinated with S1157 (O7). Approximately 85.8% of bacteria were positive. (D) Wild-type S. Newport was incubated with sera from mice vaccinated with S1116 (O8). Approximately 98.9% of bacteria were positive. (E) Wild-type S. Enteritidis was incubated with sera from mice co-vaccinated with S1075 (O9) and S1157 (O7). Approximately 96.1% of bacteria were positive. (F) Wild-type S. Choleraesuis was incubated with sera from mice co-vaccinated with S1075(O9) and S1157 (O7). Approximately 86.6% of bacteria were positive. The negative control (dark line) was wild-type S. Typhimurium incubated with sera from non-vaccinated mice, and the positive control (red line) was wild-type S. Typhimurium incubated with O4 single-factor rabbit antisera.
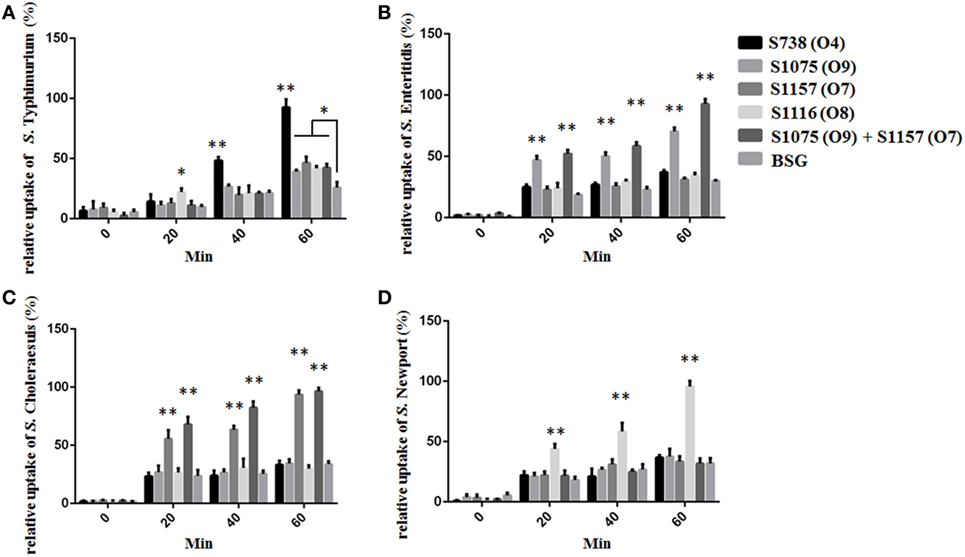
Figure 5. The differential uptake of Salmonella by RAW264.7 cells after serum opsonization. Compared to cells from control groups (Salmonella opsonized with naive serum), the uptake of S. Typhimurium (A), S. Enteritidis (B), S. Choleraesuis (C), and S. Newport (D) opsonized with sera from mice primed with S738 (O4), S1075 (O9), S1157 (O7), and S1116 (O8), respectively, was significantly higher (**, P < 0.01). The number of enumerated cells was normalized to 100% for the maximal value. Error bars represent SEs of the means.
Vaccinated mice were challenged orally on day 56 with a dose 100 times the LD50 of S. Typhimurium, S. Choleraesuis, and S. Enteritidis to evaluate protective efficacy. When challenged with S. Enteritidis and S. Choleraesuis, 100% protection was observed in mice vaccinated with S1075 (O9) (Figure 6B) and S1157 (O7) (Figure 6C), respectively. Complete protection was also observed in all vaccinated mice when challenged with S. Typhimurium (Figure 6A). As wild-type S. Newport was non-lethal in mice at an oral challenge dose of 109 CFU, the protective efficacy of vaccine S1116 (O8) or its combination with other vaccine candidates was therefore not evaluated. Most interestingly, mice vaccinated with mixed equal volumes of S1075 (O9) and S1157 (O7) were able to withstand challenges of S. Typhimurium, S. Enteritidis, and S. Choleraesuis, indicating that a S1075 (O9) and S1157 (O7) co-vaccination strategy may effectively prevent Salmonella serotype O4, O9, and O7 infections.
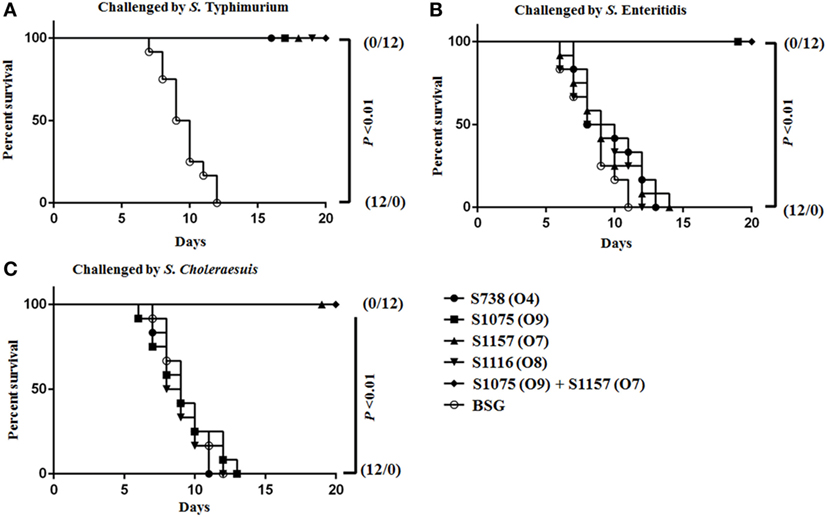
Figure 6. Survival curves after orally challenged by wild-type virulent Salmonella. Eight weeks after vaccination, BALB/c mice were challenged by about 100 times the LD50 of wild-type virulent S. Typhimurium (A), S. Choleraesuis (B), and S. Enteritidis (C). P < 0.01, for all marked group versus BSG group.
Glycoconjugate vaccines are effective against Salmonella spp. infections (10). For instance, Vi-conjugated vaccines can successfully prevent S. Typhi infections (50, 51). O-antigen polysaccharides are usually conjugated to suitable carrier proteins, such as TT, DT, the non-toxic recombinant form of DT (CRM197) (10, 11), and Salmonella proteins, to generate glycoconjugate vaccines eliciting T-cell-dependent immune responses with limited memory immunity (26, 27). Usually, this conjugated process is mediated by chemical conjugation, which is an expensive, multiple step process and influenced by various factors such as polysaccharide length and structure. Recently, the discovery of the Campylobacter jejuni N-linked and Neisseria species O-linked glycosylation pathways paved the way for in vivo glycoengineering, the expression of glycosylation machineries in combination with glycan acceptor proteins in heterologous hosts like E. coli (52–54). In these two systems, two glycosyltransferases, PglB from C. jejuni and PglL from Neisseria, are critical for functionally transferring polysaccharides to asparagine residues within the glycosylation consensus sequon D/E-X1-N-X2-S/T (X1, X2 ≠ Pro) of acceptor proteins for N-linked and to serine residues within the sequon W-P-Xn-S-Xm-P (Xn is optimal as AAA) for O-linked glycosylation (55–58). The resulting conjugate elicited a robust humoral immunogenic response in animals. Although this technology is used in industrial applications for the generation of glycoconjugated vaccine candidates, there are still some challenges to be overcome, such as limited number of glycosylation consensus sequences on the carrier protein, and poor cytotoxic T lymphocytes development as the glycoconjugates are generally inefficient at entering the class I MHC pathway of antigen presentation (59). Compared to glycoconjugate subunit vaccines, attenuated Salmonella possess many advantages in delivering O-antigen polysaccharides including the strong adjuvant effects of Salmonella LPS and porins, and the induction of mucosal, humoral, and cellular immune responses with induction of long-term protective immunity (60).
Considering that the invasive NTS are restricted to a limited number of O-antigen serogroups, including B1, D1, C1, and C2 (8, 11), we devised a novel strategy to prevent invasive NTS infections by replacing the original B1 group O-antigen polysaccharide in attenuated S. Typhimurium with heterologous O-antigen polysaccharide from Salmonella D1, C1, and C2, i.e., immunodominant O-serotype conversion. Our results demonstrated that heterologous O-antigen-specific immune responses could be effectively induced by attenuated S. Typhimurium expressing heterologous O-antigen and provide protection against heterologous wild-type virulent Salmonella challenge while maintaining homologous protection (Figures 3 and 6).
Our research was initiated by comparing the nucleotide sequences and chemical structures of the O-antigen from serovars of Groups B1, D1, C1, and C2 (Figures S1 and S2 in Supplementary Material). There was high homology in the nucleotide sequences and minor differences in chemical structures between Groups B1 and D1 (61), with an immunodominant abequose (B1) versus tyvelose (D1). Therefore, we converted O4 [group B1, α-Abe(1→3)Man] into O9 [group D1, α-Tyv(1→3)Man] by replacing the allelic abe gene with prt-tyvD1. The LPS profiles of O9 serotype-converted mutants were similar to wild-type S. Enteritidis (Figure 2A), indicating that wzxB1-flippase in S. Typhimurium was tolerant to tyvelose side-branch O-units, consistent with a previous report (62). However, the case was more complicated for group C1, as the O-antigen in group C1 serovar starts with GlcNAc initiated by WecA (63, 64). Considering that the nucleotide sequences and chemical structures of group B1 and C1 were completely different, we replaced the entire O-antigen gene cluster of Group B1 with C1 to convert O4 into the O7 serotype. For group C2 serovars, the most important difference lies in additional mannose in the main chain and the α-Abe(1→3)Rha linkages, which consequently contributed to the dominant O8 serotype. Therefore, the genes wzxB1-wbaN were replaced with wzxC2-wbaZ to convert the O4 serotype to O8. LPS silver staining and western blotting confirmed all of these O-serotype conversions (Figure 2). The LPS profile of S1124 (O7) differed from both S. Typhimurium and S. Choleraesuis, but western blotting using specific anti-O7 serum confirmed that S1124 was able to generate detectable O7 O-antigen polysaccharide, which might indicate subtle decorations on the O-polysaccharide main chain occurred in S. Typhimurium (Figure 2B).
In vitro analyses indicated that our strategy had resulted in biologically significant changes to the O-antigen. The attachment of bacteriophage P22 to Salmonella is mediated by the binding of its tailspike protein to the O-antigenic repeating units of groups B1 and D1 (65). Thus, we obtained transductants using the strain with an engineered group D1 O-antigen, S1031 (O9), while the two strains engineered to produce group C O-antigens were not infected (Table 2). O-antigen is an important factor for swimming or swarming motility on agar surfaces, by improving surface “wettability” (66). Thus, changes in O-antigen composition may have a negative impact on motility. However, our results indicate that the changes we made to O-antigen had no significant impact on motility (Table 2). There were no major changes to the cellular membrane structure, as there was no significant increase in susceptibility to the membrane-damaging agents DOC and polymyxin B (Table 2). There was a slight decrease in the growth rate of strains S1031 (O9), S1124 (O7), and S1131 (O8) compared to their parent strain (Figure S3 in Supplementary Material), and the LD50s of S1031 (O9), S1124 (O7), and S1131 (O8) were two orders of magnitude larger than S100 (O4), showing approximately 100-fold attenuation (Table 2). These negative impacts on growth and virulence are likely due to the stress imposed by synthesizing a heterologous O-antigen (67).
Enzyme-linked immunosorbent assay data showed that all the modified vaccines induced a strong heterologous O-antigen-specific serum IgG responses (Figure 3), with IgG2a dominating the anti-LPS response (Figure S6 in Supplementary Material), indicating a Th1-type immune response, consistent with our previous observations (43, 44, 49). Serum antibodies from mice immunized with S738 (O4) and S1075 (O9) were cross-reactive to LPS from S. Typhimurium and S. Enteritidis, which we ascribed to the shared glycan epitopes O1 or O12. Those from S1157 (O7)- and S1116 (O8)-vaccinated mice were specific to LPS from S. Choleraesuis and S. Newport, respectively (Figure S7 in Supplementary Material). Moreover, in vitro C3 complement deposition and opsonophagocytic assays demonstrated that the vaccine-induced antibodies were able to trigger the classical pathway of complement activation and promote the uptake of wild-type Salmonella by macrophages after serum antibody opsonization (Figures 4 and 5). These results underscored that live attenuated S. Typhimurium vaccines were able to synthesize heterologous O-antigens on the surface and elicit functional antibody responses in mice targeting the surface polysaccharide antigens of the related wild-type Salmonella serovars.
Our goal was to design and construct a S. Typhimurium vaccine to provide protection against multiple serovars of NTS infections in addition to S. Typhimurium. We observed that all mice survived challenge with 100 times dose of the LD50 of S. Typhimurium, indicating that the protective efficacy against the homologous parent strain was not compromised, consistent with a study conducted by Hormaeche et al. (33). This may be due to the robust immune responses elicited by all strains against S. Typhimurium outer membrane proteins (Figure 3E). This seems likely, as antibody responses to outer membrane proteins have been shown to play an important role in protective immunity against S. Typhimurium. For example, immunization of mice with rough S. Typhimurium mutants or mutants engineered to shut off O-antigen synthesis in vivo elicits protective immunity against lethal challenge with S. Typhimurium (22, 23, 43, 44, 49). Immunization with purified outer membrane proteins from rough S. Typhimurium elicits long-lasting protective immunity against S. Typhimurium challenge (47, 68–70). Some of these outer membrane proteins induce T-cell-mediated immune responses essential for clearance of the bacterial infection (68, 69). In our study, only mice vaccinated with S1075 (O9) or S1157 (O7) survived a challenge with 100 times the LD50 of S. Enteritidis or S. Choleraesuis, respectively, indicating that protective efficacy against heterologous Salmonella challenge was highly O-serotype related in this case.
Competition between multiple Salmonella serovars in the same host eventually results in the serovar with highest transmission success excluding the other serovars expressing the same O-serotype from the host population (71, 72). This mechanism should not impact our ability to vaccinate a single host with a mixture of our Salmonella vaccines, as strains S1075 (O9) and S1157 (O7) each expressed distinct O-antigen polysaccharides. When co-administered, they induced a high level of anti-IgG antibodies against their respective O-antigens (Figure 3). Broad protective coverage of serotypes O4, O9, and O7 was elicited by co-vaccination with S1075 (O9) and S1157 (O7) (Figure 6).
Our wild-type S. Newport strain was not virulent in mice (Table 2), so we were unable to evaluate the protective efficacy of S1116 (O8) against a S. Newport challenge. We note that the lack of virulence in mice is likely to be due to the absence of a virulence plasmid in S. Newport strains (73).
Although we only tested the protective efficacies of one serovars in each serogroup B1, D1, and C1, i.e., S. Typhimurium S100, S. Enteritidis S246, and S. Choleraesuis S340, it is reasonable to predict that vaccination with a serovar producing an immunodominant O-serotype could elicit cross-immunity against members of the same serogroup (74). In summary, we demonstrated that live attenuated S. Typhimurium vaccines based on O-serotype conversion were immunogenic and suitable for a co-vaccination strategy to provide protection against serovars S. Typhimurium (O4), S. Enteritidis (O9), S. Choleraesuis (O7), and S. Newport (O8), which account for the majority of NTS infections.
All animal studies were conducted in compliance with the Animal Welfare Act and regulations stated in the Guide for the Care and Use of Laboratory Animals, which was approved by Sichuan Agricultural University Institutional Animal Care and Use Committee (Ya’an, China; Approval No. 2011028).
PL and QK conceived and designed the experiments. PL, HL, KL, JY, YL, and YH performed the experiments. PL, QL, and YH analyzed the data. PL, QL, and QK wrote the article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Kenneth L. Roland and Roy Curtiss III for critically reading the manuscript.
This study was supported by grants 31570928 and 31472179 from the National Natural Science Foundation of China and R01AI112680 from the National Institutes of Health.
The Supplementary Material for this article can be found online at http://www.frontiersin.org/article/10.3389/fimmu.2017.01647/full#supplementary-material.
Figure S1. The O-antigen gene clusters of B1, D1, C1, and C2 serogroups. The O-antigen gene clusters of Salmonella Typhimurium (B1, O4), S. Enteritidis (D1, O9), S. Choleraesuis (C1, O7), and S. Newport (C2, O8) are shown and the immunodominant O-serotype factor was labeled in front of the gene cluster. Genes were color coded by biosynthesis pathways. Arrows represented the location and orientation of the genes. Diagrams are drawn to scale.
Figure S2. The chemical structures of B1, D1, C1, and C2 serogroup O-units. The chemical structures illustrating the sugar components and glycosidic linkages within the O-units of Salmonella Typhimurium (A), S. Choleraesuis (B), S. Enteritidis (C), and S. Newport (D). The immunodominant O-serotype factor was labeled under the structures. Sugar abbreviations: Abe, abequose; Tyv, tyvelose; l-Rha, l-rhamnose; d-Man, d-mannose; d-Gal, d-galactose; d-GlcNAc, d-acetylgalactosamine.
Figure S3. Growth curves of O-serotype converted mutants. In vitro growth rates of Salmonella Typhimurium wild-type S100, and its derivatives were determined by measurement of the optical density at OD600 at multiple time points.
Figure S4. Attachment and invasion assays in Hep-2 cells. The percentage of attachment and invasion rate of O-serotype converted mutants was evaluated as described in materials and methods. All mutants were derived from the S738 (O4) parental strain, as indicated. Error bars represented SEs of the means.
Figure S5. Colonization of murine Peyer’s patches, liver, and spleen by live attenuated Salmonella Typhimurium vaccines. All mutants were derived from the S738 (O4) parental strain, as indicated. Colonization of Peyer’s patches (A), liver (B), and spleen (C) after 4 and 8 days post-inoculation were shown. The horizontal lines represent the means, and the error bars represent SEs of the means.
Figure S6. Sera IgG1 and IgG2a responses. The responses of IgG isotype subclasses IgG2a and IgG1 were measured in vaccinated mice sera. The LPS of Salmonella Typhimurium (A), S. Enteritidis (B), S. Choleraesuis (C), and S. Newport (D) were coated in ELISA plates. The data showed the concentrations of IgG1 and IgG2a subclass in mice sera vaccinated by S738 (O4), S1075 (O9), S1157 (O7), and S1116 (O8). All vaccine candidates could induce a significantly higher amount of heterologous LPS-specific IgG2a levels than IgG1 (**, P < 0.01). The antibody concentrations were calculated through the standard curve. All concentrations of the measured samples were within the range of the standard curve. Error bars represented the SEs of the means.
Figure S7. Western-blot of Salmonella LPS treated by vaccinated mice sera. The primary antibodies used in western-blot were polyclonal antibodies from mice sera vaccinated by S738 (O4) (A), S1075 (O9) (B), S1157 (O7) (C), S1116 (O8) (D), S1075 (O9) plus S1157 (O7) (E) and BSG control (F).
1. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’brien SJ, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis (2010) 50:882–9. doi:10.1086/650733
2. Buckle GC, Walker CL, Black RE. Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010. J Glob Health (2012) 2:010401. doi:10.7189/jogh.02.010401
3. Sanderson KE, Nair S. Taxonomy and species concepts in the genus Salmonella. In: Barrow P, Methner U, editors. Salmonella in Domestic Animals. 2 ed. CAB International (2013). p. 1–19. doi:10.1079/9781845939021.0001
4. Gal-Mor O, Boyle EC, Grassl GA. Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol (2014) 5:391. doi:10.3389/fmicb.2014.00391
5. Andrews JR, Ryan ET. Diagnostics for invasive Salmonella infections: current challenges and future directions. Vaccine (2015) 33(Suppl 3):C8–15. doi:10.1016/j.vaccine.2015.02.030
6. Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet (2012) 379:2489–99. doi:10.1016/S0140-6736(11)61752-2
7. Takem EN, Roca A, Cunnington A. The association between malaria and non-typhoid Salmonella bacteraemia in children in sub-Saharan Africa: a literature review. Malar J (2014) 13:400. doi:10.1186/1475-2875-13-400
8. Fuche FJ, Sow O, Simon R, Tennant SM. Salmonella serogroup C: current status of vaccines and why they are needed. Clin Vaccine Immunol (2016) 23:737–45. doi:10.1128/CVI.00243-16
9. Kariuki S, Gordon MA, Feasey N, Parry CM. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine (2015) 33(Suppl 3):C21–9. doi:10.1016/j.vaccine.2015.03.102
10. Simon R, Levine MM. Glycoconjugate vaccine strategies for protection against invasive Salmonella infections. Hum Vaccin Immunother (2012) 8:494–8. doi:10.4161/hv.19158
11. MacLennan CA, Martin LB, Micoli F. Vaccines against invasive Salmonella disease: current status and future directions. Hum Vaccin Immunother (2014) 10:1478–93. doi:10.4161/hv.29054
12. Grabenstein JD, Pittman PR, Greenwood JT, Engler RJ. Immunization to protect the US Armed Forces: heritage, current practice, and prospects. Epidemiol Rev (2006) 28:3–26. doi:10.1093/epirev/mxj003
13. Ivanoff B, Levine MM, Lambert P. Vaccination against typhoid fever: present status. Bull World Health Organ (1994) 72:957.
14. Tennant SM, Levine MM. Live attenuated vaccines for invasive Salmonella infections. Vaccine (2015) 33(Suppl 3):C36–41. doi:10.1016/j.vaccine.2015.04.029
15. Pasetti MF, Simon JK, Sztein MB, Levine MM. Immunology of gut mucosal vaccines. Immunol Rev (2011) 239:125–48. doi:10.1111/j.1600-065X.2010.00970.x
16. Mestecky J, Nguyen H, Czerkinsky C, Kiyono H. Oral immunization: an update. Curr Opin Gastroenterol (2008) 24:713–9. doi:10.1097/MOG.0b013e32830d58be
17. Galen JE, Curtiss R. The delicate balance in genetically engineering live vaccines. Vaccine (2014) 32:4376–85. doi:10.1016/j.vaccine.2013.12.026
18. Hindle Z, Chatfield SN, Phillimore J, Bentley M, Johnson J, Cosgrove CA, et al. Characterization of Salmonella enterica derivatives harboring defined aroC and Salmonella pathogenicity island 2 type III secretion system (ssaV) mutations by immunization of healthy volunteers. Infect Immun (2002) 70:3457–67. doi:10.1128/IAI.70.7.3457-3467.2002
19. Tennant SM, Wang J-Y, Galen JE, Simon R, Pasetti MF, Gat O, et al. Engineering and preclinical evaluation of attenuated nontyphoidal Salmonella strains serving as live oral vaccines and as reagent strains. Infect Immun (2011) 79:4175–85. doi:10.1128/IAI.05278-11
20. Curtiss R, Kelly SM. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect Immun (1987) 55:3035–43.
21. Liu X, Liu Q, Xiao K, Li P, Liu Q, Zhao X, et al. Attenuated Salmonella typhimurium delivery of a novel DNA vaccine induces immune responses and provides protection against duck enteritis virus. Vet Microbiol (2016) 186:189–98. doi:10.1016/j.vetmic.2016.03.001
22. Li P, Liu Q, Huang C, Zhao X, Roland KL, Kong Q. Reversible synthesis of colanic acid and O-antigen polysaccharides in Salmonella typhimurium enhances induction of cross-immune responses and provides protection against heterologous Salmonella challenge. Vaccine (2017) 35:2862–9. doi:10.1016/j.vaccine.2017.04.002
23. Huang C, Liu Q, Luo Y, Li P, Liu Q, Kong Q. Regulated delayed synthesis of lipopolysaccharide and enterobacterial common antigen of Salmonella typhimurium enhances immunogenicity and cross-protective efficacy against heterologous Salmonella challenge. Vaccine (2016) 34:4285–92. doi:10.1016/j.vaccine.2016.07.010
24. Avci FY, Kasper DL. How bacterial carbohydrates influence the adaptive immune system. Annu Rev Immunol (2010) 28:107–30. doi:10.1146/annurev-immunol-030409-101159
25. Park BS, Lee JO. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med (2013) 45:e66. doi:10.1038/emm.2013.97
26. Svenson S, Nurminen M, Lindberg A. Artificial Salmonella vaccines: O-antigenic oligosaccharide-protein conjugates induce protection against infection with Salmonella typhimurium. Infect Immun (1979) 25:863–72.
27. Simon R, Tennant SM, Wang JY, Schmidlein PJ, Lees A, Ernst RK, et al. Salmonella enterica serovar enteritidis core O polysaccharide conjugated to H:g,m flagellin as a candidate vaccine for protection against invasive infection with S. enteritidis. Infect Immun (2011) 79:4240–9. doi:10.1128/IAI.05484-11
28. Mukkur TK, Stocker BA, Walker KH. Genetic manipulation of Salmonella serotype bovismorbificans to aromatic-dependence and evaluation of its vaccine potential in mice. J Med Microbiol (1991) 34:57–62. doi:10.1099/00222615-34-1-57
29. Nnalue NA, Stocker B. Test of the virulence and live-vaccine efficacy of auxotrophic and galE derivatives of Salmonella choleraesuis. Infect Immun (1987) 55:955–62.
30. Jörbeck HJ, Svenson SB, Lindberg AA. Immunochemistry of Salmonella O-antigens: specificity of rabbit antibodies against the O-antigen 4 determinant elicited by whole bacteria and O-antigen 4 specific saccharide-protein conjugates. J Immunol (1979) 123:1376–81.
31. Luk J, Lindberg A. Anti-Salmonella lipopolysaccharide monoclonal antibodies: characterization of Salmonella BO-, CO-, DO-, and EO-specific clones and their diagnostic usefulness. J Clin Microbiol (1991) 29:2424–33.
32. Carlin NI, Svenson SB, Lindberg AA. Role of monoclonal O-antigen antibody epitope specificity and isotype in protection against experimental mouse typhoid. Microb Pathog (1987) 2:171–83. doi:10.1016/0882-4010(87)90019-2
33. Hormaeche CE, Mastroeni P, Harrison JA, de Hormaeche RD, Svenson S, Stocker BA. Protection against oral challenge three months after iv immunization of BALBc mice with live Aro Salmonella typhimurium and Salmonella enteritidis vaccines is serotype (species)-dependent and only partially determined by the main LPS O antigen. Vaccine (1996) 14:251–9. doi:10.1016/0264-410X(95)00249-Z
34. Blomfield I, Vaughn V, Rest R, Eisenstein B. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol Microbiol (1991) 5:1447–57. doi:10.1111/j.1365-2958.1991.tb00791.x
35. Nakayama K, Kelly SM, Curtiss R. Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat Biotechnol (1988) 6:693–7. doi:10.1038/nbt0688-693
36. Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res (1988) 16:6127–45. doi:10.1093/nar/16.13.6127
37. Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. United Kingdom: Coldspring-Harbour Laboratory Press (2001).
38. Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene (1998) 207:149–57. doi:10.1016/S0378-1119(97)00619-7
39. Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods (2009) 6:343–5. doi:10.1038/nmeth.1318
40. Roland K, Curtiss R III, Sizemore D. Construction and evaluation of a Δcya Δcrp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis (1999) 43:429–41. doi:10.2307/1592640
41. Kong Q, Six DA, Roland KL, Liu Q, Gu L, Reynolds CM, et al. Salmonella synthesizing 1-dephosphorylated lipopolysaccharide exhibits low endotoxic activity while retaining its immunogenicity. J Immunol (2011) 187:412–23. doi:10.4049/jimmunol.1100339
42. Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol (1983) 154:269–77.
43. Kong Q, Liu Q, Roland KL, Curtiss R III. Regulated delayed expression of rfaH in an attenuated Salmonella enterica serovar typhimurium vaccine enhances immunogenicity of outer membrane proteins and a heterologous antigen. Infect Immun (2009) 77:5572–82. doi:10.1128/IAI.00831-09
44. Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R III. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect Immun (2011) 79:4227–39. doi:10.1128/IAI.05398-11
45. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol (1938) 27:493–7. doi:10.1093/oxfordjournals.aje.a118408
46. McKay GA, Woods DE, MacDonald KL, Poole K. Role of phosphoglucomutase of stenotrophomonasmaltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infect Immun (2003) 71:3068–75. doi:10.1128/IAI.71.6.3068-3075.2003
47. Liu Q, Liu Q, Zhao X, Liu T, Yi J, Liang K, et al. Immunogenicity and cross-protective efficacy induced by outer membrane proteins from Salmonella typhimurium mutants with truncated LPS in mice. Int J Mol Sci (2016) 17:416. doi:10.3390/ijms17030416
48. Wang S, Shi H, Li Y, Shi Z, Zhang X, Baek CH, et al. A colanic acid operon deletion mutation enhances induction of early antibody responses by live attenuated Salmonella vaccine strains. Infect Immun (2013) 81:3148–62. doi:10.1128/IAI.00097-13
49. Kong Q, Liu Q, Jansen AM, Curtiss R III. Regulated delayed expression of rfc enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Vaccine (2010) 28:6094–103. doi:10.1016/j.vaccine.2010.06.074
50. Micoli F, Rondini S, Pisoni I, Proietti D, Berti F, Costantino P, et al. Vi-CRM 197 as a new conjugate vaccine against Salmonella typhi. Vaccine (2011) 29:712–20. doi:10.1016/j.vaccine.2010.11.022
51. Kossaczka Z, Lin F-YC, Ho VA, Thuy NTT, Van Bay P, Thanh TC, et al. Safety and immunogenicity of Vi conjugate vaccines for typhoid fever in adults, teenagers, and 2-to 4-year-old children in Vietnam. Infect Immun (1999) 67:5806–10.
52. Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science (2002) 298:1790–3. doi:10.1126/science.298.5599.1790
53. Stimson E, Virji M, Makepeace K, Dell A, Morris HR, Payne G, et al. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol Microbiol (1995) 17:1201–14. doi:10.1111/j.1365-2958.1995.mmi_17061201.x
54. Faridmoayer A, Fentabil MA, Mills DC, Klassen JS, Feldman MF. Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J Bacteriol (2007) 189:8088–98. doi:10.1128/JB.01318-07
55. Kowarik M, Young NM, Numao S, Schulz BL, Hug I, Callewaert N, et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J (2006) 25:1957–66. doi:10.1038/sj.emboj.7601087
56. Wacker M, Feldman MF, Callewaert N, Kowarik M, Clarke BR, Pohl NL, et al. Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc Natl Acad Sci U S A (2006) 103:7088–93. doi:10.1073/pnas.0509207103
57. Pan C, Sun P, Liu B, Liang H, Peng Z, Dong Y, et al. Biosynthesis of conjugate vaccines using an O-linked glycosylation system. MBio (2016) 7:e443–416. doi:10.1128/mBio.00443-16
58. Ihssen J, Kowarik M, Dilettoso S, Tanner C, Wacker M, Thony-Meyer L. Production of glycoprotein vaccines in Escherichia coli. Microb Cell Fact (2010) 9:61. doi:10.1186/1475-2859-9-61
59. Nothaft H, Szymanski CM. Bacterial protein N-glycosylation: new perspectives and applications. J Biol Chem (2013) 288:6912–20. doi:10.1074/jbc.R112.417857
60. Kong W, Clark-Curtiss J, Curtiss R III. Utilizing Salmonella for antigen delivery: the aims and benefits of bacterial delivered vaccination. Expert Rev Vaccines (2013) 12:345–7. doi:10.1586/erv.13.7
61. Reeves PR, Cunneen MM, Liu B, Wang L. Genetics and evolution of the Salmonella galactose-initiated set of o antigens. PLoS One (2013) 8:e69306. doi:10.1371/journal.pone.0069306
62. Hong Y, Cunneen MM, Reeves PR. The Wzx translocases for Salmonella enterica O-antigen processing have unexpected serotype. Mol Microbiol (2012) 84:620–30. doi:10.1111/j.1365-2958.2012.08048.x
63. Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Reeves PR, et al. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol Rev (2014) 38:56–89. doi:10.1111/1574-6976.12034
64. Samuel G, Reeves P. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr Res (2003) 338:2503–19. doi:10.1016/j.carres.2003.07.009
65. Wollin R, Eriksson U, Lindberg AA. Salmonella bacteriophage glycanases: endorhamnosidase activity of bacteriophages P27, 9NA, and KB1. J Virol (1981) 38:1025–33.
66. Toguchi A, Siano M, Burkart M, Harshey RM. Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. J Bacteriol (2000) 182:6308–21. doi:10.1128/JB.182.22.6308-6321.2000
67. Liu MA, Stent TL, Hong Y, Reeves PR. Inefficient translocation of a truncated O unit by a Salmonella Wzx affects both O-antigen production and cell growth. FEMS Microbiol Lett (2015) 362. doi:10.1093/femsle/fnv053
68. Udhayakumar V, Muthukkaruppan V. Protective immunity induced by outer membrane proteins of Salmonella typhimurium in mice. Infect Immun (1987) 55:816–21.
69. Udhayakumar V, Muthukkaruppan V. An outer membrane protein (porin) as an eliciting antigen for delayed-type hypersensitivity in murine salmonellosis. Infect Immun (1987) 55:822–4.
70. Liu Q, Liu Q, Yi J, Liang K, Liu T, Roland KL, et al. Outer membrane vesicles derived from Salmonella typhimurium mutants with truncated LPS induce cross-protective immune responses against infection of Salmonella enterica serovars in the mouse model. Int J Med Microbiol (2016) 306:697–706. doi:10.1016/j.ijmm.2016.08.004
71. Santos RL, Baumler AJ. Cell tropism of Salmonella enterica. Int J Med Microbiol (2004) 294:225–33. doi:10.1016/j.ijmm.2004.06.029
72. Dorsey CW, Raffatellu M, Kingsley RA, Bäumler AJ. Mechanisms of Salmonella enterica serotype typhimurium intestinal colonization. In: Nataro JP, Cohen PS, Mobley HLT, Weiser JN, editors. Colonization of Mucosal Surfaces. Washington, DC: ASM Press (2005). p. 301–12. doi:10.1128/9781555817619.ch21
73. Roudier C, Krause M, Fierer J, Guiney D. Correlation between the presence of sequences homologous to the vir region of Salmonella dublin plasmid pSDL2 and the virulence of twenty-two Salmonella serotypes in mice. Infect Immun (1990) 58:1180–5.
Keywords: S. Typhimurium, S. Enteritidis, S. Choleraesuis, S. Newport, O-antigen, live attenuated Salmonella vaccine, cross-protection
Citation: Li P, Liu Q, Luo H, Liang K, Yi J, Luo Y, Hu Y, Han Y and Kong Q (2017) O-Serotype Conversion in Salmonella Typhimurium Induces Protective Immune Responses against Invasive Non-Typhoidal Salmonella Infections. Front. Immunol. 8:1647. doi: 10.3389/fimmu.2017.01647
Received: 15 June 2017; Accepted: 10 November 2017;
Published: 04 December 2017
Edited by:
Fabio Bagnoli, GlaxoSmithKline, ItalyReviewed by:
Axel T. Lehrer, University of Hawaii at Manoa, United StatesCopyright: © 2017 Li, Liu, Luo, Liang, Yi, Luo, Hu, Han and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingke Kong, kongqiki@163.com
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.