- 1Department of Molecular Pathology, Institute of Pathology, Comprehensive Cancer Center (CCC) Mainfranken, University of Würzburg, Würzburg, Germany
- 2Department of Pathology, Graduate School of Medicine, Dentistry and Pharmaceutical Science, Okayama University, Okayama, Japan
- 3Laboratory of Molecular Biochemistry, School of Life Science, Tokyo University of Pharmacy and Life Science, Hachioji, Tokyo, Japan
- 4Department of Gastroenterology and Gastrointestinal Oncology, University Medical Center Göttingen, Göttingen, Germany
- 5Division of Oncological Pathology, Aichi Cancer Center Research Institute, Nagoya, Japan
In lymphocytes, immune receptor signals induce the rapid nuclear translocation of preformed cytosolic NFAT proteins. Along with co-stimulatory signals, persistent immune receptor signals lead to high levels of NFATc1/αA, a short NFATc1 isoform, in effector lymphocytes. Whereas NFATc1 is not expressed in plasma cells, in germinal centers numerous centrocytic B cells express nuclear NFATc1/αA. When overexpressed in chicken DT40 B cells or murine WEHI 231 B cells, NFATc1/αA suppressed their cell death induced by B cell receptor signals and affected the expression of genes controlling the germinal center reaction and plasma cell formation. Among those is the Prdm1 gene encoding Blimp-1, a key factor of plasma cell formation. By binding to a regulatory DNA element within exon 1 of the Prdm1 gene, NFATc1/αA suppresses Blimp-1 expression. Since expression of a constitutive active version of NFATc1/αA interfered with Prdm1 RNA expression, LPS-mediated differentiation of splenic B cells to plasmablasts in vitro and reduced immunoglobulin production in vivo, one may conclude that NFATc1/αA plays an important role in controlling plasmablast/plasma cell formation.
Introduction
The maturation of peripheral B lymphocytes is a multistep process. Due to hyper-mutations and class switches in their immunoglobulin (Ig) genes during the germinal center (GC) reaction, B cell maturation results, finally, in the generation of plasma cells that produce large amounts of specific antibodies (Abs) of high affinity, and in memory B cells (1, 2). In contrast to plasma cells that lack a B cell receptor (BCR), memory B cells are able to react immediately onto further infections upon antigen contact through the BCR. Although numerous experimental studies have been devoted to elucidate the generation of plasma B cells and memory B cells, the molecular mechanisms of this important step in differentiation of adaptive immune system remained unclear to a large extent.
In this study, we investigated the role of transcription factor NFATc1 (also designated as NFAT2) in plasma cell formation. NFATc1 is expressed in most of lymphoid cells, but its expression is switched off during plasma cell formation (3). While ablation of NFATc1 did not markedly affect B cell differentiation in bone marrow and periphery, it led to a strong decrease in generation of peritoneal B1 cells (4), and in a mild reduction of BCR-mediated proliferation and increase of BCR-mediated activation-induced cell death (AICD) in B2 cells (5). Surprisingly, in mice bearing NFATc1-deficient B cells no marked defect was observed on the Ab formation upon immunization with NP-KLH, a T cell-dependent antigen, whereas the formation of Abs upon immunization with T cell-independent type II antigen was strongly affected. Moreover, in spite of the marked anti-apoptotic effect of NFATc1 in B cells, the genes targeted by NFATc1 in splenic B cells did not help to explain the phenotype typical for NFATc1-deficient B cells (5). One explanation for this discrepancy could be the opposite effects which is exerted by the various NFATc1 isoforms on AICD and, therefore, on differentiation of lymphocytes (6).
The family of NFAT transcription factors comprises five members. The Nfatc1 gene belongs to a group of mammalian genes that are expressed in multiple isoforms with opposite functions. Similar to its expression in peripheral T cells (7), due to the use of two alternate promoters (P1 and P2) and poly A sites (pA1 and pA2), and alternate splicing events the Nfatc1 gene is expressed in six isoforms in peripheral B cells (5). In splenic B cells persistent signals from the BCR and co-stimulatory receptors lead to the predominant expression of a short isoform, designated as NFATc1/αA, within 24 h. While due to the use of “basal” promoter P2 and of distal pA2 site in resting cells long NFATc1 isoforms are generated, including NFATc1/βC, activation of cells leads to the predominant synthesis of short isoform NFATc1/αA whose synthesis is directed by the proximal pA1 site and promoter P1. The induction of NFATc1/αA is strongly supported by a remote transcriptional enhancer located in the last intron of the Nfatc1 gene (8). NFATc1/αA lacks the C-terminal peptide of approximately 250 amino acid residues typical for most of the NFATc proteins. This peptide harbors two SUMOylation sites that, therefore, are present in NFATc1/C proteins. When SUMOylated, NFATc1/C was shown to recruit histone deacetylases to and, thereby, suppresses the Il2 promoter in T cells (9).
The expression of multiple isoforms with antagonistic properties from the same locus suggests that inactivating the entire locus—as in most gene targeting approaches—can lead to misleading results on the functional capacity of the inactivated gene. To circumvent this restriction, we (over-)expressed two individual NFATc1 isoforms, NFATc1/αA and NFATc1/C, in chicken DT40 B cells and murine WEHI 231 pre-B cells. In addition to their marked opposite effect on apoptosis, NFATc1/αA and NFATc1/C exerted a contrary effect on the expression of Prdm1 gene encoding Blimp-1. Whereas Blimp-1, a key factor of plasma cell differentiation (10), was suppressed by NFATc1/αA, no or a moderate stimulatory effect on Blimp-1 was observed by NFATc1/C. Expression of a constitutive active (ca) version of NFATc1/αA in splenic B cells led to a marked suppression of Blimp-1 expression and plasmablast differentiation. This indicates NFATc1 as an important transcription factor controlling terminal B cell differentiation.
Materials and Methods
Mice, Isolation, and Culture of Cells
Animal experiments were performed according to project licenses (Nr.55.2-2531.01-80/10 and 169), which were approved by the Regierung von Unterfranken, Würzburg. If not stated otherwise, 6- to 10-week-old C57BL/6 wild-type (WT) mice were used. Cd23-cre mice were described previously (11). Transgenic (tg) caNFATc1/αA mice express a mutated, ca copy of NFATc1/αA from the Rosa 26 locus upon cre-mediated removal of a “floxed” STOP sequence (12). Chicken DT40 B lymphoma cells were cultured at 39.5°C with 5% CO2 using RPMI-1640 medium supplemented with 10% FCS, 1% chicken serum, 2-mercaptoethanol (50 µM), and l-glutamine (2 mM) (13). Murine WEHI 231 cells, EL-4 thymoma cells, human Jurkat T leukemia cells and 293 HEK cells were maintained in RPMI-1640 containing 10% FCS at 37°C in 5% CO2. Splenic B cells were isolated using Miltenyi’s B cell isolation kit, cultured in X-vivo 15 medium (Lonza) and stimulated as described (5).
Inactivation of the Chicken NFATC1 Gene
Segments from the chicken genomic NFATC1 locus were amplified using PCR primers and subcloned to generate the left and right arms of target vectors. NFATC1 targeting vectors were constructed by replacing a ~3.3 kb genomic fragment encoding exons 4 and 5 with drug resistance gene cassettes. The targeting vectors were introduced into WT DT40 cells by electroporation, and cloning of the targeted cells was performed by culturing of cells in the presence of blasticidin, histidinol D, or puromycin as described (13).
Southern Blotting
Two micrograms of genomic DT40 DNA were digested by Sac I, fractionated on a 0.7% agarose gel and transferred to a Hybond N + nylon membrane (Amersham Biosciences, Buckighamshare, UK). The membrane was hybridized with a 600 bp FITC-labeled PCR-amplified genomic fragment from intron 7 of the chicken NFATC1 gene as probe.
Generation of WEHI 231 B Cells Expressing NFATc1-Bio Proteins
Full-length murine NFATc1/αA (gi:255759918 in NCBI database) and NFATc1/βC cDNAs (gi: 255759924) were amplified, fused to a bio/avidin-tag (14) and ligated into the retroviral expression vector pEGZ (15). The retroviral pMSCV-F-BirA vector was purchased from BCCM™/LMBP (Gent-Zwijnaarde, Belgium). Retroviral particles were obtained after transfection of retroviral vectors, along with the retroviral packaging plasmids pHIT60 and pHIT123, into HEK 293T cells. After infection, WEHI cells were kept under selective conditions (using zeocin or puromycin) for 14 days. Positive integration and expression of NFATc1/αA-bio, NFATc1/βC-bio and/or BirA constructs was determined by intracellular streptavidin–fluorophore labeling and flow cytometry.
Real-Time and Semi-Quantitative PCR Assays
Total RNA was isolated from DT40 B cells using the QIAamp RNA Blood kit (QIAGEN). cDNAs were synthesized from 2 µg of total RNA by using oligo dT primers and superscript reverse transcriptase (Invitrogen) and amplified by LA-Taq polymerase (Takara Bio Co. Ltd., Tokyo, Japan). To confirm the inactivation of NFATC1 gene, primers for the detection of chicken NFATC1 RNA were designed as follows:
Fw chNFATc1, exon3: 5′-GGACCTTATGAACTGCGTATTGAAGTGCAG-3′,
Fw chNFATc1, exon 4; 5′-CATGGTTACTTAGAAAGCGAGCC-3′.
Rv chNFATc1, exon 6; 5′-CTTTCGAGTCTTGCAGAAAGTTATGGCCAG-3′.
For detecting GAPDH3, the probe was labeled with the VIC reporter dye at the 5′-terminal nucleotide and the TAMRA quencher dye at the 3′-terminal nucleotide. Detection of fluorescence during the thermal cycling process was performed using the ABI Prism 7700 Sequence Detection System (ABI/PE).
For detecting Prdm1 expression RNA was extracted from splenic B cells using TRIzol reagent (Invitrogen) followed by cDNA synthesis with the iScript II Kit (BioRad). Real-time PCR assays were carried out with an ABI Prism 7700 detection system using Prdm1 and β-actin primers sequences as described previously (16).
Cloning of Human NFATc1 Isoforms and Generation of Expression Vectors
To generate the human NFATC1 expression vectors carrying a neo resistant gene cassette, full-length cDNAs encoding human NFATC1/αA (or NFATC1/αC) was subcloned into the pcDNA3.1 vector between its BamHI-KpnI sites by PCR using cDNA of human BALM-14 cells as template. The primers for subcloning the NFATc1/αA and c1/αC isoforms were the following:
c1/αA; 5′-primer: 5′-CGCGGATCCATGCCAAGCACCAGCTTTCCA-3′(+BamHI site),
c1/αA 3′-primer: 5′-CGCGGTACCTTAGAAAAAGCACCCCACGC-3′ (+KpnI site),
c1/αC; 5′-primer: 5′-CGCGGATCCATGACGGGGCTGGAGGACCAG-3 “(+BamHI site), c1/αC, 3′-primer: 5′-CGCGGTACCCTAGGAGTGGGTGCTCGTGC-3” (+KpnI site).
For constructing vectors expressing chimeric EGFP-human NFATc1/αA or c1/αC proteins, a complete fragment of EGFP was PCR-amplified from the pEGFP-C3 vector (Clontech, Palo Alto, CA, USA) using the following primers:
5′-primer: 5′-CGCAAGCTTATGGTGAGCAAGGGC-3′ (+HindIII site), and
3′-primer, 5′-CGCGGATCCTCACTTGTACAGCTCGTC-3′ (+BamHI site).
Apoptosis Assays and Flow Cytometry
Apoptosis assays were done either with the Cy-3 annexin V staining kit (MBL Co. Ltd., Nagoya, Japan) following the manufacturer’s instructions or by stainings with annexin V-APC (BD Sciences) followed by flow cytometry (FACScan, BD Sciences). 3 × 104 cells of each sample were incubated for 20–30 h with 0–50 µg/ml of anti-chicken IgM mAb M4 (#8300-01, SouthernBiotech, Birmingham, AL, USA), and annexin-V-positive cells were detected by flow cytometry. For apoptosis induction by ionomycin, 105 DT40 cells were incubated with 1 µM ionomycin for 18–30 h.
RNA Expression Profiling
Upon stimulation with 25 µg/ml of M4 anti-chicken IgM mAb M4 Ab (SouthernBiotech) for 6 h, 5 µg of total RNA was used for the preparation of Cy5-labeled cDNAs from each RNA sample. They were hybridized to Filgen’s chicken 18k arrays that contain oligonucleotide probes of 18,000 identified chicken genes (Filgen, Nagoya, Japan). After hybridization, the signals of arrays were red by GenePix 4000B to obtain fluorescence images, and signal intensity of each spot was calculated using Filgen’s software.
RNA-Seq Assays
Unstimulated WEHI 231 B cells or WEHI cells stimulated for 6, 24, and 96 h by αIgM were deep-frozen in liquid nitrogen. Their RNA was extracted using Qiagen’s RNeasy kit and Illumina’s purification beads. RNA-Seq libraries for next generation sequencing were prepared using 600 ng starting material and Illumina’s TruSeq RNA sample prep kit V2 following the manufacturer’s instructions. RNA sequencing assays were described previously (17). The data were loaded in ArrayExpress (accession no. E-MTAB-4665).
Immunoblots
Whole cell protein extracts were resolved on 8–10% SDS polyacrylamide gels. Proteins were transferred to nitrocellulose membranes and probed with the mAb 7A6 (sc-7294, Santa Cruz Biotech. Inc., CA, USA) or a polyclonal Ab (#0102-50, Immunoglobe, Himmelstadt, Germany) raised against all isoforms of human NFATc1. For the detection of NFATc1 proteins tagged with a myc epitope and expressed upon transfection into DT40 cells, the mAb 9E10 was used raised against an epitope of c-Myc (Thermofisher, Scientific). As loading control, filters were incubated with actin mAb (CHEMICON International, Temecula, CA, USA) or stained by ponceau red. Signals were developed by chemiluminescence detection using Super Signal™ (Pierce Chemical Company, CA, USA).
Electrophoretic Mobility Shift Assays (EMSAs)
Nuclear proteins from murine splenic B cells or DT40 B cells were prepared for EMSAs as described previously (7). For detecting NFAT binding, the distal NFAT site from the murine Il2 promoter was used as probe.
Chromatin Immuno Precipitation (ChIP)
Chromatin immuno precipitation assays were performed as described (18, 19) with slight modifications. In brief, 5 × 107 WEHI-231 cells were fixed, the reaction was quenched and washed cells were re-suspended in 1 ml swelling buffer on ice for 30 min. Upon adding 40 µl of 10% NP-40, cells were passed through 8× through 21 G needles, nuclei were collected and re-suspended in 0.5 ml sonication buffer. Chromatin was sheared for 10 min (30 s pulses; 35% amplitude) on ice using an ultrasonic-disintegrator (Sonicor). DNA extracts of supernatants were checked for fragment sizes and quantified using a NanoDrop device (ThermoScientific). Chromatin was pre-cleared with 3 µg unrelated Ig Ab (CellSignaling, #2729p) and 40 µl immobilized protein G beads (ImmunoPure® PIERCE, 50% slurry saturated with salmon sperm DNA, #16-157, Upstate; and 2% gelatin from cold water fish skin (FGEL), Sigma-Aldrich, #G7765). Chromatin in 300 µl sonication buffer was incubated with 25 µl of streptavidin agarose resin (ThermoScientific, #20347; 50% slurry saturated with Salmon Sperm DNA and FGEL), or 2 µl NFATc1 Ab (7A6, BD 556609), or 2 µl Ig Ab, or 2 µl H3 Ab (CellSignaling; #4620s), or 2 µl H3K9me3 Ab (abcam ab8898), or 2 µl H3K9ac Ab (Merck 07-352) at 4°C o.n. Samples with Abs were additionally incubated with saturated protein G beads for 1 h. All beads were washed, and chromatin complexes were eluted twice by incubation with 250 µl of elution buffer. Eluates were supplemented with 21 µl 5 M NaCl and 2 µl RNase (10 mg/ml) for removal of cross-links. DNA was extracted for PCR assays using the following primers:
Il2 F CCTGTGTGCAATTAGCTCA
Il2 R CTCTTCTGATGACTCTCTGGA
Rcan1 F GCGGCATAGTTCCCACTGGTA
Rcan1 R GAGTGCTGGGCTTTTCATCCA
Ppp3ca F TTTTCTTCAGGGTGCCAGTC
Ppp3ca R CAGTCTGTTCCCATCCACCT
Prdm1 F AAACGTGTGGGTACGACCTT
Prdm1 R AGCGCTGGTTTCTACTGAGG
β-actin F CGGTTCCGATGCCCTGAGGCTCTT
β-actin R CGTCACACTTCATGATGGAATTG
Prdm1 promoter F TATCTGCCACTTCCTCTTTC
Prdm1 promoter R ATGCAAATCTTCTCTGCTGT
Immunizations of Mice and ELISAs
Age-matched mice (of 8–10 weeks) were immunized with NP-KLH (100 μg/mouse, Alum precipitated; i.p.). NP-specific Ab levels were determined in sera of mice immunized with NP-(27)-KLH for 21 days (5). Samples were applied in threefold serial dilutions starting from a 1:20 initial dilution, and Ig concentrations were quantified against a reference serum by ELISA. NP-specific Abs were determined using MaxiSorp plates (Nunc) coated with 10 µg/ml NP-BSA. Isotype-specific Abs coupled to alkaline phosphatase were used for detection according to manufacturer’s instructions (SouthernBiotech).
Immunohistochemistry of Human Tonsils
Informed consents were obtained from patients for this study. Sections of formalin-fixed tonsils embedded in paraffin were automatically stained on a Ventana Bench Mark XT (Roche Diagnostics, Japan) using the ultra view DAB and/or Alp-Red detection system. Rabbit polyclonal Ab for NFATc1/α and mAbs for CD20, CD79a, and CD35 were used for double stains, 7A6 mAb was used in combination with αIgM polyclonal Ab. Alexa 488-goat αmouse IgG (green) and Cy3-goat αrabbit IgG (red) were used for double immunofluorescence.
Statistical Analysis
Statistical analyses were performed using GraphPad (Prism) software, version 6.0. Data are presented as mean ± SEM. Unpaired t-tests were performed to evaluate the statistical significance. Statistical significances were calculated and indicated (***p < 0.001, **p < 0.005, and * p < 0.05).
Results
NFATc1 Induction in Murine Splenic B Cells
When whole protein extracts from freshly prepared murine splenic B cells cultured in vitro were analyzed for NFATc1 induction in Western blots, six protein bands were detected upon α-IgM stimulation for 6 h in vitro. These bands correspond to the six prominent NFATc1 isoforms described previously for murine and human T cells (6). Extension of α-IgM stimulation to 24 h resulted in the predominant generation of short isoform NFATc1/αA (Figure 1). A similar effect was observed upon adding Abs directed against CD40 (α-CD40) to B cells that were “pulsed” for 0.5 h by α-IgM at 4°C, washed and maintained in culture for 24 h. “Pulsed” B cells stimulated by α-IgM for 0.5 h alone revealed very low NFATc1 levels, even after washing of cells and a further incubation for 24 h in culture. However, the addition of α-CD40 to the α-IgM-“pulsed” led to strong NFATc1/αA induction, whereas addition of LPS exerted a relative mild effect (Figure 1). These observations suggest that the BCR and co-stimulatory signals lead to the accumulation of short isoform NFATc1/αA that represents the most prominent nuclear NFAT protein in effector B cells (16).
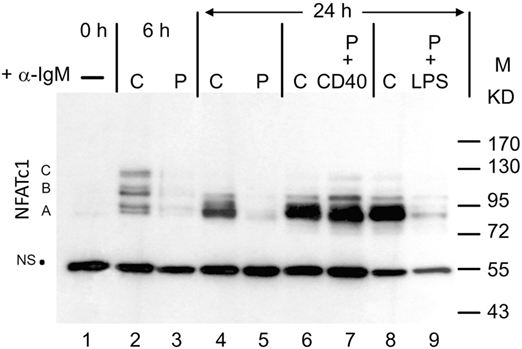
Figure 1. Induction of NFATc1 proteins in splenic B cells. Western blot using the 7A6 mAb directed against all NFATc1 proteins. B cells were left untreated (−), continuously treated (C) with α-IgM for 6 or 24 h, or “pulsed” (P) for 30 min at 4°C with α-IgM, washed and incubated for 4 h without or with α-CD40 or LPS. M, molecular weight marker; NS, non-specific band that indicates—as the ponceau red stain of the filter (not shown)—equal loading of the gel. One typical blot of more than three assays is shown. A, B, and C, NFATc1 isoforms.
Human NFATc1/αA Protects Chicken DT40 B Cells against Cell Death
To investigate the functional relevance of NFATc1/αA induction in B cells, we expressed in chicken DT40 B cells human NFATc1/αA upon inactivation of endogenous chicken NFATC1 gene. Due to the existence of three NFATC1 alleles in DT40 cells, three rounds of gene targeting had to be performed by replacing exons 4 and 5 of the chicken NFATC1 gene with gene cassettes for selection. This targeting strategy resulted in the ablation of NFATC1 transcription (Figures S1A–C in Supplementary Material). When we introduced a cDNA encoding either human NFATc1/αA or the long isoform NFATc1/αC both NFATc1 proteins were properly expressed. Upon stimulation with chicken α-IgM (M4) or ionomycin, the human NFATc1 proteins were rapidly translocated into the nuclei of chicken cells (Figures S2A–D in Supplementary Material). Testing the DNA binding activity of NFAT1/αA in EMSAs using nuclear proteins from untreated and NFATC1−/−/αA DT40 cells treated by TPA and ionomycin (T+I) for 4 h, we observed a strong increase in NFAT binding to the distal NFAT binding site from the murine Il2 promoter. Transfection of an NFAT-directed luciferase reporter gene into NFATC1−/−/αA DT40 cells showed a moderate 5- to 10-fold increase in NFAT activity compared to NFATC1−/−/− cells whereas about the same NFAT activity was measured in WT and NFATC1−/−/αA cells (Figures S2E,F in Supplementary Material).
Stimulation of DT40 cells by chicken α-IgM rapidly induces apoptosis (20). Using 30 µg/ml α-IgM for 28 h, we observed approximately 14% of annexin V-positive, apoptotic cells, in DT40 cells compared to 3% of apoptotic cells in non-stimulated cells. Stimulation of NFATC1−/−/− cells led to apoptosis in approximately 37% of cells whereas NFATC1−/−/αA cells showed a marked protection against AICD. Almost no protection was observed for NFATC1−/−/αC cells expressing the long NFATc1 isoform (Figure 2A). To identify NFATc1 target genes controlling the apoptosis of DT40 cells, we hybridized cDNA probes generated from DT40 RNAs to oligonucleotide arrays which represented 18,000 chicken genes (Filgen, Nagoya, Japan). Among the genes whose expression was strongly enhanced in α-IgM-induced NFATC1−/−/αA cells were the BAG2 and BCL6 genes. Bag-2 is a co-chaperone which, by binding to Bcl-2, could support the anti-apoptotic activity of Bcl-2 (21). Bcl-6 is highly expressed in GC B cells in which, among other effects, it suppresses cells from apoptosis induced by DNA damage (22). In real-time PCR assays using TaqMan probes, we detected a more than 20-fold increase in expression of BCL6 RNA between NFATC1−/−/− and NFATC1−/−/αA cells (Figure 2B). When we investigated the expression of Bcl-6 target genes in semi-quantitative PCR assays in non-stimulated DT40 cells, or in cells stimulated by α-IgM we observed a suppression of Blimp-1 expression and upregulation of Bach-2 in NFATC1−/−/αA cells, particularly upon stimulation (Figure 2C). Bach2 is a transcriptional repressor which is highly expressed in mature B cells, controls the GC reaction (23) and, along with Bcl-6, represses Prdm1/Blimp-1 gene expression (24). However, similar to the XBP1 gene that encodes a transcription factor required for plasma cell differentiation (25), the RNA levels of further Bcl-6 target genes, such as of CD44, ID2, and CCL3/Mip1α genes (26), remained unaffected (Figure 2C).
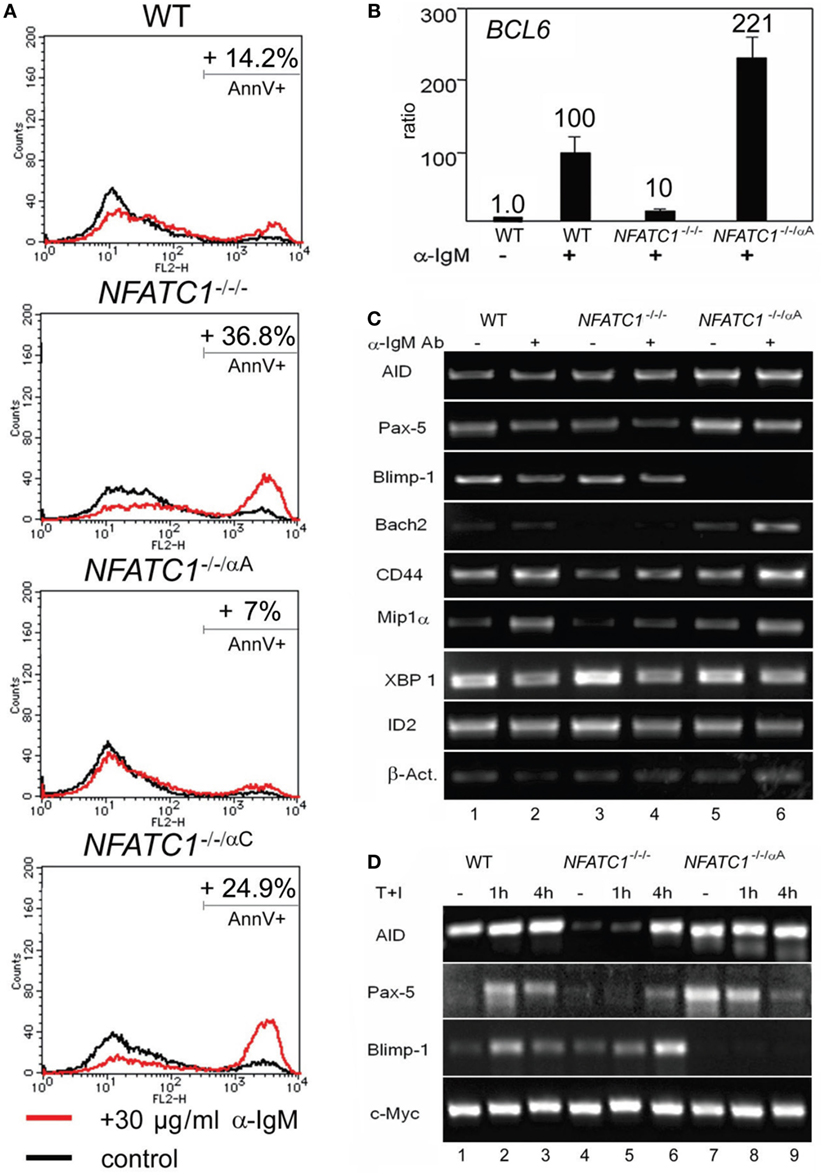
Figure 2. NFATc1/αA suppresses cell death and Prdm1/Blimp-1 expression in chicken DT40 B cells. (A) WT, NFATC1−/−/−, NFATC1−/−/αA, and NFATC1−/−/αC DT40 cells were stimulated with 30 µg/ml α-IgM M4 Ab for 28 h. Cells were stained by annexin V-Cy3. Shown is the percentage of apoptotic cells in one typical experiment from more than three. (B) Real-time qPCR assays. RNAs were isolated from either untreated DT40 B cells—or from cells treated with 30 µg/ml αIgM for 6 h (+) using a QIAamp RNA Blood kit (QIAGEN), and quantitative RT-PCR assays were performed using TaqMan probes for detecting Bcl-6 and GAPDH3 RNAs as internal control. (C,D) Human NFATc1/αA affects the gene signature of DT40 cells. RNAs from untreated DT40 B cells—or from cells treated with α-IgM for 4 h (C) or T+I for 1 and 4 h (D) were assayed in semi-quantitative PCR assays. In (C,D), one typical assay from two reactions is shown.
In a further set of PCR assays using RNAs from DT40 B cells treated with T + I for 1 and 4 h, we observed also a defective AID and Pax-5 expression in NFATC1−/−/− DT40 B cells, compared to WT cells, and again a marked suppression of Blimp-1 expression in NFATC1−/−/αA cells overexpressing NFATc1/αA (Figure 2D). These data suggest an opposite effect of NFATc1/αA on the GC reaction and plasma cell formation.
NFATc1/αA Affects Gene Expression in Murine WEHI 231 B Lymphoma Cells
To extend the findings obtained for chicken B cells to mammalian B cells, we studied murine WEHI 231 B cells stably transfected with vectors expressing the biotin ligase BirA from Escherichia coli, and either bio-tagged NFATc1/αA-bio or NFATc1/βC-bio. Due to the co-expression of BirA in these cells, bio-tagged proteins are biotinylated and can be precipitated with streptavidin beads (27). Both NFATc1-bio proteins were properly expressed in WEHI cells (Figure S3 in Supplementary Material) and translocated into the nucleus upon α-IgM stimulation (17). Similar to DT40 NFATc1−/−/αA B cells, NFATc1/αA-bio expressing WEHI cells were protected against α-IgM-mediated AICD (Figure 3A). Stimulation of WT and NFATc1/βC WEHI cells with α-IgM or αIgM + αCD40 for 48 or 96 h led to a significant increase in cell death compared to cells (over-) expressing NFATc1/αA (Figure 3A).
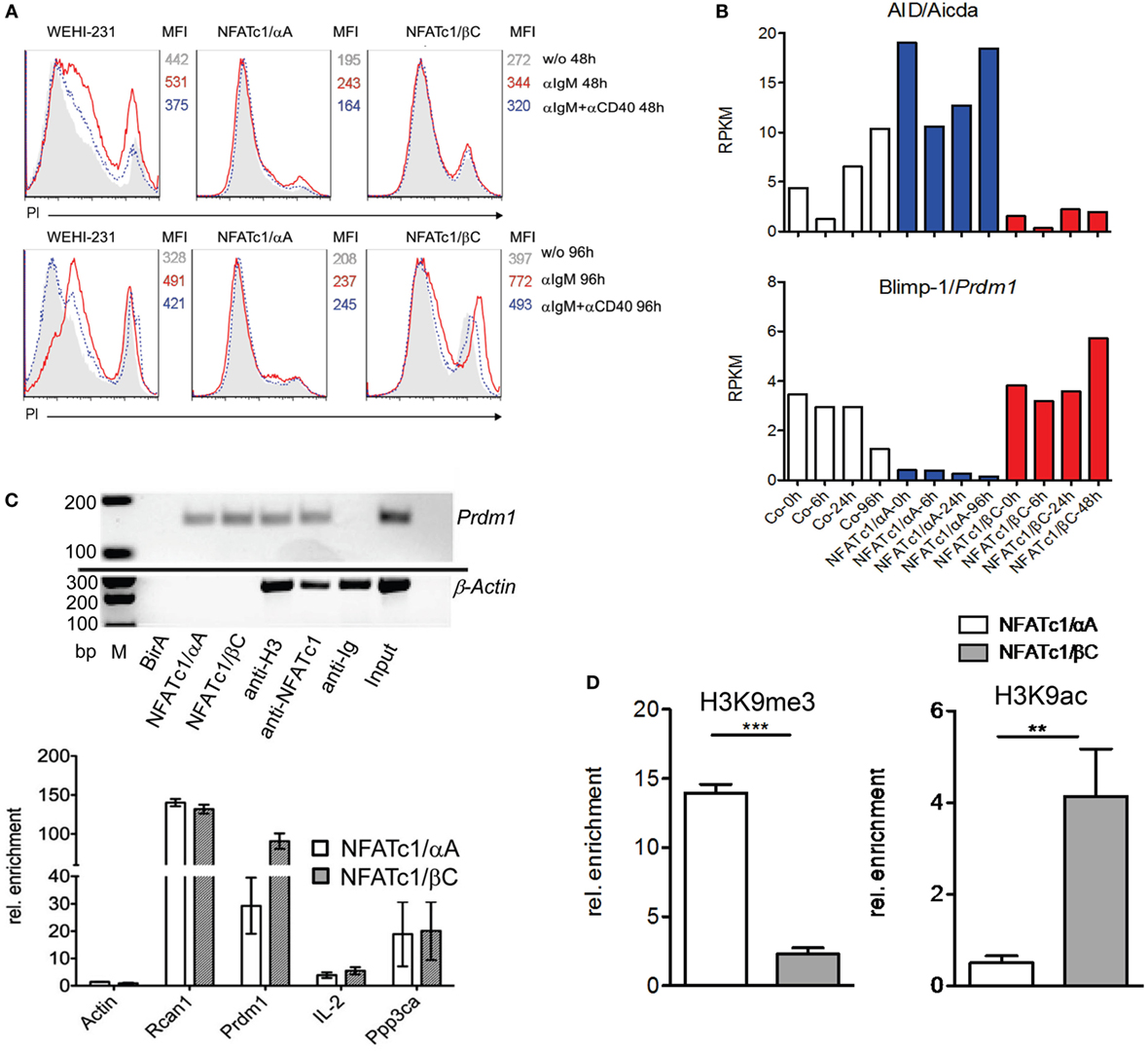
Figure 3. Effect of NFATc1/αA-bio and NFATc1/βC-bio proteins on cell death and the expression of Aicda and Prdm1 genes in murine WEHI 231 B lymphoma cells. (A) WEHI 231 B cells stably infected with retroviral vectors expressing BirA (WEHI-231), Bir A and NFATc1/αA-bio (NFATc1/αA), or BirA and NFATc1/βC-bio (NFATc1/βC) were stimulated with α-IgM or α-IgM α-IgM + αCD40 for 48 or 96 h. Apoptosis was determined by PI staining. MFI: Mean fluorescence intensity. (B) Wild-type (WT) WEHI cells (Co) or cells expressing NFATc1/αA-bio (blue) or NFATc1/βC-bio (red) were left unstimulated or stimulated for 6, 24, or 96 h with α-IgM. RNA was isolated and converted to cDNA libraries. DNA stretches of 50 bp were sequenced on a Illumina HiSeq2500 platform using the Truseq SBS kit-HS V3. Shown are the RNA reads (RPKM) from the Aicda and Prdm1 genes in the three types of WEHI cells. Results of one from two assays are shown. (C) Chromatin immuno precipitation (ChIP) assays for the binding of NFATc1-bio proteins to the Prdm1 gene in WEHI cells stimulated with T+I for 6 h. In the upper panel semi-quantitative PCR assays are shown for the detection of Prdm1 (and β-Actin) DNA in chromatin precipitations. In the first three lanes, chromatin from WEHI cells transfected with BirA alone, with NFATc1/αA-bio (+BirA) or NFATc1/βC-bio (+BirA) was precipitated with streptavidin-agarose beads. In the next lanes, chromatin was precipitated with Abs specific for histone H3, NFATc1 (7A6), and immunoglobulin. In the last two lanes, DNA input and H2O controls are shown. One typical assay from three assays is shown. In the lower panel the enrichment of β-Actin, Rcan1, Prdm1, Il2, and Ppp3ca DNAs precipitated with streptavidin beads from WEHI cells expressing either NFATc1/αA-bio or NFATc1/βC-bio is shown. Mean values of three assays are shown. (D) ChIP assays indicating histone modifications at the Prdm1 promoter. ChIP assays were performed with chromatin from WEHI cells overexpressing NFATc1/αA-bio (+BirA) (open bars) or NFATc1/βC-bio (+BirA) proteins (gray bars) using Abs directed the histone modifications H3K9me3 and H3K9ac, respectively. In semi-quantitative PCR assays, primers detecting the Prdm1 promoter (28) were used. Mean values of three assays are shown.
Using RNA from WEHI cells stably transfected with NFAT-expressing vectors for genome-wide RNA-Seq assays, we detected more than 2,000 genes whose RNA levels were changed more than twofold by NFATc1/αA-bio, and somewhat less genes whose RNAs were either affected by NFATc1/βC-bio, or by both NFATc1-bio proteins. Among the genes affected by both NFAT factors were the Aicda and Prdm1 genes encoding AID and Blimp-1, respectively. While NFATc1/αA-bio enhanced Aicda expression 2- to 5-fold, it suppressed almost 10-fold Prdm1 transcription in WEHI cells. By contrast, NFATc1/βC-bio suppressed Aicda expression and exerted a moderate stimulatory effect on the transcription of Prdm1 gene (Figure 3B). Using the same cells in ChIP assays, we observed the specific binding of both NFATc1/αA-bio or βC-bio to a regulatory site within exon 1 of Prdm1 gene which was described to be highly conserved between mouse and human (29). In real-time PCR assays, we determined a 100-fold enrichment of NFATc1/βC-bio and a 30-fold enrichment of NFATc1/αA-bio binding to this site, compared to a control site from the β-actin gene. This is comparable to the binding of NFATc1-bio proteins to Rcan1, a well-known NFATc1 target gene in B cells (5), whereas a markedly weaker binding was detected to the Ppp3ca gene encoding calcineurin’s catalytic A subunit, and the Il2 gene that is inactive in B cells. No NFATc1-bio binding to the Prdm1 gene was detected in WEHI cells expressing BirA alone, and no specific precipitation of β-actin chromatin was observed with the NFATc1 mAb 7A6 which has frequently been used in ChIP assays (Figure 3C).
The expression of the NFATc1 isoforms and their effect on Prdm1 gene expression correlated well with the occurrence of specific histone modifications at the Prdm1 promoter. While in WEHI cells overexpressing NFATc1/αA a strong increase in H3K9me3, a mark for inactive heterochromatin (30), was detected, a decrease was observed for H3K9ac, a mark for active transcription (31). Opposite chromatin modifications were determined for the Prdm1 promoter in WEHI cells overexpressing NFATc1/βC (Figure 3D).
NFATc1/αA Suppresses Plasmablast Formation and Ab Production
Stimulation of murine splenic B cells with α-IgM resulted in a more than fivefold decrease of Prdm1 RNA reads within 24 h whereas α-CD40 or LPS treatment increased the number of reads approximately twofold and more than fivefold, respectively (Figure 4A). Prolongation of LPS treatment led to a strong increase of Prdm1 RNA levels within 48–72 h (Figure 4B) and in massive production of IgM, a typical sign of differentiation of B cells to plasmablasts in vitro. However, when splenic B cells were pre-treated with α-IgM followed by LPS, or co-stimulated with α-IgM and LPS, a reduction in Prdm1 RNA levels and plasmablast formation was observed (16). To show whether the suppression of plasmablast differentiation by α-IgM and, therefore, BCR signals—which induce NFATc1/αA—are indeed due to NFATc1/αA induction we investigated plasmablast differentiation of splenic B cells from tg mice which express a ca version of NFATc1/αA (caNFATc1/αA) from the Rosa26 locus upon cre-mediated deletion of a STOP sequence. Upon crossing those mice with Cd23-cre mice, the splenic B cells of caNFATc1/αA × Cd23-cre mice express caNFATc1/αA (Figure S4 in Supplementary Material), and upon treatment of cells with LPS for 1–3 days in vitro a strong reduction in Prdm1 RNA expression was observed (Figure 4B). When such caNFATc1/αA × Cd23-cre mice were immunized with NP-KLH, a T cell dependent antigen, the secretion of IgG2a Abs into the serum was abolished and those of IgG1 and IgG2b markedly impaired, similar to IgM Ab secretion (Figure 4C). Immunization with NP-Ficoll, a T cell-independent antigen, resulted in a strong decrease in IgM, IgG2b, and IgG3 secretion, and a more moderate in IgG2a secretion (Figure 4D). This suggests that overexpressing caNFATc1/αA suppresses plasma cell formation in vivo.
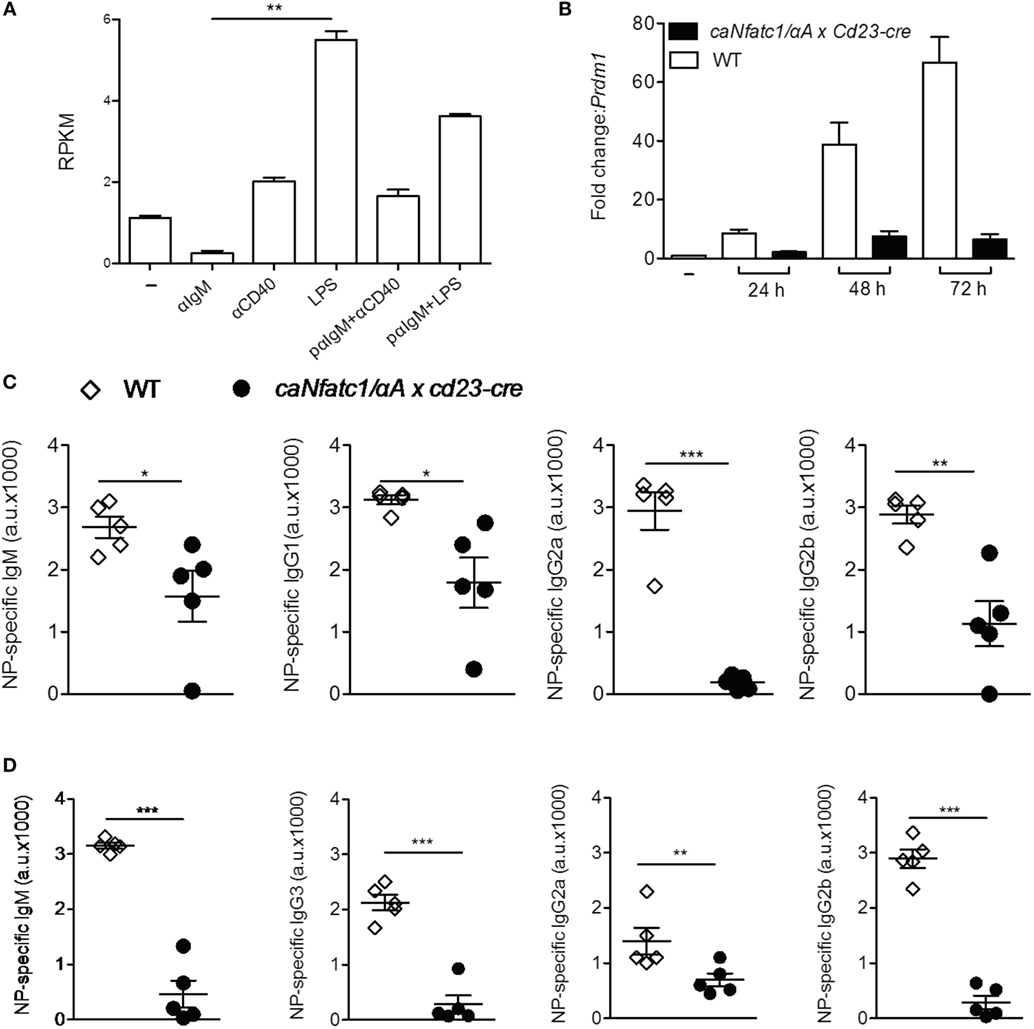
Figure 4. Suppression of plasmablast differentiation and of IgG class switch by caNFATc1/αA. (A) Changes in Prdm1 RNA levels in splenic B cells upon stimulation with α-IgM, α-CD40, or LPS for 24 h. pαIgM: cells were “pulsed” for 30 min at 4°C with α-IgM, washed and then treated with α-CD40 or LPS for 24 h. Shown are mean values of Prdm1 RNA reads (RPKM) from two assays. (B) Splenic B cells from wild-type (WT) mice (open columns) or caNfatc1/αA × Cd23-cre mice (black) were treated for 24–72 h with LPS. Real-time PCR assays for detecting Prdm1 RNA levels. Mean values of three assays are shown. (C,D) WT and caNfatc1/αA × Cd23-cre mice were immunized with NP-KLH (C) or NP-Ficoll (D) for 21 days, and the serum levels of immunoglobulins were determined in ELISAs. One dot and square indicates one mouse, respectively.
The NFATc1/αA-mediated control of genes expressed during the GC reaction and plasma cell formation prompted us to investigate whether NFATc1/αA is indeed expressed in nuclei of GC B cells. To this end, we raised a polyclonal Ab against the N-terminal α-peptide of human NFATc1 spanning 42 amino acids, and in histochemical stains of splenic GCs in mice immunized with sheep red blood cells for 7 days, we detected a strong nuclear staining for NFATc1/αA in numerous GC cells (Figure 5A). Similar results were obtained by α-NFATc1/αA staining of human tonsils from patients with chronic tonsillitis showing the nuclear expression of NFATc1/α in numerous GC cells (Figure 5B). By double-staining with Abs directed against CD79a or CD20, several of these GC cells could be identified as B cells (Figures 5C,D). Double-stainings with an Ab specific for CD35, a marker of follicular dendritic cells, show the nuclear localization of NFATc1/αA in GC cells of light zone (Figure 5E), and further double-stainings with Abs directed against various Ig proteins indicated a co-expression of NFATc1/αA with IgM (Figure 5F), but not with IgG or IgA proteins (not shown).
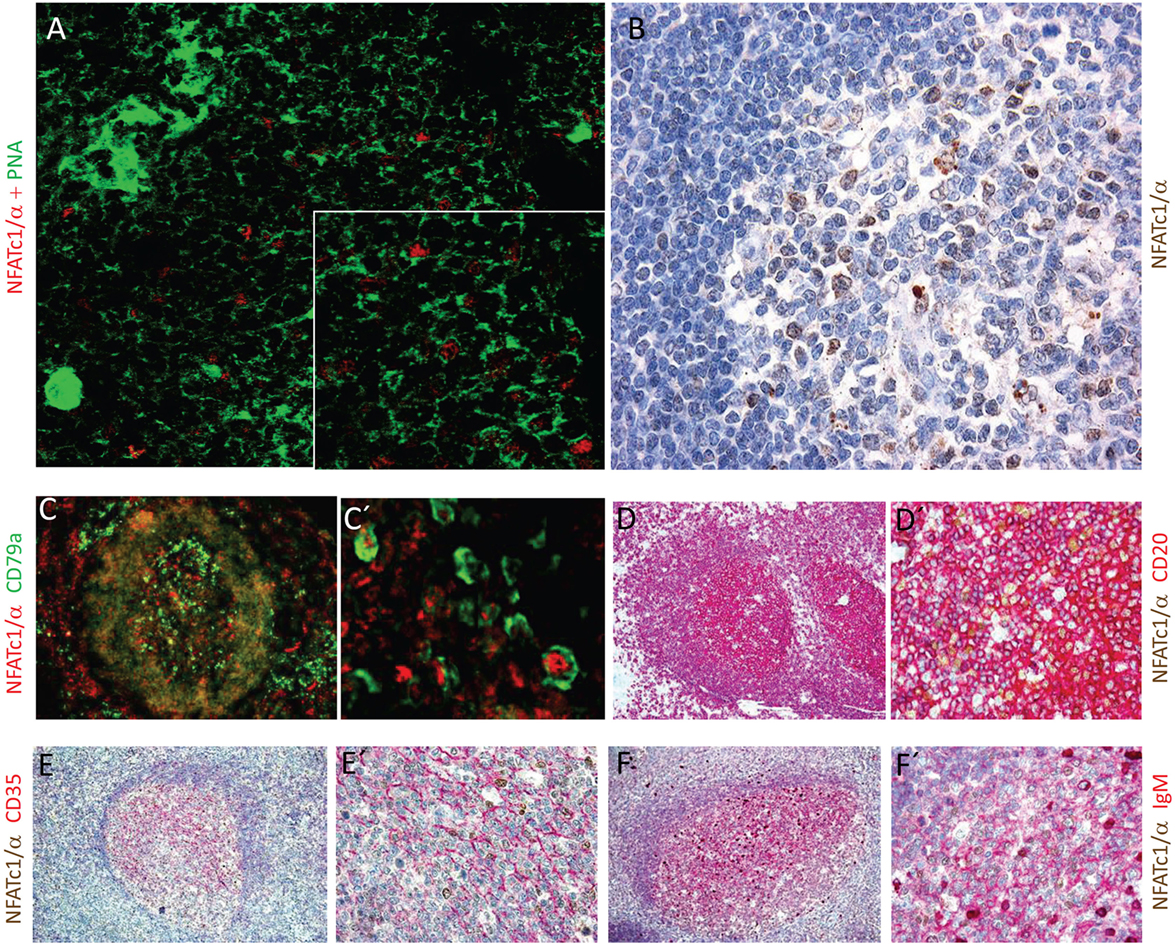
Figure 5. Nuclear expression of NFATc1/αA in germinal center (GC) B cells. (A) Histochemical co-stains of GC cells with peanut agglutinin and an antibody (Ab) raised against the N-terminal α-peptide of NFATc1. A spleen section from a mouse immunized with sheep red blood cells for 7 days is shown. Magnification, 20-fold, and insert 40-fold. (B–F) Cross sections of tonsils from tonsilitis patients. (B) Co-stains of hematoxylin/eosin and an Ab specific for the NFATc1-α peptide. (C–F) Co-stains of NFATc1-α-specific Ab with Abs specific for CD79a (C,C′), CD20 (D,D′), CD35 (E,E′), and IgM (F,F′) and at various magnifications (C vs. C′ etc.) are shown. Magnifications are 20-fold (B), 10-fold (C–F), 40-fold (D′–F′), or 50-fold (C′).
Discussion
The data presented here and those of a former report (16) show that in primary splenic B lymphocytes BCR-stimulation leads to rapid but transient appearance of nuclear NFAT complexes that consist predominantly of NFATc1. Albeit less reflected at the level of Nfatc1 transcription, persistent BCR and co-stimulatory signals induce high nuclear NFATc1/αA concentrations which, supported by the NFATc1-mediated auto-regulation of the Nfatc1 P1 promoter (7), remain high for days. By contrast, BCR signals alone lead to accumulation of cytosolic NFATc1/αA that is kept in cytosol without further (co-) induction. Although primary B cells stimulated by α-IgM alone proliferate and accumulate high levels of cytosolic NFATc1/αA, they die after 3–4 days of in vitro culture.
Published data on the strong expression of NFATc1 in follicular TFH cells (32) and our data indicate that NFATc1 is highly expressed in subsets of GC cells. The data presented here show that NFATc1/αA occurs in nuclei of numerous centrocytes within the light zone of a GC that are closely associated with the network of follicular dendritic cells. On the surface of follicular dendritic cells intact antigens are presented which, thereby, help to select GC B cells (2). It is likely that the contact between follicular dendritic cells and centrocytic B cells leads to the high levels of nuclear NFATc1/αA induction which help B cells to survive. The positive effect of NFATc1/αA on Bcl-6 and AID expression suggests that, in analogy to T cells (33), NFAT factors and, in particular, NFATc1/αA support the maturation of GC B cells to memory B cells. The occurrence of nuclear NFATc1/αA in IgM+ centrocytes suggests a role for NFATc1/αA in the survival of IgM+ memory B cells that constitute a substantial part of memory B cells upon primary immunization with complex antigens (34, 35) but further experiments are necessary to substantiate this view. In contrast to genes that control the GC reaction, NFATc1/αA suppresses the expression of the Prdm1 gene encoding the key factor of plasma cell development, Blimp-1. The binding of bio-tagged NFATc1/αA to a regulatory region of the Prdm1 gene indicates Prdm1 as a direct target of NFATc1/αA which, when overexpressed in centrocytic B cells impairs plasma cell formation.
Ethics Statement
Informed consents were obtained from patients for this study. Animal experiments were performed according to project licenses (Nr.55.2-2531.01-80/10 and 169), which are approved and controlled by the Regierung von Unterfranken, Würzburg.
Author Contributions
KM, RR, DP, SK-H, KT, and NM performed experiments; VE provided important material; EK designed and performed experiments, analyzed data, and supported the preparation of the manuscript; ES led the investigation and wrote the manuscript along with KM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very much indebted to Doris Michel for excellent technical support. For a gift of BALM-14 cells, we wish to thank Hayashibara Biochemistry Laboratories Co. Ltd., Japan, and for α-IgM hybridoma supernatants Jürgen Wienands (Göttingen). For many fold advice and support, we wish to thank Meinrad Busslinger (Vienna). For transcriptiome assays by next generation sequencing, we thank Valesca and Thomas Bukur, and Ugur Sahin (Mainz). This work was supported by the Transregio TRR52 of the DFG, the Wilhelm-Sander-Stiftung, and the Scheel-Stiftung (to ES).
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/articles/10.3389/fimmu.2018.00032/full#supplementary-material.
References
1. Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol (2010) 11(8):681–8. doi:10.1038/ni.1900
2. Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol (2008) 8(1):22–33. doi:10.1038/nri2217
3. Akimzhanov A, Krenacs L, Schlegel T, Klein-Hessling S, Bagdi E, Stelkovics E, et al. Epigenetic changes and suppression of the nuclear factor of activated T cell 1 (NFATC1) promoter in human lymphomas with defects in immunoreceptor signaling. Am J Pathol (2008) 172(1):215–24. doi:10.2353/ajpath.2008.070294
4. Berland R, Wortis HH. Normal B-1a cell development requires B cell-intrinsic NFATc1 activity. Proc Natl Acad Sci U S A (2003) 100(23):13459–64. doi:10.1073/pnas.2233620100
5. Bhattacharyya S, Deb J, Patra AK, Thuy Pham DA, Chen W, Vaeth M, et al. NFATc1 affects mouse splenic B cell function by controlling the calcineurin – NFAT signaling network. J Exp Med (2011) 208(4):823–39. doi:10.1084/jem.20100945
6. Serfling E, Chuvpilo S, Liu J, Hofer T, Palmetshofer A. NFATc1 autoregulation: a crucial step for cell-fate determination. Trends Immunol (2006) 27(10):461–9. doi:10.1016/j.it.2006.08.005
7. Chuvpilo S, Jankevics E, Tyrsin D, Akimzhanov A, Moroz D, Jha MK, et al. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity (2002) 16(6):881–95. doi:10.1016/S1074-7613(02)00329-1
8. Klein-Hessling S, Rudolf R, Muhammad K, Knobeloch KP, Maqbool MA, Cauchy P, et al. A threshold level of NFATc1 activity facilitates thymocyte differentiation and opposes notch-driven leukaemia development. Nat Commun (2016) 7:11841. doi:10.1038/ncomms11841
9. Nayak A, Glockner-Pagel J, Vaeth M, Schumann JE, Buttmann M, Bopp T, et al. Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase. J Biol Chem (2009) 284(16):10935–46. doi:10.1074/jbc.M900465200
10. Kallies A, Nutt SL. Terminal differentiation of lymphocytes depends on Blimp-1. Curr Opin Immunol (2007) 19(2):156–62. doi:10.1016/j.coi.2007.01.003
11. Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity (2008) 28(6):751–62. doi:10.1016/j.immuni.2008.04.014
12. Baumgart S, Chen NM, Siveke JT, Konig A, Zhang JS, Singh SK, et al. Inflammation-induced NFATc1-STAT3 transcription complex promotes pancreatic cancer initiation by KrasG12D. Cancer Discov (2014) 4(6):688–701. doi:10.1158/2159-8290.CD-13-0593
13. Yamamoto K, Hirano S, Ishiai M, Morishima K, Kitao H, Namikoshi K, et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol Cell Biol (2005) 25(1):34–43. doi:10.1128/MCB.25.1.34-43.2005
14. McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, et al. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J (2011) 30(12):2388–404. doi:10.1038/emboj.2011.140
15. Berberich-Siebelt F, Klein-Hessling S, Hepping N, Santner-Nanan B, Lindemann D, Schimpl A, et al. C/EBPbeta enhances IL-4 but impairs IL-2 and IFN-gamma induction in T cells. Eur J Immunol (2000) 30(9):2576–85. doi:10.1002/1521-4141
16. Hock M, Vaeth M, Rudolf R, Patra AK, Pham DA, Muhammad K, et al. NFATc1 induction in peripheral T and B lymphocytes. J Immunol (2013) 190(5):2345–53. doi:10.4049/jimmunol.1201591
17. Alrefai H, Muhammad K, Rudolf R, Pham DA, Klein-Hessling S, Patra AK, et al. NFATc1 supports imiquimod-induced skin inflammation by suppressing IL-10 synthesis in B cells. Nat Commun (2016) 7:11724. doi:10.1038/ncomms11724
18. Kolodziej KE, Pourfarzad F, de Boer E, Krpic S, Grosveld F, Strouboulis J. Optimal use of tandem biotin and V5 tags in ChIP assays. BMC Mol Biol (2009) 10:6. doi:10.1186/1471-2199-10-6
19. Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science (2002) 295(5561):1901–4. doi:10.1126/science.1068356
20. Winding P, Berchtold MW. The chicken B cell line DT40: a novel tool for gene disruption experiments. J Immunol Methods (2001) 249(1–2):1–16. doi:10.1016/S0022-1759(00)00333-1
21. Lee MY, Kim SY, Shin SL, Choi YS, Lee JH, Tsujimoto Y. Reactive astrocytes express bis, a bcl-2-binding protein, after transient forebrain ischemia. Exp Neurol (2002) 175(2):338–46. doi:10.1006/exnr.2002.7903
22. Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature (2004) 432(7017):635–9. doi:10.1038/nature03147
23. Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature (2004) 429(6991):566–71. doi:10.1038/nature02596
24. Ochiai K, Muto A, Tanaka H, Takahashi S, Igarashi K. Regulation of the plasma cell transcription factor Blimp-1 gene by Bach2 and Bcl6. Int Immunol (2008) 20(3):453–60. doi:10.1093/intimm/dxn005
25. Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature (2001) 412(6844):300–7. doi:10.1038/35085509
26. Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity (2000) 13(2):199–212. doi:10.1016/S1074-7613(00)00020-0
27. de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, et al. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A (2003) 100(13):7480–5. doi:10.1073/pnas.1332608100
28. Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, et al. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity (2013) 38(5):918–29. doi:10.1016/j.immuni.2013.04.009
29. Nishikawa K, Nakashima T, Hayashi M, Fukunaga T, Kato S, Kodama T, et al. Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc Natl Acad Sci U S A (2010) 107(7):3117–22. doi:10.1073/pnas.0912779107
30. Becker JS, Nicetto D, Zaret KS. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet (2016) 32(1):29–41. doi:10.1016/j.tig.2015.11.001
31. Gates LA, Shi J, Rohira AD, Feng Q, Zhu B, Bedford MT, et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem (2017) 292(35):14456–72. doi:10.1074/jbc.M117.802074
32. Rasheed AU, Rahn HP, Sallusto F, Lipp M, Muller G. Follicular B helper T cell activity is confined to CXCR5(hi)ICOS(hi) CD4 T cells and is independent of CD57 expression. Eur J Immunol (2006) 36(7):1892–903. doi:10.1002/eji.200636136
33. Dienz O, Eaton SM, Krahl TJ, Diehl S, Charland C, Dodge J, et al. Accumulation of NFAT mediates IL-2 expression in memory, but not naive, CD4+ T cells. Proc Natl Acad Sci U S A (2007) 104(17):7175–80. doi:10.1073/pnas.0610442104
34. Dogan I, Bertocci B, Vilmont V, Delbos F, Megret J, Storck S, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol (2009) 10(12):1292–9. doi:10.1038/ni.1814
Keywords: B cells, DT40 cells, germinal center, NFATc1, plasmablasts, plasma cells
Citation: Muhammad K, Rudolf R, Pham DAT, Klein-Hessling S, Takata K, Matsushita N, Ellenrieder V, Kondo E and Serfling E (2018) Induction of Short NFATc1/αA Isoform Interferes with Peripheral B Cell Differentiation. Front. Immunol. 9:32. doi: 10.3389/fimmu.2018.00032
Received: 27 September 2017; Accepted: 04 January 2018;
Published: 24 January 2018
Edited by:
Harry W. Schroeder, University of Alabama at Birmingham, United StatesReviewed by:
Lee Ann Garrett-Sinha, University at Buffalo, United StatesPaolo Casali, The University of Texas Health Science Center San Antonio, United States
Copyright: © 2018 Muhammad, Rudolf, Pham, Klein-Hessling, Takata, Matsushita, Ellenrieder, Kondo and Serfling. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edgar Serfling, serfling.e@mail.uni-wuerzburg.de
†Present address: Eisaku Kondo, Division of Molecular and Cellular Pathology, Niigata University Graduate School of Medical and Dental Sciences, Niigata City, Japan
‡The two last authors share the senior authorship of this article.
 Khalid Muhammad
Khalid Muhammad Ronald Rudolf1
Ronald Rudolf1 Katsuyoshi Takata
Katsuyoshi Takata Eisaku Kondo
Eisaku Kondo Edgar Serfling
Edgar Serfling