- CNRS, INSERM, CHU Lille, U1019, UMR8204, Centre d’Infection et d’Immunité de Lille (CIIL), Institut Pasteur de Lille, Université de Lille, Lille, France
Intracellular bacteria are responsible for many infectious diseases in humans and have developed diverse mechanisms to interfere with host defense pathways. In particular, intracellular vacuoles are an essential niche used by pathogens to alter cellular and organelle functions, which facilitate replication and survival. Mycobacterium tuberculosis (Mtb), the pathogen causing tuberculosis in humans, is not only able to modulate its intraphagosomal fate by blocking phagosome maturation but has also evolved strategies to successfully prevent clearance by immune cells and to establish long-term survival in the host. Mass spectrometry (MS)-based proteomics allows the identification and quantitative analysis of complex protein mixtures and is increasingly employed to investigate host–pathogen interactions. Major challenges are limited availability and purity of pathogen-containing compartments as well as the asymmetric ratio in protein abundance when comparing bacterial and host proteins during the infection. Recent advances in purification techniques and MS technology helped to overcome previous difficulties and enable the detailed proteomic characterization of infected host cells and their pathogen-containing vacuoles. Here, we summarize current findings of the proteomic analysis of Mycobacterium-infected host cells and highlight progress that has been made to study the protein composition of mycobacterial vacuoles. Current investigations focus on the pathogenicity during Mtb infection, which will allow to better understand pathogen-induced changes and immunomodulation of infected host cells. Consequently, future research in this field will have important implications on host response, pathogen survival, and persistence, induced adaptive immunity and metabolic changes of immune cells promoting the development of novel host-directed therapies in tuberculosis.
Introduction
Phagocytic cells, such as macrophages, dendritic cells, and neutrophils, represent the first line of defense against invading pathogens, such as Legionella pneumophila, Coxiella burnetti, and Leishmania donovani, by engulfing and eliminating them by phagocytosis. Phagocytosis is a complex process divided in several steps, which is initiated by the innate recognition of microbial patterns, leading to the formation of microbe-containing vesicles. These vesicles fuse successively with different endocytic compartments [early endosome (EE) and late endosome (LE), and lysosomes (LYSs)] to form a microbicidal organelle, the phagolysosome (1). This process results in the formation of an acidic, oxidative, and degradative environment, which is additionally influenced by immune signals inside and outside of the phagocyte that determine phagosomal fate (2). However, pathogen-containing vacuoles (PCVs) are also occupied and altered by intracellular pathogens providing a successful niche to survive and replicate within phagocytes. Therefore, it is not surprising that numerous microbes developed a wide variety of strategies to adapt and manipulate the phagocytic process (3).
Among these strategies induced by intracellular pathogens, the phagosome maturation arrest initiated by Mycobacterium tuberculosis (Mtb) infection allows the colonization of phagocytes and long-term survival within host cells (4). Mtb is causing tuberculosis (TB) in humans with more than 1.3 million deaths each year and an increasing incidence of drug-resistant strains (5). In addition to the lack of an efficient vaccine, tuberculosis remains together with malaria and HIV/AIDS one of the deadliest infectious diseases worldwide. The inter- and intracellular modifications of the host environment induced by Mtb infection are diverse and not fully understood (6). Briefly, after phagocytic uptake, mycobacteria-containing vacuoles (MCVs) are formed, which were shown to interact and exchange material with early endosomal compartments but are devoid of, or only interact transiently, with molecules of LEs and LYSs and are able to retain a pH at around 6.5 within MCVs. The lack of phagosomal acidification is mostly due to a defective retention of the vacuolar proton ATPase (V-ATPase) complex at the phagosome (7, 8). Moreover, it was also considered for long that MCVs are sealed, but recent findings demonstrated that Mtb is able to induce vacuolar rupture, thereby allowing direct interactions of bacterial components with host cytosolic proteins (9–11). Despite numerous studies analyzing the host machinery that controls spatiotemporal vacuolar acidification, progression of phagosome maturation, and intracellular survival of different Mycobacterium species, the Mtb-induced delay of phagosome maturation is not fully characterized. Furthermore, we also only begin to understand the impact of the different virulence factor secreted upon Mtb infection, which additionally interfere with host pathways to promote Mtb survival and growth (12). Together with the fact that the existing BCG vaccine is inefficient and needs further improvement and alternatives (13), it is clear that we need more knowledge of mycobacterial pathogenesis and the host factors manipulated by Mtb allowing to establish its intracellular niche. In particular, a better understanding of host immune responses and the involved mechanisms controlling the infection are needed, because they affect disease outcome as well as TB pathology. The possibility of modulating host immunity to maximize mycobacterial killing while minimizing inflammatory tissue damage has received increasing attention in recent years and is applied as novel host-directed therapy against drug-resistant Mtb strains (14).
The development of powerful and sensitive mass spectrometry (MS)-based methods during the past decade allows now the accurate, spatiotemporal identification, quantification, and modification of almost any expressed protein (15). These robust methodologies are increasingly applied to study host–pathogen interactions and to elucidate the proteome of the pathogen itself, of infected cells and of subcellular compartments. In the case of Mtb infection, protein profiling of different mycobacterial strains as well as of clinical, drug-resistant isolates increased tremendously our knowledge about their proteome (16, 17), while MS methods also helped to identify biomarkers of Mtb-infected patients (18). Many previous MS studies on host infection were carried out with non-pathogenic mycobacteria strains or were performed with beads coated with single mycobacterial virulence factors, while data on virulent Mtb strains were scarce. Here, we want to focus on the host side of mycobacterial infection and summarize current findings of the proteomic analysis of Mycobacterium-infected cells and the progress that has been made in the purification of PCVs that helped to overcome previous difficulties and will allow the detailed and reproducible proteomic characterization of MCVs to better understand Mtb pathogenicity. In this review, we aim to point out the potential of current MS technology to increase our knowledge of the host response during Mtb infection including pathogen-driven immunomodulation.
Proteome Analysis of Mycobacteria-Infected Cells
The intracellular niche protects mycobacteria from cellular and humoral component of the immune system. To overcome host cell defense mechanisms, Mtb subverts the normal passage through the endocytic pathway to form a distinct replicative membranous compartment. Furthermore, Mtb is also able to induce vacuolar rupture to reach nutrients in the host cytosol and to escape host defense pathways, both favoring Mtb growth. Therefore, several proteome studies of mycobacteria-infected cells were initiated to collect an inventory of host cell factors required to establish mycobacterial colonization at the cellular level. By using liquid chromatography–tandem MS (LC–MS/MS) approaches for proteome analysis, high identification rates were achieved, which also allow the measurement of the quantitative state of cellular proteomes at any given time of infection (19). In addition, the introduction of stable isotope labeling of proteins prior MS analysis further improved the relative quantification of complex proteomic samples (20). This approach has gained success due to its high proteome coverage, quantification reliability, and high-throughput format. Over the past years, a few MS studies addressed infection settings with virulent and avirulent strains of the Mtb complex, which we have summarized in Table 1A that gave insight on the host environment during mycobacterial colonization.
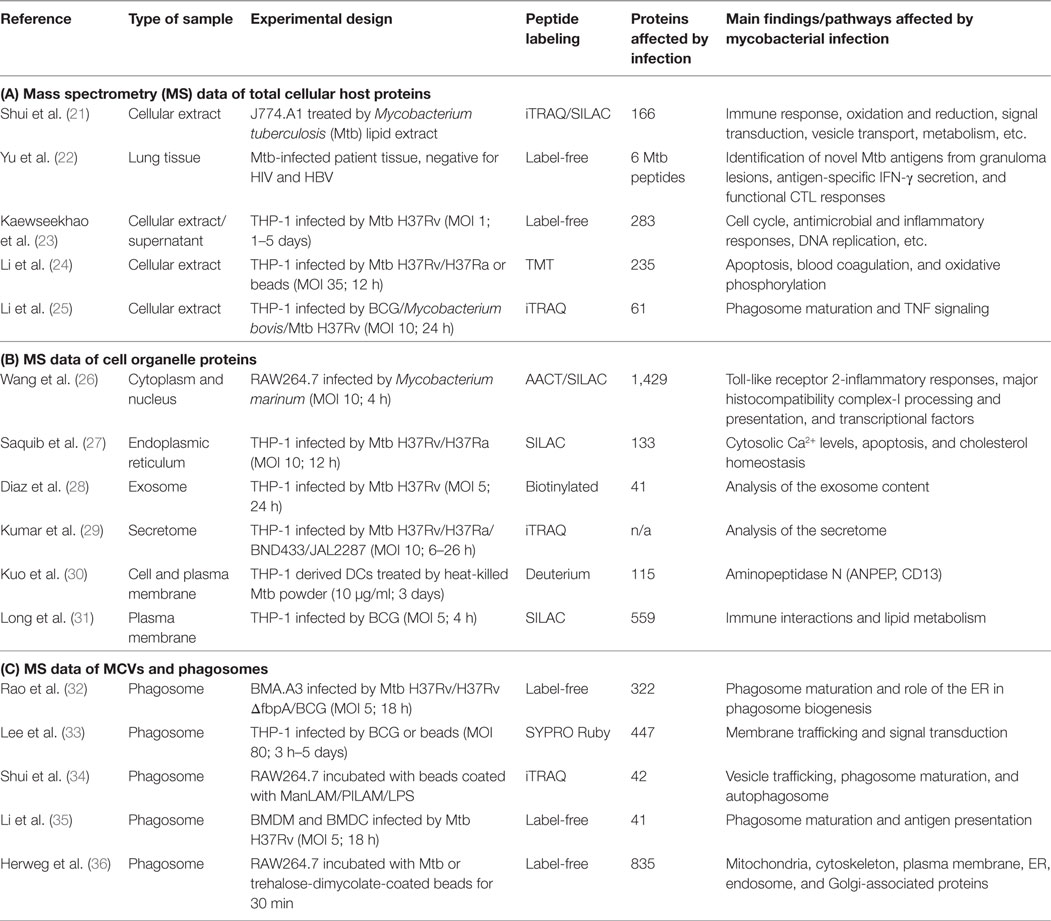
Table 1. List of host proteomic studies of mycobacterial infection performed (A) on total cellular extracts, (B) on isolated cell organelles of infected cells, and (C) specifically on mycobacteria-containing vacuoles (MCVs) and bead-containing phagosomes.
In one of the first studies, Shui et al. published a list of 166 murine macrophage proteins, which showed a differential expression after Mtb lipid exposure (21). Lipids of the mycobacterial cell wall represent approximately 60% of the total bacterial dry weight (37) and were shown to be actively trafficked out of the MCV (38). After both metabolic stable isotope labeling by amino acids in cell culture and chemical isobaric tagging (iTRAQ), the authors found that Mtb lipids act on diverse cellular processes, such as immune response, apoptosis, metabolism, vesicle transport, and signal transduction (21). In addition, also Fc gamma receptor type IIb is upregulated upon Mtb lipid exposure, which is known to block calcium influx and to inhibit phagocytosis and inflammatory responses (39). Moreover, also three proteins implicated in the oxidative burst, which help macrophages to kill intracellular pathogens, were found upregulated upon Mtb lipid exposure: p67phox, p47phox (both subunits of NADPH oxidase), and macrophage migration inhibitory factor. At the cellular level, a recent work compared human THP-1 cells infected by either the BCG vaccine strain or two virulent strains (Mtb H37Rv and Mycobacterium bovis) (25). The authors identified 61 host proteins differentially regulated depending on the used infection model with seven of them specifically upregulated upon virulent strain exposure, including CCL20 and ICAM1 involved in TNF signaling. Mtb H37Rv infection also induced upregulation of two interferon-induced transmembrane proteins (Ifitm1 and Ifitm3), which is in agreement with previously published results (40). This study has shown the importance of using appropriate infection models to identify host factors influencing the establishment of a pathogen. In line with this, another recent study identified in human macrophages the downregulation of 235 host proteins, when they compared their profile between macrophages infected with the virulent Mtb H37Rv and the avirulent H37Ra strain (24). The main host cell processes found altered by Mtb infection were blood coagulation, apoptosis, and oxidative phosphorylation. Among these candidates, some proteins showed differential expression levels between both models of infection suggesting the existence of different immunity mechanisms that influence immune responses. For example, the expression of HLA class I, a major histocompatibility complex antigen chain specific to humans, was downregulated after Mtb H37Rv infection, providing evidence that the cross-presentation pathway is affected by Mtb colonization. In another study, Mtb-specific antigens from granulomatous lesions of TB patients could be identified (22), demonstrating the potential of MS-based approaches to study pathogen-driven immune responses.
Although the proteome analysis performed on infected cells and tissue is able to give insight into the host defense mechanisms affected by Mtb colonization, it does not provide specific information about the occupied niche, the MCV, and how its composition and molecular function is altered by the pathogen to allow intracellular growth and replication. Therefore, several labs aim to purify intact cell organelles and PCVs, including those containing different mycobacteria species, to further analyze these compartments by quantitative MS approaches to better understand host–pathogen interactions.
Current State and Pitfalls of PCV Purification
Traditional organelle enrichment techniques, such as differential centrifugation in (dis-)continuous density gradients and biochemical fractionation, often resulted in preparations of low purity and contained contaminants, such as other cell organelles and molecular constituents. In recent years, additional techniques have been developed, which allow better purification of PCVs enabling organelle proteomics at higher accuracy and reproducibility. The selective enrichment of PCVs is crucial to increase both specificity and quality of sample preparations and can be achieved by immuno-affinity purification, flow cytometry-based single organelle enrichment, and subcellular fractionation in regard of the physicochemical characteristics of the PCVs of interest. Hilbi and coworkers provide a comprehensive overview of different techniques developed to purify vacuoles containing Legionella, Salmonella, Chlamydia, Simkania, and a Mycobacterium trehalose-dimycolate (TDM) bead model (36). It is essential to control all experimental steps carefully to avoid the presence of contaminating cell organelles and remnants, which will result in the identification of artifacts during MS analysis. In particular, the use of immunomagnetic separation techniques combined with fractionation and density gradient centrifugation has been used successfully to purify vacuoles of intracellular pathogens that allowed the MS-based identification of host proteins recruited to PCVs, such as IRG1, a catalytic enzyme shown to regulate an antimicrobial host response against Legionella pneumophila in vitro and in vivo (41).
There is a long-standing interest in MCV isolation to identify and characterize pathogen-specific virulence and persistence mechanisms during infection. The first protocols of MCV purification were reported by the Russell laboratory (42, 43) and Pieters laboratory (44) more than 20 years ago and were based on sucrose and/or Ficoll gradient centrifugation. More recently, these approaches have been either modified, e.g., by combining them with iso-osmotic iodixanol density gradients (33), or were replaced by separation techniques, where mycobacteria are magnetically labeled prior infection (45, 46). Currently, the latter methods seem to be superior to purify MCVs since they allow the selective enrichment of pathogen-containing compartments. In all cases, homogenous MCV isolation with a high degree of purity remains technically challenging, especially if one aims to perform it on virulent Mtb strains at BSL3 conditions. However, in combination with methods developed for other intracellular pathogens, for example, the immunomagnetic separation of PCVs using antibodies against species-specific virulence factors (47), will further boost the efforts to unravel the Mtb-specific phagosomal proteome during different times of infection.
Proteome Analysis of Mycobacteria-Containing Vacuoles
During the last years, several MS studies were published that analyzed different cell organelles of mycobacteria-infected cells (48), such as cytoplasm and nucleus (26), the ER (27), secreted exosomes (28), plasma and cell organelle membranes (30, 31), as well as phagosomes (32–36), which provide detailed insight in the cellular mechanisms of mycobacterial infection. We have listed these findings in Tables 1B,C. For example, in a quantitative MS approach, Kuo et al. performed membrane profiling on human dendritic cells and identified 115 proteins that were upregulated in response to heat-killed Mtb (30). Among those host proteins, aminopeptidase N was found largely overexpressed, and the authors could demonstrate that membranous aminopeptidase N is capable of binding live bacteria and is involved in antigen presentation that impaired T cell activation to facilitate Mtb pathogenesis.
The different MS studies on the phagosomal proteome (Table 1C) were performed in immune cells infected by either Mtb H37Rv and M. bovis BCG or beads coated with Mtb cell wall components (ManLAM, PILAM, and TDM) and identified phagosomal host proteins that are altered during mycobacterial infection. Together with the findings of non-MS studies investigating MCVs at different time points postinfection, we start to better understand the phagosome maturation block and other features of Mtb infection, which we have illustrated in Figure 1. While molecules such as transferrin receptor and Rab5 (33–36) are commonly found in MCVs, suggesting fusion with EEs, other EE markers are not found or only at much lower quantities, such as phosphatidyl-inositol-3-phosphate and early endosomal antigen 1 (32), confirming previous findings on MCV biogenesis (49–51). The different MS studies (33–35) also confirm the notion that mycobacterial infection actively suppresses the recruitment of the V-ATPase to the MCV membrane (7) to avoid phagosomal acidification due to the secreted Mtb protein tyrosine phosphatase (8). A recent study from us has also shown that the V-ATPase is targeted for ubiquitination and proteasomal degradation during Mtb infection due to the activity of cytokine-inducible SH2-containing protein (Cish) (52). Moreover, Mtb infection also blocks fusion with late endosomal and lysosomal compartments, which excludes molecules such as cathepsin D and S and lysosome-associated membrane protein 2 from MCVs (33–35, 76). Only late endosomal proteins, such as Rab7 and Nramp-1, were shown to be acquired transiently to the MCV membrane (10, 53, 54), the latter being involved in vacuolar rupture and access of Mtb to the cytosol. Recently, the mannose receptor was found to be involved in blocking phagolysosomal fusion by its recruitment of SHP-1 (55). Also two other Rab GTPases, Rab14 and Rab20, were shown to play important roles in MCV biogenesis (34, 53, 56, 57), while other molecules, such as coronin-1A, are actively retained on MCVs containing live Mtb (58). The block of phagosomal acidification, and therefore hydrolase activity, is also supported by the activity of ion channels, such as CFTR (59) and others, which lead to, e.g., high levels of chloride ions in early MCVs (60–62). Finally, there are also other cellular mechanisms, such as autophagy, necrosis, apoptosis, and impairment of antigen presentation, involved in the outcome of mycobacterial infections, which is beyond the scope of this review. However, all these different findings demonstrate an impressive body of evidence, how Mtb interferes with host defense to survive and replicate in different cell types, and how MS analysis of infected cells and of MCVs provide a valuable tool to better understand host–pathogen interactions.
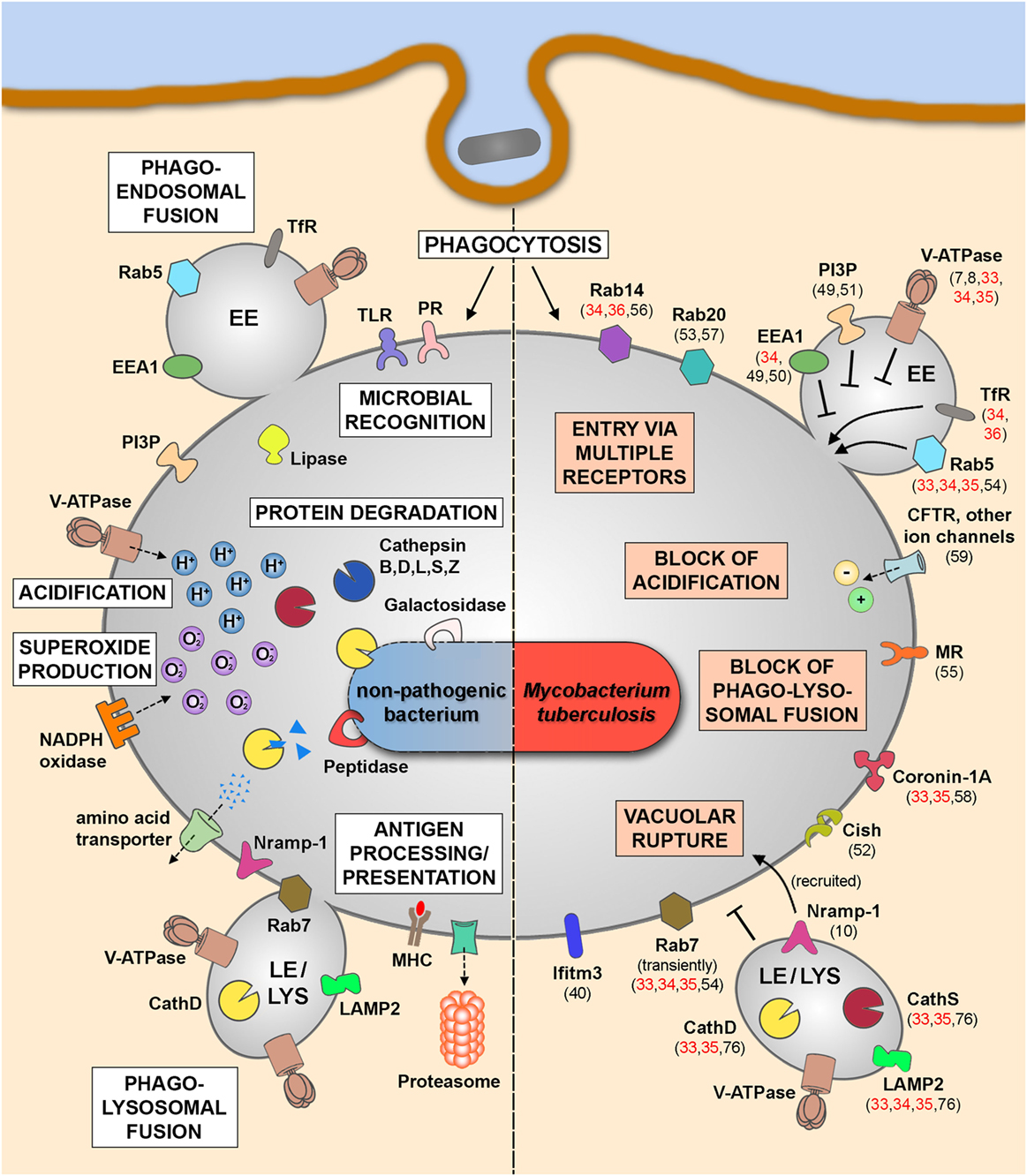
Figure 1. Phagosomal functions after internalization of non-pathogenic bacteria (left panel) and in the context of Mtb infection (right panel). Schematic representation of the key players and main functional features after the uptake of non-pathogenic bacteria leading to the clearance of the internalized cargo (left panel). Upon Mtb infection, the pathogen is internalized into mycobacteria-containing vacuoles (MCVs), which are delayed in phagosome maturation (right panel). Some of the altered phagosomal functions are indicated here together with involved host molecules that were identified by MS approaches (references shown in red) or by non-MS techniques (references shown in black). Abbreviations: CathD, cathepsin D; CathS, cathepsin S; Cish, cytokine-inducible SH2-containing protein; EE, early endosome; EEA1, early endosomal antigen 1; Ifitm3, interferon-induced transmembrane protein 3; LAMP2, lysosome-associated membrane protein 2; LE, late endosome; LYS, lysosome; MHC, major histocompatibility complex; MR, mannose receptor; MS, mass spectrometry; Mtb, Mycobacterium tuberculosis; Nramp-1, natural resistance-associated macrophage protein 1; PI3P, phosphatidyl-inositol-3-phosphate; PR, phagocytic receptor; TfR, transferrin receptor; TLR, toll-like receptor; V-ATPase, vacuolar proton ATPase.
Conclusion and Outlook
Research on mycobacterial infections includes an increasing number of MS-based approaches, which refine our understanding of the molecular mechanisms underlying Mtb pathogenesis. Together with improvements on purity and reproducibility of sample preparations, in particular of MCVs containing virulent Mtb strains, it is now feasible to combine these methods with other techniques analyzing dynamics and impact of host–pathogen interactions. In addition, current advances in quantitative MS as well as recently emerging targeting MS-based techniques, such as selected reaction monitoring and SWATH MS (17), enable accurate and very sensitive protein measurements. Combined with systems biology-driven workflows, future studies on Mtb infection will allow a better understanding of pathogen-induced changes and immunomodulation of infected host cells.
Future research should focus on the diversity of innate immune cells and their distinct functions that cooperate to control Mtb infection, such as the contribution of myeloid cells and lymphocyte populations (63–65). MS approaches could help to uncover the underlying mechanisms by identifying the involved players. Effective immunity against Mtb requires balanced adaptive immune responses while avoiding damages by immune activation, which determines the persistence of chronic TB infection (66). We now begin to understand the different mechanisms of immune manipulation induced by Mtb infection that favor disease progression, and current interventions aim to reverse these effects by altering the frequency of specific immune cell populations (67). Therefore, novel immunotherapies include depletion strategies targeting regulatory T cells (68, 69) and myeloid-derived suppressor cells (70, 71) to restrict mycobacterial replication. Finally, current findings also demonstrate the importance of metabolic remodeling of immune cells during infection, and how these processes influence effector functions, inflammatory responses, and the outcome of disease (72). How precise changes in metabolites are beneficial for the host during Mtb infection and contribute to protection from TB remains to be uncovered, but the inflammatory state of certain immune cell populations appears to be crucial (73). Without doubt, current MS technology allows the identification of metabolic adaptation during Mtb infection (74) and will allow further insight how immune responses are regulated, for example, the role of different interferons in TB pathogenesis (75). Consequently, future research in these fields will have important implications on host response, pathogen survival, and TB persistence and will promote the development of novel host-directed therapies, particularly against emergent drug-resistant Mtb strains.
Author Contributions
All the authors participated in the concept, preparation, and writing of the manuscript. EH conceived the content and edited the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Gareth Griffiths and Edouard Yeramian for their comments on the manuscript. The authors apologize to all colleagues whose work could not be cited due to space limitations. Financial support of our lab is provided by the European Research Council (ERC-STG INTRACELLTB Grant no. 260901, MM4TB Grant no. 260872, and CycloNHit no. 608407), the EMBO YIP program, the Agence Nationale de la Recherche (ANR-10-EQPX-04-01, ANR-14-CE14-0024, ANR-14-CE08-0017, and ANR-16-CE35-0009), the Projet Transversal de Recherche de l’Institut Pasteur (PTR441 and PTR22-16), the Feder [12001407 (D-AL) Equipex Imaginex BioMed], the Région Nord Pas de Calais (convention no. 12000080). AM is supported by a fellowship of the Fondation pour la Recherche Médicale (SPF20170938709).
References
1. Levin R, Grinstein S, Canton J. The life cycle of phagosomes: formation, maturation, and resolution. Immunol Rev (2016) 273:156–79. doi:10.1111/imr.12439
2. Pauwels AM, Trost M, Beyaert R, Hoffmann E. Patterns, receptors, and signals: regulation of phagosome maturation. Trends Immunol (2017) 38:407–22. doi:10.1016/j.it.2017.03.006
3. Uribe-Querol E, Rosales C. Control of phagocytosis by microbial pathogens. Front Immunol (2017) 8:1368. doi:10.3389/fimmu.2017.01368
4. Stanley SA, Cox JS. Host-pathogen interactions during Mycobacterium tuberculosis infections. Curr Top Microbiol Immunol (2013) 374:211–41. doi:10.1007/82_2013_332
6. Schorey JS, Schlesinger LS. Innate immune responses to tuberculosis. Microbiol Spectr (2016) 4:TBTB2-0010-2016. doi:10.1128/microbiolspec.TBTB2-0010-2016
7. Sturgill-Koszycki S, Schaible UE, Russell DG. Mycobacterium-containing phagosomes are accessible to early endosomes and reflect a transitional state in normal phagosome biogenesis. EMBO J (1996) 15:6960–8.
8. Wong D, Bach H, Sun J, Hmama Z, Av-Gay Y. Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc Natl Acad Sci U S A (2011) 108:19371–6. doi:10.1073/pnas.1109201108
9. Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog (2012) 8:e1002507. doi:10.1371/journal.ppat.1002507
10. Simeone R, Sayes F, Song O, Groschel MI, Brodin P, Brosch R, et al. Cytosolic access of Mycobacterium tuberculosis: critical impact of phagosomal acidification control and demonstration of occurrence in vivo. PLoS Pathog (2015) 11:e1004650. doi:10.1371/journal.ppat.1004650
11. van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell (2007) 129:1287–98. doi:10.1016/j.cell.2007.05.059
12. Philips JA, Ernst JD. Tuberculosis pathogenesis and immunity. Annu Rev Pathol (2012) 7:353–84. doi:10.1146/annurev-pathol-011811-132458
13. Kaufmann SHE, Dockrell HM, Drager N, Ho MM, McShane H, Neyrolles O, et al. TBVAC2020: advancing tuberculosis vaccines from discovery to clinical development. Front Immunol (2017) 8:1203. doi:10.3389/fimmu.2017.01203
14. Tobin DM. Host-directed therapies for tuberculosis. Cold Spring Harb Perspect Med (2015) 5:a021196. doi:10.1101/cshperspect.a021196
15. Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature (2016) 537:347–55. doi:10.1038/nature19949
16. Banaei-Esfahani A, Nicod C, Aebersold R, Collins BC. Systems proteomics approaches to study bacterial pathogens: application to Mycobacterium tuberculosis. Curr Opin Microbiol (2017) 39:64–72. doi:10.1016/j.mib.2017.09.013
17. Schubert OT, Aebersold R. Microbial proteome profiling and systems biology: applications to Mycobacterium tuberculosis. Adv Exp Med Biol (2015) 883:235–54. doi:10.1007/978-3-319-23603-2_13
18. Walzl G, Haks MC, Joosten SA, Kleynhans L, Ronacher K, Ottenhoff TH. Clinical immunology and multiplex biomarkers of human tuberculosis. Cold Spring Harb Perspect Med (2014) 5:a018515. doi:10.1101/cshperspect.a018515
19. Schubert OT, Rost HL, Collins BC, Rosenberger G, Aebersold R. Quantitative proteomics: challenges and opportunities in basic and applied research. Nat Protoc (2017) 12:1289–94. doi:10.1038/nprot.2017.040
20. Rauniyar N, Yates JR III. Isobaric labeling-based relative quantification in shotgun proteomics. J Proteome Res (2014) 13:5293–309. doi:10.1021/pr500880b
21. Shui W, Gilmore SA, Sheu L, Liu J, Keasling JD, Bertozzi CR. Quantitative proteomic profiling of host-pathogen interactions: the macrophage response to Mycobacterium tuberculosis lipids. J Proteome Res (2009) 8:282–9. doi:10.1021/pr800422e
22. Yu Y, Jin D, Hu S, Zhang Y, Zheng X, Zheng J, et al. A novel tuberculosis antigen identified from human tuberculosis granulomas. Mol Cell Proteomics (2015) 14:1093–103. doi:10.1074/mcp.M114.045237
23. Kaewseekhao B, Naranbhai V, Roytrakul S, Namwat W, Paemanee A, Lulitanond V, et al. Comparative proteomics of activated THP-1 cells infected with Mycobacterium tuberculosis identifies putative clearance biomarkers for tuberculosis treatment. PLoS One (2015) 10:e0134168. doi:10.1371/journal.pone.0134168
24. Li H, Wei S, Fang Y, Li M, Li X, Li Z, et al. Quantitative proteomic analysis of host responses triggered by Mycobacterium tuberculosis infection in human macrophage cells. Acta Biochim Biophys (2017) 49:835–44. doi:10.1093/abbs/gmx080
25. Li P, Wang R, Dong W, Hu L, Zong B, Zhang Y, et al. Comparative proteomics analysis of human macrophages infected with virulent Mycobacterium bovis. Front Cell Infect Microbiol (2017) 7:65. doi:10.3389/fcimb.2017.00065
26. Wang H, Dong D, Tang S, Chen X, Gao Q. PPE38 of Mycobacterium marinum triggers the cross-talk of multiple pathways involved in the host response, as revealed by subcellular quantitative proteomics. J Proteome Res (2013) 12:2055–66. doi:10.1021/pr301017e
27. Saquib NM, Jamwal S, Midha MK, Verma HN, Manivel V. Quantitative proteomics and lipidomics analysis of endoplasmic reticulum of macrophage infected with Mycobacterium tuberculosis. Int J Proteomics (2015) 2015:270438. doi:10.1155/2015/270438
28. Diaz G, Wolfe LM, Kruh-Garcia NA, Dobos KM. Changes in the membrane-associated proteins of exosomes released from human macrophages after Mycobacterium tuberculosis infection. Sci Rep (2016) 6:37975. doi:10.1038/srep37975
29. Kumar A, Jamwal S, Midha MK, Hamza B, Aggarwal S, Yadav AK, et al. Dataset generated using hyperplexing and click chemistry to monitor temporal dynamics of newly synthesized macrophage secretome post infection by mycobacterial strains. Data Brief (2016) 9:349–54. doi:10.1016/j.dib.2016.08.055
30. Kuo CP, Chang KS, Hsu JL, Tsai IF, Lin AB, Wei TY, et al. Analysis of the immune response of human dendritic cells to Mycobacterium tuberculosis by quantitative proteomics. Proteome Sci (2016) 14:5. doi:10.1186/s12953-016-0095-8
31. Long J, Basu Roy R, Zhang YJ, Antrobus R, Du Y, Smith DL, et al. Plasma membrane profiling reveals upregulation of ABCA1 by infected macrophages leading to restriction of mycobacterial growth. Front Microbiol (2016) 7:1086. doi:10.3389/fmicb.2016.01086
32. Rao PK, Singh CR, Jagannath C, Li Q. A systems biology approach to study the phagosomal proteome modulated by mycobacterial infections. Int J Clin Exp Med (2009) 2:233–47.
33. Lee BY, Jethwaney D, Schilling B, Clemens DL, Gibson BW, Horwitz MA. The Mycobacterium bovis bacille Calmette-Guerin phagosome proteome. Mol Cell Proteomics (2010) 9:32–53. doi:10.1074/mcp.M900396-MCP200
34. Shui W, Petzold CJ, Redding A, Liu J, Pitcher A, Sheu L, et al. Organelle membrane proteomics reveals differential influence of mycobacterial lipoglycans on macrophage phagosome maturation and autophagosome accumulation. J Proteome Res (2011) 10:339–48. doi:10.1021/pr100688h
35. Li Q, Singh CR, Ma S, Price ND, Jagannath C. Label-free proteomics and systems biology analysis of mycobacterial phagosomes in dendritic cells and macrophages. J Proteome Res (2011) 10:2425–39. doi:10.1021/pr101245u
36. Herweg JA, Hansmeier N, Otto A, Geffken AC, Subbarayal P, Prusty BK, et al. Purification and proteomics of pathogen-modified vacuoles and membranes. Front Cell Infect Microbiol (2015) 5:48. doi:10.3389/fcimb.2015.00048
37. Kolattukudy PE, Fernandes ND, Azad AK, Fitzmaurice AM, Sirakova TD. Biochemistry and molecular genetics of cell-wall lipid biosynthesis in mycobacteria. Mol Microbiol (1997) 24:263–70. doi:10.1046/j.1365-2958.1997.3361705.x
38. Beatty WL, Rhoades ER, Ullrich HJ, Chatterjee D, Heuser JE, Russell DG. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic (2000) 1:235–47. doi:10.1034/j.1600-0854.2000.010306.x
39. Maglione PJ, Xu J, Casadevall A, Chan J. Fc gamma receptors regulate immune activation and susceptibility during Mycobacterium tuberculosis infection. J Immunol (2008) 180:3329–38. doi:10.4049/jimmunol.180.5.3329
40. Ranjbar S, Haridas V, Jasenosky LD, Falvo JV, Goldfeld AE. A role for IFITM proteins in restriction of Mycobacterium tuberculosis infection. Cell Rep (2015) 13:874–83. doi:10.1016/j.celrep.2015.09.048
41. Naujoks J, Tabeling C, Dill BD, Hoffmann C, Brown AS, Kunze M, et al. IFNs modify the proteome of legionella-containing vacuoles and restrict infection via IRG1-derived itaconic acid. PLoS Pathog (2016) 12:e1005408. doi:10.1371/journal.ppat.1005408
42. Chakraborty P, Sturgill-Koszycki S, Russell DG. Isolation and characterization of pathogen-containing phagosomes. Methods Cell Biol (1994) 45:261–76. doi:10.1016/S0091-679X(08)61856-7
43. Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, et al. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science (1994) 263:678–81. doi:10.1126/science.8303277
44. Hasan Z, Schlax C, Kuhn L, Lefkovits I, Young D, Thole J, et al. Isolation and characterization of the mycobacterial phagosome: segregation from the endosomal/lysosomal pathway. Mol Microbiol (1997) 24:545–53. doi:10.1046/j.1365-2958.1997.3591731.x
45. Lönnbro P, Nordenfelt P, Tapper H. Isolation of bacteria-containing phagosomes by magnetic selection. BMC Cell Biol (2008) 9:35. doi:10.1186/1471-2121-9-35
46. Steinhäuser C, Dallenga T, Tchikov V, Schaible UE, Schütze S, Reiling N. Immunomagnetic isolation of pathogen-containing phagosomes and apoptotic blebs from primary phagocytes. Curr Protoc Immunol (2014) 105:14361–26. doi:10.1002/0471142735.im1436s105
47. Hoffmann C, Finsel I, Hilbi H. Pathogen vacuole purification from legionella-infected amoeba and macrophages. Methods Mol Biol (2013) 954:309–21. doi:10.1007/978-1-62703-161-5_18
48. He Y, Li W, Liao G, Xie J. Mycobacterium tuberculosis-specific phagosome proteome and underlying signaling pathways. J Proteome Res (2012) 11:2635–43. doi:10.1021/pr300125t
49. Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J Cell Biol (2001) 154:631–44. doi:10.1083/jcb.200106049
50. Vergne I, Chua J, Deretic V. Mycobacterium tuberculosis phagosome maturation arrest: selective targeting of PI3P-dependent membrane trafficking. Traffic (2003) 4:600–6. doi:10.1034/j.1600-0854.2003.00120.x
51. Vergne I, Chua J, Lee HH, Lucas M, Belisle J, Deretic V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci U S A (2005) 102:4033–8. doi:10.1073/pnas.0409716102
52. Queval CJ, Song OR, Carralot JP, Saliou JM, Bongiovanni A, Deloison G, et al. Mycobacterium tuberculosis controls phagosomal acidification by targeting CISH-mediated signaling. Cell Rep (2017) 20:3188–98. doi:10.1016/j.celrep.2017.08.101
53. Schnettger L, Rodgers A, Repnik U, Lai RP, Pei G, Verdoes M, et al. A Rab20-dependent membrane trafficking pathway controls M. tuberculosis replication by regulating phagosome spaciousness and integrity. Cell Host Microbe (2017) 21:619–628e5. doi:10.1016/j.chom.2017.04.004
54. Via LE, Deretic D, Ulmer RJ, Hibler NS, Huber LA, Deretic V. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J Biol Chem (1997) 272:13326–31. doi:10.1074/jbc.272.20.13326
55. Rajaram MVS, Arnett E, Azad AK, Guirado E, Ni B, Gerberick AD, et al. M. tuberculosis-initiated human mannose receptor signaling regulates macrophage recognition and vesicle trafficking by FcRgamma-Chain, Grb2, and SHP-1. Cell Rep (2017) 21:126–40. doi:10.1016/j.celrep.2017.09.034
56. Kyei GB, Vergne I, Chua J, Roberts E, Harris J, Junutula JR, et al. Rab14 is critical for maintenance of Mycobacterium tuberculosis phagosome maturation arrest. EMBO J (2006) 25:5250–9. doi:10.1038/sj.emboj.7601407
57. Seto S, Tsujimura K, Koide Y. Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic (2011) 12:407–20. doi:10.1111/j.1600-0854.2011.01165.x
58. Ferrari G, Langen H, Naito M, Pieters J. A coat protein on phagosomes involved in the intracellular survival of mycobacteria. Cell (1999) 97:435–47. doi:10.1016/S0092-8674(00)80754-0
59. Di A, Brown ME, Deriy LV, Li C, Szeto FL, Chen Y, et al. CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat Cell Biol (2006) 8:933–44. doi:10.1038/ncb1456
60. Neyrolles O, Wolschendorf F, Mitra A, Niederweis M. Mycobacteria, metals, and the macrophage. Immunol Rev (2015) 264:249–63. doi:10.1111/imr.12265
61. Soldati T, Neyrolles O. Mycobacteria and the intraphagosomal environment: take it with a pinch of salt(s)! Traffic (2012) 13:1042–52. doi:10.1111/j.1600-0854.2012.01358.x
62. Wagner D, Maser J, Lai B, Cai Z, Barry CE III, Honer Zu Bentrup K, et al. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J Immunol (2005) 174:1491–500. doi:10.4049/jimmunol.174.3.1491
63. Mayer-Barber KD, Barber DL. Innate and adaptive cellular immune responses to Mycobacterium tuberculosis infection. Cold Spring Harb Perspect Med (2015) 5:a018424. doi:10.1101/cshperspect.a018424
64. Srivastava S, Grace PS, Ernst JD. Antigen export reduces antigen presentation and limits T cell control of M. tuberculosis. Cell Host Microbe (2016) 19:44–54. doi:10.1016/j.chom.2015.12.003
65. Bénard A, Sakwa I, Schierloh P, Colom A, Mercier I, Tailleux L, et al. B cells producing type I interferon modulate macrophage polarization in tuberculosis. Am J Respir Crit Care Med (2017). doi:10.1164/rccm.201707-1475OC
66. Brighenti S, Ordway DJ. Regulation of immunity to tuberculosis. Microbiol Spectr (2016) 4:TBTB2-0006-2016. doi:10.1128/microbiolspec.TBTB2-0006-2016
67. Jayashankar L, Hafner R. Adjunct strategies for tuberculosis vaccines: modulating key immune cell regulatory mechanisms to potentiate vaccination. Front Immunol (2016) 7:577. doi:10.3389/fimmu.2016.00577
68. Parkash O, Agrawal S, Madhan Kumar M. T regulatory cells: Achilles’ heel of Mycobacterium tuberculosis infection? Immunol Res (2015) 62:386–98. doi:10.1007/s12026-015-8654-0
69. Bhattacharya D, Dwivedi VP, Kumar S, Reddy MC, Van Kaer L, Moodley P, et al. Simultaneous inhibition of T helper 2 and T regulatory cell differentiation by small molecules enhances bacillus Calmette-Guerin vaccine efficacy against tuberculosis. J Biol Chem (2014) 289:33404–11. doi:10.1074/jbc.M114.600452
70. Dorhoi A, Kaufmann SH. Versatile myeloid cell subsets contribute to tuberculosis-associated inflammation. Eur J Immunol (2015) 45:2191–202. doi:10.1002/eji.201545493
71. Gupta S, Cheung L, Pokkali S, Winglee K, Guo H, Murphy JR, et al. Suppressor cell-depleting immunotherapy with denileukin diftitox is an effective host-directed therapy for tuberculosis. J Infect Dis (2017) 215:1883–7. doi:10.1093/infdis/jix208
72. Olenchock BA, Rathmell JC, Vander Heiden MG. Biochemical underpinnings of immune cell metabolic phenotypes. Immunity (2017) 46:703–13. doi:10.1016/j.immuni.2017.04.013
73. Mishra BB, Lovewell RR, Olive AJ, Zhang G, Wang W, Eugenin E, et al. Nitric oxide prevents a pathogen-permissive granulocytic inflammation during tuberculosis. Nat Microbiol (2017) 2:17072. doi:10.1038/nmicrobiol.2017.72
74. Eoh H, Wang Z, Layre E, Rath P, Morris R, Moody DB, et al. Metabolic anticipation in Mycobacterium tuberculosis. Nat Microbiol (2017) 2:17084. doi:10.1038/nmicrobiol.2017.84
75. Travar M, Petkovic M, Verhaz A. Type I, II, and III Interferons: regulating immunity to Mycobacterium tuberculosis infection. Arch Immunol Ther Exp (2016) 64:19–31. doi:10.1007/s00005-015-0365-7
Keywords: host–pathogen interactions, immunometabolism, mass spectrometry, mycobacterial infection, Mycobacterium tuberculosis, phagocytosis, phagosome maturation, proteomics
Citation: Hoffmann E, Machelart A, Song O-R and Brodin P (2018) Proteomics of Mycobacterium Infection: Moving towards a Better Understanding of Pathogen-Driven Immunomodulation. Front. Immunol. 9:86. doi: 10.3389/fimmu.2018.00086
Received: 02 November 2017; Accepted: 11 January 2018;
Published: 30 January 2018
Edited by:
Yoann Rombouts, UMR5089 Institut de Pharmacologie et de Biologie Structurale (IPBS), FranceReviewed by:
Roland Lang, Universitätsklinikum Erlangen, GermanyShashank Gupta, Brown University, United States
Copyright: © 2018 Hoffmann, Machelart, Song and Brodin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eik Hoffmann, eik.hoffmann@ibl.cnrs.fr
 Eik Hoffmann
Eik Hoffmann Arnaud Machelart
Arnaud Machelart Ok-Ryul Song
Ok-Ryul Song