- Department of Microbiology, Immunology and Pathology, Colorado State University, Fort Collins, CO, United States
In the current quest for a complete cure for HIV/AIDS, highly sensitive HIV-1 latency detection methods are critical to verify full viral eradication. Until now, the in vitro quantitative viral outgrowth assays (qVOA) have been the gold standard for assessing latent HIV-1 viral burden. However, these assays have been inadequate in detecting the presence of ultralow levels of latent virus in a number of patients who were initially thought to have been cured, but eventually showed viral rebound. In this context, new approaches utilizing in vivo mouse-based VOAs are promising. In the murine VOA (mVOA), large numbers of CD4+ T cells or PBMC from aviremic subjects are xenografted into immunodeficient NSG mice, whereas in the humanized mouse-based VOA (hmVOA) patient CD4+ T cell samples are injected into BLT or hu-hematopoetic stem cells (hu-HSC) humanized mice. While latent virus could be recovered in both of these systems, the hmVOA provides higher sensitivity than the mVOA using a fewer number of input cells. In contrast to the mVOA, the hmVOA provides a broader spectrum of highly susceptible HIV-1 target cells and enables newly engrafted cells to home into preformed human lymphoid organs where they can infect cells in situ after viral activation. Hu-mice also allow for both xenograft- and allograft-driven cell expansions with less severe GvH providing a longer time frame for potential viral outgrowth from cells with a delayed latent viral activation. Based on these advantages, the hmVOA has great potential in playing an important role in HIV-1 latency and cure research.
Introduction
Since the beginning of the deadly HIV/AIDS epidemic, major research emphasis has been placed on developing effective vaccines for prevention and potent drugs to control the infection. Since HIV-1 is a retrovirus which integrates into the host cell genome and can establish viral latency, a complete cure was thought not to be possible until the case of the “Berlin patient” (1, 2). This HIV-1+ patient had undergone allogenic bone marrow (BM) transplantation from a homozygous CCR5Δ32 donor to treat acute myeloid leukemia. No HIV-1 could be detected during later years in this individual even after extensive testing thus confirming his HIV-1 negative status and a complete cure. Following this example additional cases of possible HIV-1 cure generated excitement.
Two individuals known as the “Boston patients,” (A and B) underwent allogeneic hematopoietic stem cell transplant (HSCT), in this case with wild-type CCR5+ donor cells to treat lymphoma (3). For 4.3 years after the transplant both patients were treated with ART (4). During this time no proviral DNA or replication-competent virus could be detected in PBMC, plasma or rectal tissues by using the most sensitive methods including the gold standard quantitative viral outgrowth assays (qVOA)(3, 4). After the cessation of ART however, virus rebounded within patient A by 12 weeks and patient B by 32 weeks (5). In the case of the “Mississippi baby,” ART was started 30 h after birth and continued for the first 18 months of life (6). After the cessation of ART, the “Mississippi baby” controlled viremia for 2 years and was antibody negative (7). No HIV-1 could be detected with PCR tests or qVOA using 22 million resting CD4+ T cells (6, 7), which led to the speculation that she could be another example of a complete HIV-1 cure. However, the virus eventually rebounded. Both of these cases exemplified “potential cures,” wherein all the tests including the gold standard qVOA (see below) could not detect the ultralow levels of latently infected cells thus necessitating the search for more sensitive HIV-1 latency detection methods.
Current Assays for Measuring the Latent Viral Reservoir and Limitations
Since the latent HIV-1 is transcriptionally silent and the minuscule number of latently infected cells (0.1–10 infectious units per million (IUPM) resting CD4+ T cells) are distributed over difficult to reach anatomical sites measuring the quiescent viral reservoir poses challenges (8–12). Many sensitive viral DNA, RNA or protein detection methods are currently employed to determine the viral burden (13). However, they overestimate the reservoir size as they cannot distinguish between the defective viral genomes and replication-competent virus. More recent advanced assays could simultaneously assess viral RNA, proteins and cell markers enabling the detection of viral induced cells (14–18). However, limitations remain to distinguish and accurately measure the true replication-competent latent virus.
The most accurate approach in determining the full efficacy of HIV-1 cure strategies is analytic treatment interruption (ATI) also known as monitored antiretroviral pause. However, this is impractical for routine application and poses unnecessary risk. The long-standing qVOA is considered as the “gold standard” in the HIV-1 latency field to measure the replication-competent virus and employs a co-culturing method to amplify the induced virus from rare latent cells (19–21). Serial dilution of test cells allows for quantitation, expressed as IUPM (22). Besides being time-consuming, a major drawback with this method is its tendency to underestimate the viral reservoir size since not all latent cells are induced during the assay period (23). New versions of the qVOA have been developed that use reporter cells and/or cell lines to amplify the virus with significantly increased sensitivity in a shorter time-span (13–17, 24, 25). Importantly, the stochastic aspect observed during in vitro viral activation wherein repeated stimulation of cells over time results in release of virus from previously non-responding cells (23) suggesting that approaches such as the in vivo methods described below that allow for long-term viral outgrowth may capture these late responder cells.
Non-Human Primate (NHP) Models of Latency Detection
Many aspects of HIV-1 pathogenesis and latent reservoirs distributed in different anatomical sites are difficult to directly assess if not impossible to study in a human subject. In this context, the simian immunodeficiency (SIV)-macaque model of AIDS has been extremely useful in gathering relevant data on viral persistence and latency (26–28). In NHP studies, latent virus was successfully recovered from naïve macaques that underwent adoptive transfer of resting CD4+ T cells obtained from virally suppressed SIV-infected macaques (as determined by all standard tests) undergoing intensive ART (29). These findings showed that ultralow levels of otherwise undetectable latently infected cells could be induced and detected with an in vivo system using adoptive transfer of test cells. More recently, Avalos et al. assessed viral persistence in brain macrophages of five ART-suppressed SIV-infected pig-tailed macaques using a newly developed macrophage quantitative viral outgrowth assay (Mϕ-VOA) (30). In one macaque, latency reversing agents (LRAs) ingenol-B (protein kinase C agonist) and vorinostat (HDAC inhibitor) reactivated latent viral genomes that were genetically distinct from virus circulating in the plasma. This data demonstrated the utility of the Mϕ-VOA for latency detection in macrophages.
Non-Humanized Mouse Models for Latent Viral Outgrowth (mVOA)
Immunodeficient mice permit the transplantation of human cells such as PBMCs without rejection which led to the development of the hu-PBL-SCID mouse model (31, 32). Infection of these mice with HIV-1 gives rise to viremia and the engraftment of PBMC from HIV-1+ subjects resulted in viral outgrowth. With this as a background, Metcaf Pate et al. recently developed a latent HIV-1 murine viral out growth assay (mVOA) (Figure 1) (33). The mVOA assay is based on the principle that engrafted human cells undergo xenograft-mediated expansion leading to consequent latent viral induction. Immunodeficient NSG mice were injected with large number of cells (66 million PBMC or 10–26 million resting CD4+ T cells) from 11 HIV-1+ subjects, including six elite controllers. All of these samples had undetectable viral loads by qRT-PCR (<50 copies/mL) but were positive for viral outgrowth by qVOA except for one elite controller. The engrafted mice were treated with anti-CD8 antibody to deplete the human CD8+ T cells and with anti-CD3/CD28 antibodies for the activation of T cells. Viral outgrowth was detected in all 11 patient samples in these mice, including the one elite controller negative for viral outgrowth in the qVOA. This study also evaluated latent cells from ART-suppressed SIV-infected pig-tailed macaques. NSG mice were injected with 40 million PBMC or 6.8 million resting CD4+ T cells. All inoculated mice had detectable SIV RNA in the plasma after 7 days.
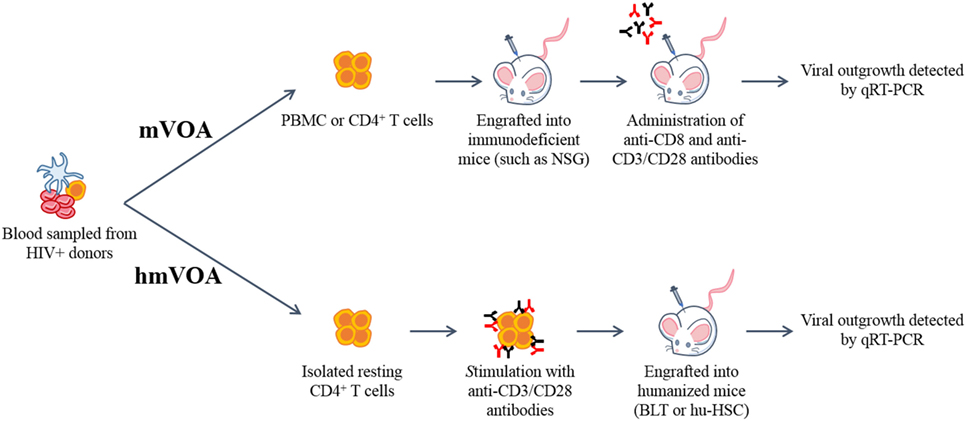
Figure 1. Schematic representation of the murine (mVOA) and humanized mouse-based viral outgrowth assay (hmVOA) for HIV-1. Previously frozen unfractionated PBMC or isolated resting CD4+ T cells obtained from HIV-1 infected donors on ART with undetectable viral loads were used for both the mVOA and hmVOA. In the mVOA, cells were either clonally expanded or directly xenografted into immunodeficient mice (such as NSG). CD8+ T cells were depleted and T cell activation was prolonged using anti-CD3/CD28 antibodies in vivo. In the hmVOA, resting CD4+ T cells were purified using PBMC and stimulated with anti-CD3/CD28 antibodies. Stimulated cells were xenografted into either hu-HSC or BLT mice. Viral outgrowth was detected by qRT-PCR.
Another recent mVOA study evaluated cells from two HIV-1 infected subjects enrolled in a PrEP program who were treated soon after infection (participants A and B treated within 10 and 12 days, respectively) (34). During the following 2-year period, participant A had undetectable HIV RNA and/or DNA in both the blood and tissue whereas participant B showed a low level of intermittent HIV RNA and/or DNA in various CD4+ T cell subsets, but not in tissue samples. The qVOA results were negative. To test these patients’ cells for latent viral detection by mVOA, 530 million peripheral CD4+ T cells from participant A (53 million per mouse, 10 mice total) and 379 million cells from participant B (50 million per mouse, eight mice total), were injected intraperitoneally into NSG mice. Approximately 5.5 weeks post-inoculation, mice were treated with anti-CD3 antibody to stimulate T cells in vivo and reactivate latent virus. One out of ten mice injected with CD4+ T cells from participant A became borderline positive (201 copies per ml) at only one time point. Terminal mouse spleen tissue sample was negative for viral detection and both RNA and DNA sequencing efforts for viral identification by an independent laboratory were unsuccessful. In contrast, three out of eight mice injected with CD4+ T cells from participant B became strongly virus positive with high viral loads (1,000, 5,000 and 11,000 copies per ml). While the sample sizes of the qVOA negative subjects are small in the above two studies, it is apparent that the mVOA could recover latent virus to a certain extent (2 out of 3 samples).
In a different twist to the mVOA, Yuan et al. utilized cells from a single aviremic subject which were positive for viral outgrowth by in vitro qVOA (0.518 IUPM) (35). First, the subject’s CD4+ T cells were clonally expanded in vitro and then split into two groups: qVOA negative or positive. NSG mice were then injected with resting or clonally expanded CD4+ T cells from each group. The clonally expanded cells that appeared qVOA positive and used to inoculate mice displayed detectable HIV-1 within 4 weeks while the qVOA negative cells used to inject mice became positive by week 10. Utilization of split portions of clonally expanded cells with a potentially uneven distribution of qVOA positive cells in the test samples, sample size of a single patient and lack of details on how many mice were used are limitations of this study.
In a recent report by Salgado et al., CD4+ T cells isolated from four HIV-1+ subjects that underwent allogenic BM stem cell transplantation to treat hematalogic malignancies were evaluated for the presence of any residual latent virus (36). Five immunodeficient NSG mice per each donor were xenografted with 10–50 million cells to detect possible viral outgrowth. However, none of these xenografted mice showed positive viral outgrowth by week 13. Since it is unlikely that these four individuals are fully cured based on previous examples like the “Boston patients,” and mVOA was not able to recover any latent virus from these, caution needs to be exercised about the reliability of mVOA for ultra-sensitive latency detection.
Several other limitations also exist for mVOA in its current form (Table 1). These include variable levels of donor cell engraftment, the need for CD8+ T cell depletion through injection of anti-CD8 antibodies and the administration of anti-CD3/CD28 antibodies for prolonged T cell activation. Most importantly, since a very large number of donor cells are xenografted, rapid GvH is a major drawback often resulting in untimely/unpredictable loss of engrafted mice thus not permitting longer assay periods to allow for the detection of delayed latent virus outgrowth.
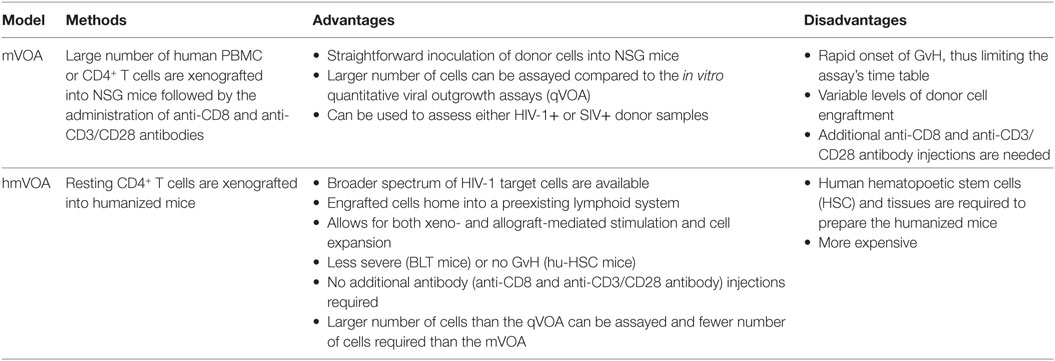
Table 1. The advantages and disadvantages of the mVOA and humanized mouse VOA (hmVOA) for HIV-1 latency detection.
Humanized Mouse Model-Based Latent Viral Outgrowth Assay (hmVOA)
New generation humanized mouse models have now become integral tools in many aspects of HIV research. The advent of highly immunodeficient mice incorporating the IL-2 receptor common gamma chain (IL2Rγc) mutation together with others, such as SCID, NOD, RAG1, or RAG2 gene mutations permitted far superior human tissue/cell engraftment (31, 37). Among these are the Rag1−/− γc−/−, Rag2−/−γc−/−, NOD/Shi-scid/γc−/− null (NOG), NOD/SCIDγc−/− (NSG), NOD.Rag1KO.IL2RγcKO (DRAG), and NOD.HLA-A2.HLA-DR4.RagKO.IL2RγcKO (DRAGA) (38, 39). Two current leading hu-mouse models are the hu-HSC and BLT mice. Hu-HSC mice are prepared by intrahepatic injection of CD34+ HSC into irradiated newborn RAG1, RAG2, NSG or NOG mice (40–43). Engraftment of these mice seeds the BM and gives rise to de novo multilineage human hematopoiesis. BLT mice are prepared by surgical implantation of human fetal liver and thymic tissue under the kidney capsule in addition to reconstitution with autologous HSC (40, 42, 44, 45). In both these models, there is de novo production of human T cells, B cells, monocytes/macrophages, dendritic cells and NK cells, as well as successful mucosal compartment engraftment (40–42, 45). While both the models permit human immune responses, the presence of an autologous human thymus in BLT mice allows for human T cell education and HLA restricted responses (40, 42–47). Thus, these hu-mice offer an excellent in vivo system for the engraftment and long-term maintenance of exogenous latently infected cells and potential outgrowth of the latent virus from these. Another potentially suitable hu-mouse model currently available employs HLA class II (DR4) transgenic mice (DRAG mice) reconstituted with HLA-matched HSC (38, 39).
In a recent study, we systematically evaluated humanized mice for developing an ultra-sensitive latent viral detection system (Figure 1) (48). First, resting CD4+ T cells from HIV-1+ subjects on ART with low, but detectable plasma HIV-1 RNA levels were tested by in vitro qVOA to measure the extent of the latent viral reservoir. These samples were positive for viral outgrowth showing a broad range of IUPM levels from 0.102 to 4.468. The CD4+ T cells either unstimulated or stimulated in vitro with PHA or anti-CD3/CD28 antibodies were injected into humanized mice. Positive viral outgrowth was observed in all of these samples within 1–3 weeks demonstrating the capacity of hu-mice to detect latently infected cells. In some of the patient samples, viral outgrowth was seen with a lesser number of input cells than in the standard qVOA. Stimulation of cells was found to give better viral outgrowth than no stimulation and anti-CD3/CD28 antibody stimulation yielded higher numbers of viable cells for testing compared to that of PHA. To determine if the hmVOA is more sensitive than conventional qVOA, five patient samples that were qVOA negative were tested using a range of CD4+ T cells (2–10 million cells/mouse) injected into mice. Of the five qVOA negative patient samples evaluated, four yielded unequivocal positive viral outgrowth in the hmVOA. The earliest time point of viral detection was 2 weeks, whereas the latest time point was 6 weeks. The negative sample did not show any viral outgrowth by 8 weeks, the last time point tested. These observations showed that the hmVOA can detect replication-competent latent HIV-1 when the standard qVOA is unable to do so thus demonstrating the higher sensitivity of this assay. The higher sensitivity of hmVOA over than the in vitro qVOA could be attributed to the provision of a more physiological in vivo setting for long-term maintenance and expansion of the engrafted cells permitting latency reactivation when compared to the short-term culture of 2 weeks employed in vitro.
Advantages of the hmVOA over the mVOA for Detecting Latent HIV-1
The hmVOA is endowed with higher sensitivity over the mVOA since it was able to detect latent HIV-1 from a higher number of qVOA negative samples and with a fewer number of input cells based on the data published so far (33–36, 48) (Table 1). The higher sensitivity of the hmVOA is likely due to the humanized mice being able to provide more optimal conditions for latent viral outgrowth for several reasons. First, hu-mice generate fresh human HIV-1 targets cells de novo (CD4+ T cells, monocytes/macrophages, and dendritic cells), including the highly susceptible immature thymocytes thus providing a much broader spectrum of susceptible cells conducive for virus outgrowth. Second, the latently infected cells have the opportunity to home into preformed human lymphoid organs where they can infect cells in situ after activation and amplifying the viral signal. Third, hu-mice provide an environment for both xenograft- and allograft-driven cell expansions. Fourth, GvH is almost non-existent in hu-HSC mice and less severe, occurring later in onset, with the BLT mice thus providing a longer time frame (2–3 months) for viral outgrowth. Furthermore, compared to mVOA, no expensive anti-CD8 or anti-CD3 antibody injections are needed after donor cell engraftment.
Limitations of mVOA and hmVOA and Future Prospects
As discussed above, the in vivo mouse-based VOA assays are more sensitive than in vitro qVOAs in detecting low levels of HIV-1 latent cells with the hmVOA being the most sensitive. However, these are limitations for these assays to be of wider use. They are not capable of a high-throughput screening, require special animal facilities and are expensive. Nevertheless, due to their higher sensitivity than any in vitro tests, they will play an important role in viral latency studies and in “kick/shock and kill” approaches toward a complete cure for HIV/AIDS. These tests will be of utmost benefit in lieu of ATI in guiding future curative drug development. Further improvements can be foreseen in the hmVOA and mVOA models with additional research. One approach would be to increase the sensitivity by using HIV-1 LRAs either alone or in various combinations. Thus far, the hmVOA has primarily focused on HIV-1 latency in CD4+ T cells. With the recent attention on viral latency in other cell types such as macrophages and work done with SIV latency detection, it is apparent that hmVOA can also be put to good use in evaluating viral outgrowth from HIV-1 latent macrophages as well. Streamlining the hu-mouse generation on a larger scale with increased efficiency should help reduce the overall costs of hmVOA permitting its wider application.
Author Contributions
Both KS and RA contributed equally in writing and editing this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Laurén Kinner-Bibeau for generating artwork in Figure 1.
Funding
Work on hmVOA research in RA’s laboratory is supported by NIH, USA grant RO1 AI120021.
References
1. Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature (2012) 487:482–5. doi:10.1038/nature11286
2. Krim M, Johnston R. AIDS: the final chapter? AIDS Res Hum Retroviruses (2014) 30:5–7. doi:10.1089/aid.2013.1503
3. Henrich TJ, Hu Z, Li JZ, Sciaranghella G, Busch MP, Keating SM, et al. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis (2013) 207:1694–702. doi:10.1093/infdis/jit086
4. Henrich TJ, Hanhauser E, Sirignano M, Davis B, Lee TH, Keating S, et al. In depth investigation of peripheral and gut HIV-1 reservoirs, HIV-1 specific cellular immunity, and host microchimerism following allogeneic hematopoetic stem cell transplantation. 7th IAS Conference on HIV Pathogenesis, Treatment and Prevention. Kuala Lumpur, Malaysia (2013).
5. Hyden EC. Hopes for HIV cure in ‘Boston patients’ dashed. Nat News (2013). doi:10.1038/nature.2013.14324
6. Persaud D, Gay H, Ziemniak C, Chen YH, Piatak M Jr, Chun TW, et al. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N Engl J Med (2013) 369:1828–35. doi:10.1056/NEJMoa1302976
7. Shah SK, Persaud D, Wendler DS, Taylor HA, Gay H, Kruger M, et al. Research on very early ART in neonates at risk of HIV infection. Lancet Infect Dis (2014) 14:797. doi:10.1016/S1473-3099(14)70893-X
8. Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol (2006) 80:6441–57. doi:10.1128/JVI.00591-06
9. Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med (1999) 5:512–7. doi:10.1038/8394
10. Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med (2003) 9:727–8. doi:10.1038/nm880
11. Bruner KM, Hosmane NN, Siliciano RF. Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol (2015) 23:192–203. doi:10.1016/j.tim.2015.01.013
12. Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog (2013) 9:e1003174. doi:10.1371/journal.ppat.1003174
13. Henrich TJ, Deeks SG, Pillai SK. Measuring the size of the latent human immunodeficiency virus reservoir: the present and future of evaluating eradication strategies. J Infect Dis (2017) 215:S134–41. doi:10.1093/infdis/jiw648
14. Prasad VR, Kalpana GV. FISHing out the hidden enemy: advances in detecting and measuring latent HIV-infected cells. MBio (2017) 8:e1433–1417. doi:10.1128/mBio.01433-17
15. Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Charlebois R, et al. Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell Host Microbe (2016) 20:368–80. doi:10.1016/j.chom.2016.07.015
16. Martrus G, Niehrs A, Cornelis R, Rechtien A, Garcia-Beltran W, Lutgehetmann M, et al. Kinetics of HIV-1 latency reversal quantified on the single-cell level using a novel flow-based technique. J Virol (2016) 90:9018–28. doi:10.1128/JVI.01448-16
17. Grau-Exposito J, Serra-Peinado C, Miguel L, Navarro J, Curran A, Burgos J, et al. A novel single-cell FISH-flow assay identifies effector memory CD4(+) T cells as a Major Niche for HIV-1 transcription in HIV-infected patients. MBio (2017) 8:e01433-17. doi:10.1128/mBio.00876-17
18. Descours B, Petitjean G, Lopez-Zaragoza JL, Bruel T, Raffel R, Psomas C, et al. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature (2017) 543:564–7. doi:10.1038/nature21710
19. Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature (1997) 387:183–8. doi:10.1038/387183a0
20. Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol (2005) 304:3–15. doi:10.1385/1-59259-907-9:003
21. Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog (2013) 9:e1003398. doi:10.1371/journal.ppat.1003398
22. Rosenbloom DI, Elliott O, Hill AL, Henrich TJ, Siliciano JM, Siliciano RF. Designing and interpreting limiting dilution assays: general principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect Dis (2015) 2:ofv123. doi:10.1093/ofid/ofv123
23. Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell (2013) 155:540–51. doi:10.1016/j.cell.2013.09.020
24. Sanyal A, Mailliard RB, Rinaldo CR, Ratner D, Ding M, Chen Y, et al. Novel assay reveals a large, inducible, replication-competent HIV-1 reservoir in resting CD4(+) T cells. Nat Med (2017) 23:885–9. doi:10.1038/nm.4347
25. Fun A, Mok HP, Wills MR, Lever AM. A highly reproducible quantitative viral outgrowth assay for the measurement of the replication-competent latent HIV-1 reservoir. Sci Rep (2017) 7:43231. doi:10.1038/srep43231
26. Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature (2014) 512:74–7. doi:10.1038/nature13594
27. Dinoso JB, Rabi SA, Blankson JN, Gama L, Mankowski JL, Siliciano RF, et al. A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J Virol (2009) 83:9247–57. doi:10.1128/JVI.00840-09
28. Policicchio BB, Pandrea I, Apetrei C. Animal models for HIV cure research. Front Immunol (2016) 7:12. doi:10.3389/fimmu.2016.00012
29. Okoye A. Early Antiretroviral Therapy Limits Viral Reservoir in SIV-Infected Macaques, in NIH Meeting: Strategies for an HIV Cure. Besthesda, MD (2014).
30. Avalos CR, Abreu CM, Queen SE, Li M, Price S, Shirk EN, et al. Brain macrophages in simian immunodeficiency virus-infected, antiretroviral-suppressed macaques: a functional latent reservoir. MBio (2017) 8(4):e1186–1117. doi:10.1128/mBio.01186-17
31. Shultz LD, Brehm MA, Bavari S, Greiner DL. Humanized mice as a preclinical tool for infectious disease and biomedical research. Ann N Y Acad Sci (2011) 1245:50–4. doi:10.1111/j.1749-6632.2011.06310.x
32. Mosier DE. Viral pathogenesis in hu-PBL-SCID mice. Semin Immunol (1996) 8:255–62. doi:10.1006/smim.1996.0032
33. Metcalf Pate KA, Pohlmeyer CW, Walker-Sperling VE, Foote JB, Najarro KM, Cryer CG, et al. A murine viral outgrowth assay to detect residual HIV type 1 in patients with undetectable viral loads. J Infect Dis (2015) 212:1387–96. doi:10.1093/infdis/jiv230
34. Henrich TJ, Hatano H, Bacon O, Hogan LE, Rutishauser R, Hill A, et al. HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: an observational study. PLoS Med (2017) 14:e1002417. doi:10.1371/journal.pmed.1002417
35. Yuan Z, Kang G, Lu W, Li Q. Reactivation of HIV-1 proviruses in immune-compromised mice engrafted with human VOA-negative CD4+ T cells. J Virus Erad (2017) 3:61–5.
36. Salgado M, Kwon M, Galvez C, Nijuis M, Vilaplana C, Bandera A, et al., editors. Murine model to predict viral rebound in HIV-1+ allotransplanted subjects. Conference on Retroviruses and Opportunistic Infections (CROI). Seattle, WA (2017).
37. Berges BK, Rowan MR. The utility of the new generation of humanized mice to study HIV-1 infection: transmission, prevention, pathogenesis, and treatment. Retrovirology (2011) 8:65. doi:10.1186/1742-4690-8-65
38. Akkina R, Allam A, Balazs AB, Blankson JN, Burnett JC, Casares S, et al. Improvements and limitations of humanized mouse models for HIV research: NIH/NIAID “Meet the Experts” 2015 Workshop Summary. AIDS Res Hum Retroviruses (2016) 32(2):109–19. doi:10.1089/AID.2015.0258
39. Allam A, Majji S, Peachman K, Jagodzinski L, Kim J, Ratto-Kim S, et al. TFH cells accumulate in mucosal tissues of humanized-DRAG mice and are highly permissive to HIV-1. Sci Rep (2015) 5:10443. doi:10.1038/srep10443
40. Akkina R. New generation humanized mice for virus research: comparative aspects and future prospects. Virology (2013) 435:14–28. doi:10.1016/j.virol.2012.10.007
41. Ito R, Takahashi T, Katano I, Ito M. Current advances in humanized mouse models. Cell Mol Immunol (2012) 9:208–14. doi:10.1038/cmi.2012.2
42. Traggiai E, Chicha L, Mazzucchelli L, Bronz L, Piffaretti JC, Lanzavecchia A, et al. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science (2004) 304:104–7. doi:10.1126/science.1093933
43. Lan P, Tonomura N, Shimizu A, Wang S, Yang YG. Reconstitution of a functional human immune system in immunodeficient mice through combined human fetal thymus/liver and CD34+ cell transplantation. Blood (2006) 108:487–92. doi:10.1182/blood-2005-11-4388
44. Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol (2012) 12:786–98. doi:10.1038/nri3311
45. Wege AK, Melkus MW, Denton PW, Estes JD, Garcia JV. Functional and phenotypic characterization of the humanized BLT mouse model. Curr Top Microbiol Immunol (2008) 324:149–65. doi:10.1007/978-3-540-75647-7
46. Melkus MW, Estes JD, Padgett-Thomas A, Gatlin J, Denton PW, Othieno FA, et al. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat Med (2006) 12:1316–22. doi:10.1038/nm1431
47. Seung E, Tager AM. Humoral immunity in humanized mice: a work in progress. J Infect Dis (2013) 208(Suppl 2):S155–9. doi:10.1093/infdis/jit448
48. Charlins P, Schmitt K, Remling-Mulder L, Hogan LE, Hanhauser E, Hobbs KS, et al. A humanized mouse-based HIV-1 viral outgrowth assay with higher sensitivity than in vitro qVOA in detecting latently infected cells from individuals on ART with undetectable viral loads. Virology (2017) 507:135–9. doi:10.1016/j.virol.2017.04.011
Keywords: HIV-1 latent viral outgrowth assay using humanized mice, humanized mouse-based HIV-1 latency outgrowth assay, comparison of quantitative viral outgrowth assays with humanized mouse-based viral outgrowth assay, comparison of mVOA with humanized mouse-based viral outgrowth assay, non-human primate-based latent simian immunodeficiency viral outgrowth assay, sensitivity of humanized mouse-based viral outgrowth assay over mVOA, ultra-sensitive HIV-1 latent viral outgrowth assay in hu-mice, mouse-based HIV-1 viral outgrowth assays
Citation: Schmitt K and Akkina R (2018) Ultra-Sensitive HIV-1 Latency Viral Outgrowth Assays Using Humanized Mice. Front. Immunol. 9:344. doi: 10.3389/fimmu.2018.00344
Received: 14 December 2017; Accepted: 07 February 2018;
Published: 05 March 2018
Edited by:
Jeffrey K. Actor, University of Texas Health Science Center at Houston, United StatesReviewed by:
Mangala Rao, United States Military HIV Research Program, United StatesJames Di Santo, Institut Pasteur, France
Copyright: © 2018 Schmitt and Akkina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ramesh Akkina, akkina@colostate.edu
 Kimberly Schmitt
Kimberly Schmitt Ramesh Akkina
Ramesh Akkina