- 1GSK, Research and Development Center, Siena, Italy
- 2Linköping University, Linköping, Sweden
- 3DTU Nanotech, Technical University of Denmark, Copenhagen, Denmark
- 4GSK, Research and Development Center, Rockville, MD, United States
Despite advancements in immunotherapeutic approaches, influenza continues to cause severe illness, particularly among immunocompromised individuals, young children, and elderly adults. Vaccination is the most effective way to reduce rates of morbidity and mortality caused by influenza viruses. Frequent genetic shift and drift among influenza-virus strains with the resultant disparity between circulating and vaccine virus strains limits the effectiveness of the available conventional influenza vaccines. One approach to overcome this limitation is to develop a universal influenza vaccine that could provide protection against all subtypes of influenza viruses. Moreover, the development of a novel or improved universal influenza vaccines may be greatly facilitated by new technologies including virus-like particles, T-cell-inducing peptides and recombinant proteins, synthetic viruses, broadly neutralizing antibodies, and nucleic acid-based vaccines. This review discusses recent scientific advances in the development of next-generation universal influenza vaccines.
Introduction
Seasonal influenza viruses circulate worldwide, spread easily from person to person, and result in the hospitalization of three to five million individuals worldwide each year (1, 2). These infections are responsible for 250,000–500,000 deaths, mainly among those with immature or compromised immunity, e.g., young children, elderly adults, and critically ill patients (2). However, all age groups can be affected, and the impact can increase significantly with an emergent human influenza-virus strain during a pandemic (3). Influenza viruses are constantly evolving through genome mutation and reassortment. During the past 100 years, new emergent influenza-virus strains have regularly appeared in human populations (“Spanish flu” in 1918 caused by the H1N1 subtype, “Asian flu” in 1957 by H2N2, “Hong Kong flu” in 1968 by H3N2, “Russian flu” in 1977 by H1N1, and “swine flu” in 2009 by H1N1). In addition, limited outbreaks of avian influenza strains in humans threaten the evolution of one or more of these viruses to the point of sustained human-to-human transmission, for example, H5N1 (“bird flu”), H7N9, H5N6 as well as virus variants transmitted from pigs to human (H1N1v, H1N2v, and H3N2v) (4–8). Pandemic influenza has claimed millions of lives globally; the 1918–1920 H1N1 pandemic alone claimed 50–100 million lives (4, 9).
The genome of the influenza virus consists of 8 single-stranded RNA segments encoding 11 proteins, including the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA). The human influenza virus is classified into three distinct types A, B, and C, on the basis of major antigenic differences. Influenza A and B viruses are responsible for annual human epidemics, whereas the influenza C virus is known to infect both humans and pigs and causes very mild upper respiratory tract disease in humans (10, 11). The influenza A virus is classified into 18 HA (H1–H18) and 11 NA (N1–N11) subtypes on the basis of HA and NA glycoproteins (3, 9, 12). On the basis of antigenic properties and structural features, influenza A virus HA subtypes can be further classified into groups 1 (H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17, and H18) and 2 (H3, H4, H7, H10, H14, and H15), which comprise different clades (A clade is a group of influenza viruses share homologous features and evolved from a common ancestor) (13, 14). Only a single subtype of influenza B virus has been identified, and this subtype comprises two distinct antigenic lineages (B/Victoria and B/Yamagata) that diverged more than 40 years ago and co-circulate at variable levels in humans (3, 15).
Vaccination is an effective approach for the control and prevention of influenza. Currently, trivalent inactivated-virus (TIV) vaccines against seasonal influenza viruses are the most frequently used influenza vaccines with a steady migration to tetravalent or quadrivalent vaccines (QIV). TIV vaccines are composed of three influenza-virus strains (2 A subtypes, H3N2, H1N1, and 1 B type) selected primarily on the basis of forecasted prevalence during the targeted influenza season. QIV vaccines include the second B lineage (16). TIV vaccines come in three different formulations; the whole virus, split virus, and subunit. Whole-virus vaccines are prepared from embryonated chicken eggs, inoculated with virus, followed by chemical inactivation and purification steps. Split-virus vaccines are prepared by treatment of influenza-virus particles by diethyl ether or detergent (e.g., ammonium deoxycholate), which dissociates the viral lipid envelope exposing all viral proteins (17, 18). Subunit vaccines contain HA and NA proteins and are prepared by applying further purification steps with detergents or diethyl ether (19, 20). A recombinant HA (rHA)-based subunit vaccine has been approved recently; and this vaccine showed higher seroconversion rates in healthy adults including elderly adults, compared with a non-recombinant TIV vaccine (21).
Inactivated vaccines primarily induce protective antibodies against epitopes on HA. Split and subunit formulations are used more frequently than the other formulations and both induce comparable immunity. Whole-virus formulation has been less preferred because of a potential association with increased reactogenicity (22). In the USA, inactivated vaccines are approved from 6 months of age, and rHA formulations are approved for those 18 years of age and older.
Live-attenuated influenza vaccines (LAIV) mimic aspects of natural influenza-virus infection, but without virus pathogenicity, and induce both humoral and cell-mediated immunity (23). Intranasal administration of LAIV induces local mucosal immunity (24) by inducing the secretion of IgA. LAIV is approved in the USA for use in children and adults between 2 and 49 years old (23). However, recently, the Center for Disease Control and Prevention (CDC) Advisory Committee on Immunization Practices (ACIP) recommended that LAIV should not be used during 2017–2018 flu season (25). The ACIP committee made this recommendation because of the lower efficacy of LAIV against A(H1N1)pdm09 viruses during the 2013–2014 and 2015–2016 seasons. Small and Cronin (26) argued that the decision of ACIP was based on Test Negative Case Controlled (TNCC) study design which measures direct rather than indirect protection and just an observational approach (25, 27). Potential causes of reduced effectiveness of A(H1N1)pdm09 in LAIV are evidenced by lower replication in alveolar cell lines and reduced binding to sialic acid receptors (receptors for influenza-virus binding) (28).
The benefits of QIV over TIV are encouraging, and an increasing proportion of influenza vaccination programs are using QIV (29). Nevertheless, both TIV and QIV vaccines require reformulation with new influenza strains each influenza season, and their effectiveness is not guaranteed because it is primarily dependent on the match between the forecasted strains in the vaccines and the actual prevalent strains in circulation.
Is there a need for a Universal Influenza Vaccine?
The leading challenge in influenza prevention and treatment is identifying effective strategies to deal with the rapidity in which the virus can evolve to evade its host’s immune system or develop resistance to drug treatments. Influenza-virus evolution is typically considered in terms of antigenic drift—the occurrence of minor changes in the virus genotype within the same virus type—and antigenic shift—emergence of new and potentially pandemic strains by the reassortment of viral genomes (13, 30, 31).
Seasonal antigenic drifts usually consist of minor amino acid mutations in the HA globular head domain that may result in some changes to the pattern of glycosylation in this domain (32, 33). These changes in the HA-glycosylation pattern affect the infectivity of the virus and its ability to escape from antibodies elicited by previous strains or vaccines (33). As a result, current influenza vaccines, which provide antibody-dominated subtype-specific protection against influenza viruses, need to be updated and produced every year before each influenza season (fall to winter period in the northern hemisphere) because of mismatches between the vaccine strains and the prevalent circulating virus subtypes.
The adult human population has some levels of cross-protective antibodies against circulating seasonal influenza strains due to previous virus exposures or vaccinations, therefore, developing only mild disease symptoms upon infection (34, 35). By contrast, the absence of preexisting immunity to an emerging pandemic strain can lead to severe pulmonary infections and death. Infants are particularly at risk as they are immunologically naïve to influenza and may develop serious symptoms even from seasonal strains, as soon as the protection from maternal antibody wanes.
Adjuvants are formulated with the antigenic component of the vaccine to increase immunogenicity. Seasonal and pandemic influenza vaccines adjuvanted with oil-in-water emulsions AS03 or MF59, in comparison with non-adjuvanted vaccines, elicit higher titers of neutralizing antibodies with higher affinities and broader cross-reactive specificities to other influenza types and promote the persistence of long-lasting memory B cells in subjects from varied age groups (36–39).
Compared with existing cell-derived and recombinant influenza vaccines, conventional egg-based influenza vaccines are usually effective and have some limitations: a complex and slow production system, limitations in production capacity (production of single dose needed two eggs), allergic responses to egg components in some individuals, and weak cross-reactivity to other influenza subtypes (40–43). Studies have shown that cultivation of influenza virus in fertilized chicken eggs often results in adapting mutations in HA and that can alter virus antigenicity and may sometimes decrease efficacy of influenza vaccines (44–46). Mammalian cell-derived influenza vaccine provide several advantages over egg-based vaccines including influenza virus propagated in mammalian cell culture system remain unchanged, provide better or comparable protection, fast production and does not require extensive advance planning, availability of controlled production system involving bio-reactors, higher yield, faster production cycle and production can be easily scaled up by adding bioreactors (43, 47, 48). A recent study has shown comparable yield of virus from 1,000-l bioreactor, 12,000 roller bottles or 30,800 chicken eggs (47). These collective advantages increase cost-effectiveness of cell-based influenza vaccines. Despite several benefits, cell culture system also have some limitations including, scaling-up different cell lines is biggest challenge, obligation of expensive new facilities and extensive adventitious virus testing required (43, 49). These collective constraints call for new design and development of unique universal influenza vaccines that could provide long-lasting immunity against all subtypes of influenza viruses, and significantly reduce the disease burden associated with influenza-virus infections.
Properties of Target Antigens for Vaccine Development
Antibodies that neutralize influenza-virus entry into host epithelial cells typically target HA, the main surface glycoprotein of the virion. HA is composed of globular and stalk domains and is expressed as a trimer at the surface of the virion. At the initial stage of infection, when influenza virus enters the respiratory tract, HA binds to sialic acid residues present on the surface of epithelial cells and allows the virus to be engulfed by the cells (50). The globular domain is highly variable across subtypes, whereas the stalk domain is much more conserved. Hence, the stalk domain is a potential target for the development of a universal influenza vaccine (51, 52). However, HA-head-specific antibodies have a greater neutralizing capacity than cross-reactive HA-stalk-specific antibodies, which are found at very low frequency (53, 54).
Neuraminidase is a tetrameric glycoprotein present on the surface of influenza viruses (55, 56). NA is important at the pre-infection, post-infection, and re-infections stages. NA is involved in the release of newly produced viruses from host cells and prevents the aggregation of virus particles by cleavage of sialic acids from respiratory tract mucins (57–59). NA may also participate in the fusion of viral and cell membranes (60) and facilitates budding of new virions by restricting their aggregation (60). NA is generally less immunogenic and lacking monodominant properties and therefore a less attractive target than HA (61). Despite this, NA has been used as a vaccine candidate in various vaccine formulations and as a target of various antiviral drugs, because of a lower rate of antigenic drift than HA (57, 59, 62–65).
Apart from HA and NA surface glycoproteins, the matrix proteins 1 and 2 (M1 and M2, respectively), are encoded by influenza-virus genes with partially overlapping reading frames (66, 67). M1 is a major constituent of the viral capsids and M2 functions as a proton channel in viral envelope. Studies suggest that the amino acid sequence of M2 extracellular domain (M2e) is conserved in influenza A viruses (IAV) and is, therefore, a target for the development of a universal influenza vaccine (68, 69). M2e-containing vaccines have shown encouraging results in animal models (68–71). Antibody responses to M2e protein provide protection by (1) binding to virus-infected cells and mediating effectors functions by antibody-dependent cell cytotoxicity, (2) killing infected cells by activating a complement cascade or by recruiting cells of the innate immune system, (3) preventing the release of virus particles by binding to the cell surface, and (4) phagocytosis of viral particles via Fcγ receptors (66, 70, 71).
Nucleoprotein (NP) is an influenza-virus protein that is associates with viral RNA and essential for viral assembly (3). NP is highly conserved across influenza subtypes and is a potential antigen for a universal influenza vaccine (72). After infection, NP is the main antigen recognized by influenza-specific cytotoxic CD8+ T lymphocytes (CTLs) that have the potential to kill virus-infected cells (73).
Virus-Like Particles (VLPs)
Virus-like particles are non-infectious particles of protein multimers and are devoid of genetic material (Figure 1A). VLPs mimic certain aspects of the conformational, structural and antigenic properties of native influenza viruses, making VLPs a useful platform for vaccine development (74, 75). The development of certain universal influenza vaccines have utilized the VLP platform because VLPs have self-assembly properties, VLPs can comprise more than one protein or protein chimeras (cVLPs) and produced in a heterologous expression system, and VLPs can be formulated with adjuvants (74, 76). VLPs activate innate immunity by stimulating antigen presenting cells and this can lead to the induction of effective B- and T-cell immunity specific to those antigens present in the VLP (75, 77–79). It is reported that highly organized and repetitive protein epitopes can directly activate B-cells by cross-linking B-cell receptors (80). Furthermore, VLPs have also been shown to provoke CD8+ T-cells mediated immune responses through cross-presentation mechanism (81).
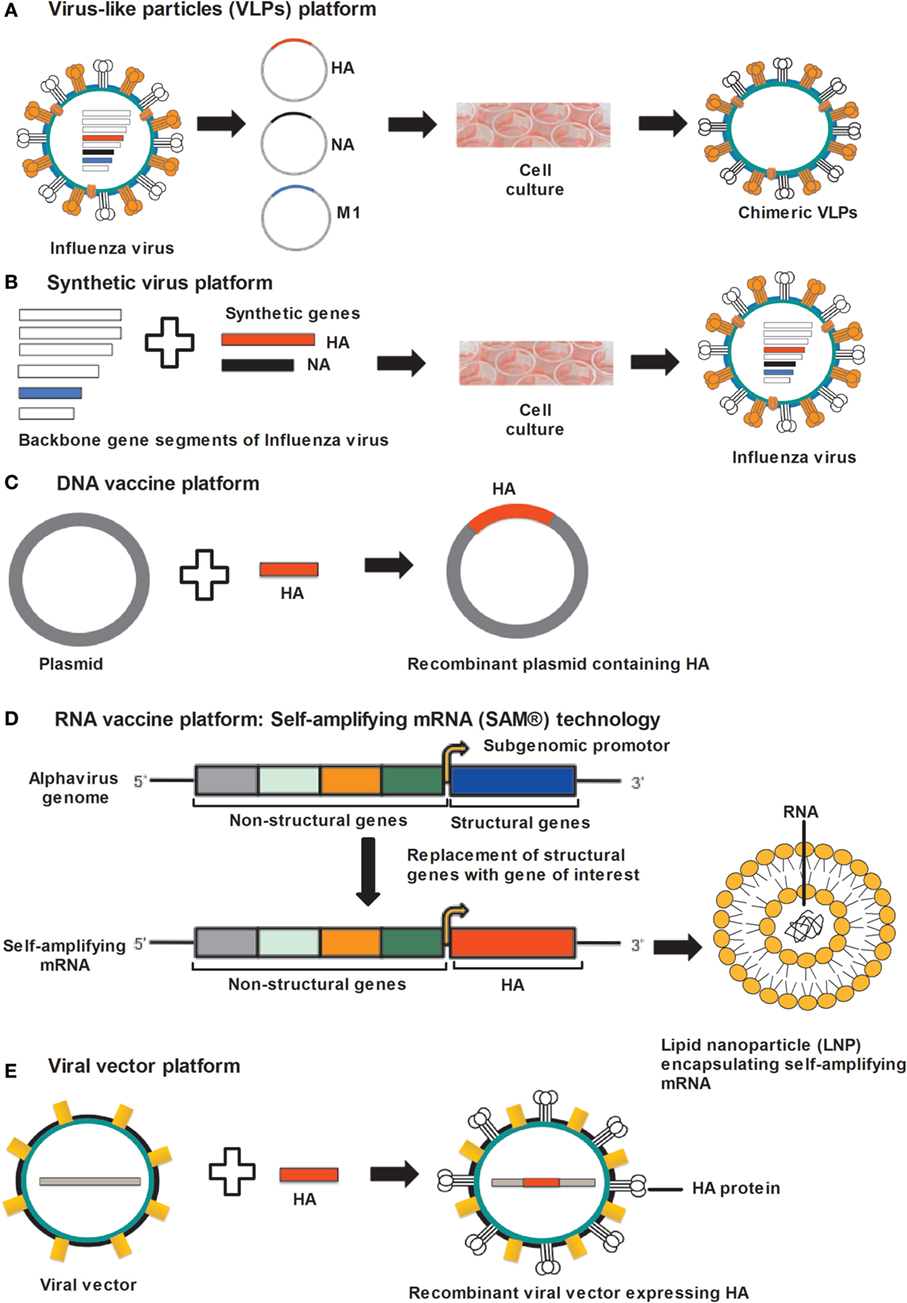
Figure 1. Approaches for universal influenza vaccine development. (A) VLP platform: VLPs produced by cloning of HA, NA, and M1 gene sequences of influenza virus into the expression vector followed by transfection in to insect cells. Co-expression of HA, NA, and M1 proteins allows self-assembly of VLPs. (B) Synthetic-virus platform: MDCK cells are transfected with plasmid DNA encoding influenza-virus backbone genes and error-free HA and NA gene segments, synthesized by an enzymatic and cell-free assembly technique. After transfection, vaccine viruses are rescued from MDCK cells. (C) DNA-vaccine platform: influenza genes encoding antigenic proteins are inserted into a DNA plasmid and delivered to the host cells, where DNA vaccines express antigenic protein. (D) RNA-vaccine platform: self-amplifying RNA expressed from an alphavirus genome in which structural genes are replaced by genes supporting the amplification of the RNA and the gene encoding the antigen. Self-amplifying mRNA (SAM) composed of a 5′ cap, genes encoding non-structural genes (NSP 1–4), a subgenomic promoter, the antigen-encoding gene, and a 3′ poly(A) tail. Diagrammatic representation of a lipid nanoparticle (LNP) encapsulating SAM. (E) Viral-vector platform: viral vector-based influenza vaccine uses a non-influenza “carrier” virus to express antigenic protein. Influenza genes encoding HA protein are placed in to the carrier virus vector to express HA protein on the virus surface. Abbreviations: HA, hemagglutinin; NA, neuraminidase; M1, matrix protein 1; VLP, virus-like particle; MDCK, Madin–Darby kidney cells.
Virus-like particles have been used in strategies for presenting epitopes from numerous different virus subtypes and engineered epitopes derived from conserved regions of viral proteins (82–85). Most VLP-based vaccines are formulated with individual VLPs containing HA, NA, or matrix proteins, or a combination of these proteins (82, 86, 87). However, VLPs containing both HA and NA tend to predominantly induce HA-specific antibodies (41) due to the characteristic immunodominance associated with HA (88). In a recent mouse study, Schwartzman et al. evaluated a vaccine consisting of a mixture of HA-derived VLPs, from influenza subtypes H1 (H1N1; A/South Carolina/1/1918), H3 (H3N8; A/pintail/Ohio/339/1987), H5 (H5N1; A/mallard/Maryland/802/2007), and H7 (H7N3; A/Environment/Maryland/261/2006) (82). After intranasal vaccination, mice were protected against lethal influenza-virus challenge from homologous (same strains as in the vaccine; H1N1 and H7N1), intrasubtypic (antigenically different strains of the same subtypes; H5N1 and H7N9), and heterosubtypic (subtypes not included in the vaccine; H2N1, H6N1, H10N1, and H11N1) (82). Hence these results suggested that this approach was suitable for developing an effective clinical vaccine against currently circulating influenza strains and potential pandemic influenza strains. In other studies, the highly conserved HA-stalk domain was used in VLPs to induce broadly cross-reactive protective immunity (84, 88, 89). In one study, VLP formation was aided by co-expression of the HA-stalk domain with the HIV gag protein in mammalian cells (84). VLPs composed of M1 and NA from A/Puerto Rico/8/1934 (H1N1) induced cross-reactive antibody responses to an H3N2 subtype and substantial levels of protection against homologous and heterologous A subtypes, although loss of body weight was observed after lethal challenge with H3N2 (65).
Although viral-surface-protein-based VLPs have appeared promising tool for developing universal vaccine, some of the strategies have also included M2e protein. M2e is a very promising target for universal vaccine development because of the conserved nature of the protein. M2e has been included in VLPs as a genetically engineered fusion protein (e.g., with the hepatitis B virus core particle (HBc), phage derived VLPs, or with human papillomavirus L1), and vaccines from these VLPs have been shown to induce high levels of M2e-specific antibodies in mice (69, 90, 91). M2e has also been included in VLPs as a tandem repeat of five M2e variants from human, swine and avian IAV (the M2e5x vaccine) (85, 92). In mice, M2e5x protected against a lethal challenge from distinct IAV (H3N2 or H5N1) (92). Finally, in a recent study, a vaccine containing a combination of three M2e, NP, and HBc VLPs induced potent humoral and cell-mediated immunity (5).
Although the abovementioned VLP strategies have shown promise in animal models, some of the vaccine formulations have required adjuvants to enhance VLP immunogenicity. Some of these adjuvants have been based on toll-like receptor (TLR) ligands. TLRs are structurally conserved molecules that recognize ligands of microbial origins. Engagement of TLRs on innate cells results in the production of pro-inflammatory cytokines and chemokines, and an enhanced ability to eliminate the pathogens (93). Wang et al. developed chimeric VLPs containing A/Puerto Rico/8/1934 influenza virus HA and M1 and a modified flagellin protein of Salmonella that engages TLR5 (94). Compared with VLPs containing only HA and M1 protein, cVLPs containing TLR5 ligand flagellin was more immunogenic and protected 67% mice against lethal challenge with a heterosubtypic A/Philippines (H3N2) influenza-virus strain (94).
The advantages of VLP-based vaccines are that the immune system of the host recognizes VLPs in a similar way to the original virus particles, and chimeric VLPs induce highly effective cross-reactive heterosubtypic immune responses (94). The existence of several licensed prophylactic VLP-based vaccines (e.g., for HBV and for HPV) (95), further suggest that a VLP-based approach is a promising approach for the development of a universal influenza vaccine.
Broadly Reactive Antibody- and T-Cell-Inducing Strategies
Antibody- and T-cell-mediated immune responses are the keys components of adaptive immune system. B cells are the source of antibodies and antibody-based immunotherapy is a very efficient approach in the treatment of various diseases caused by microbial infections (96). Most current seasonal and pandemic influenza vaccines induce high-titers of functional and neutralizing antibodies against HA and NA proteins.
Because the HA-head domain tends to contain epitopes that are immunodominant over those in the stalk, Krammer et al. investigated a strategy in which three chimeric antigens comprising a stalk domain from H1 fused with a head domain from three different subtypes, respectively, were injected sequentially in mice (97). The first vaccination with cH6/1 HA (combination of H6 head and H1 stalk) induced a very week response against the head and stalk domains, whereas a second vaccination with cH5/1 HA (combination of H5 head and H1 stalk) boosted the antibody responses against the stalk domain and induced only primary responses against the head domain (97, 98). A third vaccination with cH8/1 HA (combination of H8 head and H1 stalk) again boosted the intensity of the immune response against the stalk domain and induced a primary response against the head domain. Repeated vaccinations with chimeric HA constructs containing the same stalk domain but different heads, induced stalk-directed antibodies that mediated heterotypic immunity (98). These initial results with IAV group 1 stalk chimeric HA vaccines have since been replicated with IAV group 2 and B chimeric HA vaccines (99, 100). In addition, an inactivated split-virion vaccine adjuvanted with AS03 derived from a recombinant virus expressing an IAV group 1 chimeric HA has been evaluated in mice and shown to induce protective H1 stalk-reactive antibodies (101). Ferrets are excellent experimental animal models for the investigation of influenza-virus pathogenicity and immunobiology because of their susceptibility to influenza-virus infection and ability to develop disease symptoms similar to humans (102–104). Sequential immunization studies with chimeric HA (generated by exchanging head domains and retaining same HA-stalk domain), successfully induced broadly reactive antibody responses in ferrets (104–106).
An alternative strategy based on broadly reactive monoclonal antibodies could confer cross-protective and long-lasting immunity against influenza-virus infections. Studies on monoclonal antibodies have shown that conserved regions exist within the HA head domain. Whittle et al. isolated a human monoclonal antibody, CH65, directed against the globular head region that was able to neutralize the infectivity of about 30 H1N1 strains (107). Lee et al. demonstrated that an antibody fragment, derived from the S139/1 monoclonal IgG antibody, targets highly conserved residues in the receptor binding site of the head domain. This antibody fragment neutralized H1N1 and H3N2 strains. However, the parental antibody neutralized many more strains from H1, H2, H13, and H16 subtypes suggesting that the avidity associated with bivalent interactions of the antibody and HA molecules contributed to enhancing cross-reactivity (108).
As discussed earlier, the stalk region of HA is highly conserved and stalk-specific neutralizing antibodies cross-react with different virus subtypes. Okuno et al. isolated a monoclonal antibody, C179, from A/Okuda/1957 (H2N2)-immunized mice (51). C179 inhibited the fusion activity of HA and neutralized influenza subtypes H1, H2, H5, and H6 (51, 52). Throsby et al. isolated a monoclonal antibody, CR6261, from human IgM+ memory B cells that neutralized antigenically diverse H1, H5, H6, H8, and H9 influenza subtypes (109). Corti et al. isolated a monoclonal antibody, F16, by screening human plasma cells from peripheral blood. This monoclonal antibody was able to recognize HA of all 16 subtypes of influenza virus and efficiently neutralized groups 1 and 2 viruses (110). Recently, Dreyfus et al. isolated from a phage display library, a monoclonal antibody—CR9114—that recognized the HA-stalk domain of antigenically distinct influenza A and B viruses (111). This antibody protected against lethal challenge from both influenza A and B viruses (111). In most of the broadly cross-reactive stalk antibodies identified, the VH1-69 gene of the heavy-chain variable region is responsible for binding to the HA stalk. Recently, Pappas et al. identified the pathway for the generation of VH1-69 antibody and developed broadly neutralizing antibodies by mutation (112).
Innovative approaches based on optimal antigen design, have been implemented for the induction of cross-protective antibodies. In these approaches, antigens contain only cross-protective epitopes. Recently, Giles et al. designed vaccine antigens based on HA of H5N1 subtype by a computational approach, computational optimized broadly reactive antigen, and these antigens induced broadly reactive antibodies in mice and ferrets after challenge with H5N1 (113, 114). In another approach, mutations were incorporated in HA2 (HA1 and HA2 are subunits of HA, enzymatic cleavage product of HA0 precursor protein) of HA antigen and expressed in Escherichia coli. The resulting protein was highly immunogenic and protected mice against a lethal challenge by homologous and intrasubtypic viral strains (115).
These promising studies may be helpful for the development of an antibody-based first line of defense in the case of an emerging pandemic.
Historically, the induction of neutralizing antibodies has been the main focus in the development of influenza vaccines. However, recent data suggest that the induction of T cells with cross-reactive properties is highly important (116–118). These T cells are accountable for providing heterosubtypic immunity, wherein memory T cells generated against conserved epitopes of previous influenza virus can cross-react against multiple influenza subtypes. Both CD4+ and CD8+ T cells induce heterotypic immunity against internal and surface proteins (119–125). CTLs are the main effector cells that take part in antiviral immunity. Numerous studies have shown that CTLs orchestrate heterosubtypic effector functions against influenza virus infected cells by direct cytotoxic inhibitory mechanisms (101–105). A recent study by Sridhar et al. demonstrated a significant role of cross-protective CD8+ T cells, specific for conserved core protein epitopes, in individuals lacking cross-neutralizing antibodies (126). A significant positive correlation was found between higher frequencies of preexisting CD8+ T cells to conserved epitopes and the low grade of clinical illness among healthy individuals lacking protective antibodies against pH1N1 virus (126). These observations may help in the development of a universal influenza vaccine either by improving currently available vaccines or by designing T-cell epitopes targeting all subtypes of influenza viruses (126).
CD8+ T lymphocytes specific for conserved influenza epitopes has been identified in several studies (121–123). Kreijtz et al. showed that in humans, CTLs specific for H3N2 influenza virus display cross-reactivity with H5N1 influenza virus suggesting that a certain number of common epitopes are present in both viruses (121). Furthermore, liposomes containing full-length recombinant H5N1 NP and M2 antigens induced CTLs and showed high efficacy in mice (122). Another promising adjuvant is ISCOMATRIX (IMX), which immunomodulates humoral and cellular immune responses by inducing cytokines, enhancing antigen presentation by dendritic cells, inducing innate immune responses and cross-presentation of exogenous antigens to CD8+ T cells (123, 127, 128).
The role of CD4+ T-cells in mediating heterosubtypic immunity is less well understood. CD4+ T cells confer cross-protection by cytolytic functions (perforin-mediated) or helping B or T cells (129, 130). It has been shown that M1- and NP-specific memory CD4+ and CD8+ T cells generated by seasonal influenza A virus infection in humans could cross-react with an avian H5N1 strain (120). HA2 (stem domain) subunit is very conserved across the different influenza HA subtypes and also a promising vaccine candidate (131, 132). Some cross-reactive CD4+ T cell epitopes have been identified in this subunit (124, 131). Lee at al generated a recombinant HA2 polypeptide from influenza A/EM/Korea/W149/06 (H5N1) and reported homologous and heterologous CD4+ T cell responses in mice, immunized with HA2 protein (132).
As published reports suggested that the majority of T cell-based vaccines focus on conserved T-cell epitopes in NP and M proteins and are helpful in reducing disease progression and mortality against heterologous virus infections (133, 134). Furthermore, T-cell inducing approaches are unable to prevent infections. Therefore, a combination of antibody and CTL-inducing strategies may be required for an effective universal influenza vaccine development (133). Ongoing clinical trials based on coadministration of seasonal influenza vaccine with MVA-NP + M1 (viral vectored vaccine based on modified vaccinia virus Ankara expressing influenza NP and M1 proteins) and prime-boost strategies induced both T-cells and antibody responses (133, 135, 136).
Synthetic Influenza-Virus Platform
A recent technology platform that utilizes reverse genetics technology for the development of synthetic vaccine viruses was set up after the 2009 H1N1 pandemic (137). Dormitzer et al. developed an error-free gene assembly system for the synthesis of influenza-virus vaccine strains (137). In the first step, accurate and error-free HA and NA gene sequences are synthesized with the help of an enzymatic cell-free gene assembly technique with enzymatic error correction, followed by transfection in to the Madin–Darby canine kidney (MDCK) cells along with plasmid DNA coding viral backbone genes (137) (Figure 1B). In the final step, viruses for use in vaccines were rescued from MDCK cells. It was possible with this method to maintain HA and NA sequences identical to the original viruses (137, 138). As a proof-of-concept, artificially synthesized and wild-type H7N9 viruses were used and showed comparable immunogenicity (137). This breakthrough technology avoids the use of millions of eggs needed for virus propagation and prevents the mutations usually occurring during this step. Presently, the synthetic-virus technology is limited to the preparation of viruses for vaccine manufacturing (139). However, by applying this technology, error-free universal vaccines could be produced in a very short period of time in the future.
Nucleic Acid Platform
The feasibility of developing vaccines based on nucleic acids has been investigated for a long time. Nucleic acid vaccines have attributes for being safe and effective, and combine the positive attributes of live-attenuated and subunit vaccines. Nucleic acid vaccines are composed of DNA or RNA sequences encoding the antigen. These vaccines are delivered by viral particles competent for entry in host cell, by formulation with lipids or emulsions, or by means of electroporation (140).
DNA Platform
DNA vaccination is a technology developed about two decades ago (119–121) (Figure 1C). The ease of production in a very short period of time, stability and a very cost-effective production process makes this technology attractive for vaccine development (141). DNA vaccines are non-infectious, non-replicating, and do not induce vector-specific immunity (141). However, no DNA vaccine has been approved for human use to date.
DNA vaccines are delivered by various methods including the gene-gun approach and electroporation. The gene-gun approach involves the intracellular delivery of DNA in epidermal cells by bombarding the skin with DNA coated gold particles (142). This approach has not been explored for human use. In the electroporation method, DNA molecules are delivered by combined electric pulses that transiently permeabilize the cell membrane and promote DNA movement toward the membrane (143). Efforts are ongoing to deliver DNA vaccines efficiently, e.g., needle-free delivery and mucosal delivery (144, 145). Although DNA vaccines induce potent immune responses in animal models, they have appeared ineffective in humans because of suboptimal immunogenicity (145). There are other drawbacks of DNA vaccines including the degradation of DNA by host enzymes, the requirement for the DNA to enter the nucleus, and the possible risk of DNA integration into the vaccine recipient’s DNA (145).
The immunogenicity of various influenza virus DNA-vaccine candidates has been assessed in animal and human models (146–148). Smith et al. conducted a phase 1 study with an H5 HA-based DNA vaccine adjuvanted with Vaxfectin, and the results showed protective responses to the vaccine in about 67% of the recipients (147). A polyvalent DNA vaccine, also expressing H5 HA antigens (genes from H5N1 viruses A/Hongkong/156/97, A/Vietnam/1203/04, A/Anhui/1/2005, and A/Indonesia/5/2005) has been evaluated in rabbits and induced cross-protective antibodies against different clades (149). Results from this study in mice suggested that plasmid DNAs encoding NA from H3N2 virus provided cross-protection against lethal challenge with antigenically different viruses within the same subtype, but not against a different, H1N1, subtype (150). DNA vaccines expressing conserved NP and M1 antigens, or the expression of fusion proteins (HA and NA; or NP and M1) have been evaluated in mouse models of influenza-challenge (122, 128, 129). These strategies conferred broad protection against homologous and drifted viruses (151).
Although DNA is a flexible platform that has shown cross-protective efficacy in preclinical challenge models, it has yet to show potency in human clinical trials (152). Improvements are in progress for a broadly protective vaccine including the implementation of heterologous prime/boost strategies, where the second encounter with the heterologous influenza strain (boost) also induces cross-protective immunity by expansion of a preexisting pool of memory cells against conserved epitopes of the viral proteins (153). In an experimental setup, the sequential immunization of BALB/c mice with DNA coding for HA from H3N2 viruses arising approximately 10 years apart (prime: A/Hong Kong/1/1968, A/Alabama/1/1981 and A/Beijing/47/1992, and boost A/Wyoming/3/2003) elicited the production of antibodies with broad cross-reactivity against multiple H3N2 viruses (34).
RNA Platform
RNA-based vaccines are the most recent version of nucleic acid-based vaccines and possesses several benefits over DNA vaccines. In 1990, Wolff et al. (154) demonstrated that the direct injection of messenger RNA (mRNA) intramuscularly resulted in the expression of the encoded protein in a mouse model. In contrast to DNA vaccines, which function by DNA entering the nucleus of the vaccine recipient’s cell, mRNA vaccines function by the translation of mRNA in the cytoplasm (140, 155). Currently, there are two kinds of mRNA-based vaccines being developed: (1) conventional non-amplifying mRNA and (2) self-amplifying mRNA (SAM)-based vaccines (156).
The ease of construction, the small size (about 2–3 kb), and the absence of additional protein-encoded sequences are qualities of non-amplifying mRNA vaccines, although a shorter half-life and low protein expression make non-amplifying mRNA less attractive over SAM vaccines (157). However, nucleotide optimization may increase the potency and stability of the non-amplifying mRNA vaccines (158). Efficient delivery of mRNA in to the cytoplasm of the cell is the key challenge. Various delivery technologies have been explored for conventional non-amplifying mRNA, including administration of naked mRNA by injection through different routes (intradermal, intranodal, subcutaneous, and intramuscular) (159–162), or administration mRNA premixed with liposomes or lipopolyplexes (163, 164). Lipid–mRNA complexes have been shown to enhance the immunogenicity of various mRNA vaccines (165, 166). Preclinical and clinical studies suggest that non-amplifying mRNA vaccines are effective in mice, non-human primates and humans (159, 167). Recently, Moderna Therapeutics published the results of a phase 1 clinical trial study of a modified mRNA vaccine encoding HA protein of avian H10N8 formulated in lipid nanoparticles (LNPs) and demonstrated robust immunogenicity in humans (167). Furthermore, an HA-encoded mRNA vaccine conferred protection in new born and old BALB/c mice (18 months of age), suggesting that mRNA vaccine are capable of reducing influenza disease burden at the extremes of age (159). This latter aspect is relevant because the disease burden of influenza virus is greater in children and the elderly. Cross-protective T-cell responses to heterologous virus strains were also evident when mice were immunized with an NP-encoded mRNA vaccine. The combination of multiple antigens may be more effective to induce broadly protective immune responses, as reported with VLP-based vaccines.
Self-amplifying mRNA vaccines of sizes around 9–10 kb, have been generated using alphavirus genome as a vector, where alphavirus genes encoding structural proteins were replaced by the gene(s) of interest, therefore disabling the production of infectious viral particles (157, 168) (Figure 1D). Delivery of mRNA into the cell cytoplasm allows the translation of open reading frames encoding for non-structural proteins (nsP 1–4), which function as RNA-dependent RNA polymerase (RDRP) and produce a negative-sense copy of the genome (157). With the help of RDRP, this negative-sense RNA serves as template for positive-strand genomic mRNA and shorter subgenomic mRNA. The subgenomic mRNA is transcribed at a very high copy number resulting in high level of protein-antigen expression. The auto-amplification properties of SAM vaccines allow high-level protein expression of the vaccine antigen(s).
The effective delivery of mRNA vaccines, within acceptable limits of tolerability, is a key aspect for improving the vaccine efficacy. LNPs and cationic nano-emulsions synthetic non-viral delivery systems were used successfully with SAM in recent studies (103, 168–171). Geall et al. evaluated a synthetic LNP formulation of SAM as a way to increase the efficiency of antigen production and immunogenicity in vivo without the need of a competent viral delivery system (169). This novel vaccine technology was found to elicit broad, potent and protective immunity that was comparable to viral delivery, but without the inherent limitations of viral vectors.
Self-amplifying mRNA (HA) vaccines induce potent functional immune responses in mice and ferrets, comparable to those elicited by a licensed influenza subunit vaccine preparation (168). In mice, SAM (HA) derived from influenza A/California/7/2009 (H1N1) strain protected from a lethal challenge with a heterologous influenza strain A/Puerto Rico/8/1934 (H1N1) (103). This cross-protection may have been mediated by CD4+ and CD8+ T cells, because A/Puerto Rico/8/1934-specific CD4+ and CD8+ T cells were induced by the vaccine but not A/Puerto Rico/8/1934-specific antibodies (103). Recently, SAM vaccines encoding NP and M1 antigens, separately or in combination, delivered with LNP induced CTLs in mice (170). The expansion of central memory and effector memory CD4+ and CD8+ T-cell populations was also observed. The coadministration of monovalent inactivated influenza vaccine (MIIV) along with SAM vaccine also enhanced MIIV efficacy against heterologous challenge and enhanced the frequency of HA-specific T-cells (170).
In epidemics or disease outbreaks, the rapid development of vaccines is essential, and this is potentially feasible with SAM technology. Within 7 days, SAM (HA) vaccine against H7N9 (A/Shanghai/2/2013) was developed (168). The SAM vaccine technology is still in a preclinical development; nevertheless, present data suggest that the SAM technology has the potential to provide a platform for rapid and cost-effective vaccines.
Viral Vectors
Viral vectors with the capability of replication within the cells of the vaccine recipient are being evaluated as vaccines in the form of viral delivery of nucleic acids encoding the gene of interest (Figure 1E). The viral vector is non-infectious for the host but can express the antigen over a certain period of time.
Viral-vector-based influenza vaccines have been evaluated using double-stranded DNA (adenovirus and poxvirus) and single-stranded RNA (alphavirus, parainfluenza virus 5) viruses (172–175). Recently, by exploiting MVA vector and in silico mosaic approach, a recombinant vaccine construct, MVA-H5M (HA of H5N1), was developed (176). Mosaic technology allows the selection of potential epitopes by using a genetic algorithm (177). Immunization of BALB/c mice, MVA-H5M induced broadly protective neutralizing antibodies against clade 0–2 (avian influenza) and highly pathogenic A/Puerto Rico/8/1934 (H1N1) virus and induced cellular immunity (176). An adenovirus vector expressing NP and M2 proteins conferred protection from highly virulent H5N1, H1N1, and H3N2 influenza viruses (178) as well as reduced viral transmission from vaccinated and infected mice to unvaccinated mice (178, 179). Furthermore, Kim et al. employed a novel approach to inducing cross-reactive immune responses against HA protein by generating recombinant human adenovirus 5 vector encoding full length HA (from H5N1) and four tendem copies of M2e domain (rAdH5/M2e) (180). Adenovirus vaccine induced antibody-mediated cross-protection against heterosubtypic viruses in mice (180).
Viral-vector-based vaccines possess several advantages including the capability of inducing both humoral- and cell-mediated immunity, a high level of protein expression and long-term stability (181–184). Some of these viral-vector systems can mimic aspects of natural influenza-virus infection because of their direct delivery to the mucosa (3). Vector-based vaccines also possess several limitations including reduced immunogenicity in the presence of preexisting immunity (185) and risks of pathogenesis in certain individuals (186).
Conclusion
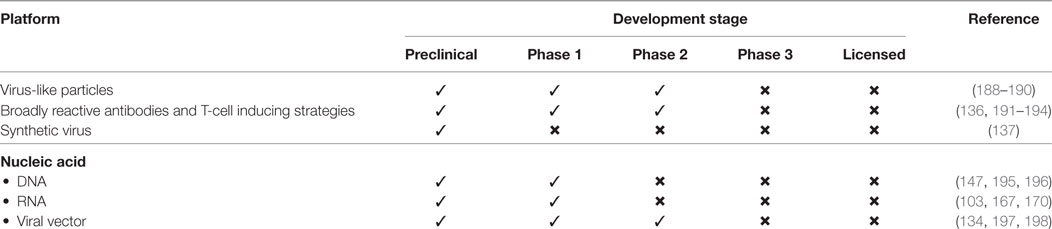
An optimal universal influenza vaccine should provide cross-protection against all subtypes of influenza viruses, particularly historic and circulating strains. Many of the universal influenza vaccines in development target conserved domains of surface and internal proteins. Numerous approaches are being applied for the development of universal flu vaccines. Proof-of-concept and initial results support the promise of these approaches (168, 187). Novel platforms focused on influenza vaccine development are currently in preclinical and early clinical development (Table 1). Applying these novel platforms may offer additional benefits for the development of a universal influenza vaccine.
Author Contributions
All authors were involved in drafting the manuscript and revising it critically for important intellectual content.
Conflict of Interest Statement
Presently, SB is an employee of the GSK group of companies while AK and TSM were employed at the time of study conduct. AK and TSM are now employed with Linköping University, Sweden and Technical University of Denmark, Denmark, respectively. All the authors declare that they have no other conflicts.
Acknowledgments
This article was sponsored by GlaxoSmithKline Biologicals SA. All the authors were involved in drafting the manuscript and revising it critically for intellectual content. AK and TM were supported by the Marie-Curie IEF grant SAMUFLU FP7-PEOPLE-2013-IEF (626283) and Marie-Curie ITN grant HOMIN FP7-PEOPLE-2012-ITN (626283), respectively. AK and TM participated in postdoctoral and postgraduate studentship program, respectively, at GSK. SB is an employee of the GSK group of companies. The authors thank Dr. Corey Mallett for his critical reading and suggestions for this manuscript.
References
1. Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine (2007) 25:5086–96. doi:10.1016/j.vaccine.2007.03.046
2. WHO. Influenza Fact Sheet (2018). Available from: http://www.who.int/mediacentre/factsheets/fs211/en (accessed January, 2018).
3. Wong SS, Webby RJ. Traditional and new influenza vaccines. Clin Microbiol Rev (2013) 26:476–92. doi:10.1128/CMR.00097-12
4. Taubenberger JK, Morens DM. 1918 influenza: the mother of all pandemics. Emerg Infect Dis (2006) 12:15–22. doi:10.3201/eid1209.05-0979
5. Gao X, Wang W, Li Y, Zhang S, Duan Y, Xing L, et al. Enhanced influenza VLP vaccines comprising matrix-2 ectodomain and nucleoprotein epitopes protects mice from lethal challenge. Antiviral Res (2013) 98:4–11. doi:10.1016/j.antiviral.2013.01.010
6. Dormitzer PR. Rapid production of synthetic influenza vaccines BT. In: Oldstone MBA, Compans RW, editors. Influenza Pathogenesis and Control – Volume II. Cham: Springer International Publishing p. 237–73.
7. Kilbourne ED. Perspectives on pandemics: a research agenda. J Infect Dis (1997) 176(Suppl):S29–31. doi:10.1086/514171
8. Sonnberg S, Webby RJ, Webster RG. Natural history of highly pathogenic avian influenza H5N1. Virus Res (2013) 178:63–77. doi:10.1016/j.virusres.2013.05.009
9. Lambert LC, Fauci AS. Influenza vaccines for the future. N Engl J Med (2010) 363:2036–44. doi:10.1056/NEJMra1002842
10. Carroll KC, Hobden JA, Miller S, Morse SA, Mietzner TA, Detrick B, et al. Chapter 39. Orthomyxoviruses (influenza viruses). In: Weitz M, Kearns B, editors. Jawetz, Melnick, & Adelberg’s Medical Microbiology, 27e. New York, NY: McGraw-Hill Education.
11. Thielen BK, Friedlander H, Bistodeau S, Shu B, Lynch B, Martin K, et al. Detection of influenza C viruses among outpatients and patients hospitalized for severe acute respiratory infection, Minnesota, 2013–2016. Clin Infect Dis (2017). doi:10.1093/cid/cix931
12. Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog (2013) 9:e1003657. doi:10.1371/journal.ppat.1003657
13. Medina RA, García-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol (2011) 9:590–603. doi:10.1038/nrmicro2613
14. Lee PS, Wilson IA. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol (2015) 386:323–41. doi:10.1007/82_2014_413
15. Vijaykrishna D, Holmes EC, Joseph U, Fourment M, Su YCF, Halpin R, et al. The contrasting phylodynamics of human influenza B viruses. Elife (2015) 4:e05055. doi:10.7554/eLife.05055
16. Ambrose CS, Levin MJ. The rationale for quadrivalent influenza vaccines. Hum Vaccin Immunother (2012) 8:81–8. doi:10.4161/hv.8.1.17623
17. Laver WG, Webster RG. Preparation and immunogenicity of a purified influenza virus haemagglutinin and neuraminidase subunit vaccine. Postgrad Med J (1976) 52:373–8. doi:10.1136/pgmj.52.608.373
18. Kon TC, Onu A, Berbecila L, Lupulescu E, Ghiorgisor A, Kersten GF, et al. Influenza vaccine manufacturing: effect of inactivation, splitting and site of manufacturing. Comparison of influenza vaccine production processes. PLoS One (2016) 11:e0150700. doi:10.1371/journal.pone.0150700
19. Bachmayer H, Liehl E, Schmidt G. Preparation and properties of a novel influenza subunit vaccine. Postgrad Med J (1976) 52:360–7. doi:10.1136/pgmj.52.608.360
20. Brady MI, Furminger IG. A surface antigen influenza vaccine. 1. Purification of haemagglutinin and neuraminidase proteins. J Hyg (Lond) (1976) 77:161–72. doi:10.1017/S0022172400024591
21. Baxter R, Patriarca PA, Ensor K, Izikson R, Goldenthal KL, Cox MM. Evaluation of the safety, reactogenicity and immunogenicity of FluBlok® trivalent recombinant baculovirus-expressed hemagglutinin influenza vaccine administered intramuscularly to healthy adults 50–64 years of age. Vaccine (2011) 29:2272–8. doi:10.1016/j.vaccine.2011.01.039
22. Gross PA, Ennis FA, Gaerlan PF, Denson LJ, Denning CR, Schiffman D. A controlled double-blind comparison of reactogenicity, immunogenicity, and protective efficacy of whole-virus and split-product influenza vaccines in children. J Infect Dis (1977) 136:623–32. doi:10.1093/infdis/136.5.623
23. Carter NJ, Curran MP. Live attenuated influenza vaccine (FluMist; Fluenz): a review of its use in the prevention of seasonal influenza in children and adults. Drugs (2011) 71:1591–622. doi:10.2165/11206860-000000000-00000
24. Barria MI, Garrido JL, Stein C, Scher E, Ge Y, Engel SM, et al. Localized mucosal response to intranasal live attenuated influenza vaccine in adults. J Infect Dis (2013) 207:115–24. doi:10.1093/infdis/jis641
25. Grohskopf LA, Sokolow LZ, Broder KR, Walter EB, Bresee JS, Fry AM, et al. Prevention and control of seasonal influenza with vaccines: recommendations of the advisory committee on immunization practices—United States, 2017–18 influenza season. MMWR Recomm Rep (2017) 66:1–20. doi:10.15585/mmwr.rr6602a1
26. Small PA, Cronin BJ. The advisory committee on immunization practices’ controversial recommendation against the use of live attenuated influenza vaccine is based on a biased study design that ignores secondary protection. Vaccine (2017) 35:1110–2. doi:10.1016/j.vaccine.2017.01.022
27. Belongia EA, Karron RA, Reingold A, Walter EB, Bennett NM. The advisory committee on immunization practices recommendation regarding the use of live influenza vaccine: a rejoinder. Vaccine (2018) 36:343–4. doi:10.1016/j.vaccine.2017.06.017
28. Ambrose CS, Bright H, Mallory R. Letter to the editor: potential causes of the decreased effectiveness of the influenza A(H1N1)pdm09 strain in live attenuated influenza vaccines. Euro Surveill (2016) 21:30394. doi:10.2807/1560-7917.ES.2016.21.45.30394
29. Tisa V, Barberis I, Faccio V, Paganino C, Trucchi C, Martini M, et al. Quadrivalent influenza vaccine: a new opportunity to reduce the influenza burden. J Prev Med Hyg (2016) 57:E28–33.
30. Foll M, Poh YP, Renzette N, Ferrer-Admetlla A, Bank C, Shim H, et al. Influenza virus drug resistance: a time-sampled population genetics perspective. PLoS Genet (2014) 10:e1004185. doi:10.1371/journal.pgen.1004185
31. van der Vries E, Schutten M, Fraaij P, Boucher C, Osterhaus A. Influenza virus resistance to antiviral therapy. Adv Pharmocol (2013) 67:217–46. doi:10.1016/B978-0-12-405880-4.00006-8
32. Das SR, Puigbo P, Hensley SE, Hurt DE, Bennink JR, Yewdell JW. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog (2010) 6:e1001211. doi:10.1371/journal.ppat.1001211
33. Medina RA, Stertz S, Manicassamy B, Zimmermann P, Sun X, Albrecht RA, et al. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci Transl Med (2013) 5:187ra70. doi:10.1126/scitranslmed.3005996
34. Wang TT, Tan GS, Hai R, Pica N, Petersen E, Moran TM, et al. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog (2010) 6:e1000796. doi:10.1371/journal.ppat.1000796
35. Skountzou I, Koutsonanos DG, Kim JH, Powers R, Satyabhama L, Masseoud F, et al. Immunity to pre-1950 H1N1 influenza viruses confers cross-protection against the pandemic swine-origin 2009 A (H1N1) influenza virus. J Immunol (2010) 185:1642–9. doi:10.4049/jimmunol.1000091
36. Galli G, Hancock K, Hoschler K, DeVos J, Praus M, Bardelli M, et al. Fast rise of broadly cross-reactive antibodies after boosting long-lived human memory B cells primed by an MF59 adjuvanted prepandemic vaccine. Proc Natl Acad Sci U S A (2009) 106:7962–7. doi:10.1073/pnas.0903181106
37. Khurana S, Chearwae W, Castellino F, Manischewitz J, King LR, Honorkiewicz A, et al. Vaccines with MF59 adjuvant expand the antibody repertoire to target protective sites of pandemic avian H5N1 influenza virus. Sci Transl Med (2010) 2:15ra5. doi:10.1126/scitranslmed.3000624
38. Khurana S, Verma N, Yewdell JW, Hilbert AK, Castellino F, Lattanzi M, et al. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med (2011) 3:85ra48. doi:10.1126/scitranslmed.3002336
39. Lofano G, Kumar A, Finco O, Del Giudice G, Bertholet S. B cells and functional antibody responses to combat influenza. Front Immunol (2015) 6:336. doi:10.3389/fimmu.2015.00336
40. Du L, Zhou Y, Jiang S. Research and development of universal influenza vaccines. Microbes Infect (2010) 12:280–6. doi:10.1016/j.micinf.2010.01.001
41. Johansson BE, Matthews JT, Kilbourne ED. Supplementation of conventional influenza A vaccine with purified viral neuraminidase results in a balanced and broadened immune response. Vaccine (1998) 16:1009–15. doi:10.1016/S0264-410X(97)00279-X
42. Widjaja L, Ilyushina N, Webster RG, Webby RJ. Molecular changes associated with adaptation of human influenza A virus in embryonated chicken eggs. Virology (2006) 350:137–45. doi:10.1016/j.virol.2006.02.020
43. Manini I, Trombetta CM, Lazzeri G, Pozzi T, Rossi S, Montomoli E. Egg-independent influenza vaccines and vaccine candidates. Vaccines (Basel) (2017) 5:18. doi:10.3390/vaccines5030018
44. Skowronski DM, Janjua NZ, De Serres G, Sabaiduc S, Eshaghi A, Dickinson JA, et al. Low 2012–13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS One (2014) 9:e92153. doi:10.1371/journal.pone.0092153
45. Schild GC, Oxford JS, de Jong JC, Webster RG. Evidence for host-cell selection of influenza virus antigenic variants. Nature (1983) 303:706. doi:10.1038/303706a0
46. Ping J, Lopes TJS, Nidom CA, Ghedin E, Macken CA, Fitch A, et al. Development of high-yield influenza A virus vaccine viruses. Nat Commun (2015) 6:8148. doi:10.1038/ncomms9148
47. Tree JA, Richardson C, Fooks AR, Clegg JC, Looby D. Comparison of large-scale mammalian cell culture systems with egg culture for the production of influenza virus A vaccine strains. Vaccine (2001) 19:3444–50. doi:10.1016/S0264-410X(01)00053-6
48. Perdue ML, Arnold F, Li S, Donabedian A, Cioce V, Warf T, et al. The future of cell culture-based influenza vaccine production. Expert Rev Vaccines (2011) 10:1183–94. doi:10.1586/erv.11.82
49. Minor PD, Engelhardt OG, Wood JM, Robertson JS, Blayer S, Colegate T, et al. Current challenges in implementing cell-derived influenza vaccines: implications for production and regulation, July 2007, NIBSC, Potters Bar, UK. Vaccine (2009) 27:2907–13. doi:10.1016/j.vaccine.2009.02.064
50. Wilks S, de Graaf M, Smith DJ, Burke DF. A review of influenza haemagglutinin receptor binding as it relates to pandemic properties. Vaccine (2012) 30:4369–76. doi:10.1016/j.vaccine.2012.02.076
51. Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol (1993) 67:2552–8.
52. Sakabe S, Iwatsuki-Horimoto K, Horimoto T, Nidom CA, Le M, Takano R, et al. A cross-reactive neutralizing monoclonal antibody protects mice from H5N1 and pandemic (H1N1) 2009 virus infection. Antiviral Res (2010) 88:249–55. doi:10.1016/j.antiviral.2010.09.007
53. Corti D, Suguitan AL, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest (2010) 120:1663–73. doi:10.1172/JCI41902
54. Jang YH, Seong BL. Options and obstacles for designing a universal influenza vaccine. Viruses (2014) 6:3159–80. doi:10.3390/v6083159
55. Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem (2010) 285:28403–9. doi:10.1074/jbc.R110.129809
56. Webster RG, Laver WG. Preparation and properties of antibody directed specifically against the neuraminidase of influenza virus. J Immunol (1967) 99:49–55.
57. Sylte MJ, Suarez DL. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol (2009) 333:227–41. doi:10.1007/978-3-540-92165-3_12
58. Moscona A. Neuraminidase inhibitors for influenza. N Engl J Med (2005) 353:1363–73. doi:10.1056/NEJMra050740
59. Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol (2004) 78:12665–7. doi:10.1128/JVI.78.22.12665-12667.2004
60. Shtyrya YA, Mochalova LV, Bovin NV. Influenza virus neuraminidase: structure and function. Acta Naturae (2009) 1:26–32.
61. Wiersma L, Rimmelzwaan G, de Vries R. Developing universal influenza vaccines: hitting the nail, not just on the head. Vaccines (Basel) (2015) 3:239–62. doi:10.3390/vaccines3020239
62. Cardoso FM, Ibanez LI, Van den Hoecke S, De Baets S, Smet A, Roose K, et al. Single-domain antibodies targeting neuraminidase protect against an H5N1 influenza virus challenge. J Virol (2014) 88:8278–96. doi:10.1128/JVI.03178-13
63. Mizuno T, Mizuno S, Kanda T. Effects of vaccination and the new neuraminidase inhibitor, laninamivir, on influenza infection. PLoS One (2014) 9:e92601. doi:10.1371/journal.pone.0092601
64. Wohlbold TJ, Krammer F. In the shadow of hemagglutinin: a growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses (2014) 6:2465–94. doi:10.3390/v6062465
65. Quan FS, Kim MC, Lee BJ, Song JM, Compans RW, Kang SM. Influenza M1 VLPs containing neuraminidase induce heterosubtypic cross-protection. Virology (2012) 430:127–35. doi:10.1016/j.virol.2012.05.006
66. Ebrahimi SM, Tebianian M. Influenza A viruses: why focusing on M2e-based universal vaccines. Virus Genes (2011) 42:1–8. doi:10.1007/s11262-010-0547-7
67. Helenius A. Unpacking the incoming influenza virus. Cell (1992) 69:577–8. doi:10.1016/0092-8674(92)90219-3
68. Zhou D, Wu TL, Lasaro MO, Latimer BP, Parzych EM, Bian A, et al. A universal influenza A vaccine based on adenovirus expressing matrix-2 ectodomain and nucleoprotein protects mice from lethal challenge. Mol Ther (2010) 18:2182–9. doi:10.1038/mt.2010.202
69. Neirynck S, Deroo T, Saelens X, Vanlandschoot P, Jou WM, Fiers W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med (1999) 5:1157–63. doi:10.1038/13484
70. Fiers W, De Filette M, El Bakkouri K, Schepens B, Roose K, Schotsaert M, et al. M2e-based universal influenza A vaccine. Vaccine (2009) 27:6280–3. doi:10.1016/j.vaccine.2009.07.007
71. Schotsaert M, De Filette M, Fiers W, Saelens X. Universal M2 ectodomain-based influenza A vaccines: preclinical and clinical developments. Expert Rev Vaccines (2009) 8:499–508. doi:10.1586/erv.09.6
72. Yewdell JW, Bennink JR, Smith GL, Moss B. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc Natl Acad Sci U S A (1985) 82:1785–9. doi:10.1073/pnas.82.6.1785
73. Zheng M, Luo J, Chen Z. Development of universal influenza vaccines based on influenza virus M and NP genes. Infection (2014) 42:251–62. doi:10.1007/s15010-013-0546-4
74. Roldao A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM. Virus-like particles in vaccine development. Expert Rev Vaccines (2010) 9:1149–76. doi:10.1586/erv.10.115
75. Kumar A, Kantele A, Jarvinen T, Chen TT, Kavola H, Sadeghi M, et al. Trichodysplasia spinulosa-associated polyomavirus (TSV) and merkel cell polyomavirus: correlation between humoral and cellular immunity stronger with TSV. PLoS One (2012) 7:e0045773. doi:10.1371/journal.pone.0045773
76. Jegerlehner A, Zabel F, Langer A, Dietmeier K, Jennings GT, Saudan P, et al. Bacterially produced recombinant influenza vaccines based on virus-like particles. PLoS One (2013) 8:e0078947. doi:10.1371/journal.pone.0078947
77. Lahtinen A, Kivela P, Hedman L, Kumar A, Kantele A, Lappalainen M, et al. Serodiagnosis of primary infections with human parvovirus 4, Finland. Emerg Infect Dis (2011) 17:79–82. doi:10.3201/eid1701.100750
78. Buonaguro L, Tornesello ML, Tagliamonte M, Gallo RC, Wang LX, Kamin-Lewis R, et al. Baculovirus-derived human immunodeficiency virus type 1 virus-like particles activate dendritic cells and induce ex vivo T-cell responses. J Virol (2006) 80:9134–43. doi:10.1128/JVI.00050-06
79. Hemann EA, Kang SM, Legge KL. Protective CD8 T cell-mediated immunity against influenza A virus infection following influenza virus-like particle vaccination. J Immunol (2013) 191:2486–94. doi:10.4049/jimmunol.1300954
80. Bachmann MF, Zinkernagel RM. Neutralizing antiviral B cell responses. Annu Rev Immunol (1997) 15:235–70. doi:10.1146/annurev.immunol.15.1.235
81. Paliard X, Liu Y, Wagner R, Wolf H, Baenziger J, Walker CM. Priming of strong, broad, and long-lived HIV type 1 p55(gag)-specific CD8+ cytotoxic T cells after administration of a virus-like particle vaccine in rhesus macaques. AIDS Res Hum Retroviruses (2000) 16:273–82. doi:10.1089/088922200309368
82. Schwartzman LM, Cathcart AL, Pujanauski LM, Qi L, Kash JC, Taubenberger JK. An intranasal virus-like particle vaccine broadly protects mice from multiple subtypes of influenza A virus. MBio (2015) 6:e1044. doi:10.1128/mBio.01044-15
83. Kang SM, Song JM, Quan FS, Compans RW. Influenza vaccines based on virus-like particles. Virus Res (2009) 143:140–6. doi:10.1016/j.virusres.2009.04.005
84. Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio (2010) 1:e00018–10. doi:10.1128/mBio.00018-10
85. Kim MC, Song JM, Eunju O, Kwon YM, Lee YJ, Compans RW, et al. Virus-like particles containing multiple M2 extracellular domains confer improved cross-protection against various subtypes of influenza virus. Mol Ther (2013) 21:485–92. doi:10.1038/mt.2012.246
86. Hu C-MJ, Chien C-Y, Liu M-T, Fang Z-S, Chang S-Y, Juang R-H, et al. Multi-antigen avian influenza a (H7N9) virus-like particles: particulate characterizations and immunogenicity evaluation in murine and avian models. BMC Biotechnol (2017) 17:2. doi:10.1186/s12896-016-0321-6
87. Park YC, Song JM. Preparation and immunogenicity of influenza virus-like particles using nitrocellulose membrane filtration. Clin Exp Vaccine Res (2017) 6:61–6. doi:10.7774/cevr.2017.6.1.61
88. Kang SM, Kim MC, Compans RW. Virus-like particles as universal influenza vaccines. Expert Rev Vaccines (2012) 11:995–1007. doi:10.1586/erv.12.70
89. Krystal M, Elliott RM, Benz EW Jr, Young JF, Palese P. Evolution of influenza A and B viruses: conservation of structural features in the hemagglutinin genes. Proc Natl Acad Sci U S A (1982) 79:4800–4. doi:10.1073/pnas.79.15.4800
90. Bessa J, Schmitz N, Hinton HJ, Schwarz K, Jegerlehner A, Bachmann MF. Efficient induction of mucosal and systemic immune responses by virus-like particles administered intranasally: implications for vaccine design. Eur J Immunol (2008) 38:114–26. doi:10.1002/eji.200636959
91. Ionescu RM, Przysiecki CT, Liang X, Garsky VM, Fan J, Wang B, et al. Pharmaceutical and immunological evaluation of human papillomavirus viruslike particle as an antigen carrier. J Pharm Sci (2006) 95:70–9. doi:10.1002/jps.20493
92. Kim MC, Lee YN, Hwang HS, Lee YT, Ko EJ, Jung YJ, et al. Influenza M2 virus-like particles confer a broader range of cross protection to the strain-specific pre-existing immunity. Vaccine (2014) 32:5824–31. doi:10.1016/j.vaccine.2014.08.030
93. Steinhagen F, Kinjo T, Bode C, Klinman DM. TLR-based immune adjuvants. Vaccine (2011) 29:3341–55. doi:10.1016/j.vaccine.2010.08.002
94. Wang BZ, Quan FS, Kang SM, Bozja J, Skountzou I, Compans RW. Incorporation of membrane-anchored flagellin into influenza virus-like particles enhances the breadth of immune responses. J Virol (2008) 82:11813–23. doi:10.1128/JVI.01076-08
95. Zhao Q, Li S, Yu H, Xia N, Modis Y. Virus-like particle-based human vaccines: quality assessment based on structural and functional properties. Trends Biotechnol (2013) 31:654–63. doi:10.1016/j.tibtech.2013.09.002
96. Saylor C, Dadachova E, Casadevall A. Monoclonal antibody-based therapies for microbial diseases. Vaccine (2009) 27(Suppl):G38–46. doi:10.1016/j.vaccine.2009.09.105
97. Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol (2013) 87:6542–50. doi:10.1128/JVI.00641-13
98. Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov (2015) 14:167–82. doi:10.1038/nrd4529
99. Margine I, Hai R, Albrecht RA, Obermoser G, Harrod AC, Banchereau J, et al. H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice. J Virol (2013) 87:4728–37. doi:10.1128/JVI.03509-12
100. Ermler M, Kirkpatrick E, Sun W, Hai R, Amanat F, Chromikova V, et al. Chimeric hemagglutinin constructs induce broad protection against influenza B virus challenge in the mouse model. J Virol (2017) 91:e286–217. doi:10.1128/JVI.00286-17
101. Nachbagauer R, Kinzler D, Choi A, Hirsh A, Beaulieu E, Lecrenier N, et al. A chimeric haemagglutinin-based influenza split virion vaccine adjuvanted with AS03 induces protective stalk-reactive antibodies in mice. NPJ Vaccines (2016) 1:16015.
102. Matsuoka Y, Lamirande EW, Subbarao K. The ferret model for influenza. Current Protocols in Microbiology (2009) 14:G:15G.2.1-15G.2.29. doi:10.1002/9780471729259.mc15g02s13
103. Brazzoli M, Magini D, Bonci A, Buccato S, Giovani C, Kratzer R, et al. Induction of broad-based immunity and protective efficacy by self-amplifying mRNA vaccines encoding influenza virus hemagglutinin. J Virol (2015) 90:332–44. doi:10.1128/JVI.01786-15
104. Krammer F, Hai R, Yondola M, Tan GS, Leyva-Grado VH, Ryder AB, et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J Virol (2014) 88:3432–42. doi:10.1128/JVI.03004-13
105. Chen C-J, Ermler ME, Tan GS, Krammer F, Palese P, Hai R. Influenza A viruses expressing intra- or intergroup chimeric hemagglutinins. J Virol (2016) 90:3789–93. doi:10.1128/JVI.03060-15
106. Nachbagauer R, Liu W-C, Choi A, Wohlbold TJ, Atlas T, Rajendran M, et al. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines (2017) 2:26. doi:10.1038/s41541-017-0026-4
107. Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A (2011) 108:14216–21. doi:10.1073/pnas.1111497108
108. Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, et al. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A (2012) 109:17040–5. doi:10.1073/pnas.1212371109
109. Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One (2008) 3:e3942. doi:10.1371/journal.pone.0003942
110. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science (2011) 333:850–6. doi:10.1126/science.1205669
111. Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, et al. Highly conserved protective epitopes on influenza B viruses. Science (2012) 337:1343–8. doi:10.1126/science.1222908
112. Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature (2014) 516:418–22. doi:10.1038/nature13764
113. Giles BM, Ross TM. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine (2011) 29:3043–54. doi:10.1016/j.vaccine.2011.01.100
114. Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, Wiley CA, et al. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis (2012) 205:1562–70. doi:10.1093/infdis/jis232
115. Bommakanti G, Citron MP, Hepler RW, Callahan C, Heidecker GJ, Najar TA, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci U S A (2010) 107:13701–6. doi:10.1073/pnas.1007465107
116. Laidlaw BJ, Decman V, Ali MAA, Abt MC, Wolf AI, Monticelli LA, et al. Cooperativity between CD8+ T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PLoS Pathog (2013) 9:e1003207. doi:10.1371/journal.ppat.1003207
117. Sambhara S, Kurichh A, Miranda R, Tumpey T, Rowe T, Renshaw M, et al. Heterosubtypic immunity against human influenza A viruses, including recently emerged avian H5 and H9 viruses, induced by FLU-ISCOM vaccine in mice requires both cytotoxic T-lymphocyte and macrophage function. Cell Immunol (2001) 211:143–53. doi:10.1006/cimm.2001.1835
118. Mohn KGI, Zhou F, Brokstad KA, Sridhar S, Cox RJ. Boosting of cross-reactive and protection-associated T cells in children after live attenuated influenza vaccination. J Infect Dis (2017) 215:1527–35. doi:10.1093/infdis/jix165
119. Boon ACM, de Mutsert G, Graus YMF, Fouchier RAM, Sintnicolaas K, Osterhaus ADME, et al. Sequence variation in a newly identified HLA-B35-restricted epitope in the influenza A virus nucleoprotein associated with escape from cytotoxic T lymphocytes. J Virol (2002) 76:2567–72. doi:10.1128/jvi.76.5.2567-2572.2002
120. Lee LYH, Ha DLA, Simmons C, De Jong MD, Chau NVV, Schumacher R, et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest (2008) 118:3478–90. doi:10.1172/JCI32460
121. Kreijtz JH, de Mutsert G, van Baalen CA, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J Virol (2008) 82:5161–6. doi:10.1128/JVI.02694-07
122. Thueng-in K, Maneewatch S, Srimanote P, Songserm T, Tapchaisri P, Sookrung N, et al. Heterosubtypic immunity to influenza mediated by liposome adjuvanted H5N1 recombinant protein vaccines. Vaccine (2010) 28:6765–77. doi:10.1016/j.vaccine.2010.07.065
123. Wilson NS, Yang B, Morelli AB, Koernig S, Yang A, Loeser S, et al. ISCOMATRIX vaccines mediate CD8+ T-cell cross-priming by a MyD88-dependent signaling pathway. Immunol Cell Biol (2012) 90:540–52. doi:10.1038/icb.2011.71
124. Duvvuri VRSK, Moghadas SM, Guo H, Duvvuri B, Heffernan JM, Fisman DN, et al. Original article: highly conserved cross-reactive CD4+ T-cell HA-epitopes of seasonal and the 2009 pandemic influenza viruses. Influenza Other Respi Viruses (2010) 4:249–58. doi:10.1111/j.1750-2659.2010.00161.x
125. Babon JAB, Cruz J, Orphin L, Pazoles P, Co MDT, Ennis FA, et al. Genome-wide screening of human T-cell epitopes in influenza A virus reveals a broad-spectrum of CD4(+) T cell responses to internal proteins, hemagglutinins and neuraminidases. Hum Immunol (2009) 70:711–21. doi:10.1016/j.humimm.2009.06.004
126. Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med (2013) 19:1305–12. doi:10.1038/nm.3350
127. Morelli AB, Becher D, Koernig S, Silva A, Drane D, Maraskovsky E. ISCOMATRIX: a novel adjuvant for use in prophylactic and therapeutic vaccines against infectious diseases. J Med Microbiol (2012) 61:935–43. doi:10.1099/jmm.0.040857-0
128. Cox MA, Kahan SM, Zajac AJ. Anti-viral CD8 T cells and the cytokines that they love. Virology (2013) 435:157–69. doi:10.1016/j.virol.2012.09.012
129. Brown DM, Lee S, Garcia-Hernandez Mde L, Swain SL. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J Virol (2012) 86:6792–803. doi:10.1128/JVI.07172-11
130. Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol (2006) 177:2888–98. doi:10.4049/jimmunol.177.5.2888
131. Cross KJ, Langley WA, Russell RJ, Steinhauer JJS, Steinhauer DA. Composition and functions of the influenza fusion peptide. Protein Pept Lett (2009) 16:766–78. doi:10.2174/092986609788681715
132. Lee J-S, Chowdhury MYE, Moon H-J, Choi Y-K, Talactac MR, Kim J-H, et al. The highly conserved HA2 protein of the influenza A virus induces a cross protective immune response. J Virol Methods (2013) 194:280–8. doi:10.1016/j.jviromet.2013.08.022
133. Rajão DS, Pérez DR. Universal vaccines and vaccine platforms to protect against influenza viruses in humans and agriculture. Front Microbiol (2018) 9:123. doi:10.3389/fmicb.2018.00123
134. Lillie PJ, Berthoud TK, Powell TJ, Lambe T, Mullarkey C, Spencer AJ, et al. Preliminary assessment of the efficacy of a T-cell-based influenza vaccine, MVA-NP+M1, in humans. Clin Infect Dis (2012) 55:19–25. doi:10.1093/cid/cis327
135. Antrobus RD, Berthoud TK, Mullarkey CE, Hoschler K, Coughlan L, Zambon M, et al. Coadministration of seasonal influenza vaccine and MVA-NP+M1 simultaneously achieves potent humoral and cell-mediated responses. Mol Ther (2014) 22:233–8. doi:10.1038/mt.2013.162
136. Atsmon J, Caraco Y, Ziv-Sefer S, Shaikevich D, Abramov E, Volokhov I, et al. Priming by a novel universal influenza vaccine (multimeric-001)—a gateway for improving immune response in the elderly population. Vaccine (2014) 32:5816–23. doi:10.1016/j.vaccine.2014.08.031
137. Dormitzer PR, Suphaphiphat P, Gibson DG, Wentworth DE, Stockwell TB, Algire MA, et al. Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci Transl Med (2013) 5:185ra68. doi:10.1126/scitranslmed.3006368
138. Settembre EC, Dormitzer PR, Rappuoli R. Bringing influenza vaccines into the 21st century. Hum Vaccin Immunother (2014) 10:600–4. doi:10.4161/hv.27600
139. Dormitzer PR. Rapid production of synthetic influenza vaccines. Curr Top Microbiol Immunol (2014) 386:237–73. doi:10.1007/82_2014_399
140. Ulmer JB, Mason PW, Geall A, Mandl CW. RNA-based vaccines. Vaccine (2012) 30:4414–8. doi:10.1016/j.vaccine.2012.04.060
141. Kim JH, Jacob J. DNA vaccines against influenza viruses. Curr Top Microbiol Immunol (2009) 333:197–210. doi:10.1007/978-3-540-92165-3_10
142. Haynes JR, McCabe DE, Swain WF, Widera G, Fuller JT. Particle-mediated nucleic acid immunization. J Biotechnol (1996) 44:37–42.
143. Mir LM, Moller PH, André F, Gehl J. Electric pulse-mediated gene delivery to various animal tissues. Adv Genet (2005) 54:83–114.
144. Rao SS, Gomez P, Mascola JR, Dang V, Krivulka GR, Yu F, et al. Comparative evaluation of three different intramuscular delivery methods for DNA immunization in a nonhuman primate animal model. Vaccine (2006) 24:367–73. doi:10.1016/j.vaccine.2005.07.072
145. Saade F, Petrovsky N. Technologies for enhanced efficacy of DNA vaccines. Expert Rev Vaccines (2012) 11:189–209. doi:10.1586/erv.11.188
146. Borggren M, Nielsen J, Bragstad K, Karlsson I, Krog JS, Williams JA, et al. Vector optimization and needle-free intradermal application of a broadly protective polyvalent influenza A DNA vaccine for pigs and humans. Hum Vaccin Immunother (2015) 11:1983–90. doi:10.1080/21645515.2015.1011987
147. Smith LR, Wloch MK, Ye M, Reyes LR, Boutsaboualoy S, Dunne CE, et al. Phase 1 clinical trials of the safety and immunogenicity of adjuvanted plasmid DNA vaccines encoding influenza A virus H5 hemagglutinin. Vaccine (2010) 28:2565–72. doi:10.1016/j.vaccine.2010.01.029
148. Gorres JP, Lager KM, Kong WP, Royals M, Todd JP, Vincent AL, et al. DNA vaccination elicits protective immune responses against pandemic and classic swine influenza viruses in pigs. Clin Vaccine Immunol (2011) 18:1987–95. doi:10.1128/CVI.05171-11
149. Wang S, Hackett A, Jia N, Zhang C, Zhang L, Parker C, et al. Polyvalent DNA vaccines expressing HA antigens of H5N1 influenza viruses with an optimized leader sequence elicit cross-protective antibody responses. PLoS One (2011) 6:e0028757. doi:10.1371/journal.pone.0028757
150. Chen Z, Kadowaki S, Hagiwara Y, Yoshikawa T, Matsuo K, Kurata T, et al. Cross-protection against a lethal influenza virus infection by DNA vaccine to neuraminidase. Vaccine (2000) 18:3214–22. doi:10.1016/S0264-410X(00)00149-3
151. Chen Z, Matsuo K, Asanuma H, Takahashi H, Iwasaki T, Suzuki Y, et al. Enhanced protection against a lethal influenza virus challenge by immunization with both hemagglutinin- and neuraminidase-expressing DNAs. Vaccine (1999) 17:653–9. doi:10.1016/S0264-410X(98)00247-3
152. Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet (2008) 9:776–88. doi:10.1038/nrg2432
153. Medina RA, Manicassamy B, Stertz S, Seibert CW, Hai R, Belshe RB, et al. Pandemic 2009 H1N1 vaccine protects against 1918 Spanish influenza virus. Nat Commun (2010) 1:28. doi:10.1038/ncomms1026
154. Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, et al. Direct gene transfer into mouse muscle in vivo. Science (1990) 247:1465–8. doi:10.1126/science.1690918
155. Rodriguez-Gascon A, del Pozo-Rodriguez A, Solinis MA. Development of nucleic acid vaccines: use of self-amplifying RNA in lipid nanoparticles. Int J Nanomedicine (2014) 9:1833–43. doi:10.2147/IJN.S39810
156. Deering RP, Kommareddy S, Ulmer JB, Brito LA, Geall AJ. Nucleic acid vaccines: prospects for non-viral delivery of mRNA vaccines. Expert Opin Drug Deliv (2014) 11:885–99. doi:10.1517/17425247.2014.901308
157. Brito LA, Kommareddy S, Maione D, Uematsu Y, Giovani C, Berlanda Scorza F, et al. Self-amplifying mRNA vaccines. Adv Genet (2015) 89:179–233. doi:10.1016/bs.adgen.2014.10.005
158. Sahin U, Kariko K, Tureci O. mRNA-based therapeutics – developing a new class of drugs. Nat Rev Drug Discov (2014) 13:759–80. doi:10.1038/nrd4278
159. Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat Biotechnol (2012) 30:1210–6. doi:10.1038/nbt.2436
160. Scheel B, Teufel R, Probst J, Carralot JP, Geginat J, Radsak M, et al. Toll-like receptor-dependent activation of several human blood cell types by protamine-condensed mRNA. Eur J Immunol (2005) 35:1557–66. doi:10.1002/eji.200425656
161. Kreiter S, Selmi A, Diken M, Koslowski M, Britten CM, Huber C, et al. Intranodal vaccination with naked antigen-encoding RNA elicits potent prophylactic and therapeutic antitumoral immunity. Cancer Res (2010) 70:9031–40. doi:10.1158/0008-5472.CAN-10-0699
162. Van Lint S, Goyvaerts C, Maenhout S, Goethals L, Disy A, Benteyn D, et al. Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy. Cancer Res (2012) 72:1661–71. doi:10.1158/0008-5472.CAN-11-2957
163. Hecker JG. Nonviral, cationic lipid-mediated delivery of mRNA. Methods Mol Biol (2013) 969:73–88. doi:10.1007/978-1-62703-260-5_5
164. Uchida S, Itaka K, Uchida H, Hayakawa K, Ogata T, Ishii T, et al. In vivo messenger RNA introduction into the central nervous system using polyplex nanomicelle. PLoS One (2013) 8:e0056220. doi:10.1371/journal.pone.0056220
165. Pollard C, Rejman J, De Haes W, Verrier B, Van Gulck E, Naessens T, et al. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol Ther (2013) 21:251–9. doi:10.1038/mt.2012.202
166. Perche F, Benvegnu T, Berchel M, Lebegue L, Pichon C, Jaffres PA, et al. Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumor antigen messenger RNA. Nanomedicine (2011) 7:445–53. doi:10.1016/j.nano.2010.12.010
167. Bahl K, Senn JJ, Yuzhakov O, Bulychev A, Brito LA, Hassett KJ, et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol Ther (2017) 25:1316–27. doi:10.1016/j.ymthe.2017.03.035
168. Hekele A, Bertholet S, Archer J, Gibson DG, Palladino G, Brito LA, et al. Rapidly produced SAM (R) vaccine against H7N9 influenza is immunogenic in mice. Emerg Microbes Infect (2013) 2:e52. doi:10.1038/emi.2013.54
169. Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci U S A (2012) 109:14604–9. doi:10.1073/pnas.1209367109
170. Magini D, Giovani C, Mangiavacchi S, MacCari S, Cecchi R, Ulmer JB, et al. Self-amplifying mRNA vaccines expressing multiple conserved influenza antigens confer protection against homologous and heterosubtypic viral challenge. PLoS One (2016) 11:e0161193. doi:10.1371/journal.pone.0161193
171. Pepini T, Pulichino A-M, Carsillo T, Carlson AL, Sari-Sarraf F, Ramsauer K, et al. Induction of an IFN-mediated antiviral response by a self-amplifying RNA vaccine: implications for vaccine design. J Immunol (2017) 198:4012–24. doi:10.4049/jimmunol.1601877
172. Hoelscher MA, Garg S, Bangari DS, Belser JA, Lu X, Stephenson I, et al. Development of adenoviral-vector-based pandemic influenza vaccine against antigenically distinct human H5N1 strains in mice. Lancet (2006) 367:475–81. doi:10.1016/S0140-6736(06)68076-8
173. Kreijtz JH, Suezer Y, de Mutsert G, van den Brand JM, van Amerongen G, Schnierle BS, et al. Preclinical evaluation of a modified vaccinia virus Ankara (MVA)-based vaccine against influenza A/H5N1 viruses. Vaccine (2009) 27:6296–9. doi:10.1016/j.vaccine.2009.03.020
174. Mooney AJ, Li Z, Gabbard JD, He B, Tompkins SM. Recombinant parainfluenza virus 5 vaccine encoding the influenza virus hemagglutinin protects against H5N1 highly pathogenic avian influenza virus infection following intranasal or intramuscular vaccination of BALB/c mice. J Virol (2013) 87:363–71. doi:10.1128/JVI.02330-12
175. Yang SG, Wo JE, Li MW, Mi FF, Yu CB, Lv GL, et al. Construction and cellular immune response induction of HA-based alphavirus replicon vaccines against human-avian influenza (H5N1). Vaccine (2009) 27:7451–8. doi:10.1016/j.vaccine.2009.05.014
176. Kamlangdee A, Kingstad-Bakke B, Anderson TK, Goldberg TL, Osorio JE. Broad protection against avian influenza virus by using a modified vaccinia Ankara virus expressing a mosaic hemagglutinin gene. J Virol (2014) 88:13300–9. doi:10.1128/JVI.01532-14
177. Barouch DH, O’Brien KL, Simmons NL, King SL, Abbink P, Maxfield LF, et al. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med (2010) 16:319–23. doi:10.1038/nm.2089
178. Price GE, Soboleski MR, Lo CY, Misplon JA, Quirion MR, Houser KV, et al. Single-dose mucosal immunization with a candidate universal influenza vaccine provides rapid protection from virulent H5N1, H3N2 and H1N1 viruses. PLoS One (2010) 5:e13162. doi:10.1371/journal.pone.0013162
179. Price GE, Lo CY, Misplon JA, Epstein SL. Mucosal immunization with a candidate universal influenza vaccine reduces virus transmission in a mouse model. J Virol (2014) 88:6019–30. doi:10.1128/JVI.03101-13
180. Kim EH, Han G-Y, Nguyen H. An adenovirus-vectored influenza vaccine induces durable cross-protective hemagglutinin stalk antibody responses in mice. Viruses (2017) 9:234. doi:10.3390/v9080234
181. Draper SJ, Moore AC, Goodman AL, Long CA, Holder AA, Gilbert SC, et al. Effective induction of high-titer antibodies by viral vector vaccines. Nat Med (2008) 14:819–21. doi:10.1038/nm.1850
182. Rollier CS, Reyes-Sandoval A, Cottingham MG, Ewer K, Hill AV. Viral vectors as vaccine platforms: deployment in sight. Curr Opin Immunol (2011) 23:377–82. doi:10.1016/j.coi.2011.03.006
183. Lambe T. Novel viral vectored vaccines for the prevention of influenza. Mol Med (2012) 18:1153–60. doi:10.2119/molmed.2012.00147
184. Geiben-Lynn R, Greenland JR, Frimpong-Boateng K, Letvin NL. Kinetics of recombinant adenovirus type 5, vaccinia virus, modified vaccinia ankara virus, and DNA antigen expression in vivo and the induction of memory T-lymphocyte responses. Clin Vaccine Immunol (2008) 15:691–6. doi:10.1128/CVI.00418-07
185. Sekaly R-P. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J Exp Med (2008) 205:7–12. doi:10.1084/jem.20072681
186. Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature (2011) 473:523–7. doi:10.1038/nature10003
187. Geall AJ, Mandl CW, Ulmer JB. RNA: the new revolution in nucleic acid vaccines. Semin Immunol (2013) 25:152–9. doi:10.1016/j.smim.2013.05.001
188. Low JGH, Lee LS, Ooi EE, Ethirajulu K, Yeo P, Matter A, et al. Safety and immunogenicity of a virus-like particle pandemic influenza A (H1N1) 2009 vaccine: results from a double-blinded, randomized phase I clinical trial in healthy Asian volunteers. Vaccine (2014) 32:5041–8. doi:10.1016/j.vaccine.2014.07.011
189. Valero-Pacheco N, Pérez-Toledo M, Villasís-Keever MÁ, Núñez-Valencia A, Boscó-Gárate I, Lozano-Dubernard B, et al. Antibody persistence in adults two years after vaccination with an H1N1 2009 pandemic influenza virus-like particle vaccine. PLoS One (2016) 11(2):e0150146. doi:10.1371/journal.pone.0150146
190. Pillet S, Aubin É, Trépanier S, Bussière D, Dargis M, Poulin J-F, et al. A plant-derived quadrivalent virus like particle influenza vaccine induces cross-reactive antibody and T cell response in healthy adults. Clin Immunol (2016) 168:72–87. doi:10.1016/j.clim.2016.03.008
191. Lim JJ, Deng R, Derby MA, Larouche R, Horn P, Anderson M, et al. Two phase 1, randomized, double-blind, placebo-controlled, single-ascending-dose studies to investigate the safety, tolerability, and pharmacokinetics of an anti-influenza A virus monoclonal antibody, MHAA4549A, in healthy volunteers. Antimicrob Agents Chemother (2016) 60:5437–44. doi:10.1128/AAC.00607-16
192. Wollacott AM, Boni MF, Szretter KJ, Sloan SE, Yousofshahi M, Viswanathan K, et al. Safety and upper respiratory pharmacokinetics of the hemagglutinin stalk-binding antibody VIS410 support treatment and prophylaxis based on population modeling of seasonal influenza A outbreaks. EBioMedicine (2016) 5:147–55. doi:10.1016/j.ebiom.2016.02.021
193. Ramos EL, Mitcham JL, Koller TD, Bonavia A, Usner DW, Balaratnam G, et al. Efficacy and safety of treatment with an anti-M2e monoclonal antibody in experimental human influenza. J Infect Dis (2015) 211:1038–44. doi:10.1093/infdis/jiu539
194. Stoloff GA, Caparros-Wanderley W. Synthetic multi-epitope peptides identified in silico induce protective immunity against multiple influenza serotypes. Eur J Immunol (2007) 37:2441–9. doi:10.1002/eji.200737254
195. Ledgerwood JE, Wei C-J, Hu Z, Gordon IJ, Enama ME, Hendel CS, et al. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis (2018) 11:916–24. doi:10.1016/S1473-3099(11)70240-7
196. DeZure AD, Coates EE, Hu Z, Yamshchikov GV, Zephir KL, Enama ME, et al. An avian influenza H7 DNA priming vaccine is safe and immunogenic in a randomized phase I clinical trial. NPJ Vaccines (2017) 2:15. doi:10.1038/s41541-017-0016-6
197. Van Kampen KR, Shi Z, Gao P, Zhang J, Foster KW, Chen D-T, et al. Safety and immunogenicity of adenovirus-vectored nasal and epicutaneous influenza vaccines in humans. Vaccine (2005) 23:1029–36. doi:10.1016/j.vaccine.2004.07.043
198. Gurwith M, Lock M, Taylor EM, Ishioka G, Alexander J, Mayall T, et al. Safety and immunogenicity of an oral, replicating adenovirus serotype 4 vector vaccine for H5N1 influenza: a randomised, double-blind, placebo-controlled, phase 1 study. Lancet Infect Dis (2013) 13:238–59. doi:10.1016/S1473-3099(12)70345-6
Keywords: influenza, hemagglutinin, virus-like particles, universal flu vaccine, neutralizing antibodies, vaccination strategies, functional antibody responses
Citation: Kumar A, Meldgaard TS and Bertholet S (2018) Novel Platforms for the Development of a Universal Influenza Vaccine. Front. Immunol. 9:600. doi: 10.3389/fimmu.2018.00600
Received: 24 November 2017; Accepted: 09 March 2018;
Published: 23 March 2018
Edited by:
Rashika El Ridi, Cairo University, EgyptReviewed by:
Karin Loré, Karolinska Institute (KI), SwedenRandy A. Albrecht, Icahn School of Medicine at Mount Sinai, United States
Copyright: © 2018 Kumar, Meldgaard and Bertholet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sylvie Bertholet, sylvie.c.bertholet-girardin@gsk.com
†Present address: Arun Kumar, Department of Clinical and Experimental Medicine, Linköping University, Linköping, Sweden;
Trine Sundebo Meldgaard, DTU Nanotech, Technical University of Denmark, Copenhagen, Denmark
 Arun Kumar
Arun Kumar Trine Sundebo Meldgaard
Trine Sundebo Meldgaard Sylvie Bertholet
Sylvie Bertholet