- 1Department of Biology, Virginia Commonwealth University, Richmond, VA, United States
- 2Department of Biochemistry and Molecular Biology, Virginia Commonwealth University, Richmond, VA, United States
Mast cells are tissue resident, innate immune cells with heterogenous phenotypes tuned by cytokines and other microenvironmental stimuli. Playing a protective role in parasitic, bacterial, and viral infections, mast cells are also known for their role in the pathogenesis of allergy, asthma, and autoimmune diseases. Here, we review factors controlling mast cell activation, with a focus on receptor signaling and potential therapies for allergic disease. Specifically, we will discuss our work with FcεRI and FγR signaling, IL-4, IL-10, and TGF-β1 treatment, and Stat5. We conclude with potential therapeutics for allergic disease. Much of these efforts have been influenced by the work of Bill Paul. With many mechanistic targets for mast cell activation and different classes of therapeutics being studied, there is reason to be hopeful for continued clinical progress in this area.
Introduction
Mast cells were first described by Paul Ehrlich in 1878. The future Nobel Laureate identified them based on their unique staining characteristics with aniline dyes and their position in all tissues of the body, particularly at interfaces with the external environment (1). As with T cells and macrophages, mast cells are a heterogeneous population, consisting of at least two major subsets (2). Although the origin of these cells remained elusive, seminal work in the 1970s and 1980s established that mast cells originate from hematopoetic stem cells in the bone marrow, spleen, fetal liver, and peripheral blood (3–5). Subsequent work showed that mast cells can be differentiated and expanded in vitro with relative ease, which greatly increased interest and progress in the field. What followed was detailed work describing how mast cells bind and respond to IgE, providing evidence for the role of mast cells in allergic disease (6, 7).
However, our understanding of mast cell biology changed drastically in the late 1980s with work by Bill Paul and colleagues. Bill Paul’s career centered on understanding T cell function and cytokine biology, contributing to the discovery, and understanding of T cell MHC-restriction, the B cell receptor mIg, IL-4, and Th2 polarization, as he eloquently described in a review of his life’s work (8). Following the discovery of IL-4, Bill Paul’s group showed that transformed and non-transformed mast cells express IL-4 in response to PMA and ionomycin (9) and that mast cells secrete a Th2-like panel of cytokines, including IL-4, in response to IgE receptor cross-linking (10). These were tectonic shifts in our fundamental understanding of mast cells, providing evidence that in addition to granule release, mast cells produce cytokine mediators that influence adaptive immunity and have a broader role in allergic disease. It is in keeping with Bill Paul’s visionary abilities that he could abruptly cast a broad light on field tangential to his primary interests. He would go on to publish two dozen mast cell-related articles, including one that initiated our group’s focus on Stat5 in mast cell biology (11). Furthermore, Bill trained many researchers who have gone on to have productive careers in the field of mast cell biology and allergic disease, including the senior author of this article, Takashi Saito, Fred Finkleman, Melissa Brown, Achsah Keegan, and Joshua Milner, many of whom have work cited here. In this review, we will cover several areas of mast cell activation and homeostasis, all of which are of great interest to our lab and have been impacted by Bill Paul’s intellect and productivity.
Mast Cell Growth, Survival, and Apoptosis
Mast cells are long-living tissue-resident immune cells that migrate to and differentiate within the tissue. Development, migration, and survival are shaped by two growth factors, in particular, SCF and IL-3, which are included in Figure 1. In healthy tissue, mast cells are maintained in constant numbers, while the mast cell population increases dramatically in chronically allergic tissue (12). This section will summarize findings on mast cell survival and death. Prior to discovery of the c-Kit receptor and its ligand SCF, mice with double mutations at the ckit-encoding W loci (W/Wv mice) or scf-encoding Sl loci (Sl/Sld mice) were known to exhibit hypoplastic, macrocytic anemia, sterility, and a lack of cutaneous melanocytes (13–15). Importantly, these mice were found to have a defect of mast cells in W/Wv mice due to lineage abnormality and a defect of mast cells in Sl/Sld mice due to an abnormality in the microenvironment (4, 16). A decade later, two groups reported that the W gene product encodes the c-Kit tyrosine kinase receptor (17, 18), while in 1990, eight groups described and identified the ligand for c-Kit: SCF/MGF/steel factor, encoded by the SI locus [prefaced in Ref. (19)]. These papers clarified the complementary receptor–ligand relationship yielding the similar phenotypes of W/Wv and Sl/Sld mice and suggested a role for c-Kit and SCF in mast cell development.
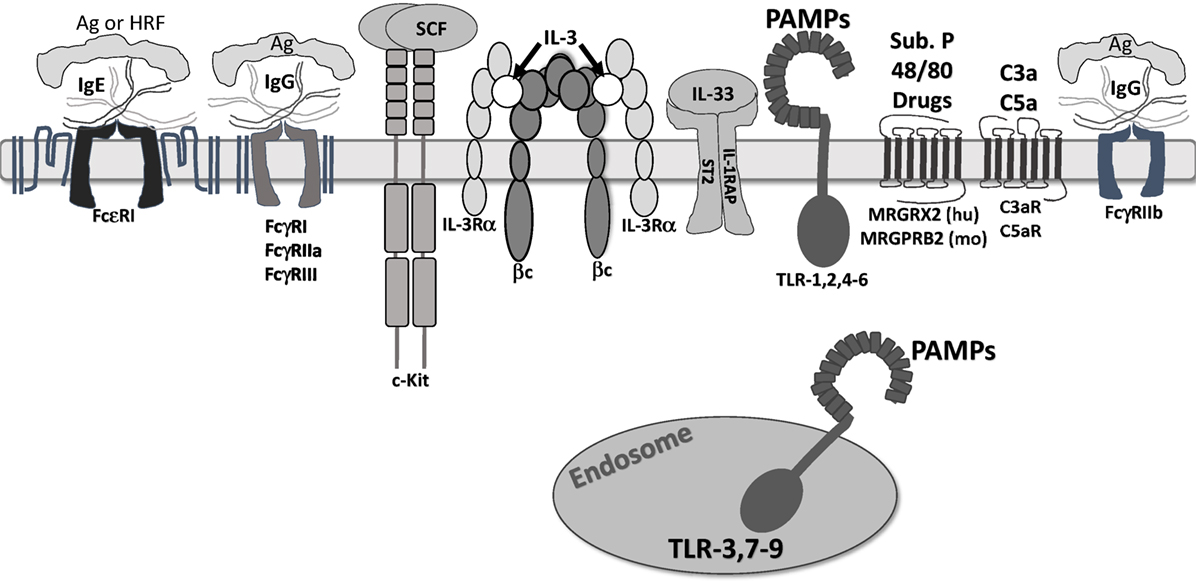
Figure 1. Receptors that regulate mast cell function. The receptors shown are confirmed to regulate mast cell function. They are depicted at approximate scale. All except FcγRIIb are known to induce mast cell degranulation and/or cytokine secretion. FcγRIIb activates SHIP-1 and SHP-1, suppressing inositol and tyrosine kinase activity. c-Kit is a weak mast cell activator, but augments signals by other receptors. IL-3 receptor is modeled after work by Broughton et al. (20). Note that ligands for Mas-related G protein-coupled receptor-X2 (MRGPRX2)/B2 are not fully known, but include drug classes discussed in the accompanying text. In addition, we do not show “cytokinergic” IgE molecules. These form aggregates in the absence of antigen and elicit FcεRI signaling.
c-Kit is a tyrosine kinase growth factor receptor, with a large extracellular domain of five Ig-like domains, a single transmembrane span, and a long cytosolic tail, containing a tyrosine kinase domain and tyrosine phosphorylation sites (21). Dimerization initiates phosphorylation of at least eight tyrosine residues, serving as a docking site for Src homology 2 domains present on signaling proteins such as Grb2, Gab2, Lyn, Fyn, PI3K, phospholipase Cɣ, and the negative regulator, SHP-1 (22).
SCF is best known for eliciting mast cell survival and inhibiting apoptosis (23). SCF-mediated activation of the PI3K–AKT cascade is important for mast cell survival, inactivating the pro-apoptotic proteins Bad and Bim, and increasing expression of the pro-survival proteins, such as Bcl-2 and Bcl-XL (24, 25). Interestingly, somatic and germline ckit gain-of-function mutations are present in mastocytosis patients (26, 27), suggesting regulatory control by the c-Kit pathway.
In addition to its role in survival, SCF has other important effects. These include inducing mast cell migration, adhesion, and IL-6 secretion (28–30). SCF also augments mast cell activation by FcεRI, ST2, and TLR4, receptors which will be further reviewed below (31–33). c-Kit signaling enhances mast cell degranulation and cytokine production by inducing calcium influx and transcriptional activity (34). These abilities make SCF arguably the most critical factor controlling mast cell biology.
IL-3 is also an important factor for mouse mast cell precursor survival, with an unclear role in human mast cells. Mouse mast cells can differentiate in response to IL-3 (mucosal tissue type) or IL-3 + SCF (connective tissue type), while human mast cells typically require SCF + IL-6 for differentiation (35–38). Contradictory studies suggest that IL-3 plays no role in human mast cell progenitor differentiation (38), while a recent study demonstrated that IL-3 alone is sufficient to drive differentiation and survival of human mast cell progenitors (39). Similar to SCF, withdrawal of IL-3 from cultured mast cells induces apoptosis and appears to play an important role in mast cell survival and development in at least mouse mast cells (23, 40). Lantz et al. showed that IL-3-deficient mice have normal numbers of mast cells in the naïve state, but fail to properly expand intestinal mast cells in response to parasite infection (41).
To balance cell growth, mast cells have pathways for both apoptosis and autophagy. The Fas and TRAIL death receptors are expressed on primary mouse and human mast cells and various mast cell lines (42, 43). Mast cells are susceptible to Fas- or TRAIL-mediated apoptosis via caspase activation (1, 43). However, they can overcome Fas death signals by upregulating Fas-associated death domain-like IL-1-converting enzyme-inhibitory protein, a caspase-8 inhibitor that lacks the cysteine domain (42). While transformed and healthy mast cells use this mechanism to bypass Fas-mediated cell death, making this an unreliable and weak apoptosis inducer (43), our group has shown that that BMMCs are more responsive to Fas/FasL-induced apoptosis in the presence of IL-4 and IL-10 (44). Furthermore, rather than acting solely as an apoptosis signal, Fas functions in mast cell development and maturation (45).
In addition to apoptosis, mast cells also undergo autophagy, which controls the clearance and reuse of intracellular organelles and proteins and is essential for eukaryotic cell survival. Light chain (LC)-3 is localized to autophagosomes through conversion of LC3-I to LC3-II, which requires Atg5 and -7. LC3-II-expressing autophagosomes are delivered to lysosomes, where the auto-cargo is degraded (46). Conversion of LC3-I to IC3-II has been shown to be constitutive in BMMC, in which LC3-II associated with secretory granules (47). The same study showed that BMMC lacking Atg7 or 12 have normal granule formation, but defective IgE-mediated degranulation, demonstrating the importance of autophagic machinery in granule movement and release. Furthermore, dysregulation of autophagy in mast cells has been shown in various disease states such as systemic sclerosis, chronic rhinosinusitis, and asthma (48–50). Overall, research on mast cell survival, apoptosis, and autophagy pathways suggests that these pathways are important for both maintenance and function in both health and disease.
Activating Ligands and Receptors Regulating Mast Cell Function
Mast cells respond to myriad signals consistent with their role in defense against pathogens, while contributing to their effects in allergy, asthma, and autoimmunity. In addition, mast cells are a heterogeneous population, particularly susceptible to different tissue microenvironments during development and maturation. Residence in different tissues is linked to distinct mast cell protease and receptor expression (51–53). Moreover, mast cell phenotype is highly “tunable” based on short-term modulation by inflammatory stimuli, growth factors, cytokines, and metabolites (52, 54–56). We will first describe several activating receptors and their stimuli, which are depicted in Figure 1.
IgE and FcεRI
FcεRI, the receptor for IgE, has been the most commonly studied mast cell receptor. While new studies suggest a role for many mediators in allergic disease, IgE remains the best-understood mechanism of mast cell activation in allergic disease (57). The interaction between mast cells and IgE was first shown in 1970 (6, 7). A previously unknown component found in the serum of allergic patients, IgE played a role in the classic Prausnitz–Küstner reaction. In addition, these studies showed that IgE bound to mast cells and basophils. Further characterization found the interaction between IgE and FcεRI, showing that monomeric IgE binds to a singular unit of FcεRI with very high affinity (58). Subsequent studies showed that FcεRI consists of three subunits: one IgE-binding α subunit, one β subunit, and a dimer of disulfide-linked γ subunits (59). FcεRI-mediated activation typically occurs when IgE, bound by its Fc portion (specifically the CH3 domain), interacts with antigen via the Fab portion, driving receptor aggregation. This initiates signaling cascades dependent on tyrosine phosphorylation, leading to a biphasic response. The first phase is the immediate degranulation. The second phase is the production of other mediators such as prostaglandins, leukotrienes, cytokines, chemokines, and growth factors (60).
Many of the pathways controlling FcεRI-induced mast cell activation have been described in detail, and we direct the reader to other recent reviews (61–63). However, some aspects of FcεRI activation warrant special attention here. In addition to signaling induced by receptor aggregation, monomeric IgE induces mast cell survival and activation (64–66). Prior to this, there was knowledge that IgE greatly increases FcεRI expression (67, 68), but its ability to induce signals in the absence of antigen was unexpected and controversial. Subsequent studies revealed that the clone of IgE molecule used greatly impacted its effects, with different IgE clones being designated “highly cytokinergic” or “poorly cytokinergic.” The latter type has since been shown to form large aggregates through Fv–Fv interactions in the absence of antigen, and hence signals much like IgE + antigen (69). In a similar way, histamine-releasing factor (also called translationally controlled tumor protein or fortillin) binds the Fab region on approximately 25% of IgE molecules tested, allowing for clonally restricted mast cell activation that may be more common among atopic patients (59). These unexpected and clinically important effects of IgE subtypes emphasize the importance of fundamentally understanding receptor–ligand interactions. They also support the approach of suppressing IgE–FcεRI interaction, which is proving effective with the drug omalizumab, discussed in the “Potential Therapies” section below.
IgG and FcγR
Mast cells can be activated by IgG immune complexes binding pro-inflammatory FcγRI, FcγRIIA, or FcγRIII, which are variably expressed on mouse and human mast cells. These receptors induce a signaling cascade resembling IgE–FcεRI activation that elicits cytokine secretion, arachidonic acid metabolism, and degranulation (70). Our lab has published work suggesting that like FcεRI, FcγR induces Fyn, Lyn, Akt, Erk, p38, and JNK phosphorylation (71–73). Opposingly, the ITIM-containing receptor FcγRIIb activates SHIP-1 and SHP-1, suppressing IgG-induced signals by reducing PI3K and tyrosine kinase activities (74). IgG-mediated mast cell function is less understood than its IgE counterpart, including its direct clinical roles. Various studies show associations for FcγR-induced mast cell function in rheumatoid arthritis, Sjögren’s syndrome, multiple sclerosis, bullous pemphigoid, thyroiditis, systemic sclerosis, and glomerulonephritis (74). Thus far, much of these data are correlative. For example, mast cells and their mediators are increased in the rheumatoid synovium (75). Moreover, the results of some studies are contradictory. While there were initial reports of the inflammatory role of mast cells in experimental autoimmune encephalitis [EAE; (76)], there are contradictory reports on the role of mast cells in the disease, potentially due to differences in the severity of the EAE protocols or the mast cell-deficient mouse strains used (77, 78). While many studies have made use of W/Wv and Wsh/Wsh c-Kit-deficient mice, newer mast cell knockout models, such as the CPA3-cre/+ mice cited above and the CPA3-Cre; Mcl-1fl/fl, do not have the same changes in neutrophils observed with mutations in c-Kit. Therefore, future studies may clarify the role of mast cells in IgG-associated pathologies such as multiple sclerosis and arthritis (77, 79). This area of research may provide high-impact outcomes, since IgG-induced inflammatory diseases involve pathological processes overlapping with the IgE response and may therefore be responsive to similar therapies.
Complement Receptors
The complement system is made up of proteolytic pro-enzymes and non-enzymatic proteins that form functional complexes, co-factors, regulators, and receptors (80). Larger fragments derived from C3 and C4 regulate opsonization, phagocytosis, and immunomodulation. The smaller fragments C3a and C5a are anaphylatoxins that mediate inflammatory reactions. Anaphylatoxins can activate mast cells but, like other signals, the outcome depends on factors such as location and microenviroment (81). A few studies have examined the role of complement in mast cell activation. C3a enhances mast cell degranulation in the presence of FcγRI signaling (82), while C5a induces mast cell migration, adhesion, and mast cell mediator production (83).
Pathogen-Associated Molecular Patterns and Toll-Like Receptors (TLRs)
Toll-like receptors are a part of the pattern-recognition receptor family interacting with a multitude of pattern-associated molecular patterns as well as host-derived damage associated molecular patterns. This family consists of 10 reported receptors (TLR 1–10) (84). Mast cells can express 9 TLRs, including TLR-1, 2, and 4–6 on the cell surface and TLR-3, and 7–9 intracellularly (85). However, some caveats are worth noting: TLR8 has not been detected on human mast cells; receptor distribution varies with mast cell location; and some receptors have only been shown at the mRNA level (86).
Toll-like receptor activation induces cytokine secretion, which can proceed through a DAP12-independent signaling cascade (87). TLR-induced cytokine profiles overlap but have some distinctions. For example, TLR-2 activation led to the production of TNF, IL-6, IL-13, IL-4, and IL-5, while TLR-4 induced TNF, IL-6, IL-13, and IL-1b (88). Interestingly, pre-exposure to TLR ligands suppressed IgE-induced mast cell responses in two mouse models, possibly by transiently reducing FcεRI expression (89, 90). By contrast, simultaneous exposure of human mast cells to various TLR ligands and FcεRI stimulation yielded increased cytokine secretion without altering degranulation (91). This is an area that warrants further study and clarification, since environmental and even laboratory exposure to allergens (e.g., house dust mite extract) is often in the context of TLR ligands.
Compound 48/80, Substance P, and Mas-Related G Protein-Coupled Receptor-X2 (MRGPRX2)
Mas-related G protein-coupled receptor-X2 has drawn increased interest recently. Human MRGPRX2 is one of approximately 50 7-transmembrane domain proteins in the larger Mas-related gene family. It is unique in its apparently selective expression on human mast cells in the MCTC subtype, outside of the dorsal root ganglion (92). While the MRGPRX gene family is restricted to humans and other primates, a mouse ortholog, MRGPRB2 has recently been described (93). Using transgene and knockout approaches, this group showed that MRGPRB2 is similarly restricted in expression to connective tissue mast cells. Both orthologs are functional receptors for compound 48/80 and the neuropeptide substance P. While of interest to those using these well-known mast cell-activating stimuli, the more important point is that MRGPRX2 (and likely its mouse ortholog) responds to other peptides and drugs. For example, MRGPRX2 binds the host defense protein LL-37 (94). Three classes of drugs have been shown to activate MRGPRX2: fluoriquinone antibiotics such as ciprofloxacin, neuromuscular-blocking drugs such as rocurinium, and the bradykinin B2 receptor antagonist icatibant (95). Although much remains to be done in this area, it appears that MRGPRX2 has an important role in the pseudoallergic reactions induced by some drugs. Inhibiting this receptor may therefore be clinically important.
Chemokines and Their Receptors
Chemokines are cytokines known to induce cellular locomotion. These are particularly important in the migration of cells to areas of inflammation. All chemokine receptors described are seven-transmembrane-spanning G protein receptors (96). Mast cells have been shown to express multiple chemokine receptors, including CCR1, CCR3–5, CXCR1–4, and CX3CR1 (97). These play a significant role in directing mast cell progenitors to the tissues where they mature, a process that is altered by on-going inflammation (98–100). In addition to migration, recent studies show that chemokines can induce partial degranulation (100–102).
Cytokines and Their Receptors
Cytokines act as messengers and modulate many functions including growth, proliferation, and migration. The importance of cytokines to mast cell biology was first shown in the 1980s, when a method to culture mast cells in vitro was being determined. Several groups showed that mast cells could be cultured in media from concanavalin A-activated T cells, cloned Ly + 2 inducer T cells, or WEHI-3B tumor cells. Analysis of this media showed the presence of numerous cytokines such as IL-3, IL-4, IL-9, IL-10, and nerve growth factor (1). The precise role of each cytokine is still a topic of research, partly because mast cells are a heterogenous population due to the microenvironment determining mature phenotype (103). This plasticity allows mast cells to alter their phenotype throughout their lifespan, with the phenotypic profile shaped by the cytokine and growth factor milieu they encounter (54). Our lab has been specifically interested in IL-4, IL-10, and TGF-β, which will be addressed below. In addition, the cytokines IL-33 and TSLP deserve specific attention here, due to their ability to activate mast cells and their known role in allergic disease.
IL-33 is an unusual cytokine, in that it is constitutively produced as a pro-form and localized to the nucleus of barrier cells such as keratinocytes, epithelial cells, endothelial cells, and fibroblasts. Its cleavage and release is stimulated by cell damage or inflammation, supporting its classification as an alarmin (104). Mast cells can also secrete IL-33 upon activation with signals such as IgE stimulation (105). IL-33 interacts with a receptor complex composed of T1/ST2 and IL-1RAcP (106, 107), triggering a MyD88-dependent NFκB-activating cascade resembling TLR signaling. Mast cells were among the first lineages shown to express T1/ST2 (108), 7 years before IL-33 was identified (109). IL-33 is a potent mast cell activator, eliciting survival, maturation, adhesion, and cytokine production (15, 106, 107, 110, 111). IL-33 also enhances mast cell responses to IgE (112) and IgG (113). Furthermore, IL-33 injections induce a rapid peritoneal neutrophil influx that requires mast cell-derived TNF secretion (114). IL-33 is now thought to play a significant role in mast cell-associated diseases such allergy, although precise mast cell-restricted functions are not clear and remain to be elucidated (115).
TLSP has some similarities to IL-33, including expression by epithelial and other barrier cell types and constitutive production among some lineages. In 2015, Bill Paul’s group published a ZsGreen TSLP reporter mouse, which showed TSLP expression not only in epithelial cells and keratinocytes but also in dendritic cells, basophils, and mast cells (116). TSLP secretion is induced by TLR-type signals, allergen and air irritant exposure, viral and bacterial infection, and trauma (115). TSLP interacts with a complex of TSLP-R and IL-7Rα. TSLP KO mice have reduced mast cell numbers (117), which is consistent with data showing TSLP induces mast cell proliferation through a Stat6- and MDM2-dependent pathway (117). TSLP does not induce mast cell degranulation and alone is a poor inducer of cytokine secretion. However, in combination with IL-1β + TNF, TSLP elicits the release of many cytokines, including IL-5, IL-6, and IL-13 from human CD34+ progenitor-derived mast cells (110, 111). It should be noted that TSLP-induced cytokine secretion has not been shown using mouse mast cells. Hence, while TSLP is clearly relevant to mast cell development and function, further studies should examine differences between mice and humans, which may be important caveats for experimental design.
IL-4, IL-10, and TGFβ1 Regulate Mast Cell Function and Homeostasis
Regarding other cytokine effects, our lab has specific interest and experience studying IL-4, IL-10, and TGFβ effects on mast cell function and homeostasis. These cytokines augment or impair activation by the mechanisms introduced above, but do not directly induce mast cell activation alone.
IL-4
IL-4, originally termed B cell stimulatory factor-1, is a cytokine primarily known for its role in antibody driven-allergic disease and protection from parasite infections (118). IL-4 was first discovered to induce B cell proliferation during anti-IgM stimulation and to promote isotype switching to IgG1 and IgE (119–121). In addition, Bill Paul’s lab and others showed that IL-4 elicits Th2 differentiation from naïve T cells in vitro, which subsequently release IL-4 in a positive feedback loop (122, 123). IL-4 signals through IL-4Rα, as part of a heterodimer containing either the common gamma chain (124) or IL13Rα (125). These receptors allow IL-4 to act on non-hematopoietic cells such as intestinal and bronchial epithelial cells and the vasculature to facilitate the protective expulsion of parasites. For details on IL-4 signaling pathways, we direct readers to reviews (126–128).
Unfortunately, IL-4 is also a major contributor to the symptoms observed with allergy and asthma (129–131). IL-4 was the first cytokine shown to be produced by mast cell lines (9), later confirmed to be secreted in response to IgE and lectin activation in human mast cells (132) as well as IL-33 in mouse mast cells (105). Mast cells also respond to IL-4, first reported to increase proliferation of mast cell lines costimulated with IL-3 by Bill Paul’s group (133). This work has been supported with data from other mast cell lines, human gut mast cells, BMMC, and in a mouse model of food allergy (9, 134, 135). Culture with IL-4 + IgE for 4–21 days has been shown to enhance FcεRI receptor expression compared with IgE alone on human cord blood, fetal liver-derived mast cells, and BMMC. It also enhances histamine, PGD2, and LTC4, and IL-5 production following IgE receptor cross-linking (134, 136–139). In addition, IL-4 differentially affected mediator release, augmenting Th2-type cytokines (IL-3, IL-5, and IL-13), and downregulating pro-inflammatory cytokines (IL-6 and TNF) in response to IgE receptor cross-linking and Gram-negative bacterial activation (137).
In contrast to its stimulatory effects on mast cells, IL-4 has been reported to suppress c-Kit expression and mast cell development in human fetal liver-derived mast cells grown in SCF in two studies (138, 140). Similarly, we showed IL-4-mediated inhibition of FcεRI and c-Kit expression on BMMC and PMC following 4–21 days of treatment, an effect dependent on Stat6 (141, 142). IL-4 suppressed IL-4, IL-5, IL-6, and IL-13 secretion induced by IgE crosslinkage, and TNF and IL-13 secretion induced downstream of SCF. Interestingly, our lab subsequently found that IL-4 increases IgG-mediated degranulation and cytokine production in mouse BMMC, involving Stat6 and increased FcγRIIIA protein expression (143). In addition to the role of IL-4 on cellular activation, we found that IL-4 induces apoptosis in developing mouse or human mast cell precursors derived from bone marrow or umbilical cord blood, respectively (144, 145).
The different pro- and anti-inflammatory effects observed by IL-4 are intriguing. Maturation, phenotype, and culture conditions likely play a role in these IL-4 responses, which we also discussed in a recent review (146). Mouse BMMC is considered less mature than human skin, human intestinal, or mouse peritoneal mast cells, which likely contributes to different experimental outcomes. For example, BMMC attain responsiveness to endothelin-1 when cultured in IL-4, while peritoneal mast cells respond to endothelin-1 without IL-4 (147). Similarly, we found that IL-4 induces apoptosis in developing mouse or human mast cell precursors (144, 145), while mature mouse and human mast cells receive survival and proliferation signals from IL-4, which also promotes the MCT (tryptase-positive) phenotype in human intestinal mast cells (137). Hence, IL-4 effects on mast cells vary with stage of differentiation, with suppressive signals being most overt on developing or less mature mast cells. Future research should experimentally clarify these observations, examining cells at different maturation stages and following both IL-3 and IL-3/SCF differentiation to examine the effects of mast cell phenotype.
IL-10
IL-10, originally termed cytokine synthesis inhibitory factor, is a homeostatic mediator in many inflammatory diseases. Secreted by macrophages, Th1 and Th2 cells, regulatory T and B cells, and cytotoxic T cells (148), IL-10 is traditionally considered an anti-inflammatory cytokine. It suppresses monocyte MHC II expression (149), dendritic cell maturation (150), and reduces inflammatory cytokine production from monocytes and neutrophils (151, 152). However, IL-10 also has stimulatory effects. It enhances B cell antibody class switching and plasma cell development (153, 154) and increases IL-2-induced proliferation and cytotoxic activity in NK cells (155). These effects correlate with clinical data, as IL-10 therapy has induced platelet loss in RA patients (156) and promoted IFNγ production in sepsis (157) and Crohn’s patients (158). In agreement with this, anti-IL-10 therapy has improved SLE measures in a clinical trial (159). While it is likely that anti-IL-10 therapy impacts many cell types in vivo, we have studied both the pro- and anti-inflammatory roles of IL-10 in mast cells.
Similar to IL-4, IL-10 is produced by BMMC and affects mast cell survival, proliferation, and function. IL-10 enhances IL-3-mediated growth of mouse mast cells and their progenitors (160–162). Interestingly, when co-cultured with IL-3 and IL-4, IL-10 induces BMMC apoptosis by diminishing Bcl-2 and Bcl-xL expression, in a Stat6-dependent manner (44), and induces apoptosis following IgE receptor cross-linking, a known pro-survival pathway (64).
Early work showed variable responses to IL-10 treatment. IL-10 was shown to induce BMMC expression of mouse mast cell protease (MCPT)-2 (163). Several studies showed inhibition or no change in TNF, IL-6, and histamine secretion following IgE receptor cross-linking and LPS-induced activation in HMC-1, rat peritoneal mast cells, BMMC, and human cord blood-derived mast cells (164–167). Our lab showed that 4-day IL-10 treatment inhibited FcεRI beta chain expression and IgE-induced TNF production in BMMC (168, 169). Recently, we found that while TNF is diminished, IgE-induced degranulation and secretion of other inflammatory cytokines were increased by IL-10 after 24-h treatment, through a Stat3–miR-155 cascade that inhibits the negative regulator, suppressor of cytokine signaling-1 (170). These effects were consistent in mouse and human mast cells and in a model of passive systemic anaphylaxis in our study, as well as a mouse model of food allergy used by Clinton Mathias’s group (160). How IL-10 suppresses TNF while enhancing other pro-inflammatory cytokines is unknown and may be important for understanding how mast cell function can be tuned. Our interpretation of these data is that while IL-10 has well-established inhibitory roles, it is not monolithic. Instead, stimulatory effects are clear from both clinical and basic research outcomes.
TGF-β
Our group has also studied the role of TGF-β in mast cell homeostasis. Similar to IL-10, TGF-β is primarily known for its immunosuppressive effects, but pleotropic activities have been reported based on environmental and differentiation factors (171). TGF-β suppresses T cell proliferation and induces Treg differentiation (172, 173), suppresses B cell proliferation and IgG antibody class switching (172, 174), and inhibits macrophage nitric oxide release and TNF translation (175, 176).
TGF-β also alters mast cell development and function. It enhances early mast cell precursor differentiation and increases protease expression, while antagonizing survival of late stage precursors and mature mast cells (177–179). TGF-β elicits mast cell chemotaxis, but can also suppress migration toward SCF (180–182). Our lab has published that TGF-β-1, -2, and -3 inhibit the expression of FcεRI subunits, c-Kit, T1/ST2, and Fcγ receptor chains in BMMC, peritoneal mast cells, and human skin mast cells. In addition, granule formation, degranulation, and IgE-induced cytokine production were reduced by TGF-β (177, 183, 184). Recently, we found that TGF-β-1, -2, and -3 also inhibit IL-33-induced TNF, IL-6, IL-13, and MCP-1 secretion in mouse and human mast cells and suppress IL-33-induced cytokine production in vivo (185). Interestingly, a mouse model of lung inflammation suggests TGF-β enhances LPS-induced mast cell IL-6 production, ultimately inducing neutrophil apoptosis and controlling neutrophilic inflammation (186).
As with IL-4 and IL-10, TGF-β effects on mast cells are altered by microenvironment and genetic background. For example, IL-4 and TGFβ1 block the expression and function of the other’s receptor, with IL-4 inhibiting TGFβ1-mediated migration and vice versa (187). TGF-β1 has divergent effects on C57Bl6/J versus 129/SvJ mast cells. Not only do 129/SvJ BMMCs resist TGF-β1-mediated suppression of IgE-induced cytokine secretion, these BMMCs show enhanced SCF-induced migration in the presence of TGF-β1 (180). We found that matched C57BL/6J and 129/SvJ BMMC cultures have no difference in TGF-β receptor expression, but 129/SvJ BMMC express twofold to threefold greater levels of Fyn and Stat5 proteins. Since inhibiting the Fyn–Stat5 cascade appears to be important for TGF-β1-mediated suppression, this may convey resistance. In keeping with these BMMC data, we found that human skin mast cells show considerable donor-to-donor variability in TGF-β1-mediated suppression, when measuring IgE-induced cytokine secretion. These donors also showed variable Fyn and Stat5 expression (180). An additional explanation for TGF-β1 resistance is polymorphic TGF-β receptor variation. C57BL/6J and 129/SvJ strains have known polymorphic variations in TGFβR1 between, and similar variations are tied to human cancers (188).
In summary, mast cell development, survival, and function are greatly altered by IL-4, IL-10, and TGF-β. These effects are impacted by microenvironment, since cytokines can act in opposition; by stage of differentiation, since precursors and mature mast cells can respond differently; and by genetic background, with some inbred mouse strains and human donors showing complete resistance or even opposite responses. Understanding how these signals are integrated will provide a coherent approach to mast cell-associated diseases.
The Role of Stat5 in Mast Cell Biology
The transcription factor Stat5 is expressed ubiquitously and activated by many growth factors and cytokines, including IL-2, IL-3, GM-CSF, prolactin, erythropoietin, thrombopoietin, and growth hormone (189). Stat5 is implicated in immune homeostasis and inflammation, as mice lacking the 110-kb Stat5A/B locus had perinatal lethality and severely compromised immune systems, similar to mice lacking the proteins γc, Jak3, or IL-7R (190).
Our lab was the first to show SCF-induced Stat5-DNA binding activity in mast cells (11). Utilizing the Stat5DN mouse, mutated to have truncated Stat5A and B lacking the N-terminus and able to form teramers but not dimers (191), we showed that BMMC from Stat5DN-expressing mice exhibited increased apoptosis and delayed cell cycle progression when cultured in either IL-3 or SCF alone (192). Specifically, we observed reduced expression of the anti-apoptotic proteins Bcl-2, Bcl-(x)l, reduced expression of the cell cycle regulators cyclin A2 and cyclin B1, reduced mitochondrial membrane potential, and greater activation of caspases-9 and -3. In vivo, Stat5DN-expressing mice were born with normal mast cell distribution, but had near complete loss of tissue mast cells by 12 weeks of age, indicating that Stat5 tetramer formation is essential for regulating mast cell survival. Furthermore, neoplastic mast cells transformed by mutant c-Kit have constitutive Stat5 activation, which can be successfully targeted to inhibit proliferation and survival (193, 194). Together, these results suggest that Stat5 plays a critical role in mast cell survival.
We later reported direct and transient Stat5 activation downstream of FcεRI-mediated mast cell stimulation (195). Stat5-deficient BMMC showed impaired immediate and late phase mediator release in response to IgE-induced stimulation, indicating a critical role for Stat5 in mast cell function. A subsequent study found that Stat5 activation depends on Fyn kinase expression, and that Fyn and Stat5 are physically associated in resting mast cells (72, 73). We also noted that Stat5 co-localizes with FcεRI in antigen-activated mast cells. Interestingly, the absence of Lyn kinase, Gab2, or SHP-1 enhanced FcεRI-mediated Stat5 phosphorylation (72, 73).
In addition to FcεRI, Stat5 has been implicated in other signaling cascades controlling mast cell function. While IL-33 does not appear to activate Stat5, Bill Paul’s group showed that IL-33-induced IL-13 secretion required IL-3-mediated Stat5 activation (196). Furthermore, IL-33 elicits a complex between its receptor, ST2/IL-1RAcP, and c-Kit, supporting the finding that IL-33 signaling is enhanced by SCF in mast cells (31). Since c-Kit is a strong Stat5 activator, Stat5 may similarly contribute to IL-33 signaling via this pathway. In a separate line of work, Toshio Kawakami’s group reported enhanced Stat5 activity tied to increased mast cell numbers in animal models of atopic dermatitis (AD) and lesions in the skin of AD patients (197), suggesting that Stat5 contributes to this disease phenotype.
Stat5A and B are encoded by distinct genes (198–200). While murine Stat5A and Stat5B exhibit 96% sequence similarity and a very similar expression pattern, these isoforms are not completely redundant and have unique biological activity (199). For example, Stat5A is critical for murine mast cell proliferation and survival (201). Our own targeting of Stat5A or Stat5B using siRNA has found that Stat5B has a selective influence over IgE-mediated mast cell cytokine release and SCF-induced migration, with Stat5A being dispensable (72, 73, 180). The idea that Stat5 can directly promote allergic disease is supported by recent work from Joshua Milner’s group, who linked a gain-of-function Stat5b mutation to eosinophilia, urticaria, and dermatitis (202). Although no further work was done to study the specific role and functionality of mast cells in these patients, the Stat5 mutation resulted in greater Th2 cytokine production by CD4 T cells, potentially skewing the immune response toward a Th2 response. It should also be noted that increased STAT5b activity, as observed in these patients, was associated with atopic-like skin inflammation that typically involves mast cell activation. Collectively, these findings show that Stat5 is one of the central factors controlling mast cell survival and function. With evidence that Stat5 can be targeted pharmacologically, we see this as a productive avenue for addressing mast cell-associated diseases.
Therapeutics Targeting Mast Cells
Due to the variety of inflammatory diseases in which mast cells participate, targeting mast cell function and survival can be broadly effective. This section will briefly discuss current and potential mast cell-directed therapies. We also refer the reader to several recent reviews focused on this topic (203–206).
There are several therapeutics on the market that are FDA approved for allergic disease or asthma, targeting mast cell activation and/or mediators. These include H1 inhibitors that prevent histamine signaling (207) and antagonists of leukotriene synthesis or signaling via the CysLT1 receptor (208). While these drugs are effective in targeting select mediators, broader inhibition of the many mast cell-derived inflammatory factors can be needed for clinical efficacy. We should note here that the newer generation antihistamines have better binding affinity for histamine receptors, reduced adverse side effects, and often have mast cell stabilizing and anti-inflammatory properties in addition to their effects on histamine, which may provide better relief for patients (209). Corticosteroids such as dexamethasone are effective inhibitors of mast cell activation and a proven treatment for mast cell-associated diseases (210, 211). In fact, we recently found that dexamethasone inhibits not only IgE- but also IL-33-mediated mast cell function (212). However, steroid medications have many adverse effects and reduced efficacy in viral-induced exacerbations, asthmatics who smoke, and in more severe forms of the disease (213, 214). The mast cell stabilizer disodium cromoglycate is approved for some mast cell proliferative and activation diseases (215), although the mechanism of action is poorly understood (216), effects are slower than drugs like antihistamines, and there is evidence that mast cell stabilizers are less efficacious than inhaled corticosteroids for asthma (209, 217). More recently, a humanized anti-IgE antibody, omalizumab, has been developed to prevent binding of circulating IgE to FcεRI and the downstream effects of cross-linking (218, 219). Omalizumab is a common and preferred treatment for chronic urticaria (220, 221) and shows efficacy for the treatment of asthma (218, 219); however, like many drugs, there appear to be responders and non-responders (222). These drugs represent both progress toward suppressing mast cell function and shortcomings supporting further development.
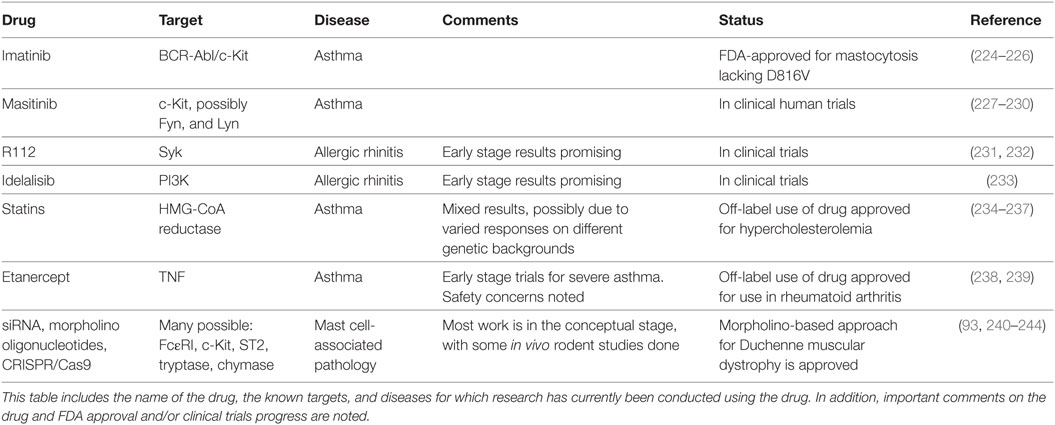
Repurposing FDA-Approved Drugs
There are several FDA-approved therapeutics with potential to treat diseases caused or exacerbated by mast cell activation (Table 1). For instance, the tricyclic antidepressant doxepin was found to be a potent H1 receptor inhibitor and is prescribed to treat chronic urticaria and AD (223). The repurposing approach to targeting mast cells represents a potentially rapid avenue for clinical progress and includes several drug classes.
Kinase Inhibitors
Imatinib is a chemotherapeutic agent designed to target the BCR–ABL tyrosine kinase common in chronic myeloid lymphoma (224). Although imatinib was designed to target the ABL tyrosine kinase domain, it also inhibits c-Kit kinase activity. Due to this off-target effect, imatinib has been used to treat mastocytosis cases lacking the c-Kit D816V mutation (225). More recently, imatinib has been tested in a clinical trial to treat severe refractory asthma. Patients treated with imatinib had reduced airway hyperresponsiveness and decreased serum tryptase levels compared to placebo (226), supporting broader use of this drug. Unlike imatinib, masitinib was designed as a c-Kit kinase inhibitor and has been used therapeutically to treat canine mast cell tumors (227, 228). It has since entered clinical trials for human mastocytosis and asthma. In a phase 2a clinical trial, masitinib improved the quality of life in 14 of 25 mastocytosis patients for at least 60 weeks (229). It is also in a phase 3 clinical trial to treat severe and persistent asthma in conjunction with corticosteroids (227). An additional kinase inhibitor capable of suppressing c-Kit, toceranib phosphate, is being tested in canines (236). Kinase targeting in mast cell-associated diseases is not limited to c-Kit. The Syk kinase inhibitor R112 has shown mixed results for allergic rhinitis in two trials (231, 232). Similarly, the phosphatidyl inositol 3′-kinase inhibitor idelalisib has shown progress in a phase 1 trial for allergic rhinitis (233). These studies collectively support the approach of targeting kinases activated early in signaling cascades controlling mast cell function.
Statins
Statin drugs are HMG-CoA reductase (HMGCR) inhibitors designed to reduce cholesterol synthesis (234). These drugs are primarily approved to treat hypercholesterolemia and reduce cardiovascular disease, but have been beneficial in asthma and atopic diseases, albeit with mixed results. For example, a 1-month trial of simvastatin as a monotherapy for asthma showed little benefit (245), but subsequent studies demonstrated positive effects. Most have employed simvastatin or atorvastatin as an adjuvant therapy. Simvastatin has been shown to decrease eosinophils, improve lung function, and promote Treg development in mild asthmatics (235, 246). Similarly, atorvastatin decreased sputum inflammatory cytokine levels, suppressed LTB4 production, and improved quality of life scores among mild-to-moderate asthmatics (247–249). A retrospective study found that among severe asthmatics, statins in combination with inhaled therapies had better asthma control in comparison to patients who were not currently taking statins (236). Despite these encouraging findings, meta-analysis studies show that the overall effects of statins in asthma are at the best modest (249–251).
These conflicting outcomes prompted us to study statin effects on mast cells. Our lab found that one drug in particular, fluvastatin, blocked FcεRI-mediated mast cell activation in human and mouse mast cells and reduced passive systemic anaphylaxis in mice (237). However, these effects showed strong genetic influences: BMMC derived from C57BL/6J mice were most sensitive, BALB/c showed intermediate responses, and 129/SvJ mice were completely resistant. Human mast cells cultured from multiple donors showed similar variation. Our data showed that statin resistance was not tied to HMGCR coding polymorphisms, but did correlate with drug-induced HMGCR upregulation. More importantly, geranylgeranyl transferase (GGT), downstream of HMGCR in the cholesterol pathway, appears to be critical for FcεRI-mediated function. These findings suggest that statin efficacy in mast cell-associated disease might be predicted by measuring drug-induced HMGCR expression, and that targeting GGT may be a better means of disrupting mast cell function.
Targeting Gene Expression
Another promising means of inhibiting mast cell activation is by selectively suppressing gene expression, including FcεRI, c-Kit, histadine decarboxylase, or other mast cell receptors and mediators. There are several approaches that hold promise, with two decades of clinical trials supporting progress. Morpholino oligomers bind mRNA and either block translation or modify pre-mRNA splicing and induce exon skipping (252). This approach has recently been demonstrated in mice showing that morpholinos targeting the FcεRI β-subunit decreased IgE receptor expression and function on mast cells and basophils (240) and was beneficial in treating a mouse model of allergic dermatitis. Morpholino-based therapy is approved for Duchenne muscular dystrophy (243, 244), suggesting that this approach can succeed.
siRNAs are another gene targeting tool, explored in various clinical trials and on the cusp of FDA approval (253). Similar to morpholinos, siRNAs base pair with mRNAs, inhibiting translation or decreasing half-life. Several studies have used siRNAs to diminish mast cell activation or mediator production in vitro (93, 241, 242, 254). A critical step for any nucleotide-based approach is customizing targeting and improving cellular uptake. Several approaches are under study, including nanoparticles and lipid-based carriers (255). Progress in this area may also come from excitement surrounding the CRISPR/Cas9 system. Although less vetted in clinical trials than morpholinos and siRNA, the specificity and efficacy of CRISPR/Cas9 elicits great hope for molecular-based therapies in many fields, including mast cell-associated disease.
Potential Targets for New Inhibitors
Although several mast cell mediators and receptors are targeted by existing therapies, others warrant consideration. Tryptase is a mast cell protease that is expressed by all mast cells and contributes to inflammation in atopy and several autoimmune diseases by causing smooth muscle contraction and fibrosis (205, 206). Several beta-tryptase inhibitors have entered clinical trials, with APC 366 moving as far as phase 2a for asthma. However, issues with target selectivity, formulation, and reproductive toxicity have thus far prevented these inhibitors from gaining FDA approval (256). There is potential for antibodies targeting tryptase to be used for mastocytosis and atopic diseases (257). Chymase is another pro-inflammatory protease made in abundance by mast cells with the potential to be targeted by chemical inhibitors or antibodies (258). Several small molecule inhibitors such as ONO-WH-236 have been developed to selectively inhibit chymase, but none of these drugs have been clinically tested in patients with mast cell-associated diseases (205, 259).
As stated above, IL-33 activates mast cells (106, 107, 260) and is elevated in patients with asthma and AD (106, 107, 261, 262). Since it also activates Th2 cells, targeting IL-33 or its receptor ST2 could be effective. An anti-ST2 human monoclonal antibody, MSTT1041A, is in phase 2 of clinical trials to treat severe asthma (http://Clinicaltrials.gov, NCT02918019). In addition, an anti-IL-33 monoclonal antibody is in a phase 2 clinical trial for peanut allergy (http://Clinicaltrials.gov, NCT02920021) and also in phase 2 trials for AD (EU Clinical Trials Register number 2016-002539-14). With varied mechanistic targets for mast cell activation and different classes of therapeutics currently being studied, there is reason to be hopeful for progress in this area.
Conclusion
By demonstrating that mast cells produce a Th2-type profile of cytokines, Bill Paul’s group allowed those of us fortunate to work in this field to expand our horizons and our definition of what the mast cell is. The current view is of a long-lived innate immune cell with considerable plasticity that responds to its microenvironment through a range of surface receptors, allowing the mast cell to greatly alter the course of immunity. It is therefore an ideal target for therapeutic intervention and a lineage that still yields novel insights, more than a century after its discovery.
Author Contributions
All authors wrote and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
We gratefully acknowledge funding support from the U.S. National Institutes of Health [1R01AI59638 and 1R01AI101153 (to JR)].
References
1. Hartmann K, Wagelie-Steffen AL, von Stebut E, Metcalfe DD. Fas (CD95, APO-1) antigen expression and function in murine mast cells. J Immunol (1997) 159(8):4006–14.
2. Enerback L. Mast cells in rat gastrointestinal mucosa. 2. Dye-binding and metachromatic properties. Acta Pathol Microbiol Scand (1966) 66:303–12. doi:10.1111/apm.1966.66.3.303
3. Sonoda T, Ohno T, Kitamura Y. Concentration of mast-cell progenitors in bone marrow, spleen, and blood of mice determined by limiting dilution analysis. J Cell Physiol (1982) 112(1):136–40. doi:10.1002/jcp.1041120120
4. Kitamura Y, Go S. Decreased production of mast cells in S1/S1 anemic mice. Blood (1979) 53(3):492–7.
5. Kitamura Y, Yokoyama M, Matsuda H, Ohno T, Mori KJ. Spleen colony-forming cell as common precursor for tissue mast cells and granulocytes. Nature (1981) 291(5811):159–60. doi:10.1038/291159a0
6. Ishizaka K, Tomioka H, Ishizaka T. Mechanisms of passive sensitization. I. Presence of IgE and IgG molecules on human leukocytes. J Immunol (1970) 105(6):1459–67. doi:10.1007/BF00258552
7. Tomioka H, Ishizaka K. Mechanisms of passive sensitization. II. Presence of receptors for IgE on monkey mast cells. J Immunol (1971) 107(4):971–8.
8. Paul WE. Endless fascination. Annu Rev Immunol (2014) 32:1–24. doi:10.1146/annurev-immunol-032713-120247
9. Brown MA, Pierce JH, Watson CJ, Falco J, Ihle JN, Paul WE. B cell stimulatory factor-1/interleukin-4 mRNA is expressed by normal and transformed mast cells. Cell (1987) 50(5):809–18. doi:10.1016/0092-8674(87)90339-4
10. Plaut M, Pierce JH, Watson CJ, Hanley-Hyde J, Nordan RP, Paul WE. Mast cell lines produce lymphokines in response to cross-linkage of FcεRI or to calcium ionophores. Nature (1989) 339(6219):64–7. doi:10.1038/339064a0
11. Ryan JJ, Huang H, McReynolds LJ, Shelburne C, Hu-Li J, Huff TF, et al. Stem cell factor activates STAT-5 DNA binding in IL-3-derived bone marrow mast cells. Exp Hematol (1997) 25(4):357–62.
12. Crimi E, Chiaramondia M, Milanese M, Rossi GA, Brusasco V. Increased numbers of mast cells in bronchial mucosa after the late-phase asthmatic response to allergen. Am Rev Respir Dis (1991) 144(6):1282–6. doi:10.1164/ajrccm/144.6.1282
13. Sarvella PA, Russell LB. Steel, a new dominant gene in the house mouse. J Hered (1956) 47:123–8. doi:10.1093/oxfordjournals.jhered.a106620
14. Mintz B, Russell ES. Gene-induced embryological modifications of primordial germ cells in the mouse. J Exp Zool (1957) 134:207–37. doi:10.1002/jez.1401340202
15. Iikura M, Suto H, Kajiwara N, Oboki K, Ohno T, Okayama Y, et al. IL-33 can promote survival, adhesion and cytokine production in human mast cells. Lab Invest (2007) 87(10):971–8. doi:10.1038/labinvest.3700663
16. Kitamura Y, Go S, Hatanaka K. Decrease of mast cells in W/Wv mice and their increase by bone marrow transplantation. Blood (1978) 52(2):447–52.
17. Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature (1988) 335(6185):88–9. doi:10.1038/335088a0
18. Geissler EN, Ryan MA, Housman DE. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell (1988) 55(1):185–92. doi:10.1016/0092-8674(88)90020-7
19. Witte ON. Steel locus defines new multipotent growth factor. Cell (1990) 63(1):5–6. doi:10.1016/0092-8674(90)90280-R
20. Broughton S, Hervus T, Hardy M, McClure B, Nero T, Dottore M, et al. Dual mechanism of interleukin-3 receptor blockade by an anti-cancer antibody. Cell Rep (2014) 8:410–9. doi:10.1016/j.celrep.2014.06.038
21. Reber L, Da Silva CA, Frossard N. Stem cell factor and its receptor c-Kit as targets for inflammatory diseases. Eur J Pharmacol (2006) 533(1–3):327–40. doi:10.1016/j.ejphar.2005.12.067
22. Linnekin D. Early signaling pathways activated by c-Kit in hematopoietic cells. Int J Biochem Cell Biol (1999) 31(10):1053–74. doi:10.1016/S1357-2725(99)00078-3
23. Iemura A, Tsai M, Ando A, Wershil BK, Galli SJ. The c-kit ligand, stem cell factor, promotes mast cell survival by suppressing apoptosis. Am J Pathol (1994) 144(2):321–8.
24. Hartmann K, Artuc M, Baldus SE, Zirbes TK, Hermes B, Thiele J, et al. Expression of Bcl-2 and Bcl-xL in cutaneous and bone marrow lesions of mastocytosis. Am J Pathol (2003) 163(3):819–26. doi:10.1016/S0002-9440(10)63442-6
25. Möller C, Alfredsson J, Engström M, Wootz H, Xiang Z, Lennartsson J, et al. Stem cell factor promotes mast cell survival via inactivation of FOXO3a-mediated transcriptional induction and MEK-regulated phosphorylation of the proapoptotic protein Bim. Blood (2005) 106(4):1330–6. doi:10.1182/blood-2004-12-4792
26. Aichberger KJ, Gleixner KV, Mirkina I, Cerny-Reiterer S, Peter B, Ferenc V, et al. Identification of proapoptotic Bim as a tumor suppressor in neoplastic mast cells: role of KIT D816V and effects of various targeted drugs. Blood (2009) 114(26):5342–51. doi:10.1182/blood-2008-08-175190
27. Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Kshimizu U, et al. Identification of mutations in the coding sequence of the pro-oncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product. J Clin Invest (1993) 92:1736–44. doi:10.1172/JCI116761
28. Dastych J, Metcalfe DD. Stem cell factor induces mast cell adhesion to fibronectin. J Immunol (1994) 152(1):213–9.
29. MacNeil AJ, Junkins RD, Wu Z, Lin T-J. Stem cell factor induces AP-1-dependent mast cell IL-6 production via MAPK kinase 3 activity. J Leukoc Biol (2014) 95(6):903–15. doi:10.1189/jlb.0713401
30. Meininger CJ, Yano H, Rottapel R, Bernstein A, Zsebo KM, Zetter BR. The c-kit receptor ligand functions as a mast cell chemoattractant. Blood (1992) 79(4):958–63.
31. Drube S, Heink S, Walter S, Lohn T, Grusser M, Gerbaulet A, et al. The receptor tyrosine kinase c-Kit controls IL-33 receptor signaling in mast cells. Blood (2010) 115(19):3899–906. doi:10.1182/blood-2009-10-247411
32. Iwaki S, Tkaczyk C, Satterthwaite AB, Halcomb K, Beaven MA, Metcalfe DD, et al. Btk plays a crucial role in the amplification of FcϵRI-mediated mast cell activation by kit. J Biol Chem (2005) 280(48):40261–70. doi:10.1074/jbc.M506063200
33. Wei JJ, Song CW, Sun LC, Yuan Y, Li D, Yan B, et al. SCF and TLR4 ligand cooperate to augment the tumor-promoting potential of mast cells. Cancer Immunol Immunother (2012) 61(3):303–12. doi:10.1007/s00262-011-1098-z
34. Hundley TR, Gilfillan AM, Tkaczyk C, Andrade MV, Metcalfe DD, Beaven MA. Kit and FcεRI mediate unique and convergent signals for release of inflammatory mediators from human mast cells. Blood (2004) 104(8):2410–7. doi:10.1182/blood-2004-02-0631
35. da Silva EZM, Jamur MC, Oliver C. Mast cell function: a new vision of an old cell. J Histochem Cytochem (2014) 62:698–738. doi:10.1369/0022155414545334
36. Kinoshita T, Sawai N, Hidaka E, Yamashita T, Koike K. Interleukin-6 directly modulates stem cell factor-dependent development of human mast cells derived from CD34(+) cord blood cells. Blood (1999) 94(2):496–508.
37. Okayama Y, Kawakami T. Development, survival, and function of mast cells. Immunol Res (2006) 34(2):97–115. doi:10.1385/IR:34:2:97
38. Shimizu Y, Matsumoto K, Okayama Y, Kentaro S, Maeno T, Suga T. Interleukin-3 does not affect the differentiation of mast cells derived from human bone marrow progenitors. Immunol Invest (2008) 37(1):1–17. doi:10.1080/08820130701741742
39. Dahlin JS, Ekoff M, Grootens J, Löf L, Amini RM, Hagberg H, et al. KIT signaling is dispensable for human mast cell progenitor development. Blood (2017) 130(16):1785–94. doi:10.1182/blood-2017-03-773374
40. Mekori YA, Oh CK, Metcalfe DD. IL-3-dependent murine mast cells undergo apoptosis on removal of IL-3. Prevention of apoptosis by c-kit ligand. J Immunol (1993) 151(7):3775–84.
41. Lantz C, Boesiger J, Song C, Mach N, Kobayashi T, Mulligan R, et al. Role for interleukin-3 in mast cell and basophil development and in immunity to parasites. Nature (1998) 392(6671):90–3. doi:10.1038/32190
42. Yoshikawa H, Nakajima Y, Tasaka K. Enhanced expression of Fas-associated death domain-like IL-1-converting enzyme (FLICE)-inhibitory protein induces resistance to Fas-mediated apoptosis in activated mast cells. J Immunol (2000) 165(11):6262–9. doi:10.4049/jimmunol.165.11.6262
43. Berent-Maoz B, Piliponsky AM, Daigle I, Simon H-U, Levi-Schaffer F. Human mast cells undergo TRAIL-induced apoptosis. J Immunol (2006) 176(4):2272–8. doi:10.4049/jimmunol.176.4.2272
44. Yeatman CF II, Jacobs-helber SM, Mirmonsef P, Gillespie SR, Bouton LA, Collins HA, et al. Combined stimulation with the T helper cell type 2 cytokines interleukin (IL)-4 and IL-10 induces mouse mast cell apoptosis. J Exp Med (2000) 192(8):1093–104. doi:10.1084/jem.192.8.1093
45. Berent-Maoz B, Gur C, Vita F, Soranzo MR, Zabucchi G, Levi-Schaffer F. Influence of FAS on murine mast cell maturation. Ann Allergy Asthma Immunol (2011) 106(3):239–44. doi:10.1016/j.anai.2010.12.001
46. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature (2008) 451(7182):1069–75. doi:10.1038/nature06639
47. Ushio H, Ueno T, Kojima Y, Komatsu M, Tanaka S, Yamamoto A, et al. Crucial role for autophagy in degranulation of mast cells. J Allergy Clin Immunol (2011) 127(5):1267–76. doi:10.1016/j.jaci.2010.12.1078
48. Choi GE, Yoon S-Y, Kim J-Y, Kang D-Y, Jang YJ, Kim HS. Autophagy deficiency in myeloid cells exacerbates eosinophilic inflammation in chronic rhinosinusitis. J Allergy ClinImmunol (2017) 141(3):938–50.e12. doi:10.1016/j.jaci.2017.10.038
49. Frech T, De Domenico I, Murtaugh MA, Revelo MP, Li DY, Sawitzke AD, et al. Autophagy is a key feature in the pathogenesis of systemic sclerosis. Rheumatol Int (2014) 34(3):435–9. doi:10.1007/s00296-013-2827-8
50. Pham DL, Kim S, Losol P, Yang E, Shin YS, Ye Y, et al. Association of autophagy related gene polymor-phisms with neutrophilic airway inflammation in adult asthma. Korean J Intern Med (2015) 31:375–85. doi:10.3904/kjim.2014.390
51. Dwyer DF, Barrett NA, Austen KF, Immunological Genome Project Consortium. Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat Immunol (2016) 17(7):878–87. doi:10.1038/ni.3445
52. Kitamura Y. Heterogeneity of mast cells and phenotypic change between subpopulations. Annu Rev Immunol (1989) 7(8):59–76. doi:10.1146/annurev.iy.07.040189.000423
53. Xing W, Austen KF, Gurish MF, Jones TG. Protease phenotype of constitutive connective tissue and of induced mucosal mast cells in mice is regulated by the tissue. Proc Natl Acad Sci U S A (2011) 108(34):14210–5. doi:10.1073/pnas.1111048108
54. da Silva EZM, Jamur MC, Oliver C. Mast cell function: a new vision of an old cell. J Histochem Cytochem (2014) 62(10):698–738. doi:10.1369/0022155414545334
55. Galli SJ, Kalesnikoff J, Grimbaldeston MA, Piliponsky AM, Williams CMM, Tsai M. Mast cells as tunable effector and immunoregulatory cells: recent advances. Annu Rev Immunol (2005) 23(1):749–86. doi:10.1146/annurev.immunol.21.120601.141025
56. Ryan JJ, Kashyap M, Bailey D, Kennedy S, Speiran K, Brenzovich J, et al. Mast cell homeostasis: a fundamental aspect of allergic disease. Crit Rev Immunol (2007) 27(1):15–32. doi:10.1615/CritRevImmunol.v27.i1.20
57. Saito H, Ishizaka T, Ishizaka K. Mast cells and IgE: from history to today. Allergol Int (2013) 62(1):3–12. doi:10.2332/allergolint.13-RAI-0537
58. Kulczycki BYA, Isersky C, Metzger H. The interaction of IgE with rat basophilic leukemia cells. J Exp Med (1974) 139:600–16. doi:10.1084/jem.139.3.600
59. Kawakami T, Blank U. From IgE to omalizumab. J Immunol (2016) 197(11):4187–92. doi:10.4049/jimmunol.1601476
60. Holowka D, Sil D, Torigoe C, Baird B. Insights into immunoglobulin E receptor signaling from structurally definined ligands. Immunol Rev (2007) 217:269–79. doi:10.1111/j.1600-065X.2007.00517.x
61. Gonzalez-Espinosa C, Medina-Tamayo J, Sanchez-Miranda E, Benitez-Garrido JP, Avila-Hernandez AM, Padilla A, et al. Signaling through the high affinity IgE receptor and conditions able to modify IgE-antigen responsiveness of mast cells. Signal Transduction (2007) 7(5–6):402–14. doi:10.1002/sita.200700142
62. Manikandan J, Kothandaraman N, Hande MP, Pushparaj PN. Deciphering the structure and function of FceRI/mast cell axis in the regulation of allergy and anaphylaxis: a functional genomics paradigm. Cell Mol Life Sci (2012) 69:1917–29. doi:10.1007/s00018-011-0886-0
63. Metcalfe DD, Peavy RD, Gilfillan AM. Mechanisms of mast cell signaling in anaphylaxis. J Allergy Clin Immunol (2009) 124(4):639–48. doi:10.1016/j.jaci.2009.08.035
64. Xiang Z, Ahmed AA, Möller C, Nakayama K, Hatakeyama S, Nilsson G. Essential role of the prosurvival bcl-2 homologue A1 in mast cell survival after allergic activation. J Exp Med (2001) 194(11):1561–9. doi:10.1084/jem.194.11.1561
65. Asai K, Kitaura J, Kawakami Y, Yamagata N, Tsai M, Carbone DP, et al. Regulation of mast cell survival by IgE. Immunity (2001) 14(6):791–800. doi:10.1016/S1074-7613(01)00157-1
66. Kalesnikoff J, Huber M, Lam V, Damen JE, Zhang J, Siraganian RP, et al. Monomeric IgE stimulates signaling pathways in mast cells that lead to cytokine production and cell survival. Immunity (2001) 14(6):801–11. doi:10.1016/S1074-7613(01)00159-5
67. Hsu C, MacGlashan D. IgE antibody up-regulates high affinity IgE binding on murine bone marrow-derived mast cells. Immunol Lett (1996) 52(2–3):129–34. doi:10.1016/0165-2478(96)02599-0
68. Yamaguchi M, Lantz CS, Oettgen HC, Katona IM, Fleming T, Miyajima I, et al. IgE enhances mouse mast cell Fc(epsilon)RI expression in vitro and in vivo: evidence for a novel amplification mechanism in IgE-dependent reactions. J Exp Med (1997) 185(4):663–72. doi:10.1084/jem.185.4.663
69. Bax HJ, Bowen H, Dodev TS, Sutton BJ, Gould HJ. Mechanism of the antigen-independent cytokinergic SPE-7 IgE activation of human mast cells in vitro. Sci Rep (2015) 5(1):9538. doi:10.1038/srep09538
70. Futosi K, Fodor S, Mócsai A. Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int Immunopharmacol (2013) 17(3):638–50. doi:10.1016/j.intimp.2013.06.034
71. Falanga YT, Chaimowitz NS, Charles N, Finkelman FD, Pullen NA, Barbour S, et al. Lyn but not Fyn kinase controls IgG-mediated systemic anaphylaxis. J Immunol (2012) 188(9):4360–8. doi:10.4049/jimmunol.1003223
72. Pullen NA, Barnstein BO, Falanga YT, Wang Z, Suzuki R, Tamang TDL, et al. Novel mechanism for Fc{epsilon}RI-mediated signal transducer and activator of transcription 5 (STAT5) tyrosine phosphorylation and the selective influence of STAT5B over mast cell cytokine production. J Biol Chem (2012) 287(3):2045–54. doi:10.1074/jbc.M111.311142
73. Pullen NA, Falanga YT, Morales JK, Ryan JJ. The Fyn-STAT5 pathway: a new frontier in IgE- and IgG-mediated mast cell signaling. Front Immunol (2012) 3:117. doi:10.3389/fimmu.2012.00117
74. Malbec O, Daëron M. The mast cell IgG receptors and their roles in tissue inflammation. Immunol Rev (2007) 217:206–21. doi:10.1111/j.1600-065X.2007.00510.x
75. Rivellese F, Nerviani A, Rossi FW, Marone G, Matucci-Cerinic M, de Paulis A, et al. Mast cells in rheumatoid arthritis: friends or foes? Autoimmun Rev (2017) 16(6):557–63. doi:10.1016/j.autrev.2017.04.001
76. Robbie-Ryan M, Tanzola MB, Secor VH, Brown MA. Cutting edge: both activating and inhibitory Fc receptors expressed on mast cells regulate experimental allergic encephalomyelitis disease severity. J Immunol (2003) 170(4):1630–4. doi:10.4049/jimmunol.170.4.1630
77. Feyerabend TB, Weiser A, Tietz A, Stassen M, Harris N, Kopf M, et al. Cre-mediated cell ablation contests mast cell contribution in models of antibody- and T cell-mediated autoimmunity. Immunity (2011) 35(5):832–44. doi:10.1016/j.immuni.2011.09.015
78. Costanza M, Colombo MP, Pedotti R. Mast cells in the pathogenesis of multiple sclerosis and experimental autoimmune encephalomyelitis. Int J Mol Sci (2012) 13(11):15107–25. doi:10.3390/ijms131115107
79. Lilla JN, Chen CC, Mukai K, BenBarak MJ, Franco CB, Kalesnikoff J, et al. Reduced mast cell and basophil numbers and function in Cpa3-Cre; Mcl-1fl/fl mice. Blood (2011) 118(26):6930–8. doi:10.1182/blood-2011-03-343962
80. Sarma JV, Ward PA. The complement system. Cell Tissue Res (2011) 343(1):227–35. doi:10.1007/s00441-010-1034-0
81. Erdei A, Andrásfalvy M, Péterfy H, Tóth G, Pecht I. Regulation of mast cell activation by complement-derived peptides. Immunol Lett (2004) 92(1–2):39–42. doi:10.1016/J.IMLET.2003.11.019
82. Woolhiser MR, Brockow K, Metcalfe DD. Activation of human mast cells by aggregated IgG through FcγRI: additive effects of C3a. Clin Immunol (2004) 110(2):172–80. doi:10.1016/j.clim.2003.11.007
83. Pundir P, MacDonald CA, Kulka M. The novel receptor C5aR2 is required for C5a-mediated human mast cell adhesion, migration, and proinflammatory mediator production. J Immunol (2015) 195(6):2774–87. doi:10.4049/jimmunol.1401348
84. Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol (2001) 2(8):675–80. doi:10.1038/90609
85. Sandig H, Bulfone-Paus S. TLR signaling in mast cells: common and unique features. Front Immunol (2012) 3:185. doi:10.3389/fimmu.2012.00185
86. Yu Y, Blokhuis BR, Garssen J, Redegeld FA. Non-IgE mediated mast cell activation. Eur J Pharmacol (2016) 778:33–43. doi:10.1016/j.ejphar.2015.07.017
87. Smrž D, Iwaki S, McVicar DW, Metcalfe DD, Gilfillan AM. TLR-mediated signaling pathways circumvent the requirement for DAP12 in mast cells for the induction of inflammatory mediator release. Eur J Immunol (2010) 40(12):3557–69. doi:10.1002/eji.201040573
88. Agier J, Zelechowska P, Kozłowska E, Brzezińska-Błaszczyk E. Expression of surface and intracellular toll-like receptors by mature mast cells. Cent Eur J Immunol (2016) 41(4):333–8. doi:10.5114/ceji.2016.65131
89. Laiño J, Wangorsch A, Blanco F, Wolfheimer S, Krause M, Flaczyk A, et al. Targeting of immune cells by dual TLR2/7 ligands suppresses features of allergic Th2 immune responses in mice. J Immunol Res (2017) 2017:7983217. doi:10.1155/2017/7983217
90. Wang N, McKell M, Dang A, Yamani A, Waggoner L, Vanoni S, et al. Lipopolysaccharide suppresses IgE-mast cell mediated reactions. Clin Exp Allergy (2017) 47(12):1574–85. doi:10.1111/cea.13013
91. Suurmond J, Dorjée AL, Knol EF, Huizinga TWJ, Toes REM. Differential TLR-induced cytokine production by human mast cells is amplified by FcεRI triggering. Clin Exp Allergy (2015) 45(4):788–96. doi:10.1111/cea.12509
92. Sarah C. G protein-coupled receptors: novel probe for MRGPRX2. Nat Rev Drug Discov (2017) 16(5):314–314. doi:10.1038/nrd.2017.77
93. McNeil BD, Pundir P, Meeker S, Han L, Undem BJ, Kulka M, et al. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature (2015) 519(7542):237–41. doi:10.1038/nature14022
94. Subramanian H, Gupta K, Guo Q, Price R, Ali H. Mas-related gene X2 (MrgX2) is a novel G protein-coupled receptor for the antimicrobial peptide LL-37 in human mast cells. J Biol Chem (2011) 286(52):44739–49. doi:10.1074/jbc.M111.277152
95. Subramanian H, Gupta K, Ali H. Roles of MAS-related G protein coupled receptor-X2 (MRGPRX2) on mast cell-mediated host defense, pseudoallergic drug reactions and chronic inflammatory diseases. J Allergy Clin Immunol (2016) 138(3):700–10. doi:10.1016/j.jaci.2016.04.051
96. Quackenbush EJ, Wershil BK, Aguirre V, Gutierrez-Ramos JC. Eotaxin modulates myelopoiesis and mast cell development from embryonic hematopoietic progenitors. Blood (1998) 92(6):1887–97.
97. Juremalm M, Nilsson G. Chemokine receptor expression by mast cells. Chem Immunol Allergy (2005) 87:130–44. doi:10.1159/000087640
98. Hallgren J, Gurish MF. Mast cell progenitor trafficking and maturation. Adv Exp Med Biol (2011) 716:14–28. doi:10.1007/978-1-4419-9533-9_2
99. Juremalm M, Hjertson M, Olsson N, Harvima I, Nilsson K, Nilsson G. The chemokine receptor CXCR4 is expressed within the mast cell lineage and its ligand stromal cell-derived factor-1alpha acts as a mast cell chemotaxin. Eur J Immunol (2000) 30(12):3614–22. doi:10.1002/1521-4141(200012)30:12<3614::AID-IMMU3614>3.0.CO;2-B
100. Juremalm M, Olsson N, Nilsson G. Selective CCL5/RANTES-induced mast cell migration through interactions with chemokine receptors CCR1 and CCR4. Biochem Biophys Res Commun (2002) 297(3):480–5. doi:10.1016/S0006-291X(02)02244-1
101. Migalovich-Sheikhet H, Friedman S, Mankuta D, Levi-Schaffer F. Novel identified receptors on mast cells. Front Immunol (2012) 3:238. doi:10.3389/fimmu.2012.00238
102. Willox I, Mirkina I, Westwick J, Ward SG. Evidence for PI3K-dependent CXCR3 agonist-induced degranulation of human cord blood-derived mast cells. Mol Immunol (2010) 47(14):2367–77. doi:10.1016/j.molimm.2010.05.005
103. Jamur MC, Oliver C. Origin, maturation and recruitment of mast cell precursors. Front Biosci (2011) 3(7):1390–406. doi:10.2741/231
104. Cayrol C, Girard JP. IL-33: an alarmin cytokine with crucial roles in innate immunity, inflammation and allergy. Curr Opin Immunol (2014) 31:31–7. doi:10.1016/j.coi.2014.09.004
105. Hsu CL, Neilsen CV, Bryce PJ. IL-33 is produced by mast cells and regulates IgE-dependent inflammation. PLoS One (2010) 5(8):e11944. doi:10.1371/journal.pone.0011944
106. Saluja R, Ketelaar ME, Hawro T, Church MK, Maurer M, Nawijn MC. The role of the IL-33/IL-1RL1 axis in mast cell and basophil activation in allergic disorders. Mol Immunol (2015) 63(1):80–5. doi:10.1016/J.MOLIMM.2014.06.018
107. Saluja R, Khan M, Church MK, Maurer M. The role of IL-33 and mast cells in allergy and inflammation. Clin Transl Allergy (2015) 5(1):1–8. doi:10.1186/s13601-015-0076-5
108. Moritz DR, Rodewald HR, Gheyselinck J, Klemenz R. The IL-1 receptor-related T1 antigen is expressed on immature and mature mast cells and on fetal blood mast cell progenitors. J Immunol (1998) 161(9):4866–74.
109. Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity (2005) 23(5):479–90. doi:10.1016/j.immuni.2005.09.015
110. Allakhverdi Z, Comeau MR, Jessup HK, Yoon B-RP, Brewer A, Chartier S, et al. Thymic stromal lymphopoietin is released by human epithelial cells in response to microbes, trauma, or inflammation and potently activates mast cells. J Exp Med (2007) 204(2):253–8. doi:10.1084/jem.20062211
111. Allakhverdi Z, Smith DE, Comeau MR, Delespesse G. Cutting edge: the ST2 ligand IL-33 potently activates and drives maturation of human mast cells. J Immunol (2007) 179(4):2051–4. doi:10.4049/jimmunol.179.4.2051
112. Ho LH, Ohno T, Oboki K, Kajiwara N, Suto H, Iikura M, et al. IL-33 induces IL-13 production by mouse mast cells independently of IgE-Fc RI signals. J Leukoc Biol (2007) 82(6):1481–90. doi:10.1189/jlb.0407200
113. Kaieda S, Wang JX, Shnayder R, Fishgal N, Hei H, Lee RT, et al. Interleukin-33 primes mast cells for activation by IgG immune complexes. PLoS One (2012) 7(10):e47252. doi:10.1371/journal.pone.0047252
114. Enoksson M, Möller-Westerberg C, Wicher G, Fallon PG, Forsberg-Nilsson K, Lunderius-Andersson C, et al. Intraperitoneal influx of neutrophils in response to IL-33 is mast cell-dependent. Blood (2013) 121(3):530–6. doi:10.1182/blood-2012-05-434209
115. Saluja R, Zoltowska A, Ketelaar ME, Nilsson G. IL-33 and thymic stromal lymphopoietin in mast cell functions. Eur J Pharmacol (2016) 778:68–76. doi:10.1016/j.ejphar.2015.04.047
116. Dewas C, Chen X, Honda T, Junttila I, Linton J, Udey MC, et al. TSLP expression: analysis with a ZsGreen TSLP reporter mouse. J Immunol (2015) 194(3):1372–80. doi:10.4049/jimmunol.1400519
117. Han NR, Oh HA, Nam SY, Moon PD, Kim DW, Kim HM, et al. TSLP induces mast cell development and aggravates allergic reactions through the activation of MDM2 and STAT6. J Invest Dermatol (2014) 134(10):2521–30. doi:10.1038/jid.2014.198
119. Howard MC, Farrar J, Hilfiker M, Johnson B, Takatsu K, Hamaoka T, et al. Identification of a T cell-derived B cell growth factor distinct from interleukin 2. J Exp Med (1982) 155(3):914–23. doi:10.1084/jem.155.3.914
120. Sudowe S, Arps V, Vogel T, Kölsch E. The role of interleukin-4 in the regulation of sequential isotype switch from immunoglobulin G1 to immunoglobulin E antibody production. Scand J Immunol (2000) 51(5):461–71. doi:10.1046/j.1365-3083.2000.00705.x
121. Vitetta ES, Ohara J, Myers CD, Layton JE, Krammer PH, Paul WE. Serological, biochemical, and functional identity of B cell-stimulatory factor 1 and B cell differentiation factor for IgG1. J Exp Med (1985) 162(5):1726–31. doi:10.1084/jem.162.5.1726
122. Le Gros G, Ben-Sasson SZ, Seder R, Finkelman FD, Paul WE. Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4-producing cells. J Exp Med (1990) 172(3):921–9. doi:10.1017/CBO9781107415324.004
123. Swain SL, Weinberg AD, English M, Huston G. IL-4 directs the development of Th2-like helper effectors. J Immunol (1990) 145(11):3796–806.
124. Russell SM, Keegan AD, Harada N, Nakamura Y, Noguchi M, Leland P, et al. Interleukin-2 receptor gamma chain: a functional component of the interleukin-4 receptor. Science (1993) 262(5141):1880–3. doi:10.1126/science.8266078
125. Hilton DJ, Zhang JG, Metcalf D, Alexander WS, Nicola NA, Willson TA. Cloning and characterization of a binding subunit of the interleukin 13 receptor that is also a component of the interleukin 4 receptor. Proc Natl Acad Sci U S A (1996) 93(1):497–501. doi:10.1073/pnas.93.1.497
126. Keegan AD, Zamorano J. Regulation of gene expression, growth, and cell survival by IL-4: contribution of multiple signaling pathways. Cell Res (1998) 8(1):1–13. doi:10.1038/cr.1998.1
127. Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol (1999) 17:701–38. doi:10.1146/annurev.immunol.17.1.701
128. Kelly-Welch A, Hanson EM, Keegan AD. Interleukin-4 (IL-4) pathway. Sci STKE (2005) 2005(293):cm9. doi:10.1126/stke.2932005cm9
129. Galietta LJV, Pagesy P, Folli C, Caci E, Romio L, Costes B, et al. IL-4 is a potent modulator of ion transport in the human bronchial epithelium in vitro. J Immunol (2002) 168(2):839–45. doi:10.4049/jimmunol.168.2.839
130. Madden KB, Whitman L, Sullivan C, Gause WC, Urban JF, Katona IM, et al. Role of STAT6 and mast cells in IL-4- and IL-13-induced alterations in murine intestinal epithelial cell function. J Immunol (2002) 169:4417–22. doi:10.4049/jimmunol.169.8.4417
131. Strait RT, Morris SC, Smiley K, Urban JF, Finkelman FD. IL-4 exacerbates anaphylaxis. J Immunol (2003) 170(7):3835–42. doi:10.4049/jimmunol.170.7.3835
132. Hoffmann HJ, Dahl C, Schiøtz PO, Berglund L, Dahl R. Lectins interact differentially with purified human eosinophils, cultured cord blood-derived mast cells and the myeloid leukaemic cell line AML14.3D10: induction of interleukin-4 secretion is conserved among granulocytes, but is not proportional to agglut. Clin Exp Allergy (2003) 33(7):930–5. doi:10.1046/j.1365-2222.2003.01625.x
133. Mossman TR, Bond MW, Coffman RL, Ohara J, Paul WE. T cell and mast cell lines respond to B-cell stimulatory factor 1. Proc Natl Acad Sci U S A (1986) 83(15):5654–8. doi:10.1073/pnas.83.15.5654
134. Bischoff SC, Sellge G, Lorentz A, Sebald W, Raab R, Manns MP. IL-4 enhances proliferation and mediator release in mature human mast cells. Proc Natl Acad Sci U S A (1999) 96(14):8080–5. doi:10.1073/pnas.96.14.8080
135. Burton OT, Darling AR, Zhou JS, Noval-Rivas M, Jones TG, Gurish MF, et al. Direct effects of IL-4 on mast cells drive their intestinal expansion and increase susceptibility to anaphylaxis in a murine model of food allergy. Mucosal Immunol (2013) 6(4):740–50. doi:10.1038/mi.2012.112
136. Karimi K, Redegeld FA, Heijdra B, Nijkamp FP. Stem cell factor and interleukin-4 induce murine bone marrow cells to develop into mast cells with connective tissue type characteristics in vitro. Exp Hematol (1999) 27(4):654. doi:10.1016/S0301-472X(98)00083-6
137. Lorentz A, Bischoff SC. Regulation of human intestinal mast cells by stem cell factor and IL-4. Immunol Rev (2001) 179:57–60. doi:10.1034/j.1600-065X.2001.790106.x
138. Xia HZ, Du Z, Craig S, Klisch G, Noben-Trauth N, Kochan JP, et al. Effect of recombinant human IL-4 on tryptase, chymase, and Fc epsilon receptor type I expression in recombinant human stem cell factor-dependent fetal liver-derived human mast cells. J Immunol (1997) 159(6):2911–21.
139. Yamaguchi M, Sayama K, Yano K, Lantz CS, Noben-Trauth N, Ra C, et al. IgE enhances Fc epsilon receptor I expression and IgE-dependent release of histamine and lipid mediators from human umbilical cord blood-derived mast cells: synergistic effect of IL-4 and IgE on human mast cell Fc epsilon receptor I expression and mediator release. J Immunol (1999) 162(9):5455–65.
140. Nilsson G, Miettinen U, Ishizaka T, Ashman LK, Irani AM, Schwartz LB. Interleukin-4 inhibits the expression of Kit and tryptase during stem cell factor-dependent development of human mast cells from fetal liver cells. Blood (1994) 84(5):1519–27.
141. Mirmonsef P, Shelburne CP, Yeatman CF, Chong HJ, Ryan JJ. Inhibition of kit expression by IL-4 and IL-10 in murine mast cells: role of STAT6 and phosphatidylinositol 3’-kinase. J Immunol (1999) 163(5):2530–9.
142. Ryan JJ, DeSimone S, Klisch G, Shelburne CP, McReynolds LJ, Han K, et al. IL-4 inhibits mouse mast cell Fc epsilonRI expression through a STAT6-dependent mechanism. J Immunol (1998) 161(12):6915–23.
143. Chong HJ, Bouton LA, Bailey DP, Wright H, Ramirez C, Gharse A, et al. IL-4 selectively enhances FcγRIII expression and signaling on mouse mast cells. Cell Immunol (2003) 224(2):64–72. doi:10.1016/j.cellimm.2003.08.004
144. Bailey DP, Kashyap M, Bouton LA, Murray PJ, Ryan JJ. Interleukin-10 induces apoptosis in developing mast cells and macrophages. J Leukoc Biol (2006) 80(3):581–9. doi:10.1189/jlb.0405201
145. Speiran K, Bailey DP, Fernando J, Macey M, Barnstein B, Kolawole M, et al. Endogenous suppression of mast cell development and survival by IL-4 and IL-10. J Leukoc Biol (2009) 85(5):826–36. doi:10.1189/jlb.0708448
146. McLeod JJA, Baker B, Ryan JJ. Mast cell production and response to IL-4 and IL-13. Cytokine (2015) 75(1):57–61. doi:10.1016/j.cyto.2015.05.019
147. Egger D, Geuenich S, Denzlinger C, Schmitt E, Mailhammer R, Ehrenreich H, et al. IL-4 renders mast cells functionally responsive to endothelin-1. J Immunol (1995) 154(4):1830–7.
148. Wolk K, Kunz S, Asadullah K, Sabat R. Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol (2002) 168(11):5397–402. doi:10.4049/jimmunol.168.11.5397
149. Chadban SJ, Tesch GH, Foti R, Lan HY, Atkins RC, Nikolic-Paterson DJ. Interleukin-10 differentially modulates MHC class II expression by mesangial cells and macrophages in vitro and in vivo. Immunology (1998) 94(1):72–8. doi:10.1046/j.1365-2567.1998.00487.x
150. Corinti S, Albanesi C, la Sala A, Pastore S, Girolomoni G. Regulatory activity of autocrine IL-10 on dendritic cell functions. J Immunol (2001) 166(7):4312–8. doi:10.4049/jimmunol.166.7.4312
151. Kasama T, Strieter RM, Lukacs NW, Burdick MD, Kunkel SL. Regulation of neutrophil-derived chemokine expression by IL-10. J Immunol (1994) 152(7):3559–69.
152. Wang P, Wu P, Siegel MI, Egan RW, Billah MM. IL-10 inhibits transcription of cytokine genes in human peripheral blood mononuclear cells. J Immunol (1994) 153(2):811–6.
153. Malisan F, Brière F, Bridon JM, Harindranath N, Mills FC, Max EE, et al. Interleukin-10 induces immunoglobulin G isotype switch recombination in human CD40-activated naive B lymphocytes. J Exp Med (1996) 183(3):937–47. doi:10.1084/jem.183.3.937
154. Rousset F, Peyrol S, Garcia E, Vezzio N, Andujar M, Grimaud JA, et al. Long-term cultured CD40-activated B lymphocytes differentiate into plasma cells in response to IL-10 but not IL-4. Int Immunol (1995) 7(8):1243–53. doi:10.1093/intimm/7.8.1243
155. Carson BWE, Lindemann MJ, Baiocchi R, Linett M, Tan JC, Chou C, et al. The functional characterization of interleukin-l0 receptor expression on human natural killer cells. Blood (1995) 85(12):3577–86.
156. van Roon J, Wijngaarden S, Lafeber FPJG, Damen C, van deWinkel J, Bijlsma JWJ. Interleukin 10 treatment of patients with rheumatoid arthritis enhances Fc γ receptor expression on monocytes and responsiveness to immune complex stimulation. J Rheumatol (2003) 30(4):648–51.
157. Lauw FN, Pajkrt D, Hack CE, Kurimoto M, van Deventer SJH, van der Poll T. Proinflammatory effects of IL-10 during human endotoxemia. J Immunol (2000) 165(5):2783–9. doi:10.4049/jimmunol.165.5.2783
158. Tilg H. Treatment of Crohn’s disease with recombinant human interleukin 10 induces the proinflammatory cytokine interferon gamma. Gut (2002) 50(2):191–5. doi:10.1136/gut.50.2.191
159. Claret E, Llorente L, Richaud-patin Y, Garci C, Jakez-ocampo J, Cardiel MH, et al. Clinical and biologic effects of anti-interleukin-10 monoclonal antibody administration in systemic lupus erythematosus. Arthritis Rheum (2000) 43(8):1790–800. doi:10.1002/1529-0131(200008)43:8<1790::AID-ANR15>3.0.CO;2-2
160. Polukort SH, Rovatti J, Carlson L, Thompson C, Ser-Dolansky J, Kinney SRM, et al. IL-10 enhances IgE-mediated mast cell responses and is essential for the development of experimental food allergy in IL-10-deficient mice. J Immunol (2016) 196(12):4865–76. doi:10.4049/jimmunol.1600066
161. Rennick D, Hunte B, Holland G, Thompson-Snipes L. Cofactors are essential for stem cell factor-dependent growth and maturation of mast cell progenitors: comparative effects of interleukin-3 (IL-3), IL-4, IL-10, and fibroblasts. Blood (1995) 85(1):57–65.
162. Thompson-Snipes L, Dhar V, Bond MW, Mosmann TR, Moore KW, Rennick DM. Interleukin 10: a novel stimulatory factor for mast cells and their progenitors. J Exp Med (1991) 173(2):507–10. doi:10.1084/jem.173.2.507
163. Ghildyal N, Mcneil HP, Gurish MF, Austen KF, Stevens RL. Transcriptional regulation of the mucosal mast cell-specific protease gene, MMCP-8. Mol Biol (1992) 267(12):8473–7.
164. Arock M, Zuany-Amorim C, Singer M, Benhamou M, Pretolani M. Interleukin-10 inhibits cytokine generation from mast cells. Eur J Immunol (1996) 26(1):166–70. doi:10.1002/eji.1830260126
165. Conti P, Kempuraj D, Kandere K, Di Gioacchino M, Barbacane RC, Castellani ML, et al. IL-10, an inflammatory/inhibitory cytokine, but not always. Immunol Lett (2003) 86(2):123–9. doi:10.1016/S0165-2478(03)00002-6
166. Lin TJ, Befus AD. Differential regulation of mast cell function by IL-10 and stem cell factor. J Immunol (1997) 159(8):4015.
167. Marshall JS, Leal-Berumen I, Nielsen L, Glibetic M, Jordana M. Interleukin (IL)-10 inhibits long-term IL-6 production but not preformed mediator release from rat peritoneal mast cells. J Clin Invest (1996) 97(4):1122–8. doi:10.1172/JCI118506
168. Gillespie SR, DeMartino RR, Zhu J, Chong HJ, Ramirez C, Shelburne CP, et al. IL-10 inhibits Fc epsilon RI expression in mouse mast cells. J Immunol (2004) 172(5):3181–8. doi:10.4049/jimmunol.172.5.3181
169. Millat G, Marc C, Tomasetto C, Chikh K, Fensom AH, Harzer K, et al. Niemann-pick C1 disease: correlations between NPC1 mutations, levels of NPC1 protein, and phenotypes emphasize the functional significance of the putative sterol-sensing domain and of the cysteine-rich luminal loop. Am J Hum Genet (2001) 68(6):1373–85. doi:10.1086/320606
170. Qayum AA, Paranjape A, Abebayehu D, Kolawole EM, Haque TT, McLeod JJA, et al. IL-10-induced miR-155 targets SOCS1 to enhance IgE-mediated mast cell function. J Immunol (2016) 196(11):4457–67. doi:10.4049/jimmunol.1502240
171. Letterio JJ, Roberts AB. Regulation of immune responses by TGF-beta. Annu Rev Immunol (1998) 16(1):137–61. doi:10.1146/annurev.immunol.16.1.137
172. Kehrl JH. Production of transforming growth factor beta by human T lymphocytes and its potential role in the regulation of T cell growth. J Exp Med (1986) 163(5):1037–50. doi:10.1084/jem.163.5.1037
173. Rao PE, Petrone AL, Ponath PD. Differentiation and expansion of T cells with regulatory function from human peripheral lymphocytes by stimulation in the presence of TGF-β. J Immunol (2005) 174(3):1446–55. doi:10.4049/jimmunol.174.3.1446
174. Petit-Koskas E, Génot E, Lawrence D, Kolb JP. Inhibition of the proliferative response of human B lymphocytes to B cell growth factor by transforming growth factor-beta. Eur J Immunol (1988) 18(1):111–6. doi:10.1002/eji.1830180117
175. Bogdan C, Paik J, Vodovotz Y, Nathan C. Contrasting mechanisms for suppression of macrophage cytokine release by transforming growth factor-B and interleukin-10. J Biol Chem (1992) 267(32):23301–8.
176. Vodovotz Y. Mechanisms of suppression of macrophage nitric oxide release by transforming growth factor beta. J Exp Med (1993) 178(2):605–13. doi:10.1084/jem.178.2.605
177. Kashyap M, Bailey DP, Gomez G, Rivera J, Huff TF, Ryan JJ. TGF-β1 inhibits late-stage mast cell maturation. Exp Hematol (2005) 33(11):1281–91. doi:10.1016/j.exphem.2005.07.001
178. Miller HR, Wright SH, Knight PA, Thornton EM. A novel function for transforming growth factor-beta1: upregulation of the expression and the IgE-independent extracellular release of a mucosal mast cell granule-specific beta-chymase, mouse mast cell protease-1. Blood (1999) 93(10):3473–86.
179. Norozian F, Kashyap M, Ramirez CD, Patel N, Kepley CL, Barnstein BO, et al. TGFβ1 induces mast cell apoptosis. Exp Hematol (2006) 34(5):579–87. doi:10.1016/j.exphem.2006.02.003
180. Fernando J, Faber TW, Pullen NA, Falanga YT, Kolawole EM, Oskeritzian CA, et al. Genotype-dependent effects of TGF-β1 on mast cell function: targeting the Stat5 pathway. J Immunol (2013) 191(9):4505–13. doi:10.4049/jimmunol.1202723
181. Gruber BL, Marchese MJ, Kew RR. Transforming growth factor-b-1 mediates mast cell chemotaxis. J Immunol (1994) 152:5860–7.
182. Olsson N, Piek E, ten Dijke P, Nilsson G. Human mast cell migration in response to members of the transforming growth factor-beta family. J Leukoc Biol (2000) 67(3):350–6. doi:10.1002/jlb.67.3.350
183. Gomez G, Ramirez CD, Rivera J, Patel M, Norozian F, Wright HV, et al. TGF-β1 inhibits mast cell FcεRI expression. J Immunol (2005) 174(10):5987–93. doi:10.4049/jimmunol.174.10.5987
184. Zhao W, Gomez G, Yu S, Ryan JJ, Schwartz LB. TGF-β1 attenuates mediator release and de novo kit expression by human skin mast cells through a Smad-dependent pathway. J Immunol (2008) 181(10):7263–72. doi:10.4049/jimmunol.181.10.7263
185. Ndaw VS, Abebayehu D, Spence AJ, Paez PA, Kolawole EM, Taruselli MT, et al. TGF-β1 suppresses IL-33-induced mast cell function. J Immunol (2017) 199(3):866–73. doi:10.4049/jimmunol.1601983
186. Ganeshan K, Johnston LK, Bryce PJ. TGF-1 limits the onset of innate lung inflammation by promoting mast cell-derived IL-6. J Immunol (2013) 190(11):5731–8. doi:10.4049/jimmunol.1203362
187. Macey MR, Sturgill JL, Morales JK, Falanga YT, Morales J, Norton SK, et al. IL-4 and TGF-1 counterbalance one another while regulating mast cell homeostasis. J Immunol (2010) 184(9):4688–95. doi:10.4049/jimmunol.0903477
188. Liu L, Wei Y, Xu H, Huang Y, Luo F, Huang Z. A systematic review and meta-analysis of the association of transforming growth factor β receptor I 6A/9A gene polymorphism with ovarian cancer risk. J Recept Signal Transduct Res (2014) 34(4):313–6. doi:10.3109/10799893.2014.885051
189. Schindler C, Darnell JE. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem (1995) 64(1):621–52. doi:10.1146/annurev.bi.64.070195.003201
190. Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A (2006) 103(4):1000–5. doi:10.1073/pnas.0507350103
191. Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell (1998) 93(5):841–50. doi:10.1016/S0092-8674(00)81444-0
192. Shelburne CP, McCoy ME, Piekorz R, Sexl V, Roh KH, Jacobs-Helber SM, et al. Stat5 expression is critical for mast cell development and survival. Blood (2003) 102(4):1290–7. doi:10.1182/blood-2002-11-3490
193. Baumgartner C, Cerny-Reiterer S, Sonneck K, Mayerhofer M, Gleixner KV, Fritz R, et al. Expression of activated STAT5 in neoplastic mast cells in systemic mastocytosis. Am J Pathol (2009) 175(6):2416–29. doi:10.2353/ajpath.2009.080953
194. Peter B, Bibi S, Eisenwort G, Wingelhofer B, Berger D, Stefanzl G, et al. Drug-induced inhibition of phosphorylation of STAT5 overrides drug resistance in neoplastic mast cells. Leukemia (2018) 32(4):1016–22. doi:10.1038/leu.2017.338
195. Barnstein BO, Li G, Wang Z, Kennedy S, Chalfant C, Nakajima H, et al. Stat5 expression is required for IgE-mediated mast cell function. J Immunol (2006) 177(5):3421–6. doi:10.4049/JIMMUNOL.177.5.3421
196. Junttila IS, Watson C, Kummola L, Chen X, Hu-Li J, Guo L, et al. Efficient cytokine-induced IL-13 production by mast cells requires both IL-33 and IL-3. J Allergy Clin Immunol (2013) 132(3):704–12.e10. doi:10.1016/j.jaci.2013.03.033
197. Ando T, Xiao W, Gao P, Namiranian S, Matsumoto K, Tomimori Y, et al. Critical role for mast cell Stat5 activity in skin inflammation. Cell Rep (2014) 6(2):366–76. doi:10.1016/j.celrep.2013.12.029
198. Azam M, Erdjument-Bromage H, Kreider BL, Xia M, Quelle F, Basu R, et al. lnterleukin-3 signals through multiple isoforms of Stat5. EMBO J (1995) 14(7):1402–11.
199. Liu X, Robinson GW, Gouilleuxt F, Gronert B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue (Janus kinase/signal transducers and activators of transcription/mammary gland factor/milk protein). Dev Biol (1995) 92:8831–5.
200. Mui AL, Wakao H, O’Farrell AM, Harada N, Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. EMBO J (1995) 14(6):1166–75.
201. Ikeda K, Nakajima H, Suzuki K, Watanabe N, Kagami S, Iwamoto I. Stat5a is essential for the proliferation and survival of murine mast cells. Int Arch Allergy Immunol (2005) 137(Suppl 1):45–50. doi:10.1159/000085431
202. Ma CA, Liqiang X, Cauff B, DeZure A, Freeman AF, Hambleton S, et al. Somatic Stat5b gain-of-function mutations in early onset nonclonal eosinophilia, urticaria, and dermatitis. Blood (2017) 129:650–3. doi:10.1182/blood-2016-09-737817
203. Harvima IT, Levi-schaffer F, Draber P, Friedman S. Molecular targets on mast cells and basophils for novel therapies. J Allergy ClinImmunol (2014) 134(3):530–44. doi:10.1016/j.jaci.2014.03.007
204. Molderings GJ, Haenisch B, Brettner S, Homann J, Menzen M, Dumoulin FL, et al. Pharmacological treatment options for mast cell activation disease. Naunyn Schmiedebergs Arch Pharmacol (2016) 389(7):671–94. doi:10.1007/s00210-016-1247-1
205. Reber LL, Frossard N. Targeting mast cells in inflammatory diseases. Pharmacol Ther (2014) 142(3):416–35. doi:10.1016/j.pharmthera.2014.01.004
206. Siebenhaar F, Redegeld FA, Bischoff SC, Gibbs BF, Maurer M. Mast cells as drivers of disease and therapeutic targets. Trends Immunol (2017) 39(2):151–62. doi:10.1016/j.it.2017.10.005
207. Simons FER, Simons KJ. Histamine and H1-antihistamines: celebrating a century of progress. J Allergy Clin Immunol (2011) 128(6):1139–50.e4. doi:10.1016/j.jaci.2011.09.005
208. Tantisira KG, Drazen JM. Genetics and pharmacogenetics of the leukotriene pathway. J Allergy Clin Immunol (2009) 124(3):422–7. doi:10.1016/j.jaci.2009.06.035
209. Bielory L, Lien KW, Bigelsen S. Efficacy and tolerability of newer antihistamines in the treatment of allergic conjunctivitis. Drugs (2005) 65(2):215–28. doi:10.2165/00003495-200565020-00004
210. Oppong E, Flink N, Cato ACB. Molecular mechanisms of glucocorticoid action in mast cells. Mol Cell Endocrinol (2013) 380(1–2):119–26. doi:10.1016/j.mce.2013.05.014
211. Yamaguchi M, Hirai K, Komiya A, Miyamasu M, Furumoto Y, Teshima R, et al. Regulation of mouse mast cell surface Fc epsilon RI expression by dexamethasone. Int Immunol (2001) 13(7):843. doi:10.1093/intimm/13.7.843
212. Paranjape A, Chernushevich O, Qayum AA, Spence AJ, Taruselli MT, Abebayehu D, et al. Dexamethasone rapidly suppresses IL-33-stimulated mast cell function by blocking transcription factor activity. J Leukoc Biol (2016) 100(6):1395–404. doi:10.1189/jlb.3A0316-125R
213. Holgate ST, Polosa R. Treatment strategies for allergy and asthma. Nat Rev Immunol (2008) 8(3):218–30. doi:10.1038/nri2262
214. Gupta R, Fonacier LS. Adverse effects of nonsystemic steroids (inhaled, intranasal, and cutaneous): a review of the literature and suggested monitoring tool. Curr Allergy Asthma Rep (2016) 16(6):44. doi:10.1007/s11882-016-0620-y
215. Zhang T, Finn DF, Barlow JW, Walsh JJ. Mast cell stabilisers. Eur J Pharmacol (2016) 778:158–68. doi:10.1016/j.ejphar.2015.05.071
216. Oka T, Kalesnikoff J, Starkl P, Tsai M, Galli SJ. Evidence questioning cromolyns effectiveness and selectivity as a mast cell stabilizer in mice. Lab Invest (2012) 92(10):1472–82. doi:10.1038/labinvest.2012.116
217. Guevara JP, Ducharme FM, Keren R, Nihtianova S, Zorc J. Inhaled corticosteroids versus sodium cromoglycate in children and adults with asthma. Cochrane Database Syst Rev (2006) 19(2):CD003558. doi:10.1002/14651858.CD003558.pub2
218. Lai T, Wang S, Xu Z, Zhang C, Zhao Y, Hu Y, et al. Long-term efficacy and safety of omalizumab in patients with persistent uncontrolled allergic asthma: a systematic review and meta-analysis. Sci Rep (2015) 5:8191. doi:10.1038/srep08191
219. Pennington LF, Tarchevskaya S, Brigger D, Sathiyamoorthy K, Graham MT, Nadeau KC, et al. Structural basis of omalizumab therapy and omalizumab-mediated IgE exchange. Nat Commun (2016) 7:11610. doi:10.1038/ncomms11610
220. Tonacci A, Billeci L, Pioggia G, Navarra M, Gangemi S. Omalizumab for the treatment of chronic idiopathic urticaria: systematic review of the literature. Pharmacotherapy (2017) 37(4):464–80. doi:10.1002/phar.1915
221. Goldstein S, Gabriel S, Kianifard F, Ortiz B, Skoner DP. Clinical features of adolescents with chronic idiopathic or spontaneous urticaria: review of omalizumab clinical trials. Ann Allergy Asthma Immunol (2017) 118(4):500–4. doi:10.1016/j.anai.2017.02.003
222. Suzukawa M, Matsumoto H, Ohshima N, Tashimo H, Asari I, Tajiri T, et al. Baseline serum CXCL10 and IL-12 levels may predict severe asthmatics’ responsiveness to omalizumab. Respir Med (2018) 131:95–102. doi:10.1016/j.rmed.2017.12.002
223. Adhya Z, Karim Y. Doxepin may be a useful pharmacotherapeutic agent in chronic urticaria. Clin Exp Allergy (2015) 45(8):1370–1370. doi:10.1111/cea.12580
224. Cohen MH, Williams G, Johnson JR, Duan J, Gobburu J, Rahman A, et al. Approval summary for imatinib mesylate capsules in the treatment of chronic myelogenous leukemia approval summary for imatinib mesylate capsules in the treatment of chronic myelogenous leukemia. Clin Cancer Res (2002) 8(5):935–42.
225. Growney JD, Clark JJ, Adelsperger J, Stone R, Fabbro D, Griffin JD, et al. Activation mutations of human c-KIT resistant to imatinib mesylate are sensitive to the tyrosine kinase inhibitor PKC412. Blood (2005) 106(2):721–4. doi:10.1182/blood-2004-12-4617
226. Cahill KN, Katz HR, Cui J, Lai J, Kazani S, Crosby-Thompson A, et al. KIT inhibition by Imatinib in patients with severe refractory asthma. N Engl J Med (2017) 376(20):1911–20. doi:10.1056/NEJMoa1613125
227. Dubreuil P, Letard S, Ciufolini M, Gros L, Humbert M, Castéran N, et al. Masitinib (AB1010), a potent and selective tyrosine kinase inhibitor targeting KIT. PLoS One (2009) 4(9):e7258. doi:10.1371/journal.pone.0007258
228. Marech I, Patruno R, Zizzo N, Gadaleta C, Introna M, Zito AF, et al. Masitinib (AB1010), from canine tumor model to human clinical development: where we are? Crit Rev Oncol Hematol (2014) 91(1):98–111. doi:10.1016/j.critrevonc.2013.12.011
229. Paul C, Sans B, Suarez F, Casassus P, Barete S, Lanternier F, et al. Masitinib for the treatment of systemic and cutaneous mastocytosis with handicap: a phase 2a study. Am J Hematol (2010) 85(12):921–5. doi:10.1002/ajh.21894
230. Humbert M, de Blay F, Garcia G, Prud’homme A, Leroyer C, Magnan A, et al. Masitinib, a c-kit/PDGF receptor tyrosine kinase inhibitor, improves disease control in severe corticosteroid-dependent asthmatics. Allergy (2009) 64(8):1194–201. doi:10.1111/j.1398-9995.2009.02122.x
231. Meltzer EO, Berkowitz RB, Grossbard EB. An intranasal Syk-kinase inhibitor (R112) improves the symptoms of seasonal allergic rhinitis in a park environment. J Allergy Clin Immunol (2005) 115(4):791–6. doi:10.1016/j.jaci.2005.01.040
232. Guyer BJ, Shimamoto SR, Bradhurst AL, Grossbard EB, Dreskin SC, Nelson HS. Mast cell inhibitor R112 is well tolerated and affects prostaglandin D2 but not other mediators, symptoms, or nasal volumes in a nasal challenge model of allergic rhinitis. Allergy Asthma Proc (2006) 27(3):208–13. doi:10.2500/aap.2006.27.2861
233. Horak F, Puri KD, Steiner BH, Holes L, Xing G, Zieglmayer P, et al. Randomized phase 1 study of the phosphatidylinositol 3-kinase δ inhibitor idelalisib in patients with allergic rhinitis. J Allergy Clin Immunol (2016) 137(6):1733–41. doi:10.1016/j.jaci.2015.12.1313
234. Ross S, Allen I, Connelly J, Korenblat B, Smith M, Bishop D, et al. Clinical outcomes in statin treatment trials: a meta-analysis. Arch Intern Med (1999) 159(15):1793–802. doi:10.1001/archinte.159.15.1793
235. Cowan DC, Cowan JO, Palmay R, Williamson A, Taylor DR. Simvastatin in the treatment of asthma: lack of steroid-sparing effect. Thorax (2010) 65(10):891–6. doi:10.1136/thx.2010.138990
236. Zeki AA, Oldham J, Wilson M, Fortenko O, Goyal V, Last M, et al. Statin use and asthma control in patients with severe asthma. BMJ Open (2013) 3(8). doi:10.1136/bmjopen-2013-003314
237. Kolawole EM, McLeod JJA, Ndaw V, Abebayehu D, Barnstein BO, Faber T, et al. Fluvastatin suppresses mast cell and basophil IgE responses: genotype-dependent effects. J Immunol (2016) 196(4):1461–70. doi:10.4049/jimmunol.1501932
238. Howarth PH, Babu KS, Arshad HS, Lau L, Buckley M, McConnell W, et al. Tumour necrosis factor (TNFalpha) as a novel therapeutic target in symptomatic corticosteroid dependent asthma. Thorax (2005) 60(12):1012–8. doi:10.1136/thx.2005.045260
239. Bathon JM, Martin RW, Fleischmann RM, Tesser JR, Schiff MH, Keystone EC, et al. A comparison of etanercept and methotrexate in patients with early rheumatoid arthritis. N Engl J Med (2000) 343(22):1586–93. doi:10.1056/NEJM200011303432201
240. Cruse G, Yin Y, Fukuyama T, Desai A, Arthur GK, Bäumer W, et al. Exon skipping of FcεRIβ eliminates expression of the high-affinity IgE receptor in mast cells with therapeutic potential for allergy. Proc Natl Acad Sci U S A (2016) 113(49):14115–20. doi:10.1073/pnas.1608520113
241. Cruse G, Beaven MA, Music SC, Bradding P, Gilfillan AM, Metcalfe DD. The CD20 homologue MS4A4 directs trafficking of KIT toward clathrin-independent endocytosis pathways and thus regulates receptor signaling and recycling. Mol Biol Cell (2015) 26(9):1711–27. doi:10.1091/mbc.E14-07-1221
242. Woska JR, Gillespie ME. SNARE complex-mediated degranulation in mast cells. J Cell Mol Med (2012) 16(4):649–56. doi:10.1111/j.1582-4934.2011.01443.x
243. Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet (2011) 378(9791):595–605. doi:10.1016/S0140-6736(11)60756-3
244. Lim KRQ, Maruyama R, Yokota T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des Devel Ther (2017) 11:533–45. doi:10.2147/DDDT.S97635
245. Menzies D, Nair A, Meldrum KT, Fleming D, Barnes M, Lipworth BJ. Simvastatin does not exhibit therapeutic anti-inflammatory effects in asthma. J Allergy Clin Immunol (2007) 119(2):328–35. doi:10.1016/j.jaci.2006.10.014
246. Maneechotesuwan K, Ekjiratrakul W, Kasetsinsombat K, Wongkajornsilp A, Barnes PJ. Statins enhance the anti-inflammatory effects of inhaled corticosteroids in asthmatic patients through increased induction of indoleamine 2,3-dioxygenase. J Allergy Clin Immunol (2010) 126(4):754–62. doi:10.1016/j.jaci.2010.08.005
247. Braganza G, Chaudhuri R, McSharry C, Weir CJ, Donnelly I, Jolly L, et al. Effects of short-term treatment with atorvastatin in smokers with asthma—a randomized controlled trial. BMC Pulm Med (2011) 11(1):16. doi:10.1186/1471-2466-11-16
248. Hothersall EJ, Chaudhuri R, McSharry C, Donnelly I, Lafferty J, McMahon AD, et al. Effects of atorvastatin added to inhaled corticosteroids on lung function and sputum cell counts in atopic asthma. Thorax (2008) 63(12):1070–5. doi:10.1136/thx.2008.100198
249. Thomson NC, Charron CE, Chaudhuri R, Spears M, Ito K, McSharry C. Atorvastatin in combination with inhaled beclometasone modulates inflammatory sputum mediators in smokers with asthma. Pulm Pharmacol Ther (2015) 31:1–8. doi:10.1016/j.pupt.2015.01.001
250. Bhattacharjee D, Chogtu B, Magazine R. Statins in asthma: potential beneficial effects and limitations. Pulm Med (2015) 2015:835204. doi:10.1155/2015/835204
251. Silva D, Couto M, Delgado L, Moreira A. A systematic review of statin efficacy in asthma. J Asthma (2012) 49(9):885–94. doi:10.3109/02770903.2012.721433
252. Subbotina E, Koganti S, Hodgson-Zingman D, Zingman L. Morpholino-driven gene editing: a new horizon for disease treatment and prevention. Clin Pharmacol Ther (2016) 99(1):21–5. doi:10.1002/cpt.276
253. Beierlein JM, McNamee LM, Ledley FD. As technologies for nucleotide therapeutics mature, products emerge. Mol Ther Nucleic Acids (2017) 9:379–86. doi:10.1016/j.omtn.2017.10.017
254. Safaralizadeh R, Soheili ZS, Deezagi A, Pourpak Z, Samiei S, Moin M. FcεRI-α siRNA inhibits the antigen-induced activation of mast cells. Iran J Allergy Asthma Immunol (2009) 8(4):177–83. doi:08.04/ijaai.177183
255. Kaczmarek JC, Kowalski PS, Anderson DG. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med (2017) 9(1):60. doi:10.1186/s13073-017-0450-0
256. Cairns JA. Inhibitors of mast cell tryptase beta as therapeutics for the treatment of asthma and inflammatory disorders. Pulm Pharmacol Ther (2005) 18(1):55–66. doi:10.1016/j.pupt.2004.09.032
257. Fukuoka Y, Schwartz LB. The B12 anti-tryptase monoclonal antibody disrupts the tetrameric structure of heparin-stabilized-tryptase to form monomers that are inactive at neutral pH and active at acidic pH. J Immunol (2006) 176(5):3165–72. doi:10.4049/jimmunol.176.5.3165
258. Pejler G, Abrink M, Ringvall M, Wernersson S. Mast cell proteases. Adv Immunol (2007) 95:167–255. doi:10.1016/S0065-2776(07)95006-3
259. Nabe T, Kijitani Y, Kitagawa Y, Sakano E, Ueno T, Fujii M, et al. Involvement of chymase in allergic conjunctivitis of guinea pigs. Exp Eye Res (2013) 113:74–9. doi:10.1016/j.exer.2013.05.015
260. Liew FY, Pitman NI, McInnes IB. Disease-associated functions of IL-33: the new kid in the IL-1 family. Nat Rev Immunol (2010) 10(2):103–10. doi:10.1038/nri2692
261. Prefontaine D, Lajoie-Kadoch S, Foley S, Audusseau S, Olivenstein R, Halayko AJ, et al. Increased expression of IL-33 in severe asthma: evidence of expression by airway smooth muscle cells. J Immunol (2009) 183(8):5094–103. doi:10.4049/jimmunol.0802387
Keywords: mast cell, IgE, IgG, IL-4, IL-10, TGF-β, Stat5, allergy
Citation: Caslin HL, Kiwanuka KN, Haque TT, Taruselli MT, MacKnight HP, Paranjape A and Ryan JJ (2018) Controlling Mast Cell Activation and Homeostasis: Work Influenced by Bill Paul That Continues Today. Front. Immunol. 9:868. doi: 10.3389/fimmu.2018.00868
Received: 24 January 2018; Accepted: 09 April 2018;
Published: 26 April 2018
Edited by:
Jinfang Zhu, National Institute of Allergy and Infectious Diseases (NIAID), United StatesReviewed by:
Avery August, Cornell University, United StatesGunnar Nilsson, Karolinska Institute (KI), Sweden
Copyright: © 2018 Caslin, Kiwanuka, Haque, Taruselli, MacKnight, Paranjape and Ryan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John J. Ryan, jjryan@vcu.edu
 Heather L. Caslin1
Heather L. Caslin1 Kasalina N. Kiwanuka
Kasalina N. Kiwanuka John J. Ryan
John J. Ryan