- 1INSERM, Centre d’Etude des Pathologies Respiratoires (CEPR), UMR 1100, Tours, France
- 2Université de Tours, Tours, France
- 3Service de Médecine Intensive Réanimation, Centre Hospitalier Régional Universitaire de Tours, Tours, France
- 4Division of Radiotherapy and Imaging, Targeted Therapy Team, The Institute of Cancer Research, London, United Kingdom
- 5Department of Biochemistry and Molecular Genetics, Faculty of Medicine, American University of Beirut, Beirut, Lebanon
γδT cells comprise a unique T cell sublineage endowed with a wide functional repertoire, which allow them to play important—sometimes opposite—roles in many immune responses associated with infection, cancer, and inflammatory processes. This is largely dependent on the existence of pre-programmed discrete functional subsets that differentiate within the thymus at specific temporal windows of life. Since they represent a major early source of interleukin-17A in many models of immune responses, the γδT17 cell population has recently gained considerable interest. Thus, a better dissection of the developmental program of this effector γδT subset appears critical in understanding their associated immune functions. Several recent reports have provided new exciting insights into the developmental mechanisms that control γδT cell lineage commitment and differentiation. Here, we review the importance of thymic cues and intrinsic factors that shape the developmental program of γδT17 cells. We also discuss the potential future areas of research in γδT17 cell development especially in regards to the recently provided data from deep RNA sequencing technology. Pursuing our understanding into this complex mechanism will undoubtedly provide important clues into the biology of this particular T cell sublineage.
Interleukin-17 (IL-17) is a highly conserved cytokine in vertebrates that plays a critical role in host homeostasis and immune response to pathogens especially at barrier sites (1, 2). Recent evidence indicates that IL-17 also emerges as a key contributor in immunity beyond the scope of infection, such as inflammation and cancer (3, 4). Given the pivotal role of lymphoid cell-derived IL-17 in orchestrating immune responses, its cellular sources have been extensively searched over the last decade. Initially believed to be mainly produced by conventional CD4+ T (Th17) cells (5, 6), the discovery of innate and innate-like lymphocytes endowed with potent capacities to produce IL-17 (7) suggests that this cytokine is well poised at the border between innate and adaptive immunity. These populations include γδT cells (8), natural killer T (NKT) cells (9), mucosal-associated invariant T (MAIT) cells (10), and group 3 innate lymphoid cells (ILC3) (11). Among those, γδT cells have been demonstrated to be the main contributors in IL-17 production in many settings, such as infection, autoimmunity, and cancer. Here, we discuss the recent advances on our understanding of IL-17-producing γδT (γδT17) cell biology with a particular emphasis on the transcriptional road maps that drives their innately “pre-programmed” effector fate.
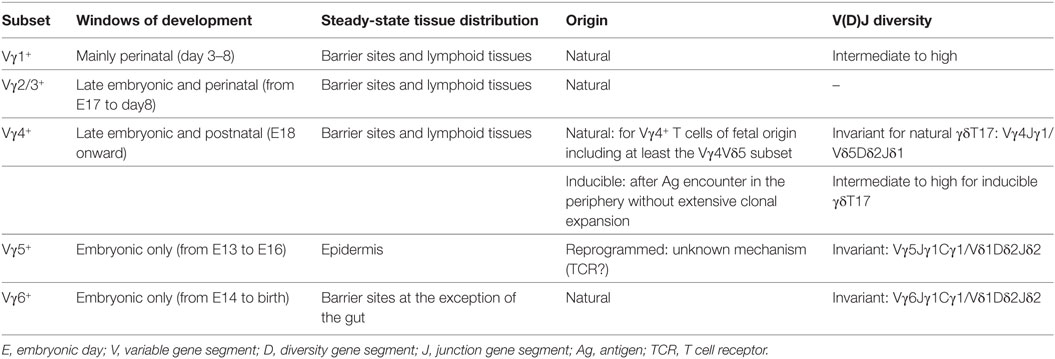
Characteristics of γδT17 Cells
Mouse γδT cells consist of a heterogeneous population of thymus-derived T lymphocytes characterized by distinct functional properties (e.g., cytokine profile and/or cytotoxic properties) and tissue distribution (12). Within this subset diversity, γδT17 cells can be defined based on their T cell receptor (TCR) repertoire usage and surface markers. Thus, γδT17 cells are almost exclusively restricted to γδT cells expressing either a Vγ6 or a Vγ4 TCR (N.B.: The Heilig and Tonegawa’s nomenclature (13) has been used in this review) (Table 1). In addition, many cell surface antigens (Ags) have been shown to distinguish γδT17 cells and can be defined as CD27−, NK1.1−, IL-7Rαhigh, IL-18Rhigh, CD122−, and CCR6+ cells (14–16). γδT17 cells mainly establish residency at barrier sites including lung, skin, vagina, and oral cavity. However, they can recirculate in particular pathological situations including infections and cancer (17). The migratory capacity of γδT17 cells is regulated by the chemokine receptors CCR2 (during inflammation) and CCR6 (at homeostasis) (18). This preferential location at barrier sites might indicate a preferential interplay between γδT17 cells and the endogenous flora (19) as exemplified by the strong reduction in frequency of lung resident γδT17 cells in germ-free mice (20). Interestingly, while Vγ4+ γδT17 cells can be detected in the gastrointestinal tract (21), we and others failed to detect Vγ6+ γδT17 cells in this tissue in adult mice under steady-state condition (20, 22). This might suggest that the nature and/or diversity of commensals in the various mucosa could differentially influence the maintenance of γδT17 cell subsets.
γδT17 cells are characterized by their ability to promptly produce copious amounts of IL-17A/F, IL-22, IL-21, and GM-CSF (8, 23–25). This rapid capacity to produce these cytokines can be mainly attributed to their innate-like feature. Despite expressing a fully functional rearranged TCR, γδT17 cells can respond to activating cytokines (IL-1β, IL-23, and IL-18) even in absence of concomitant TCR engagement (8, 16). However, TCR ligation on naive γδT17 cells has been shown to license them by increasing activating cytokine receptor expression (e.g., IL-1R1 and IL-23R) and thus rendering them permissive to “innate” stimulation (26). The nature of these Ags is yet to be determined. Notably, the unprocessed form of the red algae protein phycoerythrin has been shown to interact with a small proportion of naive γδT cell TCRs irrespectively of their TCR repertoire (26). Even if the physiological relevance of phycoerythrin in the biology of mammalian cells is difficult to conceive, it is tempting to speculate that structurally related Ags could be relevant in the general selection and licensing of γδT cells. In addition, the fact that a single Ag can be recognized by various γδTCRs harboring distinct CDR3 regions is reminiscent with the NKT cell biology (27) and can suggest the existence of a restricted conformational “hot-spot” comprising few amino acid residues in CDR3 regions responsible for γδT cell antigenicity in mice. This structural basis for Ag recognition by innate-like T cells might have been conserved all through the evolution from jawless vertebrates (28).
Despite leaving the thymus with a pre-programmed effector fate, γδT17 cells have been shown to conserve a certain degree of plasticity in the periphery. This characteristic originates from an epigenetic regulation program for specific genes in γδT17 cells such as Dickkopf-related protein 3 (29). Thus, along with IL-17, γδT17 cells can also produce interferon (IFN)-γ under inflammatory conditions. The biological relevance of this plasticity has been revealed in various settings including Listeria monocytogenes infection (22). In this later model, long-lasting accumulation of Vγ6+ IFN-γ/IL-17 double producers was observed within intestinal lamina propria (22, 30). Since Vγ6+ γδT17 cells were reported to be absent from the gastrointestinal tract at steady-state (20), it is possible that the combination of the γδT17 cell epigenome and local environment modifications under this inflammatory condition favors their homing and survival in the gut tissue. On the other hand, it is interesting to mention that IFN-γ-producing pre-programmed γδT cells (γδT1) do not possess the capacity to produce IL-17 (31). However, a small proportion of epidermal γδT1 (e.g., Vγ5+) cells has been demonstrated to produce IL-17 in vivo upon skin wounding (32). The molecular determinants involved in giving rise to this cytokine production capacity are currently unknown but seem to rely on TCR signaling (33).
The existence of γδT17 cells in humans is still a matter of debate (34). Actually, the thymic program of γδT cells in humans seems to differ from the one described in mice (35). Most of the data available suggest that human γδT cells might not be “innately” programmed to produce IL-17 during their thymic development but rather acquire this capacity under inflammatory conditions once in the periphery akin to CD4+ Th17 cells. Thus, circulating Vγ9Vδ2+ [using Lefranc’s nomenclature (36)] T cells from adult healthy donors produce no or little IL-17 (37) except under complex stimulatory protocol including both activating cytokines and TCR engagement (38). However, it is important to mention that purified Vγ9+ γδT cells from cord blood seem more prone to produce IL-17 (37, 39, 40) that could suggest an embryonic origin for human γδT17 cells similar to the murine situation. It is, therefore, possible that these putative γδT17 cells occupy particular niches of the body that render them difficult to assess under homeostatic conditions. Murine pre-committed γδT17 cells are often characterized by the expression of an almost clonal TCR (41). Thanks to next-generation sequencing, the existence of clonal TCR-expressing γδT cell subsets in humans has recently emerged. Upon cytomegalovirus reactivation, a recent study demonstrated the massive proliferation of diverse γδT cell clones in patients after allogeneic-hematopoietic-stem-cell transplantation (42). However, the TRG and TRD sequences of these clones were not shared among individuals at the nucleotide level (42). In addition, analysis of the human Vδ1+ T cells in healthy adults indicates that this repertoire is dominated by few private clonotypes (43). Determining the cytokine profile of these clones will be helpful to better appreciate the existence and origins of γδT17 cells in humans. Whatever the mechanisms that drive their emergence in humans, the capacity of human γδT cells to produce IL-17 has been demonstrated in various immune responses including infection, cancer, and autoimmunity (38, 44–46).
Development of γδT17 Cells
Dealing With the Concept of “Innate/Natural” vs “Adaptive/Inducible” Origins of γδT17 Cells
Mouse γδT cells develop in a standardized manner by sequential waves that can be conveniently followed based on their Vγ chain usage (47). This process starts during embryonic life from day 13 (E13) onward. The first wave is exclusively constituted of the IFN-γ-producing Vγ5+ cells and lasts for about 4 days. This is shortly followed by a developmental wave of “natural” IL-17-producers comprising both canonical Vγ6+ (from E14 to birth) and restricted subsets of Vγ4+ (E18 onward) (48, 49). Around birth, IFN-γ-producing Vγ1+ and Vγ4+ subsets start to develop along with the IL-4/IFN-γ-double producers Vγ1+Vδ6+ subset. After birth, developing γδT cells mainly exhibit a naive uncommitted profile (47).
According to this scheme, natural γδT17 cell development is believed to be restricted to the gestational period. To support this notion, Haas and colleagues demonstrated that transplantation of bone marrow from IL-17-competent mice into lethally irradiated Il17af-deficient adult recipients failed to induce γδT17 cell development (49). Partial γδT17 cell development in the thymus could be achieved in reconstituted Il17af-deficient neonate recipients but failed to give rise to γδT17 cells in the periphery. In addition, inducible expression of Rag1 in T cell precursors in adult mice did not restore de novo generation of γδT17 cells (49). Likewise, CCR6+ IL-17-producing dermal γδT cells failed to reconstitute 8 weeks after bone marrow transplantation unless the mice received an additional transfer of neonatal thymocytes (33). Surprisingly, the same lab reported the presence of Vγ4+ CCR6+ (but not Vγ4− CCR6+) γδT17 cells in lymph nodes of recipient TCRδ−/− mice following bone marrow transplantation in absence of neonatal thymocytes at 12 weeks post-grafting (50). The basis for this difference remains unclear but one can argue that, in the 12 weeks model, authors have reconstituted “inducible” γδT17 cells only. Several studies have also reported that the peripheral pool of γδT17 cells decreased with age (51, 52), which further support the embryonic origin of natural γδT17 cells.
Despite suggesting a strict favorable temporal window for natural γδT17 cell development during fetal life, these studies also raised some additional interrogations. Is this developmental model imposed by intrinsic (nature of γδT precursors) or extrinsic (embryonic vs adult thymic environment) factors? Zúñiga-Pflücker’s team recently started to provide an answer to this question. Culture of γδTCR-transduced fetal or adult hematopoietic precursors with OP9-Delta-like protein 4 (Dll4) cells led to the development of γδT cells with IL-17 production capacity in a similar manner (53). Thus, these data suggest that adult progenitors conserve their intrinsic capacity to develop as γδT17 cells once in an appropriate environment. However, since the authors used a clonal TCR (Vγ4Vδ5) (54) for transfection, it cannot be excluded that in addition to the favorable environment, the nature of the TCR expressed plays a role in the IL-17 effector fate, in particular regarding the recent discovery of clonal Vγ4Jγ1/Vδ5Dδ2Jδ1 T cells with a strongly biased IL-17-producing profile (55). In addition, recent studies have shown that adult peripheral γδT cells (from adult bone marrow-derived precursors) can convert into “induced” γδT17 cells upon inflammatory conditions (56, 57). Both studies highlighted a critical role for the cytokines IL-23 and IL-1β, and to a lesser extent TCR signaling, in this process (56, 57). Importantly, the potential to give rise to inducible γδT17 cells seems to be restricted to the IL-2Rβ− γδT cell subset (57). To further illustrate this peripheral polarization capacity, Buus and colleagues recently provided a RNA-Seq analysis of adult γδ thymocytes indicating IL-17 potential in certain subsets including notably IL-2Rα+ Clec12A+ Vγ1+ and Vγ4+ cells (58).
This situation illustrates the recent concept of “natural” vs “inducible” γδT17 cells (48). While “natural” (Vγ6+ and Vγ4+ subsets) γδT17 cells are committed to this effector fate during their embryonic/perinatal thymic developmental program, “inducible” γδT17 cells stem from naive (Vγ1+ and Vγ4+ subsets) γδT cells within the periphery upon inflammatory conditions through cytokine, and/or Ag recognition akin to conventional CD4+ Th17 cells (26). Thus, it is tempting to view “natural” γδT17 cells as innate-like T cells whereas “inducible” γδT17 cells can rather be considered adaptive. This is reminiscent with the situation in the αβ lineage comprising innate-like T17 (NKT17 and MAIT17) cells and adaptive conventional Th17 cells.
Thymic Molecular Determinants of γδT17 Cell Effector Fate
In this section, we will review the thymic determinants that drive γδT cell differentiation into an IL-17 effector fate by deciphering how “natural” IL-17-committed γδT cells emerge from the rest of the γδT cell compartment. Before commitment into γδT17 cell sublineage, thymic precursors have to first undergo a bifurcation into αβ or γδ lineages. Mechanisms driving this initial dichotomy are beyond the scope of this review, but it is worth mentioning that the strength of TCR engagement in thymocytes emerges as a driving force in this process [see Ref. (53, 59) for reviews]. However, requirement for TCR ligation in IL-17-commited γδT cell differentiation is still an intense matter of debates that will be discussed later. Commitment toward the γδT lineage happens at double negative (DN)2 and DN3 stages (60, 61). Interestingly, the effector fate of “natural” γδT cell subsets (IFN-γ- vs IL-17A-producing) appears to be already predetermined at this stage. Thus, commitment to “natural” γδT17 cells exclusively arises from the late DN2 stage in a B cell leukemia/lymphoma 11b-dependent manner (46).
Further differentiation into the IL-17 effector fate is a complex and highly dynamic process involving multiple molecular and cellular interactions. For the salve of clarity, we distinguish here the (1) extrinsic factors (e.g., thymic environmental cues) and (2) intrinsic factors [e.g., intracellular signaling pathways and transcription factors (TFs)] that tune γδT17 precursors into mature γδT17 cells.
Extrinsic Factors
Molecular and cellular players within the thymic microenvironment are crucial for acquisition of γδT cell effector fate (Figure 1). Anatomically, precursors migrate from the cortex to the medulla during this process, where they receive multiple signals that dictate their differentiation. In this three-dimensional environment, it is important to keep in mind that time should be considered as a fourth dimension when deciphering the “natural” γδT17 cells ontogeny.
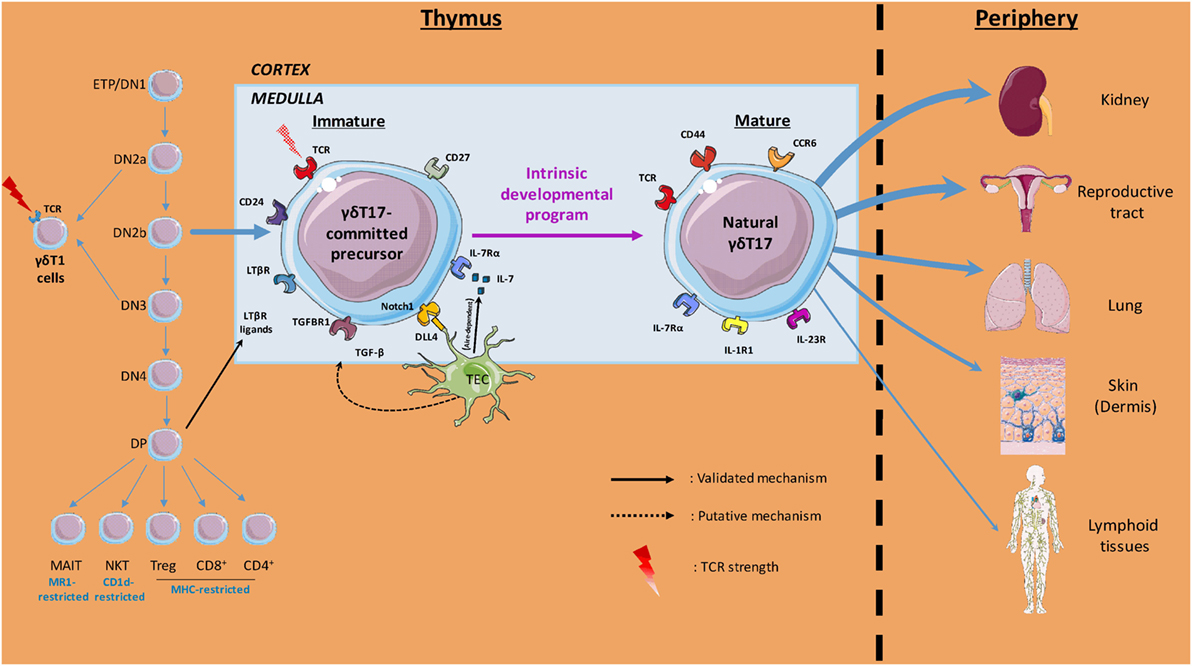
Figure 1. Overview of thymic γδT17 cell ontogeny. Initial intrathymic pathways leading to pre-committed γδT17 cells divergence from other T cell lineages are illustrated. The thymic (cortical and medullary) environmental cues involved in γδT17 cell effector fate and the preferential peripheral niches of mature γδ17 cells are also depicted. Labels indicate the cells, soluble factors, proteins, surface markers, and T cell receptor (TCR) signal strength involved in the γδT17 cell program of differentiation.
Cytokines
Many cytokines have been reported to directly or indirectly regulate thymic γδT17 cell differentiation/development.
IL-7 is a critical and non-redundant cytokine in lymphopoiesis (62). IL-7Rα-deficient mice completely lack γδT cells (63, 64) partly due to the role of IL-7 in V-J recombination of TCRγ genes (63, 65). Its specific role in survival and proliferation of γδT17 cells in peripheral tissues under homeostatic and pathological situations has been demonstrated (37, 66, 67). Even if this question has never been directly addressed, many studies imply a requirement for IL-7 in the proper natural γδT17 cell development. First, in a model of conditional abrogation (RBP-Jκ) that precludes IL-7Rα (CD127) expression, but in which the initial generation of γδ precursors (DN2 and DN3 stages) is maintained, the pool of both thymic and peripheral γδT17 cells is markedly reduced (68). Second, addition of recombinant IL-7 in fetal thymic organ culture from E16 thymus promotes γδT17 cell expansion over IFN-γ-producing γδT cells (37). Understanding whether IL-7 directly participates in the differentiation program or solely in the post-differentiation expansion of γδT17 cells will require further investigations. In line with the favorable temporal window for γδT17 cell development around birth, it is important to mention that thymus is particularly enriched for IL-7 [especially in thymic epithelial cells (TECs)] in neonates and its presence declines with age (69, 70).
Akin to conventional αβ Th17 cells, TGF-β1 has also been proposed to participate in γδT17 cell development (71). In addition, this report indicates that some other Th17-driving cytokines, such as IL-23 and IL-21, appear to be dispensable. However, since the authors have performed their investigation in 11-day-old mice, a time at which natural γδT17 cells already egressed the thymus, it is difficult to precisely evaluate the contribution of TGF-β1 in natural γδT17 cell differentiation.
The role of IL-6 in γδT17 cell development is somehow controversial. While several studies indicated that IL-6 deficiency did not influence γδT17 cell homeostasis and cytokine production capacity (71, 72), others reported a defect in γδT17 cells in both thymus (73) and peripheral organs (74). Thus, the contribution of IL-6 in natural γδT17 cell differentiation remains unclear. In addition, mRNAs for IL-6Rα (CD126) are barely expressed on both immature and mature thymic natural γδT17 cells (from the ImmGen database). However, it cannot be firmly excluded that IL-6 influences the thymic microenvironment to support γδT17 cell differentiation as previously hypothesized (73).
Thus, this indicates that the cytokine network within the thymic microenvironment has to be tightly regulated in a time-dependent manner to allow natural γδT17 cell differentiation.
Thymic Epithelial Cells
Akin to other thymocytes, interaction with TEC is likely critical in γδT17 cell differentiation, even though few experimental data are currently available. Thus, medullary (m)TEC has been implicated in regulating Vγ6+ γδT17 cell development through the TF autoimmune regulator (Aire) (69). In Aire−/− mice, IL-7 production is up-regulated in mTEC, and this is accompanied by an overproduction of Vγ6+ γδT17 thymocytes. Interestingly, other subsets of natural γδT17 cells, especially Vγ4+ subsets were not affected by specific Aire deletion in mTEC (69). This feature also indicates that the various natural γδT17 cell subsets probably require different signals to develop.
Cortical (c)TEC has also been recently shown to control γδT17 cell development. Using a mouse model with specific cTEC ablation, Nitta and colleagues observed a strong dysregulation in the proportion of natural γδT17 cell subsets (75). Specifically, absence of cTEC skewed the γδT17 TCR repertoire toward Vγ6 expression at the expense of the Vγ4+ T cell subset during the postnatal period while the proportions of Vγ6+ and Vγ4+ γδT17 cells remained normal during embryonic life. Authors hypothesize that in their mouse model, the postnatal thymic microenvironment resembles to the fetal microenvironment, which in turn favors the Vγ6+ subset. In addition, since they have been proposed to be a prime source of TGFβ (76), cTEC could also participate in thymic development of γδT17 cells through this mechanism (71).
The importance of Dll4, a Notch ligand expressed by TEC (77) has also been proposed in γδT17 cell differentiation (78). In a co-culture model of E15 thymocytes with stromal cells, absence of Dll4 expression by stromal cells led to an abrogation in γδT17 cell development (78). This phenotype indicates that Dll4 is likely to be a common factor for the differentiation of all natural γδT17 cell subsets.
Finally, a recent study proposed that signaling through NF-κB-inducing kinase (NIK) in TEC is essential for the generation of a fully functional pool of γδT17 cells (79). However, the molecular factors regulated by NIK in TEC are yet to be determined. Understanding the NIK-dependent pathways in TEC will certainly provide important clues in the TEC-γδT17 precursor interaction mechanisms that drive γδT17 cell effector fate.
Altogether, the fetal and perinatal thymic environment offers a temporal window of opportunity for γδT17 cell differentiation. However, the available literature indicates the requirement for differential factors according to the subset of naturally occurring γδT17 cells. This might somewhat rely on the intrinsic nature of the γδT17 precursors. It can also be hypothesized that these precursors (Vγ6+ and Vγ4+) require timely expressed TCR ligands in the thymic environment. However, no host-derived Ags have been proposed to date to participate in γδT17 cell differentiation.
Intrinsic Factors
A Requirement for TCR Ligation: Still an Open Question?
Beyond its importance into γδ lineage commitment, TCR signal strength is also involved in the functional maturation of γδ-committed thymocytes. Specifically, TCR signal strength drives the IL-17- vs IFN-γ-producing γδT cell dichotomy. However, it appears difficult to clearly attribute a specific strength to a specific effector fate. While the literature tends to demonstrate consensually that a strong TCR signaling in γδ thymocytes drives their commitment toward a Th1-like effector fate (31, 80–82), the situation in γδT17 cell differentiation remains highly debated.
Chronologically, a first set of data suggested that γδT17 cell differentiation occurred in the absence of TCR cognate ligands (80). However, (1) this study used adult thymocytes and focused on peripheral organs that are weakly if not populated with natural γδT17 cells and (2) it cannot be excluded that ligand-independent TCR signaling plays a part in this model. Therefore, these results are likely to provide specific information about the requirement of TCR signals for inducible γδT17 cells. In this sense, these data perfectly fit with the concept that inducible γδT17 cells egress the thymus with a naive uncommitted profile and need further encounter with peripheral Ags to gain their capacity to produce IL-17. Few years later, the lab of Adrian Hayday highlighted the butyrophilin-like molecule Skint-1 as a molecular determinant in Th1-like effector fate of Vγ5+ T cells (31). Interestingly, in absence of Skint-1, the differentiation of Vγ5+ T cells resulted in the generation of cells displaying a phenotype of natural γδT17 cells (31). Indeed, Skint-1 engagement in Vγ5+ thymocytes induces the upregulation of TCR-dependent genes that subsequently repress the transcriptional differentiation program of natural γδT17 cells. In line, Pennington and colleagues recently demonstrated that differentiation of E15 γδ thymocytes in presence of an anti-TCRδ mAb (GL3) blunted their commitment toward a γδT17 cell profile (82). Altogether, weak or no TCR signals seem required to allow proper γδT17 cell development. Thus, the developmental program of natural γδT17 cells appears to be a TCR Ag-free process acquired by “neglect.”
On the other hand, mice presenting a reduced function in the TCR proximal signaling kinase ZAP-70 displayed a reduced pool of both IL-17A-producing Vγ6+ and to a lesser extent Vγ4+ T cells in neonate thymocytes (83). In the same line, Silva-Santos and colleagues observed a reduction in the frequency of IL-17A-producing Vγ6+ subset using double-heterozygous mice for the CD3 subunits γ and δ in which TCR signaling is attenuated (81).
These apparently contradictory results may have multiple explanations. Notably, it is assumable that the different subsets of γδT17 precursors may require a specific and fine-tuned TCR signals to engage in their differentiation program. Specifically, Vγ6+ may require an “intermediate” TCR signals while Vγ4+ subsets may need weak or no signals. Moreover, intensity of TCR signaling can be hardly compared from one experimental setting to another, making any generalization risky. In this context, the importance of TCR signaling in programming γδT17 cell differentiation is still an open question. This also raises the putative existence of TCR self-ligands for natural γδT17 cells. The recent discovery of butyrophilin-like molecules as Ags for mouse γδT cells (84, 85) opens a new exciting avenue of research in the field. Identification of the enlarged butyrophilin family in both mouse and human γδT cell biology might offer an interesting anchoring point for future translational studies.
Costimulatory Molecules
On top of the TCR, its accessory receptors have been proposed to participate in γδT cell differentiation. In addition to be a convenient marker to distinguish γδT functional subsets, the costimulatory receptor CD27 has been shown to participate in γδT cell development (14). Indeed, γδ thymocytes from Cd27−/− mice presented altered expression of ifng. Although CD27 deficiency did not influence the pool of γδT17 cells, CD27 gain of function in thymic cultures resulted in lower IL-17 transcripts by CD27− γδ thymocytes (14). Thus, CD27 appears as a thymic regulator in γδT cell effector fate.
Inducible T cell co-stimulator (ICOS) signaling pathway has also recently emerged as a possible determinant in γδT17 cell (at least for the Vγ4+ subset) development (86). Agonistic activity of anti-ICOS mAb in fetal thymic organ culture significantly impaired Vγ4+ γδT17 development. In line, genetic ablation of ICOS tends to increase the pool of thymic Vγ4+ γδT17 cells (86). Thus, this study indicates that ICOS-dependent intracellular pathways in thymocytes controls γδT17 cell effector fate.
Besides, one must keep in mind, that, alongside with TCR and costimulation receptor signaling, multiple other signals have to be integrated by embryonic thymocytes to, in fine, engage toward the γδT17 effector fate.
Soluble Mediator Receptor Signaling Pathways
γδT17 precursors express specific receptors for various soluble factors produced in the thymic environment by hematopoietic and non-hematopoietic cells. Signals provided by these mediators have to be integrated by γδT cells to fully develop.
For instance, TGFβ receptor (TGFβR) signaling pathway in developing γδT17 cells could be important in their effector fate. Mice deficient for Smad3, a critical component of the TGFβR signaling pathway presented a striking defect in frequency of thymic γδT17 cells compared with littermate controls (71). However, since the authors did not provide direct evidence (bone marrow chimera and OP-9 models) for an intrinsic role of the TGFβR signaling pathway, it cannot be excluded that this pathway is indirectly linked to γδT17 cell development.
In addition, the targeting of lymphotoxin-β receptor through double positive thymocytes-derived ligands (87) also controls the generation of γδT17 cells by regulating the expression of TFs from the NF-κB family namely RelA and RelB (88).
As mentioned above, the IL-7/IL-7Rα axis is important to generate a normal pool of γδT17 cells. However, a better understanding of the downstream molecular cascade involved will be helpful to understand whether this axis controls the differentiation program or the homeostasis of γδT17 cells.
On the other hand, IL-15Rα signaling disruption favors the development of γδT17 cells in thymus of neonates (65). The molecular mechanisms responsible for this are currently unknown but might rely on the activation of repressing factors in the IL-15Rα signaling pathway or could be indirect by reducing the competition with γδT17-driving γ-chain-dependent cytokines such as IL-7.
The expression of the prostacyclin (PGI2) receptor (IP) on thymocytes has also been demonstrated to control γδT17 cell development. This was evidenced by a failure to generate γδT17 cells in the thymus of IP−/− mice (89). However, the molecular determinants involved in this process are yet to be defined.
The γδT17 Cell Transcriptional Program
The dynamic integration of these multiple signals leads to the implementation of a complex transcriptional program that dictates γδT17 cell effector fate (Figure 2). The aim of this program is ultimately to maintain and/or to favor the expression of Rorc (encoding RORγt), the cardinal TF for IL-17-secreting cells (90) including γδT cells (72). The recent advances in RNA deep sequencing analysis allowed a better understanding of the transcriptional regulation involved in γδT17 development.
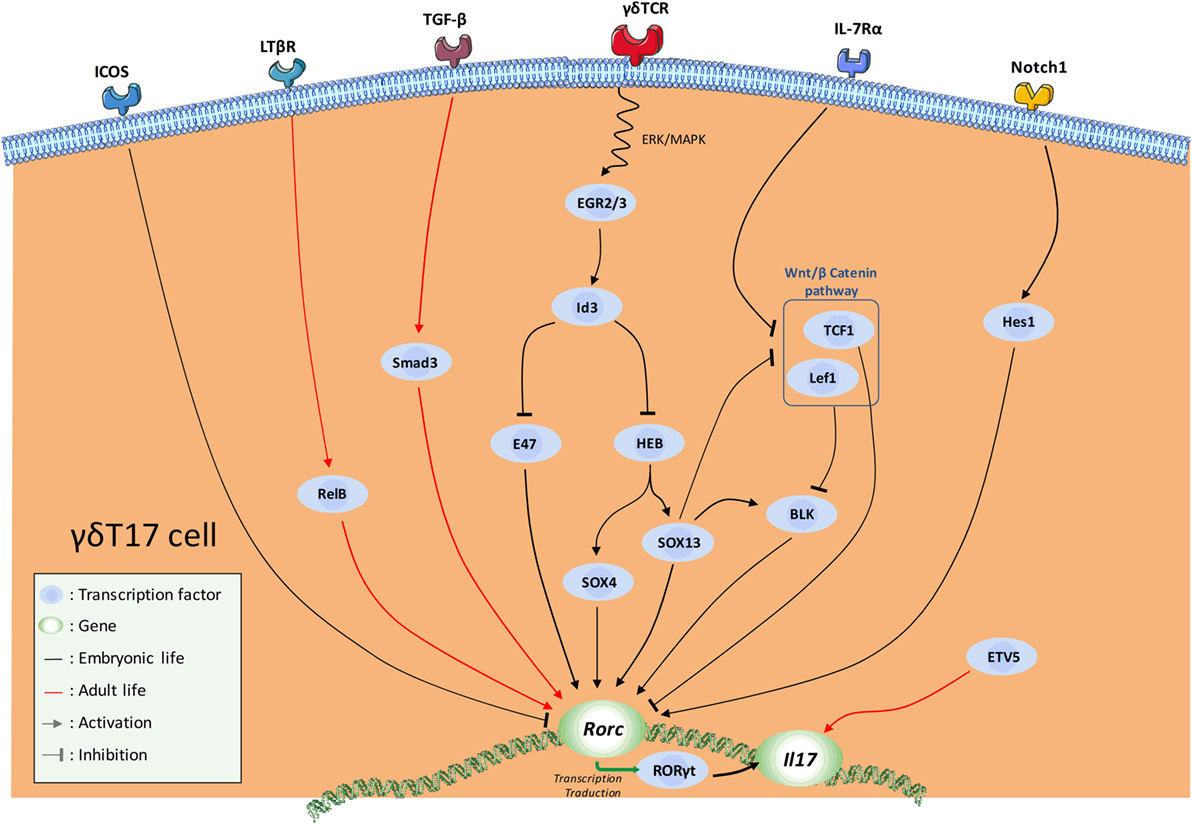
Figure 2. Schematic overview of the current knowledge in the transcription factor (TF) network involved in natural γδT17 cell effector fate. Major activating and repressing pathways implicated during γδ17 effector fate acquisition are depicted.
Thus, SRY-related HMG-box (SOX) 4 and SOX13, two members of the high mobility group box TF family constitute a central node of regulation in this program (50, 91, 92). SOX4/13 are paramount in the acquisition of the γδT17 effector fate by (1) directly controlling Rorc transcription, (2) possibly enhancing important γδT17 cell-driving pathways such as IL-7Rα signaling, and (3) possibly inhibiting Rorc-repressing TFs (31, 92). In this later mechanism, SOX13 was suggested to inhibit the Rorc-repressing activity of two downstream mediators of the Wnt/β-catenin signaling pathway namely lymphoid enhancer-binding factor 1 (Lef1) and transcription factor 1 (TCF1) (92). In this context, Tcf7 (encoding for TCF1) deficiency leads to an aberrant high proportion of γδT17 cells (92). At this stage, it is also important to mention that this pathway also regulates the development of the IFN-γ-producing Vγ1Vδ6.3+ and Vγ5+ cells (92). The mechanisms by which the TCF1–Lef1 axis counteracts the γδT17 transcriptional program are not fully understood. First, it is possible that TCF1 and Lef1 control Rorc expression through epigenetic (histone deacetylase) activity as suggested in conventional T cells (93). Alternatively, this axis could also indirectly repress Rorc expression by inhibiting the transcription of B lymphocyte kinase (Blk) (92), an important signal transducer in γδT17 cell development (94). However, how Blk controls RORγt expression is currently unknown. It is noteworthy that, using blk−/− mice, Vγ6+ T cells were shown to be more Blk-dependent than Vγ4+ (94). As SOX13 was shown to regulate Blk expression and to control Vγ4+, but minimally Vγ6+, subset development (92), these results appear somewhat contradictory. However, Vγ4 and Vγ6 subsets develop at different temporal windows; therefore, they are likely to integrate different thymic signals. As a result, this might modulate the relative importance of a same regulatory axis, and eventually leading to different effects on their respective transcriptional program. Thus, regulatory network required for Vγ4+ ontogeny may be more SOX13-dependent than Vγ6+ subset. Regarding the differential contribution of the TCR signaling in the γδT17 effector fate of these populations, Blk may play a role at this stage. According to its regulatory activity on TCR signaling, Blk could act as a “rheostat” to fine-tune signals delivered by the thymic γδT cell ligands. Thus, Blk deficiency might affect more Vγ6+ ontogeny as TCR signaling has been proposed to control the development of these latter but not Vγ4+. Of note, Blk overexpression has been shown to enhance IL-7 responsiveness in B cells (95).
As stated earlier, the TCR signaling pathway strongly influences the acquisition of the γδT17 effector fate. Mechanistically, TCR engagement induces upregulation of proteins of the early growth response (Egr) family namely Egr2 and Egr3 (31, 81). These two TFs positively regulate the DNA-binding protein inhibitor Id3. Thus, Id3 impairs γδT17 cell differentiation through (1) inhibition of HeLa E-box binding protein (HEB)-dependent Sox4 and Sox13 expressions (96) and (2) inhibition of the Rorc promoter E47 (91). This scheme is also in line with the differential requirement for TCR strength in γδT17 cell effector fate of Vγ6+ and Vγ4+ further emphasizing the differences in the developmental programs of Vγ6+ vs Vγ4+ γδT17 cells.
In addition, the promyelocytic leukemia zinc finger (PLZF) protein is a key TF in the development of some innate and innate-like lymphocytes that dictates their acquisition of a Th-like effector program (97–99). PLZF was shown to control Vγ6+ differentiation into γδT17 cells (100). The molecular mechanisms that govern PLZF activity in Vγ6+ development are currently unknown and require further investigations. PLZF contribution in Vγ4+ γδT17 cell development has not been assessed yet. However, it is noteworthy that PLZF does not appear to be expressed in neonates Vγ4+ precluding a role of this TF for this particular subset (100).
Among the intracellular pathways that dictate the γδT17 effector fate, Notch signaling contributes to the generation of γδT17 cells through the helix-loop-helix protein Hes1 (78). Since it mainly exerts transcriptional repressing activities, it is possible that Hes1 acts in one of the pathways discussed above. Unrevealing the Hes1 interactome in developing γδT17 cells will be informative to get insight into the molecular factors that regulate this mechanism. An additional pathway by which the Notch signaling pathway could influence γδT17 cell development is by promoting IL-7Rα expression through the RBP-Jκ pathway (68). However, this pathway was only described in peripheral γδT17 cells of adult mice to control their homeostasis and self-renewal.
In addition to its role in proliferation/survival of γδT17 cells during development, the IL-7Rα signaling pathway is also likely to contribute to their transcriptional program. Indeed, IL-7/IL-7Rα signaling in fetal thymocytes was demonstrated to blunt both Lef1 and Tcf7 expression (101).
In silico analyses have also been fruitful to understand the transcriptional program of γδT17 cells. Using an algorithm that predicts important regulators across various lineages, the TF ETV5, along with SOX13 was proposed as a master regulator in Vγ4+ γδT17 cell differentiation (102). Conditional ablation of ETV5 in T cells confirmed the role of this TF in Vγ4+ γδT17 effector fate (102). Absence of ETV5 in developing Vγ4+ slightly reduced RORγt expression but severely impaired IL-17 secretion. This is reminiscent with the situation for Th17 cell differentiation in which ETV5 directly promotes il17a and il17f expressions but has no influence on Rorc (103). The role of ETV5 in Vγ6+ development remains to be determined; however, ETV5 is highly expressed on immature fetal Vγ6+ and strongly repressed upon maturation (http://www.immgen.org/databrowser/index.html).
Originally thought to be acquired by “neglect,” this literature underlines that the γδT17 effector fate is under the control of a very active process in which the SOX4/13 axis acts as a guardian for proper RORγt expression. In addition, the discrepancies in the phenotypes observed for Vγ4+ and Vγ6+ γδT17 cells imply different programs for natural γδT17 cell development. Thus, further transcriptomic analyses at single cell resolution are clearly required to better decipher the overlapping and/or specific developmental “trajectories” that drive the effector fate of these subsets.
What Can We Learn From Transcriptomic Analysis of Developing Natural γδT17 Cells?
As stated above, the recent advances in the quality of whole genome analyses allowed to validate and/or to predict the involvement of numerous genes in the transcriptional signature of many cell populations including γδT cells. In addition, normalized and comparative analysis of various gene sets among lymphocyte lineages led to the identification of conserved and/or distinct signature pathways in their effector program including Th17(-like) effector fate (104).
However, in silico analysis of the transcriptional program of γδT17 cells has been mainly discussed regarding the maturation of the Vγ4+ cell subset in adult mice (91, 92, 104). Thus, this population comprises both “inducible” γδT17 cells as well as non-IL-17-producing subsets. To focus on “natural” γδT17 cells that develop during embryonic life, we reanalyzed the datasets of developing Vγ6+ T cells (immature/CD24hi vs mature/CD24low) (GSE37448). Bioinformatic analysis generated a gene set of the top 1,000 transcripts significantly regulated during the effector fate acquisition of this subset (Table S1 in Supplementary Material). Interestingly, while similar analysis on developing fetal Vγ5+ T cells (GSE15907) indicated that 87.8% of the 1,000 top regulated genes were up-regulated, only 48.7% did so in the Vγ6+ dataset (Figure 3). This emphasizes the fact that, unlike γδT1, γδT17 cell effector fate is rather acquired using a repressing model.
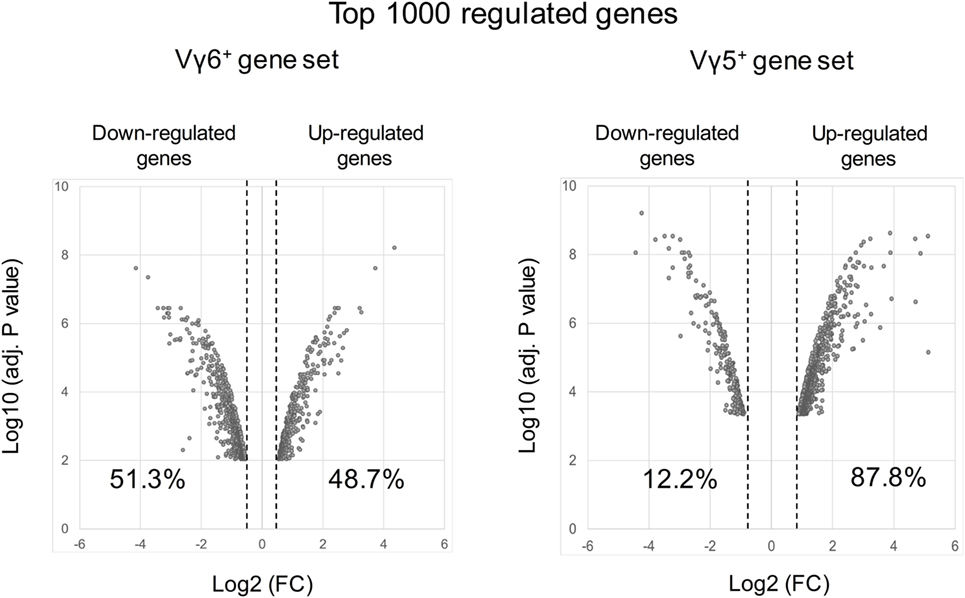
Figure 3. Volcano plots of the top 1,000 regulated genes during Vγ6 and Vγ5 maturation. Raw data were extracted from datasets (GSE37448 and GSE15907) downloaded from the NCBI’s data repositories. Vγ6+ and Vγ5+ gene sets were generated by comparing gene expression in immature (CD24high) vs mature (CD24low) thymocytes at E17 for both populations (2–3 replicates/subset). The top 1,000 regulated genes (P < 0.05) were used to constitute the two gene sets (Table S1 in Supplementary Material). Volcano plots represent either positively or negatively regulated genes (as fold change) according to their respective P value. Labels indicate the percentage of genes that are either positively or negatively regulated in each dataset. FC, fold change.
As expected, among the list of gene generated, we found many up-regulated genes shared with other innate(-like) or adaptive IL-17-producing lymphocytes including Rorc, Il17a, and Il17f and the cytokine/chemokine receptors Il1r1, Il2rb, Il7r, Il17rc, Il17re, Il23r, Il18r1, Ccr6, and Cxcr6. This is paralleled by a silencing of genes involved in Th1 and Th2 differentiation, such as Il2ra, Il12rb2, Lck, Gata3, and Maml2.
In addition, we noted numerous genes involved in TCR signaling including Lck, Nck2, Pak1, Plcg, Prkcq, Ptpn22, and Nfkbie. Notably, all these transcripts were down-modulated during Vγ6+ maturation. Moreover, genes involved in costimulation, such as Cd27, Cd28, Icos, Themis, Slamf1, Slamf6, and Pik3r2, were also repressed upon differentiation. In line, it is noteworthy that the transcript encoding for the nuclear receptor Nur77 (Nr4a1), a faithful marker of TCR strength (105) is strongly repressed during Vγ6+ maturation. Given the controversy discussed previously, these observations clearly suggest that the TCR signaling pathway has to be maintained under tight regulation to allow Vγ6+ T cell differentiation.
More importantly, using advanced pathway analysis, we pinpointed, in the Vγ6+ T cell gene set, multiple family of genes involved in biological processes and molecular pathways that might be involved in their developmental program (Figure 4).
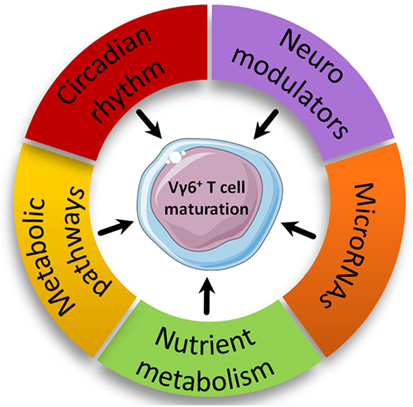
Figure 4. Proposed new biological pathways involved in intrathymic Vγ6+ maturation. Biological pathways with enriched modulated genes in the Vγ6+ dataset are represented based on advanced pathway analysis using the trial version of iPathwayGuide (©Advaita Corporation).
Nutrient Metabolism
Along with Nr4a1, other genes encoding for nuclear receptors such as receptors of vitamin A (Rarg) and D (Vdr), two vitamins reported to participate in ILC, γδT and NKT (25, 106) homeostasis and development are up-regulated during Vγ6+ maturation. In addition, genes encoding for vitamin transporters (Slc23a2 and Slc2a3) were also modulated in the Vγ6+ gene set. This could imply an important role for vitamins in γδT17 cell development. Somewhat related, expression of gpr183, a sensor of oxysterols was strongly up-regulated (3.6 fold) in developing Vγ6+ T cells. Interestingly, GPR183 has recently emerged as a critical player in the control of ILC3 homeostasis (107). Moreover, oxysterols are ligands for RORγt and drive Th17 cell differentiation (108). The influence of nutrient-derived metabolites on lymphocyte immunity including early development has recently gained considerable attention (2). The recent discovery of vitamin B2 metabolites as Ags for MAIT cells (109) will certainly reinforce the interest of immunologists for nutrient metabolism. The availability of vitamins in utero has also been shown to control the quality of the immune system in later life (110). According to these arguments and the temporal window of development for γδT17 cells, investigating the nutrient metabolism in γδT17 cell biology will be likely to provide new interesting data on the influence of maternal diet in shaping immunity.
Neuroimmunology
In line with the emerging concept of neuroimmunology, numerous members of the “neuroactive ligand-receptor interaction” pathway were present in the Vγ6+ gene set including genes encoding for neuroactive substance receptors of neuropeptide (Gpr83), hormones (Sstr2, Rxfp1, and Calcrl), prostanoids (Ptger4, Ptgfrn, and Ptgir), leukotrienes (Cysltr2 and Ltb4r1), nucleotides (P2rx7, Adora2a, and Lpar4), and amino acids (Gabbr1, Gabbr2, and Gria3). Interestingly, the above-mentioned genes encoding for prostanoids, hormones, and nucleotide receptors are down-regulated during Vγ6+ T cell maturation, while those encoding for leukotrienes are up-regulated.
The relationship between neuromodulators and immune cell development is largely unexplored. However, there are evidence for an expression of neuromodulators and their associated-receptors in TEC and on thymocytes, respectively (111). Furthermore, ex vivo somatostatin (ligand for Sstr2) addition in FTOC increased thymocyte numbers and maturation. By contrast, both neuropeptide Y (ligand for Gpr83) and calcitonin (ligand for Calcrl) reduced thymocyte numbers (112). Last, Ptgir (encoding for the prostaglandin I2 receptor) has already been biologically validated to participate in natural γδT17 cell development (89). Regarding this, a broad analysis of the eicosanoid family in γδT17 cell development should be encouraged.
Circadian Rhythm
Among highly regulated genes, our analysis also retrieved genes related to circadian rhythm. Thus, we found that Nr1d1, Nr1d2, and Bhlhe40, which encodes for REV-ERBα, REV-ERBβ, and Dec1 proteins, respectively, were all up-regulated in mature Vγ6+ T cells. These proteins are critical repressors of Arntl (encoding for Bmal1), Npas2, and Clock, three master clock genes (113, 114). Of note, Arntl, Npas2, and Clock were also significantly regulated in developing Vγ6+ T cells. Recent literature has emphasized the importance of circadian rhythms in fine-tuning immune responses (113, 115). Interestingly, RORγt expression has been shown to be under circadian regulation through a REV-ERBα-dependent mechanism (116). Thus, these data may suggest a role for the circadian rhythm in Vγ6+ T cell biology. Of note, NFIL3 (E4BP4), a repressor of the circadian clock, is implicated in the differentiation/development of ILCs and NK cells (117–119). In addition, we can speculate that the regulation of these clock genes helps mature Vγ6+ T cells to integrate and to regulate circadian cues once in the periphery, as demonstrated for many other cellular actors of innate immunity (113).
Immunometabolism
Regarding the growing interest for understanding immunometabolic pathways implicated in leukocyte biology, we searched for genes involved in metabolic pathways. Our analysis revealed that many genes involved in the six major metabolic pathways specifically glycolysis (Aldh2, Ldhb, Acss1, and Acss2), tricarboxylic acid cycle (Idh1, Idh2, and Aco1), pentose phosphate pathway (Fbp1), fatty acid oxidation (Cpt1a, Nr4a3, Peci, Auh, Pex5, and Ivd), fatty acid synthesis (Fads2 and Slc45a3), and amino acid metabolism (Aco1 and Bcat1) are down-regulated in mature Vγ6+ T cells.
Interestingly, fatty acid oxidation is associated with regulatory T cell differentiation while glycolysis is a major metabolic pathway in effector T cell differentiation (120, 121). Of note, expression of Cpt1a was reduced in Th17 cells compared with regulatory T cells (120). Enhanced fatty acid synthesis and glycolysis in immune cells, especially T cells, have been regarded as markers of inflammatory cells required for acquisition of effector functions upon inflammatory conditions (122). This adds a metabolic argument into the fact that Vγ6+ T cells are “preset” cells with metabolic programing occurring during development, to be immediately and fully functional once in peripheral tissues.
MicroRNAs
To date, there is limited literature on the role of microRNAs in leukocyte development. In our gene set, we detected the presence of five microRNAs (mir15b, mir181a-1, mir181a-2, mir181b-1, and mir181b-2), all of them being down-regulated in mature Vγ6+ T cells. Of note, miR-181 was reported to be essential in NKT cell development (123, 124). Interestingly, the group of Immo Prinz studied the impact of miR-181a/b-1 deficiency on γδT cell development. While thymic Vγ1+ and Vγ4+ T cell subsets were unaltered in the absence of miR-181a/b-1, authors reported a higher frequency of thymic, but not peripheral Vγ6+ cells in miR-181a/b-1-deficient mice (125). The reason that underlines this feature is currently unknown. However, since miR-181 is a well-known positive regulator of the TCR signal strength (126), it is tempting to hypothesize that a reduced TCR signaling confers an advantage for Vγ6+ T cell differentiation. Thus, defining the miRome of developing γδT17 cells will potentially bring a novel layer of complexity in their developmental program.
Even if these in silico analyses suggest a role for scantily explored biological pathways in γδT17 cell development and/or maintenance, supportive experimental data are clearly required to explore these predictive hypotheses. It is noteworthy that these proposed biological pathways are not meant to be Vγ6+ cell-specific and, therefore, it does not preclude that other cell types including other γδT cell subsets may rely on similar biological pathways to develop. In addition, instruction that drives γδT17 effector fate is likely to start early in thymocyte development [even before TCR rearrangement (49)]; therefore, comparing immature vs mature populations probably induced an important bias in our analysis.
Concluding Remarks
Despite the considerable body of work performed in the field of γδT17 cell ontogeny, many questions remain unsolved and sometimes appear more complex than initially thought. Even if revealing bulk transcriptomes have been informative to predict the developmental program of γδT17 cells, they present a major drawback since this kind of analysis does not reflect the dynamic aspect of the development. In this context, the recent and rapid evolution in single cell deep RNA sequencing and bioinformatics technologies will undoubtedly help to reveal the developmental “trajectories” that dictate γδT17 cell effector fate. In addition, other layers of regulation such as post-transcriptional (especially epigenetic) regulation and implication of microRNAs deserve further investigations and will have to be integrated in order to better decipher the general mechanism(s) driving γδT17 cell development. Given the critical role of γδT17 cells in major health concerns such as infections and cancer, advances in these fundamental biological processes are clearly mandatory.
Author Contributions
YJ, EP, MH, MS-T, TB, and CP prepared and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
YJ is a recipient of a PhD scholarship (poste d’acceuil) from INSERM. MS-T and CP are supported by INSERM. TB is supported by the University of Tours. We apologize to colleagues whose works could not be cited due to space constraints. This work benefited from data assembled by the ImmGen consortium. Some of the figures were created using the vector image bank of Servier Medical Art (http://smart.servier.com/). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/).
Funding
This work was supported by the recurrent annual financial support from INSERM and by a grant from the “Institut National du Cancer” (INCa, PLBIO14-155) as well as a grant from the Région Centre-Val-de-Loire (project 7UP).
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fimmu.2018.00981/full#supplementary-material.
Abbreviations
Ag, antigen; APC, Ag presenting cells; Blk, B lymphocyte kinase; DETC, dendritic epidermal γδT cell; IL, interleukin; IFN, interferon; ILC, innate lymphoid cells; Lef1, lymphoid enhancer-binding factor; MAIT, mucosal-associated invariant T; NKT, Natural Killer T; TCR, T cell receptor; TEC, thymic epithelial cells; TF, transcription factor.
References
1. Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol (2014) 14:585–600. doi:10.1038/nri3707
2. Veldhoen M. Interleukin 17 is a chief orchestrator of immunity. Nat Immunol (2017) 18:612–21. doi:10.1038/ni.3742
3. Zou W, Restifo NP. T(H)17 cells in tumour immunity and immunotherapy. Nat Rev Immunol (2010) 10:248–56. doi:10.1038/nri2742
4. Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov (2012) 11:763–76. doi:10.1038/nrd3794
5. Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol (2005) 6:1123–32. doi:10.1038/ni1254
6. Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med (2005) 201:233–40. doi:10.1084/jem.20041257
7. Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol (2010) 10:479–89. doi:10.1038/nri2800
8. Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KHG. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity (2009) 31:331–41. doi:10.1016/j.immuni.2009.08.001
9. Michel M-L, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, et al. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J Exp Med (2007) 204:995–1001. doi:10.1084/jem.20061551
10. Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood (2011) 117:1250–9. doi:10.1182/blood-2010-08-303339
11. Crellin NK, Trifari S, Kaplan CD, Satoh-Takayama N, Di Santo JP, Spits H. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by toll-like receptor 2. Immunity (2010) 33:752–64. doi:10.1016/j.immuni.2010.10.012
12. Nielsen MM, Witherden DA, Havran WL. γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nat Rev Immunol (2017) 17:733–45. doi:10.1038/nri.2017.101
13. Heilig JS, Tonegawa S. Diversity of murine gamma genes and expression in fetal and adult T lymphocytes. Nature (1986) 322:836–40. doi:10.1038/322836a0
14. Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, et al. CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat Immunol (2009) 10:427–36. doi:10.1038/ni.1717
15. Haas JD, González FHM, Schmitz S, Chennupati V, Föhse L, Kremmer E, et al. CCR6 and NK1.1 distinguish between IL-17A and IFN-gamma-producing gammadelta effector T cells. Eur J Immunol (2009) 39:3488–97. doi:10.1002/eji.200939922
16. Lalor SJ, Dungan LS, Sutton CE, Basdeo SA, Fletcher JM, Mills KHG. Caspase-1-processed cytokines IL-1beta and IL-18 promote IL-17 production by gammadelta and CD4 T cells that mediate autoimmunity. J Immunol (2011) 186:5738–48. doi:10.4049/jimmunol.1003597
17. Papotto PH, Ribot JC, Silva-Santos B. IL-17+ γδ T cells as kick-starters of inflammation. Nat Immunol (2017) 18:604–11. doi:10.1038/ni.3726
18. McKenzie DR, Kara EE, Bastow CR, Tyllis TS, Fenix KA, Gregor CE, et al. IL-17-producing γδ T cells switch migratory patterns between resting and activated states. Nat Commun (2017) 8:15632. doi:10.1038/ncomms15632
19. Duan J, Chung H, Troy E, Kasper DL. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe (2010) 7:140–50. doi:10.1016/j.chom.2010.01.005
20. Paget C, Chow MT, Gherardin NA, Beavis PA, Uldrich AP, Duret H, et al. CD3bright signals on γδ T cells identify IL-17A-producing Vγ6Vδ1+ T cells. Immunol Cell Biol (2015) 93:198–212. doi:10.1038/icb.2014.94
21. Muzaki ARBM, Soncin I, Setiagani YA, Sheng J, Tetlak P, Karjalainen K, et al. Long-lived innate IL-17-producing γ/δ T cells modulate antimicrobial epithelial host defense in the colon. J Immunol (2017) 199:3691–9. doi:10.4049/jimmunol.1701053
22. Sheridan BS, Romagnoli PA, Pham Q-M, Fu H-H, Alonzo F, Schubert W-D, et al. γδ T cells exhibit multifunctional and protective memory in intestinal tissues. Immunity (2013) 39:184–95. doi:10.1016/j.immuni.2013.06.015
23. Barros-Martins J, Schmolka N, Fontinha D, Pires de Miranda M, Simas JP, Brok I, et al. Effector γδ T cell differentiation relies on master but not auxiliary Th cell transcription factors. J Immunol (2016) 196:3642–52. doi:10.4049/jimmunol.1501921
24. Lukens JR, Barr MJ, Chaplin DD, Chi H, Kanneganti T-D. Inflammasome-derived IL-1β regulates the production of GM-CSF by CD4(+) T cells and γδ T cells. J Immunol (2012) 188:3107–15. doi:10.4049/jimmunol.1103308
25. Mielke LA, Jones SA, Raverdeau M, Higgs R, Stefanska A, Groom JR, et al. Retinoic acid expression associates with enhanced IL-22 production by γδ T cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med (2013) 210:1117–24. doi:10.1084/jem.20121588
26. Zeng X, Wei Y-L, Huang J, Newell EW, Yu H, Kidd BA, et al. γδ T cells recognize a microbial encoded B cell antigen to initiate a rapid antigen-specific interleukin-17 response. Immunity (2012) 37:524–34. doi:10.1016/j.immuni.2012.06.011
27. Scott-Browne JP, Matsuda JL, Mallevaey T, White J, Borg NA, McCluskey J, et al. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol (2007) 8:1105–13. doi:10.1038/ni1510
28. Deng L, Velikovsky CA, Xu G, Iyer LM, Tasumi S, Kerzic MC, et al. A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey. Proc Natl Acad Sci U S A (2010) 107:13408–13. doi:10.1073/pnas.1005475107
29. Schmolka N, Serre K, Grosso AR, Rei M, Pennington DJ, Gomes AQ, et al. Epigenetic and transcriptional signatures of stable versus plastic differentiation of proinflammatory γδ T cell subsets. Nat Immunol (2013) 14:1093–100. doi:10.1038/ni.2702
30. Hamada S, Umemura M, Shiono T, Hara H, Kishihara K, Tanaka K, et al. Importance of murine Vdelta1gammadelta T cells expressing interferon-gamma and interleukin-17A in innate protection against Listeria monocytogenes infection. Immunology (2008) 125:170–7. doi:10.1111/j.1365-2567.2008.02841.x
31. Turchinovich G, Hayday AC. Skint-1 identifies a common molecular mechanism for the development of interferon-γ-secreting versus interleukin-17-secreting γδ T cells. Immunity (2011) 35:59–68. doi:10.1016/j.immuni.2011.04.018
32. MacLeod AS, Hemmers S, Garijo O, Chabod M, Mowen K, Witherden DA, et al. Dendritic epidermal T cells regulate skin antimicrobial barrier function. J Clin Invest (2013) 123:4364–74. doi:10.1172/JCI70064
33. Gray EE, Suzuki K, Cyster JG. Cutting edge: identification of a motile IL-17-producing gammadelta T cell population in the dermis. J Immunol (2011) 186:6091–5. doi:10.4049/jimmunol.1100427
34. Deknuydt F, Scotet E, Bonneville M. Modulation of inflammation through IL-17 production by gammadelta T cells: mandatory in the mouse, dispensable in humans? Immunol Lett (2009) 127:8–12. doi:10.1016/j.imlet.2009.08.003
35. Ribot JC, Ribeiro ST, Correia DV, Sousa AE, Silva-Santos B. Human γδ thymocytes are functionally immature and differentiate into cytotoxic type 1 effector T cells upon IL-2/IL-15 signaling. J Immunol (2014) 192:2237–43. doi:10.4049/jimmunol.1303119
36. LeFranc MP, Forster A, Baer R, Stinson MA, Rabbitts TH. Diversity and rearrangement of the human T cell rearranging gamma genes: nine germ-line variable genes belonging to two subgroups. Cell (1986) 45:237–46. doi:10.1016/0092-8674(86)90388-0
37. Michel M-L, Pang DJ, Haque SFY, Potocnik AJ, Pennington DJ, Hayday AC. Interleukin 7 (IL-7) selectively promotes mouse and human IL-17-producing γδ cells. Proc Natl Acad Sci U S A (2012) 109:17549–54. doi:10.1073/pnas.1204327109
38. Caccamo N, La Mendola C, Orlando V, Meraviglia S, Todaro M, Stassi G, et al. Differentiation, phenotype, and function of interleukin-17-producing human Vγ9Vδ2 T cells. Blood (2011) 118:129–38. doi:10.1182/blood-2011-01-331298
39. Ness-Schwickerath KJ, Jin C, Morita CT. Cytokine requirements for the differentiation and expansion of IL-17A- and IL-22-producing human Vgamma2Vdelta2 T cells. J Immunol (2010) 184:7268–80. doi:10.4049/jimmunol.1000600
40. Moens E, Brouwer M, Dimova T, Goldman M, Willems F, Vermijlen D. IL-23R and TCR signaling drives the generation of neonatal Vgamma9Vdelta2 T cells expressing high levels of cytotoxic mediators and producing IFN-gamma and IL-17. J Leukoc Biol (2011) 89:743–52. doi:10.1189/jlb.0910501
41. O’Brien RL, Roark CL, Jin N, Aydintug MK, French JD, Chain JL, et al. gammadelta T-cell receptors: functional correlations. Immunol Rev (2007) 215:77–88. doi:10.1111/j.1600-065X.2006.00477.x
42. Ravens S, Schultze-Florey C, Raha S, Sandrock I, Drenker M, Oberdörfer L, et al. Human γδ T cells are quickly reconstituted after stem-cell transplantation and show adaptive clonal expansion in response to viral infection. Nat Immunol (2017) 18:393–401. doi:10.1038/ni.3686
43. Davey MS, Willcox CR, Joyce SP, Ladell K, Kasatskaya SA, McLaren JE, et al. Clonal selection in the human Vδ1 T cell repertoire indicates γδ TCR-dependent adaptive immune surveillance. Nat Commun (2017) 8:14760. doi:10.1038/ncomms14760
44. Fenoglio D, Poggi A, Catellani S, Battaglia F, Ferrera A, Setti M, et al. Vdelta1 T lymphocytes producing IFN-gamma and IL-17 are expanded in HIV-1-infected patients and respond to Candida albicans. Blood (2009) 113:6611–8. doi:10.1182/blood-2009-01-198028
45. Wu P, Wu D, Ni C, Ye J, Chen W, Hu G, et al. γδT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity (2014) 40:785–800. doi:10.1016/j.immuni.2014.03.013
46. Shibata K, Yamada H, Nakamura M, Hatano S, Katsuragi Y, Kominami R, et al. IFN-γ-producing and IL-17-producing γδ T cells differentiate at distinct developmental stages in murine fetal thymus. J Immunol (2014) 192:2210–8. doi:10.4049/jimmunol.1302145
47. Prinz I, Silva-Santos B, Pennington DJ. Functional development of γδ T cells. Eur J Immunol (2013) 43:1988–94. doi:10.1002/eji.201343759
48. Chien Y, Zeng X, Prinz I. The natural and the inducible: IL-17 producing gamma delta T cells. Trends Immunol (2013) 34:151–4. doi:10.1016/j.it.2012.11.004
49. Haas JD, Ravens S, Düber S, Sandrock I, Oberdörfer L, Kashani E, et al. Development of interleukin-17-producing γδ T cells is restricted to a functional embryonic wave. Immunity (2012) 37:48–59. doi:10.1016/j.immuni.2012.06.003
50. Gray EE, Ramírez-Valle F, Xu Y, Wu S, Wu Z, Karjalainen KE, et al. Deficiency in IL-17-committed Vγ4(+) γδ T cells in a spontaneous Sox13-mutant CD45.1(+) congenic mouse substrain provides protection from dermatitis. Nat Immunol (2013) 14:584–92. doi:10.1038/ni.2585
51. Roark CL, Aydintug MK, Lewis J, Yin X, Lahn M, Hahn Y-S, et al. Subset-specific, uniform activation among V gamma 6/V delta 1+ gamma delta T cells elicited by inflammation. J Leukoc Biol (2004) 75:68–75. doi:10.1189/jlb.0703326
52. Shibata K, Yamada H, Nakamura R, Sun X, Itsumi M, Yoshikai Y. Identification of CD25+ gamma delta T cells as fetal thymus-derived naturally occurring IL-17 producers. J Immunol (2008) 181:5940–7. doi:10.4049/jimmunol.181.9.5940
53. Zarin P, Chen ELY, In TSH, Anderson MK, Zúñiga-Pflücker JC. Gamma delta T-cell differentiation and effector function programming, TCR signal strength, when and how much? Cell Immunol (2015) 296:70–5. doi:10.1016/j.cellimm.2015.03.007
54. Bonneville M, Ito K, Krecko EG, Itohara S, Kappes D, Ishida I, et al. Recognition of a self major histocompatibility complex TL region product by gamma delta T-cell receptors. Proc Natl Acad Sci U S A (1989) 86:5928–32. doi:10.1073/pnas.86.15.5928
55. Kashani E, Föhse L, Raha S, Sandrock I, Oberdörfer L, Koenecke C, et al. A clonotypic Vγ4Jγ1/Vδ5Dδ2Jδ1 innate γδ T-cell population restricted to the CCR6+CD27− subset. Nat Commun (2015) 6:6477. doi:10.1038/ncomms7477
56. Papotto PH, Gonçalves-Sousa N, Schmolka N, Iseppon A, Mensurado S, Stockinger B, et al. IL-23 drives differentiation of peripheral γδ17 T cells from adult bone marrow-derived precursors. EMBO Rep (2017) 18:1957–67. doi:10.15252/embr.201744200
57. Muschaweckh A, Petermann F, Korn T. IL-1β and IL-23 promote extrathymic commitment of CD27+CD122-γδ T cells to γδT17 cells. J Immunol (2017) 199:2668–79. doi:10.4049/jimmunol.1700287
58. Buus TB, Ødum N, Geisler C, Lauritsen JPH. Three distinct developmental pathways for adaptive and two IFN-γ-producing γδ T subsets in adult thymus. Nat Commun (2017) 8:1911. doi:10.1038/s41467-017-01963-w
59. Ciofani M, Zúñiga-Pflücker JC. Determining γδ versus αß T cell development. Nat Rev Immunol (2010) 10:657–63. doi:10.1038/nri2820
60. Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat Immunol (2010) 11:666–73. doi:10.1038/ni.1887
61. Hayday AC, Saito H, Gillies SD, Kranz DM, Tanigawa G, Eisen HN, et al. Structure, organization, and somatic rearrangement of T cell gamma genes. Cell (1985) 40:259–69. doi:10.1016/0092-8674(85)90140-0
62. Fry TJ, Mackall CL. The many faces of IL-7: from lymphopoiesis to peripheral T cell maintenance. J Immunol (2005) 174:6571–6. doi:10.4049/jimmunol.174.11.6571
63. Maki K, Sunaga S, Ikuta K. The V-J recombination of T cell receptor-gamma genes is blocked in interleukin-7 receptor-deficient mice. J Exp Med (1996) 184:2423–7. doi:10.1084/jem.184.6.2423
64. He YW, Malek TR. Interleukin-7 receptor alpha is essential for the development of gamma delta + T cells, but not natural killer cells. J Exp Med (1996) 184:289–93. doi:10.1084/jem.184.1.289
65. Appasamy PM, Kenniston TW, Weng Y, Holt EC, Kost J, Chambers WH. Interleukin 7-induced expression of specific T cell receptor gamma variable region genes in murine fetal liver cultures. J Exp Med (1993) 178:2201–6. doi:10.1084/jem.178.6.2201
66. Patin EC, Soulard D, Fleury S, Hassane M, Dombrowicz D, Faveeuw C, et al. Type I IFN receptor signaling controls IL7-dependent accumulation and activity of protumoral IL17A-producing γδT cells in breast cancer. Cancer Res (2018) 78:195–204. doi:10.1158/0008-5472.CAN-17-1416
67. Rei M, Gonçalves-Sousa N, Lança T, Thompson RG, Mensurado S, Balkwill FR, et al. Murine CD27(-) Vγ6(+) γδ T cells producing IL-17A promote ovarian cancer growth via mobilization of protumor small peritoneal macrophages. Proc Natl Acad Sci U S A (2014) 111:E3562–70. doi:10.1073/pnas.1403424111
68. Nakamura M, Shibata K, Hatano S, Sato T, Ohkawa Y, Yamada H, et al. A genome-wide analysis identifies a notch-RBP-Jκ-IL-7Rα axis that controls IL-17-producing γδ T cell homeostasis in mice. J Immunol (2015) 194:243–51. doi:10.4049/jimmunol.1401619
69. Fujikado N, Mann AO, Bansal K, Romito KR, Ferre EMN, Rosenzweig SD, et al. Aire inhibits the generation of a perinatal population of interleukin-17A-producing γδ T cells to promote immunologic tolerance. Immunity (2016) 45:999–1012. doi:10.1016/j.immuni.2016.10.023
70. Alves NL, Richard-Le Goff O, Huntington ND, Sousa AP, Ribeiro VSG, Bordack A, et al. Characterization of the thymic IL-7 niche in vivo. Proc Natl Acad Sci U S A (2009) 106:1512–7. doi:10.1073/pnas.0809559106
71. Do J, Fink PJ, Li L, Spolski R, Robinson J, Leonard WJ, et al. Cutting edge: spontaneous development of IL-17-producing gamma delta T cells in the thymus occurs via a TGF-beta 1-dependent mechanism. J Immunol (2010) 184:1675–9. doi:10.4049/jimmunol.0903539
72. Lochner M, Peduto L, Cherrier M, Sawa S, Langa F, Varona R, et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J Exp Med (2008) 205:1381–93. doi:10.1084/jem.20080034
73. Hayes SM, Laird RM. Genetic requirements for the development and differentiation of interleukin-17-producing γδ T cells. Crit Rev Immunol (2012) 32:81–95. doi:10.1615/CritRevImmunol.v32.i1.50
74. Petermann F, Rothhammer V, Claussen MC, Haas JD, Blanco LR, Heink S, et al. γδ T cells enhance autoimmunity by restraining regulatory T cell responses via an interleukin-23-dependent mechanism. Immunity (2010) 33:351–63. doi:10.1016/j.immuni.2010.08.013
75. Nitta T, Muro R, Shimizu Y, Nitta S, Oda H, Ohte Y, et al. The thymic cortical epithelium determines the TCR repertoire of IL-17-producing γδT cells. EMBO Rep (2015) 16:638–53. doi:10.15252/embr.201540096
76. Takahama Y, Letterio JJ, Suzuki H, Farr AG, Singer A. Early progression of thymocytes along the CD4/CD8 developmental pathway is regulated by a subset of thymic epithelial cells expressing transforming growth factor beta. J Exp Med (1994) 179:1495–506. doi:10.1084/jem.179.5.1495
77. Ribeiro AR, Rodrigues PM, Meireles C, Di Santo JP, Alves NL. Thymocyte selection regulates the homeostasis of IL-7-expressing thymic cortical epithelial cells in vivo. J Immunol (2013) 191:1200–9. doi:10.4049/jimmunol.1203042
78. Shibata K, Yamada H, Sato T, Dejima T, Nakamura M, Ikawa T, et al. Notch-Hes1 pathway is required for the development of IL-17-producing γδ T cells. Blood (2011) 118:586–93. doi:10.1182/blood-2011-02-334995
79. Mair F, Joller S, Hoeppli R, Onder L, Hahn M, Ludewig B, et al. The NFκB-inducing kinase is essential for the developmental programming of skin-resident and IL-17-producing γδ T cells. Elife (2015) 4:e10087. doi:10.7554/eLife.10087
80. Jensen KDC, Su X, Shin S, Li L, Youssef S, Yamasaki S, et al. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity (2008) 29:90–100. doi:10.1016/j.immuni.2008.04.022
81. Muñoz-Ruiz M, Ribot JC, Grosso AR, Gonçalves-Sousa N, Pamplona A, Pennington DJ, et al. TCR signal strength controls thymic differentiation of discrete proinflammatory γδ T cell subsets. Nat Immunol (2016) 17:721–7. doi:10.1038/ni.3424
82. Sumaria N, Grandjean CL, Silva-Santos B, Pennington DJ. Strong TCRγδ signaling prohibits thymic development of IL-17A-secreting γδ T cells. Cell Rep (2017) 19:2469–76. doi:10.1016/j.celrep.2017.05.071
83. Wencker M, Turchinovich G, Di Marco Barros R, Deban L, Jandke A, Cope A, et al. Innate-like T cells straddle innate and adaptive immunity by altering antigen-receptor responsiveness. Nat Immunol (2014) 15:80–7. doi:10.1038/ni.2773
84. Vantourout P, Laing A, Woodward MJ, Zlatareva I, Apolonia L, Jones AW, et al. Heteromeric interactions regulate butyrophilin (BTN) and BTN-like molecules governing γδ T cell biology. Proc Natl Acad Sci U S A (2018) 115:1039–44. doi:10.1073/pnas.1701237115
85. Di Marco Barros R, Roberts NA, Dart RJ, Vantourout P, Jandke A, Nussbaumer O, et al. Epithelia use butyrophilin-like molecules to shape organ-specific γδ T cell compartments. Cell (2016) 167:203–218.e17. doi:10.1016/j.cell.2016.08.030
86. Buus TB, Schmidt JD, Bonefeld CM, Geisler C, Lauritsen JPH. Development of interleukin-17-producing Vγ2+ γδ T cells is reduced by ICOS signaling in the thymus. Oncotarget (2016) 7:19341–54. doi:10.18632/oncotarget.8464
87. Silva-Santos B, Pennington DJ, Hayday AC. Lymphotoxin-mediated regulation of gammadelta cell differentiation by alphabeta T cell progenitors. Science (2005) 307:925–8. doi:10.1126/science.1103978
88. Powolny-Budnicka I, Riemann M, Tänzer S, Schmid RM, Hehlgans T, Weih F. RelA and RelB transcription factors in distinct thymocyte populations control lymphotoxin-dependent interleukin-17 production in γδ T cells. Immunity (2011) 34:364–74. doi:10.1016/j.immuni.2011.02.019
89. Jaffar Z, Ferrini ME, Shaw PK, FitzGerald GA, Roberts K. Prostaglandin I2 promotes the development of IL-17-producing γδ T cells that associate with the epithelium during allergic lung inflammation. J Immunol (2011) 187:5380–91. doi:10.4049/jimmunol.1101261
90. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell (2006) 126:1121–33. doi:10.1016/j.cell.2006.07.035
91. Narayan K, Sylvia KE, Malhotra N, Yin CC, Martens G, Vallerskog T, et al. Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat Immunol (2012) 13:511–8. doi:10.1038/ni.2247
92. Malhotra N, Narayan K, Cho OH, Sylvia KE, Yin C, Melichar H, et al. A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity (2013) 38:681–93. doi:10.1016/j.immuni.2013.01.010
93. Xing S, Li F, Zeng Z, Zhao Y, Yu S, Shan Q, et al. Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat Immunol (2016) 17:695–703. doi:10.1038/ni.3456
94. Laird RM, Laky K, Hayes SM. Unexpected role for the B cell-specific Src family kinase B lymphoid kinase in the development of IL-17-producing γδ T cells. J Immunol (2010) 185:6518–27. doi:10.4049/jimmunol.1002766
95. Tretter T, Ross AE, Dordai DI, Desiderio S. Mimicry of pre-B cell receptor signaling by activation of the tyrosine kinase Blk. J Exp Med (2003) 198:1863–73. doi:10.1084/jem.20030729
96. In TSH, Trotman-Grant A, Fahl S, Chen ELY, Zarin P, Moore AJ, et al. HEB is required for the specification of fetal IL-17-producing γδ T cells. Nat Commun (2017) 8:2004. doi:10.1038/s41467-017-02225-5
97. Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity (2008) 29:391–403. doi:10.1016/j.immuni.2008.07.011
98. Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature (2014) 508:397–401. doi:10.1038/nature13047
99. Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol (2008) 9:1055–64. doi:10.1038/ni.1641
100. Lu Y, Cao X, Zhang X, Kovalovsky D. PLZF controls the development of fetal-derived IL-17+Vγ6+ γδ T cells. J Immunol (2015) 195:4273–81. doi:10.4049/jimmunol.1500939
101. Yu Q, Erman B, Park J-H, Feigenbaum L, Singer A. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORgammat: impact on thymocyte development. J Exp Med (2004) 200:797–803. doi:10.1084/jem.20032183
102. Jojic V, Shay T, Sylvia K, Zuk O, Sun X, Kang J, et al. Identification of transcriptional regulators in the mouse immune system. Nat Immunol (2013) 14:633–43. doi:10.1038/ni.2587
103. Pham D, Sehra S, Sun X, Kaplan MH. The transcription factor Etv5 controls TH17 cell development and allergic airway inflammation. J Allergy Clin Immunol (2014) 134:204–14. doi:10.1016/j.jaci.2013.12.021
104. Lee YJ, Starrett GJ, Lee ST, Yang R, Henzler CM, Jameson SC, et al. Lineage-specific effector signatures of invariant NKT cells are shared amongst γδ T, innate lymphoid, and Th cells. J Immunol (2016) 197:1460–70. doi:10.4049/jimmunol.1600643
105. Sekiya T, Kashiwagi I, Yoshida R, Fukaya T, Morita R, Kimura A, et al. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat Immunol (2013) 14:230–7. doi:10.1038/ni.2520
106. Yu S, Bruce D, Froicu M, Weaver V, Cantorna MT. Failure of T cell homing, reduced CD4/CD8alphaalpha intraepithelial lymphocytes, and inflammation in the gut of vitamin D receptor KO mice. Proc Natl Acad Sci U S A (2008) 105:20834–9. doi:10.1073/pnas.0808700106
107. Emgård J, Kammoun H, García-Cassani B, Chesné J, Parigi SM, Jacob J-M, et al. Oxysterol sensing through the receptor GPR183 promotes the lymphoid-tissue-inducing function of innate lymphoid cells and colonic inflammation. Immunity (2018) 48:120–132.e8. doi:10.1016/j.immuni.2017.11.020
108. Soroosh P, Wu J, Xue X, Song J, Sutton SW, Sablad M, et al. Oxysterols are agonist ligands of RORγt and drive Th17 cell differentiation. Proc Natl Acad Sci U S A (2014) 111:12163–8. doi:10.1073/pnas.1322807111
109. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature (2012) 491:717–23. doi:10.1038/nature11605
110. van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature (2014) 508:123–7. doi:10.1038/nature13158
111. Silva AB, Palmer DB. Evidence of conserved neuroendocrine interactions in the thymus: intrathymic expression of neuropeptides in mammalian and non-mammalian vertebrates. Neuroimmunomodulation (2011) 18:264–70. doi:10.1159/000329493
112. Solomou K, Ritter MA, Palmer DB. Somatostatin is expressed in the murine thymus and enhances thymocyte development. Eur J Immunol (2002) 32:1550–9. doi:10.1002/1521-4141(200206)32:6<1550::AID-IMMU1550>3.0.CO;2-W
113. Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science (2016) 354:994–9. doi:10.1126/science.aah4965
114. Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature (2002) 419:841–4. doi:10.1038/nature01123
115. Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol (2013) 13:190–8. doi:10.1038/nri3386
116. Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, et al. TH17 cell differentiation is regulated by the circadian clock. Science (2013) 342:727–30. doi:10.1126/science.1243884
117. Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJM, et al. Differential requirement for Nfil3 during NK cell development. J Immunol (2014) 192:2667–76. doi:10.4049/jimmunol.1302605
118. Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol (2009) 10:1118–24. doi:10.1038/ni.1787
119. Xu W, Domingues RG, Fonseca-Pereira D, Ferreira M, Ribeiro H, Lopez-Lastra S, et al. NFIL3 orchestrates the emergence of common helper innate lymphoid cell precursors. Cell Rep (2015) 10:2043–54. doi:10.1016/j.celrep.2015.02.057
120. Gerriets VA, Kishton RJ, Nichols AG, Macintyre AN, Inoue M, Ilkayeva O, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest (2015) 125:194–207. doi:10.1172/JCI76012
121. Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol (2011) 186:3299–303. doi:10.4049/jimmunol.1003613
122. O’Neill LAJ, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol (2016) 16:553–65. doi:10.1038/nri.2016.70
123. Henao-Mejia J, Williams A, Goff LA, Staron M, Licona-Limón P, Kaech SM, et al. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity (2013) 38:984–97. doi:10.1016/j.immuni.2013.02.021
124. Ziętara N, Łyszkiewicz M, Witzlau K, Naumann R, Hurwitz R, Langemeier J, et al. Critical role for miR-181a/b-1 in agonist selection of invariant natural killer T cells. Proc Natl Acad Sci U S A (2013) 110:7407–12. doi:10.1073/pnas.1221984110
125. Sandrock I, Ziętara N, Łyszkiewicz M, Oberdörfer L, Witzlau K, Krueger A, et al. MicroRNA-181a/b-1 Is Not Required for Innate γδ NKT Effector Cell Development. PLoS One (2015) 10:e0145010. doi:10.1371/journal.pone.0145010
Keywords: γδT cells, innate immunity, interleukin-17A, thymus, transcription factor, development
Citation: Jouan Y, Patin EC, Hassane M, Si-Tahar M, Baranek T and Paget C (2018) Thymic Program Directing the Functional Development of γδT17 Cells. Front. Immunol. 9:981. doi: 10.3389/fimmu.2018.00981
Received: 22 February 2018; Accepted: 20 April 2018;
Published: 08 May 2018
Edited by:
Pierre Vantourout, King’s College London, United KingdomReviewed by:
Melanie Wencker, UMR5308 Centre International de Recherche en Infectiologie (CIRI), FranceMarie-Laure Michel, INRA Centre Jouy-en-Josas, France
Julie Ribot, Instituto de Medicina Molecular (IMM), Portugal
Copyright: © 2018 Jouan, Patin, Hassane, Si-Tahar, Baranek and Paget. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christophe Paget, christophe.paget@inserm.fr
 Youenn Jouan1,2,3
Youenn Jouan1,2,3 Maya Hassane
Maya Hassane Mustapha Si-Tahar
Mustapha Si-Tahar Thomas Baranek
Thomas Baranek Christophe Paget
Christophe Paget