- 1Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Harbin, China
- 2National and Local Joint Engineering Laboratory for Freshwater Fish Breeding, Harbin, China
- 3Key Laboratory of Aquatic Genomics, Ministry of Agriculture, Chinese Academy of Fishery Sciences, Beijing, China
- 4Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 5College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China
- 6College of Fisheries and Life Science, Dalian Ocean University, Dalian, China
- 7The Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing, China
Anti-disease breeding is becoming the most promising solution to cyprinid herpesvirus-3 (CyHV-3) infection, the major threat to common carp aquaculture. Virus challenging studies suggested that a breeding strain of common carp developed resistance to CyHV-3 infection. This study illustrates the immune mechanisms involved in both sensitivity and anti-virus ability for CyHV3 infection in fish. An integrative analysis of the protein-coding genes and long non-coding RNAs (lncRNAs) using transcriptomic data was performed. Tissues from the head kidney of common carp were extracted at days 0 (the healthy control) and 7 after CyHV-3 infection (the survivors) and used to analyze the transcriptome through both Illumina and PacBio sequencing. Following analysis of the GO terms and KEGG pathways involved, the immune-related terms and pathways were merged. To dig out details on the immune aspect, the DEGs were filtered using the current common carp immune gene library. Immune gene categories and their corresponding genes in different comparison groups were revealed. Also, the immunological Gene Ontology terms for lncRNA modulation were retained. The weighted gene co-expression network analysis was used to reveal the regulation of immune genes by lncRNA. The results demonstrated that the breeding carp strain develops a marked resistance to CyHV-3 infection through a specific innate immune mechanism. The featured biological processes were autophagy, phagocytosis, cytotoxicity, and virus blockage by lectins and MUC3. Moreover, the immune-suppressive signals, such as suppression of IL21R on STAT3, PI3K mediated inhibition of inflammation by dopamine upon infection, as well as the inhibition of NLRC3 on STING during a steady state. Possible susceptible factors for CyHV-3, such as ITGB1, TLR18, and CCL4, were also revealed from the non-breeding strain. The results of this study also suggested that Nramp and PAI regulated by LncRNA could facilitate virus infection and proliferation for infected cells respectively, while T cell leukemia homeobox 3 (TLX3), as well as galectin 3 function by lncRNA, may play a role in the resistance mechanism. Therefore, immune factors that are immunogenetically insensitive or susceptible to CyHV-3 infection have been revealed.
Introduction
Cyprinid herpesvirus-3 (CyHV-3) infection is a major threat to common carp aquaculture (1), leading to widespread mortality and substantial economic loss. CyHV-3 is thought to cause death by weakening the host’s immune system, resulting in susceptibility to pathogenic microbes (1). In common carp, clinical signs of the disease develop rapidly and may induce morbidity and mortality within a period of 6 to 10 days following infection (2).
Carp that survive a primary infection with CyHV-3 can be resistant to future infection with this virus. Since latency and persistent carrying of CyHV-3 exist in carp (2–4), genetic backgrounds are crucial in developing an understanding of resistance against the virus. Experimental infections of carp from pure lines or crosses have indicated the existence of a genetic background of resistance by divergent survival rates (2). For example, a markedly higher expression of immune-related genes involved in pathogen recognition, complement activation, major histocompatibility complex class I (MHC I)-restricted antigen presentation, and the development of adaptive mucosal immunity was noted in the more resistant R3 line. Higher activation of CD8+ T cells was also observed (5). The diallelic cross of four European carp lines, including Polish ‘K’ and ‘R6’, Hungarian ‘R7’ and French ‘F’ also has been done to select the resistant fish, and then found that MH class II B genes of carp can affect immunity against CyHV-3 infection (6). Additionally, carp strains of Asian origin, particularly Amur wild carp, were shown to be more resistant to CyHV-3 than strains originating from Europe, such as the Prerov scale carp or koi carp from a breed in the Czech Republic (7).
The immune response of carp to CyHV-3 involves both innate and adaptive aspects with the outcome of the disease largely depending on whether the balance is tipped in favor of the host’s immune response or virus’s evasion strategy (2). In general, transcriptomic analysis has revealed that three immune-related pathways, namely the mitogen-activated protein kinase (MAPK) signaling pathway, the innate immune response, and the cytokine-mediated signaling pathway, were highly involved in the infection with CyHV-3 (8). In red common carp × koi, the expression of interleukin 12 (IL12) p35, interferon (IFN) αβ, and toll-like receptor 9 (TLR9) may provide potential genes related to resistance against KHV (another term for CyHV-3) (9). However, the magnitude of type I IFN response did not correlate with a higher resistance in CyHV-3-infected carp, during the challenge test among different strains, although CyHV-3 infection can induce type I IFNs (7, 10). Regarding the innate resistance in carp, the mapped CyHV-3 survival quantitative trait loci have been reported mainly in IL10 and MHC II (11). Recently, by quantitative trait locus mapping and genome-wide association study, tumor necrosis factor-alpha (tnfa), hypoxia inducible factor 1 subunit alpha (hif1a), galectin-8 (LGALS8), rootletin, and paladin, have also been related to resistance against CyHV-3 (12). Adaptive immunity through both cytotoxicity and immunoglobulin (Ig) secretion may be involved in resistance. Matthew et al. revealed that, in the anterior kidney, Ig secretion plays an important role in the resistance during the persistent infection or reactivation phases of CyHV-3 (1). In addition, CyHV-3 profoundly influences the expression of host miRNA, although the regulation of immune processes by miRNA in the clinical and latent phases differs (4). This suggests an important role of non-coding RNAs in anti-CyHV-3 immunity.
Transcriptomic studies have been a widely used tool to reveal molecular pathology (13, 14) and even pathogen discovery (15) during fish viral infection. This could facilitate understanding the pathogenic mechanisms of diseases and the immune system of fish (16). For CyHV-3 infection, the first transcriptional analysis of carp anterior kidney mainly pointed out the important role of humoral immune responses, especially those related to immunoglobulin (1). Spleen transcriptomic analyses comparing the susceptible and resistant common carp revealed that the susceptible fish elicited a typical anti-viral interferon response, while the upregulated IL-8 attracted innate immune cells and related response may play an essential role in resistant fish (17). Recent studies have demonstrated that lncRNAs are widely modulated during fish viral or bacterial infections (18–21). As for virus infection, lncRNAs could be involved in regulating the host response during ISAV infection in salmon (18). In zebrafish, SVCV could induce both immune and antiviral processes related to LncRNA (21). The widespread differential expression of lncRNAs in response to infections with different types of pathogens suggested that lncRNAs are pivotal players during immune responses in fish. However, the specific LncRNA modulation of the immune response during CyHV-3 infection has not been accessed in carp.
German mirror carp selection G4 is a strain of common carp cultivated widely in China, yet with high mortality caused by the CyHV-3 virus. Based on our previous study, a strain of common carp from German mirror carp selection G4 has already shown a higher survival rate after breeding for three generations. Among fish from the G3 generation, 1,000 individuals were genotyped by four SNP loci, including carp065309, carp070076, carp183811 and carp160380, and then four main groups, with genotypes of GG/GT/GG/AA, GG/TT/TT/AA, AG/TT/AG/AT and GG/GG/GG/AA, whose survival rates were 89.9, 94.7, 88.0 and 92.7% respectively, were propagated (22). While the unselected mirror carp was 62.6%. Since decreased viral load in tissues directly indicates resistance to CyHV-3 (23), the fact that it has displayed a better immunological index as well as a reduced virus load in immune organs, such as the kidneys and spleen (24, 25), strongly suggests resistance to CyHV-3 in current breeding strain. In detail, acid phosphatase in the spleen, glutathione and total antioxidation capacity in the kidney, lysozyme and immunoglobulin M in the serum, and alkaline phosphatase in the spleen and kidney also showed significant differences between G1 and G3 or G1 and G2/G3. Meanwhile, the survival rate after the CyHV-3 challenge increased generation by generation. The strong resistance to CyHV-3 has been stable for G3 (24). Recently, Sun has revealed that virus genes TK and ORF72 in the G4 were expressed at levels significantly lower than those in the non-breeding strain (25). Thus, the resistance to CyHV-3 in G4 was strongly deduced.
However, there is a lack of systemic studies focusing on a detailed network of the immune system for the anti-CyHV-3 immune mechanism in this resistant strain. The third-generation sequencing method, such as PacBio, can decode the genetic sequences that are markedly longer compared with the second-generation method, such as Illumina (26). Therefore, a higher quality assembly of transcripts, including both mRNA and non-coding RNAs, may provide a more detailed understanding of the mechanism of genetic resistance.
In this study, to reveal the immune mechanisms involved in anti-CyHV3 breeding for common carp, integrative transcriptomic analysis was performed for both mRNA and long non-coding RNA (lncRNA) during the CyHV-3 challenge, using the strategy shown in Figure 1. Immune-related transcripts, from both survivors and healthy controls of either breeding or non-breeding strains, were analyzed. By comparing data from different groups for different strains obtained through both Illumina (New England Biolabs, Ipswich, MA, USA) and Pacific Biosciences (PacBio) sequencing, both immune-related differentially expressed genes (DEG) and lncRNA have been revealed for the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and/or Gene Ontology (GO) terms involved. The immune gene-related lncRNA was also explored. This study may shed some light on molecular breeding for virus-resistant fish strains.
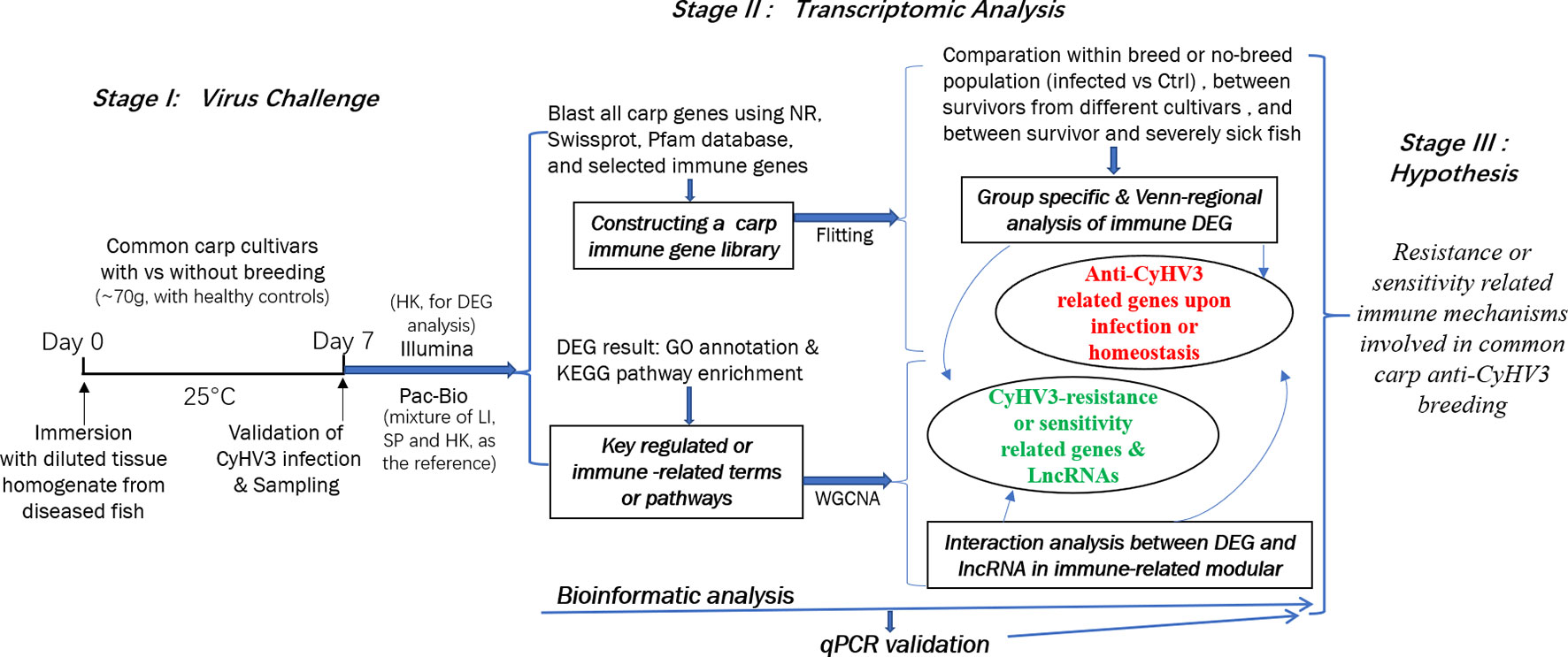
Figure 1 The strategy of integrative transcriptomic analysis for revealing immune mechanisms involved for resistance or sensitivity for current anti-CyHV3 breeding in common carp. During stage I, the disease modelling for CyHV3 infection was established using immersion of virus-containing tissue homogenate and then validated by checking the expression of virus genes. The transcriptomic sampling was done on day 7 post-infection for sequencing using both Illumina and Pac-Bio platforms. In stage II, the bioinformatic analysis was carried out for the transcriptomic data. DEG was generated from five comparison groups, including SvC-B, SvC-N, SvS-BvN-D7, CvC-BvN-D0, and SvI-B&N. The fish with severe illness swam very slowly or even floated on the water. The revealed DEGs were annotated by GO terms and KEGG pathways. Further, a common carp immune gene library was constructed for further immune-related analysis. Additionally, to reveal the LncRNA regulation, WGCNA was used to reveal the interaction between differential expressed transcripts and LncRNA in immune-related module. Thereafter, the resistance or sensitivity related immune mechanisms involved in common carp anti-CyHV3 breeding could be enlightened accordingly.
Materials and Methods
Ethics Statement
All procedures involving animals in this study were conducted according to the guidelines for the care and use of laboratory animals of Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences (Harbin, China). The studies involving animals were reviewed and approved by the Committee for the Welfare and Ethics of Laboratory Animals of Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences.
Animals and Virus Challenge
German mirror carp selection G4 used in this study were obtained from Kuandian Research Base of Heilongjiang River Fisheries Research Institute (Liaoning, China). The breeding strain was the fourth generation after the selection of resistance to CyHV-3 (25). Two G4 groups with survival rates of 94.7 and 92.7% were mixed with the ratio 1:1 and used as the experimental populations in this study. Both the breeding strain G4 and non-breeding strain were used for the virus challenge. Since CyHV-3 has a mucosal route of infection mainly via the skin rather than the gut (2), this study induced infection with CyHV-3 by adding the homogenate solution of internal organs from sick fish into the water of the tanks following the previously published methods (24, 25, 27). The homogenate solution was prepared using organs from 10 severely sick fish, which swam very slowly or floated on the water. Before using the homogenate to infect fish, the head kidney tissues from those sick fish were checked for the CyHV-3 infection by PCR of virus genes TK and Sph following the method described in the industry-standard SC/T 7212.1-2011 (28). The CyHV-3 that contained internal organs from sick fish was homogenized to generate 100 ml homogenate, which contained 1.5 × 107 copy CyHV-3 per mg homogenate by PCR-checking virus gene Sph. Then the homogenate was used to infect the experimental fish, 17 ml homogenate was used for each tank. For either the breeding or non-breeding strains, the virus challenging group (using diseased fish tissue’s homogenate) was paralleled with the control group (using saline instead). For each treatment groups, three replicates have been done. In each tank (1.6 m × 1.2 m × 0.6 m), which contained around 1 m3 water, healthy juvenile common carp (~70 g; N = 100 fish) were used. The water temperature during the experiment was maintained at 25 ± 1°C.
Sampling and Pathological Analysis
A selection of common carp, from either the breeding or non-breeding strains, were sacrificed. Head kidney samples were obtained at days 0 and 7 after challenge, representing the control and survivor carp. For each tissue sample, three fish were used. For transcriptomic analysis, the samples (N = 3) were collected immediately and soaked in 10 volumes of RNAlater (Qiagen, Hilden, Germany), for sequencing using Illumina (New England Biolabs). Further, to better sequence and generate the lncRNAs, PacBio sequencing was applied to analyze the mixture of liver, spleen, and kidney, with two sample replicates. Additionally, for the validation of transcriptomic data, the qPCR samples (N = 3) were also collected using RNAlater. The number of dead fish during the experiment was counted daily to calculate the mortality rate. Meanwhile, to analyze the degree of swelling of inner organs, the proportion of trunk kidney to the whole trunk area was calculated by ImageJ (https://imagej.en.softonic.com/). In detail, the area of both trunk kidney and trunk regions were selected and measured, and the percentages between them were calculated based on eight survivors of breeding or non-breeding strain. T-tests were applied to test the significance of the difference.
RNA Extraction
RNA was isolated using the AllPrep DNA/RNA FFPE Kit (Qiagen, Hilden, Germany), according to the instructions provided by the manufacturer. RNA degradation and contamination were monitored on 1% agarose gels. RNA purity was checked using the NanoPhotometer spectrophotometer (Implen Inc., Westlake Village, CA, USA). RNA concentration was measured using the Qubit RNA Assay Kit in a Qubit 2.0 Flurometer (Life Technologies, Carlsbad, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA).
Library Preparation and Sequencing for Transcriptomic Analysis
Optimized RNA-Seq strategies, including both PacBio and Illumina (New England Biolabs) sequencing (29), were used to more precisely resolve the sequence of transcripts. Firstly, the total RNA isolated from the head kidney was used to construct the cDNA library. Subsequently, the library was sequenced on a PacBio RS II platform (Biomarker Technologies, Beijing, China). For the PacBio Long Read Processing, raw reads were processed into error-corrected reads of insert using Iso-seq pipeline with minFullPass = 0 and minPredictedAccuracy = 0.90. Next, full-length, non-chemiric transcripts were determined by searching for the polyA tail signal and the 5’ and 3’ cDNA primers in reads of the insert. Iterative Clustering for Error Correction (ICE) was used to obtain consensus isoforms, and full-length consensus sequences from ICE were polished using Quiver. High-quality transcripts with post-correction accuracy of >99% were retained for further analysis.
The illustration of transcripts obtained from the results of PacBio could provide reference transcriptional sequences for the assembly of Illumine sequencing data, to improve sequencing quality. Therefore, Illumina (New England Biolabs) sequencing was performed on all head kidney samples. The procedure used for the preparation of the gene library and sequencing of the transcriptome followed previously published methods (30). Briefly, sequencing libraries were generated using the NEBNext UltraTM RNA Library Prep Kit for Illumina (New England Biolabs), and the library quality was assessed on the Agilent Bioanalyzer 2100 system. The library preparations were sequenced on an Illumina platform, and 150 bp paired-end reads were generated.
Annotation and Functional Analysis of Transcripts by Public Databases
All reads in the transcriptome data were mapped on the common carp genome (https://asia.ensembl.org/Cyprinus_carpio_german_mirror/Info/Index) (31) for annotation. In detail, GMAP (Genomic Mapping and Alignment Program) and BLAST (version 2.2.26) were applied for mapping the transcriptome data to carp genome and obtaining annotation respectively. The data from the Illumina platform were also used to check and replace errors by proovread (version 2.14.1) in the data from the PacBio platform. Both the annotation of genes and lncRNAs were subsequently generated. The protein-coding transcripts were annotated by NR, swissprot and Pfam database. DEGs were detected from five comparison groups, including intra-strain comparison groups SvC-B (survivors vs. controls in the breeding strain) and SvC-N (survivors vs. controls in the non-breeding strain), inter-strain comparison groups SvS-BvN-D7 (the comparison between survivors at day 7 for the two strains to compare survivors from the breeding to non-breeding strain) and CvC-BvN-D0 (the comparison between control fish at day 0 for the two strains to compare controls from the breeding to non-breeding strain), and group SvI-B&N. The vital genes for survival were investigated through group SvI-B&N, in which the DEGs with no significant differences (p >0.05) were compared between the survivors from the two strains with the transcripts from fish with severe illness. The revealed DEGs were annotated by GO terms and KEGG pathways, following a previously published protocol (30, 32). In brief, functional annotation and the classification of genes were determined by both employing local genes blasts against GO Consortium (http://geneontology.org/) and KEGG (https://www.kegg.jp/kegg/pathway.html). Enrichment of the KEGG pathways was carried out for both upregulated and downregulated genes for all comparison groups. Then to demonstrate the immune DEGs involved pathways more clearly, the gene list of the current construct common carp immune gene library was used as follows. In addition, the LncRNAs were also annotated by GO terms.
Construction of the Common Carp Immune Gene Library
The common carp immune library was constructed by following our previously published method, which was applied to grass carp (30) and tilapia (32), with some adjustment for viral infection-related immune genes. The modifications were based on gene information obtained by blasting each sequence to databases, including NCBI NR database, as well as Swiss-Prot and Pfam databases. The common carp immune gene library contained information for immune genes at two levels. For the first level, nine categories of immune processes, namely “acute phase reactions”, “pattern recognition”, “antigen processing and regulators”, “complement system”, “inflammatory cytokines and receptors”, “adapters, effectors and signal transducers”, “innate immune cells related”, “T/B cell antigen activation”, and “other genes related to immune response”, were proposed. Subsequently, many categories of immune genes for each immune process (detailed in Table S1) were applied for the second level. The library was used to filter transcriptome data and obtain details of the immune processes and particular immune genes for each comparison group, during the GO term and KEGG pathway enrichment.
Statistical Analysis
The DEG was generated by comparing the RPKM (Reads Per Kilobase of transcript, per Million mapped reads) using the DESeq2 R package (1.16.1). The resulting p values were adjusted using the Benjamini and Hochberg approach for controlling the false discovery rate. Genes with an adjusted p <0.05 found by DESeq2 were assigned as being differentially expressed. The following bioinformatics analysis was performed to select immune-related transcripts from the common carp immune gene library and construct the bar plots for the major immune processes and immune categories. The t-test was used to assess differences, with a false discovery rate adjusted p <0.05. Qualitative comparisons were performed between samples by counting the number of DEG. The data were rearranged in Microsoft EXCEL, and applied to plot charts by ggplot2 (2.2.1) using R language.
Correlation Analysis Between LncRNA and Genes Involved in Immune-Related GO Terms
The weighted gene co-expression network analysis (WGCNA) was performed using the R package “WGCNA” (33). Specifically, all genes with an expression variance ranked in the top 75 percentile of the data set were retained (34). The R package WGCNA was used to construct the weighted gene co-expression network (35). A matrix of signed Pearson correlations between all gene pairs was computed, and the transformed matrix was used as input for linkage hierarchical clustering. All transcripts in current transcriptome data with similar expression patterns were clustered together as one module. Subsequently, using the R package clusterProfiler (36), enriched GO terms for lncRNA-related protein-coding genes were generated for the DET list of every module. The p values of enriched GO terms were produced from the Kolmogorov–Smirnov test. To elucidate the detailed lncRNA-mRNA network, the immune-related GO term containing module was selected, and the relationship of involved top 100 transcripts and related lncRNAs were shown by the cystoscope software. In addition, the immune DETs (differentially expressed transcripts) involved in the immune-related module were classified into comparison groups SvC-B, SvC-N and SvS-BvN-D7, to reveal the LncRNA modulation of immunity at the aspect of surviving from CyHV-3 infection.
Quantitative Reverse Transcription Polymerase Chain Reaction (qPCR)
The mRNA samples used for transcriptome sequencing were also subjected to qPCR validation (n = 3), using SYBR Green PCR master mix (Bio-Rad) and CFX real-time PCR detection system (Bio-Rad), following a previously published protocol (37, 38). After RNA isolation, reverse transcription and qPCR experiments were carried out for 11 genes. Gene-specific primers (Table S4) were designed with the Primer Premier 5.0 software. Housekeeping gene 18s RNA was used to normalize data, and 2-DDCt formula was used to calculate the relative expression.
Results
Virus Identification, Mortality Rate and Pathological Appearance
The successful infection of CyHV-3 has been validated using PCR, to check the expression of virus genes (TK and Sph), in randomly sampled severely sick fish (N = 10) at day 7 after challenge (Figure 2A). Further, to reveal the mortality, the number of dead fish was recorded from days 1 to 15 after challenge (Table S1), the mortality rate was then calculated for all groups (Table S2). For the challenged non-breeding strain (Figure 1B), the mortality rate increased daily, except for a decrease observed on day 5. Mortality peaked at day 7 (69%) and decreased to a markedly low level thereafter. For the challenged breeding strain (Figure 2B), the mortality rate remained at a markable low level, with the highest value (11%) recorded at day 7. The mortality rates of the two challenged groups were very significantly different (p <0.01). The number of dead fish for each day is listed in Table 1 for the breeding or non-breeding strain, while in the two unchallenged controls, there were no dead fish (data not shown). Compared with the organs in survivors of the breeding strain, pathological swelling and hyperemia of the internal organs (particularly the kidney) were obvious in survivors from the non-breeding strain on day 7 (Figure 2C). In detail, the proportion of kidney area to the whole trunk area in the breeding strain was significantly different from that of the non-breeding strain (p <0.01) (Figure 2D). The reduced immune organ swelling was reflected by the ratio of 0.44 comparing the proportions between the breeding and non-breeding strains (Table S2).
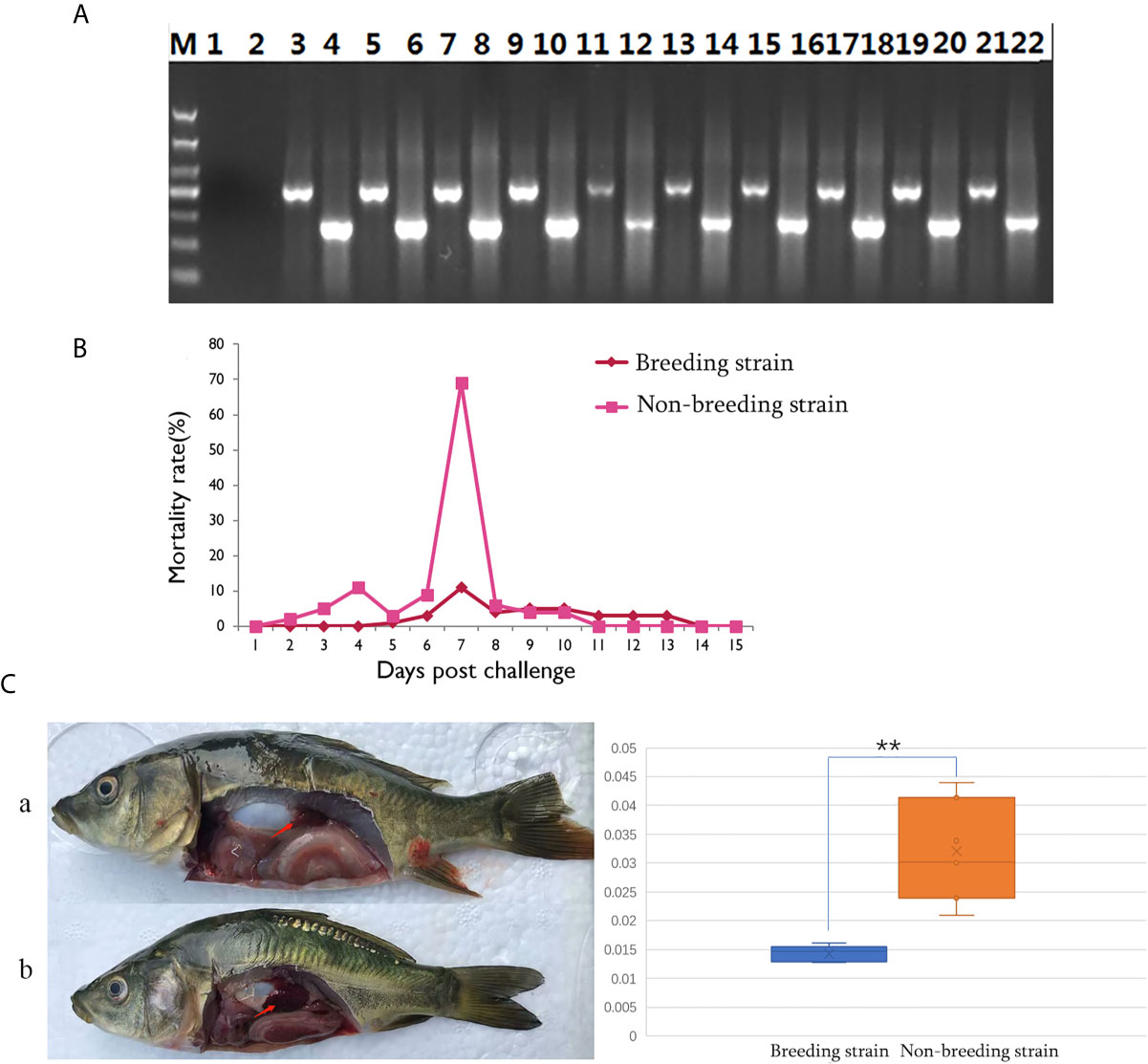
Figure 2 Differential appearance of general mortality and pathology between fish from the breeding and non-breeding strains. (A) The PCR validation of the CyHV-3 virus genes TK and Sph. Lane 1-2 represents the negative control, and afterwards, lane 3-22 represents the result for tested 10 virus infected fish. The lanes of odd and even numbers showed the PCR result of TK and Sph genes, respectively. M is the abbreviation of “marker”. (B) Comparison of daily mortality between the breeding and non-breeding strains. The mortality was monitored within 15 days post challenge. N=300 per group. (C) The degree of swelling trunk kidney was limited in the survivors from the breeding strain (a) compared with the markedly enlarged trunk kidney observed in fish from the non-breeding strain (b). The arrow indicates the trunk kidney region. “**” means the very significant difference (p < 0.01) between current compared two groups.

Table 1 The number of dead fish for each day during the CyHV-3 challenging for breeding or non-breeding stain.
Quality and Validation of Transcriptomic Data
Sequencing was performed with both the PacBio RS II and Illumina platforms to analyze the gene information of the common carp. Through PacBio sequencing, 19.49 G data were obtained. Of the 258,346 ccs, 79.79% were full-length sequences, and 17,769 polished high-quality isoforms were also revealed (Table S3, sheet 1). Meanwhile, the Illumina data were also characterized by high quality, and the information for each sample is provided in Table S3 (sheet 2). To validate the transcriptomic data, qPCR was conducted. A total of 24 reactions were performed to validate the transcripts of 11 genes, most of which were immune genes. The correlation analysis showed that fold changes between transcriptome and qPCR results correlated well (R2 = 0.929718564). The gene ID, annotation, primers, and fold-change information are listed in Table S4.
GO Analysis of Immune-Related DEGs Among Tested Fish
According to the enriched general GO terms (Figure 3) of immune-related DEGs, there are “immune system process” and “antioxidant activity” for all comparison groups, “chemoattractant activity” only in comparison group SvC-B, “rhythmic process” only in comparison group SvC-N, “cell killing” in both comparison groups SvC-B and SvS-BvN-D7, and “virion” in both comparison groups SvC-N and SvS-BvN-D7. The detailed significant enriched GO terms involved in biological processes (BP), cellular components (CC), and molecular functions (MF) were detailed in Tables S5–S9 for all comparison groups. The immune-related significant regulated BP terms (p <0.01) were in the largest number (Table 2), for example, “positive regulation of cell migration” in group SvC-B; “activation of MAPK activity”, “regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process”, “oxication-reduction process” and “positive regulation of cell migration” in group SvC-N; “activation of MAPK activity” and “dopaminergic neuron differentiation” in group SvS-BvN-D7. Besides, the CC terms (p <0.01) includes “Pre-autophagosomal structure membrane” in group SvC-B, SvC-N, CvC-BvN-D0 and SvI-B&N, and the MF terms (p <0.01) includes “NF-KappaB binding” and “lysozyme activity” in group SvC-N; “cytokine activity” in group SvS-BvN-D7; “Lysozyme activity”, “MAP kinase activity” and “ubiquitin-like protein binding” in group CvC-BvN-D0.
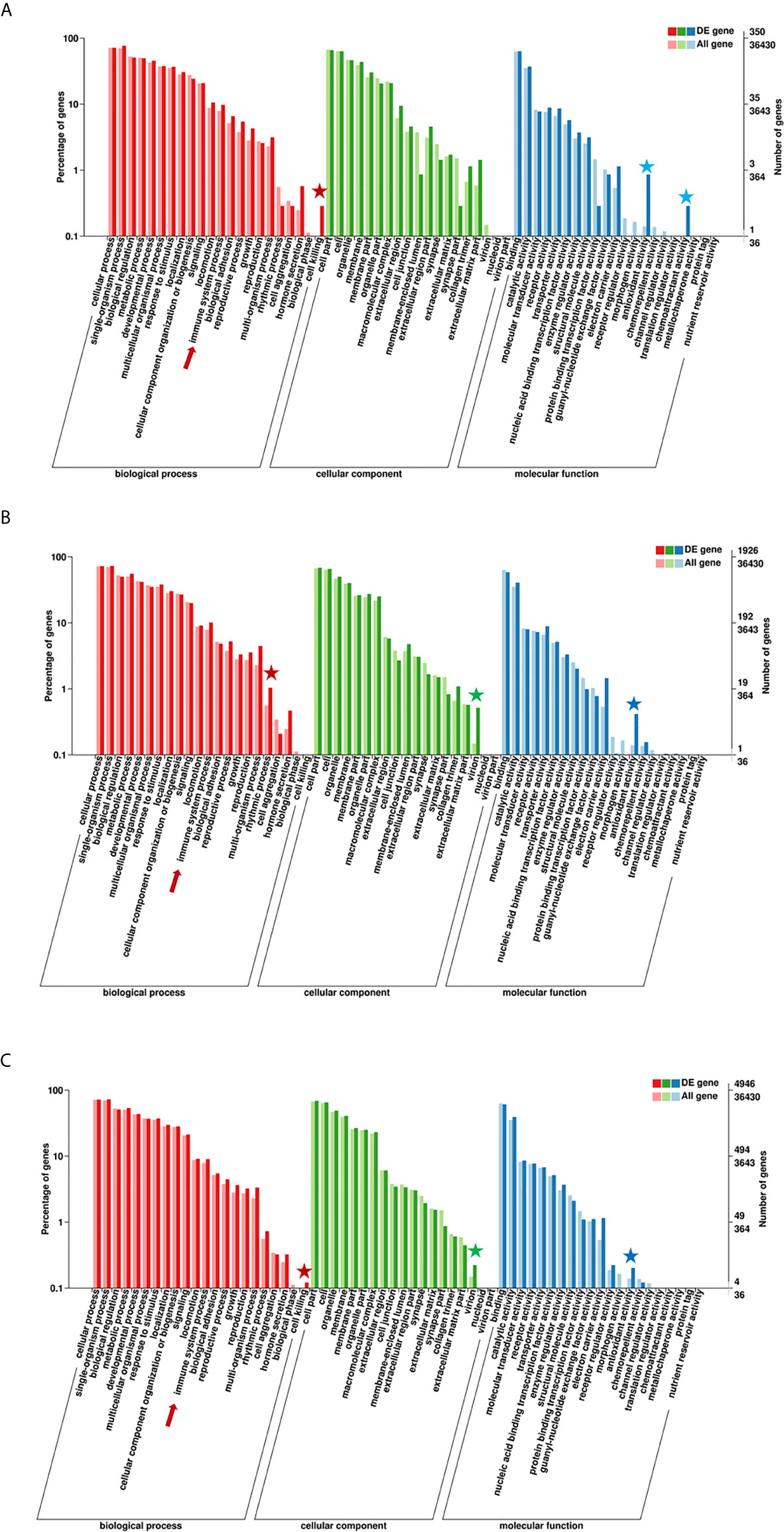
Figure 3 GO (Gene Ontology) terms of comparison groups SvC-B, SvC-N, and SvS-BvN-D7. The different colors of the bar indicate the comparison of the GO terms between all genes and immune-related genes. In detail, red and pink bars represent all genes and immune-related genes respectively for the biological process (BP, in part A), green and light green bars represent those for the cellular component (CC, in part B), blue and light blue bars represent those for the molecular function (MF, in part C). The immune-related terms are labeled with stars.
KEGG Analysis of DEGs Among Tested Fish
Compared with group SvC-B, there were markedly more genes involved in immune and related pathways in group SvC-N. Current revealed immune pathways in group SvC-B were “phagosome”, “regulation of autophagy”, “ubiquitin mediated proteolysis”, and “plant-pathogen interaction” (Figure 4Aa). Meanwhile, there are four immune pathways, including “endocytosis”, “phagosome”, “FoxO signaling pathway”, “ubiquitin mediated proteolysis”, “DNA replication”, “fructose and mannose metabolism”, “oxidative phosphorylation”, and “plant–pathogen interaction” in group SvC-N (Figure 4Ab). To get a better understanding of the different survival mechanism, the pathways revealed in group SvS-BvN-D7 includes “endocytosis”, “phagosome”, “ubiquitin mediated proteolysis”, “DNA replication”, “fructose and mannose metabolism”, and “oxidative phosphorylation” (Figure 4Ac, left).
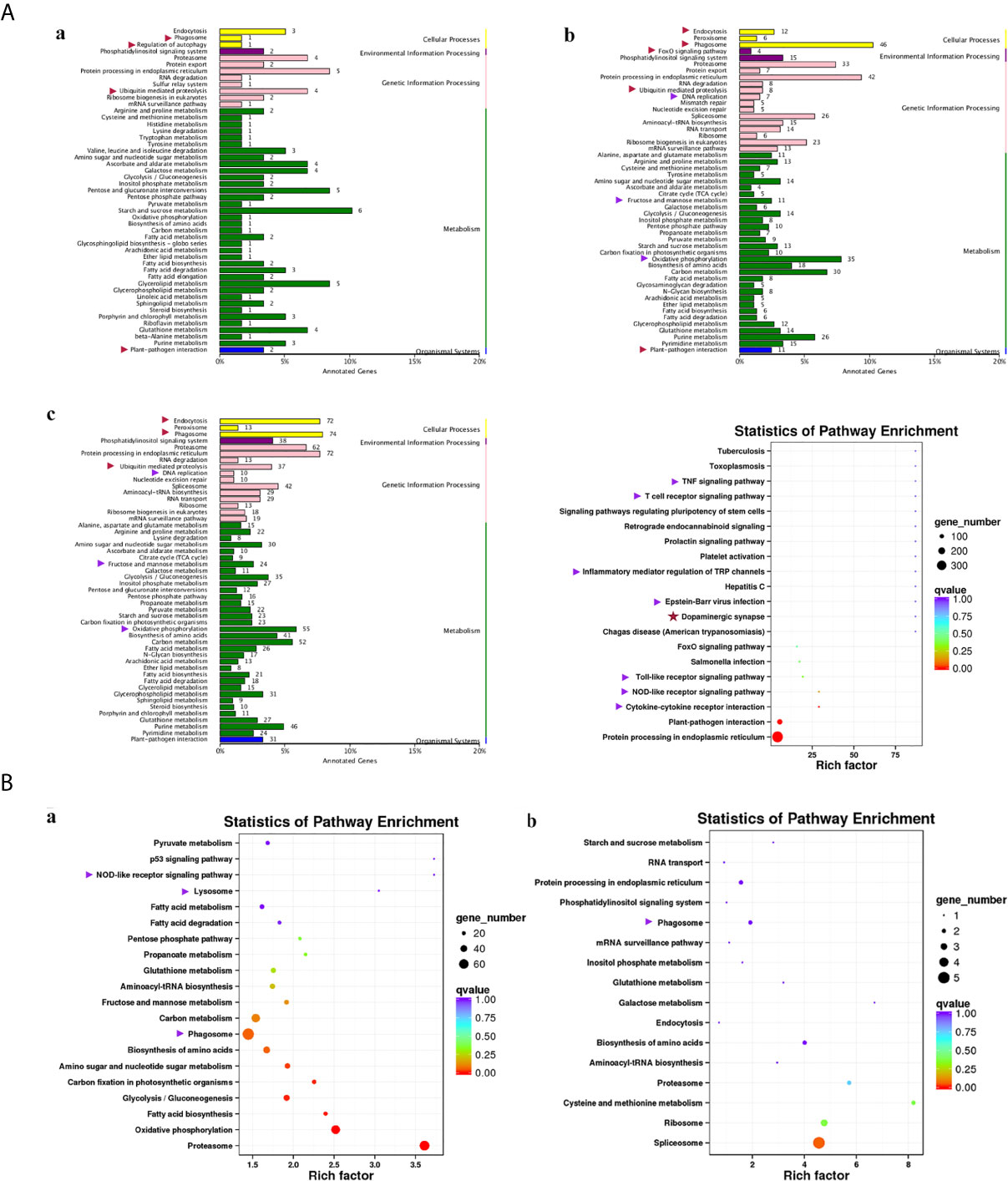
Figure 4 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways revealed for all comparison groups. In part (A), the enriched pathways for comparison group SvC-B (a), SvC-N (b) and SvS-BvN-D7 (c) were shown mainly in bar plots. In addition, for the comparison group SvS-BvN-D7, the babble plot of pathways for immune-related DEG was also given in the right to illustrate the rich factor. In part (B), the pathways of immune-related DEG for both comparison group CvC-BvN-D0 (a), which compared controls of both stains, and group SvI-B&N (b), which has compared similar genes of survivors from both strains with protein-coding transcripts of severely sick fish, were demonstrated in babble plots. The immune-related terms are labeled with arrows. Red and purple arrows indicate the immune and related pathways representatively.
To better illustrate the immune and related pathways more clearly, filtering of DEG was conducted using the gene list of the common carp immune gene library (Table S1), and then a bubble chart was used to clarify both the enrichment factors and gene numbers for revealed immune pathways. For group SvS-BvN-D7, which reflected differential surviving mechanism [Figure 4A (c, right)], the revealed immune pathways include “TNF signaling pathway”, “T cell receptor signaling pathway”, “inflammatory mediator regulation of TRP channels”, “Epstein–Barr virus infection”, “Toll-like receptor signaling pathway”, “NOD-like receptor signaling pathway” and “cytokine-cytokine receptor interaction”. In addition, “dopaminergic synapse” was also found. While, groups CvC-BvN-D0 and SvI-B&N, revealed the difference and similarity for maintaining basic homeostasis, respectively. The most enriched immune pathways were the “p53 signaling pathway”, “NOD-like receptor signaling pathway”, “lysosome”, “oxidative phosphorylation”, and “proteasome” in group CvC-BvN-D0 (Figure 4Ba), yet only “phagosome” in group SvI-B&N (Figure 4Bb).
Classification of DEG From Different Comparison Groups Into Immune Process and Immune Gene Category
The current construction of the common carp immune gene library (Table S4) was used to classify the revealed immune transcripts and refine the involved immune processes and immune gene categories. The details of DEG involved in all comparison groups were provided in Tables S5–S9. At the level of immune processing, in group SvC-B (Figure 5A, Tables 3A and S5), most immune mRNAs were upregulated in the immune processes of “pattern recognition”, “inflammatory cytokines and receptors”, and “T/B cell antigen activation”, while downregulated in the immune process of “inflammatory cytokines and receptors”, “complement system”, “adapters, effectors and signal transducers”, and “pattern recognition”. Interestingly, there were no genes in “antigen processing and regulators” for group SvC-B. In addition, there were only downregulated genes in “acute phase reactions”, as well as only upregulated genes in “innate immune cells related” in group SvC-B. In group SvC-N (Figure 5A, Tables 3A and S6), immune genes were upregulated in the immune processes, such as “other genes related to immune response”, “inflammatory cytokines and receptors”, and “adapters, effectors and signal transducers”. Meanwhile, the downregulated genes were involved in the immune processes, such as “pattern recognition”, “inflammatory cytokines and receptors” and “other genes related immune response”. The largest number of immune mRNAs was observed in group SvS-BvN-D7 (Figure 5A, Tables 3A, and S7). The upregulated immune DEGs (BS-advantage) were mainly involved in “inflammatory cytokines and receptors”, “other genes related to immune response”, “T/B cell antigen activation”, “pattern recognition”, and “adapters, effectors and signal transducers”, whereas the downregulated immune DEGs (NBS-advantage) were mainly involved in “other genes related to immune response”, “adapters, effectors and signal transducers”, “inflammatory cytokines and receptors”, “T/B cell antigen activation”, “pattern recognition”, and “antigen processing and regulators”. Meanwhile, in group CvC-BvN-D0 (Figure 5A, Tables 3B and S8), the upregulated immune DEGs (BS-advantage), were found mostly in “adapters, effectors and signal transducers”, and noted in “antigen processing and regulators” and “T/B cell antigen activation”, while downregulated immune DEGs (NBS-advantage), were mostly involved in “inflammatory cytokines and receptors”, and also noted in “pattern recognition” and “complement system”. In group SvI-B&N (Figure 5B, Tables 3B and S9), among the revealed six processes, “T/B cell antigen activation” and “other genes related to immune response” exhibited the greatest DEG number. To illustrate the immune process, the involved immune gene categories of the immune-related DEG in all comparisons were listed in Table 3.
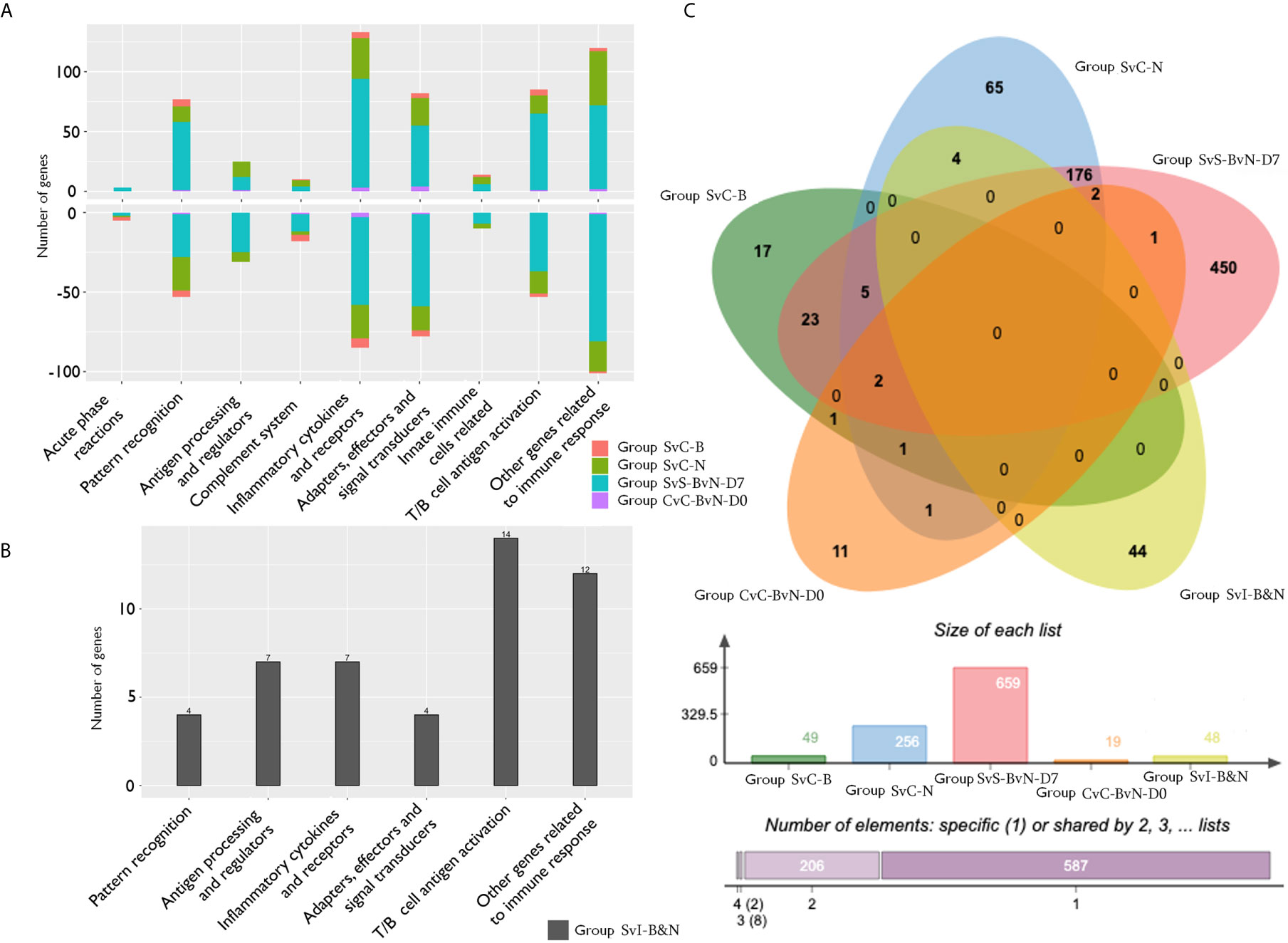
Figure 5 The analysis of immune processes of DEG among different comparison groups. (A) The bar plots of gene number for the involved immune processes of up or down-regulated DEGs in groups SvC-B, SvC-N, SvS-BvN-D7, and CvC-BvN-D0; (B) The barplot of gene number for the involved immune processes in group SvI-B&N; (C) The Venn diagram of gene number for all comparison groups. DEG, differentially expressed gene.
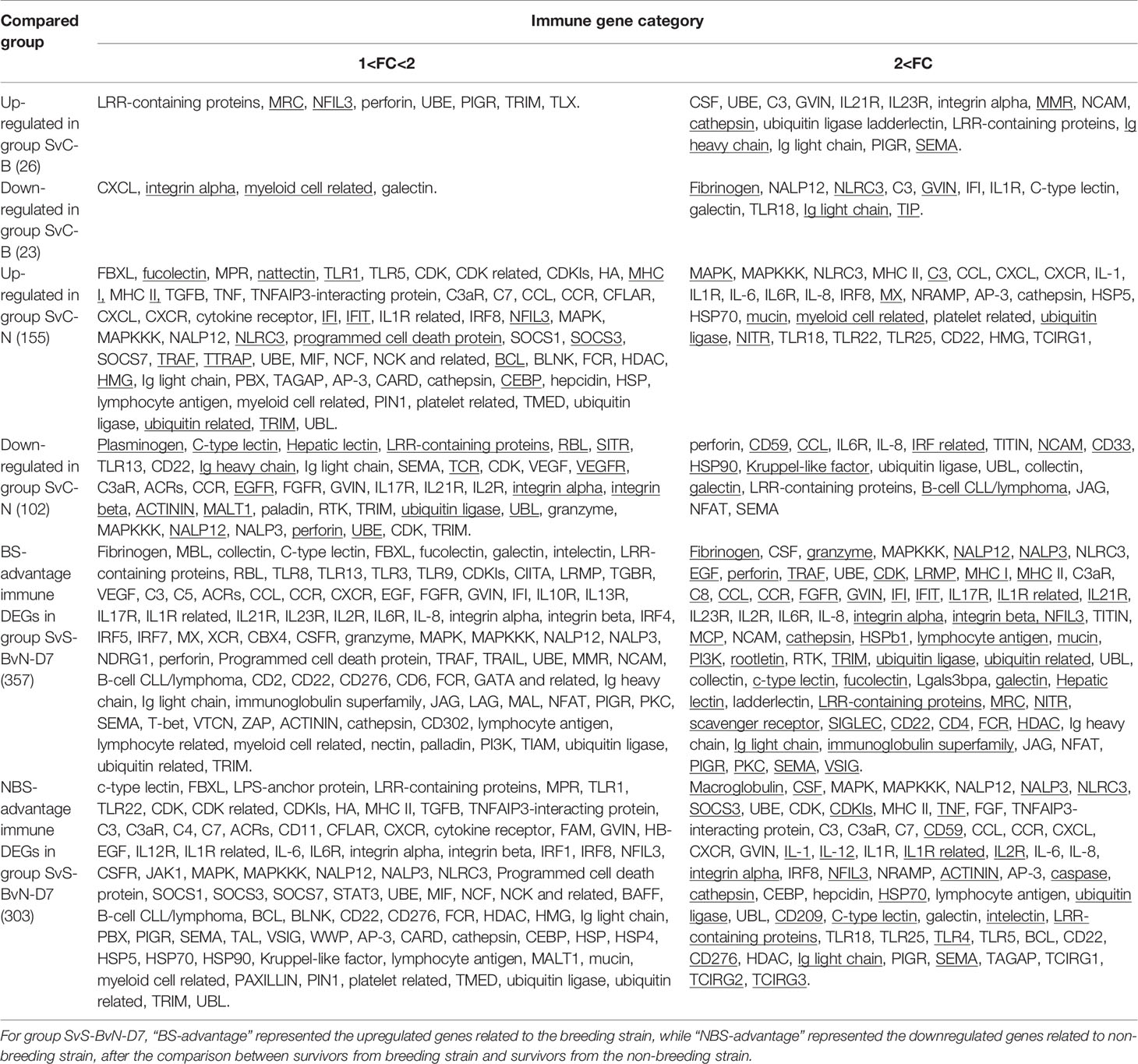
Table 3A Immune gene categories involved in differentially expressed mRNA for comparison groups SvC-B, SvC-N, and SvS-BvN-D7.
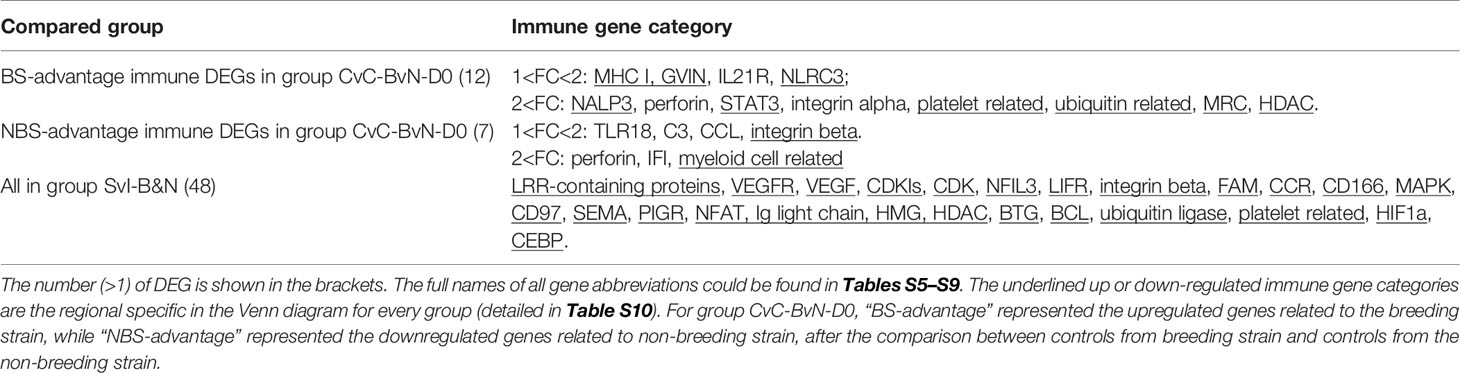
Table 3B Immune gene categories involved in differentially expressed mRNA each comparison group CvC-BvN-D0 and SvI-B&N.
Venn-Regional Analysis of Immune-Related DEG From Different Comparison Groups
A Venn diagram was created to demonstrate the relationship between the immune-related DEG among all comparison groups (Figure 5C). The details for all regions were in Table S10. Though the region-specific DEGs accounted for a large proportion of each comparison group, with immune gene categories for region-specific DEG underlined in Table 3, the overlapping DEGs (Table 4), which contained important genes, were listed with the corresponding immune process and immune gene category.
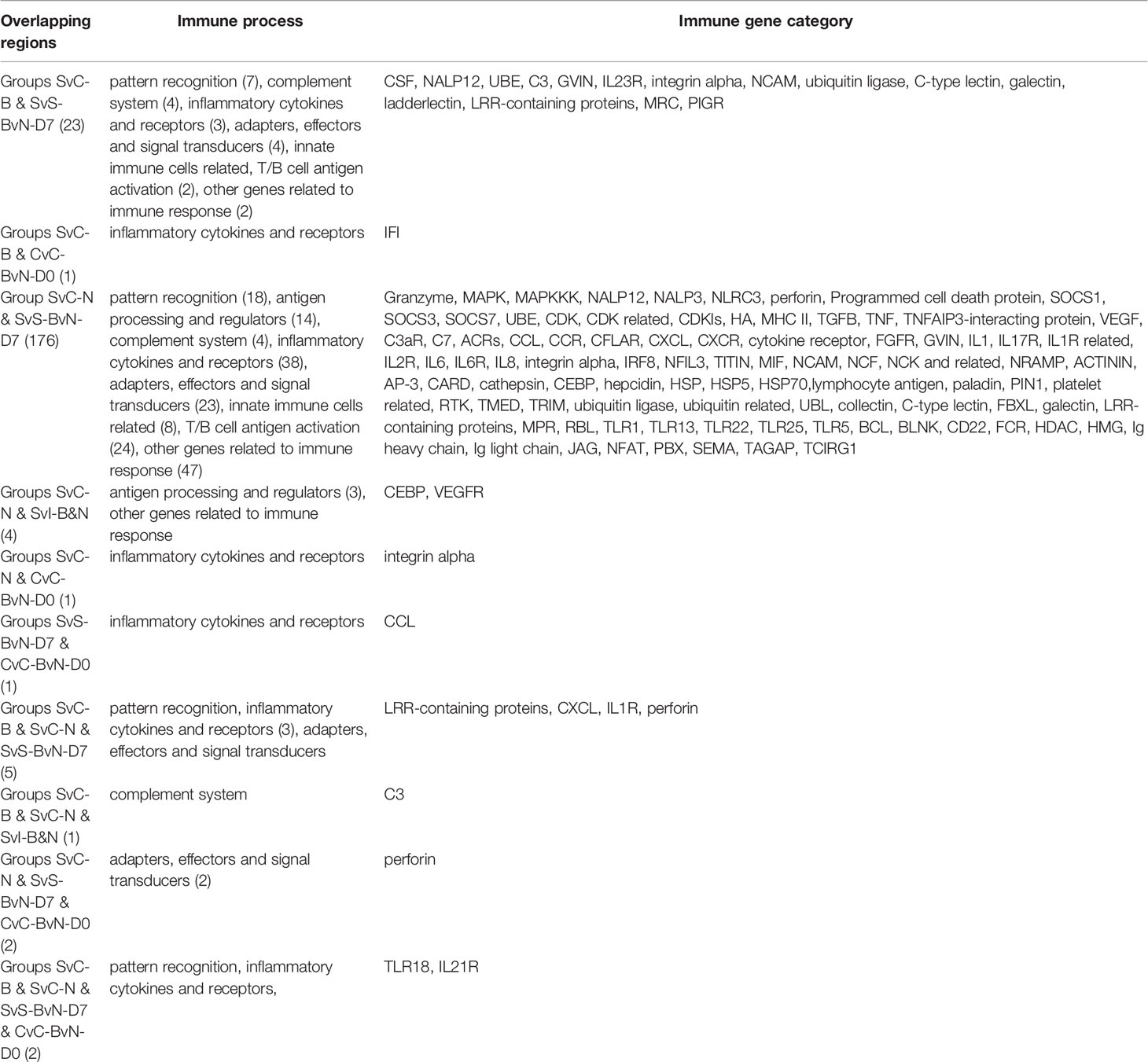
Table 4 The immune process and immune gene category for overlapping genes in the Venn regional analysis.
Immune-Related GO Terms as Well as Network Correlation Between LncRNAs and Genes in Immune-Related Module
The results of “WGCNA” were selected to determine the biological processes involved in CyHV-3-induced modulation of genes by lncRNAs, from an immune perspective. Among the currently divided 35 gene modules (Figure 6A), 12 modules were found with immune-related GO terms (Figure S1). Generally, “double-stranded DNA binding”, “receptor-mediated endocytosis”, “response to bacterium”, “lysosome”, “defense response to fungus”, “killing of cells of other organism”, “oxygen carrier activity”, “oxygen binding”, “defense response to bacterium”, “killing of cells of other organism”, “NuA4 histone acetyltransferase complex”, “defense response to gram-negative bacterium”, “defense response to gram-positive bacterium”, “immune response”, “regulation of cytokine secretion”, “cytolysis”, “execution phase of apoptosis”, “cellular response to gamma radiation”, “nucleic acid metabolic process”, “TOR signaling”, “DNA replication”, “ubiquitin-dependent protein catabolic process”, “granulocyte differentiation”, and “macrophage differentiation” were revealed after the “WGCNA” analysis. The top five immune-related modules containing most immune-related GO terms (Figure 6B) were analyzed for the relationship between LncRNA and transcripts by PPI networks in cystoscope (Figure 6C, with details in Table S11). The LncRNA regulated genes, which upregulated in group SvS-BvN-D7, such as Semaphorin-4E in the blue module, can be the factor associated with the resistance in the breeding strain. The LncRNA regulated genes, which downregulated in group SvS-BvN-D7, such as Lgals3l in the grey60 module, can be the factor associated with less accessibility to CyHV-3 in the breeding strain. The LncRNA regulated genes, which was upregulated in group SvC-N and downregulated in group SvS-BvN-D7, such as natural resistance-associated macrophage protein (Nramp), plasminogen activator inhibitor (PAI) in the brown module, could be the key clues for susceptibility of CyHV-3 in the non-breeding strain. Specifically, for detailed correlations of lncRNAs and DEG in groups SvC-B, SvC-N, and SvS-BvN-D7 (Table S12), T cell leukemia homeobox 3 (TLX3) and LGALS3 were revealed, respectively, in the survivors of the breeding strain.
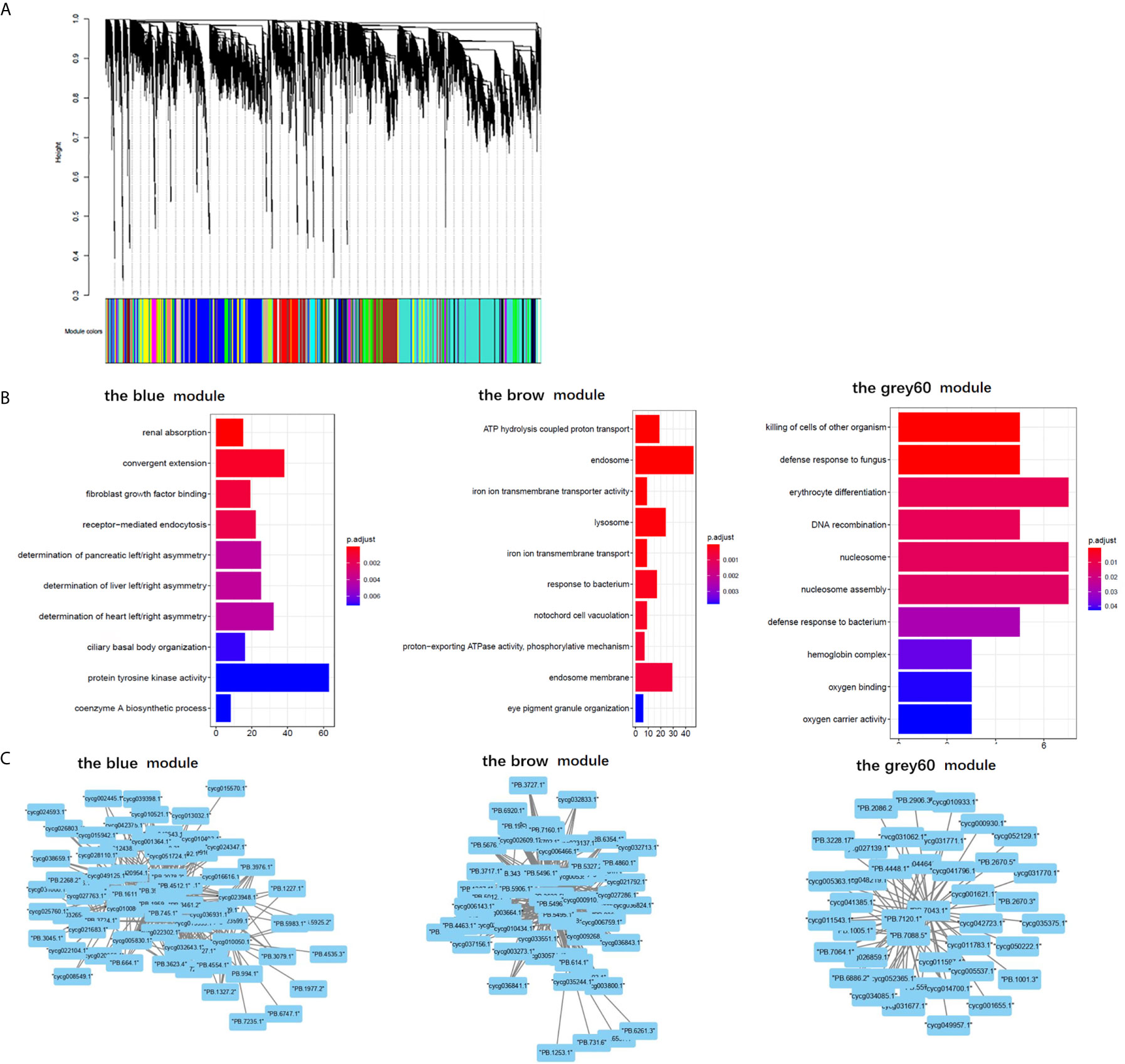
Figure 6 Gene ontology analysis and cystoscopes of key matched lncRNA and transcripts in the immune-related module. (A) the cluster dendrogram of all gene module. (B) The GO terms involved in the immune-related module, including the blue, brow, and the grey60 module, which contain the matched lncRNA and transcripts involved in current analyzed comparison groups. The star, triangle, and arrow represent the revealed GO term for comparison groups SvC-B, SvC-N, and SvS-BvN-D7, respectively. (C) Cystoscopes of key matched lncRNA and transcripts in the immune-related module, as shown for blue, brow and grey60 module, respectively. The PPI network here just showed the pattern of closely related genes and LncRNAs, and to access the detailed information of genes and LncRNAs, please refer to Table S11.
Discussion
This study demonstrated that the anti-CyHV-3 immune mechanisms of a breeding strain of common carp. This demonstrated dramatically decreased levels of mortality and less inner tissues swelling together with the previously revealed reduced virus load of tissues (24, 25), proving resistance to CyHV-3. The current studies of biological processes and pathways discovered through transcriptomic analysis have been merged to improve clarity. Afterwards, by comparing the survivors and healthy fish in either breeding or non-breeding strains, key genes and related lncRNAs involved in immune processes were also revealed. Accordingly, the immunogenetically insensitive or susceptible factors to CyHV-3 infection were determined.
From the aspect of cell surface receptors, which could participate in virus-host interaction, current comparison results suggested potential components involved in virus entry and downstream proinflammatory signaling. Firstly, integrin acts as a herpesvirus receptor (39), and the integrin-dependent signalosome in herpesvirus-infected cells mediated or coactivated numerous inflammatory responses and signaling transductions (39). Therefore, the finding that there was more expression of integrin beta 1 (ITGB1) in the survivors from non-breeding strain compared with those from the breeding strain, even during the steady status, may indicate that ITGB1 facilitates the binding of herpes virus glycoprotein for entry (40). This coincides with the finding that integrin signaling promotes the release of intracellular calcium stores and contributes to viral entry and cell-to-cell spreading via glycoprotein H, during herpes simplex virus infection in humans (41). Recent reports have also demonstrated that some integrins on lymphocytes, such as B cells, could facilitate mucosa-specific homing (42). Secondly, higher TLR expression in survivors of the non-breeding strain compared with that of the breeding strain also indicated its role in facilitating the virus infection. Of note, TLR4 signaling leads to the production of proinflammatory cytokines in human lymphatic endothelial cells (39). In addition, fish-specific and virus-responding TLR18 (43, 44) (the most overlapping gene among groups) was downregulated in the comparison group SvC-B, whereas it was upregulated in group SvC-N. Also, TLR18 was higher in steady-state in the nonbreeding strain. This suggested its possibility to enhance the severity of CyHV-3 infection. MAPK signaling, as the downstream process of pattern recognition receptors, could facilitate the tnf-alpha-induced suppressor of cytokine signaling 3 (SOCS3) expression. This can lead to both pro-inflammatory immune response and failure in growth (45), according to the upregulated DEGs upon infection observed in the group SvC-N.
To reveal the breeding driven improvement for the resistance from CyHV-3 infection, by comparing the survivors from different strains, DEGs of the inflammatory status also provide clues for how to block the virus. Non-specific binding of the virus by lectins played a protective role in preventing virus entry. Both upregulated ladderlectins and higher galectin expression were detected in survivors of breeding strains compared with non-breeding strains. This is suggestive of their blocking ability for viral proteins (46, 47), such as glycoprotein (48). Moreover, as the head kidney is one of the major reticuloendothelial systems in fish (49), mucin was also found to be upregulated as the anti-virus barrier. This was shown in the comparison group SvC-N as the gel-forming mucin 5B (MUC5B) (50) was upregulated in the survivors of the non-breeding strain. However, higher expression levels of membrane-bound MUC3 (51) were found in survivors of the breeding strain versus the non-breeding strain. Additionally, as shown in the comparison group SvS-BvN-D7, the complement system functioned differently. BS-advantage complement components C8 indicate the formation of terminal complement complex (52), while NBS-advantage CD59, which is the inhibitor of complement membrane attack complex (MAC) (52), suggesting the inhibition of f complement-dependent cytotoxicity (CDC). Thus, the effective CDC may be more helpful for surviving. These findings suggested that more mucus was secreted, thereby causing tissue swelling upon CyHV-3 infection in the non-breeding strain. For the survivors of the breeding strain, the membrane-bound mucin could effectively bind the virus with no gel.
For the anti-viral biological process for the breeding strain, the lectin complement pathway has been involved in the clearance of apoptotic cells, reflected by the GO term “cell killing” in both comparison groups SvC-B and SvS-BvN-D7, as well as more types of lectins in survivors from breeding strains. Hence, in survivors of the breeding strain, the complement components reported in the acute phase in common carp during CyHV-3 infection (53), could facilitate the phagocytic process via binding of MRC (Mannose-Receptor C) in fish (54). The interleukin 21 receptor (IL21R), which was found regulated in the survivors of the breeding strain for group SvC-B and the healthy control of the non-breeding strain for group SvC-N, indicated the control of inflammation by suppressing STAT3 signaling (55). The upregulated IL23R in the survivors of the breeding strain for group SvC-B can promote cytotoxic ability (56), which may immediately kill infected cells. The lack of antigen presentation in comparison group SvC-B suggested that the infection was overcome before the amplification of CyHV-3 in the breeding strain. As to the BS-advantage interferon related genes in group SvS-BvN-D7, the activator of IRF7 (57) as well as MX, the inhibitor of virus replication, may elicit the interferon antiviral response. Meanwhile, for proinflammatory cytokine, echoed with upregulated IL-1, IL-6, and IL-8 in group SvC-N, the NBS-advantage proinflammatory signaling dominated in the survivors from non-breeding strain, and also numerous upregulated genes were involved in antigen presentation in the non-breeding strain in group SvC-N. Therefore, the survivors from breeding strains overcame the infection mainly through phagocytosis and cytotoxicity at the cellular level and may elicit IFN response, without activating the typical process of proinflammation as in survivors from the non-breeding strain.
Thus, there are different signatures between the two strains, regarding the survival strategy. For the survivors in the breeding strain, self-repairing related autophagy was detected as both the KEGG pathways “regulation of autophagy” and autophagy-related fish antiviral tripartite motif (TRIM) protein (58) were found in group SvC-B. These findings were in line with TRIMs, which were found regulated in the survivors of breeding strain for group SvC-B and in the healthy control of non-breeding strain for group SvC-N. The above finding of TRIM is coincident with one of the recently revealed CyHV-3 resistant related DEGs with identified QTLs (17). In the survivors from the breeding strain, the higher PI3K also suggests higher autophagy, since the PI3K/AKT/mTOR pathway enhances this process (59). In the breeding strain, the suppressed IFN activation was also suggested, for that fish TRIM may inhibit the activation of IFN and attenuate IFN regulatory factor (IRF) (60, 61). The factor that there was more expression of TRAF6 in survivors of breeding strain compared with those of non-breeding strain, is in line with the resistant related SNP on TRAF6 (62) and suggests the possible repression on the production of type I IFN (63). In addition, nuclear factor, interleukin 3 regulated (NFIL3) can control Treg cell function via directly binding to and negatively regulates the expression of Foxp3 (64), and has been revealed stimulating both proinflammatory (e.g., NF-kappa B [NF-κB]) and anti-inflammatory factors (e.g., IL10) in carp (65). Thus, the upregulated NFIL3 in the survivors of the breeding strain for group SvC-B may suggest the extensive activation of immune cells, with diminished immune regulation. The directed lymphocytes response could be present as there was upregulation of IL23R, which significantly enhances the expression of cytotoxic mediators (56), as well as cathepsin L (a component of lysosomes) (66). This indicated an enhanced activation of the cytotoxic ability of T cells in survivors from the breeding strain. For B cells, the secretion of Ig-related genes (e.g., polymeric immunoglobulin receptor [PIGR], Ig heavy chain and light chain) was also upregulated in the survivors of breeding strain for group SvC-B. To fight the virus, the survivors from the non-breeding strain developed typical inflammatory cascades, including pro-inflammatory cytokines (e.g., IL-1, IL-6, and IL-8), as well as MAPK signaling, which is profoundly involved in cell survival functions during viral infection (67). For the control of inflammation, the survivors from the common strain exhibited suppression of the IFN response, which was also reflected by the upregulated IRF1 and IRF8, the inhibitor of the MYD88-mediated NF-κB signaling pathway (68, 69). Downstream hypoxia was also found, as indicated by the “p53 signaling pathway” and “oxidative phosphorylation”. The hypoxic status could also protect survivors of the non-breeding strain from death since p53 suppresses cell apoptosis (70), and HIF1A regulates virus-induced glycolytic genes (71).
Furthermore, the DEGs generated by comparing the survivors from the two strains in comparison group SvS-BvN-D7, which can provide clues for how to develop the resistance to CyHV-3 infection. The BS-advantage semaphorin, which was also regulated by LncRNA, is related to immunoregulation (72, 73). This indicated a possible tolerance of viral replication or latency after primary infection in the survivors (2), while had no negative effects on the proliferation of host cells (74). There was less cyclin-dependent kinase inhibitor 1D (CDKN1D) in survivors from the breeding strain compared with the non-breeding strain. This indicated that there was an interrupted circadian cell-cycle timing in the survivors from non-breeding strains (75). The higher levels of STAT3 and possible its upregulated integrins (76) in survivors from the non-breeding strain may facilitate the effect of enhanced IL6 signaling. Apart from the typical immune signals in comparison group SvS-BvN-D7, there was relatively higher PI3K activity, which was involved in the KEGG pathway “dopaminergic synapse”. This indicated that dopamine inhibited inflammation (77) in survivors from the breeding strain. This is because PI3K is dependent on the accumulation of DOPA decarboxylase, the enzyme involved in the production of dopamine, which is reduced by a viral infection (78). Additionally, genes responsive to the secretion of immunoglobulin (e.g., PIGR and Ig light chain), which is important for anti-virus immunity, were common (in comparison group SvI-B&N) between the two strains.
Additionally, the comparison of two strains with health status in group CvC-BvN-D0 suggested the differential basal properties for immune homeostasis. Among BS-advantage immune DEGs, which was related to the healthy control of breeding strain, the expression of NACHT, LRR, and PYD domains-containing protein 3 (NALP3) indicated a stronger ability to form the inflammasome in the breeding strain. The higher levels of NLR family CARD domain containing 3 (NLRC3), which is a negative regulator of the DNA sensor STING, suggest less sensitivity to the DNA virus in the breeding strain. The higher levels of MHC I indicate greater potential to activate response by cytotoxic CD8+ T cells upon herpes virus infection, as previously revealed in the resistant strain R3 (79). For the potential T helper cell differentiation-related JAK/STAT pathway, the upregulation of both STAT3 and histone deacetylase (HDAC) indicated an easier differentiation of both T helper 17 and regulatory T directions (80), respectively, in the breeding strain even in the steady state (81, 82). Upregulation of HDAC in the healthy control of breeding strain for group CvC-BvN-D0, which could inhibit the function of macrophages in fish (83), also suggests a more delicate immune regulation during the steady state in the breeding strain. The macrophage related MRC (Mannose-Receptor C) in fish could potentially induce the phagocytic process if necessary (54). However, among NBS-advantage immune DEGs, which indicated higher expression in the non-breeding stain at a steady state, myeloid-associated differentiation marker-like protein 2 (myadml2) suggested the basic more differentiation of monocyte cell types (84) As the surface receptor, TLR18 and ITGB1 suggested susceptibility to virus binding. Moreover, C-C motif chemokine ligand 4 (CCL4), which can protect infected cells with the viral burden (85), may be a risk factor for the non-breeding strain. In addition, the downregulated DEGs in group SvC-B and SvC-N also provided clues for how to maintain immune homeostasis, which can prevent the virus challenge. Among the downregulated DEGs in group SvC-B, both the existence of C3 and IFI suggested a restricting effector on the virus, since that C3b can bind directly to purified glycoprotein C of herpesvirus (86), and interferon-induced protein 44 (IFI44) may restrict virus replication (87). These factors were also revealed in NBS-advantage immune DEGs in group CvC-BvN-D0. Therefore, upon steady state, C3b and IFI44 also served as the basic defense function even in the virus carrier state, which could be especially important for the non-breeding strain.
The revealed regulation of resistance to CyHV-3 by lncRNA involved numerous biological processes. Among the revealed GO terms in comparison group SvC-N, “DNA replication” could indicate virus proliferation in the non-breeding strain. In survivors from the non-breeding strain, the lncRNA-regulated genes were mainly involved in innate immune cell function (e.g., macrophage protein and cathepsin, as components of lysosomes) and cell apoptosis. Nramp and PAI were found regulated by LncRNA in comparison groups SvC-N (up) and SvS-BvN-D7 (down). This finding indicates the more expression of Nramp and PAI in the survivors of non-breeding strain, could facilitate virus infection and proliferation for infected cell respectively, since that Nramp may serve as a virus receptor (88), meanwhile, PAI can inhibit apoptosis in cell lines infected with viruses (89). While, in survivors from the breeding strain, the only lncRNA-modulated gene, TLX3, is strongly methylated. This indicates that TLX3 expression was suppressed during hepatitis B virus-related cancer (90). This suggests that TLX3 in survivors from the breeding strain may play a protective role, and participate in lymphocyte proliferation in the head kidney. Meanwhile, echoing with the recent finding of the participation of zebrafish galectin proteins in immunity against viral infection (48, 91), in comparison group SvS-BvN-D7 the LncRNA regulated transcripts Lgals3l (down) and Lgals3bpa (up), as well as BS-advantage Lgals3bpa and galectin 3 suggested the regulation of galectin-3 related biological activities could be related to reduce the viral attachment for survivors of the breeding strain.
Therefore, based on the present findings, a hypothesis has been proposed for the immune mechanisms involved in both healthy controls and survivors from infection in both strains (Figure 7). In conclusion, the breeding strain of common carp showed a better ability to maintain immune homeostasis in both steady and inflammatory states and displayed enhanced blockage of CyHV-3 infection compared with the non-breeding strain. Thus, this strain could be termed as a resistant strain accordingly. Since the modulation of mRNA and lncRNA expression dynamics currently remains a hypothesis, further molecular evidence is needed to elucidate the mechanism of both resistance and susceptibility. In addition, the finding that both the inhibition of inflammation by dopamine in the breeding strain and the disrupted bio-clock in the non-breeding strain upon CyHV-3 infection suggested better growth performance for the breeding strain. Therefore, the possible advantage of this resistant strain for growth performance warrants further study.
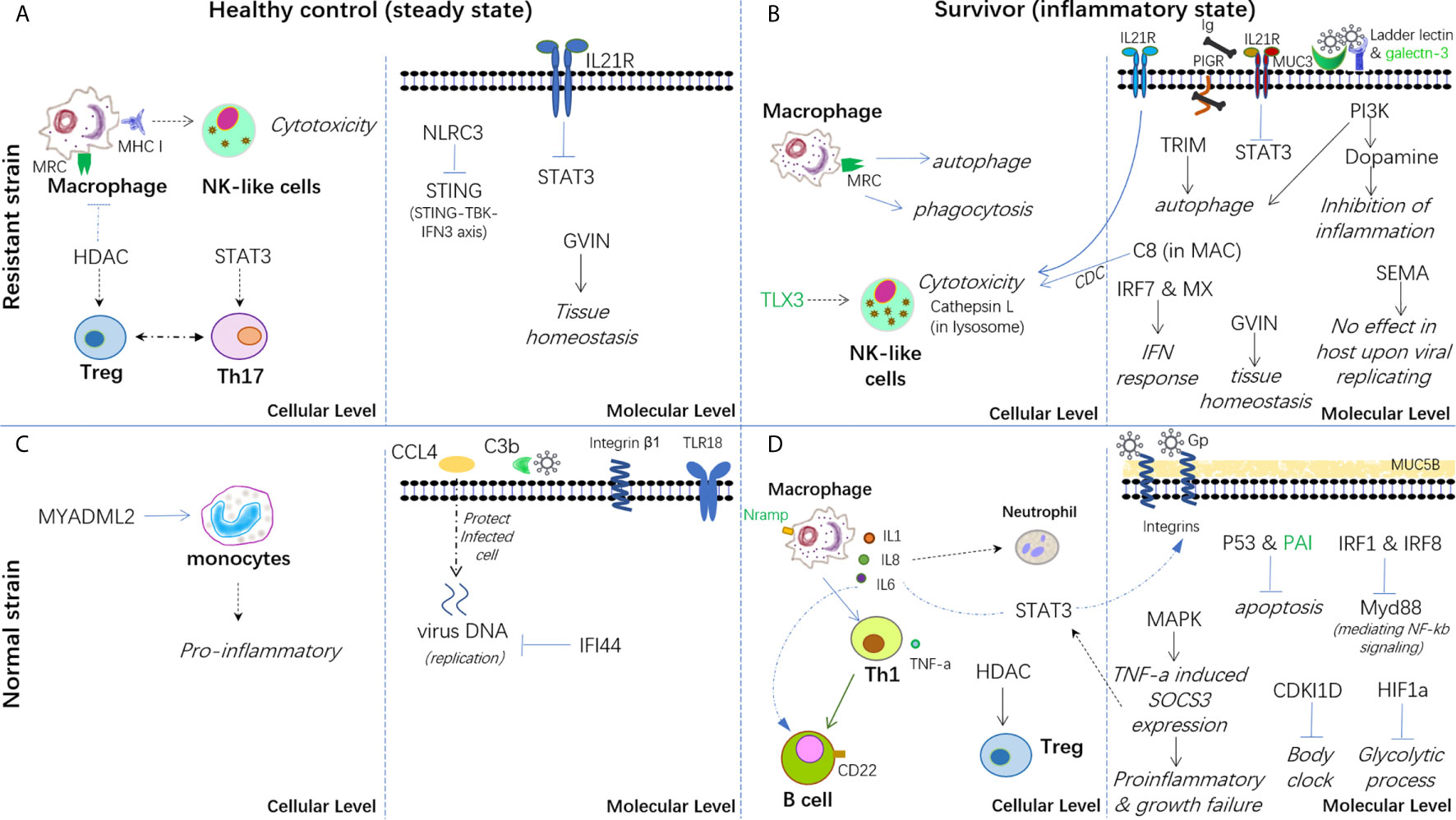
Figure 7 Hypothesis of the carp immune mechanism in both breeding (resistant) and non-breeding (normal) strains, either at the steady-state or upon surviving for CyHV-3 infection. (A) Healthy control in the steady state in the breeding strain. (B) Survivors in an inflammatory state in the breeding strain. (C) Healthy control in the steady state in non-breeding strain. (D) Survivors in an inflammatory state in the non-breeding strain. The words in italics, bold, and green represent the biological process, cell type, and lncRNA-modulated immune genes, respectively. The arrow denotes facilitation or promotion, and --- denotes inhibition. The dashed line indicates a possible correlation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by The Committee for the Welfare and Ethics of Laboratory Animals, Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences.
Author Contributions
LS and ZJ conceived the project and designed the experiments. NW and ZJ wrote the manuscript. ZJ performed the experiments. ZJ and JXS conducted the CyHV-3 infection experiment and collected samples. CL, XH, and YG performed fish propagation and culture. XJ conducted the RT-PCR experiment. NW and X-QX coordinated the data analysis tasks. NW, HL, MS, WY, YT, JWS, and YC analyzed the data. The manuscript was revised and approved by X-QX and LS. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by grants from the National Key R&D Program of China (2018YFD0900302-6), The Natural Science Foundation of Heilongjiang Province (TD2019C004), China Aquaculture Research System (CARS-45-06), and Central Public-interest Scientific Institutional Basal Research Fund (2020TD31).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Bruno Unger from the University of Canterbury in New Zealand for language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.687151/full#supplementary-material
References
1. Neave MJ, Sunarto A, McColl KA. Transcriptomic Analysis of Common Carp Anterior Kidney During Cyprinid Herpesvirus 3 Infection: Immunoglobulin Repertoire and Homologue Functional Divergence. Sci Rep (2017) 7:41531. doi: 10.1038/srep41531
2. Adamek M, Steinhagen D, Irnazarow I, Hikima J, Jung TS, Aoki T. Biology and Host Response to Cyprinid Herpesvirus 3 Infection in Common Carp. Dev Comp Immunol (2014) 43:151–9. doi: 10.1016/j.dci.2013.08.015
3. Sunarto A, McColl KA. Expression of Immune-Related Genes of Common Carp During Cyprinid Herpesvirus 3 Infection. Dis Aquat Organ (2015) 113:127–35. doi: 10.3354/dao02824
4. Reichert M, Lukasik A, Zielenkiewicz P, Matras M, Maj-Paluch J, Stachnik M, et al. Host microRNA Analysis in Cyprinid Herpesvirus-3 (CyHV-3) Infected Common Carp. BMC Genomics (2019) 20:46. doi: 10.1186/s12864-018-5266-9
5. Rakus KL, Irnazarow I, Adamek M, Palmeira L, Kawana Y, Hirono I, et al. Gene Expression Analysis of Common Carp (Cyprinus Carpio L.) Lines During Cyprinid Herpesvirus 3 Infection Yields Insights Into Differential Immune Responses. Dev Comp Immunol (2012) 37:65–76. doi: 10.1016/j.dci.2011.12.006
6. Rakus KL, Wiegertjes GF, Adamek M, Siwicki AK, Lepa A, Irnazarow I. Resistance of Common Carp (Cyprinus Carpio L.) to Cyprinid Herpesvirus-3 is Influenced by Major Histocompatibility (MH) Class II B Gene Polymorphism. Fish Shellfish Immun (2009) 26:737–43. doi: 10.1016/j.fsi.2009.03.001
7. Adamek M, Matras M, Dawson A, Piackova V, Gela D, Kocour M, et al. Type I Interferon Responses of Common Carp Strains With Different Levels of Resistance to Koi Herpesvirus Disease During Infection With CyHV-3 or SVCV. Fish Shellfish Immunol (2019) 87:809–19. doi: 10.1016/j.fsi.2019.02.022
8. Lee X, Yi Y, Weng S, Zeng J, Zhang H, He J, et al. Transcriptomic Analysis of Koi (Cyprinus Carpio) Spleen Tissue Upon Cyprinid Herpesvirus 3 (CyHV3) Infection Using Next Generation Sequencing. Fish Shellfish Immunol (2016) 49:213–24. doi: 10.1016/j.fsi.2015.12.007
9. Hwang JA, Kim JE, Kim HS, Lee JH. Immune Response to Koi Herpesvirus (KHV) of Koi and Koi X Red Common Carp (Cyprinus Carpio). Dev Reprod (2017) 21:361–70. doi: 10.12717/DR.2017.21.4.361
10. Adamek M, Rakus KL, Brogden G, Matras M, Chyb J, Hirono I, et al. Interaction Between Type I Interferon and Cyprinid Herpesvirus 3 in Two Genetic Lines of Common Carp (Cyprinus Carpio). Dis Aquat Organ (2014) 111:107–18. doi: 10.3354/dao02773
11. Tadmor-Levi R, Hulata G, David L. Multiple Interacting QTLs Affect Disease Challenge Survival in Common Carp (Cyprinus Carpio). Heredit (Edinb) (2019) 123:565–78. doi: 10.1038/s41437-019-0224-0
12. Jia ZY, Chen L, Ge YL, Li SW, Peng WZ, Li CT, et al. Genetic Mapping of Koi Herpesvirus Resistance (KHVR) in Mirror Carp (Cyprinus Carpio) Revealed Genes and Molecular Mechanisms of Disease Resistance. Aquaculture (2020) 519:1–10. doi: 10.1016/j.aquaculture.2019.734850
13. Ren X, Liu P, Li J. Comparative Transcriptomic Analysis of Marsupenaeus Japonicus Hepatopancreas in Response to Vibrio Parahaemolyticus and White Spot Syndrome Virus. Fish Shellfish Immunol (2019) 87:755–64. doi: 10.1016/j.fsi.2019.02.030
14. Liu X, Qin Z, Babu VS, Zhao L, Li J, Zhang X, et al. Transcriptomic Profiles of Striped Snakehead Cells (SSN-1) Infected With Snakehead Vesiculovirus (SHVV) Identifying IFI35 as a Positive Factor for SHVV Replication. Fish Shellfish Immunol (2019) 86:46–52. doi: 10.1016/j.fsi.2018.11.031
15. Tengs T, Rimstad E. Emerging Pathogens in the Fish Farming Industry and Sequencing-Based Pathogen Discovery. Dev Comp Immunol (2017) 75:109–19. doi: 10.1016/j.dci.2017.01.025
16. Sudhagar A, Kumar G, El-Matbouli M. Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review. Int J Mol Sci (2018) 19(1):245. doi: 10.3390/ijms19010245
17. Tadmor-Levi R, Doron-Faigenboim A, Marcos-Hadad E, Petit J, Hulata G, Forlenza M, et al. Different Transcriptional Response Between Susceptible and Resistant Common Carp (Cyprinus Carpio) Fish Hints on the Mechanism of CyHV-3 Disease Resistance. BMC Genomics (2019) 20(1):1019. doi: 10.1186/s12864-019-6391-9
18. Boltana S, Valenzuela-Miranda D, Aguilar A, Mackenzie S, Gallardo-Escarate C. Long Noncoding RNAs (lncRNAs) Dynamics Evidence Immunomodulation During ISAV-Infected Atlantic Salmon (Salmo Salar). Sci Rep (2016) 6:22698. doi: 10.1038/srep22698
19. Tarifeno-Saldivia E, Valenzuela-Miranda D, Gallardo-Escarate C. In the Shadow: The Emerging Role of Long Non-Coding RNAs in the Immune Response of Atlantic Salmon. Dev Comp Immunol (2017) 73:193–205. doi: 10.1016/j.dci.2017.03.024
20. Sun W, Feng J. Differential lncRNA Expression Profiles Reveal the Potential Roles of lncRNAs in Antiviral Immune Response of Crassostrea Gigas. Fish Shellfish Immunol (2018) 81:233–41. doi: 10.1016/j.fsi.2018.07.032
21. Valenzuela-Munoz V, Pereiro P, Alvarez-Rodriguez M, Gallardo-Escarate C, Figueras A, Novoa B. Comparative Modulation of lncRNAs in Wild-Type and Rag1-Heterozygous Mutant Zebrafish Exposed to Immune Challenge With Spring Viraemia of Carp Virus (SVCV). Sci Rep (2019) 9:14174. doi: 10.1038/s41598-019-50766-0
22. Jia Z, Shi L, Shi J, Li C, Hu X, Ge Y, et al. Identification Methods of Carp Germplasm Resources. ZL201610945256.9, the National Invention Patent in China.
23. Tadmor-Levi R, Asoulin E, Hulata G, David L. Studying the Genetics of Resistance to CyHV-3 Disease Using Introgression From Feral to Cultured Common Carp Strains. Front Genet (2017) 8:24. doi: 10.3389/fgene.2017.00024
24. Jia ZY, Wang SH, Bai SS, Ge YL, Li CT, Hu XS, et al. Survival Rate and Immunological Responses of Mirror Carp Selective Breeding Generations to CyHV-3. J World Aquacult Soc (2018) 49:388–95. doi: 10.1111/jwas.12511
25. Sun JX, Shi LY, Jiang XN, Li CT, Ge YL, Hu XS, et al. Research on Expression of Virus in Cyprinid Herpesvirus 3 (CyHV-3) F4 Disease Resistant Strains. J Shanghai Ocean Univ (2020) 30(02):258–65. doi: CN 31-2024/S
26. Conte MA, Gammerdinger WJ, Bartie KL, Penman DJ, Kocher TD. A High Quality Assembly of the Nile Tilapia (Oreochromis Niloticus) Genome Reveals the Structure of Two Sex Determination Regions. BMC Genomics (2017) 18:341. doi: 10.1186/s12864-017-3723-5
27. Adamek M, Syakuri H, Harris S, Rakus KL, Brogden G, Matras M, et al. Cyprinid Herpesvirus 3 Infection Disrupts the Skin Barrier of Common Carp (Cyprinus Carpio L.). Vet Microbiol (2013) 162:456–70. doi: 10.1016/j.vetmic.2012.10.033
28. Tan A, Ye X, Jiang L, Zhao F, Zou W, Lin Z, et al. Detection Methods of Cyprinid Herpesvirus (CyHv). Beijing: China MoAaRAotPsRo (2011).
29. Zhang X, Li G, Jiang H, Li L, Ma J, Li H, et al. Full-Length Transcriptome Analysis of Litopenaeus Vannamei Reveals Transcript Variants Involved in the Innate Immune System. Fish Shellfish Immunol (2019) 87:346–59. doi: 10.1016/j.fsi.2019.01.023
30. Wu N, Wang B, Cui ZW, Zhang XY, Cheng YY, Xu X, et al. Integrative Transcriptomic and Micrornaomic Profiling Reveals Immune Mechanism for the Resilience to Soybean Meal Stress in Fish Gut and Liver. Front Physiol (2018) 9:1154. doi: 10.3389/fphys.2018.01154
31. Xu P, Zhang X, Wang X, Li J, Liu G, Kuang Y, et al. Genome Sequence and Genetic Diversity of the Common Carp. Cyprinus Carpio Nat Genet (2014) 46:1212–9. doi: 10.1038/ng.3098
32. Wu N, Song YL, Wang B, Zhang XY, Zhang XJ, Wang YL, et al. Fish Gut-Liver Immunity During Homeostasis or Inflammation Revealed by Integrative Transcriptome and Proteome Studies. Sci Rep (2016) 6:36048. doi: 10.1038/srep36048
33. Wang J, Fu L, Koganti PP, Wang L, Hand JM, Ma H, et al. Identification and Functional Prediction of Large Intergenic Noncoding RNAs (lincRNAs) in Rainbow Trout (Oncorhynchus Mykiss). Mar Biotechnol (NY) (2016) 18:271–82. doi: 10.1007/s10126-016-9689-5
34. Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, et al. Large-Scale Prediction of Long Non-Coding RNA Functions in a Coding-Non-Coding Gene Co-Expression Network. Nucleic Acids Res (2011) 39:3864–78. doi: 10.1093/nar/gkq1348
35. Langfelder P, Horvath S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinf (2008) 9:559. doi: 10.1186/1471-2105-9-559
36. Yu GC, Wang LG, Han YY, He QY. Clusterprofiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics (2012) 16:284–7. doi: 10.1089/omi.2011.0118
37. Zhang XJ, Zhang XY, Zhang N, Guo X, Peng KS, Wu H, et al. Distinctive Structural Hallmarks and Biological Activities of the Multiple Cathelicidin Antimicrobial Peptides in a Primitive Teleost Fish. J Immunol (2015) 194:4974–87. doi: 10.4049/jimmunol.1500182
38. Wu N, Zhang XY, Huang B, Zhang N, Zhang XJ, Guo X, et al. Investigating the Potential Immune Role of Fish NCAMs: Molecular Cloning and Expression Analysis in Mandarin Fish. Fish Shellfish Immun (2015) 46:765–77. doi: 10.1016/j.fsi.2015.08.006
39. Campadelli-Fiume G, Collins-McMillen D, Gianni T, Yurochko AD. Integrins as Herpesvirus Receptors and Mediators of the Host Signalosome. Annu Rev Virol (2016) 3:215–36. doi: 10.1146/annurev-virology-110615-035618
40. Azab W, Gramatica A, Herrmann A, Osterrieder N. Binding of Alphaherpesvirus Glycoprotein H to Surface Alpha4beta1-Integrins Activates Calcium-Signaling Pathways and Induces Phosphatidylserine Exposure on the Plasma Membrane. mBio (2015) 6:e01552–01515. doi: 10.1128/mBio.01552-15
41. Cheshenko N, Trepanier JB, Gonzalez PA, Eugenin EA, Jacobs WR Jr., Herold BC. Herpes Simplex Virus Type 2 Glycoprotein H Interacts With Integrin Alphavbeta3 to Facilitate Viral Entry and Calcium Signaling in Human Genital Tract Epithelial Cells. J Virol (2014) 88:10026–38. doi: 10.1128/JVI.00725-14
42. Delecluse S, Tsai MH, Shumilov A, Bencun M, Arrow S, Beshirova A, et al. Epstein-Barr Virus Induces Expression of the LPAM-1 Integrin in B Cells. Vitro Vivo J Virol (2019) 93(5):e01618–18. doi: 10.1128/JVI.01618-18
43. Huang WJ, Shen Y, Xu XY, Hu MY, Li JL. Identification and Characterization of the TLR18 Gene in Grass Carp (Ctenopharyngodon Idella). Fish Shellfish Immunol (2015) 47:681–8. doi: 10.1016/j.fsi.2015.09.052
44. Lee PT, Zou J, Holland JW, Martin SA, Collet B, Kanellos T, et al. Identification and Characterisation of TLR18-21 Genes in Atlantic Salmon (Salmo Salar). Fish Shellfish Immunol (2014) 41:549–59. doi: 10.1016/j.fsi.2014.10.006
45. Jiang X, He M, Bai J, Chan CB, Wong AOL. Signal Transduction for TNFalpha-Induced Type II SOCS Expression and Its Functional Implication in Growth Hormone Resistance in Carp Hepatocytes. Front Endocrinol (Lausanne) (2020) 11:20. doi: 10.3389/fendo.2020.00020
46. Wang W, Cole AM, Hong T, Waring AJ, Lehrer RI. Retrocyclin, an Antiretroviral Theta-Defensin, Is a Lectin. J Immunol (2003) 170:4708–16. doi: 10.4049/jimmunol.170.9.4708
47. Zhu D, Fu P, Huang R, Xiong L, Wang Y, He L, et al. Molecular Characterization, Tissue Distribution and Functional Analysis of Galectin 1-Like 2 in Grass Carp (Ctenopharyngodon Idella). Fish Shellfish Immunol (2019) 94:455–63. doi: 10.1016/j.fsi.2019.09.041
48. Ghosh A, Banerjee A, Amzel LM, Vasta GR, Bianchet MA. Structure of the Zebrafish Galectin-1-L2 and Model of its Interaction With the Infectious Hematopoietic Necrosis Virus (IHNV) Envelope Glycoprotein. Glycobiology (2019) 29:419–30. doi: 10.1093/glycob/cwz015
49. Wu C, Zhao X, Babu VS, Yuan G, Wang W, Su J, et al. Distribution of Mannose Receptor in Blunt Snout Bream (Megalobrama Amblycephala) During the Embryonic Development and its Immune Response to the Challenge of Aeromonas Hydrophila. Fish Shellfish Immunol (2018) 78:52–9. doi: 10.1016/j.fsi.2018.03.049
50. Schneider JR, Shen X, Orlandi C, Nyanhete T, Sawant S, Carias AM, et al. A MUC16 IgG Binding Activity Selects for a Restricted Subset of IgG Enriched for Certain Simian Immunodeficiency Virus Epitope Specificities. J Virol (2020) 94(5):e01246–19.94. doi: 10.1128/JVI.01246-19
51. Tang MLK. The Physiological Induction of Tolerance to Allergens, Allergy. In: Allergy, Immunity and Tolerance in Early Childhood. Academic Press, Elsevier Inc (2016).
52. Li L, Yang W, Shen Y, Xu X, Li J. Fish Complement C8 Evolution, Functional Network Analyses, and the Theoretical Interaction Between C8 Alpha Chain and CD59. Mol Immunol (2020) 128:235–48. doi: 10.1016/j.molimm.2020.10.013
53. Pionnier N, Adamek M, Miest JJ, Harris SJ, Matras M, Rakus KL, et al. C-Reactive Protein and Complement as Acute Phase Reactants in Common Carp (Cyprinus Carpio) During CyHV-3 Infection. Dis Aquat Organ (2014) 109:187–99. doi: 10.3354/dao02727
54. Magnadottir B, Gudmundsdottir S, Lange S. A Novel Ladder-Like Lectin Relates to Sites of Mucosal Immunity in Atlantic Halibut (Hippoglossus Hippoglossus L.). Fish Shellfish Immunol (2019) 87:9–12. doi: 10.1016/j.fsi.2018.12.034
55. Ryu JG, Lee J, Kim EK, Seo HB, Park JS, Lee SY, et al. Treatment of IL-21r-Fc Control Autoimmune Arthritis via Suppression of STAT3 Signal Pathway Mediated Regulation of the Th17/Treg Balance and Plasma B Cells. Immunol Lett (2015) 163:143–50. doi: 10.1016/j.imlet.2014.09.007
56. Moens E, Brouwer M, Dimova T, Goldman M, Willems F, Vermijlen D. IL-23R and TCR Signaling Drives the Generation of Neonatal Vgamma9Vdelta2 T Cells Expressing High Levels of Cytotoxic Mediators and Producing IFN-Gamma and IL-17. J Leukoc Biol (2011) 89:743–52. doi: 10.1189/jlb.0910501
57. Zhang YB, Gui JF. Molecular Regulation of Interferon Antiviral Response in Fish. Dev Comp Immunol (2012) 38:193–202. doi: 10.1016/j.dci.2012.06.003
58. Langevin C, Levraud JP, Boudinot P. Fish Antiviral Tripartite Motif (TRIM) Proteins. Fish Shellfish Immunol (2019) 86:724–33. doi: 10.1016/j.fsi.2018.12.008
59. Shanware NP, Bray K, Abraham RT. The PI3K, Metabolic, and Autophagy Networks: Interactive Partners in Cellular Health and Disease. Annu Rev Pharmacol (2013) 53:89–106. doi: 10.1146/annurev-pharmtox-010611-134717
60. Wu M, Dan C, Gui JF, Zhang YB. Fish Species-Specific TRIM Gene FTRCA1 Negatively Regulates Interferon Response Through Attenuating IRF7 Transcription. Fish Shellfish Immunol (2019) 90:180–7. doi: 10.1016/j.fsi.2019.04.297
61. Wu M, Zhao X, Gong XY, Wang Y, Gui JF, Zhang YB. FTRCA1, a Species-Specific Member of finTRIM Family, Negatively Regulates Fish IFN Response Through Autophage-Lysosomal Degradation of TBK1. J Immunol (2019) 202:2407–20. doi: 10.4049/jimmunol.1801645
62. Kongchum P, Palti Y, Hallerman EM, Hulata G, David L. SNP Discovery and Development of Genetic Markers for Mapping Innate Immune Response Genes in Common Carp (Cyprinus Carpio). Fish Shellfish Immunol (2010) 29:356–61. doi: 10.1016/j.fsi.2010.04.013
63. Wei JG, Zang SQ, Xu M, Zheng QJ, Chen XL, Qin QW. TRAF6 is a Critical Factor in Fish Immune Response to Virus Infection. Fish Shellfish Immun (2017) 60:6–12. doi: 10.1016/j.fsi.2016.11.008
64. Kim HS, Sohn H, Jang SW, Lee GR. The Transcription Factor NFIL3 Controls Regulatory T-Cell Function and Stability. Exp Mol Med (2019) 51:80. doi: 10.1038/s12276-019-0280-9
65. Yu H, Shen Y, Sun J, Xu X, Wang R, Xuan Y, et al. Molecular Cloning and Functional Characterization of the NFIL3/E4BP4 Transcription Factor of Grass Carp, Ctenopharyngodon Idella. Dev Comp Immunol (2014) 47:215–22. doi: 10.1016/j.dci.2014.07.019
66. Szymczak M. Distribution of Cathepsin D Activity Between Lysosomes and a Soluble Fraction of Marinating Brine. J Food Sci (2016) 81:E1966–70. doi: 10.1111/1750-3841.13375
67. Kumar R, Khandelwal N, Thachamvally R, Tripathi BN, Barua S, Kashyap SK, et al. Role of MAPK/MNK1 Signaling in Virus Replication. Virus Res (2018) 253:48–61. doi: 10.1016/j.virusres.2018.05.028
68. Xuan MH, Yan XL, Liu XZ, Xu TJ. IRF1 Negatively Regulates NF-Kappa B Signaling by Targeting MyD88 for Degradation in Teleost Fish. Dev Comp Immunol (2020) 110:103709. doi: 10.1016/j.dci.2020.103709
69. Yan X, Zhao X, Huo R, Xu T. IRF3 and IRF8 Regulate NF-kappaB Signaling by Targeting MyD88 in Teleost Fish. Front Immunol (2020) 11:606. doi: 10.3389/fimmu.2020.00606
70. Feng X, Liu X, Zhang W, Xiao W. P53 Directly Suppresses BNIP3 Expression to Protect Against Hypoxia-Induced Cell Death. EMBO J (2011) 30:3397–415. doi: 10.1038/emboj.2011.248
71. Godoy-Lugo JA, Miranda-Cruz MM, Rosas-Rodriguez JA, Adan-Bante NP, Icedo-Garcia R, Sonanez-Organis JG. Hypoxia Inducible Factor -1 Regulates WSSV-Induced Glycolytic Genes in the White Shrimp Litopenaeus Vannamei. Fish Shellfish Immunol (2019) 92:165–71. doi: 10.1016/j.fsi.2019.05.040
72. Lepelletier Y, Moura IC, Hadj-Slimane R, Renand A, Fiorentino S, Baude C, et al. Immunosuppressive Role of Semaphorin-3A on T Cell Proliferation is Mediated by Inhibition of Actin Cytoskeleton Reorganization. Eur J Immunol (2006) 36:1782–93. doi: 10.1002/eji.200535601
73. Alamri A, Soussi Gounni A, Kung SKP. View Point: Semaphorin-3E: An Emerging Modulator of Natural Killer Cell Functions? Int J Mol Sci (2017) 18:2337. doi: 10.3390/ijms18112337
74. Yan Y, Cui H, Guo C, Wei J, Huang Y, Li L, et al. Singapore Grouper Iridovirus-Encoded Semaphorin Homologue (SGIV-Sema) Contributes to Viral Replication, Cytoskeleton Reorganization and Inhibition of Cellular Immune Responses. J Gen Virol (2014) 95:1144–55. doi: 10.1099/vir.0.060608-0
75. Laranjeiro R, Tamai TK, Peyric E, Krusche P, Ott S, Whitmore D. Cyclin-Dependent Kinase Inhibitor P20 Controls Circadian Cell-Cycle Timing. Proc Natl Acad Sci USA (2013) 110:6835–40. doi: 10.1073/pnas.1217912110
76. Rivera-Soto R, Dissinger NJ, Damania B. Kaposi’s Sarcoma-Associated Herpesvirus Viral Interleukin-6 Signaling Upregulates Integrin Beta3 Levels and Is Dependent on STAT3. J Virol (2020) 94(5):e01384–19. doi: 10.1128/JVI.01384-19
77. Kalkman HO, Feuerbach D. Antidepressant Therapies Inhibit Inflammation and Microglial M1-Polarization. Pharmacol Ther (2016) 163:82–93. doi: 10.1016/j.pharmthera.2016.04.001
78. Frakolaki E, Kalliampakou KI, Kaimou P, Moraiti M, Kolaitis N, Boleti H, et al. Emerging Role of L-Dopa Decarboxylase in Flaviviridae Virus Infections. Cells (2019) 8(8):837. doi: 10.3390/cells8080837
79. Rakus KL, Irnazarow I, Adamek M, Palmeira L, Kawana Y, Hirono I, et al. Gene Expression Analysis of Common Carp (Cyprinus Carpio L.) Lines During Cyprinid Herpesvirus 3 Infection Yields Insights Into Differential Immune Responses. Dev Comp Immunol (2012) 37:65–76. doi: 10.1016/j.dci.2011.12.006
80. Zhang H, Xiao Y, Zhu Z, Li B, Greene MI. Immune Regulation by Histone Deacetylases: A Focus on the Alteration of FOXP3 Activity. Immunol Cell Biol (2012) 90:95–100. doi: 10.1038/icb.2011.101
81. Jin H, Guo X. Valproic Acid Ameliorates Coxsackievirus-B3-Induced Viral Myocarditis by Modulating Th17/Treg Imbalance. Virol J (2016) 13:168. doi: 10.1186/s12985-016-0626-z
82. Sugimoto K, Itoh T, Takita M, Shimoda M, Chujo D, SoRelle JA, et al. Improving Allogeneic Islet Transplantation by Suppressing Th17 and Enhancing Treg With Histone Deacetylase Inhibitors. Transpl Int (2014) 27:408–15. doi: 10.1111/tri.12265
83. Moreira JD, Koch BEV, van Veen S, Walburg KV, Vrieling F, Guimaraes TMPD, et al. Functional Inhibition of Host Histone Deacetylases (HDACs) Enhances In Vitro and In Vivo Anti-Mycobacterial Activity in Human Macrophages and in Zebrafish. Front Immunol (2020) 11:36. doi: 10.3389/fimmu.2020.00036
84. Wang QQ, Li N, Wang XJ, Shen JG, Hong XJ, Yu H, et al. Membrane Protein hMYADM Preferentially Expressed in Myeloid Cells Is Up-Regulated During Differentiation of Stem Cells and Myeloid Leukemia Cells. Life Sci (2007) 80:420–9. doi: 10.1016/j.lfs.2006.09.043
85. Parekh NJ, Krouse TE, Reider IE, Hobbs RP, Ward BM, Norbury CC. Type I Interferon-Dependent CCL4 Is Induced by a cGAS/STING Pathway That Bypasses Viral Inhibition and Protects Infected Tissue, Independent of Viral Burden. PloS Pathog (2019) 15(10):e1007778. doi: 10.1371/journal.ppat.1007778
86. Eisenberg RJ, Ponce de Leon M, Friedman HM, Fries LF, Frank MM, Hastings JC, et al. Complement Component C3b Binds Directly to Purified Glycoprotein C of Herpes Simplex Virus Types 1 and 2. Microb Pathog (1987) 3:423–35. doi: 10.1016/0882-4010(87)90012-x
87. Busse DC, Habgood-Coote D, Clare S, Brandt C, Bassano I, Kaforou M, et al. Interferon-Induced Protein 44 and Interferon-Induced Protein 44-Like Restrict Replication of Respiratory Syncytial Virus. J Virol (2020) 94(18):e00297–20. doi: 10.1128/JVI.00297-20
88. Rose PP, Hanna SL, Spiridigliozzi A, Wannissorn N, Beiting DP, Ross SR, et al. Natural Resistance-Associated Macrophage Protein Is a Cellular Receptor for Sindbis Virus in Both Insect and Mammalian Hosts. Cell Host Microbe (2011) 10:97–104. doi: 10.1016/j.chom.2011.06.009
89. Jensen P. Structure and Function of Plasminogen Activator Inhibitor-2. Int J Oncol (1997) 11:557–70. doi: 10.3892/ijo.11.3.557
90. Tao R, Li J, Xin J, Wu J, Guo J, Zhang L, et al. Methylation Profile of Single Hepatocytes Derived From Hepatitis B Virus-Related Hepatocellular Carcinoma. PloS One (2011) 6:e19862. doi: 10.1371/journal.pone.0019862
91. Nita-Lazar M, Mancini J, Feng C, Gonzalez-Montalban N, Ravindran C, Jackson S, et al. The Zebrafish Galectins Drgal1-L2 and Drgal3-L1 Bind In Vitro to the Infectious Hematopoietic Necrosis Virus (IHNV) Glycoprotein and Reduce Viral Adhesion to Fish Epithelial Cells. Dev Comp Immunol (2016) 55:241–52. doi: 10.1016/j.dci.2015.09.007
Keywords: transcriptome, lncRNA, CyHV-3, resistant immune mechanism, common carp
Citation: Jia Z, Wu N, Jiang X, Li H, Sun J, Shi M, Li C, Ge Y, Hu X, Ye W, Tang Y, Shan J, Cheng Y, Xia X-Q and Shi L (2021) Integrative Transcriptomic Analysis Reveals the Immune Mechanism for a CyHV-3-Resistant Common Carp Strain. Front. Immunol. 12:687151. doi: 10.3389/fimmu.2021.687151
Received: 29 March 2021; Accepted: 14 June 2021;
Published: 05 July 2021.
Edited by:
Maria Del Mar Ortega-Villaizan, Miguel Hernández University of Elche, SpainReviewed by:
Paulina Schmitt, Pontificia Universidad Católica de Valparaíso, ChileJing Xing, Ocean University of China, China
Copyright © 2021 Jia, Wu, Jiang, Li, Sun, Shi, Li, Ge, Hu, Ye, Tang, Shan, Cheng, Xia and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lianyu Shi, sly2552@aliyun.com; Xiao-Qin Xia, xqxia@ihb.ac.cn
†These authors share first authorship
 Zhiying Jia
Zhiying Jia Nan Wu
Nan Wu Xiaona Jiang1,2
Xiaona Jiang1,2 Yingyin Cheng
Yingyin Cheng Xiao-Qin Xia
Xiao-Qin Xia