Insights on nitrate respiration by Shewanella
- 1State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
- 2State Key Laboratory of Ocean Engineering, Shanghai Jiao Tong University, Shanghai, China
Shewanellae are well known for their ability to utilize a number of electron acceptors and are therefore considered to have important roles in element cycling in the environment, such as nitrogen cycling through dissimilatory nitrate reduction to ammonia (DNRA) and denitrification. Possessing two periplasmic nitrate reductase systems (NAP-α and NAP-β) is a special trait of the Shewanella genus, and both enzymes are likely to provide selective advantage to the host. This review relates the current knowledge and aspects of the nitrate respiration system of Shewanella. Specifically, the potential physiological functions and regulation mechanisms of the duo-NAP system are discussed in addition to the evolution of anaerobic respiration systems of Shewanella.
Introduction
Shewanella species are widespread in diverse environments and have major environmental relevance (Fredrickson et al., 2008). They are commonly found in food, freshwater, hydrothermal vents, cold water, deep-sea sediments, and even fields contaminated with arsenic, chromium (VI), and explosives (Viamajala et al., 2004; Saltikov et al., 2005; Zhao et al., 2005; Gao et al., 2006). Shewanella utilize a number of electron acceptors for energy sources, such as nitrate, nitrite, thiosulfate, elemental sulfur, iron oxide and manganese oxide (Fredrickson et al., 2008). Among these electron acceptors, nitrate is considered to be the preferred acceptor for anaerobic respiration by microorganisms due to its relatively high standard redox potential, with a powerful ability to generate energy, and its abundance in aquatic environments (Richardson, 2000).
Briefly, the microbial nitrogen cycle involves biotic chain reactions comprising nitrogen fixation, dissimilatory nitrate reduction to ammonia (DNRA), denitrification, nitrification and anammox (anaerobic ammonium oxidation) (Figure 1). Denitrification has been intensely studied in recent decades and is largely assumed to be the major pathway for nitrate removal (Wallenstein et al., 2006; Kraft et al., 2011). In contrast to denitrification, knowledge on DNRA is mostly lacking.
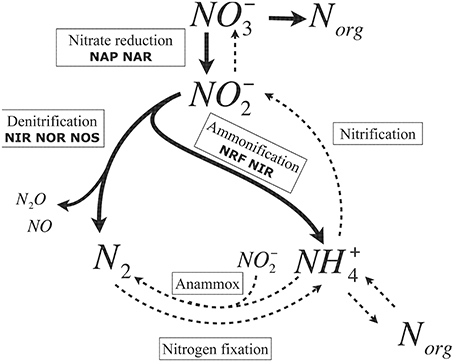
Figure 1. The major pathways of the microbial nitrogen cycle. Thick lines highlight the processes in which Shewanella are involved. Dashed lines represent the pathway catalyzed by other organisms. Denitrification comprises three enzymatic steps: nitrite to nitric oxide (copper nitrite reductase, NirK; or cytochrome cd1 nitrite reductase, NirS), nitric oxide to nitrous oxide (nitric oxide reductase, NOR), and nitrous oxide to dinitrogen (nitrous oxide reductase, NOS). Anammox (anaerobic ammonium oxidation) involves microorganisms participating in oxidation: one producing NO−2 from ammonium and another reducing NO−2 to N2 (Strous and Jetten, 2004).
To the best of our knowledge all Shewanella species are able to reduce nitrate. Most species reduce nitrate to nitrite and then to ammonia via the DNRA pathway, though a few species, such as S. denitrificans and S. amazonesis, carry out respiratory denitrification where nitrate is reduced to dinitrogen via nitrite, nitric oxide and nitrous oxide (Venkateswaran and Dollhopf, 1998; Brettar et al., 2002). Moreover, a notable number of Shewanellae are amenable to genetic manipulation, which makes detailed genetic studies on nitrate reducing pathways feasible. At the time of the writing of this review, 25 complete genomes of Shewanella were available at NCBI.
Many genes of the nitrate respiration system in Shewanella are identified as having multiple copies. The reduction of nitrate to nitrite is catalyzed by one of two distinct enzyme systems: the periplasmic nitrate reductase system (NAP) and the membrane-bound nitrate reductase system (NAR). Most Shewanella have duo-NAP (Nap-α and Nap-β) systems, which have been documented (Simpson et al., 2010b). Additionally, five strains (S. halifaxensis HAW-EB4, S. sediminis HAW-EB3, S. sp. MR-4, S sp. MR-7 and S. woodyi ATC51908) have both NAP and NAR systems, as listed in Table 1. Moreover, many genomes of Shewanella contain several nitrite reductase genes; these will be discussed below.
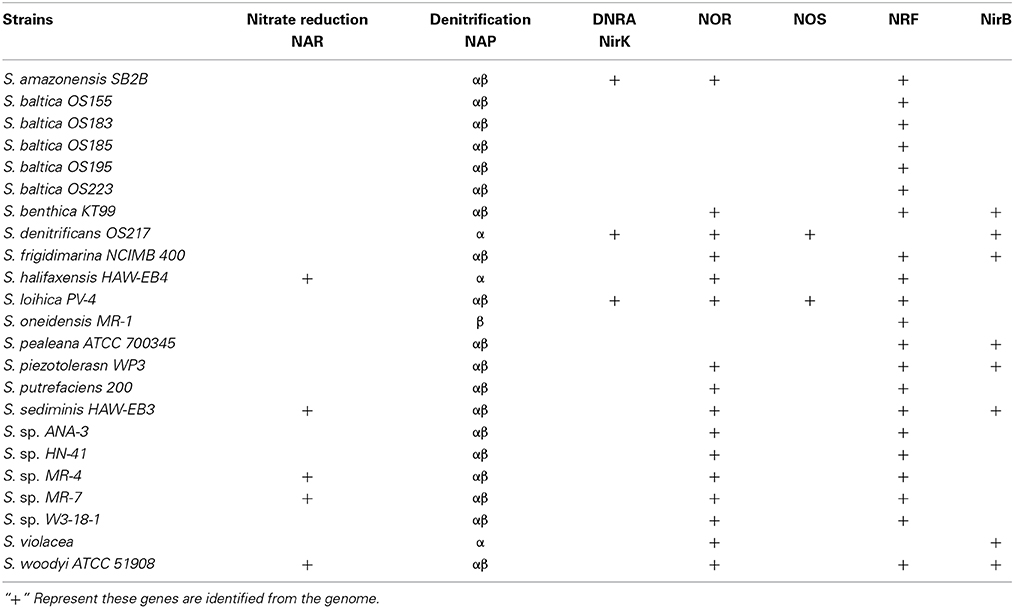
Table 1. Membrane-bound nitrate reductase (NarG), periplasmic nitrate reductase (NAP) and nitrite reductase (denitrification, NirK; ammonification, NrfA, NirB) in the genomes of Shewanella species.
At the genetic level, horizontal gene transfer (HGT), gene duplication and loss of gene function are common strategies by microorganisms during evolution to adapt to environmental stress. Previous studies have suggested that Shewanella might acquire many new adaptive strategies through step-wise processes: the acquisition of mobile islands, selection of islands carrying ecologically important genes and loss of mobile and ecologically unimportant genes (Konstantinidis et al., 2009). Many genes of the nitrate respiration system in Shewanella, e.g., duo-NAP (Nap-α and Nap-β) systems, are identified as multi-copy (Simpson et al., 2010b), and the additional gene clusters that encode the same enzymes are likely to confer selective advantages to the host rather than be functionally redundant (reviewed and discussed below).
In this review, the current knowledge of the Shewanella nitrate respiration system will be reviewed with a specific focus on physiological functions, regulation mechanisms and evolutionary origin. This knowledge will certainly help in the better understanding of nitrate reduction pathways and the impact of HGT on the evolution of anaerobic respiration.
Denitrification and Ammonification by the Shewanella Genus
Denitrification is a respiratory process in which nitrite is first reduced to nitric oxide by nitrite reductase (NIR), nitric oxide reductase (NOR) then catalyzes nitric oxide reduction to nitrous oxide, and dinitrogen is synthesized from the reduction of nitrous oxide by nitrous oxide reductase (NOS). This process occurs in three separate reactions, and a proton-motive force is independently generated by each reaction. S. denitrificans possesses all sets of genes involved in denitrification in its genome (Genebank accession: NC_007954.1), and its capability of denitrification was confirmed by physiological analysis (Brettar et al., 2002). In addition to S. denitrificans, two other Shewanella species are potentially capable of denitrification. Although no growth on nitrate or nitrite was observed, S. loihica possesses the entire set of genes for denitrification (Gao et al., 2006). S. amazonesis was reported to denitrify nitrate to dinitrogen (Venkateswaran and Dollhopf, 1998); however, the genome of S. amazonesis lacks the gene encoding the NOS enzyme, which catalyzes the last step of denitrification, reducing nitrous oxide to dinitrogen. As the enzymatic mechanisms of dinitrogen synthesis in S. amazonesis have not yet been established, S. amazonesis is not a typical denitrifying bacterium. In addition, the nitrite reductase gene nrfA within DNRA pathway has been discovered in the genomes of S. amazonesis and S. loihica. Advanced studies on the function of the nrf gene in these two potential denitrifying bacteria will be interesting because to the best of our known, no strain reported so far is able to perform both denitrification and DNRA.
In DNRA, nitrite is reduced directly to ammonia by nitrite reductase (NrfA or NirB) without any intermediate; energy is also obtained by generating an electrochemical proton potential across the membrane. The pentaheme cytochrome NrfA is a membrane-associated respiratory enzyme that couples to membrane-associated formate-oxidizing enzymes via quinones to generate the membrane potential (Simon, 2002). NirB is an iron-sulfur protein that carries FAD and a siroheme cofactor (Jackson et al., 1981). The function of NirB was assigned primarily to the detoxification of nitrite produced by cytoplasmically oriented membrane-bound nitrate reductase (NarGHI) (Cole, 1996). However, the NirB enzyme may function as a nitrite-scavenger rather than a respiratory enzyme because energy is not conserved during nitrite reduction by NADH (Simon, 2002). For example, in E. coli, the nrfA operon is only induced under low-nitrate concentrations, whereas the NirB nitrite reductase is optimally synthesized only when nitrate or nitrite is in excess (Wang and Gunsalus, 2000). Most shewanellae carry out DNRA because their genomes have either the nrfA or nirB gene (or both) and lack genes for denitrification.
Notably, there are some indications that nitrous oxide is also generated as a byproduct of DNRA (Cruz-García et al., 2007; Vermeiren et al., 2009), though the mechanisms by which non-denitrifying bacteria generate nitrous oxide have not been revealed. One reasonable interpretation is that nitrous oxide is generated by nitrate-dependent iron reduction because the abiotic reduction of nitrite to nitrous oxide was observed in S. putrefaciens when both nitrate and iron (III) were available (Coby and Picardal, 2005).
Periplasmic and Membrane-bound Nitrate Reductase (NAR) in Shewanella
Although the subsequent reactions involve different enzymes and intermediates, the first steps of DNRA and denitrification are identical and can be carried out by the same enzyme: periplasmic nitrate reductase (NAP) or membrane-bound nitrate reductase (NAR). All the available Shewanella genomes contain NAP, and only 5 strains out of 23 (S. halifaxensis HAW-EB4, S. sediminis HAW-EB3, S. sp. MR-4, S sp. MR-7 and S. woodyi ATC51908) possess both the NAP and NAR systems (Table 1). Although not well understood, NAR and NAP may also be of physiological significance in addition to nitrate reduction. In Shewanella species, the understanding of the NAP system is more comprehensive and therefore is selected for discussion in greater detail in this review.
Nap genes are widely distributed in gram-negative bacteria, and our understanding on the NAP system of Shewanella is mainly derived from research in model organisms such as E. coli. The current understanding of the subunits involved in the NAP system of the Shewanella genus is that the NAP system comprises seven compounds: NapA-H (except for NapF). NapA is the catalytic subunit where nitrate is reduced to nitrite, whereas NapB serves as the direct electron transporter, without a catalytic function. NapC is a quinol dehydrogenase protein, transporting electrons from both ubiquinol and menaquinol to terminal reductase NapAB (Simon and Kern, 2008). When NapC is missing, protein CymA performs the function of NapC, which will be discussed later. The electron transport model of NapABC and CymA is shown in Figure 3. NapGH couples electron transfer from the quinol pool via NapB to NapA in E. coli (Brondijk et al., 2002, 2004). However, it seems functionally redundant to retain NapGH in the few species of Shewanella that also possess NapC and/or CymA. It was suggested that NapGH might mediate the alternative pathway for nitrate reduction in addition to NapC or CymA (Gao et al., 2009), though the physiological significance of this alternative pathway is unclear.
Gene Arrangement and Protein Structure Differences Between NAP-α and NAP-β
One of the apparent differences between NAP-α and NAP-β is the arrangement of their gene clusters. Briefly, NAP-α (napEDABC) could be defined as a napC-dependent system, with NAP-β (napDAGHB) being napC independent. In S. oneidensis, which lacks NapC, the NapC homolog CymA mediates electrons from the quinone pool to both NapAB-β and NrfA (Cruz-García et al., 2007; Gao et al., 2009). In S. piezotolerans, which possesses both NAP-α and NAP-β, CymA is involved in both the reduction of nitrate to nitrite and nitrite ammonification (Chen et al., 2011). Furthermore, the different gene arrangements indicate the structural distinction between NAP-α and NAP-β proteins, which will be discussed below.
NAP could be divided into two types: monomeric and heterodimeric NAP. Both NAP-α and NAP-β are components of heterodimeric NAP, which is considered more evolved than monomeric NAP. In heterodimeric NAP, all NapA monomers form a heterodimeric complex with NapB. In contrast, monomeric NapA does not form a complex with the redox partner NapB; instead, a small tetraheme cytochrome C (~10 kDa), called NapM, may be the direct electron donor for NapA (Marietou et al., 2005). The monomeric class of NapA is considered to be the evolutionary link between monomeric NAS (assimilatory nitrate reductase) and heterodimeric NAP and thus a more ancient protein compared with heterodimeric NAP (Stolz and Basu, 2002; Jepson et al., 2006).
Physiological Significance of the duo-NAP System of Shewanella
The existence of multiple nitrate reductases within Shewanella is not as functionally redundant as it appears. By investigating the NAP system of Shewanella and other nitrate reducers, we can find some clues to the functional importance of duo-NAP, especially when the host is under environmental stress.
Scavenging a Limited Concentration of Nitrate
Despite the fact that the majority of organic matter degradation in marine shelf sediments is coupled to sulfate reduction, the reduction of nitrate yields more energy than that of sulfate, and the former is therefore consumed preferentially (Jørgensen, 1982). It has been proposed that NAP may assist bacteria in scavenging nitrate in environments in which the amount of nitrate is extremely limited. For example, in E. coli, which possesses two NAR (NarGHJI and NarZYWV) and one NAP (NapFDAGHBC) enzyme, the NAP enzyme is primarily expressed at low nitrate concentrations and may assist nitrate respiration via NarGHI when the bioenergetic efficiency of nitrate respiration via the NAR enzyme is limiting (Potter and Cole, 1999; Stewart et al., 2002). In addition to E. coli, five species of Shewanella harbor both NAP and NAR, as mentioned above (Table 1), and similar mechanisms may be utilized by shewanellae.
Furthermore, NapB and NapC are suggested to support the scavenging of nitrate by transferring electrons specifically to NapA. For example, S. oneidensis MR-1 cells have only one NAP (NAP-β), and the function of NapB has been proposed as the scavenging of nitrate by routing electrons to NapA only, therefore gaining a benefit during a competition assay test (Gao et al., 2009). Another example is S. piezotolerans WP3, which harbors a duo-NAP system (NAP-α and NAP-β). The NapC of NAP-α in WP3 transports electrons to NapAB exclusively, specifically providing a growth advantage to the host (Chen et al., 2011). Overall, both NapB and NapC may support the host in coping with nitrate-limiting conditions by routing electrons exclusively to NapA.
Maintaining Redox Balance
During chemoheterotrophic growth on reduced carbon compounds, the substrate must be oxidized to a level at which it can be assimilated (Richardson, 2000). During the oxidation process of carbon compounds, reductants are released and partially utilized for ATP synthesis. However, when reductants are in excess, a deposition system must be available. The NAP-α of the duo-NAP system is thought to help maintain this redox balance (Simpson et al., 2010a,b), as implicated in early studies on denitrifying bacteria. For example, in Paracoccus denitrificans, Paracoccus pantotrophus and Rhodobacter capsulatus, the expression of NapABC was found to be stimulated when reduced carbon compounds (such as butyrate), rather than oxidized carbon compounds (such as succinate and malate), were supplied as a carbon source (Sears et al., 2000; Gavira et al., 2002). Thus, the NAP-α system controls the redox balance by using nitrate as an ancillary oxidant to dissipate any excess reductant.
Factors Regulating the duo-NAP System
The distinct physiological functions of the NAP system indicate that NAP-α and NAP-β may have multiple regulation mechanisms. Studies with S. oneidensis suggest that anaerobic respiration systems containing NAP are regulated by the electron transport regulator (EtrA) and cyclic AMP receptor protein (CRP), which control anaerobic respiratory gene expression individually (Maier and Myers, 2001; Beliaev et al., 2002; Saffarini et al., 2003).
EtrA is a global transcriptional regulator involved in the regulation of aerobic and anaerobic metabolism, and it shares 73.6% amino acid sequence similarity with the oxygen-sensing regulator Fnr in E. coli (Saffarini and Nealson, 1993). In mutant strains of S. oneidensis with etrA gene deletion, the expression of nap genes (nap-β) was significantly repressed (Cruz-García et al., 2011). CRP is a well-known regulator of carbon metabolism (Botsford and Harman, 1992), but CRP is essential for anaerobic growth in S. oneidensis, and a crp gene-deleted strain is defective in nitrate reduction (Saffarini et al., 2003; Gao et al., 2008). According to previous studies, CRP lacks an apparent redox-sensing domain, and it is still unclear which CRP is activated under anaerobic conditions (Fredrickson et al., 2008). Moreover, the nap, nar, nrf, and nir gene operons are further controlled by the NarQ-P two-component system. NarQ senses the presence of nitrate/nitrite, the response regulator NarP is then phosphorylated; finally, phospho-NarP activates the transcription of nap gene operons (Darwin and Stewart, 1995). Other environmental factors that may also influence duo-NAP system regulation include the carbon source, temperature, and oxygen concentration.
Carbon Source
We investigated the upstream regions of nap gene operons from 23 genomes of Shewanella and confirmed that there were no apparent Crp sites in the upstream region of nap-α (Figure 2). In all cases, the upstream control regions of napDAGHB (nap-β) contain NarP, EtrA, and Crp sites, whereas napEDABC (nap-α) contain sites for NarP and EtrA. The above findings suggest that NarP-Q, CRP, and EtrA are likely to be involved in the regulation of the duo-NAP system and that the nap-α operon may not belong to the CRP regulation system, which is a well-known regulator associated with carbon metabolism (Botsford and Harman, 1992). Therefore, the transcription of the nap-β gene operon may respond to different types of carbon sources, such as the reduced carbon compounds and oxidized carbon compounds mentioned above.
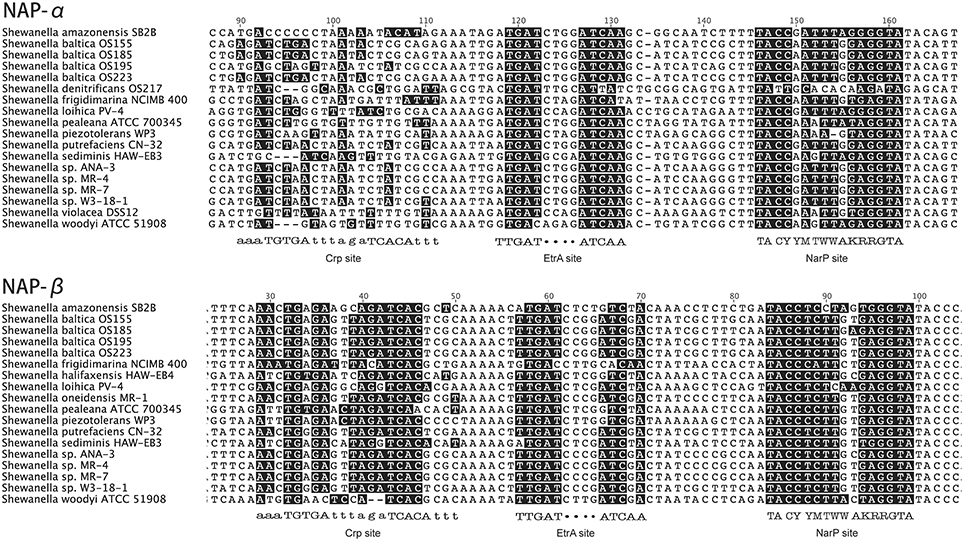
Figure 2. Upstream regions of the NAP-α and NAP-β operons from many Shewanella species. The binding sites for Crp EtrA and NarP were analyzed. NAP-α contains sites for EtrA and NarP, whereas NAP-β has sites for Crp EtrA and NarP. The nucleotides that match the consensus sequences are shaded in black (Crp, EtrA, and NarP site); Y, C or T; M, A or C; K, G or T; and W, A or T. The upper case letters in the binding sites refer to the conserved nucleotides; while the lower case letters are not conserved.
Low Temperature
Almost all species of Shewanella can thrive under cold temperature (2~4°C), which is one of the hallmark features of this genus. Recently, the influence of low temperatures on the duo-NAP system was studied in S. piezotolerans WP3 (Chen et al., 2011). Under conditions of 4°C anaerobic nitrate respiration, the transcription of napDABC (nap-α) was fivefold higher relative to its transcription at 20°C. Moreover, the transcription of napDAB (nap-β) was dramatically reduced compared to that at 20°C. However, no typical cold-shock box sequence was found in the potential control regions of the nap gene operons (Chen et al., 2011). A gene encoding a DEAD box RNA helicase (HrpA) is located upstream of the nap-α operon in almost all Shewanella species (excluding S. denitrificans and S. frigidimarina) (Chen et al., 2011). RNA helicases have been observed to be overexpressed at low temperature in many psychrophilic microorganisms and help to unwind RNA secondary structures for efficient translation under cold conditions (Cartier et al., 2010; Piette et al., 2011). Therefore, the HrpA protein of Shewanella may be involved in the regulation and mRNA processing of NAP-α at low temperatures. Under low temperature, the rates of biochemical reactions usually decrease. And the NAP-α is proposed a more efficient system on electron transporting (Chen et al., 2011). As a consequence, up-regulation of NAP-α may confer advantages to Shewanella in the cold environment.
Oxygen
Most Shewanella are facultative anaerobes, and oxygen levels are proposed to be another factor regulating the expression of the duo-NAP system (Simpson et al., 2010b). Studies of Shewanella genomes show that most Shewanella species have both ubiquinones and menaquinones. Ubiquinones are considered to be “aerobic quinol” (Soballe and Poole, 1999), and menaquinones are suggested to be “anaerobic quinol” (Maier and Myers, 2001) (Simpson et al., 2010b). S. oneidensis mutants deficient in menaquinone and methylmenaquinone, but having wild-type levels of ubiquinone, lose the ability to utilize nitrate, iron(III) and fumarate as electron acceptors (Myers and Myers, 1993). These results indicate that the single NAP-β of S. oneidensis may draw electrons from menaquinone. Aerobic nitrate reduction via NAP is of physiological significance. It could be a self defense mechanism for the cell converting nitrate to nitrite that is very toxic. And it also may provide poorly coupled route for the oxidation of highly reduced carbon sources in order to poise the redox balance (Sears et al., 2000). Studies on the sources of electrons and the transporter of electrons in NAP-α and NAP-β may further elucidate the influence of oxygen on the activation of the duo-NAP system.
In summary, the duo-NAP system in Shewanella has distinct physiological functions, would confer the cells advantages in scavenging nitrate when it's limited. Various environmental factors including carbon sources, temperature, and oxygen might influence the regulation of duo-NAP system. It's generally understood that these regulations are required to maintain/control the redox balance within the cells, such as using nitrate as an ancillary oxidant to dissipate any excess reductant. The detail molecular mechanisms are still waiting to be elucidated.
The Origin of the duo-NAP System
As discussed above, intensive studies have focused on the regulating and operating mechanisms of the duo-NAP system. However, little is known about the origin of this system; for example, how did the duo-NAP system of the Shewanella genus arise during evolution? Based on current understanding, the duo-NAP system is more likely to have resulted from HGT as opposed to gene duplication. Indeed, on the phylogenetic tree constructed with NapA protein sequences, NapA-β of Shewanella forms a relatively independent branch, while NapA-α are more closely related to the NapA of Vibrio species and Photobacterium profundum, indicating that the duo-NAP system is the result of HGT rather than gene duplication (Chen et al., 2011).
To understand why most Shewanella species have acquired NAP-α, we should take a close look at NapC and CymA. Both NapC and CymA are tetraheme c-type cytochromes belonging to the NapC/NirT family of proteins. The distribution of NapC/NirT proteins is relatively less widespread (they are found only in several subgroups of proteobacterial), which suggests that they may be evolutionarily more recent (Sharma et al., 2010). Members of this protein family play a role in the mediation of electrons in several anaerobic respiration pathways: NrfH and NirT for nitrite reduction, TorC for TMAO reduction, DorC for DMSO reduction, and FccC for methacrylate reduction (Simon and Kern, 2008). The proposed electron transport network of CymA and other specific transfers is illustrated in Figure 3. Sequences among the members of this family have a high degree of similarity, and they are suggested to have a common ancestor prior to recruitment into their specific anaerobic respiration system (Bergmann et al., 2005). This common ancestor is likely to function in the most ancient microbial metabolism, such as iron (III) reduction because iron reduction is widely agreed as one of the most ancient microbial metabolisms on the early Earth (Vargas et al., 1998). Alternatively, the ancestor may be a universal component of many electron transport pathways. If this is correct, CymA may have developed various functions at the time, and CymA may have also inherited multi-function capacities from the common ancestor. In addition, E. coli NapC has been reported to show ferric reductase activity (Gescher et al., 2008), suggesting that the specified electron transport proteins still share a commonality with their ancestor.
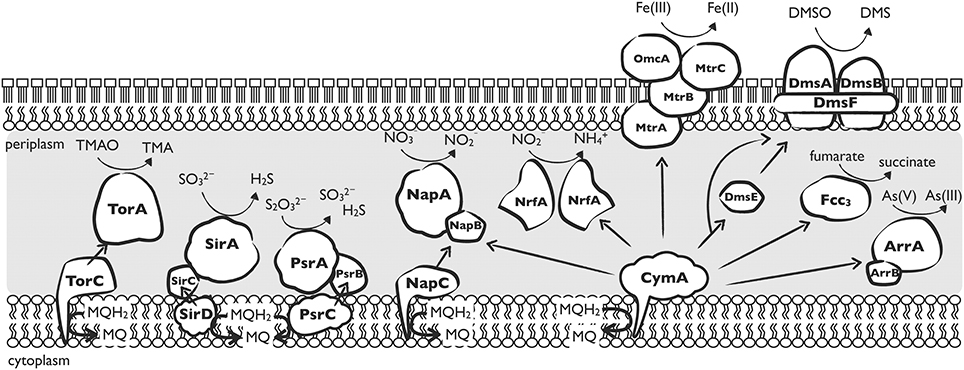
Figure 3. Depiction of the nine anaerobic electron transfer pathways reported in Shewanella. Arrows represent proposed electron flows. CymA-independent respiration systems include the TMAO reductase TorA (Schwalb et al., 2003), sulfite reductase SirA (Shirodkar et al., 2011), thiosulfate reductase PsrA (Shirodkar et al., 2011), periplasmic nitrate reductase NapA, and associated subunits TorC, SirCD, PsrBC, and NapC. CymA-dependent respiration systems: nitrite reductase NrfA (Gao et al., 2009); dissimilatory iron reductase MtrABC and associated outer membrane protein OmcA (Fredrickson et al., 2008); DMSO reductase DmsAB located at the outer membrane and associated subunits DmsF and DmsE (Gralnick et al., 2006); flavocytochrome C3 catalyzing the reduction of fumarate to succinate (Schwalb et al., 2003); and arsenate reductase ArrAB (Murphy and Saltikov, 2007).
Based on multiple lines of evidence, we speculate that the origin of the duo-NAP system is the consequence of the co-evolution of microorganisms and environments. At the very beginning of its origin, Shewanella may have used metal as its sole electron acceptor. During evolution on Earth when extensive substrates became available, CymA may have developed its enzyme functions specifically to utilize alternative electron acceptors. By making use of the multifunctional protein CymA, the organism benefits by not increasing the burden of an expanding genome when selection pressure limits genome size (Huberts and Van Der Klei, 2010). However, for a protein that has multiple related functions, the additional roles might result in a low specificity of the active site (Jeffery, 1999). This model of respiration may have become less competitive as the environment changed. To cope with this deficiency, Shewanella, had to recruit specific electron-transport proteins (such as NapC, PsrC, SirC, and TorC, see Figure 3) by HGT. Nitrate is the most preferred electron acceptor in the natural environment for Shewanella because the cells can synthesize more energy for growth. Therefore, to primarily use nitrate, it is conceivable that Shewanella recruited the specific electron transport proteins NapB and NapC and routed electrons to NapA only.
Moreover, HGT was analyzed in several Shewanella baltica strains (Caro-Quintero et al., 2011), and the results indicate that the exchange rate of the genes, including nap, is rapid and that this gene-exchange contributes to successful adaptation to specific physicochemical conditions.
Concluding Remarks
According to genetic and physiological studies, most Shewanella species are ammonifiers, whereas only a few are denitrifiers. Genes for nitrate reductase (NAP and NAR) and nitrite reductase (NRF and NIR) are identified as multi-copies in the genomes of most shewanellae. The duo-NAP system (NAP-α and NAP-β) was probably derived through horizontal gene transfer and environmental adaptation, with NAP-α having apparent advantages over NAP-β with regard to nitrate scavenging. Moreover, environmental factors, such as carbon source, temperature and oxygen levels, may influence the regulation of the duo-NAP system of Shewanella via different mechanisms. Based on available evidence, a model for the evolutionary course of the anaerobic respiration system in Shewanella is proposed here (Figure 4). At the early point of their emergence, Shewanella species may have used CymA as a common electron transfer that served multiple pathways of respiration; afterward, the specific transport protein NapC was acquired via gene transfer, which provided competitive advantages to the host under stressed conditions.
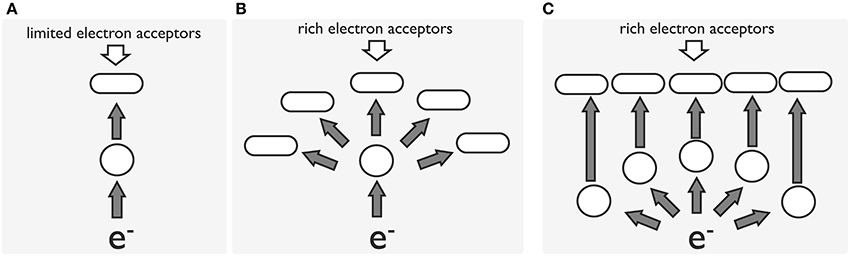
Figure 4. A model for the evolution of the electron transport pathway in Shewanellae. In all cases, e− represents an electron source, a blank cycle represents an electron-transporting protein, an oblong represents a terminal reductase, and a gray arrow represents electron flow. Initially, the transporting protein and terminal reductase functioned for a limited number of acceptors (A). As the environment changed, substrates accessible to the cell became enriched and more diverse, requiring the bacteria to develop a specialized system to utilize different types of electron acceptors (B,C). Model (C) is proposed to have advantage over models (A,B) with regard to electron transfer efficiency when rich electron acceptors are available.
Challenges and Future Work
Recently, DNAR was suggested to play an important role in N metabolism (Kraft et al., 2011), which has been long ignored in previous investigations, as it was generally assumed that most of the nitrate in nature is respired via denitrification. It would be important and challenging to quantify the ecological contributions, diversity, and controlling factors of different nitrate respiration processes (DNAR and denitrification) in different environments. Shewanellae are believed to be important players in environmental N cycling, though additional detail and quantitative evaluation on their contribution are still lacking. Possessing multiple genes in nitrogen metabolism pathways is believed to confer Shewanella with environmental adaptation flexibility and advantages, though these assumptions need to be tested in the future. In addition, the mechanisms by which the duo NAP system benefits the host need to be elucidated. Understanding the regulation and evolution of the NAP systems in Shewanella would help in the understanding of the adaptation and evolution of this environmentally important bacterial genus and their application in bioremediation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was support by National Natural Science Foundation of China (Grant No. 31290232, 41306129) and China Ocean Mineral Resources R & D Association (Grant No. DY125-15-T-04).
References
Beliaev, A. S., Thompson, D. K., Fields, M. W., Wu, L., Lies, D. P., Nealson, K. H., et al. (2002). Microarray transcription profiling of a Shewanella oneidensis etrA mutant. J. Bacteriol. 184, 4612–4616. doi: 10.1128/JB.184.16.4612-4614.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bergmann, D. J., Hooper, A. B., and Klotz, M. G. (2005). Structure and sequence conservation of hao cluster genes of autotrophic ammonia-oxidizing bacteria: evidence for their evolutionary history. Appl. Environ. Microbiol. 71, 5371–5382. doi: 10.1128/AEM.71.9.5371-5382.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Botsford, J. L., and Harman, J. G. (1992). Cyclic AMP in prokaryotes. Microbiol. Rev. 56, 100–122. doi: 10.1146/annurev.mi.28.100174.002033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brettar, I., Christen, R., and Höfle, M. G. (2002). Shewanella denitrificans sp. nov., a vigorously denitrifying bacterium isolated from the oxic-anoxic interface of the Gotland Deep in the central Baltic Sea. Int. J. Syst. Evol. Microbiol. 52, 2211–2217. doi: 10.1099/ijs.0.02255-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brondijk, T. H., Fiegen, D., Richardson, D. J., and Cole, J. A. (2002). Roles of NapF, NapG and NapH, subunits of the Escherichia coli periplasmic nitrate reductase, in ubiquinol oxidation. Mol. Microbiol. 44, 245–255. doi: 10.1046/j.1365-2958.2002.02875.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brondijk, T. H. C., Nilavongse, A., Filenko, N., Richardson, D. J., and Cole, J. A. (2004). NapGH components of the periplasmic nitrate reductase of Escherichia coli K-12: location, topology and physiological roles in quinol oxidation and redox balancing. Biochem. J. 379, 47–55. doi: 10.1042/BJ20031115
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Caro-Quintero, A., Deng, J., Auchtung, J., Brettar, I., Höfle, M. G., Klappenbach, J., et al. (2011). Unprecedented levels of horizontal gene transfer among spatially co-occurring Shewanella bacteria from the Baltic Sea. ISME J. 5, 131–140. doi: 10.1038/ismej.2010.93
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cartier, G., Lorieux, F., Allemand, F., Dreyfus, M., and Bizebard, T. (2010). Cold adaptation in DEAD-box proteins. Biochemistry 49, 2636–2646. doi: 10.1021/Bi902082d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, Y., Wang, F., Xu, J., Mehmood, M. A., and Xiao, X. (2011). Physiological and evolutionary studies of NAP systems in Shewanella piezotolerans WP3. ISME J. 5, 843–855. doi: 10.1038/ismej.2010.182
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Coby, A. J., and Picardal, F. W. (2005). Inhibition of NO3- and NO2- reduction by microbial Fe(III) reduction: evidence of a reaction between NO2- and cell surface-bound Fe2+. Appl. Environ. Microbiol. 71, 5267–5274. doi: 10.1128/AEM.71.9.5267-5274.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cole, J. (1996). Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol. Lett. 136, 1–11. doi: 10.1111/j.1574-6968.1996.tb08017.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cruz-García, C., Murray, A. E., Klappenbach, J. A., Stewart, V., and Tiedje, J. M. (2007). Respiratory nitrate ammonification by Shewanella oneidensis MR-1. J. Bacteriol. 189, 656–662. doi: 10.1128/JB.01194-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cruz-García, C., Murray, A. E., McCue, L. A., Romine, M. F., and Tiedje, J. M. (2011). Fnr (EtrA) acts as a fine-tuning regulator of anaerobic metabolism in Shewanella oneidensis MR-1. BMC Microbiol. 11:64. doi: 10.1186/1471-2180-11-64
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Darwin, A. J., and Stewart, V. (1995). Nitrate and nitrite regulation of the Fnr-dependent aeg-46.5 promoter of Escherichia coli K-12 is mediated by competition between homologous response regulators (NarL and NarP) for a common DNA-binding site. J. Mol. Biol. 251, 15–29. doi: 10.1006/jmbi.1995.0412
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fredrickson, J. K., Romine, M. F., Beliaev, A. S., Auchtung, J. M., Driscoll, M. E., Gardner, T. S., et al. (2008). Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6, 592–603. doi: 10.1038/nrmicro1947
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gao, H., Obraztova, A., Stewart, N., Popa, R., Fredrickson, J. K., Tiedje, J. M., et al. (2006). Shewanella loihica sp. nov., isolated from iron-rich microbial mats in the Pacific Ocean. Int. J. Syst. Evol. Microbiol. 56, 1911–1916. doi: 10.1099/Ijs.0.64354-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gao, H., Wang, X., Yang, Z., Palzkill, T., and Zhou, J. (2008). Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics 9:42. doi: 10.1186/1471-2164-9-42
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gao, H., Yang, Z. K., Reed, S. B., Romine, M. F., Nealson, K. H., Fredrickson, J. K., et al. (2009). Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME J. 3, 966–976. doi: 10.1038/ismej.2009.40
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gavira, M., Roldán, M. D., Castillo, F., and Moreno-Vivián, C. (2002). Regulation of nap gene expression and periplasmic nitrate reductase activity in the phototrophic bacterium Rhodobacter sphaeroides DSM158. J. Bacteriol. 184, 1693–1702. doi: 10.1128/JB.184.6.1693-1702.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gescher, J. S., Cordova, C. D., and Spormann, A. M. (2008). Dissimilatory iron reduction in Escherichia coli: identification of CymA of Shewanella oneidensis and NapC of E. coli as ferric reductases. Mol. Microbiol. 68, 706–719. doi: 10.1111/j.1365-2958.2008.06183.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gralnick, J., Vali, H., Lies, D., and Newman, D. (2006). Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. U.S.A. 103, 4669–4674. doi: 10.1073/Pnas.0505959103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huberts, D. H. E. W., and Van Der Klei, I. J. (2010). Moonlighting proteins: an intriguing mode of multitasking. Biochim. Biophys. Acta 1803, 520–525. doi: 10.1016/j.bbamcr.2010.01.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jackson, R. H., Cornish-Bowden, A., and Cole, J. A. (1981). Prosthetic groups of the NADH-dependent nitrite reductase from Escherichia coli K12. Biochem. J. 193, 861–867.
Jepson, B. J. N., Marietou, A., Mohan, S., Cole, J. A., Butler, C. S., and Richardson, D. J. (2006). Evolution of the soluble nitrate reductase: defining the monomeric periplasmic nitrate reductase subgroup. Biochem. Soc. Trans. 34, 122–126. doi: 10.1042/BST0340122
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jørgensen, B. B. (1982). Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature 296, 643–645. doi: 10.1038/296643a0
Konstantinidis, K. T., Serres, M. H., Romine, M. F., Rodrigues, J. L. M., Auchtung, J., McCue, L. A., et al. (2009). Comparative systems biology across an evolutionary gradient within the Shewanella genus. Proc. Natl. Acad. Sci. U.S.A. 106, 15909–15914. doi: 10.1073/pnas.0902000106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kraft, B., Strous, M., and Tegetmeyer, H. E. (2011). Microbial nitrate respiration – genes, enzymes and environmental distribution. J. Biotechnol. 155, 104–117. doi: 10.1016/j.jbiotec.2010.12.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maier, T. M., and Myers, C. R. (2001). Isolation and characterization of a Shewanella putrefaciens MR-1 electron transport regulator etrA mutant: reassessment of the role of EtrA. J. Bacteriol. 183, 4918–4926. doi: 10.1128/JB.183.16.4918-4926.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marietou, A., Richardson, D., Cole, J., and Mohan, S. (2005). Nitrate reduction by Desulfovibrio desulfuricans: a periplasmic nitrate reductase system that lacks NapB, but includes a unique tetraheme c-type cytochrome, NapM. FEMS Microbiol. Lett. 248, 217–225. doi: 10.1016/j.femsle.2005.05.042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Murphy, J. N., and Saltikov, C. W. (2007). The cymA gene, encoding a tetraheme c-type cytochrome, is required for arsenate respiration in Shewanella species. J. Bacteriol. 189, 2283–2290. doi: 10.1128/JB.01698-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Myers, C., and Myers, J. (1993). Role of menaquinone in the reduction of fumarate, nitrate, iron(III) and manganese(IV) by Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 114, 215–222. doi: 10.1111/j.1574-6968.1993.tb06576.x
Piette, F., Struvay, C., and Feller, G. (2011). The protein folding challenge in psychrophiles: facts and current issues. Environ. Microbiol. 13, 1924–1933. doi: 10.1111/j.1462-2920.2011.02436.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Potter, L. C., and Cole, J. A. (1999). Essential roles for the products of the napABCD genes, but not napFGH, in periplasmic nitrate reduction by Escherichia coli K-12. Biochem. J. 344, 69.
Richardson, D. J. (2000). Bacterial respiration: a flexible process for a changing environment. Microbiology 551–571.
Saffarini, D. A., and Nealson, K. H. (1993). Sequence and genetic characterization of etrA, an fnr analog that regulates anaerobic respiration in Shewanella putrefaciens MR-1. J. Bacteriol. 175, 7938–7944.
Saffarini, D. A., Schultz, R., and Beliaev, A. (2003). Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J. Bacteriol. 185, 3668–3671. doi: 10.1128/JB.185.12.3668-3671.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saltikov, C. W., Wildman, R. A., and Newman, D. K. (2005). Expression dynamics of arsenic respiration and detoxification in Shewanella sp. strain ANA-3. J. Bacteriol. 187, 7390–7396. doi: 10.1128/JB.187.21.7390-7396.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schwalb, C., Chapman, S., and Reid, G. (2003). The tetraheme cytochrome CymA is required for anaerobic respiration with dimethyl sulfoxide and nitrite in Shewanella oneidensis. Biochemistry 42, 9491–9497. doi: 10.1021/Bi034456f
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sears, H. J., Sawers, G., Berks, B. C., Ferguson, S. J., and Richardson, D. J. (2000). Control of periplasmic nitrate reductase gene expression (napEDABC) from Paracoccus pantotrophus in response to oxygen and carbon substrates. Microbiology 146, 2977–2985.
Sharma, S., Cavallaro, G., and Rosato, A. (2010). A systematic investigation of multiheme c-type cytochromes in prokaryotes. J. Biol. Inorg. Chem. 15, 559–571. doi: 10.1007/s00775-010-0623-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shirodkar, S., Reed, S., Romine, M., and Saffarini, D. (2011). The octahaem SirA catalyses dissimilatory sulfite reduction in Shewanella oneidensis MR-1. Environ. Microbiol. 13, 108–115. doi: 10.1111/j.1462-2920.2010.02313.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Simon, J. (2002). Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol. Rev. 26, 285–309. doi: 10.1111/j.1574-6976.2002.tb00616.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Simon, J., and Kern, M. (2008). Quinone-reactive proteins devoid of haem b form widespread membrane-bound electron transport modules in bacterial respiration. Biochem. Soc. Trans. 36, 1011–1016. doi: 10.1042/BST0361011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Simpson, P. J. L., McKinzie, A. A., and Codd, R. (2010a). Resolution of two native monomeric 90kDa nitrate reductase active proteins from Shewanella gelidimarina and the sequence of two napA genes. Biochem. Biophys. Res. Commun. 398, 13–18. doi: 10.1016/j.bbrc.2010.06.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Simpson, P. J. L., Richardson, D. J., and Codd, R. (2010b). The periplasmic nitrate reductase in Shewanella: the resolution, distribution and functional implications of two NAP isoforms, NapEDABC and NapDAGHB. Microbiology 156, 302–312. doi: 10.1099/mic.0.034421-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Soballe, B., and Poole, R. K. (1999). Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145, 1817–1830.
Stewart, V., Lu, Y., and Darwin, A. J. (2002). Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184, 1314–1323. doi: 10.1128/JB.184.5.1314-1323.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stolz, J., and Basu, P. (2002). Evolution of nitrate reductase: molecular and structural variations on a common function. Chembiochem 3, 198–206. doi: 10.1002/1439-7633(20020301)3:2/3<198::AID-CBIC198>3.0.CO;2-C
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Strous, M., and Jetten, M. S. (2004). Anaerobic oxidation of methane and ammonium. Annu. Rev. Microbiol. 58, 99–117. doi: 10.1146/annurev.micro.58.030603.123605
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vargas, M., Kashefi, K., Blunt-Harris, E., and Lovley, D. (1998). Microbiological evidence for Fe(III) reduction on early Earth. Nature 395, 65–67. doi: 10.1038/25720
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Venkateswaran, K., and Dollhopf, M. (1998). Shewanella amazonensis sp. nov., a novel metal-reducing facultative anaerobe from Amazonian shelf muds. Int. J. Syst. Bacteriol. 48, 965–972.
Vermeiren, J., Van De Wiele, T., Verstraete, W., Boeckx, P., and Boon, N. (2009). Nitric oxide production by the human intestinal microbiota by dissimilatory nitrate reduction to ammonium. J. Biomed. Biotechnol. 2009:284718. doi: 10.1155/2009/284718
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Viamajala, S., Peyton, B. M., Sani, R. K., Apel, W. A., and Petersen, J. N. (2004). Toxic effects of chromium(VI) on anaerobic and aerobic growth of Shewanella oneidensis MR-1. Biotechnol. Prog. 20, 87–95. doi: 10.1021/bp034131q
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wallenstein, M. D., Myrold, D. D., Firestone, M., and Voytek, M. (2006). Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol. Appl. 16, 2143–2152. doi: 10.1890/1051-0761(2006)016[2143:ECODCA]2.0.CO;2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, H., and Gunsalus, R. P. (2000). The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. J. Bacteriol. 182, 5813–5822. doi: 10.1128/JB.182.20.5813-5822.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, J., Manno, D., Beaulieu, C., Paquet, L., and Hawari, J. (2005). Shewanella sediminis sp. nov., a novel Na+-requiring and hexahydro-1,3,5-trinitro-1,3,5-triazine-degrading bacterium from marine sediment. Int. J. Syst. Evol. Microbiol. 55, 1511–1520. doi: 10.1099/ijs.0.63604-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Shewanella, NAP, nitrate
Citation: Chen Y and Wang F (2015) Insights on nitrate respiration by Shewanella. Front. Mar. Sci. 1:80. doi: 10.3389/fmars.2014.00080
Received: 14 August 2014; Accepted: 16 December 2014;
Published online: 13 January 2015.
Edited by:
Andrew Stanley Mount, Clemson University, USAReviewed by:
Stanley Chun Kwan Lau, The Hong Kong University of Science and Technology, ChinaMatthew James Neave, King Abdullah University of Science and Technology/Woods Hole Oceanographic Institution, Saudi Arabia
Copyright © 2015 Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengping Wang, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, 800 Dongchuan Road, Shanghai 200240, China e-mail: fengpingw@sjtu.edu.cn
†Present address: Ying Chen, Department of Biochemistry and Molecular Biology, Guilin Medical University, Guilin, China
 Ying Chen
Ying Chen Fengping Wang
Fengping Wang