Estimating Symbiont Abundances and Gill Surface Areas in Specimens of the Hydrothermal Vent Mussel Bathymodiolus puteoserpentis Maintained in Pressure Vessels
- 1Sorbonne Universités, UMR7208 Laboratoire Biologie des Organismes et Écosystèmes Aquatiques (UPMC, CNRS, MNHN, IRD, UCBN), Université Pierre et Marie Curie, Paris, France
- 2Institut Universitaire de France, Paris, France
The hydrothermal vent mussel Bathymodiolus puteoserpentis hosts gill-associated sulfur- and methane-oxidizing bacteria which sustain host nutrition and allow it to reach high densities at various sites along the northern Mid-Atlantic Ridge. Previous studies have demonstrated that in similar dual symbioses, relative abundances of each bacterial type could change following variations in symbiont substrate availabilities. In this study, pressurized recovery and incubations in pressure vessels were used to test whether B. puteoserpentis symbionts displayed similar behavior in the presence of symbiont substrates. The relative abundances of both types of symbionts were analyzed using fluorescence in situ hybridization (FISH) and group-specific gene copy numbers were assessed using qPCR. Specimens sampled using isobaric and non-isobaric recovery contained similar relative proportions (in surface coverage) of sulfur- and methane-oxidizing bacteria indicating that recovery type did not have impact on measured relative areas. Similarly, pressurized incubations with different substrates did lead to significant differences in the relative surface coverage of the two types of bacteria, although slight variations were measured with qPCR, suggesting changes in relative abundances of gene copy numbers but not in relative areas covered. Total gill surface areas and total bacterial numbers in specimens were estimated for the first time. Symbiont bearing-mussels display exchange surfaces about 20-fold higher than those found in similar-sized coastal mussels, and mean bacterial numbers of 2.5*1012 per specimen were estimated. This emphasizes that symbiotic mussels are a major reservoir of bacteria in vent ecosystems.
Introduction
The large mussels (Mytilidae: Bathymodiolinae) thriving at deep-sea hydrothermal vents and cold seeps owe their success to the symbiotic chemosynthetic bacteria they harbor in their gill tissues. These bacteria benefit from the oxic-anoxic interface to gain access to both electron donors and oxygen, and the derived energy is used to fix carbon from inorganic carbon or methane and ultimately ensure nutrition of the host (Fisher et al., 1993; Duperron, 2010). Dual symbioses involving the simultaneous presence of sulfur- and methane-oxidizing bacteria (SOX and MOX) within the same host cells are a pervasive feature of bathymodiolins from vents and seeps in the Atlantic Ocean (Distel et al., 1995; Fiala-Médioni et al., 2002). The flexibility of these environmentally-acquired associations theoretically allows the symbiotic system to adapt to the reported spatial and temporal heterogeneities of symbiont substrate availability, and contributes to the ecological success of bathymodiolins (Won et al., 2003a; Halary et al., 2008).
In the species Bathymodiolus azoricus, from Mid-Atlantic Ridge (MAR) vents, and B. aff. boomerang from seeps in the Gulf of Guinea, symbiont relative abundances were shown to reflect the composition of surrounding fluid at the time of sampling (Halary et al., 2008; Duperron et al., 2011). Recent in vivo experiments have demonstrated that even a relatively short exposure to sulfide or methane could promote a shift in the balance between symbiont types, in favor of the symbiont using that particular substrate (Kadar et al., 2005; Halary et al., 2008; Riou et al., 2008). Although most of these experiments were conducted at ambient pressure on B. azoricus from the shallow vent site Menez Gwen (800 m), a recent study employed isobaric recovery and pressurized vessels to perform exposure experiments on specimens from the 2300 m deep Rainbow site. Despite that only two specimens were used for each treatment, exposure to bicarbonate ions and to a combination of sulfur and bicarbonate caused a spectacular increase in the relative abundance of sulfur-oxidizers measured as their volume within bacteriocytes (Szafranski et al., 2015).
These in vivo studies emphasized the relevance of isobaric recovery and use of pressurized vessels for the investigation of deep-sea symbiotic systems (Shillito et al., 2008). However, they raised several important questions that need to be addressed. One is about the universality of symbiosis flexibility in dual symbiotic mussels. Some species occur only at depths greater than B. azoricus, for example B. puteoserpentis, which lives at MAR vents situated between 2900 and 3700 m (O'Mullan et al., 2001; Desbruyères et al., 2006; van der Heijden et al., 2012). For these species, whether symbiont abundances also reflect habitat characteristics and are as flexible, and even whether maintenance and similar exposure experiments are tractable using available tools remain unknown (Le Bris and Duperron, 2010). Another issue is with the quantification method. The aim of most studies to date was to identify and characterize symbionts or host features from a limited number of host specimens, so they did not quantify symbiont numbers in any way. Among the few studies which attempted quantification, the methods used were indirect. Indeed, due to the structural complexity of gill tissue, they could not rely on direct cell counts. They included measurements of bacterial-specific compounds, rRNA content, fractions of the total bacterial volume occupied by each symbiont type, or qPCR on symbiont-specific genes (Yamamoto et al., 2002; Duperron et al., 2007; Halary et al., 2008; Guezi et al., 2014). Such approaches however provide relative abundances at best. The limitations of the methods were not always discussed, and the various methods were never compared directly. Reliable estimation of total numbers of bacteria in symbiotic mussels is also lacking. This information would be of prime importance to evaluate the significance of symbiotic metazoans as reservoirs of bacteria in comparison to other deep-sea habitats such as seafloor sediment, water column, exposed surfaces, or subsurface sediment. Obtaining these estimates from mussels requires measuring several parameters that have been poorly investigated to date, including gill surface area and bacterial content. Not only would this data be important for scientists investigating symbiosis, but it would also contribute to a better understanding of deep-sea microbial ecology.
In this study, specimens of B. puteoserpentis from the Snake Pit vent site were investigated. B. puteoserpentis is a sister-species to the well-documented B. azoricus and shares the same symbionts based on their 16S rRNA gene sequence (Won et al., 2003b; Duperron et al., 2006; Faure et al., 2009). We thus tested two hypotheses. The first was that B. puteoserpentis from the 3500 meter-deep site displayed a higher relative abundance of sulfur- vs. methane-oxidizers compared to the data available for B. azoricus, as can be predicted based on the higher sulfide-to-methane ratio observed at Snake Pit compared to other MAR sites previously investigated (Halary et al., 2008; Le Bris and Duperron, 2010). The second was that symbionts displayed similar dynamics in both species when live specimens are exposed to the substrates used by bacteria, given that B. puteoserpentis habitats are reported to be highly fluctuant (Zielinski et al., 2011). For this we performed isobaric recoveries and in vivo experiments in pressurized vessels and quantified symbiont relative abundances by fluorescence in situ hybridization (FISH) and image analysis (Halary et al., 2008), and by qPCR. In an attempt to bridge the gap between focused symbiosis case studies and more quantitative ecological investigations, we also estimated total gill surface area and bacterial numbers in individual specimens. Results represent a first step toward integrating the dynamics of mussel symbioses into the broader picture of vent ecosystem functioning, and provides a basis for the future evaluation of potential impacts of deep-sea exploitation on endemic symbiotic metazoans (Moskvitch, 2014).
Materials and Methods
Specimen Recovery
Specimens of B. puteoserpentis with shell lengths between 41 and 125 mm (mean: 80.8 mm) were sampled during the BICOSE cruise aboard RV Pourquoi Pas? using ROV Victor 6000 (2014, chief scientists: MA. Cambon-Bonavita; M. Zbinden). Specimens were sampled on the hydrothermal vent site Snake Pit, close to the “Elan” marker (23°22′54″N, 44°55′48″W, 3520 m depth). Five specimens (specimens 101–105) collected during dive 1 were stored in insulated closed boxes and surfaced within 2 h of sampling (non-isobaric recovery). Seventeen and ten mussels were sampled using the PERISCOP pressure-maintaining device (Shillito et al., 2008) during dives 2 and 5 (specimens 201–217 and 501–511, respectively, isobaric recovery, Table 1) and recovered 70 min and 100 min respectively after the PERISCOP had been closed and released from the bottom. When the PERISCOP reached the ship's deck, the data from the pressure recorder (SP2T4000, NKE Instruments, Hennebont, France) were downloaded before pressure was released in order to proceed with further experiments (see below).
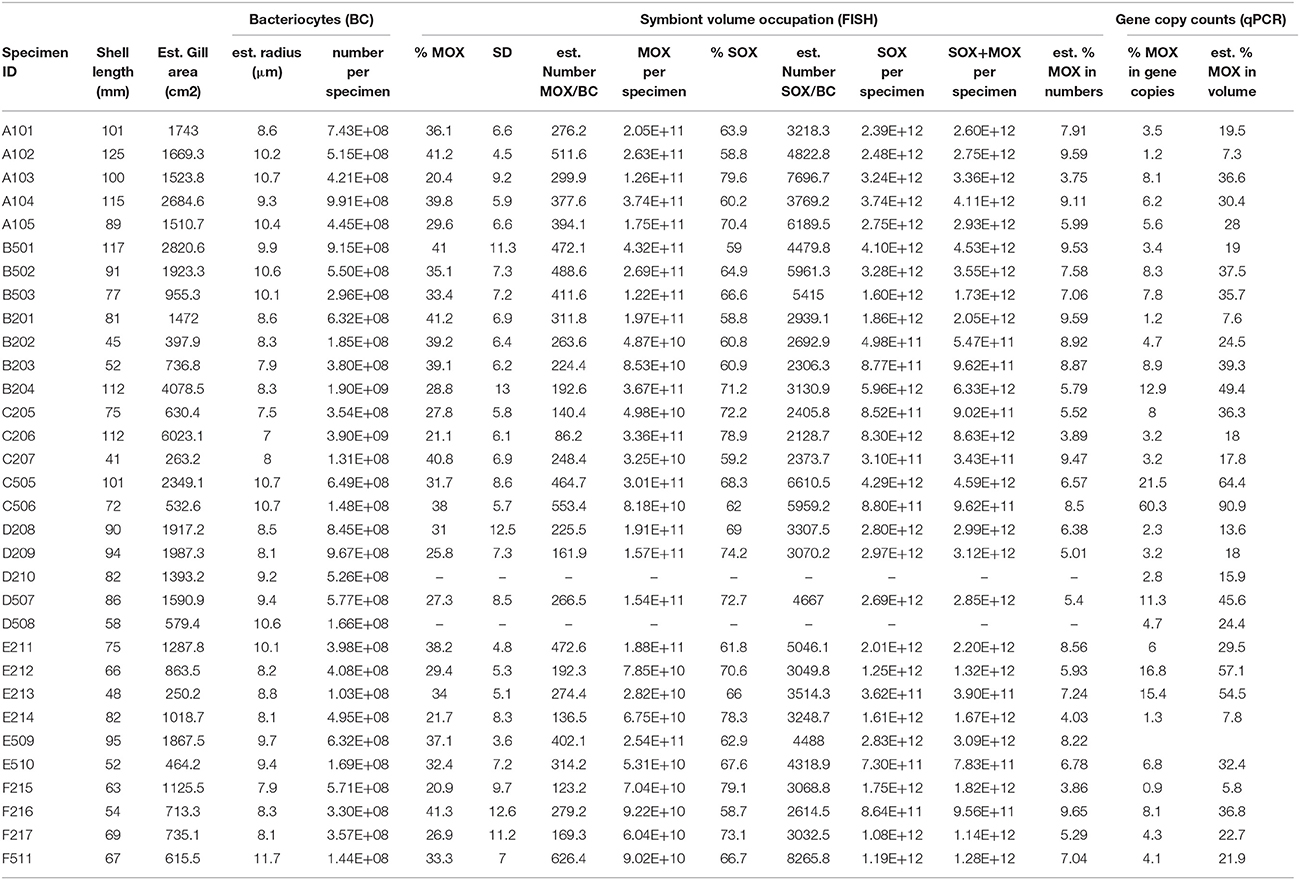
Table 1. Specimens from this study, their shell length, estimated gill area, bacteriocyte radius and number, symbiont area occupation as percentage of the total area occupied by bacteria on 2D sections, standard deviation (SD), estimated numbers of symbionts per bacteriocyte and per whole specimen, and estimated percentages of MOX based on gene copies and derived volumes (percentages of SOX correspond to (100-%MOX)%).
Live Experiments and Sample Fixation
All five specimens from non-isobaric recovery (treatment A, Table 1) and seven from isobaric recovery (treatment B) were dissected and fixed immediately upon recovery onboard (see below). Other specimens from isobaric recoveries were transferred to impermeable 1L Nalgene bottles containing seawater (treatment C, five specimens), seawater saturated with methane (treatment D, five specimens), seawater containing 5.9 μM (treatment E, six specimens), and seawater with 5.9 μM and 36.4 μM Na2S (treatment F, four specimens). Chosen concentrations of sulfide and bicarbonate are within the range of concentrations reported in the vicinity of mussels at MAR vent sites, while methane is almost absent in end-member fluids at Snake Pit, but present at other MAR vents (Le Bris and Duperron, 2010). Bottles were closed, placed into the pressure vessel IPOCAMP (Shillito et al., 2014), and incubated for 5 h at 30 Mpa (the maximum capacity of IPOCAMP) and 8°C (the lowest temperature we could achieve in the system given the constraints of operating on a ship). Specimens were then dissected and fixed within 5 min after the end of the experiment. Gill tissue was split. The most anterior half of the gill was fixed for FISH as described previously (Duperron, 2015), and another fragment for DNA.
Estimation of Gill Surface Area
Shell length (SL), height and width were measured using a caliper. To avoid tissue RNA and DNA degradation, gills had to be dissected, subsampled and fixed within minutes after opening the pressure vessels. For this reason, gill length could not be measured directly. To estimate gill length, we instead used published values and measurements made on fresh specimens (not used for the present study) which indicate that gill length represents ~80% of SL in B. puteoserpentis, with a highly significant correlation (Von Cosel et al., 1999; Von Cosel and Marshall, 2003). Numbers of filaments per millimeter gill length were measured on FISH-fixed gills along the antero-posterior axis on one side of the specimen (filament density Dfil). Two filaments were then individually extracted from the median part of the FISH-fixed gill of each individual and photographed under a SZX12 dissecting microscope, showing no sign of tissue shrinkage (Olympus, Japan). The surface (Sfil) of one side of individual “W-shaped” filaments was measured on calibrated images using ImageJ (Abramoff et al., 2004). Assuming that all gill filaments have similar surface (i.e., that the gill is rectangular in shape), the total gill surface area Sgill was then estimated using the following formula:
Although gills are roughly rectangular in shape, their anterior and posterior parts are more rounded, with filaments being shorter along their dorso-ventral axis, so this formula leads to a slight overestimation of the total gill surface, but certainly not by orders of magnitude.
Fluorescence In situ Hybridization and Relative Area Occupation by SOX and MOX Bacteria in the Gills
A fragment of FISH-fixed gill tissue was embedded in Steedman wax and 8 μm-thick transverse sections were obtained following the protocol in Duperron (2015). FISH was performed on these sections, as well as on gill filaments spread flat onto glass slides. Gill tissues were hybridized for 3 h at 46°C using the same probes and a formamide concentration of 30% as described previously (Halary et al., 2008; Szafranski et al., 2015). Three-channel RGB (red-green-blue) image stacks were acquired at 400x magnification under a BX61 epifluorescence microscope (Olympus Japan). The red, green and blue channels were used simultaneously and attributed to the Eub-338, ImedM-138, and BangT-642 probes, specific respectively for Eubacteria, methane- and sulfur-oxidizers (Table 2). Probe Non-338 was employed on separate sections as a negative control to confirm the absence of non-specific binding, and did not yield any signal (not shown). Image stacks representing volumes obtained from the 8 μm-thick sections were summed using the “Extended Depth of Field, Local Contrast Composite” function of ImagePro to produce the 2D images that were subsequently analyzed, as this makes following analyses faster without altering quality of estimations (Szafranski et al., 2015). Within each field of view, the numbers of pixels corresponding to the eubacterial (bacterial area), MOX, and SOX signals were computed from gill filament cross-section images using the SymbiontJ plugin implemented in ImageJ as previously described (Halary et al., 2008). The fraction of the bacterial area actually occupied by SOX and MOX was then determined across the entire field of view. For each specimen, percentages of MOX and standard deviation were calculated from a total of 4–10 images (one image consisting in a whole field of view) of good quality, from 4 to 5 distinct transverse gill sections. Image quality was verified by careful eye-examination of the areas actually detected by SymbiontJ.
Estimation of Gill Bacteriocyte Densities and Numbers of Gill-Associated Bacteria
For each specimen, five FISH images were acquired of the surface of whole-mount hybridized gill filaments. Bacteriocyte densities (DBC, the number of bacteriocytes per μm2 of gill surface area) were measured based on bacteriocytes counted using the “cell counter” module of ImageJ. To estimate bacterial number, mean bacteriocyte diameter was inferred by assuming a circle shape on flat images, and the volume of the corresponding sphere (VolBC) was calculated. Based on previous studies indicating bacteriocyte volume occupation of 55–58% in Thyasiridae (Dufour and Felbeck, 2006) and 52–55% in Vesicomyidae (Decker, pers. comm.), we assumed that bacteria roughly occupied 50% of the bacteriocyte volume in Mytilidae. The FISH-calculated fraction %SOX and %MOX of this volume corresponding to SOX and MOX allowed estimating the total volume occupied by each symbiont within a single bacteriocyte. Due to the high density of bacteria, diameters of SOX and MOX could not be measured directly on images. To estimate numbers of SOX and MOX per bacteriocyte, typical diameters of symbionts reported in previous studies were used (Fiala-Médioni et al., 2002). The volume occupied by SOX within a single bacteriocyte was thus divided by the volume of a typical spherical 0.8 μm diameter bacterium (VolSOX), and that occupied by MOX was divided by the volume of a spherical 1.5 μm diameter bacterium (VolMOX).
Total numbers nSOX and nMOXof SOX and MOX bacteria were then estimated by the following formulas:
qPCR Analysis of Bacterial Genes
DNA was extracted from frozen gill fragments using a QiaQuick Kit (Qiagen, USA), and re-suspended at 20 ng/μL. Four gene-specific primer sets were employed, targeting fragments of genes encoding host H3 histone, eubacterial 16S rRNA (i.e., both symbiont types), SOX 16S rRNA, and MOX 16S rRNA (Table 2). Primer specificity was tested by sequencing cloned PCR products. The qPCR was run in triplicate for each gene and mussel specimen, and any replicate with CT differing by more than 0.5 were discarded. A dilution series of an equimolar mixture of DNA from all specimens was used to evaluate amplification efficiency. Results for each of the three bacterial genes were normalized and expressed as “-folds” vs. host histone by the ΔCT method (Table S1). We compared the sum of SOX and MOX quantifications vs. that of eubacteria to detect potentially aberrant results. The effect of isobaric vs. non-isobaric recovery on SOX and MOX gene copy numbers was evaluated by calculating mean “-fold” abundance of specimens in treatment A vs. the mean obtained for isobaric recovery (B) by the ΔΔCT method. Similarly, “-fold” values of incubation treatments D, E, and F were normalized to that of specimens kept for 5 h in seawater (treatment C). In order to test the correlation between qPCR and FISH results, percentages of SOX and MOX vs. the sum of SOX and MOX in gene copy numbers were calculated. Corresponding percentages in volume occupied were computed assuming that all bacteria contained the same number of 16S rRNA gene copies, and using the aforementioned VolSOX and VolMOX.
Statistical Analyses
All statistical analyses were performed using the R software and the Vegan package (Oksanen et al., 2007; R Development Core Team, 2013).
Results
Sampling and Live Experiments
The lowest pressures experienced during the two PERISCOP recoveries were respectively 70.4 and 70.5% of the pressure at the seafloor where mussels were sampled. Only tightly closed specimens with unbroken shells were used for further treatments or fixation. A pressure of 30 Mpa was maintained during the incubations in IPOCAMP, which is lower than in situ pressure (about 35 MPa), and corresponds to the maximum working pressure of the instrument. Specimens were still closed tightly when recovered after 5 h, indicating that they were alive. Due to the constraint of processing several bottles within a single IPOCAMP, no video recording of mussel activity (including valve opening) could be obtained.
Gill Surface Areas and Bacteriocyte Densities
In the specimens investigated, gill filament densities ranged from 8 to 13 mm−1 (mean: 9.9) and bacteriocyte densities from 2340.8 to 6470.1 mm−2 (mean: 3986.5, Table 1; Figures 2A,B). Neither measurements correlated with shell length (t = −1.23, df = 30; p = 0.23 and t = −1.20, df = 28, p = 0.24). Estimated gill filament numbers per specimen ranged from 330.5 to 1173.5 (mean: 641.2), and gill surface areas ranged from 250.2 to 6023.1 cm2 (mean: 1491.4 cm2). Total estimated gill surface area correlated positively with shell length and best fit was obtained when applying a power function (R2 = 0.81, equation was Sgill = 0.0674 * SL2.24; Table 1, Figure 1).
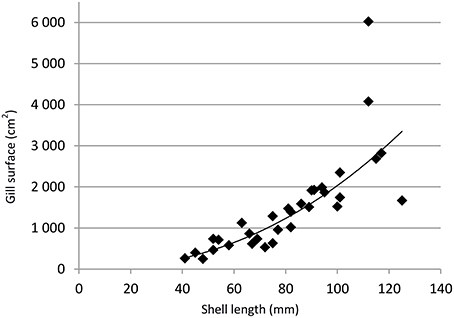
Figure 1. Total estimated gill surface (Sgill) area in cm2 vs. shell length (SL) in mm. The line represents to the best fitting power function (R2 = 0.81) and follows the equation Sgill = 0.0674.SL2.24.
Relative Areas Covered by SOX and MOX Symbionts and Effect of Recovery and Incubations
Images of FISH hybridizations on gill sections displayed clear non-ambiguous signals (Figures 2A,B, Figure S1). Bacteria labeled with the MOX-targeting probe were larger than those labeled with the SOX-targeting probe and displayed the donut-shape typical of methanotrophic symbionts reported in previous studies under high magnification (Duperron et al., 2006; Halary et al., 2008). No overlap existed between signals of the two probes. The signal from the Eubacteria-targeting probe fully overlapped with both MOX and SOX signals, suggesting very low abundance of other bacterial types. For this reason, percentages of SOX and MOX were calculated vs. the sum of SOX and MOX (SOX + MOX = 100%) and analyses were performed on %MOX. Percentages of MOX ranged between 20.4 and 41.3% of total bacterial area per field of view (Table 1). After Arcsine transformation (Halary et al., 2008), distributions of percentages did not follow normal distributions in at least some of the treatments (Shapiro-Wilk test, p < 0.05), and variances were unequal among treatments (Levene test, p < 0.05). A non-parametric Kruskal Wallis test revealed that differences existed among some treatments (KW test, p = 0.0005). Treatments were compared using paired Mann-Whitney-Wilcoxon (MWW) tests with a Bonferroni correction for multiple testing when needed. Percentages of MOX in specimens from non-isobaric (A, mean = 33.4%, Figure 3) and isobaric (B, mean = 36.8%) recoveries were not statistically different (MWW test, p = 0.15). Among the 5 h treatments, the only significant difference was between specimens exposed to methane (D, mean = 28.0%) and those exposed to bicarbonate (E, mean = 32.1%, MWW test with a Bonferroni correction, p = 0.03). A redundancy analysis investigating the contributions of treatment, individual, dive of origin, shell length, and bacteriocyte density was conducted. Inter-individual variation explained 34.8% of observed variations. Treatment explained 6.9% of the variance, followed by bacteriocyte density (3.6%). Shell length and dive had no significant effect (permutation test, n = 999 permutations).
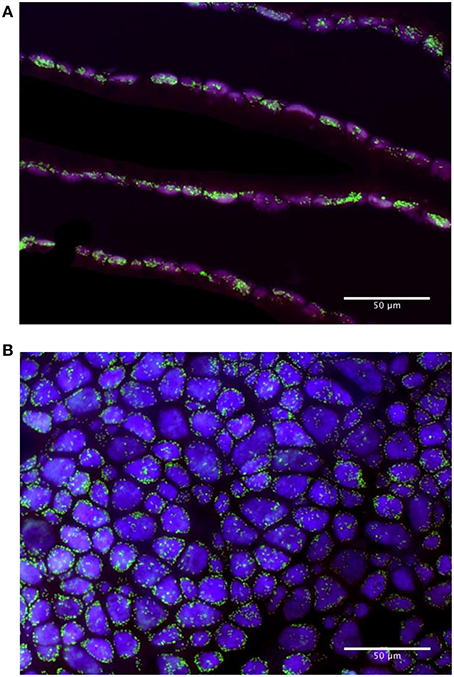
Figure 2. (A) Fluorescence in situ hybridization on transverse sections of gill filaments used for FISH estimation of symbiont relative abundances; (B) FISH on the apical surface of whole filaments spread on a slide, used for estimation of bacteriocyte densities. Sulfur-oxidizers, labeled with the BangT-642 probe appear in purple, methane-oxidizers labeled with probe ImedM-138 appear in green.
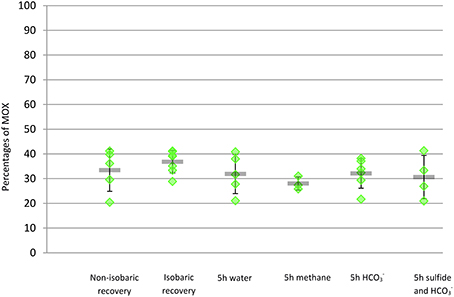
Figure 3. Percentage of area occupied by methane-oxidizers vs. total bacterial area (comprising methane- and sulfide-oxidizers) in transverse sections of gill filaments of individual mussel specimen from each treatment (green diamonds). Mean value and standard deviation within each treatment are provided (gray line and error bars).
Variations in SOX and MOX 16S rRNA-Encoding Gene Copy Numbers
Primer pairs used were specific based on sequences obtained after cloning PCR products. The first observation was that although intra-specimen replicate qPCRs yielded similar values, a high variability existed among the different mussel specimens (Table S1). As a result, the percentages of the total bacterial gene copy numbers represented by MOX genes varied widely (Table 1). Gene copy numbers were considered to be significantly different when “-fold” values between two treatments were below 0.5 or above 2, as usually considered in gene expression studies (Bustin et al., 2009). Using these thresholds, average SOX gene abundance in specimens from non-isobaric recovery (A) was higher than in specimens from isobaric recovery (B, 2.03 vs. 1), but MOX was not (1.35 vs. 1). Among incubation treatments, SOX gene copy numbers were higher in the methane exposure (by 2.0 fold) and in the bicarbonate and sulfur treatment (by 2.5 fold) than in the seawater treatment (Figure 4). For MOX, the significance threshold value was reached in the methane and bicarbonate treatments, with a decrease in gene copy numbers compared to seawater (Figure 4). These results however need to be interpreted with great caution, as inter-individual variability was high, as shown in Figure 4. Volumes inferred based on percentages of MOX and SOX gene copy numbers estimated by qPCR did not correlate significantly with percentages measured with FISH in the same specimens (Table 1, Figure 5, t = 0.40; df = 27; p = 0.69).
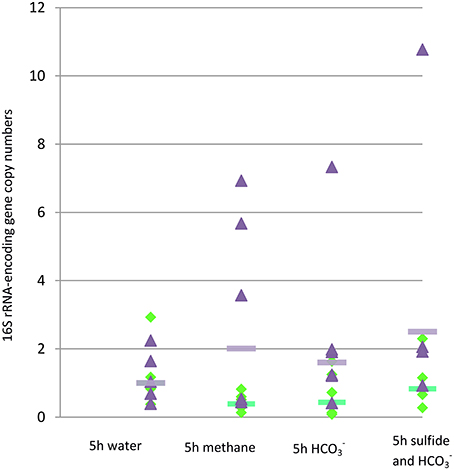
Figure 4. Number of copies of genes encoding 16S rRNA from methane- and sulfur-oxidizers (green diamonds and purple triangles, respectively) in gill tissue of individual mussel specimens from treatments C to F estimated by qPCR normalized vs. average value of the control 5 h exposure to seawater. Average values for MOX and SOX of the specimens from each treatment, normalized vs. average value of the control 5 h exposure to seawater, are represented by green and purple lines, respectively.
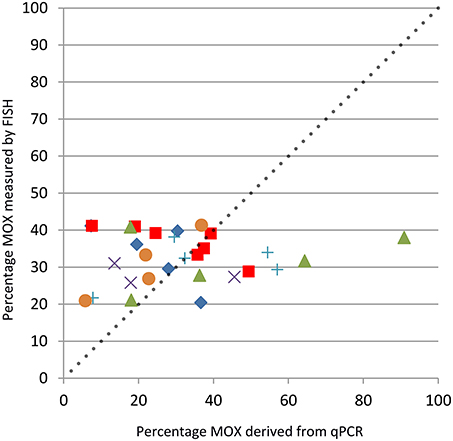
Figure 5. Percentage of the total bacterial area occupied by methane-oxidizers derived from qPCR results (x-axis) vs. percentages measured by FISH on gill filament sections (y-axis). Non-isobaric recovery, blue diamond; isobaric recovery, red square; 5 h water, green triangle; 5 h methane, violet “ × ”; 5 h , blue “+”; 5 h sulfide and , orange circle. The dotted line with slope 1 corresponds to the relationship that would be expected if every symbiont contained a single copy of their 16S rRNA-encoding gene, and thus each copy would correspond to a bacterium.
Estimation of Total Bacterial Numbers in Gills
Based on FISH-measured percentages of MOX and SOX, on a model assuming spherical bacteriocytes and bacteria, and a 50% bacteriocyte volume occupation by symbionts, bacteriocytes contained on average 312 MOX and 4126 SOX symbionts (7 and 93% of the symbiont population in cell numbers, respectively, Table 1). Any given specimen contained between 2.8 * 1010 and 4.3 * 1011 MOX (mean = 1.7 * 1011), and between 3.1 * 1011 and 8.3 * 1012 SOX symbionts (mean = 2.3 * 1012). The total estimated number of bacterial symbionts in a specimen ranged between 3.4 * 1011 and 8.6 * 1012 (mean = 2.5 * 1012). Total symbiont numbers positively correlated with shell length (nSYMB = 6 * 107 * SL2.39; R2 = 0.84; Figure 6).
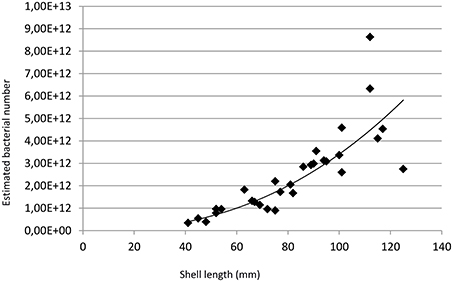
Figure 6. Total estimated bacterial numbers (nSYMB) vs. shell length (SL) in mm. The line represents to the best fitting power function (R2 = 0.84) and follows the equation nSYMB = 6.107* SL2.39.
Discussion
Distel et al. initially confirmed dual symbiosis involving sulfur and methane-oxidizing bacteria using 16S rRNA gene sequencing and FISH in B. puteoserpentis from Snake Pit. They formulated the hypothesis that dual symbiosis may be “providing the host with greater flexibility to exploit carbon and energy sources in the environment” (Distel et al., 1995: p. 199). Twenty years later we had the opportunity to perform isobaric recovery and in vivo experiments in pressurized vessels on this species, the deepest on which this type of approach has been applied to date (Shillito et al., 2008). In the present study, we measured relative area occupation of gill by symbionts based on 2D images, a method recently shown to yield proportions of MOX and SOX similar to those obtained by 3D FISH-based approaches (Szafranski et al., 2015). Relative area occupation by sulfur- and methane oxidizers is not statistically different between specimens from isobaric and non-isobaric recoveries. This indicates that the recovery type has limited impact on the relative area occupied by each type of bacteria as measured in this study. A similar lack of impact was previously observed on the sister species B. azoricus from the shallower Menez Gwen and Rainbow vent sites situated at 850 and 2300 meter depth, respectively, but our study extends this result to an even deeper site (3500 m) (Szafranski et al., 2015). MOX 16S rRNA gene copy numbers estimated by qPCR were also comparable between the two recovery methods, although SOX gene copy number was higher in non-isobaric recovery, again very close to the significance limit (2.03-fold). The fact that gene copy numbers and areas are unaffected by non-isobaric recovery is probably due to the stability of DNA and of bacterial cell structure in general. However, this does not preclude major potential effect on physiological and functional traits which were not specifically investigated here, and isobaric recovery is certainly to be recommended for further studies on these topics, and this is why we used isobaric-recovery specimens for in vivo experiments. Of course, recovery from even greater depths might involve significant changes in qPCR and proportion areal coverage data.
Symbiont Relative Abundances and Relation with Hydrothermal Fluid Composition on the MAR
Methane-oxidizers account for 36.8% of the bacterial area in specimens from the isobaric recovery. This is less than the recently investigated PERISCOP-recovered B. azoricus specimens from the Menez Gwen and Rainbow vent sites, which displayed 44.3 and 56.3% MOX, respectively (Szafranski et al., 2015). This is also less than values of 39.4 and 53.1% reported in other previous works using a slightly different approach (Halary et al., 2008; Le Bris and Duperron, 2010). Available data in other previous studies on the chemical composition of end-member hydrothermal fluids at Snake Pit indicate that reduced sulfur concentrations are higher than reported at the two other sites. They range between 2.7 and 6.1 mmol.l−1, compared to less than 1.5 and 2.5 mmol.l−1, respectively. Methane is on the other hand below 0.06 mmol.l−1 at Snake Pit, while it is present at Menez Gwen and Rainbow (1.35–2.63 mmol.l−1) (Desbruyères et al., 2000; Le Bris and Duperron, 2010). Overall, the fluid at Snake Pit is thus higher in sulfur and much lower in methane than the two others. It explains the higher relative abundance of SOX in gills of mussels compared to B. azoricus, and one can even wonder where the MOX obtain their methane. It must be noted that another closely-related mussel, namely B. aff. boomerang (Lorion et al., 2013), is reported to harbor up to 88.4% methanotrophs in volume at a methane-rich cold seep site in the Gulf of Guinea, indicating that dual symbioses can adapt to a broad range of substrate availabilities (Duperron et al., 2011).
The Limited Impact of Exposure to Symbiont Substrates and its Potential Causes
Depending on the incubation treatment, methane-oxidizers occupied on average between 28.0 and 32.1% of the total bacterial area (Figure 3). The only treatment that was different from the others was the methane exposure in which, paradoxically, methanotrophs displayed a significantly lower area occupation (based on three specimens only). In similar experiments and using the same FISH-based approach, the proportion of SOX was shown to increase from 38.5 to 90.1% in B. azoricus specimens from Rainbow upon 5 h-exposure to bicarbonate and sulfur, and to 76.0% with bicarbonate alone (Szafranski et al., 2015). No methane experiment was done in this previous study. Other exposure experiments at atmospheric pressure, including some with methane, also triggered marked changes in symbiont volume occupations (Kadar et al., 2005; Halary et al., 2008; Riou et al., 2008). Herein, treatments explained <7% of the variance observed among specimens, indicating a very limited effect on symbiont area occupation compared to the previous study under pressure (68.5% in Szafranski et al., 2015). Results from qPCR indicate on the other hand that SOX gene copy numbers increased during exposure to bicarbonate and sulfur, suggesting their presence may have promoted genome replication. Unexpectedly, an increase was also recorded in the methane treatment. As observed in FISH, qPCR indicated a decrease in numbers of 16S rRNA gene copies of methanotrophs in the methane treatment. However, inter-individual variability was high in qPCR results and the trends showing an increase in SOX gene copy numbers are the consequence of a limited number of specimens within each of the treatments (1–3, see Figure 4), thus more specimens would be needed to confirm our results.
Overall, neither direct results from qPCR nor volume inferred based on qPCR results correlated significantly with area percentages observed using FISH (Figure 5). This is an interesting finding because these two methods have never been used in tandem before. It indicates that the dynamics of gene copy numbers, and thus genomes, and bacterial volume are not directly coupled. This is not unreasonable, as bacterial growth and division are not necessarily correlated, and correspond to distinct time frames. This is illustrated by a study on SOX symbionts of Lucinidae bivalves which revealed the existence of distinct groups of symbionts differing by the number of genome copies they harbored (Caro et al., 2007). Authors suggested that the host did prevent cell division in this case, but did not rule out the possibility that symbionts are preparing for division. It can be assumed that genomes respond differently to variations than the whole bacterial volume or area. Genome amplification without cytokinesis was also documented in Medicago-associated Rhozobium symbionts (Mergaert et al., 2006). This could explain why we find significant increase in SOX gene copy numbers without seeing corresponding increase in their area. In another study, symbiont gene expression patterns were suggested to respond faster to variations than symbiont abundances in B. puteoserpentis (Wendeberg et al., 2012). In the present case, one could hypothesize that symbionts start replicating their genome under favorable conditions in order to prepare for cell division. We may indeed be seeing the first step of a slower response to variations than what was observed in B. azoricus in at least some specimens. Yet the lack of clear-cut changes in symbiont areas is still intriguing given that environmental fluctuations are reported around B. puteoserpentis (most documented at the Logatchev site; Zielinski et al., 2011). Although it may be that B. puteoserpentis or its symbionts actually have a slower response, several experimental constraints might provide an alternative explanation. The transfer of PERISCOP-recovered mussels to IPOCAMP involved a short, yet brutal, depressurization. Furthermore, the incubation pressure (30 Mpa) did not match exactly that naturally experienced by the animals (35 Mpa). Finally, experiments in closed bottles may have led to oxygen limitation and thus limited the use of substrates by symbionts. Mussels were not dead as the muscles still kept valves strongly stuck together at the end of the experiments, however the stress associated with these events may have caused mussels to change their behavior. For example, they may have opened their valves for shorter periods than what would have happened in normal conditions, limiting exchanges with the surrounding water. This would have isolated symbionts from the substrates that were added to the water, and could explain the very limited variations we observe. The fact that B. puteoserpentis is a deeper-dwelling species compared to B. azoricus, and is never found shallower than 2900 m, suggests that it could be less tolerant to depressurization, and would need to be treated even more carefully (O'Mullan et al., 2001). Further work may benefit from instruments which allow isobaric recovery and subsequent transfer toward an incubator without decompression. Such devices were successfully employed with other deep vent fauna but cannot yet be used if several treatments have to be tested simultaneously (Ravaux et al., 2013).
Estimates of Gill Surface Area and Number Of Bacteriocytes and Bacteria in B. puteoserpentis
To our knowledge, this study provides the first estimates of total gill surface areas in Bathymodiolinae. On average, mussels from this study had a shell length of 80.8 mm, harbored 641 gill filaments (9.9 mm−1 gill length), and had a total gill surface area of 1491.4 cm2, ~2.4 times the surface of an A4 sheet. Adult Bathymodiolus gills are roughly rectangular in shape, with relatively constant dorso-ventral elongation of the filaments over most of the gills' antero-posterior axis. However, filaments from the most anterior and posterior regions are less elongated dorso-ventrally. Because our method assumed a rectangular shape for the gill, it thus overestimates gill surface, but certainly not by orders of magnitude. Gill surface areas in coastal non-symbiotic mussels were estimated to be around 25 cm2 for a 55 mm-long specimen (Petersen et al., 2004). Jones et al. using a method similar to ours, obtained total gill areas around 35 cm2 and 13.5 filaments per mm gill length for 65 mm-long Mytilus edulis (Jones et al., 1992). These areas are around 20 times lower than those reported here for mussels comparable in size. For example, the smallest specimen in our study (C207, Table 1) had a surface of 263.2 cm2. This difference is well above what could result from the aforementioned overestimation bias from our method, and clearly indicates that Bathymodiolus gills display much higher gill surface areas than the coastal non-symbiotic species. Gill filament densities being lower in B. puteoserpentis, the difference is thus linked to the elongation of the lateral zone of gill filaments, which considerably increases the surface of individual filaments compared with non-symbiotic mussels (authors pers. obs.). Even the gill surface areas of larger non-symbiotic bivalves are much lower those reported here. They reach up to 120 cm2 in large specimens of the pearl oyster Pinctada margaritifera (120 mm shell length; Pouvreau et al., 1999), and 237 cm2 in 20 cm-long Acesta excavata (Jarnegren and Altin, 2006). Few estimates of surface areas of symbiont-containing gills are available from chemosymbiotic bivalves. In Solemya velum, Scott estimated gill surfaces of 107 cm2.g−1 and 276 cm2.g−1 total and gill wet weight, respectively (Scott, 2005). With gills between 6 and 80 mg and two gills per specimen, total surface areas between 3.3 and 44.2 cm2 can be extrapolated in S. velum. Shell lengths of specimens were not reported in this study, but they usually reach only around 25 mm (http://eol.org). In this regard, Bathymodiolus mussels display unprecedented exchange surfaces for their size, even among chemosymbiotic bivalves, although other large chemosymbiotic bivalves such as vesicomyids may also have comparatively large gill surfaces. Estimation should be improved for example using total imaging of gills in dedicated mussels (i.e., specimens that do not need to be subsampled quickly as we did).
Gill surface area in B. puteoserpentis increased with shell length following a power law. If gill surface area did display isometric growth with shell length, a scaling exponent of 2 would be expected. The aforementioned A. excavata for example displays a scaling exponent of gill area vs. shell length of 1.94, close to this value (Jarnegren and Altin, 2006). The value of 2.24 reported herein indicates positive allometric growth in B. puteoserpentis, with gill area increasing faster than the square of shell length.
We estimated that each mussel contained between 1.0 * 108 and 3.9 * 109 bacteriocytes, and overall between 3.4 * 1011 and 8.6 * 1012 bacteria. Very few estimates of actual bacterial numbers are available for chemosymbiotic organisms, because reliable estimates are really hard to obtain from tissues with complex shapes. Here, we assumed that the whole surface of each gill filament was paved with bacteriocytes. Although most of the surface is indeed occupied by bacteriocytes, there are small areas that are not, such as the ciliary junctions or the ciliated area in the frontal zone of gill filaments. For this reason, bacteriocytes and bacterial numbers are probably slightly overestimated, yet comparable among specimens. Interestingly, estimates of 1010–1011 cells per gram gill tissue were reported based on Q-8 quinone measurements in Bathymodiolus sp. (possibly brevior) and Calyptogena laubieri. These are within an order of magnitude of our estimates, and not far from the highest bacterial densities recorded in some mats of photosynthetic or methanotrophic prokaryotes (1012 bacteria per mL; Amaral-Zettler et al., 2010). Recent results from the census of marine microbes provided typical estimates of 108–109 bacteria per liter seawater and 108–109 per gram marine surface sediment (Amaral-Zettler et al., 2010), which means that a single mussel may harbor more bacteria than 1000 liters seawater or 1 kg seafloor surface sediment. Around hydrothermal vents, 108–1010 bacteria per kg chimney (Harmsen et al., 1997; Takai et al., 2001) and 107–109 per liter in the hydrothermal plume (Cowen et al., 1999; Takai et al., 2004; Anderson et al., 2013) were reported. Although numbers including those reported herein are only rough estimates, they emphasize the significance of symbiotic mussels as bacterial habitats around hydrothermal vents. They confirm that symbionts are probably amongst the most abundant bacteria at vents which display high densities of Bathymodiolus mussels. Associated with host density data, these estimations open the possibility of integrating chemosymbiotic organisms into the energy budget of hydrothermal vents. This would help bridge the gap between basic symbiosis characterization and evaluation of their ecological impact. It may also be of interest in future evaluations of the impact of human mining activities.
Conclusions
B. puteoserpentis from Snake Pit harbors the same symbionts as the shallower sister-species B. azoricus from which it diverged 0.76 MYA (Duperron et al., 2006; Faure et al., 2009), with a higher relative abundance of sulfur-oxidizers than observed in the latter, probably due to differences in habitat chemistry. Its association displays signs of flexibility, but it may be that isobaric transfer from the recovery device to the incubation aquarium is mandatory to maintain mussels in good condition to study the holobiont's response to habitat variation. Future studies should employ appropriate equipment to test this hypothesis, and when possible monitor the chemical characteristics of the water around mussels during the recovery and the experiments. The gill surface per length is much higher than in other documented bivalves, with the consequence that symbiont abundances in mussel gills are high when compared to other bacterial habitats around vents. This clearly points to symbiotic metazoans as major reservoirs of bacterial numbers at hydrothermal vents. Future work should integrate these findings into ecological approaches to better understand the ecosystem as a whole.
Author Contributions
SD designed the study, analyzed the results and wrote the manuscript. AQ performed the FISH experiments and analyzed the results. KS sampled the mussels, performed the live experiments, and analyzed the results. NL performed the qPCR experiments and analyzed the results. BS sampled the mussels, performed live experiments, and supervised the work at sea. All authors read and agreed on the latest version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the captain, crew and pilots of RV Pourquoi Pas? and ROV Victor6000 as well as scientists onboard for their help during the cruise BICOSE, in particular chief scientist M-A Cambon-Bonavita. We also thank E. Roussel for his input. We thank the reviewers for constructive comments which helped improve the manuscript. The research leading to these results has received funding from UPMC, from the European Union Seventh Framework Programme (FP7/2007-2013) under the MIDAS project, grant agreement n° 603418, and through an excellence grant to SD from Institut Universitaire de France. KS was funded through a Ph.D. grant from the Marie Curie Actions Initial Training Network (ITN) SYMBIOMICS (contract number 264774). This study did not involve endangered species and no specific permit was required for sampling at the Snake Pit site.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fmars.2016.00016
Table S1. Specimens from this study, qPCR results obtained for SOX-, MOX-, and Eubacteria-specific target genes expressed as “-folds” vs. the host reference gene (histone). Each qPCR was run in triplicates.
Figure S1. Fluorescence in situ hybridization on transverse sections of gill filaments of Bathymodiolus puteoserpentis displaying probe-labeled bacterial symbionts. On the left: composite image displaying the overlapped signals from probes Eub-338 (FITC, blue), BangT-642 (Cy5, red, sulfur-oxidizers thus appear purple) and ImedM-138 (Cy3, green). The three black and white images display individual channels, from left to right: BangT-642, ImedM-138, and Eub-338. Scale bar is 10 μm.
References
Abramoff, M. D., Magelhaes, P. J., and Ram, S. J. (2004). Image processing with ImageJ. Biophotonics Int. 11, 36–42.
Amann, R., Binder, B. J., Olson, R. J., Chisholm, S. W., Devereux, R., and Stahl, D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analysing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925.
Amaral-Zettler, L., Artigas, L. F., Baross, J., Bharathi, P. A., Boetius, A., Chandramohan, D., et al. (2010). “A global census of marine microbes,” in Life in the World's Oceans, ed A. D. McIntyre (Aberdeen: Wiley-Blackwell), 221–245.
Anderson, R. E., Beltrán, M. T., Hallam, S. J., and Baross, J. A. (2013). Microbial community structure across fluid gradients in the Juan de Fuca Ridge hydrothermal system. FEMS Microbiol. Ecol. 83, 324–339. doi: 10.1111/j.1574-6941.2012.01478.x
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Kubista, M., Mueller, R., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Caro, A., Gros, O., Got, P., De Wit, R., and Trousselier, M. (2007). Characterization of the population of the sulfur-oxidizing symbiont of Codakia orbicularis (Bivalvia, Lucinidae) by single-cell analyses. Appl. Environ. Microbiol. 73, 2102–2109. doi: 10.1128/AEM.01683-06
Colgan, D. J., McLauchlan, A., Wilson, G. D. F., Livingston, S. P., Edgecombe, G. D., Macaranas, J., et al. (1998). Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Aust. J. Zool. 46, 419–437. doi: 10.1071/ZO98048
Cowen, J. P., Shackelford, R., McGee, D., Lam, P., Baker, E. T., and Olson, E. (1999). Microbial biomass in the hydrothermal plumes associated with the 1998 axial volcano eruption. Geophys. Res. Lett. 26, 3637–3640. doi: 10.1029/1999GL002343
Desbruyères, D., Almeida, A., Biscoito, M., Khripounoff, A., Le Bris, N., Sarradin, P. M., et al. (2000). A review of the distribution of hydrothermal vent communities along the northern Mid-Atlantic Ridge: dispersal vs. environmental controls. Hydrobiologia 440, 201–216. doi: 10.1023/A:1004175211848
Desbruyères, D., Segonzac, M., and Bright, M. (2006). Handbook of Deep-Sea Hydrothermal Vent Fauna. Denisia 18.
Distel, D. L., Lee, H. K. W., and Cavanaugh, C. M. (1995). Intracellular coexistence of methano- and thioautotrophic bacteria in a hydrothermal vent mussel. Proc. Natl. Acad. Sci. U.S.A. 92, 9598–9602. doi: 10.1073/pnas.92.21.9598
Dufour, S. C., and Felbeck, H. (2006). Symbiont abundance in thyasirids (Bivalvia) is related to particulate food and sulphide availability. Mar. Ecol. Prog. Ser. 320, 185–194. doi: 10.3354/meps320185
Duperron, S. (2010). “The diversity of deep-sea mussels and their bacterial symbioses,” in The Vent and Seep Biota, ed S. Kiel (Dordrecht: Springer), 137–167.
Duperron, S. (2015). “Characterization of bacterial symbionts in deep-sea fauna: protocols for sample conditioning, fluorescence in situ hybridization, and image analysis,” in Hydrocarbon and Lipid Microbiology Protocols, Springer Protocols Handbooks, eds T. J. McGenity, K. N. Timmis, and B. Nogales (Berlin; Heidelberg: Springer-Verlag).
Duperron, S., Bergin, C., Zielinski, F., McKiness, Z. P., DeChaine, E. G., Cavanaugh, C. M., et al. (2006). A dual symbiosis shared by two mussel species, Bathymodiolus azoricus and B. puteoserpentis (Bivalvia: Mytilidae), from hydrothemal vents along the northern Mid-Atlantic Ridge. Environ. Microbiol. 8, 1441–1447. doi: 10.1111/j.1462-2920.2006.01038.x
Duperron, S., Guezi, H., Gaudron, S. M., Pop Ristova, P., Wenzöfer, F., and Boetius, A. (2011). Relative abundances of methane- and sulphur-oxidizing symbionts in the gills of a cold seep mussel and link to their potential energy sources. Geobiology 9, 481–491. doi: 10.1111/j.1472-4669.2011.00300.x
Duperron, S., Halary, S., Lorion, J., Sibuet, M., and Gaill, F. (2008). Unexpected co occurence of 6 bacterial symbionts in the gill of the cold seep mussel Idas sp. (Bivalvia: Mytilidae). Environ. Microbiol. 10, 433–445. doi: 10.1111/j.1462-2920.2007.01465.x
Duperron, S., Nadalig, T., Caprais, J. C., Sibuet, M., FIala-Médioni, A., Amann, R., et al. (2005). Dual symbiosis in a Bathymodiolus mussel from a methane seep on the Gabon continental margin (South East Atlantic): 16S rRNA phylogeny and distribution of the symbionts in the gills. Appl. Environ. Microbiol. 71, 1694–1700. doi: 10.1128/AEM.71.4.1694-1700.2005
Duperron, S., Sibuet, M., MacGregor, B. J., Luypers, M. M., Fisher, C. R., and Dubilier, N. (2007). Diversity, relative abundance, and metabolic potential of bacterial endosymbionts in three Bathymodiolus mussels (Bivalvia: Mytilidae) from cold seeps in the Gulf of Mexico. Environ. Microbiol. 9, 1423–1438. doi: 10.1111/j.1462-2920.2007.01259.x
Faure, B., Jollivet, D., Tanguy, A., Bonhomme, F., and Bierne, N. (2009). Speciation in the deep sea: multi-locus analysis of divergence and gene flow between two hybridizing species of hydrothermal vent mussels. PLoS ONE 4:e6485. doi: 10.1371/journal.pone.0006485
Fiala-Médioni, A., McKiness, Z. P., Dando, P., Boulègue, J., Mariotti, A., Alayse-Danet, A. M., et al. (2002). Ultrastructural, biochemical and immunological characterisation of two populations of the Mytilid mussel Bathymodiolus azoricus from the Mid Atlantic Ridge: evidence for a dual symbiosis. Mar. Biol. 141, 1035–1043. doi: 10.1007/s00227-002-0903-9
Fisher, C. R., Brooks, J. M., Vodenichar, J. S., Zande, J. M., Childress, J. J., and Burke, R. A. (1993). The co-occurence of methanotrophic and chemoautotrophic sulfur oxydixing bacterial symbionts in a deep-sea mussel. Mar. Ecol. 14, 277–289. doi: 10.1111/j.1439-0485.1993.tb00001.x
Guezi, H., Boutet, I., Andersen, A. C., Lallier, F. H., and Tanguy, A. (2014). Comparative analysis of symbiont ratios and gene expression in natural populations of two Bathymodiolus mussel species. Symbiosis 63, 19–29. doi: 10.1007/s13199-014-0284-0
Halary, S., Riou, V., Gaill, F., Boudier, T., and Duperron, S. (2008). 3D FISH for the quantification of methane- and sulphur-oxidising endosymbionts in bacteriocytes of the hydrothermal vent mussel Bathymodiolus azoricus. ISME J. 2, 284–292. doi: 10.1038/ismej.2008.3
Harmsen, H. J. M., Prieur, D., and Jeanthon, C. (1997). Distribution of microorganisms in deep-sea hydrothermal vent chimneys investigated by whole-cell hybridization and enrichment culture of thermophilic subpopulations. Appl. Environ. Microbiol. 63, 2876–2883.
Jarnegren, J., and Altin, D. (2006). Filtration and respiration of the deep living bivalve Acesta excavata (J.C. Fabricius, 1779) (Bivalvia; Limidae). J. Exp. Mar. Biol. Ecol. 334, 122–129. doi: 10.1016/j.jembe.2006.01.014
Jones, H. D., Richards, O. G., and Southern, T. A. (1992). Gill dimensions, water pumping rate and body size in the mussel Mytilus edulis L. J. Exp. Mar. Biol. Ecol. 155, 213–237. doi: 10.1016/0022-0981(92)90064-H
Kadar, E., Bettencourt, R., Costa, V., Serrao Santos, R., Lobo-da-Cunha, A., and Dando, P. R. (2005). Experimentally induced endosymbiont loss and re-acquirement in the hydrothermal vent bivalve Bathymodiolus azoricus. J. Exp. Mar. Biol. Ecol. 318, 99–110. doi: 10.1016/j.jembe.2004.12.025
Le Bris, N., and Duperron, S. (2010). “Chemosynthetic communities and biogeochemical energy pathways along the MAR: the case of Bathymodiolus azoricus,” in Diversity of Hydrothermal Systems on Slow Spreading Ocean Ridges, eds P. A. Rona, C. W. Devey, J. Dyment, et al. (Washington, DC: American Geophysical Union), 409–429.
Lorion, J., Kiel, S., Faure, B., Kawato, M., Ho, S. Y. W., Marshall, B., et al. (2013). Adaptive radiation of chemosymbiotic deep-sea mussels. Proc. Biol. Sci. 280:20131243. doi: 10.1098/rspb.2013.2972
Mergaert, P., Uchiumi, T., Alunni, B., Evano, G. G., Cheron, A., Catrice, O., et al. (2006). Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 5230–5235. doi: 10.1073/pnas.0600912103
Moskvitch, K. (2014). MARINE SCIENCE Health check for deep-sea mining. Nature 512, 122–123. doi: 10.1038/512122a
O'Mullan, G. D., Maas, P. A. Y., Lutz, R. A., and Vrijenhoek, R. C. (2001). A hybrid zone between hydrothermal vent mussels (Bivalvia : Mytilidae) from the Mid-Atlantic Ridge. Mol. Ecol. 10, 2819–2831. doi: 10.1046/j.0962-1083.2001.01401.x
Oksanen, J., Kindt, R., Legendre, P., O'Hara, B., Stevens, M. H., Oksanen, M. J., et al. (2007). The Vegan Package. Community Ecol Package (Oulu).
Petersen, J. K., Bougrier, S., Smaal, A. C., Garen, P., Robert, S., Larsen, J. E. N., et al. (2004). Intercalibration of mussel Mytilus edulis clearance rate measurements. Mar. Ecol. Prog. Ser. 267, 187–194. doi: 10.3354/meps267187
Pouvreau, S., Jonquieres, G., and Buestel, D. (1999). Filtration by the pearl oyster, Pinctada margaritifera, under conditions of low seston load and small particle size in a tropical lagoon habitat. Aquaculture 176, 295–314. doi: 10.1016/S0044-8486(99)00102-7
Ravaux, J., Hamel, G., Zbinden, M., Tasiemski, A. A., Boutet, I., Léger, N., et al. (2013). Thermal limit for metazoan life in question: in vivo heat tolerance of the pompeii worm. PLoS ONE 8:e64074. doi: 10.1371/journal.pone.0064074
Riou, V., Halary, S., Duperron, S., Bouillon, S., Elskens, M., Bettencourt, R., et al. (2008). Influence of CH4 and H2S availability on symbiont distribution, carbon assimilation and transfer in the dual symbiotic vent mussel Bathymodiolus azoricus. Biogeosciences 5, 1681–1691. doi: 10.5194/bg-5-1681-2008
Scott, K. M. (2005). Allometry of gill weights, gill surface areas, and foot biomass d13C values of the chemoautotroph-bivalve symbiosis Solemya velum. Mar. Biol. 147, 935–941. doi: 10.1007/s00227-005-1630-9
Shillito, B., Gaill, F., and Ravaux, J. (2014). The ipocamp pressure incubator for deep-sea fauna. J. Mar. Sci. Technol. 22, 97–102. doi: 10.6119/JMST-013-0718-3
Shillito, B., Hamel, G., Duchi, C., Cottin, D., Sarrazin, J., Sarradin, P. M., et al. (2008). Live capture of megafauna from 2300 m depth, using a newly designed pressurized recovery device. Deep Sea Res. Part I Oceanogr. Res. Pap. 55, 881–889. doi: 10.1016/j.dsr.2008.03.010
Szafranski, K. M., Piquet, B., Shillito, B., Lallier, F. H., and Duperron, S. (2015). Relative abundances of methane- and sulfur-oxidizing symbionts in gills of the deep-sea hydrothermal vent mussel Bathymodiolus azoricus under pressure. Deep Sea Res. Part I Oceanogr. Res. Pap. 101, 7–13. doi: 10.1016/j.dsr.2015.03.003
Takai, K., Komatsu, T., Inagaki, F., and Horikoshi, K. (2001). Distribution of Archaea in a black smoker chimney structure. Appl. Environ. Microbiol. 67, 3618–3629. doi: 10.1128/AEM.67.8.3618-3629.2001
Takai, K., Oida, H., Suzuki, Y., Hirayama, H., Nakagawa, S., Nunoura, T., et al. (2004). Spatial distribution of marine crenarchaeota group I in the vicinity of deep-sea hydrothermal systems. Appl. Environ. Microbiol. 70, 2404–2413. doi: 10.1128/AEM.70.4.2404-2413.2004
van der Heijden, K., Petersen, J. M., Dubilier, N., and Borowski, C. (2012). Genetic connectivity between North and South mid-atlantic ridge chemosynthetic bivalves and their symbionts. PLoS ONE 7:e39994. doi: 10.1371/journal.pone.0039994
Von Cosel, R., Comtet, T., and Krylova, E. M. (1999). Bathymodiolus (Bivalvia: Mytilidae) from hydrothermal vents on the Azores Triple junction and the Logatchev hydrothermal field, Mid-Atlantic Ridge. Veliger 42, 218–248.
Von Cosel, R., and Marshall, B. A. (2003). Two new species of large mussels (Bivalvia: Mytilidae) from active submarine volcanoes and a cold seep off the eastern North Island of New Zealand, with description of a new genus. Nautilus 117, 31–46.
Wang, Y., and Qian, P.-Y. (2009). Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 4:e7401. doi: 10.1371/journal.pone.0007401
Wendeberg, A., Zielinski, F. U., Borowski, C., and Dubilier, N. (2012). Expression patterns of mRNAs for methanotrophy and thiotrophy in symbionts of the hydrothermal vent mussel Bathymodiolus puteoserpentis. ISME J. 6, 104–112. doi: 10.1038/ismej.2011.81
Won, Y. J., Hallam, S. J., O'Mullan, G. D., Pan, I. L., Buck, K. R., and Vrijenhoek, R. C. (2003a). Environmental acquisition of thiotrophic endosymbionts by deep-sea mussels of the genus Bathymodiolus. Appl. Environ. Microbiol. 69, 6785–6792. doi: 10.1128/AEM.69.11.6785-6792.2003
Won, Y. J., Hallam, S. J., O'Mullan, D., and Vrijenhoek, R. C. (2003b). Cytonuclear disequilibrium in a hybrid zone involving deep-sea hydrothermal vent mussels of the genus Bathymodiolus. Mol. Ecol. 12, 3185–3190. doi: 10.1046/j.1365-294X.2003.01974.x
Yamamoto, H., Fujikura, K., Hiraishi, A., Kato, K., and Maki, Y. (2002). Phylogenetic characterization and biomass estimation of bacterial endosymbionts associated with invertebrates dwelling in chemosynthetic communities of hydrothermal vents and cold seeps. Mar. Ecol. Prog. Ser. 245, 61–67. doi: 10.3354/meps245061
Zielinski, F. U., Gennerich, H.-H., Borowski, C., Wenzhöfer, F., and Dubilier, N. (2011). In situ measurements of hydrogen sulfide, oxygen, and temperature in diffuse fluids of an ultramafic-hosted hydrothermal vent field (Logatchev, 14 degrees 45′N, Mid-Atlantic Ridge): Implications for chemosymbiotic bathymodiolin mussels. Geochem. Geophys. Geosyst. 12, Q0AE04. doi: 10.1029/2011GC003632
Keywords: symbiosis, in vivo experiments, pressurized recovery, bivalve, Mid-Atlantic Ridge
Citation: Duperron S, Quiles A, Szafranski KM, Léger N and Shillito B (2016) Estimating Symbiont Abundances and Gill Surface Areas in Specimens of the Hydrothermal Vent Mussel Bathymodiolus puteoserpentis Maintained in Pressure Vessels. Front. Mar. Sci. 3:16. doi: 10.3389/fmars.2016.00016
Received: 02 November 2015; Accepted: 29 January 2016;
Published: 16 February 2016.
Edited by:
Joerg Graf, University of Connecticut, USAReviewed by:
Roxanne Beinart, Harvard University, USASuzanne Dufour, Memorial University of Newfoundland, Canada
Copyright © 2016 Duperron, Quiles, Szafranski, Léger and Shillito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sébastien Duperron, sebastien.duperron@upmc.fr
†Present Address: Kamil M. Szafranski, Unité d'Ecologie, Systématique et Evolution, Centre National de la Recherche Scientifique UMR 8079, Université Paris-Sud, Orsay, France
 Sébastien Duperron
Sébastien Duperron Adrien Quiles1
Adrien Quiles1  Kamil M. Szafranski
Kamil M. Szafranski