The Ferrojan Horse Hypothesis: Iron-Virus Interactions in the Ocean
- College of Marine Science, University of South Florida, St. Petersburg, FL, USA
Iron is an essential nutrient and the sub-nanomolar concentrations of iron in open ocean surface waters are often insufficient to support optimal biological activity. More than 99.9% of dissolved iron in these waters is bound to organic ligands, yet determining the identity of these ligands in seawater remains a major challenge. Among the potential dissolved organic ligands in the colloidal fraction captured between a 0.02 and a 0.2 μm filter persists an extremely abundant biological candidate: viruses, most of which are phages (viruses that infect bacteria). Recent work in non-marine model systems has revealed the presence of iron ions within the tails of diverse phages infecting Escherichia coli. Based on these findings and the presence of conserved protein motifs in marine phages, here we present several lines of evidence to support the hypothesis that phages are organic iron-binding ligands in the oceans. With average concentrations of 107 phages per milliliter surface seawater, we predict that phages could contain up to 0.7 pM iron, a value equivalent to as much as 70% of the colloidal fraction of organically complexed dissolved iron in the surface ocean. Additionally, the production and uptake of siderophores, a strategy that bacteria have developed for assimilating iron, renders cells vulnerable to phage infection due to the dual function of these cell surface receptors. Iron ions present in phage tails enable phages to exploit their bacterial host's iron-uptake mechanism via the “Ferrojan Horse Hypothesis” proposed herein, where the apparent gift of iron leads to cell lysis. Finally, if host iron stores are recycled during the assembly of progeny phages, as much as 14% of the cellular iron released into the water column upon lysis would already be incorporated into new phage tails. The potential role of phages as iron-binding ligands has significant implications for both oceanic trace metal biogeochemistry and marine phage-host interactions.
The Ferrojan Horse Hypothesis
Iron is an essential micronutrient for microbial life. To thrive in the ocean, where iron concentrations are astonishingly low (Gordon et al., 1982), bacteria have evolved specialized iron-acquisition strategies, such as the production of siderophores (Neilands, 1957; Sandy and Butler, 2009). Siderophores are exceedingly strong organic iron-binding ligands produced by bacteria to outcompete other ligands for the binding and uptake of iron via a specific “trap door” receptor on the cell surface (Pawelek et al., 2006; Sandy and Butler, 2009; Vraspir and Butler, 2009; Hider and Kong, 2010). In non-marine systems, it has long been demonstrated that bacteriophages (phages; viruses that infect bacteria) take advantage of their bacterial hosts' vulnerabilities by utilizing the siderophore-bound iron receptors on the bacterial cell surface membrane for infection, directly competing with siderophore-bound iron uptake (Neilands, 1979; Braun, 2009). These siderophore-bound iron receptors, collectively referred to as TonB-dependent receptors, are essential for bacterial iron acquisition (Luria and Delbruck, 1943; Braun et al., 1973; Hantke and Braun, 1975; Luckey et al., 1975; Braun and Endriss, 2007). Mutations in critical regions of these surface-exposed receptors are therefore selected against, providing an advantage to phages in the ongoing phage-host evolutionary arms race (Van Valen, 1973; Stern and Sorek, 2011; Samson et al., 2013).
A series of recent studies demonstrating the presence of iron ions in the tail proteins of several well-studied phages that infect enteric bacteria (Bartual et al., 2010; Figure 1) leads us to propose the “Ferrojan Horse Hypothesis” (Ferrum = Latin for iron; Figure 2). If marine phages, which are present at concentrations of ~10 million colloidal-sized (0.02–0.2 μm) particles per milliliter of surface seawater, contain similar amounts of iron, then a significant amount of the dissolved iron in the oceans could be complexed within phages. In this model, phages use the essential micronutrient iron in the manner of a Trojan horse: iron within the phage tail fibers is recognized by the host siderophore-bound iron receptor, enabling the phage to attach to the bacterial cell, puncture the cell membrane and inject its nucleic acid into the host for infection (Figure 2). Furthermore, we hypothesize that cellular iron stocks are recycled for incorporation into the tail fibers of new phage progeny during phage protein synthesis and assembly, which occurs inside the host cell prior to lysis (King, 1968; Chuprov-Netochin et al., 2010; Rohwer et al., 2014). Following cell lysis, host cell materials are remineralized via the viral shunt and new phages (containing a substantial fraction of the cellular iron) are released into the environment where they proceed to infect new hosts. Here we present the lines of evidence that support the plausibility for phages to act as organically complexed dissolved iron in the oceans and discuss the implications on oceanic biogeochemistry and phage-host interactions.
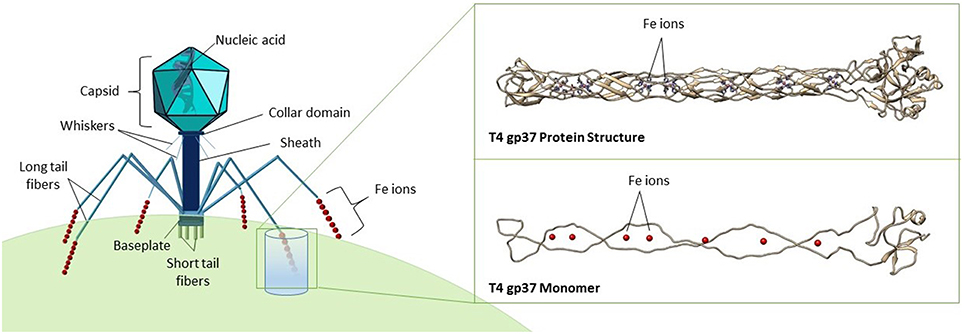
Figure 1. Structure of a typical bacteriophage belonging to the Myoviridae family. The expanded inset shows a model of the gp37 tail fiber protein of phage T4 (PDB ID code 2XGF), visualized in Chimera (Pettersen et al., 2004), and VMD (Humphrey et al., 1996). Seven iron ions (red spheres) are coordinated octahedrally by histidine residues, forming a trimer as shown in the top image, while the bottom image shows a gp37 monomer.
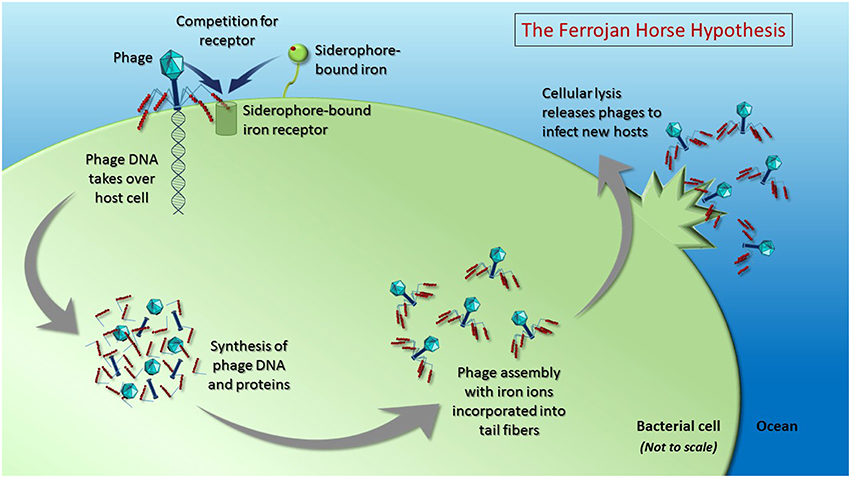
Figure 2. The Ferrojan Horse Hypothesis depicts the use of iron ions (red spheres) within phage tail fibers to compete with siderophore-bound iron for access to the same receptor on the surface of the host cell. Once a phage docks on the cell surface, it can pierce the cell membrane and inject its nucleic acid, which takes over the host cell, instructing the cell to produce phage DNA and proteins. Cellular mechanisms are used to fold the proteins and assemble them into progeny phages, with iron from within the cell incorporated into the tail fibers. Upon cell lysis, new phages are released to infect hosts using the iron that has been pre-packaged within their tail fibers. This proposed recycling of cellular iron during phage production results in a depletion of the amount of iron available for remineralization upon lysis (i.e., via the viral shunt). Figure is not drawn to scale.
Oceanic Iron Biogeochemistry
Bioavailable iron is scarce in open ocean surface waters due to its low solubility in oxic seawater at a pH of 8 and the vast distance from continental iron sources (Kuma et al., 1996; Johnson et al., 1997). This poses a challenge for marine microbes, since iron is essential for multiple biological functions, including the production of enzymes necessary for photosynthesis and respiration (Raven et al., 1999). As such, the iron quotas (Fe:C) for these cells are high relative to the requirements for any other trace metal (Bruland et al., 1991; Tortell et al., 1999; Morel and Price, 2003; Twining and Baines, 2013). In order to satisfy such a high iron requirement, marine bacteria have developed specialized mechanisms for iron acquisition. The synthesis and uptake of siderophores, which are high-affinity iron-chelating compounds, is a common strategy that bacteria use to secure iron from the marine environment (Butler, 2005). The process of organic complexation protects dissolved iron from precipitation (Kuma et al., 1996), stabilizing the pool of bioavailable dissolved iron so that bacteria may then take up the entire complex via a siderophore-bound iron receptor (Sandy and Butler, 2009; Shaked and Lis, 2012). Furthermore, some bacteria have developed strategies to acquire the iron bound to siderophores produced by other bacteria, called “xenosiderophores” (Matzanke et al., 1997; Sandy and Butler, 2009; Cornelis, 2010; Gauglitz et al., 2014).
Iron bioavailability serves as a bottom-up control on primary productivity in much of the modern surface ocean, making characterization of iron speciation of vital importance to understanding the carbon cycle and global climate (e.g., Martin et al., 1989). The concentration of dissolved iron in seawater is largely regulated by organic complexation, with >99.9% of dissolved iron complexed by organic iron-binding ligands as measured by competitive ligand exchange-adsorptive cathodic stripping voltammetry (CLE-ACSV: Gledhill and van den Berg, 1994; Rue and Bruland, 1995). Most recently, applications of CLE-ACSV to basin-scale surveys of the Atlantic Ocean as part of the GEOTRACES program have reported widespread organic complexation, with iron-binding ligands present in excess of dissolved iron concentrations through most of the Atlantic water column (Buck et al., 2015; Gerringa et al., 2015).
Numerous studies have sought to identify the organic iron-binding ligands measured by CLE-ACSV throughout the oceans. The strongest known iron-binding ligands are siderophores, which are typically small, soluble molecules (<10 kDa or <0.02 μm), though some marine siderophores are colloidal in size (>0.02 μm but <0.2 μm) due to long fatty acid chains presumably used to tether the ligand to the bacterial cell that produced it (Martinez et al., 2003). Siderophores are the best-characterized strong iron-binding ligand class, yet the concentration of siderophores measured in the Atlantic Ocean is only 3–20 pM (Mawji et al., 2008)—though this is likely an underestimate due to the challenges of isolating siderophores from seawater (Vraspir and Butler, 2009; Gledhill and Buck, 2012). Iron-binding humic substances have been increasingly recognized as an important component of a weaker iron-binding ligand pool in the oceans, particularly in the deep sea and in coastal environments (Laglera et al., 2007, 2011; Laglera and van den Berg, 2009). Similar to humic substances, other weaker iron-binding ligands most likely also originate from the remineralization of organic matter in the oceans (Hunter and Boyd, 2007; Boyd and Ellwood, 2010; Boyd et al., 2010). These may include exopolymeric substances, transparent exopolymers, nanofibrils, and even the biotoxin domoic acid (Figure 3; Rue and Bruland, 2001; Mancuso Nichols et al., 2004; Stolpe and Hassellov, 2010; Hassler et al., 2011).
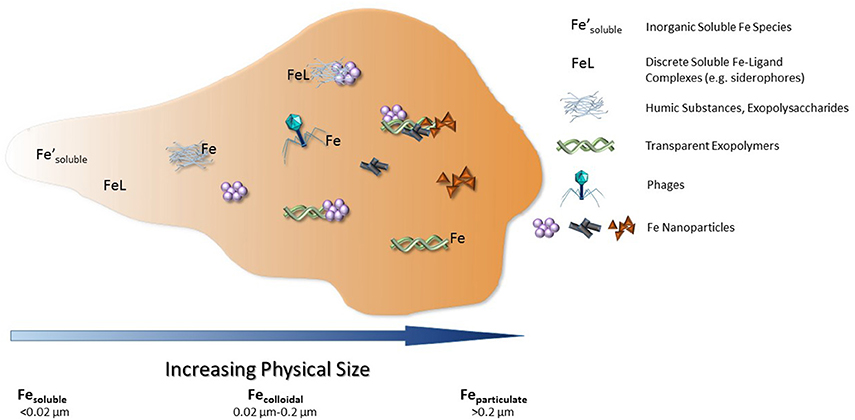
Figure 3. Current dissolved iron-binding ligands identified within seawater, with the addition of phages within the colloidal size range. Figure modified from Gledhill and Buck (2012).
To date, few studies have combined size fractionation with iron-binding ligand measurements (Cullen et al., 2006; Thuroczy et al., 2010; Fitzsimmons et al., 2015). These studies have all measured the largest excess of strong iron-binding ligands within the soluble (<0.02 μm) size fraction, consistent with the traditional siderophore model. However, strong iron-binding ligands were also detected within the colloidal fraction (0.02–0.2 μm), though at concentrations closer to that of dissolved iron with little to no excess ligands found in this size fraction (Cullen et al., 2006; Thuroczy et al., 2010; Fitzsimmons et al., 2015). Since measuring organic ligands that are already saturated with iron is particularly challenging by CLE-ACSV (e.g., Gledhill and Buck, 2012), the persistence of colloidal iron in these studies was suggested to result from recent dust deposition of inorganic colloidal iron or an unknown but organically-stabilized colloidal iron complex. Size fractionation studies of dust dissolution from aerosols in the Atlantic Ocean have shown that iron in dust proceeds from particulate to colloidal to soluble size fractions in the presence of strong organic iron-binding ligands like siderophores (Aguilar-Islas et al., 2010; Fishwick et al., 2014). In the absence of strong ligands, on the other hand, the iron remains in the colloidal size fraction, re-aggregates and is lost (Aguilar-Islas et al., 2010; Fishwick et al., 2014). The same may be expected of other particulate iron sources such as hydrothermal vents or shelf sediments, and the inorganic colloidal iron remains difficult to characterize. Recently, the ability of iron-oxidizing bacteria to produce iron oxides from hydrothermal vents has been proposed to contribute to the dissolved iron pool as a source of inorganic iron colloids (Emerson, 2016). While these studies highlight the complexity of iron speciation in seawater and the likelihood that inorganic iron colloids are an important dissolved iron species upon iron addition, the majority of the dissolved iron pool in the oceans is still thought to be organically complexed. Indeed, UV oxidation of seawater samples is commonly employed to release ligand-bound iron in seawater samples (Rue and Bruland, 1997; van den Berg, 2005; Biller and Bruland, 2012). Intriguingly, UV oxidation also reduces the infectivity of viruses and has been shown to destroy viral particles (Wilhelm et al., 1998); such treatment of phages in seawater would presumably also release any iron sequestered in phage tails.
The contribution of biologically sequestered iron to the organically complexed dissolved iron fraction has been largely ignored to date. The “biogenic” iron incorporated into the cells of phytoplankton and bacteria (typically >0.2 μm in diameter) can be a substantial portion of total (unfiltered) iron in the oceans, and when the fraction of dissolved iron is operationally defined as <0.45 μm, many marine bacteria may contribute to the dissolved iron pool (Tortell et al., 1999). However, to our knowledge, no studies have explored the potential for viruses, the most abundant biological entities in the ocean, to contribute to the iron-binding ligand pool in seawater, where they may account for a significant proportion of dissolved, and specifically colloidal (0.02–0.2 μm), organically complexed iron.
Marine Viruses
With average concentrations of 10 million particles per milliliter, viruses (the majority of which are phages that infect bacteria) are the most abundant biological entities in the oceans (Wommack and Colwell, 2000; Breitbart, 2012). The impact of viruses on ocean biogeochemistry is often evoked purely through the act of host lysis (Fuhrman, 1999; Suttle, 2007), but very few studies have considered the chemical contributions of the viral particles themselves. Recent modeling work suggests that marine virus particles contain a significant amount of macronutrients (carbon, nitrogen, and phosphorus), and that the stoichiometric mismatch (i.e., the relative difference in elemental contents) between phages and their bacterial hosts has the potential to drive the differential release of nutrients upon cell lysis (Jover et al., 2014). However, no studies have considered the potential impact of trace elements within the structure of marine viral particles. If viruses contain or interact with trace metals such as iron, their small size and sheer abundance in the oceans would translate into a major influence for biogeochemical cycling of this vital micronutrient. Since phages dominate marine viral communities and the field of marine viral ecology is relatively young compared to the century of knowledge of phage biology (Rohwer and Segall, 2015; Salmond and Fineran, 2015), here we present the lines of evidence from non-marine model systems that suggest critical linkages may exist between phages and oceanic iron cycling.
Evidence from Non-Marine Model Systems
Iron within Phages Contributes to Colloidal Organically Complexed Dissolved Iron
Phage particles are composed of nucleic acids contained within a protein shell (capsid) with or without a proteinaceous tail, and occasionally surrounded by a lipid membrane (Figure 1; Hershey and Chase, 1952; Rohwer et al., 2014). A recent X-ray crystallography study demonstrated the presence of seven iron ions within the receptor-binding tip of each of the six long tail fibers (gp37) of phage T4, one of the most extensively studied Escherichia coli phages (Bartual et al., 2010). These iron ions, assumed to be Fe2+, are coordinated through paired histidine residues (HxH domains) in the phage's long tail fiber protein (Figure 1; Bartual et al., 2010). The octahedral coordination of iron ions by the paired histidine residues in phage tails suggests that the iron ions are a stable part of the phage particle which would contribute to the colloidal organically complexed iron pool (i.e., FeL). This is consistent with recent evidence that colloidal and particulate Fe(II) is more abundant in the oceans than previously considered and that the majority of this Fe(II) is associated with organic carbon (von der Heyden et al., 2014). The HxH domains (PFAM 03335; Figure 4) are present in the tail fibers of several model phages that infect E. coli such as T4 (family Myoviridae) and lambda (family Siphoviridae), where they are repeated a variable number of times (Finn et al., 2014). From the marine environment, we used BLAST similarity to T4 gp37 to identify HxH domains in putative tail fibers from several cyanophages infecting Prochlorococcus (phage P-SSM2) and Synechococcus (phages ACG2014f_1 and KBS-2A), as well as in an uncultured phage with an unknown host from the Mediterranean Sea (uvMED; Figure 4). The conservation of HxH motifs known to facilitate iron binding in the putative tail fibers of several marine phages suggests that these pathways documented in well-studied phages infecting E. coli may also occur within diverse phages in the oceans. Although the receptors for the vast majority of marine phages are yet unknown (Breitbart, 2012; Silva et al., 2016), if some marine phages also utilize siderophore-bound iron receptors for host infection, this mechanism would have major implications for our current understanding of marine phage-host interactions in the iron-limited surface ocean.
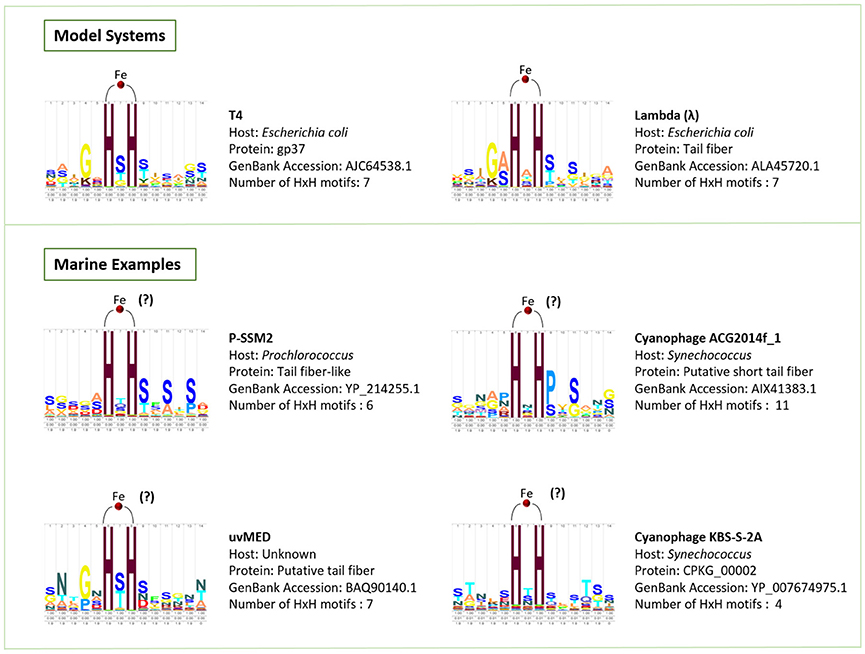
Figure 4. HMM logos created using Skylign (Wheeler et al., 2014) for the HxH motifs that bind iron ions in the model phages T4 and lambda, as well as conserved HxH motifs identified here in the putative tail fiber proteins of marine phages, which we hypothesize play a role in binding iron.
In addition to the tail fibers, conserved HxH domains within other phage structural proteins have also been shown to bind iron ions. For example, the baseplate assembly proteins of phages P2 and Φ92 (family Myoviridae), which form a membrane piercing spike, have recently been shown to contain iron ions coordinated octahedrally by paired histidine residues (Yamashita et al., 2011; Browning et al., 2012). Although the P2 and Φ92 proteins share conserved HxH motifs and the same function, the proteins that comprise the structure only share 19% amino acid identity. Since most phages with contractile tails (Myoviridae) utilize the same cell-puncturing mechanism (Leiman and Shneider, 2012), it is likely that distantly related Myoviridae may also contain iron-loaded tail spikes, even if these proteins cannot be identified through sequence homology. In addition, the tail tip protein (gpL) of phage N15, a member of the Siphoviridae family of phages with non-contractile tails, binds an iron-sulfur cluster (Tam et al., 2013). While the function of this cluster remains unknown, it has been speculated to play a role in stabilizing the tail tip protein or in conformational changes that this protein undergoes during assembly or DNA injection (Tam et al., 2013), implying that the incorporation of metals into phage tails may broadly be utilized by diverse phages for strengthening and stabilizing host-piercing proteins.
In order to estimate the proportion of colloidal dissolved iron that could be bound within the tail fibers of oceanic phages, we calculated the number of iron atoms present in phage tail fibers based on knowledge from E. coli phage T4. According to Bartual et al. (2010), seven iron ions are coordinated within each of the six long tail fibers of phage T4 (Leiman et al., 2010), equaling 42 iron ions per phage. Using the average of 107 viruses per milliliter of seawater (Wommack and Colwell, 2000; Breitbart, 2012; Parsons et al., 2012; Wommack et al., 2015) and assuming all viruses are tailed phages containing the same number of iron ions as T4, there are potentially a total of 4.2 × 108 iron ions bound to phage tail fibers in a milliliter of seawater, accounting for up to 0.7 pM dissolved iron.
However, several caveats accompany these assumptions, which may significantly affect the relative impact of phages on dissolved iron concentrations. First, the presence of iron has not been demonstrated in any marine phage tails to date, so the numbers used here are based on well-studied model systems and the detection of conserved HxH motifs in marine phages. Since strong evolutionary relationships exist between marine and non-marine phages, with many phage proteins conserved across ecosystems, it is likely that successful infection strategies documented in non-marine model systems will also be present in the oceans (Rohwer et al., 2000; Sullivan et al., 2005). Second, the number of iron ions per phage tail fiber is likely to vary, as demonstrated by the variable number of HxH motifs seen in marine phage sequences (Figure 4). While our preliminary analyses have identified many HxH motifs among metagenomic sequences of uncultured marine viral communities (e.g., uvMED in Figure 4 and additional data not shown), the limited database of marine phage genomes and limited levels of sequence similarity between phage tail proteins despite their conserved function makes a complete analysis of the prevalence and abundance of conserved iron-binding motifs in phage tail fibers challenging. Third, marine viral communities are extremely diverse, containing viruses with a myriad of particle sizes and morphologies that infect hosts across all domains of life (Breitbart et al., 2007; Suttle, 2007; Rohwer and Thurber, 2009; Breitbart, 2012; Wommack et al., 2015). While tailed phages are consistently present in the oceans, the proportion of the viral community that they comprise can vary widely (Brum et al., 2013) and still requires further evaluation. A quantitative transmission electron microscopy study performed across the world's oceans demonstrated that tailed phages comprised between 8 and 49% of the total viral communities sampled (Brum et al., 2013). Repeating the above calculations using the lower bound of 8% tailed phages, phage tail fibers would account for 0.06 pM dissolved iron. In addition, phage tails may not be the only structural proteins that interact with iron. For example, Daughney et al. (2004) showed that iron could also adsorb to phage capsids, creating the possibility that non-tailed phages (and perhaps eukaryotic or archaeal viruses) could also play a role in iron cycling. Since these experiments used iron oxide concentrations far exceeding typical marine concentrations, future work is needed to explore the relevance of these studies to natural systems and test the ability of marine phage capsids to bind iron.
With average dissolved iron concentrations for open ocean surface waters of 0.01–0.2 nM (Johnson et al., 1997; Mawji et al., 2015) and the colloidal fraction accounting for 10–25% of the total dissolved iron at the surface (Nishioka et al., 2001; Bergquist et al., 2007; Fitzsimmons and Boyle, 2014), the proportion of iron bound to the tail fibers of phages in 1 ml of seawater could thus account for ~5.6–70% of the colloidal fraction of dissolved iron, depending on the proportion of tailed phages. When considering that siderophore-bound iron concentrations in the Atlantic Ocean may comprise 0.2–4.6% (likely an underestimate) of the soluble fraction of dissolved iron (Mawji et al., 2008), phages potentially constitute a similarly substantial proportion of ligand-bound iron within the colloidal fraction.
Phage Utilization of Outer Membrane Siderophore-Bound Iron Receptors
In 1943, Luria and Delbruck launched the field of bacterial genetics with their seminal work identifying mutants of E. coli strain B that were resistant to infection by a specific phage (Luria and Delbruck, 1943). Although not named in the original study, the phage used was T1 and one of the resistant mutants was designated tonA. It wasn't until 1973 that researchers were able to ascribe the phage resistance to a single monomeric protein, TonA (named for T one resistance), which was also shown to bind phage T5 and colicin M (Braun et al., 1973). The exciting discovery in 1975 that TonA was capable of transporting iron bound to the siderophore ferrichrome (Hantke and Braun, 1975; Luckey et al., 1975) later prompted a change in the protein's name from TonA to FhuA (named for f erric hydroxamate uptake; Kadner et al., 1980). It is now known that the outer membrane protein FhuA functions as a receptor for numerous phages (T1, T5, Φ80, UC-1, H8; Lundrigan et al., 1983; Poon and Dhillon, 1987; Rabsch et al., 2007; Braun, 2009; Silva et al., 2016), as well as in the transport of ferrichrome, the peptide toxins colicin M, and microcin 25 (Braun et al., 1976), and the antibiotics albomycin and rifamycin CGP 4832 (which contain ferrichrome attached to an antimicrobial agent; Braun et al., 2001). Finally, the broad host range phage H8 achieves infection via adsorption to a different siderophore-bound iron receptor, the outer membrane porin protein FepA, suggesting that utilizing TonB-dependent siderophore-bound iron receptors (including FhuA and FepA) is a highly effective infection strategy (Rabsch et al., 2007).
Iron is so critical for growth that bacteria must rely upon specialized systems for the uptake of siderophore-bound iron, even though these strategies render them vulnerable to phage infection (Neilands, 1979) This places bacterial host cells in a precarious position because mutations in critical regions of these surface-exposed receptors that could provide resistance against phage infection may compromise their ability to acquire the iron they need for growth (Rabsch et al., 2007), thus providing an advantage to phages in the ongoing phage-host evolutionary arms race (Van Valen, 1973; Stern and Sorek, 2011; Samson et al., 2013). Additionally, the dual function of these cell surface receptors in siderophore-bound iron uptake and phage infection creates competition for receptor binding. In early in vitro studies, several phages (T5, T1, Φ80, UC-1) were shown to compete with ferrichrome for the FhuA receptor on the outer membrane of E. coli (Luckey et al., 1975; Wayne and Neilands, 1975; Hantke and Braun, 1978; Lundrigan et al., 1983; Bonhivers et al., 1996). Likewise, addition of iron bound to the siderophore enterobactin inhibits the binding of phage H8 to its receptor FepA (Rabsch et al., 2007).
Iron Incorporated into Phage Particles Prior to Host Lysis
Marine heterotrophic bacteria, which are highly abundant and contain significantly more iron per unit biomass than eukaryotic phytoplankton, have been estimated to account for up to half of the total iron in the marine environment (Tortell et al., 1996, 1999). While grazing of bacteria transfers cellular contents to higher trophic levels, phage infection plays an important role in regeneration of carbon, nutrients, and other elements through the viral shunt (Fuhrman, 1999; Wilhelm and Suttle, 1999). A growing body of literature has documented the critical roles of viral activity on iron cycling and bioavailability in the surface oceans (Gobler et al., 1997; Poorvin et al., 2004; Mioni et al., 2005). Experimental studies have demonstrated that organic iron-ligand complexes generated through phage lysis of infected bacterial cells are highly bioavailable and rapidly assimilated by heterotrophic bacteria (Poorvin et al., 2004; Mioni et al., 2005). In high nutrient, low chlorophyll (HNLC) systems, iron regeneration via phage lysis is critical for recycling organically complexed iron and speculated to regenerate enough dissolved iron to support some phytoplankton growth (Poorvin et al., 2004).
Although the source of the iron in phage tails has not yet been experimentally determined in any system, the fact that phage progeny fully assemble within the host cell prior to lysis, combined with the much higher iron concentrations inside bacterial cells compared to seawater, suggests that iron reserves in bacterial cells are the most likely source of the iron in phage tails (King, 1968; Tortell et al., 1999; Chuprov-Netochin et al., 2010; Rohwer et al., 2014). The incorporation of iron into phage tails assembled within the bacterial cell has significant implications for understanding the biogeochemistry of dissolved organic matter and trace metals released as a result of bacterial host cell lysis. An average marine heterotrophic bacterial cell has an iron quota of 7.5 μmol Fe per mol C (Tortell et al., 1996, 1999). Using an estimate of 20 fg C per cell (Ducklow, 2000), this translates to 7518 iron ions per heterotrophic bacterial cell. If marine phages contain similar amounts of iron in their tails as phage T4 (42 iron irons/phage; Bartual et al., 2010), and assuming an average burst size (number of progeny phages released through a lysis event) of 25 phages per infection (Wommack and Colwell, 2000; Parada et al., 2006), then ~14% of the bacterial iron previously assumed to be released into the water column as dissolved iron for remineralization would already be incorporated into new phage particles. The magnitude of this impact could vary widely as the calculations required several assumptions, including the iron quota of marine bacterial cells, which has been reported to range between 6.05 and 112 μmol Fe per mol C, depending on bioavailable iron concentrations (Brand, 1991; Tortell et al., 1996, 1999; Maldonado and Price, 1999; Boyd et al., 2010; Shire and Kustka, 2015), and the average burst size, which ranges from 10 to 500 for marine viruses (Wommack and Colwell, 2000; Parada et al., 2006). Nevertheless, if phage-bound iron originates from cellular iron reserves, this form of recycling and subsequent depletion of the iron available for remineralization upon lysis will be important to consider in modeling ocean biogeochemistry.
Implications and Future Work
Here we have detailed several lines of evidence supporting the Ferrojan Horse Hypothesis. The three facets of this hypothesis are: (1) Phages constitute important iron-binding ligands in the marine environment, where they can account for a substantial portion of the colloidal fraction of organically complexed dissolved iron; (2) Marine phages compete with siderophore-bound iron for uptake receptors on the bacterial cell surface, effectively acting as a Trojan horse where the apparent gift of iron leads to cell lysis; (3) The iron incorporated into marine phage tails originates from bacterial cellular iron reserves, which reduces the amount of iron available for remineralization upon lysis. All the calculations described in this manuscript are based on knowledge from the structure of E. coli phage T4 as a model system (Bartual et al., 2010) and combined with average published values for viral abundance and dissolved iron concentrations within the surface oceans; therefore, a great deal of work still needs to be performed to validate this hypothesis and quantify its impact on ocean biogeochemistry and phage-host interactions. In addition, it is still unknown how strongly phages bind iron, or how this binding strength compares to known organic ligands such as siderophores within the marine environment. Yet if marine phages incorporate iron into their tail fibers as a mechanism to gain access to bacterial hosts through receptors developed for iron acquisition and uptake, as seen in the model system of E. coli and T4, this infection strategy could answer fundamental questions regarding marine phage-host specificity and infection dynamics in the marine environment.
Notably, the competition for iron is not unique to the marine environment and the topics discussed here have broad implications for many ecosystems. For example, biomedical studies have demonstrated the importance of iron regulation within the human blood stream, with both pathogens and the human immune system manipulating iron as a tool for survival (Barasch and Mori, 2004; Drakesmith and Prentice, 2012; Barber and Elde, 2014). Analogous to the manner in which phages exploit bacterial siderophore-bound iron receptors, several mammalian viruses also utilize the cell-surface transferrin receptor for gaining entry to mammalian cells (Demogines et al., 2013). Additionally, although the mechanisms are largely unknown, recent phage therapy studies have demonstrated that the addition of iron-doped apatite nanoparticles to phages in solution enhances phage infection, resulting in an increase in the number of plaques forming on bacterial lawns by up to 128% (Andriolo et al., 2014). Given similarities in ionic strength and pH between seawater (Byrne and Breland, 1989) and blood (May et al., 1977), insights gained from human systems can aid in our understanding of the interactions between iron, bacteria, and viruses within the marine environment and vice versa.
Finally, although the Ferrojan Horse Hypothesis focuses on iron, the same concepts may be applicable to other essential nutrients found within the protein structure of phages, including metals like zinc and macronutrients like phosphorus. Zinc was found to be bound by histidines to one of the short tail fibers (gp12) of E. coli phage T4 in a manner similar to the iron ions in the T4 long tail fibers (Thomassen et al., 2003; Bartual et al., 2010). In addition, recent work by Jover et al. (2014) has estimated that viral particles can account for 0.01–24 nM of dissolved organic phosphorus in the oceans. Phages have a skewed C:N:P of ~20:7:1 (Jover et al., 2014), which is highly enriched in phosphorous and nitrogen compared to the Redfield ratio of cellular life in the oceans, 106:16:1 (Redfield, 1958). This stoichiometric mismatch suggests that the chemical composition of phages should be further investigated to determine elements for which they can make disproportionately large contributions to marine chemistry.
Future research will test the Ferrojan Horse Hypothesis by documenting the ability of phages to act as iron-binding ligands within the oceans and exploring the role of iron exploitation in governing phage-host interactions. In the meantime, however, the linkages made in this manuscript clearly demonstrate the importance of considering the potential contributions of phages, as well as other biological entities, to the concentrations and biogeochemical cycling of dissolved trace metals and macronutrients in the ocean.
Author Contributions
MB and KNB jointly conceived this hypothesis. CB took the lead in assembling evidence, compiling manuscript content, and creating graphics. All three authors contributed equally to writing the manuscript text and approved the manuscript for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Leon Hardy at the University of South Florida Saint Petersburg for help visualizing the structure of phage T4 gp37 presented in Figure 1 and Dr. Robert A. Edwards at San Diego State University for bioinformatics support. This work was supported by National Science Foundation grants from the Division of Integrative Organismal Systems (IOS-1456301) to MB and from the Division of Ocean Sciences (Chemical Oceanography, OCE-1446327) to KNB, as well as a donation from George H. Lorton. We thank three reviewers for their helpful contributions that improved this manuscript.
References
Aguilar-Islas, A. M., Wu, J., Rember, R., Johansen, A. M., and Shank, L. M. (2010). Dissolution of aerosol-derived iron in seawater: leach solution chemistry, aerosol type, and colloidal iron fraction. Mar. Chem. 120, 25–33. doi: 10.1016/j.marchem.2009.01.011
Andriolo, J. M., Hensleigh, R. M., McConnell, C. A., Pedulla, M., Hailer, K., Kasinath, R., et al. (2014). Iron-doped apatite nanoparticles for improvement of phage therapy. J. Vac. Sci. Technol. B 32:06FD01. doi: 10.1116/1.4894460
Barasch, J., and Mori, K. (2004). Cell biology - iron thievery. Nature 432, 811–813. doi: 10.1038/432811a
Barber, M. F., and Elde, N. C. (2014). Escape from bacterial iron piracy through rapid evolution of transferrin. Science 346, 1362–1366. doi: 10.1126/science.1259329
Bartual, S. G., Otero, J. M., Garcia-Doval, C., Llamas-Saiz, A. L., Kahn, R., Fox, G. C., et al. (2010). Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc. Natl. Acad. Sci. U.S.A. 107, 20287–20292. doi: 10.1073/pnas.1011218107
Bergquist, B. A., Wu, J., and Boyle, E. A. (2007). Variability in oceanic dissolved iron is dominated by the colloidal fraction. Geochim. Cosmochim. Acta 71, 2960–2974. doi: 10.1016/j.gca.2007.03.013
Biller, D. V., and Bruland, K. W. (2012). Analysis of Mn, Fe, Co, Ni, Cu, Zn, Cd, and Pb in seawater using the Nobias-chelate PA1 resin and magnetic sector inductively coupled plasma mass spectrometry (ICP-MS). Mar. Chem. 130, 12–20. doi: 10.1016/j.marchem.2011.12.001
Bonhivers, M., Ghazi, A., Boulanger, P., and Letellier, L. (1996). FhuA, a transporter of the Escherichia coli outer membrane, is converted into a channel upon binding of bacteriophage T5. EMBO J. 15, 1850–1856.
Boyd, P. W., and Ellwood, M. J. (2010). The biogeochemical cycle of iron in the ocean. Nat. Geosci. 3, 675–682. doi: 10.1038/ngeo964
Boyd, P. W., Ibisanmi, E., Sander, S. G., Hunter, K. A., and Jackson, G. A. (2010). Remineralization of upper ocean particles: implications for iron biogeochemistry. Limnol. Oceanogr. 55, 1271–1288. doi: 10.4319/lo.2010.55.3.1271
Brand, L. E. (1991). Minimum iron requirements of marine-phytoplankton and the implications for the biogeochemical control of new productuon. Limnol. Oceanogr. 36, 1756–1771.
Braun, V. (2009). FhuA (TonA), the career of a protein. J. Bacteriol. 191, 3431–3436. doi: 10.1128/JB.00106-09
Braun, V., Bos, C., Braun, M., and Killmann, H. (2001). Outer membrane channels and active transporters for the uptake of antibiotics. J. Infect. Dis. 183, S12–S16. doi: 10.1086/318840
Braun, V., and Endriss, F. (2007). Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals 20, 219–231. doi: 10.1007/s10534-006-9072-5
Braun, V., Hancock, R. E. W., Hantke, K., and Hartmann, A. (1976). Functional organization of outer membrane of Escherichia coli - phage and colicin receptors as components of iron uptake systems. J. Supramol. Struct. 5, 37–58. doi: 10.1002/jss.400050105
Braun, V., Schaller, K., and Wolff, H. (1973). Common receptor protein for phage T5 and colicin M in outer membrane of Escherichia coli B. Biochim. Biophys. Acta 323, 87–97. doi: 10.1016/0005-2736(73)90433-1
Breitbart, M. (2012). Marine viruses: truth or dare. Annu. Rev. Mar. Sci. 4, 425–448. doi: 10.1146/annurev-marine-120709-142805
Breitbart, M., Thompson, L. R., Suttle, C. A., and Sullivan, M. B. (2007). Exploring the vast diversity of marine viruses. Oceanography 20, 135–139. doi: 10.5670/oceanog.2007.58
Browning, C., Shneider, M. M., Bowman, V. D., Schwarzer, D., and Leiman, P. G. (2012). Phage pierces the host cell membrane with the iron-loaded spike. Structure 20, 326–339. doi: 10.1016/j.str.2011.12.009
Bruland, K. W., Donat, J. R., and Hutchins, D. A. (1991). Interactive influences of bioactive trace-metals on biological production in oceanic waters. Limnol. Oceanogr. 36, 1555–1577. doi: 10.4319/lo.1991.36.8.1555
Brum, J. R., Schenck, R. O., and Sullivan, M. B. (2013). Global morphological analysis of marine viruses shows minimal regional variation and dominance of non-tailed viruses. ISME J. 7, 1738–1751. doi: 10.1038/ismej.2013.67
Buck, K. N., Sohst, B., and Sedwick, P. N. (2015). The organic complexation of dissolved iron along the US GEOTRACES (GA03) North Atlantic Section. Deep-Sea Res. Part II-Top. Stud. Oceanogr. 116, 152–165. doi: 10.1016/j.dsr2.2014.11.016
Butler, A. (2005). Marine siderophores and microbial iron mobilization. Biometals 18, 369–374. doi: 10.1007/s10534-005-3711-0
Byrne, R. H., and Breland, J. A. (1989). High- precision multiwavelength pH determinations in seawater using cresol red. Deep-Sea Res. Part A-Oceanogr. Res. Pap. 36, 803–810. doi: 10.1016/0198-0149(89)90152-0
Chuprov-Netochin, R. N., Faizullina, N. M., Sykilinda, N. N., Simakova, M. N., Mesyanzhinov, V. V., and Miroshnikov, K. A. (2010). The beta-helical domain of bacteriophage T4 controls the folding of the fragment of long tail fibers in a chimeric protein. Russ. J. Bioorganic Chem. 36, 172–178. doi: 10.1134/S1068162010020056
Cornelis, P. (2010). Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86, 1637–1645. doi: 10.1007/s00253-010-2550-2
Cullen, J. T., Bergquist, B. A., and Moffett, J. W. (2006). Thermodynamic characterization of the partitioning of iron between soluble and colloidal species in the Atlantic Ocean. Mar. Chem. 98, 295–303. doi: 10.1016/j.marchem.2005.10.007
Daughney, C. J., Chatellier, X., Chan, A., Kenward, P., Fortin, D., Suttle, C. A., et al. (2004). Adsorption and precipitation of iron from seawater on a marine bacteriophage (PWH3A-P1). Mar. Chem. 91, 101–115. doi: 10.1016/j.marchem.2004.06.003
Demogines, A., Abraham, J., Choe, H., Farzan, M., and Sawyer, S. L. (2013). Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol. 11:e1001571. doi: 10.1371/journal.pbio.1001571
Drakesmith, H., and Prentice, A. M. (2012). Hepcidin and the iron-infection axis. Science 338, 768–772. doi: 10.1126/science.1224577
Ducklow, H. (2000). “Bacterial production and biomass in the oceans,” in Microbial Ecology of the Oceans, ed D. L. Kirchman (New York, NY: Wiley-Liss Inc.), 85–120.
Emerson, D. (2016). The irony of iron - biogenic iron oxides as an iron source to the ocean. Front. Microbiol. 6:1502. doi: 10.3389/fmicb.2015.01502
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Fishwick, M. P., Sedwick, P. N., Lohan, M. C., Worsfold, P. J., Buck, K. N., Church, T. M., et al. (2014). The impact of changing surface ocean conditions on the dissolution of aerosol iron. Global Biogeochem. Cycles 28, 1235–1250. doi: 10.1002/2014GB004921
Fitzsimmons, J. N., and Boyle, E. A. (2014). Both soluble and colloidal iron phases control dissolved iron variability in the tropical North Atlantic Ocean. Geochim. Cosmochim. Acta 125, 539–550. doi: 10.1016/j.gca.2013.10.032
Fitzsimmons, J. N., Bundy, R. M., Al-Subiai, S. N., Barbeau, K. A., and Boyle, E. A. (2015). The composition of dissolved iron in the dusty surface ocean: an exploration using size-fractionated iron-binding ligands. Mar. Chem. 173, 125–135. doi: 10.1016/j.marchem.2014.09.002
Fuhrman, J. (1999). Marine viruses: biogeochemical and ecological effects. Nature 399, 541–548. doi: 10.1038/21119
Gauglitz, J. M., Inishi, A., Ito, Y., and Butler, A. (2014). Microbial tailoring of acyl peptidic siderophores. Biochemistry 53, 2624–2631. doi: 10.1021/bi500266x
Gerringa, L. J. A., Rijkenberg, M. J. A., Schoemann, V., Laan, P., and de Baar, H. J. W. (2015). Organic complexation of iron in the West Atlantic Ocean. Mar. Chem. 177, 434–446. doi: 10.1016/j.marchem.2015.04.007
Gledhill, M., and Buck, K. N. (2012). The organic complexation of iron in the marine environment: a review. Front. Microbiol. 3:69. doi: 10.3389/fmicb.2012.00069
Gledhill, M., and van den Berg, C. M. G. (1994). Determination of complexation of iron (III) with natural organic complexing ligands in seawater using cathodic stripping voltammetry. Mar. Chem. 47, 41–54. doi: 10.1016/0304-4203(94)90012-4
Gobler, C. J., Hutchins, D. A., Fisher, N. S., Cosper, E. M., and Saňudo-Wilhelmy, S. A. (1997). Release and bioavailability of C, N, P Se, and Fe following viral lysis of a marine chrysophyte. Limnol. Oceanogr. 42, 1492–1504. doi: 10.4319/lo.1997.42.7.1492
Gordon, R. M., Martin, J. H., and Knauer, G. A. (1982). Iron in Northeast Pacific waters. Nature 299, 611–612. doi: 10.1038/299611a0
Hantke, K., and Braun, V. (1975). Membrane receptor dependent iron transport in Escherichia coli. FEBS Lett. 49, 301–305. doi: 10.1016/0014-5793(75)80771-X
Hantke, K., and Braun, V. (1978). Functional interaction of TonA-TonB receptor system in Escherichia-coli. J. Bacteriol. 135, 190–197.
Hassler, C. S., Alasonati, E., Nichols, C. A. M., and Slaveykova, V. I. (2011). Exopolysaccharides produced by bacteria isolated from the pelagic Southern Ocean - role in Fe binding, chemical reactivity, and bioavailability. Mar. Chem. 123, 88–98. doi: 10.1016/j.marchem.2010.10.003
Hershey, A. D., and Chase, M. (1952). Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36, 39–56. doi: 10.1085/jgp.36.1.39
Hider, R. C., and Kong, X. (2010). Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637–657. doi: 10.1039/b906679a
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: visual molecular dynamics. J. Mol. Graphics 14, 33–38. doi: 10.1016/0263-7855(96)00018-5
Hunter, K. A., and Boyd, P. W. (2007). Iron-binding ligands and their role in the ocean biogeochemistry of iron. Environ. Chem. 4, 221–232. doi: 10.1071/EN07012
Johnson, K. S., Gordon, R. M., and Coale, K. H. (1997). What controls dissolved iron concentrations in the world ocean? Mar. Chem. 57, 137–161.
Jover, L. F., Effler, T. C., Buchan, A., Wilhelm, S. W., and Weitz, J. S. (2014). The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat. Rev. Microbiol. 12, 519–528. doi: 10.1038/nrmicro3289
Kadner, R. J., Heller, K., Coulton, J. W., and Braun, V. (1980). Genetic control of hydroxamate-mediated iron uptake in Escherichia coli. J. Bacteriol. 143, 256–264.
King, J. (1968). Assembly of tail of bacteriophage T4. J. Mol. Biol. 32, 231–262. doi: 10.1016/0022-2836(68)90007-7
Kuma, K., Nishioka, J., and Matsunaga, K. (1996). Controls on iron(III) hydroxide solubility in seawater: the influence of pH and natural organic chelators. Limnol. Oceanogr. 41, 396–407. doi: 10.4319/lo.1996.41.3.0396
Laglera, L. M., Battaglia, G., and van den Berg, C. M. G. (2007). Determination of humic substances in natural waters by cathodic stripping voltammetry of their complexes with iron. Anal. Chim. Acta 599, 58–66. doi: 10.1016/j.aca.2007.07.059
Laglera, L. M., Battaglia, G., and van den Berg, C. M. G. (2011). Effect of humic substances on the iron speciation in natural waters by CLE/CSV. Mar. Chem. 127, 134–143. doi: 10.1016/j.marchem.2011.09.003
Laglera, L. M., and van den Berg, C. M. G. (2009). Evidence for geochemical control of iron by humic substances in seawater. Limnol. Oceanogr. 54, 610–619. doi: 10.4319/lo.2009.54.2.0610
Leiman, P. G., Arisaka, F., van Raaij, M. J., Kostyuchenko, V. A., Aksyuk, A. A., Kanamaru, S., et al. (2010). Morphogenesis of the T4 tail and tail fibers. Virol. J. 7, 1–28. doi: 10.1186/1743-422X-7-355
Leiman, P. G., and Shneider, M. M. (2012). Contractile tail machines of bacteriophages. Viral Mol. Mach. 726, 93–114. doi: 10.1007/978-1-4614-0980-9_5
Luckey, M., Wayne, R., and Neilands, J. B. (1975). In vitro competition between ferrichrome and phage for outer membrane T5 receptor complex of Escherichia coli. Biochem. Biophys. Res. Commun. 64, 687–693. doi: 10.1016/0006-291X(75)90375-7
Lundrigan, M. D., Lancaster, J. H., and Earhart, C. F. (1983). UC-1, a new bacteriophage that uses the TonA polypeptide as its receptor. J. Virol. 45, 700–707.
Luria, S. E., and Delbruck, M. (1943). Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511.
Maldonado, M. T., and Price, N. M. (1999). Utilization of iron bound to strong organic ligands by plankton communities in the subarctic Pacific Ocean. Deep-Sea Res. Part II-Top. Stud. Oceanogr. 46, 2447–2473. doi: 10.1016/S0967-0645(99)00071-5
Mancuso Nichols, C. A., Garon, S., Bowman, J. P., Raguenes, G., and Guezennec, J. (2004). Production of exopolysaccharides by Antarctic marine bacterial isolates. J. Appl. Microbiol. 96, 1057–1066. doi: 10.1111/j.1365-2672.2004.02216.x
Martin, J. H., Gordon, R. M., Fitzwater, S., and Broenkow, W. W. (1989). VERTEX - Phytoplankton iron studies in the Gulf of Alaska. Deep-Sea Res. Part A-Oceanogr. Res. Pap. 36, 649–680. doi: 10.1016/0198-0149(89)90144-1
Martinez, J. S., Carter-Franklin, J. N., Mann, E. L., Martin, J. D., Haygood, M. G., and Butler, A. (2003). Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. Proc. Natl. Acad. Sci. U.S.A. 100, 3754–3759. doi: 10.1073/pnas.0637444100
Matzanke, B. F., Bohnke, R., Mollmann, U., Reissbrodt, R., Schunemann, V., and Trautwein, A. X. (1997). Iron uptake and intracellular metal transfer in mycobacteria mediated by xenosiderophores. Biometals 10, 193–203. doi: 10.1023/A:1018351728081
Mawji, E., Gledhill, M., Milton, J. A., Tarran, G. A., Ussher, S., Thompson, A., et al. (2008). Hydroxamate siderophores: occurrence and importance in the Atlantic Ocean. Environ. Sci. Technol. 42, 8675–8680. doi: 10.1021/es801884r
Mawji, E., Schlitzer, R., Dodas, E. M., Abadie, C., Abouchami, W., Anderson, R. F., et al. (2015). The GEOTRACES intermediate data product 2014. Mar. Chem. 177, 1–8. doi: 10.1016/j.marchem.2015.04.005
May, P. M., Linder, P. W., and Williams, D. R. (1977). Computer simulation of metal-ion equilibria in biofluids: models for the low-molecular-weight complex distribution of calcium(II), magnesium(II), manganese(II), iron(III), copper(II), zinc(II), and lead(II) ions in human blood plasma. J. Chem. Soc. Dalton Trans. 6, 588–595. doi: 10.1039/dt9770000588
Mioni, C. E., Poorvin, L., and Wilhelm, S. W. (2005). Virus and siderophore-mediated transfer of available Fe between heterotrophic bacteria: characterization using an Fe-specific bioreporter. Aquat. Microb. Ecol. 41, 233–245. doi: 10.3354/ame041233
Morel, F. M. M., and Price, N. M. (2003). The biogeochemical cycles of trace metals in the oceans. Science 300, 944–947. doi: 10.1126/science.1083545
Neilands, J. B. (1979). Ironic function of bacteriophage receptors. Trends Biochem. Sci. 4, 115–118. doi: 10.1016/0968-0004(79)90396-7
Nishioka, J., Takeda, S., Wong, C. S., and Johnson, W. K. (2001). Size-fractionated iron concentrations in the northeast Pacific Ocean: distribution of soluble and small colloidal iron. Mar. Chem. 74, 157–179. doi: 10.1016/S0304-4203(01)00013-5
Parada, V., Herndl, G. J., and Weinbauer, M. G. (2006). Viral burst size of heterotrophic prokaryotes in aquatic systems. J. Mar. Biol. Assoc. U.K. 86, 613–621. doi: 10.1017/S002531540601352X
Parsons, R. J., Breitbart, M., Lomas, M. W., and Carlson, C. A. (2012). Ocean time-series reveals recurring seasonal patterns of virioplankton dynamics in the northwestern Sargasso Sea. ISME J. 6, 273–284. doi: 10.1038/ismej.2011.101
Pawelek, P. D., Croteau, N., Ng-Thow-Hing, C., Khursigara, C. M., Moiseeva, N., Allaire, M., et al. (2006). Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312, 1399–1402. doi: 10.1126/science.1128057
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Poon, A. P. W., and Dhillon, T. S. (1987). The attachment sites of T5-host range temperate coliphages. Virology 158, 431–434. doi: 10.1016/0042-6822(87)90215-7
Poorvin, L., Rinta-Kanto, J., Hutchins, D., and Wilhelm, S. (2004). Viral release of iron and its bioavailability to marine plankton. Limnol. Oceanogr. 49, 1734–1741. doi: 10.4319/lo.2004.49.5.1734
Rabsch, W., Ma, L., Wiley, G., Najar, F. Z., Kaserer, W., Schuerch, D. W., et al. (2007). FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. J. Bacteriol. 189, 5658–5674. doi: 10.1128/JB.00437-07
Raven, J. A., Evans, M. C. W., and Korb, R. E. (1999). The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosyn. Res. 60, 111–149. doi: 10.1023/A:1006282714942
Redfield, A. C. (1958). The biological control of chemical factors in the environment. Am. Sci. 46, 205–221.
Rohwer, F., Segall, A., Steward, G., Seguritan, V., Breitbart, M., Wolven, F., et al. (2000). The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with nonmarine phages. Limnol. Oceanogr. 45, 408–418. doi: 10.4319/lo.2000.45.2.0408
Rohwer, F., and Segall, A. M. (2015). In retrospect: a century of phage lessons. Nature 528, 46–48. doi: 10.1038/528046a
Rohwer, F., and Thurber, R. V. (2009). Viruses manipulate the marine environment. Nature 459, 207–212. doi: 10.1038/nature08060
Rohwer, F., Youle, M., Maughan, H., and Hisakawa, N. (2014). Life in Our Phage World: A Centennial Field Guide to the Earth's Most Diverse Inhabitants. San Diego, CA: Wholon.
Rue, E., and Bruland, K. (2001). Domoic acid binds iron and copper: a possible role for the toxin produced by the marine diatom Pseudo-nitzschia. Mar. Chem. 76, 127–134. doi: 10.1016/S0304-4203(01)00053-6
Rue, E. L., and Bruland, K. W. (1995). Complexation of iron(III) by natural organic-ligands in the central north Pacific as determined by a new competitive ligand equilibration adsorptive cathodic stripping voltammetric method. Mar. Chem. 50, 117–138. doi: 10.1016/0304-4203(95)00031-L
Rue, E. L., and Bruland, K. W. (1997). The role of organic complexation on ambient iron chemistry in the equatorial Pacific Ocean and the response of a mesoscale iron addition experiment. Limnol. Oceanogr. 42, 901–910. doi: 10.4319/lo.1997.42.5.0901
Salmond, G. P. C., and Fineran, P. C. (2015). A century of the phage: past, present and future. Nat. Rev. Microbiol. 13, 777–786. doi: 10.1038/nrmicro3564
Samson, J. E., Magadan, A. H., Sabri, M., and Moineau, S. (2013). Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687. doi: 10.1038/nrmicro3096
Sandy, M., and Butler, A. (2009). Microbial iron acquisition: marine and terrestrial siderophores. Chem. Rev. 109, 4580–4595. doi: 10.1021/cr9002787
Shaked, Y., and Lis, H. (2012). Disassembling iron availability to phytoplankton. Front. Microbiol. 3:123. doi: 10.3389/fmicb.2012.00123
Shire, D. M., and Kustka, A. B. (2015). Luxury uptake, iron storage and ferritin abundance in Prochlorococcus marinus (Synechococcales) strain MED4. Phycologia 54, 398–406. doi: 10.2216/14-109.1
Silva, J. B., Storms, Z., and Sauvageau, D. (2016). Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 363:fnw002. doi: 10.1093/femsle/fnw002
Stern, A., and Sorek, R. (2011). The phage-host arms race: shaping the evolution of microbes. Bioessays 33, 43–51. doi: 10.1002/bies.201000071
Stolpe, B., and Hassellov, M. (2010). Nanofibrils and other colloidal biopolymers binding trace elements in coastal seawater: significance for variations in element size distributions. Limnol. Oceanogr. 55, 187–202. doi: 10.4319/lo.2010.55.1.0187
Sullivan, M. B., Coleman, M. L., Weigele, P., Rohwer, F., and Chisholm, S. W. (2005). Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:e144. doi: 10.1371/journal.pbio.0030144
Suttle, C. A. (2007). Marine viruses - major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
Tam, W., Pell, L. G., Bona, D., Tsai, A., Dai, X. X., Edwards, A. M., et al. (2013). Tail tip proteins related to bacteriophage lambda gpL coordinate an iron-sulfur cluster. J. Mol. Biol. 425, 2450–2462. doi: 10.1016/j.jmb.2013.03.032
Thomassen, E., Gielen, G., Schutz, M., Schoehn, G., Abrahams, J. P., Miller, S., et al. (2003). The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J. Mol. Biol. 331, 361–373. doi: 10.1016/S0022-2836(03)00755-1
Thuroczy, C. E., Gerringa, L. J. A., Klunder, M. B., Middag, R., Laan, P., Timmermans, K. R., et al. (2010). Speciation of Fe in the Eastern North Atlantic Ocean. Deep-Sea Res. Part I-Oceanogr. Res. Pap. 57, 1444–1453. doi: 10.1016/j.dsr.2010.08.004
Tortell, P. D., Maldonado, M. T., Granger, J., and Price, N. M. (1999). Marine bacteria and biogeochemical cycling of iron in the oceans. FEMS Microbiol. Ecol. 29, 1–11. doi: 10.1111/j.1574-6941.1999.tb00593.x
Tortell, P. D., Maldonado, M. T., and Price, N. M. (1996). The role of heterotrophic bacteria in iron-limited ocean ecosystems. Nature 383, 330–332. doi: 10.1038/383330a0
Twining, B. S., and Baines, S. B. (2013). The trace metal composition of marine phytoplankton. Annu. Rev. Mar. Sci. 5, 191–215. doi: 10.1146/annurev-marine-121211-172322
van den Berg, C. M. G. (2005). Organic iron complexation is real, the theory is used incorrectly. Comment on 'Measuring marine iron(III) complexes by CLE-AdSV'. Environ. Chem. 2, 88–89. doi: 10.1071/EN05029
von der Heyden, B. P., Hauser, E. J., Mishra, B., Martinez, G. A., Bowie, A. R., Tyliszczak, T., et al. (2014). Ubiquitous presence of Fe(II) in aquatic colloids and its association with organic carbon. Environ. Sci. Technol. Lett. 1, 387–392. doi: 10.1021/ez500164v
Vraspir, J. M., and Butler, A. (2009). Chemistry of marine ligands and siderophores. Annu. Rev. Mar. Sci. 1, 43–63. doi: 10.1146/annurev.marine.010908.163712
Wayne, R., and Neilands, J. B. (1975). Evidence for common binding-sites for ferrichrome compounds and bacteriophage Phi-80 in cell-envelope of Escherichia coli. J. Bacteriol. 121, 497–503.
Wheeler, T. J., Clements, J., and Finn, R. D. (2014). Skylign. BMC Bioinform. 15:7. doi: 10.1186/1471-2105-15-7
Wilhelm, S. W., and Suttle, C. A. (1999). Viruses and nutrient cycles in the sea. Bioscience 49, 781–783. doi: 10.2307/1313569
Wilhelm, S. W., Weinbauer, M. G., Suttle, C. A., and Jeffrey, W. H. (1998). The role of sunlight in the removal and repair of viruses in the sea. Limnol. Oceanogr. 43, 586–592. doi: 10.4319/lo.1998.43.4.0586
Wommack, K. E., and Colwell, R. R. (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64, 69–114. doi: 10.1128/MMBR.64.1.69-114.2000
Wommack, K. E., Nasko, D. J., Chopyk, J., and Sakowski, E. G. (2015). Counts and sequences, observations that continue to change our understanding of viruses in nature. J. Microbiol. 53, 181–192. doi: 10.1007/s12275-015-5068-6
Keywords: iron, ligand, phage, marine, virus, siderophore, infection, bacteria
Citation: Bonnain C, Breitbart M and Buck KN (2016) The Ferrojan Horse Hypothesis: Iron-Virus Interactions in the Ocean. Front. Mar. Sci. 3:82. doi: 10.3389/fmars.2016.00082
Received: 16 February 2016; Accepted: 17 May 2016;
Published: 08 June 2016.
Edited by:
Toshi Nagata, The University of Tokyo, JapanReviewed by:
Arvind Singh, Physical Research Laboratory, IndiaElvira Pulido-Villena, Mediterranean Institute of Oceanography, France
Dave Kirchman, University of Delaware, USA
Copyright © 2016 Bonnain, Breitbart and Buck. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kristen N. Buck, kristenbuck@usf.edu
 Chelsea Bonnain
Chelsea Bonnain Mya Breitbart
Mya Breitbart Kristen N. Buck
Kristen N. Buck