Quantifying Tradeoffs for Marine Viruses
- 1Bigelow Laboratory for Ocean Sciences, East Boothbay, ME, USA
- 2Department of Earth, Atmosphere and Planetary Sciences, Massachusetts Institute of Technology, Cambridge, MA, USA
- 3Department of Biology, Hjort Centre for Marine Ecosystem Dynamics, University of Bergen, Bergen, Norway
The effects of viruses on marine microbial communities are myriad. The high biodiversity of viruses and their complex interactions with diverse hosts makes it a challenge to link modeling work with experimental work. In various trophic groups, trait-based approaches have helped to simplify this complexity, as traits describe organism properties in terms of taxon-transcending units, allowing for easier identification of generic, underlying principles. By predicting large-scale biogeography of different plankton functional types based on key sets of traits and their associated tradeoffs, these approaches have made major contributions to our understanding of global biogeochemistry and ecology. This review addresses the question of how a trait-based approach can make contributions toward understanding marine virus ecology. We review and synthesize current knowledge on virus traits with a focus on quantifying the associated tradeoffs. We use three case studies—virulence, host range, and cost of resistance—to illustrate how quantification of tradeoffs can help to explain observed patterns, generate hypotheses, and improve our theoretical understanding of virus ecology. Using a nutrient-susceptible-infected-virus model as a framework, we discuss tradeoffs as a link between model building (theory) and experimental design (practice). Finally, we address how insights from virus ecology can contribute back to the trait-based ecology community.
1. Introduction
“We live in a dancing matrix of viruses”
Linking modeling and experimental work is an ongoing challenge in ecology. For marine viruses, ecologists are beginning to model the role of viruses in shaping biogeography, biogeochemistry, macroecology, and climate in the oceans. As bearers of life and death, viruses influence growth and mortality rates within microbial communities, mediating biogeochemical processes, and driving adaptation and evolution (Fuhrman, 1999; Brussaard, 2004; Suttle, 2007; Weitz and Wilhelm, 2012). Lytic viruses may account for up to 50% of bacterial mortality in the pelagic ecosystem (Suttle, 1994; Fuhrman and Noble, 1995; Fuhrman, 1999), and they can abruptly terminate eukaryotic algae blooms (Bratbak et al., 1993; Nagasaki et al., 1994). Lysogenic viruses, embedded in the host genome, may on the other hand have effects on hosts ranging from parasitic to mutualistic (Weinbauer, 2004). Viruses contribute to both top-down and bottom-up control of the microbial community (Weitz et al., 2016). They are thus key components of ocean ecosystem models.
Virus-mediated mortality has multiple, often contrasting, and globally significant biogeochemical effects. For example, there are competing hypotheses on whether viral lysis increases or decreases carbon export from the euphotic zone. If dissolved matter from lysis is typically remineralized, this would lead to increased regenerate production in the euphotic zone (Fuhrman, 1999; Wilhelm and Suttle, 1999; Weinbauer, 2004) and reduce export by shortcutting transfer to higher trophic levels and sinking fecal pellets (De La Rocha and Passow, 2007). On the other hand, transparent exopolymeric particles released during lysis may actually enhance particle aggregation and sinking (Proctor and Fuhrman, 1991). Furthermore, by stimulating regenerate production, viruses may sustain higher microbial biomass (Weinbauer, 2004), which may affect export. Virus infection can also alter cell stoichiometry and uptake rates, thereby altering biogeochemical pathways (Weitz and Wilhelm, 2012; Jover et al., 2014).
Viruses are also important drivers of microbial diversity (Thingstad, 2000; Weinbauer, 2004; Thingstad et al., 2015). A consequence of the strong top-down control by viruses on the microbial community is selection for host cell structures and functions rendering resistance. Such structural adaptations may be physiologically costly, which allows coexistence of both susceptible and resistant hosts. Typically, structural changes are such that virus-host interactions become highly specific (Lima-Mendez et al., 2015). The result is that viruses can regulate strain diversity within species (Thingstad et al., 2014), or species diversity within microbial communities, depending on the specificity of the virus-host interactions. In a dynamic world, co-evolutionary arms races—where hosts try to escape virus infections and viruses evolve to infect resistant hosts—provide an important mechanism to build and maintain microbial diversity (Martiny et al., 2014).
The increasing recognition of the importance of viruses has motivated work to incorporate them into ecosystem models—particularly those representing biogeochemical processes (Weitz et al., 2015; Middleton et al., in press). Virus ecology is complex, however, and representing the diversity of processes and interactions described above remains a challenge for modelers. Most ecosystem models approach ecological problems either at the species level or at the level of trophic groups. This strategy is reasonable, given the fundamental importance of species as a taxonomic unit and of trophic interactions. However, this approach has limitations when it comes to certain objectives. Challenges include: (1) connecting community structure with ecological function, (2) drawing causal links between organism properties and macroecological patterns, and (3) making diversity tractable in ecosystem models. Each of these challenges is key both to understanding basic ecology as well as to predicting the ocean's response to changing climate and other pressures.
An alternative to species-centric models is the trait-based approach to ecology, which has seen a recent resurgence in marine ecology (Barton et al., 2016). The trait-based approach to ecology has some early roots in marine systems (Sheldon et al., 1972), but much of the foundational work comes from terrestrial ecosystems, where global trait databases have facilitated breakthroughs (Wright et al., 2005; Violle et al., 2007; Kattge et al., 2011). The approach has gained traction in marine ecosystems over the past 10 years, primarily for phytoplankton (Follows et al., 2007; Litchman et al., 2007; Litchman and Klausmeier, 2008; Edwards et al., 2012; Barton et al., 2013), but also for zooplankton (Kiørboe et al., 2010; Litchman et al., 2013; Record et al., 2013b) and fish (Claudet et al., 2010; Marras et al., 2013; Opdal and Jørgensen, 2015; Pimentel et al., 2016). While ecologists have not agreed upon a single rigorous definition of “trait,” the objective is to define characteristics of organisms that link processes at the individual level to population-, community-, or ecosystem-level processes. Traits are typically taxon-transcending properties that describe growth, ontogeny, reproduction, and defense, and ultimately determine fitness (Litchman et al., 2013). When successful, the trait-based approach can explain mechanistically the organization and function of an ecosystem (Hansen et al., 1994; Follows et al., 2007; Rose et al., 2007; Banas, 2011; Record et al., 2012, 2013a; Zhang et al., 2013; Andersen and Beyer, 2015). The notion of a trait often goes hand-in-hand with modeling, as traits can be represented as parameters in systems of dynamical equations. As such, they provide a useful link between models, which require values for these parameters, and experiments, which measure them.
Many of the parameters found in virus models have been reviewed and described in the context of traits (Weitz et al., 2016, Ch. 2). To link traits with the underlying ecology, we also require an understanding of tradeoffs associated with sets of traits. Tradeoffs occur when an increase in fitness associated with one trait comes at a fitness cost associated with another trait. Identification of tradeoffs has recently been proposed as a key step toward linking the work of modelers and empiricists (Barton et al., 2016). The concept of tradeoffs is commonly invoked in the virus literature (Bohannan et al., 2002; De Paepe and Taddei, 2006; Jessup and Bohannan, 2008; Pradeep Ram and Sime-Ngando, 2010; Keen, 2014), and with the recent reemergence of the trait-based approach in marine ecology, it is timely to review our understanding of marine virus tradeoffs in order to help incorporate it into the broader trait-based dialogue. Here we provide a review of marine virus traits and attempt to characterize and quantify the tradeoffs associated with some of the most important traits. We include viruses that infect pelagic bacteria, archaea, and eukaryotes, though there is considerably more literature on bacteriophages. We use three case studies and simple illustrative model simulations to demonstrate how quantification of tradeoffs can help to explain observed patterns, generate hypotheses, and link modeling work and empirical work.
2. A Framework for Trait-Based Virus Models
The first step in a trait-based approach to the ecology of a group of organisms is to build a framework that includes the traits involved in important tradeoffs (Litchman et al., 2013). Here we use a dynamical systems model as the core of our framework. A recent review of marine virus-host models (Middleton et al., in press) found a wide variety in model equations and state variables used to describe marine virus-host interactions. While there was some commonality among models, nearly one third did not include a state variable tracking virion abundance (e.g., Beltrami and Carroll, 1994; Chattopadhyay and Pal, 2002; Singh et al., 2004; Rhodes and Martin, 2010). This follows the convention in biogeochemical models of using a linear mortality term to capture all background mortality including viral lysis. Other notable state variables in marine virus-host models included a separate infected host population (Bratbak et al., 1998), a virus population within host cells (Siekmann and Malchow, 2008), a virus inhibitor (Thyrhaug et al., 2003), and a resistant population (Middelboe, 2000). Model design is often dictated by available data or by hypotheses, or, for marine virus-host models, based on other standard forms such as susceptible-infected-removed (SIR) models of epidemics (Kermack and McKendrick, 1927).
Following Litchman et al. (2013), we have classified virus traits based on ecological function and trait type (Table 1). This exercise itself illustrates some of the challenges around building a general trait-based framework for marine pelagic ecology. Some of the trait categories in the Litchman classification needed to be changed to apply to viruses. Additionally, not all of these traits convert easily into model parameters. Some traits that have been studied and reviewed (Weitz et al., 2016) translate directly to model parameters (e.g., burst size). Those that do not (e.g., size) generally have one or more underlying relationships that translate indirectly into model parameters. We constructed a set of equations based on the common components in marine virus-host models (Middleton et al., in press) designed to incorporate the quantitative traits outlined in Table 1. The full model includes all traits discussed in this review so that the associated tradeoffs can be explored mathematically. The model can be reduced based on simplifying assumptions into various forms that are used in other modeling studies (detailed in Supplementary Material). A chemostat is simulated, where hosts are partitioned into those that are susceptible and those that are infected. Encounter between free living viruses and susceptible hosts creates infected hosts. The full model is a Nutrient-Susceptible-Infected-Virus (NSIV) model, representing virus infection of a generic host population. The equations are written:
where N is the nutrient concentration supplied at an input concentration Nin and dilution rate δ, S is the population of susceptible hosts, I is the population of infected hosts, and V is the population of viruses (Table 2). The parameter ϕ represents an interaction kernel between viruses and hosts, μ is a growth function for hosts, and c is the host nutrient quota. Host and virus losses, ω and ψ respectively, include losses due to dilution, as well as other forms of mortality or decay. The parameter γ determines the virulence strategy, representing the lysogenic proportion of the virus. Hosts are lysed at rate λ, and each lysis event results in β free living virions. Each parameter represents a trait explicitly or can be expressed as a function of a trait. In some contexts, a parameter can combine multiple traits, such as the interaction kernel, which includes processes like encounter (dependent upon size) as well as structure or entry mechanism. In such cases, as we understand more about these traits, we can replace the parameter with a more refined function that captures multiple traits. We will use this model, or its reduced versions (detailed in Supplementary Material), to illustrate the tradeoffs associated with the reviewed traits.
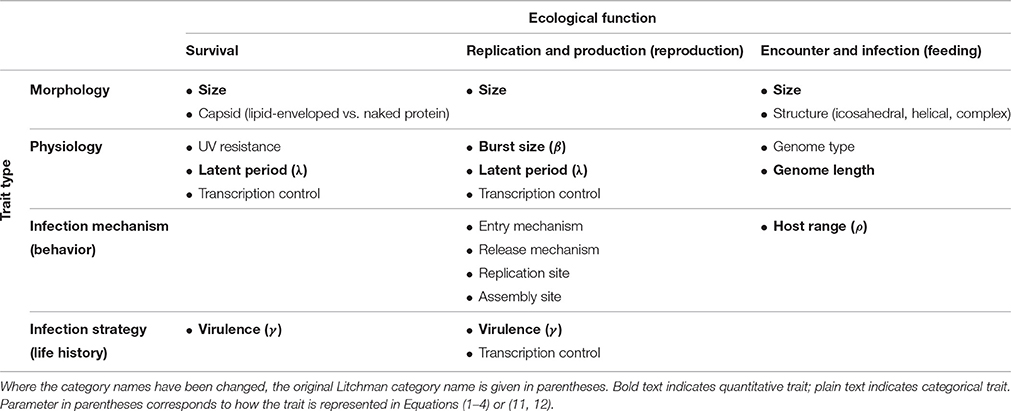
Table 1. Overview of virus traits, following the classification scheme of Litchman et al. (2013).
3. Identifying and Quantifying Tradeoffs
Each of the traits listed in Table 1 is hypothetically involved in one or more tradeoffs. Here we focus on four traits and their associated tradeoffs: size, virulence, host range, and resistance. For the latter three cases, we use the NSIV model framework (Equations 1–7) to evaluate the tradeoff associated with the trait and compare predictions to observations. The fourth case—resistance—illustrates the interaction between virus traits and host traits.
3.1. Size
Ocean ecosystems are strongly size-structured. Organism size is a first-order determinant of many aspects of ecosystem structure and function, including production and metabolism (López-Urrutia et al., 2006), reproduction (Sheldon et al., 1972), predator-prey interactions (Banas, 2011; Golet et al., 2015), and species richness (Record et al., 2012) to name a few. As a trait, it is often viewed as a primary axis for describing marine ecosystems (Barton et al., 2013; Litchman et al., 2013), with a strong body of theory explaining how size structures marine ecosystems from bacteria to whales (Andersen et al., 2016). It is unknown whether these allometric relationships extend to the viruses.
3.1.1. Survival
Important sources of virus loss include ultraviolet light (Murray and Jackson, 1993; Wilhelm et al., 2003), grazing (Deng et al., 2014), adsorption, and genome size and density (De Paepe and Taddei, 2006). These processes have strong allometric dependencies in general, but it is an open question whether virus loss in the marine environment is strongly size- dependent.
3.1.2. Encounter and Infection
As non-motile entities, extracellular virus particles depend on Brownian motion for random encounter with hosts. As such, diffusive transport is likely to be a primary determinant of host-virus encounter. Larger virus particles diffuse more slowly, and are thus expected to be at a disadvantage in terms of contact rates (Murray and Jackson, 1992). Simple physical arguments may also be used to understand which hosts are more susceptible to encounter with virus particles. Probability of virus attachment is greater for larger organisms with higher surface area. Furthermore, larger organisms generally swim faster, clearing a larger volume of water and enhancing the probability of virus encounter (Murray and Jackson, 1992). Simple physical controls on encounter do not necessarily manifest in clear relations between virus particle size and the size of the host it is able to infect. For example, coccolithoviruses with capsids ~170–200 nm in diameter infect haptophyte Emiliania huxleyi (5–8 μm diameter) (Schroeder et al., 2002), while the significantly smaller Rhizosolenia setigera-virus (~30 nm diameter) infects the much larger diatom R. setigera (2–50 μm diameter and 0.1–1 mm length) (Nagasaki et al., 2004). Furthermore, the sizes of virus and host genomes appear to be unrelated (Brown et al., 2006). While these examples do not point toward a simple, canonical virus:host size ratio, to the best of our knowledge, no one has systematically addressed this question in a broad range of marine virus-host systems.
3.1.3. Virus Production
Over sufficiently large ranges, growth rate tends to decline with organism size in eukaryotes, while the opposite is true for prokaryotes (Kempes et al., 2012). Very little is known about the relation between virus particle size and production rate. Since viruses utilize host metabolic machinery for replication, virus production rate generally follows growth rates of the host and varies depending on host growth conditions and physiology (Van Etten et al., 1983; Moebus, 1996; Bratbak et al., 1998; Middelboe, 2000; Baudoux and Brussaard, 2008). On the other hand, replication rate may depend more directly on virus particle size. Virus replication rate is also likely to depend on morphological variation across diverse phylogenies. A proxy often used to infer virus accumulation rate is burst size. There is evidence that burst size is related to the ratio between host and virus genome lengths (Weitz et al., 2015), and that number of base pairs is related to capsid size (Jover et al., 2014), suggesting that a smaller virus can make more copies of itself than a large virus, all other factors being equal. However, burst size is not a direct reflection of the metabolic efficiency of virus replication. It says nothing about the time required for replication, which in part may be reflected in the duration of latent period. Few direct measurements of virus replication rates are available, making it difficult to know the allometric scaling of virus production.
3.2. Virulence: Tradeoffs with Lysogeny and Lysis
Pathogenic virulence influences microbial fitness in complex and interesting ways. Lysogenic viruses can enhance host fitness by increasing the growth of infected hosts (Edlin et al., 1975), and providing protection against closely related viruses (Ptashne, 1967). Lysogeny is associated with horizontal gene transfer (Chiura, 1997; Lawrence and Ochman, 1998; Ochman et al., 2000), and bacterial genomes contain a high portion of dormant or repurposed phage genes (Casjens et al., 2000; Ochman et al., 2000; Hayashi et al., 2001; Daubin et al., 2003). Due to its importance for public health and epidemiology, there is a sizeable literature on factors controlling the switch from lysogeny to lysis (Brüssow et al., 2004; Ptashne, 2004), as well as some marine examples (Paul and Jiang, 2001; Brum et al., 2016). The switch may be related to molecular interactions between host and virus (Ptashne, 2004; Zeng et al., 2010), or environmental cues that relate, for example, to host production (Koudelka et al., 1988; Wilson and Mann, 1997; Williamson et al., 2002; Weinbauer et al., 2003). Relatively few studies (Levin and Lenski, 1983; Stewart and Levin, 1984; Koudelka et al., 1988; Williamson et al., 2002; Weinbauer et al., 2003) have explored the ecological factors that may ultimately govern when and where lysogenic vs. lytic replication modes are selected. Here, we briefly review observed relationships between environmental variables and prevalence of lysogeny. We then use Equations (1–4) to explore a tradeoff that may govern observed trends.
3.2.1. Temperature
Wilson and Mann (1997) concluded that the only studies relating temperature to the switch between lysogeny and lysis were with enteric hosts (Edgar and Lielausis, 1964; Gough, 1968), and that evidence in marine systems was limited. Since then, correlations between temperature and lysogeny have been found in a range of environments (Cochran and Paul, 1998; Williamson et al., 2002; Maurice et al., 2010; Payet and Suttle, 2013). The general pattern shows low temperatures favoring lysogenic phages, and high temperatures favoring lytic phages. Temperature has been used to provoke prophage induction in natural seawater samples (Jiang and Paul, 1996), but laboratory experiments testing the dependence of the switch on temperature have been equivocal (Williamson and Paul, 2006). Early in their experiments, Williamson and Paul (2006) observed elevated temperature leading to higher production rates and reduced lysogeny. Later in the experiments however, high temperature cultures reduced overall production of free phage by both lytic and lysogenic infections.
3.2.2. Light
Tests of the influence of light intensity on the switch between lysogeny and lysis can be divided into those that tested effects of UV radiation, and those that tested whether direct sunlight could induce lysis. In general, the results evidence that induction response to light varies among host species (possibly even at the strain level) and specific lysogens. For example, Vibrio lysogens could be induced by sunlight (Faruque et al., 2000) and Synechococcus phages could be induced by continuous, high light (McDaniel et al., 2006). In contrast, other studies failed to induce natural assemblages with sunlight (Wilcox and Fuhrman, 1994; Jiang and Paul, 1996) while UV radiation has been proven effective to revert lysogenic to lytic infections within natural assemblages (Jiang and Paul, 1996; Weinbauer and Suttle, 1996).
3.2.3. Nutrients
Field and laboratory observations suggest nutrient status is likely to influence the prevalence of lysogeny. There is ample evidence, from a range of studies in different oceanic regions, that the proportion of hosts in natural assemblages carrying inducible prophages varies as a function of trophic status. The abundance of hosts with lysogenic infections is often lower in more productive than in less productive waters and also lower in highly productive summer months than in low productive winter months (Jiang and Paul, 1994; McDaniel et al., 2002; Weinbauer et al., 2003; Lymer and Lindström, 2010; Payet and Suttle, 2013). Additionally, it has been empirically shown that nutrient enrichment of natural samples may induce lysogens in some bacterial assemblages (Wilson et al., 1998; Williamson et al., 2002), but not in others, e.g., Synechococcus populations (McDaniel and Paul, 2005), while nutrient starvation can lead to the establishment of lysogeny in the heterotrophic bacteria Pseudomonas aeruginosa (Kokjohn et al., 1991). Similarly, phosphate depletion in cultures of the photoautotrophic bacteria Synechococcus sp. (Wilson et al., 1996) has been shown to decrease burst size and lysis rate of infected host populations, suggesting cyanophages entered a lysogenic state in response to nutrient starvation.
3.2.4. The Virulence Tradeoff
There must be a tradeoff associated with the switch between lysis and lysogeny, otherwise one would always be favored over the other. In low productive systems, encounter between hosts and extracellular virus particles may be low, and insufficient to overcome losses, rendering lysogeny the favorable strategy for production. In relatively productive systems, when encounter is high, virus replication could be maximized by frequently lysing hosts. This amounts to a tradeoff with the maximum host production potential. The above literature review suggests increases in light, temperature, and nutrients are all correlated with diminished lysogeny, with arguably the strongest support for nutrient related control. We can examine this hypothesized tradeoff using the NSIV model (Equations 1–4), where the parameter γ determines the virulence strategy of the virus. When γ = 1, the virus is lysogenic. Infected hosts grow at the same rate as susceptible hosts (a simplifying assumption that can potentially be relaxed), and there is no lysis. When γ = 0, infected hosts do not grow, and lysis proceeds at rate λ. With the system solved in steady state (see Supplementary Material), we can explore two scenarios exemplifying contrasting virulence strategies. The following two expressions are the total equilibrium virus biomass when γ = 1 and when γ = 0, respectively.
Note that these expressions assume the rate of lysis, λ, is significantly greater than other parameters in the model (Supplementary Material). The above expressions are both linear functions of the nutrient input concentration Nin (Figure 1A), and give a guide to when lytic replication may be favorable over lysogeny. Interestingly, the minimal nutrient input required for lytic viruses to exist is always greater than the nutrient input required for lysogenic viruses to exist:
Since c, ω, ψ, ϕ, δ, and β are all positive, we have . The prediction is that lytic viruses may only be present at a nutrient input concentration that is higher than that required to sustain host biomass. Thus, low nutrient supply may naturally favor lysogenic viruses, since lytic viruses may not survive as free-existing entities. If viruses are assumed to switch from lysogeny to lysis once nutrient supply is sufficient to support a free living population of viruses, then the overall prevalence of lysogeny would decrease as a function of nutrient enrichment, perhaps replaced by predominately lytic viruses (Figure 1B). Thus, lysogeny may be favored under low nutrient conditions, and lytic production favored under relatively high nutrient conditions (Knowles et al., 2016; Weitz et al., 2016, cf).
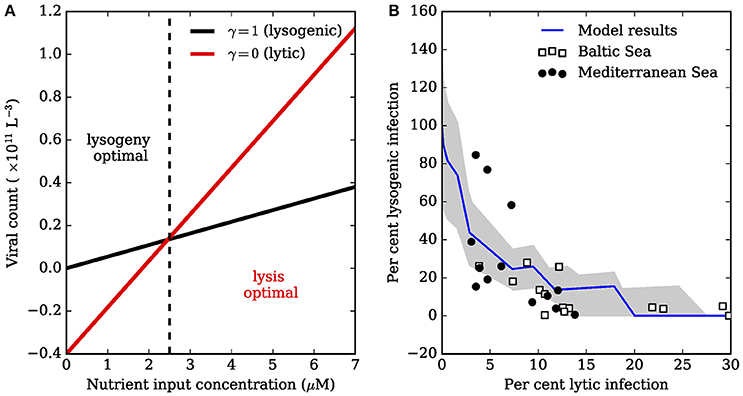
Figure 1. Tradeoff between lysogenic and lytic infection. (A) Lines were generated using Equations (5) and (6) with δ, μ, c, β, ϕ, ω, and ψ equal to 0.1, 1 × 10−9, 2.3 × 10−12, 50, 1 × 10−11, 0.4, 0.1, respectively. The lysogenic and lytic populations can sustain non-negative biomass when , and , respectively (Equations 13, 15). Low nutrient input enables lysogeny to survive when the lytic strategy would fail. High nutrient input can lead to survival of free-existing lytic phage. (B) The model in (A) can be used to explain the relationship between fraction of hosts that are lysogenic (FLC), and fraction of hosts with lytic infections (FIC). Data are of bacterial assemblages in a range of trophic conditions, ranging from the relatively oligotrophic Mediterranean Sea, to the more productive Baltic Sea (Weinbauer et al., 2003). Total lysogenic infection was inferred by treatment with mitomycin C. The decline in FLC as a function of FIC is thought to be due to nutrient enrichment. The jagged blue line averages total lysogenic (FIC) and lytic infection (FLC) for a community of phages, each modeled with Equations (1–7) where parameters β and ϕ were drawn randomly from uniform distributions ±50% of the values in (A), and no tradeoffs were imposed between parameters. Each virus in the community was assumed to switch from lysis to lysogeny when . The jagged line arises because each community member has a unique determined by the randomly drawn β and ϕ. The gray shaded region is one standard deviation from the mean for FLC and FIC.
The analysis presented here examines the potential for lytic viruses to exist in a range of nutrient conditions, but says nothing about the biological mechanisms that directly control virus replication strategy. Host metabolic state is associated with changes in replication strategy (Ptashne, 2004; Ghosh et al., 2009), and is expected to improve as nutrient supply increases. Host metabolic state may therefore provide a direct, mechanistic link between environmental conditions and virus replication strategy. Better understanding of the transition from lysogeny to lysis thus depends on our mechanistic understanding of metabolic cues, as well as the trade-offs governing feasibility of the different strategies, shown here to include host-virus encounter, burst, virus replication rate, host, and virus mortality (Equations 5, 6, Figure 1).
3.3. Host Range: Tradeoffs with Specificity and Generality
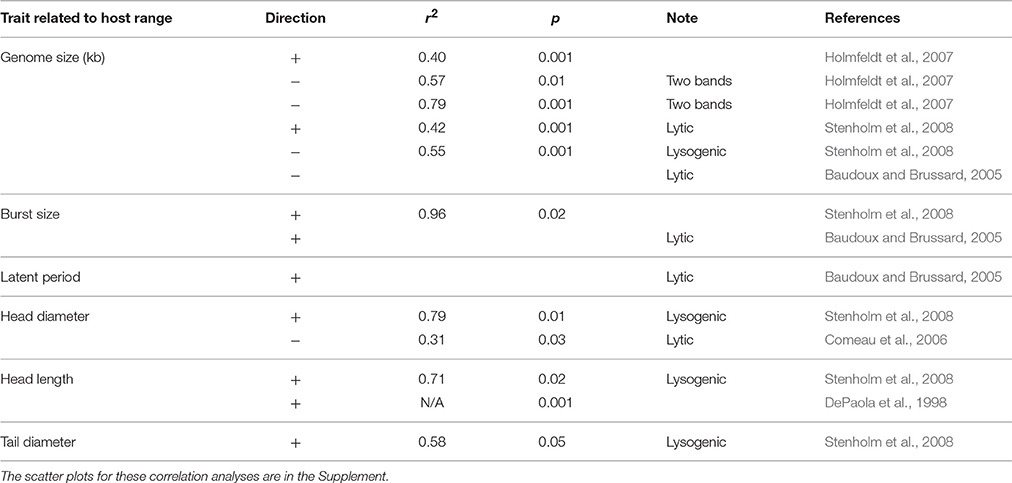
The prevalent view of high host specificity among marine viruses has been changing recently as wide and variable host ranges are observed among marine viruses (Holmfeldt et al., 2007; Flores et al., 2011). Host ranges can be variable across host strains (DePaola et al., 1998; Liu et al., 2001; Allen et al., 2007; Holmfeldt et al., 2007; Stenholm et al., 2008; Martínez et al., 2015) as well as across host phyla (Malki et al., 2015). A broad host range (generalist) must come with some cost; otherwise we would expect no host specialists. Coexistence between generalists and specialists is a topic of broader interest in ecology (Egas et al., 2004; Ma and Levin, 2006). In other ecological contexts, tradeoffs associated with generalism versus specialism are associated with foraging strategy (Wilson, 1994), scale of temporal variability in environment or resource (Gilchrist, 1995), and habitat fragmentation (Marvier et al., 2004) to name a few. For viruses, there has been some analytical work examining possible tradeoffs (Jover et al., 2013). By revisiting data in published studies, we found evidence for possible tradeoffs with host range size, including virus genome size, burst size, and morphology (Table 3, data shown in Supplementary Material). There are other sources of information that suggest possible tradeoffs but without sufficient measurements to show statistical significance (e.g., Mojica and Brussaard, 2014).
3.3.1. Genome Size
A broader host range could imply that the virus strain has some increased ability to overcome a wider range of host defenses. The hypothesized tradeoff would be that this requires a longer genome with larger functional potential, which requires more cell resources to assemble, ultimately slowing virus population increase rates. In a diverse host community, access to a greater number of hosts would outweigh this cost for a generalist, and in less diverse host communities, the more rapid population increase rate of specialists would be favored. There is some evidence that host range size correlates positively with genome size for lytic viruses (Table 3). These relationships are driven by the extremes (i.e., generalists and specialists), with higher variance for intermediate host ranges. The relationship is also reversed for lysogenic viruses (Table 3) (e.g., Stenholm et al., 2008). In some cases there is no relationship (Comeau et al., 2006).
3.3.2. Burst Size
There is at least one example of evidence for a strong relationship between host range and burst size, though this relationship is based on a small sample size (Table 3) (Stenholm et al., 2008). The relationship is also in the reverse direction from what one would expect from a tradeoff: a higher host range is associated with a higher burst size, so generalists would hypothetically have an advantage over specialists in both cases. However, greater burst size is also associated with greater latency period (Wang, 2006), and greater burst size, as with host range, is associated with larger genomes (Brown et al., 2006; Weitz et al., 2016). A tradeoff might therefore involve more than two traits, with a greater host range requiring extra genetic machinery and longer latency time, producing a cost with the potential to offset the advantages of generalism.
3.3.3. Morphology
A number of morphological traits associated with size relate positively with host range size, namely head length, head diameter, and tail diameter (Table 3). Additionally, DePaola et al. (1998) found among Vibrio phages a distinct short-tailed, long capsid morphotype that had a markedly wider host range than the other phages examined. Again, the tradeoff would likely have to do with the extra resources required for larger sizes.
3.3.4. Interaction Networks
The host range trait can be categorized into generalists, specialists, and intermediate cases, and the overall prevalence of these different strategies manifest in distinct network structures (Proulx et al., 2005). Nestedness—that is, the specialist virus infects the host that is infected most commonly—appears to be common among closely related host-virus groups, though other configurations occur (Flores et al., 2011). Interestingly, these network structures appear limited to more closely related phylogentic host-virus networks. Broad phylogenetic groups are characterized by a nested-modular network structure, whereby nestedness only occurs in modules, and viruses within each module are generally unable to infect more distantly related hosts belonging to separate modules (Beckett and Williams, 2013). Marine viruses follow these patterns as well, though an aggregation of the reviewed data shows a disproportionately high number of generalists and specialists as compared to intermediate cases (Figure 2A).
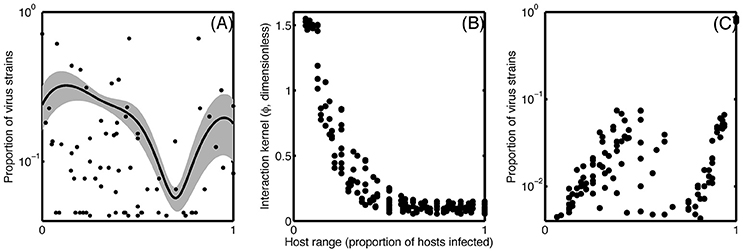
Figure 2. (A) Smoothed histogram showing the proportion of virus strains with each host range strategy from 0 (infecting no tested hosts) to 1 (infecting all tested hosts), averaged across 11 published experiments (Suttle and Chan, 1993; DePaola et al., 1998; Wichels et al., 1998; Sullivan et al., 2003; Comeau et al., 2005, 2006; Allen et al., 2007; Holmfeldt et al., 2007; Stenholm et al., 2008; Middelboe et al., 2009; Martínez et al., 2015). Gray shading shows variance around the mean. (B) Input relationship between host range and interaction kernel, representing a hypothetical tradeoff. (C) Resulting output, after convergence, from simulation examining a tradeoff associated with host range using the tradeoff depicted in (B).
3.3.5. The Host Range Tradeoff
To examine the potential tradeoffs associated with host range, we use a simplified version of the NSIV model (Equations 1–7), where we consider only the lytic case without an explicit population of infected hosts (Supplementary Material Section 1.3). If we assume a nutrient replete environment, we can approximate host population growth rate with a constant α, and write the system as two equations:
Simplifications like this allow for focusing on a single tradeoff. To understand the host range dimension, we include multiple hosts and viruses, with different combinations of infection:
where i and j subscripts represent hosts and viruses respectively, and ρij is a binary parameter that describes whether virus j infects host i. In this configuration, different host-virus interaction matrices (Flores et al., 2011) can be input as binary matrices of ρ values.
In this system, a diversity of viruses and hosts can coexist if host range trades off with ϕiβi/ψi (Jover et al., 2013). We tested a tradeoff between host range and the interaction kernel ϕ, which encapsulates the above reviewed traits that likely tradeoff with host range. We ran an ensemble of simulations with varying numbers of hosts and viruses, nested host range structures, and a forced tradeoff between host range and ϕ (Figure 2B). The simulation reproduced the observed bimodal pattern of higher abundance of generalists and specialists as compared to the intermediate cases (Figure 2C) when two conditions were met: the interaction network has a nested structure, and the tradeoff relationship (Figure 2B) is concave up. If the tradeoff is concave down, then increasing host range toward the extreme comes at a very high cost in terms of the interaction kernel (and vice versa), shifting the resulting distribution to a unimodal one with an intermediate host range. This example shows how the trait-based approach gives us a quantitative hypothesis about the shape of a tradeoff and its ability to describe observed patterns.
Host range is not just a function of virus traits, however. Defense mechanisms of hosts can vary across strains as well. Because the life histories of viruses and hosts are intimately coupled, tradeoffs with host range can occur across the virus-host relationship. We discuss this idea more generally in the next section.
3.4. Resistance to Infection: Tradeoffs Relating to Host Traits
Because viruses rely on hosts for their genetic material, tradeoffs associated with virus traits can be intimately tied to host traits as well. Tradeoffs around competitive and defensive traits in hosts (in the following referred to as cost of resistance, COR) are an intriguing example of such a link between virus and host traits.
3.4.1. COR in Natural Communities
Natural virus-host communities provide indirect evidence for COR. The apparent paradox between laboratory experiments, where host resistance readily evolves and eventually excludes viruses, and the large abundance of viruses in the pelagic environment (Weinbauer, 2004) gets resolved when considering COR. Specifically, resource limitation in natural environments may render expensive defense less viable. There is ample evidence of coexistence of susceptible and resistant hosts in natural communities (Waterbury and Valois, 1993; Tarutani et al., 2000; Holmfeldt et al., 2007; Middelboe et al., 2009), supporting the idea that COR prevents resistant types to outcompete susceptible types. Further support for COR is found in the observation that dominant host types tend to be both resistant (Rosenzweig, 1973; Lenski and Levin, 1985; Suttle and Chan, 1993; Waterbury and Valois, 1993; Malmstrom et al., 2004; Suttle, 2007; Middelboe et al., 2009; Campbell et al., 2011; Våge et al., 2013; Thingstad et al., 2014) and slow growing (Malmstrom et al., 2004; Suttle, 2007; Campbell et al., 2011; Samo et al., 2014; Thingstad et al., 2014). Finally, it appears that nested infection networks are widespread in natural communities (Chao et al., 1977; Flores et al., 2011; Jover et al., 2013; Koskella and Brockhurst, 2014; Martiny et al., 2014). In these networks, COR prevents defense-specialized hosts infected by generalist viruses only to outcompete competition-specialized hosts that are infected by most viruses. Supporting this idea, there is evidence that hosts infected by specialist viruses have faster growth rates than hosts infected by generalist viruses (Chao et al., 1977).
3.4.2. COR in Experimental Communities
Observing COR directly in experiments can be difficult, since COR may be small and dependent on the environment (Bohannan et al., 2002). Nevertheless, a number of studies have measured COR. The most prevalent expression of COR is a reduced growth rate in resistant types, as observed experimentally in prokaryotes including Escherichia coli (Lenski and Levin, 1985; Lenski, 1988; Bohannan et al., 1999; Bohannan and Lenski, 2000; Harcombe and Bull, 2005), Pseudomonas (Lythgoe and Chao, 2003), Synechococcus (Waterbury and Valois, 1993; Lennon et al., 2007), Flavobacteria (Middelboe et al., 2009), Prochlorococcus (Avrani et al., 2011) and eukaryotic Phaeocystis pouchetti (Haaber and Middelboe, 2009) and Ochromonas tauri (Thomas et al., 2011). Besides reduced growth rates, increased susceptibility to other viruses (Avrani et al., 2011, Prochlorococcus) and reduced abilities to form biofilms (Buckling and Rainey, 2002; Brockhurst et al., 2005, Pseudomonas) are other known expressions of COR.
3.4.3. Defense Mechanisms with Varying COR
Besides environmental conditions that may influence the expression of COR (Lennon et al., 2007), different defense mechanisms probably have varying COR, both in quality and quantity. Changes in surface receptors that hamper virus adsorption (Middelboe, 2000; Middelboe et al., 2001; Buckling and Rainey, 2002; Mizoguchi et al., 2003; Stoddard et al., 2007; Middelboe et al., 2009; Pagarete et al., 2009; Avrani et al., 2011; Bidle and Vardi, 2011) often hamper uptake of limiting nutrients as well, which can explain reduced growth rates in resistant hosts. On the other hand, internal defense mechanisms such as the CRISPR-Cas (Barrangou et al., 2007; Sorek et al., 2008; Levin, 2010; Makarova et al., 2011) and restriction enzymes (Wilson and Murray, 1991; Labrie et al., 2010) prevent take-over of the host by the virus after adsorption, which does not influence nutrient uptake dynamics directly. Instead, costs for these internal defense systems may arise from resource allocation and maintenance of the enzymatic machinery. It is conceivable that extending resistance to new viruses with these internal defense systems (e.g., by adding an additional recognition sequence in the CRISPR system) may be relatively inexpensive, but we lack quantitative evidence. Other mechanisms rendering resistance to various degrees, whose specific COR are poorly understood, include prophage incorporation (Stoddard et al., 2007; Martiny et al., 2014), chronic infection (Fuhrman, 1999; Thomas et al., 2011), immunization through viral lysate (Bidle and Vardi, 2011) and quorum sensing allowing a regulated expression of surface receptors (Høyland-Kroghsbo et al., 2013). The most drastic defense is abortive infection leading to induced cell death (Bidle and Vardi, 2011; Berngruber et al., 2013; Refardt et al., 2013).
3.4.4. Linking Host and Virus Traits through COR
An intriguing aspect of virus ecology is the strong link between virus and host traits. A resistance trait in the host leads to adaptive and evolutionary changes in the virus to sustain infectivity. Interestingly, however, COR also affects viruses in a more direct way. In a review on marine viruses, Suttle (2007) hypothesized inverse rank-abundance distributions of hosts and their associated viruses, where the most abundant hosts are low-active and infected by rare and low virulent viruses, while the rare hosts are active and infected by abundant and highly virulent viruses. Findings of abundant low-active and defensive hosts (Rosenzweig, 1973; Lenski and Levin, 1985; Suttle and Chan, 1993; Waterbury and Valois, 1993; Malmstrom et al., 2004; Suttle, 2007; Middelboe et al., 2009; Campbell et al., 2011; Våge et al., 2013; Samo et al., 2014; Thingstad et al., 2014) and inverse rank-abundance distributions emerging in virus-host interaction models when assuming COR (Våge et al., 2013; Thingstad et al., 2014) support Suttle's hypothesis. The positive correlation between host activity and virulence in this scenario provides a direct link between COR and viral infectivity.
Inverse rank abundance distributions also impose a tradeoff between host abundance and virulence, which becomes apparent when considering the model for host infection rates used earlier:
where R is the infection rate for Sj by Vi. Given inverse rank-abundance distributions as described by Suttle (2007), infection rates for highly competitive hosts should be reduced due to the low host abundance, despite high adsorption coefficients of their viruses, whereas infection rates for highly defensive hosts should be reduced due to the low adsorption coefficient of their viruses, despite the high host abundance. The consequence is that highest infection rates may occur at intermediate virulence and host defensiveness (Figure 3). This tradeoff between host abundance and virulence of the virus can only be corroborated once we overcome the major challenge of quantifying abundances of viruses associated with defensive vs. competitive hosts (Jover et al., 2013). A step in that direction can be made through methods that identify single virus-host pairs in natural communities, such as phageFish (e.g., Martiny et al., 2014).
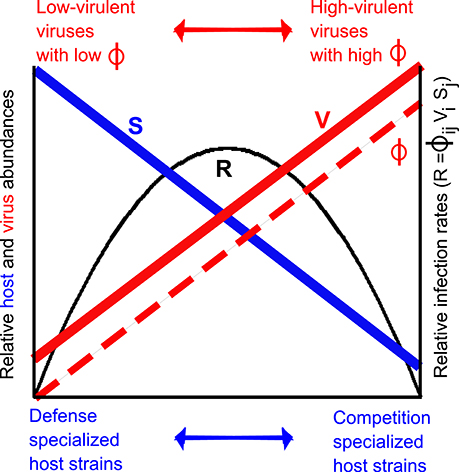
Figure 3. Illustration of virus tradeoff in inverse rank-abundance scenario. Defense specialized host strains are more abundant than competition specialized host strains (blue line), whereas low-virulent viruses (here illustrated by small interaction kernel ϕ) associated with defense specialists are less abundant than high-virulent viruses (large interaction kernel ϕ) associated with competition specialists (red line). Relative interaction kernel for pairs of host and associated viruses are shown in dashed red line. Relative infection rate R (Equation 13) is shown in black.
We also note that “host abundance” needs to be treated with care, as analysis of SSU rRNA used to quantify host abundances in the field resolves “species” level diversity, whereas virus-host interactions typically take place on the more strain-specific level. Investigations of virus-host interactions in natural communities will therefore rely on refinements in sequence-based methods, such as CRISPR spacer similarities and single-cell sequencing (Roux et al., 2016).
3.5. Summary
In this section, we have reviewed different virus traits and discussed quantification of potential tradeoffs by means of a dynamic NSIV model framework. We have shown that tradeoffs fundamentally influence community structure, and we have pointed out areas where more knowledge regarding tradeoffs would better inform our understanding. In the case studies we have discussed, we used trait-based tradeoffs to generate hypotheses, to explain observed patterns, and to link empirical and modeling work. In the virulence example, a steady-state analysis provided a hypothesized relationship between nutrient concentration and the fractions of hosts that are lytic and lysogenic. This hypothesis held up well to a small dataset of field measurements, and with additional support of field measurements could have important implications for the biogeography of lysogeny and lysis at basin and global scales. In the host range example, we explored a particular tradeoff between host range and interaction kernel. The simulation provided an explanation for the observed pattern where host range tends toward the extremes. Because host range can tradeoff with other traits as well (Jover et al., 2013), this is just one candidate explanation for the observed pattern, and it could be that the host range tradeoff involves multiple traits. Our review of the literature data (Table 3, Supplementary Material) provides strong evidence for host range tradeoffs, but no clear single trait involved in this tradeoff. In this case, analysis shows how a tradeoff can explain an observed pattern, and more experimental work is required to resolve the specifics of tradeoff and the traits involved. In these examples, the close interaction between virus traits and host traits is inescapable. Both the lysis-lysogeny switch and the host range tradeoff depend on traits such as μ-the host growth rate. The interaction between virus traits and host traits is perhaps most pronounced with COR, where this interaction can drive evolutionary dynamics and shape the rank-abundance structure of the community. Understanding these interactions and their dynamics would have value beyond virus ecology, to the field of trait-based ecology as a whole.
4. Future Directions
“The best theory is inspired by practice. The best practice is inspired by theory.”
One of the current challenges identified by the trait-based ecology community is bridging the gap between empiricists (i.e., practice) and modelers (i.e., theory) (Barton et al., 2016). This challenge is a common vein that runs through much of the history of science (Knuth, 1991), but has become particularly pronounced with the increased specialization required in both ecological modeling and experimentation. The quantification of ecological tradeoffs, as we have presented in this review, is an objective that provides a useful nexus of modeling and experimental work and an avenue that can cut across subdisciplines. The use of traits and tradeoffs to organize ecological information has the potential to offer a robust theory for understanding a system. Here we briefly discuss a few of the central questions in marine virus ecology, with an eye toward how the study of tradeoffs can inform these questions. We also emphasize points where the study of virus ecology can contribute new insights to trait-based ecology in general.
4.1. What Portion of Primary and Bacterial Production Goes to Viruses vs. to Grazers?
In some ways, viruses and grazers compete for the same resource. Additionally, virus infection could alter grazing rates. This question is key to understanding when and under what conditions production is exported or recycled (Fuhrman, 1999; Brussaard, 2004; Suttle, 2007; Weitz and Wilhelm, 2012). It is a major challenge and one for which models are indispensable (Weitz et al., 2015). To tackle this problem with models, traits that control consumption of bacteria and phytoplankton must be understood, but there is currently a lack of clarity on how to compare zooplankton traits with virus traits. For decades, zooplankton ecologists have evaluated grazing competitiveness by comparing parameters that control the shape of the curve defining zooplankton consumption as a function of prey concentration (Holling, 1959; Gentleman et al., 2003; Jeschke et al., 2004; Kiørboe, 2008; Prowe et al., 2012).
Typically, zooplankton consumption increases linearly with prey density at low prey concentrations and saturates toward a handling time limit—for example,
where H is the total prey concentration and Kg is the prey concentration at which predator consumption rate is half the maximal value, gmax. The parameters that define the shape of this curve, gmax and Kg, can be a useful guide for zooplankton ecologists to evaluate consumption of prey biomass. This raises the question of whether virus consumption of prey biomass can be described in a comparable way and, if so, how do the different rate constants compare.
If we assume that the total concentration of prey can be approximated by the total concentration of susceptible hosts, we can write an analogous expression in terms of virus abundance and traits (Supplementary Material),
Evaluating the parameters that control curve shapes may be a useful way to understand competition between viruses and grazers. To the best of our knowledge, very little work has been done to standardize and compare virus and zooplankton Holling curve parameters. Most virus models that explicitly resolve free-existing viruses assume a linear relation between virus production and host density, where the slope of the line is defined by adsorption and burst size (Thingstad, 2000; Beckett and Williams, 2013; Weitz et al., 2015). Yet, it is conceivable that at a sufficiently high host density, the burst becomes independent of encounter, and depends instead on the duration of the latent phase. Understanding tradeoffs among key parameters λ and ϕ will help evaluate whether losses due to viral infection saturate as a function of host cell density, with implications for transfer to higher trophic levels.
4.2. What Regulates Virus to Host Ratios?
Virus to host ratios have been measured as a proxy for the importance of viruses in aquatic systems for over two decades (Ogunseitan et al., 1990; Wommack and Colwell, 2000; Wigington et al., 2016). Numbers vary greatly (Wigington et al., 2016), but the ratio is typically considered to be roughly 10:1. What regulates this number still remains an important question, as the ratio has far reaching biogeochemical consequences (Fuhrman, 1999; Weinbauer, 2004; Suttle, 2007). A recent analysis of an idealized microbial food web model revealed intricate links between mechanisms within the bacterial host community and between different plankton functional types, providing a framework for how virus to host ratios may emerge in the bacterial community (Våge et al., 2016). Briefly, assuming total host abundance to be controlled by micrograzers as a result of their quick response to bacterial production (Azam et al., 1983) and assuming grazing to be non-selective, viral lysis compensates for differences in growth rates of the host strains. Virus abundance is thus positively correlated to the width of the growth rate spectrum in the host community. Interestingly, the width of the host growth rate spectrum and thus virus to bacteria ratios as well as food web structure at the level of plankton functional types are highly sensitive to COR. This suggests that mechanistically understanding and quantifying COR will be key to better understand aquatic microbial ecology and evolutionary dynamics. In trait-based ecology in general, the emphasis has been on tradeoffs within a species or trophic group, so an understanding of how traits can tradeoff between viruses and hosts has the potential to bring new knowledge to how traits can tradeoff across trophic groups more generally.
4.3. How Can the Trait-Based Approach Better Inform us on the Role of Marine Viruses in Regulating Climate?
Understanding climate and the cycling of carbon is one of the most pressing earth science challenges of the day. The role of marine viruses is complex and nuanced, and answering this question relies on incorporating knowledge of virus ecology into climate system models. Global and ocean scale climate models are state variable limited because of computational costs. Adding the level of complexity required to resolve food web dynamics, let alone virus ecology, is often too computationally burdensome at the global scale. Particular ecological components, if they are included at all, are typically simplified to a single state variable with fixed parameters. As computational restrictions are gradually overcome, tradeoffs offer one potentially useful technique for capturing some of the ecological complexity while minimizing state variable increases. If we relax the notion that parameter values (or traits) are fixed for species, but rather change and respond to other traits, we can capture some aspects of adaptation and diversity within modeled ecosystems. For example, virus infections can drive changes in host traits, such as growth rate, rapidly (Avrani and Lindell, 2015). By allowing parameter values to be flexible and depend on each other based on tradeoffs, we can capture some of the community dynamics without adding state variables. Virus-host dynamics are rapid enough to allow for studying these dynamics in detail and for revealing their underlying principles. Incorporating this information into climate models is a promising approach both to answer questions about the role of marine viruses in climate and to introduce ecological processes more generally into climate models.
5. Conclusion
Virus ecology and trait-based ocean ecology have new insights to offer each other. There are some ways in which trait-based approaches to ocean life can inform virus ecology. For example, the strongly size structured character of the ocean ecosystem has been explored in depth across trophic levels (cf. Andersen et al., 2016), but there is limited work on the allometry of marine viruses. A detailed investigation of the marine virus size spectrum could facilitate the incorporation of virus ecology into models. However, as we have discussed, there are ways in which virus allometry might deviate from pelagic food web allometry in general. Because of differences like this, the exchange of knowledge works in both directions: the study of virus ecology can also provide new insights to trait-based ecology. For example, the role of adaptation and evolution in altering traits dynamically has been a challenge to measure at higher trophic levels. This type of dynamic has been explored in models (Record et al., 2013a; Sauterey et al., 2014), but it is difficult to link with empirical work. The short time scales at which virus-host interactions take place makes it possible to quantify and study these dynamics, and the insights gained can help guide ecology at higher trophic levels. Along a similar vein, trait-based studies typically focus on the traits of the focal taxa, while ignoring the traits of adjacent trophic groups. The intimate interaction between virus replication and host reproduction at the genetic level forces us to consider both virus and host traits together. Because of these perspectives, virus ecology is uniquely poised to offer new insights to the broader field of trait-based ecology.
Author Contributions
NR, DT, and SV contributed equally to this manuscript.
Funding
This publication was supported by the Ocean Carbon and Biogeochemistry program (NSF, NASA). NR was supported by Bigelow Laboratory institutional funds. DT was supported by NSF grant OCE-1536521 and the Gordon and Betty Moore Foundation through grant GBMF3778 to M.J. Follows. SV was supported by the University of Bergen.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript is a product of a collaboration at the “Trait-based Approaches to Ocean Life” Scoping Workshop, which was supported by the Ocean Carbon and Biogeochemistry group (NSF, NASA), the Simons Foundation, and the Gordon and Betty Moore Foundation. We thank two reviewers for their input and Joaquín Martínez Martínez for valuable comments that greatly improved the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fmars.2016.00251/full#supplementary-material
References
Allen, J., Somerfield, P., and Gilbert, F. (2007). Quantifying uncertainty in high-resolution coupled hydrodynamic-ecosystem models. J. Mar. Syst. 64, 3–14. doi: 10.1016/j.jmarsys.2006.02.010
Andersen, K. H., Berge, T., Gonçalves, R. J., Hartvig, M., Heuschele, J., Hylander, S., et al. (2016). Characteristic sizes of life in the oceans, from bacteria to whales. Mar. Sci. 8, 217–241. doi: 10.1146/annurev-marine-122414-034144
Andersen, K. H., and Beyer, J. E. (2015). Size structure, not metabolic scaling rules, determines fisheries reference points. Fish Fish. 16, 1–22. doi: 10.1111/faf.12042
Avrani, S., and Lindell, D. (2015). Convergent evolution toward an improved growth rate and a reduced resistance range in prochlorococcus strains resistant to phage. Proc. Natl. Acad. Sci. U.S.A. 112, E2191–E2200. doi: 10.1073/pnas.1420347112
Avrani, S., Wurtzel, O., Sharon, I., Sorek, R., and Lindell, D. (2011). Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 474, 604–608. doi: 10.1038/nature10172
Azam, F., Fenchel, T., Field, J., Gray, J., Meyer-Reil, L., and Thingstad, F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Banas, N. S. (2011). Adding complex trophic interactions to a size-spectral plankton model: emergent diversity patterns and limits on predictability. Ecol. Model. 222, 2663–2675. doi: 10.1016/j.ecolmodel.2011.05.018
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). Crispr provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Barton, A., Finkel, Z., Ward, B., Johns, D., and Follows, M. (2013). On the roles of cell size and trophic strategy in North Atlantic diatom and dinoflagellate communities. Limnol. Oceanogr. 58, 254–266. doi: 10.4319/lo.2013.58.1.0254
Barton, A. D., Dutkiewicz, S., Andersen, K. H., Fiksen, Ø. Ø., Follows, M. J., Mouw, C. B., et al. (2016). Report on the “Trait-Based Approaches to Ocean Life” Scoping Workshop. Waterville Valley, NH: U.S. Ocean Carbon and Biogeochemistry Program, p. 35. doi: 10.1575/1912/8017
Baudoux, A.-C., and Brussaard, C. P. D. (2008). Influence of irradiance on virus-algal host interactions. J. Phycol. 44, 902–908. doi: 10.1111/j.1529-8817.2008.00543.x
Baudoux, A.-C., and Brussard, C. P. D. (2005). Characterization of different viruses infecting the marine harmful algal bloom species phaeocystis globosa. Virology 341, 80–90. doi: 10.1016/j.virol.2005.07.002
Beckett, S., and Williams, H. (2013). Coevolutionary diversification creates nested-modular structure in phage bacteria interaction networks. Interface Focus 3:20130033. doi: 10.1098/rsfs.2013.0033
Beltrami, E., and Carroll, T. (1994). Modeling the role of viral disease in recurrent phytoplankton blooms. J. Math. Biol. 32, 857–863. doi: 10.1007/BF00168802
Berngruber, T. W., Lion, S., and Gandon, S. (2013). Evolution of suicide as a defence strategy against pathogens in a spatially structured environment. Ecol. Lett. 16, 446–453. doi: 10.1111/ele.12064
Bidle, K., and Vardi, A. (2011). A chemical arms race at sea mediates algal host-virus interactions. Curr. Opin. Microbiol. 14, 449–457. doi: 10.1016/j.mib.2011.07.013
Bohannan, B., and Lenski, R. (2000). Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3, 362–377. doi: 10.1046/j.1461-0248.2000.00161.x
Bohannan, B. J., Kerr, B., Jessup, C. M., Hughes, J. B., and Sandvik, G. (2002). Trade-offs and coexistence in microbial microcosms. Antonie Van Leeuwenhoek 81, 107–115. doi: 10.1023/A:1020585711378
Bohannan, B. J. M., Travisano, M., and Lenski, R. E. (1999). Epistatic interactions can lower the cost of resistance to multiple consumers. Evolution 53, 292–295.
Bratbak, G., Egge, J., and Heldal, M. (1993). Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar. Ecol. Prog. Ser. 93, 39–48.
Bratbak, G., Jacobsen, A., Heldal, M., Nagasaki, K., and Thingstad, F. (1998). Virus production in Phaeocystis pouchetii and its relation to host cell growth and nutrition. Aquat. Microb. Ecol. 16, 1–9.
Brockhurst, M. A., Buckling, A., and Rainey, P. B. (2005). The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc. Biol. Sci. 272, 1385–1391. doi: 10.1098/rspb.2005.3086
Brown, C., Lawrence, J., and Campbell, D. (2006). Are phytoplankton population density maxima predictable through analysis of host and viral genomic DNA content? J. Mar. Biol. Assoc. U.K. 86:491. doi: 10.1017/s0025315406013397
Brum, J. R., Hurwitz, B. L., Schofield, O., Ducklow, H. W., and Sullivan, M. B. (2016). Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 10, 437–449. doi: 10.1038/ismej.2015.125
Brussaard, C. (2004). Viral control of phytoplankton populations - A review. J. Eukaryot. Microbiol. 51, 125–138. doi: 10.1111/j.1550-7408.2004.tb00537.x
Brüssow, H., Canchaya, C., and Hardt, W. D. (2004). Phages and the evolution of bacterial pathogens : from genomic rearrangements to lysogenic conversion phages and the evolution of bacterial pathogens : from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602. doi: 10.1128/MMBR.68.3.560-602.2004
Buckling, A., and Rainey, P. B. (2002). Antagonistic coevolution between a bacterium and a bacteriophage. Proc. Biol. Sci. 269, 931–936. doi: 10.1098/rspb.2001.1945
Campbell, B. J., Yu, L., Heidelberg, J. F., and Kirchman, D. L. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. U.S.A. 108, 12776–12781. doi: 10.1073/pnas.1101405108
Casjens, S., Palmer, N., Van Vugt, R., Huang, W. M., Stevenson, B., Rosa, P., et al. (2000). A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35, 490–516. doi: 10.1046/j.1365-2958.2000.01698.x
Chao, L., Levin, B. R., and Stewart, F. M. (1977). A complex community in a simple habitat: an experimental study with bacteria and phage. Ecology 58, 369–378. doi: 10.2307/1935611
Chattopadhyay, J., and Pal, S. (2002). Viral infection on phytoplankton-zooplankton system - a mathematical model. Ecol. Model. 151, 15–28. doi: 10.1016/S0304-3800(01)00415-X
Chiura, H. X. (1997). Generalized gene transfer by virus-like particles from marine bacteria. Aquat. Microb. Ecol. 13, 75–83.
Claudet, J., Osenberg, C. W., Domenici, P., Badalamenti, F., Milazzo, M., Falcón, J., et al. (2010). Marine reserves: fish life history and ecological traits matter. Ecol. Appl. 20, 830–839. doi: 10.1890/08-2131.1
Cochran, P. K., and Paul, J. H. (1998). Seasonal abundance of lysogenic bacteria in a subtropical estuary. Appl. Environ. Microbiol. 64, 2308–2312.
Comeau, A. M., Buenaventura, E., and Suttle, C. A. (2005). A persistent, productive, and seasonally dynamic vibriophage population within pacific oysters (Crassostrea gigas). Appl. Environ. Microbiol. 71, 5324–5331. doi: 10.1128/AEM.71.9.5324-5331.2005
Comeau, A. M., Chan, A. M., and Suttle, C. A. (2006). Genetic richness of vibriophages isolated in a coastal environment. Environ. Microbiol. 8, 1164–1176. doi: 10.1111/j.1462-2920.2006.01006.x
Daubin, V., Lerat, E., and Perrière, G. (2003). The source of laterally transferred genes in bacterial genomes. Genome Biol. 4:R57. doi: 10.1186/gb-2003-4-9-r57
De La Rocha, C., and Passow, U. (2007). Factors influencing the sinking of POC and the efficiency of the biological carbon pump. Deep Sea Res. II 54, 639–658. doi: 10.1016/j.dsr2.2007.01.004
De Paepe, M., and Taddei, F. (2006). Viruses' life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 4:e193. doi: 10.1371/journal.pbio.0040193
Deng, L., Krauss, S., Feichtmayer, J., Hofmann, R., Arndt, H., and Griebler, C. (2014). Grazing of heterotrophic flagellates on viruses is driven by feeding behaviour. Environ. Microbiol. Rep. 6, 325–330. doi: 10.1111/1758-2229.12119
DePaola, A., Motes, M. L., Chan, A. M., and Suttle, C. A. (1998). Phages infecting vibrio vulnificus are abundant and diverse in oysters (Crassostrea virginica) collected from the gulf of mexico. Appl. Environ. Microbiol. 64, 346–351.
Edgar, R., and Lielausis, I. (1964). Temperature-sensitive mutants of bacteriophage T4D: their isolation and genetic characterization. Genetics 49, 649–662.
Edlin, G., Lin, L., and Kudrna, R. (1975). Lambda lysogens of E. coli reproduce more rapidly than non-lysogen. Nature 255, 735–737.
Edwards, K., Thomas, M., Klausmeier, C., and Litchman, E. (2012). Allometric scaling and taxonomic variation in nutrient utilization traits and maximum growth rate of phytoplankton. Limnol. Oceanogr. 57, 554–566. doi: 10.4319/lo.2012.57.2.0554
Egas, M., Dieckmann, U., and Sabelis, M. W. (2004). Evolution restricts the coexistence of specialists and generalists: the role of trade-off structure. Am. Nat. 163, 518–531. doi: 10.1086/382599
Faruque, S., Asadulghani, Rahman, M. M., Waldor, M. K., and Sack, D. A. (2000). Sunlight-induced propagation of the lysogenic phage encoding cholera toxin. Infect. Immun. 68, 4795–4801. doi: 10.1128/IAI.68.8.4795-4801.2000
Flores, C. O., Meyer, J. R., Valverde, S., Farr, L., and Weitz, J. S. (2011). Statistical structure of host-phage interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 288–297. doi: 10.1073/pnas.1101595108
Follows, M., Dutkiewicz, S., Grant, S., and Chisholm, S. W. (2007). Emergent biogeography of microbial communities in a model ocean. Science 315, 1843–1846. doi: 10.1126/science.1138544
Fuhrman, J. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548.
Fuhrman, J. A., and Noble, R. T. (1995). Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol. Oceanogr. 40, 1236–1242.
Gentleman, W., Leising, A., Frost, B., Strom, S., and Murray, J. (2003). Functional responses for zooplankton feeding on multiple resources: a review of assumptions and biological dynamics. Deep. Res. II Top. Stud. Oceanogr. 50, 2847–2875. doi: 10.1016/j.dsr2.2003.07.001
Ghosh, D., Roy, K., Williamson, K. E., Srinivasiah, S., Wommack, K. E., and Radosevich, M. (2009). Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl. Environ. Microbiol. 75, 7142–7152. doi: 10.1128/AEM.00950-09
Gilchrist, G. W. (1995). Specialists and generalists in changing environments. I. fitness landscapes of thermal sensitivity. Am. Nat. 146, 252–270.
Golet, W. J., Record, N. R., Lehuta, S., Lutcavage, M., Galuardi, B., Cooper, A. B., et al. (2015). The paradox of the pelagics: why bluefin tuna can go hungry in a sea of plenty. Mar. Ecol. Prog. Ser. 527, 181–192. doi: 10.3354/meps11260
Gough, M. (1968). Second locus of bacteriophage P22 necessary for the maintenance of lysogeny. J. Virol. 2, 992–998.
Haaber, J., and Middelboe, M. (2009). Viral lysis of Phaeocystis pouchetii: implications for algal population dynamics and heterotrophic c, n and p cycling. ISME J. 3, 430–441. doi: 10.1038/ismej.2008.125
Hansen, B., Gaard, E., and Reinert, J. (1994). “Physical effects on recruitment of faroe plateau cod,” in ICES Marine Science Symposia. Copenhagen [ICES MAR. SCI. SYMP.]., eds J. Jakobsson, O. Astthorsson, R. Beverton, B. Bjoernsson, N. Daan, K. Frank, J. Meincke, B. Rothschild, S. Sundby, and S. Tilseth, International Counc. for the Exploration of the Sea (Copenhagen).
Harcombe, W. R., and Bull, J. J. (2005). Impact of phages on two-species bacterial communities. Appl. Environ. Microbiol. 71, 5254–5259. doi: 10.1128/AEM.71.9.5254-5259.2005
Hayashi, T., Makino, K., Ohnishi, M., Kurokawa, K., Ishii, K., Yokoyama, K., et al. (2001). Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8, 11–22. doi: 10.1093/dnares/8.1.11
Holling, C. (1959). The Components of predation as revealed by a study of small-mammal predation of the European pine sawfly. Can. Entomol. 91, 293–320.
Holmfeldt, K., Middelboe, M., Nybroe, O., and Riemann, L. (2007). Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their flavobacterium hosts. Appl. Environ. Microbiol. 73, 6730–6739. doi: 10.1128/AEM.01399-07
Høyland-Kroghsbo, N. M., Mærkedahl, R. B., and Svenningsen, S. L. (2013). A quorum-sensing-induced bacteriophage defense mechanism. MBio 4, e00362-e00312. doi: 10.1128/mBio.00362-12
Jeschke, J. M., Kopp, M., and Tollrian, R. (2004). Consumer-food systems: why type I functional responses are exclusive to filter feeders. Biol. Rev. 79, 337–349. doi: 10.1017/S1464793103006286
Jessup, C. M., and Bohannan, B. J. (2008). The shape of an ecological trade-off varies with environment. Ecol. Lett. 11, 947–959. doi: 10.1111/j.1461-0248.2008.01205.x
Jiang, S., and Paul, J. (1994). Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine environment. Mar. Ecol. Prog. Ser. 104, 163–172.
Jiang, S., and Paul, J. (1996). Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar. Ecol. Prog. Ser. 142, 27–38.
Jover, L. F., Effler, T. C., Buchan, A., Wilhelm, S. W., and Weitz, J. S. (2014). The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat. Rev. Microbiol. 12, 519–528. doi: 10.1038/nrmicro3289
Jover, L. F., Cortez, M. H., and Weitz, J. S. (2013). Mechanisms of multi-strain coexistence in host–phage systems with nested infection networks. J. Theor. Biol. 332, 65–77. doi: 10.1016/j.jtbi.2013.04.011
Kattge, J., Ogle, K., Bönisch, G., Díaz, S., Lavorel, S., Madin, J., et al. (2011). A generic structure for plant trait databases. Methods Ecol. Evol. 2, 202–213. doi: 10.1111/j.2041-210X.2010.00067.x
Keen, E. C. (2014). Tradeoffs in bacteriophage life histories. Bacteriophage 4:e28365. doi: 10.4161/bact.28365
Kempes, C. P., Dutkiewicz, S., and Follows, M. J. (2012). Growth, metabolic partitioning, and the size of microorganisms. Proc. Natl. Acad. Sci. U.S.A. 109, 495–500. doi: 10.1073/pnas.1115585109
Kermack, W. O., and McKendrick, A. G. (1927). A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. A 115, 700–721.
Kiørboe, T. (2008). A mechanistic Approach to Plankton Ecology. Princeton, NJ: Princeton University Press.
Kiørboe, T., Andersen, A., Langlois, V. J., and Jakobsen, H. H. (2010). Unsteady motion: escape jumps in planktonic copepods, their kinematics and energetics. J. R. Soc. Interface 7, 1591–1602. doi: 10.1098/rsif.2010.0176
Knowles, B., Silveira, C. B., Bailey, B. A., Barott, K., Cantu, V. A., Cobián-Güemes, A., et al. (2016). Lytic to temperate switching of viral communities. Nature 531, 466–470. doi: 10.1038/nature17193
Kokjohn, T., Sayler, G., and Miller, R. (1991). Attachment and replication of Pseudomonas aeruginosa bacteriophages under conditions simulating aquatic environments. J. Gen. Microbiol. 137, 661–666.
Koskella, B., and Brockhurst, M. A. (2014). Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 38, 916–931. doi: 10.1111/1574-6976.12072
Koudelka, G. B., Harbury, P., Harrison, S. C., and Ptahsne, M. (1988). Dna twisting and the affinity of bacteriophage 434 operator for bacteriophage 434 represso. Proc. Natl. Acad. Sci. U.S.A. 85, 4633–4637.
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Lawrence, J. G., and Ochman, H. (1998). Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. U.S.A. 95, 9413–9417.
Lennon, J., Khatana, S., Marston, M., and Martiny, J. (2007). Is there a cost of virus resistance in marine cyanobacteria? ISME J. 1, 300–312. doi: 10.1038/ismej.2007.37
Lenski, R. E. (1988). Experimental studies of pleiotropy and epistasis in Escherichia coli. I. variation in competitive fitness among mutants resistant to virus T4. Evolution 42, 425–432.
Lenski, R. E., and Levin, B. R. (1985). Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am. Nat. 125, 585–602.
Levin, B. R., and Lenski, R. E. (1983). “Coevolution in bacteria and their viruses and plasmids,” in Coevolution, eds D. J. Futuyma and M. Slatkin (Sunderland, MA: Sinauer Associates Inc.), 99–127.
Levin, B. R. (2010). Nasty viruses, costly plasmids, population dynamics, and the conditions for establishing and maintaining crispr-mediated adaptive immunity in bacteria. PLoS Genet. 6:e1001171. doi: 10.1371/journal.pgen.1001171
Lima-Mendez, G., Faust, K., Henry, N., Decelle, J., Colin, S., Carcillo, F., et al. (2015). Determinants of community structure in the global plankton interactome. Science 348:1262073. doi: 10.1126/science.1262073
Litchman, E., and Klausmeier, C. (2008). Trait-based community ecology of phytoplankton. Annu. Rev. Ecol. Evol. Syst. 39, 615–639. doi: 10.1146/annurev.ecolsys.39.110707.173549
Litchman, E., Klausmeier, C. A., Schofield, O. M., and Falkowski, P. G. (2007). The role of functional traits and trade-offs in structuring phytoplankton communities: scaling from cellular to ecosystem level. Ecol. Lett. 10, 1170–1181. doi: 10.1111/j.1461-0248.2007.01117.x
Litchman, E., Ohman, M. D., and Kiørboe, T. (2013). Trait-based approaches to zooplankton communities. J. Plankton Res. 35, 473–484. doi: 10.1093/plankt/fbt019
Liu, H., Laws, E., Villareal, T., and Buskey, E. (2001). Nutrient-limited growth of Aureoumbra lagunensis (Pelagophyceae), with implications for its capability to outgrow other phytoplankton species in phosphate-limited environments. J. Phycol. 37, 500–508. doi: 10.1046/j.1529-8817.2001.037004500.x
López-Urrutia, Á., San Martin, E., Harris, R. P., and Irigoien, X. (2006). Scaling the metabolic balance of the oceans. Proc. Nat. Acad. Sci. U.S.A. 103, 8739–8744. doi: 10.1073/pnas.0601137103
Lymer, D., and Lindström, E. (2010). Changing phosphorus concentration and subsequent prophage induction alter composition of a freshwater viral assemblage. Freshw. Biol. 55, 1984–1996. doi: 10.1111/j.1365-2427.2010.02435.x
Lythgoe, K., and Chao, L. (2003). Mechanisms of coexistence of a bacteria and a bacteriophage in a spatially homogeneous environment. Ecol. Lett. 6, 326–334. doi: 10.1046/j.1461-0248.2003.00433.x
Ma, J., and Levin, S. A. (2006). The evolution of resource adaptation: how generalist and specialist consumers evolve. Bull. Math. Biol. 68, 1111–1123. doi: 10.1007/s11538-006-9096-6
Makarova, K. S., Wolf, Y. I., Snir, S., and Koonin, E. V. (2011). Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 193, 6039–6056. doi: 10.1128/JB.05535-11
Malki, K., Kula, A., Bruder, K., Sible, E., Hatzopoulos, T., Steidel, S., et al. (2015). Bacteriophages isolated from lake michigan demonstrate broad host-range across several bacterial phyla. Virol. J. 12, 164. doi: 10.1186/s12985-015-0395-0
Malmstrom, R. R., Kiene, R. P., Cottrell, M. T., and Kirchman, D. L. (2004). Contribution of sar11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic Ocean. Appl. Environ. Microbiol. 70, 4129–4135. doi: 10.1128/AEM.70.7.4129-4135.2004
Marras, S., Killen, S. S., Domenici, P., Claireaux, G., and McKenzie, D. J. (2013). Relationships among traits of aerobic and anaerobic swimming performance in individual european sea bass Dicentrarchus labrax. PLoS ONE 8:e72815. doi: 10.1371/journal.pone.0072815
Martínez, J. M., Boere, A., Gilg, I., van Lent, J. W., Witte, H. J., van Bleijswijk, J. D., et al. (2015). New lipid envelope-containing dsdna virus isolates infecting micromonas pusilla reveal a separate phylogenetic group. Aquat. Microbiol. Ecol. 74, 17–28. doi: 10.3354/ame01723
Martiny, J. B. H., Riemann, L., Marston, M. F., and Middelboe, M. (2014). Antagonistic coevolution of marine planktonic viruses and their hosts. Mar. Sci. 6, 393–414. doi: 10.1146/annurev-marine-010213-135108
Marvier, M., Kareiva, P., and Neubert, M. G. (2004). Habitat destruction, fragmentation, and disturbance promote invasion by habitat generalists in a multispecies metapopulation. Risk Anal. 24, 869–878. doi: 10.1111/j.0272-4332.2004.00485.x
Maurice, C. F., Bouvier, T., Comte, J., Guillemette, F., and Del Giorgio P. (2010). Seasonal variations of phage life strategies and bacterial physiological states in three northern temperate lakes. Environ. Microbiol. 12, 628–641. doi: 10.1111/j.1462-2920.2009.02103.x
McDaniel, L., DelaRosa, M., and Paul, J. (2006). Temperate and lytic cyanophages from the Gulf of Mexico. J. Mar. Biol. Assoc. U.K. 86, 517–527. doi: 10.1017/S0025315406013427
McDaniel, L., Houchin, L. A., Williamson, S. J., and Paul, J. H. (2002). Lysogeny in marine Synechococcus. Nature 415:496. doi: 10.1038/415496a
McDaniel, L., and Paul, J. (2005). Effect of nutrient addition and environmental factors on prophage induction in natural populations of marine Synechococcus species. Appl. Environ. Microbiol. 71, 842–850. doi: 10.1128/AEM.71.2.842-850.2005
Middelboe, M. (2000). Bacterial growth rate and marine virus–host dynamics. Microb. Ecol. 40, 114–124. doi: 10.1007/s00248-001-0012-1
Middelboe, M., Hagström, A., Blackburn, N., Sinn, B., Fischer, U., Borch, N., et al. (2001). Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microb. Ecol. 42, 395–406. doi: 10.1007/s00248-001-0012-1
Middelboe, M., Holmfeldt, K., Riemann, L., Nybroe, O., and Haaber, J. (2009). Bacteriophages drive strain diversification in a marine flavobacterium: implications for phage resistance and physiological properties. Environ. Microbiol. 11, 1971–1982. doi: 10.1111/j.1462-2920.2009.01920.x
Middleton, J., Martínez Martínez, J., Wilson, W., and Record, N. (in press). Functional dynamics of Emiliania huxleyi virus-host interactions across multiple spatial scales. Limnol. Oceanogr.
Mizoguchi, K., Morita, M., Fischer, C. R., Yoichi, M., Tanji, Y., and Unno, H. (2003). Coevolution of bacteriophage pp01 and Escherichia coli o157: H7 in continuous culture. Appl. Environ. Microbiol. 69, 170–176. doi: 10.1128/AEM.69.1.170-176.2003
Moebus, K. (1996). Marine bacteriophage reproduction under nutrient-limited growth of host bacteria. II. investigations with phage-host system [H3:H3/1]. Mar. Ecol. Prog. Ser. 144, 13–22.
Mojica, K. D., and Brussaard, C. P. (2014). Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol. Ecol. 89, 495–515. doi: 10.1111/1574-6941.12343
Murray, A., and Jackson, G. (1992). Viral dynamics : a model of the effects of size, shape, motion and abundance of single-celled planktonic organisms and other particles. Mar. Ecol. Prog. Ser. 89, 103–116.
Murray, A. G., and Jackson, G. A. (1993). Viral dynamics II: a model of the interaction of ultraviolet light and mixing processes on virus survival in seawater. Mar. Ecol. Prog. Ser. 102, 105–114.
Nagasaki, K., Ando, M., Imai, I., Itakura, S., and Ishida, Y. (1994). Virus-like particles in Heterosigma akashiwo (raphidophyceae): a possible red tide disintegration mechanism. Mar. Biol. 119, 307–312.
Nagasaki, K., Tomaru, Y., Katanozaka, N., Nishida, K., Itakura, S., Shirai, Y., et al. (2004). Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl. Environ. Microbiol. 70, 704–711. doi: 10.1128/AEM.70.2.704-711.2004
Ochman, H., Lawrence, J. G., and Groisman, E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304. doi: 10.1038/35012500
Ogunseitan, O. A., Sayler, G. S., and Miller, R. V. (1990). Dynamic interactions of Pseudomonas aeruginosa and bacteriophages in lake water. Microb. Ecol. 19, 171–185.
Opdal, A. F., and Jørgensen, C. (2015). Long-term change in a behavioural trait: truncated spawning distribution and demography in northeast arctic cod. Glob. Change Biol. 21, 1521–1530. doi: 10.1111/gcb.12773
Pagarete, A., Allen, M. J., Wilson, W. H., Kimmance, S. A., and de Vargas, C. (2009). Host-virus shift of the sphingolipid pathway along an Emiliania huxleyi bloom: survival of the fattest. Environ. Microbiol. 11, 2840–2848. doi: 10.1111/j.1462-2920.2009.02006.x
Paul, J. H., and Jiang, S. C. (2001). Lysogeny and transduction. Method Microbiol. 30, 105–125. doi: 10.1016/S0580-9517(01)30042-9
Payet, J., and Suttle, C. (2013). To kill or not to kill: the balance between lytic and lysogenic viral infection is driven by trophic status. Limnol. Oceanogr. 58, 465–474. doi: 10.4319/lo.2013.58.2.0465
Pimentel, M. S., Faleiro, F., Marques, T., Bispo, R., Dionísio, G., Faria, A. M., et al. (2016). Foraging behaviour, swimming performance and malformations of early stages of commercially important fishes under ocean acidification and warming. Clim. Change 137, 495–509. doi: 10.1007/s10584-016-1682-5
Pradeep Ram, A. S., and Sime-Ngando, T. (2010). Resources drive trade-off between viral lifestyles in the plankton: evidence from freshwater microbial microcosms. Environ. Microbiol. 12, 467–479. doi: 10.1111/j.1462-2920.2009.02088.x
Proctor, L., and Fuhrman, J. (1991). Roles of virus infection in organic particle flux. Mar. Ecol. Prog. Ser. 69, 133–142.
Proulx, S. R., Promislow, D. E., and Phillips, P. C. (2005). Network thinking in ecology and evolution. Trends Ecol. Evol. 20, 345–353. doi: 10.1016/j.tree.2005.04.004
Prowe, A., Pahlow, M., Dutkiewicz, S., Follows, M., and Oschlies, A. (2012). Top-down control of marine phytoplankton diversity in a global ecosystem model. Prog. Oceanogr. 101, 1–13. doi: 10.1016/j.pocean.2011.11.016
Ptashne, M. (1967). Specific binding of the lambda phage repressor to lambda DNA. Nature 214, 232–234.
Ptashne, M. (2004). A Genetic Switch: Phage Lambda Revisited. Palo Alto, CA: Cold Spring Harbour Laboratory Press.
Record, N., Pershing, A., and Maps, F. (2012). First principles of copepod development help explain global marine diversity pattern. Oecologia 170, 289–295. doi: 10.1007/s00442-012-2313-0
Record, N. R., Pershing, A. J., and Maps, F. (2013a). Emergent copepod communities in an adaptive trait-structured model. Ecol. Model. 260, 11–24. doi: 10.1016/j.ecolmodel.2013.03.018
Record, N. R., Pershing, A. J., and Maps, F. (2013b). The paradox of the “paradox of the plankton”. ICES J. Mar. Sci. 71, 236–240. doi: 10.1093/icesjms/fst049
Refardt, D., Bergmiller, T., and Kümmerli, R. (2013). Altruism can evolve when relatedness is low: evidence from bacteria committing suicide upon phage infection. Proc. R. Soc. Lond. B 280:20123035. doi: 10.1098/rspb.2012.3035
Rhodes, C. J., and Martin, A. P. (2010). The influence of viral infection on a plankton ecosystem undergoing nutrient enrichment. J. Theor. Biol. 265, 225–237. doi: 10.1016/j.jtbi.2010.04.022
Rose, K. A., Megrey, B. A., Werner, F. E., and Ware, D. M. (2007). Calibration of the NEMURO nutrient-phytoplankton-zooplankton food web model to a coastal ecosystem: evaluation of an automated calibration approach. Ecol. Model. 202, 38–51. doi: 10.1016/j.ecolmodel.2006.08.016
Roux, S., Brum, J. R., Dutilh, B. E., Sunagawa, S., Duhaime, M. B., Loy, A., et al. (2016). Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537, 689–693. doi: 10.1038/nature19366
Samo, T. J., Smriga, S., Malfatti, F., Sherwood, B. P., and Azam, F. (2014). Broad distribution and high proportion of protein synthesis active marine bacteria revealed by click chemistry at the single cell level. Front. Mar. Sci. 1:48. doi: 10.3389/fmars.2014.00048
Sauterey, B., Ward, B. A., Follows, M. J., Bowler, C., and Claessen, D. (2014). When everything is not everywhere but species evolve: an alternative method to model adaptive properties of marine ecosystems. J. Plankton Res. 37, 28–47. doi: 10.1093/plankt/fbu078
Schroeder, D. C., Oke, J., Malin, G., and Wilson, W. H. (2002). Coccolithovirus (Phycodnaviridae): characterisation of a new large dsDNA algal virus that infects Emiliana huxleyi. Arch. Virol. 147, 1685–1698. doi: 10.1007/s00705-002-0841-3
Sheldon, R., Prakash, A., and Sutcliffe, W. (1972). The size distribution of particles in the ocean. Limnol. Oceanogr. 17, 327–340.
Siekmann, I., and Malchow, H. (2008). An extension of the beretta-kuang model of viral disease. Math. Biosci. Eng. 5, 549–565. doi: 10.3934/mbe.2008.5.549
Singh, B. K., Chattopadhyay, J., and Sinha, S. (2004). The role of virus infection in a simple phytoplankton zooplankton system. J. Theor. Biol. 231, 153–166. doi: 10.1016/j.jtbi.2004.06.010
Sorek, R., Kunin, V., and Hugenholtz, P. (2008). Crispr—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6, 181–186. doi: 10.1038/nrmicro1793
Stenholm, A. R., Dalsgaard, I., and Middelboe, M. (2008). Isolation and characterization of bacteriophages infecting the fish pathogen flavobacterium psychrophilum. Appl. Environ. Microbiol. 74, 4070–4078. doi: 10.1128/AEM.00428-08
Stewart, F. M., and Levin, B. R. (1984). The population biology of bacterial viruses: why be temperate. Theor. Popul. Biol. 26, 93–117.
Stoddard, L. I., Martiny, J. B., and Marston, M. F. (2007). Selection and characterization of cyanophage resistance in marine Synechococcus strains. Appl. Environ. Microbiol. 73, 5516–5522. doi: 10.1128/AEM.00356-07
Sullivan, M. B., Waterbury, J. B., and Chisholm, S. W. (2003). Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424, 1047–1051. doi: 10.1038/nature01929
Suttle, C. (2007). Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
Suttle, C., and Chan, A. (1993). Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Mar. Ecol. Prog. Ser. 92, 99–109.
Suttle, C. A. (1994). The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 28, 237–243.
Tarutani, K., Nagasaki, K., and Yamaguchi, M. (2000). Viral impacts on total abundance and clonal composition of the harmful bloom-forming phytoplankton Heterosigma akashiwo. Appl. Environ. Microbiol. 66, 4916–4920. doi: 10.1128/AEM.66.11.4916-4920.2000
Thingstad, T. (2000). Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45, 1320–1328. doi: 10.4319/lo.2000.45.6.1320
Thingstad, T. F., Våge, S., Storesund, J. E., Sandaa, R. A., and Giske, J. (2014). A theoretical analysis of how strain-specific viruses can control microbial species diversity. Proc. Natl. Acad. Sci. U.S.A. 111, 7813–7818. doi: 10.1073/pnas.1400909111
Thingstad, T. F., Pree, B., Giske, J., and Våge, S. (2015). What difference does it make if viruses are strain-, rather than species-specific? Front. Microbiol. 6:320. doi: 10.3389/fmicb.2015.00320
Thomas, R., Grimsley, N., Escande, M. L., Subirana, L., Derelle, E., and Moreau, H. (2011). Acquisition and maintenance of resistance to viruses in eukaryotic phytoplankton populations. Environ. Microbiol. 13, 1412–1420. doi: 10.1111/j.1462-2920.2011.02441.x
Thyrhaug, R., Larsen, A., Thingstad, T., and Bratbak, G. (2003). Stable coexistence in marine algal host-virus systems. Mar. Ecol. Prog. Ser. 254, 27–35. doi: 10.3354/meps254027
Våge, S., Pree, B., and Thingstad, T. F. (2016). Linking internal and external bacterial community control gives mechanistic framework for pelagic virus-to-bacteria ratios. Environ. Microbiol. 18, 3932–3948. doi: 10.1111/1462-2920.13391
Våge, S., Storesund, J. E., and Thingstad, T. F. (2013). Adding a cost of resistance description extends the ability of virus-host model to explain observed patterns in structure and function of pelagic microbial communities. Environ. Microbiol. 15, 1842–1852. doi: 10.1111/1462-2920.12077
Van Etten, J. L., Burbank, D. E., Kia, Y., and Meints, R. H. (1983). Growth cycle of a virus, PBCV-1, that infects clorella-like algae. Virology 126, 117–125.
Violle, C., Navas, M., Vile, D., Kazakou, E., Fortunel, C., Hummel, I., et al. (2007). Let the concept of trait be functional! Oikos 116, 882–892. doi: 10.1111/j.0030-1299.2007.15559.x
Wang, I.-N. (2006). Lysis timing and bacteriophage fitness. Genetics 172, 17–26. doi: 10.1534/genetics.105.045922
Waterbury, J. B., and Valois, F. W. (1993). Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59, 3393–3399.
Weinbauer, M. (2004). Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181. doi: 10.1016/j.femsre.2003.08.001
Weinbauer, M., Brettar, I., and Höfle, M. (2003). Lysogeny and virus-induced mortality of bacterioplankton in surface, deep, and anoxic marine waters. Limnol. Oceanogr. 48, 1457–1465. doi: 10.4319/lo.2003.48.4.1457
Weinbauer, M. G., and Suttle, C. A. (1996). Potential significance of lysogeny to bacteriophage production and bacterial mortality in coastal waters of the gulf of Mexico. Appl. Environ. Microbiol. 62, 4374–4380.
Weitz, J., Beckett, S., Brum, J., Cael, B., and Dushoff, J. (2016). Lysis, lysogeny, and virus-microbe ratios. bioRxiv 1–5. doi: 10.1101/051805
Weitz, J. S., and Wilhelm, S. W. (2012). Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol. Rep. 4:17. doi: 10.3410/b4-17
Weitz, J. S., Stock, C. A., Wilhelm, S. W., Bourouiba, L., Coleman, M. L., Buchan, A., et al. (2015). A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J. 9, 1352–1364. doi: 10.1038/ismej.2014.220
Wichels, A., Biel, S. S., Gelderblom, H. R., Brinkhoff, T., Muyzer, G., and Schütt, C. (1998). Bacteriophage diversity in the north sea. Appl. Environ. Microbiol. 64, 4128–4133.
Wigington, C. H., Sonderegger, D., Brussard, C. P. D., Buchan, A., Finke, J. F., Fuhrman, J. A., et al. (2016). Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 1:15024. doi: 10.1038/nmicrobiol.2015.24
Wilcox, R., and Fuhrman, J. (1994). Bacterial viruses in coastal seawater: lytic rather than lysogenic production. Mar. Ecol. Prog. Ser. 114, 35–46.
Wilhelm, S. W., and Suttle, C. A. (1999). Viruses and nutrient cycles in the sea viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49, 781–788.
Wilhelm, S. W., Jefferey, W. H., Dean, A. L., Meador, J., Pakulski, J. D., and Mitchell, D. L. (2003). UV radiation induced DNA damage in marine viruses along a latitudinal gradient in the southereastern pacific ocean. Aquat. Microb. Ecol. 31, 1–8. doi: 10.3354/ame031001
Williamson, S. J., Houchin, L., McDaniel, L., and Paul, J. H. (2002). Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl. Environ. Microbiol. 68, 4307–4314. doi: 10.1128/AEM.68.9.4307-4314.2002
Williamson, S. J., and Paul, J. H. (2006). Environmental factors that influence the transition from lysogenic to lytic existence in the phiHSIC/Listonella pelagia marine phage-host system. Microb. Ecol. 52, 217–225. doi: 10.1007/s00248-006-9113-1
Wilson, G. G., and Murray, N. E. (1991). Restriction and modification systems. Annu. Rev. Genet. 25, 585–627.
Wilson, W., Carr, N., and Mann, N. (1996). The effect of phosphate status on the kinetic of cyanophage infection in the oceanic cyanobacterium Synechococcus sp. WH7803. J. Phycol. 32, 506–516.
Wilson, W., and Mann, N. (1997). Lysogenic and lytic viral production in marine microbial communities. Aquat. Microb. Ecol. 13, 95–100.
Wilson, W., Turner, S., and Mann, N. (1998). Population dynamics of phytoplankton and viruses in a phosphate-limited mesocosm and their effect on DMSP and DMS production. Estuar. Coast. Shelf Sci. 46, 49–59.
Wommack, K. E., and Colwell, R. R. (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64, 69–114. doi: 10.1128/MMBR.64.1.69-114.2000
Wright, I. J., Reich, P. B., Cornelissen, J. H., Falster, D. S., Garnier, E., Hikosaka, K., et al. (2005). Assessing the generality of global leaf trait relationships. New Phytol. 166, 485–496. doi: 10.1111/j.1469-8137.2005.01349.x
Zeng, L., Skinner, S. O., Zong, C., Sippy, J., Feiss, M., and Golding, I. (2010). Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell 141, 682–691. doi: 10.1016/j.cell.2010.03.034
Keywords: marine virus, host, tradeoff, trait, model
Citation: Record NR, Talmy D and Våge S (2016) Quantifying Tradeoffs for Marine Viruses. Front. Mar. Sci. 3:251. doi: 10.3389/fmars.2016.00251
Received: 31 July 2016; Accepted: 18 November 2016;
Published: 15 December 2016.
Edited by:
Susanne Menden-Deuer, University of Rhode Island, USAReviewed by:
Kristina Dee Anne Mojica, Oregon State Univeristy, USAUrania Christaki, Ministry of Research and Higher Education, France
Copyright © 2016 Record, Talmy and Våge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicholas R. Record, nrecord@bigelow.org
 Nicholas R. Record
Nicholas R. Record David Talmy
David Talmy Selina Våge
Selina Våge