Colonial Tube-Dwelling Ciliates Influence Methane Cycling and Microbial Diversity within Methane Seep Ecosystems
- 1Integrative Oceanography Division, Center for Marine Biodiversity and Conservation, Scripps Institution of Oceanography, San Diego, CA, USA
- 2Division of Geological and Planetary Sciences, California Institute of Technology, Pasadena, CA, USA
- 3Department of Biology, Occidental College, Los Angeles, CA, USA
- 4Marine Biology Research Division, Center for Marine Biodiversity and Conservation, Scripps Institution of Oceanography, San Diego, CA, USA
In a variety of marine ecosystems, microbial eukaryotes play important ecological roles; however, our knowledge of their importance in deep-sea methane seep ecosystems is limited. Microbial eukaryotes have the potential to influence microbial community composition and diversity by creating habitat heterogeneity, and may contribute to carbon cycling through grazing or symbiotic associations with microorganisms. In this study, we characterized the distribution, substrate variability and ecology of a particular group of microbial eukaryotes, known as folliculinid ciliates, at methane seeps along the eastern Pacific margin. Folliculinid ciliates were recently recognized as an abundant and ecologically important component of hydrothermal vent ecosystems, but their ecology in methane seeps has not been examined. Folliculinid ciliates inhabited methane seeps from Costa Rica to Oregon, suggesting a broad distribution in the eastern Pacific. Using phylogenetic analyses of the 18S rRNA gene, two different species of folliculinid were identified and are formally described here. Folliculinids occupied a range of physical substrates, including authigenic carbonate rocks, shells of dead vesicomyid clams, polychaete tubes and gastropod shells. Molecular analysis of the folliculinid-associated microorganisms (16S rRNA and particular methane monoxygenase) revealed that these ciliates not only influence overall microbial diversity, but also have a specific relationship with bacteria in the “Deep sea-2” methanotroph clade. Natural δ13C isotope signatures of the folliculinids (−35‰) and their 13C-enrichment patterns in shipboard 13CH4 stable isotope-probing experiments indicated these ciliates and their associated microbes are involved in cycling methane-derived carbon. Folliculinids were significantly enriched in 13C after the addition of 13CH4 over short-term (3–8 day) incubations. Together, these results suggest that folliculinid ciliates represent a previously overlooked contributor to the ecology and biogeochemical cycling at deep-sea methane seep ecosystems.
Introduction
Methane seeps are geomorphologic features and novel ecosystems common to most of the world's continental margins (German et al., 2011). Due to their novel metabolic activities, methane seep bacteria and archaea have extremely high rates of in situ primary production (relative to the surrounding deep sea), perform important roles in biogeochemical cycling, and provide trophic support to unique, dense animal assemblages which contribute to regional biodiversity patterns along continental margins (Levin, 2005; Cordes et al., 2010; Grupe et al., 2015). While the bacterial, archaeal and metazoan communities living at methane seeps are well-studied (e.g., Orphan et al., 2001; Levin, 2005; Knittel and Boetius, 2009; Cordes et al., 2010; Valentine, 2011), research on the importance of microbial eukaryotes is limited. Recent culture-independent surveys of eukaryotic SSU rRNA genes have revealed a great diversity of microbial eukaryotes (commonly referred to as heterotrophic protists) living at deep-sea methane seeps (Takishita et al., 2007, 2010; Pasulka et al., 2016) and hydrothermal vents (e.g., López-García et al., 2007; Sauvadet et al., 2010; Coyne et al., 2013). In other marine ecosystems, heterotrophic protists greatly influence energy flow and biogeochemical cycling by consuming bacteria and remineralizing nutrients (Azam et al., 1983; Sherr and Sherr, 1994, 2002). However, we are just beginning to investigate how these marine protists influence the structure and function of chemosynthetic ecosystems (e.g., Edgcomb et al., 2010, 2011).
Ciliates are one of the most conspicuous groups of protists, particularly the sessile ciliates, as they can often be large in size and can form dense colonies (Dovgal, 2001; Lynn, 2008). Folliculinids are a particular group of sessile ciliates within the Family Folliculinidae (class Heterotrichea) easily recognized by their vase-shaped chitin tubes (lorica) and peristomal lobes (arm-like extensions typical to folliculinid ciliates), which are thought to be used for feeding (e.g., Andrews, 1920, 1923; Mathews, 1964, 1968; Lynn, 2008). These ciliates are found in non-chemosynthetic coastal marine habitats (e.g., Fauré-Fremiet, 1936; Das, 1949; Mathews, 1964, 1968; Ji et al., 2004) and are known to colonize a variety of substrates including oyster and mussel shells (Mathews, 1968; Toupoint et al., 2012), serpulid polychaete worm tubes (Mathews, 1964), crayfish (Jaszczolt and Szaniawsk, 2007), wood (Mathews, 1964), coral skeletons (Cróquer et al., 2006), and even artificial substrates (Ji et al., 2004). Their presence in chemosynthetic ecosystems was first discovered at hydrothermal vents in the 1980's (Tunnicliffe et al., 1985; Van Dover et al., 1988), but their abundance and ecological importance was only more recently recognized (Kouris et al., 2007, 2010). Folliculinids form dense “blue mat” colonies within hydrothermal vent fields, sometimes occupying 70% of mineralized substrate, influencing community composition and food web structure within these ecosystems (Kouris et al., 2010). Folliculinids have also been collected and documented from a methane seep in the Gulf of California (Lobban et al., 2007), but their ecological role in seep ecosystems has not been explored.
Folliculinids have the potential to influence microbial community composition and diversity by creating habitat heterogeneity, and may contribute to carbon cycling through grazing or symbiotic associations with microorganisms. As such, a more complete analysis of their distribution and microbial associations is important for characterizing the ecology and energy flows within these chemosynthetic habitats. In this study we determined the distribution of folliculinids inhabiting a variety of methane seep ecosystems along the Pacific margin, characterized folliculinid phylogenetic relationships and substrate variability within methane seep habitats, identified their associated microorganisms, and assessed their potential influence on carbon cycling in these ecosystems. Folliculinids were collected from five methane seep sites along the eastern Pacific margin (spanning a distance of 5560 km and ranging in depth from 779 to 1565 m). The phylogenetic diversity of these folliculinids was assessed using molecular systematics of the 18S rRNA gene. Microscopy, sequencing (particulate methane monooxygenase and 16S rRNA gene), fluorescence in situ hybridization (FISH), stable isotope analyses and short-term stable isotope probing experiments with carbon isotope tracers (13CH4 and H13CO3) were used to assess the diversity and metabolic potential of folliculinid-associated microorganisms as well as the potential ecological and biogeochemical roles of folliculinid ciliates in methane seep ecosystems.
Methods
Sample Collection
Authigenic carbonate rocks and other substrates covered in folliculinid ciliates were collected from five active seep sites along the Pacific coast of North and Central America (Figure 1). Samples were collected from Costa Rica (AT 15-59, Mound 12; 989 m) and Hydrate Ridge, OR (South HR; 779 m) in 2010 (AT15-68) using the HOV Alvin (WHOI) and in 2011 (AT18-10) using the ROV JASON (WHOI), from Guaymas Basin in 2010 (1565 m, MBAR DR399) using the ROV Doc Ricketts (Monterey Bay Aquarium Research Institute, MBARI), from Split Ridge in 2013 (MBARI DR466, 865 m) using the ROV Doc Ricketts (MBARI) and from the Del Mar Seep (1022 m) in 2012 (MV12-17) and 2013 (MBARI DR470/472) using the ROV Trident (SIO) and ROV Doc Ricketts (MBARI), respectively (details provided in Table 1). Upon recovery immediately following the dive, the substrates containing folliculinid ciliates were subsampled for various analyses. Samples for DNA extraction and sequencing were preserved (in 95% ethanol) or frozen at −80°C until processed in the laboratory. Samples for microscopy were fixed in 2% paraformaldehyde for 24 h, washed with phosphate-buffered saline (PBS; 1X) and stored at −20°C in 1:1 1X PBS:ethanol. Samples for stable isotope incubation experiments (Hydrate Ridge and Split Ridge) were stored at 4°C in mylar bags with chilled 0.22-μm filtered bottom water and N2/air (80:20%) headspace for ~1–2 days until experiments were set up at sea. In addition to the samples that were collected, we also did non-quantitative visual surveys of folliculinid-covered substrates in situ at the Del Mar Seep using high-definition video recorded by the Monterey Bay Aquarium Research Institute's ROV Doc Ricketts operated on the R/V Western Flyer.
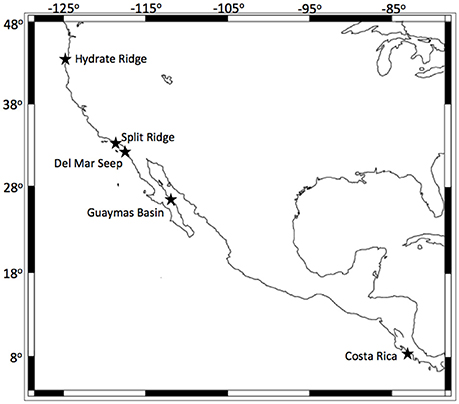
Figure 1. Map of folliculinid ciliate collection sites along the Pacific coast of North and Central America. Each site is indicated with a black star. Coastline created using NOAA's GEODAS-NG software.
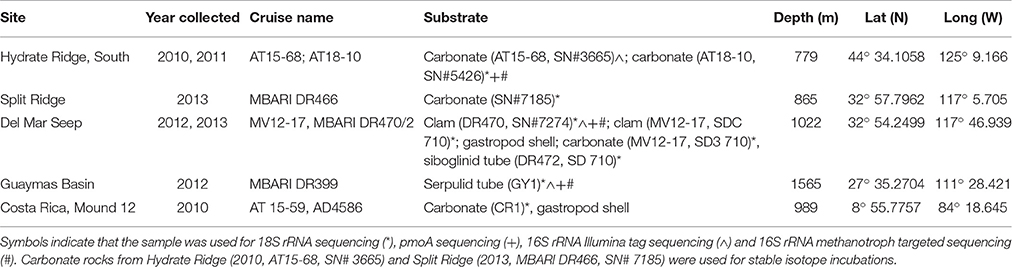
Table 1. Information about sampling locations including the latitude and longitude of each site, the year and cruise name as well as the depth and type of substrate that was collected (ordered from north to south).
Folliculinid DNA Extraction, Primer Design, and PCR Amplification
DNA was extracted from folliculinid samples using a Power Soil DNA Extraction Kit (MO BIO Laboratories, Carlsbad, CA) for samples associated with seep carbonates or a DNeasy Blood and Culture Kit (Qiagen, Valencia, CA) for the remaining samples using the manufacturer recommendations. Samples used for 18S rRNA sequencing are indicated in Table 1. Folliculinids were removed gently from carbonate rocks under a microscope with ultra-fine forceps. However, folliculinids from Hydrate Ridge were embedded in the carbonate rocks to a greater extent than at the other sites and therefore the cells could not be completely separated from all carbonate fragments without damage to the cell. Folliculinid-specific primers were designed using Geneious (Biomatters, http://www.geneious.com) based on available 18S rRNA gene sequences from folliculinids and several closely related heterotrich (GenBank accession nos: AY630405, EU583992, AY630405, AM713187, U47620, EF688408, AM713184, EU583990). Regions that were highly conserved within the heterotrichs and not within common carbonate-associated metazoan meiofauna (e.g., nematodes) were targeted for primer selection. Primer sets were tested in silico using NCBI's Primer-BLAST feature. Based on the results from Primer-BLAST, primer set 1F_follic (5′-TGCTGGTTGCGCTGTGCGTA-3′) and 1R_follic (5′-AGGCGAGAGCCTGCCTGGAA-3′) was optimized using a culture of the heterotrich Blepharisma sp. (Flinn Scientific, Inc., LM1073), using a gradient of annealing temperatures. The optimum thermal protocol was 3 min at 95°C followed by 35 cycles of: 30 s at 95°C (denaturation), 30 s at 60.5°C (annealing), and 2 min at 72°C (extension), with a final elongation step of 7 min at 72°C. All PCR amplifications were done using PuReTaq Ready-To-Go PCR beads (GE Healthcare). PCR products (537 bp) were directly purified with ExoSAP-IT (USB Corporation, Ohio, USA) and sequenced with PCR primers. All sequencing was carried out by Eurofins MWG Operon. Sequences were deposited in GenBank (KX012915-KX012922).
Phylogenetic Analysis of Folliculinid Ciliates
In order to determine the phylogenetic relationships among folliculinids collected during this study and previously identified folliculinids, we compared 18S rRNA sequences in relation to available sequences that were representative of the ciliate class Heterotrichea (obtained from GenBank). The root of the phylogenetic analysis was placed by using representatives of Karyorelictea as an outgroup (Schmidt et al., 2007). Sequences were aligned using MUSCLE (Edgar, 2004). A maximum parsimony analysis was carried out with PAUP*4.0 (Swofford, 2002; Swofford and Sullivan, 2003) using the heuristic search option with 100 random additions. A strict consensus tree was generated, and clade supports were determined using 100 jackknife replicates (Farris et al., 1996). A maximum likelihood analysis was completed using RAxML GUI v. 0.93 (Stamatakis, 2006; Silvestro and Michalak, 2010) with the GTRGAMMA model based on author recommendations (Stamatakis, 2008). A “thorough” bootstrap analysis was carried out with 1000 pseudoreplicates using the same model.
Isotope Labeling Experiments and Analysis
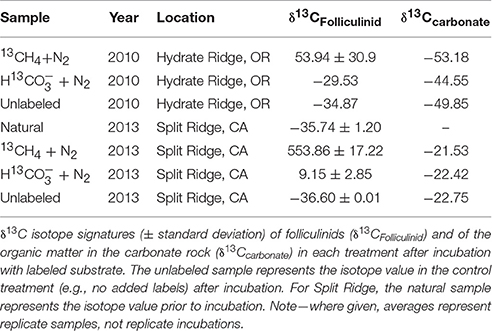
In order to evaluate potential interactions with methanotrophs and other carbon-fixing microorganisms, short-term stable isotope incubation experiments with 13C-labeled methane (13CH4) and bicarbonate (H13CO3) were set up at sea using folliculinid-colonized carbonates from Split Ridge, CA and Hydrate Ridge, OR. Controls amended with unlabeled substrates, were set up as part of each incubation series. All treatments were incubated at 4°C in sealed mylar bags with 0.2-μm filtered bottom seawater from the respective collection site (initial concentrations of labeled substrates and incubation times as given in Table 2). All labeled substrates were at least 99% pure, and the initial conditions given for the gases assumed complete equilibration of the labeled gas between the headspace and seawater inside the mylar bag. Isotopic analyses of the Hydrate Ridge incubation experiments were conducted using a Costech ECS 4010 elemental analyzer with a continuous flow Micromass Isoprime isotope-ratio mass spectrometer at Washington State University. Isotopic analyses of the Split Ridge incubation experiments were analyzed at California Institute of Technology using Thermo Scientific Delta V Plus isotope ratio mass spectrometer with a Costech ECS 4010 elemental analyzer. The carbon isotope ratios are expressed using δ-notation where δ13C ‰ = [(13C/12Csample)/(13C/12Cstandard) − 1] × 1000. Enrichment in 13C is expressed as Δδ13C, indicating a relative increase in δ13C in the sample compared with the background (e.g., unlabeled folliculinid ciliates), and calculated as Δ δ13C ‰ = δ13Csample − δ13Cbackground. Bulk organic carbon isotopic composition was determined for carbonate rocks without folliculinids from each incubation treatment. Details of sample processing and background δ13C folliculinid values are provided in the Supplemental Materials.
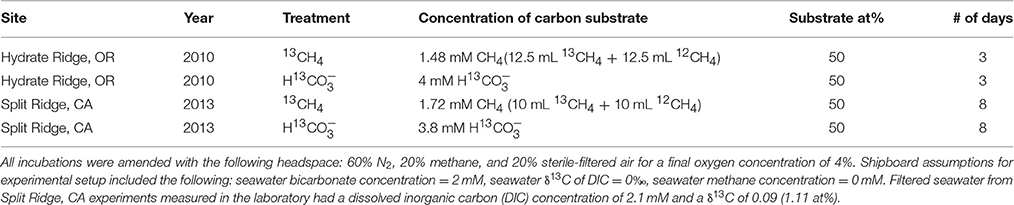
Table 2. Initial conditions of labeled substrates (concentration and the 13C atom percent; at%) at the start of each isotope incubation experiment.
Identification of Folliculinid-Associated Microorganisms: Sequencing and Microscopy
Short-read, high-throughput 16S rRNA sequencing (iTAG) was used to survey the diversity of bacteria and archaea occurring in association with the folliculinid ciliates. The same DNA extracts from the Del Mar Seep (DR470, SN#7274) and Guaymas Basin (DR399, GY1) that were used for folliculinid 18S rRNA sequencing were used for this analysis. However, for Hydrate Ridge, DNA for 16S rRNA iTAG sequencing was extracted from folliculinds on a different piece of carbonate that had been paraformaldehyde-fixed in preparation for microscopy (AT15-68, SN#3665). These folliculinids were more easily separated from the carbonate than the folliculinids used for 18S rRNA sequencing (AT18-10, SN#5426), which was important for identifying only the folliculinid-associated microbial community. DNA from these fixed samples was extracted using the modifications outlined in Trembath-Reichert et al. (2013) to reverse the DNA cross-linking that occurs during fixation. To obtain information about the microbial community associated with the carbonate rock, DNA was extracted from a piece of this same carbonate (AT15-68, SN#3665) that was not covered in folliculinids. Sequencing of the V4 region of the 16S rRNA gene was performed according to the Earth Microbiome Project (EMP; Gilbert et al., 2011) recommended protocol (http://www.earthmicrobiome.org/emp-standard-protocols/16s/) (Caporaso et al., 2011, 2012), with two alterations (see Supplemental Material and Mason et al., 2015, for details). Paired-end 2 × 250 basepair sequencing was performed at Laragen, Inc (Los Angeles, CA) on an Illumina MiSeq platform. The sequence data from the folliculinds are available in the Sequence Read Archive under BioProject number PRJNA315164. The full dataset for the carbonate rock microbial community was published by Case et al. (2015; PRJNA275905). The sequences were processed in-house through QIIME1.8.0 (Caporaso et al., 2010) as in Mason et al. (2015) with operational taxonomic unit (OTU) binning (similarity = 97%), singleton OTU removal, taxonomy assignment, and contaminant removal from negative PCR controls (Salter et al., 2014). Each sample was normalized to its sequence total (McMurdie and Holmes, 2014), OTUs appearing at less than 0.01% relative abundance were removed in order to minimize spurious sequencing products, and the samples were renormalized. OTU richness (# of OTUs per sample), Shannon Diversity (H′) and Pielous' evenness (J′) were calculated with PRIMER v6 with PERMANOVA+ add-on software (PRIMER-E Ltd.).
Targeted 16S rRNA and particulate methane monooxygenase (pmoCAB) sequencing was done to assess the presence and phylogenetic identity of bacterial aerobic methanotrophs associated with folliculinids. The same DNA extracts that were used for 18S rRNA (Table 1) were used. Short read iTAG sequences within the Methylococcales were used to identify the main clade with which the methanotrophs sequences were affiliated (see Table S1 of Methylococcales OTUs). Most of the diversity of aerobic methanotrophs fell into the “Deep sea-2” group (Lüke and Frenzel, 2011; Li et al., 2014). Therefore, in order to obtain longer sequences for better phylogenetic resolution of this group, clade-specific primers were subsequently designed in ARB following the recommendations of Hugenholtz et al. (2001). The following oligonucleotide primer was designed: Mg849-5′-ACGTTAGCTCCACCACTAAG-3′ (16S rRNA position 849-868 from E. coli as in Brosius et al., 1981). This primer was then used in combination with a modified 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGYTACCTTGTTACGACT-3′) (Lane, 1991) to obtain full-length folliculinid-associated Methylococcales 16S rRNA sequences. Amplicons were directly sequenced by Laragen (Culver City, CA) using an AB3730XL instrument with amplification primers. Sequences were manually trimmed, edited and assembled in Sequencher (Gene Codes Corporation, MI). A maximum likelihood analysis was completed using RAxML in ARB with the GTRGAMMA model based on author recommendations (Stamatakis, 2006, 2008). A rapid bootstrap analysis was carried out with 1000 pseudoreplicates using the same model (Stamatakis et al., 2008). 16S rRNA sequences from this study have been deposited in GenBank with Accession Numbers KU896787 to KU896789.
For pmo sequencing, extracts from cleaned individual folliculinid cells (Del Mar Seep, Guaymas Basin) were subjected to 20 cycles of PCR as previously described (pmoA strategy 2, Tavormina et al., 2008) with minor modifications (see Supplemental Materials for more details). In short, this nested PCR approach included an initial amplification with forward primer spacer_pmoC599f and pmoA682r (Holmes et al., 1995) followed by amplification with primers spacer_pmoC626f and mb_661r (Costello and Lidstrom, 1999). Based on microscopy and initial clone library results, the PCR-amplified pmoC-pmoA fragment from cleaned individual folliculinid cells (Del Mar Seep, Guaymas) was expected to contain predominantly folliculinid-associated methanotrophs. As a result of this expectation, the amplified product was sequenced directly, using the spacer_pmoC626f primer.
Slightly different amplification procedures were used for pmo sequencing of Hydrate Ridge samples, as these folliculinids were more-tightly associated with the carbonate rock and therefore were anticipated to reveal both folliculinid-associated as well as carbonate-associated methanotrophs. The amplification strategy for this specimen targeting the pmoC-pmoA region of the pmo operon has been previously described (Tavormina et al., 2010), and was used here with minor primer modifications to capture additional pmo diversity. Nested PCR was done with an initial amplification with primers spacer_pmoC599f and spacer_pmoA192r, followed by amplification with spacer_pmoC626f and spacer_pmoA189r. The resulting PCR-amplified DNA fragment was cloned into the pSMART-GC-LK cloning vector (Lucigen) following manufacturer's guidelines. Unique clones (as determined by restriction fragment length polymorphism; RFLP) were sequenced. Particulate methane monooxygenase (pmo) sequences have been deposited in GenBank with Accession Numbers KX021875-KX021877. Unique pmo sequences, from the Hydrate Ridge clone library, and from direct sequence data from folliculinids from Guaymas Basin and Del Mar seep, were translated in silico to pMMO-A sequence and aligned. A maximum likelihood tree was constructed from this alignment using PhyML available at phylogeny.fr (Dereeper et al., 2008, 2010).
Additional microscopy was completed to visualize the presence and spatial orientation of bacteria associated with individual folliculinid ciliates. Fluorescence in situ hybridization (FISH) was done on paraformaldehyde-fixed folliculinids from Hydrate Ridge. Individual folliculinids were picked and embedded in Technovit 8100 (Electron Microscopy Sciences) according to the manufacturers recommendations with the exception that ethanol was used instead of acetone for dehydration. Embedded samples were cut on a microtome into 1-μm thick sections and mounted onto Poly-L-Lysine coated slides (Teckdon Incorporated). Slides were then stored at room temperature prior to FISH. In order to detect the presence of either bacteria or archaea in association with the folliculinids, FAM-labeled general bacterial and archaeal probes, EUB338 (EUB I, II, III mix) and Arch915, respectively, were used as previously described (Stahl and Amann, 1991; Amann et al., 1995; Daims et al., 1999). Hybridization was conducted at 46°C for 2 h with a hybridization volume of 40 μl followed by a 15-min wash step in 50 ml at 48°C. 35% formamide concentrations were used for both probes. Sections were counter-stained using DAPI (1 μg/ml) and examined using an Olympus BX51 fluorescence microscope and 100X objective. One non-sectioned folliculinid specimen was examined using light sheet fluorescence microscopy (Zeiss Z.1, see Supplemental Material for details). Folliculinid samples embedded in Durocopan ACM resin and cut into ultrathin sections (70–80 nm thick) were imaged using either a FEI Tecnai Spirit or T11 and acquired using TVIPS F224 (with Spirit) and Gatan 2 × 2 k (with T12) CCD camera (see Supplemental Material for details).
Results
Folliculinid Distribution and Phylogenetic Relationships
Folliculinid ciliates were found at all five of the seep sites visited along the Pacific coast (Table 1, Figure 1). Phylogenetic analyses of the 18S rRNA gene were congruent with previous phylogenetic analyses of heterotrich ciliates (Miao et al., 2005, 2009; Schmidt et al., 2007). The results revealed two well-supported clades of folliculinids from seep habitats (Figure 2) that grouped with the other folliculinids as a well-supported Folliculinidae clade. Each of these seep folliculinid clades had closest folliculinid relatives that were not collected from seeps. The low variability of sequences within each of the seep folliculinid clades suggests that each can be regarded as a species with different distributions along the eastern Pacific margin. We did not observe any clear patterns between folliculinid species and the depth of collection or the type of substrate upon which they were collected. The two species of seep folliculinids both contained multiple macronuclei (beaded nuclei, Figure S3). This morphologically informative trait (e.g., Fauré-Fremiet, 1936) was used to designate the two species from this study in the genera Eufolliculina. We propose to designate these seep species as Eufolliculina methanicola n. sp. and Eufolliculina caerulea n. sp. (Figures S4, S5; see Supplemental Material for a more detailed description of folliculinid ciliate phylogenetic designation and taxonomy; http://zoobank.org/NomenclaturalActs/48877B40-44B9-4ACB-B7FD-4F6CB6EDEE3D, http://zoobank.org/NomenclaturalActs/25319BB9-008C-4AA0-BE00-C0F22A777E1B). Eufolliculina methanicola n. sp. was found at Hydrate Ridge (Oregon) and the Del Mar Seep (California), whereas Eufolliculina caerulea n. sp. was found at all the sites, except Hydrate Ridge. The sequences of the Eufolliculina caerulea n. sp. were 99% similar to sequences obtained from folliculinids recently recovered from authigenic carbonates in Guaymas Basin (Lobban et al., 2007, indicated by accession number EF688408 in Figure 2). The Del Mar Seep was the only site where both species were collected together.
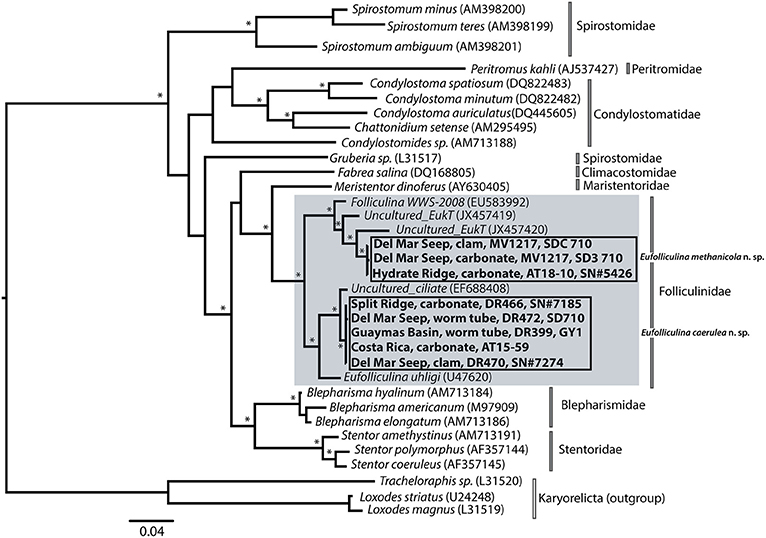
Figure 2. 18S rRNA maximum likelihood tree of ciliates within the class Heterotrichea with emphasis on folliculinid ciliates, highlighted in gray. Sequences from this study are in bold. Jackknife (parsimony) and bootstrap (likelihood) support of greater than 70 are indicated with an asterisk (*). Direct distance calculations among two clades of folliculinid sequences collected in this study were calculated using PAUP*4.0 (Swofford, 2002) and found to be 94% identical. Details about sampling location can be found in Table 1; the substrate and cruise name provided in the tree correspond with starred substrates in Table 1.
Folliculinid Substrate Variability and Colony Morphology
Folliculinids were observed living on a variety of substrates including authigenic carbonates, shells of dead vesicomyid clams, serpulid (Neovermilia sp.) and siboglinid polychaete (Lamellibrachia sp., Escarpia sp.) tubes, and gastropod (Provanna sp., Kanoia myronfeinbergi sp.) shells (Warén and Rouse, 2016; Figure 3). Folliculinid density and colony morphology varied across the different substrates and seep sites. For example, densely clumped colonies were found on a carbonate rock from Costa Rica (Eufolliculina caerulea n. sp.; Figure 3E), but more loosely dispersed cells were found on a gastropod shell from the same seep (Figure 3D). The same species, found in dense clumps on a siboglinid tube from the Del Mar Seep (Figure 3I), had narrow lorica with longer necks than the colonies found at Costa Rica. Similar variability in lorica neck length was observed in Eufolliculina methanicola n. sp. found on a carbonate rock at Hydrate Ridge (Figure 3B) and on a dead vesicomyid clam shell at the Del Mar Seep (Figure 3C). In addition, the lorica of individual folliculinids from Hydrate Ridge were more intertwined and built on top of one another than those at the Del Mar Seep. The colonies also differed in color. Under a light microscope, folliculinids from the Del Mar Seep were purplish-red (Figure 3I), while folliculinids from Costa Rica were blue-green (Figure 3E). Folliculinids were also patchily distributed on each substrate. For example, folliculinids did not cover entire gastropod shells, but rather were concentrated in the grooves of the shell (Figure 3H). The same was true for carbonate rocks; folliculinids occurred in dense clumps on parts of the carbonate rock often in depressions, but they were absent from other parts of the same carbonate rock (Figure 3A). Based on in situ observations from the Del Mar Seep, folliculinid colonies were also patchily distributed within seep habitats. For example, folliculinids were often found living in dense colonies on one patch of dead clam shells, siboglinid tube worms or carbonate rocks, but not on adjacent patches (Figure S1).

Figure 3. Images of folliculinids on substrates from various collection sites. (A) Carbonate rock from Hydrate Ridge covered with folliculinid ciliates (photo credit: L. Levin); (B) Close-up of Hydrate Ridge folliculinids embedded in carbonate rock (photo credit: G. Rouse); (C) Folliculinid ciliates on a dead vesicomyid clam shell from the Del Mar Seep (photo credit: A. Pasulka); (D) Folliculinid ciliates on a gastropod shell (Kanoia myronfeinbergi sp.) collected from Costa Rica (photo credit: L. Levin); (E) Folliculinid ciliates on a carbonate rock from Costa Rica (photo credit: G. Rouse); (F) Guaymas Basin folliculinids on serpulid worm tube (photo credit: G. Rouse); (G) Close-up of folliculinids on serpulid worm tube from Guaymas (photo credit: A. Pasulka); (H) Folliculinid ciliates on a gastropod shell (Provanna sp.) collected from the Del Mar Seep (photo credit: B. Grupe); (I) Folliculinid ciliates on a siboglinid tube from the Del Mar Seep with in set photo showing zoomed in view of the ciliates living in their tubes (photo credit: G. Rouse); (J–L) ROV frame grabs of folliculinids in situ at the Del Mar Seep on Lamellibrachia sp. tubes (L) and surrounding carbonate (J) and a dead vesicomyid clam shell (K) (photo credit: MBARI). Folliculinids in images (A–C) are Eufolliculina methanicola n. sp. and (E–G,I–L) are Eufolliculina caerulea n. sp. The species shown in (D,H) was not assessed with molecular methods. Black and white arrows point to folliculinids and scale bars are indicated on each image.
Stable Isotopes—Natural Abundance and Tracer Experiments
The natural abundance δ13C signature of folliculinid ciliates from the Split Ridge methane seep off southern California was −35.74 ± 1.2‰. This value was similar to the Hydrate Ridge seep folliculinids (δ13C = −34.87‰) recovered from a 3-day control incubation supplied with unlabeled methane (Table 3). Analysis of the carbonate substrates colonized by the folliculinids revealed that the natural carbon isotope value of the carbonate rock-associated organic carbon from Hydrate Ridge (−49.85‰) was lighter than the carbonate-associated organic carbon from Split Ridge (−22.75‰).
Folliculinids became highly enriched in 13C when incubated shipboard with 13C-labeled methane (13CH4) (Figure 4, Tables 2, 3). Parallel incubations amended with 13C-labeled-bicarbonate (H13CO3), averaged only 7% of the enrichment observed in the 13C-labeled-methane treatment. Enrichment after incubation with 13CH4 was significantly greater than enrichment after incubation with H13CO3 (unpaired one-tailed t-test P-value = 0.07 and 0.002 for Hydrate Ridge and Split Ridge, respectively). There was no significant 13C-enrichment observed in folliculinids in the unlabeled control treatments (CH4 or HCO3; Table 3). Similar enrichment patterns were observed in folliculinid incubations from both Hydrate Ridge and Split Ridge, even though the magnitudes of carbon uptake varied. Bulk-carbonate associated organic carbon did not show 13C-enrichment in the short-term incubation treatments (Table 3).
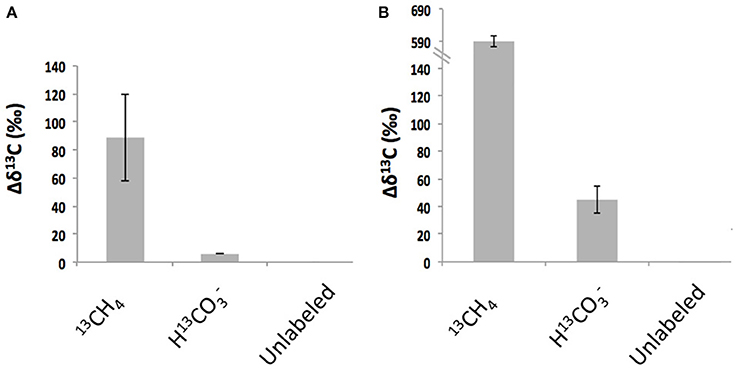
Figure 4. Enrichment of 13C in folliculinid ciliates from Hydrate Ridge (A) and Split Ridge (B) after incubation with different 13C-labeled substrates indicated on the x-axis. Note y-axis scale change in B. These results correspond to data shown in Table 3.
Identification of Folliculinid-Associated Microorganisms
The iTAG sequencing recovered a variety of microorganisms in association with the folliculinids and were dominated by Gammaproteobacteria (Figure 5). The folliculinid-associated Gammaproteobacteria recovered from three different geographic regions (Hydrate Ridge, Del Mar Seep and Guaymas Basin) all contained sequences from the orders Oceanospirillales and Methylococcales (Figure 5). Oceanospirillales was the dominant group recovered in two of the three folliculinid samples (Figure 5), with diversity in many different clades (see Supplemental Material for Oceanospirillales OTUs; Table S2). Most of the Methylococcales clustered with the “Deep sea-2” clade (see Supplemental Material for Methylococcales OTUs; Table S1). Members of the Oceanospirillales were completely absent from the carbonate rock community and sequences associated with Methylococcales occurred at a lower abundance in the carbonate rock (0.67%) relative to the folliculinid ciliates (9.7, 3.2, and 1.7% for Guaymas Basin, the Del Mar Seep and Hydrate Ridge, respectively). Lineages from the order Campylobacterales were also found in association with all three ciliates (constituting between 1.6 and 17% of the 16S rRNA gene sequences), but representatives from this lineage were also found in high abundance on the carbonate rock (4% of the 16S rRNA gene sequences). The dominant sequences of Epsilonproteobacteria fell within the sulfur-oxidizing genera Arcobacter, Sulfurimonas, and Sulfurovum (see Supplemental Material for Campylobacterales OTUs; Table S3). The only archaeal sequences found in association with the folliculinids were closely related to the Thaumarchaeota Marine Group 1. It is important to note that we are not able to confirm whether the microbial sequences associated with the folliculinids are from remnant DNA of prey they have consumed, lorica-associated microorganisms, or from ecto- and endosymbionts.
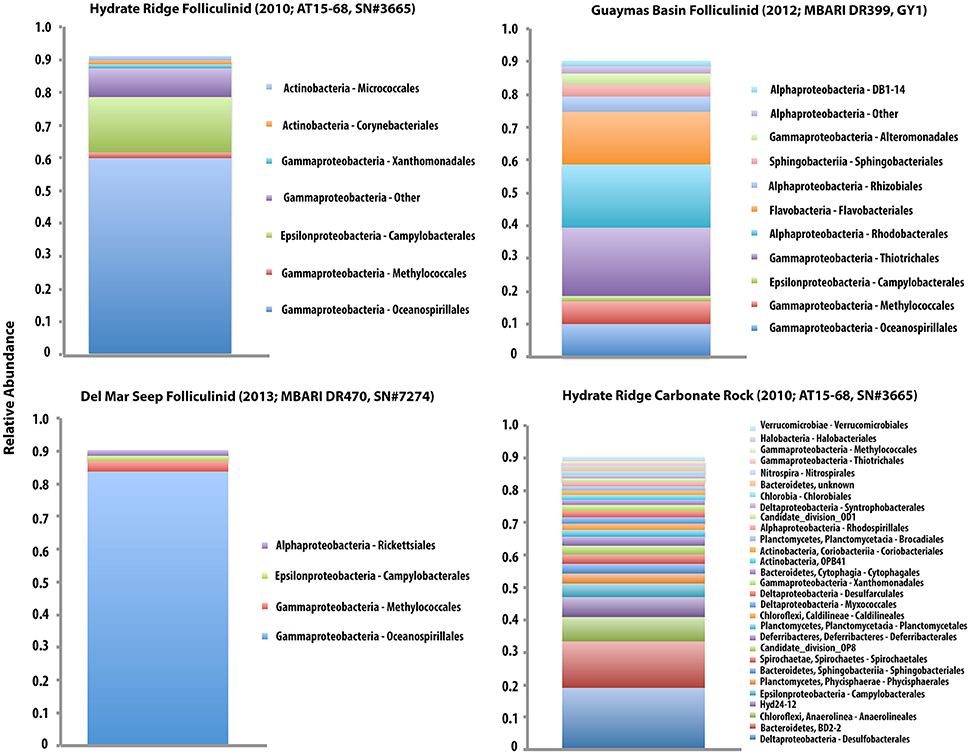
Figure 5. Bar chart displaying the community composition of three folliculinid samples (cell+lorica; Hydrate Ridge, Guaymas Basin, and Del Mar Seep) and a carbonate rock from Hydrate Ridge (HR AT15-68; SN#3665). Data is displayed as relative abundance (at the order level) with the taxa that make up 90% of the community. Note that the legends within each sample correspond with the order of the stacked bars within that sample, but the colors are not consistent across samples. The Hydrate Ridge folliculinid (shown in the top left) was isolated from the same rock displayed in the bottom right, but the portion of the carbonate used for this analysis was absent of folliculinids. OTU richness was 72, 196, 191 for folliculinids from Hydrate Ridge, Guaymas Basin and the Del Mar Seep, respectively and 457 in the carbonate rock from Hydrate Ridge.
Overall, folliculinid-associated microbial communities were less diverse than carbonate-associated microbial communities (Figure 5). OTU richness and Shannon Diversity [H'] were 72[1.8], 196[3.8], and 191[1.3] for folliculinids from Hydrate Ridge, Guaymas Basin and the Del Mar Seep, respectively and 457[4.4] in the carbonate rock from Hydrate Ridge. The folliculinid from Guaymas Basin had the highest diversity of associated microbes (at the OTU level and at the order level). Microbial communities associated with folliculinids from Hydrate Ridge and the Del Mar Seep also had higher dominance than communities associated with the carbonate rock (J' = 0.44, 0.72, 0.24 for folliculinids from Hydrate Ridge, Guaymas Basin and the Del Mar Seep, respectively and 0.72 in the carbonate rock from Hydrate Ridge).
Targeted PCR amplification and sequencing of the particulate methane monooxygenase gene (pmoA) and clade-specific 16S rRNA genes was used to better characterize bacterial methanotrophs in association with folliculinids that may potentially be responsible for the observed host enrichment from 13C-methane. Folliculinids from three different geographic regions (Hydrate Ridge, Del Mar Seep, and Guaymas Basin) had phylogenetically similar pmoA sequences (Figure 6A) that grouped with the “Deep sea-2” clade (Lüke and Frenzel, 2011). These pmoA sequences were closely related to a pmoA sequence recovered from Jaco Scar, on the Costa Rica margin (Tavormina et al., 2013). This sample was collected near the base of a large tubeworm bush (Lamellibrachia barhami) and had elevated temperatures (3.6–5.2°C) and higher methane concentrations (4300 nM) compared to the background (see Levin et al., 2012 for more details). The “Deep sea-2” clade is a sister clade to the “Deep sea-1/symbiont-like” clade, in which the pmo of the bathymodiolin endosymbiont Methyloprofundus sedimenti “WF1” (Tavormina et al., 2015) is found on both trees. Both “Deep sea-1” and “Deep sea-2” lineages are “Type 1a” methanotrophs as described by Lüke and Frenzel (2011). Targeted 16S rRNA amplification and sequencing of the “Deep sea-2” clade revealed a phylogenetically similar 16S rRNA sequence in all three was folliculinids (Figure 6B). These sequences were most closely related to a 16S rRNA sequence recovered from the Guaymas Basin deep-sea hydrothermal plume (Dick and Tebo, 2010).
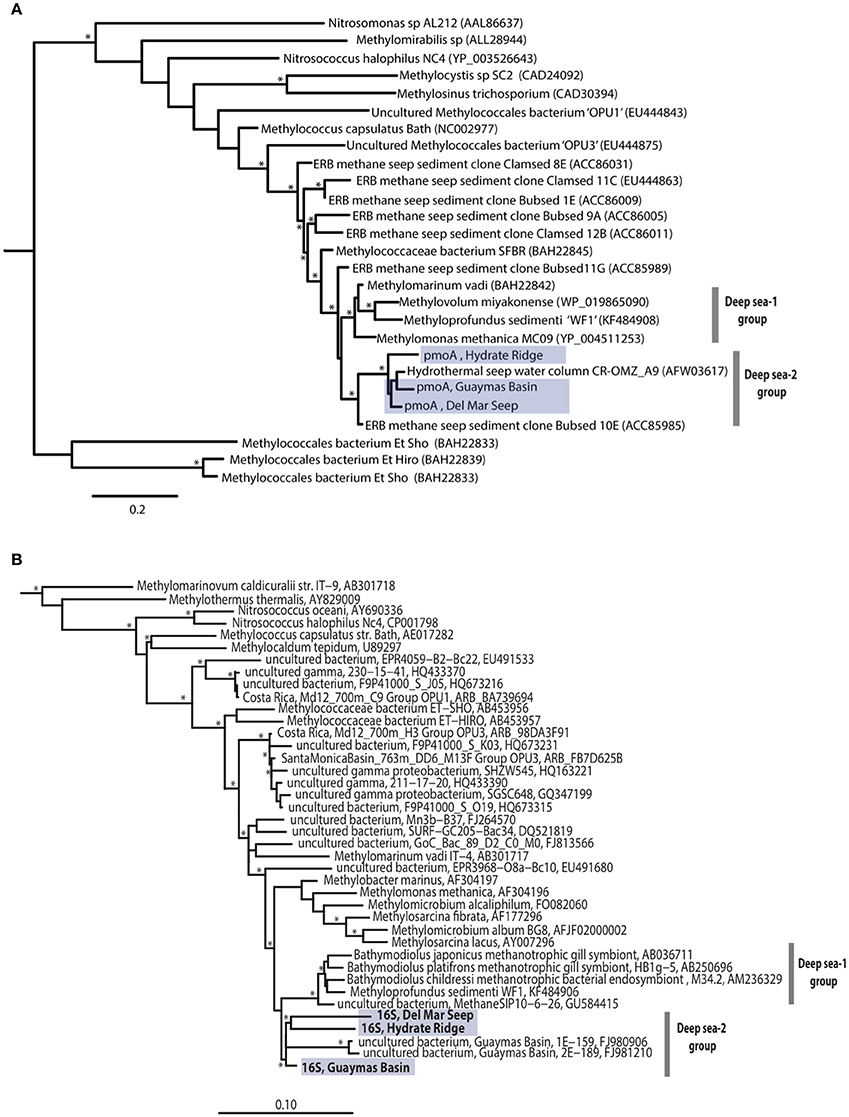
Figure 6. Maximum likelihood tree of unique pmoA sequences (amino acid residues 82–208, Methylococcus capsulatus Bath numbering) from three folliculinid samples (A). SAR324 was used as an outgroup. 16S rRNA maximum likelihood tree showing phylogenetic relationships of three folliculinid-associated methanotrophic sequences within order Methylococcales (B). The following sequences were used as an outgroup: Hydrogenobacter thermophiles AP011112, Aquifex pyrophilus M83548, Hydrogenobaculum acidophilum D16296. Nodes with bootstrap value >70 are indicated with an asterisk (*). Folliculinid-associated sequences highlighted in blue.
Epifluorescence microscopy and TEM revealed an abundance of microorganisms associated with the ciliates and their lorica (Figure 7). FISH microscopy using Domain-targeted probes (Eub338 and Arch915) on folliculinid thin sections revealed bacteria, but not archaea (see Supplemental Material Figures S1–S3) inside and attached to folliculinid cells (Figures 7D,E). This finding is consistent with the iTAG sequencing results, as Thaumarchaeota would not hybridize with Arch915. Bacteria that positively hybridized with the Eub 338 probe were visible on the outside edge of the wings within the cilia and in the center of each wing, whereas bacteria were not detected within the area between these two regions (Figure 7D). Within the wings, some FISH-stained bacteria were clearly located in vacuoles, while others were not. TEM revealed the presence of vacuolated bodies as well as bacteria into food vacuoles inside the cytoplasm (Figures 7G,H). Some bacteria were located inside the cytoplasm, while some were located between the pellicle (cell covering) and the cytoplasm (Figure 7I). TEM observations also revealed many bacterial morphotypes associated with the interior of the lorica (Figures 7J–M), some of which exhibited methanotroph-like stacked membranes.
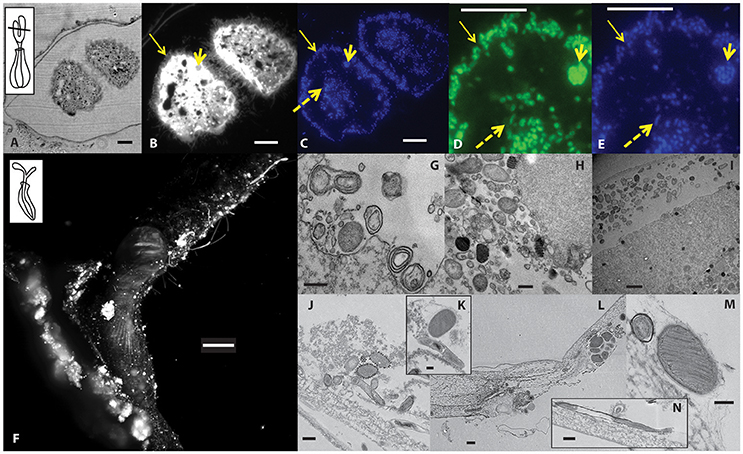
Figure 7. Microscopy images showing the abundance and diversity of microorganisms associated with folliculinid ciliates at Hydrate Ridge. (A–E) Images of a thin section through folliculinid ciliate peristomal lobes. (A) Bright field image with inset of folliculinid schematic showing approximate location of cross section and epifluorescence microscopy images of (B) ciliate autofluorescence and visible cilia (650 nm/670 nm; Cy5) and (C) DAPI-stained cell (350 nm/450 nm). (D) FAM-labeled EUB338-prob and (E) and DAPI-stained cells corresponding to the same section in D (Arch915 probe on similar section shown in Figure S1). Yellow arrows pointing to epibiotic bacteria that are located within cilia (thin arrow), internal bacteria (dashed arrow) and bacteria within a vacuole (short thick arrow). Scale bars = 10 μm. (F) Epifluorescent image of a whole DAPI-stained folliculinid ciliate inside its tube imaged by light sheet fluorescence microscopy (Zeiss, Lightsheet Z.1). Inset showing the general orientation of the cell, however the imaged cell is compacted down into its chitin tube with its peristomal lobes bent over. Scale bar = 500 μm. (G–N) Transmission electron microscopy (TEM) images of microorganisms and vacuolated bodies inside the folliculinid cytoplasm (G–I). TEM images of microorganisms attached to the interior of the folliculinid chitin tube (J–N). Scale bars: G,H,J,L,N = 0.5 μm; I = 2 μm; K,M = 0.2 μm.
Discussion
Distribution and Substrate Variability of Seep-Associated Folliculinid Ciliates
This survey revealed that folliculinid ciliates not only have a broad distribution across seep habitats in the eastern Pacific, but also occupy a range of authigenic and biogenic hard substrates within methane seeps. Using 18S rRNA gene phylogeny, two different seep-associated folliculinid species were found across the five Pacific seep sites. One species (Eufolliculina caerulea n. sp.) inhabited seeps from San Diego (Split Ridge and Del Mar Seep) to Costa Rica. The other species (Eufolliculina methanicola n. sp.) was recovered only at Hydrate Ridge and the Del Mar Seep. There is evidence for the connectivity of animals between vents and seeps across this entire geographic range (e.g., Stiller et al., 2013). Therefore, the absence of Eufolliculina methanicola n. sp. from most of the southern seeps represents either a limited geographical distribution for this species or missed presence due to non-exhaustive sampling.
We cannot yet determine if the two species are endemic to seeps because the current sequence data for folliculinids from different locations and/or habitats is limited. Only 2 of ~70 known folliculinid species (Hadzi, 1951; Hammerschmidt et al., 1996; Miao et al., 2009; Warren, 2013) have been both morphologically described and also sequenced for the 18S rRNA genes. Based on the 18S rRNA gene in this study, the two seep species are phylogenetically distinct from each other and from the two morphologically described and sequenced species (Eufolliculina uhligi and Folliculina producta/simplex). They are most closely related to several undescribed species with available 18S rRNA sequences. Eufolliculina caerulea n. sp. was most closely related to a species (Uncultured_ciliate EF688408, Figure 2) recovered from the seafloor in the Gulf of California (1567 m, cold seep environment; Lobban et al., 2007). Whereas, Eufolliculina methanicola was related to a species (Uncultured_EukT JX457420, Figure 2) recovered from the shell of a mussel in a shallow marine habitat (~3 m; Toupoint et al., 2012). Additional sequences within the Folliculinidae would assist in establishing the evolutionary relationships among seep and non-seep folliculinids.
Authigenic carbonates (carbonates created during the anaerobic oxidation of methane; Aloisi et al., 2002; Teichert et al., 2005), as well as other hard substrates of biogenic origin such as clam shells and polychaete worm tubes, are ubiquitous within seep ecosystems, but have recently been identified as important habitat for seep organisms (Marlow et al., 2014a,b; Case et al., 2015; Levin et al., 2015; Pasulka et al., 2016). Within each seep, folliculinids were observed living on a variety of these types of hard substrates (Figure 3). This finding is consistent with the types of substrates that shallow-water folliculinid species have been observed to inhabit including, but not limited to, the calcium carbonate shells of pearl oysters (Mathews, 1968) and gastropod larvae (Scheltema, 1973) and the chitinous skeletons of crayfish (Jaszczolt and Szaniawsk, 2007).
It is not yet understood what controls colonization, growth and morphological diversity of folliculinids in seep habitats, or why they only occur on some rocks or some bivalve shells, despite the fact that they all appear to afford the same attachment potential. We hypothesize that the various scales of folliculinid patchiness (e.g., within substrate and within seep) result from a combination of habitat selection and folliculinid life history. The fact that folliculinids were commonly found in substrate depressions suggests an important link to increased folliculinid success. This success may be related to higher access to chemical-laden fluids (via substrate induced turbulence) or perhaps even protection from predation. Alternatively, folliculinids may be more likely to get caught in substrate depressions while dispersing.
Folliculinid life histories may enable them to disperse short distances and contribute to their patchy distributions within seep habitats. The swarmer, non-feeding motile stage, has been reported to last one to several hours before the ciliate settles down to secrete its lorica and become sessile (Andrews, 1920; Mulisch et al., 1993). In some instances, folliculinids have been observed to alternate between sessile and motile forms several times within a 24-h period (Andrews, 1920). This short-lived motile stage is likely a mechanism for habitat selection, offering the folliculinids the ability to locate and colonize new suitable substrates in response to spatially and temporally variable seep activity (e.g., MacDonald et al., 1989; Tryon et al., 2002; Levin et al., 2003; Treude et al., 2003). In shallow water environments, folliculinids rapidly colonized artificial substrates within 10 days (Ji et al., 2004). In a hydrothermal vent ecosystem, while a few individuals colonized recruitment arrays within 26 days, they became the most abundant eukaryotic organisms on the arrays after 3.3 years (>1000 individuals; Van Dover et al., 1988). While we did not explicitly test colonization rates of folliculinids at seeps, out of 56 artificial substrates (including wood, bone and carbonate rocks) that were deployed for 1 year at Hydrate Ridge, only one of the whale bones was colonized by folliculinids (Rouse, unpublished observation). Folliculinids also appeared on polypropylene-line and whale bone associated with 2-year old colonization experiments at the Del Mar seep (Levin, Grupe unpublished observations). Therefore, microhabitat variability, not just the presence of available hard substrate, is likely important for folliculinid colonization and subsequent growth in chemosynthetic ecosystems.
Influence of Folliculinid Ciliates on Microbial Composition and Diversity in Seep Ecosystems
The high abundance of folliculinid ciliates observed on a variety of hard substrates at methane seeps along the Pacific margin suggest that these ciliates are an ecologically successful, yet poorly understood, member of methane seep ecosystems. Ecological theory predicts an increase in diversity with increasing habitat heterogeneity (e.g., Simpson, 1949; Connor and McCoy, 1979; Tews et al., 2004). Therefore, the habitat structure created by individual folliculinid tubes and larger colonies has the potential to influence both microbial and meiofaunal diversity. Preliminary microscopy surveys revealed a variety of organisms attached to (e.g., filamentous bacteria and other sessile ciliates) and nestled between (e.g., nematodes) folliculinid lorica. These observations are consistent with the observed utilization of “blue mat” habitat by micro- and meiofaunal organisms at vents (Kouris et al., 2010). TEM observations also revealed a diversity of microorganisms attached to the folliculinid lorica. This finding is consistent with microscopic analyses of Thioploca sp. sheaths from cold seeps in Monterey Bay and Santa Barbara which also host a diverse bacterial and protistan assemblage associated with interstitial spaces between trichomes (Buck et al., 2014), including methanotrophs (Ding and Valentine, 2008). Together, these findings support the notion that small-scale heterogeneity created by lorica, sheaths and the folliculinids themselves can provide habitat niches for other organisms and may influence seep community structure.
The iTAG data revealed two bacterial orders that were more abundant in association with the folliculinid relative to the carbonate rock. Lineages from the order Oceanospirillales (which constituted between 10 and 84% of the 16S rRNA gene sequences recovered from the ciliates) have been found in symbiotic association with a variety of deep-sea chemosynthetic marine invertebrates such as bathymodiolin mussels, marine siboglinid polychaetes of the genus Osedax, and gastropod snails of the genus Alviniconcha (e.g., Goffredi et al., 2005; Zielinski et al., 2009; Beinart et al., 2014), but they have also been found as free living organisms. Oceanospirillales became a dominant component the bacterial community following the Deepwater Horizon spill (Yang et al., 2016). Oceanospirillales are known to be heterotrophic (not chemoautotrophic) and can degrade complex organic compounds (Garrity et al., 2005; Goffredi et al., 2005; Mason et al., 2012).
Lineages from the order Methylococcales were also abundant within the ciliates (constituting 2–7% of the 16S rRNA gene sequences recovered from the ciliates). Many of these sequences were assigned to the “Deep sea-2” clade of methanotrophs. Methanotrophs within this clade have not been described as symbionts, but have been found in the water column within hydrothermal plumes (Dick and Tebo, 2010), in the water column adjacent to a large tubeworm bush (Lamellibrachia barhami) within a hydrothermal seep (Levin et al., 2012; Tavormina et al., 2013), and in response to spilled methane in the Gulf of California (Kessler et al., 2011). A sister clade of methanotrophs (“Deep sea-1”) includes known symbionts of a few seep and vent marine invertebrate taxa (e.g., Cavanaugh et al., 1987; Schmaljohann et al., 1990; DeChaine and Cavanaugh, 2006; Wendeberg et al., 2012) as well as a ciliate from a low-oxygen environment (Edgcomb et al., 2011). The vent-associated folliculinids were reported to have intracellular microorganisms that were morphologically similar to methanotrophs (Kouris et al., 2007), however, morphology alone was not able to confirm whether these features were methanotrophs or pigmented granules.
The recovery of phylogenetically similar methanotroph pmo and 16S rRNA gene sequences (Figure 6) from two different folliculinid species across three distinct geographic regions suggests a specific association between folliculinid ciliates and “Deep sea-2” clade methanotrophs. Folliculinids may provide a specific niche for the “Deep sea-2” clade of methanotrophs; however, at this stage it is not clear if this association is obligate or facultative. Overall, our data demonstrate that these conspicuous single-celled eukaryotes have the ability to alter microbial community structure. Since microbial community structure is important for methane seep ecosystem function (Orphan et al., 2004; Knittel et al., 2005; Russ et al., 2013), enhancing our understanding of how these micro-scale community interactions influence ecosystem function within these unique habitats is of critical importance.
Biogeochemical Influence of Folliculinid Ciliates in Seep Ecosystems: Insights from Stable Isotopes
Natural abundance carbon isotope signatures coupled with isotope tracer experiments reveal that folliculinids and their associated microorganisms have the ability to directly assimilate methane-derived carbon. The isotopically light carbon composition of methane provides a natural isotope tracer for methane-derived biomass (Kastner et al., 1998; Suess et al., 1999; Orphan et al., 2001; Levin and Michener, 2002; Van Dover, 2007); in the absence of a chemosynthetic source, we might have expected the natural carbon isotope signature to be more similar to marine phytoplankton (−18 to −24‰; Fry and Sherr, 1984; Peterson and Fry, 1987). The depleted carbon isotope signature of the folliculinids (−34.87‰ for Hydrate Ridge, −35.74‰ for Split Ridge) indicates that a portion of their energy is derived from chemosynthetically fixed carbon. The 13C-enrichment observed with the addition of 13CH4 over short-term incubations (3–8 days; Figure 4A) further confirms that methanotrophy occurs in association with the folliculinids and may be an important nutritional source for these ciliates in seep habitats. While autotrophy coupled to CO2 uptake is also thought to be common in these habitats (Treude et al., 2007), the 13C-enrichment with H13CO3 during our incubations was a small fraction (~7%) of that in the 13CH4 treatment. The lack of enrichment in the carbonate-associated organic carbon during the short-term incubations demonstrates that the folliculinid-associated microorganisms, not the carbonate-associated microorganisms, were assimilating labeled substrates. This enrichment pattern was consistent in two independent stable isotope tracer experiments with folliculinids from seep regions that were 1480 kilometers apart. Folliculinids from hydrothermal vents also have a depleted carbon isotope signature (−32.8 ± 1.7‰ at Juan de Fuca, Kouris et al., 2010; −34‰ at Gorda Ridge, Goffredi and Orphan, unpublished) indicating the importance of folliculinid-associated chemoautotrophs may be more widespread across many types of chemosynthetic environments. While further work needs to be done to better understand the exact nature of this association, based on the natural carbon isotope signature of the folliculinids and 13C-enrichment patterns in the isotope tracer experiments, we hypothesize that this specific association between folliculinid ciliates and “Deep sea-2” aerobic methanotrophs enables the ciliate to participate in ecosystem-level methane cycling.
The relative importance of different metabolic substrates can be difficult to resolve since in many cases organisms have a mixed diet. Assuming that methanotrophy is a dominant source of chemosynthetic carbon for folliculinids, a simple two-source linear mixing model (Fm = (δF- δPOC)/(δm- δPOC); Fry and Sherr, 1984), can be used to estimate the magnitude of methane-derived vs. photosynthesis-derived carbon that would result in the observed folliculinid carbon isotope signature at Hydrate Ridge. Where Fmis the fraction of methane derived carbon, δF, δPOC, and δm are the δ13C signature of the folliculinid, particulate organic carbon and methane, respectively. For δPOC, we used the isotope signature of non-seep macrofauna (−21‰; Levin and Michener, 2002), and for δm, we used methane δ13C signatures measured from the Oregon margin (−65‰; Suess and Whiticar, 1989). Using this equation with the above values, folliculinid ciliates would need to assimilate 34% of methane-derived carbon in order to obtain the observed natural carbon isotope signature of −34.87‰. At Split Ridge, this same approach revealed that folliculinid ciliates would need to assimilate 24% methane-derived carbon (see Supplemental Materials for details). These estimates are within the range of estimates of methane-derived carbon consumption for macrofauna on the Oregon margin (Levin and Michener, 2002). This type of nutritional mixotrophy could occur if seep folliculinids have maintained their ability to feed on surface-derived production (e.g., heterotrophic bacteria or detrital material) in addition to interacting with chemosynthetic microorganisms (e.g., through consumption or symbiotic associations). The presence of food vacuoles within the cytoplasm as indicated by microscopy (this study, Uhlig et al., 1965) suggests that this type of nutritional mixotrophy is possible.
Characterizing the trophic structure and key nutritional sources that support deep-sea methane seep ecosystems is fundamental for understanding methane biogeochemical cycling and the trophic fate of methane-derived production in the ocean. We are just beginning to understand the specific roles of protists in chemosynthetic ecosystems. Our study reveals a previously unrecognized, but potentially ecologically important group of protists that contributes to the fate of methane-derived carbon in seep food webs. We have demonstrated that these colonial lorica-dwelling ciliates not only influence seep community structure and diversity by creating a unique and patchy habitat, but are also involved in methane cycling through their interactions with methane-oxidizing bacteria. Continuing to explore how protists influence community structure and methane cycling rates within chemosynthetic ecosystems is of critical importance for developing a complete understanding of ecosystem function within these unique habitats.
Funding
Support for this research was provided by grants OCE 0825791, OCE 0826254 and OCE 0939557 from the US National Science Foundation (NSF). This work was funded in part by the Gordon and Betty Moore Foundation through Grant GBMF3780 to VO.
Author Contributions
AP, LL, GR, VO contributed to the research design. AP, SG, LL, KD, GR, PT, VO performed the research. AP, PT, KD analyzed the data. AP, SG, and VO wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the captains and crew members of the R/V Atlantis, R/V Melville, and R/V Western Flyer as well as the pilots of Doc Ricketts and JASON II who helped make sampling possible. UC Ship Funds provided the ship time for the original discovery and sampling of the Del Mar Seep. We thank B. Vrijenhoek and P. Brewer (MBARI) for inviting GR to Guaymas, and GR, AP, KD, SG, LL, and VO to Split Ridge and the Del Mar Seep. We also thank P. Brewer and R. Vrijenhoek for use of video observations and collected specimens. Work aboard the R/V Western Flyer was partially supported by the David and Lucile Packard Foundation's funding of MBARI. We are particularly grateful to J. Marlow and J. Steele for their help setting up incubation experiments, D. Case for help with carbonate isotope measurements, S. McGlynn and G. Chadwick for help with TEM, A. Sessions and F. Wu for access to the EA-IRMS and for help with measurements, B. Grupe for his help at sea, in the laboratory and for many insightful conversations and S. Connon for help with ARB. J. Gonzalez assisted with the processing of isotope samples in the laboratory, R. Lee ran some of the isotope samples, R. Hatzenpichler helped with literature translations, and M. Summers and J. Carvajal assisted with folliculinid phylogenetic analyses. We would also like to thank A. Collazo at the Biological Imaging Facility at Caltech for support with Light Sheet Microscopy as well as T. Deerinck and M. Ellisman from the National Center for Microscopy and Imaging Research for support with TEM.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fmars.2016.00276/full#supplementary-material
Table S1. Table of OTUs within the class Gammaproteobacteria and Order Methylococcales. The OTU name, taxonomic designtation and representative sequence are provided.
Table S2. Table of OTUs within the class Gammaproteobacteria and Order Oceanospirillales. The OTU name, taxonomic designtation and representative sequence are provided.
Table S3. Table of OTUs within the class Epsilonproteobacteria and Order Campylobacterales. The OTU name, taxonomic designtation and representative sequence are provided.
References
Aloisi, G., Bouloubassi, I., Heijs, S. K., Pancost, R. D., Pierre, C., Damsté, J. S. S., et al. (2002). CH4-consuming microorganisms and the formation of carbonate crusts at cold seeps. Earth Planet. Sci. Lett. 203, 195–203. doi: 10.1016/S0012-821X(02)00878-6
Amann, R. I., Ludwig, W., and Schleifer, K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169.
Andrews, E. A. (1923). Folliculina: case making, anatomy and transformation. J. Morphol. 38, 207–278. doi: 10.1002/jmor.1050380202
Azam, F., Fenchel, T., Field, J. G., Gray, J. S., Meyer-Reil, L. A., and Thingstad, F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Beinart, R. A., Nyholm, S. V., Dubillier, N., and Girguis, P. R. (2014). Intracellular Oceanospirillales inhabit the gills of the hydrothermal vent snail Alviniconcha with chemosynthetic, γ-Proteobacterial symbionts. Environ. Microbiol. Rep. 6, 656–664. doi: 10.1111/1758-2229.12183
Brosius, J., Dull, T. J., Sleeter, D. D., and Noller, H. F. (1981). Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148, 107–127. doi: 10.1016/0022-2836(81)90508-8
Buck, K. R., Barry, J. P., and Hallam, S. J. (2014). Thioploca spp. sheaths as niches for bacterial and protistan assemblages. Mar. Ecol. 35, 395–400. doi: 10.1111/maec.12076
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinberg, J., Bushman, F. D., Costello, E. L., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high- throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Case, D. H., Pasulka, A. L., Marlow, J. J., Grupe, B. M., Levin, L. A., and Orphan, V. J. (2015). Methane seep carbonates host distinct, diverse, and dynamic microbial assemblages. mBio 6:e01348-15. doi: 10.1128/mBio.01348-15
Cavanaugh, C. M., Levering, P. R., Maki, J. S., Mitchell, R., and Lidstrom, M. E. (1987). Symbiosis of methylotrophic bacteria and deep-sea mussels. Nature 325, 346–347. doi: 10.1038/325346a0
Connor, E. F., and McCoy, E. D. (1979). The statistics and biology of the species-area relationship. Am. Nat. 113, 791–833. doi: 10.1086/283438
Cordes, E. E., Cunha, M. R., Galeron, J., Mora, C., Roy, K. O., Myriam, S., et al. (2010). The influence of geological, geo- chemical, and biogenic habitat heterogeneity on seep bio- diversity. Mar. Ecol. 31, 51–65. doi: 10.1111/j.1439-0485.2009.00334.x
Costello, A. M., and Lidstrom, M. E. (1999). Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65, 5066–5074.
Coyne, K. J., Countway, P. D., Pilditch, C. A., Lee, C. K., Caron, D. A., and Cary, S. C. (2013). Diversity and distributional patterns of ciliates in Guaymas Basin hydrothermal vent sediments. J. Eukaryot. Microbiol. 60, 433–447. doi: 10.1111/jeu.12051
Cróquer, A., Bastidas, C., and Lipscomb, D. (2006). Folliculinid ciliates: a new threat to Caribbean corals? Dis. Aquat. Org. 69, 75–78. doi: 10.3354/dao069075
Daims, H., Brühl, A., Amann, R., Schleifer, K. H., and Wagner, M. (1999). The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444. doi: 10.1016/S0723-2020(99)80053-8
Das, S. M. (1949). British Folliculinidae (Ciliata, Heterotrichea). J. Mar. Biol. Assoc. UK. 28, 381–393. doi: 10.1017/S0025315400023286
DeChaine, E. G., and Cavanaugh, C. M. (2006). “Symbioses of methanotrophs and deep-sea mussels (Mytilidae: Bathymodiolinae),” in Molecular Basis for Symbiosis: Progress in Molecular and Subcellular Biology, ed O. Jörg (Heidelberg: Springer), 227–249.
Dereeper, A., Audic, S., Claverie, J. M., and Blanc, G. (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. doi: 10.1186/1471-2148-10-8
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr, robust phylogenetic analysis for the non-specialist. Nucleic. Acids. Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Dick, G. J., and Tebo, B. M. (2010). Microbial diversity and biogeochemistry of the Guaymas Basin deep-sea hydrothermal plume. Environ. Microbiol. 12, 1334–1347. doi: 10.1111/j.1462-2920.2010.02177.x
Ding, H., and Valentine, D. L. (2008). Methanotrophic bacteria occupy benthic microbial mats in shallow marine hydrocarbon seeps, Coal Oil Point, California. J. Geophys. Res. 113, G01015. doi: 10.1029/2007JG000537
Dovgal, I. V. (2001). Some regularities in sessile protists evolution: study and protection of the animal world on the end of the century. Baku Elm, -P 111–115.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic. Acids. Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgcomb, V. P., Breglia, S. A., Yubuki, N., Beaudoin, D., Patterson, D. J., Leander, B. S., et al. (2010). Identity of epibiotic bacteria on symbiontid euglenozoans in O2-depleted marine sediments: evidence for symbiont and host co-evolution. ISME J. 5, 231–243. doi: 10.1038/ismej.2010.121
Edgcomb, V. P., Leadbetter, E. R., Bourland, W., Deaudoin, D., and Bernhard, J. M. (2011). Structured multiple endosymbiosis of bacteria and archaea in a ciliate from marine sulfidic sediments: a survival mechanism in low oxygen, sulfidic sediments? Front. Microbiol. 2:55. doi: 10.3389/fmicb.2011.00055
Farris, J. S., Albert, V. A., Källersjö, M., Lipscomb, D., and Kluge, A. G. (1996). Parsimony jackknifing outperforms neighbor-joining. Cladistics 12, 99–124. doi: 10.1111/j.1096-0031.1996.tb00196.x
Fauré-Fremiet, E. (1936). The Folliculinidae (Infusoria heterotricha) of the Breton Coast. Biol. Bull. 70, 353–360. doi: 10.2307/1537293
Fry, B., and Sherr, E. B. (1984). δ13C measurements as indicators of carbon flow in marine and freshwater systems. Contrib. Mar. Sci. 27, 13–46.
Garrity, G. M., Bell, J. A., and Lilburn, T. (2005). “Oceanospirillales ord. nov.,” in Bergey's Manual of Systematic Bacteriology, eds D. J. Brenner, N. R. Krieg, G. M. Garrity, D. R. Boone, P. De Vos et al. (Springer: New York, NY), 270–323. doi: 10.1007/0-387-28022-7_8
German, C. R., Ramirez-Llodra, E., Baker, M. C., Tyler, P. A., and ChEss Scientific Steering C.ommittee (2011). Deep- water chemosynthetic ecosystem research during the census of marine life decade and beyond: a proposed deep-ocean road map. PLoS ONE 6:e23529. doi: 10.1371/journal.pone.0023259
Gilbert, J. A., Meyer, F., Jansson, J., Gordon, J., Pace, N., Tiedje, J., et al. (2011). The Earth Microbiome Project: meeting report of the “1st EMP meeting on sample selection and acquisition” at Argonne National Laboratory October 6th 2010. Stand. Genomic. Sci. 3, 249–253. doi: 10.4056/aigs.1443528
Goffredi, S. K., Orphan, V. J., Rouse, G. W., Jahnke, L., Embaye, T., Turk, K., et al. (2005). Evolutionary innovation: a bone-eating marine symbiosis. Environ. Microbiol. 7, 1369–1378. doi: 10.1111/j.1462-2920.2005.00824.x
Grupe, M. B., Krach, M. L., Pasulka, A. L., Maloney, J. M., Levin, L. A., and Frieder, C. A. (2015). Methane seep ecosystem functions and services from a recently discovered southern California seep. Mar. Ecol. 36, 91–108. doi: 10.1111/maec.12243
Hammerschmidt, B., Schlegel, M., Lynn, D. H., Leipe, D. D., Sogin, M. L., and Raiko, I. G. (1996). Insights into the evolution of nuclear dualism in the ciliates revealed by phylogenetic analysis of rRNA sequences. J. Euk. Microbiol. 43, 225–230. doi: 10.1111/j.1550-7408.1996.tb01396.x
Holmes, A. J., Costello, A., Lidstrom, M. E., and Murrell, J. C. (1995). Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132, 203–208. doi: 10.1111/j.1574-6968.1995.tb07834.x
Hugenholtz, P., Tyson, G. W., and Blackall, L. L. (2001). “Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization,” in Methods in Molecular Biology Gene Probes, eds M. Aquino de Muro and R. Rapley (Totowa, NJ: Humana Press, Inc.), 179, 29–42.
Jaszczolt, J., and Szaniawsk, A. (2007). Folliculinids - epibiotic ciliates from Puck Bay. Int. Oceanogr. Hydrobiol. 36, 165–168. doi: 10.2478/v10009-007-0028-z
Ji, D., Lin, X., and Song, W. (2004). Complementary notes on “well-known” marine heterotrichous ciliate, Folliculinopsis producta (Wright 1859) Frauré-Fremiet, 1936 (Protozoa, Ciliaphora) J. Ocean. Univ. China 3, 65–69. doi: 10.1007/s11802-004-0011-1
Kastner, M., Kvenvolden, K. A., and Lorenson, T. D. (1998). Chemistry, isotopic composition and origin of a methane-hydrogen sulfide hydrate at the Cascadia subduction zone. Earth Planet. Sci. Lett. 156, 173–183. doi: 10.1016/S0012-821X(98)00013-2
Kessler, J. D., Valentine, D. L., Redmond, M. C., Du, M., Chan, E. W., Mendes, S. D., et al. (2011). A persistent oxygen anomaly reveals the fate of spilled methane in the deep Gulf of Mexico. Science 331, 312–315. doi: 10.1126/science.1199697
Knittel, K., and Boetius, B. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Knittel, K., Lösekann, T., Boetius, A., Kort, R., and Amann, R. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479. doi: 10.1128/AEM.71.1.467-479.2005
Kouris, A., Juniper, S. K., Frébourg, G., and Gaill, F. (2007). Protozoan-bacterial symbiosis in a deep-sea hydrothermal vent folliculinid ciliate (Folliculinopsis sp.) from the Juan de Fuca Ridge. Mar. Ecol. 28, 63–71. doi: 10.1111/j.1439-0485.2006.00118.x
Kouris, A., Limén, H., Stevens, C. J., and Juniper, S. K. (2010). Blue mats: faunal composition and food web structure in colonial ciliate (Folliculinopsis sp.) mats in Northeast Pacific hydrothermal vents. Mar. Ecol. Prog. Ser. 412, 93–101. doi: 10.3354/meps08675
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, NY: Wiley), 115–175.
Levin, L. A. (2005). Ecology of cold seep sediments: interactions of fauna with flow, chemistry and microbes. Oceanogr. Mar. Biol. Annu. Rev. 43, 1–46. doi: 10.1201/9781420037449.ch1
Levin, L. A., Mendoza, G. F., Grupe, B. M., Gonzalez, J. P., Jellison, B., Rouse, G., et al. (2015). Biodiversity on the rocks: macrofauna inhabiting authigenic carbonate at Costa Rica methane seeps. PLoS ONE 10:e0131080. doi: 10.1371/journal.pone.0131080
Levin, L. A., and Michener, R. (2002). Isotopic evidence of chemosynthesis-based nutrition of macrobenthos: the lightness of being at Pacific methane seeps. Limnol. Oceanogr. 47, 1336–1345. doi: 10.4319/lo.2002.47.5.1336
Levin, L. A., Orphan, V. J., Rouse, G. W., Rathburn, A. E., Ussler, W. III, Cook, G. S., et al. (2012). A hydrothermal seep on the Costa Rica margin: middle ground in a continuum of reducing ecosystems. Proc. R. Soc. B. 279, 2580–2588. doi: 10.1098/rspb.2012.0205
Levin, L. A., Ziebis, W., Mendoza, G., Growney, V., Tryon, M., Brown, K., et al. (2003). Spatial heterogeneity of macrofauna at northern California methane seeps: the influence of sulfide concentration and fluid flow. Mar. Ecol. Prog. Ser. 265, 123–139. doi: 10.3354/meps265123
Li, M., Jain, S., Baker, B. J., Taylor, C., and Dick, G. J. (2014). Novel hydrocarbon monooxygenase genes in the metatranscriptome of a natural deep-sea hydrocarbon plume. Environ. Microbiol. 16, 60–71. doi: 10.1111/1462-2920.12182
Lobban, C. S., Hallam, S. J., Mukherjee, P., and Petrich, J. W. (2007). Photophysics and multifunctionality of hypericin-like pigments in heterotrich ciliates: a phylogenetic perspective. Photochem. Photobiol. 83, 1074–1094. doi: 10.1111/j.1751-1097.2007.00191.x
López-García, P., Vereshchaka, A., and Moreira, D. (2007). Eukaryotic diversity associated with carbonates and fluid-seawater interface in Lost City hydrothermal field. Environ. Microbiol. 9, 546–554. doi: 10.1111/j.1462-2920.2006.01158.x
Lüke, C., and Frenzel, P. (2011). Potential of pmoA amplicon pyrosequencing for methanotroph diversity studies. Appl. Environ. Microbiol. 77, 6305–6309. doi: 10.1128/AEM.05355-11
Lynn, D. H. (2008). The Ciliated Protozoa: Characterization, Classification and Guide to Literature. Dordrecht: Springer Science.
MacDonald, I. R., Boland, G. S., Baker, J. S., Brooks, J. M., Kennicutt, M. C., and Bidigare, R. R. (1989). Gulf of Mexico hydrocarbon seep communities II. Spatial distribution of seep organisms and hydrocarbons at Bush Hill. Mar. Biol. 101, 235–247. doi: 10.1007/BF00391463
Marlow, J. J., Steele, J. A., Case, D. H., Connon, S. A., Levin, L. A., and Orphan, V. J. (2014a). Microbial abundance and diversity patterns associated with sediments and carbonates from the methane seep environments of Hydrate Ridge, OR. Front. Mar. Sci. 1:16. doi: 10.3389/fmars.2014.00044
Marlow, J. J., Steele, J. A., Ziebis, W., Thurber, A. R., Levin, L. A., and Orphan, V. J. (2014b). Carbonate-hosted methanotrophy represents an unrecognized methane sink in the deep sea. Nat. Commun. 5, 5094. doi: 10.1038/ncomms6094
Mason, O. U., Case, D. H., Naehr, T. H., Lee, R. W., Thomas, R. B., Bailey, J. V., et al. (2015). Comparison of archaeal and bacterial diversity in methane seep carbonate nodules and host sediments, Eel River Basin and Hydrate Ridge, USA. Microb. Ecol. 70, 766–784. doi: 10.1007/s00248-015-0615-6
Mason, O. U., Hazen, T. C., Borglin, S., Chain, P. S. G., Dubinsky, E. A., Fortney, J. L., et al. (2012). Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deeepwater Horizon oil spill. ISME J. 6, 1715–1727. doi: 10.1038/ismej.2012.59
Mathews, D. C. (1964). Recent observations on neck extensions in folliculinids (Protozoa). Pac. Sci. 18, 229–235.
Mathews, D. C. (1968). The folliculinids (protozoa) of Ago Bay, Japan, and their relation to the epifauna of the pearl oyster (Pinctada martensii). Pac. Sci. 22, 232–250.
McMurdie, P. J., and Holmes, S. (2014). Waste no, want not, why rarefying microbiome data is inadmissible. PLoS Comp. Biol. 10:e1003531. doi: 10.1371/journal.pcbi.1003531
Miao, M., Song, W., Clamp, J. C., Al-Rasheid, A. S., Al-Khedhairy, A. A., and Al-Arifi, S. (2009). Further consideration of the phylogeny of some “traditional” heterotrichs (Protista, Ciliaphora) of uncertain affinities, based on new sequences of the small subunit rRNA gene. J. Eukaryot. Microbiol. 56, 244–250. doi: 10.1111/j.1550-7408.2009.00391.x
Miao, W., Simpson, A. G. B., Fu, C., and Lobban, C. S. (2005). The giant zooxanthellae-bearing ciliate Maristentor dinoferus (Heterotrichea) is closely related to Folliculinidae. J. Eukaryot. Microbiol. 52, 11–16. doi: 10.1111/j.1550-7408.2005.3311r.x
Mulisch, M., Smock, U., and Uhlig, G. (1993). Life cycle and ultrastructure of Lagotia minor Dons, 1948 (Ciliaphora, Heterotrichia) revealing new characters for folliculinid classification. Europ. J. Protistol. 29, 144–154. doi: 10.1016/S0932-4739(11)80267-1
Orphan, V. J., House, C. H., Hinrichs, K. U., McKeegan, K. D., and DeLong, E. F. (2001). Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293, 484–487. doi: 10.1126/science.1061338
Orphan, V. J., Ussler, W. III, Naehr, T. H., House, C. H., Hinrichs, K. U., and Paull, C. K. (2004). Geological, geochemical, and microbiological heterogenetic of the seafloor around methane vents in the Eel River Basin, offshore California. Chem. Geol. 205, 265–289. doi: 10.1016/j.chemgeo.2003.12.035
Pasulka, A. L., Levin, L. A., Steele, J. A., Case, D. H., Landry, M. R., and Orphan, V. J. (2016). Microbial eukaryotic distributions and diversity patterns in a deep-sea methane seep ecosystem. Environ. Microbiol. 18, 3022–3043. doi: 10.1111/1462-2920.13185
Peterson, B. J., and Fry, B. (1987). Stable isotopes in ecosystem studies. Ann. Rev. Ecol. Syst. 18, 293–320. doi: 10.1146/annurev.es.18.110187.001453
Russ, L., Kartal, B., op den Camp, H. J. M., Sollai, M., Le Bruche, J., Caprais, J. C., et al. (2013). Presence and diversity of anammox bacteria in cold hydrocarbon-rich seeps and hydrothermal vent sediments of the Guaymas Basin. Front. Microbiol. 4:219. doi: 10.3389/fmicb.2013.00219
Salter, S., Cox, M., Turek, E., Calus, S. T., Cookson, W. O., Moffatt, M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12:87. doi: 10.1186/s12915-014-0087-z
Sauvadet, A. L., Gobet, A., and Guillou, L. (2010). Comparative analysis between protist communities from the deep-sea pelagic ecosystem and specific deep hydrothermal habitats. Environ. Microbiol. 12, 2946–2964. doi: 10.1111/j.1462-2920.2010.02272.x
Scheltema, R. S. (1973). On the unusual means by which the sessile marine ciliate Folliculina simplex maintains its widespread geographical distribution. Neth. J. Sea. Res. 7, 122–125. doi: 10.1016/0077-7579(73)90038-0
Schmaljohann, R., Faber, E., Whiticar, M. J., and Dando, P. R. (1990). Co-existence of methane- and sulphur-based endosymbiosis between bacteria and invertebrates at a site in the Skagerrak. Mar. Ecol. Prog. Ser. 51, 119–124. doi: 10.3354/meps061119
Schmidt, S. L., Foissner, W., Schlegel, M., and Bernhard, D. (2007). Molecular phylogeny of the Heterotrichea (Ciliaphora, Postciliodesmatophora) based on small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 54, 358–363. doi: 10.1111/j.1550-7408.2007.00269.x
Sherr, E. B., and Sherr, B. F. (1994). Bacterivory and herbivory: key roles of phagotrophic protists in pelagic food webs. Microb. Ecol. 28, 223–235. doi: 10.1007/BF00166812
Sherr, E. B., and Sherr, B. F. (2002). Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81, 293–308. doi: 10.1023/A:1020591307260
Silvestro, D., and Michalak, I. (2010). raxmlGUI: a graphical front-end for RAxML. Org. Divers. Evol. 12, 335–337. doi: 10.1007/s13127-011-0056-0
Stahl, D. A., and Amann, R. (1991). “Development and application of nucleic acid probes,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester: John Wiley and Sons Ltd, Chichester), 205–248.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stamatakis, A (2008). The RAxML 7.0.4 Manual [WWW Document]. Available online at: http://sco.h-its.org/exelixis/oldPage/RAxML-Manual.7.0.4.pdf.
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Stiller, J., Roussett, V., Pleijel, F., Chevaldonné, P., Vrijenhoek, R. C., and Rouse, G. W. (2013). Phylogeny, biogeography and systematics of hydrothermal vent and methane seep Amphisamytha (Ampharetidae, Annelida), with descriptions of three new species. Syst. Biodiv. 11, 35–65. doi: 10.1080/14772000.2013.772925
Suess, E., Torres, M. E., Bohrmann, G., Collier, R. W., Greinert, J., Linke, P., et al. (1999). Gas hydrate destabilization: enhanced dewatering, benthic material turnover and large methane plumes at the Cascadia convergent margin. Earth Planet. Sci. Lett. 170, 1–15. doi: 10.1016/S0012-821X(99)00092-8
Suess, E., and Whiticar, M. J. (1989). Methane-derived CO2 in pore water fluids expelled from the Oregon subduction zone. Palaeogeogr. Palaeoclimatol. Palaeoecol. 71, 119–136. doi: 10.1016/0031-0182(89)90033-3
Swofford, D. (2002). PAUP 4.0 B10: Phylogenetic Analysis Using Parsimony. Sunderland, MA: Sinauer Associates.
Swofford, D., and Sullivan, J. (2003). “Phylogeny inference based on parsimony and other methods using PAUP*,” in The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny, eds P. Lemey, M. Salemi, and A. M. Vandamme (Cambridge, UK: Cambridge University Press), 160–206.
Takishita, K., Kakizoe, N., Yoshida, T., and Maruyama, T. (2010). Molecular evidence that phylogenetically diverged ciliates are active in microbial mats of deep-sea cold-seep sediment. J. Eukaryot. Microbiol. 57, 76–86. doi: 10.1111/j.1550-7408.2009.00457.x
Takishita, K., Yubuki, B., Kakizoe, N., Inagaki, Y., and Maruyama, T. (2007). Diversity of microbial eukaryotes in sediment at a deep-sea methane cold seep: surveys of ribosomal DNA libraries from raw sediment samples and two enrichment cultures. Extremophiles 11, 563–576. doi: 10.1007/s00792-007-0068-z
Tavormina, P. L., Hatzenpichler, R., McGlynn, S., Chadwick, G., Dawson, K. S., Connon, S. A., et al. (2015). Methyloprofundus sediment gen. nov., sp. nov., an obligate methanotroph from ocean sediment belonging to the “deep sea-1” clade of marine methanotrophs. Int. J. Syst. Evol. Microbiol. 65, 251–259. doi: 10.1099/ijs.0.062927-0
Tavormina, P. L., Ussler, W. III, Joyem, S. B., Harrison, B. K., and Orphan, V. J. (2010). Distributions of putative aerobic methanotrophs in diverse pelagic marine environments. ISME J. 4, 700–710. doi: 10.1038/ismej.2009.155
Tavormina, P. L., Ussler, W. III, and Orphan, V. J. (2008). Planktonic and sediment-associated aerobic methanotrophs in two seep systems along the North American Margin. Appl. Environ. Microbiol. 74, 3985–3995. doi: 10.1128/AEM.00069-08
Tavormina, P. L., Ussler, W. III, Steele, J. A., Connon, S. A., Klotz, M. G., and Orphan, V. J. (2013). Abundance and distribution of diverse membrane-bound monooxygenase (Cu-MMO) genes within the Costa Rica oxygen minimum zone. Environ. Microbiol. Rep. 5, 414–423. doi: 10.1111/1758-2229.12025
Teichert, B. M. A., Bohrmann, G., and Suess, E. (2005). Chemoherms on Hydrate Ridge – unique microbially-mediated carbonate build-ups growing into the water column. Palaeogeogr. Palaeoclimatol. Palaeoecol. 227, 67–85. doi: 10.1016/j.palaeo.2005.04.029
Tews, J., Brose, U., Grimm, V., Tielbörger, K., Wichmann, M. C., Schwager, M., et al. (2004). Animal species diversity driven by habitat heterogeneity/diversity: the importance of keystone structures. J. Biogeogr. 31, 79–92. doi: 10.1046/j.0305-0270.2003.00994.x
Toupoint, N., Mohit, V., Linossier, I., Bourgougnon, N., Myrand, B., Olivier, F., et al. (2012). Effect of biofilm age on settlement of Mytilus edulis. Biofouling 28, 985–1001. doi: 10.1080/08927014.2012.725202
Trembath-Reichert, E., Green-Saxena, A., and Orphan, V. J. (2013). Whole cell immunomagnetic enrichment of environmental microbial consortia using rRNA-targeted magneto-FISH. Method. Enzymol. 531, 21–44. doi: 10.1016/B978-0-12-407863-5.00002-2
Treude, T., Boetius, A., Knittel, K., Wallmann, K., and Jøregensen, B. (2003). Anaerobic oxidation of methane above gas hydrate at Hydrate Ridge, NE Pacific Ocean. Mar. Ecol. Prog. Ser. 264, 1–14. doi: 10.3354/meps264001
Treude, T., Orphan, V., Knittle, K., Gieseke, A., House, C. H., and Boetius, A. (2007). Consumption of methane and CO2 by methanotrophic microbial mats from gas seeps of the anoxic black sea. Appl. Environ. Microbiol. 73, 2271–2283. doi: 10.1128/AEM.00806-07
Tryon, M. D., Brown, K. M., and Torres, M. E. (2002). Fluid and chemical flux in and out of sediments hosting methane hydrate deposits on Hydrate Ridge, OR, II: hydrological processes. Earth Planet. Sci. Lett. 201, 541–557. doi: 10.1016/S0012-821X(02)00732-X
Tunnicliffe, V., Juniper, S. K., and de Burgh, M. E. (1985). The hydrothermal vent community on axial seamount, Juan de Fuca Ridge. Biol. Soc. Wash. Bull. 6, 434–464.
Uhlig, G., Komnick, H., and Wohlfarth-bottermann, K. E. (1965). Intrazelluläre zellzotten in nahrungsvakuolen von ciliaten. Helgol. Wissenschaftliche Meeresunters. 12, 61–77. doi: 10.1007/BF01612094
Valentine, D. L. (2011). Emerging topics in marine methane biogeochemistry. Ann. Rev. Mar. Sci. 3, 147–171. doi: 10.1146/annurev-marine-120709-142734
Van Dover, C. L. (2007). “Stable isotope studies in marine chemoautrotrophically based ecosystems: an update,” in Stable Isotopes in Ecology and Environmental Science, eds R. Michener and K. Lajtha (Malden, MA: Wiley-Blackwell), 202–237.
Van Dover, C. L., Berg, C. J., and Turner, R. D. (1988). Recruitment of marine invertebrates to hard substrates at deep-sea hydrothermal vents on the East Pacific Rise and Galapagos spreading center. Deep-Sea Res. 35, 1833–1849. doi: 10.1016/0198-0149(88)90052-0
Warén, A., and Rouse, G. W. (2016). A new genus and species of Cataegidae (Gastropoda; Seguenzioidea) from eastern Pacific Ocean methane seeps. Novaplex 17, 59–66.
Warren, A (2013). World Ciliaphora Database. Accessed through: World Register of Marine Species. Available online at: http://www.marinespecies.org.
Wendeberg, A., Zielinski, F. U., Borowski, C., and Dubilier, N. (2012). Expression patterns of mRNAs for methanotrophy and thiotrophy in symbionts of the hydrothermal vent mussel Bathymodiolus puteoserpentis. ISME J. 6, 104–112. doi: 10.1038/ismej.2011.81
Yang, T., Nigro, L. M., Gutierrez, T., D'Ambrosio, L., Joye, S. B., Highsmith, R., et al. (2016). Pulsed blooms and persistent oil-degrading bacterial populations in the water column during and after the Deepwater Horizon blowout. Deep Sea Res. II Top. Stud. Oceanogr. 129, 282–291. doi: 10.1016/j.dsr2.2014.01.014
Keywords: folliculinid ciliate, methane cycling, microbial ecology, methane seep, microbial diversity
Citation: Pasulka AL, Goffredi SK, Tavormina PL, Dawson KS, Levin LA, Rouse GW and Orphan VJ (2017) Colonial Tube-Dwelling Ciliates Influence Methane Cycling and Microbial Diversity within Methane Seep Ecosystems. Front. Mar. Sci. 3:276. doi: 10.3389/fmars.2016.00276
Received: 22 July 2016; Accepted: 13 December 2016;
Published: 04 January 2017.
Edited by:
Télesphore Sime-Ngando, Centre National de la Recherche Scientifique, FranceCopyright © 2017 Pasulka, Goffredi, Tavormina, Dawson, Levin, Rouse and Orphan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexis L. Pasulka, apasulka@caltech.edu
 Alexis L. Pasulka
Alexis L. Pasulka Shana K. Goffredi
Shana K. Goffredi Patricia L. Tavormina
Patricia L. Tavormina Katherine S. Dawson
Katherine S. Dawson Lisa A. Levin
Lisa A. Levin Greg W. Rouse
Greg W. Rouse Victoria J. Orphan
Victoria J. Orphan