Lactic Acid Bacteria Selection for Biopreservation as a Part of Hurdle Technology Approach Applied on Seafood
- 1Laboratoire Ecosystèmes Microbiens et Molécules Marines pour les Biotechnologies, Ifremer, Nantes, France
- 2UMR1014 SECALIM, Institut National de la Recherche Agronomique, Oniris, Nantes, France
As fragile food commodities, microbial, and organoleptic qualities of fishery and seafood can quickly deteriorate. In this context, microbial quality and security improvement during the whole food processing chain (from catch to plate), using hurdle technology, a combination of mild preserving technologies such as biopreservation, modified atmosphere packaging, and superchilling, are of great interest. As natural flora and antimicrobial metabolites producers, lactic acid bacteria (LAB) are commonly studied for food biopreservation. Thirty-five LAB known to possess interesting antimicrobial activity were selected for their potential application as bioprotective agents as a part of hurdle technology applied to fishery products. The selection approach was based on seven criteria including antimicrobial activity, alteration potential, tolerance to chitosan coating, and superchilling process, cross inhibition, biogenic amines production (histamine, tyramine), and antibiotics resistance. Antimicrobial activity was assessed against six common spoiling bacteria in fishery products (Shewanella baltica, Photobacterium phosphoreum, Brochothrix thermosphacta, Lactobacillus sakei, Hafnia alvei, Serratia proteamaculans) and one pathogenic bacterium (Listeria monocytogenes) in co-culture inhibitory assays miniaturized in 96-well microtiter plates. Antimicrobial activity and spoilage evaluation, both performed in cod and salmon juice, highlighted the existence of sensory signatures and inhibition profiles, which seem to be species related. Finally, six LAB with no unusual antibiotics resistance profile nor histamine production ability were selected as bioprotective agents for further in situ inhibitory assays in cod and salmon based products, alone or in combination with other hurdles (chitosan, modified atmosphere packing, and superchilling).
Introduction
Benefiting from a healthy image, as a source of valuable nutrients (proteins, vitamins, minerals, omega-3 fatty acids etc.), seafood and fishery products contribute to an important part of our alimentation with an average world consumption value of 20.1 kg/per capita in 2014 (FAO, 2016). As the demand is increasing with the world population, total fisheries and aquaculture production for human consumption is expected to grow from 146.3 million tons (for a trade value of US$ 148 billion) in 2014 up to 181.1 million tons in 2022 (Lem et al., 2014; FAO, 2016).
Fishery products are very fragile commodities with a short shelf-life not exceeding 1–2 weeks for fresh products to 3–4 weeks for lightly preserved ones. This is mainly due to a high post-mortem pH often superior to 6.0, combined with a high non-protein nitrogen fraction including trimethylamine oxide (TMAO) and amino acids such as methionine and cysteine which are related to strong off-odors and off-flavors molecules production (Gram and Huss, 1996). Thus, this intrinsic flesh composition makes an appropriate growth environment for specific spoilage microorganisms (SSO; Dalgaard, 1995) involved in sensory degradation. Fresh and lightly preserved fish products are more likely spoiled by psychrotrophic Gram negative bacteria such as Shewanella sp. (S. putrefaciens, S. baltica), Aeromonas sp., Pseudomonas sp. (P. fragi, P. fluorescens, P. putida, P. lundensis, etc.), Photobacterium sp. (P. phosphoreum, P. illiopiscarium), Enterobacteriaceae (Serratia proteomaculans, Hafnia alvei, etc.), or Brochothrix thermosphacta (Gram and Huss, 1996; Gram and Dalgaard, 2002; Cortesi et al., 2009; Leroi, 2014; Løvdal, 2015). Lactic acid bacteria (LAB), mainly Carnobacterium sp. (C. maltaromaticum, C. divergens), Lactobacillus sp. (L. curvatus, L. sakei, L. farciminis, L. plantarum) can also be found in high proportion and may contribute to seafood spoilage (Cortesi et al., 2009; Leroi, 2010, 2014; Pilet and Leroi, 2011).
Seafood products can also be important vectors for human illnesses, as 10–20% of food-borne diseases are attributed to fish consumption (Pilet and Leroi, 2011). The main microbial risk in seafood products is related to Listeria monocytogenes, Vibrio sp., Salmonella sp., Staphylococcus aureus, which are indigenous to the aquatic environment, or resulting from post-contamination during manufacturing processes (Huss et al., 1995, 2000; Pilet and Leroi, 2011; Leroi, 2014; Løvdal, 2015).
Thus, microbial food spoilage and pathogens growth control are currently representing a crucial challenge, as they have been evaluated to be responsible for the loss of 25% of all post-harvesting food production (Gram and Dalgaard, 2002). In addition to industrial traditional technologies and to face the consumers' demand for minimally processed food, new trends such as biopreservation, high hydrostatic pressure, pulsed electric fields, superchilling, chitosan coating, and active packagings are promising complementary ways to extend food shelf-life and reduce microbial risks (Devlieghere et al., 2004; Cortesi et al., 2009).
Biopreservation corresponds to the use of bacteria (or their metabolites) with antimicrobial properties to prevent undesirable bacteria growth (Stiles, 1996). LAB are excellent candidates as they are naturally present in many food commodities (Rodgers, 2001; Ghanbari et al., 2013). They possess the ability to produce a wide range of broad spectrum antimicrobial compounds such as bacteriocins, organic acids, fatty acids, diacetyl, acetaldehyde, H2O2, reuterin (Caplice and Fitzgerald, 1999). They are generally recognize as harmless for human consumption and benefiting from a healthy and natural image from consumers (Holzapfel et al., 1995; Ghanbari et al., 2013). In food, biopreservation is mainly studied to control pathogens and many authors succeeded to inhibit L. monocytogenes or S. aureus growth in fresh or lightly processed products. On the contrary only few studies showed sensory quality improvement by targeting SSO (Devlieghere et al., 2004; Pilet and Leroi, 2011; Ghanbari et al., 2013).
This study aimed to select protective cultures (PC), from a collection of 35 LAB strains with interesting antimicrobial profiles, to be combined with others hurdles (MAP, chitosan, superchilling) for cod and salmon based products microbial quality and safety improvement. LAB strains were screened according to seven criteria (Figure 1). Five criteria are commonly used for antimicrobial or functional cultures assessment and safety issue for human consumption (Holzapfel et al., 1995; Ammor and Mayo, 2007; Jones et al., 2011; Ghanbari et al., 2013; Leroi et al., 2015) and two specific criteria, chitosan and freezing tolerance, were especially designed for this study. Unlike conventional semi-quantitative antimicrobial methodologies, mainly based on agar diffusion tests (Jones et al., 2011), a miniaturized method in fish juice model was performed to quantify LAB inhibitory activity. To keep a bacterial diversity through the whole screening process and future experiments, at least one strain was selected per LAB species.
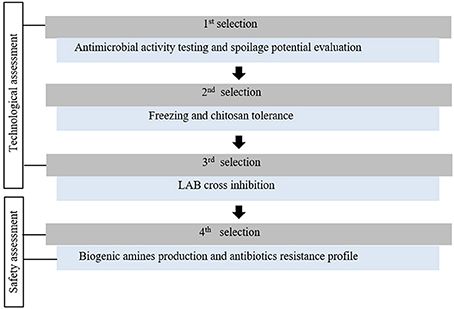
Figure 1. Protective cultures selection strategy for seafood products microbial safety and quality improvement.
Materials and Methods
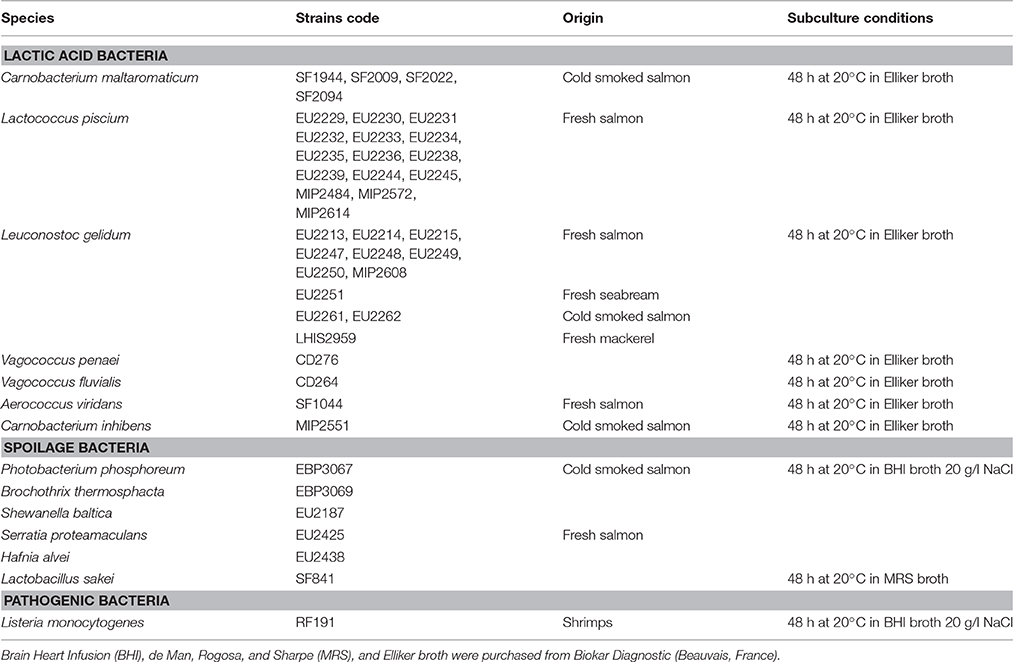
Bacterial Strains and Subculture Conditions
Bacterial strains were isolated from seafood matrices through previous collaborative projects between Ifremer and Oniris/UMR1014 SECALIM, INRA. Bacterial strains subculture conditions are listed in Table 1. All strains were then stored at −80°C in their growth respective medium supplemented with 10% of sterile glycerol (Sigma-Aldrich, Steinheim, Germany).
Fish Juice Preparation
Salmon and cod juices were prepared according to an adapted method from Dalgaard (1995) and Leroi et al. (1998). Five hundred grams of fresh cod or salmon filet obtained from local retailer (Carrefour, Nantes, France) were added with 1 l of distilled water prior to a blending step using a blendor (Waring, Torrington, Connecticut, USA). The mixture was then boiled for 2 min before being filtered through a pleated filter (Whatman, Maldstone, England). Fish juice extracts was sterilized at 100°C for 30 min and stored at −20°C.
Co-culture Inhibitory Assay in 96-Well Plates
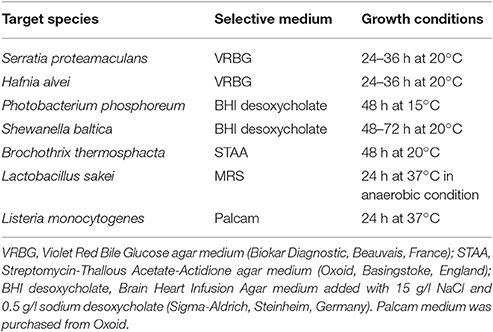
Bacteria were precultured in their respective broth medium for 48 h at 15°C and then diluted in tryptone salt (Biokar Diagnostic, Beauvais, France) to reach, respectively, 108 CFU/ml for PC and 104 for the target bacteria. Prior to use, fish juices were thawed at room temperature, 90 ml were supplemented with 10 ml of 1 M K2HPO4/KH2PO4(Merck, Darmstadt, Germany) buffer solution at pH 6.7, 1 g of D-glucose (Merck) and 1.5 g of NaCl (Merck). Enriched fish juices were then sterilized on 0.45 μm PTFE filter (Merck). Co-cultures were performed in a 96-well plate containing 196 μl of fish juice and 2 μl of both PC and target diluted suspension. Thus, initial bacterial concentration in co-cultures was 106 CFU/ml for PC and 102 CFU/ml for targets. After 96 h at 15°C, co-culture products were diluted following a 10-fold serial dilution in tryptone salt in a 96-well plate. Five microliters of each dilution were then platted on target selective agar medium in 12 cm2 Petri dishes. Selective media and growth conditions are summarized in Table 2. After incubation period, colonies were enumerated for each spot with a maximum of fifty colonies to avoid overlap. Inhibition was then quantified by comparing target concentration in co-culture with pure culture. According to the deposit volume (5 μl) for enumeration, the method threshold was 2.30 log CFU/ml. A principal component analysis (PCA) was firstly performed on PC inhibition scores against the seven targets. Then a Ward's hierarchical clustering method with squared Euclidian distance was performed on PCA components to separate the 35 PC into clusters according to their antimicrobial profile (R, Vienna, Austria).
Spoilage Potential Evaluation
PC strains spoilage potential was evaluated both in cod and salmon juice prepared as described in Co-Culture Inhibitory Assay in 96-Well Plates and supplemented with 0.1 g/l of L-cysteine, 0.1 g/l of L-methionine, and 0.1 g/l of TMAO (Sigma-Aldrich). Four hundred fifty microliters of PC preculture (Elliker 48 h at 15°C) were inoculated into 45 ml of fish juice and incubated at 15°C for 96 h. For each PC strain, 12 trained panelists, experienced in seafood sensory evaluation and spoilage assessment (Macé et al., 2012, 2013, 2014), carried out a conventional profiling test (ISO 13299, 2003) on seven relevant descriptors (spoilage intensity, fish, pungent acid, sour, acid/lemon, feet/banana, and sulfur). After a sniffing step, descriptors were scored on a continuous scale anchored by low intensity (score 0) up to high intensity (score 10). Sessions were performed in individual partitioned booths, as described in the procedure NF V-09-105 (ISO 8598, 2010) and equipped with a computerized system (Fizz, Biosystèmes, Couternon, France). All the inoculated juices were kept frozen at −80°C until sensory sessions and thawed 1 h in cold water prior to use. During sensory evaluation, panelists received 5 ml from samples. For each session, a sample of non-inoculated cod or salmon juice was set as a reference of non-spoiled sample. This sample was not scored but served as a baseline for sensory evaluation. The 35 samples per fish juice (cod and salmon) were divided in four profiling sessions, with nine samples per session. Strains choice was balanced all over the sensory evaluation by presenting the same bacterial groups diversity at each session. Samples were assigned with three digit numbers and randomized for the order presentation within panelists. PCA with standardization was performed on scores mean for each sensory descriptor.
Freezing Impact on Cells Viability
Freezing tolerance test was performed in salmon fish juice as prepared in Co-culture Inhibitory Assay in 96-Well Plates. PC were precultured in Elliker broth 48 h at 15°C and then 48 h at 15°C in salmon juice. For each bacterial strain, 1 ml of preculture was frozen in eppendorf tube at −80°C for 3 h and thawed at room temperature. Bacterial enumeration was performed prior and after the freezing period using the micro-enumeration method described above in Co-culture Inhibitory Assay in 96-Well Plates. All strains were enumerated after 48–72 h at 20°C on Elliker agar medium, except for Leuconostoc gelidum strains, which were instead counted on BHI 20 g/l NaCl agar medium to avoid exopolysaccharides production.
Chitosan Treatment Tolerance
PC chitosan tolerance was evaluated at two different pH values (6.0 and 6.6) in 10-fold diluted salmon juice prepared as described in Fish Juice Preparation and supplemented with less sugar and salt, 0.15% (w/v) and 0.30% (w/v) respectively, to avoid chitosan precipitation. pH was adjusted with NaOH or HCl solution addition (Grosseron, Nantes, France) and was then filter-sterilized. Prior to use, juices were supplemented with 0.02% (v/v) of chitosan D (Primex, Siglufjordu, Iceland). PC strains were pre-cultivated twice successively in Elliker broth 48 h at 15°C followed by 48 h at 15°C in salmon juice (as prepared in Co-culture Inhibitory Assay in 96-Well Plates). One hundred and ninety-eight microliters of diluted salmon juice containing chitosan were inoculated in 96-well plates with 2 μl of PC preculture. Microtiter plates were incubated 3 h at 15°C. PC concentration was determined just after inoculation and every hour with micro-enumeration method described in Co-Culture Inhibitory Assay in 96-Well Plates.
Lactic Acid Bacteria Cross Inhibition Assay
The cross inhibition was evaluated by the double layer method used by Matamoros et al. (2009a). PC were precultured 48 h at 15°C in Elliker broth. Ten microliters were spotted onto Elliker agar Petri dish. Plates were incubated 48 h at 15°C under anaerobic conditions. A second preculture was performed at 15°C for 48 h. Bacterial suspensions were 100-fold diluted in physiological water. One milliliters of bacterial dilution was transferred into 15 ml of molten Elliker medium containing 1% agar, which were quickly spread to form a double layer onto previously inoculated Elliker plates containing PC spots. Plates were incubated 96 h at 20°C and cross inhibition was quantified by measuring inhibition diameters from the colonies center.
Biogenic Amines Production
After preculture (Elliker 48 h at 15°C), each PC was cultivated 96 h at 15°C in salmon juice as prepared in Co-culture Inhibitory Assay in 96-Well Plates and supplemented with histidine or tyrosine (Sigma-Aldrich) at a final concentration of 350 μg/ml. Histamine and tyramine were quantified following a protocol adapted from Duflos et al. (1999). Ten milliliters from sample were added with 5 ml of TCA (Panreac, Darmstadt, Germany) at 12% and kept frozen at −20°C until analysis. After thawing, samples were spiked with 100 μl of intern standard (1,7 diaminoheptane dihydrochloride at 2 mg/ml) and centrifuged 15 min at 7,000 g. One hundred microliters of supernatant were mixed with 300 μl of satured Na2CO3(Sigma-Aldrich) solution, 400 μl of dansyl chloride solution at 7.5 mg/ml (Sigma-Aldrich) and incubated in obscurity in water bath at 40°C for 45 min. Tubes were left at room temperature for 10 min before the addition of 100 μl of ammoniac solution (Carlo Erba Reagents, Val de Reuil, France) at 25% and kept in obscurity for 30 min. Biogenic amines were then recovered as following Duflos et al. (1999) methodology. For each sample, 20 μl were injected in a HPLC chain (Shimadzu, France) equipped with a C18 Kinetexcolumn (5 μm, 100A, 250 × 4.6 mm, Phenomenex, Le Pecq, France). Elution gradient started with 60% acetonitrile (Fisher Chemical, Loughborough, UK) and 40% water and ended with 95% acetronitrile and 5% water after 18 min. Biogenic amines were detected with UV detector. Chromatograms were analyzed with LabSolutions software (Shimadzu).
Antibiotics Resistance Profile
PC antimicrobial profile was evaluated according to the standard method procedure ISO 10932 (2010). A minimum inhibitory concentration (MIC) was determined for the 9 following antimicrobial agents as recommended by EFSA (2012). For each agent, the concentration range expressed in μg/ml is given in brackets: gentamicin (0.5–256), kanamycin (2–1024), streptomycin (0.5–256), tetracycline (0.125–64), erythromycin (0.016–8), clindamycin (0.032–16), chloramphenicol (0.125–64), ampicillin (0.032–16), vancomycin (0.25–128). MICs were determined for all strains after 48 h in anaerobic conditions at 28°C, except for L. gelidum strains, incubated at 26°C. Lactobacillus plantarum ATCC® 14917 was used as reference strain for quality control (ISO 10932, 2010).
Results
First Selection
Antimicrobial Activity
The 35 LAB strains antimicrobial activity was evaluated in co-culture in fish juice (cod and salmon) miniaturized in 96-well plates against seven targets frequently isolated from seafood (Shewanella baltica, Photobacterium phosphoreum, B. thermosphacta, Lactobacillus sakei, H. alvei, Serratia proteamaculans, and L. monocytogenes).
All LAB strains demonstrated an antagonist effect against at least one target in fish juice model. Inhibitory results are summarized in Table 3.
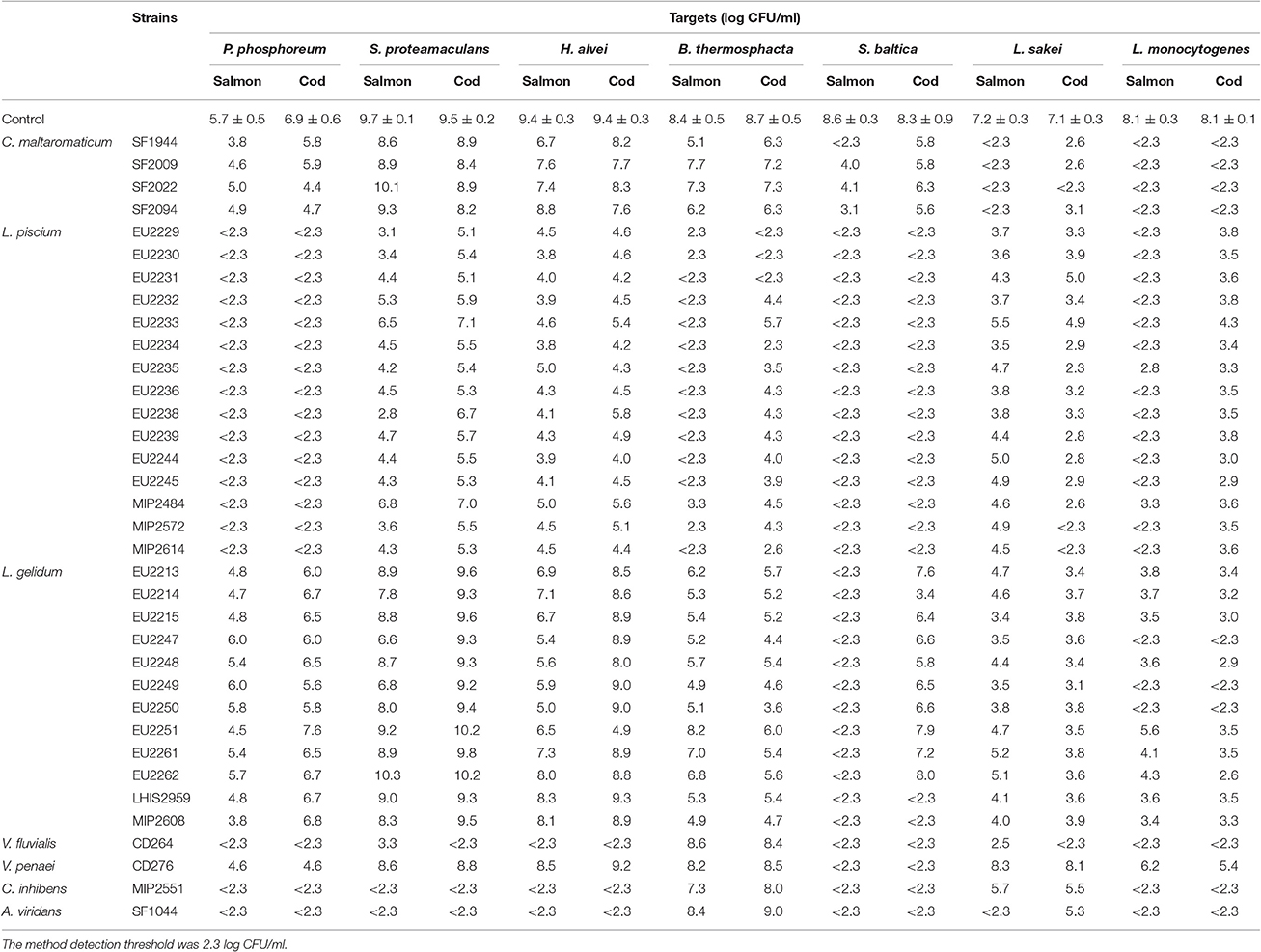
Table 3. Growth of the seven targets strains in pure culture (control) and in co-culture with the 35 LAB in cod and salmon juice after 96 h at 15°C.
L. piscium isolates showed the widest antimicrobial spectrum by demonstrating inhibition activity for all target bacteria in both fish juices. P. phosphoreum, B. thermosphacta, S. baltica, and L. monocytogenes, were strongly inhibited (count < 2.3 log CFU/ml). S. proteamaculans, H. alvei and L. sakei were also broadly inhibited with 2.5–7.0 log CFU/ml reduction depending on the L. piscium strain.
Carnobacterium inhibens MIP2551, Vagococcus fluvialis CD264, and Aerococcus viridans SF1044 were the most active, by almost totally inhibiting all target bacteria (count < 2.3 log CFU/ml). There were exceptions such as B. thermosphacta which was never inhibited, L. sakei which was not inhibited by C. inhibens MIP2551 and A. viridans SF1044 (only in cod juice) or strongly inhibited (4.7 log CFU/ml reduction) by V. fluvialis CD264 only in salmon juice. Among these three strains, V. fluvialis CD264 did not totally inhibit S. proteamaculans in salmon juice, but still displayed a strong activity with 6.4 log CFU/ml reduction.
Vagoccoccus penaei strain CD272 exhibited the weakest inhibition properties. This isolate inhibited only S. baltica (count < 2.3 log CFU/ml), and was slightly active against P. phosphoreum and L. monocytogenes with 2.0 log CFU/ml reduction.
In presence of C. maltaromaticum strains, both in cod and salmon juice, L. sakei and L. monocytogenes were strongly inhibited with, respectively, 4.0–4.5 and 5.5 log CFU/ml reduction. C. maltaromaticum strains also demonstrated a strong antagonist activity against S. baltica in salmon juice (4.5–6.3 log CFU/ml reduction), but not in cod juice, which was drastically lower (2.0 log CFU/ml). P. phosphoreum and H. alvei were only slightly inhibited (1.0–2.0 log CFU/ml reduction), while S. proteamaculans growth was not significantly reduced in comparison with the control culture (< 1 log CFU/ml reduction).
L. gelidum strains showed contrasting inhibitory activities against S. baltica, S. proteamaculans and H. alvei according to the fish juice and the strain. For instance, all strains strongly inhibited S. baltica (count < 2.3 log CFU/ml) in salmon juice, but lost their activity in cod juice, except EU2214, LHIS2959, and MIP2608. L. monocytogenes was highly inhibited with more than 4.0 log CFU/ml reduction. Especially in the presence of the strains EU2247, EU2249 and EU2250, L. monocytogenes bacterial count was under the detection threshold. B. thermosphacta and L. sakei were only slightly inhibited. According to the fish juice and for some L. gelidum strains, P. phosphoreum was also slightly inhibited with < 1.9 log CFU/ml reduction.
Figure 2, representing Ward's hierarchical classification based on PCA components of the 35 PC inhibition scores against the seven targets, showed that PC strains were strongly clustered together according to the species. Fish juice type played minor effect with antimicrobial activity generally slightly lower in cod juice compared to salmon juice. V. fluvialis CD264, C. inhibens MIP2551, and A. viridans SF1044 antimicrobial spectra were found to be closed to L. piscium strains activity, while L. gelidum EU2247, EU2249, EU2250 antimicrobial activity were closed to C. maltaromaticum strains antimicrobial activity.
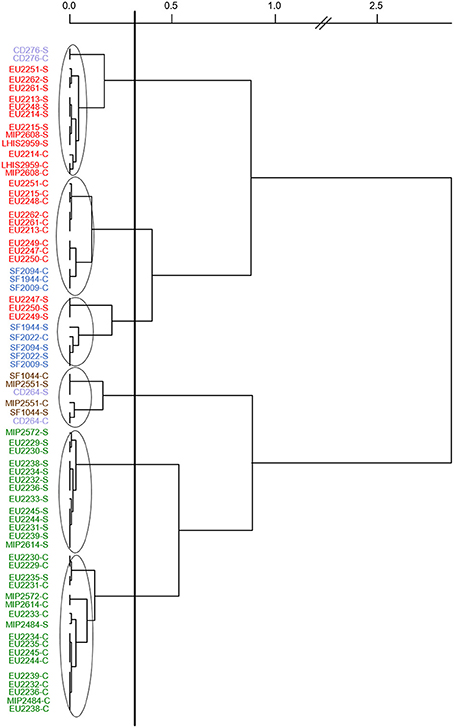
Figure 2. Ward's hierarchical classification realized with squared Euclidian distance on PCA components of the 35 protective cultures inhibition scores against the 7 targets in both salmon and cod juices. Strain code followed by a letter S: Salmon juice, a letter C: cod juice. Strains in red: L. gelidum, in blue: C. maltaromaticum, in green: L. piscium, in brown: C. inhibens MIP2551 and A. viridans SF1044, in violet: V. fluvialis CD264 and V. penaei CD276.
Spoilage Potential Evaluation
The 35 PC spoilage potential was investigated in cod and salmon juice supplemented with 0.1 g/l of L-cysteine, 0.1 g/l of L-methionine, and 0.1 g/l of TMAO, precursors of bad smell molecules related to seafood spoilage. For each strain, sensory descriptors were scored from 0 to 10 according to their intensity during a profiling test.
For both juices, sensory results are synthetized in Figure 3, which shows normalized PCA, performed on sensory descriptors mean scores. Reference samples were mainly characterized by fresh fish, dried fish, fish flour, or crustacean odors. According to their sensory profile, all PC strains were found to be distinctly clustered according to the species.
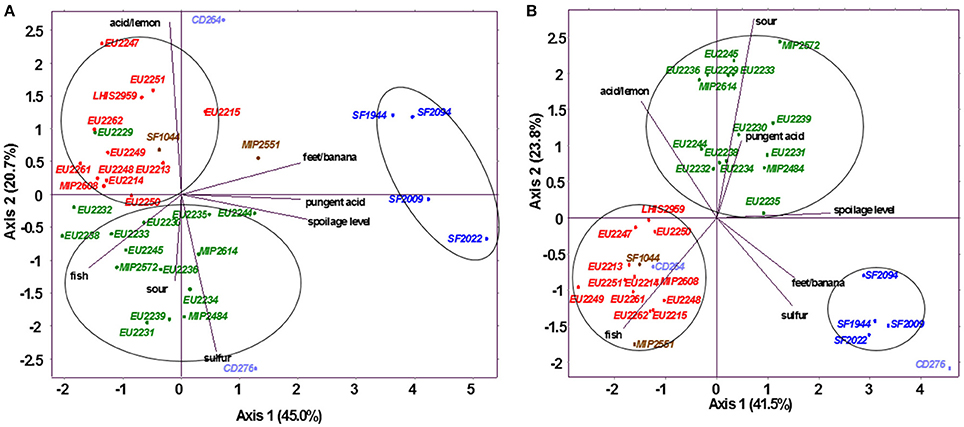
Figure 3. Normalized principal component analysis representation of inoculated salmon juice (A) and cod juice (B) based on odor descriptors, on dimension 1–2. Strains in red: L. gelidum, in blue: C. maltaromaticum, in green: L. piscium, in brown: C. inhibens MIP2551 and A. viridans SF1044, in violet: V. fluvialis CD264 and V. penaei CD276.
C. maltaromaticum strains were considered as the most spoiling bacteria group in both fish juices. They were able to reach a spoilage level of 3.0 to 4.5 in salmon juice, and even exceeding 5.0 for strains SF1994 and SF2009 in cod juice. C. maltaromaticum spoilage was mainly characterized by pungent acid and feet/banana odor production, also described as malt or rhubarb by some panelists.
L. piscium strains spoilage level were found to be slightly lower in salmon juice (0.5–2.0) in comparison with cod juice (2.0–3.5). In salmon juice, these strains produced weak sour and fish odors, also associated with a slight sulfur smell for isolates EU2231, EU2234, EU2239, EU2244, EU2245, and MIP2484. L. piscium EU2229 was the only exception with acid/lemon odor production. In cod juice, no L. piscium strains produced sulfur, but were associated with a higher sour and pungent acid smell.
In both juices, L. gelidum strains constitute the LAB group identified to possess the weakest alteration potential, by never exceeding 1.5 as spoilage score. In salmon juice, they were mostly related to fish odor close to the reference, and for some isolates such as EU2215, EU2247, EU2249, EU2251, and LHIS2959, with a small acid/lemon smell. In cod juice, L. gelidum strains were the most neutral, with same sensory profile than the non-inoculated control.
As for L. gelidum, C. inhibens MIP2551, V. fluvialis CD264, and A. viridans SF1044 were not different from the reference in cod juice. On the contrary, V. penaei CD276 was considered as the most spoiling bacteria with a high sulfur smell and a spoilage level reaching 6.6. In salmon juice, C. inhibens MIP2551 was associated with slight feet/banana odor, V. fluvialis CD264 and A. viridans SF1044 to a small acid/lemon smell, while V. penaei CD276 was still related to a sulfur smell, but in much less proportion than in cod juice.
Although almost all PC produced odors non-specific to seafood or fishery products, it was always in very small proportions and not always perceived by panelists as unpleasant smells related to spoilage, as for instance for acid/lemon odor production. According to these two first screening steps and by maintaining species diversity, 14 LAB strains were selected as the most interesting LAB in terms of antimicrobial activity with the less spoilage potential (per species): 2 strains of C. maltatomaticum (SF1944, SF2094), five strains of L. piscium (EU2229, EU2231, EU2234, EU2238, MIP2614), four strains of L. gelidum (EU2247, EU2249, EU2250, MIP2608), C. inhibens MIP2551, A. viridans SF1044 and V. fluvialis CD264.
Second Selection
Freezing Tolerance Assay
The 14 previously selected PC were enumerated before and after 3 h at −80°C. The bacterial count revealed no impact of the freezing step on strains viability, as the bacterial concentration was identical in salmon juice before and after the freezing period (data not shown).
Chitosan Tolerance Assay
The 14 PC tolerance to chitosan D treatment was evaluated at a concentration of 0.02% at pH 6.0 and 6.6 in salmon juice during 3 h. Bacterial enumeration was realized after each hour of treatment.
Chitosan D at 0.02% showed a variable antimicrobial activity against the fourteen LAB at pH 6.0 (Figure 4).
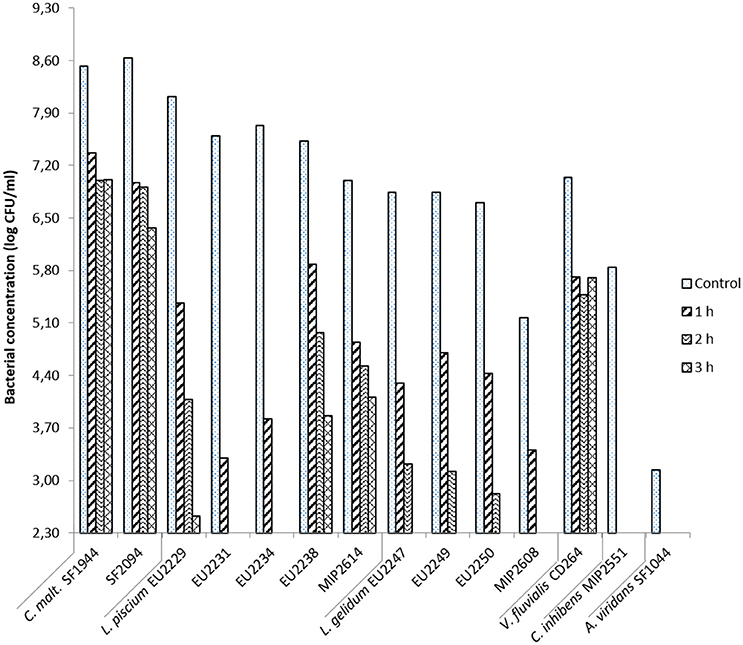
Figure 4. Chitosan D antimicrobial activity at 0.02% in salmon juice at pH 6.0 toward protective cultures strains after 1, 2, and 3 h of treatment.
C. inhibens MIP2551 and A. viridans SF1044, were the two most sensitive strains to chitosan treatment. Their count was below the detection threshold after only 1 h. In the same way L. gelidum isolates, were also highly sensitive but still persistent after 2 h of treatment, with an exception for the strain MIP2608 inhibited after only 1 h. All L. gelidum counts were below the detection threshold after 3 h of chitosan treatment.
C. maltaromaticum strains and V. fluvialis CD264 were the most tolerant PC. They were slightly inhibited after 1 h of treatment, with 1.2–1.7 log CFU/ml reduction, then no differences in microbial count was observable after 2 and 3 h.
L. piscium tolerance to chitosan was heterogeneous. Isolates EU2231 and EU2234 were drastically inhibited after 1 h of treatment, with respectively 4.3 and 3.9 log CFU/ml reduction in comparison with the initial bacterial load. After 2 h, the bacterial concentrations were below 2.3 log CFU/ml. In contrast, L. piscium EU2229, EU2238 and MIP2614 bacterial concentration were respectively 2.5, 3.9, and 4.1 log CFU/ml after 3 h of treatment.
At pH 6.6 chitosan sensitivity was identical to pH 6.0 for all PC, with an exception for L. gelidum EU2249 and EU2250 which were less sensitive at pH 6.6 (data not shown).
The chitosan tolerance assay allowed to refine the LAB screening by eliminating L. piscium strains EU2231, EU2234, L. gelidum EU2247, and MIP2608, as the most sensitive LAB. Although very sensitive to chitosan treatment, C. inhibens MIP2551 and A. viridans SF1044 were retained for further screening steps as they showed the broadest and the most interesting antimicrobial profiles.
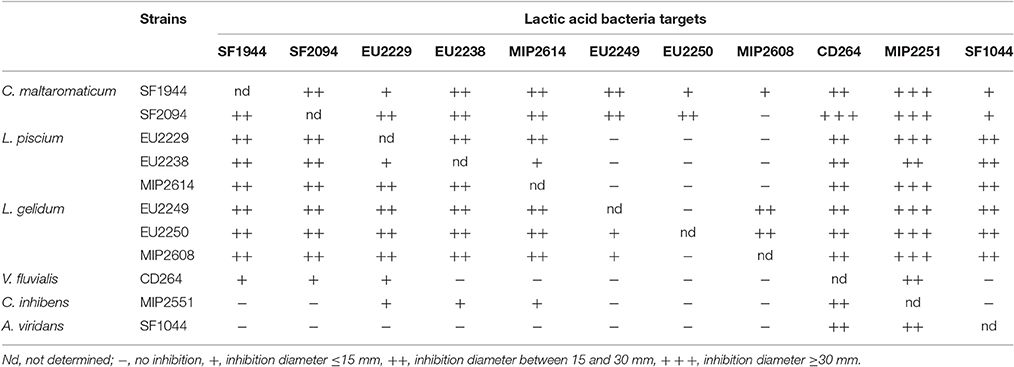
PC Cross Inhibition Activity
PC antagonist activity between them was evaluated for potential association for food biopreservation. Inhibitory results are exposed in Table 4.
All PC demonstrated inhibitory activity against the others, including strains from the same species. This was the case for L. piscium, C. maltaromaticum, and L. gelidum strains, which displayed the strongest activity characterized by large inhibition halos with diameters < 15 to 30 mm.
On the contrary, V. fluvialis CD264, C. inhibens MIP2551, and A. viridans SF1044 showed the weakest antagonist activity. V. fluvialis CD264 displayed only small inhibition diameters (< 15 mm) against C. maltaromaticum strains, L. piscium EU2229 and a strong inhibition against C. inhibens MIP2551. C. inhibens MIP2551, was only lightly active against L. piscium strains and V. fluvialis CD264, while A. viridans SF1044 only displayed strong inhibition against C. inhibens MIP2551 and V. fluvialis CD264.
L. gelidum strains were the less inhibited PC, being only slightly sensitive between them and to C. maltaromaticum strains, while C. inhibens MIP2551 was the most sensitive one, with inhibition halos almost all superior to 30 mm.
According to this criterion, for each species the strain displaying the lowest inhibition activity toward LAB was selected. Thus, C. maltaromaticum SF1994, L. piscium EU2229, L. gelidum EU2249, V. fluvialis CD264, C. inhibens MIP2551, and A. viridans SF1044 were selected to be screened further on risk assessment criteria.
LAB Strains Safety Assessment
Biogenic Amines Production Evaluation
The six PC were assessed on their ability to produce histamine and tyramine in fish juice model containing precursors. No PC produced histamine and tyramine in these experimental conditions, with an exception for C. maltaromaticum SF1994 and V. fluvialis CD264 which produced respectively 52 ± 2 and 164 ± 1 mg/l of tyramine.
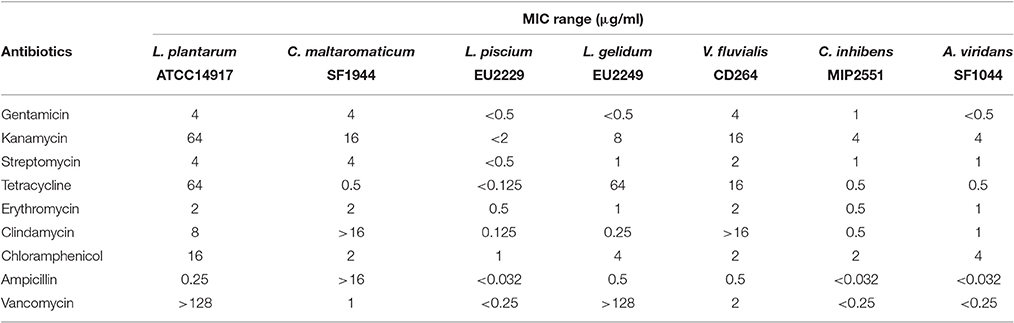
Antibiotic Resistance Profile
PC resistance profile was assessed following the ISO procedure 10932: (2010). MICs for nine antimicrobials against PC strains are summarized in Table 5. The MICs for the quality control L. plantarum ATCC® 14917 were similar to the results expressed in the procedure ISO 10932 (2010). There was an exception for streptomycin, where the MIC was slightly lower than in the standard procedure (4 μg/ml vs. 16–256 μg/ml), but also for tetracycline whose MIC was somewhat higher (64 μg/ml vs. 8–32 μg/ml). Overall, MICs ranges for all antibiotics were variable depending on the strain. Some strains, such as L. piscium EU2229, C. inhibens MIP2551, and A. viridans SF1044 were more sensitive to all antimicrobials. On the contrary C. maltaromaticum SF1944 was less sensitive to antibiotics, especially to clindamycin and ampicillin (not inhibited in the concentration range tested).
Discussion
This study presents a new strategy, based on seven criteria, implemented to finally select PC of interest for cod and salmon based products biopreservation. A collection of 35 isolates with antimicrobial properties and represented by seven species was screened. To our knowledge, C. inhibens, V. fluvialis, A. viridans, and V. penaei have never been tested for food preservation. Although, C. maltaromaticum, L. piscium, L. gelidum antimicrobial activity has been studied (Pilet and Leroi, 2011; Ghanbari et al., 2013), the screening of several isolates at the same time (respectively 4, 15, and 12) allowed to determine their intra-species biodiversity. The possibility for combining PC with other hurdles was also investigated.
Technological Assessment
The first selection was made on two crucial aspects for food biopreservation: presence of antimicrobial activity without sensory adverse effects when added to food commodities (Jones et al., 2011; Leroi et al., 2015). Instead of usual semi-quantitative method to study LAB antimicrobial activity, based on commercial culture media, a quantitative miniaturized method was developed in two fish juices, allowing a rapid and precise comparison of several isolates.
According to these two first criteria, all PC strains could be clustered distinctly per species, based on their specific sensory signature and antimicrobial profile. Intra-species variability was quite weak.
C. maltaromaticum were strongly active against L. sakei, L. monocytogenes, and S. baltica (in salmon juice) with more than 4.5–5.0 log CFU/ml reduction, but also to a lesser extent against P. phosphoreum and H. alvei. Antilisterial activity is largely described among Carnobacterium species, as they are known to produce a wide range of bacteriocins targeting more specifically Gram positive bacteria (Leisner et al., 2007). Thus, many studies reported C. maltaromaticum (formerly C. piscicola), C. divergens and C. alterfunditum strong inhibitory activity against L. monocytogenes, in vitro and in seafood (Duffes et al., 1999a,b; Brillet et al., 2004, 2005; Nilsson et al., 2004; Tahiri et al., 2004, 2009; Vescovo et al., 2006). C. maltaromaticum and C. divergens were also found to inhibit, in supernatant spot assays, several other Gram positive bacteria, such as L. innocua, and LAB (L. sakei, E. faecalis, C. maltaromaticum, and C. divergens; Duffes et al., 1999b). On the contrary, Carnobacterium sp. are not or weakly active toward Gram negative bacteria. When inoculated at 8.0 log CFU/g in cold smoked salmon (CSS), Carnobacterium spp. 10 and 39 did not show any H2S-producing bacteria inhibition (Leroi et al., 1996). C. maltaromaticum V1 and C. divergens V41 did not demonstrate antimicrobial activity against Escherichia coli or S. putrefaciens (Duffes et al., 1999b). C. alterfunditum EU2257 is the only exception, which exhibited a broad-spectrum antimicrobial profile by inhibiting in double layer assay C. sporogenes, L. monocytogenes, B. thermosphacta, L. farciminis, but also Gram negative bacteria such as Pseudomonas sp., S. liquefaciens, Psychrobacter sp., S. putrefaciens, and P. phosphoreum (Matamoros et al., 2009a). When inoculated in cooked and peeled shrimps stored under vacuum packaging, this strain decreased the total Enterobacteriaceae count by 2 log CFU/g in comparison with control (Matamoros et al., 2009b). In our screening approach C. maltaromaticum was considered as the most spoiling bacterium in both fish juices by producing feet/banana, pungent acid, malt or rhubarb off-odor. However, the spoilage score remained acceptable in all cases and the 4 C. maltaromaticum strains might be suitable in salmon and cod based products. As for all LAB, Carnobacterium sp. spoilage activity is food matrices and strains dependent (Leisner et al., 2007; Leroi, 2010). Some authors did not find spoilage evidence in seafood products (Leroi et al., 1996; Duffes et al., 1999a; Brillet et al., 2005; Joffraud et al., 2006; Laursen et al., 2006; Vescovo et al., 2006; Matamoros et al., 2009a; Jaffrès et al., 2011). On the other hand, Macé et al. (2013, 2014) showed that C. maltaromaticum strains, isolated from spoiled seafood products, were related to fresh salmon and cooked peeled shrimps alteration, with strong feet/cheese, bitter, sour, and acid smell. Malt, burnt, herring, oxidized, sweet/nauseous, nutty, chlorine, grass/hay, butter, and plastic were also described for C. maltaromaticum and C. divergens (Leroi et al., 1998; Paludan-Müller et al., 1998; Truelstrup Hansen and Huss, 1998; Duffes et al., 1999a; Joffraud et al., 2001; Stohr et al., 2001; Brillet et al., 2005; Laursen et al., 2006; Saraoui et al., 2017), but never at a level that could jeopardize their use in seafood.
L. piscium showed the broadest antimicrobial activity toward targets in both fish juices. L. monocytogenes, B. thermosphacta, S. baltica, and P. phosphoreum were totally inhibited, while S. proteamaculans, H. alvei, L. sakei bacterial count were reduced from 2.5 to 7.0 log CFU/ml, depending on the strain. In double layer assays on Elliker medium, L. piscium EU2229 (also screened in this study) and CNCM I-4031 (formerly EU2241) displayed such wide antimicrobial spectrum by inhibiting C. sporogenes, L. monocytogenes, Pseudomonas sp., S. liquefaciens, B. thermosphacta, S. aureus, Psychrobacter sp., S. putrefaciens, P. phosphoreum, E. coli, and Salmonella sp. (Matamoros et al., 2009a; Fall et al., 2010a). When inoculated in MAP cooked peeled shrimps and CSS under vacuum packaging, L. piscium CNCM I-4031 also reduced L. monocytogenes by 2–4 log CFU/g and totally inhibited B. thermosphacta, and Enterobacteriaceae (Matamoros et al., 2009b; Fall et al., 2010a,b; Leroi et al., 2015). L. piscium strains did not exhibit a strong spoilage potential in both juices. Only very slight sour, acid/lemon and for some strains very light sulfur odors were produced. L. piscium is not considered as spoiler in seafood product (Matamoros et al., 2009b; Fall et al., 2010a; Macé et al., 2012; Leroi et al., 2015; Saraoui et al., 2016), although some strains have sometimes been related to light butter/fatty fish, floor cloth, sour, and seaweed/iodine-like odors (Matamoros et al., 2009b; Macé et al., 2013).
L. gelidum strains did not display a strong inhibitory activity, except against S. baltica (only in salmon juice) and L. monocytogenes. B. thermosphacta, L. sakei, H. alvei, and S. proteamaculans were also slightly inhibited by some strains. In in vitro conditions, Matamoros et al. (2009a) demonstrated similar inhibition spectra for L. gelidum EU2213, EU2247 and EU2262 (also screened in this study). Harding and Shaw (1990) demonstrated for the strain L. gelidum IN139, in double layer and supernatant assays, an antagonist activity only against L. monocytogenes on a pool of 21 targets including pathogenic or spoilage bacteria such as B. thermosphacta, H. alvei, and S. liquefaciens. In challenge test in CSS and cooked peeled shrimps, L. gelidum was also able to slightly inhibit P. phosphoreum (1.5 log CFU/g), B. thermosphacta (4.0 log CFU/g), S. proteamaculans (2.5 log CFU/g), without inducing any sensory adverse effect (Matamoros et al., 2009b; Leroi et al., 2015) and in some cases, leading to sensory shelf-life extension. In the same way, in our study, L. gelidum strains were the most neutral LAB, with for some strains an appreciated acid/lemon smell.
C. inhibens MIP2551, V. fluvialis, and A. viridans SF1044 did not induce spoilage off-odor and showed the strongest antimicrobial activity among LAB strains. Regardless the fish juice type, except B. thermosphacta, all target bacteria were inhibited. No data are available concerning their antimicrobial activity or spoilage potential, thus, this study constitutes the first report for these three species. In contrast to V. fluvialis CD264, V. penaei CD276 displayed the lowest antimicrobial activity by inhibiting only S. baltica and induced a strong spoilage of cod juice by producing an important sulfur odor. V. penaei, is a recently described species (Jaffrès et al., 2010) which was not associated with spoilage in sterile cooked and peeled shrimps (Jaffrès et al., 2011).
PC cross inhibition activity was also investigated for mixed culture applications in food biopreservation. LAB antimicrobial activity is not commonly determined against other LAB, except for some species which might be related to spoilage such as L. farciminis and L. sakei (Stohr et al., 2001; Joffraud et al., 2006; Leroi et al., 2015). In this study, the 10 selected PC displayed antagonist activity between them, including strains from the same species. This might act as a brake for their use in combination. These results are not surprising for LAB that can be bacteriocins or antimicrobial peptides producers such as Carnobacterium sp. (Leisner et al., 2007). Bacteriocins are known to be active against closely-related species (Thonart and Dortu, 2009). Moreover, many authors observed LAB strains possessing antimicrobial activity toward a wide range of other LAB (Harding and Shaw, 1990; Jöborn et al., 1999; Duffes et al., 1999b; Fall et al., 2010a). According to our results, C. maltaromaticum strains could only be used in combination with A. viridans SF1044. Among L. gelidum strains, EU2249 and EU2250 can only be combined together, while L. piscium strains, V. fluvialis CD264, and C. inhibens MIP2551 cannot be combined with any other PC strains. However, as Saraoui et al. (2017) improved cooked and peeled shrimp quality and safety using in combination C. divergens V41 with L. piscium CNCM I-4031 without any adverse effect on their respective growth, antagonist activity in model conditions might be moderated.
In order to be combined with chitosan coating and superchilling, PC were also screened for their chitosan and freezing tolerance, as part of an entire hurdle technology strategy. A rapid freezing step of 1 ml of subculture in salmon juice at −80°C, used to mimic superchilling conditions (−37/−40°C for 2 min to rapidly decrease internal fish flesh temperature at −1.5 to −2°C) did not affect PC viability. Thus, although superchilling tolerance in seafood still needs to be confirmed, the freezing tolerance was not a discriminant criterion in our screening approach. Several hypotheses could explain these results: fish juice may contain cryoprotectants (in addition to 1% glucose added in our study) and 3 h at −80°C may not be sufficient to induce bacterial viability decrease. 15°C as preculture temperature may induce cold-shock proteins (CSP) production. Indeed, LAB are generally naturally tolerant to freezing. Several studies demonstrated a correlation between CSP (CspL and CspP) produced after a cold-shock at suboptimal temperatures and cryotolerance (Kim and Dunn, 1997; Derzelle et al., 2003; Song et al., 2014).
Chitosan tolerance among PC was both linked to the species and to the strain. Chitosan and derivatives (metal complexes or oligomers) are well-known for their antimicrobial activity against a broad range of bacteria and fungi (Kong et al., 2010). The activity may depend on numerous factors like microorganism species, environmental pH, molecular weight, deacetylation degree, pKa, and presence of metal cations (No et al., 2002; Wang et al., 2004; Kong et al., 2010). No et al. (2002) observed that chitosans and chitosans oligomers antimicrobial activity, within a wide range of molecular weights, was highly variable depending on the target bacteria. For instance, a 746 kDa chitosan molecule (0.1% in MHB or MRS broth, pH 5.9, 24 h at 37°C) was able to strongly inhibit L. plantarum (8.0 log CFU/ml), while other LAB such as L. brevis and L. bulgaricus were much less sensitive. Likewise, S. aureus, B. cereus and L. monocytogenes were totally inhibited, while E. coli, P. fluorescens, S. typhimurium, V. parahaemolyticus, Bacillus megaterium growth was variably inhibited from 3.4 to 7.2 log CFU/ml.
Safety Assessment
Despite the lack of strict regulations regarding the use of microbial cultures as protective agents in food for human consumption (Zagorec and Christieans, 2013), LAB must fulfill several safety requirements (Wessels et al., 2004). Based on existing safety regulation for the use of microbial cultures in animal feed or food fermentation and on the Quality Presumption of Safety status of species (EFSA, 2007), LAB should not produce any toxins such as biogenic amines (tyramine, histamine) and should not possess enterotoxic activity nor transmissible antibiotic resistance (Holzapfel et al., 1995; Holzapfel, 2002; Ammor and Mayo, 2007; Jones et al., 2011; Vogel et al., 2011; Bourdichon et al., 2012). Among the six LAB strains selected so far, no histamine production was detected, and only C. maltaromaticum SF1944 and V. fluvialis CD264 were able to produce tyramine. Carnobacterium spp. are known to be important tyramine producers (Masson et al., 1996; Bover-Cid and Holzapfel, 1999; Duffes et al., 1999a; Brillet et al., 2005; Leisner et al., 2007). When inoculated in sterile CSS, C. maltaromaticum V1, SF668 and C. divergens V41 produced 37, 63, and 122 mg/kg after 4 weeks of storage at 4–8°C (Brillet et al., 2005). Higher concentrations were even observed (up to 260 mg/kg) by Duffes et al. (1999a). Tyramine-producing microbial cultures addition to food might be a safety concern, as it may cause headaches and hypertensive effects at concentration superior to 100–800 mg/kg (ten Brink et al., 1990; Halász et al., 1994). Nevertheless, there is no legal regulation for tyramine in food. Unlike tyramine, only few LAB species were associated with histamine production (Masson et al., 1996; Bover-Cid and Holzapfel, 1999).
The use of microbial cultures as active agents in food and feed might represent a risk for antimicrobials resistance genes transfer to gut bacterial population through the food chain. Thus, it is important to assess the absence of acquired antibiotics resistance genes for each LAB strain intended for use in food (EFSA, 2012). A standardized procedure already exists for antimicrobials susceptibility testing (ISO 10932, 2010) and data are available for results interpretation based on MICs cut-off values (Klare et al., 2007; EFSA, 2012). Nevertheless, these guidelines mainly focus on LAB genera or species found and/or intended for use in dairy or fermented dairy products. The species used in the present study have never been extensively studied for antibiotic resistance, in consequence, no data are available. Only L. gelidum EU2249 could be compared with Leuconostoc sp. MICs cut-off values from EFSA (2012). For the nine antimicrobials tested, L. gelidum EU2249 MICs values were below the recommended values, except for tetracycline where the MIC was well-above (64 vs. 8 μg/ml). A deeper look at the genetic level may be necessary to determine whether this resistance might be acquired or intrinsic to the species. Concerning V. fluvialis, Teixeira et al. (1997) investigated the susceptibility of seven isolates to 18 antibiotics. MICs values were similar to those found in our study with V. fluvialis CD264. Martín et al. (2007) tested 58 A. viridans isolates for their susceptibility to 12 antimicrobials using the disk diffusion method. They found that A. viridans isolates were slightly resistant to streptomycin, erythromycin, clindamycin and tetracycline. On the contrary, in our study, A. viridans SF1044 growth was inhibited with very low concentrations. The other PC (L. piscium EU2229, C. inhibens MIP2551, and C. maltaromaticum SF1994) were sensitive to all antibiotics with very low MICs values, except C. maltaromaticum which was not inhibited by the highest concentrations in clindamycin and ampicillin.
As no adverse health effects have been reported so far for the consumption of LAB added into food commodities (Wessels et al., 2004) and as L. piscium, L. gelidum, and C. maltaromaticum are commonly isolated from meat and seafood products (Leisner et al., 2007; Leroi, 2010; Saraoui et al., 2016), our strains seems to be suitable for biopreservation strategy. Moreover, one strain of C. maltaromaticum is already commercialized by Sacco S.r.l (Italy). However, caution must be taken in the case of V. fluvialis CD264 and A. viridans SF1044. Indeed, in our study, V. fluvialis CD264 was found to be able to grow at 37°C and showed a β hemolytic activity on sheep blood Columbia agar medium (data not shown). Pot et al. (1994) and Teixeira et al. (1997) highlighted similar phenotype results for V. fluvialis isolates from various environment including clinical sources. This character may not lead to the species rejection but should be investigated deeper. A. viridans SF1044, although not resistant to antibiotics, was able to grow at 37°C and showed α hemolytic activity (data not shown). Kerbaugh and Evans (1968) also underlined such phenotype for A. viridans isolated from hospital environment. Moreover, A. viridans is a crustacean pathogen, causing the lobster's gaffkemia disease, and has been associated with different human infections such as endocarditis, urinary tract infections, arthritis, or meningitis (Kerbaugh and Evans, 1968; Martín et al., 2007).
Conclusion
According to this screening approach, 6 out of 35 LAB strains were selected for being suitable as bioprotective agents in a hurdle technology strategy applied to cod and salmon based products. Nevertheless, for some strains, more investigations are needed to entirely assess their safety. Strains whole genome sequencing will provide more information on this aspect. This screening approach also permitted to highlight the existence of species related sensory signatures and antimicrobial profiles. Consequently, we finally selected one strain per species in order to ensure a bacterial diversity for future experiments. C. maltaromaticum SF1994, L. piscium EU2229, L. gelidum EU2249, and V. fluvialis CD264 seem to be suitable for combining with chitosan coating treatment, freezing or superchilling, while C. inhibens, MIP2551 and A. viridans SF1044 should not be combined with chitosan.
Author Contributions
NW: Ph.D. student, in charge of experimentations, data processing and writing of the article. JC and MC in charge of sensory analyses and data processing. MP, DP, and FL: Co-Directors of the Ph.D. student.
Funding
This study was supported by a public grant from the French Agence Nationale de la Recherche within the context of ERANET COFASP program (reference ANR-14-COFA-0001, project SAFEFISHDISH).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Christine Chopin and Claire Donnay-Moreno for their kind assistance with biogenic amines determination and Frédérique Chevalier for fish juice preparation.
References
Ammor, M. S., and Mayo, B. (2007). Selection criteria for lactic acid bacteria to be used as functional starter cultures in dry sausage production: an update. Meat Sci. 76, 138–146. doi: 10.1016/j.meatsci.2006.10.022
Bourdichon, F., Casaregola, S., Farrokh, C., Frisvad, J. C., Gerds, M. L., Hammes, W. P., et al. (2012). Food fermentations: microorganisms with technological beneficial use. Int. J. Food Microbiol. 154, 87–97. doi: 10.1016/j.ijfoodmicro.2011.12.030
Bover-Cid, S., and Holzapfel, W. H. (1999). Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 53, 33–41. doi: 10.1016/S0168-1605(99)00152-X
Brillet, A., Pilet, M.-F., Prevost, H., Bouttefroy, A., and Leroi, F. (2004). Biodiversity of Listeria monocytogenes sensitivity to bacteriocin-producing Carnobacterium strains and application in sterile cold-smoked salmon. J. Appl. Microbiol. 97, 1029–1037. doi: 10.1111/j.1365-2672.2004.02383.x
Brillet, A., Pilet, M.-F., Prevost, H., Cardinal, M., and Leroi, F. (2005). Effect of inoculation of Carnobacterium divergens V41, a biopreservative strain against Listeria monocytogenes risk, on the microbiological, chemical and sensory quality of cold-smoked salmon. Int. J. Food Microbiol. 104, 309–324. doi: 10.1016/j.ijfoodmicro.2005.03.012
Caplice, E., and Fitzgerald, G. F. (1999). Food fermentations: role of microorganisms in food production and preservation. Int. J. Food Microbiol. 50, 131–149. doi: 10.1016/S0168-1605(99)00082-3
Cortesi, M. L., Panebianco, A., Giuffrida, A., and Anastasio, A. (2009). Innovations in seafood preservation and storage. Vet. Res. Commun. 33(Suppl. 1), 15–23. doi: 10.1007/s11259-009-9241-4
Dalgaard, P. (1995). Qualitative and quantitative characterization of spoilage bacteria from packed fish. Int. J. Food Microbiol. 26, 319–333. doi: 10.1016/0168-1605(94)00137-U
Derzelle, S., Hallet, B., Ferain, T., Delcour, J., and Hols, P. (2003). Improved adaptation to cold-shock, stationary-phase, and freezing stresses in Lactobacillus plantarum overproducing cold-shock proteins. Appl. Environ. Microbiol. 69, 4285–4290. doi: 10.1128/AEM.69.7.4285-4290.2003
Devlieghere, F., Vermeiren, L., and Debevere, J. (2004). New preservation technologies: possibilities and limitations. Int. Dairy J. 14, 273–285. doi: 10.1016/j.idairyj.2003.07.002
Duffes, F., Corre, C., Leroi, F., Dousset, X., and Boyaval, P. (1999a). Inhibition of Listeria monocytogenes by in situ produced and semipurified bacteriocins of Carnobacterium spp. on vacuum-packed, refrigerated cold-smoked salmon. J. Food Prot. 62, 1394–1403. doi: 10.4315/0362-028X-62.12.1394
Duffes, F., Leroi, F., Boyaval, P., and Dousset, X. (1999b). Inhibition of Listeria monocytogenes by Carnobacterium spp. strains in a simulated cold smoked fish system stored at 4°C. Int. J. Food Microbiol. 47, 33–42. doi: 10.1016/S0168-1605(98)00206-2
Duflos, G., Dervin, C., Malle, P., and Bouquelet, S. (1999). Use of biogenic amines to evaluate spoilage in plaice (Pleuronectes platessa) and whiting (Merlangus merlangus). J. AOAC Int. 82, 1357–1363.
EFSA (2007). Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA- Opinion of the Scientific Committee. EFSA J. 587, 1–16. doi: 10.2903/j.efsa.2007.587
EFSA (2012). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 10:2740. doi: 10.2903/j.efsa.2012.2740
Fall, P. A., Leroi, F., Cardinal, M., Chevalier, F., and Pilet, M. F. (2010a). Inhibition of Brochothrix thermosphacta and sensory improvement of tropical peeled cooked shrimp by Lactococcus piscium CNCM I-4031. Lett. Appl. Microbiol. 50, 357–361. doi: 10.1111/j.1472-765X.2010.02801.x
Fall, P. A., Leroi, F., Chevalier, F., Guérin, C., and Pilet, M.-F. (2010b). Protective effect of a non-bacteriocinogenic Lactococcus piscium CNCM I-4031 strain against Listeria monocytogenes in sterilized tropical cooked peeled shrimp. J. Aquat. Food Prod. Technol. 19, 84–92. doi: 10.1080/10498850.2010.486910
FAO (2016). The State of World Fisheries and Aquaculture (SOFIA) FAO Food and Agriculture Organization of the United Nations. Available online at: http://www.fao.org/publications/sofia/2016/en/ (Accessed December 7, 2016).
Ghanbari, M., Jami, M., Domig, K. J., and Kneifel, W. (2013). Seafood biopreservation by lactic acid bacteria – a review. LWT Food Sci. Technol. 54, 315–324. doi: 10.1016/j.lwt.2013.05.039
Gram, L., and Dalgaard, P. (2002). Fish spoilage bacteria – problems and solutions. Curr. Opin. Biotechnol. 13, 262–266. doi: 10.1016/S0958-1669(02)00309-9
Gram, L., and Huss, H. H. (1996). Microbiological spoilage of fish and fish products. Int. J. Food Microbiol. 33, 121–137. doi: 10.1016/0168-1605(96)01134-8
Halász, A., Baráth, Á., Simon-Sarkadi, L., and Holzapfel, W. (1994). Biogenic amines and their production by microorganisms in food. Trends Food Sci. Technol. 5, 42–49. doi: 10.1016/0924-2244(94)90070-1
Harding, C. D., and Shaw, B. G. (1990). Antimicrobial activity of Leuconostoc gelidum against closely related species and Listeria monocytogenes. J. Appl. Bacteriol. 69, 648–654. doi: 10.1111/j.1365-2672.1990.tb01558.x
Holzapfel, W. H. (2002). Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 75, 197–212. doi: 10.1016/S0168-1605(01)00707-3
Holzapfel, W. H., Geisen, R., and Schillinger, U. (1995). Biological preservation of foods with reference to protective cultures, bacteriocins and food-grade enzymes. Int. J. Food Microbiol. 24, 343–362. doi: 10.1016/0168-1605(94)00036-6
Huss, H. H., Embarek, P. K. B., and Jeppesen, V. F. (1995). Control of biological hazards in cold smoked salmon production. Food Control 6, 335–340. doi: 10.1016/0956-7135(95)00043-7
Huss, H. H., Reilly, A., and Karim Ben Embarek, P. (2000). Prevention and control of hazards in seafood. Food Control 11, 149–156. doi: 10.1016/S0956-7135(99)00087-0
ISO 10932 (2010). Milk and Milk Products. Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB). Geneva.
ISO 13299 (2003). Sensory Analysis, Methodology. General Guidance for Establishing a Sensory Profile. Geneva: ISO.
Jaffrès, E., Lalanne, V., Macé, S., Cornet, J., Cardinal, M., Sérot, T., et al. (2011). Sensory characteristics of spoilage and volatile compounds associated with bacteria isolated from cooked and peeled tropical shrimps using SPME-GC-MS analysis. Int. J. Food Microbiol. 147, 195–202. doi: 10.1016/j.ijfoodmicro.2011.04.008
Jaffrès, E., Prévost, H., Rossero, A., Joffraud, J.-J., and Dousset, X. (2010). Vagococcus penaei sp. nov., isolated from spoilage microbiota of cooked shrimp (Penaeus vannamei). Int. J. Syst. Evol. Microbiol. 60, 2159–2164. doi: 10.1099/ijs.0.012872-0
Jöborn, A., Dorsch, M., Olsson, J. C., Westerdahl, A., and Kjelleberg, S. (1999). Carnobacterium inhibens sp. nov., isolated from the intestine of Atlantic salmon (Salmo salar). Int. J. Syst. Bacteriol. 49(Pt 4), 1891–1898. doi: 10.1099/00207713-49-4-1891
Joffraud, J. J., Leroi, F., Roy, C., and Berdagué, J. L. (2001). Characterisation of volatile compounds produced by bacteria isolated from the spoilage flora of cold-smoked salmon. Int. J. Food Microbiol. 66, 175–184. doi: 10.1016/S0168-1605(00)00532-8
Joffraud, J.-J., Cardinal, M., Cornet, J., Chasles, J.-S., Léon, S., Gigout, F., et al. (2006). Effect of bacterial interactions on the spoilage of cold-smoked salmon. Int. J. Food Microbiol. 112, 51–61. doi: 10.1016/j.ijfoodmicro.2006.05.014
Jones, R. J., Wescombe, P. A., and Tagg, J. R. (2011). “Identifying new protective cultures and culture components for food biopreservation,” in Protective Cultures, Antimicrobial Metabolites and Bacteriophages for Food and Beverage Biopreservation, ed C. Lacroix (Cambridge: Woodhead Publishing), 3–26.
Kerbaugh, M. A., and Evans, J. B. (1968). Aerococcus viridans in the hospital environment. Appl. Microbiol. 16, 519–523.
Kim, W. S., and Dunn, N. W. (1997). Identification of a cold shock gene in lactic acid bacteria and the effect of cold shock on cryotolerance. Curr. Microbiol. 35, 59–63. doi: 10.1007/s002849900212
Klare, I., Konstabel, C., Werner, G., Huys, G., Vankerckhoven, V., Kahlmeter, G., et al. (2007). Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J. Antimicrob. Chemother. 59, 900–912. doi: 10.1093/jac/dkm035
Kong, M., Chen, X. G., Xing, K., and Park, H. J. (2010). Antimicrobial properties of chitosan and mode of action: a state of the art review. Int. J. Food Microbiol. 144, 51–63. doi: 10.1016/j.ijfoodmicro.2010.09.012
Laursen, B. G., Leisner, J. J., and Dalgaard, P. (2006). Carnobacterium species: effect of metabolic activity and interaction with Brochothrix thermosphacta on sensory characteristics of modified atmosphere packed shrimp. J. Agric. Food Chem. 54, 3604–3611. doi: 10.1021/jf053017f
Leisner, J. J., Laursen, B. G., Prévost, H., Drider, D., and Dalgaard, P. (2007). Carnobacterium: positive and negative effects in the environment and in foods. FEMS Microbiol. Rev. 31, 592–613. doi: 10.1111/j.1574-6976.2007.00080.x
Lem, A., Bjorndal, T., and Lappo, A. (2014). Economic Analysis of Supply and Demand for Food Up to 2030 – Special Focus on Fish and Fishery Products. FAO Fisheries and Aquaculture Circular No. (1089), 106.
Leroi, F. (2010). Occurrence and role of lactic acid bacteria in seafood products. Food Microbiol. 27, 698–709. doi: 10.1016/j.fm.2010.05.016
Leroi, F. (2014). “Role of bacteria in seafood products,” in Seafood Science: Advances in Chemistry, Technology and Applications, ed S. K. Kim (Boca Raton: CRC Press), 458–482.
Leroi, F., Arbey, N., Joffraud, J.-J., and Chevalier, F. (1996). Effect of inoculation with lactic acid bacteria on extending the shelf-life of vacuum-packed cold smoked salmon. Int. J. Food Sci. Technol. 31, 497–504. doi: 10.1046/j.1365-2621.1996.00366.x
Leroi, F., Cornet, J., Chevalier, F., Cardinal, M., Coeuret, G., Chaillou, S., et al. (2015). Selection of bioprotective cultures for preventing cold-smoked salmon spoilage. Int. J. Food Microbiol. 213, 79–87. doi: 10.1016/j.ijfoodmicro.2015.05.005
Leroi, F., Joffraud, J.-J., Chevalier, F., and Cardinal, M. (1998). Study of the microbial ecology of cold-smoked salmon during storage at 8°C. Int. J. Food Microbiol. 39, 111–121. doi: 10.1016/S0168-1605(97)00126-8
Løvdal, T. (2015). The microbiology of cold smoked salmon. Food Control 54, 360–373. doi: 10.1016/j.foodcont.2015.02.025
Macé, S., Cardinal, M., Jaffrès, E., Cornet, J., Lalanne, V., Chevalier, F., et al. (2014). Evaluation of the spoilage potential of bacteria isolated from spoiled cooked whole tropical shrimp (Penaeus vannamei) stored under modified atmosphere packaging. Food Microbiol. 40, 9–17. doi: 10.1016/j.fm.2013.11.018
Macé, S., Cornet, J., Chevalier, F., Cardinal, M., Pilet, M.-F., Dousset, X., et al. (2012). Characterisation of the spoilage microbiota in raw salmon (Salmo salar) steaks stored under vacuum or modified atmosphere packaging combining conventional methods and PCR–TTGE. Food Microbiol. 30, 164–172. doi: 10.1016/j.fm.2011.10.013
Macé, S., Joffraud, J.-J., Cardinal, M., Malcheva, M., Cornet, J., Lalanne, V., et al. (2013). Evaluation of the spoilage potential of bacteria isolated from spoiled raw salmon (Salmo salar) fillets stored under modified atmosphere packaging. Int. J. Food Microbiol. 160, 227–238. doi: 10.1016/j.ijfoodmicro.2012.10.013
Martín, V., Vela, A. I., Gilbert, M., Cebolla, J., Goyache, J., Domínguez, L., et al. (2007). Characterization of Aerococcus viridans isolates from swine clinical specimens. J. Clin. Microbiol. 45, 3053–3057. doi: 10.1128/JCM.00156-07
Masson, F., Talon, R., and Montel, M. C. (1996). Histamine and tyramine production by bacteria from meat products. Int. J. Food Microbiol. 32, 199–207. doi: 10.1016/0168-1605(96)01104-X
Matamoros, S., Leroi, F., Cardinal, M., Gigout, F., Kasbi Chadli, F., Cornet, J., et al. (2009b). Psychrotrophic lactic acid bacteria used to improve the safety and quality of vacuum-packaged cooked and peeled tropical shrimp and cold-smoked salmon. J. Food Prot. 72, 365–374. doi: 10.4315/0362-028X-72.2.365
Matamoros, S., Pilet, M. F., Gigout, F., Prévost, H., and Leroi, F. (2009a). Selection and evaluation of seafood-borne psychrotrophic lactic acid bacteria as inhibitors of pathogenic and spoilage bacteria. Food Microbiol. 26, 638–644. doi: 10.1016/j.fm.2009.04.011
Nilsson, L., Ng, Y. Y., Christiansen, J. N., Jørgensen, B. L., Grótinum, D., and Gram, L. (2004). The contribution of bacteriocin to inhibition of Listeria monocytogenes by Carnobacterium piscicola strains in cold-smoked salmon systems. J. Appl. Microbiol. 96, 133–143. doi: 10.1046/j.1365-2672.2003.02129.x
No, H. K., Young Park, N., Ho Lee, S., and Meyers, S. P. (2002). Antibacterial activity of chitosans and chitosan oligomers with different molecular weights. Int. J. Food Microbiol. 74, 65–72. doi: 10.1016/S0168-1605(01)00717-6
Paludan-Müller, C., Dalgaard, P., Huss, H. H., and Gram, L. (1998). Evaluation of the role of Carnobacterium piscicola in spoilage of vacuum- and modified-atmosphere-packed cold-smoked salmon stored at 5°C. Int. J. Food Microbiol. 39, 155–166. doi: 10.1016/S0168-1605(97)00133-5
Pilet, M.-F., and Leroi, F. (2011). “Applications of protective cultures, bacteriocins and bacteriophages in fresh seafood and seafood products,” in Protective Cultures, Antimicrobial Metabolites and Bacteriophages for Food and Beverage Biopreservation, ed C. Lacroix (Cambridge: Woodhead Publishing), 324–347. doi: 10.1533/9780857090522.3.324
Pot, B., Devriese, L. A., Hommez, J., Miry, C., Vandemeulebroecke, K., Kersters, K., et al. (1994). Characterization and identification of Vagococcus fluvialis strains isolated from domestic animals. J. Appl. Bacteriol. 77, 362–369. doi: 10.1111/j.1365-2672.1994.tb03436.x
Rodgers, S. (2001). Preserving non-fermented refrigerated foods with microbial cultures—a review. Trends Food Sci. Technol. 12, 276–284. doi: 10.1016/S0924-2244(01)00093-0
Saraoui, T., Cornet, J., Guillouet, E., Pilet, M. F., Chevalier, F., Joffraud, J.-J., et al. (2017). Improving simultaneously the quality and safety of cooked and peeled shrimp using a cocktail of bioprotective lactic acid bacteria. Int. J. Food Microbiol. 241, 69–77. doi: 10.1016/j.ijfoodmicro.2016.09.024
Saraoui, T., Leroi, F., Björkroth, J., and Pilet, M. F. (2016). Lactococcus piscium: a psychrotrophic lactic acid bacterium with bioprotective or spoilage activity in food-a review. J. Appl. Microbiol. 121, 907–918. doi: 10.1111/jam.13179
Song, S., Bae, D.-W., Lim, K., Griffiths, M. W., and Oh, S. (2014). Cold stress improves the ability of Lactobacillus plantarum L67 to survive freezing. Int. J. Food Microbiol. 191, 135–143. doi: 10.1016/j.ijfoodmicro.2014.09.017
Stiles, M. E. (1996). Biopreservation by lactic acid bacteria. Antonie Van Leeuwenhoek 70, 331–345. doi: 10.1007/BF00395940
Stohr, V., Joffraud, J. J., Cardinal, M., and Leroi, F. (2001). Spoilage potential and sensory profile associated with bacteria isolated from cold-smoked salmon. Food Res. Int. 34, 797–806. doi: 10.1016/S0963-9969(01)00101-6
Tahiri, I., Desbiens, M., Benech, R., Kheadr, E., Lacroix, C., Thibault, S., et al. (2004). Purification, characterization and amino acid sequencing of divergicin M35: a novel class IIa bacteriocin produced by Carnobacterium divergens M35. Int. J. Food Microbiol. 97, 123–136. doi: 10.1016/j.ijfoodmicro.2004.04.013
Tahiri, I., Desbiens, M., Kheadr, E., Lacroix, C., and Fliss, I. (2009). Comparison of different application strategies of divergicin M35 for inactivation of Listeria monocytogenes in cold-smoked wild salmon. Food Microbiol. 26, 783–793. doi: 10.1016/j.fm.2009.05.003
Teixeira, L. M., Carvalho, M. G., Merquior, V. L., Steigerwalt, A. G., Brenner, D. J., and Facklam, R. R. (1997). Phenotypic and genotypic characterization of Vagococcus fluvialis, including strains isolated from human sources. J. Clin. Microbiol. 35, 2778–2781.
ten Brink, B., Damink, C., Joosten, H. M., and Huis in't Veld, J. H. (1990). Occurrence and formation of biologically active amines in foods. Int. J. Food Microbiol. 11, 73–84.
Thonart, P., and Dortu, C. (2009). Les bactériocines des bactéries lactiques : caractéristiques et intérêts pour la bioconservation des produits alimentaires. Base. Available online at: http://popups.ulg.ac.be/1780-4507/index.php?id=3626 (Accessed December 15, 2016).
Truelstrup Hansen, L., and Huss, H. H. (1998). Comparison of the microflora isolated from spoiled cold-smoked salmon from three smokehouses. Food Res. Int. 31, 703–711. doi: 10.1016/S0963-9969(99)00049-6
Vescovo, M., Scolari, G., and Zacconi, C. (2006). Inhibition of Listeria innocua growth by antimicrobial-producing lactic acid cultures in vacuum-packed cold-smoked salmon. Food Microbiol. 23, 689–693. doi: 10.1016/j.fm.2005.12.002
Vogel, R. F., Hammes, W. P., Habermeyer, M., Engel, K.-H., Knorr, D., Eisenbrand, G., et al. (2011). Microbial food cultures–opinion of the Senate Commission on Food Safety (SKLM) of the German Research Foundation (DFG). Mol. Nutr. Food Res. 55, 654–662. doi: 10.1002/mnfr.201100010
Wang, X., Du, Y., and Liu, H. (2004). Preparation, characterization and antimicrobial activity of chitosan–Zn complex. Carbohydr. Polym. 56, 21–26. doi: 10.1016/j.carbpol.2003.11.007
Keywords: antimicrobial activity, spoilage, screening, chitosan, fish juice, safety assessment
Citation: Wiernasz N, Cornet J, Cardinal M, Pilet M-F, Passerini D and Leroi F (2017) Lactic Acid Bacteria Selection for Biopreservation as a Part of Hurdle Technology Approach Applied on Seafood. Front. Mar. Sci. 4:119. doi: 10.3389/fmars.2017.00119
Received: 02 February 2017; Accepted: 13 April 2017;
Published: 19 May 2017.
Edited by:
Ioannis S. Boziaris, University of Thessaly, GreeceReviewed by:
Spiros Paramithiotis, Agricultural University of Athens, GreeceCarla C. C. R. de Carvalho, Universidade de Lisboa, Portugal
Copyright © 2017 Wiernasz, Cornet, Cardinal, Pilet, Passerini and Leroi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Françoise Leroi, fleroi@ifremer.fr
 Norman Wiernasz1,2
Norman Wiernasz1,2  Marie-France Pilet
Marie-France Pilet Delphine Passerini
Delphine Passerini Françoise Leroi
Françoise Leroi