Seriatopora Diversity Preserved in Upper Mesophotic Coral Ecosystems in Southern Japan
- 1Sesoko Station, Tropical Biosphere Research Center, University of the Ryukyus, Okinawa, Japan
- 2Global Change Institute, The University of Queensland, St Lucia, QLD, Australia
Coral reefs worldwide are facing increasing stress due to drastic changes in their environment. Mesophotic coral ecosystems (MCEs) have been considered as a potential refuge from several major stressors, such as warm-water bleaching events. However, their role as a subsequent source of larvae remains unclear for many species, particularly as genetic differentiation of corals over depth has frequently been observed. In 1998 and 2001, two severe bleaching events around Okinawa Island in Japan resulted in major changes to the shallow reefs, including the local “extinction” of species such as Seriatopora hystrix at Sesoko Island. However, recently this species was found to be present in abundance at mesophotic depths in the area, despite no clear signs of recovery being observed in the adjacent shallow waters. Here, we assessed the genetic diversity of Seriatopora from this deep population and provide a comparison with populations from shallow to mesophotic depths in other parts of the Ryukyu archipelago, to understand their depth specificity and their importance in genetic diversity conservation of affected shallow populations. High levels of genetic diversity were observed in both shallow and mesophotic Seriatopora populations for both the nuclear (internal transcribed spacer 2, ITS2) and the mitochondrial (hypervariable open reading frame, ORF) markers, with no clear partitioning of haplotypes over depth or across locations in the archipelago. Both ITS2 and ORF suggest the presence of potential cryptic species and the discrepancy between the markers could reflect hybridization or incomplete lineage sorting. Although associated endosymbionts (Symbiodinium spp.) all shared the same mitochondrial haplotype (cytochrome oxidase subunit 1, COI), the nuclear ribosomal ITS2 revealed slight potential habitat partitioning between the genotypes, with a small decrease of C59 Symbiodinium types below 10 m depth and a mirrored increase in C1/C78a-related types. The relative absence of depth-specific host lineages and the substantial overlap between shallow and deep Symbiodinium types indicate that Okinawan MCEs may act as a refuge preserving genotypic diversity of bleaching-sensitive coral such as Seriatopora, and on the long-term may have the potential to contribute to shallow-water recolonization of this species.
Introduction
Over the last decades, major concerns have arisen from the increasing amount of stress sustained by coral reefs because of climate change and other human impacts (reviewed by Hoegh-Guldberg et al., 2007; Fabricius, 2011). Especially, the occurrence of mass-bleaching events driven by elevated temperatures and high solar irradiance has led to a dramatic decrease of coral coverage worldwide (Hoegh-Guldberg, 1999). In this context, certain reef environments have been identified that have the potential to act as a thermal refuge, such as areas of upwelling, offshore banks and areas at moderate depth (Glynn, 1996; Riegl and Piller, 2003). Particularly, the latter has gained much attention as it could provide a “local refuge” adjacent to threatened shallow communities, given that many coral reef systems (e.g., fringing, barrier and atolls) extend across a wide depth range.
Although these deeper sections of coral reefs have remained poorly studied, recent advancements in underwater technologies are now facilitating access to these otherwise logistically challenging environments (Kahng et al., 2010, 2014). This has sparked a recent interest and led to a definition of Mesophotic Coral Ecosystems (MCEs) as coral reef habitat occurring at depths greater than 30 m (Hinderstein et al., 2010). The role of MCEs as refuges against episodic disturbances, referred to as the Deep Reef Refuge Hypothesis (DRRH) (sensu Bongaerts et al., 2010a, 2017), has played a central part in the motivation of mesophotic studies (Slattery et al., 2011; van Oppen et al., 2011; Bridge et al., 2012; Smith et al., 2014). Based on the original hypothesis that deeper reefs could act as a thermal refuge (Glynn, 1996; Riegl and Piller, 2003), the DRRH was then extended to also include other disturbances (e.g., storms) and the subsequent potential of deep reef habitat to act as source of larvae for shallow reef areas post-disturbance (Bongaerts et al., 2010a). Although, models of coral spawning and larval settlement suggest a strong potential for some upper mesophotic corals to act as a source of larvae for shallow environments (Holstein et al., 2016), initial genetic studies on both hosts and symbiotic algae, Symbiodinium, showed differences between coral populations at different depths as well as specific symbiont associations in mesophotic corals (Frade et al., 2008; van Oppen et al., 2011; Serrano et al., 2014; Pochon et al., 2015; Bongaerts et al., 2015a,b). A recent study demonstrated that the potential for vertical connectivity can differ greatly between species within a single reef location indicating that reseeding potential may be relevant for individual species rather than representing a broader ecosystem-wide phenomenon (Bongaerts et al., 2017).
In the Northwest Pacific, the number of mesophotic studies is extremely limited. Despite the high coral diversity found in the Ryukyu Archipelago and the large number of studies on Japanese shallow coral reefs, only a few MCEs have been reported in the upper part of the mesophotic range (between 30 and 55 m; Yamazato, 1972; Kimura et al., 2011; Sinniger et al., 2013; White et al., 2013). Located near relatively densely populated islands, Okinawan reefs are subject to heavy anthropogenic stresses, such as terrestrial runoffs (Hongo and Yamano, 2013) in addition to the strong tropical typhoons that frequently hit this region (Hongo et al., 2012). Consequently, major changes in shallow reef coral communities have been observed in this region (Loya et al., 2001; van Woesik et al., 2011; Hongo and Yamano, 2013; Muko et al., 2013; Harii et al., 2014). While it remains largely unknown how these stressors have impacted mesophotic reefs, deep reefs may not be affected in the same way as shallow reefs (Bak et al., 2005).
Following bleaching events in 1998 and 2001, Seriatopora hystrix, a coral species that was commonly found in shallow reefs, completely disappeared from the shallow reefs surrounding Sesoko Island in the north part of Okinawa (Loya et al., 2001; van Woesik et al., 2011). While presumed locally extinct for more than a decade, recent exploration of nearby mesophotic communities showed a dense population of this species between 38 and 47 m depth (Sinniger et al., 2013). Although, this observation appears to support the existence of a deep refuge (in line with the first stipulation of the DRRH), it remains unclear whether the deep populations of S. hystrix are genetically similar to and could aid in the recovery of shallow S. hystrix populations (second stipulation of the DRRH). For example, on the northern Great Barrier Reef, deeper sections of the reef harbored S. hystrix populations genetically distinct from those in shallow reef habitats, potentially representing distinct species (Bongaerts et al., 2010b, 2011). Similar depth-partitioning was observed in Western Australia, with the difference that there was evidence of shallow-deep connectivity over intermediate depth ranges (0–30 m), indicating a potential role of populations deeper on the slope to aid in the recovery of shallow populations. Here, we use a set of two independent nuclear and mitochondrial markers to examine both the host and Symbiodinium genotypic diversity of the mesophotic refuge populations in Okinawa. By comparing genotypic diversity to other shallow and mesophotic populations in the archipelago (e.g., Kume Island and the Yaeyama Islands), we assess whether this refuge population represents an isolated depth-adapted population and/or to what extent it harbors genotypes also found in shallow water.
Methods
Sample Collection and DNA Extraction
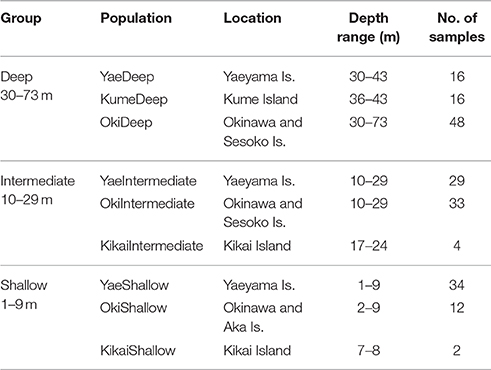
For genetic analyses, a total of 194 Seriatopora specimens exhibiting S. hystrix external morphology were sampled using SCUBA diving between 3 and 50 m at various locations in the Ryukyu archipelago: Okinawa Island (Shigeo's Reef, offshore of Sesoko and Okinawa Islands, 26°40′ N, 127°52′ E), in Kume Island (26°18′ N, 126°45′ E and 26°20′ N, 126°43′ E), and in the Yaeyama Islands in Taketomi Island (24°20′ N, 124°06′ E), Ishigaki Island (Nagura Bay, 24°24′ N, 124°06′ E), Iriomote Island (Amitori Bay and Cape Sabazaki, 24°20′ N, 123°41′ E and 24°21′ N, 123°42′ E) (Figure 1). In addition, 4 samples were obtained using a Smith-McIntyre grab between 68 and 73 m depth offshore of Sesoko Island, 2 samples were obtained from less than 2 m depth in Aka Island, near Okinawa Island and 6 samples were sampled further north in Kikai Island (28°20′ N, 130°00′ E) between 7 and 24 m depth. The depth ranges were classified as follows: “shallow” 1–9 m depth, “intermediate” from 10–29 m depth and “deep” 30–73 m depth. Samples were fixed and preserved in 95% ethanol and DNA was extracted using the guanidine extraction method: a fragment of a few mm3 was digested in 100–300 μl of guanidine solution without EDTA. After at least 1 h digestion, 100 μl of the solution were transferred to a new tube and the extraction process was continued as described in Sinniger et al. (2010).
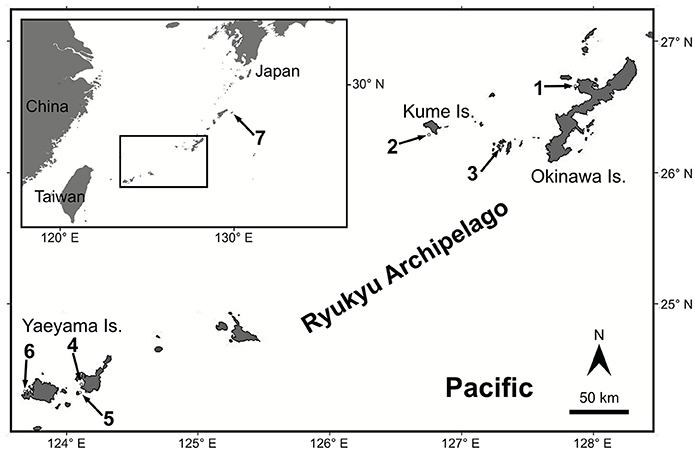
Figure 1. Map of the studied area. Numbered arrows point at the sampling locations. 1. Shigeo's Reef, offshore Sesoko and Okinawa Island, 2. Kume Island, 3. Aka Island, 4. Nagura Bay in Ishigaki Island, 5. Taketomi Island, 6. Amitori Bay and Cape Sabazaki in Iriomote Island, 7. Kikai Island.
Genetic Analyses of the Coral Host
The host genotypes were identified by sequencing the internal transcribed spacer 2 (ITS2) of the nuclear ribosomal RNA gene (n = 192) and the putative mitochondrial open reading frame region (mtORF) (n = 194). The target DNA fragments were amplified using the primer pairs ITS2-5c/R28S1 (Flot and Tillier, 2006) and FATP6.1/RORF (Flot et al., 2008b), respectively. The thermal cycling conditions of the PCR for both DNA regions were according to the publications referring for each primer pair. The PCR products were purified using ExoSAP-IT (USB Corp., USA) and Sanger sequencing was performed by Macrogen Japan (http://www.macrogen-japan.co.jp/) using the same primer pairs as for the PCR. Chromatograms of Seriatopora ITS2 showing double peaks were sorted using the online software Champuru v1.0 (Flot, 2007) and manual phase determination (Flot et al., 2006) using Geneious 10.1.2 (https://www.geneious.com, Kearse et al., 2012). In a few situations, ambiguities were left in the sequence when a site could not be reliably assigned to a specific base pair. All sequences obtained are deposited on GenBank (Accession numbers KY987652-KY988112). Sequences were aligned in Geneious 10.1.2 (https://www.geneious.com, Kearse et al., 2012) and Bayesian phylogenetic analyses were conducted using MrBayes 3.2.6 (Huelsenbeck and Ronquist, 2001). A GTR substitution model was used with a 6 gamma categories and Stylophora sp. as outgroup with ITS2 and ORF sequences from the specimen 04Oki195 (JN559077 and JN558888, Flot et al., 2011). In addition the 6 Seriatopora sequences from Nakajima et al. (2017) were included in the ORF tree for comparison. Median-joining networks (Bandelt et al., 1999) were drawn using PopART (http://popart.otago.ac.nz). Analyses of molecular variance (AMOVA) on the ORF results were performed using ARLEQUIN v.3.5.2 (Excoffier and Lischer, 2010). To test the correlation between ITS and ORF haplotypes, the ORF “populations” were determined based on ITS group to which each sample belonged. To test depth distribution of ORF haplotypes, the populations were defined based on depth (shallow, intermediate, deep) and geographic origin (Okinawa Island, Kume Island and Yaeyama Islands including Taketomi, Ishigaki and Iriomote Islands), although shallow specimens were not found in Kume and none of the 6 samples collected in Kikai Island were found below 24 m depth (Table 1). In this analysis, populations were then grouped according to the depth category (e.g., the group “Deep” included the following three populations: YaeyamaDeep, KumeDeep and OkinawaDeep). In total, the groups “Deep,” “Intermediate,” and “Shallow” comprised 80, 66, and 48 samples, respectively. The rarity or absence of samples from specific depths and locations may create an artificial structure due to incomplete sampling, therefore we limited the comparisons to populations/groups including sufficient amount of samples.
Genetic Analyses of the Associated Symbiodinium
A fragment of the mitochondrial cytochrome c oxidase subunit I gene (COI) was amplified and sequenced for selected samples (n = 35) representing the main locations (Yaeyama Islands, Kume and Okinawa Islands) and all depths using the primer pair CoxI_for2 (Pochon et al., 2015)/CoxI_r1 (Pochon et al., 2012) with the thermal cycling conditions described in Pochon et al. (2012). The PCR products of COI were subsequently purified and sequenced as mentioned above. Sequence data were edited and aligned as for the host DNA sequences. Symbiodinium types associated with S. hystrix colonies (n = 115) were identified by denaturing gradient gel electrophoresis (DGGE) of the nuclear ITS2 region following the method of LaJeunesse and Trench (2000). The ITS2 region was amplified by PCR using the primer pair ITSint-for2/ITS2CLAMP (LaJeunesse and Trench, 2000) and amplification conditions described by LaJeunesse et al. (2003). The amplified ITS2 PCR products were separated by DGGE using a Bio-Rad DCode System (Bio-Rad Laboratories, Inc., USA) as described by LaJeunesse and Trench (2000) with slight modifications. The denaturing gradient concentrations, running time, and voltage were modified for optimal resolution of the samples that were used in this study: 8% polyacrylamide gels with an internal gradient of 20–75% denaturants (urea and formamide) were run at 90 V for 15 h at a constant temperature of 60°C. Representative DGGE profiles of 10 samples were selected and dominant bands were sequenced to identify the ITS sequences associated with the harbored Symbiodinium spp. The target DGGE bands were cut out from gels and dissolved in 10 μl of pure water. The DNA fragments in the excised DGGE bands were amplified using primers ITSint-for2 and ITS2 rev (reverse primer lacking the GC clamp) with following thermal cycle profile: initial denaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 45 s, annealing at 52°C for 45 s, and extension at 72°C for 60 s; final extension at 72°C for 10 min. The PCR products of the DGGE bands were purified and sequenced using the primers pair ITSint-for2/ITS2 rev. In addition, samples representing each DGGE pattern were amplified with the above-mentioned primers for ITS2 and PCR products were cloned using the pGEM-T. Easy Vector system (Promega) and competent DH5-α cells (Toyobo). Sixteen clones were sequenced for each pattern. ITS2 sequences were identified using BLAST against the GeoSymbio database (Franklin et al., 2012), which provides alignments of all Symbiodinium ITS2 sequences that have been published from 1982 to 2012. As for the host sequences, a median joining network was drawn with the ITS sequences obtained both from cloning and DGGE band sequencing. Unlike the procedure applied to insertion and deletion in the host sequences, due to the amount of single nucleotide deletions in Symbiodinium ITS2, these sequences were kept unmodified for drawing networks. All sequences obtained are deposited on GenBank (Accession numbers KY988113-KY988299).
Results
Seriatopora Host Genotypic Diversity
The ITS2 sequences obtained for 192 out of 194 Seriatopora samples ranged from 535 to 561 bp length. Two out of the four deepest samples (from between 69 and 73 m) failed to sequence with this marker. The different genotypes could be separated into 4 large clusters mainly because of the presence/absence of large insertions/deletion (indels), whereas single nucleotide polymorphisms (SNPs) and small indels were responsible for the intra-cluster diversity (Figure S1). As expected for this marker, heterozygosity was observed in several instances with a total of 247 sequences recovered for 192 samples. Yet, the ITS2 haploweb clearly shows the different genotypes found in one sample always grouped within the same cluster (Figure 2). Within three of the four clusters (II, III, and IV), the haploweb distinguish haplotypes that appear independent (indicated by dashed lines in Figure 2), however, based on the close phylogenetic relationship of these haplotypes with others haplotypes in clusters II or III, the limited specimen number and in the absence of other elements justifying to distinguish these haplotypes as separate clusters, we deviated from a strict interpretation of the haploweb and considered them as belonging to the clusters II and III respectively. Interestingly, except of cluster IV that contains only a few samples (n = 3), clusters I, II, and III were found at all depths ranges and at all of the main locations (Figure 2).
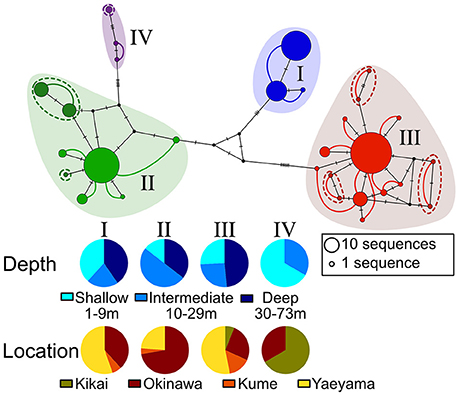
Figure 2. Haploweb made from the Median-Joining haplotype network of the host nuclear ITS2. Colors correspond to the different clusters and size of the circles is proportional to the frequency of the haplotypes. Colored lines indicate haplotypes shared by the same individual and thickness of the lines is proportional to the frequency. Dashed lines indicate small independent clusters that are considered to belong to the larger clusters. Marks on the lines of the haplotype network indicate the number of evolutionary steps between haplotypes.
The mtORF sequences ranged between 824 and 1,014 bp in length. As observed for ITS2, several haplotypes (12) were identified and named from S to Z with a number distinguishing closely related haplotypes (e.g., Z and Z2, Figure 3A). Except for the three samples possessing both the ORF haplotype S and forming the ITS cluster IV, no strict correlation was observed between the two markers as haplotypes from the main ITS groups are present in each of the ORF clusters (Figure 3A). Large indels and several SNPs separated ORF haplotypes into three distinct clusters called here “α”, “β,” and “γ.” However, these groups match only partially to the three groups previously identified using microsatellites and ORF in Nakajima et al. (2017) (Figure 3B and Figure S2). Most haplotypes were found at all depths ranges except for a few haplotypes (Figure 3B). This limitation is easily explained by these haplotypes being represented by only one to three samples. Similarly, clusters α and γ were present in all main locations (Figure S3) and the absence of cluster β from Kume and Kikai Islands may be a consequence of the low number of samples from these locations (n = 16 and n = 6 respectively).
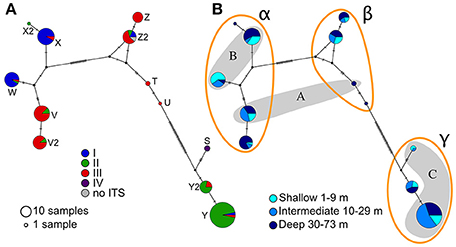
Figure 3. Median-Joining haplotype networks for the host mitochondrial ORF. Size of the circles is proportional to the frequency of the haplotypes. As in Figure 2, marks on the lines of the network indicate evolutionary steps; letters by the haplotypes correspond to the letters used in the text to refer to specific haplotypes. (A) Colors correspond to the clusters of ITS2 (see Figure 2), (B) colors correspond to the depth range and capital letters in the shaded areas correspond to the haplotype groups in Nakajima et al. (2017).
AMOVA results on the ORF sequences (Table 2) suggest significant structure between both markers (FST 0.651, p = 0.000). On the other hand, results were not significant when testing for depth partitioning (FCT: −0.065, p = 0.652) suggesting an absence of significant structure among depth groups and most of the variation was observed within location-depth groups.
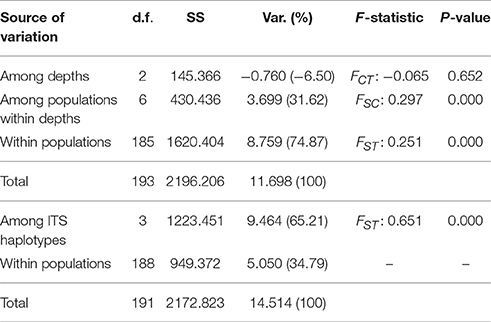
Table 2. Analyses of molecular variance (AMOVA) of Seriatopora ORF according to depth of ITS haplotypes.
Symbiodinium Diversity
All the samples analyzed for Symbiodinium COI (n = 35) possessed identical sequences regardless of the depth or geographical origin of the colonies. However, the ITS2 DGGE data (n = 119) showed that Seriatopora hosted only clade C Symbiodinium with two types of communities composed of different subtypes. One community was composed of Symbiodinium types with dominant sequences related to C1/C78a (in this context “related” means that C1/C78a were the closest BLAST matches although the sequences diverged from up to 2 bp/gaps from the reference sequences). Colonies with this pattern (n = 37) were found at depths ranging from 3 to 47 m and in Okinawa Island, Kume Island and the Yaeyama Islands. The other group consisted of dominant Symbiodinium belonging to C59 or related sequences. This group (n = 82) was found from 3 to 40 m depth and from Okinawa Island to the Yaeyama Islands. The C59-related communities were predominant in the shallow areas and decreased at both intermediate and deep depths in comparison with the C1/C78a-related community (Figure 4). However, contrasting with the C1/C78a-related group, single SNPs could split the C59 related group into several subgroups recognizable by different DGGE patterns. One subgroup composed of Seriatopora hosting mainly Symbiodinium C59 (n = 28) was found from 3 to 38 m in Okinawa, Kume Island, and the Yaeyama Islands. This subgroup contributed to most of the symbionts found in shallow waters (15 out of 17 samples). Another group hosting a different C59-variant (n = 35) was found at all depths, but mainly below 10 m in Okinawa and the Yaeyama Islands, while the two other subgroups, less common, composed of different variants of C59, appeared to have more restricted geographic distributions, either Nagura Bay (Ishigaki Island, Yaeyama Islands) and Kume Island between 10 and 39 m depth (n = 6) or Okinawa Island between 37 and 45 m depth (n = 13) (Figure S4). In addition to the C59 variant, the latter group also contained C1 and C3 Symbiodinium subclades in the dominant DGGE bands.
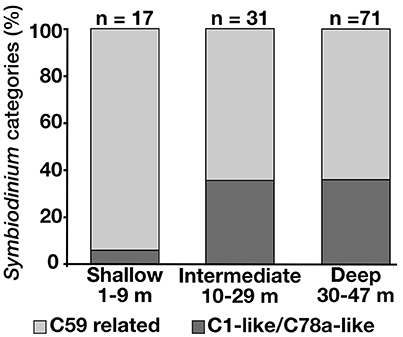
Figure 4. Proportions of the C59-related Symbiodinium assemblages and of the C1-like/C78a-like Symbiodinium in Seriatopora at different depths.
DGGE allows to rapidly identifying dominant Symbiodinium types in a sample, however cloning is more sensitive than DGGE to symbiont types present in lower densities in the host (Thornhill et al., 2007; Sampayo et al., 2008). As expected, cloning produced a large diversity of variants spreading around C1 and C59 with little structure visible between the different DGGE-groups on the haplotype network (Figure S5). Most groups included C1 sequences, the only exception being the first group that showed several close variants (1–2 bp) of C1 but no exact C1 sequences. Likewise, this group is the only one in which we did not recover exact C59 sequences. Although the cloning results suggest more intricate structuring, it supports the simplification of the symbiont communities described above, with two main groups, one containing C59 and the other composed mainly of C1 variants.
Discussion
Seriatopora hystrix, which was reported to have gone locally extinct in some the shallow reef areas such as around Sesoko Island following the past two bleaching events (van Woesik et al., 2011), represents an ideal model to test the potential of mesophotic coral populations to act as a refuge for shallow corals exposed to stressful environmental disturbances due to climate change.
Both ITS2 and ORF markers analyzed independently separate specimens into 3 distinct clusters and suggest the presence of cryptic lineages/species of this coral. Haplowebs based on ITS2 have been proposed as an efficient way to delineate species (Adjeroud et al., 2014; Fontaneto et al., 2015), on the other hand, the mitochondrial mtORF was suggested to be an efficient proxy to detect cryptic species of Seriatopora in Australia (Warner et al., 2015). Here, ORF and ITS structuration appear significantly consistent (Table 2). However, the presence of a few samples from most ITS clusters within each of the ORF groups clearly illustrates some genetic exchange between these groups (Figure 3). This marker discrepancy could be the result of hybridization or incomplete lineage sorting as observed for several other scleractinian corals (Richards et al., 2008). In the absence of unambiguous evidence of clear genetic isolation between specific clusters, further morphological and ecological studies should assess whether these clusters represent lineages or species.
Molecular analyses at population levels will also help understanding the connectivity between the different clusters discussed above. In this context, a recent microsatellite study on shallow and intermediate Seriatopora samples collected around Okinawa also distributed specimens into 3 groups, with 6 different ORF haplotypes (Nakajima et al., 2017). Here 12 distinct ORF haplotypes were found (Figure 3B and Figure S3) covering nearly the same geographic region although with a larger depth range. Interestingly, 4 out of the 6 additional haplotypes reported here were collected at depths investigated in Nakajima et al. (2017). Despite nearly 200 samples analyzed in each study, the differences between both studies still show the effect of sampling on haplotypes diversity (e.g., the haplotype Ser-1 that was found only on the east coast of Okinawa Island in Nakajima et al. (2017), while here we found this haplotype only on the west coast of Ishigaki Island). Such sampling bias will contribute to artificially creating a structure when there is none. Limited diversity found despite a reasonable sampling effort could results from the patchy geographical distribution of haplotypes within one sampling site. This distribution of mitochondrial haplotypes could result from the rapid settlement of Seriatopora planulae (Atoda, 1951; Prasetia et al., 2017) leading to patches of identical mitochondrial haplotypes nearby a mother colony.
Flot et al. (2008a) examined Seriatopora specimens from New Caledonia, Philippines and Okinawa with two mitochondrial markers and showed no relation between the mitochondrial genetic signal and morphology, although some potential biogeographic signal could be detected with the absence in New Caledonia of a group of haplotypes found in Philippines and Okinawa (named “cluster 4”) and the presence of “clusters 1 and 3” only in New Caledonia. However, they also proposed that the absence of haplotypes belonging to clusters 1 and 3 in Okinawa and Philippines could be due to the lower sampling intensity in those locations rather than a biogeographic pattern, however, in our relatively extensive survey of Okinawan Seriatopora, we also did not recover haplotypes assigned to the clusters 1 and 3 in Flot et al. (2008a). Additionally, one haplotype found in both Okinawa Island and Yaeyama Islands across all depth categories was identical to one of the haplotypes observed by van Oppen et al. (2011) on the shallow reef (upper slope, between 5 and 7 m) in Eastern Australia and a deeper site (between 25 and 40 m) in Western Australia (referred to as “HostB”). Overall, the genetic diversity observed in Seriatopora indicates that although this species is particularly prone to bleaching under high temperatures, it benefits from a large and diverse gene pool that may facilitate adaptation to new conditions or the colonization of new ecological niches.
In this study, independently of the taxonomic status of the different clusters of haplotypes, all haplotypes present in more than three samples were found at all depth ranges and no significant structure with depth could be detected with the ORF data (Table 2). This overlap between shallow and deep sites suggests the absence of depth-specific Seriatopora and, at least on the long term, some levels of connectivity between shallow and deep communities. This absence of a clear depth zonation contrasts with results from East Australia where a clear partitioning of S. hystrix mitochondrial lineages was observed (Bongaerts et al., 2010b, 2011; van Oppen et al., 2011), and that of observations from Western Australia, as at this location, all depths were largely dominated by a single mitochondrial haplotype (van Oppen et al., 2011). In the Atlantic, the brooding coral Porites astreoides also showed genetic patterns suggesting deep-shallow connectivity in several locations although in this case the symbionts exhibited depth-zonation in some cases (Serrano et al., 2016). Strong vertical connectivity was also detected in the Atlantic broadcasting species, Stephanocoenia intersepta, while another brooding coral, Agaricia fragilis, showed strong genetic structure with depth in the same region (Bongaerts et al., 2017).
Symbiodinium plays an important role in the depth distribution of scleractinian corals (Bongaerts et al., 2015b) and the adaptation of corals to the mesophotic environment (Lesser et al., 2010; Cooper et al., 2011). High diversity of Symbiodinium was detected using COI in Hawaiian Leptoseris (Pochon et al., 2015), however, no variation for this marker was observed among all the samples analyzed. In our study, ITS2 is potentially more informative. While both C1/C78-related and C59-related Symbiodinium types were found at all depths investigated, the frequency shifts from C59-related to C1/C78-related types below 10 m depth (Figure 4). This change of symbiont association may reflect metabolic transition and adaptation to depth. Similarly, change of symbiont association with depth was correlated with change in photosynthesis/respiration ratio for S. hystrix in Western Australia (Cooper et al., 2011). In the Caribbean, zonation in associated Symbiodinium was found to be common in depth-generalist species (Bongaerts et al., 2015b), with putative deep-specialist types observed in association with mesophotic corals (Lesser et al., 2010; Bongaerts et al., 2015a; Pochon et al., 2015). Nonetheless, despite the increase of frequency in the C1/C78a-related type with increasing depth, more than half of the sampled intermediate and mesophotic colonies associated with the C59-like Symbiodinium types that dominate shallower depths. Given the maternal symbiont acquisition mode (vertical transmission) of S. hystrix (Atoda, 1951), larvae from mesophotic colonies hosting the C1/C78-related type may be less likely to successfully recruit to and survive in shallow reef areas, whereas those hosting the C59-related types are more likely given the dominance of these types in shallow-water. However, more shallow samples would be required to confirm the trends observed as the vast majority of the samples analyzed here originated from mesophotic depths.
The persistence of bleaching-sensitive species at mesophotic depths in Okinawa, despite localized “extinction” of corals such as Seriatopora on the adjacent shallow reef, demonstrates the potential of MCEs as a thermal refuge during warm-water bleaching episodes. Given that most host and Symbiodinium genotypes identified for mesophotic Seriatopora in Okinawa are not unique to that depth zone but are also found on shallow reefs elsewhere in the archipelago, the refuge potential appears to extend beyond just the species and includes safeguarding at least a proportion of shallow-water genotypic diversity (of both host and associated endosymbionts).
Author Contributions
FS and SH planned the experiments; FS conducted the experiments; RP, MY, PB, and SH contributed to the experiments; FS, RP, MY, and SH analyzed the data; FS and SH wrote the manuscript; RP, MY, and PB critically reviewed the manuscript.
Funding
This research was supported by a grant from the Mitsubishi Foundation to SH (No. 24133), a University of the Ryukyus Research Project Promotion Grant for Foreign Researchers, a Sasakawa Scientific Research Grant from the Japan Science Society to FS (No. 24-748), two Grants-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science to FS (Nos. 26870917 and 17K15176) and a Grant-in-Aid for Scientific Research (A) of the Japan Society for the Promotion of Science to SH (No. 16H02490).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Some samples from Nagura Bay were collected with the collaboration of Prof. H. Kan. Samples from Kikai Island were collected with the collaboration of Dr. T. Watanabe, Dr. A. Yamazaki and the Kikai Institute for Coral Reef Sciences. We are grateful for the field assistance from Dr. M. Morita, Mr. M. Jinza, Mr. S. Kadena, Mr. M. Sunagawa, Ms. A. Izeki, Mr. I. Nakayoshi, Mr. T. Iwamoto and Mr. N. Saeki and for the laboratory assistance of Mr. Y. Nakatsuji and Mr. M. Jinza. The authors also want to thank Dr. J. F. Flot and the three reviewers for their constructive comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fmars.2017.00155/full#supplementary-material
References
Adjeroud, M., Guérécheau, A., Vidal-Dupiol, J., Flot, J.-F., Arnaud-Haond, S., and Bonhomme, F. (2014). Genetic diversity, clonality and connectivity in the scleractinian coral Pocillopora damicornis: a multi-scale analysis in an insular, fragmented reef system. Mar. Biol. 161, 531–541. doi: 10.1007/s00227-013-2355-9
Atoda, K. (1951). The larva and postlarval development of the reef-building corals. V. Seriatopora hystrix Dana. Sci. Rep. Tohoku Univ. 4, 33–39.
Bak, R. P., Nieuwland, G., and Meesters, E. H. (2005). Coral reef crisis in deep and shallow reefs: 30 years of constancy and change in reefs of Curacao and Bonaire. Coral Reefs 24, 475–479. doi: 10.1007/s00338-005-0009-1
Bandelt, H.-J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bongaerts, P., Carmichael, M., Hay, K. B., Tonk, L., Frade, P. R., and Hoegh-Guldberg, O. (2015a). Prevalent endosymbiont zonation shapes the depth distributions of scleractinian coral species. R. Soc. Open Sci. 2:140297. doi: 10.1098/rsos.140297
Bongaerts, P., Frade, P. R., Hay, K. B., Englebert, N., Latijnhouwers, K. R., Bak, R. P., et al. (2015b). Deep down on a Caribbean reef: lower mesophotic depths harbor a specialized coral-endosymbiont community. Sci. Rep. 5:7652. doi: 10.1038/srep07652
Bongaerts, P., Ridgway, T., Sampayo, E. M., and Hoegh-Guldberg, O. (2010a). Assessing the ‘deep reef refugia’ hypothesis: focus on Caribbean reefs. Coral Reefs 29, 309–327. doi: 10.1007/s00338-009-0581-x
Bongaerts, P., Riginos, C., Brunner, R., Englebert, N., Smith, S. R., and Hoegh-Guldberg, O. (2017). Deep reefs are not universal refuges: reseeding potential varies among coral species. Sci. Adv. 3:e1602373. doi: 10.1126/sciadv.1602373
Bongaerts, P., Riginos, C., Hay, K. B., Van Oppen, M. J. H., Hoegh-Guldberg, O., and Dove, S. (2011). Adaptive divergence in a scleractinian coral: physiological adaptation of Seriatopora hystrix to shallow and deep reef habitats. BMC Evol. Biol. 11:303. doi: 10.1186/1471-2148-11-303
Bongaerts, P., Riginos, C., Ridgway, T., Sampayo, E. M., Van Oppen, M. J. H., Englebert, N., et al. (2010b). Genetic divergence across habitats in the widespread coral Seriatopora hystrix and its associated Symbiodinium. PLoS ONE 5:10871. doi: 10.1371/journal.pone.0010871
Bridge, T., Beaman, R., Done, T., and Webster, J. (2012). Predicting the location and spatial extent of submerged coral reef habitat in the Great Barrier Reef World Heritage Area, Australia. PLoS ONE 7:48203. doi: 10.1371/journal.pone.0048203
Cooper, T. F., Ulstrup, K. E., Dandan, S. S., Heyward, A. J., Kuhl, M., Muirhead, A., et al. (2011). Niche specialization of reef-building corals in the mesophotic zone: metabolic trade-offs between divergent Symbiodinium types. Proc. R. Soc. B Biol. Sci. 278, 1840–1850. doi: 10.1098/rspb.2010.2321
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fabricius, K. E. (2011). “Factors determining the resilience of coral reefs to eutrophication: a review and conceptual model,” in Coral Reefs: An Ecosystem in Transition, eds Z. Dubinsky and N. Stambler (New York, NY: Springer), 493–505.
Flot, J.-F. (2007). Champuru 1.0: a computer software for unraveling mixtures of two DNA sequences of unequal lengths. Mol. Ecol. Notes 7, 974–977. doi: 10.1111/j.1471-8286.2007.01857.x
Flot, J.-F., Blanchot, J., Charpy, L., Cruaud, C., Licuanan, W. Y., Nakano, Y., et al. (2011). Incongruence between morphotypes and genetically delimited species in the coral genus Stylophora: phenotypic plasticity, morphological convergence, morphological stasis or interspecific hybridization? BMC Ecol. 11:22. doi: 10.1186/1472-6785-11-22
Flot, J.-F., Licuanan, W., Nakano, Y., Payri, C., Cruaud, C., and Tillier, S. (2008a). Mitochondrial sequences of Seriatopora corals show little agreement with morphology and reveal the duplication of a tRNA gene near the control region. Coral Reefs 27, 789–794. doi: 10.1007/s00338-008-0407-2
Flot, J.-F., Magalon, H., Cruaud, C., Couloux, A., and Tillier, S. (2008b). Patterns of genetic structure among Hawaiian corals of the genus Pocillopora yield clusters of individuals that are compatible with morphology. C. R. Biol. 331, 239–247. doi: 10.1016/j.crvi.2007.12.003
Flot, J.-F., Tillier, A., Samadi, S., and Tillier, S. (2006). Phase determination from direct sequencing of length-variable DNA regions. Mol. Ecol. Notes 6, 627–630. doi: 10.1111/j.1471-8286.2006.01355.x
Flot, J.-F., and Tillier, S. (2006). “Molecular phylogeny and systematics of the scleractinian coral genus Pocillopora in Hawaii,” in Proceedings of the 10th International Coral Reef Symposium (Okinawa), 24–29.
Fontaneto, D., Flot, J.-F., and Tang, C. Q. (2015). Guidelines for DNA taxonomy, with a focus on the meiofauna. Marine Biodiv. 45, 433–451. doi: 10.1007/s12526-015-0319-7
Frade, P. R., Bongaerts, P., Winkelhagen, A. J. S., Tonk, L., and Bak, R. P. M. (2008). In situ photobiology of corals over large depth ranges: a multivariate analysis on the roles of environment, host, and algal symbiont. Limnol. Oceanogr. 53, 2711–2723. doi: 10.4319/lo.2008.53.6.2711
Franklin, E. C., Stat, M., Pochon, X., Putnam, H. M., and Gates, R. D. (2012). GeoSymbio: a hybrid, cloud-based web application of global geospatial bioinformatics and ecoinformatics for Symbiodinium-host symbioses. Mol. Ecol. Resour. 12, 369–373. doi: 10.1111/j.1755-0998.2011.03081.x
Glynn, P. W. (1996). Coral reef bleaching: facts, hypotheses and implications. Glob. Chang. Biol. 2, 495–509. doi: 10.1111/j.1365-2486.1996.tb00063.x
Harii, S., Hongo, C., Ishihara, M., Ide, Y., and Kayanne, H. (2014). Impacts of multiple disturbances on coral communities at Ishigaki Island, Okinawa, Japan, during a 15 year survey. Marine Ecol. Progr. Ser. 509, 171–180. doi: 10.3354/meps10890
Hinderstein, L. M., Marr, J. C. A., Martinez, F. A., Dowgiallo, M. J., Puglise, K. A., Pyle, R. L., et al. (2010). Theme section on “Mesophotic Coral Ecosystems: characterization, Ecology, and Management”. Coral Reefs 29, 247–251. doi: 10.1007/s00338-010-0614-5
Hoegh-Guldberg, O. (1999). Climate change, coral bleaching and the future of the world's coral reefs. Marine Freshw. Res. 50, 839–866. doi: 10.1071/MF99078
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Holstein, D. M., Paris, C. B., Vaz, A. C., and Smith, T. B. (2016). Modeling vertical coral connectivity and mesophotic refugia. Coral Reefs 35, 23–37. doi: 10.1007/s00338-015-1339-2
Hongo, C., Kawamata, H., and Goto, K. (2012). Catastrophic impact of typhoon waves on coral communities in the Ryukyu Islands under global warming. J. Geophys. Res. Biogeosci. 117:G02029. doi: 10.1029/2011jg001902
Hongo, C., and Yamano, H. (2013). Species-specific responses of corals to bleaching events on anthropogenically turbid reefs on Okinawa Island, Japan, over a 15-year period (1995–2009). PLoS ONE 8:e60952. doi: 10.1371/journal.pone.0060952
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Kahng, S. E., Copus, J. M., and Wagner, D. (2014). Recent advances in the ecology of mesophotic coral ecosystems (MCEs). Curr. Opin. Environ. Sustain. 7, 72–81. doi: 10.1016/j.cosust.2013.11.019
Kahng, S. E., Garcia-Sais, J. R., Spalding, H. L., Brokovich, E., Wagner, D., Weil, E., et al. (2010). Community ecology of mesophotic coral reef ecosystems. Coral Reefs 29, 255–275. doi: 10.1007/s00338-010-0593-6
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kimura, T., Shimoike, K., Suzuki, G., Nakayoshi, I., Shioiri, A., Tabata, A., et al. (2011). Large scale communities of Acropora horrida in the mesophotic zone off Kume Island, Okinawa. J. Jpn. Coral Reef Soc. 13, 43–45. doi: 10.3755/jcrs.13.43
LaJeunesse, T. C., Loh, W. K., Van Woesik, R., Hoegh-Guldberg, O., Schmidt, G. W., and Fitt, W. K. (2003). Low symbiont diversity in southern Great Barrier Reef corals, relative to those of the Caribbean. Limnol. Oceanogr. 48, 2046–2054. doi: 10.4319/lo.2003.48.5.2046
LaJeunesse, T., and Trench, R. (2000). Biogeography of two species of Symbiodinium (Freudenthal) inhabiting the intertidal sea anemone Anthopleura elegantissima (Brandt). Biol. Bull. 199, 126–134. doi: 10.2307/1542872
Lesser, M. P., Slattery, M., Stat, M., Ojimi, M., Gates, R. D., and Grottoli, A. (2010). Photoacclimatization by the coral Montastraea cavernosa in the mesophotic zone: light, food, and genetics. Ecology 91, 990–1003. doi: 10.1890/09-0313.1
Loya, Y., Sakai, K., Yamazato, K., Nakano, Y., Sambali, H., and Van Woesik, R. (2001). Coral bleaching: the winners and the losers. Ecol. Lett. 4, 122–131. doi: 10.1046/j.1461-0248.2001.00203.x
Muko, S., Arakaki, S., Nagao, M., and Sakai, K. (2013). Growth form-dependent response to physical disturbance and thermal stress in Acropora corals. Coral Reefs 32, 269–280. doi: 10.1007/s00338-012-0967-z
Nakajima, Y., Nishikawa, A., Iguchi, A., Nagata, T., Uyeno, D., Sakai, K., et al. (2017). Elucidating the multiple genetic lineages and population genetic structure of the brooding coral Seriatopora (Scleractinia: Pocilloporidae) in the Ryukyu Archipelago. Coral Reefs 36, 415–426. doi: 10.1007/s00338-017-1557-x
Pochon, X., Forsman, Z., Spalding, H., Padilla-Gami-o, J., Smith, C., and Gates, R. (2015). Depth specialization in mesophotic corals (Leptoseris spp.) and associated algal symbionts in Hawai'i. R. Soc. Open Sci. 2:140351. doi: 10.1098/rsos.140351
Pochon, X., Putnam, H. M., Burki, F., and Gates, R. D. (2012). Identifying and characterizing alternative molecular markers for the symbiotic and free-living Dinoflagellate genus Symbiodinium. PLoS ONE 7:29816. doi: 10.1371/journal.pone.0029816
Prasetia, R., Sinniger, F., Hashizume, K., and Harii, S. (2017). Reproductive biology of the deep brooding coral Seriatopora hystrix: implications for shallow reef recovery. PLoS ONE 12:e0177034. doi: 10.1371/journal.pone.0177034
Richards, Z. T., Van Oppen, M. J. H., Wallace, C. C., Willis, B. L., and Miller, D. J. (2008). Some rare indo-pacific coral species are probable hybrids. PLoS ONE 3:e3240. doi: 10.1371/journal.pone.0003240
Riegl, B., and Piller, W. E. (2003). Possible refugia for reefs in times of environmental stress. Int. J. Earth Sci. 92, 520–531. doi: 10.1007/s00531-003-0328-9
Sampayo, E. M., Ridgway, T., Bongaerts, P., and Hoegh-Guldberg, O. (2008). Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. Proc. Natl. Acad. Sci. U.S.A. 105, 10444–10449. doi: 10.1073/pnas.0708049105
Serrano, X., Baums, I. B., O'reilly, K., Smith, T. B., Jones, R. J., Shearer, T. L., et al. (2014). Geographic differences in vertical connectivity in the Caribbean coral Montastraea cavernosa despite high levels of horizontal connectivity at shallow depths. Mol. Ecol. 23, 4226–4240. doi: 10.1111/mec.12861
Serrano, X. M., Baums, I. B., Smith, T. B., Jones, R. J., Shearer, T. L., and Baker, A. C. (2016). Long distance dispersal and vertical gene flow in the Caribbean brooding coral Porites astreoides. Sci. Rep. 6:21619. doi: 10.1038/srep21619
Sinniger, F., Morita, M., and Harii, S. (2013). “Locally extinct” coral species Seriatopora hystrix found at upper mesophotic depths in Okinawa. Coral Reefs 32, 153–153. doi: 10.1007/s00338-012-0973-1
Sinniger, F., Reimer, J. D., and Pawlowski, J. (2010). The Parazoanthidae (Hexacorallia: Zoantharia) DNA taxonomy: description of two new genera. Marine Biodiversity 40, 57–70. doi: 10.1007/s12526-009-0034-3
Slattery, M., Lesser, M. P., Brazeau, D., Stokes, M. D., and Leichter, J. J. (2011). Connectivity and stability of mesophotic coral reefs. J. Exp. Mar. Biol. Ecol. 408, 32–41. doi: 10.1016/j.jembe.2011.07.024
Smith, T. B., Glynn, P. W., Maté, J. L., Toth, L. T., and Gyory, J. (2014). A depth refugium from catastrophic coral bleaching prevents regional extinction. Ecology 95, 1663–1673. doi: 10.1890/13-0468.1
Thornhill, D. J., LaJeunesse, T. C., and Santos, S. R. (2007). Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol. Ecol. 16, 5326–5340. doi: 10.1111/j.1365-294X.2007.03576.x
van Oppen, M. J. H., Bongaerts, P., Underwood, J. N., Peplow, L. M., and Cooper, T. F. (2011). The role of deep reefs in shallow reef recovery: an assessment of vertical connectivity in a brooding coral from west and east Australia. Mol. Ecol. 20, 1647–1660. doi: 10.1111/j.1365-294X.2011.05050.x
van Woesik, R., Sakai, K., Ganase, A., and Loya, Y. (2011). Revisiting the winners and the losers a decade after coral bleaching. Mar. Ecol. Prog. Ser. 434, 67–76. doi: 10.3354/meps09203
Warner, P. A., Oppen, M. J., and Willis, B. L. (2015). Unexpected cryptic species diversity in the widespread coral Seriatopora hystrix masks spatial-genetic patterns of connectivity. Mol. Ecol. 24, 2993–3008. doi: 10.1111/mec.13225
Keywords: deep reef refuge hypothesis, reef recovery, diversity, Seriatopora, NW pacific
Citation: Sinniger F, Prasetia R, Yorifuji M, Bongaerts P and Harii S (2017) Seriatopora Diversity Preserved in Upper Mesophotic Coral Ecosystems in Southern Japan. Front. Mar. Sci. 4:155. doi: 10.3389/fmars.2017.00155
Received: 20 December 2016; Accepted: 08 May 2017;
Published: 23 May 2017.
Edited by:
Hajime Kayanne, University of Tokyo, JapanReviewed by:
Aldo Cróquer, Simón Bolívar University, VenezuelaAnnika Noreen, Inter-Research, Germany
Zac H. Forsman, Hawaii Institute of Marine Biology, United States
Copyright © 2017 Sinniger, Prasetia, Yorifuji, Bongaerts and Harii. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frederic Sinniger, fredsinniger@hotmail.com
 Frederic Sinniger
Frederic Sinniger Rian Prasetia
Rian Prasetia Makiko Yorifuji
Makiko Yorifuji Pim Bongaerts2
Pim Bongaerts2  Saki Harii
Saki Harii