Quorum Sensing and Quorum Quenching in the Mediterranean Seagrass Posidonia oceanica Microbiota
- 1UPMC Univ Paris 06, Laboratoire de Biodiversité et Biotechnologies Microbiennes, Centre National De La Recherche Scientifique (CNRS), Observatoire Océanologique, Sorbonne Universités, Banyuls-sur-Mer, France
- 2Unité Molécules de Communication et Adaptation des Micro-organismes (UMR 7245), Sorbonne Universités, Muséum National d'Histoire Naturelle, Centre National De La Recherche Scientifique (CNRS), Paris, France
- 3Toxicology Laboratory, Pharmaceutical Science Course, Federal University of Amapá, Macapa, Brazil
- 4UPMC Univ Paris 06, Centre National De La Recherche Scientifique (CNRS), Bio2MAR, Observatoire Océanologique, Sorbonne Universités, Banyuls-sur-Mer, France
- 5UPMC Univ Paris 06, Laboratory of Cancer Biology and Therapeutics, Centre de Recherche Saint-Antoine, Institut National De La Santé Et De La Recherche Médicale, Sorbonne Universités, UMR_S 938, Paris, France
Posidonia oceanica (L.) Delile is an endemic Mediterranean seagrass angiosperm. The taxonomic diversity of the Posidonia-associated microbiota has previously been inventoried using culture-based and culture-independent approaches. However, little is known about the type of chemical interactions that occur between the cells within this microbiota. In this study, we isolated 60 bacterial strains from the leaves and rhizomes of P. oceanica and evaluated their capacity to communicate through quorum sensing. We found 6 strains able to emit 8 different types of quorum sensing acyl homoserine lactones, identified through UHPLC-HRMS/MS after bioguided fractionation of large cultures. We also identified 19 strains able to produce furanosyl diester borate, another type of quorum sensing autoinducer. Also, 5 strains were found to produce quorum quenching compounds. Finally, as quorum sensing is known to be involved in biofilm production, we also tested whether strains positive to our biotests were able to produce biofilms. Collectively, these data reveal the importance of examining chemical interactions within this bacterial consortium associated with P. oceanica that could have an impact on the physiology of this key Mediterranean seagrass.
Introduction
Seagrasses are major underwater coastal ecosystems that are usually dominated by one of the 60 species of marine angiosperms living in seashore environments (Hemminga and Duarte, 2000; Orth et al., 2006). These ecosystems provide important ecosystem services, including acting as a fish nursery, biodiversity protection, carbon sequestration, nutrient recycling, and limiting coastal erosion by sediment stabilization (Orth et al., 2006). The genus Posidonia is the evolutionarily oldest genus of seagrasses, having colonized the marine environment during the Cretaceous (Den Hartog, 1970). This genus is composed of 9 species. Among them, Posidonia oceanica (L.) Delile, a Mediterranean endemic species abundant along the coasts, occupies ~50,000 km2 (Garcias-Bonet et al., 2012).
Only a few works have examined seagrass angiosperm microbiotas, while such studies on terrestrial plants are more common (Turner et al., 2013). However, a few pioneering studies revealed that the seagrass-associated bacteria are phylogenetically diverse. The rhizobiome of the seagrasses Zostera and Cyrnodocea contains a large bacterial diversity (Cúcio et al., 2016). Garcias-Bonet et al. (2012) have examined the diversity of prokaryotic communities in surface-sterilized tissues. They revealed 34 distinct operational taxonomic units (OTUs) that are affiliated with diverse Proteobacteria and Bacteroidetes. They also reported that bacterial communities depended on the seagrass tissues and on the locations where the seagrass were collected.
Many chemical interactions between microbial cells occur in plant-associated microbiotas. Also, in recent decades, chemical ecologists have provided many experimental clues indicating that the exchange of semiochemicals is a widespread pattern in prokaryotic communities (Fuqua et al., 1994; Miller and Bassler, 2001). However, such an exchange of semiochemicals has never been examined within the microbiota of P. oceanica, where it could regulate important physiological traits (Turner et al., 2013; Vandenkoornhuyse et al., 2015) and thus have important implications for the whole Posidonia-based seagrass ecosystem.
Quorum sensing (QS) constitutes a widespread mode of chemical communication in prokaryotic communities. Cells engaged in quorum sensing liberate small compounds, named autoinducers, and are able to detect their presence in the nearby environment. At low cell densities, few autoinducers are present, and prokaryotes display individual phenotypes and behaviors. When the cell density increases, the concentration of autoinducers also increases, causing bacteria to switch their genetic and physiological program and display collective phenotypes and comportments (Fuqua et al., 1994). Quorum-sensing processes induce many transduction cascades, and the regulation of the expression of many genes influencing bacterial metabolism and phenotypes (Miller and Bassler, 2001). The establishment of biofilms (Miller and Bassler, 2001), the emission of bioluminescence (Nealson, 1977), virulence (Smith and Iglewski, 2003), and many other bacterial phenotypes (Smith and Iglewski, 2003) are examples that have been shown to be controlled by quorum sensing. Two major types of autoinducers are involved in Gram negative marine bacteria quorum sensing: homoserine lactones (AHL, or autoinducer type 1, or AI-1), involved in intraspecies communication, and furanosyl diester borate (AI-2), which might plays a role in interspecies communication (Hmelo, 2017). However, to date, the prevalence and the nature of the quorum sensing that occur in the P. oceanica microbiota remain unknown.
Quorum quenching mechanisms are engaged by competitors to limit the growth and the coordination of bacteria engaged in QS communication (Defoirdt et al., 2004; Rasmussen and Givskov, 2006). These processes rely on the secretion of enzymes leading to QS-signal cleavage including acylases or lactonases or secretion of metabolites acting as QS inhibitors (Dong and Zhang, 2005). Again, whether quorum quenching compounds that are expressed by bacteria are associated with the P. oceanica microbiota remains unknown.
In this work, we examined quorum sensing and quorum quenching to determine if these processes are widespread phenotypes of the prokaryotic communities associated with P. oceanica and we evaluated the nature of the secreted autoinducers (AI-1 and/or AI-2). Collectively, our data reveal for the first time the nature of the quorum-based chemical exchanges occurring in the prokaryotic communities of the seagrass P. oceanica, and will serve as a starting point for future studies that will focus on the microbial interactions in seagrass microbiotas.
Materials and Methods
Elaboration of a Bacterial Culture Collection Associated with Posidonia oceanica
P. oceanica plants were collected in June 2014 in front of the port of Banyuls-sur-mer, south of France (42°28′54.12″N, 3° 8′11.99″E; 2 m deep). To maximize the recovery of the bacterial strains associated with P. oceanica (epiphytes and endophytes strains), we combined two protocols of isolation. The first protocol consisted of cutting a clean piece of leaves or rhizomes, after observations of epibionts presence under the stereomicroscope, to isolate epiphytic strains. A second protocol was used to isolate endophytic strains (Zinniel et al., 2002). This protocol included a series of surface sterilizations with first 99% ethanol, second 3.125% NaOCl, and third a second 99% ethanol step before a final and gentle washing with sterile water.
The different pieces of Posidonia (rhizomes and leaves) were ground with a blender. Serial dilutions (1 to 10−3) in presterilized seawater were performed, and 100 μL was plated on various media. All these media were reconstituted with 75% seawater: BG11 medium, R2A medium and Starch Casein Agar (SCA) medium. Petri plates were incubated at 25°C until growth of the colonies occurred and new colonies stopped appearing. Colonies were isolated and purified based on their different morphotypes on their respective media. Each bacterial strain was then stored at −80°C in a 35% glycerol solution and in a 5% DMSO solution in Marine Broth (MB, Difco™ BD). The strains were deposited in the Banyuls Bacterial Culture Collection (WDCM911) http://collection.obs-banyuls.fr/.
Bacterial Identification by 16S rRNA Sequencing and Phylogenetic Analysis
The bacterial identification was based on their partial 16SrRNA gene sequencing. All the sequences were deposited in Genbank (see Supplementary Table 1). The obtained 16S rRNA gene sequences were compared using the BLAST algorithm against the sequences of the EzTaxon server (http://eztaxon-e.ezbiocloud.net) database (Kim et al., 2012). Phylogenetic analyses were conducted using MEGA 6 software (Kim et al., 2012). All alignments were performed with ClustalW (Larkin et al., 2007) and adjusted manually. Phylogenetic trees were constructed using the Neighbor-Joining (NJ) method with p-distance correction and 1,000 bootstrap replicates.
Quorum Sensing Bioassay: Detection of Al-1
Bacterial culture supernatants were tested in the AHL biosensor assay following previously described protocols using Pseudomonas putida and Escherichia coli-based biosensors. Briefly, P. putida F117 (pRK-C12; Kmr; ppuI:mpt) was used for the detection of long-chain AHLs (>8 carbons in the acyl side chain) (Andersen et al., 2001; Girard et al., 2017), and E. coli MT102 (pJBA132) for the detection of short-chain AHLs (≤8 carbons in the acyl side chain) (Riedel et al., 2001; Girard et al., 2017). E. coli MT102 and P. putida F117 were cultivated in Luria-Bertani (LB) Broth (Sigma L3022) overnight with continuous shaking (200 rpm) at 37°C supplemented with tetracycline (25 μL/mL) and at 30°C supplemented with gentamicin (20 μL/mL), respectively. An overnight culture of each biosensor strain was inoculated in fresh LB medium (dilution 1/50). This fresh biosensor culture was dispensed into 96-well microplates (150 μL/well for bacterial supernatant evaluation and 180 μL/well for extracts or fractions). To evaluate bacteria for their ability to produce Al-1, they were grown on MB medium 24 h at 25°C with agitation (120 rpm). Culture supernatants were collected after centrifugation (10 min at 10,000 rpm) and added to the microplate wells containing the biosensor strains (50 μL/well, in triplicate). Extracts or fractions were dissolved in DMSO/LB (25:75, v/v) and added to each well in triplicate (20 μL/well). Microplates were incubated at 30°C and 37°C, depending of optimum growth temperature of the selected biosensor strain, without shaking. After 0, 5, and 24 h of incubation, fluorescence was determined with a Victor1420 Multilabel Counter (Perkin-Elmer) at an excitation wavelength of 485 nm and a detection wavelength of 535 nm. The optical density (OD) at 620 nm was also measured to monitor biosensor cell growth. Negative controls were biosensor cultures with sterile LB medium and without extract. Biosensor cultures with added commercial AHLs (C6-HSL for E. coli MT102 and oxo-C10-HSL for P. putida F117) were used as positive controls.
Quorum Sensing Bioassay: Detection of AI-2
A mutant strain of Vibrio harveyi, BB170, was used to detect the presence of AI-2 in the culture supernatants (Bassler et al., 1997; Taga, 2005). Briefly, the V. harveyi reporter strain was grown at 30°C in Autoinducer Bioassay (AB) medium. The overnight culture was then diluted 1:5,000 into fresh medium, and 180 μL of the diluted cells was added to wells containing 20 μL of the culture supernatants of the bacteria to be tested for AI-2 production. The 96-well plates were incubated at 30°C. Every hour, luminescence was measured using a Victor1420 Multilabel Counter (Perkin-Elmer) until the medium control sample produced the least amount of light. The Al-2 production is reported as the fold induction of the reporter strain divided by the background when only medium was added to the reporter. All assays were performed in duplicate.
Quorum Quenching Bioassay
The AHL biosensor E. coli MT102 was also used to highlight the quorum quenching ability of the bacterial strains. The biosensor was cultivated in Luria-Bertani (LB) Broth (Sigma L3022) overnight with continuous shaking (200 rpm) in the same conditions (temperature, antibiotics) used for the quorum-sensing bioassay. An overnight culture of the biosensor strain was inoculated into fresh LB medium (dilution 1/25). This fresh biosensor culture was dispensed into 96-well microplates (150 μL/well). Bacteria were grown on MB medium supplemented with an AHL (100 μL of C6-AHL or oxo-C10-HSL (both 1 mM) in 100 mL of MB for the assay with E. coli MT102 and P. putida F117, respectively) for 24 h at 25°C with agitation (120 rpm). Culture supernatants were collected after centrifuging for 10 min at 10,000 rpm and added to the microplate containing the biosensor strains (50 μL/well in triplicate). Extracts or fractions were dissolved in DMSO/LB (25:75, v/v) and added to each well in triplicate (20 μL/well). The microplates were analyzed as previously describe above (detection of AI 1 bioassay). The negative controls were biosensor cultures with sterile MB medium supplemented with AHL. Two strains of the Banyuls Bacterial Culture Collection (BBCC116 and BBCC2153) were used as positive controls.
Biofilm Formation Assay
The ability of strains to produce biofilm has been evaluated using a crystal violet-based protocol previously published (Stepanovic et al., 2000; O'Toole, 2011). Strains from fresh agar plates were inoculated in 5 mL of Marine Broth and incubated for 24 h at 25°C. Biofilm formation was conducted in 96-well flat bottom plates. To increase biofilm formation (Moreira et al., 2013), culture strains were diluted with fresh medium until a final OD630 nm of 0.2 was reached. The microplate wells were filled with 100 μL of diluted culture and 100 μL of a 4.5% glucose solution, whereas only broth with glucose served as a control to check the sterility of the medium and to test for non-specific attachment. Ruegeria mobilis (strain MOLABBCC1144, Banyuls Culture Collection) was used as a negative control, and Sulfitobacter brevis (strain BBCC1436, Banyuls Culture Collection) was used as a positive control. Each inoculated plate was incubated for 48 h at 25°C under static conditions. After incubation, the medium was gently removed, and the wells were washed with 200 μl of sterile phosphate-buffered saline (PBS, pH 7.2) to remove all planktonic cells while preserving the integrity of the biofilm. The remaining bacteria that were attached to the bottom of wells were stained for 15 min at room temperature with 100 μL of 0.2% crystal violet solution prepared in 20% ethanol (v/v). Subsequently, the unbound crystal violet stain was removed, and the wells were rinsed three times with sterile water before adding 200 μL of a solution of 10% acetic acid, 50% ethanol, and 40% distilled water (v/v/v) to each well. The absorbance was measured at 540 nm using a microplate reader [Victor1420 Multilabel Counter (Perkin-Elmer)]. We classified isolates according to their ability to form biofilms, and following Stepanovic et al. (2000), a cut-off value was calculated for the negative control using the following formula: ODc = Average OD of negative control + 3 × SD (Standard Deviation) of negative control. Based on the calculated cut-off OD, strains were classified into the following categories: non-biofilm producers: (OD < ODc), weak biofilm producers: (0Dc < OD < 2 × 0Dc), moderate biofilm producers: (2 × ODc < OD < 4 × 0Dc), and strong biofilm producers: (OD > 4 × 0Dc) (Stepanovic et al., 2000).
Bacterial Culture for AHL Characterization
The bacterial strains that demonstrated positive results for the production of AHLs were grown in MB broth at 25°C with shaking (120 rpm). Cultures were realized in 6 1-L Erlenmeyer flasks, each containing 500 mL of Marine Broth (MB) 2216 (BD Difco, Sparks, MD). After 48 h, ethyl acetate (EtOAc, 200 mL) was added to each flask, and the ground mixture was shaken for 12 h at room temperature (150 rpm). After dehydration on anhydrous Na2SO4 and filtration, the organic phase was evaporated to dryness. The ethyl acetate extracts were treated prior to analysis using solid-phase extraction (Phenomenex Strata C18, 55 1.1m). The stationary SPE phase was activated with 100 mL of acetonitrile. The cartridge was loaded with 100 mg of each extract, which was solubilized in 2 mL of acetonitrile. The elution was performed with 50 mL of acetonitrile to eliminate the most lipophilic metabolites. The eluates were dried and dissolved in DMSO.
HPLC Fractionation
The treated extracts were then fractionated by liquid chromatography on a Phenomenex Luna C18 (5 mm, 250 × 21.20 mm, 100 Å) column with a separation HPLC system with 2 Varian Prep Star pumps, a manual injector, a Dionex Ultimate 3000 RS variable wavelength detector and a Dionex Ultimate 3000 fraction collector. The solvent was made of gradient grade H2O and CH3CN (70:30 for 3 min, followed by a 12-min linear gradient from 70:30 to 0:100, followed by 100% CH3CN for 10 min) and pumped at a flow rate of 20 mL.min−1. The eluents were monitored at 214, 254, 274, and 280 nm and were collected between 3 and 25 min (1 fraction/min, 22 fractions). The fractions were dried with a genevac HT-4X system and were dissolved in 100 μL DMSO to evaluate the presence of AHL with the biosensors and diluted.
HPLC-HRMS/MS Analyses
UHPLC-MS analyses were performed with a Dionex Ultimate 3000 UHPLC-HESI HRMS Q Exactive focus (Thermo Scientific) system controlled by Thermo Xcalibur software (Thermo scientific). Elution was performed on a Hypersil gold C18 (2.1 × 150 mm) column with 1.9 μm particle size (Thermo Scientific) maintained at 50°C; solvent A consisted of water with 0.1% formic acid, and solvent B was acetonitrile with 0.1% formic acid. The gradient profile for chromatography was as follows: 100% solvent A for 5 min, a linear increase in solvent B to 100% over 5 min, hold at 100% solvent B for 5 min, and then equilibrate with 100% solvent A for 2 min. The flow rate was maintained at 0.8 mL.min−1, and the injection volume was 5 μL of the fractions diluted to 1 mg.mL−1. The detection using ESI was performed in positive mode. The ion source parameters were: capillary voltage 3 kV, aux gas flow rate 20, sheath gas flow rate 75, spray current 4 μA, capillary temperature 350°C, heater temperature 450°C, and S-lens RF 40. Nitrogen was used as the nebulizing gas in the ion trap source.
The MS/MS method was designed to perform an MS1 full-scan (scan range m/z 130–900) alternating with a MS/MS scan [AIF, all-ion fragmentation, scan range 60–600 m/z, normalized-collision energy (NCE) 25]. The mass resolution was set at 35,000, the AGC targets were 1 × 106 and 5 × 104 for the full scan and AIF modes, respectively, and injection time was 40 ms.
Standard AHLs (0.1 mg/mL in DMSO, 2 μL injected), were analyzed to determine the fragmentation patterns and retention time under our chromatographic conditions. As previous studies reported (Morin et al., 2003; Patel et al., 2016), we observed some characteristic fragments arising from fragmentation of the lactone ring. A daughter ion with m/z 102.055 is always observed, and three others ions with m/z 56.050, 74.061, and 84.045 are usually detected. These four daughter ions were chosen as the specific ions to indicate the presence of HSL-type compounds. To confirm the AHL identification, a second step of MS analyses was performed using SIM (resolution 3,500) and dd-ms2 (resolution 17,500) modes. Each ion of interest was isolated using an isolation width of 3.0 m/z and fragmented with a resolution of 17 500 and a collision energy of 20 eV. The presence of the daughter ion with 102.055 m/z and at least one other daughter ion that is characteristic of the lactone ring were needed to confirm AHL identification. This confirmation has also been conducted using a combination of 25 commercially available AHL standards with acyl chains that vary in their length, the nature of substituent on their third carbon (hydroxy or oxo group) and the presence of unsaturation (see more details of our methods in Girard et al., 2017 and Supplementary Table 2).
Results
Bacterial Strains Isolated from Posidonia oceanica
Sixty bacterial strains were isolated from P. oceanica (16SrRNA genes Accession numbers KY787128 to KY787185, KY697665 and KY697666, see Supplementary Table 1). Thirty-nine were from leaves (only epiphytic strains), and 21 were from rhizomes (both epiphytic and endophytic strains). The identified strains are affiliated with distinct groups of bacteria representing a broad spectrum of diversity, including Alphaproteobacteria (72%), Gammaproteobacteria (20%), Flavobacteriia (3%) Bacilli, and Actinobacteria (5%). Rhodobacteraceae-affiliated isolates were dominate among the Alphaproteobacteria fraction (55% of Alphaproteobacterial isolates), while isolates affiliated with the genus Marinomonas sp. dominated the Gammaproteobacteria (14%) (Figures 1, 2). With the second isolation protocol, we focused on the endophytic bacteria that were present in the Posidonia rhizomes. A similarly broad diversity pattern was observed with the endophytic strains, which belong to Alphaproteobacteria (70%), Gammaproteobacteria (20%), and Flavobacteriia (10%). The collected bacteria were also affiliated with a large diversity of genera, including Labrenzia sp. (17 strains), Cohaesibacter sp. (3 strains), Ruegeria sp. (3 strains), Pseudophaeobacter sp. (4 strains), Nisaea sp. (4 strains), Marinomonas sp. (4 strains), Vibrios sp. (3 strains), Micrococcus sp. (2 strains), Aquimarina sp. (2 strains), Microbulbifer sp. (1 strain), Alteromonas sp. (1 strain) Pseudovibrio sp. (1 strain), Flavivirga sp. (1 strain), Sphingopyxis sp. (3 strains), Sphingorhabdus sp. (1 strain), Kordiimonas sp. (1 strain), and Erythrobacter sp. (1 strain). Eight strains were found to be phylogenetically too distant from the known isolates to be affiliated with a specific genus with enough confidence (Figure 1).
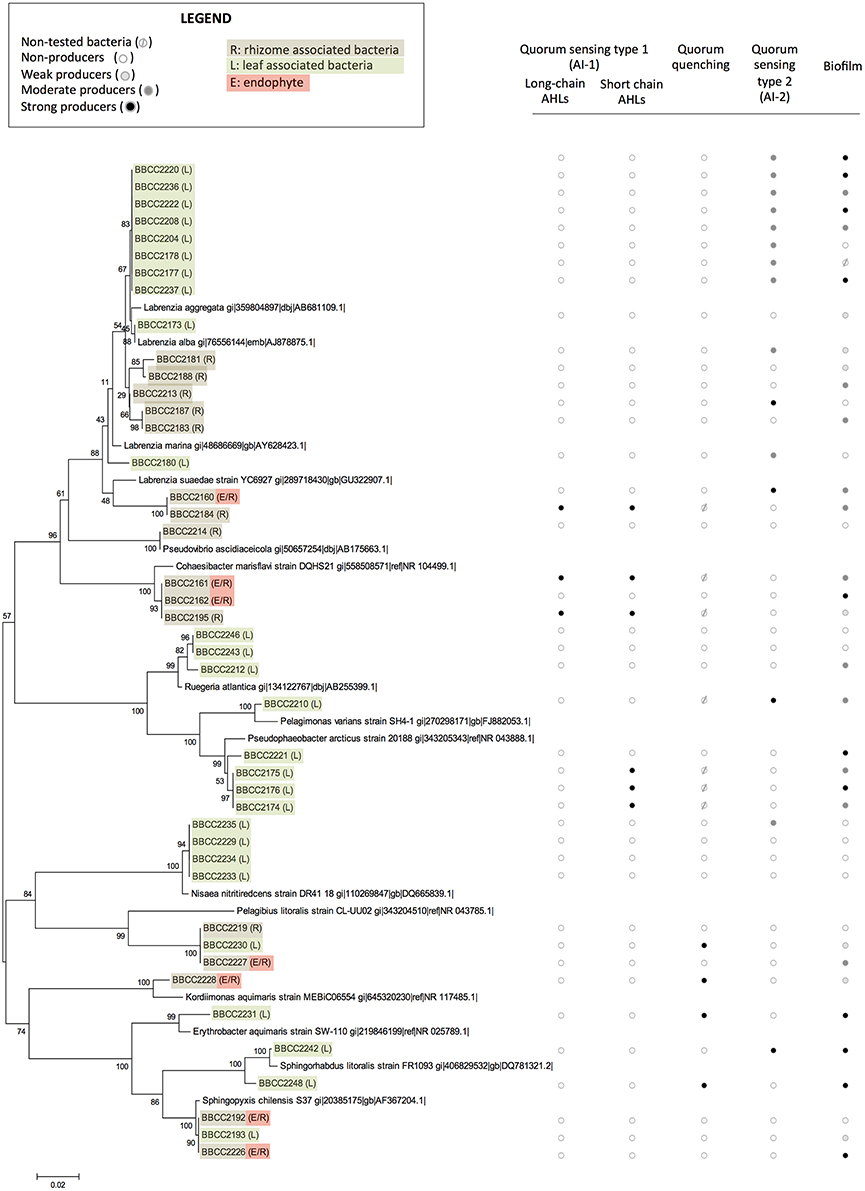
Figure 1. The phylogenetic affiliations of sequences of Alphaproteobacteria-related strains associated with Posidonia oceanica and their abilities to express quorum sensing, quorum quenching, or biofilm formation. The values at each node represent the bootstrap values generated using MEGA 6.0 [Neighbor-joining method with bootstrap (1,000 replications)]. Non-tested bacteria ( ), non-producers (
), non-producers ( ), weak producers (
), weak producers ( ), moderate producers (
), moderate producers ( ), and strong producers (
), and strong producers ( ) R: rhizome associated bacteria, L: leaf associated bacteria, and E: endophyte.
) R: rhizome associated bacteria, L: leaf associated bacteria, and E: endophyte.
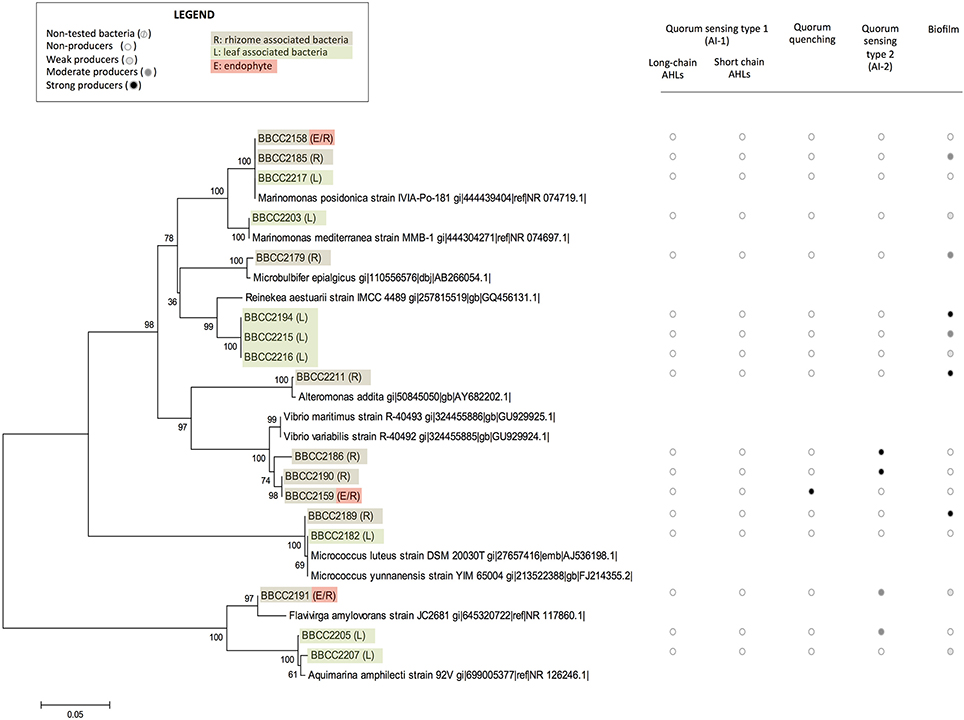
Figure 2. The phylogenetic affiliations of sequences of non-Alphaproteobacteria-related strains associated with Posidonia oceanica and their abilities to express quorum sensing, quorum quenching or biofilm formation. The values at each node represent the bootstrap values generated using MEGA 6.0 [Neighbor-joining method with bootstrap (1,000 replications)]. Non-tested bacteria ( ), non-producers (
), non-producers ( ), weak producers (
), weak producers ( ), moderate producers (
), moderate producers ( ), and strong producers (
), and strong producers ( ) R: rhizome associated bacteria, L: leaf associated bacteria, and E: endophyte.
) R: rhizome associated bacteria, L: leaf associated bacteria, and E: endophyte.
Diversity of Isolates Involved in Quorum Sensing
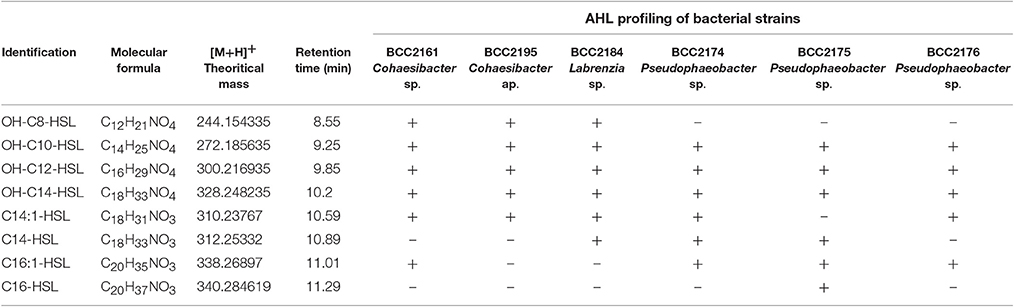
Among the 60 isolated strains, 6 were identified as AHL-producing isolates: 3 strains were double positive (responded to the P. putida F117 and the E.coli MT102 reporter strains), suggesting the production of long and short chain AHLs, and 3 strains gave rise only to a positive signal with the E. Coli MT102 reporter strain, revealing the production of short chain AHLs (Figure 1, and Datasheet S1). All six AHL-producing strains were isolated from both the rhizomes and leaves of P. oceanica and were affiliated with Alphaproteobacteria: 3 strains are related to to Pseudophaeobacter sp., 2 strains appeared phylogenetically related to the genus Cohaesibacter sp. and 1 strain of Labrenzia sp.
Among the whole isolates, the AI-2 biosensor V. harveyi BB170 responded positively to the supernatant of 19 isolated strains (Figures 1, 2, and Datasheet S1). The autoinducer secretion was measured by calculating the increased induction of bioluminescence using data collected at the optimal time point, i.e., the moment at which the control sample produced the least amount of light (Taga, 2005). A total of 6 strains were found to be strong producers of AI-2 and exhibited a >10-fold induction of bioluminescence, while 13 strains were weak producers with an induction between 5- and 10-fold. The AI-2 producers were found among isolates from Alphaproteobacteria (78%), Gammaproteobacteria (11%), and Flavobacteriia (11%). Among the Alphaproteobacteria, the genus Labrenzia seemed to be a large producer of AI-2, with 12 strains able to activate the AI-2 biosensor among the 17 that were isolated. Five of these strains were isolated from rhizomes, and 14 were associated with the leaves.
Diversity of Isolates Involved in Quorum Quenching
The bacterial ability to interfere in AHL-based quorum sensing was evaluated with the E. Coli MT102 reporter strain. Five strains (9%) produced quorum quenching signals. Four strains were related to Alphaproteobacteria (affiliated with the genera Kordiimonas, Erythrobacter, Sphingorhabdus, and an unknown Rhodospirillaceae). Additionally, one strain affiliated with Gammaproteobacteria (related to the genus Vibrio) was positive in our quorum quenching bioassay. Three of these strains were isolated in the leaves of Posidonia, and two were sampled from the rhizomes (Figures 1, 2 and Datasheet S1).
Capacity of Biofilm Formation
A crystal violet-based microplate assay was used to evaluate the capacity of microbial strains to attach to abiotic surfaces. We classified our isolates based on the calculated cut-off OD, resulting in the following categories: non-biofilm producers (OD < 3), weak biofilm producers (0.3 < OD < 0.6), moderate biofilm producers (0.6 < OD < 1.2), and strong biofilm producers (OD >1.4). A total of 67% of the P. oceanica-associated strains were able to produce biofilm (75% related to Alphaproteobacteria). Among them 11 strains are weak biofilm producers, 15 are moderate producers and 14 are strong biofilm producers (Figures 1, 2 and Datasheet S1). In contrast, 19 strains did not produce a biofilm. We observed that closely phylogenetically related strains responded differently to this biotest. For example, among the16 isolated strains related to the Labrenzia genus, 3 did not produce biofilm, while 3, 6, and 4 of the isolated strains related to the Labrenzia genus were either weak, moderate or strong biofilm producers, respectively. Interestingly, OD values above 1 were observed for the 4 strains that are related to Pseudophaeobacter sp. (and isolated from P. oceanica leaves), while the 4 strains related to Nisaea sp. were unable to produce biofilm. All strains identified as AHL producers were also, according to our bioassays, able to produce biofilms, while 58% of the strains involved in AI-2-based quorum sensing were able to synthesize a biofilm (Figures 1, 2 and Datasheet S1).
Diversity of the Emitted Acylhomoserine Lactones
We combined HPLC fractionation and UHPLC-HRMS analyses to identify the diversity of the AHLs secreted by the 6 bacterial strains that our biosensors identified as positive. Our method consists of a non-targeted UHPLC-MS/MS analysis to identify AHLs in microbial samples. Our chromatographic method yielded good separation, and the mass accuracy for all standards was within 3 ppm (parts per million). The analytical method includes full-scan MS with successive all-ion fragmentation (AIF) MS/MS scan allowing the acquisition high resolution data of the parent ion and daughter ion masses in a single chromatographic run. As described by Patel et al. (2016), four characteristic fragments originating from the lactone ring in AHLs were detected in almost all of the MS/MS spectra of the AHL standards (Girard et al., 2017). One daughter ion was abundant and always present in fragmentation patterns: the 102.055 m/z ion, which corresponds to the lactone ring itself; three other ions that arose from the fragmentation of the lactone ring (the 56.050, 74.061, and 84.045 m/z fragments) were usually observed. These daughter ions were sought in the MS/MS scans of bacteria fractions to detect HSL-type compounds. All AHLs were also confirmed with commercially available standards.
Thus, we determine that the 3 strains related to Pseudophaeobacter sp. (BBCC2174, 2175, and 2176) produced 5–6 different AHLs with acyl side chains harboring 10–16 carbons with unsaturated regions or/and hydroxyl substitution (Table 1). The two Cohaesibacter strains, BBCC2161 and BBCC2195, as well as the Labrenzia strain BBCC2184 produce respectively 6, 5, and 6 AHLs with acyl chain harboring between 8 and 16 carbons; in which unsaturated regions or/and hydroxyl substitution could be found too. Because of the use of our non-targeted UHPLC-MS/MS method allowing the detection of known and novel AHLs and because of the presence in MS/MS spectra of the daughter ions characteristic of the lactone ring, we also suspect the presence of many other HSL-type compounds. However, the confirmation of such result is highly difficult. Especially, the Nuclear Magnetic Resonance step requires large amounts of pure compounds, while AHLs are emitted at very low concentrations (Table 1).
Discussion
In this study, we identified a cultivable bacterial diversity associated with the collected Mediterranean P. oceanica, largely dominated by Alphaproteobacteria and Gammaproteobacteria. This observation correlates with previous studies, which found a dominance of Proteobacteria and Bacteroidetes in endophytic communities of P. oceanica using DNA-based inventories of bacterial diversity (Garcias-Bonet et al., 2012). Many hypotheses could be raised to explain our difficulty to cultivate some Bacteroidetes. One possibility is the light conditions which were not appropriate in our experiments, as it has been previously shown that at least some of these lineages are proteorhodopsins-containing cells (Fernandez-Gomez et al., 2013). However, similar bacteria lineages were also found in epiphytic bacterial communities of seaweeds (Egan et al., 2013) and in the core microbiota of seagrass roots, which appears dominated by Gammaproteobacteria (Egan et al., 2013) represented in our study by three strains affiliated with Marinomonas posidonica, and one strain affiliated with M. mediterranea. Previous studies revealed that microorganisms within the genus Marinomonas can be readily isolated from the different tissues of P. oceanica (Espinosa et al., 2010; Lucas-Elio et al., 2011), including Marinomonas balnearica, pollencensis, mediterranea, and posidonica. Marinomonas posidonica is frequently detected in the microbiota of P. oceanica, and some authors suggest an ancient relationship between these two species (Lucas-Elio et al., 2011), including the bacteria promoting the growth of the angiosperm (Celdran et al., 2012).
In these bacterial communities, we detected many strains that are able to produce AHLs. These cells are phylogenetically related to Alphaproteobacteria and, in particular, to the Rhodobacteraceae and the Cohaesibacteraceae families. Such observation is in line with previous studies, as many Rhodobacteraceae strains are known to be involved in quorum sensing (Gram et al., 2002; Cude and Buchan, 2013; Zan et al., 2014) but it is the first time that Cohaesibacter sp. strains are shown to be involved in quorum sensing. In our culture collection, we also detected 19 isolates capable of an AI-2-based quorum sensing, including many strains affiliated with the genus Labrenzia sp., which appears to be an original observation for this taxonomic group (Pereira et al., 2013). However, this result must be used cautiously, especially within the Alphaproteobacteria group, as AI-2 could simply be a side product of the methyl cycle and not a molecular signal in some strains (Rezzonico and Duffy, 2008; Hmelo, 2017). We also reported two Vibrio sp. strains that were identified as positive by our AI-2 bioassay (BBCC2186 and BBCC2190), in line with previous studies mentioning the existence of quorum sensing regulations in this genus (Bassler et al., 1997; Bassler, 1999; Milton, 2006). The role of AI-2 signaling remains unclear, but this semiochemical is hypothesized to be involved in interspecies communication (Hmelo, 2017).
The percentage of bacteria able to AHL-based quorum sensing in our strain collection could appear low (10% of total strains), despite the fact that we used a combination of biosensors that are able to detect a wide range of AHLs (Wagner-Döbler et al., 2005; Steindler and Venturi, 2007). However, we probably underestimated the real number of AHL-producing strains, since culture conditions may influence the expression of quorum sensing (Bhedi et al., 2017). However, despite these limitations, our data clearly indicate that a noticeable fraction of bacteria associated with the P. oceanica microbiota are able to communicate using an AHL- or AI-2-based quorum sensing.
Our UHPLC-HRMS analyses revealed the secretion of a large variety of AHLs by the isolated strains. For example, the three strains affiliated with Pseudophaeobacter produced 3-OH-C10-HSL, as detected in previous studies that focused on Phaeobacter gallaeciensis (Berger et al., 2011). Similarly, the C14:1-HSL that was detected in some of our isolated Alphaproteobacteria was also detected in Rhodobacter sphaeroides (Puskas et al., 1997). Interestingly, our results are consistent with previous reports showing that marine Alphaproteobacteria tend to preferentially produce long-acyl chain rather than short-acyl chain AHLs (Wagner-Döbler et al., 2005). Additionally, many of the detected AHLs were found to frequently possess −OH groups. Two reasons for these observations may be that long chains are known to increase HSL half-lives in saltwater environments (Hmelo and Van Mooy, 2009) and OH substitution might favor AHL solubility. These features might reveal an adaptation to marine environments.
In addition to the production of a 3-OH-C10-AHL, the three strains affiliated with Pseudophaeobacter were also identified as producers of 3-0H-C12-HSL, 3-0H-C14-HSL, and C16:1-HSL. However, the BBCC2175 strain, also affiliated with Pseudophaeobacter, was readily identified as being able to produce C16-HSL and the only one to not produce C14:1-HSL. Previous studies revealed that the patterns of AHL production can differ between phylogenetically closely related strains (Zimmer et al., 2014; Rajput et al., 2016). Such patterns may result from the selective pressure that occurs when related organisms share the same ecological niche (Steidle et al., 2001). Conversely, bacteria from different genera producing similar types of AHL compounds may promote interspecies communication in the mixed communities of a natural environment (Viswanath et al., 2015).
Interestingly, we also detected a few strains that are capable of quorum quenching. A total of 9% of the strains in our collection produced compounds that led to a reduction of quorum sensing signals, including four strains affiliated with Alphaproteobacteria and one affiliated with Gammaproteobacteria. Such quorum quenching activity has previously been found in strains isolated from various types of marine ecological niches. It is well-established that quorum quenching relies on many different types of biological processes, including the enzymatic degradation of AHLs by lactonases or acylases or analog production mimicking the signaling compounds without inducing the biological activity (Rasmussen et al., 2000). We suspect that this latter mechanism is involved in the quorum quenching signals of our strains, as we executed our tests with extracts prepared in ethyl acetate, thus denaturing proteins and avoiding any kind of enzyme-based activity. The report of quorum quenching activities in P. oceanica-associated bacteria also opens interesting perspectives based on these biochemical compounds, once their structures are elucidated, that could represent important biotechnological potential (Uroz et al., 2009).
From a wider perspective, our data raise the question of the ecological role of quorum sensing and quorum quenching in the microbiota of P. oceanica. It is known that quorum sensing is involved in biofilm formation mechanisms in many model strains and that this biofilm colonization is a physiological trait that may favor niche colonization (Rolland et al., 2016). Thus, we evaluated the capacity of our isolates to produce biofilms using the classic crystal violet-based biotest. Interestingly, all the strains identified as being involved in AHL-based quorum sensing were also able to produce biofilms. Such results do not imply a role for quorum sensing in biofilm development for these strains, as we also detected many other non-AHL producing strains that are able to biofilm formation. However, the regulation of biofilm production by quorum sensing, if demonstrated in future studies, might increase the fitness of bacteria inhabiting the Posidonia microbiota.
Collectively, our data constitute the first report of quorum sensing and quorum quenching mechanisms in bacterial strains associated with the seagrass P. oceanica. Our data reveals a large diversity of AHLs involved in the quorum sensing mechanisms and the existence of AI-2-based communication systems. Our study underscores the need to consider bacterial interactions in the Posidonia-associated microbiota, with the ultimate goal of better understanding Posidonia-bacteria relationships and the physiology of a key angiosperm species in the seagrass ecosystem.
Author Contributions
EB, JO, YF, NB, and LI performed the experiments. SP, AE, and RL designed the experiments. All authors wrote the MS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sorbonne Universités (ENDOQUO project) for funding, as well as the ANR (Agence Nationale de la Recherche, project SECIL, ANR-15-CE21-0016). We also thank the BIO2MAR platform and Karine Escoubeyrou for assistance, as well as Jeanne Naudet and Austin Showalter for technical help.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fmars.2017.00218/full#supplementary-material
References
Andersen, J. B., Heydorn, A., Hentzer, M., Eberl, L., Geisenberger, O., Christensen, B. B., et al. (2001). gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67, 575–585. doi: 10.1128/AEM.67.2.575-585.2001
Bassler, B. L. (1999). How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2, 582–587. doi: 10.1016/S1369-5274(99)00025-9
Bassler, B. L., Greenberg, E. P., and Stevens, A. M. (1997). Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179, 4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997
Berger, M., Neumann, A., Schulz, S., Simon, M., and Brinkhoff, T. (2011). Tropodithietic acid Production in Phaeobacter gallaeciensis is regulated by N-acyl homoserine lactone-mediated quorum sensing. J. Bacteriol. 193, 6576–6585. doi: 10.1128/JB.05818-11
Bhedi, C. D., Prevatte, C. W., Lookadoo, M. S., Waikel, P. A., Gillevet, P. M., Sikaroodi, M., et al. (2017). Elevated temperature enhances short to medium chain acyl homoserine lactone production by black band disease associated vibrios. FEMS Microbiol. Ecol. 93:fix005. doi: 10.1093/femsec/fix005
Celdran, D., Espinosa, E., Sanchez-Amat, A., and Marin, A. (2012). Effects of epibiotic bacteria on leaf growth and epiphytes of the seagrass Posidonia oceanica. Mar. Ecol. Prog. Ser. 456, 21–27. doi: 10.3354/meps09672
Cúcio, C., Engelen, A. H., Costa, R., and Muyzer, G. (2016). Rhizosphere microbiomes of european+ seagrasses are selected by the plant, but are not species specific. Front. Microbiol. 7:440. doi: 10.3389/fmicb.2016.00440
Cude, W. N., and Buchan, A. (2013). Acyl-homoserine lactone-based quorum sensing in the Roseobacter clade: complex cell-to-cell communication controls multiple physiologies. Front. Microbiol. 4:336. doi: 10.3389/fmicb.2013.00336
Defoirdt, T., Boon, N., Bossier, P., and Verstraete, W. (2004). Disruption of bacterial quorum sensing: an unexplored strategy to fight infections in aquaculture. Aquaculture 240, 69–88. doi: 10.1016/j.aquaculture.2004.06.031
Dong, Y.-H., and Zhang, L.-H. (2005). Quorum sensing and quorum-quenching enzymes. J. Microbiol. 43, 101–109.
Egan, S., Harder, T., Burke, C., Steinberg, P., Kjelleberg, S., and Thomas, T. (2013). The seaweed holobiont: understanding seaweed-bacteria interactions. FEMS Microbiol. Rev. 37, 462–476. doi: 10.1111/1574-6976.12011
Espinosa, E., Marco-Noales, E., Gómez, D., Lucas-Elío, P., Ordax, M., Garcías-Bonet, N., et al. (2010). Taxonomic study of Marinomonas strains isolated from the seagrass Posidonia oceanica, with descriptions of Marinomonas balearica sp. nov. and Marinomonas pollencensis sp. nov. Int. J. Syst. Evol. Microbiol. 60, 93–98. doi: 10.1099/ijs.0.008607-0
Fernandez-Gomez, B., Richter, M., Schuler, M., Pinhassi, J., Acinas, S. G., Gonzalez, J. M., et al. (2013). Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 7, 1026–1037. doi: 10.1038/ismej.2012.169
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275. doi: 10.1128/jb.176.2.269-275.1994
Garcias-Bonet, N., Arrieta, J., De Santana, C., Duarte, C., and Marbà, N. (2012). Endophytic bacterial community of a Mediterranean marine angiosperm (Posidonia oceanica). Front. Microbiol. 3:342. doi: 10.3389/fmicb.2012.00342
Girard, L., Blanchet, E., Intertaglia, L., Baudart, J., Stien, D., Suzuki, M., et al. (2017). Characterization of N-Acyl Homoserine Lactones in Vibrio tasmaniensis LGP32 by a biosensor-based UHPLC-HRMS/MS method. Sensors 17:906. doi: 10.3390/s17040906
Gram, L., Grossart, H. P., Schlingloff, A., and Kiorboe, T. (2002). Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68, 4111–4116. doi: 10.1128/AEM.68.8.4111-4116.2002
Hemminga, M. A., and Duarte, C. M. (2000). Seagrass Ecology. Cambridge, UK: Cambridge University Press.
Hmelo, L. R. (2017). Quorum sensing in marine microbial environments. Ann. Rev. Mar. Sci. 9, 257–281. doi: 10.1146/annurev-marine-010816-060656
Hmelo, L., and Van Mooy, B. A. S. (2009). Kinetic constraints on acylated homoserine lactone-based quorum sensing in marine environments. Aquat. Microb. Ecol. 54, 127–133. doi: 10.3354/ame01261
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lucas-Elio, P., Marco-Noales, E., Espinosa, E., Ordax, M., Lopez, M. M., Garcias-Bonet, N., et al. (2011). Marinomonas alcarazii sp. nov., M. rhizomae sp. nov., M. foliarum sp. nov., M. posidonica sp. nov. and M. aquiplantarum sp. nov., isolated from the microbiota of the seagrass Posidonia oceanica. Int. J. Syst. Evol. Microbiol. 61, 2191–2196. doi: 10.1099/ijs.0.027227-0
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Milton, D. L. (2006). Quorum sensing in vibrios: complexity for diversification. Int. J. Med. Microbiol. 296, 61–71. doi: 10.1016/j.ijmm.2006.01.044
Moreira, J. M. R., Gomes, L. C., Araujo, J. D. P., Miranda, J. M., Simoes, M., Melo, L. F., et al. (2013). The effect of glucose concentration and shaking conditions on Escherichia coli biofilm formation in microtiter plates. Chem. Eng. Sci. 94, 192–199. doi: 10.1016/j.ces.2013.02.045
Morin, D., Grasland, B., Vallee-Rehel, K., Dufau, C., and Haras, D. (2003). On-line high-performance liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J. Chromatogr. A 1002, 79–92. doi: 10.1016/S0021-9673(03)00730-1
Nealson, K. H. (1977). Autoinduction of bacterial luciferase. Occurrence, mechanism and significance. Arch. Microbiol. 112, 73–79. doi: 10.1007/BF00446657
Orth, R. J., Carruthers, T. J. B., Dennison, W. C., Duarte, C. M., Fourqurean, J. W., Heck, K. L., et al. (2006). A global crisis for seagrass ecosystems. Bioscience 56, 987–996. doi: 10.1641/0006-3568(2006)56[987:AGCFSE]2.0.CO;2
O'Toole, G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. e2437. doi: 10.3791/2437
Patel, N. M., Moore, J. D., Blackwell, H. E., and Amador-Noguez, D. (2016). Identification of unanticipated and novel N-acyl L-homoserine lactones (AHLs) using a sensitive non-targeted LC-MS/MS method. PLoS ONE 11:e0163469. doi: 10.1371/journal.pone.0163469
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Puskas, A., Greenberg, E. P., Kaplan, S., and Schaefer, A. L. (1997). A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J. Bacteriol. 179, 7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997
Rajput, A., Kaur, K., and Kumar, M. (2016). SigMol: repertoire of quorum sensing signaling molecules in prokaryotes. Nucleic Acids Res. 44, D634–D639. doi: 10.1093/nar/gkv1076
Rasmussen, T. B., and Givskov, M. (2006). Quorum sensing inhibitors: a bargain of effects. Microbiology 152, 895–904. doi: 10.1099/mic.0.28601-0
Rasmussen, T. B., Manefield, M., Andersen, J. B., Eberl, L., Anthoni, U., Christophersen, C., et al. (2000). How Delisea pulchra furanones affect quorum sensing and swarming motility in Serratia liquefaciens MG1. Microbiology 146, 3237–3244. doi: 10.1099/00221287-146-12-3237
Rezzonico, F., and Duffy, B. (2008). Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 8:154. doi: 10.1186/1471-2180-8-154
Riedel, K., Hentzer, M., Geisenberger, O., Huber, B., Steidle, A., Wu, H., et al. (2001). N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147, 3249–3262. doi: 10.1099/00221287-147-12-3249
Rolland, J. L., Stien, D., Sanchez-Ferandin, S., and Lami, R. (2016). Quorum sensing and quorum quenching in the phycosphere of phytoplankton: a case of chemical interactions in ecology. J. Chem. Ecol. 42, 1201–1211. doi: 10.1007/s10886-016-0791-y
Smith, R. S., and Iglewski, B. H. (2003). P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6, 56–60. doi: 10.1016/S1369-5274(03)00008-0
Steidle, A., Sigl, K., Schuhegger, R., Ihring, A., Schmid, M., Gantner, S., et al. (2001). Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67, 5761–5770. doi: 10.1128/AEM.67.12.5761-5770.2001
Steindler, L., and Venturi, V. (2007). Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 266, 1–9. doi: 10.1111/j.1574-6968.2006.00501.x
Stepanovic, S., Vukovic, D., Dakic, I., Savic, B., and Svabic-Vlahovic, M. (2000). A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40, 175–179. doi: 10.1016/S0167-7012(00)00122-6
Taga, M. E. (2005). Methods for analysis of bacterial autoinducer-2 production. Curr. Protoc. Microbiol. Chapter 1, Unit 1C.1. doi: 10.1002/9780471729259.mc01c01s00
Turner, T. R., James, E. K., and Poole, P. S. (2013). The plant microbiome. Genome Biol. 14:209. doi: 10.1186/gb-2013-14-6-209
Uroz, S., Dessaux, Y., and Oger, P. (2009). Quorum sensing and quorum quenching: the yin and yang of bacterial communication. Chembiochem 10, 205–216. doi: 10.1002/cbic.200800521
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., and Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Viswanath, G., Jegan, S., Baskaran, V., Kathiravan, R., and Prabavathy, V. R. (2015). Diversity and N-acyl-homoserine lactone production by Gammaproteobacteria associated with Avicennia marina rhizosphere of South Indian mangroves. Syst. Appl. Microbiol. 38, 340–345. doi: 10.1016/j.syapm.2015.03.008
Wagner-Döbler, I., Thiel, V., Eberl, L., Allgaier, M., Bodor, A., Meyer, S., et al. (2005). Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine alphaproteobacteria. Chembiochem 6, 2195–2206. doi: 10.1002/cbic.200500189
Zan, J., Liu, Y., Fuqua, C., and Hill, R. T. (2014). Acyl-homoserine lactone quorum sensing in the Roseobacter clade. Int. J. Mol. Sci. 15, 654–669. doi: 10.3390/ijms15010654
Zimmer, B. L., May, A. L., Bhedi, C. D., Dearth, S. P., Prevatte, C. W., Pratte, Z., et al. (2014). Quorum sensing signal production and microbial interactions in a polymicrobial disease of corals and the coral surface mucopolysaccharide layer. PLoS ONE 9:e108541. doi: 10.1371/journal.pone.0108541
Keywords: microbiota, quorum sensing, quorum quenching, acyl-homoserine lactone, seagrass, Posidonia oceanica
Citation: Blanchet E, Prado S, Stien D, Oliveira da Silva J, Ferandin Y, Batailler N, Intertaglia L, Escargueil A and Lami R (2017) Quorum Sensing and Quorum Quenching in the Mediterranean Seagrass Posidonia oceanica Microbiota. Front. Mar. Sci. 4:218. doi: 10.3389/fmars.2017.00218
Received: 24 April 2017; Accepted: 23 June 2017;
Published: 21 July 2017.
Edited by:
Russell T. Hill, Institute of Marine and Environmental Technology, University of Maryland Center for Environmental Science, United StatesReviewed by:
Ilana Kolodkin-Gal, Weizmann Institute of Science, IsraelJulie L. Meyer, University of Florida, United States
Copyright © 2017 Blanchet, Prado, Stien, Oliveira da Silva, Ferandin, Batailler, Intertaglia, Escargueil and Lami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raphaël Lami, raphael.lami@obs-banyuls.fr
 Elodie Blanchet
Elodie Blanchet Soizic Prado2
Soizic Prado2  Didier Stien
Didier Stien Laurent Intertaglia
Laurent Intertaglia Alexandre Escargueil
Alexandre Escargueil Raphaël Lami
Raphaël Lami