Metabolic Remodeling of Membrane Glycerolipids in the Microalga Nannochloropsis oceanica under Nitrogen Deprivation
- 1Center for Microalgal Biofuels and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 2SDIC Microalgae Biotechnology Center, China Electronics Engineering Design Institute, State Development and Investment Corporation, Beijing, China
- 3CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing, China
- 4Laboratory for Algae Research and Biotechnology, Department of Applied Biological Sciences, Arizona State University, Mesa, AZ, United States
- 5CAS Key Laboratory of Biofuels and Shandong Key Laboratory of Energy Genetics and BioEnergy Genome Center, Single-Cell Center, Qingdao Institute of BioEnergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, China
- 6Key Laboratory of Algal Biology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
HIGHLIGHTS
• An electrospray ionization mass spectrometry-based lipidomics method was developed and integrated with transcriptomics to elucidate metabolic remodeling and turnover of microalgal membrane lipids by using Nannochloropsis oceanica as a model.
The lack of lipidome analytical tools has limited our ability to gain new knowledge about lipid metabolism in microalgae, especially for membrane glycerolipids. An electrospray ionization mass spectrometry-based lipidomics method was developed for Nannochloropsis oceanica IMET1, which resolved 41 membrane glycerolipids molecular species belonging to eight classes. Changes in membrane glycerolipids under nitrogen deprivation and high-light (HL) conditions were uncovered. The results showed that the amount of plastidial membrane lipids including monogalactosyldiacylglycerol, phosphatidylglycerol, and the extraplastidic lipids diacylglyceryl-O-4′-(N, N, N,-trimethyl) homoserine and phosphatidylcholine decreased drastically under HL and nitrogen deprivation stresses. Algal cells accumulated considerably more digalactosyldiacylglycerol and sulfoquinovosyldiacylglycerols under stresses. The genes encoding enzymes responsible for biosynthesis, modification and degradation of glycerolipids were identified by mining a time-course global RNA-seq data set. It suggested that reduction in lipid contents under nitrogen deprivation is not attributable to the retarded biosynthesis processes, at least at the gene expression level, as most genes involved in their biosynthesis were unaffected by nitrogen supply, yet several genes were significantly up-regulated. Additionally, a conceptual eicosapentaenoic acid (EPA) biosynthesis network is proposed based on the lipidomic and transcriptomic data, which underlined import of EPA from cytosolic glycerolipids to the plastid for synthesizing EPA-containing chloroplast membrane lipids.
Introduction
Microalgae are emerging as a promising feedstock for biofuels and non-fuel lipids owing to their rapid growth and high cellular lipid content, rich particularly in triacylglycerol (TAG) (Hu et al., 2008; Wijffels and Barbosa, 2010). However, the lipid productivity is inevitably compromised by the impaired photosynthesis and retarded growth when the typical stress conditions [e.g., high-light (HL) and nitrogen (N) deprivation] are imposed to induce TAG formation. This is largely attributable to the breakdown and remodeling of polar glycerolipid-based cell membranes, particularly thylakoid membranes under stress conditions. Numerous previous studies have shown that membrane glycerolipids are susceptible to HL or N deprivation in microalgae (Khotimchenko and Yakovleva, 2005; Yoon et al., 2012; Goncalves et al., 2013; Simionato et al., 2013), but the molecular mechanisms underlying biosynthesis and turnover of membrane lipids have remained largely unexplored.
Nannochloropsis is a genus of heterokont unicellular microalgae that have been intensively explored for fuel and animal feed applications. Nannochloropsis species can accumulate substantial amounts of eicosapentaenoic acid (EPA), a high-value polyunsaturated fatty acid, when environmental conditions are favorable for vegetative growth, but synthesize large quantities of TAG under stress (Sukenik, 1991; Zittelli et al., 1999; Rodolfi et al., 2009; Bondioli et al., 2012; Wang et al., 2012). As one of the few production species with their genomes sequenced and genetic tools developed, Nannochloropsis is also being utilized as a model species for investigation of lipid metabolism (Kilian et al., 2011; Radakovits et al., 2012; Vieler et al., 2012; Corteggiani Carpinelli et al., 2013; Liang et al., 2013; Simionato et al., 2013; Xiao et al., 2013; Li et al., 2014; Wang et al., 2014; Jia et al., 2015). Recently, gene annotation led to the discovery that the number of genes involved in lipid metabolism is expanded in this organism (Wang et al., 2014), as compared with the well-established model organisms, like the green alga Chlamydomonas reinhardtii, the diatom Phaeodactylum tricornutum and the brown alga Ectocarpus siliculosus (Radakovits et al., 2012; Jinkerson et al., 2013). Genome-wide gene expression analyses of Nannochloropsis species/strains were conducted in several independent studies with a focus on identification of key regulatory components controlling fatty acid and TAG biosynthesis (Vieler et al., 2012; Corteggiani Carpinelli et al., 2013; Liang et al., 2013; Hu et al., 2014; Li et al., 2014; Wang et al., 2014). Recently, an integrative omics approach was used to investigate remodeling of membrane lipids of Nannochloropsis under changing light regimes (Alboresi et al., 2016). The results showed that HL induced accumulation of extraplastidic lipid diacylglyceryl-O-4′-(N, N, N,-trimethyl) homoserine (DGTS) and caused substantial breakdown of chloroplast polar lipids, which correlated with up-regulation of a number of genes encoding cytosolic central carbon metabolism components (e.g., fatty acid biosynthesis) and the parallel inhibition of their chloroplast counterpart. Regarding their biological and biotechnical significance, metabolic remodeling of microalgal membrane lipids in response to a variety of environmental stimuli (e.g., nutrient starvation and excess light) remains to be investigated in a more broader context.
Despite the transcriptomic and proteomic studies that have provided valuable insights into lipid metabolism and regulation in microalgae (Corteggiani Carpinelli et al., 2013; Dong et al., 2013; Liang et al., 2013; Simionato et al., 2013; Li et al., 2014), changes at mRNA and protein level are not necessarily always reflective of the metabolic flux. Therefore, direct measurements of the end products and intermediates in lipid biosynthesis pathways are necessary to develop a better understanding of the function and global regulation of lipid metabolism. The most commonly used method for algal lipid analysis is gas chromatography-mass spectrometry (GC-MS), which resolves the fatty acid composition of a given lipid class. This method involves separation of lipids based on polarity by using thin-layered liquid chromatography (TLC), followed by recovery of the separated lipid classes for GC-MS analysis. However, application of this approach in high-throughput studies is limited by the relatively time-consuming and labor-intensive preparation processes. In addition, changes in those minor constituent lipids and intermediates among treatments could rarely be detected by employing this approach, due to possible sample loss and incomplete lipid separation/detection in the TLC. Moreover, GC-MS cannot been employed to measure the cellular content of a specific glycerolipid molecule, and thus offers few advantages in complementing the efforts in functional genomics studies.
Mass spectrometry (MS)-based lipidomics is a powerful tool for quantitative measurement of lipids and assists in describing the dynamics of lipid metabolism at the molecule species level (Han and Gross, 2003, 2005; Ejsing et al., 2009; Li et al., 2014). This technique has been attempted to study lipid compositions of microalgae (Vieler et al., 2007; Xu et al., 2010; Lee et al., 2013; Liu et al., 2013), but absolute quantification of individual lipid molecular species was rarely reported. The aim of this study is to develop an advanced MS/MS-based lipidomics platform for quantifying the glycerolipids in microalgae, using Nannochloropsis oceanica IMET1 as a model system. To gain better insights into the regulation and physiological role of membrane glycerolipids synthesis, particularly under N deprivation conditions, lipidomic and mRNA-seq-based transcriptomic analyses were conducted. It revealed that the membrane glycerolipids, especially galactolipids, underwent considerable degradation and remodeling under N deprivation condition, accompanied by the biosynthesis pathway-specific regulation at the gene expression level, making the cells more tolerant to stress.
Materials and Methods
Nannochloropsis Strain and Culture Conditions
Cells of N. oceanica IMET1 were inoculated in 800 mL modified f/2 medium in a 1000 mL glass column, with the initial biomass concentration of 1.3 g L−1. Algal cell cultures were supplied with 1.5% CO2 (v/v) and 50 μmol photons m−2 s−1 light for 4 days prior to N-depletion or HL treatments. For N-depletion experiment, cells were harvested by centrifugation at 3,000 g for 5 min and resuspended in N-free f/2 medium followed by 4-day cultivation under the same light conditions. For HL experiments, cells were exposed into an irradiance of 200 μmol photons m−2 s−1 and were grown for 4 days. The temperature for all the cultures was maintained at 25°C.
Shotgun Mass Spectrometry for Glycerolipid Identification
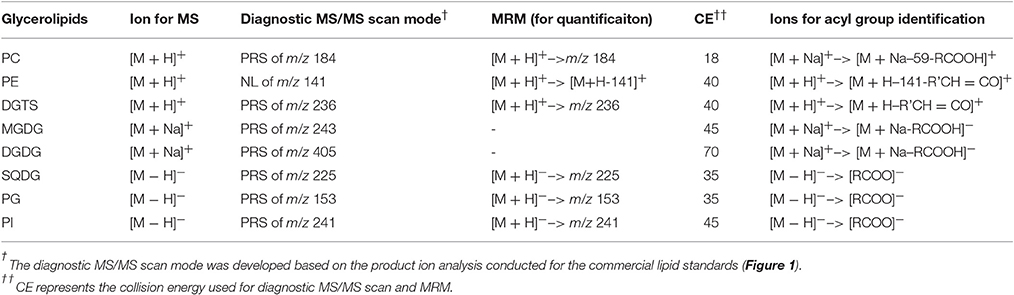
Lipids were extracted from 10 mg lypholized cells and recovered in 2 mL of chloroform:methanol (1:1, v:v) according to the previously described method (Yoon et al., 2012). Lipid extracts were diluted by 10-fold in chloroform:methanol (1:1, v/v) and were analyzed on an electrospray triple-quadruple mass spectrometry equipped with a high performance LC (Agilent, USA). For shotgun analysis, samples were directly infused into a mass spectrometer by loop injections with methanol as a mobile phase at a flow rate of 0.1 mL min−1. Two types of tandem MS, i.e., precursor-ion and neutral loss scan, were employed to identify lipid molecular species belonging to a given class. The diagnostic ions and scan mode for identification of each class of lipids are listed in Table 1. Product ion scan mode was used to identify acyl groups.
Quantitative LC-MS/MS
MGDG 36:0 (18:0/18:0), DGDG 36:0 (18:0/18:0), PE 31:1 (14:1/17:0), and PG 37:4 (17:0/20:4) were used as internal standards (ITSD) for MGDG, DGDG, PE, and PG quantification, respectively. PC 37:4 (17:0/20:4) was used as ITSD for both PC and DGTS quantification, and PI 37:4 (17:0/20:4) for both PI and SQDG quantification. MGDG 34:6 (16:3/18:3), DGDG 36:3 (18:3/18:3), PE 36:1 (18:0/18:1), PG 36:1 (18:0/18:1), PC 36:2 (18:1/18:1), and DGTS 32:0 (16:0/16:0) were used as calibration standards for their corresponding lipid class. All the standards for phospholipids and DGTS standards were purchased from Avanti Polar Lipids (Atlanta, USA). SQDG were ordered from Indofine Chemical (New Jersey, USA). ITSD and calibration standards of MGDG and DGDG were ordered from Matreya (Pennsylvania, USA) and Avanti Polar Lipids, respectively.
Positive ion mode ESI-MS was applied for detection of MGDG, and DGDG in the form of [M + NH4]+; PC, PE, and DGTS in the form of [M+H]+, respectively with a prior separation on a ZOBARX SBC18 column (particle size 1.8 μm, internal diameter 2.1 mm, length 150 mm, Agilent). The mobile phase for this mode was composed of A: methanol: acetonitrile: H2O (19: 19: 2, v/v/v) and B: isopropanol, both containing 10 mm ammonium acetate and 0.1% (w/v) formic acid. The following gradient was used: starting from isocratic elution by 90% A: 10% B for 5 min before a linearly 20-min gradient elution to 60% A: 40% B, followed by another 35-min linearly gradient to 45% A: 55% B and held for 2 min. For PI, PG and SQDG, negative ion mode was applied and an Extend C18 column (particle size 1.8 μm, internal diameter 2.1 mm, length 150 mm, Agilent) was used for separation. For negative mode, the mobile phase consists of A: methanol: acetonitrile: water (100: 100: 32, v/v/v) and B: isopropanol. Both A and B contained 0.025% (w/v) ammonium hydroxide. The elution gradient was as follows: a linear 10-min gradient from 100 to 95% A, followed by another linear gradient from 95 to 45% A within 2 min and then an 18-min isocratic elution at 45% A in B. Finally, mobile phase A went back to the initial gradient (100%) within 2 min. The flow rate of the mobile phase for both modes was 0.2 mL min−1. The column was maintained at 40°C.
mRNA Seq Analysis
A 48 h time course experiment of N-depletion was performed to investigate changes in transcripts of the genes involved in membrane lipid metabolism. The growth curve under N-depleted conditions showed that the endogenous N in IMET1 cells was completely consumed approximately 24 h after being transferred into the N- f/2 medium, as indicated by the arrested cell division (Supplemental Figure 1). To capture the dynamics of transcripts after the depletion of N both in the growth medium and within the algal cells, cell aliquots were collected for RNA isolation after being transferred for the six time points of 3, 4, 6, 12, 24, and 48 h. Total algal RNA was extracted using Trizol reagents (Invitrogen). For mRNA-Seq, the poly (A)-containing mRNA molecules were purified using Sera-mag Magnetic Oligo (dT) Beads (Thermo Scientific) and libraries were prepared according to the Illumina protocol. Sequencing was performed on an Illumina HiSeq2000 with 90 bp paired-end reads. Raw data processing and differential gene expression analysis were performed as a previous study described (Li et al., 2014).
Statistical Tests
Student's t-test was used to compare the lipid contents between two data points (N-deprived and N-replete, n = 4~6). If the test gives a p ≤ 0.05, the difference was interpreted as being significant. For transcriptomics analysis, when the expression values showed at least 2-fold change between the control and N-deprivation conditions, with a false discovery rate (FDR)-corrected p ≤ 0.05 (Benjamini-Hochberg correction which is provided by the Cuffdiff program in the Cufflinks package version 2.0.4), the gene was considered to be significantly differentially expressed.
Results
Development of a Lipidomics Platform
To identify the molecular species of glycerolipids in IMET1 lipid extracts, we first conducted single-stage MS followed by MS/MS analysis of a collection of glycerolipid standards representing glycerolipid classes that have been previously identified in Nannochloropsis and other microalgae. The ions associated with dissociated head groups of phospholipids and galactolipids were utilized as diagnostic product for a given glycerolipid class (Table 1). For example, the dissociation of PC 17:0/20:4 (m/z 796.2, [M+H]+) gave rise to m/z184, which was deciphered as [C5H15NO4P]+, corresponding to the head group of phosphoryl choline (Figure 1A). Consequently, all the PC species in the crude lipid extract can be identified by employing product ion scanning for m/z 184. Similarly, in a positive-ion mode, DGTS, MGDG, and DGDG species of IMET1 were determined by precursor ion scanning for m/z 236 [(C10H22NO5)+], m/z 243 [(C9H16O6+Na)+], and m/z 405 [(C15H26O11+Na)+], respectively (Figures 1B–D); PG, PI and SQDG were identified by precursor scanning, in the negative-ion mode, for the ion of m/z 153 [(C15H12O7P)−], m/z 241 [(C7H14O7P)−], and m/z 225 [(C6H9O7S)−], respectively (Figures 1E–G). Many glycerolipids lose a neutral ion associated with the headgroup due to collision energy induced dissociation, and this feature can facilitate identification through the neutral loss scanning mode. For instance, PE 17:0/14:1 (m/z 576.5) subjected to collisional energy yielded a product ion of m/z 535.3, which corresponded to a neutral loss of a phosphoryl ethanolamine group (C2H8NO4P) (Figure 1H). Thus, neutral loss scanning for m/z 141 led to the identification of PE species in IMET1. Fatty acyl groups of glycerolipid molecular species were determined in the product ion scanning mode. For PE and DGTS, fragmentary ions derived from [M + H]+ were used for structural analysis (Figures 1A,D) (Figures 1H,B). For PC, MGDG, and DGDG, fragments arising from their metal adducts ([M + Na]+) allowed unambiguous structural identification (Figures 1I,C,D). In the negative-ion mode, fatty acids dissociated from PG, PI, and SQDG were directly detected and identified as the acyl groups of the precursor ions (Figures 1E–G).
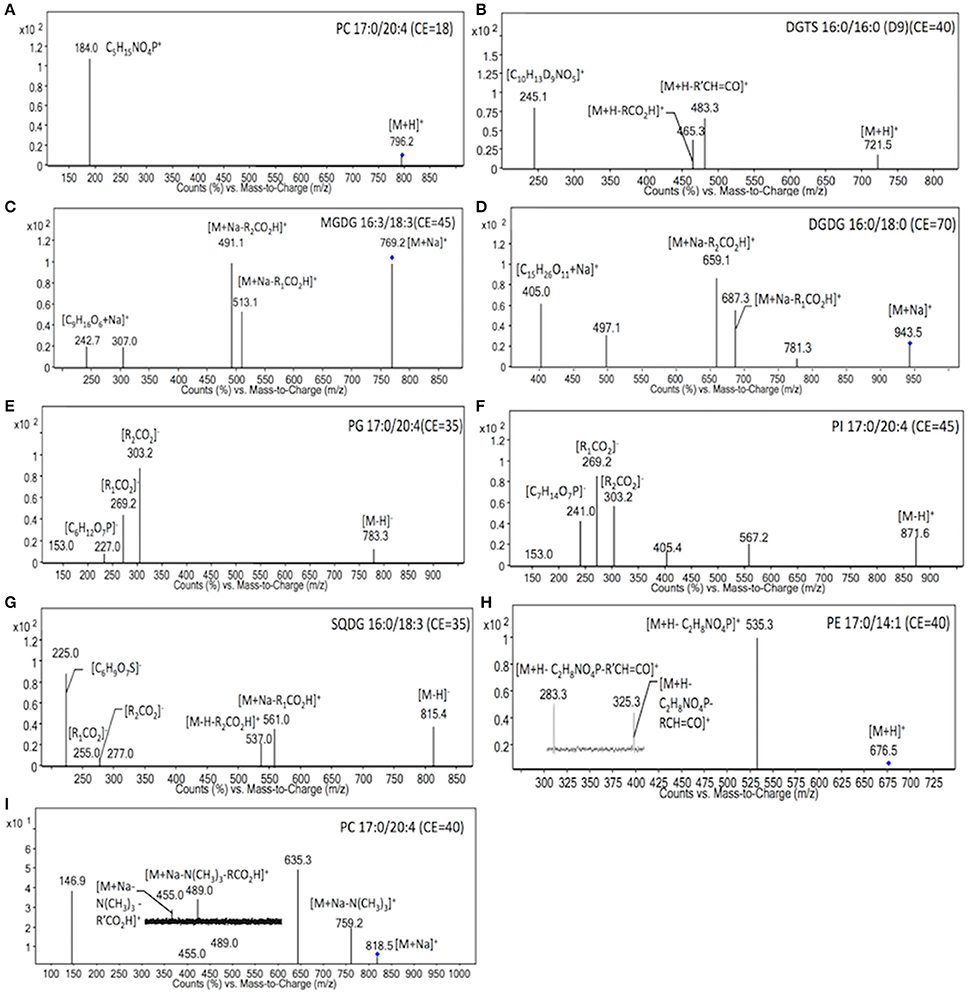
Figure 1. MS/MS spectra of glycerolipids standards. The diagnostic structural information was indicated with the respective product ions in the MS/MS spectra. Blue dots indicated the precursor ions subjected to MS/MS analysis.
For absolute quantification, calibration lipid standards were used for establishing the linear function between the abundance and concentration of lipid ions. The results showed that by using this developed LC/MS/MS method the relative responses of each calibration standard to the internal standards were strictly linearly correlated with their relative concentrations (R2 > 0.99, Supplemental Figure 2). The linear correlation is over two orders of magnitude for all of the membrane glycerolipids except for MGDG. A picomolar detection limit was achieved with high reproducibility.
Glycerolipids Profiling and Quantification
Based on the method described above, a total of 41 glycerolipid molecular species belonging to glycerolipid classes, i.e., PC, PE, PI, PG, DGTS, MGDG, DGDG, and SQDG, were identified from crude lipid extracts of IMET1 (Figures 2, 3). An MS/MS analysis indicated that a variety of fatty acids with different acyl chain lengths and desaturation degrees were unevenly distributed among the glycerolipid classes (Figure 3). For instance, C18 fatty acids were primarily associated with the extraplastidic membrane lipids including PC, PE and PI, but virtually undetectable in the chloroplast membrane glycerolipids (e.g., MGDG, DGDG, SQDG, and PG) of IMET1. Lipidome profiling also revealed the presence of both prokaryotic type (16:1/16:0) and eukaryotic type (20:5/20:5) MGDG, implying that the DAG backbones for MGDG synthesis were supplied by both the prokaryotic and cytoplasmic Kennedy pathways in IMET1. Surprisingly, a typical prokaryotic DAG backbone C16/C16 was detected in several classes of extraplastidic membrane lipids (e.g., DGTS, PC, and PI), pointing to the possible presence of a yet unknown process to transport DAG C16/C16 from the chloroplast into ER.
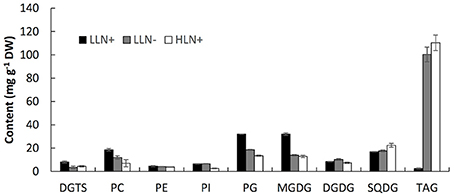
Figure 2. Content of the major glycerolipids of IMET1 quantified by using LC/MS. LLN+, low-light and N replete conditions; LLN−, low-light and N deprivation conditions; HLN+, high-light and N replete conditions. Values represent means ± SD (n = 4~6).
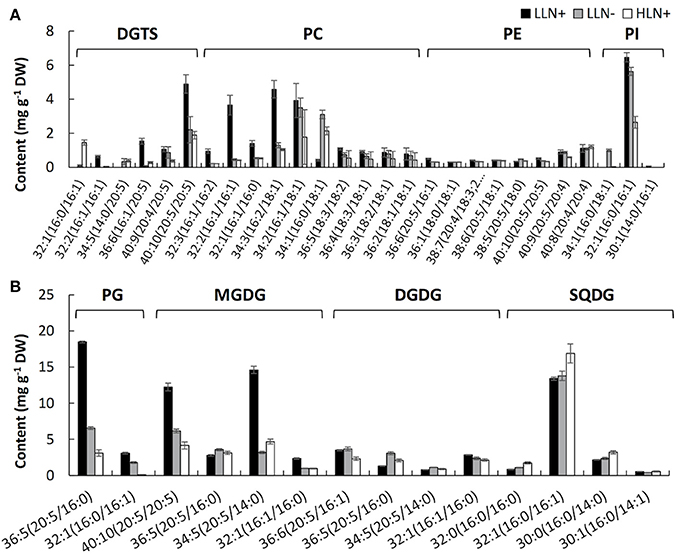
Figure 3. Content of the molecular species of extraplastidic membrane glycerolipids (A), chloroplast membrane glycerolipids (B) of IMET1. LLN+, low-light and N replete conditions; LLN−, low-light and N deprivation conditions; HLN+, high-light and N replete conditions. Acyl chains of glycerolipid molecular species were presented by “carbon number: number of double bonds.” Values represent means ± SD (n = 4~6).
As shown in Figure 2, MGDG (31.9 mg g−1 dry weight) was measured as the most abundant membrane glycerolipid (Figure 2). The chloroplast membrane lipids, including MGDG, DGDG, SQDG, and PG, accounted for up to 66.2% of total glycerolipids under normal growth conditions. PC was the most abundant extraplastidic membrane glycerolipid, and the content of PC was ca. 2.5-fold of its structural analogs–DGTS. PE and PI were detected as the minor extraplastidic membrane glycerolipids, which contributed to 3.1 and 3.9% of total glycerolipids, respectively (Figure 2).
When the algal cells were subjected to N deprivation for 4 days, the cellular content of MGDG, PG, DGTS, and PC decreased by 57.1, 67.1, 56.8, and 19.5%, respectively, whereas PE, PI, and SQDG were not significantly altered. Among the eight membrane glycerolipid classes, DGDG was the only group that increased by 20.5% in response to N deprivation. Compared to N deprivation, HL treatment caused a more profound decline in MGDG, PG, DGTS, PC, and PI, which decreased by 59.6, 85.3, 43.9, 42.2, and 59.1% (p < 0.05), respectively. Surprisingly, despite the chloroplast membrane MGDG and PG declining substantially in response to HL, the cellular content of SQDG increased by 32.1% compared to that under favorable growing conditions, whereas DGDG did not change significantly. Under favorable growth conditions (LLN+), IMET1 cells accumulate a small amount of TAG (5 mg g−1 cellular dry weight), which was greatly stimulated by both N deprivation and HL stresses, accounting 100.3 and 110.3 mg g−1 cellular dry weight under N deprivation and HL conditions, respectively (Figure 2).
As shown in Figure 3, the responses of membrane glycerolipids to N deprivation and HL stresses differed among various molecular species. Among the 41 membrane glycerolipid species detected in this study, most membrane glycerolipids were degraded to various extents under stress conditions, among which the DGTS 16:1/20:5 showed the highest fold change (decreased by ca. 16-fold) under N deprivation conditions (Figure 3A), whereas PG 16:0/16:1 was identified as the most susceptible lipid molecular species (decreased by ca. 50-fold) under HL conditions (Figure 3B). A few lipid species, like most PE molecular species and MGDG 36:5, were constitutive in IMET1 cells, irrespective of the changing environmental conditions (Figures 3A,B). Two DGTS species (i.e., DGTS 34:5 and 32:1) were induced under stress conditions (Figure 3A). PI 34:1 and PG 32:1, as well as the N-containing compounds PC 34:2 and 36:3, were specifically up-regulated by N deprivation, whereas HL stress specifically caused an increase in the content of SQDG 32:0 and 32:1 (Figures 3A,B).
Transcriptional Regulation of Membrane Glycerolipids Biosynthesis under N-Deprivation Conditions
To gain more insights into the regulation of glycerolipid metabolism in IMET1, expression dynamics of the genes involved in membrane lipids biosynthesis were analyzed by mRNA-seq. In IMET1, the monogalactosyldiacylglycerol synthase (MGD) and digalactosyldiacylglycerol synthase (DGD) are each encoded by a single-copy gene. As the galactolipid content in IMET1 was reduced under N deprivation, down-regulation of the two genes and, correspondingly, the galactolipid biosynthesis pathways would be expected. MGD (g4968) was expressed at a lower level under N deprivation conditions (vs. N replete conditions), and the fold-change gradually declined over 48 h (Figure 4). Different from MGD, the DGD exhibited an almost identical expression pattern under both growth conditions.
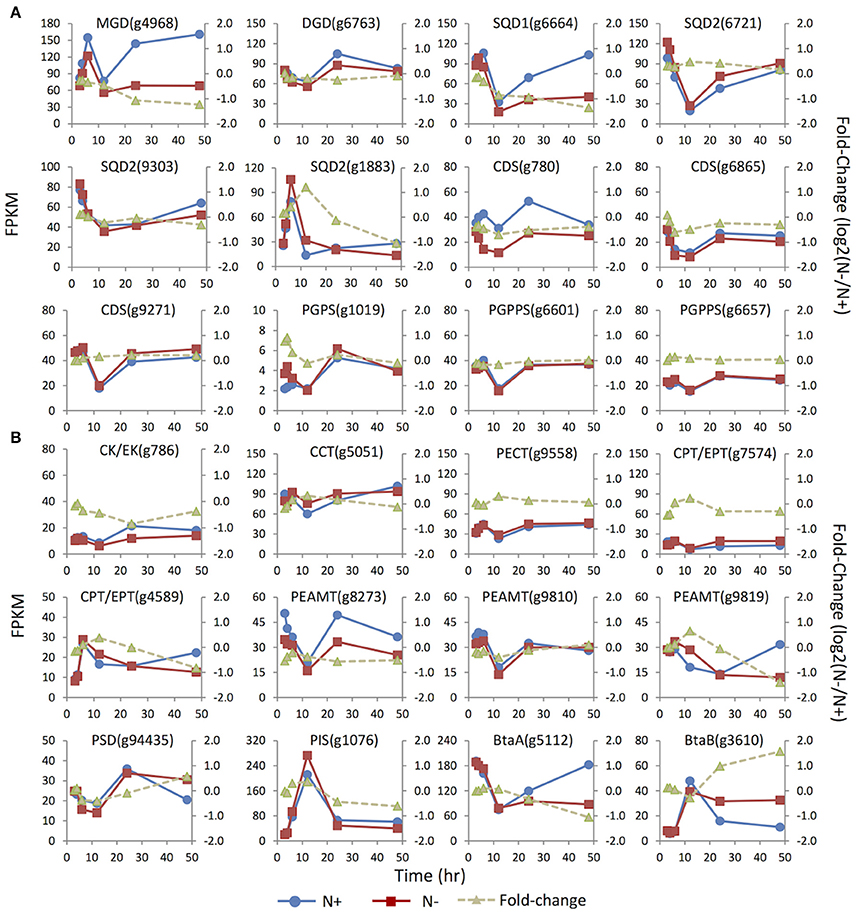
Figure 4. Expression dynamics of the genes involved in membrane glycerolipids biosynthesis under N replete (N+) and N deprivation (N−) conditions. Absolute transcript abundances (FPKM, left y-axis) and fold-change (log2 (N−/N+), right y-axis) in response to N deprivation was plotted against time (3, 4, 6, 12, 24, and 48 h) after the onset of N deprivation, respectively. (A) Genes involved in biosynthesis of the chloroplast membrane glycerolipids; (B) Genes involved in biosynthesis of the extraplastidic membrane glycerolipids. Values represent means of three independent experiments.
Three copies of the SQDG synthase genes SQD2 (i.e., g1883, g6721, and 9303) were identified. Under N replete conditions, the mRNA abundance of g6721 and g9303 was much higher than that of g1883, suggesting that the former two isoforms were likely responsible for bulk SQDG synthesis (Figure 4). The minor isoform g1883 more likely played a specific role in de novo SQDG synthesis under N-depleted conditions because its transcript level was transiently up-regulated 2.5-fold at 12 h in response to N deprivation, while the two major isoforms were down-regulated (g9303) or unaffected (g6721).
Three enzymes—CDP-DAG synthase (CDS), PG-phosphate synthase (PGPS), and PG-phosphate phosphatase (PGPPS)–were proposed to be involved in PG synthesis (Andrews and Mudd, 1985). In contrast to the absence of the candidate PGPPS in plants Arabidopsis and C. reinhardtii (Riekhof et al., 2005b), all the genes required for PG biosynthesis, including two copies of putative PGPPS, were identified in IMET1. Among the three CDS genes present in the IMET1 genome, g6865 and g9271 were the homologs of Arabidopsis ER-associated CDS2, and the third isoform (g780) was identified as the homolog of Arabidopsis plastidic CDS5, indicating that the synthesis of PG occurred in both ER and chloroplasts of this organism. Under N deprivation, the transcript level of the chloroplast CDS (g780) was down-regulated over 48 h, with the most substantial decrease (ca. 40%, p < 0.001) occurring at 24 h (Figure 4). By contrast, one of the ER isoforms (g9271) was slightly up-regulated at 48 h in response to N deprivation (p < 0.001), while transcription of the third (g6865) remained constant at the transcript level. A putative PGPS (g1019) gene in IMET1 was the homolog of Arabidopsis PGPS1, which encoded a dual targeting protein involved in PG biosynthesis in both chloroplast and mitochondria (Muller and Frentzen, 2001; Babiychuk et al., 2003). This gene was up-regulated by 2-fold at 4 h under N deprivation. No significant change in transcript was observed for the two PGPPS genes over 48 h under N deprivation.
According to previous studies on vascular plants, the phospholipids PC, PI, and PE were synthesized in ER membranes through the eukaryotic pathways (Li-Beisson et al., 2010). The first step of PC and PE synthesis begins with the phosphorylation of choline and ethanolamine by choline kinase and ethanolamine kinase, respectively, which belong to the CK/EK super family (Tasseva et al., 2004). A single-copy gene encoding putative CK/EK (g786) is present in the IMET1 genome, which may encode a bifunctional enzyme that can use both choline and ethanolamine as substrates. This gene was down-regulated by 2-fold at 24 h under N-conditions (Figure 4). The rate-limiting step in the biosynthesis of PC and PE was catalyzed by CTP:phosphorylcholinecytidyltransferase (CCT) and CTP:phosphorylethanolamine cytidyltransferase (PECT), respectively (Kent, 1997; Maheshwari et al., 2013). In IMET1, the CCT gene was slightly up-regulated at 12 h under N− conditions (p < 0.001), whereas PECT was transcriptionally unchanged. IMET1 harbors two copies of the plant-type DAG:CDP-alchohol (choline/ethanolamine) phosphotransferase (DAG-CPT/EPT), which putatively encodes a bifunctional enzyme that can incorporate a CDP-choline or a CDP-ethanolamine into the DAG backbone CDP-CPT/EPT (Dewey et al., 1994; Qi et al., 2003). Of the two genes, g4589 was transiently up-regulated at 12 h (p < 0.001), but transcripts were reduced 2-fold at 48 h following N deprivation (Figure 4). Another isoform (g7574) was immediately down-regulated at 3 h under N deprivation (p < 0.001). Three putative phosphoethanolamine N-methyltransferase (PEAMT) genes (g8273, g9810, and g9819) encoding the enzyme converting PE to PC were found in the IMET1 genome (Keogh et al., 2009). Of the three putative PEAMT genes, g9819 was slightly up-regulated at 12 h and then gradually declined thereafter, and g8273 was down-regulated over 48 h under N deprivation, whereas g9558 was essentially unchanged at the transcript level.
Interestingly, the phosphatidylserine (PS) was absent from the lipidome of IMET1, but a PS synthase (PSS) gene (g6304) was identified in the genome and transcriptomes (data not shown). In mammals, PSS can use PC or PE as the substrate for a base-exchange reaction to form PS (Kuge and Nishijima, 2003). PS can be converted to PE by a PS decarboxylase (PSD) (Nerlich et al., 2007). Therefore, we propose that the function of PS in IMET1 is likely to serve as a bridge connecting the PC and PE synthesis pathways. The single-copy PSD exhibited 50% up-regulation at 48 h following N deprivation (Figure 4).
The acidic phospholipid PI was synthesized from CDP-DAG and myo-inositol by PI synthase (PIS) in Arabidopsis (Xue et al., 2000; Lofke et al., 2008). A single-copy PIS gene was found in IMET1, and its transcript level increased by 14-fold at 12 h under both N replete and N deprivation conditions (Figure 4), with a fold change of 1.2 (p < 0.001), indicating that the gene expression was slightly induced by N deprivation but was largely stimulated by as yet unknown factors in our experimental system.
Previous studies on bacteria identified two genes involved in DGTS biosynthesis: a BtaA gene encoding an S-adenosylmethionine (AdoMet):DAG 3-amino-3-carboxypropyltransferaseleading to the formation of diacylglycerolhomoserine (DGHS). DGHS is then converted to DGTS by an AdoMet-dependent M-methylase encoded by BtaB (Hofmann and Eichenberger, 1996; Riekhof et al., 2005a). Under N replete conditions, the transcription of a single-copy BtaAin IMET1 was about 10–20-fold greater than that of BtaB (Figure 4). After the onset of N deprivation, the transcription of BtaA decreased by ca. 2-fold, whereas that of BtaB increased 4-fold, indicating that BtaB was the regulatory gene maintaining the homeostasis of DGTS under N- conditions.
Transcriptional Regulation of Membrane Glycerolipids Desaturation, Turnover and Elongation under N-Deprivation
Distinct unsaturation degrees of the fatty acyl groups of membrane glycerolipidsare derived from the action of a suite of fatty acid desaturases. Insertion of one double bond to C16:0 is mediated by highly substrate-specific desaturases, acting on different groups of lipids. For instance, FAD4 was a Δ3 desaturase that specifically inserted a trans double bond into the sn-2 C16:0 of PG. Two (g1711 and g10079) of the three putative homologs of FAD4 in IMET1 were up-regulated under N- conditions, whereas the expression of the third gene (g2551) was slightly suppressed at 12 h under N deprivation (Figure 5). FAD5, a Δ7 desaturase, was responsible for the formation of C16:1 in MGDG and DGDG in Arabidopsis (Browse et al., 1985; Heilmann et al., 2004; Gao et al., 2009). In IMET1, one putative homolog of FAD5(g2635) was up-regulated at the transcript level under N deprivation (Figure 5).
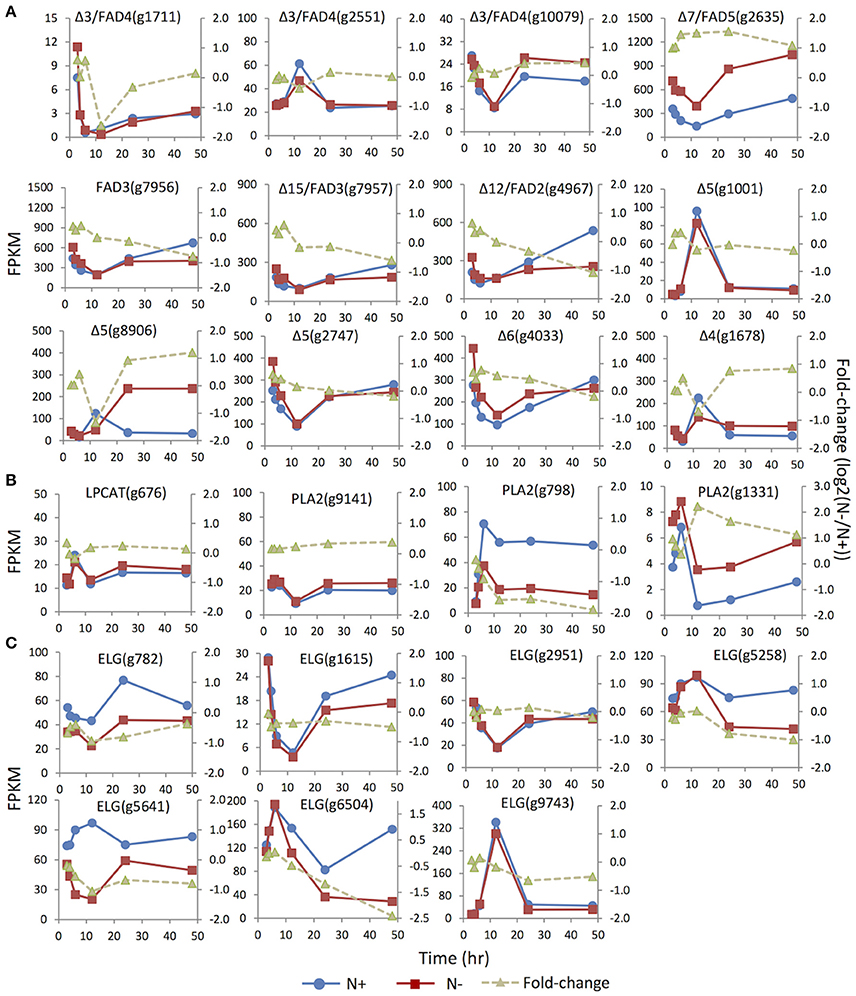
Figure 5. Expression dynamics of the genes involved in membrane glycerolipids desaturation, turnover and elongation under N replete (N+) and N deprivation (N−) conditions. Absolute transcript abundances (FPKM, left y-axis) and fold-change (log2 (N−/N+), right y-axis) in response to N deprivation was plotted against the time (3, 4, 6, 12, 24, and 48 h) after the onset of N deprivation, respectively. (A) Genes involved encoding FAD (fatty acid desaturase). (B) Genes involved in the PC acyl editing cycle, including LPCAT (lyso-phosphatidylcholine acyltransferase), PLA2 (phospholipase A2). (C) Genes encoding ELG (elongase).Values represent means of three independent experiments.
Insertion of more double bonds into C16:1 and C18:1 is catalyzed by desaturases with broad substrate specificity, includingFAD6 (Δ12 desaturase), FAD7/8 (Δ15/ω3 desaturases), FAD2 (Δ12 desaturase) and FAD 3 (Δ15/ω3 desaturase) (Miquel and Browse, 1992; Browse et al., 1993). IMET1 lacks the homologs of FAD6, FAD7, and FAD8, consistent with the absence of polyunsaturatedC16 and C18 acyl moieties in the chloroplast membrane glycerolipids of this organism. Two putative desaturases encoded by g7956 and g7957 exhibited 48 and 44% sequence similarities to FAD3, respectively, and the putative protein of g4967 showed 45% similarity to FAD2, suggesting that these proteins may be the homologs of enzymes involved in the desaturation of extraplastidic glycerolipids in IMET1. These three genes showed slight up-regulation at 3 and 4 h under N deprivation, followed by a gradual decrease (Figure 5). Moreover, up-regulation of the putative FAD2 and FAD3 pointed to a possible stimulation on the PC-mediated acyl editing cycle at the early stage after the onset of N deprivation. To further test this hypothesis, we investigated the expression of the genes encoding lysophosphatidylcholine acyltransferase (LPCAT) and phospholipase A2, the enzymes previously reported to be responsible in vascular plants for acylation and deacylation in the PC acyl editing cycle, respectively. The results indicated that one (g1331) of the three putative PLA2 genes was up-regulated by ca. 2-fold at 3 h and achieved the highest fold-change (4-fold) at 12 h under N deprivation conditions (Figure 5).
Three Δ5 desaturase genes (g1001 and g8906, g2747) and one single-copy gene for Δ6 desaturase (g782) were identified in IMET1. The Δ5 and Δ6 desaturases are known to be involved in EPA biosynthesis, of which the former one is responsible for production of arachidonic acid (C20:4, the direct precursor of EPA), and the latter catalyzes the reaction producing the intermediate C18:3. While the two Δ5 desaturase genes (g1001 and g8906) were expressed at a relatively low level under N replete conditions, the third one (g2747), along with the Δ6 desaturase gene, exhibited considerably higher basal expression (Figure 5). Moreover, under N deprivation conditions, the mRNA abundance of g2747 and g4033 decreased simultaneously with a similar expression pattern. Taken together, these results suggested g2747 and g4033 maybe participated in EPA biosynthesis under normal growth conditions. Distinct from the major isoforms, g8906, one of the minor Δ5 desaturase genes, was up-regulated by 2-fold under N deprivation, indicative of its involvement in de novo EPA synthesis during N deprivation.
A Δ4 fatty acid desaturase gene (g1678) was identified in the IMET1 genome. The homologs of this gene have previously been characterized in many other microalgae; for instance, EgΔ4FAD was involved in the formation of docosahaxaenoic acid in Euglena gracilis, and CrΔ4FAD catalyzed the formation of C16:4 in the chloroplast of C. reinhardtii (Meyer et al., 2003; Zauner et al., 2012). However, neither DHA nor C16:4 are present in the lipidome of IMET1.
A total of seven genes encoding fatty acid elongases (ELO) were identified in IMET1, and their expression dynamics are shown in Figure 5. The expressions of the putative elongase genes were overall down-regulated or unchanged under N deprivation conditions. The deduced protein sequence ofg782 showed 51% similarity to a Δ6 elongase of Phaeodactylum tricornutum (EEC47836), indicative of its possible function in EPA biosynthesis in this organism. This speculation is also supported by the observation that g782 was congruently down-regulated with the two desaturase genes (g2747 and g4033).
Discussion
Remodeling of the Membrane Glycerolipids in Response to Changing Environmental Conditions
This study addressed the impact of the adverse environmental conditions (N deprivation and HL) on glycerolipid composition and possible physiological functions of lipid remodeling. Both N deprivation and HL stresses led to drastic declines in the chloroplast lipid MGDG. MGDG is known as a non-bilayer lipid, because it possesses a smaller cross-sectional area of the head group as compared with that of the acyl chains, resulting in an overall conical shape with a preference of forming the inverted hexagonal phase (HII) (van den Brink-van der Laan et al., 2004). However, certain membrane proteins (e.g., light harvesting complexes) can act to stabilize the non-bilayer lipid MGDG in the thylakoid membranes (Simidjiev et al., 2000). Assembly of non-bilayer lipids into the bilayer membranes may lower the pressure and packing density in the lipid head-group region, and facilitates binding of peripheral membrane proteins; in addition, the increased pressure in the acyl chain region can stabilize the structure of integral membrane proteins (van den Brink-van der Laan et al., 2004). The intrinsic properties of non-bilayer lipids and their interactions with membrane proteins bestow their important roles in creating a dynamic membrane environment for multiple biological processes. It was reported that under high light conditions, MGDG in the thylakoid membranes tended to transform into the HIIphase, due to the lateral movement of LHC during the state transition (Garab et al., 2000). Although MGDG in the HIIphase can enhance photoprotection (e.g., xanthophylls cycle), extensive formation of the non-bilayer structure may cause fusion between bilayer membranes (Siegel and Epand, 1997; Epand, 1998). Therefore, reduction of MGDG under stress conditions may represent a protective mechanism to minimize the destruction of the thylakoid and chloroplast envelope membranes.
In contrast to MGDG, no substantial decrease was observed in DGDG under stress conditions. DGDG is known as a bilayer lipid, due to its cylindrical shape, which enables it always self-assembled into a lamellar phase. A stable amount of DGDG may be essential for structural and functional stabilization of the chloroplast and thus cell survival. As DGDG can be exported from the chloroplast to replace phospholipids in extraplastidial membranes under P-limited conditions (Sandelius et al., 2003; Andersson et al., 2005; Jouhet et al., 2007), we can't rule out that trafficking of DGDG from the chloroplast to other subcelluar organelles may occur under N-deprivation conditions, especially when the extraplastidic bilayerglycerolipids (e.g., PC and DGTS) are drastically decreased.
This study also revealed that IMET1 contained a high content of the anionic glycerolipid SQDG. A similar feature was reported in the diatom Cyclotella meneghiniana, in which the amount of SQDG accounted for 40% of the total lipids (Vieler et al., 2007; Goss et al., 2009). These observations suggested the presence of a highly negatively-charged microdomain in the thylakoid membrane for these heterokont microalgae derived from the secondary endosymbiosis. Additionally, SQDG has been identified in PSII of Thermosynechococcus elongatus and cytochrome b6f of C. reinhardtii, with its headgroup embedded in the photosynthetic complexes and acyl chain located in the cytoplasm side of the thylakoid membranes (Stroebel et al., 2003; Loll et al., 2005, 2007). Recently, Chlamydomonas mutants deficient in SQDG biosynthesis have been obtained and characterization of the mutant suggested that SQDG plays a structural role for the QB binding site of PSII, while maintaining a correct conformation at the PS II donor side (Minoda et al., 2003; Riekhof et al., 2003).
The anionic chloroplast glycerolipids PG was remarkably decreased in IMET1 in response to HL and N deprivation stresses. PG was identified in the cytoplasmic side of PSII, PI and LHCII from oxygenic photosynthetic organisms(Jordan et al., 2001; Liu et al., 2004; Loll et al., 2005, 2007; Standfuss et al., 2005). The function of PG in PSII was suggested to be involved in dimerization of PSII and stabilization of D1-CP43 interaction in Synechocystis (Laczko-Dobos et al., 2008). Depletion of PG in Chlamydomonas was essential for the correct insertion of D1 protein into PSII complexes and thus the thylakoid membrane (Pineau et al., 2004). PG was also involved in trimerization of PSI as suggested by studies with cyanobacteria (Sato et al., 2004). In LHCII, PG provided axial ligation for Mg2+ of one of the chlorophylls, resulting in the increased number of chlorophyll molecule arrangements within a protein-lipid framework (Liu et al., 2004; Standfuss et al., 2005). Taken together, the decrease of PG under stress conditions is likely to facilitate degradation of chlorophyll and thylakoid membranes, attenuate linear electron transport by reducing the functional PSII and PI, as well as enhancing D1 protein turnover, so as to reduce photosynthetic activities essential for the cells to survive under stress conditions.
Surprisingly, the cellular contents of the N-containing extraplastidic glycerolipids DGTS and PC decreased to a comparable level under N-deprivation and HL conditions, indicating that the decreases in PC and DGTS were not attributable to the depletion of nitrogen for their de novo biosynthesis under N-deprivation conditions, but probably due to the shunting of carbon flux from the membrane lipid biosynthesis to TAG synthesis at the ER membranes. The non-bilayer lipid PE is known to influence protein folding and functions of numerous membrane proteins, and its deficiency was shown to result in severe growth defects in many organisms (Emoto and Umeda, 2000; Birner et al., 2001; Steenbergen et al., 2005). Lipid analysis in this study indicated that PE played an indispensible functional role in cellular responses to adverse environmental conditions for IMET1.
Conceptual Network of EPA Biosynthesis
In eukaryotic organisms, EPA is thought to be synthesized from linoleic (18:2) and α-linolenic acid (18:3) via the ω-6 and ω-3 pathways, respectively (Khozin et al., 1997; Abbadi et al., 2004). Both the ω-6 and ω-3 pathways are involved in serial desaturations and elongations of FA, eventually leading to the formation of EPA (20:5). Previous studies suggested that PC and PE were involved in EPA synthesis pathways (Schneider and Roessler, 1994; Vieler et al., 2012).
Based on the results of glycerolipid profiling and prior knowledge, we propose the possible pathways for EPA biosynthesis and distribution in IMET1 (Figure 6). Given that the fatty acids 18:2 and 18:3 were particularly enriched in PC, and 20:4 was found primarily in PE, we proposed that PC is the site for the desaturation of C18 FA, while PE be the site for desaturation of C20 FA in IMET1 (Figure 4). As such, C18:3 can first be cleaved from PC molecules by a PLA2 and then activated by a long chain acyl-CoA synthase (LACS) to form C18:3-CoA.C18:3-CoA in turn can be utilized as the substrate of Δ6 elongase to produce C20:3-CoA, which is then incorporated along with C18:3-CoA into a DAG pool via the Kennedy pathway for de novo PE synthesis. In the PE pool, the molecular species 20:5/20:5 may coexist with the PE species containing one 20:5 and one C18 acyl-group, indicating that at least two EPA biosynthesis routes exist (Figure 6). One route involved the simultaneous formation of two C20:4 prior to the final desaturation step yielding C20:5, whereas the other underwent stepwise desaturation of one acyl chain. Given that there are three loci encoding Δ5 desaturase in IMET1, it remains to be determined if the two routes are catalyzed by different isoforms of desaturases.
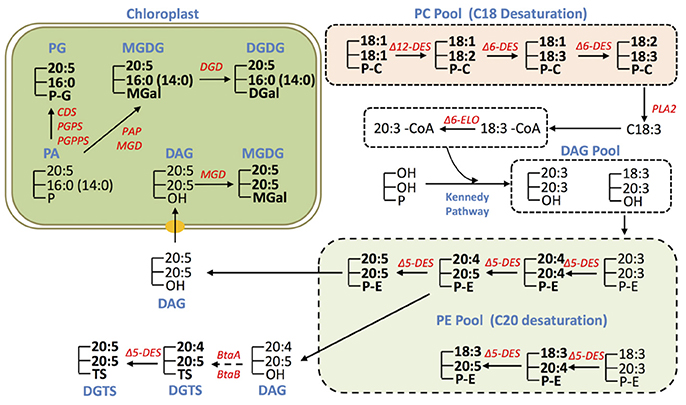
Figure 6. Conceptual EPA biosynthesis network in IMET1. The glycerolipid species detected by LC/MS analysis are marked in bold, and the unmarked lipid species represent the deduced intermediates that have yet to be validated. The genes involved in EPA biosynthesis/allocation and validated by transcriptomic analysis in this study are presented.
In addition to PC and PE, DGTS may also be a site for EPA synthesis, as both 20:4 and 20:5 were detected in DGTS. By contrast, EPA distributed in MGDG, DGDG, and seemed to be allocated from PE (or DGTS), because no intermediates for EPA biosynthesis (e.g., C20:4) were detected in these chloroplast membrane glycerolipids. For instance, DAG derived from PE by dephosphorylation could serve as a precursor for synthesis of the eukaryotic MGDG species 20:5/20:5 in the chloroplast. On the other hand the observation of a number of MGDG, DGDG, and PG species containing EPA at sn-1 and 16:0 or 14:0 at sn-2 suggested that these glycerolipid species are synthesized through a prokaryotic pathway in the chloroplast. It is plausible that EPA de-acylated from the existing EPA-containing glycerolipid species (e.g., PE, DGTS) is incorporated into the prokaryotic PA, the common precursor for synthesis of PG, MGDG and DGDG in the chloroplast Kennedy Pathway. Recently, an elongase gene was found to play a role in utilizing C16:0 for producing EPA associated with MGDG in N. gaditana, but its full function remains to be uncovered, which may provide more mechanistic insights into EPA rechanneling from the ER to the plastids (Dolch et al., 2017).
In summary, the results of glycerolipid profiling suggest that EPA was produced via serial desaturations and elongations associated with biosynthesis of the extraplastidic glycerolipids, including PC, PE, and DGTS in IMET1. EPA distributed in the chloroplast membrane glycerolipids pool is presumed to be derived from PE and probably DGTS as well. Furthermore, it can be inferred from the gene expression results that the de novo EPA biosynthesis was slightly up-regulated at the early stage of N deprivation.
Transcriptional Regulation of Membrane Lipids Biosynthesis under N-Deprivation
It was well documented that supply of N has an impact on galactolipids composition in microalgae and photosynthetic tissue of plants (Gaude et al., 2007), but little is known about the regulation of galactolipids biosynthesis and turnover under N deprivation at the gene expression level. A previous study in Arabidopsis revealed that expression of DGD1 and DGD2 were elevated under N deprivation, which correlated with the increased DGDG content (Gaude et al., 2007). Recently, expression of the genes involved in membrane lipids biosynthesis in C. reinhardtii in response to N deprivation was investigated by mRNA-seq (Boyle et al., 2012). However, due to the lack of essential controls (expression level under N replete conditions), regulation of membrane lipid biosynthesis in C. reinhardtii remained elusive, although it was observed that the absolute abundances of MGD was dramatically reduced after being subjected to N deprivation conditions for 2 h, while DGD was transiently up-regulated and peaked at 24 h under N deprivation. Based on a time-course global transcriptomics analysis with the appropriate control (N-replete), it can be concluded that in IMET1 MGD was down-regulated at the transcript level in response to N deprivation, whereas DGD was unchanged. When the changes in the content of MGDG and DGDG were taken into consideration collectively, the results suggest that the metabolic flux in galactolipids synthesis was reduced under N deprivation, and the continuous accumulation of DGDG under stress conditions is attributable to the unchanged DGD abundance inferred from the gene expression results.
Our lipidomic analysis revealed that most molecular species of SQDG and PE were not susceptible to N deprivation stress. For SQDG, three SQD loci seemed to be controlled by distinct regulatory mechanisms, since one was up-regulated while the other two were down-regulated under N deprivation. Therefore, we speculated that these three SQD genes possess distinct physiological roles in IMET1: the up-regulated one was likely specifically responsible for de novo SQDG synthesis under N deprivation, whereas the other two are likely responsible for bulk SQDG synthesis under normal growth conditions. The homeostasis of SQDG probably involves coordination of multiple loci, which are subject to different regulatory mechanisms.
The transcriptomic analysis indicated that PE is synthesized via two distinct pathways in IMET1, i.e., phosphoethanolamine transfer from CDP-ethanolamine to diacylglycerol, and decarboxylation of PS by PSD. Under N deprivation conditions, the genes involved in the synthesis of PE from ethanolamine and diacylglycerol were down-regulated at the transcript level, whereas the single copy PSD was up-regulated. Despite that the genes responsible for PS biosynthesis are present in the genome of IMET1 and were expressed under both N replete and N deprivation conditions, PS was under the detection limit when total cellular lipids were analyzed using the MS approach. These results indicated that similar to other eukaryotes IMET1 probably contains a very low amount of PS, which mainly serves as the precursor for PE synthesis. Our results implied that PSD may be primarily responsible for synthesis of PE under N deprivation.
Although dramatic decrease were observed in PG, DGTS, and PC under N deprivation, it was not attributable to the retarded biosynthesis process, at least at the gene expression level, as most genes involved in biosynthesis of PG, DGTS and PC were unaffected by N supply, yet several genes were significantly up-regulated, such as PGPS, BtaB. Therefore, de novo glycerolipid synthesis may be required not only for growth and cell division under N-replete conditions, but also for lipid turnover and remodeling to ensure lipid homeostasis for cell survival under N deprivation or other stress conditions.
In summary, lipidomes and transcriptomes characterized in this study may serve as a base upon which molecular mechanisms of membrane lipid metabolism may be further explored in IMET1 and other related microalgae. Advances in understanding lipid remodeling and gene regulation in membrane lipid biosynthesis can facilitate genetic manipulation for enhanced production of microalgae biomass, TAG and PUFA. When the information obtained from this system-level study is coalesced with future functional explorations, these efforts may eventually provide the society with a microalgae-based biomass feedstock for food, feed, nutraceuticals, and biofuels.
Author Contributions
JJ and JL contribute to the acquisition and analysis of data; JX contributes to design of the work and manuscript revising; MS contributes to revise manuscript; QH contributes to the conception of the work and manuscript revising; DH contributes to the conception of the work, interpretation of data, and manuscript drafting and revising.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by State Development & Investment Corporation (SDIC) of China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fmars.2017.00242/full#supplementary-material
Supplementary Figure1. Growth curve and nitrate consumption curve of N. oceanica IMET1 under nitrogen-replete (N+) and nitrogen-depleted (N−) conditions.
Supplementary Figure2. Dynamic quantification of membrane glycerolipids. Calibration lipid standards were titrated relative to a constant amount of internal standards for the quantitative LC/MS analysis. The x axis shows the relative concentration of calibration standard compared to the corresponding internal standard. The y axis shows the relative peak area of calibration standard compared to the corresponding internal standard. Calibration standards include DGTS 16:0/16:0, PC18:1/18:1, PE18:0/18:0, PI 18:1/18:1, PG18:0/18:1, SQDG16:0/18:3, MGDG 16:3/18:3, and DGDG18:3/18:3; internal standards include PC17:0/20:4, PE14:1/17:0, PI17:0/20:4, PG17:0/20:4, MGDG18:0/18:0 and DGDG18:0/18:0. The line indicates the linear function between relative response and relative concentration. Values derived from three independent LC/MS analyses.
References
Abbadi, A., Domergue, F., Bauer, J., Napier, J. A., Welti, R., Zahringer, U., et al. (2004). Biosynthesis of very-long-chain polyunsaturated fatty acids in transgenic oilseeds: constraints on their accumulation. Plant Cell 16, 2734–2748. doi: 10.1105/tpc.104.026070
Alboresi, A., Perin, G., Vitulo, N., Diretto, G., Block, M., Jouhet, J., et al. (2016). Light remodels lipid biosynthesis in nannochloropsis gaditana by modulating carbon partitioning between organelles. Plant Physiol. 171, 2468–2482. doi: 10.1104/pp.16.00599
Andersson, M. X., Larsson, K. E., Tjellstrom, H., Liljenberg, C., and Sandelius, A. S. (2005). The plasma membrane and the tonoplast as major targets for phospholipid- to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J. Biol. Chem. 280, 27578–27586. doi: 10.1074/jbc.M503273200
Andrews, J., and Mudd, J. B. (1985). Phosphatidylglycerol synthesis in pea chloroplasts: pathway and localization. Plant Physiol. 79, 259–265. doi: 10.1104/pp.79.1.259
Babiychuk, E., Muller, F., Eubel, H., Braun, H. P., Frentzen, M., and Kushnir, S. (2003). Arabidopsis phosphatidylglycerophosphate synthase 1 is essential for chloroplast differentiation, but is dispensable for mitochondrial function. Plant J. 33, 899–909. doi: 10.1046/j.1365-313X.2003.01680.x
Birner, R., Burgermeister, M., Schneiter, R., and Daum, G. (2001). Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 997–1007. doi: 10.1091/mbc.12.4.997
Bondioli, P., Della Bella, L., Rivolta, G., Chini Zittelli, G., Bassi, N., Rodolfi, L., et al. (2012). Oil production by the marine microalgae Nannochloropsis sp. F&M-M24 and Tetraselmis suecica F&M-M33. Bioresour. Technol. 114, 567–572. doi: 10.1016/j.biortech.2012.02.123
Boyle, N. R., Page, M. D., Liu, B., Blaby, I. K., Casero, D., Kropat, J., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287, 15811–15825. doi: 10.1074/jbc.M111.334052
Browse, J., Mcconn, M., James, D., and Miquel, M. (1993). Mutants of Arabidopsis deficient in the synthesis of apha-linolenate. Biochemical and genetic characterization of the endoplasmic-reticulum linoleoyl desaturase. J. Biol. Chem. 268, 16345–16351.
Browse, J., McCourt, P., and Somerville, C. R. (1985). A mutant of Arabidopsis lacking a chloroplast-specific lipid. Science 227, 763–765. doi: 10.1126/science.227.4688.763
Corteggiani Carpinelli, E., Telatin, A., Vitulo, N., Forcato, C., D'Angelo, M., Schiavon, R., et al. (2013). Chromosome scale genome assembly and transcriptome profiling of nannochloropsis gaditana in nitrogen depletion. Mol. Plant. 7, 323–335. doi: 10.1093/mp/sst120
Dewey, R. E., Wilson, R. F., Novitzky, W. P., and Goode, J. H. (1994). The Aapt1 gene of soybean complements a cholinephosphotransferase-deficient mutant of yeast. Plant Cell 6, 1495–1507. doi: 10.1105/tpc.6.10.1495
Dolch, L. J., Rak, C., Perin, G., Tourcier, G., Broughton, R., Leterrier, M., et al. (2017). A palmitic acid elongase affects eicosapentaenoic acid and plastidial monogalactosyldiacylglycerol levels in Nannochloropsis. Plant Physiol. 173, 742–759. doi: 10.1104/pp.16.01420
Dong, H. P., Williams, E., Wang, D. Z., Xie, Z. X., Hsia, R. C., Jenck, A., et al. (2013). Responses of Nannochloropsis oceanica IMET1 to long-term nitrogen starvation and recovery. Plant Physiol. 162, 1110–1126. doi: 10.1104/pp.113.214320
Ejsing, C. S., Sampaio, J. L., Surendranath, V., Duchoslav, E., Ekroos, K., Klemm, R. W., et al. (2009). Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 2136–2141. doi: 10.1073/pnas.0811700106
Emoto, K., and Umeda, M. (2000). An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J. Cell Biol. 149, 1215–1224. doi: 10.1083/jcb.149.6.1215
Epand, R. M. (1998). Lipid polymorphism and protein-lipid interactions. Biochim. Biophys. Acta 1376, 353–368. doi: 10.1016/S0304-4157(98)00015-X
Gao, J., Ajjawi, I., Manoli, A., Sawin, A., Xu, C., Froehlich, J. E., et al. (2009). Fatty acid desaturase 4 of Arabidopsis encodes a protein distinct from characterized fatty acid desaturases. Plant J. 60, 832–839. doi: 10.1111/j.1365-313X.2009.04001.x
Garab, G., Lohner, K., Laggner, P., and Farkas, T. (2000). Self-regulation of the lipid content of membranes by non-bilayer lipids: a hypothesis. Trends Plant Sci. 5, 489–494. doi: 10.1016/S1360-1385(00)01767-2
Gaude, N., Brehelin, C., Tischendorf, G., Kessler, F., and Dormann, P. (2007). Nitrogen deficiency in Arabidopsis affects galactolipid composition and gene expression and results in accumulation of fatty acid phytyl esters. Plant J. 49, 729–739. doi: 10.1111/j.1365-313X.2006.02992.x
Goncalves, E. C., Johnson, J. V., and Rathinasabapathi, B. (2013). Conversion of membrane lipid acyl groups to triacylglycerol and formation of lipid bodies upon nitrogen starvation in biofuel green algae Chlorella UTEX29. Planta 238, 895–906. doi: 10.1007/s00425-013-1946-5
Goss, R., Nerlich, J., Lepetit, B., Schaller, S., Vieler, A., and Wilhelm, C. (2009). The lipid dependence of diadinoxanthin de-epoxidation presents new evidence for a macrodomain organization of the diatom thylakoid membrane. J. Plant Physiol. 166, 1839–1854. doi: 10.1016/j.jplph.2009.05.017
Han, X., and Gross, R. W. (2003). Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 44, 1071–1079. doi: 10.1194/jlr.R300004-JLR200
Han, X., and Gross, R. W. (2005). Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24, 367–412. doi: 10.1002/mas.20023
Heilmann, I., Mekhedov, S., King, B., Browse, J., and Shanklin, J. (2004). Identification of the Arabidopsis palmitoyl-monogalactosyldiacylglycerol delta7-desaturase gene FAD5, and effects of plastidial retargeting of Arabidopsis desaturases on the fad5 mutant phenotype. Plant Physiol. 136, 4237–4245. doi: 10.1104/pp.104.052951
Hofmann, M., and Eichenberger, W. (1996). Biosynthesis of diacylglyceryl-N,N,N-trimethylhomoserine in Rhodobacter sphaeroides and evidence for lipid-linked N methylation. J. Bacteriol. 178, 6140–6144. doi: 10.1128/jb.178.21.6140-6144.1996
Hu, J. Q., Wang, D. M., Li, J., Jing, G. C., Ning, K., and Xu, J. (2014). Genome-wide identification of transcription factors and transcription-factor binding sites in oleaginous microalgae Nannochloropsis. Sci. Rep. 4:5454. doi: 10.1038/srep05454
Hu, Q., Sommerfeld, M., Jarvis, E., Ghirardi, M., Posewitz, M., Seibert, M., et al. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54, 621–639. doi: 10.1111/j.1365-313X.2008.03492.x
Jia, J., Han, D. X., Gerken, H. G., Li, Y. T., Sommerfeld, M., Hu, Q., et al. (2015). Molecular mechanisms for photosynthetic carbon partitioning into storage neutral lipids in Nannochloropsis oceanica under nitrogen-depletion conditions. Algal Res. 7, 66–77. doi: 10.1016/j.algal.2014.11.005
Jinkerson, R. E., Radakovits, R., and Posewitz, M. C. (2013). Genomic insights from the oleaginous model alga Nannochloropsis gaditana. Bioengineered 4, 37–43. doi: 10.4161/bioe.21880
Jordan, P., Fromme, P., Witt, H. T., Klukas, O., Saenger, W., and Krauss, N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411, 909–917. doi: 10.1038/35082000
Jouhet, J., Marechal, E., and Block, M. A. (2007). Glycerolipid transfer for the building of membranes in plant cells. Prog. Lipid Res. 46, 37–55. doi: 10.1016/j.plipres.2006.06.002
Kent, C. (1997). CTP:phosphocholine cytidylyltransferase. Biochim. Biophys. Acta 1348, 79–90. doi: 10.1016/S0005-2760(97)00112-4
Keogh, M. R., Courtney, P. D., Kinney, A. J., and Dewey, R. E. (2009). Functional characterization of phospholipid N-methyltransferases from Arabidopsis and Soybean. J. Biol. Chem. 284, 15439–15447. doi: 10.1074/jbc.M109.005991
Khotimchenko, S. V., and Yakovleva, I. M. (2005). Lipid composition of the red alga Tichocarpus crinitus exposed to different levels of photon irradiance. Phytochemistry 66, 73–79. doi: 10.1016/j.phytochem.2004.10.024
Khozin, I., Adlerstein, D., Bigongo, C., Heimer, Y. M., and Cohen, Z. (1997). Elucidation of the biosynthesis of eicosapentaenoic acid in the microalga Porphyridium cruentum.2. Studies with radiolabeled precursors. Plant Physiol. 114, 223–230. doi: 10.1104/pp.114.1.223
Kilian, O., Benemann, C. S., Niyogi, K. K., and Vick, B. (2011). High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl. Acad. Sci. U.S.A. 108, 21265–21269. doi: 10.1073/pnas.1105861108
Kuge, O., and Nishijima, M. (2003). Biosynthetic regulation and intracellular transport of phosphatidylserine in mammalian cells. J. Biochem. 133, 397–403. doi: 10.1093/jb/mvg052
Laczko-Dobos, H., Ughy, B., Toth, S. Z., Komenda, J., Zsiros, O., Domonkos, I., et al. (2008). Role of phosphatidylglycerol in the function and assembly of Photosystem II reaction center, studied in a cdsA-inactivated PAL mutant strain of Synechocystis sp. PCC6803 that lacks phycobilisomes. Biochim. Biophys. Acta 1777, 1184–1194. doi: 10.1016/j.bbabio.2008.06.003
Lee, Y. J., Leverence, R. C., Smith, E. A., Valenstein, J. S., Kandel, K., and Trewyn, B. G. (2013). High-throughput analysis of algal crude oils using high resolution mass spectrometry. Lipids 48, 297–305. doi: 10.1007/s11745-013-3757-7
Li, J., Han, D. X., Wang, D. M., Ning, K., Jia, J., Wei, L., et al. (2014). Choreography of transcriptomes and lipidomes of Nannochloropsis reveals the mechanisms of oil synthesis in microalgae. Plant Cell 26, 1645–1665. doi: 10.1105/tpc.113.121418
Liang, C. W., Cao, S. N., Zhang, X. W., Zhu, B. H., Su, Z. L., Xu, D., et al. (2013). De novo sequencing and global transcriptome analysis of Nannochloropsis sp. (Eustigmatophyceae) following nitrogen starvation. Bioenergy Res. 6, 494–505. doi: 10.1007/s12155-012-9269-0
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2010). Acyl-lipid metabolism. Arabidopsis Book 8:e0133. doi: 10.1199/tab.0133
Liu, B., Vieler, A., Li, C., Jones, A. D., and Benning, C. (2013). Triacylglycerol profiling of microalgae Chlamydomonas reinhardtii and Nannochloropsis oceanica. Bioresour. Technol. 146, 310–316. doi: 10.1016/j.biortech.2013.07.088
Liu, Z., Yan, H., Wang, K., Kuang, T., Zhang, J., Gui, L., et al. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428, 287–292. doi: 10.1038/nature02373
Lofke, C., Ischebeck, T., Konig, S., Freitag, S., and Heiliviann, I. (2008). Alternative metabolic fates of phosphatidylinositol produced by phosphatidylinositol synthase isoforms in Arabidopsis thaliana. Biochem. J. 413, 115–124. doi: 10.1042/BJ20071371
Loll, B., Kern, J., Saenger, W., Zouni, A., and Biesiadka, J. (2005). Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II. Nature 438, 1040–1044. doi: 10.1038/nature04224
Loll, B., Kern, J., Saenger, W., Zouni, A., and Biesiadka, J. (2007). Lipids in photosystem II: interactions with protein and cofactors. Biochim. Biophys. Acta 1767, 509–519. doi: 10.1016/j.bbabio.2006.12.009
Maheshwari, S., Lavigne, M., Contet, A., Alberge, B., Pihan, E., Kocken, C., et al. (2013). Biochemical characterization of Plasmodium falciparum CTP:phosphoethanolamine cytidylyltransferase shows that only one of the two cytidylyltransferase domains is active. Biochem. J. 450, 159–167. doi: 10.1042/BJ20121480
Meyer, A., Cirpus, P., Ott, C., Schlecker, R., Zahringer, U., and Heinz, E. (2003). Biosynthesis of docosahexaenoic acid in Euglena gracilis: biochemical and molecular evidence for the involvement of a Δ4-fatty acyl group desaturase. Biochemistry 42, 9779–9788. doi: 10.1021/bi034731y
Minoda, A., Sonoike, K., Okada, K., Sato, N., and Tsuzuki, M. (2003). Decrease in the efficiency of the electron donation to tyrosine Z of photosystem II in an SQDG-deficient mutant of Chlamydomonas. FEBS Lett. 553, 109–112. doi: 10.1016/S0014-5793(03)00981-5
Miquel, M., and Browse, J. (1992). Arabidopsis mutants deficient in polyunsaturated fatty-acid synthesis - biochemical and genetic-characterization of a plant oleoyl-phosphatidylcholine desaturase. J. Biol. Chem. 267, 1502–1509.
Muller, F., and Frentzen, M. (2001). Phosphatidylglycerophosphate synthases from Arabidopsis thaliana. FEBS Lett. 509, 298–302. doi: 10.1016/S0014-5793(01)03163-5
Nerlich, A., von Orlow, M., Rontein, D., Hanson, A. D., and Dormann, P. (2007). Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria. Plant Physiol. 144, 904–914. doi: 10.1104/pp.107.095414
Pineau, B., Girard-Bascou, J., Eberhard, S., Choquet, Y., Tremolieres, A., Gerard-Hirne, C., et al. (2004). A single mutation that causes phosphatidylglycerol deficiency impairs synthesis of photosystem II cores in Chlamydomonas reinhardtii. Eur. J. Biochem. 271, 329–338. doi: 10.1046/j.1432-1033.2003.03931.x
Qi, Q. G., Huang, Y. F., Cutler, A. J., Abrams, S. R., and Taylor, D. C. (2003). Molecular and biochemical characterization of an aminoalcoholphosphotransferase (AAPT1) from Brassica napus: effects of low temperature and abscisic acid treatments on AAPT expression in Arabidopsis plants and effects of over-expression of BnAAPT1 in transgenic Arabidopsis. Planta 217, 547–558. doi: 10.1007/s00425-003-1031-6
Radakovits, R., Jinkerson, R. E., Fuerstenberg, S. I., Tae, H., Settlage, R. E., Boore, J. L., et al. (2012). Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 3:686. doi: 10.1038/ncomms1688
Riekhof, W. R., Andre, C., and Benning, C. (2005a). Two enzymes, BtaA and BtaB, are sufficient for betaine lipid biosynthesis in bacteria. Arch. Biochem. Biophys. 441, 96–105. doi: 10.1016/j.abb.2005.07.001
Riekhof, W. R., Ruckle, M. E., Lydic, T. A., Sears, B. B., and Benning, C. (2003). The sulfolipids 2'-O-acyl-sulfoquinovosyldiacylglycerol and sulfoquinovosyldiacylglycerol are absent from a Chlamydomonas reinhardtii mutant deleted in SQD1. Plant Physiol. 133, 864–874. doi: 10.1104/pp.103.029249
Riekhof, W. R., Sears, B. B., and Benning, C. (2005b). Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1(Cr). Eukaryotic Cell 4, 242–252. doi: 10.1128/EC.4.2.242-252.2005
Rodolfi, L., Chini Zittelli, G., Bassi, N., Padovani, G., Biondi, N., Bonini, G., et al. (2009). Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 102, 100–112. doi: 10.1002/bit.22033
Sandelius, A. S., Andersson, M. X., Stridh, M. H., Larsson, K. E., and Lijenberg, C. (2003). Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett. 537, 128–132. doi: 10.1016/S0014-5793(03)00109-1
Sato, N., Suda, K., and Tsuzuki, M. (2004). Responsibility of phosphatidylglycerol for biogenesis of the PSI complex. Biochim. Biophys. Acta 1658, 235–243. doi: 10.1016/j.bbabio.2004.06.008
Schneider, J. C., and Roessler, P. (1994). Radiolabeling studies of lipids and fatty acids in Nannochloropsis (Eustigmatophyceae), an oleaginous marine alga. J. Phycol. 30, 594–598. doi: 10.1111/j.0022-3646.1994.00594.x
Siegel, D. P., and Epand, R. M. (1997). The mechanism of lamellar-to-inverted hexagonal phase transitions in phosphatidylethanolamine: implications for membrane fusion mechanisms. Biophys. J. 73, 3089–3111. doi: 10.1016/S0006-3495(97)78336-X
Simidjiev, I., Stoylova, S., Amenitsch, H., Javorfi, T., Mustardy, L., Laggner, P., et al. (2000). Self-assembly of large, ordered lamellae from non-bilayer lipids and integral membrane proteins in vitro. Proc. Natl. Acad. Sci. U.S.A. 97, 1473–1476. doi: 10.1073/pnas.97.4.1473
Simionato, D., Block, M. A., La Rocca, N., Jouhet, J., Maréchal, E., Finazzi, G., et al. (2013). The Response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryotic Cell. 12, 665–676. doi: 10.1128/EC.00363-12
Standfuss, J., Terwisscha van Scheltinga, A. C., Lamborghini, M., and Kuhlbrandt, W. (2005). Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. Embo J. 24, 919–928. doi: 10.1038/sj.emboj.7600585
Steenbergen, R., Nanowski, T. S., Beigneux, A., Kulinski, A., Young, S. G., and Vance, J. E. (2005). Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J. Biol. Chem. 280, 40032–40040. doi: 10.1074/jbc.M506510200
Stroebel, D., Choquet, Y., Popot, J. L., and Picot, D. (2003). An atypical haem in the cytochrome b6f complex. Nature 426, 413–418. doi: 10.1038/nature02155
Sukenik, A. (1991). Ecophysiological considerations in the optimization of eicosapentaenoic acid production by Nannochloropsis-sp. (Eustigmatophyceae). Bioresour. Technol. 35, 263–269. doi: 10.1016/0960-8524(91)90123-2
Tasseva, G., Richard, L., and Zachowski, A. (2004). Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 566, 115–120. doi: 10.1016/j.febslet.2004.04.015
van den Brink-van der Laan, E., Killian, J. A., and de Kruijff, B. (2004). Non-bilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta 1666, 275–288. doi: 10.1016/j.bbamem.2004.06.010
Vieler, A., Wilhelm, C., Goss, R., Suss, R., and Schiller, J. (2007). The lipid composition of the unicellular green alga Chlamydomonas reinhardtii and the diatom Cyclotella meneghiniana investigated by MALDI-TOF MS and TLC. Chem. Phys. Lipids 150, 143–155. doi: 10.1016/j.chemphyslip.2007.06.224
Vieler, A., Wu, G., Tsai, C. H., Bullard, B., Cornish, A. J., Harvey, C., et al. (2012). Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 8:e1003064. doi: 10.1371/journal.pgen.1003064
Wang, D. M., Lu, Y. D., Huang, H., and Xu, J. (2012). Establishing oleaginous microalgae research models for consolidated bioprocessing of solar energy. Biotechnol. China 128, 69–84. doi: 10.1007/10_2011_122
Wang, D. M., Ning, K., Li, J., Hu, J. Q., Han, D. X., Wang, H., et al. (2014). Nannochloropsis genomes reveal evolution of microalgal oleaginous traits. PLoS Genet. 10:e1004094. doi: 10.1371/journal.pgen.1004094
Wijffels, R. H., and Barbosa, M. J. (2010). An outlook on microalgal biofuels. Science 329, 796–799. doi: 10.1126/science.1189003
Xiao, Y., Zhang, J., Cui, J., Feng, Y., and Cui, Q. (2013). Metabolic profiles of Nannochloropsis oceanica IMET1 under nitrogen-deficiency stress. Bioresour. Technol. 130, 731–738. doi: 10.1016/j.biortech.2012.11.116
Xu, J., Chen, D., Yan, X., Chen, J., and Zhou, C. (2010). Global characterization of the photosynthetic glycerolipids from a marine diatom Stephanodiscus sp. by ultra performance liquid chromatography coupled with electrospray ionization-quadrupole-time of flight mass spectrometry. Anal. Chim. Acta 663, 60–68. doi: 10.1016/j.aca.2010.01.026
Xue, H. W., Hosaka, K., Plesch, G., and Mueller-Roeber, B. (2000). Cloning of Arabidopsis thaliana phosphatidylinositol synthase and functional expression in the yeast pis mutant. Plant Mol. Biol. 42, 757–764. doi: 10.1023/A:1006308909105
Yoon, K., Han, D. X., Li, Y. T., Sommerfeld, M., and Hu, Q. (2012). Phospholipid: diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 24, 3708–3724. doi: 10.1105/tpc.112.100701
Zauner, S., Jochum, W., Bigorowski, T., and Benning, C. (2012). A cytochrome b(5)-containing plastid-located fatty acid desaturase from Chlamydomonas reinhardtii. Eukaryotic Cell 11, 856–863. doi: 10.1128/EC.00079-12
Keywords: Nannochloropsis oceanica, membrane glycerolipid, lipidomics, transcriptomics, environmental stresses, eicosapentaenoic acid
Citation: Han D, Jia J, Li J, Sommerfeld M, Xu J and Hu Q (2017) Metabolic Remodeling of Membrane Glycerolipids in the Microalga Nannochloropsis oceanica under Nitrogen Deprivation. Front. Mar. Sci. 4:242. doi: 10.3389/fmars.2017.00242
Received: 30 April 2017; Accepted: 18 July 2017;
Published: 04 August 2017.
Edited by:
Jin Liu, Peking University, ChinaReviewed by:
Hong-Ye Li, Jinan University, ChinaYuan Kun Lee, National University of Singapore, Singapore
Matthew Posewitz, Colorado School of Mines, United States
Copyright © 2017 Han, Jia, Li, Sommerfeld, Xu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danxiang Han, danxianghan@ihb.ac.cn
†Co-first author
 Danxiang Han
Danxiang Han Jing Jia1,2†
Jing Jia1,2†