Small Scale Genetic Population Structure of Coral Reef Organisms in Spermonde Archipelago, Indonesia
- 1Biotechnology and Molecular Genetics, Center for Environmental Research and Sustainable Technology (UFT), University Bremen, Bremen, Germany
- 2Marine Biology, Vrije Universiteit Brussel, Brussels, Belgium
- 3Marine Science and Technology, Bogor Agricultural University, Bogor, Indonesia
- 4Marum Center for Marine Environmental Sciences, University Bremen, Bremen, Germany
Small island archipelagos with fringing and dispersed reef systems represent special marine ecosystems, providing a patchy habitat for many coral reef organisms. Although geographic distances may be short, it is still unclear if such environments are inhabited evenly with panmictic conditions or if limited connectivity between marine populations, even on small geographic scales, leads to genetic differentiation between areas within the archipelago or even single reef structures. To study diversity patterns and connectivity between reefs of the Spermonde Archipelago, Indonesia, population genetic analyses of two reef organisms were performed by using the mitochondrial control region and microsatellite markers. A vertebrate (clown anemonefish) and an invertebrate species (sea squirt) were studied in parallel to investigate if there are general patterns of connectivity in Spermonde for sessile or site attached marine species, which can be extrapolated to a larger group. The genetic population structures revealed restrictions in gene flow in the clown anemone fish (Amphiprion ocellaris), especially between near-shore reefs in the South of the archipelago. This indicates very localized genetic exchange and may also reflect the high self-recruitment typical for these fish. The northern reefs show higher connectivity despite geographic distances being larger. The filter-feeding sessile sea squirt, Polycarpa aurata, features similar population patterns, especially in the southern area. However, connectivity is generally higher in the middle and shelf edge areas of Spermonde for this species. The results underline that there are restrictions to gene flow even on very small geographic scales in the studied organisms, with many barriers to gene flow in the southern shallower shelf area. Weaker currents in this area may lead to more influence of biological factors for dispersal, such as larval behavior, motility and competition for suitable habitat. The results are discussed in the context of conservation and MPA planning in the Spermonde Archipelago.
Introduction
In recent years, marine shallow water habitats have been facing increasing rates of environmental change and impacts through industrialization, accelerated climate change, and deposition of sediments, toxins, and nutrients (Wild et al., 2011). Additionally, sensitive coastal habitats, like coral reefs, are more intensely exploited for the marine ornamental trade, food, and other products. Many species of commercial interest suffer from over-exploitation, while the use of inadequate fishery techniques and coastal developments can alter the habitat structure enormously (e.g., blast fishing, Edinger et al., 1998). Over-exploitation, paired with anthropogenic changes of environmental conditions, may lead to the depletion of populations and eventually to local extinctions, causing an inadvertent overall decrease of ecosystem resilience (Hughes et al., 2003; Hughes and Stachowicz, 2004). High genetic diversity is of crucial importance for species exposed to adverse or very variable conditions, ensuring the potential to adapt and maintain critical resilience levels (Hughes et al., 2003; Ehlers et al., 2008). Demographically relevant genetic exchange among populations helps to maintain a high degree of diversity and stabilizes local biological resources (Cowen et al., 2006). For exploited organisms, like many of the coral reef species in Indonesia, this aspect is critical to sustain populations and is therefore a major consideration in conservation and marine reserve network design (Almany et al., 2009).
Archipelagos constitute an extremely fragmented habitat for coral reef organisms, as they are comprised of small island groups and submerged reefs separated by deeper water or sandy areas. Coral reefs are “islands of bounty” in this otherwise desert marine space, which reflects in the life history of most coral reef dependent species. These species often exhibit a site attached or even sessile adult and juvenile phase, whereas their larvae are pelagic, possessing the ability to disperse over smaller or larger spatial scales. Various techniques and methods for studying larval dispersal and connectivity on different scales have been developed and used (in Jones et al., 2009). To study connectivity patterns among reefs in an archipelago, population genetic analysis can be applied for an indirect estimation of larval dispersal in site-attached organisms. By studying different taxa in parallel (e.g., vertebrate and invertebrate species) in the same habitat and area, a more general picture about common or distinguishing factors and prevailing barriers to connectivity can be revealed. This enables and improves adaptive management and conservation strategies for the studied area and similar systems.
Biological, geographical, and physical factors influence the genetic connectivity of marine populations. Biological factors include the mobility of adults, mode of reproduction (e.g., breeding, broadcast spawning), the pelagic larval duration (PLD), as well as larval development, behavior and motility. Important geographical and physical factors include prevailing currents [e.g., eddies, tidal currents, and major currents like the Indonesian Throughflow (ITF)], geographical location and history of the study area (e.g., remote islands with barriers to other suitable habitats, plate tectonics, sea level changes).
Spermonde Archipelago is an interesting environment to study small-scale connectivity patterns in coral reef organisms, as it represents a large patchy habitat. It is located in the Makassar Strait in the Southwest of Sulawesi, Indonesia (Tomascik et al., 1997). The region is part of the Coral Triangle (Briggs, 2009), which is considered to harbor the world's highest marine biodiversity (Allen and Werner, 2002; Allen, 2008; Veron et al., 2009), but also the most threatened coral reef systems worldwide (Burke et al., 2006). The Spermonde Archipelago consists of about 150 islands, as well as many submerged reefs with distances of only a few to up to 70 km between them. Islands found within the archipelago differ in size, are uninhabited to densely populated, and surrounding reefs are exploited to varying degrees and with various techniques. Reef systems are therefore differentially impacted and the health of the reefs differs quite dramatically as a result of these impacts (e.g., bomb fishing, cyanide fishing, sediments, sewage; Plass-Johnson et al., 2015). Located on the southern end of the archipelago's coastline is the city of Makassar with about 1.34 million inhabitants and an industrial harbor. The reef's distance from shore determines its accessibility for exploitation and potential impact from pollution by river runoff or other shore based pollutants (e.g., sewage, sediments, and toxins). These factors, in addition to the changing bathymetry of the shelf itself and potential current strength, may drive the persistence of coral reef populations and their genetic diversity.
To analyse the genetic connectivity among reefs and shelf areas of Spermonde, the anemonefish, Amphiprion ocellaris, and the sea squirt, Polycarpa aurata, were chosen as vertebrate and invertebrate models, respectively. The chosen species represent different organism groups with different reproductive strategies, but are both expected to exhibit small-scale population genetic patterns as a result of their short PLDs.
The clown anemonefish, A. ocellaris (Pomacentridae), lives in symbiosis with up to three different sea anemone species (Heteractis magnifica, Stichodactyla gigantea, and S. mertensii). The adults and juvenile settlers are bound to their host anemone for protection. The species lays demersal eggs at the base of the host and cares for them until the larvae hatch and enter the pelagic larval phase (Fautin and Allen, 1994). The PLD of the species is 8–10 days in captivity, which is considered relatively short for reef fishes (Gopakumar et al., 2009). Still one of the most popular marine aquarium species, A. ocellaris is of high commercial interest for the international marine ornamental trade and numbers of wild catches from Spermonde and other sites in Indonesia are very high (Wabnitz et al., 2003; Madduppa et al., 2014a; Glaser et al., 2015). Additionally, host species Heteractis magnifica and Stichodactyla spp. are also collected in Spermonde for the marine ornamental trade, reducing the availability of habitat for adults and new recruits (Madduppa et al., 2014a). The combined pressures from fisheries, host reduction and reef degradation are affecting this species, emphasizing its dependence on population re-seeding through larval dispersal. The site-attached nature of A. ocellaris juveniles and adults allows important conclusions to be made about the dispersal achieved during the pelagic larval phase, which is the main mechanism by which population connectivity and admixture is achieved. Genetic signals of connectivity or isolation among sampled sites can be used for this or similar species to inform small-scale management plans and efforts.
The solitary, broadcast-spawning sea squirt, Polycarpa aurata, is distributed on coral reefs of the Indo-West Pacific (Kott, 1985) and can be found at many study sites in Spermonde. Ascidians play an important ecological role in bentho-pelagic coupling as filter feeding re-mineralizers in coral reefs (Kaiser et al., 2005). They are simultaneous hermaphrodites, usually with external fertilization (Yund and O'Neil, 2000) and their eggs are broadcast for up to 15 h before hatching (Davis and Davis, 2007). Larvae of P. aurata are non-feeding, leading to a very short PLD of <12 h to up to 1 week (Svane and Young, 1989). The tadpole shaped ascidian larvae have a tail that allows for a certain degree of auto-locomotion, but strong currents undoubtedly play a more important role for dispersal. The species was chosen as an invertebrate model for small-scale connectivity within the archipelago because it is a reef dependent organism with an important ecological function, short PLD, and site attached adults. In addition, its pervasive presence at most sample sites allowed collecting a representative dataset for the study area.
Although both chosen model species show limited dispersal capabilities, they have different reproductive strategies as vertebrate and invertebrate representatives and may behave differently in response to prevailing environmental parameters. An additional distinguishing feature between populations of the two species from identical sites is that fishing pressure greatly affects anemonefish populations, while the sea squirt is not a species of commercial interest and therefore not affected.
The importance of considering population connectivity for conservation is increasingly recognized, playing a crucial role in population persistence and recovery from exploitation and disturbances (Planes et al., 2009). Several years ago, a network of small marine reserves was established in Spermonde (Radjawali, 2012). However, in absence of precise background data on larval dispersal and connectivity needed for marine spatial planning, the site and size selection of protected sites was instead aimed at providing a first opportunity to interact, educate, and engage island inhabitants in marine conservation and management. Following this first important step, the present study was conducted to provide the missing scientific data to local decision makers to potentially increasing the effectiveness of conservation measures in Spermonde and other small island systems.
Materials and Methods
Sampling and DNA Extraction
A total of 356 samples of the clown anemonefish, A. ocellaris, were collected at 22 sites in Spermonde Archipelago (Figure 1, Table 1). Individual fish were caught with aquarium nets and small fin clips (about 2 mm2, dependent on the individual size of the fish) were taken. Specimens were then released back into their host anemone. This sampling method was tested prior on captive fish, where normal behavior of the fish after sampling and an eventual regrowth of the clipped section was observed. For the sea squirt, P. aurata, a total of 332 samples from 16 sites in Spermonde were collected as small tissue pieces (3 mm-1 cm, dependent on the size of the individual), cut from either the oral or atrial siphon opening (Figure 1, Table 2). In most individuals, reopening after the procedure was observed during the sampling time and healing of older lesions could be detected. Sample sites for both species cover the entire shelf area (Figure 1). Samples were transferred to ethanol (>96%) and finally stored at 4°C.
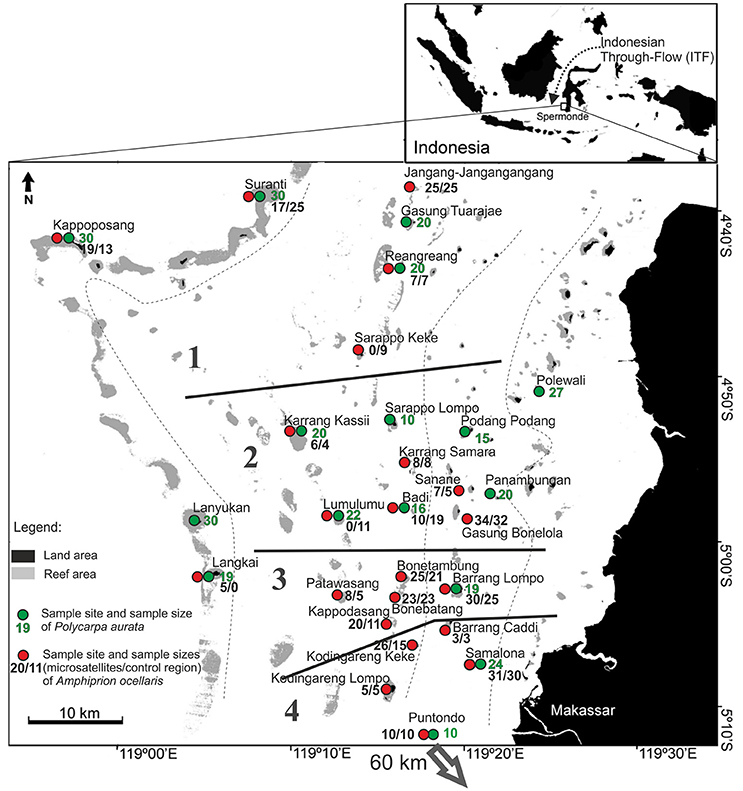
Figure 1. Sample sites for the clown anemonefish, Amphiprion ocellaris, (red circles) and the solitary sea squirt, Polycarpa aurata, (green circles) with corresponding sample sizes for each location in Spermonde Archipelago, Indonesia. Regional groups (1–4) tested for significant differences in nucleotide diversity are indicated on the map through solid black lines. Dashed lines are the major isobaths found on the shelf (simplified after Cleary et al., 2005). Puntondo is located 60 km South of Spermonde on the coast of Sulawesi.
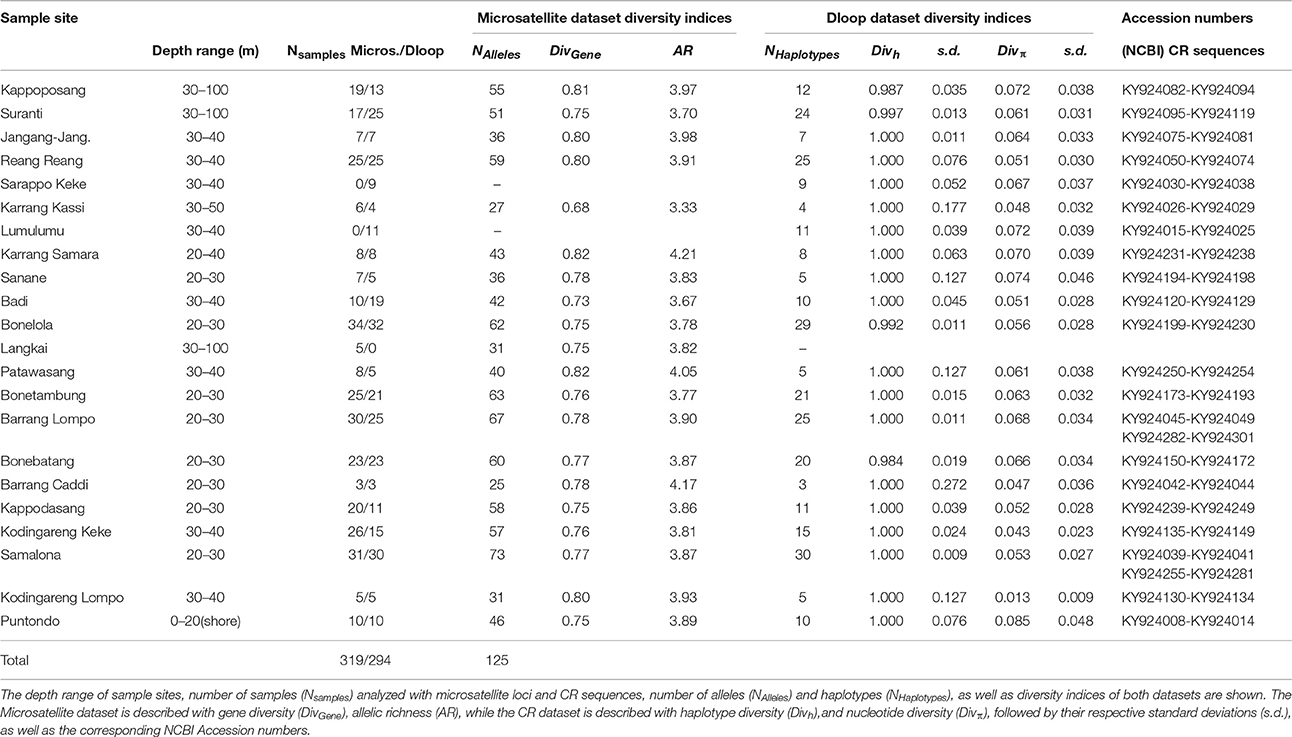
Table 1. Samples of the clown anemonefish, Amphiprion ocellaris, from sample sites in Spermonde Archipelago, Indonesia.
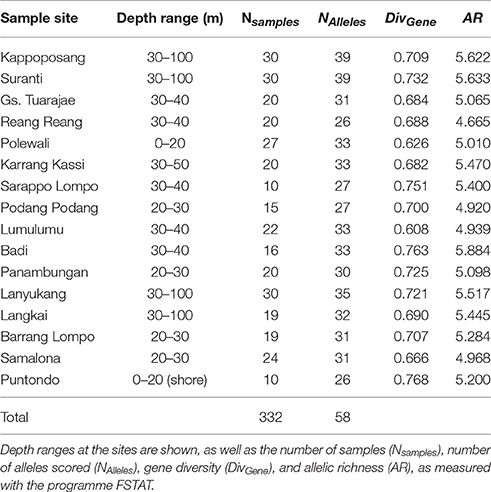
Table 2. Samples of the sea squirt, Polycarpa aurata, from sample sites in the Spermonde Archipelago, Indonesia.
For further analysis, genomic DNA was extracted using the NucleoSpin Tissue Extraction kit (Macherey-Nagel, Germany) or the DNEasy Blood and Tissue Extraction Kit (Qiagen, Germany) for A. ocellaris, following the manufacturer's protocols. The Chelex extraction procedure was used for P. aurata samples, with the following protocol: 500 μL 10% Chelex® 100 suspension, 25 μL Dithiothreitol (100 mM), and 20 μL Proteinase K (10 mg/ml) were added to the tissue sample (about 100 mg) and incubated at 54°C and 1,000 rpm in a thermoshaker for minute 5 h or overnight. After incubation, the mixture was centrifuged at 13,000 rpm for 3 min, and the supernatant, containing the DNA, was transferred to a new reaction tube.
Genetic Markers and PCR
Vertebrate Model (A. ocellaris)
In A. ocellaris a 370 base-pairs fragment of the mitochondrial control region, including a hypervariable region (CR; Lee et al., 1995), was amplified, in addition to a set of six microsatellite loci (Quenouille et al., 2004; Buston et al., 2007). Information derived from these markers was used for the genetic population analysis (Table 3). A larger number of published microsatellite loci for A. ocellaris were tested, but excluded due to either a lack of polymorphism at this small scale, stutter bands, or null alleles.
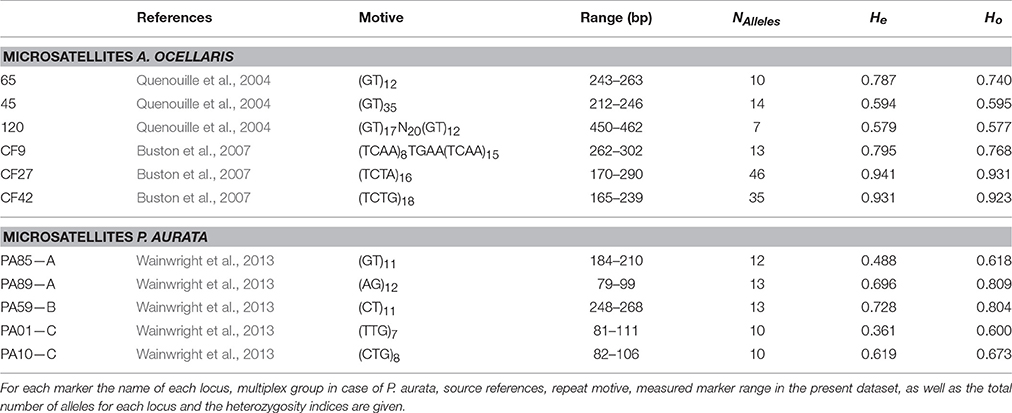
Table 3. Microsatellite markers used for the population genetic analysis of the clown anemonefish, Amphiprion ocellaris, and the sea squirt, Polycarpa aurata, in this study.
The PCR for CR amplification was conducted in an Eppendorf Ep S Master cycler (Eppendorf) and a T Professional Gradient Cycler (Biometra) in 25 μl reaction volume containing: 2.5 μl 10x PCR buffer, 3 μl 25 mM MgCl2, 1 μl 10 μM dNTP mix, 10 pmol each primer, 0.5 U Taq polymerase (Molzym, Bremen), and 10–30 ng genomic DNA. The thermal profile was as following: 95°C for 2 min, followed by 35 cycles at 95°C for 30 s, 50°C for 30 s, and 72°C for 60 s, followed by a final elongation step at 72°C for 2 min. The PCR products were purified with the QIA quick PCR purification kit (Qiagen) and both strands sequenced on an ABI Automated Sequencers (Applied Biosystems) using the PCR primers.
For the microsatellite PCRs the same thermocyclers were used as for the CR and the PCR reactions (25 μl volume) contained: 2.5 μl 10x PCR buffer, 3 μl 25 mM MgCl2, 0.5 μl 10 μM dNTP mix, 10 pmol each primer, 0.5 U Taq polymerase (Molzym, Bremen), and 10–30 ng template DNA. The thermal profile was as follows: 94°C for 2 min, then 35 cycles at 94°C for 45 s, specific annealing temperature (54–62°C; Table 3) for 45 s, followed by 72°C for 60 s, and ending with a final elongation at 72°C for 2 min. Samples for fragment length analysis were prepared by diluting to similar PCR product quantities, mixing with the internal size standard LIZ500 and formamide HiDi (Thermo-Fisher Scientific). The analysis was performed on an ABI automated sequencer (Applied Biosystems).
The A. ocellaris datasets were supplemented with CR sequences from previously published studies (Timm and Kochzius, 2008; Timm et al., 2012).
Invertebrate Model (P. aurata)
For the P. aurata dataset, five microsatellite loci (Wainwright et al., 2013) were amplified in a multiplex reaction, using the Type-it Multiplex PCR kit (Qiagen) with 25 μl reaction mix containing 12.5 μl Master Mix, 2.5 μl 2 μM primer mix A, B, or C (consisting of two primer pairs each; Table 3), and about 50 ng template DNA. Here too, a much larger number of published microsatellite loci were tested for suitability, but excluded due to various issues with amplification or scoring. For the five selected loci the thermal profile was as follows: 95°C for 5 min, 28 cycles of 95°C for 30 s, primer specific annealing temperature for 90 s, 72°C for 30 s, and a final elongation at 60°C for 30 min. The PCR products were diluted as required, mixed with the internal size standard LIZ500 and HiDi, and analyzed with an ABI automated sequencer (Applied Biosystems).
Data Analysis
The forward and reverse CR sequences were aligned with the software Seqman (ver. 4.05 DNAStar) and quality checked by eye. Multiple alignments were generated with the ClustalW algorithm implemented in Bioedit (ver. 7.0.0.1, Hall, 1999). The haplotype and nucleotide diversities were calculated with the programme Arlequin (ver.3.5, Excoffier et al., 2005), as well as the Analysis of Molecular Variance (AMOVA) and pairwise differentiation indices. To discover regional structures, a hierarchical AMOVA was run with Arlequin, testing various regional groupings. Groupings were selected based on geographical proximity of sample sites. Sample sites on possible “borders” between sub-populations were tested for both possible groups and as stand-alone groups. The highest significant value obtained for the tested groups was interpreted to indicate the most probable population subdivision in the study area for the corresponding dataset.
To investigate possible regional differences in the nucleotide diversity of the sampled populations, a one-way ANOVA was performed, comparing diversities in four larger “regional” groups (Figure 1). Puntodo was excluded from this analysis based on its distant, off-shelf location and because the high nucleotide diversity found there was not considered representative for the southern shelf sites.
For the microsatellite datasets of A. ocellaris and P. aurata, allele scoring and quality control was performed using the Programme GeneMarker (ver. 1.91) (Supplementary Material 1—A. ocellaris; Supplementary Material 2—P. aurata). Genetic diversity indices for co-dominant markers were complemented using FSTAT (ver. 2.9; Goudet, 1995).
To identify the most important gene flow barriers for both species in Spermonde, Voronoï tessellation maps were constructed in BARRIER (ver. 2.2, Manni et al., 2004). The maps consist of Voronoï tessellations generated from XY coordinates of the sampling locations. The tessellations represent individual polygonal neighborhoods for each of the included sites (population samples) and determine which sites are neighbors (i.e., adjacent). A Delaunay triangulation (Brassel and Reif, 1979) is applied to connect all neighboring sites on the map. Only one possible Delaunay triangulation can be generated for a set of known geographic locations. The Monmonier (1973) maximum difference algorithm is run on these maps to identify polygon edges where the distance (here Fst- and Φst- indices) between populations within adjacent polygons is the greatest. By generating bootstrapped distance matrices and applying them to the map, the robustness of the emerging patterns can be evaluated. The first ten barriers with the highest bootstrap support were selected for each marker individually. To identify barriers with support from both marker systems in A. ocellaris, the BARRIER maps were superimposed and barriers marked accordingly. Barriers generated by only one of the datasets (CR sequences or microsatellites) are marked in orange, while barriers supported by both marker systems are drawn in red (A. ocellaris only).
In addition, STRUCTURE (ver. 2.2., Pritchard et al., 2000) analyses were performed (default setup with 80,000 Burn-in period and 100,000 Replicates) for both species with a priori information about populations (LOCPRIOR model used), to compare the results from the different analytic approaches. For each k (number of clusters) three to five runs were performed (Supplementary Material 3). If a subdivision was found in the dataset (k > 1), the proportion of membership of each pre-defined population in each of the found clusters was visualized via a pie chart for each sample site.
Results
A total of 294 CR sequences and 319 multilocus genotypes based on six microsatellite loci of the clown anemonefish, A. ocellaris, were analyzed, while 332 multilocus genotypes were scored based on five microsatellite loci for P. aurata.
Genetic Diversity
In the A. ocellaris CR dataset 253 haplotypes were detected, resulting in high haplotype diversities at all locations (Table 1). The hypervariable region contained many nucleotide substitutions and several insertions, including a repetitive sequence stretch, as is characteristic for this genetic marker (Lee et al., 1995). This resulted in a high proportion of private haplotypes (over 70% in most sample sites, data not shown). The number of shared haplotypes was limited and haplotype frequencies were generally low due to the high number of private singleton haplotypes. The nucleotide diversity (π) showed a high variation between sites, with a trend of increasing diversity moving from the most southern sampling site of the archipelago (Kodingareng Lompo, π = 0.013) to the north and also increasing with distance from the coast (exception here is Karang Kassi). High to moderate nucleotide diversities were found in the North and the middle of the shelf area (π = 0.048–0.074) and moderate to low values were found on the most southern reefs closer to shore (π = 0.013–0.053). Puntondo, located about 60 km to the South of the archipelago, presented the highest nucleotide diversity in the dataset (π = 0.085). Testing regional group mean nucleotide diversities (groups 1–4, Figure 1) with a one-way ANOVA and Tukey HSD Post-hoc Test revealed that the nucleotide diversity of group 4 was significantly lower at the p < 0.05 level than any of the other three groups [F(3, 16) = 4,5944, p < 0.017] (Figure 2). Group 4 includes the islands Kodingareng Lompo, Kodingareng Keke, Samalona, and Barrang Caddi.
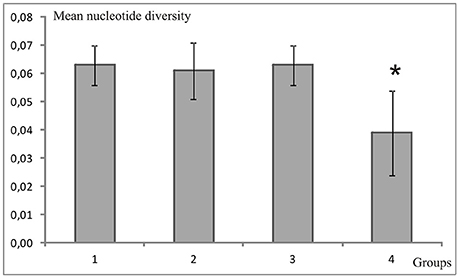
Figure 2. Mean nucleotide diversities of the clown anemonefish, Amphiprion ocellaris, for groups (1–4) sorted by shelf areas (see Figure 1) in Spermonde Archipelago. The asterisk indicates a significantly lower value (one-way ANOVA and Tukey HSD Post-hoc Test).
The microsatellite analysis of A. ocellaris could not confirm the tendency of lower diversities in the southern region of Spermonde. Diversity values were similar at all sites, ranging from 0.679 to 0.824 for gene diversity and from 3.33 to 4.208 for allelic richness, with the lowest values found at a mid-shelf location (Karrang Kassi, 0.679 and 3.33, respectively; Table 1).
For the sea squirt, P. aurata, diversity values were also within a small range (0.608–0.768 for gene diversity and 4.665–5.884 for allelic richness), showing no tendency toward lower diversities in the South of the archipelago.
Genetic Population Structure
Amphiprion ocellaris
The overall AMOVA with CR sequences of A. ocellaris revealed a significant subdivision within Spermonde (including Puntondo), with a Φst-value of 0.055 (p < 0.001). The hierarchical AMOVA showed that the most probable subdivision (Φct = 0.076; p < 0.001) resulted in nine groups, with the most northern sites forming one large group and increasing differentiation among proximate sites when moving south and closer to shore (Figure 3, dashed colored circles).
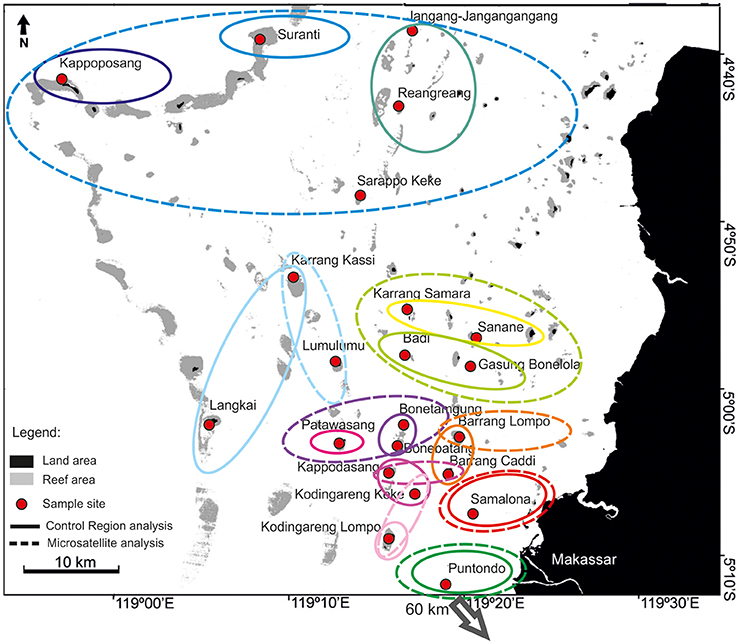
Figure 3. Results of the hierarchical AMOVA (Arlequin ver.3.5, Excoffier et al., 2005) for the clown anemonefish, A. ocellaris, in Spermonde Archipelago and Puntondo. Analysis is based on 6 microsatellite loci (solid circles; 13 groups: Fct = 0.013; p = 0.01) and control region sequences (dashed circles; 9 groups: Φct = 0.076; p < 0.001).
The microsatellite dataset showed no significant overall Fst-value, but when groups were tested with a hierarchical AMOVA, significant groupings could be detected. The highest Fct-value was found for 13 groups (Fct = 0.013; p = 0.01; Figure 3, solid colored circles). Similar to the gradient detected with CR data, differentiation among proximate sites increased when moving south and closer to shore.
The STRUCTURE analysis of A. ocellaris showed the highest probability of subdivision into 3 clusters (k = 3, Supplementary Material 3A, Figure 4). Cluster 1 (red) is most frequent in the North of the archipelago, as well as in Puntondo, which lies to the South of Spermonde archipelago, on the coast of Sulawesi. Cluster 2 (green) was dominant in the middle and southern shelf area and Cluster 3 (blue) genotypes dominated in Samalona, the site closest to the city of Makassar. To smaller proportions, Cluster 3 genotypes were also found at a few other sites, mainly in the South of Spermonde and close to Samalona (Figure 4).
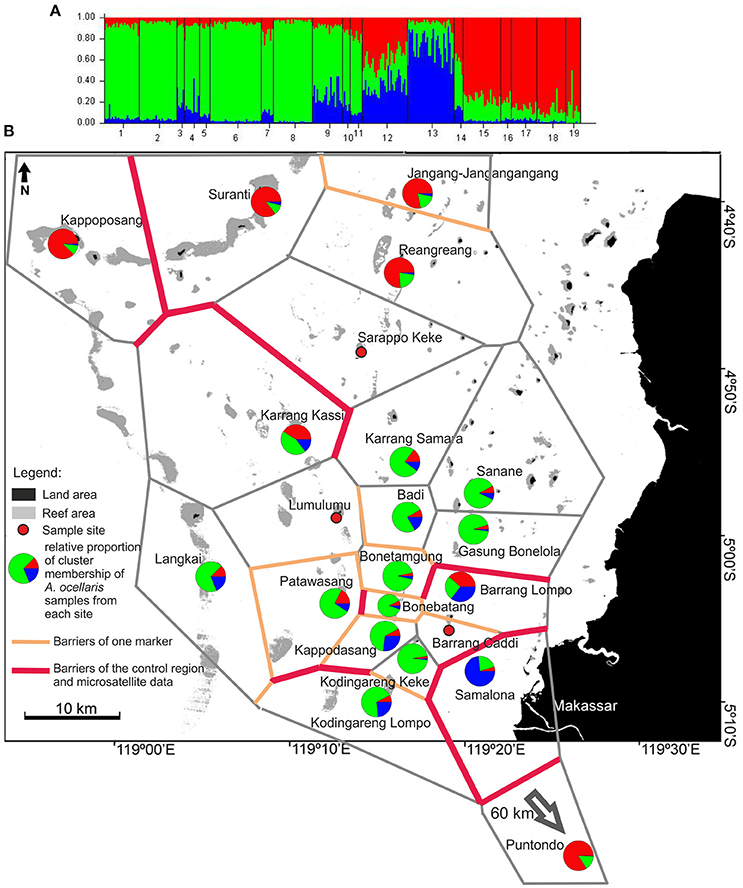
Figure 4. (A) of the figure shows the frequency diagram generated by STRUCTURE (ver. 2.2., Pritchard et al., 2000) as part of the analysis output showing the proportional cluster membership of sampled clown anemonefish, A. ocellaris, at the sites. (B) Shows the summarized results of the BARRIER analysis (Manni et al., 2004) based on two datasets (control region and microsatellites, A. ocellaris) and STRUCTURE analysis. For BARRIER, the 10 strongest/most important barriers to gene flow are shown. Red lines indicate barriers detected in both data sets, while orange lines indicate barriers detected in only one dataset. The polygon structures (gray lines/grit) are Voronoï-Tesselations (Voronoï, 1908) as implemented in the software BARRIER (Manni et al., 2004). The pie charts depict the relative proportion of membership of individuals from each site in the three genotype clusters as generated by STRUCTURE (ver. 2.2., Pritchard et al., 2000). The analysis was run for all k from 1 to 10, with the highest probability of correctly dividing the data into k = 3 groups (Supplementary Material 3, Table 1). The analysis used the LOCPRIOR model on 316 individuals, from 19 sites, using data from 6 microsatellite loci. The burn-in period was set to 80,000 and 100,000 replicates were generated.
The BARRIER analysis, conducted for both datasets of A. ocellaris, revealed a high number of barriers in the southern area (Figure 4), concordant with the high number of separate groups resolved with the hierarchical AMOVA (Figure 3) and the structure detected with STRUCTURE (Figure 4). High order barriers (i.e., strongest restrictions to gene flow) derived from both datasets (bold red lines in Figure 4) indicate a concordant barrier to gene flow to Puntondo, the most distant population, 60 km south of the archipelago, and between Samalona in the South and all other sites. In general, single marker and concordant (both markers) barriers were detected most frequently among the southern sites and between these sites and more northern locations. Barriers based on CR data alone, separate almost all islands in the southern region of the archipelago. More northern sites experience less barriers to geneflow. One prominent concordant barrier was shown running in North-South direction, separating Karrang Kassi as a northwestern location from the northern rim and sites closer to the shore in the East (Figure 4).
Polycarpa aurata
The overall AMOVA for the sea squirt, P. aurata, revealed a significant structure in the microsatellite dataset (Fst = 0.02; p = 0.001). The hierarchical AMOVA showed the highest significant value (Fct = 0.024; p = 0.001) for nine groups (Figure 5). The displayed patterns were similar to the groupings of A. ocellaris, with fine scale differentiation of only one or a few reefs. Connectivity is highest among sites in the middle of the shelf (Figure 5, light green circle), and slightly more pronounced in this species, than in A. ocellaris (light green circle, Figure 3). This indicates higher connectivity of P. aurata in central Spermonde, as well as between the shelf edge and the offshore sites in the northern part (Suranti and Gs. Tuarajae; blue oval), when comparing with results for A. ocellaris (microsatellite dataset).
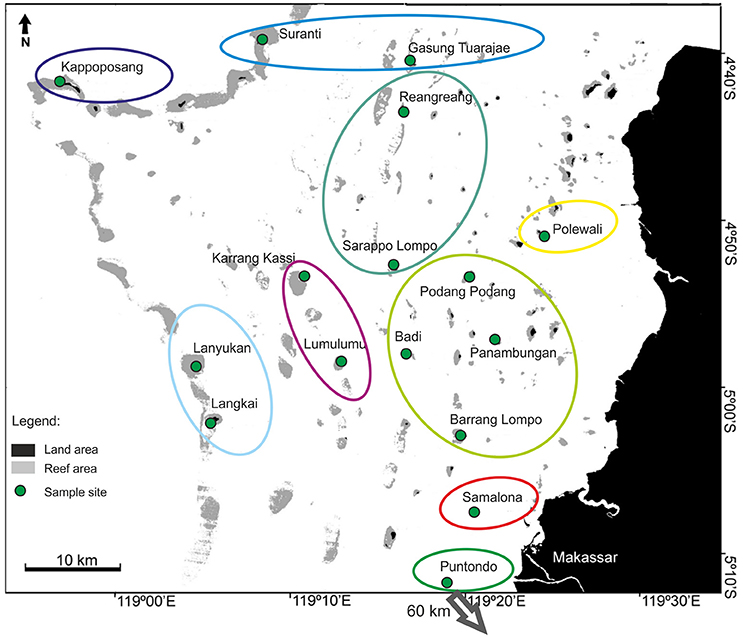
Figure 5. Results of the hierarchical AMOVA (Arlequin ver.3.5, Excoffier et al., 2005) for the sea squirt, Polycarpa aurata, in Spermonde Archipelago and Puntondo, Indonesia. Colored circles represent the highest significant Fct-value (0.024; p = 0.001), dividing the dataset into 9 groups.
The results of the STRUCTURE analysis for P. aurata revealed a most probable subdivision of the data into three clusters (k = 3, Supplementary Material 3B, Figure 6).
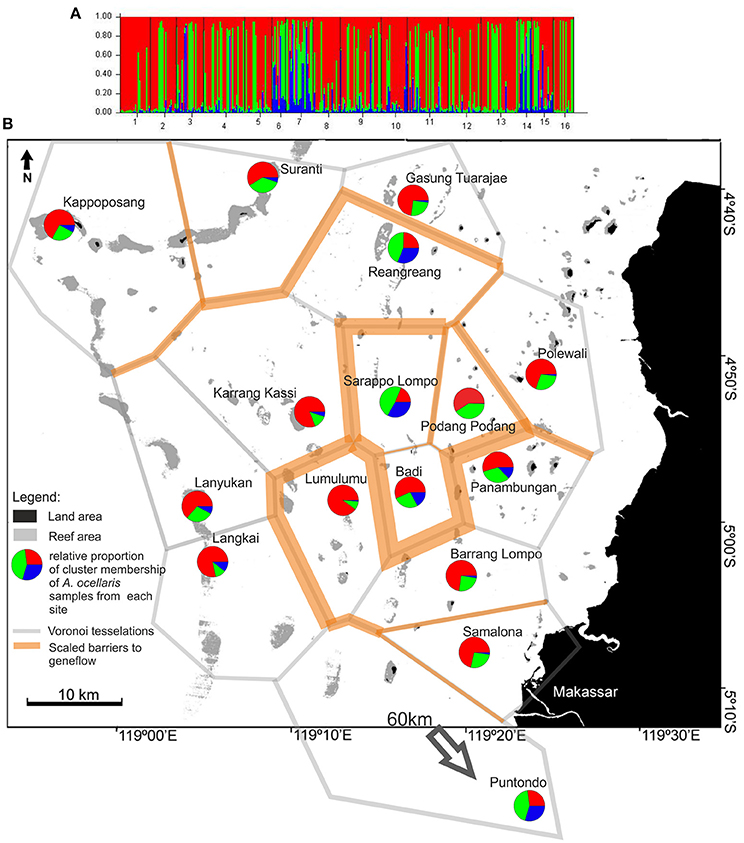
Figure 6. (A) of the figure shows the frequency diagram generated by STRUCTURE (ver. 2.2., Pritchard et al., 2000) as part of the analysis output showing the proportional cluster membership of Polycarpa aurata individuals at sites. (B) Shows the summarized results of the BARRIER analysis (Manni et al., 2004) based on two datasets (control region and microsatellites, P. aurata) and a projection of the STRUCTURE analysis. For BARRIER, the 10 strongest/most important barriers to gene flow are shown. Red lines indicate barriers detected in both data sets, while orange lines indicate barriers detected in only one dataset. The polygon structures (gray lines/grit) are Voronoï-Tesselations (Voronoï, 1908) as implemented in the software BARRIER (Manni et al., 2004). The pie charts depict the relative proportion of membership of individuals from each site in the three genotype clusters as generated by STRUCTURE (ver. 2.2., Pritchard et al., 2000). The analysis was run for all k from 1 to 10, with the highest probability of correctly dividing the data at k = 3 groups (Supplementary Material 3, Table 1). The analysis used the LOCPRIOR model on 332 individuals, from 16 sites, using data from 5 microsatellite loci. The burn-in period was set to 80,000 and 100,000 replicates were generated.
The distribution of the clusters among the sample sites in Spermonde showed no clear regional subdivision within the archipelago. In most sites “Red” was the dominant cluster and present at all sites, while “Blue” was present in larger proportions (>) only at three sites, in different parts of the study area (middle of the archipelago and Puntondo, Figure 6).
Overall, the conducted analyses with all datasets in the studied species revealed a small scale genetic population structure, especially in the southern region of the shelf. Puntondo is clearly separated from Spermonde archipelago, underlining its geographic distance. Samalona (directly offshore from the city of Makassar), was significantly differentiated in all datasets and in all analyses, although well inside the archipelago and not further apart than other sites. Similarly, Kappoposang was differentiated in part of the microsatellite analyses, but not as pronounced. The site is located at the drop-off (around 100 m depth) of the most northwestern shelf edge, directly adjacent to the Makassar Strait and exposed to the strong current of the Indonesian Throughflow (ITF).
Discussion
Spermonde Archipelago is characterized by a complex arrangement of islands, impacted by a wide range of conditions and threats. In addition to warming oceans under climate change, these ecosystems have to cope with the influx of nutrients, toxins, and sediment (Plass-Johnson et al., 2015), in addition to overexploitation, destructive fishing practices and other mechanic impacts, such as anchoring and solid waste. Understanding the vulnerability of reef organisms under these conditions is very important to ensure that measures for protection, monitoring and sustainable exploitation are put into place. This is urgently needed in order to prepare reefs and reef resource users for changing conditions. Studies that examine the genetic diversity and connectivity of local populations are very helpful in this context. By revealing general and reef specific patterns, conservation and management efforts can focus on vulnerable and/or impacted populations (e.g., identifying the most isolated populations and/or those most impacted by exploitation or other decimating mechanisms).
Genetic Diversity
Amphiprion ocellaris
The CR data analysis revealed a significantly lower genetic diversity (nucleotide diversity) in the southern shelf populations of A. ocellaris. A possible explanation for the detected diversity distribution could be the bathymetry of the study area, with reduced current mediated larval transport and limited exchange to other populations in the shallower sea basins of the southern near shore area. However, the very low genetic diversity at Kodingareng Lompo (most southern Spermonde site) indicates that other factors might have further reduced the diversity at this site (e.g., strong exploitation) and close attention should be paid to this result. Higher influx of “new” genetic material via the ITF at the shelf edge and stronger currents in the deeper northern sea basins around sites further from shore, may explain the higher diversity found there. In these site-attached organisms, larval transport is the dominant mechanism by which population connectivity can be achieved and maintained, so ocean currents are an important factor to be considered. However, this explanation is speculative and needs to be revisited when data for the local or regional shelf currents are available.
Unfortunately, the overall asymmetry of genetic diversity on the shelf was not confirmed by the microsatellite data. The inconsistencies among results from the two marker systems further prevents reliable conclusions about the detected reduction in diversity and how far this indicates a severe genetic depletion and immediate threat to the populations of the local reefs. However, the microsatellite data for A. ocellaris showed that Karrang Kassi had the lowest genetic diversity, which could be due to the high fishing pressure at that location and often used destructive fishing methods. A low nucleotide diversity in Karrang Kassi was confirmed by CR data, being lowest among northern and middle shelf sites and rather in the range of the values detected at sites further south and inshore (group 4, Figure 2). Findings of a lower diversity measured by microsatellite analysis at Barrang Lompo (middle shelf area) possibly reflect the higher fishing pressure compared to Samalona (South of Barrang Lompo, closer to shore) (Madduppa, 2012).
Generally, nucleotide diversity measures on such small scales should be interpreted with caution, as it is commonly accepted that sequence markers are mainly used to answer questions concerning evolutionary time scales and not ecological time scales (or accordingly, smaller geographic scales) due to slow mutation rates. Mutation rates in microsatellites, on the other hand, are considered fast enough to reflect processes on ecological timescale. Naturally, these scales may overlap when mutation rates in sequence markers are extremely high. The hypervariable fragment of the mitochondrial Control Region included in the present study has very high mutation rates and variability, compared to other parts of the mitochondrial DNA (Alvarado Bremer et al., 1995). The region contains insertions and many single nucleotide mutations, resulting in a high proportion of private and rare haplotypes and very low haplotype frequencies in general. These characteristics of the marker leads to uninformative values when F- statistics are calculated based on haplotype frequencies only. The genetic distances between haplotypes provide important information when the mutation rate of the marker is very high, as is the case here. Additionally, the studied species is marked by high self-recruitment rates, strong site attached behavior and a short PLD. This suggests that even short geographic distances may limit genetic exchange enough to drive differentiation of smaller sub-populations in a meta-population over small spatial and temporal scales.
Despite medium to high genetic diversity (Timm and Kochzius, 2008; Timm et al., 2012; Dohna et al., 2015) currently measured at many sites, exploitation and destructive fishing practices continue to impact the reefs of Spermonde and may further decrease genetic diversity. The effect may be particularly strong in the southern area of the archipelago, close to the shore, where access and exploitation are already high and genetic evidence suggests localized reduction in diversity.
Connectivity
Amphiprion ocellaris and Polycarpa aurata
The connectivity patterns detected for both species best describe meta-populations with restricted gene flow due to hidden barriers between distant and neighboring subpopulations (Hanski, 1998). This is best exemplified in the southern and more near shore areas of the archipelago. Similarities between the two studied species indicate that local bathymetry and current features may dictate larval dispersal to a large degree. Deeper and larger sea basins in areas further from shore (Cleary et al., 2005) lead to stronger currents supported by the ITF, moving from North to South through the Strait of Makassar while shallower basins in the southern inshore shelf areas are less exposed, causing more localized genetic exchange. Active swimming behavior plays a more important role at such distances, especially for the dispersal of fish larvae. It is known that anemonefish larvae can sense suitable reef habitat, in particular their natal reef, and actively swim toward it. This increases their chances of finding suitable settlement habitat at the end of the PLD (Jones et al., 2005; Dixson et al., 2008). Invertebrate larvae, like that of P. aurata, do not show the same level of recognition and swimming activity (Bradbury and Snelgrove, 2001), leaving them more prone to passive drifting with currents. This could explain a higher degree of connectivity between sites at the shelf edge and locations further on-shelf, as well as among sites in the deeper areas of the shelf, when compared to A. ocellaris. This emphasizes that, despite the general patterns that were detected, species specific larval behavior may need to be considered.
The most differentiated sites in all datasets were predominantly located at the periphery of the study area. Puntondo is located outside the archipelago and most distant to the other sites, therefore a restriction in connectivity was expected. The results of the STRUCTURE analysis indicate higher connectivity between Puntondo and sites at the outer edge of Spermonde, both affected by the ITF coming from the North through the Strait of Makassar. Additionally, Puntondo displayed high genetic diversity, which might be due to its geographic location South of Spermonde, close to the tip of south-western Sulawesi, adjacent to the monsoon driven major currents between the Java and Flores Seas. Therefore, the population in Puntondo might be subject to gene flow from other regions in the Indo-Malay Archipelago (Timm and Kochzius, 2008; Timm et al., 2012). Kappoposang, at the northern shelf edge, is located directly in the path of the ITF and therefore influenced by strong currents that transport larvae from upstream populations. The same should be valid for the other more eastern shelf edge sites, but contrary to expectations, Kappoposang was differentiated from these sites (microsatellite data of both species). Its prominent position on the most western and northern tip of the shelf edge may provide an explanation for this result.
Samalona, immediately offshore from the city of Makassar, was differentiated in all datasets, although geographically near to neighboring reefs (<10 km). In another study, the anemonefish population of this reef showed high proportions of self-recruitment (up to 60%; Madduppa et al., 2014b) and a high proportion of private alleles (Madduppa, 2012), explaining signals of restricted connectivity to other reefs. High self-recruitment was generally measured for A. ocellaris (Madduppa et al., 2014b) and its sibling species A. percula (Berumen et al., 2012), raising the question, if the degree of self-recruitment would differ in the more offshore and shelf edge areas of the archipelago, being influenced by stronger currents and increased larval input from other populations. This question should be investigated in a few select populations to draw important conclusions in this regard.
The sub-divisions inferred in all datasets indicate restrictions in gene flow over small geographic distances, especially in the Southeast of the study area. These results, combined with the high self-recruitment found in earlier studies for this fish species, suggest that small (enforced) MPAs for each island or submerged reef would prove effective for the persistence of populations in the studied species. The same should be valid for coral reef species with similar reproductive strategies and adult behavior, leading to similarly low dispersal potential as in the clown anemonefish and the sea squirt P. aurata. The larval exchange necessary for maintaining genetic diversity is possible over the relatively short distances between reefs in Spermonde, as long as reefs of such a network maintain a breeding stock and suitable habitat as “stepping stones” for connectivity in the form of enforced MPAs. However, with an overall decrease in the quality of reef habitat the availability of suitable habitat may well act as the bottleneck in this scenario.
Since larvae tend to avoid their natal host on their home reef when settling, protecting a part of the reef as a no-take zone would still ensure reseeding of exploited parts of the reef through “spill-over,” as was shown to be effective in earlier studies (Harrison et al., 2012), and can lead to a sustainable use of this particular resource for the ornamental trade. Protection of no-take zones by coastal law enforcement agencies and through local community schooling and participation is crucial for the functioning of a reserve network as a conservation and stock replenishment strategy (Ferse et al., 2010; Radjawali, 2012). Currently, the established system of MPAs receives no protection and much work and effort has to go into establishing and sustaining protective measures on all levels. Additionally, the amount of “spillover” that can be expected from a network of small scale no-take MPAs needs to be truly tried and tested to be able to evaluate its potential and effect for the recovery and sustenance of the stocks in the fished zones and its effective value for the local resource users.
Ethics Statement
This study was carried out in accordance with the recommendations of the Ministry of Marine Affairs and Fisheries (KKP), Jakarta, Indonesia. The species of the study are not CITES listed. Small tissue samples were taken in situ, the specimens were not extracted from their habitat, or killed for the sampling. In order to reduce long-lasting changes in regional populations or substantially reducing the number of individuals present, the sampling method was chosen was only minimally invasive and approved by scientists of the Research and Development Center for Marine, Coastal, and Small Islands (MaCSI), Hasanuddin University, Makassar and the Indonesian Ministry of Research and Technology. The fish were caught with hand nets and released back into their host anemone after sampling. No anesthetic or toxic additive was used, so neither the populations of the species, other organisms in the surrounding area, nor the habitat or ecosystem were threatened.
For the present study, no experiments were conducted on the living individuals, but small tissue samples of about 2 mm2 (specific size according to individual size of the specimens) were taken with fine scissors. This method of sampling, the so called finclipping, is well established and implies as little impact on the specimens as possible. For the sea squirt species, similarly small tissue samples were taken with scissors from the opening edge. Small injuries at the caudal fin or opening edge, respectively, occur as well naturally through predatory species in the habitat or intraspecific competition in the fish species, and usually heal quickly in healthy individuals.
For sampling and transfer of samples to Germany, a Material Transfer Agreement between German and Indonesian project partners were signed and no further approval of an ethical committee was needed for the sampling process. Sampling of coral reef organisms means sampling in sensitive habitat. To avoid physical, chemical, biological or geological changes or damage to marine habitats, hand nets were chosen as the least destructive method. Divers were well trained and equipped for sampling in coral reefs and boat staff was instructed to avoid anchor or propeller damage to the reef.
Author Contributions
JT: Study development and planning, as well as sampling, conducting of experiments, analysis and manuscript writing. TD: Conducting part of the experiments and analysis, writing and reviewing manuscript. HM: Conducting part of the sampling and experiments. AN: Sampling, standardizing, and conducting part of the experiments. MK: Study development and planning, as well as part of the sampling and writing support.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the German Federal Ministry of Education and Research (BMBF, grant no. 03F0390B, 03F0472B and 03F0643B) for funding; Prof. Dr. D. Blohm for support and sub-project coordination; the Centre for Tropical Marine Ecology (Bremen, Germany) for support, cooperation, and project coordination; R. Zelm (University of Bremen) for technical advice, A. Nuryanto (Jenderal Soedirman University, Purwokerto, Indonesia) and scientists and students of the Hasanuddin University (Makassar, Indonesia) for logistics and help during field work; the competent authorities for permits. The SPICE project was conducted and permitted under the governmental agreement between the German Federal Ministry of Education and Research (BMBF) and the Indonesian Ministry for Research and Technology (RISTEK), Indonesian Institute of Sciences (LIPI), Indonesian Ministry of Maritime Affairs and Fisheries (DKP), and Indonesian Agency for the Assessment and Application of Technology (BPPT).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fmars.2017.00294/full#supplementary-material
References
Allen, G. R. (2008). Conservation hotspots of biodiversity and endemism for Indo-Pacific coral reef fishes. Aqua. Conserv. 18, 541–556. doi: 10.1002/aqc.880
Allen, G. R., and Werner, T. B. (2002). Coral reef fish assessment in the “coral triangle” of Southeastern Asia. Environ. Biol. Fishes. 65, 209–214. doi: 10.1023/A:1020093012502
Almany, G. R., Connolly, S. R., Heath, D. D., Hogan, J. D., Jones, G. P., McCook, L. J., et al. (2009). Connectivity, biodiversity conservation and the design of marine reserve networks for coral reefs. Coral Reefs. 28, 339–351. doi: 10.1007/s00338-009-0484-x
Alvarado Bremer, J. R., Baker, A. J., and Mejuto, J. (1995). Mitochondria1 DNA control region sequences indicate extensive mixing of swordfish gladius populations in the Atlantic Ocean. Can. J. Fish. Auqat. Sci. 52, 1720–1732. doi: 10.1139/f95-764
Berumen, M. L., Almany, G. R., Planes, S., Jones, G. P., Saenz-Agudelo, P., and Thorrold, S. R. (2012). Persistence of self-recruitment and patterns of larval connectivity in a marine protected area network. Ecol. Evol. 2, 444–453. doi: 10.1002/ece3.208
Bradbury, I. R., and Snelgrove, P. V. R. (2001). Contrasting larval transport in demersal fish and benthic invertebrates: the roles of behaviour and advective processes in determining spatial pattern. Can. J. Fish. Aquat. Sci. 58, 811–823. doi: 10.1139/f01-031
Brassel, K. E., and Reif, D. (1979). A procedure to generate Thiessen polygons. Geogr. Anal. 325, 31–36. doi: 10.1111/j.1538-4632.1979.tb00695.x
Briggs, J. C. (2009). Diversity, endemism and evolution in the Coral Triangle. J. Biogeogr. 36, 2008–2010. doi: 10.1111/j.1365-2699.2009.02146.x
Burke, L., Selig, E., and Spalding, M. (2006). Reefs at Risk in Southeast Asia. Washington, DC: World Resources Institute.
Buston, P. M., Bogdanowicz, A. W., and Harrison, R. G. (2007). Are clownfish groups composed of close relatives? An analysis of microsatellite DNA variation in Amphiprion percula. Mol. Ecol. 16, 3671–3678. doi: 10.1111/j.1365-294X.2007.03421.x
Cleary, D. F. R., Becking, L. E., de Voogd, N. J., Renema, W., de Beer, M., van Soest, R. W. M., et al. (2005). Variation in the diversity and composition of benthic taxa as a function of distance offshore, depth and exposure to the Spermonde Archipelago, Indonesia. Estuar. Coast. Shelf Sci. 65, 557–570. doi: 10.1016/j.ecss.2005.06.025
Cowen, R. K., Paris, C. B., and Srinivasan, A. (2006). Scaling of connectivity in marine populations. Science 311, 522–527. doi: 10.1126/science.1122039
Davis, M. H., and Davis, M. E. (2007). The distribution of Styela clava (Tunicata, Ascidiacea) in European waters. J. Exp. Mar. Biol. Ecol. 342, 182–184. doi: 10.1016/j.jembe.2006.10.039
Dixson, D. L., Jones, G. P., Munday, P. L., Planes, S., Pratchett, M. S., Srinivasan, M., et al. (2008). Coral reef fish smell leaves to find island homes. Proc. R. Soc. B. 275, 2831–2839. doi: 10.1098/rspb.2008.0876
Dohna, T. A., Timm, J., Hamid, L., and Kochzius, M. (2015). Limited connectivity and a phylogeographic break characterize populations of the pink anemonefish, Amphiprion perideraion, in the Indo-Malay Archipelago: inferences from a mitochondrial and microsatellite loci. Ecol. Evol. 5, 1717–1733. doi: 10.1002/ece3.1455
Edinger, E. N., Jompa, J., Limmon, G. V., Widjatmoko, W., and Risk, M. J. (1998). Reef degradation and coral biodiversity in Indonesia: effects of land-based pollution, destructive fishing practices and changes over time. Mar. Poll. Bull. 36, 617–630. doi: 10.1016/S0025-326X(98)00047-2
Ehlers, A., Worm, B., and Reusch, T. B. H. (2008). Importance of genetic diversity in eelgrass Zostera marina for its resilience to global warming. Mar. Ecol. Prog. Ser. 355, 1–7. doi: 10.3354/meps07369
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50.
Ferse, S. C. A., Manez Costa, M., Schwerdtner Manez, K., Adhuri, D. S., and Glaser, M. (2010). Allies, not aliens: increasing the role of local communities in marine protected area implementation. Environ. Conserv. 37, 1–12. doi: 10.1017/S0376892910000172
Glaser, M., Breckwoldt, A., Deswandi, R., Radjawali, I., Baitoningsih, W., and Ferse, S. C. A. (2015). Of exploited reefs and fishers - A holistic view on participatory coastal and marine management in an Indonesian archipelago. Ocean Coast. Manag. 116, 193–213. doi: 10.1016/j.ocecoaman.2015.07.022
Gopakumar, G., Madhu, K., Madhu, R., Ignatius, B., Krishnan, L., and Mathew, G. (2009). Broodstock development, breeding and seed production of selected marine food fishes and ornamental fishes. Mar. Fish Inf. Serv. 201, 1–9.
Goudet, J. (1995). FSTAT (Version 1.2): a computer program to calculate F-Statistics. J. Hered. 86, 485–486. doi: 10.1093/oxfordjournals.jhered.a111627
Hall, T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Harrison, H. B., Williamson, D. H., Evans, R. D., Almany, G. R., Thorrold, S. R., Russ, G. R., et al. (2012). Larval export from marine reserves and the recruitment benefit for fish and fisheries. Curr. Biol. 22, 1023–1028. doi: 10.1016/j.cub.2012.04.008
Hughes, A. R., and Stachowicz, J. J. (2004). Genetic diversity enhances the resistance of a seagrass ecosystem to disturbance. Proc. Natl. Acad. Sci. U.S.A. 101, 8998–9002. doi: 10.1073/pnas.0402642101
Hughes, T. P., Baird, A. H., Bellwood, D. R., Card, M., Connolly, S. R., Folke, C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933. doi: 10.1126/science.1085046
Jones, G. P., Almany, G. R., Russ, G. R., Sale, P. F., Steneck, R. S., van Oppen, M. J. H., et al. (2009). Larval retention and connectivity among populations of corals and reef fishes: history, advances and challenges. Coral Reefs. 28, 307–325. doi: 10.1007/s00338-009-0469-9
Jones, G. P., Planes, S., and Thorrold, S. R. (2005). Coral reef fish larvae settle close to home. Curr. Biol. 15, 1314–1318. doi: 10.1016/j.cub.2005.06.061
Kaiser, M., Attrill, M., Jennings, S., Thomas, D., Barnes, D., Brierley, A., et al. (2005). Marine Ecology: Processes, Systems, and Impacts. Oxford: Oxford University Press.
Kott, P. (1985). The australian ascidiacea. part 1, Phlebobranchia and Stolidobranchia. Mem. Queensl. Mus. 23, 1–439
Lee, W. J., Howell, W. H., and Kocher, T. D. (1995). Structure and evolution of teleost mitochondrial control regions. J. Mol. Evol. 41, 54–66. doi: 10.1007/BF00174041
Madduppa, H. H. (2012). Self-recruitment in Anemonefish and the Impact of Marine Ornamental Fishery in Spermonde Archipelago, Indonesia: Implications for Management and Conservation. Doctoral thesis, University of Bremen, Bremen, Germany. Available online at: http://elib.suub.uni-bremen.de/peid=D00102836
Madduppa, H. H., Juterzenka, K., Syakir, M., and Kochzius, M. (2014a). Socio-economy of marine ornamental fishery and its impact on the population structure of the clown anemonefish Amphiprion ocellaris and its host anemones in Spermonde Archipelago, Indonesia. Ocean Coast. Manag. 100, 41–50. doi: 10.1016/j.ocecoaman.2014.07.013
Madduppa, H. H., Timm, J., and Kochzius, M. (2014b). Interspecific, spatial and temporal variability of self-recruitment in anemonefishes. PLoS ONE 9:e90648. doi: 10.1371/journal.pone.0090648
Manni, F., Guerard, E., and Heyer, E. (2004). Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by “Monmonier's algorithm”. Hum. Biol. 76, 173–190. doi: 10.1353/hub.2004.0034
Monmonier, M. (1973). Maximum-difference barriers: an alternative numerical regionalization method. Geogr. Anal. 3, 245–261. doi: 10.1111/j.1538-4632.1973.tb01011.x
Planes, S., Jones, G. P., and Thorrold, S. R. (2009). Larval dispersal connects fish populations in a network of marine protected areas. Proc. Natl. Acad. Sci. U.S.A. 106, 5693–5697. doi: 10.1073/pnas.0808007106
Plass-Johnson, J. G., Ferse, S. C. A., Jompo, J., Wild, C., and Teichberg, M. (2015). Fish herbivory as key ecological function in a heavily degraded coral reef system. Limnol. Oceanogr. 60, 1382–1391. doi: 10.1002/lno.10105
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Quenouille, B., Bouchenak-Khelladi, Y., Hervet, C., and Planes, S. (2004). Eleven microsatellite loci for the saddleback clownfish Amphiprion polymnus. Mol. Ecol. Notes. 4, 291–293. doi: 10.1111/j.1471-8286.2004.00646.x
Radjawali, I. (2012). Examining local conservation and development: live reef food fishing in Spermonde Archipelago, Indonesia. J. Integr. Coast. Zone Manag. 12, 545–557. doi: 10.5894/rgci337
Svane, I., and Young, C. (1989). The ecology and behaviour of ascidian larvae. Oceanogr. Mar. Biol. 27, 45–90.
Timm, J., and Kochzius, M. (2008). Geological history and oceanography of the Indo-Malay Archipelago shape the genetic population structure in the false clown anemonefish (Amphiprion ocellaris). Mol. Ecol. 17, 3999–4014. doi: 10.1111/j.1365-294X.2008.03881.x
Timm, J., Planes, S., and Kochzius, M. (2012). High similarity of genetic population structure in the false clown anemonefish (Amphiprion ocellaris) found in microsatellite and mitochondrial control region analysis. Conserv. Genet. 13, 693–706. doi: 10.1007/s10592-012-0318-1
Tomascik, T., Mah, A., Nontji, A., and Moosa, M. (1997). The Ecology of Indonesian Seas, Part II. Hong Kong: Periplus Edition Ltd.
Veron, J. E. N., Devantier, L. M., Turak, E., Green, A. L., Kininmonth, S., Stafford-Smith, M., et al. (2009). Delineating the coral triangle. Galaxea. J. Coral Reef Stud. 11, 91–100. doi: 10.3755/galaxea.11.91
Voronoï, M. G. (1908). Nouvelles application des paramètres continus à la théorie des formes quadratiques, deuxième mémoire, recherche sur le paralléloedres primitifs. J. Reine Angew. Math. 134, 198–207.
Wabnitz, C., Taylor, M., Green, E., and Razak, T. (2003). From Ocean to Aquarium: The Global Trade in Marine Ornamental Species. Cambridge: UNEP-WCMC.
Wainwright, B., Arlyza, I., and Karl, S. (2013). Isolation and characterization of twenty-one polymorphic microsatellite loci for Polycarpa aurata using third generation sequencing. Conserv. Genet. Resour. 5, 671–673. doi: 10.1007/s12686-013-9879-7
Wild, C., Hoegh-Guldberg, O., Naumann, M. S., Colombo-Pallotta, M. F., Ateweberhan, M., Fitt, W. K., et al. (2011). Climate change impedes scleractinian corals as primary reef ecosystem engineers. Mar. Freshw. Res. 62, 205–215. doi: 10.1071/MF10254
Keywords: Polycarpa aurata, clownfish, Amphiprion ocellaris, indo-malay archipelago, marine populations, population connectivity, self-recruitment, coral triangle
Citation: Timm J, Kochzius M, Madduppa HH, Neuhaus AI and Dohna T (2017) Small Scale Genetic Population Structure of Coral Reef Organisms in Spermonde Archipelago, Indonesia. Front. Mar. Sci. 4:294. doi: 10.3389/fmars.2017.00294
Received: 12 February 2017; Accepted: 28 August 2017;
Published: 25 September 2017.
Edited by:
Angel Borja, AZTI Pasaia, SpainReviewed by:
Xavier Turon, Consejo Superior de Investigaciones Científicas (CSIC), SpainAnne Chenuil, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2017 Timm, Kochzius, Madduppa, Neuhaus and Dohna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Janne Timm, jatimm@uni-bremen.de
 Janne Timm
Janne Timm Marc Kochzius
Marc Kochzius Hawis H. Madduppa
Hawis H. Madduppa Anouk I. Neuhaus
Anouk I. Neuhaus Tina Dohna
Tina Dohna