Assessing the Functional Limitations of Lipids and Fatty Acids for Diet Determination: The Importance of Tissue Type, Quantity, and Quality
- 1College of Science and Engineering, Flinders University, Bedford Park, SA, Australia
- 2CSIRO Oceans and Atmosphere, Hobart, TAS, Australia
- 3SARDI Aquatic Sciences, South Australian Research and Development Institute, West Beach, SA, Australia
- 4Griffith Centre for Coastal Management, Griffith University, Gold Coast, QLD, Australia
Lipid and fatty acid (FA) analysis is commonly used to describe the trophic ecology of an increasing number of taxa. However, the applicability of these analyses is contingent upon the collection and storage of sufficient high quality tissue, the limitations of which are previously unexplored in elasmobranchs. Using samples from 110 white sharks, Carcharodon carcharias, collected throughout Australia, we investigated the importance of tissue type, sample quantity, and quality for reliable lipid class and FA analysis. We determined that muscle and sub-dermal tissue contain distinct lipid class and FA profiles, and were not directly comparable. Muscle samples as small as 12 mg dry weight (49 mg wet weight), provided reliable and consistent FA profiles, while sub-dermal tissue samples of 40 mg dry weight (186 mg wet weight) or greater were required to yield consistent profiles. This validates the suitability of minimally invasive sampling methods such as punch biopsies. The integrity of FA profiles in muscle was compromised after 24 h at ambient temperature (~20°C), making these degraded samples unreliable for accurate determination of dietary sources, yet sub-dermal tissue retained stable FA profiles under the same conditions, suggesting it may be a more robust tissue for trophic ecology work with potentially degraded samples. However, muscle samples archived for up to 16 years in −20°C retain their FA profiles, highlighting that tissue from museum or private collections can yield valid insights into the trophic ecology of marine elasmobranchs.
Introduction
The field of trophic ecology has seen a substantial increase in the number of available techniques and applications across aquatic and terrestrial taxa within the last half century (Layman et al., 2012, 2015; Christiansen et al., 2015; Nielsen et al., 2015; Young et al., 2015; Roslin and Majaneva, 2016). More recently, there has been a growing number of studies moving from traditional stomach-content analysis, which may provide a potentially limited view due to differences in digestibility among prey species (Hyslop, 1980), to time-integrated biochemical methods (reviewed in Traugott et al., 2013; Pethybridge et al., 2018). Lipid and fatty acid (FA) analysis is one such method growing in popularity as it has the capacity to elucidate key biological and ecological aspects, such as an organism's physiology and bioenergetics (Parrish et al., 2007; Pond and Tarling, 2011), and most often, trophic relationships (e.g., Bradshaw et al., 2003; Iverson et al., 2004; Budge et al., 2006). As per the saying “you are what you eat,” certain FAs are transferred from prey to predator with minimal modification (Iverson et al., 2004; Budge et al., 2006), allowing certain functional trophic groups to be traced within a food chain. Owing to this broad applicability, more than 29,000 published studies featured FA analysis for marine and aquatic taxa alone between 1990 and 2014 (Rudy et al., 2016).
The applicability of FA analysis is especially pertinent for threatened and iconic species for which lethal sampling, which is often used to obtain stomach contents, is not possible especially for large numbers of specimens. Instead, minimally invasive biopsy techniques are often employed to obtain tissue samples for biochemical studies (e.g., Hooker et al., 2001; Carlisle et al., 2012; Hussey et al., 2012). With the development of specialized biopsy probes (Reeb and Best, 2006; Robbins, 2006; Daly and Smale, 2013), tissue samples can be obtained from free-swimming marine organisms, reducing the stress and detrimental effects of the capture and release process, and enabling the increased use of FA analyses across a number of species, including threatened elasmobranchs (Couturier et al., 2013; Rohner et al., 2013; Every et al., 2016).
The accuracy and reliability of biochemical analyses are dependent on the methods used to collect and store samples. Sampling elasmobranchs in particular poses a series of logistical challenges, due in part to the large proportion of species considered at risk of extinction (Dulvy et al., 2014), leading to samples often being difficult and expensive to obtain. As a result, these samples are often highly valuable and one needs to understand the functional limitations of collecting and storing these tissues to maximize sampling opportunities and reliability of resulting data.
The increasing use of biopsies to collect tissues from elasmobranchs has led to constraints on the type, amount, and quality of tissue collected. Beneath the epidermis, elasmobranchs contain a deep sub-dermal layer of collagen and elastin fibers, which varies in thickness between species (Motta, 1977). The underlying physiological differences between the two tissue types (muscle, a metabolically active and protein-rich tissue vs. sub-dermal tissue, a less bioactive and largely structural tissue composed of elastin and collagen) results in distinct biochemical properties, with the potential to yield different ecological data. This is evidenced by recent isotopic studies on white sharks, Carcharodon carcharias, whereby muscle and sub-dermal tissue had the same 15N isotopic signatures, but divergent 13C signatures, which was attributed to differing tissue-specific incorporation rates (Carlisle et al., 2012; Kim et al., 2012; Jaime-Rivera et al., 2013). How these tissue-specific physiological and biochemical differences manifest in FA profiles remains poorly studied, with most elasmobranch work to date focused on the FA differences between skeletal muscle and the lipid-rich liver (e.g., Schaufler et al., 2005; Pethybridge et al., 2011; Beckmann et al., 2013), myocardial tissue (Davidson et al., 2011, 2014), and blood plasma (Ballantyne et al., 1993; McMeans et al., 2012). However, Every et al. (2016) recently showed differences in FA profiles between muscle tissue and fin clips (a mixed-tissue sample, including cartilage, connective tissue, muscle, vascularization and an outer dermal layer with denticles).
The functional limitations of various biopsy methods also extend to the amount of tissue obtained. With the thick epidermal layer serving as a barrier, collecting sufficient amounts of usable muscle from large elasmobranchs in particular, has proven challenging. The sub-dermal layer of white sharks can be up to 3 cm, hindering the ability to collect the underlying muscle (Jaime-Rivera et al., 2013). Whale sharks, Rhincodon typus, sampled with a biopsy probe penetrating ~2 cm yielded exclusively sub-dermal tissue (Rohner et al., 2013), whereas the ~2 cm biopsies of bull sharks, Carcharhinus leucas yielded 5% dermis, 40% sub-dermal and 55% muscle (Daly and Smale, 2013). These differences in the thickness of the sub-dermal layer complicate the collection of elasmobranch muscle samples. Although small amounts of tissue are sufficient for genetic [1 mg dry weight (DW), Kasajima et al., 2004] and stable isotope analysis (~10 mg DW, Jaime-Rivera et al., 2013), the minimum amount of muscle or sub-dermal tissue necessary for accurate FA analysis remains relatively unknown. Although Every et al. (2016) reported that FA were detectable in fin clips as small as 20 mg and muscle biopsies >10 mg dry weight, minimum sample sizes yielding consistent results were not quantitatively assessed. Such evaluations are vital however, particularly when considering the appropriateness of various biopsy probes, and the applicability of the sampling method across smaller elasmobranch species, from which removing large amounts of tissue is not feasible.
Appropriate sample acquisition, storage and tissue preservation is vital when applying FA analysis techniques, as certain FAs (particularly long-chain(≥C20) polyunsaturated FAs, LC-PUFAs) oxidize when exposed to air, high temperatures, and direct sunlight, leading to tissue degradation and loss of information (Budge et al., 2006). This becomes particularly challenging when there are scarce opportunities for sampling (e.g., for highly mobile, rare, or cryptic species) and when working in remote and hostile field locations (e.g., hot and humid tropics, and offshore sampling sites). Furthermore, despite the growing utilization of non-lethal biopsies, many FA studies use samples taken from deceased elasmobranch carcasses obtained from fisheries bycatch (Pethybridge et al., 2011), beach strandings (Rohner et al., 2013), and shark-control measures (Davidson et al., 2011, 2014; Pethybridge et al., 2014). Given the variable condition of these carcasses, which may have spent multiple days at ambient temperature, there is the high potential for lipid and FA degradation within samples collected via these means. Additionally, FA studies often use tissue samples collected over a long period of time (e.g., 5 years—Davidson et al., 2011, 2014; 2 years—Rohner et al., 2013; 12 years—Pethybridge et al., 2014 and 3 years—Jaime-Rivera et al., 2014), providing another opportunity for unchecked FA degradation throughout these long periods of frozen storage. Several recent studies examining storage procedures have revealed significant species- and tissue-specific lipid and FA degradation over the course of several months held at −20°C (Refsgaard et al., 1998; Roldán et al., 2005; Phleger et al., 2007; Sahari et al., 2014; Paola and Isabel, 2015; Rudy et al., 2016). To date, the focus of such investigations have remained limited to highly valued commercial teleost (Roldán et al., 2005; Paola and Isabel, 2015; Rudy et al., 2016) and cephalopod species (Gullian-Klanian et al., 2017). Despite this evidence of FA degradation, it remains unassessed for the many archived elasmobranch tissues stored over the period of months to years.
Given the aforementioned lack of information regarding the functional limitations and capabilities of lipid and FA biomarkers for application to highly mobile, rare or cryptic elasmobranchs, this study seeks to assess:
1) Differences in lipid content, lipid class, and FA profiles between muscle and sub-dermal tissue from white sharks;
2) The minimum muscle and sub-dermal tissue sample size required for consistent analysis of FA profiles; and
3) The effects of handling and freezing storage time on FA degradation via a controlled experiment with shark muscle tissue left at 20°C for 5 days, and by comparing profiles of shark tissue stored over known periods of time at −20°C, up to 16 years.
The knowledge gained from addressing these functional limitations will facilitate the more effective use of lipid and FA profiling on biopsied or potentially degraded tissues, allowing them to be employed with greater confidence in a range of ecological studies.
Materials and Methods
Sample Collection and Data Compilation
Tissue samples were collected from 110 white sharks, C. carcharias from South Australia (SA), New South Wales (NSW), and Queensland (QLD), Australia between 2000 and 2016 (Table 1). Tissues were obtained through punch-biopsies of live, free-swimming white sharks from the Neptune Islands, SA, opportunistically through fisheries bycatch, the NSW Department of Primary Industries Shark Meshing Program and QLD Department of Agriculture and Fisheries Shark Control Program as part of the QLD large shark tagging research program. Samples were frozen and stored from 3 weeks to 16 years at −20°C, until freeze-drying immediately prior to lipid analysis.
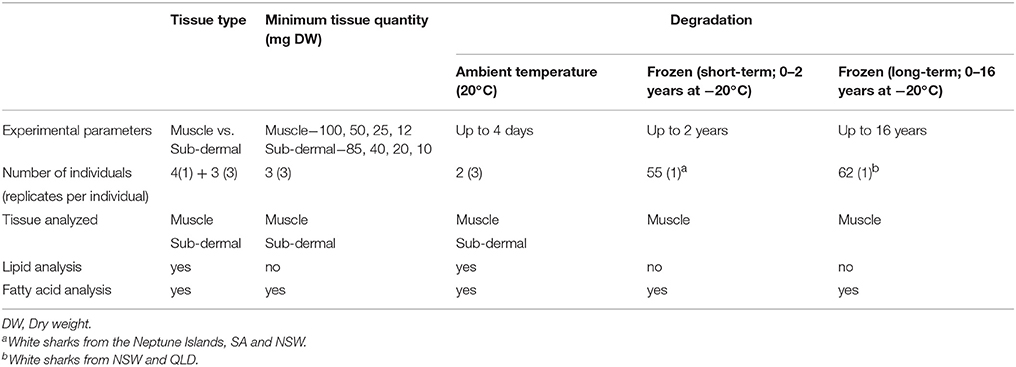
Table 1. Sample details across the three study aims, including the number of individual white sharks Carcharodon carcharias and the tissue and lipid parameter analyzed.
Ethics Statement
In South Australia, fieldwork at the Neptune Islands was carried out in accordance with ethics permit #E398, approved by The Flinders University Animal Welfare Committee, and under DEWNR permit # Q26292. In New South Wales, tissue collection under NSW DPI Scientific Collection Permit (P07/0099-3.0 and P07/0099-4) was approved by New South Wales Department of Primary Industries (NSW DPI) Animal Research Authority (ACEC 12/07). Tissue from Queensland was obtained as part of the QLD Shark Meshing Program and QLD Department of Agriculture and Fisheries Shark Control Program as part of the QLD large shark tagging research program under fisheries permit 143005 and QLD Department of Agriculture and Fisheries Shark Animal Ethics Committee approved ethics CA 2010/11/482, CA 2013/11/737, ENV 1709 AEC.
Experimental Design
Three sets of comparative lipid and FA analyses were undertaken, each addressing one of the aims; the difference between muscle and sub-dermal tissue, minimum tissue quantity for each tissue, and the effect of tissue degradation on resulting lipid and FA profiles (Table 1). To investigate the difference between the muscle and sub-dermal tissue, ~300 g sections, comprising both muscle and sub-dermal tissue were collected from three deceased white sharks (a, b, and c). Lipid class and FA profiles were assessed across triplicate subsamples from these three sharks (Table 1) to incorporate the within-individual variability. Minimum tissue quantity was also assessed in triplicate, across the three sharks, for both muscle and sub-dermal tissue using progressively smaller samples sizes. The tissue degradation analysis was performed in three parts: (i) at ambient temperature, and (ii) short term storage at −20°C (for up to 2 years), and (iii) long-term storage at −20°C (for up to 16 years). The remaining portions of sharks a and b were then held at room temperature (~20°C) for 4 days, and muscle and sub-dermal tissue were sub-sectioned in triplicate, every 24 h. Immediately prior to sub-sectioning, ~1 cm of the outermost edge was removed and discarded, allowing the sample to be taken from the interior of the tissue section. This was to minimize incidentally measuring the co-occurring effects of oxygen-contact induced FA oxidation on the samples. Only sharks a and b underwent the ambient temperature degradation trial, as there was insufficient remaining tissue from shark c.
The remaining 107 white shark muscle samples were used to assess both short- to mid-term (1 month up to 2 years) and long-term (1 month up to 16 years) FA profile degradation associated with storage at −20°C (Table 1). Forty-five samples from the Neptune Islands, SA and 10 of the 31 samples from NSW were processed within 2 years of being obtained and thus these were assessed together for short- to mid-term degradation (1 month up to 2 years). These results were grouped into 3 months bins for statistical analysis. Sixty-two muscle samples (31 from NSW, 31 from QLD) were assessed together for long-term freezer degradation (1 month up to 16 years). This excluded the 45 Neptune Islands samples included in short-term freezer degradation analysis, limiting the potential confounding factor of collection location within long-term degradation. These long-term freezer degradation results were also grouped into bins for statistical analysis, with group 1 = 0–1 years at −20°C, 2 = 1.1–2 years, 3 = 3–5 years, 4 = 6–10 years, 5 = 11–16 years.
Lipid Extraction
Total lipid was extracted using the modified Bligh and Dyer method (Bligh and Dyer, 1959). Briefly, samples were left overnight in a one-phase CH2Cl2:CH3OH:milliQ H2O mixture (10:20:8 mL) before the solution was broken into two phases by the addition of 10 mL CH2Cl2 and 10 mL of 9 g NaCL L−1 saline milliQ H2O. The lower phase containing the lipid fraction was drained into a round bottom flask and the solvent removed using a rotary evaporator. The lipid was re-suspended in CH2Cl2 and transferred to a 2 mL vial and dried under N2 gas until a constant weight was noted. The total lipid extract (TLE) was then re-suspended in 1.5 mL of CH2Cl2.
Lipid Content and Class Analysis
Water content, reported as percent of tissue wet weight, was determined for each sample by taking weights before and after freeze-drying at −82°C for 72 h and calculating the wet to dry ratio. Similarly, the lipid content was calculated by subtracting tissue dry weight prior to lipid extraction from the weight of the resulting TLE, then multiplied by the wet to dry ratio, and reported as percent of tissue wet weight.
Lipid class composition [triacylglycerols (TAG), phospholipids (PL), sterols (ST), wax esters (WE), and free fatty acids (FFA)] were measured using an Iatroscan Mark V TH10 thin layer chromatrograph coupled with a flame ion detector (TLC-FID). TLE from each sample was analyzed in triplicate. Aliquots of TLE were spotted onto chromarods and developed for 25 min in a polar solvent system [70:10:0.1 v/v/v, C6H14:(C2H5)2O:CH3COOH]. Rods were oven dried at 100°C for 10 min and analyzed immediately. SIC-480 Scientific Software was used to identify and quantify the areas of the resulting peaks.
Fatty Acid Analysis
An aliquot of the TLE was transferred into a teflon-lined screw cap glass test tube and trans-methylated with 3 mL of CH3OH: CH2Cl2:HCl (10:1:1 v/v/v) for 2 h at 80°C. The tube was then cooled in a water bath, and 1 mL MilliQ H2O was added. The resulting fatty acid methyl esters (FAME) were extracted into a 2 mL glass vial using three washes of C6H14: CH2Cl2 (4:1 v/v), each thoroughly mixed and then the tube centrifuged at 2,000 rpm for 5 min. The resulting FAME were dried under N2 gas prior to the addition of 1.0 mL of C19 internal injection standard solution in preparation for gas chromatography (GC) and GC-mass spectrometry (GC-MS) analysis.
Each FAME sample was injected into an Agilent Technologies 7890B GC (Palo Alto, California USA) equipped with an Equity-1 fused silica capillary column (15 m × 0.1 mm internal diameter and 0.1 mm film thickness), a flame ionization detector, a splitless injector, and an Agilent Technologies 7683B Series auto-sampler. At an oven temperature of 120°C, samples were injected in splitless mode and carried by helium gas. Oven temperature was raised to 270°C at a rate of 10°C per min, and then to 310°C at a rate of 5°C per min. Peaks were quantified using Agilent Technologies ChemStation software (Palo Alto, California USA). The identities of the peaks were confirmed using a Finnigan Thermoquest DSQ GC-MS system. All FAs were converted from chromatogram peak area to percentage of total area.
Statistical Analysis
Of the 50 total FAs detected, 21 (with averages >0.1% of total FAs across either tissue type, in quantities of 100 mg non-degraded muscle and 80 mg of non-degraded sub-dermal tissue) were used for multivariate analysis comparing the differences in profiles across factors. Statistical analysis was undertaken in PRIMER 7 (Plymouth Routines in Multivariate Ecological Research, Clarke et al., 2014) +PERMANOVA. We used Principal Coordinates Analysis (PCO) of Bray-Curtis similarity matrices calculated from the square-root transformed data to determine clustering of individual samples. To test the differences between factors we used permutational analysis of variance (PERMANOVA) with Monte Carlo simulations denoted as p(MC) on the unrestricted raw values to account for the small sample sizes. PERMANOVA analyses used factors nested within shark to incorporate the triplicate samples from each individual shark. Significance was determined by p < 0.05. Following significant ANOSIM tests, similarity percentage (SIMPER) analyses were undertaken to quantify the contribution of each parameter to the separation between the designated groups.
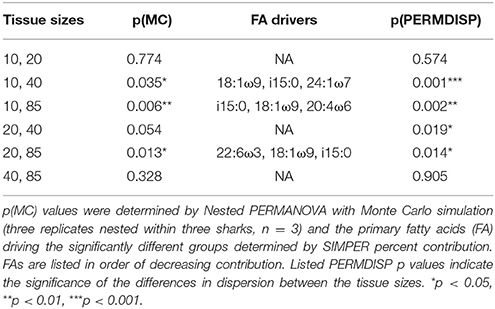
Additionally, the sum of the saturated (SFA), monounsaturated (MUFA), total polyunsaturated fatty acids (PUFA), ω3 PUFA and the ratio of ω3 PUFA:ω6 PUFA and EPA+DHA/16:0 were calculated per replicate. We used nested (factor within shark) PERMANOVA analysis with Monte Carlo simulations to assess the response of individual lipid classes, FA values, and FA metrics (aforementioned sums and ratios). Permutational analysis of multidimensional dispersion PERMDISP denoted at p(perm) was used to determine the relative amount and statistical significance level of the dispersion within factor groups.
Results
Muscle vs. Sub-dermal Tissue
White shark muscle was high in water content 82.1 ± 1.1% wet weight (WW) and low in lipid content (0.6 ± 0.1% WW), with a wet to dry ratio of 4.1 ± 0.2. Sub-dermal tissue contained even lower amounts of total lipid (0.4 ± 0.2% WW), which was on average 33% less lipid than the muscle tissue.
The lipid class profiles of both tissues were dominated by PL (Table 2) followed by ST, which were 13.5% (as % of total lipid) more abundant in sub-dermal tissue than muscle. ST contributed the greatest source of dissimilarity between the tissue types (46%) as determined by SIMPER, and when assessed individually, was the only lipid class significantly different between the tissues [p(MC) = 0.001] (Table 2).
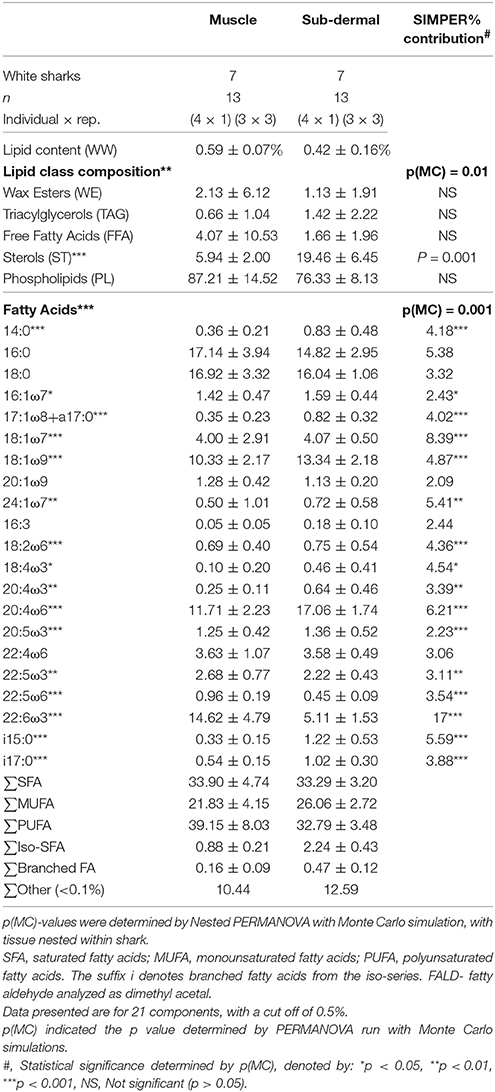
Table 2. Total lipid content, relative proportions of lipid classes and fatty acids (FA) (as percent of total lipid or FA) (mean ± standard deviation) of muscle and sub-dermal tissue (wet weight, WW) from Carcharodon carcharias.
Muscle tissue contained primarily PUFA 39.2 ± 8.0%, mostly consisting of 22:6ω3 (docosahexaenoic acid, DHA) and 20:4ω6 (arachidonic acid, ARA) (Table 2). SFA contributed 33.9 ± 4.7%, dominated by 16:0 and 18:0. MUFA contributed the remaining 21.8 ± 4.2% of the muscle tissue FA profile, nearly half of which was 18:1ω9. Sub-dermal tissue contained similar relative levels of PUFA (32.8 ± 3.5%) dominated by 20:4ω6 and 22:6ω3, and SFA (33.3 ± 3.2%) mostly 18:0 and 16:0, with MUFA (26.1 ± 2.7%) primarily consisting of 18:1ω9 (Table 2).
Muscle and sub-dermal tissue comparisons resulted in distinctly different FA profiles [Nested PERMANOVA: shark p(MC) = 0.439, tissue p(MC) = 0.001, Figure 1]. The difference was primarily driven by high levels of 22:6ω3 in the muscle (SIMPER 17% dissimilarity contribution), followed by 18:1ω7 (8.4%), 20:4ω6 (6.2%), and i15:0 (5.6%) (Table 2). Sixteen of the 21 individual FAs were found to be significantly different [p(MC) < 0.05] across the two tissue types (Table 2), with only 16:0, 18:0, 20:1ω9, 16:3, and 22:4ω6 not significantly different between the two tissues.
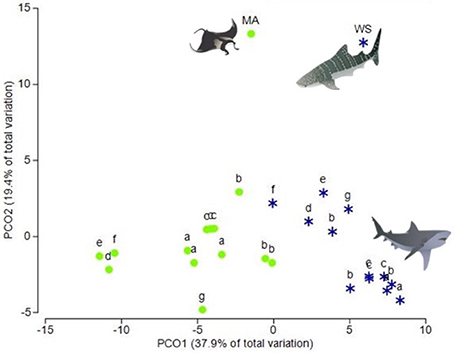
Figure 1. Principal coordinates analysis (PCO) of fatty acid profiles from the muscle and sub-dermal tissue of white sharks, reef manta rays and whale sharks. Principal coordinates analysis (PCO) of fatty acid profiles of muscle (green circles) and sub-dermal tissue (blue stars) from seven white sharks, Carcharodon carcharias (a–g) three of which (a, b, and c) were analyzed in triplicate. Mean fatty acid profiles from reef manta rays Mobula alfredi (MA) and whale sharks Rhincodon typus (WS) from Couturier et al. (2013) and Rohner et al. (2013), respectively, are also included. Eigenvalues denote the percent of variation attributed to each axis (PCO1 and PCO2).
Muscle tissue samples showed greater dispersion than the sub-dermal tissue [p(perm) = 0.030; Figure 1] across the three individual sharks. However, this difference in tissue-specific dispersion was not seen within the three triplicate samples of sharks a, b, and c [Shark a p(perm) = 0.600, Shark b p(perm) = 0.456, Shark c p(perm) = 0.812].
Minimum Sample Size
The progressively smaller muscle tissue increments (100, 50, 25, and 12 mg DW) showed no statistical difference between size groups [p(MC) = 0.28], or difference in dispersion (PERMDISP means of 5.2, 4.1, 5.3, 6.6 for the 100, 50, 25, and 12 mg samples, respectively, p > 0.05). Principal coordinates analysis showed that the clustering is not driven by tissue amount, but by individual shark (Nested PERMANOVA p(MC) = 0.28 nested within shark p(MC) = 0.001), with shark c separating from sharks a and b (Figure 2A).
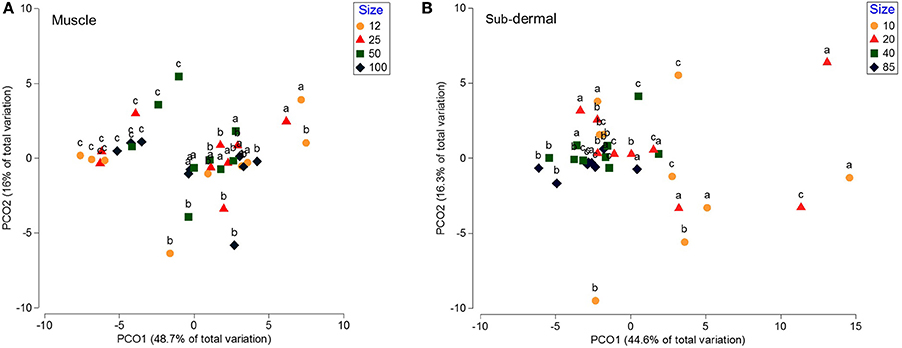
Figure 2. Principal coordinates analysis (PCO) of the fatty acid profiles from white shark muscle and sub-dermal tissue across differing tissue sizes. Principal coordinates analysis (PCO) of (A) muscle, and (B) sub-dermal tissue from three white shark Carcharodon carcharias individuals (a–c), analyzed in triplicate across differing tissue sizes in mg dry weight (DW). Eigenvalues denote the percent of variation attributed to each axis (PCO1 and PCO2).
Sub-dermal tissue increments (85, 40, 20, and 10 mg DW) revealed differing FA profiles with decreasing tissue amounts [p(MC) = 0.042 for tissue size], with the difference between the two larger (85 and 40 mg) and two smaller (20 and 10 mg) amounts driven by 18:1ω9, i15:0, 22:6ω3, and 20:4ω3 (Table 3). The difference in FA profiles is exacerbated by an increase in dispersion with decreasing tissue size (Figure 2B), particularly between the two smaller 10 and 20 mg tissue samples and the two larger 85 and 40 mg sample sizes (Table 3).
Lipid Class and FA Degradation at Ambient Temperature (20°C)
The lipid class profiles from the muscle tissue showed no differences across the 4 day period at 20°C [p(MC) = 0.127]. However, the muscle tissue showed a significant shift in FA profile over the 4 day period at 20°C [p(MC) = 0.009], with significant (p < 0.05) differences between the fresh samples and days 1, 2, and 3 (Figure 3A). This was mostly driven by changes in 18:4ω3, 22:6ω3, and 18:0 (SIMPER analysis). PERMANOVA analysis of individual FA found significant differences in 18:0, total SFA, 18:4ω3, 18:2ω6, 20:5ω3, 22:6ω3, total PUFA, total ω3 PUFA, and the ω3:ω6 ratio, but not 20:5ω3 + 22:6ω3/16:0 (EPA+DHA/16:0).
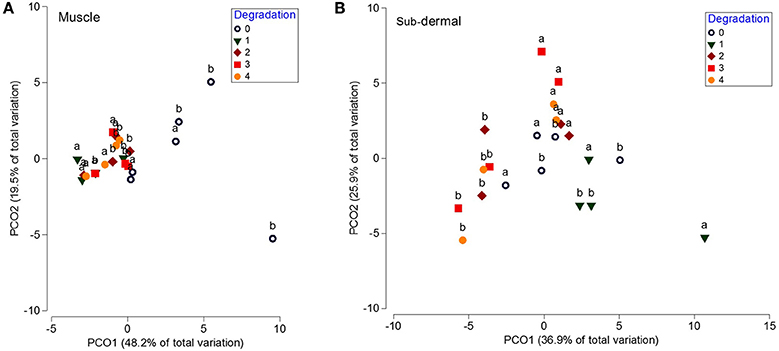
Figure 3. Principal coordinates analysis (PCO) of the fatty acid profiles from white shark muscle and sub-dermal tissue across 4 days of degradation at 20°C. Principal coordinates analysis (PCO) of fatty acid profiles from (A) muscle, and (B) sub-dermal tissue from two white shark Carcharodon carcharias individuals (a and b), analyzed in triplicate across 4 days of degradation (0, indicating fresh tissue, 1, 2, 3, and 4 indicating the number of days left at 20°C prior to analysis). Eigenvalues denote the percent of variation attributed to each axis (PCO1 and PCO2).
Additionally, PERMDISP analysis revealed a significant decrease in dispersion when the tissue was left at ambient temperature [p(perm) = 0.03]. The mean dispersion for the fresh tissue (4.9) was significantly larger than the 2.0, 2.1, and 2.0 dispersion means for days 1, 2, and 3, respectively (p < 0.05), but not significantly different than the 3.0 dispersion mean at day 4 [p(perm) = 0.104]. Similar to the muscle lipid class profile, the sub-dermal tissue did not show any significant differences across the 4 day period at 20°C [p(MC) = 0.183]. There was also no discernible shift in FA profile over the 4 day period [p(MC) = 0.141; Figure 3B]. Unlike the muscle tissue, there were no differences in the level of dispersion between the groups [overall p(perm) = 0.631].
FA Degradation of Frozen Tissue (−20°C)
The FA profiles showed distinct degradation across the 24 months spent in the −20°C freezer, regardless of the location where the sharks were caught [location p(MC) = 0.317; time in freezer nested within location p(MC) = 0.008]. Within group comparisons reveal differences primarily between group 2 (3–6 months in the freezer) and all other groups, aside from group 1. Group 1 (0–3 months in the freezer) was only different to group 7 (the 19–21 month period) (Table 4). SIMPER analysis reveal that these differences were driven largely by 18:0, 22:6ω3, 18:2ω6, 16:0, and 18:4ω3 across the groups. Similar to the unfrozen, controlled muscle degradation trial, the total FA profile degradation manifests in changes to the level of dispersion, which decreases significantly with the amount of time spent in the freezer [p(perm) = 0.001, Table 4].
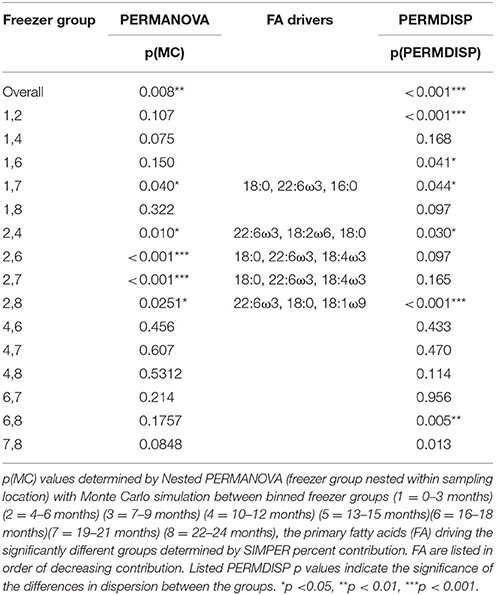
Table 4. The differences between groups of samples combined by time spent frozen at −20°C for 55 white shark Carcharodon carcharias samples from the Neptune Islands, South Australia and throughout New South Wales.
When assessing freezer-based degradation of archived samples over a long time frame (up to 16 years), there was slight discernible degradation, however, the capture location of the white sharks was more highly significant than period in the freezer [p(MC) = 0.002 vs. 0.045]. For the sharks captured in NSW, none of the group level comparisons showed significant degradation [all p(MC)-values > 0.05], and within the QLD samples, only the difference between group 2 and 4 (1.1–2 years and 5.1–10 years) was significant [p(MC) = 0.041]. Unlike the short-term freezer degradation and the unfrozen muscle degradation trial, there was no decrease in dispersion with the longer storage period [p(perm) = 0.620].
Discussion
Lipid class and FA analysis are increasingly used to describe the trophic ecology of a range of species, including elasmobranchs, necessitating greater understanding of the functional limitations of collection and storage methodologies. Here, we determined that muscle and sub-dermal tissue were not directly comparable, as they had tissue-specific lipid class and FA profiles. We also provide the first estimation of the minimum amount of muscle and sub-dermal tissue required to provide reliable FA profiles, which validated the suitability of minimally invasive sampling methods such as punch biopsies. Additionally, we determined that muscle tissue stored at ambient temperature was compromised after as little as 24 h, making muscle samples from beach strandings and fisheries bycatch potentially unreliable for accurate determination of dietary sources. Yet, sub-dermal tissue retained stable FA profiles under the same conditions, suggesting it may offer a more robust tissue for trophic ecology work with potentially compromised samples. However, muscle samples archived for up to 16 years in −20°C retain their FA profiles, highlighting that muscle tissue from museum or private collections can yield valid insights into the trophic ecology of marine elasmobranchs. Knowledge gained from addressing these functional limitations will facilitate the more effective use of lipid and FA profiling on biopsied or potentially degraded tissues for the white shark, and in addition for other species, allowing them to be employed with greater confidence in a range of ecological studies.
Muscle vs. Sub-dermal Tissue
The lipid classes of the muscle tissue, dominated by PL (87%), were consistent with previously reported values for white sharks (92 ± 5%, Pethybridge et al., 2014) whereas the sub-dermal tissue contained higher relative levels of sterols (ST), closely resembling the profile of whale shark sub-dermal tissue (21 ± 4%, Rohner et al., 2013). Regardless of ST contribution, both tissues were dominated by PL, with relatively little contribution from the neutral lipids (triacylglycerols, wax esters, FFA) responsible for metabolic energy storage (Sargent et al., 1999). This affirms the understanding that both muscle and sub-dermal tissue contain little capacity for metabolic energy storage, unlike elasmobranch livers, which are high in lipid content and dominated by triacylglycerols (Beckmann et al., 2013; Pethybridge et al., 2014).
Tissue differences across 16 of the 21 FAs (contributing >76% of total FA) are likely a reflection of divergent functions and underlying physiology. For example, 22:6ω3 and other key essential FAs including 18:2ω6, 20:4ω6 (ARA), 20:5ω3 (EPA), which serve as indicators for a range of trophic pathways differed between the two tissues. As such, the variation in FAs that accounted for the separation between muscle and sub-dermal tissue indicates that interpretation of a species' diet would be greatly affected by the tissue from which the FA profiles is derived, and thus the profiles of the different tissues are not directly comparable.
Recent studies have suggested that differences in FA profiles between muscle and sub-dermal tissue of euryhaline elasmobranches are species-specific (Every et al., 2016). However, when we include the FA profiles of manta ray muscle from Couturier et al. (2013) and whale shark sub-dermal tissue, from Rohner et al. (2013) in the PCO with our white shark samples, the manta ray and whale shark FA profiles align with the tissue-specific clusters (Figure 1). This suggests that the difference in FA profiles between muscle and sub-dermal tissues are not limited to white sharks, but extends to other species and across trophic levels.
The sub-dermal tissue serves as a key structural component, with a slower metabolic turnover rate than muscle (assessed in relation to divergent isotopic signatures by del Rio et al., 2009). As such, these tissues may therefore present complementary results, reflecting diets incorporated across different time frames (Every et al., 2016). Given the opportunity to collect both tissue types through non-lethal biopsies, further investigations comparing the tissue-specific FA incorporation rates should be undertaken. Results discerning the time-frame of both tissue's FA profiles would provide the opportunity to assess multiple temporal scales of an individual's trophic history, valuable additional information when investigating individual specialization, location specific, seasonal, or ontogenetic dietary shifts.
Minimum Samples Size
Muscle biopsies of variable forms have previously been developed to collect samples for genetic and isotopic studies, e.g., punch biopsies (Robbins, 2006; Daly and Smale, 2013) or thick-gauged needles (Baker et al., 2004). Based on the ability of samples as small as 12 mg DW (= 49 mg WW) to provide consistent FA profiles, our study shows that sufficient tissue samples are collected by standard biopsy darts (e.g., Daly and Smale, 2013; Jaime-Rivera et al., 2013) including the small dart assessed by Robbins (2006) which obtained 6.6–122 mg of total tissue. Although not stated what proportion of these biopsies were muscle, the large quantity of tissue obtained (up to 122 mg WW) suggests that sufficient muscle can be collected. Furthermore, biopsy needles (14-gauge, 4 cm long, double-barreled Tru-Cut needles), designed to collect 60 mg WW of tissue from small teleosts are also sufficient to collect tissue for FA analysis (Baker et al., 2004; Logan and Lutcavage, 2010). This ability to obtain FA profiles from small amounts of muscle validates the suitability of minimally invasive sampling methods, and allows trophic ecologists to apply FA analyses to smaller elasmobranchs than previously thought, without the need for lethal sampling. Additionally, multiple studies investigated the variation in muscle-derived FA profiles across different anatomical sites, and found no significant differences (Davidson et al., 2011; Pethybridge et al., 2014). Thus, these biopsy methods can be reliably used regardless of variation in sampling site, furthering the applicability of signature FA analyses. Furthermore, FA profiles can be obtained from the lipids extracted during standard sample preparation for isotopic analysis (Marcus et al., 2017). Therefore, the minimal tissue quantities already retrieved for SIA provide researchers with the opportunity for distinct and complementary FA analyses from the same non-lethal tissue biopsies, without the need to prioritize one of the two datasets. Considering the small amount of muscle necessary, minimally invasive biopsy methods collect sufficient muscle tissue to undertake FA analysis which can be paired with existing standard sample preparation for isotopic analysis, enhancing the method's suitability for ongoing work in trophic ecology.
In contrast to muscle tissue, the FA profiles of sub-dermal tissue smaller than 40 mg DW became highly variable, indicating a minimum reliable tissue quantity of 40 mg DW (= 184 mg WW), which is more than three times the minimal requirement for muscle. This is potentially due to the difference in PL concentration between the two types of tissue of the lipid profile. Combined with the lower lipid content, the lower relative PL contribution in the sub-dermal tissue may explain the comparatively larger minimum sub-dermal tissue quantity, as the ST, which are found in higher abundance in the sub-dermal tissue, do not contribute to the FA pool. This larger minimum tissue quantity required for sub-dermal tissue compared to muscle may limit the applicability of many aforementioned non-lethal biopsy methods. For example, the biopsy method yielding the second highest tissue volume provided only 80–172 mg WW of sub-dermal tissue (Daly and Smale, 2013), which is not sufficient for reliable FA analysis. Only the Reeb and Best's dart head (Reeb and Best, 2006) which retained an average of 0.35 cm3 of sub-dermal tissue when trialed by Jaime-Rivera et al. (2013), obtained potentially suitable tissue quantities. Furthermore, biopsies from small elasmobranchs are unlikely to yield sufficient tissue, as the thickness of the sub-dermis is greatly reduced. For example, sub-dermal tissue layers in atlantic sharpnose shark, scalloped hammerhead and dusky smooth-hound sharks ranged 0.02–0.16 cm (Motta, 1977), compared to white sharks averaging 1.1 cm (Jaime-Rivera et al., 2013) and whale sharks exceeding 2 cm (Rohner et al., 2013).
Degradation
The consistently low levels of FFA in muscle and sub-dermal tissue throughout the degradation trial contrasts with findings across marine taxa, which highlight large increases in FFA from enzymatic hydrolysis of several non-polar lipid classes (Fernández-Reiriz et al., 1992; Kaneniwa et al., 2000; Losada et al., 2005). The difference between our findings and the pervasive trends in previous studies may be attributable to species- and taxa-specific enzymatic processes. Rudy et al. (2016) and Kaneniwa et al. (2000) hypothesized that total lipid content drove the species-specific differences in the level of observed lipid class and FA degradation amongst teleost species, with the “fatty” fish most susceptible. Compared with the six teleosts assessed in Rudy et al. (2016), white sharks were orders of magnitude leaner, with muscle containing 0.6% lipid WW and sub-dermal tissue 0.4% lipid WW (vs. 10.3–2.9% WW in teleosts). The low lipid content may explain the lack of discernable lipid class degradation across both tissues and the comparative stability in FA profiles within the sub-dermal tissue. Given the aim of determining the functional limitations of using elasmobranch specimens not immediately frozen, for example from fisheries bycatch and shark mitigation measures, our results indicate that lipid classes from muscle and sub-dermal tissues are not convoluted by degradation within a 4 day period.
The lipid-poor sub-dermal tissue also showed no discernible shift in FA profile or level of dispersion through exposure to ambient temperature for 4 days. However, the FA profiles derived from muscle tissue immediately changed, with a decrease in dispersion observed after 24 h, potentially compromising the ability to distinguish between individual samples. This advocates for exploring the use of sub-dermal tissue over muscle in situations when samples have been left at ambient temperature, and should be the subject of controlled feeding trails to assess the capacity for sub-dermal tissue to reflect diet. Our earlier findings, however, highlights that such FA profiles based on sub-dermal layers cannot be directly compared to FA profiles from muscle and that this discrepancy should be accounted for.
Muscle segments stored at −20°C showed significant FA profile shifts in both assessment periods, highlighting concerns regarding the capacity to accurately use archived samples. Results in this study suggest that although there may be some level of FA degradation, the time frame at which this occurs and processes involved remains unclear. It is also plausible that the difference in the 3–6 months group is not driven by the time spent in the freezer, but by the influence of unassessed biotic factors (e.g., individual's state of maturity, sex, season of capture). The comparison of FA profiles from archived samples stored for 1–16 years did not provide further clarification and showed no clear differences in FA profiles. Furthermore, neither trial's FA profiles decreased in dispersion, a pattern characteristic of FA degradation in the ambient temperature trial. Regardless of the degradation that might be occurring through long-term storage, differences between locations (NSW vs. QLD) remained, further suggesting that frozen samples may retain viable and indicative FA signatures.
The shift in the relative proportions of individual FAs of the muscle tissue illustrates the complex nature of FA degradation at both 20 and −20°C. Our study found that SFA, driven primarily by 18:0, can remain constant during some time periods, but also decreased drastically through other periods. The MUFA, unchanged at 20°C, demonstrated some resistance to degradation, with no shifts in either individual MUFA, or the ∑MUFA. Unexpectedly, they showed variable patterns of alteration in the early month of storage, suggesting that they are prone to degradation at −20°C, consistent with findings across other taxa (Table 5, e.g., teleosts in Rudy et al., 2016 and octopus in Gullian-Klanian et al., 2017). PUFA are more reactive owing to their numerous double-bonds and are especially prone to degradation (Refsgaard et al., 1998; Paola and Isabel, 2015; Rudy et al., 2016; Gullian-Klanian et al., 2017). However, shifts in relative levels of PUFA of white sharks, including key dietary indicators 22:6ω3 (DHA) and 20:5ω3 (EPA), were only distinguishable in the ambient temperature trial, and not in either the short- or long-term −20°C analysis (with the exception of 18:2ω6). Additionally, the polyene index (EPA+DHA/16:0), a well-established metric for tissue degradation, thought to be ubiquitous across taxa (Jeong et al., 1990; Paola and Isabel, 2015), showed no decrease across any trials (Table 5). The present study shows that white shark muscle PUFA might not show the stark degradation seen in the muscle tissue of other species. Given the relative importance of PUFA, as essential FAs and key dietary markers, these findings suggest that elasmobranch samples may retain these key FAs throughout extensive storage at −20°C.
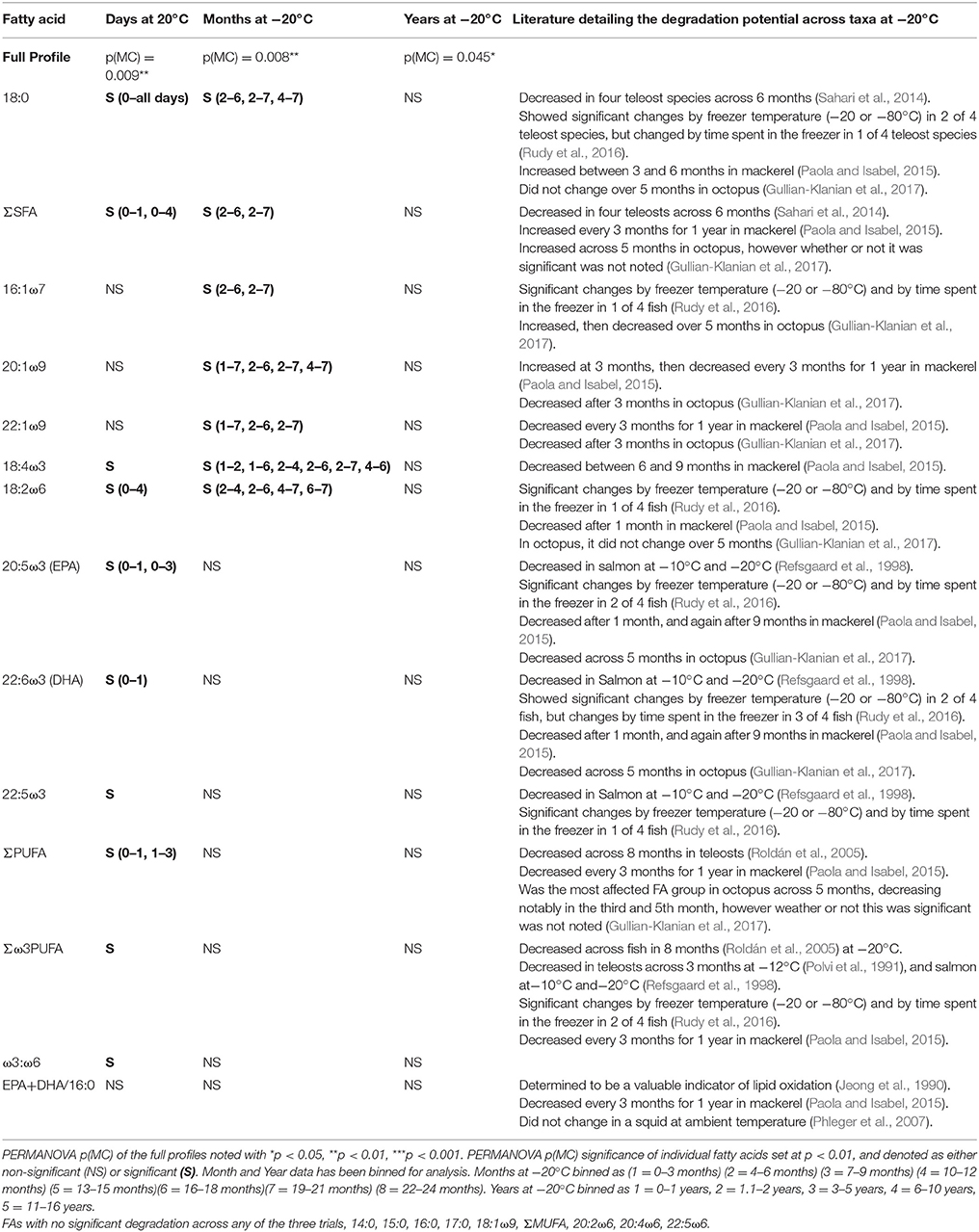
Table 5. Individual fatty acid degradation, assessed by days at −20°C, months stored at −20°C, and years stored at −20°C.
Conclusion
Our findings indicate that muscle and sub-dermal tissue contain distinct FA profiles and differing individual FAs, many of which are key trophic indicators. As such, these tissues are not directly comparable. They may, however, present complementary trophic information reflecting differing time frames, providing the opportunity to garner additional information from non-lethal biopsies. The minimum tissue amount for sub-dermal tissue was 40 mg DW (184 mg WW), whereas muscle samples as small as 12 mg DW (equating to 49 mg WW) retained consistent FA profiles. This makes FA analysis an ideal tool for elucidating trophic ecology of rare or endangered elasmobranchs for which lethal sampling is inappropriate. Degradation of muscle tissue occurred within the first 24 h at ambient temperature, unlike sub-dermal tissue, which revealed no discernible degradation across 4 days. As such, the use of deceased organisms, from shark mitigation strategies, by-catch, or beach strandings should be undertaken with caution, ensuring that preservation occurs within 24 h. Muscle tissue appears to retain viable and indicative FA signatures across long periods of frozen storage (up to 16 years), advocating for the use of archived samples, especially in cases where sampling opportunities are rare or opportunistic. Overall, lipid class and FA analysis can be reliably assessed from small tissue quantities derived from minimally invasive, non-lethal biopsies, deceased elasmobranchs preserved within 24 h and archived samples, proving a robust toolset for elucidating the trophic ecology of rare and endangered wildlife.
Author Contributions
LM, CH, CB, HP, PN, JW, and BB: conceived and designed the experiments; BB, JW, CB, and CH: provided tissue samples; LM: performed the experiments, analyzed the data with the help of CH, CB, PN, and HP; LM wrote the manuscript with the advice of CH, CB, HP, PN, JW, and BB; all authors provided editorial advice.
Funding
This work was supported by a research grant from the Save Our Seas Foundation (grant number RPF14/553).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Save Our Seas Foundation. We thank the Queensland Department of Agriculture and Fisheries Shark Control Program as part of the QLD large shark tagging research program and the QLD white shark tracking program for providing tissue samples. We also thank the New South Wales Department of Primary Industries Shark Meshing Program and the South Australian Research and Development Institute for collecting and storing tissue material. The white shark tourism operators; Andrew Fox and the Fox Shark Research Foundation, Calypso Star Charters and Adventure Bay Charters staff and crew provided ongoing support and in-kind contributions, facilitating sample collection within South Australia.
References
Baker, R. F., Blanchfield, P. J., Paterson, M. J., Flett, R. J., and Wesson, L. (2004). Evaluation of nonlethal methods for the analysis of mercury in fish tissue. Trans. Am. Fish. Soc. 133, 568–576. doi: 10.1577/T03-012.1
Ballantyne, J. S., Glemet, H. C., Chamberlin, M. E., and Singer, T. D. (1993). Plasma nonesterified fatty acids of marine teleost and elasmobranch fishes. Mar. Biol. 116, 47–52. doi: 10.1007/BF00350730
Beckmann, C. L., Mitchell, J. G., Seuront, L., Stone, D. A., and Huveneers, C. (2013). Experimental evaluation of fatty acid profiles as a technique to determine dietary composition in benthic elasmobranchs. Physiol. Biochem. Zool. 86, 266–278. doi: 10.1086/669539
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Bradshaw, C. J., Hindell, M. A., Best, N. J., Phillips, K. L., Wilson, G., and Nichols, P. D. (2003). You are what you eat: describing the foraging ecology of southern elephant seals (Mirounga leonina) using blubber fatty acids. Proc. R. Soc. Lond. 270, 1283–1292. doi: 10.1098/rspb.2003.2371
Budge, S. M., Iverson, S. J., and Koopman, H. N. (2006). Studying trophic ecology in marine ecosystems using fatty acids: a primer on analysis and interpretation. Mar. Mamm. Sci. 22, 759–801. doi: 10.1111/j.1748-7692.2006.00079.x
Carlisle, A. B., Kim, S. L., Semmens, B. X., Madigan, D. J., Jorgensen, S. J., Perle, C. R., et al. (2012). using stable isotope analysis to understand the migration and trophic ecology of northeastern pacific white sharks (Carcharodon carcharias). PLoS ONE 7:e30492. doi: 10.1371/journal.pone.0030492
Christiansen, H., Fisk, A., and Hussey, N. (2015). Incorporating stable isotopes into a multidisciplinary framework to improve data inference and their conservation and management application. Afr. J. Mar. Sci. 37, 189–197. doi: 10.2989/1814232X.2015.1039583
Clarke, K. R., Gorley, R. N., Somerfield, P. J., and Warwick, R. M. (2014). Change in Marine Communities: An Approach to Statistical Analysis and Interpretation, 3rd Edn. Plymouth: PRIMER-E.
Couturier, L. I., Rohner, C. A., Richardson, A. J., Pierce, S. J., Marshall, A. D., Jaine, F. R., et al. (2013). Unusually high levels of n-6 polyunsaturated fatty acids in whale sharks and reef manta rays. Lipids 48, 1029–1034. doi: 10.1007/s11745-013-3829-8
Daly, R., and Smale, M. J. (2013). Evaluation of an underwater biopsy probe for collecting tissue samples from bull sharks Carcharhinus leucas. Afr. J. Mar. Sci. 35, 129–132. doi: 10.2989/1814232X.2013.769910
Davidson, B. C., Nel, W., Rais, A., Namdarizandi, V., Vizarra, S., and Cliff, G. (2014). Comparison of total lipids and fatty acids from liver, heart and abdominal muscle of scalloped (Sphyrna lewini) and smooth (Sphyrna zygaena) hammerhead sharks. Springerplus 3:521. doi: 10.1186/2193-1801-3-521
Davidson, B., Sidell, J., Rhodes, J., and Cliff, G. (2011). A comparison of the heart and muscle total lipid and fatty acid profiles of nine large shark species from the east coast of South Africa. Fish Physiol. Biochem. 37, 105–112. doi: 10.1007/s10695-010-9421-8
del Rio, C. M., Wolf, N., Carleton, S. A., and Gannes, L. Z. (2009). Isotopic ecology ten years after a call for more laboratory experiments. Biol. Rev. 84, 91–111. doi: 10.1111/j.1469-185X.2008.00064.x
Dulvy, N. K., Fowler, S. L., Musick, J. A., Cavanagh, R. D., Kyne, P. M., Harrison, L. R., et al. (2014). Extinction risk and conservation of the world's sharks and rays. eL 3:e00590. doi: 10.7554/eLife.00590
Every, S. L., Pethybridge, H. R., Crook, D. A., Kyne, P. M., and Fulton, C. J. (2016). Comparison of fin and muscle tissues for analysis of signature fatty acids in tropical euryhaline sharks. J. Exp. Mar. Biol. Ecol. 479, 46–53. doi: 10.1016/j.jembe.2016.02.011
Fernández-Reiriz, M. J., Pastoriza, L., and Sampedro, G. (1992). Lipid changes in muscle tissue of Ray (Raja clavata) during processing and frozen storage. J. Agric. Food Chem. 40, 484–488. doi: 10.1021/jf00015a025
Gullian-Klanian, M., Terrats-Preciat, M., Pech-Jiménez, E. C., and Cutz De Ocampo, J. (2017). Effect of frozen storage on protein denaturation and fatty acids profile of the red octopus (Octopus maya). J. Food Process. Preserv. 41:e13072. doi: 10.1111/jfpp.13072
Hooker, S. K., Iverson, S. J., Ostrom, P., and Smith, S. C. (2001). Diet of northern bottlenose whales inferred from fatty-acid and stable-isotope analyses of biopsy samples. Can. J. Zool. 79, 1442–1454. doi: 10.1139/z01-096
Hussey, N. E., MacNeil, M. A., Olin, J. A., McMeans, B. C., Kinney, M. J., Chapman, D. D., et al. (2012). Stable isotopes and elasmobranchs: tissue types, methods, applications and assumptions. J. Fish Biol. 80, 1449–1484. doi: 10.1111/j.1095-8649.2012.03251.x
Hyslop, E. J. (1980). Stomach contents analysis—a review of methods and their application. J. Fish Biol. 17, 411–429. doi: 10.1111/j.1095-8649.1980.tb02775.x
Iverson, S. J., Field, C., Bowen, W. D., and Blanchard, W. (2004). Quantitative fatty acid signature analysis: a new method of estimating predator diets. Ecol. Monogr. 74, 211–235. doi: 10.1890/02-4105
Jaime-Rivera, M., Caravea-Patino, J., Hoyos-Padilla, M., and Galván-Magana, F. (2013). Evaluation of biopsy systems for sampling white shark Carcharodon carcharias (Lamniformes: Lamnidae) muscle for stable isotope analysis. Rev. Biol. Mar. Oceanogr. 48, 345–351. doi: 10.4067/S0718-19572013000200013
Jaime-Rivera, M., Caraveo-Patiño, J., Hoyos-Padilla, M., and Galván-Magaña, F. (2014). Feeding and migration habits of white shark Carcharodon carcharias (Lamniformes: Lamnidae) from Isla Guadalupe inferred by analysis of stable isotopes δ15N and δ13C. Rev. Biol. Trop. 62, 637–647. doi: 10.15517/rbt.v62i2.7767
Jeong, Y., Ohshima, T., Koizumi, C., and Kanou, Y. (1990). Lipid deterioration and Its inhibition of Japanese Oyster Crassostrea gigas during frozen storage. Nippon Suisan Gakkaishi 56, 2083–2091. doi: 10.2331/suisan.56.2083
Kaneniwa, M., Miao, S., and Yuan, C. (2000). Lipid components and enzymatic hydrolysis of lipids in muscle of Chinese freshwater fish. J. Am. Oil Chem. Soc. 77, 1–7. doi: 10.1007/s11746-000-0132-3
Kasajima, I., Ide, Y., Ohkama-Ohtsu, N., Hayashi, H., Yoneyama, T., and Fujiwara, T. (2004). A protocol for rapid DNA extraction from Arabidopsis thaliana for PCR analysis. Plant Mol. Biol. Rep. 22, 49–52. doi: 10.1007/BF02773348
Kim, S. L., del Rio, C. M., Casper, D., and Koch, P. L. (2012). Isotopic incorporation rates for shark tissues from a long-term captive feeding study. J. Exp. Biol. 215, 2495–2500. doi: 10.1242/jeb.070656
Layman, C. A., Araujo, M. S., Boucek, R., Hammerschlag-Peyer, C. M., Harrison, E., Jud, Z. R., et al. (2012). Applying stable isotopes to examine food-web structure: an overview of analytical tools. Biol. Rev. 87, 545–562. doi: 10.1111/j.1469-185X.2011.00208.x
Layman, C. A., Giery, S. T., Buhler, S., Rossi, R., Penland, T., Henson, M. N., et al. (2015). A primer on the history of food web ecology: fundamental contributions of fourteen researchers. Food Webs 4, 14–24. doi: 10.1016/j.fooweb.2015.07.001
Logan, J. M., and Lutcavage, M. E. (2010). Stable isotope dynamics in elasmobranch fishes. Hydrobiologia 644, 231–244. doi: 10.1007/s10750-010-0120-3
Losada, V., Pi-eiro, C., Barros-Velázquez, J., and Aubourg, S. P. (2005). Inhibition of chemical changes related to freshness loss during storage of horse mackerel (Trachurus trachurus) in slurry ice. Food Chem. 93, 619–625. doi: 10.1016/j.foodchem.2004.09.041
Marcus, L., Virtue, P., Nichols, P. D., Meekan, M. G., and Pethybridge, H. (2017). Effects of sample treatment on the analysis of stable isotopes of carbon and nitrogen in zooplankton, micronekton and a filter-feeding shark. Mar. Biol. 164, 124. doi: 10.1007/s00227-017-3153-6
McMeans, B. C., Arts, M. T., and Fisk, A. T. (2012). Similarity between predator and prey fatty acid profiles is tissue dependent in Greenland sharks (Somniosus microcephalus): implications for diet reconstruction. J. Exp. Mar. Biol. Ecol. 429, 55–63. doi: 10.1016/j.jembe.2012.06.017
Motta, Ñ. J. (1977). Anatomy and functional morphology of dermal collagen fibres in shark. Copeia 3, 454–464. doi: 10.2307/1443263
Nielsen, J. M., Popp, B. N., and Winder, M. (2015). Meta-analysis of amino acid stable nitrogen isotope ratios for estimating trophic position in marine organisms. Oecologia 178, 631–642. doi: 10.1007/s00442-015-3305-7
Paola, A. S., and Isabel, Y. M. (2015). Effect of srozen storage on biochemical changes and fatty acid composition of mackerel (Scomber japonicus) muscle. J. Food Res. 4, 135–147. doi: 10.5539/jfr.v4n1p135
Parrish, C. C., Whiticar, M., and Puvanendran, V. (2007). Is ω6 docosapentaenoic acid an essential fatty acid during early ontogeny in marine fauna?. Limnol. Oceanogr. 52, 476–479. doi: 10.4319/lo.2007.52.1.0476
Pethybridge, H. R., Choy, C. A., Polovina, J. J., and Fulton, E. A. (2018). Improving marine ecosystem models with biochemical tracers. Ann. Rev. Mar. Sci. 10:30. doi: 10.1146/annurev-marine-121916-063256
Pethybridge, H. R., Parrish, C. C., Bruce, B. D., Young, J. W., and Nichols, P. D. (2014). Lipid, fatty acid and energy density profiles of white sharks: insights into the feeding ecology and ecophysiology of a complex top predator. PLoS ONE 9:7877. doi: 10.1371/journal.pone.0097877
Pethybridge, H., Daley, R. K., and Nichols, P. D. (2011). Diet of demersal sharks and chimaeras inferred by fatty acid profiles and stomach content analysis. J. Exp. Mar. Biol. Ecol. 409, 290–299. doi: 10.1016/j.jembe.2011.09.009
Phleger, C. F., Young, J. W., Guest, M., Lansdell, M. J., and Nichols, P. D. (2007). “Signature fatty acids: a robust method for evaluating trophic relationships in open ocean ecosystems,” in Report of a GLOBEC-CLIOTOP/PFRP Workshop on the Role of Squid in Open Ocean Ecosystems, GLOBEC Report 24, eds R. J. Olson and J. W. Young (Honolulu, HI), 64–67.
Pond, D. W., and Tarling, G. A. (2011). Phase transitions of wax esters adjust buoyancy in diapausing Calanoides acutus. Limnol. Oceanogr. 56, 1310–1318. doi: 10.4319/lo.2011.56.4.1310
Polvi, S. M., Ackman, R. G., Lall, S. P., and Saunders, R. L. (1991). Stability of lipids and OMEGA-3 fatty acids during frozen storage of Atlantic salmon. J. Food Process. Preserv. 15, 167–181. doi: 10.1111/j.1745-4549.1991.tb00164.x
Reeb, D., and Best, P. B. (2006). A biopsy system for deep-core sampling of the blubber of southern right whales, Eubalaena australis. Mar. Mamm. Sci. 22, 206–213. doi: 10.1111/j.1748-7692.2006.00015.x
Refsgaard, H. H., Brockhoff, P. B., and Jensen, B. (1998). Sensory and chemical changes in farmed Atlantic Salmon (Salmo salar) during frozen storage. J. Agric. Food Chem. 46, 3473–3479. doi: 10.1021/jf980309q
Robbins, W. D. (2006). Evaluation of two underwater biopsy probes for in situ collection of shark tissue samples. Mar. Ecol. Prog. Ser. 310, 213–217. doi: 10.3354/meps310213
Rohner, C. A., Couturier, L. I. E., Richardson, A. J., Pierce, S. J., Prebble, C. E. M., Gibbons, M. J., et al. (2013). Diet of whale sharks Rhincodon typus inferred from stomach content and signature fatty acid analyses. Mar. Ecol. Prog. Ser. 493, 219–235. doi: 10.3354/meps10500
Roldán, H. A., Roura, S. I., Montecchia, C. L., Pérez Borla, O., and Crupkin, M. (2005). Lipid changes in frozen stored fillets from pre- and postspawned hake (Merluccius hubbsi Marini). J. Food Biochem. 29, 187–204. doi: 10.1111/j.1745-4514.2005.00006.x
Roslin, T., and Majaneva, S. (2016). The use of DNA barcodes in food web construction—terrestrial and aquatic ecologists unite!. Genome 59, 603–628. doi: 10.1139/gen-2015-0229
Rudy, M. D., Kainz, M. J., Graeve, M., Colombo, S. M., and Arts, M. T. (2016). Handling and storage procedures have variable effects on fatty acid content in fishes with different lipid quantities. PLoS ONE 11:e0160497. doi: 10.1371/journal.pone.0160497
Sahari, M. A., Farahani, F., Soleimanian, Y., and Javadi, A. (2014). Effect of frozen storage on fatty acid composition of the different tissues of four scombrid and one dussumeriid species. J. Appl. Ichthyol. 30, 381–391. doi: 10.1111/jai.12400
Sargent, J., Bell, G., McEvoy, L., Tocher, D., and Estevez, A. (1999). Recent developments in the essential fatty acid nutrition of fish. Aquaculture 177, 191–199. doi: 10.1016/S0044-8486(99)00083-6
Schaufler, L., Heintz, R., Sigler, M., and Hulbert, L. (2005). Fatty Acid Composition of Sleeper Shark (Somniosus pacificus) Liver and Muscle Reveals Nutritional Dependence on Planktivores. ICES CM, 5.
Traugott, M., Kamenova, S., Ruess, L., Seeber, J., and Plantegenest, M. (2013). Empirically characterising trophic networks: what emerging DNA-based methods, stable isotope and fatty acid analyses can offer. Adv. Ecol. Res. 49, 177–224. doi: 10.1016/B978-0-12-420002-9.00003-2
Keywords: biochemical tracer, elasmobranch, biopsy, trophic ecology, white shark, Carcharodon carcharias
Citation: Meyer L, Pethybridge H, Nichols PD, Beckmann C, Bruce BD, Werry JM and Huveneers C (2017) Assessing the Functional Limitations of Lipids and Fatty Acids for Diet Determination: The Importance of Tissue Type, Quantity, and Quality. Front. Mar. Sci. 4:369. doi: 10.3389/fmars.2017.00369
Received: 18 August 2017; Accepted: 31 October 2017;
Published: 24 November 2017.
Edited by:
Jeremy Kiszka, Florida International University, United StatesReviewed by:
Luis Cardona, University of Barcelona, SpainValentina Franco-Trecu, Facultad de Ciencias, Universidad de la República, Uruguay
Copyright © 2017 Meyer, Pethybridge, Nichols, Beckmann, Bruce, Werry and Huveneers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lauren Meyer, lauren.meyer@flinders.edu.au
 Lauren Meyer
Lauren Meyer Heidi Pethybridge
Heidi Pethybridge Peter D. Nichols
Peter D. Nichols Crystal Beckmann
Crystal Beckmann Barry D. Bruce2
Barry D. Bruce2  Jonathan M. Werry
Jonathan M. Werry Charlie Huveneers
Charlie Huveneers