- 1Biosciences, University of Exeter, Exeter, United Kingdom
- 2Daniel P. Haerther Center for Conservation and Research, John G. Shedd Aquarium, Chicago, IL, United States
- 3Science and Policy, Bahamas National Trust, Nassau, Bahamas
- 4Perry Institute for Marine Science, Waitsfield, VT, United States
Severe declines of endangered Nassau grouper (Epinephelus striatus) across The Bahamas and Caribbean have spurred efforts to improve their fisheries management and population conservation. The Bahamas is reported to hold the majority of fish spawning aggregations for Nassau grouper, however, the status and genetic population structure of fish within the country is largely unknown, presenting a major knowledge gap for their sustainable management. Between August 2014–February 2017, 464 individual Nassau grouper sampled from The Bahamas were genotyped using 15 polymorphic microsatellite loci to establish measures of population structure, genetic diversity and effective population size (Ne). Nassau grouper were characterized by mostly high levels of genetic diversity, but we found no evidence for geographic population structure. Microsatellite analyses revealed weak, but significant genetic differentiation of Nassau grouper throughout the Bahamian archipelago (Global FST 0.00236, p = 0.0001). Temporal analyses of changes in Ne over the last 1,000 generations provide evidence in support of a pronounced historic decline in Bahamian Nassau grouper that appears to pre-date anthropogenic fishing activities. M-ratio results corroborate significant reductions in Ne throughout The Bahamas, with evidence for population bottlenecks in three islands and an active fish spawning aggregation along with apparent signs of inbreeding at two islands. Current estimates of Ne for Nassau grouper are considerably lower compared with historic levels. These findings represent important new contributions to our understanding of the evolutionary history, demographics and genetic connectivity of this endangered species, which are of critical importance for advancing their sustainable management.
Introduction
Anthropogenic activities, most notably the over-exploitation of species and habitat degradation has led to significant global declines in biodiversity and genetic diversity (Worm et al., 2006; Allendorf et al., 2008; Paddack et al., 2009; Pinsky and Palumbi, 2014). Understanding population status, demographics and patterns of connectivity is vital from a management perspective to conserve ecologically and economically important species. Species that reproduce via fish spawning aggregations (FSAs; Domeier, 2012) are particularly susceptible to unsustainable fishing and many of these species are not only commercially valuable, but also provide important marine ecosystem services (Coleman et al., 2010; Sadovy de Mitcheson et al., 2013; Archer et al., 2015). Nassau grouper (Epinephelus striatus, Bloch, 1792) are among the top predatory reef fish species, naturally distributed throughout the Tropical Western Atlantic including Bermuda, Florida, The Bahamas and Yucatan Peninsula, the Caribbean Sea and parts of the Gulf of Mexico (Heemstra and Randall, 1993; Albins et al., 2009; Froese and Pauly, 2014). However, they have experienced severe population declines and even local extirpations throughout their native range over recent decades (Olsen and LaPlace, 1979; Sadovy and Eklund, 1999; Sadovy De Mitcheson et al., 2008).
The highly synchronous and predictable nature of FSAs is often associated with targeted fishing activity (Smith, 1972; Aguilar-Perera, 2006). FSA fishing has been identified as one of the leading drivers for significant reductions in Nassau grouper abundance (Sadovy De Mitcheson et al., 2008; Sadovy de Mitcheson et al., 2013; Cheung et al., 2013). While profitable for fishers (Cheung et al., 2013), this practice heavily reduces reproductive fish biomass and is detrimental to the integrity of Nassau grouper stocks (Sala et al., 2001; Sadovy and Domeier, 2005; Aguilar-Perera, 2006; Sadovy de Mitcheson et al., 2013). Global declines in Nassau grouper abundance and the disappearance of historic FSAs have led to IUCN-Red List and United States Endangered Species Act designations of endangered and threatened respectively (Cornish and Eklund, 2003; Albins et al., 2009). This has helped to focus conservation management efforts with fisheries management regulations now implemented in a number of countries where Nassau grouper still exist, with recent reports from the Cayman Islands and the United States Virgin Islands of population recovery (USVI; Kadison et al., 2010; Heppell et al., 2012). More generally, however, stock assessment data are deficient and marked declines in commercial landings, FSA abundance and densities on reef habitats have been documented in The Bahamas despite regulatory policies (Cheung et al., 2013; Dahlgren et al., 2016b; Sherman et al., 2016; Stump et al., 2017).
Advances in molecular biology and population genetics have proven to be extremely valuable in generating information on population status, genetic diversity and connectivity of a variety of fish species (Carvalho and Hauser, 1998; Silva-Oliveira et al., 2008; Davey et al., 2011; Adams et al., 2016; Garcia-Mayoral et al., 2016); in turn, these data have been applied to support population management and conservation efforts (Waples et al., 2008; Beldade et al., 2014; Selkoe et al., 2016). In particular, the application of genetics for stock identification (Carvalho and Hauser, 1994) and estimates of effective population size, Ne (Wright, 1931), combined with traditional fisheries stock assessment models (Hilborn and Walters, 1992) are emerging approaches for advancing conservation management of at-risk species (Luikart et al., 2010; Hare et al., 2011; Ovenden et al., 2016). Effective population size is a particularly informative measure for genetic diversity studies because it accounts for genetic drift and inbreeding. Reduced genetic diversity and Ne are often indicative of a population bottleneck, which may reduce adaptive potential and exacerbate extinction risk, especially in vulnerable species (Smith et al., 1991; Hauser et al., 2002; Luikart et al., 2010; Hare et al., 2011).
Previous assessments of the population genetic dynamics of Nassau grouper have primarily focused on the wider Caribbean (Hateley, 1995; Stevenson et al., 1998; Jackson A. M. et al., 2014; Bernard et al., 2016), with reduced representation (i.e., both in sample size and spatial coverage) from The Bahamas. Jackson A. M. et al. (2014) provided arguments for regional genetic subdivision based upon analyses using multiple molecular makers [i.e., mtDNA, microsatellites and single nucleotide polymorphisms (SNPs)] applied to samples collected throughout the Caribbean and The Bahamas. The authors also suggested that The Bahamas may be genetically distinct, with the Exumas representing a barrier to gene flow within The Bahamas, and between The Bahamas and the Caribbean. More recently, Bernard et al. (2016) explored genetic differentiation from two geographically disparate FSAs in the Caribbean and found no significant difference (FST = −0.0004) between FSAs in the Cayman Island and USVI, with low relatedness of Nassau grouper within the USVI. Based on these findings, the authors deduced that external rather than local recruitment was responsible for maintenance of the USVI FSA (Bernard et al., 2016).
The life history and ecological characteristics of Nassau grouper (Sadovy and Colin, 1995; Sadovy and Eklund, 1999) coupled with its economic value make it an ideal model species to demonstrate the value of applying population genetics to the conservation management of endangered and FSA targeted species. The Bahamas contains the majority (50–60%) of Nassau grouper FSAs globally, but the status of spawning stocks is poorly known and there is a paucity of information on the genetic composition of these fish (Sherman et al., 2016).
In the present study, we aimed to resolve whether Bahamian populations of Nassau grouper are indeed genetically distinct and to explore how anthropogenic activities may have influenced the genetic architecture of contemporary stocks. Specifically, our objectives were: to (1) assess the current status, genetic population structure and dynamics of Nassau grouper in The Bahamas; (2) estimate the effective population size of Bahamian Nassau grouper; (3) ascertain whether bottlenecks have occurred, which may compromise the genetic health of contemporary populations; and (4) based on these findings, to provide management recommendations for supporting their recovery.
Methods
Study Area and Sample Collection
The Bahamas is an archipelagic nation consisting of 700 islands and 3,000 cays located in the tropical Western Atlantic (~80.5 km/50 mi) southeast of the United States of America and north of Cuba (Buchan, 2000; Figure 1). A total of 464 Nassau grouper fin clip samples were collected from 13 islands (n = 407) and one active FSA in The Bahamas (n = 57) between August 2014–February 2017 (Figure 1, Table 1). Tissue samples from living fish (n = 186) were collected with permission from the Department of Marine Resources during 10 research cruises (see Acknowledgements). Nassau grouper were captured via fish traps or divers using hand-held nets in water depths ranging from 4.6 to 32 m. Fish were slowly brought to the surface and processed using methods described by Stump et al. (2017). Nassau grouper were Floy™ tagged, measured (standard length, SL and total length, TL to the nearest 0.1 centimeter), and a small (~5 × 5 mm) tissue sample was removed from the anal or dorsal fin. Each fish was weighed (to the nearest 0.1 kg) and allowed to recover. Fish were then individually escorted back to the reef from which they were captured and monitored for 1–2 min to ensure there were no adverse impacts due to handling. Fin clips from recently killed specimens (n = 280) were also opportunistically collected from local fishing docks and landing sites throughout The Bahamas. If the capture location for a fish was not specified or was ambiguous, these individuals were treated as unknown. Samples were preserved in 95–100% ethanol (n = 452) or DMSO (n = 14) prior to genomic DNA extraction.
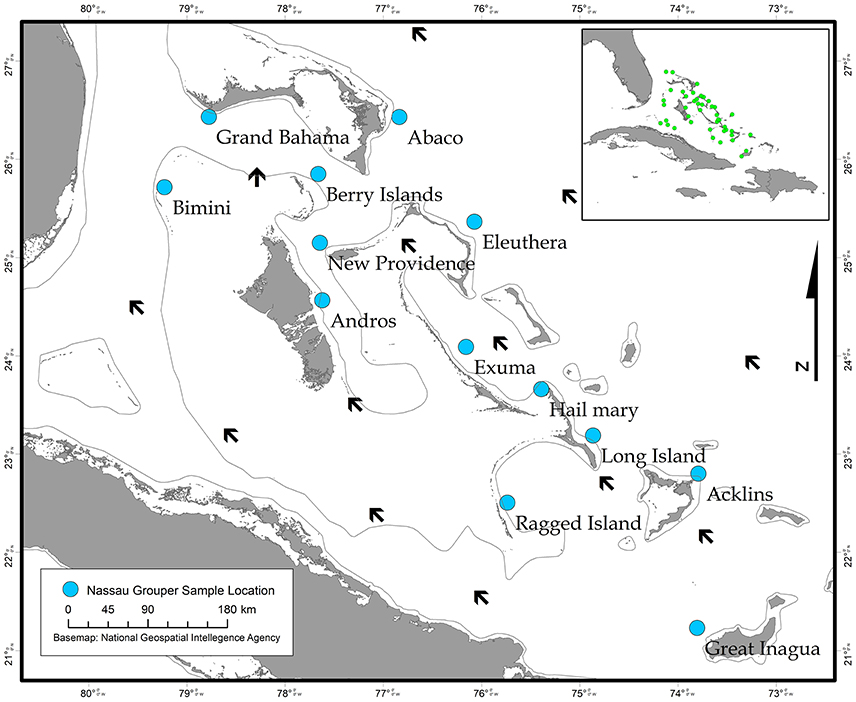
Figure 1. Map showing sampling locations of Nassau grouper collected in The Bahamas (blue circles) and prevailing currents (black arrows), with an inset map depicting approximate locations of in-country Nassau grouper fish spawning aggregations (FSAs) (green circles).
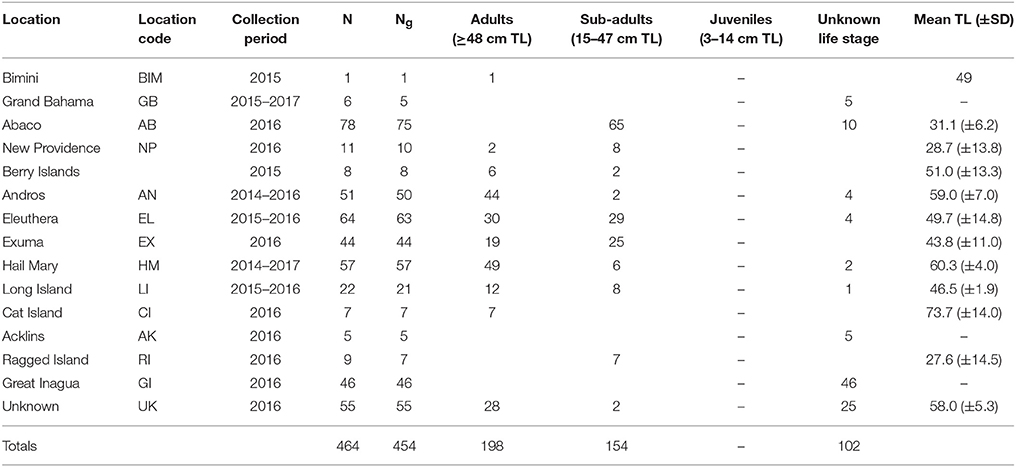
Table 1. Sampling information for Nassau grouper specimens including a breakdown of the number of fin clips = N, number of genotypes = Ng, life stages for genotyped fish, and the mean total length (TL) ± standard deviation (SD) of fish measured in each location.
DNA Extraction, PCR and Genotyping
Genomic DNA for microsatellite analysis was extracted from fin clip tissues (n = 464) using the HotSHOT method (Truett et al., 2000) and its quality assessed via NanoDrop Spectrophotometry (ND−1,000). Twelve species-specific and three cross-species grouper microsatellite loci: Est33a, Est416, Est360, Est92, Est265, Est376, Est267, Est420, Est338, Est340, Est290, Est262, EACD08, EACB6, and EACD02 developed by Bernard et al. (2012) and Jackson et al. (2012) were selected for this research. Polymerase Chain Reaction (PCR) amplification was performed using a BIO-RAD MyCycler Thermal Cycler® in a total of three multiplexes each containing 1 μl DNA, 3 μl of RNase-free water and 5 μl of Qiagen HotStarTaq plus MasterMix, and 1 μl of primer mixture under identical touchdown thermal cycling conditions (Supplementary Tables 1, 2; Hamilton and Tyler, 2008).
Amplified PCR products were visualized and scored with a Beckman Coulter CEQTM 8,000 sequencer (Fullerton, California, USA) using the manufacturer specified internal size standard. Genotypes with more than four missing loci (i.e., 10 individuals) were excluded from further analyses. Microsatellite loci were assessed for the presence of null alleles, large allele dropout, scoring error and stutter using Microchecker v.2.2.3 (van Oosterhout et al., 2004; Supplementary Table 3). The log-likelihood ratio statistic and probability test in GENEPOP v.4.2 was used to test for linkage disequilibrium and conformance to Hardy-Weinberg equilibrium (Rousset, 2008). To reveal Type I errors in both tests, the false discovery rate (FDR) correction was also applied (Storey and Tibshirani, 2003).
Genetic Diversity
To address whether patterns of genetic composition were due to temporal and/or Wahlund effects (i.e., heterozygosity deficiency due to unknown/cryptic population sub-structuring), we computed a range of genetic diversity parameters for Nassau grouper by sampling year (2014–2016) and for sub-adult and adult fish from two locations (Eleuthera and Exuma) with relatively robust sample sizes (Supplementary Table 4). Specifically, observed (HO) and expected heterozygosity (HE) were calculated using GenAlEx v.6.503 (Peakall and Smouse, 2006, 2012). The rarefaction method in HP-Rare v. 1.1 (Kalinowski, 2005) was used to calculate allelic richness (AR) and private allelic richness (PAR) for 2014, 2015, 2016, sub-adult and adult fish with minimum sample sizes of 14, six, 10, 14, and 14 genes per locus respectively. Because these measures produced similar results (Supplementary Table 4), data was pooled for subsequent analyses. Genetic diversity statistics including gene diversity and allelic richness were determined using microsatellite profiles for 454 fish (Table 2). AR and PAR for the pooled data set was computed with a minimum sample size of 14 genes for each location. Significant differences in genetic diversity parameters (HO, HE, AR, and PAR) among sample locations were calculated using the non-parametric Kruskal-Wallis rank sum test in R v.3.4.0.
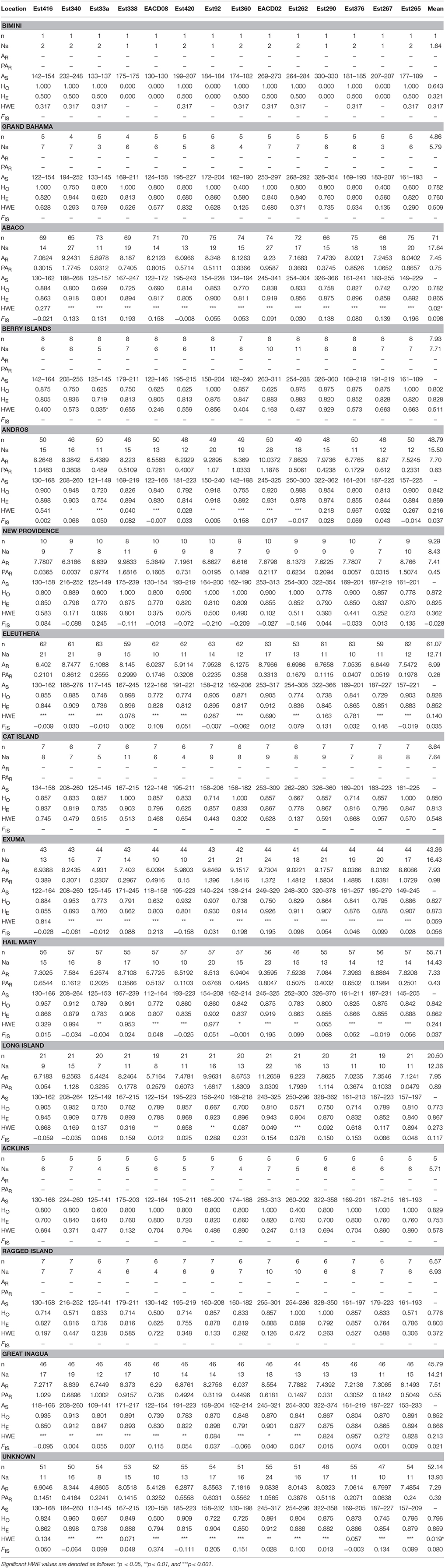
Table 2. Genetic Diversity Summary Statistics for Nassau Grouper where n, number of individuals; Na, number of effective alleles; AR, allelic richness; PAR, private allelic richness; AS, size range of alleles; HO, observed heterozygosity; HE, expected heterozygosity; HWE, probability of conforming to Hardy-Weinberg equilibrium, FIS, inbreeding coefficient.
Ascertainment bias was not a consideration in the comparison of our diversity results with those of Jackson A. M. et al., 2014. The current study had three loci in common with that of Jackson A. M. et al., 2014: EACB6, EACD02, and EACD08; these loci were originally designed for another grouper species (Gulf coney-Hyporthodus acanthistius) and successfully cross-amplified for Nassau grouper (Jackson et al., 2012). In both our study and that of Jackson A. M. et al., 2014 these loci were used with the same species (Nassau grouper).
Genetic Differentiation and Demographic Analyses
Locations with sample sizes of ≥10 individuals were used in assessments of genetic differentiation, corresponding to a total of 421 Nassau grouper sampled from Abaco in the north to Great Inagua in the south. Significant differences in genetic differentiation (i.e., global and pairwise FST values; Weir and Cockerham, 1984) were computed using 10,000 permutations of the data set in Microsatellite Analyzer (Dieringer and Schlötterer, 2003). To examine the extent of genetic variance among sampled locations, GenAlEx v.6.503 was used to perform an analysis of molecular variance (AMOVA). Statistical significance was assessed using 999 permutations of the data. Principal coordinate analysis (PoCA) plots were generated from codominant genetic distance matrices for all samples (n = 454) initially and then for locations with sample sizes of ≥10 per location (n = 421) in GenAlEx v.6.503. A discriminate analysis of principal components (DAPC) was also constructed using the adegenet package in R to expose any fine-scale genetic partitioning (Jombart et al., 2010).
To visualize genetic population structure, the admixture ancestry model using correlated allele frequencies in STRUCTURE v.2.3.4 (Pritchard et al., 2000) was implemented with a pre- and post-burn-in of 75,000 and 150,000 Monte Carlo Markov Chain iterations respectively over 10 independent runs for inferred populations, K, ranging from 1 to 17. STRUCTURE analysis was re-run using a pre- and post-burn-in of 250,000 and 1,000,000 MCMCs respectively for K 1–10. The most likely partition of the dataset for both STRUCTURE runs was determined using the delta K (ΔK) statistic (Supplementary Figures 1, 2; Evanno et al., 2005) and was calculated using the web-based Structure Harvester v. 0.6.94 programme (Earl and vonHoldt, 2012). The PopHelper web application v.1.0.10 (with a ΔK of 2) was used to create a Structure plot of aligned and merged repeats for the 10 independent runs of K 1–10. Because marine species often show weak patterns of genetic differentiation, the locprior model (Hubisz et al., 2009) was also run with parameters identical to those used for the admixture model for K 1–10. Because the ΔK was the same for both models, only the results of the more conservative admixture model (which assumes no prior information about structure) are reported.
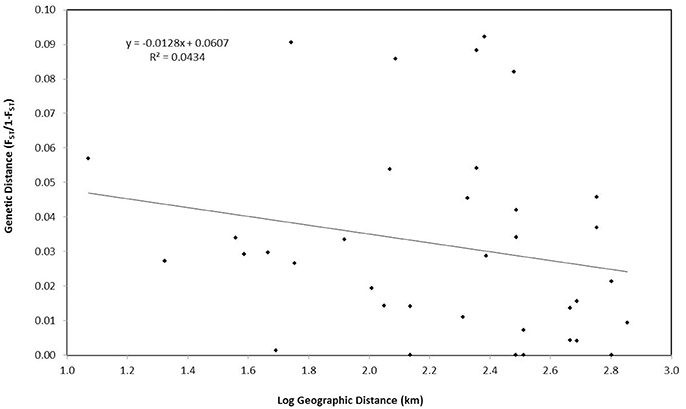
Finally, an assessment of whether genetic structure could be attributed to isolation-by-distance was performed using a Mantel test in GenAlEx v.6.503. Matrices of unbiased pairwise genetic distances FST/(1−FST) were compared with the logarithm of geographic distances between sites (km). Google Earth Pro was used to estimate the shortest straight line distance between islands. Significance was based on 999 permutations of the data.
Effective Population Size and Population Bottlenecks
Estimates of contemporary effective population size, Ne based on linkage disequilibrium were assessed using the random mating model with minimum allele frequencies of 0.05, 0.02, 0.01, and 0.005, as implemented in LDNE v.1.31 (Waples and Do, 2008). LDNE analyses were performed using sub-adult and adult fish from Eleuthera and Exuma and for the entire dataset. Additionally, we used STRUCTURE outputs for ΔK = 2 to assign individuals to the most likely genetic cluster and re-analyzed contemporary Ne using these two populations. M-ratio was used to test whether anthropogenic activities (e.g., overfishing) have led to declines in population size of Nassau grouper throughout the archipelago and within an active FSA. We calculated the M-ratio, the ratio of the number of microsatellite alleles compared to allele size range (Garza and Williamson, 2001). Critical M (Mc) values were computed using a pre-bottleneck Ne ranging from 10,000 to 15,000; corresponding to theta (θ) values between 5.6 and 120 and a mutation rate of μ = 5 × 10−4. We also tested mutation rates of 5.57 × 10−4, 2.0 × 10−3, and 1.5 × 10−4 derived from Common carp (Cyprinus carpio, Yue et al., 2007), Broadnosed pipefish (Syngnathus typhle, Jones et al., 1999) and Zebrafish (Danio rerio, Shimoda et al., 1999). The proportion of stepwise mutations was set to 0.78 and the mean size of non-stepwise mutation (δg) to 3.1 following the suggestions of Peery et al. (2012).
Temporal changes in Ne were analyzed using the VarEff package in R to determine when historical population declines occurred (Nikolic and Chevalet, 2014). The VarEff model uses the coalescent method and approximate likelihoods to derive posterior distributions of Ne over a specified past generation time. Initially, this analysis was performed assuming a two-phase model, T, with a commonly used microsatellite mutation rate, μ = 5 × 10−4 (Estoup et al., 2002), and burn-in of 10,000 over the past 1,000 generations. Additional parameters included setting the number batch to 50,000, length and space batch to 10, with an acceptance rate of 0.25 and diagonale of 0.5, following recommendations from Nikolic and Chevalet (2014). The model was also run using mutation rates of 5.57 × 10−4 (C. carpio), 2.0 × 10−3 (S. typhle) and 1.5 × 10−4 (D. rerio) respectively, under the same parameters. The procedure was re-run modifying the number of times Ne changed (JMAX 2 and 3) and assumed prior values of Ne (NBAR 10,000 and 15,000) for each of the mutation rates listed above (Supplementary Figures 3–6).
Results
Quality Control of Genotypes
Following the removal of 10 individuals with missing data for four or more loci, genotypes from 454 Nassau grouper were retained for analysis. Evidence of null alleles and homozygote excess or heterozygote deficiencies were detected in eight loci, but these were inconsistent across sample locations (Supplementary Table 3). Three occurrences of stuttering and potential scoring error were detected for loci EACB6 and Est360. However, no large allele dropout was detected, and stuttering and scoring error were negligible across populations (Supplementary Table 3). Tests for linkage disequilibrium revealed associations between 10 pairs of loci for fish collected from unknown locations within The Bahamas and Abaco, Eleuthera and Exuma. Samples at three locations, Abaco, Exuma and Long Island, showed possible deviations from Hardy Weinberg equilibrium. Locus EACB6 exhibited significant deviations from HWE after FDR correction across ≥50% of populations and 10 instances of homozygote excess, which can be indicative of null alleles. Accordingly, genotypes for this locus were excluded from subsequent analyses.
Genetic Diversity
Microsatellite loci were highly polymorphic, with the number of alleles per locus ranging from 21 (Est33a) to 42 (EACD02; Supplementary Table 5). A total of 394 alleles were detected, averaging 28.14 across all 14 loci. Measures of genetic diversity and allele size range varied among loci and across sample locations (Table 2, Supplementary Tables 4, 5). Gene diversity by sampling year and life stage ranged from HE = 0.746–0.873 and from 0.838 to 0.877 respectively (Supplementary Table 4). Estimates of allelic richness were variable, ranging from AR = 4.4–7.84 (Supplementary Table 4). Inbreeding coefficients also varied by sampling year and life stage, with higher FIS values observed for sub-adults vs. adults (Supplementary Table 4). Allelic richness for the pooled dataset (adjusted for sample size) was similar among locations, with values ranging from 6.99 to 7.95 (Table 2). Private allelic richness varied from 0.26 to 0.89. Mean allelic richness was highest in Long Island (7.95) and lowest in Eleuthera (6.99). Private alleles were identified in all locations, with more private alleles present in Exuma (mean PAR of 0.98) specimens (Table 2). Heterozygosity, HE was generally high across the sampling area (ranging from 0.753 to 0.873), with the exception of Bimini (0.321), which contained the smallest samples size (Table 2). Mean HO was lowest in Bimini (0.643) and highest in New Providence (0.872). Long Island had the highest mean inbreeding coefficient, (FIS = 0.117) and New Providence had the lowest (FIS = −0.028). Kruskal-Wallis tests showed no significant differences in HE, HO, AR, and PAR among locations (p > 0.05).
Genetic Differentiation and Demographic Analyses
PCoA, DAPC, and STRUCTURE analyses revealed no distinct clustering or definitive geographic structure in the genetic composition of Bahamian Nassau grouper when examined by island or individual (Figures 2, 3). However, there was a low but significant difference in the overall FST value across samples and loci (Global FST = 0.00236, p = 0.0001; Table 3). Genetic differentiation based on pairwise FST values ranged from −0.00097 to 0.01194 and was greatest between Eleuthera and Long Island. Pairwise FST values were significant between only 8 population pairs (Table 3). The Mantel test revealed no significant isolation-by-distance (rxy = −0.208, p = 0.255; Figure 4). AMOVA results indicated that most genetic variability, 90%, occurs within individuals, with 10% among individuals (Supplementary Table 6).
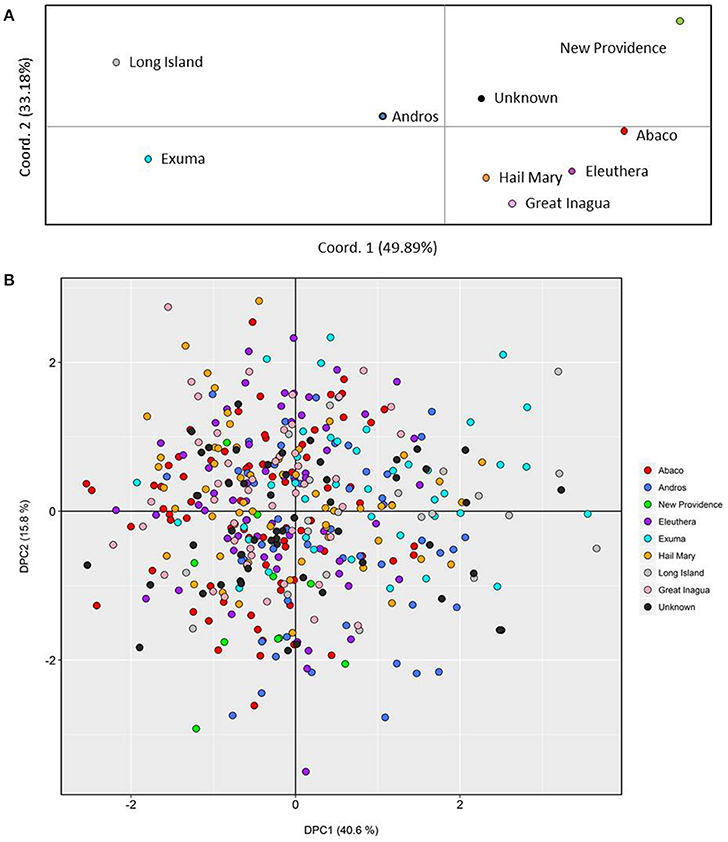
Figure 2. Genetic clustering of Nassau grouper based upon 421 genotypes and inferred from (A) Principal Coordinates Analysis (PCoA) by location and (B) individual based Discriminate Analysis of Principal Components (DAPC). The per cent variation explained by each axis is presented within brackets for both the PCoA and DAPC.
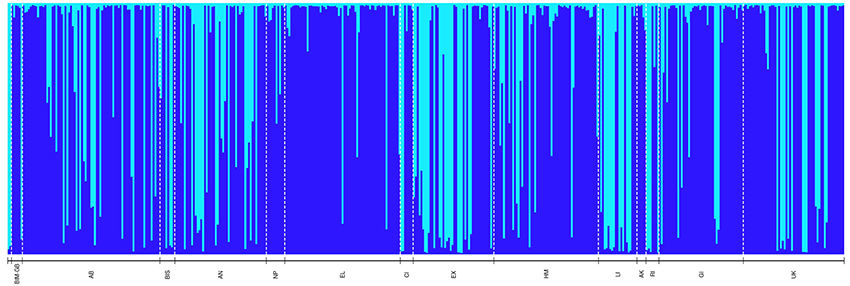
Figure 3. Bayesian-based Structure analysis depicting genetic clustering (K = 2) of 454 Nassau grouper sampled organized geographically from north to south (left-right) throughout The Bahamas (n = 454). Codes correspond to the following sampling locations: BIM-GB includes pooled samples from Bimini (n = 1) and GB = Grand Bahama (n = 5), AB, Abaco; BIS, Berry Islands; AN, Andros; NP, New Providence; EL, Eleuthera; CI, Cat Island; EX, Exuma; HM, Hail Mary FSA; LI, Long Island; AK, Acklins; RI, Ragged Island; GI, Great Inagua; UK, Unknown.
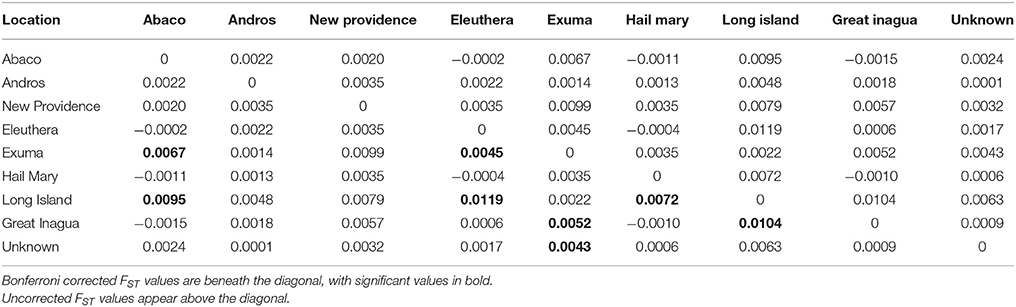
Table 3. Results of Nassau grouper population differentiation within The Bahamas based on pairwise FST values.
Effective Population Size and Population Bottlenecks
Contemporary linkage disequilibrium based estimates of Ne varied among locations and in association with the allele frequency used (Table 4, Supplementary Table 7). Ne also varied by sampling year, life stage and population (Supplementary Table 7). Values based on a critical allele frequency of 0.02 are reported (Table 4) following recommendations by Waples and Do (2010). Ne was lowest for New Providence (−888.3), but this was likely due to the small sample size. To investigate this, we randomly sub-sampled data (n = 10) from Abaco, Eleuthera and Andros respectively and re-ran LDNE analysis, which also produced negative Ne estimates (Supplementary Table 8). Excluding New Providence, Ne estimates were lowest for Abaco (~375) and highest for Andros (~2,978).
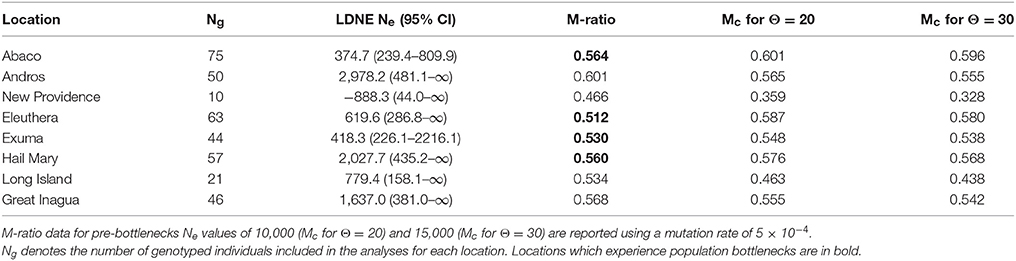
Table 4. Summary of effective population size, Ne per location with 95% confidence intervals, and M-ratio results.
The VarEff modeling approach using μ = 5 × 10−4 revealed historically very large and stable effective population sizes for Nassau grouper within The Bahamas (Figure 5). The upper estimates of median Ne ranged from ~111,000 in Long Island to ~315,000 in Andros. Ne estimates for The Bahamas peaked at ~227,000 and began to gradually decline around 400 generations ago. Similar patterns of population decline were identified for all islands in The Bahamas and Hail Mary around the same period, with a severe decline occurring within the last 150 generations (Figure 5). The upper estimates for Nassau grouper within the last 100 generations were variable, ranging from ~135,742 in Great Inagua to 5,024 Long Island (Figure 5). Lower and upper contemporary Ne estimates during the last 50 generations varied from ~830 to 1,514 in Great Inagua and ~2,672 to 4,539 in Exuma. The upper estimate for The Bahamas during this timeframe was ~2,016 (Figure 5).
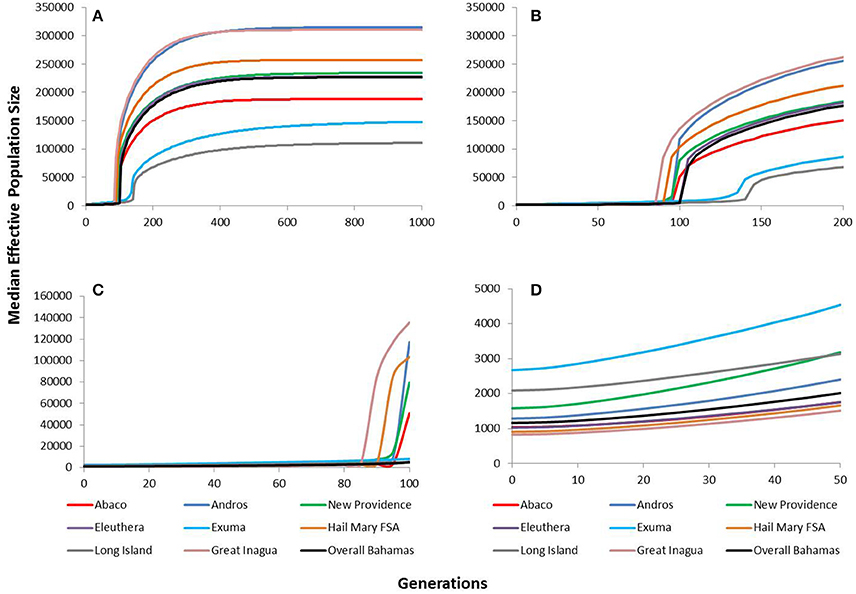
Figure 5. VarEff estimates of temporal changes in the effective population sizes, Ne of Nassau grouper from Abaco (red), Andros (blue), New Providence (green), Eleuthera (purple), Exuma (blue), Hail Mary FSA (tan), Long Island (gray), Great Inagua (pink) and overall Bahamas (black) for (A) the past 1,000 generations, (B) the past 200 generations, (C) the past 100 generations, and (D) the past 50 generations.
Tests using faster and slower mutation rates shifted the timing of genetic bottlenecks (Supplementary Figures 3–6). Under a faster rate of mutation (μ = 2 × 10−3), bottlenecks would have occurred within the last 50 generations, whereas under the slower mutation rate (μ = 1.5 × 10−4), declines occurred around 300–400 generations ago (Supplementary Figure 6). All model simulations consistently revealed a pronounced decline in Nassau grouper within the past 400 generations for all islands (Supplementary Figures 3–6). The only exceptions were Exuma and Long Island, which under the slower mutation rate, showed population declines within the last 450 generations (Supplementary Figure 5).
Dating when bottlenecks occurred varied in accordance with the generation time used. We explored this using generation times ranging from 4 to 9 years. These times encompass the earliest age of sexual maturity, 4 years (Sadovy and Colin, 1995), the suggested generation time calculated for an unexploited Nassau grouper FSA, 9 years (Sadovy and Eklund, 1999), and the approximate age at which Bahamian Nassau grouper undertake their first spawning migration, 7 years (Dahlgren et al., 2016a). Comparisons of historical vs. contemporary derived VarEff Ne estimates for The Bahamas indicate a dramatic reduction in Ne over the last 400 generations, which would have occurred between ~1,600–3,600 years ago (corresponding to generation times of 4 and 9 years respectively). M-ratio estimates also varied in relation to the mutation rate used. Generally, under the fastest mutation rate, no bottlenecks were detected, however, under more realistic mutation rates, M-ratio values were below critical M values for both pre-bottleneck thetas (Supplementary Table 9). Overall, VarEff outputs were consistent with M-ratio analyses, showing historic bottlenecks on several islands and throughout The Bahamas (Table 4, Figure 5).
Discussion
As a species at risk of extinction, developing and applying science-based conservation management policies at the appropriate scale are critical to facilitate recovery of Nassau grouper. From a conservation perspective, the preservation of genetic variation is an important component for mitigating biodiversity loss, highlighting the necessity for research in this area. We aimed to delineate population structure and establish Ne estimates for Nassau grouper throughout the Bahamian archipelago to support current conservation efforts. The highly polymorphic nature of the species-specific microsatellites employed for this study support their utilization for assessing genetic variability and population subdivision.
Genetic Diversity, Differentiation, and Demographic Analyses
Results from microsatellite analysis of genotyped Nassau grouper revealed weak genetic differentiation, lacking definitive geographical population structure. Patterns of weak or no genetic subdivision at varying spatial and temporal scales using microsatellites and other molecular markers have been reported for a number of Epinephelids, e.g., Camouflage grouper, E. polyphekadion (Rhodes et al., 2003); Hawaiian grouper, E. quernus (Rivera et al., 2004); Red grouper, E. morio (Zatcoff et al., 2004); Dusky grouper, E. marginatus (Maggio et al., 2006; Schunter et al., 2011; Buchholz-Sørensen and Vella, 2016) and Goliath grouper, E. itajara (Silva-Oliveira et al., 2008). We expected to see a stronger signal of genetic differentiation separating the northern and southern Bahamas given that the Exumas were purported to present a barrier to gene flow (Jackson A. M. et al., 2014). However, Nassau grouper sampled throughout the archipelago were overlapping in DAPC analysis and no geographical clustering was apparent in either PCoA or Bayesian-based Structure analyses. Moreover, a closer examination of fish sampled north to south across (~200 km) in the Exuma chain, revealed considerable genetic mixing, which is inconsistent with the hypothesis of reduced gene flow. In contrast, low estimates of genetic differentiation and similarly high levels of genetic diversity suggest moderate to extensive gene flow or connectivity throughout The Bahamas.
There are several plausible explanations for the patterns of genetic variation observed. Firstly, pelagic larval dispersal (PLD) and recruitment success are affected by many factors including interspecific competition, predation, larval behavior, oceanographic processes, environmental conditions (e.g., water temperature) and habitat suitability (Colin, 1992, 2012; Shenker et al., 1993; Choat, 2012; Hamner and Largier, 2012; Kough et al., 2016). The stochastic nature of these biotic and abiotic factors helps to determine population structure and demographic connectivity of adults. For example, in bonefishes (Albula spp.), the convergence of larval dispersal and ecological processes has been shown to influence genetic population structuring (Wallace, 2015; Wallace and Tringali, 2016). Nassau grouper have a protracted PLD of up to 50 days, with a mean of 42 days (Tucker and Woodard, 1991; Colin et al., 1997). In the Caribbean, in situ monitoring of sub-surface currents along with modeled trajectories of ocean currents and larval dispersal of Nassau grouper have shown varied recruitment patterns including advection from spawning sites and retention via eddies (Colin, 1992; Heppell et al., 2009, 2011). However, behavioral traits and ecology of larval Nassau grouper in tandem with local oceanographic-driven current patterns have not been explicitly investigated for The Bahamas.
Secondly, adult Nassau grouper can undertake long migrations (>300 km) to participate in annual transient FSAs (Bolden, 2000; Domeier, 2012; Dahlgren et al., 2016a; Stump et al., 2017). Telemetry studies have demonstrated that adults exhibit philopatry to specific FSAs (Starr et al., 2007; Dahlgren et al., 2016a). It is likely therefore, that the population dynamics of Nassau grouper is influenced by both adult migration to and from FSAs via continental shelf margins (Bolden, 2000; Stump et al., 2017), FSA site fidelity (Sala et al., 2001; Starr et al., 2007; Dahlgren et al., 2016a), larval dispersal (Colin, 1992; Shenker et al., 1993) and ocean currents (Colin et al., 1997; Paris, 2009; Paris et al., 2013). Ocean circulation patterns in The Bahamas show predominately northerly flowing currents (Figure 1); however, these patterns are not static and exhibit considerable complexity (Gunn and Watt, 1982; Colin, 1992, 1995). Strong winds and cross-continental shelf currents driven by winter cold fronts have been shown to facilitate larval dispersal from FSAs to recruitment habitats (e.g., Shenker et al., 1993; Colin, 1995; but see Colin et al., 1997). Additionally, mesoscale gyres have been documented in the Exuma Sound, enabling entrainment of larvae through to recruitment stage within this area (Colin, 1995). Collectively, the processes of adult migration and larval dispersal have the potential to increase gene flow, thus contributing to the apparent maintenance of genetic diversity despite ongoing anthropogenic pressures (e.g., FSA fishing).
However, the ability to detect genetic differentiation can be masked by high levels of polymorphism, genetic diversity and large Ne, which can act to retard mutation-drift equilibrium (O'Reilly et al., 2004; Hellberg, 2009). In this study, both patterns of genetic partitioning (K = 2) and a high degree of genetic diversity were found. The measures of genetic diversity reported here (HE = 0.753–0.873) using neutral microsatellite markers are comparable to those found for Nassau grouper in the Caribbean (HE = 0.608–0.966, Jackson A. M. et al., 2014; HE = 0.32–0.91, Bernard et al., 2016) and to other grouper species (HE = 0.703–0.762, Schunter et al., 2011; HE = 0.04–0.886, Buchholz-Sørensen and Vella, 2016). However, given the extent of overfishing and other anthropogenic activities, which have been shown to collapse FSAs (Aguilar-Perera, 2006; Sadovy De Mitcheson et al., 2008) and alter essential marine habitats (Buchan, 2000; Gardner et al., 2003; Pandolfi et al., 2003; Jackson J. B. C et al., 2014), the high levels of genetic diversity observed were somewhat surprising. Indeed, genetic diversity measures for Hail Mary, an active but fished Nassau grouper FSA, were similar to those for the rest of The Bahamas, suggesting no loss in genetic diversity. In other exploited grouper species, e.g., E. itajara, molecular analyses employing microsatellites have provided evidence for reduced genetic variability (Silva-Oliveira et al., 2008). However, this is not always the case, as demonstrated by Zatcoff et al. (2004) for E. morio and Scamp, Mycteroperca phenax; in their study, both species were genetically homogeneous and genetically diverse, despite intense fishing pressure.
Typically, high heterozygosity and allelic richness is beneficial because, potentially, it confers a fitness advantage and has implications for both short and long-term adaptations to both natural and anthropogenic stressors (Bouzat, 2010; Bernatchez, 2016). We found no significant difference in allelic richness in fish sampled from across the islands, but these values were low and may indicate possible loses of genetic diversity (Pinsky and Palumbi, 2014). The allelic richness results reported here are analogous to other published data for Nassau grouper (Bernard et al., 2016) and E. marginatus (Schunter et al., 2011), but higher than values reported from E. quernus (Rivera et al., 2011). In populations that undergo a bottleneck, rare or unique alleles may be lost, deleterious alleles may become fixed and inbreeding is more likely to occur (Garza and Williamson, 2001). In our study, coefficients of inbreeding were high in Abaco (FIS = 0. 098) and Long Island (FIS = 0.117), suggesting that inbreeding of Nassau grouper may be occurring at these locations. However, it is important to highlight that samples from both these islands showed possible deviations from Hardy-Weinberg Equilibrium. Additionally, sub-adults comprised the majority of Nassau grouper from Abaco and most of these fish were caught around the same area (Table 1). This is likely due to Wahlund effects (from the incorporation of mixed cohorts and/or population sub-structuring), providing some explanation for one of the highest FIS values found in this study (Table 2, Supplementary Table 4). Similar patterns may not have been observed if Nassau grouper were sampled over a broader geographic area and across different life stages. In contrast, both sub-adults and adults were sampled over a wider area in Long Island, although mostly around the northern part of the island.
Effective Population Size and Population Bottlenecks
In most instances, LDNE derived Ne estimates varied in accordance with the critical allele frequency used, with the smallest allele frequency (0.005) and smaller sample sizes generating the lowest estimates for Ne (Table 4, Supplementary Table 7). Analysing the data by sampling year, life stage, and population greatly reduced sample sizes and produced mostly negative results, which is either an artifact of sampling or suggest that Ne is indistinguishable from infinity (Waples and Do, 2010; MacBeth et al., 2013). Recently, MacBeth et al. (2013) compared empirical and simulated data of various population sizes to estimate Ne and examine the accuracy of these estimates for Spanish mackerel (Scomberomorus commerson), demonstrating how sample size and the presence of conspecific migrants from genetically differentiated populations could result in reduced or negative Ne estimates.
In the present study, contemporary values of Ne were generally low, but higher estimates were observed in Andros, Hail Mary, and Great Inagua (Table 4). M-ratio and VarEff analyses provided strong evidence in support of both recent and historic population declines. Recent bottlenecks were detected for Abaco, Eleuthera, Exuma and Hail Mary fish despite showing high levels of genetic diversity consistent with the rest of The Bahamas. This is particularly interesting for Hail Mary, which was also the location with the second highest Ne estimate (Table 4). Fishing pressure on Nassau grouper around Eleuthera and Exuma, and FSA fishing at Hail Mary are likely to have contributed to the observed bottlenecks. The majority of contemporary LDNE-based Ne values we derived are smaller than those reported (using different microsatellite markers) for E. marginatus—an endangered grouper species with similar life history characteristics to Nassau grouper (Schunter et al., 2011). However, the smallest contemporary Ne value observed (Abaco: 375) was similar to those reported for other teleosts (e.g., Santer and Slinger sea bream, Cheimerius nufar and Chrysoblephus puniceus; Coscia et al., 2016).
We recognize that most Nassau grouper used in this study were sampled over consecutive years and across life stages with a few exceptions (Table 1), thus potentially violating assumptions regarding discrete generations (Waples, 2006; Waples et al., 2014). Variability in Ne estimates has been linked to differences in reproductive success and/or the methods used in their computation (Hare et al., 2011; Waples et al., 2013, 2014; Coscia et al., 2016; Waples and Anderson, 2017). For example, Waples et al. (2014) demonstrated that Ne estimates based on a variety of age-structured samples are likely to be underestimated due to Wahlund effects. In the present study, both LDNE and VarEff produced similar estimates of contemporary Ne, providing additional confidence in modeled results. However, based on potential biases associated with Ne estimates, results should be interpreted with caution.
The timing of contemporary Ne losses (i.e., within the last 100 generations) coincides with known anthropogenic impacts, e.g., fishing, habitat degradation to Bahamian marine resources (Smith, 1972; Lang et al., 1988; Buchan, 2000; Jackson J. B. C et al., 2014). Historical estimates of Nassau grouper Ne were very large. Jue (2006) also reported a large historical Ne of ~30,000 for Gag (Mycteroperca microlepi), another exploited aggregating grouper species. M-ratio values for Bahamian Nassau grouper were below the critical M (0.68) recommended by Garza and Williamson (2001), highlighting the severity of population declines. In contrast, Bernard et al. (2016) reported a weakly bottlenecked Nassau grouper population from an FSA in the USVI.
More remarkably, if we examine historical Ne reductions in The Bahamas, assuming a generation time of 9 years, the first signs of population decline would have occurred 3,600 year ago. Although Florida is within close proximity to The Bahamas, during the 1800s most of the population resided in northern Florida (Smith, 2005). Consequently, it seems unlikely that the native Indian tribes residing there 3,600 years ago would have traveled to The Bahamas to fish with access to ample marine resources around the shallow Floridian coast. The earliest known inhabitants to populate the Bahamian archipelago were the Lucayan or Arawak Indians (900–1500 AD) who survived by engaging in low technology subsistence fishing and farming, before their eradication in the 1500s (Buchan, 2000). A decline in Ne 3,600 years ago pre-dates the existence of human occupancy in The Bahamas and implies that historical population bottlenecks were likely due to natural as opposed to anthropogenic events.
According to marine geological records, four major glacial events happened during the Pleistocene (2.6 Ma to 11,700 years. ago), resulting in massive (120 m) fluctuations in sea level, which impacted The Bahamas (Sealey, 1994). This was punctuated by smaller variations in sea level (5–6 m) above current conditions during inter-glacial periods (White et al., 1998) that also would have affected marine organisms. Unfortunately, no historical genetic material was available to establish baseline genetic variability and cross-validate modeled estimates of temporal changes in the effective population sizes and contemporary genetic diversity values of Nassau grouper. However, previous studies have shown that the demography, distribution and genetic composition of species can be strongly influenced by geological processes and associated eustatic sea level changes (e.g., Hewitt, 2000; Pellissier et al., 2014; Brüniche-Olsen et al., 2017). For example, an analysis of >6,000 fish species showed strong correlations between current species richness and distribution patterns with historic climatic events that negatively impacted reef habitats (Pellissier et al., 2014). This finding was particularly pronounced for Caribbean reefs and strongly associated reef species, which experienced drastic reductions in water temperatures during past glacial events (White et al., 1998; Hewitt, 2000; Pellissier et al., 2014). Although speculative, it is possible that historical bottlenecks experienced by Bahamian Nassau grouper ~3,600 years ago could have been associated with severe climatic perturbations. Collectively, these results suggest that both climate-driven biological processes and human exploitation have impacted the genetic composition of Nassau grouper.
Management Recommendations
The concept of management units (MUs) is based upon populations with genetically distinct population dynamics and demographics, resulting in management strategies tailored to preserve genetic variability within units (Palsbøll et al., 2007). Two putative genetic clusters were revealed by STRUCTURE analysis, but these do not appear related to geography. Possible explanations for this finding include panmixia, temporal differences in spawning between two sympatric stocks, and secondary contact from previously isolated Nassau grouper populations (e.g., Reid et al., 2016). However, our study was not designed to investigate these hypotheses, and to address it would require expanding spatial and temporal sampling of Nassau grouper throughout The Bahamas and the Caribbean. In the absence of demographic genetic structure, Ne is a viable alternative to use for conservation management. Prior to this study, no estimates of Ne were available for Nassau grouper. Genetic analyses have been successfully used to demonstrate the genetic consequences of exploitation through reductions in Ne (e.g., Hauser et al., 2002; Hoarau et al., 2005; Poulsen et al., 2006), a finding we observed in the present study. The protection of FSAs has been identified as a biodiversity target for The Bahamas (Moultrie, 2012) and has been incorporated into Marxan analyses for the establishment of new MPAs (Moultrie, 2012; Moss and Moultrie, 2014). Given the substantial reduction in Ne and the low contemporary values of allelic richness, strengthened national, regional and international relationships are recommended to tackle illegal fishing, to eliminate FSA targeted fishing and to establish MPA networks to protect marine habitats that are of critical importance to rebuild Nassau grouper stocks.
Conclusions
Given that two putative genetic stocks have been identified for The Bahamas, over-harvesting of Nassau grouper may lead to reductions and losses in genetic diversity of the least abundant stock. The apparent lack of clear geographical or spatial population structure implies the need to adopt a conservative approach for managing Nassau grouper in The Bahamas until such time as the main drivers shaping the genetic composition of these fish are fully understood. Although estimates of genetic diversity were mostly high and fairly uniform, temporal analyses of changes in Ne show that contemporary populations have dramatically reduced effective population sizes compared to historic levels. Because Ne accuracy is affected by sample size, marker numbers, and marker polymorphism (Tallmon et al., 2010; Waples et al., 2014; Wang et al., 2016), performing cohort analysis, increasing the numbers of polymorphic molecular markers, sample size, and spatial coverage throughout the archipelago would allow for enhanced assessments of the genetic population dynamics of Nassau grouper. The application of more sophisticated molecular techniques than microsatellites, for example, RAD-sequencing, which can identify thousands of single nucleotide polymorphisms (SNPs), could help to resolve potential fine-scale intraspecific genetic structure (e.g., Larson et al., 2013). SNP data coupled with oceanographic and biophysical models and in situ biological data (Miller, 2007; Kough et al., 2016), would likely provide additional insight into the mechanisms influencing the spatial distribution, source-sink dynamics and connectivity of the species. The data presented provide an important foundation upon which future changes to Ne and genetic diversity can be monitored and compared to report on the status of management practices. Overall, this research represents the most comprehensive assessment of the genetic architecture of Nassau grouper in The Bahamas to date and has yielded novel insights into the historical processes that may have shaped contemporary genetic patterns. These findings highlight the utility of molecular approaches for enhancing management of endangered species.
Author Contributions
KS designed and conducted fieldwork, coordinated sample collection, performed genomic DNA extractions and quality testing, primer optimisation, genotyping, microsatellite data analysis and wrote the manuscript. RK provided laboratory support, assisted with data analysis and contributed to writing the manuscript. CD conducted fieldwork and commented on the manuscript. SS, JS, and CT helped in the project design and to refine the manuscript. All authors reviewed and approved of the final version. We have no competing interests to declare. Feedback from two reviewers enhanced the manuscript.
Funding
Molecular research was financially supported by the University of Exeter and research cruises were funded by the John G. Shedd Aquarium. Partial funding for KS was provided by the Shirley Oakes Butler Charitable Trust, Rotary Club of East Nassau and a private donation by I. de la Rocha.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer RH and handling Editor declared their shared affiliation.
Acknowledgments
Animal handing followed established and approved ethical procedures reviewed by Shedd Aquarium's internal and external research committee. All fin clip samples were obtained with permission from The Bahamas Department of Marine Resources under scientific research permit (MAMR/FIS/17–Sherman) to KS and follow UK Home Office guidelines. Figure 1 was produced by Lindy Knowles. Special thanks to Kristine Stump, Randolph Burrows, Stephanie Buhler, Jocelyn Curtis-Quick, Candice Brittain, Tim Higgs, Jonisha Cartwright, Shenica Campbell, Andrieka Burrows, Felice Knowles, Catherine Booker, Annabelle Brooks, and Elizabeth Wallace for assisting with sample collection and to Keith Pamper and the crew of the R/V Coral Reef II for logistical support in preparation for and during research cruises. L. Knowles and A. Lundy (Bahamas National Trust), supported logistics for sample transportation and fieldwork.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2017.00393/full#supplementary-material
References
Adams, C. E., Bean, C. W., Dodd, J. A., Down, A., Etheridge, E. C., Gowans, A. R. D., et al. (2016). Inter- and intra-population phenotypic and genotypic structuring in the European whitefish Coregonus lavaretus, a rare freshwater fish in Scotland. J. Fish Biol. 88, 580–594. doi: 10.1111/jfb.12855
Aguilar-Perera, A. (2006). Disappearance of a Nassau grouper spawning aggregation off the southern Mexican Caribbean coast. Mar. Ecol. Prog. Ser. 327, 289–296. doi: 10.3354/meps327289
Albins, M. A., Hixon, M. A., and Sadovy, Y. (2009). Threatened fishes of the world: Epinephelus striatus (Bloch, 1792) (Serranidae). Environ. Biol. Fishes 86, 309–310. doi: 10.1007/s10641-009-9512-5
Allendorf, F. W., England, P. R., Luikart, G., Ritchie, P. A., and Ryman, N. (2008). Genetic effects of harvest on wild animal populations. Trends Ecol. Evol. 23, 327–337. doi: 10.1016/j.tree.2008.02.008
Archer, S. K., Allgeier, J. E., Semmens, B. X., Heppell, S. A., Pattengill-Semmens, C. V., Rosemond, A. D., et al. (2015). Hot moments in spawning aggregations: implications for ecosystem-scale nutrient cycling. Coral Reefs 34, 19–23. doi: 10.1007/s00338-014-1208-4
Beldade, R., Jackson, A. M., Cudney-Bueno, R., Raimondi, P. T., and Bernardi, G. (2014). Genetic structure among spawning aggregations of the gulf coney Hyporthodus acanthistius. Mar. Ecol. Prog. Ser. 499, 193–201. doi: 10.3354/meps10637
Bernard, A. M., Feldheim, K. A., Nemeth, R., Kadison, E., Blondeua, J., Semmens, B. X., et al. (2016). The ups and downs of coral reef fishes: the genetic characteristics of a formerly severely overfished but currently recovering Nassau grouper fish spawning aggregation. Coral Reefs 35, 273–284. doi: 10.1007/s00338-015-1370-3
Bernard, A. M., Feldheim, K. A., Richards, V. P., Nemeth, R. S., and Shivji, M. S. (2012). Development and characterization of fifteen novel microsatellite loci for the Nassau grouper (Epinephelus striatus) and their utility for cross-amplification on a suite of closely related species. Consererv. Genet. Res. 4, 983–986. doi: 10.1007/s12686-012-9688-4
Bernatchez, L. (2016). On the maintenance of genetic variation and adaptation to environmental change: considerations from population genomics in fishes. J. Fish Biol. 89, 2519–2556. doi: 10.1111/jfb.13145
Bolden, S. K. (2000). Long-distance movement of a Nassau grouper (Epinephelus striatus) to a spawning aggregation in the central Bahamas. Fish Bull. 98, 642–645. Available online at: https://www.st.nmfs.noaa.gov/spo/FishBull/983/12.pdf
Bouzat, J. L. (2010). Conservation genetics of population bottlenecks: the role of chance, selection, and history. Conserv. Genet. 11, 463–478. doi: 10.1007/s10592-010-0049-0
Brüniche-Olsen, A., Hazlitt, S. L., and Eldridge, M. (2017). Genetic evidence of range-wide population declines in an Australian marsupial prior to European settlement. Conserv. Genet. 18, 1–13. doi: 10.1007/s10592-017-0960-8
Buchholz-Sørensen, M., and Vella, A. (2016). Population structure, genetic diversity, effective population size, demographic history and regional connectivity patterns of the endangered dusky grouper, Epinephelus marginatus (Teleostei: Serranidae), within Malta's fisheries management zone. PLoS ONE 11:e0159864. doi: 10.1371/journal.pone.0159864
Carvalho, G. R., and Hauser, L. (1994). Molecular-Genetics and the stock concept in fisheries. Rev. Fish Biol. Fish. 4, 326–350.
Carvalho, G. R., and Hauser, L. (1998). Advances in the molecular analysis of fish population structure. Ital. J. Zool. 65, 21–33. doi: 10.1080/11250009809386791
Cheung, W. W. L., Sadovy, Y., Braynen, M. T., and Gittens, L. G. (2013). Are the last remaining Nassau grouper Epinephelus striatus fisheries sustainable? Status quo in the Bahamas. Endanger. Species Res. 20, 27–39. doi: 10.3354/esr00472
Choat, J. H. (2012). “Spawning aggregations in reef fishes: ecological and evolutionary processes,” in Reef Fish Spawning Aggregations: Biology, Research and Management, eds Y. Sadovy de Mitcheson and P. L. Colin (New York, NY: Springer), 85–116.
Coleman, F. C., Koenig, C. C., Scanlon, K. M., Heppell, S., Heppell, S., and Miller, M. W. (2010). Benthic Habitat Modification through Excavation by Red Grouper, Epinephelus morio, in the Northeastern Gulf of Mexico. Open Fish Sci. J. 3, 1–15. doi: 10.2174/1874401X01003010001
Colin, P. L. (1992). Reproduction of the Nassau grouper, Epinephelus striatus, (Pisces: Serranidae) and its relationship to environmental conditions. Environ. Biol. Fish. 34, 357–377. doi: 10.1007/BF00004740
Colin, P. L. (1995). Surface currents in Exuma Sound, Bahamas and adjacent areas with reference to potential larval transport. Bull. Mar. Sci. 56, 48–57.
Colin, P. L. (2012). “Aggregation spawning: biological aspects of the early life history,” in Reef Fish Spawning Aggregations: Biology, Research and Management, eds Y. Sadovy de Mitcheson and P. L. Colin (New York, NY: Springer), 191–224.
Colin, P. L., Laroche, W. A., and Brothers, E. B. (1997). Ingress and settlement in the Nassau grouper, Epinephelus striatus (Pisces: Serranidae), with relationship to spawning occurrence. Bull. Mar. Sci. 60, 656–667
Cornish, A., and Eklund, A.-M. (2003). Epinephelus Striatus. The IUCN Red List of Threatened Species. e.T7862A12858266. Available online at: www.iucnredlist.org.
Coscia, I., Chopelet, J., Waples, R. S., Mann, B. Q., and Mariani, S. (2016). Sex change and effective population size: implications for population genetic studies in marine fish. Heredity 117, 251–258. doi: 10.1038/hdy.2016.50
Dahlgren, C. P., Buch, K., Rechisky, E., and Hixon, M. A. (2016a). Multiyear tracking of Nassau grouper spawning migrations. Mar. Coast. Fish. 8, 522–535. doi: 10.1080/19425120.2016.1223233
Dahlgren, C. P., Sherman, K., Lang, J., Kramer, P. R., and Marks, K. (2016b). Bahamas Coral Reef Report Card, Vol. 1: 2011–2013. Nassau. Available online at: http://www.agrra.org/wp-content/uploads/2016/05/Bahamas-2016-Coral-Reef-Report-Card.pdf
Davey, J. W., Hohenlohe, P. A., Etter, P. D., Boone, J. Q., Catchen, J. M., and Blaxter, M. L. (2011). Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 12, 499–510. doi: 10.1038/nrg3012
Dieringer, D., and Schlötterer, C. (2003). Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes 3, 167–169. doi: 10.1046/j.1471-8286.2003.00351.x
Domeier, M. L. (2012). “Revisiting spawning aggregations: definitions and challenges,” in Reef Fish Spawning Aggregations: Biology, Research and Management, Vol. 35, eds Y. Sadovy de Mitcheson and P. L. Colin (Dordrecht; Heidelberg; London; New York, NY: Springer), 1–20.
Earl, D. A., and vonHoldt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Res. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Estoup, A., Jarne, P., and Cornuet, J.-M. (2002). Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol. Ecol. 11, 1591–1604. doi: 10.1046/j.1365-294X.2002.01576.x
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Froese, R., and Pauly, D. (eds.). (2014). FishBase. Available online at: www.fishbase.org (Accessed Oct 23, 2014).
Garcia-Mayoral, E., Olsen, M., Hedeholm, R., Post, S., Nielsen, E. E., and Bekkevold, D. (2016). Genetic structure of West Greenland populations of lumpfish Cyclopterus lumpus. J. Fish Biol. 89, 2625–2642. doi: 10.1111/jfb.13167
Gardner, T. A., Côté, I. M., Gill, J. A., Grant, A., and Watkinson, A. R. (2003). Long-term region-wide declines in Caribbean corals. Science 301, 958–960. doi: 10.1126/science.1086050
Garza, J. C., and Williamson, E. G. (2001). Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 10, 305–318. doi: 10.1046/j.1365-294x.2001.01190.x
Gunn, J. T., and Watt, D. R. (1982). On the currents and water masses north of the Antilles/Bahamas Arc. J. Mar. Res. 40, 1–48
Hamilton, P. B., and Tyler, C. R. (2008). PERMANENT GENETIC RESOURCES: identification of microsatellite loci for parentage analysis in roach Rutilus rutilus and eight other cyprinid fish by cross-species amplification, and a novel test for detecting hybrids between roach and other cyprinids. Mol. Ecol. Res. 8, 462–465. doi: 10.1111/j.1471-8286.2007.01994.x
Hamner, W. M., and Largier, J. L. (2012). “Oceanography of the planktonic stages of aggregation spawning reef fishes,” in Reef Fish Spawning Aggregations: Biology, Research and Management, eds Y. Sadovy de Mitcheson and P. L. Colin (New York, NY: Springer), 159–190.
Hare, M. P., Nunney, L., Schwartz, M. K., Ruzzante, D. E., Burford, M., Waples, R. S., et al. (2011). Understanding and estimating effective population size for practical application in marine species management. Conserv. Biol. 25, 438–449. doi: 10.1111/j.1523-1739.2010.01637.x
Hateley, J. G. (1995). Preliminary results of a protein electrophoretic analysis of genetic variation, population structure and gene flow in the Nassau grouper, Epinephelus striatus. Proc. Gulf Carib. Fish. Inst. 47, 888–905.
Hauser, L., Adcock, G. J., Smith, P. J., Bernal Ramírez, B., and Carvalho, G. R. (2002). Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proc. Nat. Acad. Sci. U.S.A. 99, 11742–11747. doi: 10.1073/pnas.172242899
Heemstra, P. C., and Randall, J. E. (1993). FAO Species Catalogue, Vol. 16. Groupers of the World (family Serranidae, Subfamily Epinephelinae): An Annotated and Illustrated Catalogue of the Grouper, Rockcod, Hind, Coral Grouper, and Lyretail Species Known to Date. FAO Fisheries Synopsis No. 125, Rome: Food and Agriculture Organization of the United Nations.
Hellberg, M. E. (2009). Gene flow and isolation among populations of marine animals. An. Rev. Ecol. Evol. Syst. 40, 291–310. doi: 10.1146/annurev.ecolsys.110308.120223
Heppell, S. A., Semmens, B. X., Archer, S. K., Pattengill-Semmens, C. V., Bush, P. G., McCoy, C. M., et al. (2012). Documenting recovery of a spawning aggregation through size frequency analysis from underwater laser callipers measurements. Biol. Conserv. 155, 119–127. doi: 10.1016/j.biocon.2012.06.002
Heppell, S. A., Semmens, B. X., Pattengill-Semmens, C. V., Bush, P. G., Johnson, B. C., McCoy, C. M., et al. (2009). Tracking potential larval dispersal patterns from Nassau grouper aggregation sites: evidence for local retention and the ‘importance of place’. Proc. Gulf Carib. Fish. Inst. 61, 325–327. Available online at: http://aquaticcommons.org/15740/1/GCFI_61-48.pdf
Heppell, S. A., Semmens, B. X., Pattengill-Semmens, C. V., Bush, P. G., Johnson, B. C., McCoy, C. M., et al. (2011). Oceanographic patterns associated with Nassau grouper aggregation spawn timing: shifts in surface currents on the nights of peak spawning. Proc. Gulf Carib. Fish. Inst. 63, 152–154. Available online at: http://aquaticcommons.org/15523/
Hewitt, G. (2000). The genetic legacy of the Quaternary ice ages. Nature 405:907–913. doi: 10.1038/35016000
Hilborn, R., and Walters, C. J. (1992). Quantitative Fisheries Stock Assessment. Choice, Dynamics and Uncertainty. London: Chapman and Hall.
Hoarau, G., Boon, E., Jongma, D. N., Ferber, S., Palsson, J., Van der Veer, H. W., et al. (2005). Low effective population size and evidence for inbreeding in an overexploited flatfish, plaice (Pleuronectes platessa L). Proc. R. Soc. Lon. 272, 497–503. doi: 10.1098/rspb.2004.2963
Hubisz, M. J., Falush, D., Stephens, M., and Pritchard, J. K. (2009). Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 9, 1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x
Jackson, A. M., Semmens, B. X., and Bernardi, G. (2012). Characterization and cross-species amplification of microsatellite markers in Nassau grouper (Epinephelus striatus). Mol. Ecol. Resour. 12, 972–974. Available online at: http://www.reef.org/db/publications/6337
Jackson, A. M., Semmens, B. X., Sadovy de Mitcheson, Y., Nemeth, R. S., Heppell, S. A., Bush, P. G., et al. (2014). Population structure and phylogeography in Nassau grouper (Epinephelus striatus), a mass-aggregating marine fish. PLoS ONE 9:e97508. doi: 10.1371/journal.pone.0097508
Jackson, J. B. C., Donovan, M. K., Cramer, K. L., and Lam, V. V. (2014). Status and Trends of Caribbean Coral Reefs: 1970–2012. Gland: Global Coral Reef Monitoring Network; IUCN.
Jombart, T., Devillard, S., and Balloux, F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. doi: 10.1186/1471-2156-11-94
Jones, A. G., Rosenqvist, G., Berglund, A., and Avise, J. C. (1999). Clustered microsatellite mutations in the pipefish Syngnathus typhle. Genetics 152, 1057–1063.
Jue, N. K. (2006). Exploring the structure of genetic variation and the influences of demography on effective population size in the gag grouper Mycteroperca microlepi (Goode and Bean). J. Fish Biol. 69, 217–224. doi: 10.1111/j.1095-8649.2006.01273.x
Kadison, E., Nemeth, R. S., Blondeau, J., Smith, T., and Calnan, J. (2010). Nassau grouper (Epinephelus striatus) in St. Thomas, US Virgin Islands, with evidence for a spawning aggregation site recovery. Proc. Gulf Carib. Fish. Inst. 62, 273–279. Available online at: http://nsgl.gso.uri.edu/flsgp/flsgpc09001/data/papers/039.pdf
Kalinowski, S. T. (2005). HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 5:187. doi: 10.1111/j.1471-8286.2004.00845.x
Kough, A. S., Claro, R., Lindeman, K. C., and Paris, C. B. (2016). Decadal analysis of larval connectivity from Cuban Snapper (Lutjanidae) spawning aggregations based on biophysical modelling. Mar. Ecol. Prog. Ser. 550, 175–190. doi: 10.3354/meps11714
Lang, J. C., Wicklund, R. I., and Dill, R. F. (1988). Depth and habitat related bleaching of zooxanthellate reef organisms near Lee Stocking Island, Exuma Cays, Bahamas. Proc. Sixth Intl. Coral Reef Symp. 3, 269–274.
Larson, W. A., Seeb, L. W., Everett, M. V., Waples, R. K., Templin, W. D., and Seeb, J. E. (2013). Genotyping by sequencing resolved shallow population structure to inform conservation of Chinook salmon (Oncorhynchus tshawytscha). Evol. Appl. 7, 355–369. doi: 10.1111/eva.12128
Luikart, G., Ryman, N., Tallmon, D. A., Schwartz, M. K., and Allendorf, F. W. (2010). Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conserv. Genet. 11, 255–373. doi: 10.1007/s10592-010-0050-7
MacBeth, G. M., Broderick, D., Buckworth, R. C., and Ovenden, J. R. (2013). Linkage disequilibrium estimation of effective population size with immigrants from divergent populations: a case study on Spanish Mackerel (Scomberomorus commerson). G3 Genes, Genomes, Genet. 3, 709–717. doi: 10.1534/g3.112.005124
Maggio, T., Andaloro, F., and Arculeo, M. (2006). Genetic population structure of Epinephelus marginatus (Pisces, Serranidae) revealed by two molecular markers. Ital. J. Zool. 73, 275–283. doi: 10.1080/11250000600804599
Miller, T. J. (2007). Contribution of individual-based coupled physical–biological models to understanding recruitment in marine fish populations. Mar. Ecol. Prog. Ser. 347, 127–138. doi: 10.3354/meps06973
Moss, S., and Moultrie, S. (2014). Ecological Gap Analysis: GEF FSP Sustainable Network of MPAs–The Bahamas. Nassau: BEST Commission, 60.
Moultrie, S. (2012). Master Plan for The Bahamas National Protected Area System. Nassau: The Nature Conservancy; Northern Caribbean Office.
Nikolic, N., and Chevalet, C. (2014). Detecting past changes of effective population size. Evol. Appl. 7, 663–681. doi: 10.1111/eva.12170
O'Reilly, P. T., Canino, M. F., Bailey, K. M., and Bentzen, P. (2004). Inverse relationship between Fst and microsatellite polymorphism in the marine fish, walleye pollock (Theragra chalcogramma): implications for resolving weak population structure. Mol. Ecol. 13, 1799–1814. doi: 10.1111/j.1365-294X.2004.02214.x
Olsen, D. A., and LaPlace, J. A. (1979). A study of a Virgin Islands grouper fishery based on a breeding aggregation. Proc. Gulf Carib. Fish. Inst. 31, 130–144.
Ovenden, J. R., Leigh, G. M., Blower, D. C., Jones, A. T., Moore, A., Bustamante, C., et al. (2016). Can estimates of genetic effective population size contribute to fisheries stock assessments? J. Fish Biol. 89, 2505–2518. doi: 10.1111/jfb.13129
Paddack, M. J., Reynolds, J. D., Aguilar, C., Appelodoorn, R. S., Beets, J., Burkett, E. W., et al. (2009). Recent region-wide declines in Caribbean reef fish abundance. Curr. Biol. 19, 590–595. doi: 10.1016/j.cub.2009.02.041
Palsbøll, P. J., Bérubé, M., and Allendorf, F. W. (2007). Identification of management units using population genetic data. Trends Ecol. Evol. 22, 11–16. doi: 10.1016/j.tree.2006.09.003
Pandolfi, J. M., Bradbury, R. H., Sala, E., Hughes, T. P., Bjorndal, K. A., Cooke, R. G., et al. (2003). Global trajectories of the long-term decline of coral reef ecosystems. Science 301, 955–958. doi: 10.1126/science.1085706
Paris, C. B. (2009). “Fate of reef fish larvae trough ontogeny: advection or true mortality? theme session t: death in the sea,” in Proceedings of the 2009 Annual Science Conference, September 21–25 2009, Berlin: ICES CM 2009/T: 13, 22.
Paris, C. B., Helgers, J., van Sebille, E., and Srinivasan, A. (2013). Connectivity modeling system: a probabilistic modeling tool for the multi-scale tracking of biotic and abiotic variability in the ocean. Environ. Model. Softw. 42, 47–54. doi: 10.1016/j.envsoft.2012.12.006
Peakall, R., and Smouse, P. E. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Peery, M. Z., Kirby, R., Reid, B. N., Stoelting, R., Doucet-Bëer, E., Robinson, S., et al. (2012). Reliability of genetic bottleneck tests for detecting recent population declines. Mol. Ecol. 21, 3403–3418. doi: 10.1111/j.1365-294X.2012.05635.x
Pellissier, L., Leprieur, F., Parravicini, V., Cowman, P. F., Kulbicki, M., Litsios, G., et al. (2014). Quaternary coral reef refugia preserved fish diversity. Science 344, 1016–1019. doi: 10.1126/science.1249853
Pinsky, M. L., and Palumbi, S. R. (2014). Meta-analysis reveal lower genetic diversity in overfished populations. Mol. Ecol. 23, 29–39. doi: 10.1111/mec.12509
Poulsen, N. A., Nielsen, E. E., Schieruo, M. H., Loeschcke, V., and Grønkjaer, P. (2006). Long-term stability and effective population size in North Sea and Baltic Sea cod (Gadus morhua). Mol. Ecol. 15, 321–331. doi: 10.1111/j.1365-294X.2005.02777.x
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Reid, K., Hoareau, T. B., Graves, J. E., Potts, W. M., Dos Santos, S. M., Klopper, A. W., et al. (2016). Secondary contact and asymmetrical gene flow in a cosmopolitan marine fish across the Benguela upwelling zone. Heredity 117, 307–315. doi: 10.1038/hdy.2016.51
Rhodes, K. L., Lewis, R. I., Chapman, R. W., and Sadovy, Y. (2003). Genetic structure of camouflage grouper, Epinephelus polyphekadion (Pisces: Serranidae), in the western central Pacific. Mar. Biol. 142, 771–776. doi: 10.1007/s00227-002-1002-7
Rivera, A. M. J., Kelley, C. D., and Roderick, G. K. (2004). Subtle population genetic structure in the Hawaiian grouper, Epinephelus quernus (Serranidae) as revealed by mitochondrial DNA analyses. Biol. J. Linn. Soc. 81, 449–468. doi: 10.1111/j.1095-8312.2003.00304.x
Rivera, M. A. J., Andrews, K. R., Kobayashi, D. R., Wren, J. L. K., Kelley, C., Roderick, G. K., et al. (2011). Genetic analyses and simulations of larval dispersal reveal distinct populations and directional connectivity across the range of the Hawaiian Grouper (Epinephelus quernus). J. Mar. Biol. 2011, 1–11. doi: 10.1155/2011/765353
Rousset, F. (2008). Genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Res. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Sadovy, Y., and Colin, P. L. (1995). Sexual development and sexuality in the Nassau grouper. J. Fish Biol. 46, 961–976. doi: 10.1111/j.1095-8649.1995.tb01401.x
Sadovy, Y., and Domeier, M. (2005). Are aggregation-fisheries sustainable? Reef fish fisheries as a case study. Coral Reefs 24, 254–262. doi: 10.1007/s00338-005-0474-6
Sadovy, Y., and Eklund, A. M. (1999). Synopsis of Biological Data on the Nassau Grouper, Epinephelus striatus (Bloch, 1792) and the Jewfish, E. itajara, Lichtenstein, 1822. Seattle, WA, NOAA; National Marine Fisheries Service, 1–64.
Sadovy de Mitcheson, Y., Craig, M. T., Bertoncini, A. A., Carpenter, K. E., Cheung, W. W. L., Choat, J. H., et al. (2013). Fishing groupers towards extinction: a global assessment of threats and extinction risks in a billion dollar fishery. Fish Fish. 14, 119–136. doi: 10.1111/j.1467-2979.2011.00455.x
Sadovy De Mitcheson, Y., Cornish, A., Domeier, M., Colin, P. L., Russell, M., and Lindeman, K. C. (2008). A global baseline for spawning aggregations of reef fishes. Conserv. Biol. 22, 1233–1244. doi: 10.1111/j.1523-1739.2008.01020.x
Sala, E., Ballesteros, E., and Starr, R. M. (2001). Rapid decline of nassau grouper spawning aggregations in belize: fishery management and conservation needs. Fisheries 26, 23–30. doi: 10.1577/1548-8446(2001)026<0023:RDONGS>2.0.CO;2
Schunter, C., Carreras-Carbonell, J., Planes, S., Sala, E., Ballesteros, E., Zabala, M., et al. (2011). Genetic connectivity patterns in an endangered species: the dusky grouper (Epinephelus marginatus). J. Exp. Mar. Biol. Ecol. 401, 126–133. doi: 10.1016/j.jembe.2011.01.021
Sealey, N. E. (1994). Bahamian Landscapes. An Introduction to the Geography of the Bahamas, 2nd Edn., Nassau: Media Enterprises, 128.
Selkoe, K. A., D'Aloia, C. C., Crandall, E. D., Iacchei, M., Liggins, L., Puritz, J. B., et al. (2016). A decade of seascape genetics: contributions to basic and applied marine connectivity. Mar. Ecol. Prog. Ser. 554, 1–19. doi: 10.3354/meps11792
Shenker, J. M., Maddox, E. D., Wishinski, E., Pearl, A., Thorrold, S. R., and Smith, N. (1993). Onshore transport of settlement-stage Nassau grouper Epinephelus striatus and other fishes in Exuma Sound, Bahamas. Mar. Ecol. Prog. Ser. 98, 31–43. doi: 10.3354/meps098031
Sherman, K. D., Dahlgren, C. P., Stevens, J. R., and Tyler, C. R. (2016). Integrating population biology into conservation management for endangered Nassau grouper Epinephelus striatus. Mar. Ecol. Prog. Ser. 554, 263–280. doi: 10.3354/meps11771
Shimoda, N., Knapik, E. W., Ziniti, J., Sim, C., Yamada, E., Kaplan, S., et al. (1999). Zebrafish genetic map with 2000 microsatellite markers. Genomics 58, 219–232. doi: 10.1006/geno.1999.5824
Silva-Oliveira, G. C., do Rêgo, P. S., Schneider, H., Sampaio, I., and Vallinoto, M. (2008). Genetic characterisation of populations of the critically endangered Goliath grouper (Epinephelus itajara, Serranidae) from the Northern Brazilian coast through analyses of mtDNA. Genet. Mol. Biol. 31, 988–994. doi: 10.1590/S1415-47572008005000013
Smith, C. L. (1972). A spawning aggregation of Nassau grouper, Epinephelus striatus (Bloch). Trans. Am. Fish. Soc. 101, 257–261. doi: 10.1577/1548-8659(1972)101<257:ASAONG>2.0.CO;2
Smith, P. J., Francis, R. I. C. C., and McVeagh, M. (1991). Loss of genetic diversity due to fishing pressure. Fish. Res. 10, 309–316. doi: 10.1016/0165-7836(91)90082-Q
Smith, S. K. (2005). Florida Population Growth: Past, Present and Future. Bureau of Economic and Business Research, University of Florida, Gainesville, FL. Available online at: https://www.bebr.ufl.edu/sites/default/files/FloridaPop2005_0.pdf (Accessed Oct 13, 2017).
Starr, R. M., Sala, E., Ballesteros, E., and Zabala, M. (2007). Spatial dynamics of the Nassau grouper Epinephelus striatus in a Caribbean atoll. Mar. Ecol. Prog. Ser. 343, 239–249. doi: 10.3354/meps06897
Stevenson, D. E., Champman, R. W., and Sedberry, G. R. (1998). Stock identification in nassau grouper, Epinephelus striatus, using microsatellite DNA analysis. Proc. Gulf Carib. Fish. Inst. 50, 727–749
Storey, J. D., and Tibshirani, R. (2003). Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. USA. 100, 9440–9445. doi: 10.1073/pnas.1530509100
Stump, K., Dahlgren, C. P., Sherman, K. D., and Knapp, C. R. (2017). Nassau grouper migrations during full moon suggest collapsed historic fish spawning aggregation and evidence of undocumented aggregation. Bull. Mar. Sci. 93, 375–389. doi: 10.5343/bms.2016.1042
Tallmon, D. A., Gregovich, D., Waples, R. S., Scott Baker, C., Jackson, J., Taylor, B. L., et al. (2010). When are genetic methods useful for estimating contemporary abundance and detecting population trends? Mol. Ecol. Res. 10, 684–692. doi: 10.1111/j.1755-0998.2010.02831.x
Truett, G. E., Heeger, P., Mynatt, R. L., Truett, A. A., Walker, J. A., and Warman, M. L. (2000). Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris. (HotSHOT). Biotechniques 29, 52–54.
Tucker, J. W. Jr., and Woodard, P. C. (1991). Growth and development of domestic juvenile Nassau groupers. Proc. Gulf Carib. Fish. Inst. 43, 389–391.
van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4:535. doi: 10.1111/j.1471-8286.2004.00684.x
Wallace, E. M. (2015). High intraspecific genetic connectivity in the Indo-Pacific bonefishes: implications for conservation and management. Environ. Biol. Fishes 98, 2173–2186. doi: 10.1007/s10641-015-0416-2
Wallace, E. M., and Tringali, M. D. (2016). Fishery composition and evidence of population structure and hybridization in the Atlantic bonefish species complex (Albula spp.). Mar. Biol. 163:142. doi: 10.1007/s00227-016-2915-x
Wang, J., Santiago, E., and Caballero, A. (2016). Prediction and estimation of effective population size. Heredity 117, 193–206. doi: 10.1038/hdy.2016.43
Waples, R. S. (2006). A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv. Genet. 7, 167–184. doi: 10.1007/s10592-005-9100-y
Waples, R. S., and Anderson, E. C. (2017). Purging putative siblings from population genetic data sets: a cautionary view. Mol. Ecol. 26, 1211–1224. doi: 10.1111/mec.14022
Waples, R. S., Antao, T., and Luikart, G. (2014). Effects of overlapping generations on linkage disequilibrium estimates of effective population size. Genetics 197, 769–780. doi: 10.1534/genetics.114.164822
Waples, R. S., and Do, C. (2008). LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Res. 8, 753–756. doi: 10.1111/j.1755-0998.2007.02061.x
Waples, R. S., and Do, C. (2010). Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol. Appl. 3, 244–262. doi: 10.1111/j.1752-4571.2009.00104.x
Waples, R. S., Luikart, G., Faulkner, J. R., and Tallmon, D. A. (2013). Simple life-history traits explain key effective population size ratios across diverse taxa. Proc. Royal Soc. 280:20131339. doi: 10.1098/rspb.2013.1339
Waples, R. S., Punt, A. E., and Cope, J. M. (2008). Integrating genetic data into the management of marine resources: how we can do it better. Fish Fish. 9, 423–449. doi: 10.1111/j.1467-2979.2008.00303.x
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370.
White, B., Curan, H. A., and Wilson, M. A. (1998). Bahamian coral reefs yield evidence of a brief sea-level lowstand during the last interglacial. Carbon. Evap. 13, 231–234.
Worm, B., Barbier, E. B., Beaumont, N., Duffy, J. E., Folke, C., Halpern, B. S., et al. (2006). Impacts of biodiversity loss on ecosystem services. Science 314, 787–790. doi: 10.1126/science.1132294
Yue, G. H., David, L., and Orban, L. (2007). Mutation rate and pattern of microsatellites in common carp (Cyprinus carpio L.). Genetica 129, 329–331. doi: 10.1007/s10709-006-0003-8
Keywords: bottleneck, connectivity, effective population size, endangered species, fish spawning aggregation, genetic diversity, microsatellites
Citation: Sherman KD, King RA, Dahlgren CP, Simpson SD, Stevens JR and Tyler CR (2017) Historical Processes and Contemporary Anthropogenic Activities Influence Genetic Population Dynamics of Nassau Grouper (Epinephelus striatus) within The Bahamas. Front. Mar. Sci. 4:393. doi: 10.3389/fmars.2017.00393
Received: 08 September 2017; Accepted: 22 November 2017;
Published: 06 December 2017.
Edited by:
Sophie von der Heyden, Stellenbosch University, South AfricaReviewed by:
Giacomo Bernardi, University of California, Santa Cruz, United StatesRomina Henriques, Stellenbosch University, South Africa
Copyright © 2017 Sherman, King, Dahlgren, Simpson, Stevens and Tyler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jamie R. Stevens, ai5yLnN0ZXZlbnNAZXhldGVyLmFjLnVr
Charles R. Tyler, Yy5yLnR5bGVyQGV4ZXRlci5hYy51aw==
 Krista D. Sherman
Krista D. Sherman