Transcriptomic Resilience of the Montipora digitata Holobiont to Low pH
- 1Department of Earth and Environmental Science, Palaeontology and Geobiology, Ludwig-Maximilians-Universität München, Munich, Germany
- 2Faculty of Biology, Munich Graduate School for Evolution, Ecology and Systematics, Ludwig-Maximilians-Universität München, Munich, Germany
- 3Bavarian State Collections of Palaeontology and Geology, Munich, Germany
- 4GeoBio-Center, Ludwig-Maximilians-Universität München, Munich, Germany
Ocean acidification is considered as one of the major threats for coral reefs at a global scale. Marine calcifying organisms, including stony corals, are expected to be the most affected by the predicted decrease of the surface water pH at the end of the century. The severity of the impacts on coral reefs remains as a matter of controversy. Although previous studies have explored the physiological response of stony corals to changes in pH, the response of the holobiont (i.e., the coral itself plus its symbionts) remains largely unexplored. In the present study, we assessed the changes in overall gene expression of the coral Montipora digitata and its microalgal symbionts after a short (3 days) and a longer (42 days) exposure to low pH (7.6). The short-term exposure to low pH caused small differences in the expression level of the host, impacting mostly genes associated with stress response in other scleractinians. Longer exposure to low pH resulted in no significant changes in gene expression of treated vs. control coral hosts. Gene expression in the eukaryotic symbionts remained unaltered at both exposure times. Our findings suggest resilience, in terms of gene expression, of the M. digitata holobiont to pH decrease, as well as capability to acclimatize to extended periods of exposure to low pH.
Introduction
Ocean acidification, the decreasing pH of ocean surface waters, is considered a global threat to coral reefs (Hoegh-Guldberg et al., 2007). A Business-as-Usual (BaU) carbon emissions scenario predicts surface water pH should reach values ranging between 8.0 and 7.8 by the end of the century, likely posing a challenging future for marine calcifying organisms (Kleypas et al., 2005; Jokiel et al., 2008; De'ath et al., 2009). Among these organisms, scleractinian corals have received particular attention due to their pivotal role as reef builders (Shinzato et al., 2011). In this regard, ocean acidification has the potential to decrease reef-accretion rates and negatively impact these important ecosystems that serve as biodiversity reservoirs, allow the development of other marine ecosystems, and provide important economic and environmental services (Moberg and Folke, 1999; Chen et al., 2015).
The mechanisms by which exposure to acidified water affect calcium carbonate accretion in corals appears to be related to the maintenance of pH-homeostasis at the tissue-skeleton interface, which allegedly makes calcification more energetically expensive (Krief et al., 2010; McCulloch et al., 2012; Venn et al., 2013; Holcomb et al., 2014; Cyronak et al., 2015; Von Euw et al., 2017). Other observed physiological changes triggered by low pH include impaired reproduction, growth and metabolic functions, and anomalies in skeletal morphology (Albright et al., 2008, 2010; Morita et al., 2010; Suwa et al., 2010; Albright and Langdon, 2011; Nakamura et al., 2011). Symbiosis can also be affected as coral bleaching can be triggered by ocean acidification (Anthony et al., 2008). In addition, decreased photosynthetic productivity has been reported in Symbiodinium (the microalgal symbiont inhabiting corals) exposed to low pH water. Also, delayed establishment of symbiosis between coral larvae and Symbiodinium has been observed as result of exposure to similar low pH conditions (Anthony et al., 2008; Crawley et al., 2010). However, the impacts of exposing corals to low pH vary across species and life stages (Kaniewska et al., 2012; Moya et al., 2012, 2015; Davies et al., 2016).
Previous findings suggest that corals can tolerate low pH by using different strategies, for example, through the reduction of their metabolic rate and oxygen consumption (in larvae) or by increasing calcification rates (in adults) (Nakamura et al., 2011; Rodolfo-Metalpa et al., 2011). Better characterization of the responses of these organisms to ocean acidification is therefore fundamental in order to better understand the potential of scleractinian corals to cope with the challenges associated with a changing environment. Within this framework, we sequenced the transcriptome of Montipora digitata, a coral commonly found in seawater aquaria and available in culture in different countries, and of its symbiotic microalgae and assessed the changes in global gene expression as result of exposing M. digitata to low (7.6) pH water for 3 and 42 days. To date, most studies have focused on the physiological response of stony corals to changes in pH, leaving the coupled transcriptional response of corals and their symbionts to ocean acidification largely unexplored. Here we show that the M. digitata holobiont is remarkably tolerant to low pH water exposure, highlighting the potential for resilience and acclimation of corals to ocean acidification.
Methods
Experimental Setup and Biological Material
The experimental setup consisted of two continuous-flow 20L tanks (control and treatment) connected to one of the (360 L) aquarium systems available at the Molecular Geo- and Palaeobiology Laboratory of the Department of Earth and Environmental Sciences, Palaeontology and Geobiology, LMU München (Supplementary Figure 1). Continuous water flow between the experimental tanks and the main 360 L tank was achieved by pumping 1.0–1.6 L/min of water from the main tank to the bottom of the experimental tanks and allowing an equal amount of water from the surface of the tank to return to the main water tank. To simulate ocean acidification conditions, a pH-electrode LE4099 Mettler Toledo connected to a pH computer was used to control the injection of CO2 into the water of the treatment tank to be kept pH at 7.6 (treatment tank). We used a water pump set to a flow rate of 300 L/h to keep the water circulating in each experimental tank and allow for better mix the CO2 injected into the treatment tank. Water returning from the treatment tank was equilibrated to pH ~8.0 by letting it flow through a 25 cm column containing limestone grains with diameters ranging from 2 to 5 mm and simultaneously injecting air (300 to 500 L/h). After this, the treated water was mixed with the remaining water in the main (360 L) tank. Both tanks were illuminated using a Mitras LX 6200-HV lamp simulating tropical light cycles; illuminance ranged between 19 and 22 kilolux at the seawater surface of both tanks. During the experiment, water pH in each experimental tank was controlled every day and was recorded every hour from the second day onwards with a pH electrode LE4099 Mettler Toledo and a pH-meter PCE-PHD 1. The pH in the control aquarium fluctuated between 7.90 and 8.26 (mean = 8.08, sd = 0.11) according to the respiration/photosynthesis cycles of coral and symbionts. Because the lowest pH value reached under control condition was close to the worst-case scenario (pH = 7.8) predicted for the end of the century (Meehl et al., 2007), we set the pH in the experimental tank to a lower value (~7.6), which stayed relatively constant (mean = 7.64, sd = 0.02).
Coral nubbins (n = 20) were obtained from branches of two adult M. digitata colonies from our coral culture stock. The colonies were originally donated by Prof. Christian Laforsch. The individual explants were cultured for over 2 months in the main (360 L) aquarium under the same light (19–22 kilolux) and temperature (average ~26°C) conditions used for the experiment. The coral nubbins were not fed either previous to or during the experiment. Approximately 1 month before starting the pH experiment, the nubbins were transferred to the control aquarium of the experimental setup for acclimation under the same light, temperature and feeding conditions as mentioned above. Afterwards, half of the corals were moved to the treatment tank and sampling was conducted after 3 and 42 days of exposure. Five corals were sampled from each tank at each sampling time.
To determine the phylogenetic affinities of the Symbiodinium sp. inhabiting in the colonies of M. digitata used for all experiments, a partial sequence (664 bp) of the ITS region was amplified using primers and PCR conditions detailed in Baillie et al. (2000). ITS sequences from Symbiodinium clades A to G were downloaded from GenBank (accession numbers in Supplementary Figure 2) and aligned with MUSCLE (Edgar, 2004). A maximum likelihood tree was then inferred using PhyML (Guindon et al., 2010) with a GTR substitution model, 100 bootstrap replicates, and optimized nucleotide equilibrium frequency, invariable sites and across-site rate variation. Both analyses were done in SeaView v4.6.2 (Gouy et al., 2009).
RNA Extraction and Sequencing
At each sampling point, the nubbins were flash-frozen in liquid nitrogen, broken in smaller pieces with mortar and pestle (always in liquid nitrogen) and stored at −80°C until further processing. For RNA extraction, samples were homogenized in TRIzol Reagent (Thermo Fisher Scientific) with a SilentCrusher M and a Dispersion Tool 8 F (Heidolph) and total RNA was extracted using a slightly modified version of the Chomczynski method (Chomczynski and Sacchi, 1987). RNA quality and concentration were controlled with a NanoDrop ND-1000 spectrometer and with a BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). An additional cleaning step with Agencourt RNAClean XP Magnetic Beads was done if required. RNA concentration and quality requirements for library preparation and sequencing were achieved for all samples obtained after the first sampling time, for the second sampling time only 6 samples (3 control + 3 treatment) met quality requirements and were further used. Library preparation and sequencing (50 bp PE; HiSeq2500) was done at the GeneCore of the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany. The reads generated for both control and treatment samples of the second sampling time were strand-specific and further used for de novo transcriptome assembly. The reads generated were uploaded to the Short Read Archive of the European Nucleotide Archive under the project accession PRJEB21531 (http://www.ebi.ac.uk/ena/data/view/PRJEB21531).
Transcriptome Assembly and Annotation
Illumina read pairs were quality controlled with FastQC v0.63 (Andrews, 2010), low quality bases and reads were removed with Trimmomatic v0.32 (Bolger et al., 2014). The surviving pairs were further processed with the program Filter Illumina v0.40 of the Agalma/Biolite suite (Dunn et al., 2013) to ensure the removal of adapters and low quality reads. Reads from putative bacterial contaminants were filtered out by aligning the surviving pairs against all bacterial genomes on RefSeq as of July 2015 (ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/bacteria) and discarding mapping pairs.
De novo transcriptome assembly was done with Trinity v2.0.6 using only strand-specific sequences (i.e., the surviving read pairs from the second sampling time from both conditions) (Grabherr et al., 2011). The assembled contigs from coral and dinoflagellates were separated with Psytrans (https://github.com/sylvainforet/psytrans) using the Acropora digitifera v0.9 (Shinzato et al., 2011) and Symbiodinium minutum Clade B1 v1.0 (Shoguchi et al., 2013) predicted proteins as references. The minicircle sequences of Symbiodinium sp. Chloroplast (Barbrook et al., 2014) were recovered manually from the contigs assigned to the coral. Coding sequences (CDS) from the assembled transcripts were predicted with TransDecoder v2.0.1 (https://transdecoder.github.io). Only transcripts with predicted proteins were used as the reference transcriptome for both coral and symbionts. Completeness of the transcriptomes was assessed by search of the sequences against the Core Eukaryotic Genes Dataset (CEGMA) (Parra et al., 2007; Francis et al., 2013). Data processing and transcriptome assembly were mostly executed on the Molecular Geo- and Palaeobiology Lab's Galaxy server (Goecks et al., 2010). Differentially expressed genes (as defined in the next section; DEGs hereafter) were annotated by BLASTN (E = 10−10) against the Non-Redundant (nr) database of the NCBI and by BLASTX (E = 10−10) against the SwissProt database (both downloaded in February 2015). The best SwissProt hit of each DEG was used to retrieve its associated Gene Ontology terms through the QuickGO online tool (https://www.ebi.ac.uk/QuickGO). In addition, predicted proteins of the DEGs were annotated with the protein identifiers of the A. digitifera v0.9 and S. minutum Clade B1 v1.2 by BLASTP (E = 10−10). A Galaxy history containing some of the steps previously mentioned as well as transcriptome annotations can be found in https://github.com/PalMuc/Montipora_digitata_resources.
Differential Gene Expression Analysis
For each sampling time, the remaining reads from each sample were mapped with Bowtie2 (Langmead et al., 2009) to the reference transcriptomes of M. digitata and Symbiodinium sp. Read counts from isoforms groups were added to derive a matrix of counts per Trinity-component. Differences between conditions and colonies were assessed with the adonis function of the vegan R library (Oksanen et al., 2013). As in other ocean acidification studies (Davies et al., 2016; Kurman et al., 2017), the sample-dependence effect caused by one single tank per condition could not be corrected (Riebesell et al., 2011; Cornwall and Hurd, 2016; but see Oksanen, 2001 and Schank and Koehnle, 2009 for a discussion on pseudoreplication). DEGs between conditions were detected using the package DESeq2 v1.8.1 (Love et al., 2014). DEGs were selected by their significant change in expression as assessed by a Wald test; P-values were adjusted with the Benjamini-Hochberg correction (Benjamini and Hochberg, 1995). Gene Ontology (GO) terms enrichment was assessed with topGO v1.0 (Alexa and Rahnenfuhrer, 2010) in R v3.2.1. Protein domains of the DEG products were identified using PfamScan and PFAM A release 30.0 (both downloaded from ftp://ftp.ebi.ac.uk/pub/databases/Pfam). The counts and the R scripts used to analyze the data can be found in the project repository (https://github.com/PalMuc/Montipora_digitata_resources).
Results
Reference Transcriptomes
The M. digitata meta-transcriptome was assembled using over 190 millions of strand-specific read pairs. The transcriptome assembly yielded 179,298 sequences, of which 123,710 (69%) were assigned to the coral host and 55,588 (31%) to the symbiont. These transcript sets resulted in 41,852 and 34,057 predicted CDS for the coral and symbionts, respectively. The N50 length was 1,101 bp for the coral and 744 bp for Symbiodinium sp. G+C content of both reference transcriptomes were 43.9 and 54.5%, consistent with previous reports for the Order Scleractinia and Symbiodinium spp., respectively (Bayer et al., 2012; Shoguchi et al., 2013; Sun et al., 2013; Shinzato et al., 2014). Summary quality statistics for both transcriptomes are summarized in Table 1. Most of the Core Eukarytic Genes (CEGs) were found in the transcriptomes, 89.5% in M. digitata and 79.4% in Symbiodinium sp. (Supplementary Table 1). In addition, the level of fragmentation of the assembled transcriptomes appears to be low as judged by the low number of CEGs that are longer than the queries.
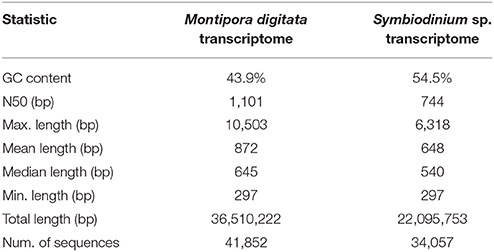
Table 1. Summary quality statistics for the reference transcriptomes of the M. digitata and Symbiodinium sp.
Only 5,541 (13.2%) out of the 41,852 genes contained in the transcriptome of M. digitata transcriptome could not be annotated with any of the databases. A large portion (48.6%) of transcripts had hits against sequences from all three databases used and about 15% of the total transcripts hit exclusively sequences of the A. digitifera genome (Figure 1). On the other hand, most of the sequences in the transcriptome of Symbiodinium sp. were annotated with genes from the S. minutum genome (85.5% of all transcripts), and almost half of them (48.4%) had hits exclusively against this genome. Similarly to the coral transcriptome, only a small fraction of the transcripts (12.9%) lacked any annotation (Figure 1).
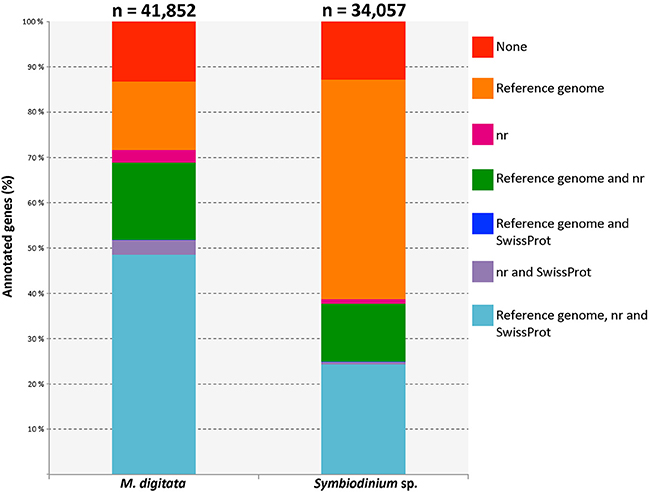
Figure 1. Bar plot displaying different fractions of the annotations for both, coral and symbiont, transcriptomes. The total number of genes for each transcriptome is shown above the corresponding bar. Each color represents an annotation category specified in the legend on the right and the lines connecting the bars indicate the position of the same category for each dataset. The category with the largest fraction in the coral transcriptome corresponds to those genes that had records from the three annotation resources. Genes with no annotation records are shown in red and represent a minor fraction of each transcriptome. Reference genomes used were Acropora digitifera (scleractinian coral) and Symbiodinium minutum (dinoflagellate symbiont).
Coral Response to Low pH
No significant differences in global expression were found with the adonis exploratory test between conditions and between colonies for neither the short (pcondition = 0.40, pcolony = 0.19) nor the longer (pcondition = 0.73, pcolony = 0.76) exposures of the coral to low pH. These findings are also supported by the variance in expression of the coral genes at both sampling times (Figure 2) and by the variance-corrected counts of mapped reads per gene per sample (Supplementary Figure 3).
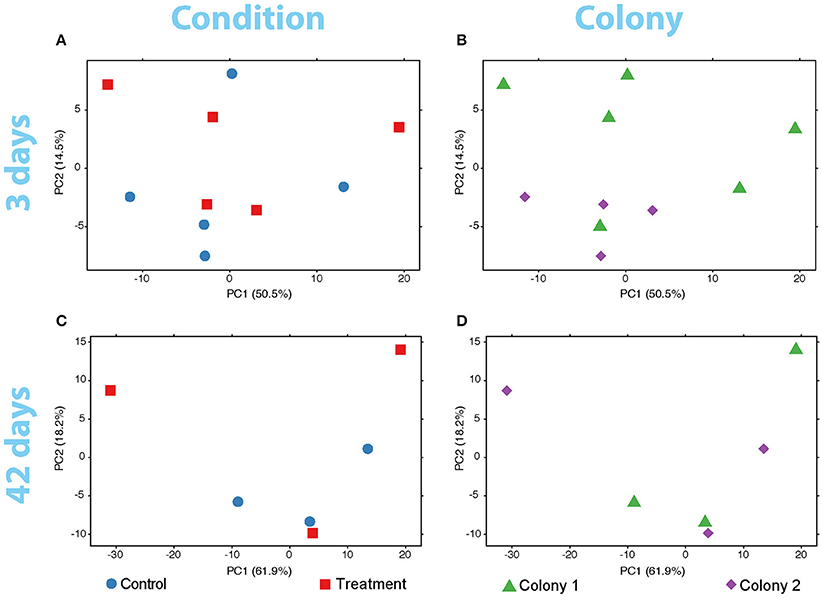
Figure 2. PCA plots of the variance of expressed genes in the coral nubbins (A) between colonies and (B) conditions after 3 days of exposure to low pH. Variance of gene expression is also plotted after the 42-days exposure comparing conditions (C) and colonies (D). No grouping patterns in any comparison can be distinguished. Dot color and shape codes are displayed at the bottom.
Still, the DeSeq2 gene expression analysis yielded 18 genes with significant (padj < 0.05) differences in expression level due to exposure to low pH after 3 days (model design: ~ colony + condition, Figure 3). The log2-fold changes for these genes ranged between −0.70 and 0.73 (Supplementary Table 2). Six up- and twelve down-regulated genes were found. From the six upregulated genes, two (including a metalloprotease) contain zinc-binding domains and others have RNA/DNA binding domains. Among the downregulated genes, proteins similar to Noelin, to an αB-crystallin and to a “platelet-derived” growth factor were found. A protein with no annotation but containing a domain of the tumor necrosis factor receptor superfamily (TNFR) was also downregulated. GO terms related to organ development, cell differentiation and cell chemotaxis were enriched in this set of genes (Supplementary Table 2). Upregulated genes were rich in GO terms related to the negative regulation of calcium ion-dependent exocytosis, regulation of extracellular matrix assembly, collagen catalysis, changes in bone morphogenesis, negative regulation of cell-substrate adhesion, transmembrane transport of zinc and response to antigenic stimulus. Additionally, the Notch-signaling pathway appeared to be positively regulated, contrary to previous findings (Kaniewska et al., 2012). No DEGs were found in the coral after 42 days of exposure to low pH.
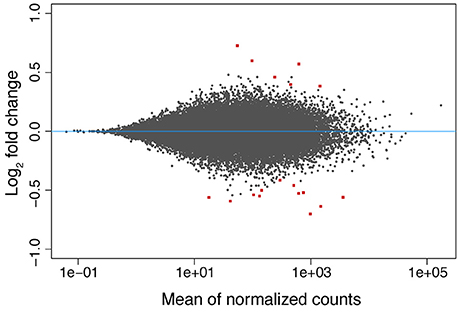
Figure 3. MA-plot of the differences in gene expression observed in the coral host between low (~7.6) and normal (~8.2) pH conditions after 3 days of exposure. The log2 fold changes in expression are shown as a function (y-axis) of the normalized read counts for each gene (x-axis). The values of the 18 DEGs found are displayed as red squares.
Symbiont Response to Low pH
The exploratory analyses of gene expression of Symbiodinium sp. suggest no change at any sampling time (Figure 4). The adonis permutation test resulted in no significant differences between conditions and colony after the first (pcondition = 0.62, pcolony = 0.78) or the second (pcondition = 0.88, pcolony = 0.99) exposure period. No DEGs were found at any time.
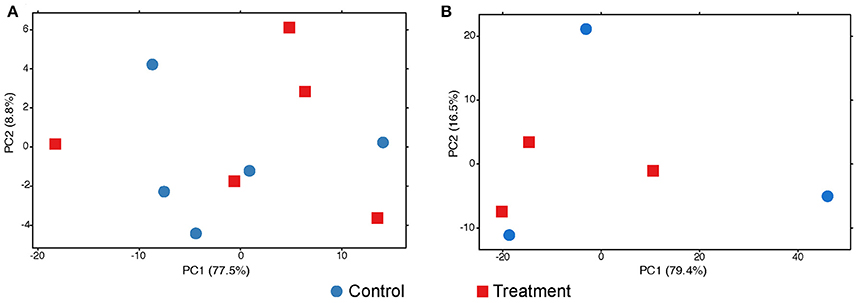
Figure 4. PCA plots of the variance of expressed genes in the symbionts after 3 (A) and 42 (B) days of exposure to low pH. No grouping patterns at any time can be distinguished. Dot color and shape codes are displayed at the bottom.
Discussion
Mild Stress Response of the Coral to Low pH
The present study used RNA-Seq to assess the coupled response of an adult stony coral (M. digitata) and its microalgal symbionts to low pH. Compared to previous investigations (Kaniewska et al., 2012; Moya et al., 2012, 2015), our results suggest a mild gene expression response of the adult coral holobiont to a 3-days exposure to acidified water in terms of number of DEGs and log2-fold change. Still, the log2-fold changes we found in this study are comparable to those observed in the resilient coral Siderastrea siderea under laboratory-induced ocean acidification conditions (Davies et al., 2016). This suggests resilience of M. digitata to low pH.
The few DEGs found after short-term exposure of M. digitata to low pH (7.6) have been shown to be involved in stress response in corals. Zinc-metalloproteases, for instance, seem to be relevant for heat tolerance in these organisms and might play a role in regulation of apoptosis and cell repair (Barshis et al., 2013). Other stress related genes were found among the significantly downregulated genes, like the αB-crystallin-like protein, an HSP20 family member. αB-crystallin seems to play a role in stress response in other corals (e.g., Orbicella annularis and O. faveolata), but its exact function is yet unknown (Downs et al., 2002). A TNFR domain-containing protein is another example, and changes in expression levels of proteins carrying this type of domain display have been found in other corals under stress conditions (Barshis et al., 2013; Seneca and Palumbi, 2015; Yuan et al., 2017). Another down-regulated gene codes for a protein containing a platelet-derived growth factor domain, though the regulation of this kind of protein upon exposure to low pH has not been previously reported in corals. However, it is already known that homologs of human bone-morphogenesis proteins (BMP2/4) participate in skeleton production of marine calcifiers, including corals (Green et al., 2013). In fact, proteins carrying this domain are currently used for bone regeneration in humans, and its use, in combination with coral skeleton, has been proposed to treat bone defects (Parizi et al., 2012).
According to previous investigations, coral larvae can acclimate to acidified water within a few days (Moya et al., 2015). Such a rapid acclimation by the adult coral holobiont would thus explain the lack of change in gene expression after 42 days of acidified water exposure in our experiments, a time by which acclimation would likely have been already completed. Our findings in the long-term response are similar to those of a previous investigation on the response of another adult scleractinian (Acropora millepora) to a 37-days exposure to low pH (elevated pCO2) that reported absence of change in the expression of target genes associated with calcification and metabolism (Rocker et al., 2015). In agreement with this, previous studies suggest that temperature stress has a greater impact in gene expression of corals than pH (Mayfield et al., 2014; Davies et al., 2016), although resilience to stress conditions varies from one species to another (Loya et al., 2001), and even among populations of the same coral species occurring in habitats with different local conditions (Poli et al., 2017).
Symbiont Response
Physiological response to stress conditions does not only depend on coral host resilience but also on the interaction with its symbionts (Hoadley et al., 2015). The Symbiodinium sp. studied here, which likely belongs to clade C and is specific to M. digitata (Supplementary Figure 2), did not seem to be affected by the decrease in pH. The more stable environment provided by the coral tissue, which acts as some kind of protection for the algae may serve as an explanation for this observation (Banaszak and Trench, 1996). Nonetheless, the study of the ex hospite transcriptomic response to heat stress of Symbiodinium in the same clade revealed distinct levels of heat-stress tolerance among different populations of this symbiont (Levin et al., 2016). These findings indicate that the tolerance level of different Symbiodinium types and populations to environmental stressors may contribute significantly to the resilience of particular coral holobiont assemblages (Yuyama et al., 2012; Putnam et al., 2017). Still, differences in response to stress between Symbiodinium spp. in hospite and isolated from the host remain largely unexplored, as does the contribution of the resilience of the symbiotic partner to the response of the holobiont.
An alternative explanation for the lack of change in overall gene expression of the symbionts in this study might lie on the constitutive expression of most genes observed in dinoflagellates (Moustafa et al., 2010) and on the reliance on post-transcriptional regulation of gene expression in these organisms (Baumgarten et al., 2013; Liew et al., 2017). However, little is known about the mechanisms of gene expression regulation in Symbiodinium and this topic needs to be more actively investigated given its potential role in holobiont resilience.
Conclusions
Here we provide baseline data in the form of a reference transcriptome of the scleractinian coral M. digitata and its dinoflagellate symbiont. We used these data to assess changes in overall gene expression of the different components of the coral holobiont, i.e., the coral host and its microalgal symbionts, after different periods of exposure to low pH (7.6) seawater. While the coral host showed an initial stress response after short-term exposure to low pH, an acclimatization of gene expression levels was indicated after longer exposure (42 days) to low pH, whereas its symbionts remained unaffected independent of exposure times. While we cannot exclude tank effects due to the experimental set-up, these findings nonetheless suggest a potential of the M. digitata holobiont for acclimatization and resilience to lowered seawater pH, and that temperature may play a greater role in coral stress than pH. Both the reference transcriptome we provide and the working hypothesis of acclimatization and resilience in this model system serve as a valuable baseline for future in-depth experiments.
Author Contributions
GW and SV conceived the study, SV coordinated the lab and analytical work, RG-P carried out the experiments, lab work and analyzed the data, WF contributed to bioinformatics analyses, GW acquired the funding.
Funding
This publication is the outcome of the Master Thesis project of the first author, who was funded by the National Council of Science and Technology of Mexico (CONACYT) to pursue his studies at LMU. The study benefited from funding by LMU Munich's Institutional Strategy LMU Excellent within the framework of the German Excellence Initiative.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SF and handling Editor declared their shared affiliation.
Acknowledgments
This study would not have been possible without the support of Gabriele Büttner, Simone Schätzle, and of Dr. Peter Naumann. Dr. Michael Eitel provided important discussions about data analysis. We would also like to thank Dr. Helmut Blum, Dr. Stefan Krebs, and Andrea Klanner, LAFUGA, Gene Center LMU München, for the support during early stages of laboratory work. We acknowledge Dr. Sylvain Foret for his help with Psytrans. SV is indebted to N. Villalobos Trigureros, M. Vargas Villalobos, S. Vargas Villalobos, and S. Vargas Villalobos for their constant support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2017.00403/full#supplementary-material
References
Albright, R., and Langdon, C. (2011). Ocean acidification impacts multiple early life history processes of the Caribbean coral Porites astreoides. Glob. Chang. Biol. 17, 2478–2487. doi: 10.1111/j.1365-2486.2011.02404.x
Albright, R., Mason, B., and Langdon, C. (2008). Effect of aragonite saturation state on settlement and post-settlement growth of Porites astreoides larvae. Coral Reefs 27, 485–490. doi: 10.1007/s00338-008-0392-5
Albright, R., Mason, B., Miller, M., and Langdon, C. (2010). Ocean acidification compromises recruitment success of the threatened Caribbean coral Acropora palmata. Proc. Natl. Acad. Sci. U.S.A. 107, 20400–20404. doi: 10.1073/pnas.1007273107
Alexa, A., and Rahnenfuhrer, J. (2010). topGO: Enrichment Analysis for Gene Ontology. R package version 2(0). doi: 10.18129/B9.bioc.topGO
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Anthony, K. R., Kline, D. I., Diaz-Pulido, G., Dove, S., and Hoegh-Guldberg, O. (2008). Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc. Natl. Acad. Sci. U.S.A. 105, 17442–17446. doi: 10.1073/pnas.0804478105
Baillie, B. K., Belda-Baillie, C. A., and Maruyama, T. (2000). Conspecificity and Indo-Pacific distribution of Symbiodinium genotypes (Dinophyceae) from giant clams. J. Phycol. 36, 1153–1161. doi: 10.1046/j.1529-8817.2000.00010.x
Banaszak, A., and Trench, R. (1996). Effects of ultraviolet (UV) radiation on marine microalgal-invertebrate symbioses. I. Response of the algal symbionts in culture and in hospite. Oceanogr. Literat. Rev. 8, 816.
Barbrook, A. C., Voolstra, C. R., and Howe, C. J. (2014). The chloroplast genome of a Symbiodinium sp. clade C3 isolate. Protist 165, 1–13. doi: 10.1016/j.protis.2013.09.006
Barshis, D. J., Ladner, J. T., Oliver, T. A., Seneca, F. O., Traylor-Knowles, N., and Palumbi, S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U.S.A. 110, 1387–1392. doi: 10.1073/pnas.1210224110
Baumgarten, S., Bayer, T., Aranda, M., Liew, Y. J., Carr, A., Micklem, G., et al. (2013). Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genomics 14:704. doi: 10.1186/1471-2164-14-704
Bayer, T., Aranda, M., Sunagawa, S., Yum, L. K., DeSalvo, M. K., Lindquist, E., et al. (2012). Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS ONE 7:e35269. doi: 10.1371/journal.pone.0035269
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chen, P.-Y., Chen, C.-C., Chu, L., and McCarl, B. (2015). Evaluating the economic damage of climate change on global coral reefs. Glob. Environ. Change 30, 12–20. doi: 10.1016/j.gloenvcha.2014.10.011
Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. doi: 10.1016/0003-2697(87)90021-2
Cornwall, C. E., and Hurd, C. L. (2016). Experimental design in ocean acidification research: problems and solutions. ICES J. Mar. Sci. 73, 572–581. doi: 10.1093/icesjms/fsv118
Crawley, A., Kline, D. I., Dunn, S., Anthony, K. E. N., and Dove, S. (2010). The effect of ocean acidification on symbiont photorespiration and productivity in Acropora formosa. Glob. Chang. Biol. 16, 851–863. doi: 10.1111/j.1365-2486.2009.01943.x
Cyronak, T., Schulz, K. G., and Jokiel, P. L. (2015). The Omega myth: what really drives lower calcification rates in an acidifying ocean. ICES J. Mar. Sci. 73, 558–562. doi: 10.1093/icesjms/fsv075
Davies, S. W., Marchetti, A., Ries, J. B., and Castillo, K. D. (2016). Thermal and pCO2 stress elicit divergent transcriptomic responses in a resilient coral. Front. Mar. Sci. 3:112. doi: 10.3389/fmars.2016.00112
De'ath, G., Lough, J. M., and Fabricius, K. E. (2009). Declining coral calcification on the Great Barrier Reef. Science 323, 116–119. doi: 10.1126/science.1165283
Downs, C., Fauth, J. E., Halas, J. C., Dustan, P., Bemiss, J., and Woodley, C. M. (2002). Oxidative stress and seasonal coral bleaching. Free Radic. Biol. Med. 33, 533–543. doi: 10.1016/S0891-5849(02)00907-3
Dunn, C. W., Howison, M., and Zapata, F. (2013). Agalma: an automated phylogenomics workflow. BMC Bioinformatics 14:330. doi: 10.1186/1471-2105-14-330
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Francis, W. R., Christianson, L. M., Kiko, R., Powers, M. L., Shaner, N. C., and Haddock, S. H. (2013). A comparison across non-model animals suggests an optimal sequencing depth for de novo transcriptome assembly. BMC Genomics 14:167. doi: 10.1186/1471-2164-14-167
Goecks, J., Nekrutenko, A., and Taylor, J. (2010). Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86. doi: 10.1186/gb-2010-11-8-r86
Gouy, M., Guindon, S., and Gascuel, O. (2009). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Green, D. W., Padula, M. P., Santos, J., Chou, J., Milthorpe, B., and Ben-Nissan, B. (2013). A therapeutic potential for marine skeletal proteins in bone regeneration. Mar. Drugs 11, 1203–1220. doi: 10.3390/md11041203
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hoadley, K. D., Pettay, D. T., Grottoli, A. G., Cai, W.-J., Melman, T. F., Schoepf, V., et al. (2015). Physiological response to elevated temperature and pCO2 varies across four Pacific coral species: understanding the unique host+symbiont response. Sci. Rep. 5:18371. doi: 10.1038/srep18371
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Holcomb, M., Venn, A., Tambutte, E., Tambutte, S., Allemand, D., Trotter, J., et al. (2014). Coral calcifying fluid pH dictates response to ocean acidification. Sci. Rep. 4:5207. doi: 10.1038/srep05207
Jokiel, P., Rodgers, K., Kuffner, I., Andersson, A., Cox, E., and Mackenzie, F. (2008). Ocean acidification and calcifying reef organisms: a mesocosm investigation. Coral Reefs 27, 473–483. doi: 10.1007/s00338-008-0380-9
Kaniewska, P., Campbell, P. R., Kline, D. I., Rodriguez-Lanetty, M., Miller, D. J., Dove, S., et al. (2012). Major cellular and physiological impacts of ocean acidification on a reef building coral. PLoS ONE 7:e34659. doi: 10.1371/journal.pone.0034659
Kleypas, J. A., Feely, R. A., Fabry, V. J., Langdon, C., Sabine, C. L., and Robbins, L. L. (2005). Impacts of Ocean Acidification on Coral Reefs and Other Marine Calcifiers: a Guide for Future Research. Report of a workshop held 18, 20.
Krief, S., Hendy, E. J., Fine, M., Yam, R., Meibom, A., Foster, G. L., et al. (2010). Physiological and isotopic responses of scleractinian corals to ocean acidification. Geochim. Cosmochim. Acta 74, 4988–5001. doi: 10.1016/j.gca.2010.05.023
Kurman, M. D., Gómez, C. E., Georgian, S. E., Lunden, J. J., and Cordes, E. E. (2017). Intra-specific variation reveals potential for adaptation to ocean acidification in a cold-water coral from the Gulf of Mexico. Front. Mar. Sci. 4:111. doi: 10.3389/fmars.2017.00111
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Levin, R. A., Beltran, V. H., Hill, R., Kjelleberg, S., McDougald, D., Steinberg, P. D., et al. (2016). Sex, scavengers, and chaperones: transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol. 33, 2201–2215. doi: 10.1093/molbev/msw119
Liew, Y. J., Li, Y., Baumgarten, S., Voolstra, C. R., and Aranda, M. (2017). Condition-specific RNA editing in the coral symbiont Symbiodinium microadriaticum. PLoS Genet. 13:e1006619. doi: 10.1371/journal.pgen.1006619
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Loya, Y., Sakai, K., Yamazato, K., Nakano, Y., Sambali, H., and van Woesik, R. (2001). Coral bleaching: the winners and the losers. Ecol. Lett. 4, 122–131. doi: 10.1046/j.1461-0248.2001.00203.x
Mayfield, A. B., Wang, Y. B., Chen, C. S., Lin, C. Y., and Chen, S. H. (2014). Compartment-specific transcriptomics in a reef-building coral exposed to elevated temperatures. Mol. Ecol. 23, 5816–5830. doi: 10.1111/mec.12982
McCulloch, M., Falter, J., Trotter, J., and Montagna, P. (2012). Coral resilience to ocean acidification and global warming through pH up-regulation. Nat. Clim. Chang. 2, 623–627. doi: 10.1038/nclimate1473
Meehl, G. A., Stocker, T. F., Collins, W. D., Friedlingstein, P., Gaye, A. T., Gregory, J. M., et al. (2007). “Global climate projections,” in Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, eds S. Salomon, D. Quin, M. Manning, Z. Chen, M. Marquis, K. B. Averyt, M. Tignor, and H.L. Miller (Cambridge; New York, NY: Cambridge University Press), 747–846.
Moberg, F., and Folke, C. (1999). Ecological goods and services of coral reef ecosystems. Ecol. Econ. 29, 215–233. doi: 10.1016/S0921-8009(99)00009-9
Morita, M., Suwa, R., Iguchi, A., Nakamura, M., Shimada, K., Sakai, K., et al. (2010). Ocean acidification reduces sperm flagellar motility in broadcast spawning reef invertebrates. Zygote 18, 103–107. doi: 10.1017/S0967199409990177
Moustafa, A., Evans, A. N., Kulis, D. M., Hackett, J. D., Erdner, D. L., Anderson, D. M., et al. (2010). Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS ONE 5:e9688. doi: 10.1371/journal.pone.0009688
Moya, A., Huisman, L., Ball, E., Hayward, D., Grasso, L., Chua, C., et al. (2012). Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2-driven acidification during the initiation of calcification. Mol. Ecol. 21, 2440–2454. doi: 10.1111/j.1365-294X.2012.05554.x
Moya, A., Huisman, L., Foret, S., Gattuso, J. P., Hayward, D. C., Ball, E., et al. (2015). Rapid acclimation of juvenile corals to CO2-mediated acidification by upregulation of heat shock protein and Bcl-2 genes. Mol. Ecol. 24, 438–452. doi: 10.1111/mec.13021
Nakamura, M., Ohki, S., Suzuki, A., and Sakai, K. (2011). Coral larvae under ocean acidification: survival, metabolism, and metamorphosis. PLoS ONE 6:e14521. doi: 10.1371/journal.pone.0014521
Oksanen, J. (2001). Logic of experiments in ecology: is pseudoreplication a pseudoissue? Oikos 94, 27–38. doi: 10.1034/j.1600-0706.2001.11311.x
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., et al. (2013). Package ‘vegan.’ Community Ecology Package 2(9).
Parizi, A. M., Oryan, A., Shafiei-Sarvestani, Z., and Bigham, A. (2012). Human platelet rich plasma plus Persian Gulf coral effects on experimental bone healing in rabbit model: radiological, histological, macroscopical and biomechanical evaluation. J. Mater. Sci. 23, 473–483. doi: 10.1007/s10856-011-4478-1
Parra, G., Bradnam, K., and Korf, I. (2007). CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067. doi: 10.1093/bioinformatics/btm071
Poli, D., Fabbri, E., Goffredo, S., Airi, V., and Franzellitti, S. (2017). Physiological plasticity related to zonation affects hsp70 expression in the reef-building coral Pocillopora verrucosa. PLoS ONE 12:e0171456. doi: 10.1371/journal.pone.0171456
Putnam, H. M., Barott, K. L., Ainsworth, T. D., and Gates, R. D. (2017). The vulnerability and resilience of reef-building corals. Curr. Biol. 27, R528–R540. doi: 10.1016/j.cub.2017.04.047
Riebesell, U., Fabry, V. J., Hansson, L., and Gattuso, J.-P. (2011). Guide to Best Practices for Ocean Acidification Research and Data Reporting. Office for Official Publications of the European Communities.
Rocker, M. M., Noonan, S., Humphrey, C., Moya, A., Willis, B. L., and Bay, L. K. (2015). Expression of calcification and metabolism-related genes in response to elevated pCO2 and temperature in the reef-building coral Acropora millepora. Mar. Genomics 24, 313–318. doi: 10.1016/j.margen.2015.08.001
Rodolfo-Metalpa, R., Houlbrèque, F., Tambutté, É., Boisson, F., Baggini, C., Patti, F. P., et al. (2011). Coral and mollusc resistance to ocean acidification adversely affected by warming. Nat. Clim. Chang. 1, 308–312. doi: 10.1038/nclimate1200
Schank, J. C., and Koehnle, T. J. (2009). Pseudoreplication is a pseudoproblem. J. Comp. Psychol. 123, 421–433. doi: 10.1037/a0013579
Seneca, F. O., and Palumbi, S. R. (2015). The role of transcriptome resilience in resistance of corals to bleaching. Mol. Ecol. 24, 1467–1484. doi: 10.1111/mec.13125
Shinzato, C., Inoue, M., and Kusakabe, M. (2014). A snapshot of a coral “holobiont”: a transcriptome assembly of the scleractinian coral, Porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS ONE 9:e85182. doi: 10.1371/journal.pone.0085182.t001
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. doi: 10.1038/nature10249
Shoguchi, E., Shinzato, C., Kawashima, T., Gyoja, F., Mungpakdee, S., Koyanagi, R., et al. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408. doi: 10.1016/j.cub.2013.05.062
Sun, J., Chen, Q., Lun, J. C., Xu, J., and Qiu, J.-W. (2013). PcarnBase: development of a transcriptomic database for the brain coral Platygyra carnosus. Mar. Biotechnol. 15, 244–251. doi: 10.1007/s10126-012-9482-z
Suwa, R., Nakamura, M., Morita, M., Shimada, K., Iguchi, A., Sakai, K., et al. (2010). Effects of acidified seawater on early life stages of scleractinian corals (Genus Acropora). Fish. Sci. 76, 93–99. doi: 10.1007/s12562-009-0189-7
Venn, A. A., Tambutté, E., Holcomb, M., Laurent, J., Allemand, D., and Tambutté, S. (2013). Impact of seawater acidification on pH at the tissue–skeleton interface and calcification in reef corals. Proc. Natl. Acad. Sci. U.S.A. 110, 1634–1639. doi: 10.1073/pnas.1216153110
Von Euw, S., Zhang, Q., Manichev, V., Murali, N., Gross, J., Feldman, L. C., et al. (2017). Biological control of aragonite formation in stony corals. Science 356, 933–938. doi: 10.1126/science.aam6371
Yuan, C., Zhou, Z., Zhang, Y., Chen, G., Yu, X., Ni, X., et al. (2017). Effects of elevated ammonium on the transcriptome of the stony coral Pocillopora damicornis. Mar. Pollut. Bull. 114, 46–52. doi: 10.1016/j.marpolbul.2016.08.036
Keywords: gene expression, ocean acidification, coral, holobiont, resilience
Citation: González-Pech RA, Vargas S, Francis WR and Wörheide G (2017) Transcriptomic Resilience of the Montipora digitata Holobiont to Low pH. Front. Mar. Sci. 4:403. doi: 10.3389/fmars.2017.00403
Received: 06 July 2017; Accepted: 28 November 2017;
Published: 12 December 2017.
Edited by:
Stefano Goffredo, Università di Bologna, ItalyReviewed by:
Aldo Nicosia, Istituto per l'Ambiente Marino Costiero (CNR), ItalySilvia Franzellitti, Università di Bologna, Italy
Adolfo Rivero-Muller, Turku Centre for Biotechnology, Finland
Copyright © 2017 González-Pech, Vargas, Francis and Wörheide. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gert Wörheide, woerheide@lmu.de
†Present Address: Raúl A. González-Pech, Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD, Australia
 Raúl A. González-Pech
Raúl A. González-Pech Sergio Vargas
Sergio Vargas Warren R. Francis
Warren R. Francis Gert Wörheide
Gert Wörheide