Single-Cell Biomolecular Analysis of Coral Algal Symbionts Reveals Opposing Metabolic Responses to Heat Stress and Expulsion
- 1School of Life Sciences, University of Technology Sydney, Ultimo, NSW, Australia
- 2Centre for Biospectroscopy, School of Chemistry, Monash University, Clayton, VIC, Australia
- 3Department of Microbiology, Faculty of Medicine, Nursing and Health Sciences, Biomedical Discovery Institute, Monash University, Clayton, VIC, Australia
The success of corals in nutrient poor environments is largely attributed to the symbiosis between the cnidarian host and its intracellular alga. Warm water anomalies have been shown to destabilize this symbiosis, yet detailed analysis of the effect of temperature and expulsion on cell-specific carbon and nutrient allocation in the symbiont is limited. Here, we exposed colonies of the hard coral Acropora millepora to heat stress and using synchrotron-based infrared microspectroscopy measured the biomolecular profiles of individual in hospite and expelled symbiont cells at an acute state of bleaching. Our results showed symbiont metabolic profiles to be remarkably distinct with heat stress and expulsion, where the two effectors elicited opposing metabolic adjustments independent of treatment or cell type. Elevated temperature resulted in biomolecular changes reflecting cellular stress, with relative increases in free amino acids and phosphorylation of molecules and a concomitant decline in protein content, suggesting protein modification and degradation. This contrasted with the metabolic profiles of expelled symbionts, which showed relative decreases in free amino acids and phosphorylated molecules, but increases in proteins and lipids, suggesting expulsion lessens the overall effect of heat stress on the metabolic signature of the algal symbionts. Interestingly, the combined effects of expulsion and thermal stress were additive, reducing the overall shifts in all biomolecules, with the notable exception of the significant accumulation of lipids and saturated fatty acids. This first use of a single-cell metabolomics approach on the coral symbiosis provides novel insight into coral bleaching and emphasizes the importance of a single-cell approach to demark the cell-to-cell variability in the physiology of coral cellular populations.
Introduction
The mutualistic association of the unicellular alga Symbiodinium with reef building corals is founded on carbon exchange. The phototrophic symbiont relies on inorganic nutrients supplied by the host and in return delivers fixed organic carbon to the host (Muscatine, 1990), supplementing the host's nutritional demands to support growth and reproduction. In the low nutrient waters of the tropics, this bilateral transfer of metabolites provides a competitive advantage for survival, and under stable conditions this host-symbiont interaction is mutually beneficial to both partners. However, during unfavorable conditions, cellular metabolism becomes compromised, causing a diminished capacity for exchange between the host and symbiont (Douglas, 2003). Today, coral reefs are under pressure from warming ocean temperatures triggering mass coral bleaching (Glynn, 1991; De'ath et al., 2009; Hoegh-Guldberg and Bruno, 2010; Hughes et al., 2018), yet despite decades of research motivated by the intensifying loss of coral cover around the globe (Hughes et al., 2017), the underlying cellular mechanisms responsible remain poorly understood.
All organisms, including corals, adapt to changes in environmental conditions by adjusting their biochemical and cellular processes with the aim of maintaining cell homeostasis (Arrigo, 2005; Moore et al., 2013). These changes result in shifts in the cellular energetics and allocation of energy stores into various macromolecules and metabolites needed to uphold systems central to the cellular machinery, such as energy production, antioxidants, compatible solutes and compounds involved in intra and inter-cell signaling. Due to the inherent complexity of these metabolic networks, many studies of the coral symbiosis now employ “omics” approaches (transcriptomics, proteomics and metabolomics) to help unravel the intricacies of these networks (Burriesci et al., 2012; Dunn et al., 2012; Oakley et al., 2016; Sogin et al., 2016; Hillyer et al., 2017a,b). These “untargeted” approaches can provide a broad-spectrum overview of the metabolic state of the coral and/or its symbionts, by simultaneously assessing the prevalence of multiple molecules relating to a wide range of metabolic processes, thus providing a “snapshot” of the current cell state. Omics studies have generated insights into the effect of the symbiosis on the proteome of the holobiont in the coral model-organism Aiptasia sp. (Oakley et al., 2016), indicating a major role of lipid metabolism in the maintenance of the symbiosis. More recently, using a metabolomics approach, Hillyer et al. (2017b) revealed how heat stress induces major changes to multiple metabolic pathways in the coral symbionts, including glycolysis, pentose phosphate, fatty acid and amino acid metabolism. Studies such as these highlight the intricacies of the coral-symbiosis including its heavy dependence on energy metabolism by the two partners and how this dependence is affected during unfavorable conditions, resulting in the demise of the symbiosis.
While “omics” methods have already become important tools to understand the coral symbiosis, their inherent requirement of relatively large amounts of tissue for analysis may conceal the finer details of the central mechanisms investigated: Coral bleaching is often described as an ecosystem wide event but is fundamentally a process occurring at the single-celled scale (Weis, 2008), with each symbiont encased within a host-derived membrane (in hospite) being released independently. During bleaching, the coral host tissue contains in hospite algal cells, as well as a large proportion of cells that are no longer encased in a host membrane (ex hospite) that are possibly in the process of being expelled (Ladrière et al., 2008). As such, the common whole tissue approach employed in “omics” studies and other conventional experiments, inevitably provides results that average across cells with potentially different pathophysiologies and/or symbiotic states. For this reason, it is evident that separating the response of the different cell-types may be important if we are to elucidate what is happening to the symbionts during the bleaching process.
Cell level experiments have proved powerful in uncovering key processes of the coral symbiosis and have been pivotal in delivering a better understanding of cellular metabolism in Symbiodinium, and the mechanism of translocation of organic carbon (Kopp et al., 2015), nitrogen (Pernice et al., 2012; Kopp et al., 2013) and lipids (Peng et al., 2011; Kopp et al., 2013) in the coral holobiont. Unlike their bulk analysis counterparts, these studies resolved processes at the individual cell level, thus removing the inherent variability in the signal that results from population level studies. Recent years also have seen a surge in the development and use of single cell “omics” to assist in differentiating cell-to-cell variability within clonal populations (Zenobi, 2013). However, due to the lack of sufficient instrument sensitivity to measure the small amounts of metabolites within a cell, single-cell metabolomics is yet to come of age (Zenobi, 2013). In the absence of fine-scale metabolomics, an alternative approach presents itself in synchrotron-based infrared microspectroscopy. This fast, non-destructive technique can detect variations in biomolecular composition at the single cell level, thus providing a snapshot of the metabolic state of individual cells (Heraud et al., 2005).
In this study, we took a single-cell approach to examine the photosynthetic viability and biomolecular composition of (1) endosymbiotic (in hospite—encased in animal cell) and (2) actively expelled Symbiodinium cells during both healthy symbiosis and heat-induced bleaching in the reef-building coral Acropora millepora. Using synchrotron-based infrared microspectroscopy we were able to create high resolution “fingerprints” of the metabolic signature of individual symbionts experiencing distinct physiological conditions, uncovering separate and combined effects of heat stress and expulsion on the biomolecular dynamics of coral symbionts.
Materials and Methods
Coral Collection and Experimental Design
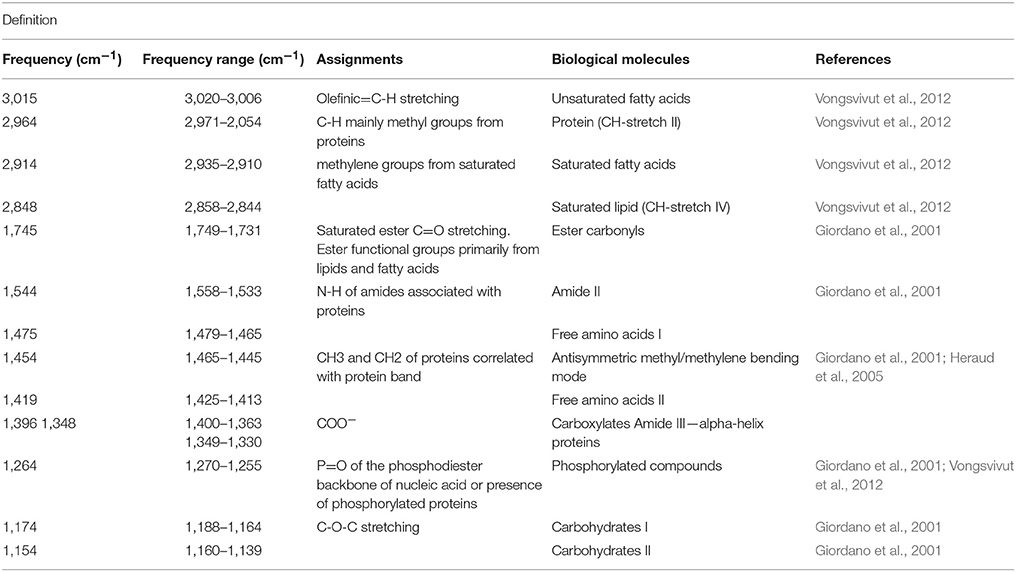
Three individual colonies of Acropora millepora were collected on the reef flat of Heron Island (< 2 m depth at low tide, a minimum of 20 m apart), Great Barrier Reef, Australia (152°06′E, 20°29′S) in February 2015. Coral colonies (~20 cm diameter) were halved and kept in aquaria (90 L) with flow through of natural seawater from the reef flat. Tanks were shaded (resulting in a daytime maximum of ~700 μmol photons m−2 s−1) for 5 days before commencement of the experiment. At commencement of the experiment, one half of each colony (n = 3) was maintained in individual aquaria at ambient summer water temperature (control; ~ 27.2°C) throughout the experiment (Figure 1). The other halves (n = 3), which were assigned to bleaching stress, were placed in individual aquaria that were exposed to temperature increases of 1°C per day until reaching 30°C and then 0.5°C per day until reaching ~32°C where they were maintained for 1 week (Figure 1). The gradual temperature increase with daily fluctuations was employed to allow for cellular adjustment in the corals and to avoid degenerative tissue loss from the animal, as often occurs during rapid temperature increase. Heating was controlled using aquarium bar heaters with continuous exchange of natural seawater (flow rate ~1 L/min) with additional water movement provided by submersible pumps in each aquarium. Water temperature was recorded every 10 min with temperature buttons (Thermochron, Australia). A proxy for coral (photosynthetic) health in situ was obtained by chlorophyll a fluorescence using pulse amplitude modulated fluorometry (Mini PAM, Walz GmbH, Effeltrich, Germany). Measurements were made in situ using a fibreoptic cable (5.5 mm diameter) held perpendicular to the coral surface with a 5 mm silicon tubing spacer. Once a stable signal was established (measuring light intensity = 2, photomultiplier gain = 12), minimum fluorescence (FO or FT) was recorded before application of a saturating pulse of light (SP width = 0.8 s, SP intensity = 12), to determine maximum fluorescence (FM or ). Measurements were made on all coral colonies twice daily, at midday and half an hour after sunset to determine light-adapted effective quantum yield of PSII (ΔF/) and the dark-adapted maximum quantum yield of PSII (FV/FM), respectively. Fluorescence data were analyzed for statistical differences over time and between treatments using rmANOVA (IBM, SPSS, Statistics v24; IBM Corporation, New York). Sampling of all colony replicates (control and heated) for single-cell fluorometry and biomolecular profiling was conducted 48 h after heat-treated corals reached an average of 31.5°C and every day for 4 consecutive days (Figure 1). This daily sampling regime was performed to ensure that the day of maximal expulsion of cells for the heated corals was captured, which occurred 96 h after reaching temperature (day 11).
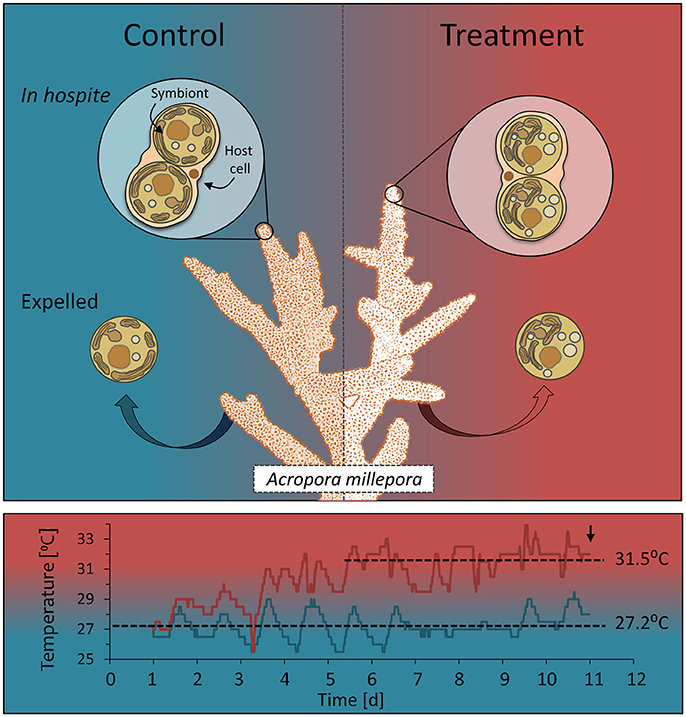
Figure 1. Sampling design showing cell types and temperature treatments. Colonies (n = 3) of the reef building coral Acropora millepora were divided in two and the two halves subjected to one of two temperature regimes: ambient summer temperatures (average 27.2°C), and warmer temperatures (average 31.5°C), following a 1-week, incremental increase. Samples from each colony and treatment of endosymbiotic (in hospite) and actively expelled cells were collected over 3 consecutive days (9, 10, 11) to capture the day of maximum bleaching, day 11 (down arrow). These cells (control and heat-treated) were then used for single-celled chl a fluorescence analysis, microscope imaging and biomolecular fingerprinting.
Intact endosymbiotic cells of Symbiodinium (ITS2 type C8; Gardner et al., 2017) were extracted from each coral colony by adding a coral fragment to a 50 mL centrifuge tube containing 5 mL of 0.22 μm filtered seawater (FSW) and firmly hitting the base of the tube against a hard surface three times. The resulting cell slurry had a high percentage of intact symbiont cells encased in viable animal cells (in hospite). Once extracted, cells were gently centrifuged (150 RCF for 30 s), resulting in precipitation of the heavier symbiont containing cells. This was necessary to concentrate the cells of interest, namely those still encased in gastrodermal host tissue (in hospite), for single cell analyses. Most of the less dense non-symbiotic animal cells were removed with the supernatant and the remaining cell pellet subsequently re-suspended in 100 μL FSW and directly analyzed for in hospite chlorophyll a fluorescence and symbiont cell integrity, or fixed in 2% formalin for microspectroscopy (see below). Expelled Symbiodinium cells were collected at mid-day by placing a large fragment from each coral colony and treatment into individual beakers containing 800 mL of FSW, maintained at respective treatment temperatures with continuous air bubbling. After 3 h the coral fragments were removed, the water from the beaker gravity filtered through a 5 μm polycarbonate filter and the captured cells gently washed into 5 mL of FSW for immediate in vivo analyses (microscopy, single-cell fluorescence) or fixation.
Physiological and Morphological Condition in Vivo
Physiological condition of Symbiodinium cells (in hospite and expelled) was assessed by single-cell chlorophyll a fluorescence and light microscopy. Structural degradation of symbionts was determined visually using a light microscope (400X; Olympus BX53) where cells showing signs of vacuolation and deformation or retraction of chloroplasts, were designated “abnormal.” Counts of normal (healthy) vs. abnormal (degenerated) appearing cells (n = 40–90 individuals from three individual colony fragments) as a percentage of the total were used to assess the overall cellular condition of the in hospite or expelled populations. Variable chlorophyll a fluorescence measurements were made on randomly selected individual cells within the control and treatment populations (n = 60–122 individuals from three individual fragment extractions or captured expelled cells) using a pulse amplitude modulated fluorometer (Imaging PAM IMAG-K4, Walz GmbH, Effeltrich, Germany) mounted on a compound microscope (Axiostar plus, Zeiss, Germany). Measurements were made at 200X magnification employing blue excitation light (440 nm) and collected using the Imaging Win software (V2.32 FW Multi RGB; Walz GmbH, Effeltrich, Germany). After 10 min dark-adaptation, minimum fluorescence (FO) was recorded before application of a saturating pulse of light (saturating pulse width = 0.8 s; saturating pulse intensity = 10; using the Special SP-routine), where maximum fluorescence (FM) was determined. From these two parameters FV/FM was calculated as (FM-FO)/FM. Cells were deemed to be in hospite based on the positive identification of an animal membrane surrounding the cell. Pooled populations of single cell FV/FM (from three individual coral fragments) of in hospite and expelled cells were tested for non-equal distributions between control and heated corals using Kolmogorov Smirnov and medians compared for statistical difference. Differences were considered significant at P < 0.05 (IBM SPSS Statistics v.24; IBM Corporation, New York). Single-cell data were pooled once the difference in the spread across individual populations had been shown to be non-significant between the extracts from the three coral fragments.
Coral Health and Biomolecular Composition
Coral fragments from each colony were excised and immediately frozen in liquid N2 for later determination of symbiont density and chlorophyll a and c2 concentrations. Briefly, tissue was released from coral fragments using a high pressure air-gun into 5 mL of FSW. For cell density measurements the resulting slurry was homogenized and concentration determined by manual counting using a haemocytometer. For chlorophyll a and c2 determination, the cell slurry was centrifuged (8000 RCF for 5 min), the supernatant discarded and the remaining pellet subsequently resuspended and extracted overnight in 3 mL of chilled 90% acetone. Samples were centrifuged prior to spectrophotometric determination of chlorophyll a and c2 (Ritchie, 2006). Cell densities and chlorophyll concentrations were normalized to surface area, which was determined for each coral fragment using a standardized wax technique (Stimson and Kinzie, 1991) and analyzed for statistical significance using ANOVA (IBM SPSS Statistics v.24; IBM Corporation, New York).
Synchrotron Infrared Microspectroscopy
Formalin-fixed cells were loaded into a micro compression cell (modified compression cell, Spectratech Inc., Oak Ridge, USA; Tobin et al., 2010) with 1 mm thick CaF2 window. Measurements were made on hydrated cells, a method shown to limit light scattering effects (Bambery et al., 2012; Sackett et al., 2014). Spectra were acquired over the measurement range 4,000–800 cm−1 with a Vertex 80v Fourier Transform Infra-Red (FTIR) spectrometer (Bruker Optics, Ettlingen, Germany) in conjunction with an IR microscope (Hyperion 3000, Bruker) fitted with a liquid nitrogen cooled mercury cadmium telluride detector. The microscope was connected to a computer-controlled microscope stage contained within a box purged with dehumidified air. Co-added interferograms (n = 64) were collected at a wavenumber resolution of 6 cm−1 s. Spectral measurements were made on individual Symbiodinium cells using transmission mapping mode with an aperture size of 4 μm × 4 μm. Spectral acquisition and instrument control was performed using Opus 6.5 software (Bruker).
As with the single celled fluorometry, positive visual identification of an animal membrane surrounding the symbiont was required, ensuring that each spectral measurement collected for an in hospite cell was representative of an intact endosymbiont. Multiple spectra (3–5) from each cell were averaged and exported for multivariate analysis using The Unscrambler software package (v10.4, Camo Inc., Oslo, Norway). The regions that contained the major biological bands were selected by excluding the non-biological wavenumbers from the spectra and collating the spectral regions from 3,050 to 2,820, 1,770 to 1,700, and 1,640 to 1,122 cm−1 (Supplementary Figure 1). Data were smoothed (4 pts either side) and second derivative (3rd order polynomial) transformed using the Savitzky-Golay algorithm and then normalized using the Single Normal Variate (SNV) before performing Principal Component Analysis (PCA). To ensure minimal influence from the animal tissue encasing the endosymbiotic cells, animal spectra from both treatment groups (n = 7) were pre-treated as described above and the averaged spectrum subtracted from each in hospite spectrum. The animal host tissue produced a considerably weaker absorption spectrum than that of the symbionts (Supplementary Figure 1). Moreover, in corals, the animal cell is tightly stretched around the symbionts, resulting in a thin layer (~0.5–1 μm) of cell membrane and cytosol encasing the algae (Supplementary Figure 2), thus further reducing the overall contribution of the animal to the spectrum of the endosymbiotic algal cells. As absorbance is a direct function of path-length and thus tissue thickness, the contribution of the animal signal was calculated based on the average path-length through the host tissue of the endoderm cell relative to the spectrum measured on individual host cells (Supplementary Figure 2).
Biomolecular contents were estimated based on the area under the assigned peaks according to the Beer—Lambert Law describing a direct relationship between absorbance and analyte concentration (Wagner et al., 2010). Second derivative spectra were used for individual biomolecule peak area integration for each cell type (in hospite and expelled) and treatment (control and heated). Integrated peak areas of populations of individual cell profiles were tested for non-equal distributions using a Kolmogorov Smirnov test at α = 0.05 (IBM SPSS Statistics v.24; IBM Corporation, New York). A heat map of the relative changes in peak areas was generated using the free software package R (R Core Team, 2013) and plugins gplots (Warnes et al., 2016) and pheatmap (Kolde, 2015).
Results
Coral fragments showed a significant decline in the effective quantum yield of PSII (ΔF/) with heat treatment on days 10 and 11 [rmANOVA, F(10,40) = 32.89, P < 0.001] and a significant drop in maximum quantum yield (FV/FM) 48 h after reaching 32°C [day 9; rmANOVA, F(9, 36) = 26.36, P = 0.006], with a further decline on the following days (Figures 2A,B). Cell density and chlorophyll a and c2 concentrations also declined significantly with elevated temperature [ANOVA F(1, 4) = 20.89, P < 0.001; F(1, 4) = 43.33, P = 0.003; F(1, 4) = 81.56, P = 0.001, respectively] by day 11 (96 h after reaching 32°C; Figures 2C–E). Corals kept at 27°C (controls) showed no changes in ΔF/, FV/, chlorophyll or cell density.
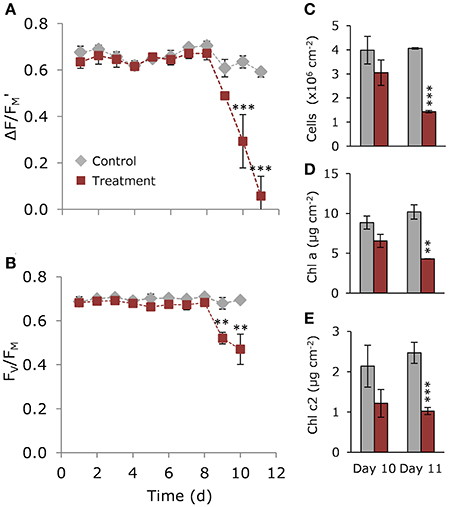
Figure 2. Coral physiological condition over time. (A) Effective quantum yield (ΔF/) and (B) maximum quantum yield (FV/FM) of PSII over time for control (gray) and heated (red) corals. (C) Cell density (D) chlorophyll a and (E) chlorophyll c2 (μg cm−1) in control (gray) and heated (red) corals on days 10 and 11. Data represent means ± SE. (n = 3). Asterisks denote treatment is significantly different from the control over time: **P < 0.01; ***P < 0.001.
On day 11, the proportion of healthy to degraded cells varied with both cell type and treatment (Figure 3A), with degraded cells making up ~2% of in hospite cells from the control compared with ~67% for in hospite cells exposed to thermal stress (Figure 3B). When compared with data from day 10, where only 28% of the population showed signs of degradation (Supplementary Figure 3), these data demonstrate the rapidity of the decline in population viability and health under thermal stress. For expelled symbionts, 26% of the cells showed signs of degradation in the control compared with 93% of cells expelled under thermal stress on day 11 (Figure 3B). The data from day 10 showed that the proportion of degraded cells was unchanged in the control, and made up only 54% of the expelled population under thermal stress (Supplementary Figure 3).
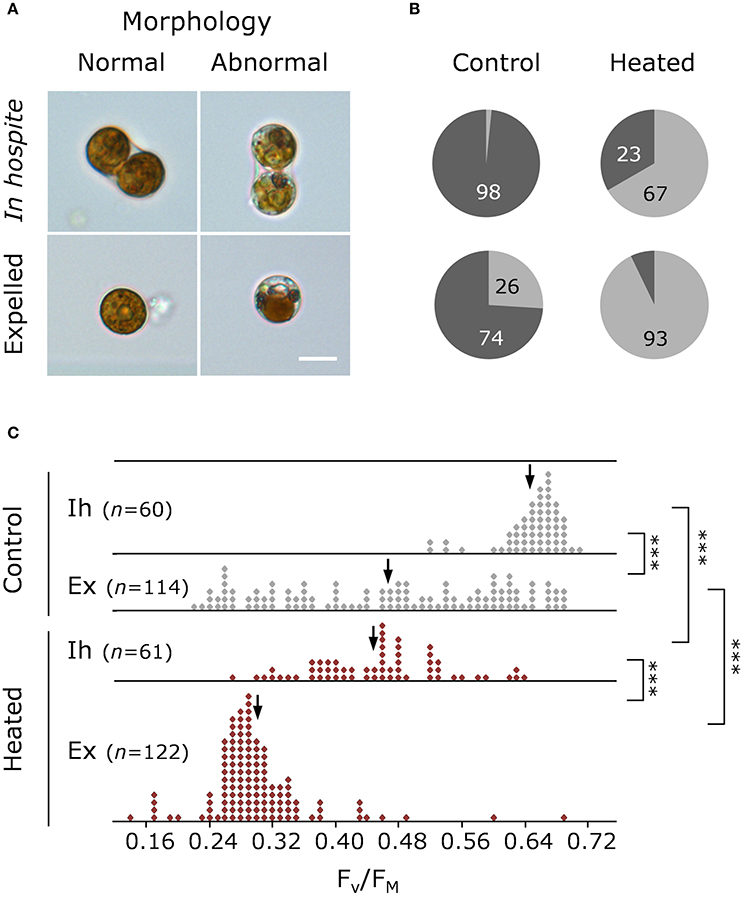
Figure 3. Health and morphology of Symbiodinium on day 11. (A) Representative light microscopy images of normal and abnormal morphologies, and (B) percentage of normal (dark gray) and abnormal (light gray) in hospite and expelled cells from control and heated corals. (C) Single-cell maximum quantum yield of PSII (FV/FM) of in hospite and expelled cell populations from three individual coral fragments from control (gray) and heated (red) corals. Scale bar = 10 μm. Down arrows denote population median. Vertical brackets indicate significant differences in population distributions (Kolmogorov Smirnoff test) and medians, and asterisks denote significance level ***P < 0.001.
The patterns observed using light microscopy are reflected in the single-celled fluorescence data, where under control (27°C) conditions, the FV/FM of in hospite cells ranged from 0.486 to 0.743 with a population mean of 0.659 (Figure 3C), while in expelled cells the range was much broader, from 0.232 to 0.679, with a significantly lower median ( = 0. 480; P < 0.001). The effect of temperature by day 11 resulted in an overall broadening of FV/FM (0.246–0.641) in the endosymbiotic population and a significantly lower median ( 0.462; P < 0.001). In the heated expelled cells, the large spread in FV/FM in cells expelled a day earlier (day 10; Supplementary Figure 3) condensed to form a unimodal distribution (0.139–0.689) around a median of 0.289 (Figure 3C) by day 11, resulting in a significant shift in the health of the population between the 2 days (PKS < 0.001).
Infrared absorbance spectra showed typical bands associated with microalgal samples with clear discrimination of profiles based on temperature and cell type (Supplementary Figure 1). The normalized 2nd derivative spectra of the selected biologically relevant regions revealed the specific absorbance bands representing key molecules of interest (Figure 4A), from which a total of 14 bands with unequivocal band assignments were selected for comparison across cell type and treatment (Figure 4A; Table 1).
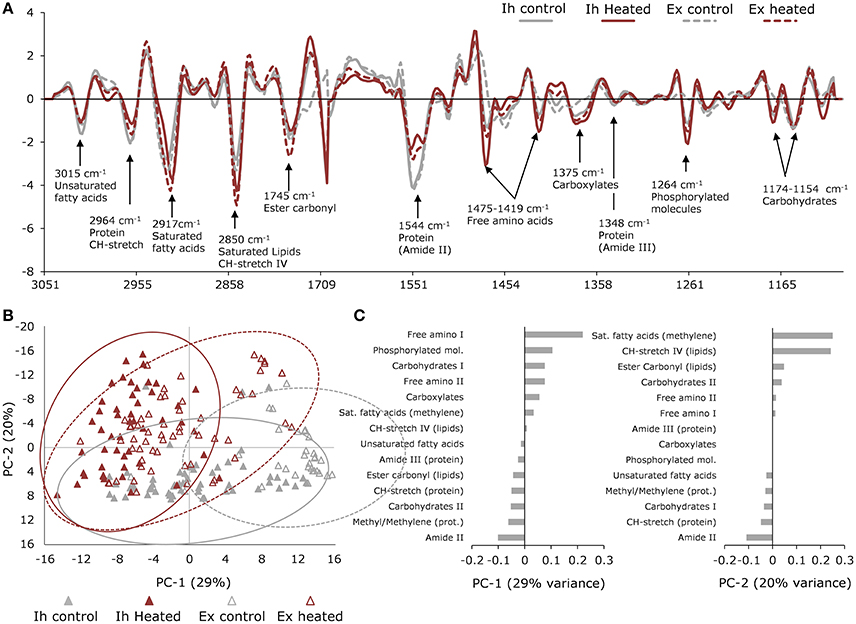
Figure 4. Synchrotron-based IR microspectroscopy data. (A) Normalized average 2nd derivative spectra of in hospite (solid line) and expelled (broken line) cells from control (gray) and heated (red) corals showing major biomolecular peaks. (B) Principle Component Analysis (PCA) scores plot showing differentiation in macromolecular profiles of in hospite (closed triangles) and expelled (open triangles) cells under control (gray) and heated (red) conditions. Ellipses represent the 95% confidence intervals of the data. (C) PCA loadings showing the contributions of each biomolecule to PC1 and PC2 variation, where negatively loaded bands have the strongest absorbance in positively scored samples and positively loaded bands will have the strongest absorbance in negatively scored samples. Pre-treatment and sorting of spectra resulted in a total of n = 55 (in hospite control); n = 48 (in hospite heated); n = 29 (expelled control); n = 59 (expelled heated) collected across three individual coral colony fragments.
Principal Component Analysis (PCA) of the biologically relevant spectra revealed the dominant effect was associated with temperature, explaining 29% of the variation, while PC2 was more closely associated with the expulsion effect, explaining 20% of the variation (Figure 4B). The most pronounced response to elevated temperature was seen in the free amino acids (bands at 1,475 and 1,419 cm−1), which were highest in the heated samples, as were the phosphorylated molecules (band at 1,264 cm−1; Figure 4A). The PC1 loadings showed an opposite trend between saturated lipids and proteins, where the heat-treated samples were high in saturated lipids (bands at 1,753, 2,848 and 2,914 cm−1) and low in proteins (bands at 1,550 and 1,348 cm−1). This trend was reversed for unsaturated lipids (band at 3,015 cm−1), which were higher in the control samples (Figure 4C). The next major direction of influence along PC2 (Figure 4B), showed the effect of expulsion without the dominating influence of temperature. The PC2 loadings showed that the expelled symbionts were higher in saturated lipids (~1,740, 2,848, 2,914 cm−1) and carbohydrates (1,154 cm−1), compared with non-expelled endosymbionts (in hospite) which showed relatively more protein (1,550 cm−1) and unsaturated lipid (3,015 cm−1) content (Figure 4C).
To further differentiate temperature from expulsion effects, we used integrated peak areas to calculate the relative change compared to controls and a heat map was generated with statistical clustering of molecules based on similarity and treatment (Figure 5). The cluster analysis grouped the heat-treated spectra to one side and expelled spectra to the other, with the heat-treated expelled cells forming an intermediary column (Figure 5). Free amino acids (FAA), carboxylates and phosphorylated molecules showed the greatest relative increase with temperature (0.6–1.7; PKS < 0.001) in both the in hospite and expelled cells. Temperature also resulted in a significant relative decrease in proteins (methyl/methylene, amide II and III, CH-stretch II peaks) in both in hospite and expelled heat-treated cells (PKS < 0.01) and a significant decline in unsaturated fatty acids in the heat-treated in hospite cells (PKS < 0.001; Figure 5). The most significant biomolecular changes associated with expulsion were increases in methyl/methylene ratio (0.4–1.1; PKS < 0.001) and a decrease in FAAs, carboxylates and phosphorylated molecules (up to −0.5; PKS < 0.01). The results were more variable for the saturated lipids, where both temperature and expulsion effects showed significant relative increases (0.2–0.6) in most instances (see Figure 5; PKS < 0.05). The heat-treated expelled cells (middle column) showed the additive effects of temperature and expulsion, with their fingerprint reflecting an intermediary response in most biomolecular changes (Figure 5). The one notable exception was the significant increase in saturated lipids and fatty acids (0.6; PKS < 0.001) that showed a cumulative effect with temperature and expulsion (Figure 5).
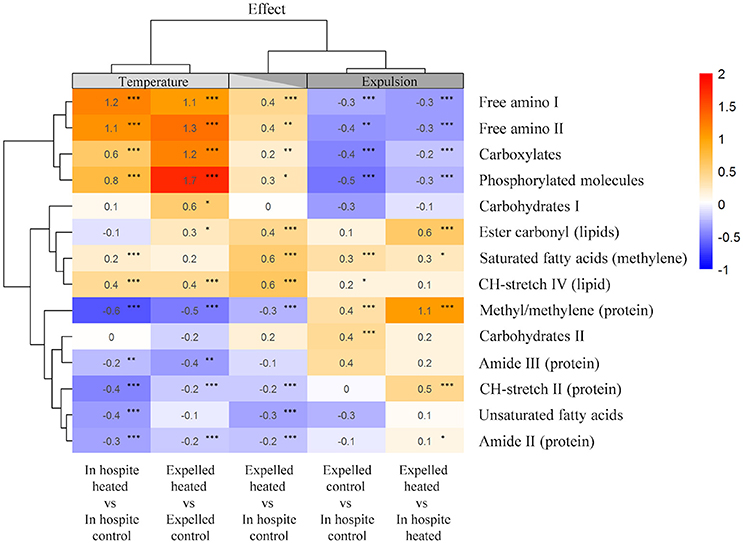
Figure 5. Heat map of the mean relative changes in peak area of key biomolecules for in hospite and expelled coral symbionts compared with their respective controls. Columns are organized based on statistical clustering of temperature (LHS) and expulsion (RHS) effects, rows are grouped according to similarity of relative change in biomolecule. Numbers show the mean relative change, asterisks denote significant differences in populations from each treatment group (Kolmogorov Smirnoff test) *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
During 2015–2016, record ocean temperatures triggered mass coral bleaching, resulting in post-bleaching mortality and a decline in the integrity of reefs across the globe (Hughes et al., 2017). The imminent risk of substantial loss of coral reef ecosystems from the increasing frequency and severity of bleaching events highlights the importance of understanding the symbiotic interactions of the two coral partners. This intimate partnership supports the metabolism and growth of healthy corals, where the maintenance of a mutually beneficial transfer of nutrients and biomolecules is essential for sustaining homeostasis and resilience. In the last decade, “omics” studies have proven useful to uncover key pathways and molecules of importance (Lehnert et al., 2014; Shinzato et al., 2014; Pinzón et al., 2015; Rosic et al., 2015; Oakley et al., 2016; Hillyer et al., 2017a,b), but given the bias inherent with averaging across the multiple cell types that make up the coral holobiont, constraining responses to that of the symbiotic system requires single-cell analyses.
In this study, we saw strong shifts in the photophysiology and morphology of coral symbionts at the individual and population level. Single-cell fluorescence clearly separated the photosynthetic performance of the different cell types with temperature stress and expulsion. Cells expelled by Acropora millepora under ambient conditions showed no discrimination toward less physiologically fit symbionts, instead they exhibited a broad variety of photo-physiological states ranging from values matching in hospite cells to cells exhibiting minimal variable fluorescence. This non-biased expulsion would not have been detected using bulk analyses; rather a mean value (~0.45) would have been reported, implying that only compromised cells were expelled. However, the spread of fluorescence viability we observed tells a different story and supports the hypothesis that non-stress related expulsion is driven simply by lack of space in the host tissue (Jones and Yellowlees, 1997). Interestingly, this pattern was reversed in heat stressed corals, revealing an expulsion bias toward low performing cells, possibly driven by a need to maintain a minimum level of productivity, whilst removing the most compromised symbionts.
Light microscopy revealed a high proportion of in hospite cells exhibiting signs of vacuolation, as well as deformation and retraction of chloroplasts with heat treatment, corroborating a previous study in which degenerated endosymbionts made up ~30–40% of the population under heat stress in Stylophora pistillata (Franklin et al., 2004). In the case of expelled cells, earlier work found 40% of the population severely degenerated with high levels of chlorosis under heat stress, from which the authors concluded that all cells were photosynthetically dysfunctional and dead (Franklin et al., 2004). In the present study, however, expelled cells remained photosynthetically viable under heat stress, despite signs of cellular degradation (chloroplast break-up and lipid globule formation; Salih et al., 1998) in up to 93% of the population. Indeed, photosynthetic efficiency of the expelled cells grouped tightly around a mean of 0.3, a measure still indicative of viability, albeit not functionally optimal, supporting previous findings (Hill and Ralph, 2007). Also, cells showed no signs of chlorosis, but rather a clumping or aggregation of chloroplasts, possibly as a form of photoprotection through self-shading (Wada, 2013), which may account for the retention of photosynthetic activity post-expulsion.
Our single-cell metabolomics approach revealed significant differentiation between the biomolecular profiles of coral algal symbionts in response to heat stress compared with expulsion. In isolating the effects of both temperature and expulsion, the data revealed that these effectors resulted in opposing metabolic adjustments by the cells. Interestingly, the biomolecular profile of thermally stressed expelled cells (i.e., those collected from actively bleaching corals) relative to in hospite control cells (middle column Figure 5) clustered as an intermediate between the temperature and expulsion effects, suggesting an additive effect of these opposing metabolic adjustment to expulsion and temperature.
Elevated temperature resulted in parallel shifts in biomolecular profiles of the in hospite and expelled cells, suggesting that the relative increases in free amino acids (FAAs), carboxylates, phosphorylated molecules and saturated lipids, with a concomitant loss of cellular proteins, is a conserved response to heat stress in Symbiodinium. In Symbiodinium, elevated temperature in conjunction with light commonly results in photoinhibition, which has been shown to be associated with a loss in the photosystem II reaction center protein D1, antenna pigment proteins, and the suppression of protein de novo synthesis (Takahashi et al., 2008). Support for such a shift can be found in this study via the concomitant thermal-induced reduction in cellular proteins, decline in photosynthetic efficiency and changes in cell morphology of the Symbiodinium, where these changes may be attributed, at least in part, to degradation of both the reaction center and key antenna proteins, known to have the majority of the light-harvesting pigments associated with them (Takahashi et al., 2008). Furthermore, the observed accumulation of FAAs likely resulted from protein degradation or from a decline in the de novo synthesis of proteins, such as the D1 or antenna proteins.
The increase in phosphorylated molecules in the heat-treated cells in hospite strongly supports the notion that these symbionts experienced significant physiological stress under the experimental conditions used. Phosphorylation is used by cells to influence transcription factors that can elicit changes in gene expression (Benschop et al., 2007), and as such play a crucial role in adjusting physiological parameters under changing environmental conditions (Evrard et al., 2013). Recent research has suggested phosphorylation events are conserved across kingdoms and important in regulating stress responses through their involvement in DNA repair, protein refolding or degradation and heat shock protein regulation (Evrard et al., 2013), all of which are common pathophysiological responses to heat stress.
In corals, lipid synthesis is primarily performed by Symbiodinium, whereby acetate derived from the host is used to synthesize lipids that are transferred back to the host in the form of triglyceride fatty acids (Wang and Douglas, 1997; Biel et al., 2007). This acetate-lipid recycling is considered a key process in host-symbiont-host energy exchange (Patton et al., 1977; Wang and Douglas, 1997; Biel et al., 2007). Consequently, the relative increase in carboxylates, lipids and fatty acids with elevated temperature measured in this study, would suggest an alteration in symbiont lipid metabolism and translocation, a finding supported by recent studies using transcriptomics and pathway activity profiling, which revealed a reduction in the turnover of glycerol, lipid and fatty acid metabolism in heat stressed algal symbionts (Lehnert et al., 2014; Hillyer et al., 2017b). In this study, the accumulation of carboxylates (such as acetate) could reflect a decline in the de novo biosynthesis of lipids by the symbiont, due to limitations in photosynthetically-derived ATP (reduced photosynthetic activity). In support of this, work on the anemone Aiptasia showed an accumulation of lipogenesis precursors in its symbionts during heat-stress, which was suggested to be the result of on-going ATP limitation (Hillyer et al., 2017a). Similarly, the strong accumulation of lipids measured; corroborate recent work that found a 5–10-fold increase in the cellular fatty acid content in heat-stressed coral symbionts (Hillyer et al., 2017b). Taken together, the strong accumulation of carboxylates, lipids and fatty acids, could be explained by reduced cellular energy causing a decline in symbiont biosynthesis and translocation of lipids back to the host.
The analogous changes in the biomolecular profiles of expelled cells, independent of temperature stress (i.e., the expulsion effect), revealed a common metabolic shift in the expelled symbionts and strongly suggest that, while the cause of the deterioration of the symbiotic relationship may differ, the expulsion results in the same short-term metabolic readjustments in symbiont metabolism. Unfortunately, because of the inability to accurately predict which cells will be expelled, we are unable to differentiate whether these metabolic shifts occurred prior to or post-expulsion, thereby limiting our possibility for discussion on cause and effect. The shift toward relatively fewer phosphorylated molecules and FAAs suggests that protein phosphorylation and possibly degradation may cease with expulsion and repair mechanisms become activated. The expulsion effect also resulted in a relative decline in carboxylates, which supports a loss of acetate delivery from the host and/or an increased consumption via the glyoxylate cycle, which enables the cell to create complex carbohydrates and energy from acetate. The measured relative increase in proteins with expulsion could be due to alleviation of host-mediated nitrogen limitation, which has been proposed to be important for the maintenance of coral symbiosis (Muscatine et al., 1989; Falkowski et al., 1993). However, a more likely explanation for this increase is the utilization of the accumulated FAAs in de novo synthesis of proteins such as D1 and pigments (Takahashi et al., 2008), which would explain the observed decline in FAAs and phosphorylated molecules, as well as the sustained photosynthetic capacity. These metabolic changes seem compatible with the idea of the “selfish symbiont,” whereby the symbiont favors its own fitness over that of its host. Whether these changes happen within the endosymbiosis thus causing its expulsion, or they occur post-expulsion in order to adjust to the new environment, is worthy of further investigation.
The biomolecular profile of the cells expelled from actively bleaching corals (i.e., temperature + expulsion effect), were intermediate in their metabolic changes, except for the strong accumulation of saturated fatty acids and lipids. These were the only compounds to increase cumulatively with both effectors, supporting a potentially important role for such molecules in the bleaching response. Given that lipids are the principal energy stores in cnidarians and considered key photosynthetically derived products transferred from the symbiont to the host (Latyshev et al., 1991), it is not surprising that thermal stress resulted in dramatic changes to the production, storage and availability of these compounds for symbiosis. It has long been shown that during stable endosymbiosis, cell-cell transport is tightly regulated and the translocation of metabolites to the host results in minimal lipid accumulation in the symbiont (Trench, 1971; Wang and Douglas, 1997; Biel et al., 2007). In contrast, destabilization of the symbiosis, such as that which occurs during coral bleaching, blocks or reduces the flow of traffic and results in the accumulation of lipids in the symbiont (Luo et al., 2009). As such, the additive effect of temperature and expulsion on lipid accumulation seen here strongly supports previous work that has implicated the production and transport of saturated lipids as key processes in the coral symbiosis. To better understand these changes, future work should include detailed metabolomics on expelled Symbiodinium cells.
The growing risk to coral reefs from the increasing frequency and severity of mass bleaching events impresses the need to understand the intricate dynamics of the coral metabolism and how they are affected by environmental perturbation. However, the difficulty in separating the symbiotic partners has stemmed our progress toward understanding the mechanisms leading to breakdown in the symbiosis. Therefore, to ensure complete separation of cell types and effectors we took a single-cell metabolomics approach to assess a broad range of metabolic components. Our results revealed a remarkable distinction in the changes to the symbiont metabolic profiles that resulted from heat stress and expulsion, with the two effectors eliciting opposing metabolic adjustments. Elevated temperature resulted in biomolecular changes reflecting cellular stress, including protein modification and degradation, whereas expulsion was found to lessen the overall effect of heat stress on the metabolic signature of coral algal symbionts. Also, in line with previous studies, we were able to confirm major shifts in fatty acid and lipid metabolism in symbionts under thermal stress. The clear separation of effectors in these findings emphasizes the importance of single cell analyses to clarify and delineate the variability in the physiology of coral cellular populations. While this methodology is unable to identify discrete metabolites, it allowed for fast, non-destructive, and quantitative assessment of the cellular composition of individual cells, delivering original insight into the biomolecular dynamics of the coral symbiosis.
Author Contributions
Conceptualization, Methodology, Investigation, Visualization: KP, and DN; Formal Analysis: KP and PH; Writing—Original draft: KP; Writing—Review and Editing: KP, DN, and PH; Funding Acquisition, Resources: KP.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AAS and handling Editor declared their shared affiliation.
Acknowledgments
We would like to thank Dr. Sackett and Australian Synchrotron Principal Beamline Scientist Dr. Tobin for technical support in synchrotron IR microspectroscopy data acquisition. Part of this work was funded by the Australian Synchrotron through merit-based beamtime awarded on the Infrared Microscopy (IRM) beamline (AS151_IRM_9051). Thanks also to the staff at Heron Island Research Station. Corals were collected under GBRMPA permit G14/36977.1 granted to KP and DN.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00110/full#supplementary-material
References
Arrigo, K. R. (2005). Marine microorganisms and global nutrient cycles. Nature 437, 349–355. doi: 10.1038/nature04159
Bambery, K. R., Wood, B. R., and McNaughton, D. (2012). Resonant Mie scattering (RMieS) correction applied to FTIR images of biological tissue samples. Analyst 137, 126–132. doi: 10.1039/C1AN15628D
Benschop, J. J., Mohammed, S., O'Flaherty, M., Heck, A. J., Slijper, M., and Menke, F. L. (2007). Quantitative phosphoproteomics of early elicitor signalling in Arabidopsis. Mol. Cell Prot. 6, 1198–1214. doi: 10.1074/mcp.M600429-MCP200
Biel, K. Y., Gates, R. D., and Muscatine, L. (2007). Effects of free amino acids on the photosynthetic carbon metabolism of symbiotic dinoflagellates. Russ. J. Plant Phys. 54, 365–371. doi: 10.1134/S1021443707020033
Burriesci, M. S., Raab, T. K., and Pringle, J. R. (2012). Evidence that glucose is the major transferred metabolite in dinoflagellate–cnidarian symbiosis. J. Exp. Biol. 215, 3467–3477. doi: 10.1242/jeb.070946
De'ath, G., Lough, J. M., and Fabricius, K. E. (2009). Declining coral calcification on the great barrier reef. Science 323, 116–119. doi: 10.1126/science.1165283
Douglas, A. E. (2003). Coral bleaching—how and why? Mar. Pollut. Bull. 46, 385–392. doi: 10.1016/S0025-326X(03)00037-7
Dunn, S. R., Thomas, M. C., Nette, G. W., and Dove, S. G. (2012). A lipidomic approach to understanding free fatty acid lipogenesis derived from dissolved inorganic carbon within cnidarian-dinoflagellate symbiosis. PLoS ONE 7:e46801. doi: 10.1371/journal.pone.0046801
Evrard, A., Kumar, M., Lecourieux, D., Lucks, J., von Koskull-Döring, P., and Hirt, H. (2013). Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. Peer J. 1:e59. doi: 10.7717/peerj.59
Falkowski, P. G., Dubinsky, Z., Muscatine, L., and McCloskey, L. (1993). Population control in symbiotic corals. Bioscience 43:606. doi: 10.2307/1312147
Franklin, D. J., Hoegh-Guldberg, O., Jones, R. J., and Berges, J. A. (2004). Cell death and degeneration in the symbiotic dinoflagellates of the coral Stylophora pistillata during bleaching. Mar. Ecol. Prog. Ser. 272, 117–130. doi: 10.3354/meps272117
Gardner, S. G., Raina, J. B., Nitschke, M. R., Nielsen, D. A., Stat, M., Motti, C. A., et al. (2017). A multi-trait systems approach reveals a response cascade to bleaching in corals. BMC Biol. 15:117. doi: 10.1186/s12915-017-0459-2
Giordano, M., Kansiz, M., Heraud, P., Beardall, J., Wood, B., and McNaughton, D. (2001). Fourier transform infrared spectroscopy as a novel tool to investigate changes in intracellular macromolecular pools in the marine microalgal Chaetoceros muellerii (Bacillariophyceae) J. Phycol. 37, 271–279. doi: 10.1046/j.1529-8817.2001.037002271.x
Glynn, P. W. (1991). Coral-reef bleaching in the 1980s and possible connections with global warming. Trends Ecol. Evol. 6, 175–179. doi: 10.1016/0169-5347(91)90208-F
Heraud, P., Wood, B. R., Tobin, M. J., Beardall, J., and McNaughton, D. (2005). Mapping of nutrient-induced biochemical changes in living algal cells using synchrotron infrared microspectroscopy. FEMS Microbiol. Lett. 249, 219–225. doi: 10.1016/j.femsle.2005.06.021
Hill, R., and Ralph, P. J. (2007). Post-bleaching viability of expelled zooxanthellae from the scleractinian coral Pocillopora damicornis. Mar. Ecol. Prog. Ser. 352, 137–144. doi: 10.3354/meps07159
Hillyer, K. E., Dias, D. A., Lutz, A., Roessner, U., and Davy, S. K. (2017a). Mapping carbon fate during bleaching in a model cnidarian symbiosis: the application of 13C metabolomics. New Phytol. 214, 1551–1562. doi: 10.1111/nph.14515
Hillyer, K. E., Dias, D. A., Lutz, A., Wilkinson, S. P., Roessner, U., and Davy, S. K. (2017b). Metabolite profiling of symbiont and host during thermal stress and bleaching in the coral Acropora aspera. Coral Reefs 36, 105–118. doi: 10.1007/s00338-016-1508-y
Hoegh-Guldberg, O., and Bruno, J. F. (2010). The Impact of Climate Change on the World's Marine Ecosystems. Science 328, 1523–1528. doi: 10.1126/science.1189930
Hughes, T. P., Anderson, K. D., Connolly, S. R., Heron, S. F., Kerry, J. T., Lough, J. M., et al. (2018). Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83. doi: 10.1126/science.aan8048
Hughes, T. P., Kerry, J. T., Álvarez-Noriega, M., Álvarez-Romero, J. G., Anderson, K. D., Baird, A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
Jones, R. J., and Yellowlees, D. (1997). Regulation and control of intracellular algae (= zooxanthellae) in hard corals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 352, 457–468.
Kopp, C., Domart-Coulon, I., Escrig, S., Humbel, B. M., Hignette, M., and Meibom, A. (2015). Subcellular investigation of photosynthesis-driven carbon assimilation in the symbiotic reef coral Pocillopora damicornis. MBio 6, e02299–e02214. doi: 10.1128/mBio.02299-14
Kopp, C., Pernice, M., Domart-Coulon, I., Djediat, C., Spangenberg, J. E., Alexander, D. T. L., et al. (2013). Highly dynamic cellular-level response of symbiotic coral to a sudden increase in environmental nitrogen. mBio 4:e00052-13. doi: 10.1128/mBio.00052-13
Ladrière, O., Compere, P., Decloux, N., Vandewalle, P., and Poulicek, M. (2008). Morphological alterations of zooxanthellae in bleached cnidarian hosts. Cahairs de Biol. Mar. 49, 215–227. Available online at: http://hdl.handle.net/2268/6020
Latyshev, N. A., Naumenko, N. V., Svetashev, V. I., and Laypov, Y. Y. (1991). Fatty acids of reef-building corals. Mar. Ecol. Prog. Ser. 76, 295–301. doi: 10.3354/meps076295
Lehnert, E. M., Mouchka, M. E., Burriesci, M. S., Gallo, N. D., Schwarz, J. A., and Pringle, J. R. (2014). Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. Genes Geneomes Genet. 4, 277–295. doi: 10.1534/g3.113.009084
Luo, Y. J., Want, L. H., Chen, W. N. U., Peng, S. E., Tzen, J. T. C., Hsiao, Y. Y., et al. (2009). Ratiometric imaging of gastrodermal lipid bodies in coral-dinoflagellate endosymbiosis. Coral Reefs 28, 289–301. doi: 10.1007/s00338-008-0462-8
Moore, C. M., Mills, M. M., Arrigo, K. R., Berman-Frank, I., Bopp, L., Boyd, P. W., et al. (2013). Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710. doi: 10.1038/ngeo1765
Muscatine, L. (1990). The role of symbiotic algae in carbon and energy flux in reef corals. Ecosyst. World 25, 75–87.
Muscatine, L., Falkowski, P. G., Dubinsky, Z., Cook, P. A., and McCloskey, L. R. (1989). The effect of external nutrient resources on the population dynamics of zooxanthellae in a reef coral. Proc. R. Soc. Lond. B Biol. Sci. 236, 311–324. doi: 10.1098/rspb.1989.0025
Oakley, C. A., Ameismeier, M. F., Peng, L., Weis, V. M., Grossman, A. R., and Davy, S. K. (2016). Symbiosis induces widespread changes in the proteome of the model cnidarian Aiptasia. Cell. Microbiol. 18, 1009–1023. doi: 10.1111/cmi.12564
Patton, J. S., Abraham, S., and Benson, A. A. (1977). Lipogenesis in the intact coral Pocillopora capitate and its isolated zooxanthellae: evidence for a light-driven carbon cycle between symbiont and host. Mar. Biol. 44, 235–247. doi: 10.1007/BF00387705
Peng, S.-E., Chen, W.-N. U., Chen, H.-K., Lu, C.-Y., Mayfield, A. B., Fang, L.-S., et al. (2011). Lipid bodies in coral–dinoflagellate endosymbiosis: Proteomic and ultrastructural studies. Proteomics 11, 3540–3555. doi: 10.1002/pmic.201000552
Pernice, M., Meibom, A., Van Den Heuvel, A., Kopp, C., Domart-Coulon, I., Hoegh-Guldberg, O., et al. (2012). A single-cell view of ammonium assimilation in coral-dinoflagellate symbiosis. ISME J. 6, 1314–1324. doi: 10.1038/ismej.2011.196
Pinzón, J. H., Kamel, B., Burge, C. A., Harvell, C. D., Medina, M., Weil, E., et al. (2015). Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. R. Soc. Open Sci. 2:140214. doi: 10.1098/rsos.140214
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing Available online at: http://www.R-project.org/
Ritchie, R. (2006). Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosyn. Res. 89, 27–41. doi: 10.1007/s11120-006-9065-9
Rosic, N., Ling, E. Y. S., Chan, C. K. K., Lee, H. C., Kaniewska, P., Edwards, D., et al. (2015). Unfolding the secrets of coral-algal symbiosis. ISME J. 9, 844–856. doi: 10.1038/ismej.2014.182
Sackett, O., Armand, L., Beardall, J., Hill, R., Doblin, M., Connelly, C., et al. (2014). Taxon-specific responses of Southern Ocean diatoms to Fe enrichment revealed by synchrotron radiation FTIR microspectroscopy. Biogeoscience 11, 5795–5808. doi: 10.5194/bg-11-5795-2014
Salih, A., Hoegh-Guldberg, O., and Cox, G. (1998). “Photoprotection of symbiotic dinoflagellates by fluorescent pigments in reef corals,” in Proceedings of the Australian Coral Reef Society 75th Anniversary Conference (Brisbane: School of Marine Science, University of Queensland, Brisbane, Australia), 217–230.
Shinzato, C., Inoue, M., and Kusakabe, M. (2014). Wide variety of genes from both the host and symbiotic zooxanthellae. PLoS ONE 9:e85182. doi: 10.1371/journal.pone.0085182
Sogin, E. M., Putnam, H. M., Anderson, P. E., and Gates, R. D. (2016). Metabolomic signatures of increases in temperature and ocean acidification from the reef-building coral, Pocillopora damicornis. Metabolomics 12, 1–12 doi: 10.1007/s11306-016-0987-8
Stimson, J., and Kinzie, R. L. (1991). The temporal pattern and rate of release of Zooxanthellae from the Reef Coral Pocillopora-damicornis (Linnaeus) under nitrogen-enrichment and control conditions. J. Exp. Bio. Aquat. Bot. 153, 63–74. doi: 10.1016/S0022-0981(05)80006-1
Takahashi, S., Whitney, S., Itoh, S., Maruyama, T., and Badger, M. (2008). Heat stress causes inhibition of the de novo synthesis of antenna proteins and photobleaching in cultured Symbiodinium. Proc. Nalt. Acad. Sci. U.S.A. 105, 4203–4208. doi: 10.1073/pnas.0708554105
Tobin, M. J., Puskar, L., Barber, R. L., Harvey, E. C., Heraud, P., Wood, B. R., et al. (2010). FTIR spectroscopy of single live cells in aqueous media by synchrotron IR microscopy using microfabricated sample holders. Vibrat. Spectrosc. 53, 34–38. doi: 10.1016/j.vibspec.2010.02.005
Trench, R. K. (1971). The physiology and biochemistry of zooxanthellae symbiotic with marine coelenterates. III. The effect of homogenates of host tissues on the excretion of photosynthetic products in vitro by zooxanthellae from two marine coelenterates. Proc. R. Soc. B 177, 251–264. doi: 10.1098/rspb.1971.0026
Vongsvivut, J., Heraud, P., Zhang, W., Kralovec, J. A., McNaughton, D., and Barrow, C. J. (2012). Quantitative determination of fatty acid compositions in micro-encapsulated fish-oil supplements using Fourier Transform Infrared (FTIR) spectroscopy. Food Chem. 135, 603–609. doi: 10.1016/j.foodchem.2012.05.012
Wagner, H., Liu, Z., Langner, U., Stehfest, K., and Wilhelm, C. (2010). The use of FTIR spectroscopy to assess quantitative changes in the biochemical composition of microalgae. J. Biophotonics 3, 557–566. doi: 10.1002/jbio.201000019
Wang, J. T., and Douglas, A. E. (1997). Nutrients, signals and photosynthates release by symbiotic algae: the impact of taurine on the dinoflagellate alga Symbiodinium from the sea anemone Aiptasia pulchella. Plant Physiol. 114, 631–636. doi: 10.1104/pp.114.2.631
Warnes, G. R., Bolker, B., Bonebakker, L., Gentleman, R., Liaw, W. H. A., Lumley, T., et al. (2016). gplots: Various R Programming Tools for Plotting Data. R package version 3.0.1.
Weis, V. M. (2008). Cellular mechanisms of cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Bio. 211, 3059–3066. doi: 10.1242/jeb.009597
Keywords: coral bleaching, Symbiodinium, biomolecules, FTIR microspectroscopy, Acropora millepora, symbiosis
Citation: Petrou K, Nielsen DA and Heraud P (2018) Single-Cell Biomolecular Analysis of Coral Algal Symbionts Reveals Opposing Metabolic Responses to Heat Stress and Expulsion. Front. Mar. Sci. 5:110. doi: 10.3389/fmars.2018.00110
Received: 21 November 2017; Accepted: 15 March 2018;
Published: 29 March 2018.
Edited by:
Iliana B. Baums, Pennsylvania State University, United StatesReviewed by:
Raquel Peixoto, Universidade Federal do Rio de Janeiro, BrazilAndrew A. Shantz, Pennsylvania State University, United States
Copyright © 2018 Petrou, Nielsen and Heraud. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katherina Petrou, katherina.petrou@uts.edu.au
†These authors have contributed equally to this work.
 Katherina Petrou
Katherina Petrou Daniel A. Nielsen1†
Daniel A. Nielsen1†