Liquid Biopsy in Non-Small Cell Lung Cancer
- 1Laboratory of Oncology, Pangaea Biotech, Quirón Dexeus University Hospital, Barcelona, Spain
- 2Instituto Oncológico Dr Rosell, Quirón Dexeus University Hospital, Barcelona, Spain
- 3Cancer Biology and Precision Medicine Program, Catalan Institute of Oncology, Germans Trias i Pujol Health Sciences Institute and Hospital, Badalona, Spain
Liquid biopsy analyses are already incorporated in the routine clinical practice in many hospitals and oncology departments worldwide, improving the selection of treatments and monitoring of lung cancer patients. Although they have not yet reached its full potential, liquid biopsy-based tests will soon be as widespread as “standard” biopsies and imaging techniques, offering invaluable diagnostic, prognostic, and predictive information. This review summarizes the techniques available for the isolation and analysis of circulating free DNA and RNA, exosomes, tumor-educated platelets, and circulating tumor cells from the blood of cancer patients, presents the methodological challenges associated with each of these materials, and discusses the clinical applications of liquid biopsy testing in lung cancer.
Introduction
The so-called “liquid biopsy” is quickly moving from research into clinical practice in lung cancer, as well as in other human malignancies. Although its full potential has not yet been reached, the “liquid biopsy” is no longer a promise but a reality that is allowing a better treatment selection and monitoring of lung cancer patients in hospitals and oncology departments worldwide. We can already foresee a day when “liquid biopsy”-based tests will be as widespread and useful as “standard” biopsies and imaging techniques, offering invaluable diagnostic, prognostic, predictive, and monitoring information. In this mini review, we will summarize the state of the art in this exciting area, placing a particular emphasis on the clinical utility of the “liquid biopsy” and the variety of applications, methodologies, and results that can be derived from it.
“Liquid biopsies” are usually defined as tests done in blood samples or other body fluids. In the case of cancer patients, the objective of those tests is to detect materials originated in the tumor. Although the term “liquid biopsy” is universally used, many pathologists argue that it is incorrect. The so-called “liquid biopsies,” they claim, are not true biopsies. A “true” biopsy is usually performed by a surgeon or a pneumologist and involves the extraction of sample cells or tissues that are subsequently examined by a pathologist under a microscope, commonly after some kind of fixation and staining. Paraffin embedding is also widespread. In contrast, “liquid biopsies” are not obtained by surgeons; involve the extraction of blood or other fluids and not of solid tissues, pathologists only occasionally intervene and fixation, embedding, or staining are equally infrequent. In addition to the “biopsy” half, the “liquid” half in the term “liquid biopsy” can also be misleading. The materials originated in the tumor that are to be detected in such “biopsies” are never liquid. Some of them are cells or fragments of cells, such as circulating tumor cells (CTCs), exosomes, or tumor-educated platelets (TEPs); others are nucleic acids dissolved in the blood, such as circulating tumor DNA or RNA (ctDNA, ctRNA). Each of these materials offers unique opportunities to test different biomarkers and analyze particular characteristics of the tumors (Table 1).
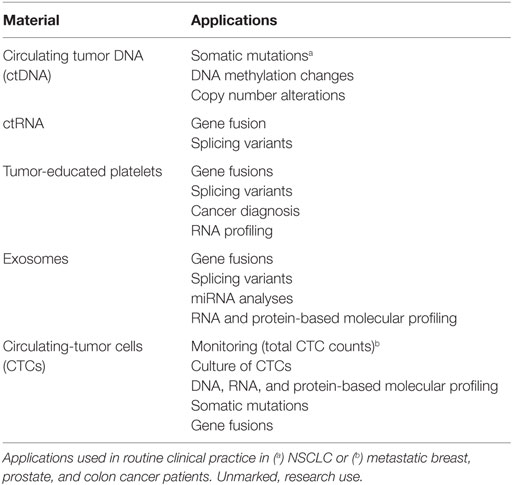
Table 1. Biological materials that can be isolated from liquid biopsies and their applications in lung cancer.
The differences between a “real” and a “liquid” biopsy—or “liquid sample,” as the pathologists would probably prefer to call them—explain the advantages of the latter. “Liquid” biopsies will never replace real biopsies, which are irreplaceable sources of information that cannot be obtained by any other means, such as tumor type and histology. However, they offer all sorts of additional data that cannot be obtained in any other way. In patients who cannot be biopsied, or where biopsies do not have enough tissue, “liquid biopsy” is the only alternative to perform genetic testing for targeted therapy. Also, in patients with advanced disease, it is not feasible to obtain biopsies of every metastasic site. But blood reaches both the primary tumor and the metastases, and materials coming from all can be found in a “liquid biopsy.” Finally, unlike “real” biopsies, blood can be repeatedly obtained without the risk of comorbidities and used to monitor the course of the disease, including early detection of response and relapse or emergence of resistance to a particular therapy.
Circulating Tumor DNA
Circulating free DNA (cfDNA) can be found dissolved in plasma and serum, at variable amounts. In the case of cancer patients, a fraction of the cfDNA is tumor derived, and ctDNA represents from less than 0.1% to more than 10% of the total cfDNA. This percentage has been shown to depend on stage, tumor burden, vascularization of the tumor, biological features like apoptotic rate and metastatic potential of the cancer cells, and factors affecting the blood volume of the patient (1, 2). In addition, variations on the relative abundance of ctDNA correlate with response to therapy (3–5). ctDNA is released by passive mechanisms, such as lysis of apoptotic and necrotic cells or digestion of tumor cells by macrophages, and also by active mechanisms. In this respect, cfDNA shows and enrichment in 150–180 bp fragments typical of the nucleosomal pattern of DNA fragmentation during apoptosis (6–9). The ctDNA carries the same somatic alterations as the tumor itself and can be used to detect clinically relevant mutations such as those in the epidermal growth factor (EGFR) or KRAS genes. This is particularly useful when no biopsy is available for genetic analyses and, in this setting, the European Medicine Agency recommends EGFR testing in liquid biopsies to select patients for tyrosine kinase inhibitor (TKI) therapy (10). However, many standard techniques for mutation detection are not useful for ctDNA analyses due to an insufficient sensitivity. Since ctDNA often represents a small percentage of the total cfDNA, somatic mutations coming from the tumor can be present at allele fractions as low as 0.01%. Highly sensitive methodologies, or variations of preexisting methodologies, have been developed in order to detect low abundance mutations in cfDNA (6, 11).
Modified real-time PCR techniques have been widely used to identify genetic alterations in the cfDNA of cancer patients. They include amplification-refractory mutation system [ARMS (12),], Scorpion-ARMS (13), and peptide nucleic acid (PNA) or locked nucleic acid (LNA) mutant-enriched PCR (14–17). The diagnostic sensitivity of these techniques, when compared to tumor tissue, ranges from 43 to more than 90%, while the specificity is usually close to 100%; and the two commercially available methods to determine EGFR mutations in the cfDNA of cancer patients (Therascreen Plasma from Qiagen and COBAS Blood from Roche Diagnostics) are based on them. In our group, we have developed a quantitative PCR technique in the presence of PNA to detect EGFR, KRAS, and BRAF mutations in the cfDNA of advanced lung, colon, and cancer patients that achieves 75–80% sensitivity with 100% specificity (18, 19). Digital PCR, droplet digital PCR, and beads, emulsion, amplification, and magnetics (BEAMing) system constitute further refinements of the PCR-based techniques and have also been used to determine mutations in cfDNA (14, 20–26) (Table 2).
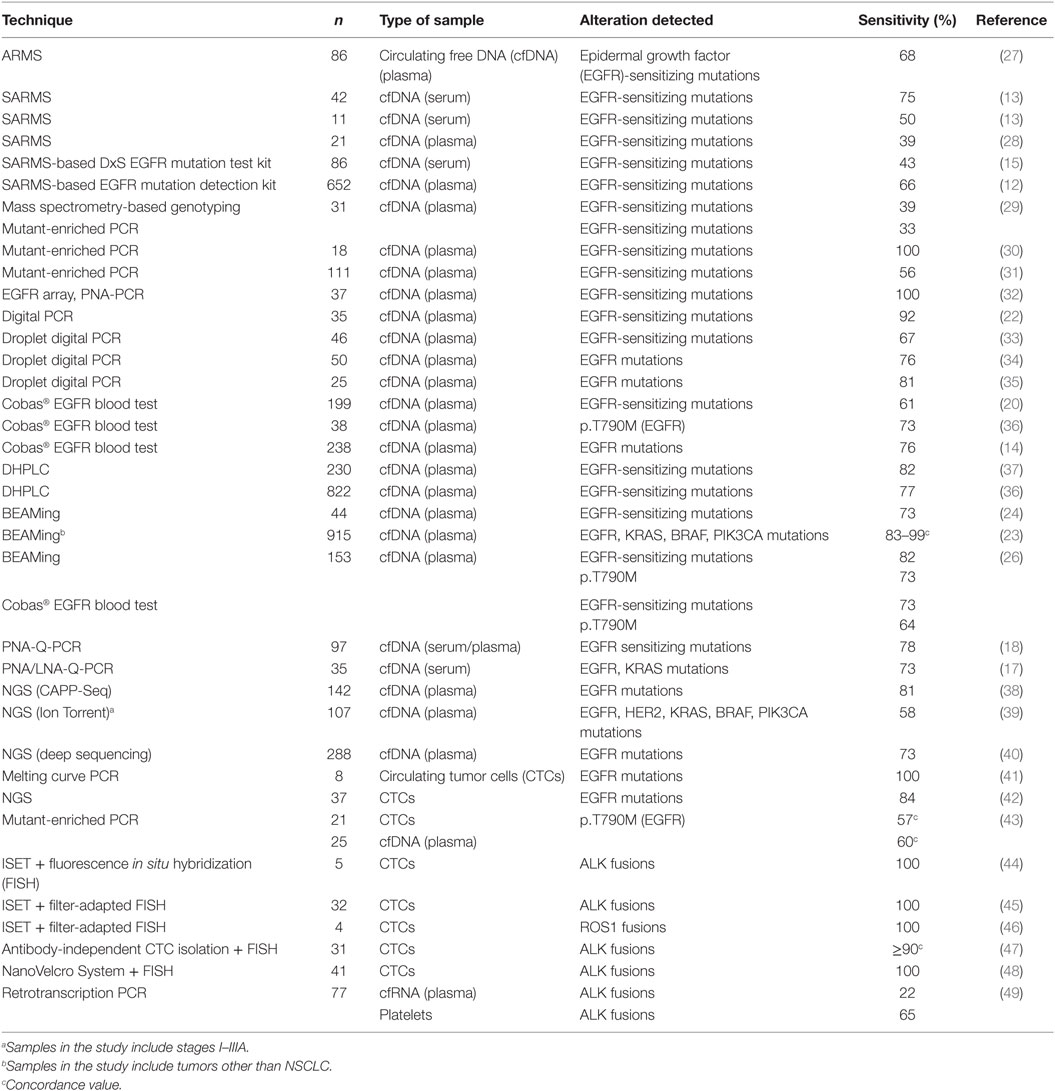
Table 2. Summary of reports on detection of genetic alterations in liquid biopsy materials from advanced NSCLC patients.
Most modified PCR techniques are easy, comparatively unexpensive, and have a quick turnaround time (19), but have the disadvantage that can only detect mutations in a limited number of loci, usually within a single gene. Next-generation sequencing methodologies can overcome these limitations but, while tissue-based NGS genotyping is already well established, the application of NGS technologies to liquid biopsies is challenging and an ultra-deep sequencing approach is commonly used in order to improve sensitivity. In this approach, the gene panels are limited so that each read is sequenced thousands of times (39, 50, 51). However, this requirement of a high sensitivity may easily lead to false-positive results and requires a careful validation of the whole testing process. Examples of NGS protocols specifically developed for ctDNA analysis include TAm-Seq (tagged-amplicon deep sequencing), which combines site-specific primers with universal tails (52, 53); Safe-SeqS (Safe-Sequencing System) (54), and CAPP-seq (capture based sequencing), which relies on hybridization-based capture of target regions followed by amplification (38, 55) (Table 2).
The detection of mutations in cfDNA by modified PCR or NGS techniques is not only useful in lung cancer patients at presentation. It has also been successfully employed for patient monitoring, including early evaluation of response and relapse, which are associated with changes in the EGFR or KRAS mutational burden in cfDNA; and for early detection of acquired resistance to EGFR TKIs, associated in many patients with the emergence of the p.T790M mutation in blood (26, 56). In this respect, p.T790M testing in cfDNA has been recently recommended in patients eligible for osimertinib treatment, in order to avoid unnecessary rebiopsies (33, 36, 56).
Circulating Tumor RNA
Similar to ctDNA, RNA derived from tumor cells (ctRNA) is present in the plasma of cancer patients and can be used for detection of the clinically relevant ALK, ROS1, and RET fusion genes and METΔ14 splicing variant. However, genetic analyses in cfRNA present specific challenges and have not been widely used. Unlike cfDNA, cfRNA degrades very quickly and needs to be purified rapidly after blood extraction. The alternative is adding a preservative such as Trizol and freezing the sample at −80°C, but this procedure is not easily accessible to many clinical sites. Despite these limitations, our group has a 5-year experience in detection of EML4-ALK fusion transcripts in plasma cfRNA by retrotranscription PCR (RT-PCR) (49) and, using improved processing and purification methods, we have demonstrated that the sensitivity of the technique can be significantly improved.
Tumor-Educated Platelets
Platelets have been recently demonstrated to sequester tumor RNA by a microvesicle dependent mechanism, and the so-called TEPs (57, 58) can be used as a source of tumor RNA for genetic analysis. Platelets can be isolated from blood by simple centrifugation steps, and its RNA content easily purified and used for the detection of gene fusions and splicing variants. Using a RT-PCR approach, our group has detected EML4-ALK fusion transcripts in TEP RNA from advanced lung cancer patients with 65% sensitivity and 100% specificity (49). In addition, we have demonstrated that the disappearance of fusion transcripts in platelets correlates with response to crizotinib treatment. Regarding splicing variants, the clinical relevance of METΔ14 in lung cancer was only described in 2015 (59–61), and there are no reports in the literature about detection of METΔ14 transcripts in liquid biopsy. However, we have recently detected the alteration in the TEP RNA of a NSCLC patient positive in tumor tissue, who attained a partial response to crizotinib (unpublished data).
Platelet RNA can also be analyzed by multiplexing techniques, and a recent report has demonstrated the diagnostic potential of this approach. Using mRNA sequencing and surrogate TEP RNA profiles of 283 samples, 228 cancer patients of six different origins were discriminated from 55 healthy individuals with 96% accuracy. Tumors with specific genetic alterations, such as KRAS or EGFR mutations, were also distinguished and the location of the primary tumor identified with 71% accuracy (58).
Exosomes
Exosomes are small vesicles present in blood and other body fluids (62–64). With a 30–100 nm diameter, they are constitutively released through exocytosis by many cells, including tumor cells, in physiological and pathological conditions. Exosomes contain lipids, proteins, mRNA, several types of non-coding RNAs, and double-stranded DNA; and their composition partly reflects that of the parental cells (65). In addition, being generated by the cell secretion pathway, all exosomes carry some common proteins independent of their origin, such as ALIX, CD63, or TSG-101 (66). Exosomes are generally isolated by sucrose gradient ultracentrifugation or immune-bead isolation techniques (such as magnetic activated cell sorting), and there are commercial kits available. Once isolated, exosomes are characterized by transmission electron microscopy, Western blot, FACS, or other methodologies (67).
Although being more difficult to purify than other materials, the lipid bilayer of exosomes makes their cargo particularly stable, theoretically allowing the identification of the tumor of origin, genetic alterations or resistances to treatments. In this respect, EML4-ALK fusion transcripts have been recently identified in the exosomal RNA of NSCLC patients (68). In addition, some studies indicate that micro RNA (miRNA) analysis of exosomes might be useful for the diagnosis of lung adenocarcinoma (69–71) and that particular miRNAs can offer prognostic information in advanced NSCLC. For example, downregulation of miRNA-373 and miRNA-512 has been associated with a poor prognosis (72), miR-208a and miR-1246 with resistance to radiotherapy (73, 74), and miR-221-3p and 222-3p with good response to osimertinib in EGFR mutated patients (75).
Circulating Tumor Cells
Together with ctDNA, CTCs are the most widely investigated material in liquid biopsies of cancer patients. First observed in 1869 (76), they are cancer cells detached from the solid tumor mass that circulate in the blood and lymphatic system (77) as single cells or as aggregates, the so-called circulating tumor microemboli (78–80). In advanced NSCLC patients, CTCs are relatively rare, 1–10 per mL against a background of 106–107 peripheral blood mononuclear cells. This low abundance poses formidable challenges for the development of robust and sensitive enrichment protocols (81).
Some CTC capture methods are label dependent, based on specific epithelial cell surface markers, such as epithelial cell adhesion molecule (EpCAM) for positive selection or CD45 for negative depletion. One of such techniques, the CellSearch® system (Veridex), has been approved by the FDA for monitoring some type of tumors (82–84), but not lung cancer. In advanced NSCLC, CellSearch® has shown a limited detection efficiency, with CTCs detectable in only 20–40% of patients (85–87). Label-dependent methods do not select CTCs that have undergone epithelial to mesenchymal transition (88) or those with stem cell characteristics that have not started epithelial differentiation. Label-independent techniques, which are based on physical characteristics such as size, can overcome this limitation. Isolation by Size of Epithelial Tumor cells (ISET®, Rarecells), based on filtration and cytological characterization, has shown an increased sensitivity in NSCLC (89–92) with an 80% detection rate of CTCs in blood from 40 stage IIIA–IV patients compared with 23% using CellSearch® (85). Another technology based on size, ScreenCell®, allows not only the detection but also the isolation of CTCs, which can be subjected to further morphological studies and used for genetic testing. Isolated CTCs can be cultured or injected into mice to generate xenografts (93–96) and CTC-derived tumor cells can thus be obtained in enough numbers for molecular and pharmacological profiling.
CTC counts have been extensively researched as a prognostic factor in NSCLC (97). In early-stage patients, the decrease or disappearance of CTCs after surgery has been reported to correlate with better clinical outcomes (98, 99), while its persistence was associated with shorter progression-free survival (PFS) (100). Regarding advanced NSCLC, some studies have reported that a higher CTC count at presentation correlates with advanced stage and shorter PFS and overall survival (85, 101). Also, the decrease or disappearance of CTCs after chemotherapy or targeted therapy has been consistently associated with better outcomes (102–104).
Finally, CTCs have also been investigated as a tool to identify clinically relevant genetic alterations in NSCLC (Table 2). Using NGS and modified PCR techniques, EGFR-sensitizing mutations and the p.T790M resistance mutation have been detected in the CTCs of EGFR-positive patients at presentation and after progression to TKI treatments, respectively (28, 41, 42, 105). The sensitivities reported range from 47 to 100%. However, unlike cfDNA, CTCs are not used for EGFR testing in the routine clinical practice. EML4-ALK fusions have been identified by fluorescence in situ hybridization (FISH) and immunochemistry in CTCs isolated using ISET (44) or other enrichment methodologies (47, 48). In some cases, filter-adapted FISH was employed, a methodology that has also been demonstrated to successfully identify ROS1 rearrangements in CTCs isolated by ISET (46).
Author Contributions
All the authors contributed to the writing and critical revision of the review.
Conflict of Interest Statement
The authors declare that this review was elaborated in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Kidess E, Jeffrey SS. Circulating tumor cells versus tumor-derived cell-free DNA: rivals or partners in cancer care in the era of single-cell analysis? Genome Med (2013) 5:70. doi: 10.1186/gm474
2. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med (2008) 14:985–90. doi:10.1038/nm.1789
3. Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov (2014) 4:650–61. doi:10.1158/2159-8290.CD-13-1014
4. Sozzi G, Conte D, Mariani L, Lo Vullo S, Roz L, Lombardo C, et al. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res (2001) 61:4675–8.
5. Sirera R, Bremnes RM, Cabrera A, Jantus-Lewintre E, Sanmartín E, Blasco A, et al. Circulating DNA is a useful prognostic factor in patients with advanced non-small cell lung cancer. J Thorac Oncol (2011) 6:286–90. doi:10.1097/JTO.0b013e31820189a5
6. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol (2013) 10:472–84. doi:10.1038/nrclinonc.2013.110
7. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res (2001) 61:1659–65.
8. Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A (2005) 102:16368–73. doi:10.1073/pnas.0507904102
9. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem (2015) 61:112–23. doi:10.1373/clinchem.2014.222679
10. European Medicines Agency. Summary of Product Characteristics for Iressa, Annex 1. London: European Medicines Agency (2016). 3 p.
11. Volik S, Alcaide M, Morin RD, Collins C. Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res (2016) 14(10):898–908. doi:10.1158/1541-7786.MCR-16-0044
12. Douillard JY, Ostoros G, Cobo M, Ciuleanu T, Cole R, McWalter G, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol (2014) 9:1345–53. doi:10.1097/JTO.0000000000000263
13. Kimura H, Kasahara K, Kawaishi M, Kunitoh H, Tamura T, Holloway B, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res (2006) 12(13):3915–21. doi:10.1158/1078-0432.CCR-05-2324
14. Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res (2015) 21(14):3196–203. doi:10.1158/1078-0432.CCR-14-2594
15. Goto K, Ichinose Y, Ohe Y, Yamamoto N, Negoro S, Nishio K, et al. Epidermal growth factor receptor mutations status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatine-paclitaxel in non-small cell lung cancer. J Thorac Oncol (2012) 7(1):115–21. doi:10.1097/JTO.0b013e3182307f98
16. Rosell R, Molina MA, Serrano MJ. EGFR mutations in circulating tumour DNA. Lancet Oncol (2012) 13:971–3. doi:10.1016/S1470-2045(12)70369-8
17. Kim HR, Lee SY, Hyun DS, Lee MK, Lee HK, Choi CM, et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J Exp Clin Cancer Res (2013) 32:1–8. doi:10.1186/1756-9966-32-50
18. Karachaliou N, Mayo-de las Casas C, Queralt C, de Aguirre I, Melloni B, Cardenal F, et al. Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC trial. JAMA Oncol (2015) 1(2):149–57. doi:10.1001/jamaoncol.2014.257
19. Gonzalez-Cao M, Mayo-de-Las-Casas C, Molina-Vila MA, De Mattos-Arruda L, Muñoz-Couselo E, Manzano JL, et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res (2015) 25(6):486–95. doi:10.1097/CMR.0000000000000187
20. Weber B, Meldgaard P, Hager H, Wu L, Wei W, Tsai J, et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer (2014) 14:294. doi:10.1186/1471-2407-14-294
21. Wang Z, Chen R, Wang S, Zhong J, Wu M, Zhao J, et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS One (2014) 9(11):e110780. doi:10.1371/journal.pone.0110780
22. Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res (2009) 15:2076–84. doi:10.1158/1078-0432.CCR-08-2622
23. Janku F, Angenendt P, Tsimberidou AM, Fu S, Naing A, Falchook GS, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget (2015) 6(14):12809–21. doi:10.18632/oncotarget.5663
24. Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res (2011) 17(24):7808–15. doi:10.1158/1078-0432.CCR-11-1712
25. Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer (2015) 90(3):509–15. doi:10.1016/j.lungcan.2015.10.004
26. Karlovich C, Goldman JW, Sun JM, Mann E, Sequist LV, Konopa K, et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO-1686). Clin Cancer Res (2016) 22(10):2386–95. doi:10.1158/1078-0432.CCR-15-1260
27. Liu X, Lu Y, Zhu G, Lei Y, Zheng L, Qin H, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol (2013) 66(12):1065–9. doi:10.1136/jclinpath-2013-201728
28. Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med (2008) 359(4):366–77. doi:10.1056/NEJMoa0800668
29. Brevet M, Johnson ML, Azzoli CG, Ladanyi M. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer (2011) 73(1):96–102. doi:10.1016/j.lungcan.2010.10.014
30. He C, Liu M, Zhou C, Zhang J, Ouyang M, Zhong N, et al. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer (2009) 125(10):2393–9. doi:10.1002/ijc.24653
31. Zhao X, Han RB, Zhao J, Wang J, Yang F, Zhong W, et al. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration (2013) 85:119–25. doi:10.1159/000338790
32. Yam I, Lam DC, Chan K, Chung-Man Ho J, Ip M, Lam WK, et al. EGFR array: uses in the detection of plasma EGFR mutations in non-small cell lung cancer patients. J Thorac Oncol (2012) 7(7):1131–40. doi:10.1097/JTO.0b013e3182558198
33. Oxnard GR, Thress KS, Alden RS, Lawrance R, Paweletz CP, Cantarini M, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol (2016) 34(28):3375–82. doi:10.1200/JCO.2016.66.7162
34. Wei Z, Shah N, Deng C, Xiao X, Zhong T, Li X. Circulating DNA addresses cancer monitoring in non small cell lung cancer patients for detection and capturing the dynamic changes of the disease. Springerplus (2016) 5:531. doi:10.1186/s40064-016-2141-5
35. Zheng D, Ye X, Zhang MZ, Sun Y, Wang JY, Ni J, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep (2016) 6:1–9. doi:10.1038/srep20913
36. Huang Z, Wang Z, Bai H, Wu M, Ann T, Zao J, et al. The detection of EGFR mutation status in plasma is reproducible and can dynamically predict the efficacy of EGFR-TKI. Thorac Cancer (2012) 3:334–40. doi:10.1111/j.1759-7714.2012.00133.x
37. Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol (2009) 27(16):2653–9. doi:10.1200/JCO.2008.17.3930
38. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol (2016) 34(5):547–55. doi:10.1038/nbt.3520
39. Couraud S, Vaca-Paniagua F, Villar S, Oliver J, Schuster T, Blanché H, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res (2014) 20:4613–24. doi:10.1158/1078-0432.CCR-13-3063
40. Uchida J, Kato K, Kukita Y, Kumagai T, Nishino K, Daga H, et al. Diagnostic accuracy of noninvasive genotyping of EGFR in lung cancer patients by deep sequencing of plasma cell-free DNA. Clin Chem (2015) 61(9):1191–6. doi:10.1373/clinchem.2015.241414
41. Breitenbuecher F, Hoffarth S, Worm K, Cortes-Incio D, Gauler TC, Köhler J, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS One (2014) 9(1):e85350. doi:10.1371/journal.pone.0085350
42. Marchetti A, Del Grammastro M, Felicioni L, Malatesta S, Filice G, Centi I, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One (2014) 9(8):e103883. doi:10.1371/journal.pone.0103883
43. Sundaresan TK, Sequist LV, Heymach JV, Riely GJ, Jänne PA, Koch WH, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res (2016) 22(5):1103–10. doi:10.1158/1078-0432.CCR-15-1031
44. Ilie M, Long E, Butori C, Hofman V, Coelle C, Mauro V, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol (2012) 23(11):2907–13. doi:10.1093/annonc/mds137
45. Pailler E, Adam J, Barthélémy A, Oulhen M, Auger N, Valent A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol (2013) 31(18):2273–81. doi:10.1200/JCO.2012.44.5932
46. Pailler E, Auger N, Lindsay CR, Vielh P, Islas-Morris-Hernandez A, Borget I, et al. High level of chromosomal instability in circulating tumor cells of ROS-1 rearranged non-small-cell lung cancer. Ann Oncol (2015) 26(7):1408–15. doi:10.1093/annonc/mdv165
47. Tan CL, Lim TH, Lim TK, Tan DS, Chua YW, Ang MK, et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget (2016) 7(17):23251–62. doi:10.18632/oncotarget.8136
48. He W, Xu D, Wang Z, Xiang X, Tang B, Li S, et al. Detecting ALK-rearrangement of CTC enriched by nanovelcro chip in advanced NSCLC patients. Oncotarget (2016). doi:10.18632/oncotarget.8305
49. Nilsson RJ, Karachaliou N, Berenguer J, Gimenez-Capitan A, Schellen P, Teixido C, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget (2016) 7(1):1066–75. doi:10.18632/oncotarget.6279
50. Paweletz CP, Sacher AG, Raymond CK, Alden RS, O’Connell A, Mach SL, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res (2016) 22:915–22. doi:10.1158/1078-0432.CCR-15-1627-T
51. Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol (2012) 30:1033–6. doi:10.1038/nbt.2403
52. Martinez P, McGranahan N, Birkbak NJ, Gerlinger M, Swanton C. Computational optimisation of targeted DNA sequencing for cancer detection. Sci Rep (2013) 3:3309. doi:10.1038/srep03309
53. Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DWY, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med (2012) 4(13):ra68. doi:10.1126/scitranslmed.3003726
54. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A (2011) 108:9530–5. doi:10.1073/pnas.1105422108
55. Lebofsky R, Decraene C, Bernard V, Kamal M, Blin A, Leroy Q, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol (2014) 9:783–90. doi:10.1016/j.molonc.2014.12.003
56. Rosell R, Karachaliou N. Implications of blood-based T790M genotyping and beyond in epidermal growth factor receptor-mutant non-small-cell lung cancer. J Clin Oncol (2016) 34(28):3361–2. doi:10.1200/JCO.2016.68.3458
57. Nilsson RJ, Balaj L, Hulleman E, van Rijn S, Pegtel DM, Walraven M, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood (2011) 118(13):3680–3. doi:10.1182/blood-2011-03-344408
58. Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell (2015) 28(5):666–76. doi:10.1016/j.ccell.2015.09.018
59. Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov (2015) 5(8):850–9. doi:10.1158/2159-8290.CD-15-0285
60. Schrock AB, Frampton GM, Suh J, Chalmers ZR, Rosenzweig M, Erlich RL, et al. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J Thorac Oncol (2016) 11(9):1493–502. doi:10.1016/j.jtho.2016.06.004
61. Paik PK, Drilon A, Fan PD, Yu H, Rekhtman N, Ginsberg MS, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov (2015) 5(8):842–9. doi:10.1158/2159-8290.CD-14-1467
62. Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol (2012) 10(12):e1001450. doi:10.1371/journal.pbio.1001450
63. Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, Valle M, et al. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J Proteome Res (2008) 7(12):5157–66. doi:10.1021/pr8004887
64. Mathivanan S, Simpson RJ. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics (2009) 9(21):4997–5000. doi:10.1002/pmic.200900351
65. Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res (2014) 24(6):766–9. doi:10.1038/cr.2014.44
66. Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol (2009) 9(8):581–93. doi:10.1038/nri2567
67. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol (2015) 8:83. doi:10.1186/s13045-015-0181-x
68. Brinkmann K, Emenegger J, Hurley J, et al. Exosomal RNA-based liquid biopsy detection of EML4-ALK in plasma from NSCLC patients. Presented at: 2016 National Comprehensive Cancer Network 21st Annual Conference. Hollywood, FL (2016).
69. Rosell R, Wei J, Taron M. Circulating microRNA signatures of tumor-derived exosomes for early diagnosis of non-small-cell lung cancer. Clin Lung Cancer (2009) 10(1):8–9. doi:10.3816/CLC.2009.n.001
70. Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer (2009) 10(1):42–6. doi:10.3816/CLC.2009.n.006
71. Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, et al. MicroRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol (2013) 8(9):1156–62. doi:10.1097/JTO.0b013e318299ac32
72. Adi Harel S, Bossel Ben-Moshe N, Aylon Y, Bublik DR, Moskovits N, Toperoff G, et al. Reactivation of epigenetically silenced miR-512 and miR-373 sensitizes lung cancer cells to cisplatin and restricts tumor growth. Cell Death Differ (2015) 22(8):1328–40. doi:10.1038/cdd.2014.221
73. Yuan D, Xu J, Wang J, Pan Y, Fu J, Bai Y, et al. Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget (2016) 7(22):32707–22. doi:10.18632/oncotarget.9017
74. Tang Y, Cui Y, Li Z, Jiao Z, Zhang Y, He Y, et al. Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J Exp Clin Cancer Res (2016) 35:7. doi:10.1186/s13046-016-0285-3
75. Giallombardo M, Chacartegui J, Reclusa P, Van Meerbeeck JP, Alessandro R, Peeters M, et al. Follow up analysis by exosomal miRNAs in EGFR mutated non-small cell lung cancer (NSCLC) patients during osimertinib (AZD9291) treatment: a potential prognostic biomarker tool. J Clin Oncol (2016) 34(suppl; abstr e23035).
76. Ashworth T. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Aus Med J (1869) 14:146–9.
77. Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene (2016) 35(10):1216–24. doi:10.1038/onc.2015.192
78. Carlsson A, Nair VS, Luttgen MS, Keu KV, Horng G, Vasanawala M, et al. Circulating tumor microemboli diagnostics for patients with non-small-cell lung cancer. J Thorac Oncol (2014) 9:1111–9. doi:10.1097/JTO.0000000000000235
79. Hou JM, Krebs M, Ward T, Sloane R, Priest L, Hughes A, et al. Circulating tumor cells as a window on metastasis biology in lung cancer. Am J Pathol (2011) 178:989–96. doi:10.1016/j.ajpath.2010.12.003
80. Hou JM, Greystoke A, Lancashire L, Cummings J, Ward T, Board R, et al. Evaluation of circulating tumor cells and serological cell death biomarkers in small cell lung cancer patients undergoing chemotherapy. Am J Pathol (2009) 175:808–16. doi:10.2353/ajpath.2009.090078
81. O’Flaherty JD, Gray S, Richard D, Fennell D, O’Leary JJ, Blackhall FH, et al. Circulating tumour cells, their role in metastasis and their clinical utility in lung cancer. Lung Cancer (2012) 76(1):19–25. doi:10.1016/j.lungcan.2011.10.018
82. Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol (2005) 23:1420–30. doi:10.1200/JCO.2005.08.140
83. de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res (2008) 14:6302–9. doi:10.1158/1078-0432.CCR-08-0872
84. Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol (2008) 26:3213–21. doi:10.1200/JCO.2007.15.8923
85. Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol (2011) 29(12):1556–63. doi:10.1200/JCO.2010.28.7045
86. Tanaka F, Yoneda K, Kondo N, Hashimoto M, Takuwa T, Matsumoto S, et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res (2009) 15:6980–6. doi:10.1158/1078-0432.CCR-09-1095
87. Isobe K, Hata Y, Kobayashi K, Hirota N, Sato K, Sano G, et al. Clinical significance of circulating tumor cells and free DNA in non-small cell lung cancer. Anticancer Res (2012) 32:3339–44.
88. de Wit S, van Dalum G, Lenferink AT, Tibbe AG, Hiltermann TJ, Groen HJ, et al. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep (2015) 5:12270. doi:10.1038/srep12270
89. Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer (2011) 105:847–53. doi:10.1038/bjc.2011.294
90. Krebs MG, Hou JM, Sloane R, Lancashire L, Priest L, Nonaka D, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol (2012) 7:306–15. doi:10.1097/JTO.0b013e31823c5c16
91. Mascalchi M, Falchini M, Maddau C, Salvianti F, Nistri M, Bertelli E, et al. Prevalence and number of circulating tumour cells and microemboli at diagnosis of advanced NSCLC. J Cancer Res Clin Oncol (2016) 142:195–200. doi:10.1007/s00432-015-2021-3
92. Hofman V, Long E, Ilie M, Bonnetaud C, Vignaud JM, Fléjou JF, et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology (2012) 23:30–8. doi:10.1111/j.1365-2303.2010.00835.x
93. Kolostova K, Spicka J, Matkowski R, Bobek V. Isolation, primary culture, morphological and molecular characterization of circulating tumor cells in gynecological cancers. Am J Transl Res (2015) 7:1203–13.
94. Zhang Z, Shiratsuchi H, Lin J, Chen G, Reddy RM, Azizi E, et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget (2014) 5:12383–97. doi:10.18632/oncotarget.2592
95. Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol (2013) 31:539–44. doi:10.1038/nbt.2576
96. Maheswaran S, Haber DA. Ex vivo culture of CTCs: an emerging resource to guide cancer therapy. Cancer Res (2015) 75:2411–5. doi:10.1158/0008-5472.CAN-15-0145
97. Rolfo C, Castiglia M, Hong D, Alessandro R, Mertens I, Baggerman G, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta (2014) 1846(2):539–46. doi:10.1016/j.bbcan.2014.10.001
98. Rolle A, Gunzel R, Pachmann U, Willen B, Höffken K, Pachmann K. Increase in number of circulating disseminated epithelial cells after surgery for nonsmall cell lung cancer monitored by MAINTRAC(R) is a predictor for relapse: a preliminary report. World J Surg Oncol (2005) 3:18. doi:10.1186/1477-7819-3-18
99. Yoon SO, Kim YT, Jung KC, Jeon YK, Kim BH, Kim CW. TTF-1 mRNA-positive circulating tumor cells in the peripheral blood predict poor prognosis in surgically resected non-small cell lung cancer patients. Lung Cancer (2011) 71:209–16. doi:10.1016/j.lungcan.2010.04.017
100. Bayarri-Lara C, Ortega FG, Cueto Ladrón de Guevara A, Puche JL, Ruiz Zafra J, de Miguel-Pérez D, et al. Circulating tumor cells identify early recurrence in patients with non-small cell lung cancer undergoing radical resection. PLoS One (2016) 11(2):e0148659. doi:10.1371/journal.pone.0148659
101. Hofman V, Bonnetaud C, Ilie MI, Vielh P, Vignaud JM, Fléjou JF, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res (2011) 17:827–35. doi:10.1158/1078-0432.CCR-10-0445
102. Li J, Shi SB, Shi WL, Wang Y, Yu LC, Zhu LR, et al. LUNX mRNA-positive cells at different time points predict prognosis in patients with surgically resected nonsmall cell lung cancer. Transl Res (2014) 163:27–35. doi:10.1016/j.trsl.2013.09.010
103. Yamashita J, Matsuo A, Kurusu Y, Saishoji T, Hayashi N, Ogawa M. Preoperative evidence of circulating tumor cells by means of reverse transcriptase-polymerase chain reaction for carcinoembryonic antigen messenger RNA is an independent predictor of survival in non-small cell lung cancer: a prospective study. J Thorac Cardiovasc Surg (2002) 124:299–305. doi:10.1067/mtc.2002.124370
104. Punnoose EA, Atwal S, Liu W, Raja R, Fine BM, Hughes BG, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res (2012) 18:2391–401. doi:10.1158/1078-0432.CCR-11-3148
105. Tan DS, Yom SS, Tsao MS, Pass HI, Kelly K, Peled N, et al. The International Association for the Study of Lung Cancer consensus statement on optimizing management of EGFR mutation positive non-small cell lung cancer: status in 2016. J Thorac Oncol (2016) 11(7):946–63. doi:10.1016/j.jtho.2016.05.008
Keywords: ctDNA, ctRNA, CTCs, exosomes, tumor-educated platelets, mutations, gene fusions, lung cancer
Citation: Molina-Vila MA, Mayo-de-las-Casas C, Giménez-Capitán A, Jordana-Ariza N, Garzón M, Balada A, Villatoro S, Teixidó C, García-Peláez B, Aguado C, Catalán MJ, Campos R, Pérez-Rosado A, Bertran-Alamillo J, Martínez-Bueno A, Gil M-d, González-Cao M, González X, Morales-Espinosa D, Viteri S, Karachaliou N and Rosell R (2016) Liquid Biopsy in Non-Small Cell Lung Cancer. Front. Med. 3:69. doi: 10.3389/fmed.2016.00069
Received: 31 October 2016; Accepted: 08 December 2016;
Published: 23 December 2016
Edited by:
Umberto Malapelle, University of Naples Federico II, ItalyReviewed by:
Dario De Biase, University of Bologna, ItalyMoira Ragazzi, Azienda Sanitaria Unità Locale di Reggio Emilia, Italy
Copyright: © 2016 Molina-Vila, Mayo-de-las-Casas, Giménez-Capitán, Jordana-Ariza, Garzón, Balada, Villatoro, Teixidó, García-Peláez, Aguado, Catalán, Campos, Pérez-Rosado, Bertran-Alamillo, Martínez-Bueno, Gil, González-Cao, González, Morales-Espinosa, Viteri, Karachaliou and Rosell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miguel A. Molina-Vila, mamolina@pangaebiotech.com
 Miguel A. Molina-Vila
Miguel A. Molina-Vila Clara Mayo-de-las-Casas
Clara Mayo-de-las-Casas Ana Giménez-Capitán1
Ana Giménez-Capitán1
 Mónica Garzón
Mónica Garzón Sergi Villatoro
Sergi Villatoro Cristina Teixidó
Cristina Teixidó Cristina Aguado
Cristina Aguado María José Catalán
María José Catalán Raquel Campos
Raquel Campos María-de-los-Llanos Gil
María-de-los-Llanos Gil María González-Cao
María González-Cao Xavier González
Xavier González Daniela Morales-Espinosa
Daniela Morales-Espinosa Santiago Viteri
Santiago Viteri Rafael Rosell
Rafael Rosell