Protein Translation and Signaling in Human Eosinophils
- 1Department of Medicine, Allergy, Pulmonary, and Critical Care Medicine Division, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, United States
- 2Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, United States
We have recently reported that, unlike IL-5 and GM-CSF, IL-3 induces increased translation of a subset of mRNAs. In addition, we have demonstrated that Pin1 controls the activity of mRNA binding proteins, leading to enhanced mRNA stability, GM-CSF protein production and prolonged eosinophil (EOS) survival. In this review, discussion will include an overview of cap-dependent protein translation and its regulation by intracellular signaling pathways. We will address the more general process of mRNA post-transcriptional regulation, especially regarding mRNA binding proteins, which are critical effectors of protein translation. Furthermore, we will focus on (1) the roles of IL-3-driven sustained signaling on enhanced protein translation in EOS, (2) the mechanisms regulating mRNA binding proteins activity in EOS, and (3) the potential targeting of IL-3 signaling and the signaling leading to mRNA binding activity changes to identify therapeutic targets to treat EOS-associated diseases.
Introduction
Control of protein production is critical for the maintenance of cell and tissue homeostasis. Excessive protein production may lead to hypertrophy and an unnecessary use of energy and other resources. However, inadequate protein synthesis antagonizes cell growth, proliferation, adaptation to environmental changes, and the implementation of new cell functions. Overproduction of transcription factors or cytokines contributes to or causes transformation and cancer. Thus, a carefully controlled balance within metabolic constraints but responsive to environmental and signaling cues is essential for optimal cellular function.
Circulating eosinophils (EOS) are differentiated, non-proliferative cells, which become apoptotic within 2–3 days if lacking contact with pro-survival cytokines, such as IL-5, GM-CSF, and IL-3 (1). Therefore, resting EOS have modest needs for new protein production. Protein production is dependent on (1) the level of coding mRNA, which in turn depends on the amount of mRNA transcribed and spliced excluding the amount degraded, and (2) the translation rate of the transcripts, which is governed by ribosomal content, activity, and associated ribosomal and mRNA binding proteins. Extracellular inputs via cell surface and intracellular receptors leading to the propagation of intracellular signals control each of these steps [reviewed in Ref. (2)].
Eosinophils have the ability to differentially regulate translation. As shown in Figure 1, the presence of high levels of a specific mRNA may or may not lead to protein translation, making inference of protein expression from mRNA quantification tenuous. Cell stimulation can trigger (1) the transcription and translation of mRNA expressed at very low level under basal conditions, (2) the stabilization of mRNA contributing to its accumulation and translation, (3) the translation of mRNA constitutively present but translationally quiescent in resting cells, and (4) an increase in the activity of the machinery, contributing to increased, global protein synthesis. As these topics are far too large to be covered adequately, here we will focus on how changes of both the translation machinery activity and the content of mRNA binding proteins affect the translatability of a subset of mRNA. We will start with an overview of protein translation and its control by intracellular signaling. During this overview, we will use previously published proteomic and phospho-proteomic data from peripheral blood EOS (3) to generalize these known protein translation mechanisms in EOS. Then, we will discuss how changes in mRNA binding proteins and the IL-3-dependent translation of a group of mRNA influence the production of the pro-survival cytokine, GM-CSF, and EOS function, respectively. Finally, the last section, titled “Regulation of translation and potential therapeutic targets,” describes potential molecular drug targets that are implicated in protein translation in EOS in addition to EOS survival and activity. This review may help identify targets that are upstream of GM-CSF and downstream of IL-3 to supplement anti-IL-5 therapies, which despite their efficacy, have not totally controlled eosinophilia and EOS-related pathology. Of note, unless indicated, the observations discussed in this manuscript were obtained using human EOS.
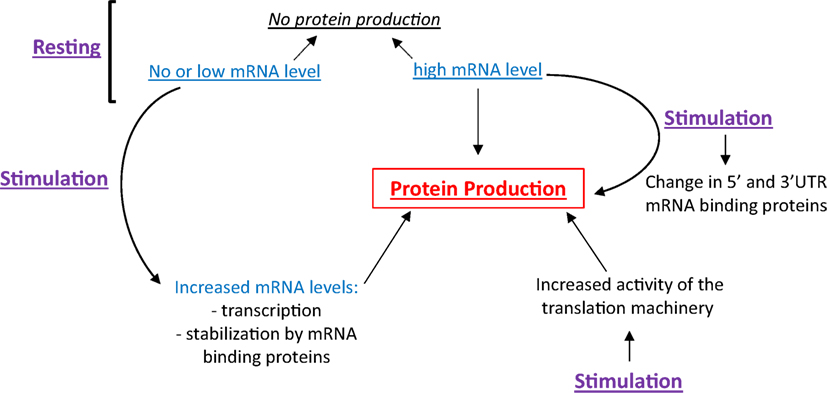
Figure 1. Protein production is a function of cellular stimulation state, mRNA expression level and RNA-binding protein functionality. In resting eosinophils (EOS), protein synthesis can be suppressed irrespective of mRNA content. Cell stimulation can trigger protein production through increased transcription, mRNA stabilization and increased translation, typically regulated by changes in RNABP function.
General Mechanisms Controlling Protein Synthesis
In eukaryotic cells, initiation, elongation, and termination are the three fundamental steps of protein translation. Some of the main proteins/mRNA interactions involved in the initiation and elongation of translation are shown in Figure 2. During translation, initiation begins with the binding of eukaryotic translation initiation factor 4E (eIF4E) to the mRNA 5′-cap. Next, eIF4E binds to eIF4G, which interacts with the other eIF4 proteins, eIF4A and eIF4B. The helicase activity of eIF4A is amplified by eIF4B, and most likely unwinds secondary GC-rich structures of the 5′-UTR, thus facilitating initiation of mRNAs possessing these structures. The interaction of eIF4G with the poly-A binding protein (PABP), which circularizes the mRNA, also increases mRNA translatability. The binding of eIF4B and eIF4G to the 43S preinitiation complex (PIC) via eIF3 links the mRNA to the ribosome. The 43S PIC is composed of the ribosomal 40S subunit, eIF3, eIF5 eIF1, eIF1A, and the complex eIF2/Met-tRNA. EIF2 binds Met-tRNA in its GTP-bound state (eIF2-GTP). The complex Met-tRNA/eIF2-GTP along with the initiation factors/40S complex scans the 5′UTR until the start codon (AUG) is recognized by complementarity with the anticodon of Met-tRNA (4). Once the start codon is reached, protein translation becomes initiated by the eIF5B-catalyzed hydrolysis of eIF2-GTP into eIF2-GDP, which frees the ribosomal 40S from eIF2 (5). The release of eIF2-GDP and other initiation factors from the 40S complex is followed by the recruitment of the 60S ribosome subunit. The newly formed 80S ribosomal complex is now ready to start elongation (6). Elongation is predominantly controlled by eukaryotic elongation factor 1 (eEF1) and eEF2. Next, eEF1A-GTP recruits the second aminoacyl (aa)-tRNA complementary to the adjacent, C-terminal codon (A-site). After the peptide bound formation between Met and the second aa at the P-site, eEF2-GTP pushes (translocates) the mRNA and allows the third aa-tRNA to become positioned on the third codon at the A-site. Simultaneously, the first Met-tRNA is removed from the P-site and is replaced by the second aa-tRNA previously on the A-site. When the ribosome reaches a stop codon, no complementary tRNA exists to fill the A-site. At that point, the release factor ERF1 (ETF1) takes position in the A-site, and along with ERF3A-B (GSPT1–2) hydrolyzes the peptide chain (protein) attached to the last t-RNA to terminate translation.
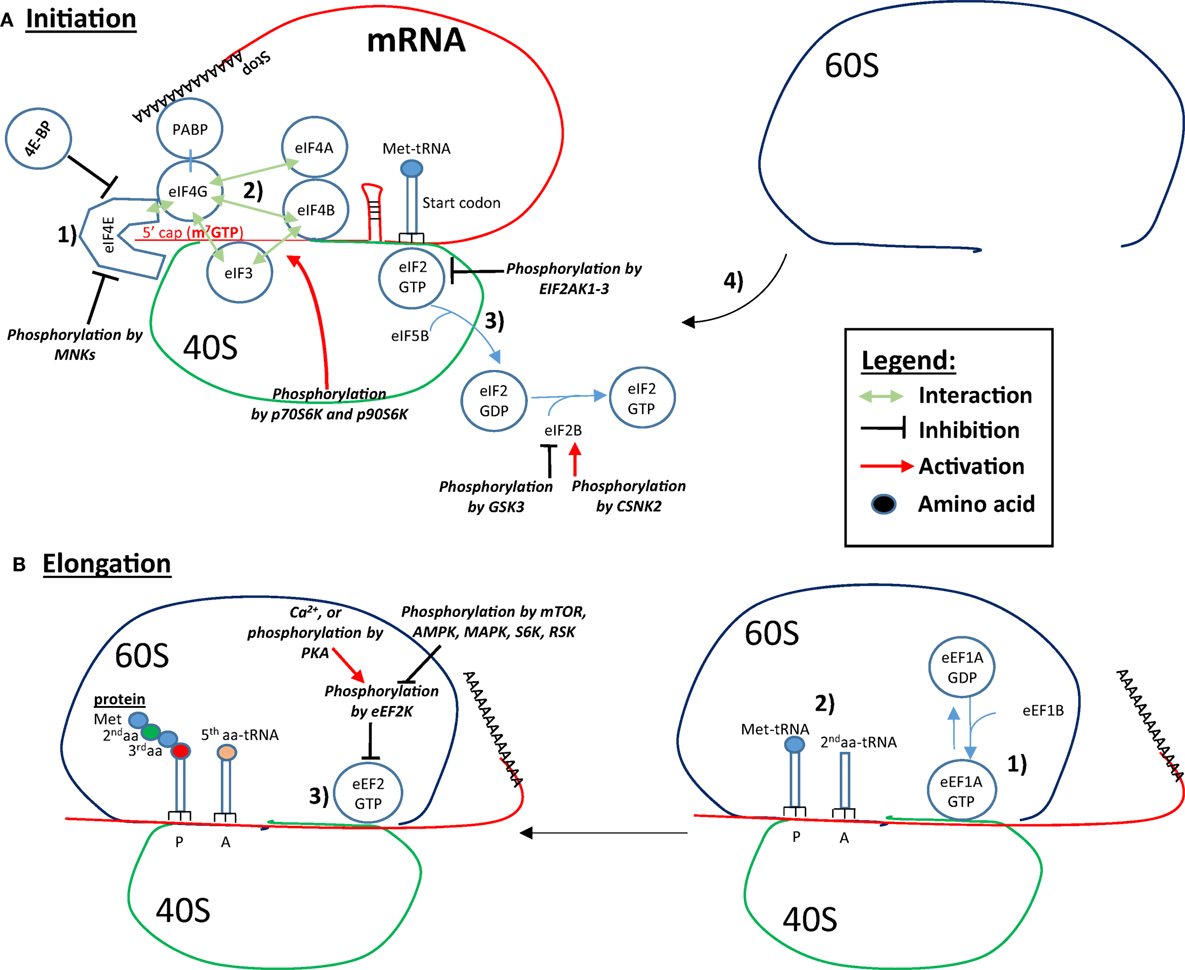
Figure 2. Controls of initiation and elongation of protein translation. (A) Initiation: (1) eukaryotic translation initiation factor 4E (eIF4E) binds to the 5′-cap structure of the mRNA and eIF4G, which interacts with eIF4A and eIF4B. eIF4G also interacts with the poly-A binding protein (PABP), which circularizes the mRNA and increases the translation rate. (2) eIF4B and eIF4G binds to eIF3, which is associated with the ribosomal 40S subunit, which forms the link between the mRNA, the ribosome and the complex eIF2-GTP/Met-tRNA. In addition, the initiation factors/40S complex scan the 5′UTR until the recognition of the start codon. (3) Protein translation is initiated by the eIF5B-catalyzed eIF2-GTP hydrolysis into GDP, which results in the freeing of the ribosomal 40S from eIF2 and other initiation factors. (4) The recruitment of the 60S ribosomal subunit forms the ribosome by binding to the 40S subunit. eIF4B interaction with eIF3 is increased by p70S6K- and p90S6K-mediated phosphorylation. Binding of eIF4E to eIF4G and to the 5′ cap can be inhibited by 4E-BP and by Mnk-mediated phosphorylation. Phosphorylation of eIF2 by the EIF2AKs inhibits eIF2 recycling. eIF2B is phosphorylated and inhibited by glycogen synthase kinase 3 (GSK3), while phosphorylation of eIF2B by CSNK2 increases its activity toward eIF2 recycling. (B) Elongation: (1) eEF1A-GTP recruits the second aminoacyl (aa)-tRNA on the A-site. (2) A peptide bond forms between Met and the second aa. (3) eEF2-GTP pushes the mRNA, Met-tRNA is removed from the P-site and is replaced by the next aa-tRNA previously on the A-site. In addition, a third aa-tRNA is placed in the now empty A-site. Eukaryotic elongation factor 2 (eEF2) is inhibited by eEF2K-mediated phosphorylation. EEF2K is inhibited by mTOR, AMP-activated protein kinase (AMPK), mitogen-activated protein kinase (MAPK), S6K, and RSK. Conversely, Ca2+ and PKA phosphorylation leads to eEF2K phosphorylation and activation, and inhibition of the elongation.
Regulation of Protein Translation
In general, in eukaryotic cells, initiation can be controlled at multiple levels. The eIF4BP proteins (4E-BP) interact with eIF4E, preventing its interaction with eIF4G and, therefore, inhibiting translation initiation (7). 4E-BP are regulated at multiple phosphorylation sites, often by the mammalian target of rapamycin (mTOR), which reduces 4E-BP interactions with eIF4E and enhances translation initiation (8). In addition, the cyclin-dependent kinase 2 (CDK2) phosphorylates 4E-BP on Thr70 leading to its release from eIF4E (9).
Eukaryotic translation initiation factor 4E (eIF4E) can be phosphorylated on Ser209, which decreases its affinity for the 5′-cap structure and, therefore, inhibits translation (10, 11). eIF4E is phosphorylated by the mitogen-activated protein kinase (MAPK) signal-integrating kinases Mnk1 and Mnk2 (MKNK1 and 2), which are downstream targets of the MAPK (ERK and p38) (10, 11). Also among the eIF4 family, eIF4B is phosphorylated by p70S6K and p90S6K (RSK) at Ser422, which increases its interaction with eIF3, enhancing translation initiation (12).
When the inactive form of eIF2, eIF2-GDP, leaves the 40S during initiation, it must be recharged with GTP for continuous translation initiation. Then, eIF2-GDP is converted to eIF2-GTP by eIF2B, which is a guanine-nucleotide-exchange factor (GEF). eIF2B is phosphorylated at multiple sites that includes two residues phosphorylated by casein kinase 2 (CSNK2A1) that are required for eIF2B/eIF2 interactions, eIF2 recycling and translation initiation (13). eIF2 is phosphorylated at Ser51 by as many as four kinases, all of which inhibit the eIF2–eIF2B interaction, demonstrating a critical role in protein synthesis (14).
The delivery of aa-tRNA required for elongation is driven by the hydrolysis of eEF1A-GTP to eEF1A-GDP. Thus, the GEF eEF1B acts on eEF1A-GDP as eIF2B does on eIF2-GDP. eEF1 is also targeted by a variety of kinases, including PKC, CSNK2, and cyclin-dependent kinase 1 (CDK1), but the role of the phosphorylation states of these elongation factors remains uncertain (15).
Phosphorylation by eEF2 kinase on Thr56 impairs eEF2’s ability to bind to the 40S subunit of the ribosome (16). Thr56 phosphorylation is enhanced if Ser595 is previously phosphorylated (17). The eEF2 kinase activity is calcium/calmodulin-dependent. Its activation after Ca2+ flux leads to the attenuation of elongation. Of note, increased eEF2 kinase activity may provide mRNA with poor translation initiation efficiency a greater chance of being synthesized (18). eEF2 kinase is itself regulated at multiple phosphorylation sites, typically by the mammalian target of rapamycin complex 1 (mTORC1) that reduces its activity (19). AMP-activated protein kinase (AMPK) and the MAPK can also phosphorylate eEF2 kinase leading to translation enhancement (18). Conversely, cAMP/PKA signaling pathway phosphorylates Ser500 (20), rending eEF2 activity independent of Ca2+ ions and activating the kinase. Figure 2 summarizes these different signaling events and control points.
General Translation in EOS
Recently, using two-dimensional liquid chromatography coupled with high-resolution mass spectrometry, 6,813 proteins were identified in unstimulated human blood EOS ((3), and see article by Mosher et al., in this issue for more details). In addition, 4,802 site-specific phosphorylation events were simultaneously identified (3). Furthermore, using RNA-Seq, ~7,981 protein-coding genes expressed by unstimulated human blood EOS were identified (21). The cellular content (mRNA and protein) and phosphorylation state of the main proteins involved in the initiation, elongation, and termination of protein translation have been extracted from the published proteome and transcriptome (shown in Table 1). Notably, Table 1 shows examples of the disconnections between mRNA and protein levels, which suggests that production of certain proteins is tightly regulated at the translational level in EOS (i.e., EIF4G2, ETF1, etc.). For instance, while ratio of protein to mRNA expression generally reached ~1,000 and above, ratios for EIF4G2 and ETF1 were only 107 and 160, respectively (Table 1), suggesting marginal translation for these two transcripts in resting EOS. With the possible exception of the inhibitor of elongation, eEF2K, resting blood EOS possess all the essential proteins involved in protein translation. However, the identification of eEF2 phosphorylation on Thr56 (Table 1) suggests the existence of eEF2K activity, preventing the eEF2/40S interaction and blockade of translation elongation (16). In addition, the lack of phosphorylation of eIF2B (Table 1) suggests a possible lack of eIF2B/eIF2 interactions and reduced recycling of eIF2 into its active form (eIF2-GTP), which would dampen translation initiation (13). Conversely, in agreement with our previous report (22), 4E-BP is phosphorylated in resting EOS (Table 1). This indicates that 4E-BP does not act as a blocker of eIF4E binding to the 5′ cap in resting EOS and, therefore, other factors are responsible for restricting protein translation in resting EOS. Thus, the combined lack of eIF2B phosphorylation with the phosphorylation of eEF2 on Thr56 suggests attenuation of both initiation and elongation of protein translation in resting EOS (22).
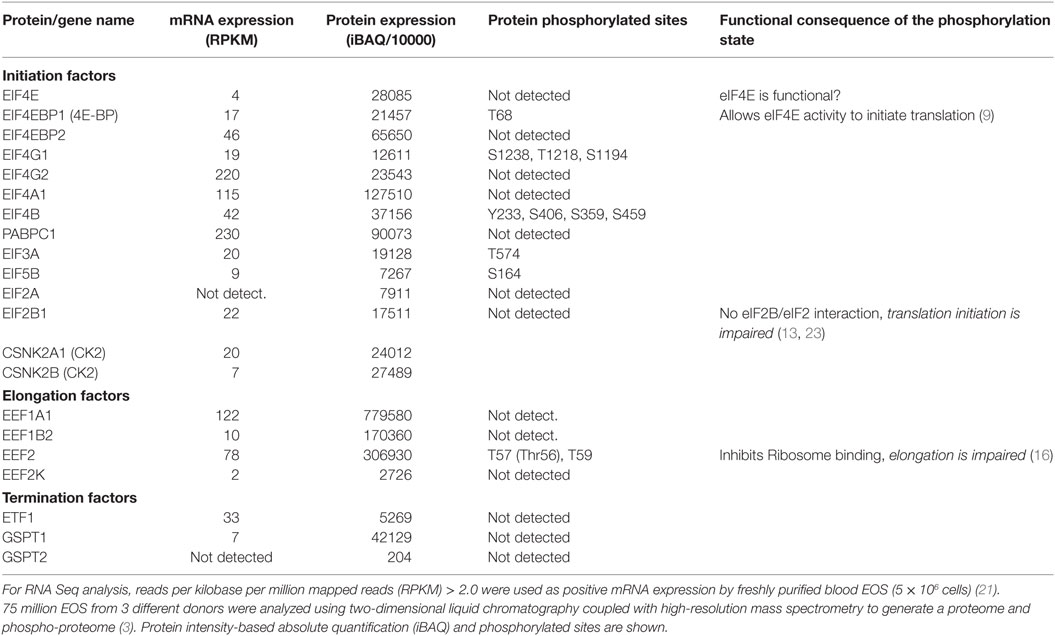
Table 1. Proteins involved in initiation, elongation, and termination, and present in fresh human blood EOS.
IL-5, GM-CSF, and IL-3 are critical cytokines for EOS development and function. Each interacts with a specific α-chain receptor and a common, associated β-chain (24, 25). Not surprisingly, these receptors can generate both common and unique signals (22, 26–28). As indicated above, we have shown that 4E-BP is highly phosphorylated in resting EOS (22). After activation with IL-3, IL-5, or GM-CSF, 4E-BP phosphorylation state remains largely unaffected (22), suggesting that the increased translation induced by these cytokines is likely 4E-BP-independent. In addition to 4E-BP, we have unpublished observations indicating that EIF4B phosphorylation at Ser422 was unaffected by GM-CSF. Therefore, as for 4E-BP, eIF4B activity cannot explain the significant enhancement of translation in GM-CSF-activated cells (22). However, a slight but significant increase in the phosphorylation of eIF4B was observed in EOS activated by IL-3 for 20 h (unpublished data). This phosphorylation on Ser422 may account for the differences in translation seen in IL-3- versus GM-CSF-activated EOS (22). The signaling accounting for regulated translation in IL-3 or GM-CSF-activated EOS remains largely unstudied.
Signaling and Protein Translation
Two major intracellular signaling pathways regulate translation in eukaryotic cells: the phosphoinositide 3-kinase (PI3K)/Akt/mTOR and the MAPK pathways. These two pathways are generally triggered by extracellular stimuli via membrane receptors but also respond to intracellular ATP levels and amino acid availability.
PI3K/Akt/mTOR Signaling
Ligation of growth factors with tyrosine kinase or G-protein coupled (GPC) receptors typically leads to phosphorylation of the membrane phospholipid, phosphatidylinositol-4,5-biphosphate (PI-4,5-P2) into phosphatidylinositol-3,4,5-triphosphate (PIP3), by the class I lipid kinase PI3K. This transformation into PIP3 is reversed by phosphatases such as the phosphatase and tensin homolog deleted on chromosome 10 (PTEN) and the SH2-domain containing inositol phosphatase (SHIP) (29). PIP3 drives the phosphorylation and activation of Akt (also called, PKB), via 3-phosphoinositol-dependent kinase 1 (PDK1) (30). Akt activity is also augmented by the mTORC2 complex, composed of mTOR and rictor (rapamycin-insensitive companion of mTOR) (29). Akt can in turn phosphorylate and inhibit the glycogen synthase kinase 3 (GSK3) leading to dephosphorylation and activation of eIF2B with translation initiation (31). In addition, Akt phosphorylates five sites leading to the inhibition of the GTPase activity of tuberous sclerosis complex 2 (TSC2), on the small GTPase Ras homolog enriched in brain (RHEB), which in its GTP form stimulates the kinase activity of mTORC1 (32, 33). Therefore, the activity of the mTORC1 complex, composed of mTOR, RHEB, the mTOR associated protein, LST8 (MLST8), and the regulatory-associated protein of mTOR (Raptor), is downregulated by unphosphorylated TSC2 that is derepressed by Akt kinase activity.
Downstream mTORC1, the TOS (target of rapamycin signaling)-containing 4E-BP and p70S6K are phosphorylated. As seen above, phosphorylated 4E-BP is inactive and allows eIF4E to bind eIF4G to initiate translation. In dividing cells, mTOR phosphorylates p70S6K at Thr389, which in turn can phosphorylate ribosomal S6 protein (RPS6), eIF4B and programmed cell death 4 (PDCD4). While the function of phosphorylated RPS6 remains largely unknown, eIF4B and PDCD4 are positive and negative regulators, respectively, of the RNA helicase, eIF4A (34). mTORC1 also downregulates the activity of the eEF2 Kinase, which then subsequently enhances the elongation step of translation by eEF2. The general protein translational capacity is also enhanced by mTOR via increased transcription (more mRNA), and stimulation of the translation of mRNAs containing a string of 5′–pyrimidines (5′TOP mRNA) (35). In addition to its activation by growth factors, mTOR also senses cellular nutrient, oxygen, and energy level (36). As its name implies, most of mTOR effects are neutralized by rapamycin. The FKBP12–Rapamycin complex quickly binds close to the kinase domain (37), leading to mTOR conformational changes, dissociation from Raptor (38, 39) and inhibition of some of mTORC1 functions (40). By binding newly produced mTOR, FKBP12–rapamycin complex also inhibits the assembly of mTORC2 (41). Rapamycin also inhibits the binding of phosphatidic acid (PA) to mTOR, reducing the stabilization of the mTORC1 and mTORC2 complexes (42). PA is synthetized during membrane phospholipid biogenesis (43), and its intracellular level modulates the amount of rapamycin required to inhibit mTOR (44). Interestingly, low doses of rapamycin inhibit mTOR-induced p70S6K phosphorylation while much higher doses are required to block mTOR-induced 4E-BP Thr37/Thr46 phosphorylation (45). As a result, other compounds that are stronger inhibitors of mTORC1 and C2 than rapamycin, such as PP242 and AZD8055, were developed.
mTOR Signaling in EOS
Surprisingly, mTOR has not been studied in EOS, although its inhibitor, rapamycin has shown effects on EOS in vitro and in vivo. As shown in Table 2, resting human blood EOS express relatively little mTOR, but very high amount of FKBP12. FKBP12 is bound by both rapamycin and FK506 and is required for these drugs to exert their inhibitory effects in cells. Interestingly, nanomolar doses of FK506 strongly inhibit calcium ionophore-induced cytokine (GM-CSF) production in EOS, while micromolar doses of rapamycin does not (46). Due to the competition between rapamycin and FK506 on FKBP12, high amount of rapamycin antagonizes the FK506-mediated inhibition of cytokine production in EOS (46). However, rapamycin is more potent than FK506 in inhibiting IL-5-induced prolonged EOS survival (46). The divergence between FK506 and rapamycin has also been described in T lymphocyte and mast cells, where rapamycin modulates proliferation rather that gene expression (47, 48). Another study (49) confirmed that rapamycin reduces IL-5-induced pro-survival signaling in EOS but the effect was modest and required high doses of drug for at least 72 h. In the same study, rapamycin also partially inhibited IL-5-induced eosinophil cationic protein (ECP) release from EOS (49). In addition, mTOR has important functions during EOS differentiation as rapamycin inhibited mouse EOS differentiation downstream of IL-5 (50). This is in agreement with the dependence of T cell proliferation and differentiation on mTOR (51). Remarkably, rapamycin has no inhibitory effect on mouse EOS recruitment into the BALF after exposure to dust-mite allergen in chronic allergic models (52), suggesting that the role of mTOR signaling is confined to development and possibly survival but not cell migration.
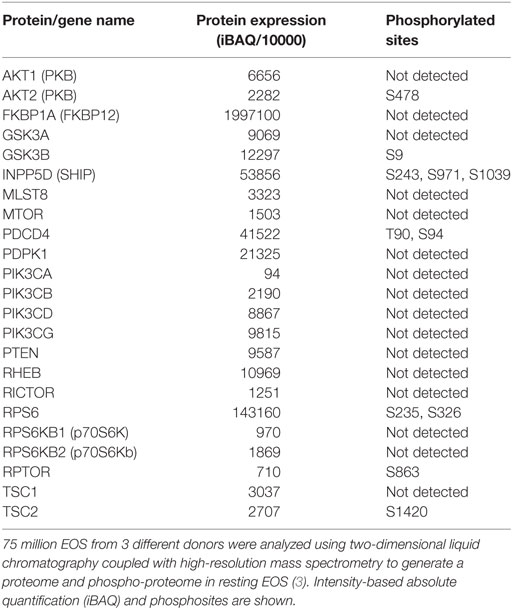
Table 2. Proteins present in human blood eosinophil (EOS) and involved in the phosphoinositide 3-kinase/mammalian target of rapamycin pathway.
In both human and mouse EOS, PI3K is required for a variety of functions. These include chemokine-induced EOS granule proteins release (53), platelet-activating factor (PAF)-induced chemotaxis but not LTC4 release (54). The PI3K/Akt pathway is also essential for IL-5-induced β2-integrin adhesion to bovine serum albumin (BSA) (55), and IL-5-induced guinea pig EOS mobilization from the bone marrow (56). In EOS, the PI3K/Akt pathway can be activated by fMLP or RANTES after priming with IL-5 or IL-3 (57). Prostaglandin E2 (PGE2) via EP4 induces PI3K/PDK1-dependent increase in Akt phosphorylation, which consequently inhibits eotaxin-induced EOS shape changes and chemotaxis (30). Therefore, the PI3K/PDK1/Akt pathway is important in EOS and regulates a variety of functions depending on its activator.
MAPK Signaling
The MAPK (ERK and p38) signaling pathways are involved in most of cellular functions, including differentiation and proliferation. ERK1, ERK2, p38α, and p38β are coded by four different genes (MAPK3, MAPK1, MAPK14, and MAPK11). Following intracellular or extracellular activation, the MAP kinase kinase kinases (MEKK) are activated, leading to phosphorylation of MAP kinase kinases (MEK) and, finally, MAPK are phosphorylated (58). ERK1/2 alone possess more than 150 substrates involved in a large variety of cell functions, including transcription, cell death, autophagy metabolism, and translation (59). Among the kinases activated by ERK or p38 are kinases involved in protein translation, including p90S6K (RSK), the MAPK-interacting kinases (Mnk), and the MAPK-activated protein kinase 2 (MK2) (2). The latter has an important role in 3′UTR directed, mRNA binding protein-dependent translation. P90S6K are activated by ERK signaling that can then phosphorylate TSC2 at Ser1798, activating mTORC1 and protein synthesis (60, 61). Of note, ERK may also directly phosphorylate and inhibit TSC2, leading to increased mTORC1 activity (62). Like p70S6K, p90S6K also phosphorylates both eIF4B and eEF2 kinase, which enhances eIF4B/eIF3 interactions and eEF2 function and, consequently, protein initiation and elongation (12, 23). While Mnk2 activity is thought to be constitutive, Mnk1 phosphorylation and activation can be triggered downstream ERK and p38 leading to eIF4E phosphorylation at Ser209 (63). Although this phosphorylation inhibits eIF4E binding to the 5′-cap, it may also control the translation of specific mRNAs (63).
MAPK Signaling in EOS
Mitogen-activated protein kinases have important roles in many critical events, including EOS survival, migration, adhesion, production of inflammatory mediators, and degranulation. In EOS, ERK and p38 are phosphorylated and active following stimulation with a variety of mediators, including the β-chain cytokines (IL-3, IL-5, and GM-CSF), chemokines, fMLP, the PAF, and matrix proteins (26, 28, 53, 57, 64–71). Table 3 shows the expression levels of ERK, and their downstream targets, RSK1–3 (p90S6K), all of which are phosphorylated at a detectable level in resting cells. However, EOS contain little Mnk1/2 (Table 3), suggesting that the MAPK activation likely does not regulate protein translation via eIF4E phosphorylation (Figure 2); and despite its phosphorylation, the low level of Mnk2 probably have little effect on eIF4E phosphorylation. Consistent with MAPK activation, upstream MEK and MEKK were also phosphorylated at multiple sites in circulating EOS (Table 3). These data suggest that such cells are not truly resting but have been partially activated or primed either in vivo or during isolation.
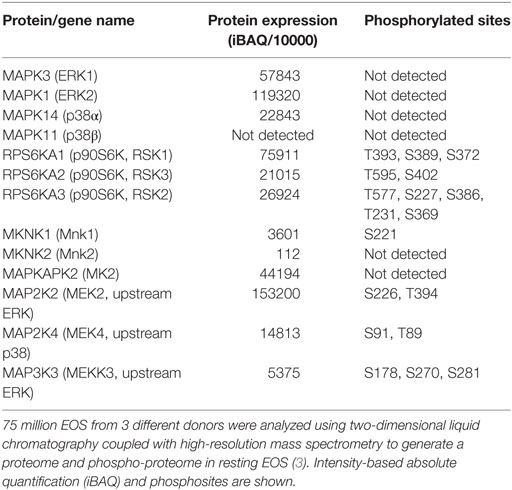
Table 3. Proteins present in human eosinophil and involved in the mitogen-activated protein kinase signaling upstream of protein translation.
Messenger RNA-Specific Protein Translation
mRNA translation is clearly not an all or nothing event. Agonists may increase or decrease ribosomal mobilization of all, the majority or subsets of mRNAs. This may occur through a slowdown of global elongation by phosphorylated eEF2 allowing poorly translated mRNAs to enter initiation and to be translated when elongation becomes derepressed (18). Alternatively, increased eIF4A helicase activity may preferentially facilitate the translation of mRNAs possessing secondary structures in their 5′-UTR that require unwinding prior to initiation.
Selective regulation requires the recognition of unique cis-elements within the mRNA by sequence-specific mRNA binding proteins. In this way, subsets of mRNAs can be selectively identified and regulated for differential translation and mRNA decay. One well-studied example is the pyrimidine-rich domain termed terminal oligopyrimidine (TOP). mRNA containing TOP usually code for elongation factors and ribosomal proteins (72) and their translation is preferably induced by the mTOR pathway (73). We will discuss additional examples below.
IL-3 Induces Translation of Semaphorin-7A mRNA in EOS
Semaphorin-7A mRNA level is relatively high in resting cells and changes only slightly in activated blood EOS. However, its translation remains almost undetectable despite GM-CSF activation (22). Surprisingly, despite similar mRNA levels, the translation rate for semaphorin-7A is more than 10-fold higher in IL-3- versus GM-CSF-activated EOS (22). Consistent with increased translation, semaphorin-7A mRNA was enriched in polyribosome fractions following IL-3 compared to GM-CSF (22). Of note, TOP mRNAs (EEF1A1 and PABP) were not enriched in the polyribosome fraction by IL-3, suggesting unique and highly selective signaling from IL-3 receptor to the translational machinery.
Freshly purified blood EOSs possess surface semaphorin-7A, which tends to decrease overtime during the first 20 h of cell culture (unpublished data). Activation with IL-5 or GM-CSF maintains or slightly increases surface semaphorin-7A over this same time span (27). On the other hand, over a broad range of doses, IL-3 significantly increased surface semaphorin-7A expression (27). Interestingly, IL-3-induced semaphorin-7A translation occurred more than 6 h after activation (unpublished data), suggesting that considerable signaling and possibly the translation of accessory proteins precedes semaphorin-7A translation initiation.
ERK/p90S6K/RPS6 Signaling Downstream from the β-Chain Cytokines in EOS
Along with RL13A (74), RPS6 is one of the rare ribosomal proteins that is phosphorylated following cellular stimulation in eukaryotic cells (75, 76). In stromal cells, RPS6 phosphorylation is directly controlled by the kinases p70S6K1 and p70S6K2, downstream of mTOR (77). In knock-in mice, genetically modified at RPS6 phospho-sites, aggregate protein synthesis was decreased in liver and embryonic fibroblasts (78). Other studies have suggested that RPS6 phosphorylation facilitated more efficient 40S ribosomal subunit assembly (79). This idea is supported by structural and biochemical data demonstrating that phosphorylated RPS6 is located at the interface between the small and the large ribosomal subunits near the tRNA-binding sites (80), and is enriched in polyribosomes (75). The correlation of RPS6 phosphorylation with cell division during mitogenic activation suggests that RPS6 participates in translation control in dividing cells (81). However, the role of phosphorylated RPS6 in non-dividing cells, such as EOS, remains unexplored.
We found that all β-chain cytokines strongly induced RPS6 phosphorylation at Ser235 and Ser236. However, while RPS6 phosphorylation persisted for only 1–4 h in EOS culture with IL-5 or GM-CSF, IL-3 induced continuous RPS6 phosphorylation for as long as IL-3 remained present in the culture medium (22). Of note, this unique feature of IL-3 to prolong RPS6 phosphorylation has also been observed in basophils (82, 83). Anti-IL-3 neutralization rapidly reversed RPS6 phosphorylation indicating that constant presence of IL-3 was required and that signaling was likely driven by a labile secondary messenger following IL-3 activation (22). Interestingly, the relatively rapid RPS6 dephosphorylation in GM-CSF-activated EOS was phosphatase 1 (PP1)-dependent, although total PP1 activity in cell lysates was the same in GM-CSF- and IL-3-activated EOS (22). This suggests that PP1 activity toward RPS6 may be negatively regulated only in IL-3-activated but not in GM-CSF-activated EOS. Of note, a 23 KDa ribosomal-associated inhibitor of PP1, termed ribosomal-associated inhibitor of phosphatase 1 (RIPP1) has been identified but remains incompletely described (84, 85).
As mentioned above, RPS6 can be phosphorylated downstream of the PI3K/Akt/mTOR/p70S6K pathway (77). However, in EOS neither rapamycin, PI3K nor p70S6K inhibitors prevented IL-3-induced RPS6 phosphorylation (22). On the contrary, p90S6K (RSK) inhibitors significantly reduced IL-3-induced, RPS6 phosphorylation on both Ser235 and Ser236 (22). GM-CSF activation of p90S6K peaked after 10 min, and p90S6K was already largely dephosphorylated by 1 h (22). Conversely, progressive phosphorylation of p90S6K occurred after IL-3, peaking, at 16–20 h and still detectable until IL-3 was removed or neutralized in the culture medium (22). P90S6K was the first RPS6-phosphorylating kinase described in Xenopus oocytes (86), but has since been implicated in cell proliferation and survival (87). P90S6K includes three isoforms (RSK1, 2, and 3), all with inducible phosphorylation-dependent activity and similar functions. P90S6K phosphorylation is downstream of ERK and phosphorylated p90S6K has been found associated with polyribosomes (88). Phosphorylation on Thr573 is sequentially followed by Thr359, Ser363, and finally Ser380. All four sites are strongly phosphorylated following IL-3-activated EOS (22). Ultimately 3′–phosphoinositol-dependent kinase-1 (PDK1) phosphorylates Ser221 leading to maximal p90S6K activation (89). In addition to RPS6, p90S6K also phosphorylates eIF4B and GSK3 (12, 90). Phosphorylated eIF4B interacts with eIF3A, enhancing translation initiation (91). P90S6K inactivates GSK3, which would in turn dephosphorylate and activate eIF2B, thus promoting eIF2 recycling and increasing translation initiation [(90); Figure 2]. While the dephosphorylation of eIF2B possibly occurs via changes in PP1 activity (90), differential activation of p90S6K by the different β-chain cytokines was not accompanied by changes in PP1 activity (22), suggesting that IL-3-induced and prolonged p90S6K activation does not affect translation via the GSK3/PP1/eIF2B pathway. As proposed above, the β-chain cytokines could differentially regulate a ribosomal specific PP1 regulatory protein (85).
Upstream, p90S6K phosphorylation is known to be regulated by the MAPK and particularly by ERK1/2 (92). Consistent with these data, a selective inhibitor of both MEK1 and MEK2 (U0126) added 3 h after IL-3, blocked the phosphorylation of p90S6K on Ser380 and RPS6 in EOS in culture. Another MAPK, p38, has also been implicated as a potential activator of p90S6K in dendritic cells (93). However, a p38 inhibitor (SB203580) had no effect on p90S6K phosphorylation in IL-3-activated EOS.
In addition to semaphorin-7A, we have more recently shown that the low-affinity IgG receptors, FCGR2B and FCGR2C (CD32B and CD32C) were upregulated at the translational level by IL-3, in a p90S6K-dependent manner (94). Therefore, we have so far identified two transcripts whose translation is exclusively enhanced by the prolonged effect of IL-3 through ERK/p90S6K signaling. MS proteomic analysis of EOS treated with IL-3 with and without ERK inhibitors will yield insight into the identity of other similarly regulated mRNA.
mRNA-Binding Proteins and Control of Protein Translation
Overview
RNA-binding proteins (RBP) regulate all aspects of RNA metabolism, including biogenesis, cellular localization and transport, stability, and translation. With the emergence of high throughput screening and quantitative proteomics, several hundred (approximately 500) potential RBP have been identified (95). Given their obvious importance, enormous effort has been directed to expand our knowledge on how RNA-protein interactions determine RNA function and cell fate. It bears reiterating that mRNA is not a rod but a complex 3-dimensional shape. As such, RBP can interact with mRNA via structure, sequence or structure, and sequence elements. A simple example is 5′-cap binding protein eIF4E. A more complex example is PABP, which interacts with poly A tails, a combination of sequence (Poly A) and structure. The iron-response binding protein (IRE-BP) interacts with a sequence presented on a stem-loop and bulge (96). Alterations in the size of the loop, the distance between the loop and bulge or the loop sequence ablates binding. Given these levels of target specificity, some RBPs will no doubt be successfully targeted with therapeutics to treat human disease.
Once transcribed from genomic DNA, newly produced pre-mRNAs are immediately covered by a number of nuclear RBP to protect from degradation by nucleases, guide splicing and prepare for cytoplasmic transport. As mature mRNA are translocated, the inventory of bound proteins are often replaced with a new set of RBP that determine intracellular location, define degradation rates as well as translatability (see above) in the cytoplasm. In response to extrinsic and intrinsic stimuli, free and bound RBP are subject to post-translational modifications (PTM) (e.g., phosphorylation, ubiquitination, acetylation, and methylation) that may induce conformational changes and alter the association between RBP and target mRNA (97, 98). Depending on stimulus and cell type, the modified RBP may associate with or dissociate from mRNA, affecting the transcript stability as well as its translation, clearly affecting protein production. RBP bind to RNA via a variety of domains, including the so-called RNA-recognition motif (RRM), zinc finger motives, K-homology domains (KH), RGG boxes, and DEAD/DEAH boxes (97). Often more than one binding domain are present allowing simultaneous interactions with multiple mRNAs, multiple sites within one mRNA target or between specific mRNA sequences and organelles such as ribosomes or stress granules. RBP can also form higher order structures through protein–protein interactions either as homo- or heterodimers/trimers, etc. As a rule, RBP that interact with 5′ or 3′ ends of mRNA often regulate translation initiation (e.g., translation initiation factors and their partners; PABP) while those that bind to coding regions can affect translation, localization, or mRNA decay (e.g., IRE-BP). 3′ UTR RBP (e.g., AUF1, HuR, TTP, TIA-1, TIAR, FMRP, PTB, KSRP, hnRNPs, nucleolin, and CUGBP) are most often involved in mRNA localization and decay (99).
Regulation of mRNA Binding Proteins in EOS and Their Potential Effect on Protein Translation
It is well known that many rapidly inducible mRNA coding for pro-inflammatory cytokines and oncoproteins are very short-lived. Inevitably, these mRNA contain cis-acting sequences into their 3′-UTR (100). The best-characterized instability determinant is composed of adenosine-uridine (AU)-rich element (ARE) repeats that are found in 3′-UTR of GM-CSF, IL-3, IL-5, IL-2, IFN-γ, and TNF-α and other cytokine mRNA. The life-span of ARE mRNA are regulated by a subset of binding proteins (AUBPs) that preferentially target the ARE and stabilize or further destabilize the transcripts. To date, approximately 20 AUBP have been identified. EOS express 7 AUBP (AUF1, hnRNP C, YB-1, nucleolin, TIA-1, HuR, and BRF1) (3) and their role in the regulation of mRNA stability has been demonstrated by many studies (101–105). In response to an exogenous pro-survival signal, Y-box binding protein 1 (YB1) and heterogeneous nuclear ribonucleoprotein C (hnRNP C) became associated with, while heterogeneous nuclear ribonucleoprotein D (hnRNP D or AUF1) dissociated from the ARE of GM-CSF mRNA (101, 106). These interactions were accompanied by the multiple phosphorylation of AUF1 (Ser83, Ser87, and Thr91) likely by ERK, CK1, GSK3β, or PKA (103, 107–109). Presumably, phosphorylation reduced the affinity of AUF1 for the ARE. AUF1 also undergoes post-transcriptional, alternative splicing events (110), yielding four AUF1 mRNAs and isoform variants (p37, p40, p42, and p45), all of which are detectable in human EOS (106). Thus, the regulatory control by AUF1 isoforms appears to be highly complex and includes their potential to form heterodimers (111) with a different affinity for ARE containing mRNAs (p37 > p42 > p45 > p40) (112, 113). While AUF1 has additional functions, its best-characterized function is to accelerate the decay of associated ARE-rich mRNAs. The p37 isoform has been shown to interact with the exosome in EOS (103) and exhibit the greatest destabilizing activity toward ARE-containing mRNAs compared to other isoforms (114).
mRNA turnover is often linked to translation (115) and the role of AUBP in RNA translation has been extensively studied in many systems. Similar mechanisms may occur in EOS although no direct evidence has yet been published. As mentioned above, PI3K/Akt/mTOR and MAPK cascades are the major signaling pathways that control global RNA translation upstream of the ribosomal machinery. These pathways have also been linked to AUBP-mediated mRNA decay in many cell types. EOS possess all translational machinery (Table 1) and can activate those kinase pathways when stimulated with various agonists (fMLP, RANTES, eotaxin, IL-5, IL-3, and PGE2) (30, 57). For example, ERK is activated by hyaluronic acid, IL-3 or IL-5, and likely drives AUF1 phosphorylation (102, 103), impacting the translation and decay of multiple ARE mRNA, including GM-CSF. These data suggest that ARE mRNA will be subject to translation control in EOS. This has been investigated by analysis of transfected mRNA, which has revealed striking differences in protein production despite similar cytosolic steady state levels of coding mRNA (116). Below, we discuss well-defined AUBP and their potential roles in target mRNA translation in EOS.
Heterogeneous Nuclear Ribonucleoprotein D
In eukaryotic cells, AUF1 (hnRNP D) is one of the best-characterized AUBPs and has a multiplicity of functions. It is a positive regulator of mRNA translation (117) but can also accelerate transcript decay. These two events may or may not be coupled. For example, AUF1 weakly targets Myc mRNAs for an accelerated decay but strongly promotes its translation by successfully competing with the cytotoxic granule-associated RNA binding protein TIA-1 and TIA-1-like 1 (TIAR) for a common binding site (118). Consistent with this observation, cells lacking AUF1 exhibited an increase binding of the translation-inhibitory TIA-1/TIAR to ARE mRNA, resulting in translation repression of the mRNA encoding TGF-β-activated kinase 1 (TAK1) and IL-10 (119, 120). Depending on the cell and its activation state, AUF1 can also assemble factors necessary for mRNA translation, including eIF4G, chaperones (hsp27 and hsp70), and PABP, thereby affecting translation (121–123). In EOS, eIF4G is phosphorylated by a brief (5 min) exposure to IL-5 (3), a condition that favors AUF1 phosphorylation and global protein translation (22). As EOS express high levels of PABP-C1 (major cytoplasmic PABP isoform) (3, 124), activated AUF1 may facilitate the displacement of TIA-1/TIAR by PABP-C1 and promote phospho-eIF4G-mediated translation initiation. Taken together, these results strongly indicate that modulation of translation efficiency by AUF1 is a common cellular event, which may not necessarily couple with ARE-mediated decay. Interestingly, AUF1 can also function as an inhibitor as was reported in EV71 virus translation. In this model, AUF1 binding to a stem–loop structure within IRES displaced HuR and Ago2, whose association promotes IRES-dependent translation and subsequent viral replication.
Y-Box Binding Protein-1
In EOS, an increase in YB-1 content led to the stabilization of GM-CSF mRNA. Binding was mediated through 3′ UTR ARE and resulted in increased GM-CSF translation and release with subsequent pro-survival signaling (101, 102). YB-1 can also stabilize non-ARE containing mRNA (125, 126), suggesting that it associates with other cis-elements or acts through another protein effector(s). Consistent with this notion, as the YB-1/mRNA ratio increases, so does the translation efficiency of the affected mRNA (125, 126). At high YB-1/mRNA ratios associated with maximal mRNA stabilization, YB-1 displaces eIF4F from the messenger ribonucleoprotein (mRNP) complex, possibly inhibiting the translation of the stabilized mRNA (127). This mechanism was not observed in EOS for GM-CSF expression, however (101, 102), which may reflect the ordinarily high basal levels of YB-1 in these cells. Thus, it is not entirely clear how endogenous AUBP such as YB-1 influence eIF4F-mRNA interactions and regulate mRNA stability and translation in these cells. In cells, at low YB-1/mRNA ratios, eIF4F is known to bind effectively to mRNA near the 5′ cap-structure and drive translation (125, 126). YB-1 can be phosphorylated at a single site (Ser316) within the C-terminal domain (CTD) by multiple kinases (Akt, ERK2, GSK3-β, and JNK) (3), which leads to increased IL-2 mRNA stability and cytokine production (128). YB-1 binds to mRNA as a monomer through the cold-shock domain (CSD) and the CTD (125), which can unfold mRNA secondary structures, likely facilitating interactions with the translation initiation machinery. Inhibition of translation is mainly attributed to the CTD. Similarly to the full length YB-1, CTD displaces eIF4G from mRNP while the CSD displaces eIF4E, eIF4A, and eIF4B by interacting with the 5′-Cap-structure or with its adjacent region (125, 126). After EOS exposure to IL-5, eIF4G (Ser1238, Thr1218, and Ser1194) and eIF4B (Tyr233, Ser406, Ser359, and Ser459) are rapidly phosphorylated (3). YB-1 can also be phosphorylated by Akt (98, 129, 130), which lowers its affinity for the 5′-cap-structure (or/and adjacent mRNA region) (130). This may also facilitate the assembly of the translation initiation complex. Of note, circadian changes of YB-1 binding to GM-CSF mRNA have been observed in circulating EOS from subjects with nocturnal asthma, with lower YB-1/GM-CSF mRNA interaction at 04.00 a.m., suggesting possible increased GM-CSF protein production and EOS activation at night (131).
Heterogeneous Nuclear Ribonucleoprotein C
Heterogeneous nuclear ribonucleoprotein C has been predominantly associated with the regulation of mRNA stability although several reports describe translational regulation through 5′ UTR interactions (132–135). This function was first identified in rabbit reticulocyte lysate supplemented with exogenous hnRNP C. Those studies revealed hnRNP C bound to a non-ARE domain, stabilizing APP mRNA and increasing its translation (132). In neurons, hnRNP C and FMRP were shown to compete for binding to a coding region element of APP mRNA that modulated APP mRNA translation in opposite directions (136). Further study clarified that increased APP translation by hnRNP C was accompanied by enhanced mRNA polyadenylation, which was mediated by a functional IRES found in the 5′ UTR of the transcript (137). Thus, the mRNA-specific translational activation by hnRNP C is generally independent of ARE and is through interactions with distinct 3′ or coding region (132, 136) target sequences, IRES (133), 5′ UTR, or heptameric U sequence in IRES (138). Whether similar mechanisms occur in EOS is unknown although hnRNP C was reported to bind GM-CSF mRNA and associated with transcript stability (103). To date, neither cytoplasmic kinases nor phosphosites on hnRNP C have been identified although several RNA-dependent protein kinases (PKA, PKC, CDK-II, and PKR) have been associated with hyperphosphorylation of hnRNP C1 (small isoform of hnRNP C) in nuclear extracts (139).
Other AUBPs
HuR (stabilizer of ARE mRNA) and TIA-1 (U-rich binding protein) bind to GM-CSF and TGF-β mRNA and are associated with transcript stability in EOS. While the role of HuR in mRNA translation has not been reported, TIA-1 is believed to repress the translation of TNF-α (140), COX-2 (141), cytochrome c (142), and 5′ TOP mRNAs (143). TIA-1 binds to the ARE of TNF-α mRNA, but has no effect on the mRNA decay. Instead, TIA-1 represses TNF-α translation by promoting its sequestration in non-polysomal mRNP complexes or the so-called stress granules (144). TIA-1 can also recruit multifunctional RBP, including PTB, La, hnRNP K, and hnRNP A1, all of which are expressed by EOS (145). However, it remains unknown whether this recruitment is associated with TIA-1-mediated translational repression. TIA-1 can be phosphorylated by FASTK but the phosphorylation sites have not been mapped (146, 147). On a similar note, the mRNA stabilizing protein, Sjögren syndrome type B antigen (SSB or La) plays a unique role in translation initiation (148–151). La is largely nuclear but acts as an RNA chaperone in the cytoplasm when translation starts. La binds in close proximity to the translation start site and unwinds second structure of mRNA to expose embedded AUG start codons. Similar actions were also observed for the translation of virus-encoded mRNA (152–154). This unique feature of La is critically important in facilitating translation initiation because the translation start sites of certain mRNA are buried in strong stem–loop or secondary structures and are not efficiently recognized by the scanning 43S ribosomal subunit. La is phosphorylated on Thr301, Ser366, and Thr389 by AKT and CK2 (151, 155, 156), which contributes to its nuclear or cytoplasmic distribution (157).
Regulation of Translation and Potential Therapeutic Targets
Endogenous GM-CSF Effects on EOS Biology and the Use of Pin-1 As a Potential Therapeutic Target
We have described above how RBP regulate mRNA stability and translatability, particularly of GM-CSF mRNA in EOS. GM-CSF plays a pivotal role in the modulation of EOS differentiation, function, and survival. The cytokine is upregulated in eosinophilic diseases and a major contributor to enhancing EOS survival in the lungs of patients during active asthma (158). Recombinant GM-CSF promotes EOS survival about five times as potently as equal concentrations of IL-5 (159). In asthmatics, GM-CSF is produced by a wide spectrum of cell types, including lung epithelial cells, lymphocytes, alveolar macrophages, EOS, endothelial cells, and fibroblasts. As EOS typically increase by 20-fold in the lung within a few days of an allergen challenge (160), autocrine GM-CSF is an important source in order to support survival. The level of endogenous GM-CSF in BAL fluid is low (161–163) compared to IL-5 in both mice and humans (164). However, intranasal delivery of Adeno-GM-CSF to the airways of OVA-sensitized mice resulted in sustained accumulation of various inflammatory cell types, most noticeably EOS, in the lung for more than 2 weeks post OVA aerosol challenge (165, 166). Conversely, neutralization of endogenous GM-CSF during aeroallergen exposure significantly inhibited eosinophilic inflammation and airway hyper-responsiveness. This suggests that small amount of endogenous GM-CSF can significantly contribute to the development and persistence of eosinophilic airway inflammation. In vitro, purified peripheral blood EOS synthesize small amounts (~1 pg/ml) of anti-apoptotic GM-CSF (167, 168), after stimulation with a variety of factors (fibronectin, hyaluronic acid, TNF-α, IL-3, IL-5, IL-15, integrins, IFN-γ, calcium ionophore, cross-linking of cell surface molecules) (67, 102, 167, 169–174). Activation-induced survival was blocked by the addition of neutralizing anti-GM-CSF even 2 days after the initiation of culture, indicating that the cells continuously release low levels of GM-CSF on which survival depends (103, 172–174). Similarly, the majority of BAL EOS obtained 48 h after segmental allergen challenge died in vitro at 6 days in the presence of neutralizing anti-GM-CSF. Both in situ hybridization (tissue EOS) (175) and qPCR (purified EOS) (67, 172, 173) analyses have demonstrated that increased GM-CSF mRNA was associated with GM-CSF protein secretion and prolonged EOS survival.
Pin1
While all ARE mRNA have relatively short half-lives (20 min-2 h), GM-CSF mRNA is extremely labile (t1/2 < 6 min) in resting EOS but show significantly increased stability (increased by fourfold to sixfold) after cell activation. This conversion likely reflects alterations in the composition of interacting AUBP (67, 103, 172, 173). Multiple AUBP, including AUF1, HuR, YB-1, and hnRNP C associate with and regulate the decay of GM-CSF mRNA in EOS (101–103). Cell activation triggered occupancy of GM-CSF mRNA by YB1, hnRNP C, and HuR, which displaced AUF1. ERK-mediated phosphorylation likely caused a decrease in affinity for GM-CSF mRNA by AUF1 (103), which led to remodeling of the GM-CSF mRNP complex. Co-immunoprecipitation and gene knockout studies have found that Pin1, a cis-trans peptidyl prolyl isomerase, interacts with multiple AUBPs, including AUF1, HuR, KSRP, SLBP, and the translation regulators eIF4E and 4E-BP1/2 (103, 176–178). Pin1 is essential for cell-cycle progression through interactions with cyclinD (179). Pin1 is the only known eukaryotic isomerase with specificity for Ser-Pro or Thr-Pro peptide bonds. Isomerization is bidirectional with cis to trans or trans to cis conversions but occurs approximately 1000-fold faster when the N-terminal Ser or Thr has been phosphorylated (180). Structurally, Pin1 has two domains, including a ~40 amino acid N-terminal WW domain and a C-terminal isomerase domain. The WW domain binds pSer/pThr-Pro motifs while the catalytic domain is responsible for substrate isomerization. Pin1-mediated isomerization has profound effects on target-protein folding, altering subsequent protein–protein and protein–nucleic acid interactions, protein stability and subcellular localization thereby altering a variety of cellular processes, including cell cycle progression, apoptosis, innate and acquired immunity, and gene regulation. We showed that Pin1 was reproducibly pulled down with AUF1 in human EOS and T cells irrespective of cell activation (103, 105). Cell activation also increased Pin1 activity, which likely isomerized phosphorylated AUF1. These events occurred with a simultaneous reduction of AUF1 binding to GM-CSF mRNA. Conversely, inhibition of Pin1 reduced isomerase activity, reconstituted the AUF1–GM-CSF mRNP complex and accelerated transcript decay. Consistent with this in vitro data, EOS obtained from the blood or BALF from patients with active asthma showed significantly elevated Pin1 isomerase activity. In vivo Pin1 blockade significantly reduced pulmonary EOS counts, GM-CSF production, and cell viability in rat models of asthma (181). These observations indicate that Pin1 is a critical regulator of GM-CSF mRNA turnover and production, which in turn controls the survival of activated EOS in the lungs of asthmatics.
In addition to its role in mRNA stabilization, Pin1 signaling amplifies or suppresses the action of kinases, phosphatases, transcription factors, cell cycle regulators, and apoptotic effectors (124, 180). This broad targeting specificity of Pin1 arises from its short consensus target (pSer/pThr-Pro) as well as the phosphorylation frequency of S/T-P sites, which are found in numerous proteins. Pin1 activity can be modulated either positively or negatively without change in protein content, in response to injury or environmental cues. Chronic activation or suppression can be pathologic as seen in immune disorders, fibrosis, cancer, and neurodegeneration (105, 182, 183). Specifically, Pin1 overexpression or amplification is highly correlated with cancer progression and metastasis while Pin1 loss is seen in evolving Alzheimer Disease. Pathology may result from loss of regulation of RBP with alterations in cytokine mRNA stability and translation. Thus, pharmacologic modulation of Pin1 activity with small molecule inhibitors may provide a novel approach to eosinophilic diseases, such as asthma. Unfortunately, current Pin1 inhibitors lack specificity or are excessively toxic.
IL-3 Signaling in EOS
TPI ASM8 is a drug in development, targeting the common β-chain receptor for all IL-5, GM-CSF, and IL-3, in the form of RNA-targeted inhaled oligonucleotide antisense phosphorotioates (184). Although TPI ASM8 seems to be well tolerated and leads to some reduction of EOS and eosinophilic hematopoietic progenitor (CD34+IL5R+), other alternative therapeutic targets more specific to each of the 3 cytokine should be developed.
So far, we have identified semaphorin-7A and FCGR2B/C (CD32) as specific genes exclusively responding to IL-3 activation via prolonged ERK/p90S6K signaling. It is probable that additional genes are similarly regulated at a translation level by IL-3/ERK/p90S6K. Likely other candidate genes may share specific mRNA cis-elements whose identity may be inferred by homology searches among IL-3 upregulated mRNA. We started analyzing how semaphorin-7A or FCGR2 affect EOS function. We found that IL-3-activated EOS adhere to the only known semaphorin-7A ligand, plexin-C1 (27). Plexin-C1 is expressed by many cell types, including lymphocytes, monocytes, dendritic cells, and neutrophils (185), and has an important role in the migration of these inflammatory cells. Plexin-C1 is also expressed by stromal cells (186), which could facilitate migration or activate EOS in fibrotic tissue. Interestingly, IL-3-activated EOS migration on plexin-C1 was largely resistant to semaphorin-7A blockade while neutralizing anti-αMβ2 integrin were far more inhibitory (187). Migration in the absence of chemotaxis indicates that a haptotaxis process is operative for plexin-C1- or periostin-mediated migration (187, 188). Semphorin-7A signaling may also skew fibroblasts toward a pro-fibrotic, more mesenchymal phenotype (27, 189, 190), although we recently demonstrated anti-fibrotic functions for endogenous semaphorin-7A expressed by lung fibroblasts (191).
The upregulation of CD32 by IL-3 on EOS has a profound impact on EOS function. EOS-driven pathology in tissue requires both EOS migration from circulating blood to the site of inflammation and the release (degranulation) of preformed toxic proteins and mediators of the inflammation. IL-3-activated EOS strongly degranulate on heat-aggregated (HA)-IgG, with extrusion of ~25% of their total cellular eosinophil-derived neurotoxin (EDN) in 6 h (94) compared to less than 10% after IL-5 (94). Degranulation on HA-IgG was CD32-dependent (94). Thus, IL-3 and its downstream intracellular effectors may be potential therapeutic targets to limit EOS degranulation and EOS-driven pathologies. The use of anti-IL-5 therapies on patients with severe eosinophilic asthma has reduced asthma exacerbations and blood eosinophilia (192, 193), see other article by Nair in this issue. However, airway EOS are still present and active despite treatment (194, 195). This partially reflects loss of the surface IL-5 receptor expression by airway EOS (22, 196, 197). Conversely, IL-3 and the surface IL-3 receptor are upregulated and highly expressed on airway EOS (27, 198). Therefore, combined targeting of the IL-3 and IL-5 pathways may provide additive or synergistic benefits.
Ribosomal S6 Protein
Whether RPS6 phosphorylation in EOS induces a unique profile of proteins (e.g., semaphorin-7A, CD32, etc.), downstream of IL-3/ERK/P90S6K signaling, is unclear. If so, phospho-RPS6 would be a possible therapeutic target to reduce EOS-related pathology. On the positive side, knock-in mice lacking the ability to phosphorylate RPS6 have modest deficits (199) and show limited changes in global protein synthesis in vivo and in embryonic fibroblasts (78). However, β-cell development may be adversely affected by RPS6 knock-in (78). RPS6 phosphorylation can be blocked in EOS by small molecules inhibitors of p90S6K, such as BI-D1870 (200). However, the consequences of p90S6K inhibition probably include transcriptional silencing, blockade of cell proliferation, and cell death (87, 201). Thus, while potentially attractive, inhibition of this pathway remains hypothetical.
Conclusion
The three β-chain cytokines, IL-3, IL-5, and GM-CSF are all present in human eosinophilic diseases and have both highly redundant and yet critically unique roles in the EOS biology. Their signaling affects differentiation, maturation, survival, migration, piecemeal release of immune-mediators, and degranulation. IL-3 is unique among the β-chain cytokines in generating prolonged intracellular signaling leading to the translation of a subset of EOS mRNA. Signaling requires ERK and p90S6K activation and culminates in the phosphorylation of RPS6. The control of both translation and decay of cytokine mRNA ultimately involves an interplay between mRNA-BP, especially those that target ARE. The AUBP in turn are often regulated by the action of Pin1, leading to multi-level control over cytokine gene expression. Critical, unanswered questions include the identification of RPS6-dependent mRNA as well as additional Pin1 RBP interactors and whether drugs can be developed to target these important pathways.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest Statement
The authors declare no conflict of interest other than an issued patent on the use of Pin1 as a drug target to treat eosinophilia.
The reviewer KWG and the handling editor declared their shared affiliation, and the handling editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
The authors would like to thank Ksenija Bernau and Andrea Noll for reading and correcting the manuscript.
Funding
This work was supported by P01 HL088594 and Clinical and Translational Research Center grant UL1 RR025011 from the National Institutes of Health.
Abbreviations
aa, aminoacyl; AMPK, AMP-activated protein kinase; ARE, adenosine-uridine (AU)-rich element; AUBP, ARE binding protein; AUF1 or hnRNP D, heterogeneous nuclear ribonucleoprotein D; CDK1, cyclin-dependent kinase 1; CSNK2A1, casein kinase 2; EDN, eosinophil-derived neurotoxin; eEF, eukaryotic elongation factor; eIF, eukaryotic translation initiation factor; EOS, eosinophil; FCGRII (CD32), receptor for Fc fragment of IgG, low affinity II; GEF, guanine-nucleotide-exchange factor; GTP, guanosine triphosphate; HA-IgG, heat-aggregated-IgG; hnRNP C, heterogeneous nuclear ribonucleoprotein C; La or SSB, Sjögren syndrome type B antigen; MAPK, mitogen-activated protein kinase; mRNP, messenger ribonucleoprotein; mTOR, mammalian target of rapamycin; mTORC1, rapamycin complex 1; p90S6K (RSK), 90-KDa ribosomal S6 kinase; PA, phosphatidic acid; PABP, poly-A binding protein; PIC, 43S preinitiation complex; RBP, RNA-binding protein; RIPP1, ribosomal-associated inhibitor of phosphatase 1; RPS6, ribosomal protein S6; TIA-1, cytotoxic granule-associated RNA binding protein; TIAR, cytotoxic granule-associated RNA binding protein like 1; TOP, terminal oligopyrimidine 5′-UTR, 5′ untranslated region; YB-1, Y-Box Binding Protein-1.
References
1. Shen ZJ, Malter JS. Determinants of eosinophil survival and apoptotic cell death. Apoptosis (2015) 20(2):224–34. doi:10.1007/s10495-014-1072-2
2. Proud CG. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J (2007) 403(2):217–34. doi:10.1042/BJ20070024
3. Wilkerson EM, Johansson MW, Hebert AS, Westphall MS, Mathur SK, Jarjour NN, et al. The peripheral blood eosinophil proteome. J Proteome Res (2016) 15(5):1524–33. doi:10.1021/acs.jproteome.6b00006
4. Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol (2010) 11(2):113–27. doi:10.1038/nrm2838
5. Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science (2016) 352(6292):1413–6. doi:10.1126/science.aad9868
6. Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev (2011) 75(3):434–67, first page of table of contents. doi:10.1128/MMBR.00008-11
7. Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem (1999) 68:913–63. doi:10.1146/annurev.biochem.68.1.913
8. Qin X, Jiang B, Zhang Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle (2016) 15(6):781–6. doi:10.1080/15384101.2016.1151581
9. Heesom KJ, Gampel A, Mellor H, Denton RM. Cell cycle-dependent phosphorylation of the translational repressor eIF-4E binding protein-1 (4E-BP1). Curr Biol (2001) 11(17):1374–9. doi:10.1016/S0960-9822(01)00422-5
10. Joshi B, Cai AL, Keiper BD, Minich WB, Mendez R, Beach CM, et al. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem (1995) 270(24):14597–603. doi:10.1074/jbc.270.24.14597
11. Scheper GC, van Kollenburg B, Hu J, Luo Y, Goss DJ, Proud CG. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem (2002) 277(5):3303–9. doi:10.1074/jbc.M103607200
12. Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J (2006) 25(12):2781–91. doi:10.1038/sj.emboj.7601166
13. Wang X, Paulin FE, Campbell LE, Gomez E, O’Brien K, Morrice N, et al. Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon-subunit and their functions in vivo. EMBO J (2001) 20(16):4349–59. doi:10.1093/emboj/20.16.4349
14. Joshi M, Kulkarni A, Pal JK. Small molecule modulators of eukaryotic initiation factor 2alpha kinases, the key regulators of protein synthesis. Biochimie (2013) 95(11):1980–90. doi:10.1016/j.biochi.2013.07.030
15. Minella O, Mulner-Lorillon O, Bec G, Cormier P, Belle R. Multiple phosphorylation sites and quaternary organization of guanine-nucleotide exchange complex of elongation factor-1 (EF-1betagammadelta/ValRS) control the various functions of EF-1alpha. Biosci Rep (1998) 18(3):119–27. doi:10.1023/A:1020140527930
16. Ryazanov AG, Davydova EK. Mechanism of elongation factor 2 (EF-2) inactivation upon phosphorylation. Phosphorylated EF-2 is unable to catalyze translocation. FEBS Lett (1989) 251(1–2):187–90. doi:10.1016/0014-5793(89)81452-8
17. Hizli AA, Chi Y, Swanger J, Carter JH, Liao Y, Welcker M, et al. Phosphorylation of eukaryotic elongation factor 2 (eEF2) by cyclin A-cyclin-dependent kinase 2 regulates its inhibition by eEF2 kinase. Mol Cell Biol (2013) 33(3):596–604. doi:10.1128/MCB.01270-12
18. Kenney JW, Moore CE, Wang X, Proud CG. Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Adv Biol Regul (2014) 55:15–27. doi:10.1016/j.jbior.2014.04.003
19. Hannan KM, Sanij E, Hein N, Hannan RD, Pearson RB. Signaling to the ribosome in cancer – it is more than just mTORC1. IUBMB Life (2011) 63(2):79–85. doi:10.1002/iub.428
20. Diggle TA, Subkhankulova T, Lilley KS, Shikotra N, Willis AE, Redpath NT. Phosphorylation of elongation factor-2 kinase on serine 499 by cAMP-dependent protein kinase induces Ca2+/calmodulin-independent activity. Biochem J (2001) 353(Pt 3):621–6. doi:10.1042/bj3530621
21. Shen ZJ, Hu J, Esnault S, Dozmorov I, Malter JS. RNA Seq profiling reveals a novel expression pattern of TGF-beta target genes in human blood eosinophils. Immunol Lett (2015) 167(1):1–10. doi:10.1016/j.imlet.2015.06.012
22. Esnault S, Kelly EA, Shen ZJ, Johansson MW, Malter JS, Jarjour NN. IL-3 maintains activation of the p90S6K/RPS6 pathway and increases translation in human eosinophils. J Immunol (2015) 195(6):2529–39. doi:10.4049/jimmunol.1500871
23. Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J (2001) 20(16):4370–9. doi:10.1093/emboj/20.16.4370
24. Clutterbuck EJ, Sanderson CJ. Regulation of human eosinophil precursor production by cytokines: a comparison of recombinant human interleukin-1 (rhIL-1), rhIL-3, rhIL-5, rhIL-6, and rh granulocyte-macrophage colony-stimulating factor. Blood (1990) 75(9):1774–9.
25. Blom M, Tool AT, Kok PT, Koenderman L, Roos D, Verhoeven AJ. Granulocyte-macrophage colony-stimulating factor, interleukin-3 (IL-3), and IL-5 greatly enhance the interaction of human eosinophils with opsonized particles by changing the affinity of complement receptor type 3. Blood (1994) 83(10):2978–84.
26. Kelly EA, Liu LY, Esnault S, Quinchia Johnson BH, Jarjour NN. Potent synergistic effect of IL-3 and TNF on matrix metalloproteinase 9 generation by human eosinophils. Cytokine (2012) 58(2):199–206. doi:10.1016/j.cyto.2012.01.009
27. Esnault S, Kelly EA, Johansson MW, Liu LY, Han S-H, Akhtar M, et al. Semaphorin 7A is expressed on airway eosinophils and upregulated by IL-5 family cytokines. Clin Immunol (2014) 150(1):90–100. doi:10.1016/j.clim.2013.11.009
28. Kelly EA, Esnault S, Johnson SH, Liu LY, Malter JS, Burnham ME, et al. Human eosinophil activin A synthesis and mRNA stabilization are induced by the combination of IL-3 plus TNF. Immunol Cell Biol (2016) 94(7):701–8. doi:10.1038/icb.2016.30
29. Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol (2015) 25(9):545–55. doi:10.1016/j.tcb.2015.06.002
30. Sturm EM, Parzmair GP, Radnai B, Frei RB, Sturm GJ, Hammer A, et al. Phosphoinositide-dependent protein kinase 1 (PDK1) mediates potent inhibitory effects on eosinophils. Eur J Immunol (2015) 45(5):1548–59. doi:10.1002/eji.201445196
31. Wang X, Janmaat M, Beugnet A, Paulin FE, Proud CG. Evidence that the dephosphorylation of Ser(535) in the epsilon-subunit of eukaryotic initiation factor (eIF) 2B is insufficient for the activation of eIF2B by insulin. Biochem J (2002) 367(Pt 2):475–81. doi:10.1042/BJ20020677
32. Manning BD, Cantley LC. United at last: the tuberous sclerosis complex gene products connect the phosphoinositide 3-kinase/Akt pathway to mammalian target of rapamycin (mTOR) signalling. Biochem Soc Trans (2003) 31(Pt 3):573–8. doi:10.1042/bst0310573
33. Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol (2005) 15(8):702–13. doi:10.1016/j.cub.2005.02.053
34. Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science (2006) 314(5798):467–71. doi:10.1126/science.1130276
35. Huo Y, Iadevaia V, Proud CG. Differing effects of rapamycin and mTOR kinase inhibitors on protein synthesis. Biochem Soc Trans (2011) 39(2):446–50. doi:10.1042/BST0390446
36. Tokunaga C, Yoshino K, Yonezawa K. mTOR integrates amino acid- and energy-sensing pathways. Biochem Biophys Res Commun (2004) 313(2):443–6. doi:10.1016/j.bbrc.2003.07.019
37. Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci U S A (1995) 92(11):4947–51. doi:10.1073/pnas.92.11.4947
38. Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer (2006) 6(9):729–34. doi:10.1038/nrc1974
39. Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature (2013) 497(7448):217–23. doi:10.1038/nature12122
40. Choo AY, Blenis J. Not all substrates are treated equally: implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle (2009) 8(4):567–72. doi:10.4161/cc.8.4.7659
41. Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell (2006) 22(2):159–68. doi:10.1016/j.molcel.2006.03.029
42. Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol Cell Biol (2009) 29(6):1411–20. doi:10.1128/MCB.00782-08
43. Foster DA. Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol Metab (2013) 24(6):272–8. doi:10.1016/j.tem.2013.02.003
44. Chen Y, Zheng Y, Foster DA. Phospholipase D confers rapamycin resistance in human breast cancer cells. Oncogene (2003) 22(25):3937–42. doi:10.1038/sj.onc.1206565
45. Yellen P, Saqcena M, Salloum D, Feng J, Preda A, Xu L, et al. High-dose rapamycin induces apoptosis in human cancer cells by dissociating mTOR complex 1 and suppressing phosphorylation of 4E-BP1. Cell Cycle (2011) 10(22):3948–56. doi:10.4161/cc.10.22.18124
46. Hom JT, Estridge T. FK506 and rapamycin modulate the functional activities of human peripheral blood eosinophils. Clin Immunol Immunopathol (1993) 68:293–300. doi:10.1006/clin.1993.1130
47. Dumont FJ, Melino MR, Staruch MJ, Koprak SL, Fischer PA, Sigal NH. The immunosuppressive macrolides FK-506 and rapamycin act as reciprocal antagonists in murine T cells. J Immunol (1990) 144(4):1418–24.
48. Hatfield SM, Mynderse JS, Roehm NW. Rapamycin and FK506 differentially inhibit mast cell cytokine production and cytokine-induced proliferation and act as reciprocal antagonists. J Pharmacol Exp Ther (1992) 261(3):970–6.
49. Meng Q, Ying S, Corrigan CJ, Wakelin M, Assoufi B, Moqbel R, et al. Effects of rapamycin, cyclosporin A, and dexamethasone on interleukin 5-induced eosinophil degranulation and prolonged survival. Allergy (1997) 52(11):1095–101. doi:10.1111/j.1398-9995.1997.tb00181.x
50. Hua W, Liu H, Xia LX, Tian BP, Huang HQ, Chen ZY, et al. Rapamycin inhibition of eosinophil differentiation attenuates allergic airway inflammation in mice. Respirology (2015) 20(7):1055–65. doi:10.1111/resp.12554
51. Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol (2011) 12(4):295–303. doi:10.1038/ni.2005
52. Mushaben EM, Brandt EB, Hershey GK, Le Cras TD. Differential effects of rapamycin and dexamethasone in mouse models of established allergic asthma. PLoS One (2013) 8(1):e54426. doi:10.1371/journal.pone.0054426
53. Shamri R, Young KM, Weller PF. PI3K, ERK, p38 MAPK and integrins regulate CCR3-mediated secretion of mouse and human eosinophil-associated RNases. Allergy (2013) 68(7):880–9. doi:10.1111/all.12163
54. Mishra RK, Scaife JE, Harb Z, Gray BC, Djukanovic R, Dent G. Differential dependence of eosinophil chemotactic responses on phosphoinositide 3-kinase (PI3K). Allergy (2005) 60(9):1204–7. doi:10.1111/j.1398-9995.2005.00845.x
55. Sano M, Leff AR, Myou S, Boetticher E, Meliton AY, Learoyd J, et al. Regulation of interleukin-5-induced beta2-integrin adhesion of human eosinophils by phosphoinositide 3-kinase. Am J Respir Cell Mol Biol (2005) 33(1):65–70. doi:10.1165/rcmb.2005-0076OC
56. Palframan RT, Collins PD, Severs NJ, Rothery S, Williams TJ, Rankin SM. Mechanisms of acute eosinophil mobilization from the bone marrow stimulated by interleukin 5: the role of specific adhesion molecules and phosphatidylinositol 3-kinase. J Exp Med (1998) 188(9):1621–32. doi:10.1084/jem.188.9.1621
57. Zhu Y, Bertics PJ. Chemoattractant-induced signaling via the Ras-ERK and PI3K-Akt networks, along with leukotriene C4 release, is dependent on the tyrosine kinase Lyn in IL-5- and IL-3-primed human blood eosinophils. J Immunol (2011) 186(1):516–26. doi:10.4049/jimmunol.1000955
58. Lake D, Correa SA, Muller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol Life Sci (2016) 73(23):4397–413. doi:10.1007/s00018-016-2297-8
59. Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors (2006) 24(1):21–44. doi:10.1080/02699050500284218
60. Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A (2004) 101(37):13489–94. doi:10.1073/pnas.0405659101
61. Rolfe M, McLeod LE, Pratt PF, Proud CG. Activation of protein synthesis in cardiomyocytes by the hypertrophic agent phenylephrine requires the activation of ERK and involves phosphorylation of tuberous sclerosis complex 2 (TSC2). Biochem J (2005) 388(Pt 3):973–84. doi:10.1042/BJ20041888
62. Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell (2005) 121(2):179–93. doi:10.1016/j.cell.2005.02.031
63. Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol (2004) 24(15):6539–49. doi:10.1128/MCB.24.15.6539-6549.2004
64. Coffer PJ, Schweizer RC, Dubois GR, Maikoe T, Lammers JW, Koenderman L. Analysis of signal transduction pathways in human eosinophils activated by chemoattractants and the T-helper 2-derived cytokines interleukin-4 and interleukin-5. Blood (1998) 91(7):2547–57.
65. Bates ME, Green VL, Bertics PJ. ERK1 and ERK2 activation by chemotactic factors in human eosinophils is interleukin 5-dependent and contributes to leukotriene C(4) biosynthesis. J Biol Chem (2000) 275(15):10968–75. doi:10.1074/jbc.275.15.10968
66. Kampen GT, Stafford S, Adachi T, Jinquan T, Quan S, Grant JA, et al. Eotaxin induces degranulation and chemotaxis of eosinophils through the activation of ERK2 and p38 mitogen-activated protein kinases. Blood (2000) 95(6):1911–7.
67. Esnault S, Malter JS. Extracellular signal-regulated kinase mediates granulocyte-macrophage colony-stimulating factor messenger RNA stabilization in tumor necrosis factor-alpha plus fibronectin-activated peripheral blood eosinophils. Blood (2002) 99(11):4048–52. doi:10.1182/blood.V99.11.4048
68. Zhu X, Jacobs B, Boetticher E, Myou S, Meliton A, Sano H, et al. IL-5-induced integrin adhesion of human eosinophils caused by ERK1/2-mediated activation of cPLA2. J Leukoc Biol (2002) 72(5):1046–53.
69. Langlois A, Chouinard F, Flamand N, Ferland C, Rola-Pleszczynski M, Laviolette M. Crucial implication of protein kinase C (PKC)-delta, PKC-zeta, ERK-1/2, and p38 MAPK in migration of human asthmatic eosinophils. J Leukoc Biol (2009) 85(4):656–63. doi:10.1189/jlb.0808492
70. Bates ME, Sedgwick JB, Zhu Y, Liu LY, Heuser RG, Jarjour NN, et al. Human airway eosinophils respond to chemoattractants with greater eosinophil-derived neurotoxin release, adherence to fibronectin, and activation of the Ras-ERK pathway when compared with blood eosinophils. J Immunol (2010) 184(12):7125–33. doi:10.4049/jimmunol.0900634
71. Burnham ME, Esnault S, Roti Roti EC, Bates ME, Bertics PJ, Denlinger LC. Cholesterol selectively regulates IL-5 induced mitogen activated protein kinase signaling in human eosinophils. PLoS One (2014) 9(8):e103122. doi:10.1371/journal.pone.0103122
72. Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci U S A (1994) 91(10):4441–5. doi:10.1073/pnas.91.10.4441
73. Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature (2012) 485(7396):109–13. doi:10.1038/nature11083
74. Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell (2003) 115(2):187–98. doi:10.1016/S0092-8674(03)00773-6
75. Thomas G, Martin-Perez J, Siegmann M, Otto AM. The effect of serum, EGF, PGF2 alpha and insulin on S6 phosphorylation and the initiation of protein and DNA synthesis. Cell (1982) 30(1):235–42. doi:10.1016/0092-8674(82)90029-0
76. Evans SW, Farrar WL. Interleukin 2 and diacylglycerol stimulate phosphorylation of 40 S ribosomal S6 protein. Correlation with increased protein synthesis and S6 kinase activation. J Biol Chem (1987) 262(10):4624–30.
77. Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell (1992) 69(7):1227–36. doi:10.1016/0092-8674(92)90643-Q
78. Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev (2005) 19(18):2199–211. doi:10.1101/gad.351605
79. Duncan R, McConkey EH. Preferential utilization of phosphorylated 40-S ribosomal subunits during initiation complex formation. Eur J Biochem (1982) 123(3):535–8. doi:10.1111/j.1432-1033.1982.tb06564.x
80. Nygard O, Nika H. Identification by RNA-protein cross-linking of ribosomal proteins located at the interface between the small and the large subunits of mammalian ribosomes. EMBO J (1982) 1(3):357–62.
81. Bandi HR, Ferrari S, Krieg J, Meyer HE, Thomas G. Identification of 40 S ribosomal protein S6 phosphorylation sites in Swiss mouse 3T3 fibroblasts stimulated with serum. J Biol Chem (1993) 268(6):4530–3.
82. Esnault S. The neglected of eosinophil biology, IL-3 finds sustenance in the basophil. J Leukoc Biol (2017) 101(3):615–6. doi:10.1189/jlb.3LT0916-383R
83. Kampfer SS, Odermatt A, Dahinden CA, Fux M. Late IL-3-induced phenotypic and functional alterations in human basophils require continuous IL-3 receptor signaling. J Leukoc Biol (2017) 101(1):227–38. doi:10.1189/jlb.2A0715-292RR
84. Beullens M, Stalmans W, Bollen M. Characterization of a ribosomal inhibitory polypeptide of protein phosphatase-1 from rat liver. Eur J Biochem (1996) 239(1):183–9. doi:10.1111/j.1432-1033.1996.0183u.x
85. Cohen PT. Protein phosphatase 1 – targeted in many directions. J Cell Sci (2002) 115(Pt 2):241–56.
86. Erikson E, Maller JL. Substrate specificity of ribosomal protein S6 kinase II from Xenopus eggs. Second Messengers Phosphoproteins (1988) 12(2–3):135–43.
87. Lara R, Seckl MJ, Pardo OE. The p90 RSK family members: common functions and isoform specificity. Cancer Res (2013) 73(17):5301–8. doi:10.1158/0008-5472.CAN-12-4448
88. Angenstein F, Greenough WT, Weiler IJ. Metabotropic glutamate receptor-initiated translocation of protein kinase p90rsk to polyribosomes: a possible factor regulating synaptic protein synthesis. Proc Natl Acad Sci U S A (1998) 95(25):15078–83. doi:10.1073/pnas.95.25.15078
89. Jensen CJ, Buch MB, Krag TO, Hemmings BA, Gammeltoft S, Frodin M. 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J Biol Chem (1999) 274(38):27168–76. doi:10.1074/jbc.274.38.27168
90. Quevedo C, Alcazar A, Salinas M. Two different signal transduction pathways are implicated in the regulation of initiation factor 2B activity in insulin-like growth factor-1-stimulated neuronal cells. J Biol Chem (2000) 275(25):19192–7. doi:10.1074/jbc.M000238200
91. Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell (2005) 123(4):569–80. doi:10.1016/j.cell.2005.10.024
92. Chen RH, Sarnecki C, Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol Cell Biol (1992) 12(3):915–27. doi:10.1128/MCB.12.3.915
93. Zaru R, Ronkina N, Gaestel M, Arthur JS, Watts C. The MAPK-activated kinase Rsk controls an acute toll-like receptor signaling response in dendritic cells and is activated through two distinct pathways. Nat Immunol (2007) 8(11):1227–35. doi:10.1038/ni1517
94. Esnault S, Johansson MW, Kelly EA, Koenderman L, Mosher DF, Jarjour NN. IL-3 up-regulates and activates human eosinophil CD32 and alphaMbeta2 integrin causing degranulation. Clin Exp Allergy (2017) 47(4):488–98. doi:10.1111/cea.12876
95. Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell (2012) 46(5):674–90. doi:10.1016/j.molcel.2012.05.021
96. Haile DJ, Rouault TA, Tang CK, Chin J, Harford JB, Klausner RD. Reciprocal control of RNA-binding and aconitase activity in the regulation of the iron-responsive element binding protein: role of the iron-sulfur cluster. Proc Natl Acad Sci U S A (1992) 89(16):7536–40. doi:10.1073/pnas.89.16.7536
97. Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett (2008) 582(14):1977–86. doi:10.1016/j.febslet.2008.03.004
98. Shen ZJ, Malter JS. Regulation of AU-rich element RNA binding proteins by phosphorylation and the prolyl isomerase Pin1. Biomolecules (2015) 5(2):412–34. doi:10.3390/biom5020412
99. Pichon X, Wilson LA, Stoneley M, Bastide A, King HA, Somers J, et al. RNA binding protein/RNA element interactions and the control of translation. Curr Protein Pept Sci (2012) 13(4):294–304. doi:10.2174/138920312801619475
100. Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene (2012) 500(1):10–21. doi:10.1016/j.gene.2012.03.021
101. Capowski EE, Esnault S, Bhattacharya S, Malter JS. Y box-binding factor promotes eosinophil survival by stabilizing granulocyte-macrophage colony-stimulating factor mRNA. J Immunol (2001) 167(10):5970–6. doi:10.4049/jimmunol.167.10.5970
102. Esnault S, Malter JS. Hyaluronic acid or TNF-alpha plus fibronectin triggers granulocyte macrophage-colony-stimulating factor mRNA stabilization in eosinophils yet engages differential intracellular pathways and mRNA binding proteins. J Immunol (2003) 171(12):6780–7. doi:10.4049/jimmunol.171.12.6780
103. Shen ZJ, Esnault S, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat Immunol (2005) 6(12):1280–7. doi:10.1038/ni1266
104. Esnault S, Shen ZJ, Whitesel E, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates granulocyte-macrophage colony-stimulating factor mRNA stability in T lymphocytes. J Immunol (2006) 177(10):6999–7006. doi:10.4049/jimmunol.177.10.6999
105. Esnault S, Shen ZJ, Malter JS. Pinning down signaling in the immune system: the role of the peptidyl-prolyl isomerase Pin1 in immune cell function. Crit Rev Immunol (2008) 28(1):45–60. doi:10.1615/CritRevImmunol.v28.i1.30
106. Shen ZJ, Esnault S, Schinzel A, Borner C, Malter JS. The peptidyl-prolyl isomerase Pin1 facilitates cytokine-induced survival of eosinophils by suppressing Bax activation. Nat Immunol (2009) 10(3):257–65. doi:10.1038/ni.1697
107. Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, et al. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol (1993) 13(12):7652–65. doi:10.1128/MCB.13.12.7652
108. Kreegipuu A, Blom N, Brunak S. PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res (1999) 27(1):237–9. doi:10.1093/nar/27.1.237
109. Wilson GM, Lu J, Sutphen K, Suarez Y, Sinha S, Brewer B, et al. Phosphorylation of p40AUF1 regulates binding to A + U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J Biol Chem (2003) 278(35):33039–48. doi:10.1074/jbc.M305775200
110. Gratacos FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA (2010) 1(3):457–73. doi:10.1002/wrna.26
111. Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J Biol Chem (2003) 278(23):20700–7. doi:10.1074/jbc.M301176200
112. Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics (1998) 48(2):195–202. doi:10.1006/geno.1997.5142
113. Misquitta CM, Iyer VR, Werstiuk ES, Grover AK. The role of 3’-untranslated region (3’-UTR) mediated mRNA stability in cardiovascular pathophysiology. Mol Cell Biochem (2001) 224(1–2):53–67. doi:10.1023/A:1011982932645
114. Sarkar B, Xi Q, He C, Schneider RJ. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol Cell Biol (2003) 23(18):6685–93. doi:10.1128/MCB.23.18.6685-6693.2003
115. Fukao A, Fujiwara T. The coupled and uncoupled mechanisms by which trans-acting factors regulate mRNA stability and translation. J Biochem (2016) 161:309–14. doi:10.1093/jb/mvw086
116. Esnault S, Malter JS. Primary peripheral blood eosinophils rapidly degrade transfected granulocyte-macrophage colony-stimulating factor mRNA. J Immunol (1999) 163(10):5228–34.
117. White EJ, Matsangos AE, Wilson GM. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip Rev RNA (2017) 8(2):e1393. doi:10.1002/wrna.1393
118. Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol (2007) 14(6):511–8. doi:10.1038/nsmb1249
119. Sarkar S, Sinsimer KS, Foster RL, Brewer G, Pestka S. AUF1 isoform-specific regulation of anti-inflammatory IL10 expression in monocytes. J Interferon Cytokine Res (2008) 28(11):679–91. doi:10.1089/jir.2008.0028
120. Sarkar S, Han J, Sinsimer KS, Liao B, Foster RL, Brewer G, et al. RNA-binding protein AUF1 regulates lipopolysaccharide-induced IL10 expression by activating IkappaB kinase complex in monocytes. Mol Cell Biol (2011) 31(4):602–15. doi:10.1128/MCB.00835-10
121. Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science (1999) 284(5413):499–502. doi:10.1126/science.284.5413.499
122. Lu JY, Bergman N, Sadri N, Schneider RJ. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA (2006) 12(5):883–93. doi:10.1261/rna.2308106
123. Sinsimer KS, Gratacos FM, Knapinska AM, Lu J, Krause CD, Wierzbowski AV, et al. Chaperone Hsp27, a novel subunit of AUF1 protein complexes, functions in AU-rich element-mediated mRNA decay. Mol Cell Biol (2008) 28(17):5223–37. doi:10.1128/MCB.00431-08
124. Kleene KC, Mulligan E, Steiger D, Donohue K, Mastrangelo MA. The mouse gene encoding the testis-specific isoform of Poly(A) binding protein (Pabp2) is an expressed retroposon: intimations that gene expression in spermatogenic cells facilitates the creation of new genes. J Mol Evol (1998) 47(3):275–81. doi:10.1007/PL00006385
125. Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Mosc) (2011) 76(13):1402–33. doi:10.1134/S0006297911130049
126. Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: functions and regulation. Wiley Interdiscip Rev RNA (2014) 5(1):95–110. doi:10.1002/wrna.1200
127. Minich WB, Ovchinnikov LP. Role of cytoplasmic mRNP proteins in translation. Biochimie (1992) 74(5):477–83. doi:10.1016/0300-9084(92)90088-V
128. Chen CY, Gherzi R, Andersen JS, Gaietta G, Jurchott K, Royer HD, et al. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev (2000) 14(10):1236–48. doi:10.1101/gad.14.10.1236
129. Coles LS, Lambrusco L, Burrows J, Hunter J, Diamond P, Bert AG, et al. Phosphorylation of cold shock domain/Y-box proteins by ERK2 and GSK3beta and repression of the human VEGF promoter. FEBS Lett (2005) 579(24):5372–8. doi:10.1016/j.febslet.2005.08.075
130. Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, et al. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol Cell Biol (2006) 26(1):277–92. doi:10.1128/MCB.26.1.277-292.2006
131. Esnault S, Fang Y, Kelly EA, Sedgwick JB, Fine J, Malter JS, et al. Circadian changes in granulocyte-macrophage colony-stimulating factor message in circulating eosinophils. Ann Allergy Asthma Immunol (2007) 98(1):75–82. doi:10.1016/S1081-1206(10)60863-0
132. Rajagopalan LE, Westmark CJ, Jarzembowski JA, Malter JS. hnRNP C increases amyloid precursor protein (APP) production by stabilizing APP mRNA. Nucleic Acids Res (1998) 26(14):3418–23. doi:10.1093/nar/26.14.3418
133. Sella O, Gerlitz G, Le SY, Elroy-Stein O. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol Cell Biol (1999) 19(8):5429–40. doi:10.1128/MCB.19.8.5429
134. Holcik M, Gordon BW, Korneluk RG. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol Cell Biol (2003) 23(1):280–8. doi:10.1128/MCB.23.1.280-288.2003
135. Schepens B, Tinton SA, Bruynooghe Y, Parthoens E, Haegman M, Beyaert R, et al. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J (2007) 26(1):158–69. doi:10.1038/sj.emboj.7601468
136. Lee EK, Kim HH, Kuwano Y, Abdelmohsen K, Srikantan S, Subaran SS, et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol (2010) 17(6):732–9. doi:10.1038/nsmb.1815
137. Beaudoin ME, Poirel VJ, Krushel LA. Regulating amyloid precursor protein synthesis through an internal ribosomal entry site. Nucleic Acids Res (2008) 36(21):6835–47. doi:10.1093/nar/gkn792
138. Kim JH, Paek KY, Choi K, Kim TD, Hahm B, Kim KT, et al. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol Cell Biol (2003) 23(2):708–20. doi:10.1128/MCB.23.2.708-720.2003
139. Fung PA, Labrecque R, Pederson T. RNA-dependent phosphorylation of a nuclear RNA binding protein. Proc Natl Acad Sci U S A (1997) 94(4):1064–8. doi:10.1073/pnas.94.4.1064
140. Phillips K, Kedersha N, Shen L, Blackshear PJ, Anderson P. Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc Natl Acad Sci U S A (2004) 101(7):2011–6. doi:10.1073/pnas.0400148101
141. Dixon DA, Balch GC, Kedersha N, Anderson P, Zimmerman GA, Beauchamp RD, et al. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J Exp Med (2003) 198(3):475–81. doi:10.1084/jem.20030616
142. Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol (2006) 26(8):3295–307. doi:10.1128/MCB.26.8.3295-3307.2006
143. Damgaard CK, Lykke-Andersen J. Translational coregulation of 5’TOP mRNAs by TIA-1 and TIAR. Genes Dev (2011) 25(19):2057–68. doi:10.1101/gad.17355911
144. Zhang T, Kruys V, Huez G, Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem Soc Trans (2002) 30(Pt 6):952–8. doi:10.1042/bst0300952
145. Wilkinson MF, Shyu AB. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays (2001) 23(9):775–87. doi:10.1002/bies.1113
146. Tian Q, Taupin J, Elledge S, Robertson M, Anderson P. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J Exp Med (1995) 182(3):865–74. doi:10.1084/jem.182.3.865
147. Izquierdo JM, Valcarcel J. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J Biol Chem (2007) 282(3):1539–43. doi:10.1074/jbc.C600198200
148. Bachmann M, Pfeifer K, Schroder HC, Muller WE. Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell (1990) 60(1):85–93. doi:10.1016/0092-8674(90)90718-T
149. Huhn P, Pruijn GJ, van Venrooij WJ, Bachmann M. Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res (1997) 25(2):410–6. doi:10.1093/nar/25.2.410
150. Martino L, Pennell S, Kelly G, Bui TT, Kotik-Kogan O, Smerdon SJ, et al. Analysis of the interaction with the hepatitis C virus mRNA reveals an alternative mode of RNA recognition by the human La protein. Nucleic Acids Res (2012) 40(3):1381–94. doi:10.1093/nar/gkr890
151. Kuehnert J, Sommer G, Zierk AW, Fedarovich A, Brock A, Fedarovich D, et al. Novel RNA chaperone domain of RNA-binding protein La is regulated by AKT phosphorylation. Nucleic Acids Res (2015) 43(1):581–94. doi:10.1093/nar/gku1309
152. Svitkin YV, Meerovitch K, Lee HS, Dholakia JN, Kenan DJ, Agol VI, et al. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J Virol (1994) 68(3):1544–50.
153. Svitkin YV, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5’ leader sequence of the human immunodeficiency virus type 1 mRNA. J Virol (1994) 68(11):7001–7.
154. Pudi R, Srinivasan P, Das S. La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation. J Biol Chem (2004) 279(29):29879–88. doi:10.1074/jbc.M403417200
155. Schwartz EI, Intine RV, Maraia RJ. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate rpL37 5’-terminal oligopyrimidine mRNA metabolism. Mol Cell Biol (2004) 24(21):9580–91. doi:10.1128/MCB.24.21.9580-9591.2004
156. Brenet F, Socci ND, Sonenberg N, Holland EC. Akt phosphorylation of La regulates specific mRNA translation in glial progenitors. Oncogene (2009) 28(1):128–39. doi:10.1038/onc.2008.376
157. Intine RV, Dundr M, Vassilev A, Schwartz E, Zhao Y, Zhao Y, et al. Nonphosphorylated human La antigen interacts with nucleolin at nucleolar sites involved in rRNA biogenesis. Mol Cell Biol (2004) 24(24):10894–904. doi:10.1128/MCB.24.24.10894-10904.2004
158. Adachi T, Motojima S, Hirata A, Fukuda T, Makino S. Eosinophil viability-enhancing activity in sputum from patients with bronchial asthma. Am J Respir Crit Care Med (1995) 151:618–23. doi:10.1164/ajrccm/151.3_Pt_1.618
159. Corrigan CJ, Hamid Q, North J, Barkans J, Moqbel R, Durham S, et al. Peripheral blood CD4 but not CD8 T-lymphocytes in patients with exacerbation of asthma transcribe and translate messenger RNA encoding cytokines which prolong eosinophil survival in the context of a Th2-type pattern: effect of glucocorticoid therapy. Am J Respir Cell Mol Biol (1995) 12:567–78. doi:10.1165/ajrcmb.12.5.7742019
160. Schneider T, van Velzen D, Moqbel R, Issekutz AC. Kinetics and quantitation of eosinophil and neutrophil recruitment to allergic lung inflammation in a brown Norway rat model. Am J Respir Cell Mol Biol (1997) 17(6):702–12. doi:10.1165/ajrcmb.17.6.2849
161. Walker C, Bauer W, Braun RK, Menz G, Braun P, Schwarz F, et al. Activated T cells and cytokines in bronchoalveolar lavages from patients with various lung diseases associated with eosinophilia. Am J Respir Crit Care Med (1994) 150:1038–48. doi:10.1164/ajrccm.150.4.7921434
162. Shaver JR, Zangrilli JG, Cho SK, Cirelli RA, Pollice M, Hastie AT, et al. Kinetics of the development and recovery of the lung from IgE-mediated inflammation: dissociation of pulmonary eosinophilia, lung injury, and eosinophil-active cytokines. Am J Respir Crit Care Med (1997) 155(2):442–8. doi:10.1164/ajrccm.155.2.9032176
163. Yamashita N, Tashimo H, Ishida H, Kaneko F, Nakano J, Kato H, et al. Attenuation of airway hyperresponsiveness in a murine asthma model by neutralization of granulocyte-macrophage colony-stimulating factor (GM-CSF). Cell Immunol (2002) 219(2):92–7. doi:10.1016/S0008-8749(02)00565-8
164. Ohkawara Y, Lei XF, Stampfli MR, Marshall JS, Xing Z, Jordana M. Cytokine and eosinophil responses in the lung, peripheral blood, and bone marrow compartments in a murine model of allergen-induced airways inflammation. Am J Respir Cell Mol Biol (1997) 16:510–20. doi:10.1165/ajrcmb.16.5.9160833
165. Lei XF, Ohkawara Y, Stampfli MR, Gauldie J, Croitoru K, Jordana M, et al. Compartmentalized transgene expression of granulocyte-macrophage colony-stimulating factor (GM-CSF) in mouse lung enhances allergic airways inflammation. Clin Exp Immunol (1998) 113(2):157–65. doi:10.1046/j.1365-2249.1998.00652.x
166. Park CS, Choi YS, Ki SY, Moon SH, Jeong SW, Uh ST, et al. Granulocyte macrophage colony-stimulating factor is the main cytokine enhancing survival of eosinophils in asthmatic airways. Eur Respir J (1998) 12(4):872–8. doi:10.1183/09031936.98.12040872
167. Moqbel R, Hamid Q, Ying S, Barkans J, Hartnell A, Tsicopoulos A, et al. Expression of mRNA and immunoreactivity for the granulocyte/macrophage colony-stimulating factor in activated human eosinophils. J Exp Med (1991) 174(3):749–52. doi:10.1084/jem.174.3.749
168. Gauvreau GM, O’Byrne PM, Moqbel R, Velazquez J, Watson RM, Howie KJ, et al. Enhanced expression of GM-CSF in differentiating eosinophils of atopic and atopic asthmatic subjects. Am J Respir Cell Mol Biol (1998) 19(1):55–62. doi:10.1165/ajrcmb.19.1.2871
169. Ohkawara Y, Lim KG, Xing Z, Glibetic M, Nakano K, Dolovich J, et al. CD40 expression by human peripheral blood eosinophils. J Clin Invest (1996) 97(7):1761–6. doi:10.1172/JCI118603
170. Kim JT, Gleich GJ, Kita H. Roles of CD9 molecules in survival and activation of human eosinophils. J Immunol (1997) 159(2):926–33.
171. Levi-Schaffer F, Temkin V, Malamud V, Feld S, Zilberman Y. Mast cells enhance eosinophil survival in vitro: role of TNF-alpha and granulocyte-macrophage colony-stimulating factor. J Immunol (1998) 160(11):5554–62.
172. Esnault S, Malter JS. Granulocyte macrophage-colony-stimulating factor mRNA is stabilized in airway eosinophils and peripheral blood eosinophils activated by TNF-alpha plus fibronectin. J Immunol (2001) 166(7):4658–63. doi:10.4049/jimmunol.166.7.4658
173. Esnault S, Malter JS. Minute quantities of granulocyte-macrophage colony-stimulating factor prolong eosinophil survival. J Interferon Cytokine Res (2001) 21(2):117–24. doi:10.1089/107999001750069980
174. Hoontrakoon R, Chu HW, Gardai SJ, Wenzel SE, McDonald P, Fadok VA, et al. Interleukin-15 inhibits spontaneous apoptosis in human eosinophils via autocrine production of granulocyte macrophage-colony stimulating factor and nuclear factor-kappaB activation. Am J Respir Cell Mol Biol (2002) 26(4):404–12. doi:10.1165/ajrcmb.26.4.4517
175. Broide DH, Firestein GS. Endobronchial allergen challenge in asthma: demonstration of cellular source of granulocyte macrophage colony-stimulating factor by in situ hybridization. J Clin Invest (1991) 88:1048–53. doi:10.1172/JCI115366
176. Stumpo DJ, Lai WS, Blackshear PJ. Inflammation: cytokines and RNA-based regulation. Wiley Interdiscip Rev RNA (2010) 1(1):60–80. doi:10.1002/wrna.1
177. Westmark PR, Westmark CJ, Wang S, Levenson J, O’Riordan KJ, Burger C, et al. Pin1 and PKMzeta sequentially control dendritic protein synthesis. Sci Signal (2010) 3(112):ra18. doi:10.1126/scisignal.2000451
178. Krishnan N, Titus MA, Thapar R. The prolyl isomerase pin1 regulates mRNA levels of genes with short half-lives by targeting specific RNA binding proteins. PLoS One (2014) 9(1):e85427. doi:10.1371/journal.pone.0085427
179. Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature (1996) 380(6574):544–7. doi:10.1038/380544a0
180. Lee TH, Pastorino L, Lu KP. Peptidyl-prolyl cis-trans isomerase Pin1 in ageing, cancer and Alzheimer disease. Expert Rev Mol Med (2011) 13:e21. doi:10.1017/S1462399411001906
181. Esnault S, Rosenthal LA, Shen ZJ, Sedgwick JB, Szakaly RJ, Sorkness RL, et al. A critical role for Pin1 in allergic pulmonary eosinophilia in rats. J Allergy Clin Immunol (2007) 120(5):1082–8. doi:10.1016/j.jaci.2007.06.024
182. Zhou CX, Gao Y. Aberrant expression of beta-catenin, Pin1 and cylin D1 in salivary adenoid cystic carcinoma: relation to tumor proliferation and metastasis. Oncol Rep (2006) 16(3):505–11. doi:10.3892/or.16.3.505
183. Driver JA, Zhou XZ, Lu KP. Pin1 dysregulation helps to explain the inverse association between cancer and Alzheimer’s disease. Biochim Biophys Acta (2015) 1850(10):2069–76. doi:10.1016/j.bbagen.2014.12.025
184. Imaoka H, Campbell H, Babirad I, Watson RM, Mistry M, Sehmi R, et al. TPI ASM8 reduces eosinophil progenitors in sputum after allergen challenge. Clin Exp Allergy (2011) 41(12):1740–6. doi:10.1111/j.1365-2222.2011.03816.x
185. Konig K, Marth L, Roissant J, Granja T, Jennewein C, Devanathan V, et al. The plexin C1 receptor promotes acute inflammation. Eur J Immunol (2014) 44(9):2648–58. doi:10.1002/eji.201343968
186. Spencer AY, Lallier TE. Mechanical tension alters semaphorin expression in the periodontium. J Periodontol (2009) 80(10):1665–73. doi:10.1902/jop.2009.090212
187. Esnault S, Kelly EA. Essential mechanisms of differential activation of eosinophils by IL-3 compared to GM-CSF and IL-5. Crit Rev Immunol (2016) 36(5):429–44. doi:10.1615/CritRevImmunol.2017020172
188. Johansson MW, Annis DS, Mosher DF. alpha(M)beta(2) integrin-mediated adhesion and motility of IL-5-stimulated eosinophils on periostin. Am J Respir Cell Mol Biol (2013) 48(4):503–10. doi:10.1165/rcmb.2012-0150OC
189. Kang HR, Lee CG, Homer RJ, Elias JA. Semaphorin 7A plays a critical role in TGF-beta1-induced pulmonary fibrosis. J Exp Med (2007) 204(5):1083–93. doi:10.1084/jem.20061273
190. De Minicis S, Rychlicki C, Agostinelli L, Saccomanno S, Trozzi L, Candelaresi C, et al. Semaphorin 7A contributes to TGF-beta-mediated liver fibrogenesis. Am J Pathol (2013) 183(3):820–30. doi:10.1016/j.ajpath.2013.05.030
191. Esnault S, Torr EE, Bernau K, Johansson MW, Kelly EA, Sandbo N, et al. Endogenous semaphorin-7A impedes human lung fibroblast differentiation. PLoS One (2017) 12(1):e0170207. doi:10.1371/journal.pone.0170207
192. Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med (2009) 360(10):973–84. doi:10.1056/NEJMoa0808991
193. Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, et al. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med (2009) 360(10):985–93. doi:10.1056/NEJMoa0805435
194. Robinson DS, Damia R, Zeibecoglou K, Molet S, North J, Yamada T, et al. CD34(+)/interleukin-5Ralpha messenger RNA+ cells in the bronchial mucosa in asthma: potential airway eosinophil progenitors. Am J Respir Cell Mol Biol (1999) 20(1):9–13. doi:10.1165/ajrcmb.20.1.3449
195. Sehmi R, Smith SG, Kjarsgaard M, Radford K, Boulet LP, Lemiere C, et al. Role of local eosinophilopoietic processes in the development of airway eosinophilia in prednisone-dependent severe asthma. Clin Exp Allergy (2016) 46(6):793–802. doi:10.1111/cea.12695
196. Liu LY, Sedgwick JB, Bates ME, Vrtis RF, Gern JE, Kita H, et al. Decreased expression of membrane IL-5R alpha on human eosinophils: II. IL-5 down-modulates its receptor via a proteinase-mediated process. J Immunol (2002) 169(11):6459–66. doi:10.4049/jimmunol.169.11.6459
197. Gregory B, Kirchem A, Phipps S, Gevaert P, Pridgeon C, Rankin SM, et al. Differential regulation of human eosinophil IL-3, IL-5, and GM-CSF receptor alpha-chain expression by cytokines: IL-3, IL-5, and GM-CSF down-regulate IL-5 receptor alpha expression with loss of IL-5 responsiveness, but up-regulate IL-3 receptor alpha expression. J Immunol (2003) 170(11):5359–66. doi:10.4049/jimmunol.170.11.5359
198. Johansson MW, Kelly EA, Busse WW, Jarjour NN, Mosher DF. Up-regulation and activation of eosinophil integrins in blood and airway after segmental lung antigen challenge. J Immunol (2008) 180(11):7622–35. doi:10.4049/jimmunol.180.11.7622
199. Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, et al. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene (2014) 33(4):474–83. doi:10.1038/onc.2012.606
200. Sapkota GP, Cummings L, Newell FS, Armstrong C, Bain J, Frodin M, et al. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem J (2007) 401(1):29–38. doi:10.1042/BJ20061088
Keywords: eosinophils, protein translation, ribosomal S6 protein, Pin-1, IL-3, intracellular signaling
Citation: Esnault S, Shen Z-J and Malter JS (2017) Protein Translation and Signaling in Human Eosinophils. Front. Med. 4:150. doi: 10.3389/fmed.2017.00150
Received: 13 April 2017; Accepted: 01 September 2017;
Published: 19 September 2017
Edited by:
Florence Emmanuelle Roufosse, Free University of Brussels, BelgiumReviewed by:
Owen McCarty, Oregon Health & Science University, United StatesKaren Willard-Gallo, Free University of Brussels, Belgium
Copyright: © 2017 Esnault, Shen and Malter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephane Esnault, sesnault@medicine.wisc.edu
 Stephane Esnault
Stephane Esnault Zhong-Jian Shen2
Zhong-Jian Shen2  James S. Malter
James S. Malter