Genetic Insights Into Frailty: Association of 9p21-23 Locus With Frailty
- 1Department of Neurology, Albert Einstein College of Medicine, Bronx, NY, United States
- 2Department of Medicine, Albert Einstein College of Medicine, Bronx, NY, United States
- 3Department of Genetics, Institute for Aging Research, Albert Einstein College of Medicine, Bronx, NY, United States
- 4Department of Biology, Faculty of Natural Science, University of Haifa, Haifa, Israel
Frailty is a complex aging phenotype associated with increased vulnerability to disability and death. Understanding the biological antecedents of frailty may provide clues to healthy aging. The genome-wide association study hotspot, 9p21-23 region, is a risk locus for a number of age-related complex disorders associated with frailty. Hence, we conducted an association study to examine whether variations in 9p21-23 locus plays a role in the pathogenesis of frailty in 637 community-dwelling Ashkenazi Jewish adults aged 65 and older enrolled in the LonGenity study. The strongest association with frailty (adjusted for age and gender) was found with the SNP rs518054 (odds ratio: 1.635, 95% CI = 1.241–2.154; p-value: 4.81 × 10−04) intergenic and located between LOC105375977 and C9orf146. The prevalence of four SNPs (rs1324192, rs7019262, rs518054, and rs571221) risk alleles haplotype in this region was significantly higher (compared with other haplotypes) in frail older adults compared with non-frail older adults (29.7 vs. 20.8%, p = 0.0005, respectively). Functional analyses using in silico approaches placed rs518054 in the CTCF binding site as well as DNase hypersensitive region. Furthermore, rs518054 was found to be in an enhancer site of NFIB gene located downstream. NFIB is a transcription factor that promotes cell differentiation during development, has antiapoptotic effect, maintains stem cell populations in adult tissues, and also acts as epigenetic regulators. Our study found novel association of SNPs in the regulatory region in the 9p21-23 region with the frailty phenotype; signifying the importance of this locus in aging.
Introduction
Frailty is a complex phenotype seen in aging, which is associated with low physiologic reserves and with increased vulnerability to adverse outcomes such as disability, hospitalization, and death (1, 2). The prevalence of frailty has been reported to range from 7 to 32% in older populations and is higher in women (3). Given the emergent aging pandemic worldwide (4), a major public health challenge is to find ways to enhance functional independence in older adults and to increase years free from disabilities. Hence, understanding the biological antecedents of frailty may provide insights into healthy aging strategies.
Frailty is a multidimensional construct involving several domains—physical, cognitive, psychological, and social domains (5–7). Even though expression and biomarker studies have pointed toward the involvement of various biological pathways in frailty (8, 9), genetic studies have not yielded consistent results. Candidate gene studies of IL6, TNF and IGF1 have shown either no association with frailty or provided contradictory results (10). This might be mainly explained by the multifactorial nature of frailty with involvement of genetic, lifestyle, and epigenetic factors (11, 12). This multidimensionality and multifactorial or complex origin of frailty is further supported by the etiological overlap between frailty and various age-related complex or multifactorial disorders (13). Prevalent frailty was a strong risk factor for cardiovascular diseases (CVD) as well as associated mortality (14). In the Cardiovascular Health Study, a cross-sectional analyses showed 38% of frail individuals had prevalent heart disease compared with 17% in non-frail individuals (15). Frailty and diabetes are strongly linked (16) with a higher incidence of type 2 diabetes seen in individuals with frailty (17). Frailty is associated with postmortem Alzheimer pathology in older adults with and without an antemortem history of dementia (18, 19). All of these point toward overlapping biological mechanism for frailty and other complex disorders. It is also possible that complex disorders may alter the frailty risk conferred by specific biological pathways.
Complex or multifactorial diseases are caused by a combination of genetic, lifestyle, and other environment factors. Genome wide association studies (GWASs) have identified a large number of genetic variants associated with complex disorders (20, 21). In particular, 9p21-23 has been shown to be a risk-associated locus with many complex disorders. For example, 9p21 has been reported to be associated with CVD (22, 23), abdominal aortic aneurysm (24), arterial stiffness (25), peripheral artery disease (26), intracranial aneurysm (27), various types of cancers (28, 29), amyotrophic lateral sclerosis (30), primary open-angle glaucoma (29), vascular dementia, and Alzheimer’s disease (31). The 9p23 region was associated with restless legs syndrome (32) and obsessive-compulsive disorder (33). Distinct haplotype blocks at the 9p21-23 region were associated with CVD and type 1 diabetes (34). This locus harbors several genes including ANRIL, a long non-coding RNA gene implicated in the pathogenesis CVD and strokes, three candidate tumor suppressor genes; CDKN2A (cyclin-dependent kinase inhibitor 2A) encoding p16 protein, CDKN2B encoding p15 protein, and p14/ARF encoding p14ARF protein (35). C9ORF72 gene was found to be associated with amyotrophic lateral sclerosis-frontotemporal dementia (36). Furthermore, protein tyrosine phosphatase receptor type delta (PTPRD) at 9p23 region was associated with restless legs syndrome (32) as well as cancers (37). While there is substantial overlap in the diseases-associated with frailty and the 9p21-23 locus, to the best of our knowledge, the association of this locus with frailty has not been specifically examined.
Discovering new biological pathways that prevent or delay frailty would increase current therapeutic options for clinicians and increase health span for individuals. Interestingly, rs2811712 located in ANRIL gene in the 9p21 locus is associated with physical function in older people with the minor allele being associated with reduced physical impairment (38). Furthermore, rs71321217 in PTPRD in the 9p23 locus is associated with gait rhythm (39). Based on these observations, we hypothesized that genetic variants in the chromosome 9p21-23 locus will increase the risk of developing frailty in older adults. To elucidate the role of the 9p21-23 locus in the pathogenesis of frailty, we conducted a preliminary cross-sectional study in 637 community-residing Ashkenazi Jewish (AJ) older adults participating in the LonGenity Study (40, 41). This population is homogenous genetically and socioeconomically (42) and allows for greater power for genetic analysis with fewer number of participants. Establishing the genetic underpinnings of frailty may provide new insights into preventive strategies to delay the occurrence of frailty and other related comorbidities as well as to promote healthy aging.
Materials and Methods
LonGenity Cohort
The LonGenity study, established in 2007, recruited a cohort of AJ adults age 65 and older, who were defined as either Offspring of Parents with Exceptional Longevity (OPEL) (having at least one parent who lived to age 95 or older) or Offspring of Parents with Usual Survival (OPUS) (neither parent survived to age 95). The goal of the LonGenity study is to identify genotypes associated with longevity and their association with successful aging. Study participants were recruited through contacts at synagogues, community organizations and advertisements in Jewish newspapers in the New York City area. Potential participants were contacted by telephone to assess interest and eligibility. They were invited to our research center for further evaluation. Exclusion criterion included diagnosis of dementia [previous physician diagnosed dementia or telephone Memory Impairment Screen scores in the dementia range (43)] as well as presence of severe visual or hearing impairments that would interfere with study assessments. Participants received detailed medical history evaluation and cognitive testing at baseline as well as at annual follow-up visits. All participants signed written informed consents for clinical assessments and genetic testing before enrollment. The Einstein institutional review board approved the study protocol.
A total of 965 older individuals were enrolled in the LonGenity study between October 2008 and August 2017. We excluded 64 individuals who did not complete frailty assessments as well as 264 who did not complete genetic testing. Hence, the eligible sample for this analysis included 637 participants, who had been genotyped and completed frailty assessments.
Frailty Syndrome
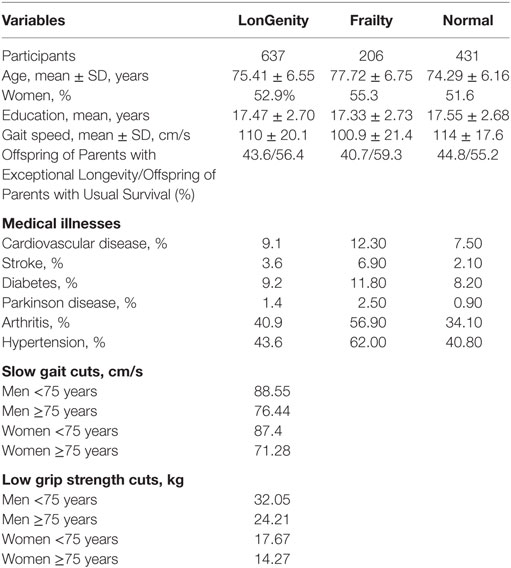
The two common approaches to defining frailty clinically are as a clinical syndrome (5) or as a cumulative deficit score (44–46). The syndromic definition of frailty (see below) is widely adopted in research and clinical practice (5). While the cumulative deficit score approach has advantages in research settings, it is less intuitive in clinical settings in the community (47). Frailty diagnosis, hence, was operationalized using the widely used Cardiovascular Health Study criteria (48) for this study. Frailty was operationally defined as meeting three or more of the following five attributes: unintentional weight loss (≥10 lb in past year), muscle weakness (objectively measured grip strength or self-report; described below), exhaustion [negative response to the question “do you feel full of energy?” on the Geriatric Depression Scale (49)], self-reported low physical activity levels [positive response to the question “Have you been less active physically?” on the Health Self-Assessment Questionnaire (5)] and slow gait (Table 1). A Jamar handgrip dynamometer was used to objectively measure dominant hand grip strength at baseline. Weakness was defined using a cut score of 1 SD or more below age and sex mean values (Table 1). Similar to previous reports (50–52), subjective grip strength (“do you feel as though your grip is weak?”) was used on follow-up waves as a frailty criterion, since objective grip strength measures were not available for all our participants on follow-up. A previous study in this same cohort showed substantial agreement between the objective and subjective grip strength rating methods (53). Gait speed (cm/s) was measured using an 8.5 m long computerized walkway with embedded pressure sensors (GAITRite; CIR Systems, PA). The GAITRite system is widely used in clinical and research settings, and excellent reliability has been reported in our and other centers (54, 55). Participants were asked to walk on the walkway at their normal pace in a quiet well-lit room wearing comfortable footwear and without any attached monitors. Slow gait was defined as 1.5 or more SD below age and sex-appropriate mean values. In total, we had 206 individuals who were diagnosed with frailty; 118 prevalent cases and 88 incident cases of frailty.
Selection of Gene Variants and Genotyping
We targeted 9p21-23 region spanning from chr9: 8743598 to 32586822 (NCBI build 37) for this analysis based on its functional significance and reported associations with major complex disorders (24, 30, 31, 34, 35). Genotyping was performed at the Center for Inherited Disease Research using Illumina HumanOmniExpress array (Illumina, San Diego, CA, USA), and the procedures have been described previously (40, 41).
Since the focus of our research was to explore complex disorder-associated alleles in this locus in regards to frailty; the few SNPs missing in the genotyping array were made available from imputation analysis. Imputation of un-genotyped autosomal SNPs were based on the 1000 Genomes data (worldwide reference panel of all 1,092 samples from the phase I integrated variant set) (v3, released March 2012) (56) using IMPUTE2, version 2.3.0. Poorly imputed SNPs with low imputation quality (info_metric < 0.3) were excluded from the analysis. For this study, we selected SNPs with minor allele frequencies of >0.10.
Statistical Analysis
Baseline characteristics of participants were compared using descriptive statistics (Table 1). The preliminary objective of this study was to identify the association of variants in the 9p21-23 region with frailty using logistic regression analysis. Prevalent and incident cases of frailty were examined together in this analysis to maximize sample size. In participants who did not have frailty at baseline or develop incident frailty, the wave at which the first non-frail status was diagnosed was used as baseline for comparing clinical characteristics. As previous studies have shown frailty to increase with age and in women (57), all analyses were adjusted for age and gender (Model 1). All SNP based association analyses were conducted using Plink v1.90.1 All other statistical analyses were carried out using SPSS software (version 24; IBM Corporation). Presence or absence of diabetes, heart failure (including myocardial infarction, angina, or congestive heart failure), hypertension, strokes, Parkinson’s disease, and arthritis was used to calculate a global health score (range 0–6) as previously described (58). To account for the LonGenity study design described above (40, 41) and health status, we conducted sensitivity analyses further adjusting the models for OPUS/OPEL status and global health score (Model 2).
A total of 5,556 variants were available for analysis in the selected region (chr9: 8743598 to 32586822) after removing SNPs that had minor allele frequencies < 0.10 (n = 1,856) and failed the Hardy–Weinberg exact test (p ≥ 0.01) (n = 79). Linkage disequilibrium (LD) plots were generated using Haploview 4.2 (59). Haplotype blocks were defined based on the Gabriel criteria (60). Haplotype analyses were performed using SNPStats software (61). Functional prediction of the associated variants was carried out using various in silico approaches. Genotype-Tissue Expression portal (GTEx)2 was used to determine the significant expression quantitative trait loci (eQTL) for SNPs associated with frailty (62). Regulome DB3 based on Encyclopedia of DNA Elements (ENCODE) project (63) was used to identify functional effects of the identified SNPs in the association and eQTL analyses. rVarBase,4 updated database for regulatory features of variants was also used to find the effect of SNP of chromatin states, interacting regulatory elements and target genes (64). Functional Single Nucleotide Polymorphism; a web-based tool that integrates 16 databases and bioinformatic tools to uncover the functional effect of the SNPs (65) and FuncPred5 were used to predict the functional effects of associated variants.
Results
Study Population
Of the 637 eligible individuals with phenotype and genotype data in the LonGenity cohort, 356 were OPUS and 281 were OPEL. Of the eligible sample, 206 individuals (32.5%) received a diagnosis of frailty at baseline (n = 118) or at various time points over the study follow-up (n = 88), and 430 individuals (67.5%) remained non-frail throughout the study follow-up. The overall median follow-up time was 3.7 years (range 0–9 years). Demographic and clinical characteristics are summarized in Table 1. The mean age of the participants was 75.41 ± 6.55 years, and 52.9% were women. The mean years of education was 17.47 ± 2.70 years. A higher percentage of OPUS individuals met frailty diagnosis (34%) compared with OPEL (30.4%). All major medical illnesses were more prevalent in individuals who had frailty compared with normal individuals (Table 1).
Association and In Silico Functional Analyses
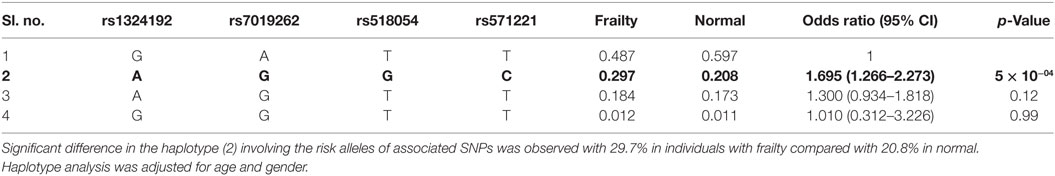
The strongest association with frailty was found with the G allele of rs518054 [odds ratio (OR): 1.635, 95% CI = 1.241–2.154; p-value: 4.81 × 10−04] (Table 2; Figure 1) (Model 1). None of the SNPs studied survive Bonferroni correction for threshold for statistical significance. The associations remained similar after adjusting for longevity status (OPEL vs. OPUS) and global health score for all of these four SNPs (Table S1 in Supplementary Material) (Model 2). We also observed modest associations of three other SNPs (rs7019262, rs571221 and rs1324192) in this region with frailty (Table 2; Figure 2). LD plot of associated SNPs showed presence of SNPs in two LD blocks (rs518054–rs571221 and rs1324192–rs7019262) in frail and single LD block in normal individuals (Figure S1 in Supplementary Material). Haplotype analysis to investigate the combined effect of associated SNPs found significant association (p-value < 0.0005) with haplotype involving risk alleles (AGGC) at four loci combination (29.7 vs. 20.8%) (Table 3).
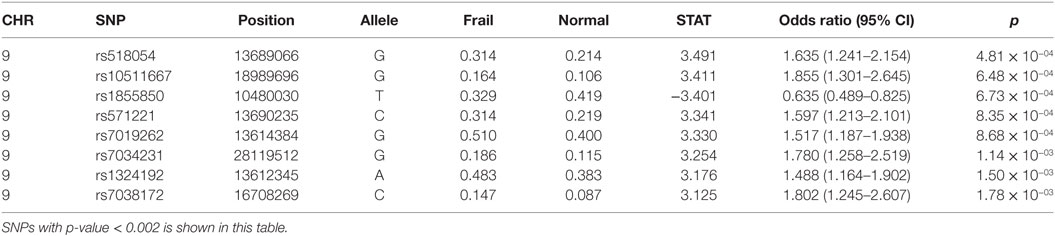
Table 2. Logistic regression analysis of 9p21-23 locus with Frailty with genotyped SNPs adjusted for age and gender (Model 1).
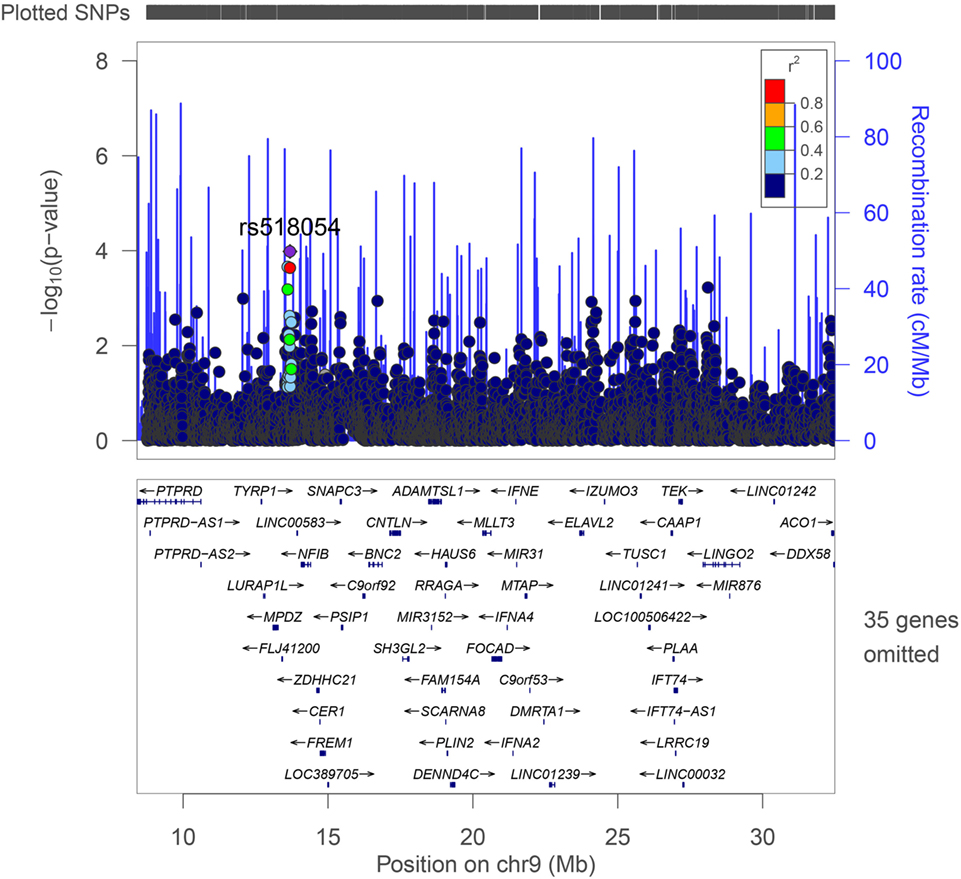
Figure 1. LocusZoom plot of the region studied with frailty on chromosome 9p21-23. Genes and ESTs within the region are shown in the lower panel, and the unbroken blue line indicates the recombination rate within the region. Each filled circle represents the p-value for one SNP, with the top SNP rs518054 shown in purple and SNPs in the region colored depending on their degree of correlation (r2) with rs518054 [as estimated internally by LocusZoom on the basis of CEU (Utah residents of Northern and Western European ancestry) HapMap haplotypes].
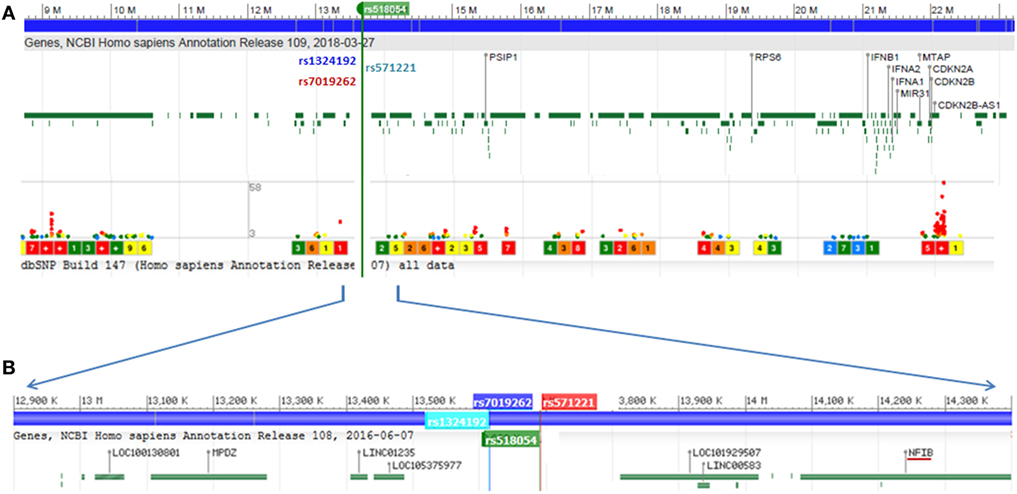
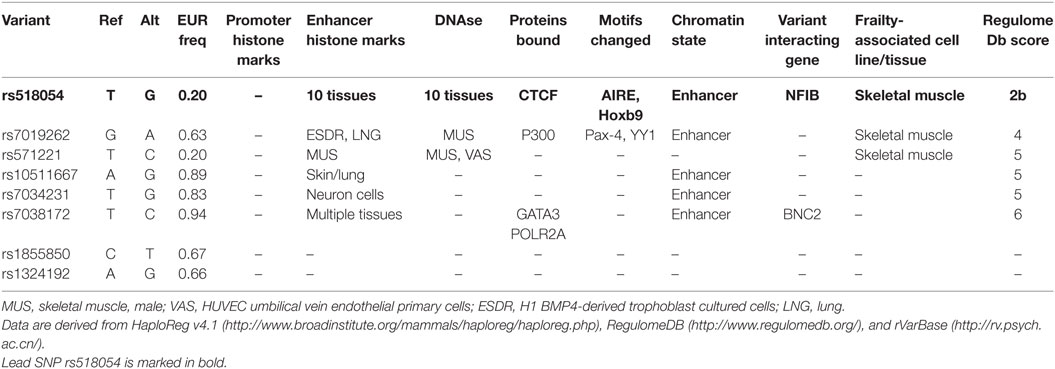
Figure 2. (A) Genome wide association study hotspot locus of 9p21-23 region screened in this study. Frailty-associated SNPs are marked in rsIDs, and lower dots indicate disease-associated SNPs in this region and level of significance. (B) Localized view of associated SNPs showing its location between LOC105375977 and LINC00583 (C9orf146). NFIB is the nearest well-characterized gene to SNP rs518054. Genomic region data adapted from NCBI dbSNP database.
All four SNPs associated with frailty in this region were intergenic and located between LOC105375977 and C9orf146 (LINC00583) (Figure 2). The nearest well-characterized gene was NFIB coding for nuclear factor 1B (Figure 2). We assessed the functional significance of the associated SNPs in our study. The lead SNP rs518054 is located in the DNase 1 hypersensitive site. ENCODE data showed a Regulome DB score of 2b for rs518054, which predicted its role as likely to affect gene expression level, and the evidence includes transcription factor binding, any motif change, DNase footprint, and DNase peak (Table 4). The data show this region to be a binding site for CTCF, a transcriptional regulator. rVarBase data further suggest this SNP to be located in the chromatin interactive region with predominantly enhancer function in most tissues including muscle in both male and female. The available ENCODE data further showed this region harboring rs518054 interacted with the NFIB gene located downstream (Table 4). The associated SNP rs518054 located in the DNase hypersensitive site might play a role in the transcriptional regulation of NFIB gene through an enhancer effect. Furthermore, considering that these SNPs were located in the regulatory regions (e.g., enhancers), we used an in silico approach to determine whether they were local eQTL. Using GTEx portal, we could not find any significant eQTLs for SNP rs518054 in studied tissues.
Though none of the SNPs survived multiple corrections in this study, rs518054 emerged to be lead SNP with functional relevance in all models studied (Table 2; Table S1 in Supplementary Material). The unadjusted association analysis results are shown in Table S3 in Supplementary Material.
Sensitivity Analyses
The next objective of our study was to find out the risk conferred by specific complex disorder-associated SNPs in this region with frailty. A number of CVD-associated SNPs were observed in the 9p21-23 locus followed by SNPs for cancers and many other complex disorders. Our analysis showed lack of association of these disease-associated SNPs with frailty (Table S2 in Supplementary Material). Interestingly, there was an increased prevalence of CVD-associated risk alleles [rs10757278 (p = 0.116), rs1333040 (p = 0.133), and rs1333049 (p = 0.116)] in frail individuals compared with non-frail individuals (Table S2 in Supplementary Material). SNPs associated with gait rhythm (rs71321217; p-value = 0.384) and physical activity (rs2811712; p-value = 0.205) in previous studies (38, 39) were not associated with frailty in our cohort (Table S2 in Supplementary Material).
Even though genetic studies have been carried out combining prevalent and incident cases (66–68), to check the possibility of survival bias arising from the possible systematic differences in allele frequencies between the prevalent and incident cases, we carried out case only analysis comparing allele frequency in incident and prevalent cases. There was slight difference in the allele frequency of rs518054 in incident and prevalent cases of frailty (p-value = 0.014) with associated G allele found to be 0.36 in prevalent cases and 0.25 in incident cases. The frequency of G allele in controls was 0.21. The overall association was mainly driven by cases of prevalent frailty (OR: 1.980, 95% CI = 1.426–2.749; p-value: 4.50 × 10−05) than incident frailty (OR: 1.198, 95% CI = 0.808–1.776; p-value: 0.368) when each of them were compared independently to controls adjusting for age and gender.
Discussion
This study attempted to delineate the role of the 9p21-23 region with frailty in a well-characterized AJ cohort as a strategy to understand healthy aging. We uncovered a novel association of SNPs at the 9p21-23 region with frailty, not implicated previously with any of the complex disorders associated with this locus. Using functional analyses, we found the lead variant to be located in the enhancer region and involved in the transcriptional regulation of the NFIB gene. The study further observed increased frequency of CVD-associated alleles in individuals with frailty though failed to reach statistical significance with frailty phenotype.
The 9p21-23 region has emerged as a genetic hotspot for complex disorder associations in recent studies. With regard to the frailty-associated SNPs discovered in our study, the nearest well-characterized gene was NFIB, coding for the transcription factor Nuclear Factor IB, which plays a key role in the transcriptional regulation of a large number of genes in which our lead SNP rs518054 was found to be located in the enhancer region of this gene. NFIB has various functions ranging from promoting cell differentiation during development to maintaining stem cell populations in adult tissues and also possess antiapoptotic effect (69–72). In vivo studies have shown a multi-potency restriction of adult hippocampal neuronal stem cells by Drosha–NFIB interactions (73). It plays an important role in lung maturation and brain development (74), mediates repression of the epigenetic factor ezh2 which regulates cortical development (75), and also has an important role in chromatin remodeling (76). NFIB alters and globally maintains hyper accessible chromatin state and an increase of chromatin accessibility at distal regulatory elements enacts a program of gene expression (76). Thus the association we observed in the enhancer region of NFIB gene seems clinically and functionally relevant. The wide spread binding of NFIB in open chromatin sites has been linked to its regulatory action in adipocyte differentiation (77) and cancer metastasis (76). NFIB is also associated with osteosarcoma (78) and sciatica (79) in GWAS. All these findings point toward possible tissue-specific as well as genome-wide effects mediated through NFIB.
There is a paucity of studies examining the role of epigenetic mechanisms in frailty (12, 80). Epigenetic mechanisms including chromatin remodeling plays a pivotal role in the aging process (81, 82). Genes in the 9p21-23 locus have an important role in chromatin remodeling (76, 83). For instance, non-coding RNA ANRIL, specifically binds two polycomb proteins: CBX7 (PRC1) and SUZ12 (PRC2) to regulate histone modification in the CDKN2A/B locus. Overexpression of this gene also causes the down regulation of several genes involved in important chromatin architecture and remodeling mechanisms in other chromosomal regions (83). These results point toward a possible role of this locus in mediating environmental factors influenced epigenetic mechanisms. This might explain why this locus is linked with various age and environmental risk-associated diseases such as CVD, strokes and diabetes (24, 27, 34). Our finding thus supports a possible role of epigenetic mechanisms in frailty pathogenesis. Though there was higher prevalence of CVDs-associated risk allele with frailty, the association was not statistically significant. This might be mainly due to the smaller sample size as well as multifactorial origin of these diseases and frailty. Larger studies need to validate the initial observations in this study. Furthermore, since dementia was an exclusion criterion for the cohort, the association of some dementia related risk alleles with frailty might have minimized.
The strengths of our study include the systematic clinical and frailty assessments as well as the well-characterized population (40, 41). Limitation of this study is inclusion of incident frailty for increasing statistical power. The allele frequency of associated rs518054 “G” allele was observed to be more in prevalent and incident cases of frailty when independently compared with individuals free from frailty during course of this study. But the association was mainly driven by the prevalent frailty. The inclusion of incident frailty in the model provides us healthy controls free from frailty throughout the course of study. The lack of significant association with incident frailty might be mainly due to smaller sample size as well as objective-subjective definition of frailty used in this study. Limitations also include the absence of functional studies to validate the effect of associated genotypes with gene expression and chromatin interaction as well as the relatively small sample size. We noted the lack of consensus regarding frailty definitions, and chose a widely used and clinically relevant syndromic definition of frailty. Further studies need to be carried out to find the relevance of these observations in case of other frailty definitions. Our findings were based in a relatively genetically homogenous AJ population with high levels of education, which was used successfully for other genetic discoveries (40–42, 84–86) that have then been cross validated in other heterogeneous cohorts. However, our findings need to be validated in other more diverse populations.
In conclusion, we found novel association of variants in the 9p21-23 locus with frailty with lead SNP located in the enhancer region of the NFIB gene. Further investigation of this region is required to gain insights into potential interventions to address biological derangements in these pathways to extend health span and to maintain functional independence in older adults. The dynamics of healthy aging are complex and maintaining functional ability in older age is multifactorial. Frailty is one of the most significant geriatric syndrome observed in elderly population. Studies have shown that complex disorders increases with age but whether aging is the cause or consequence of these diseases is controversial. Our study supports a role for the complex disorder GWAS-associated 9p21-23 locus in frailty and provides insights into healthy aging.
Ethics Statement
The study was approved by the Institutional Review Board at the Albert Einstein College of Medicine. Written informed consent was obtained from all the study participants in accordance with the Declaration of Helsinki.
Author Contributions
SS and JV contributed to the design of the study and interpretation of the data. SS, NB, SM, and EA contributed to the acquisition of data and writing of the manuscript. EA, SM, and GA contributed to the analysis of the data. SS, NB, GA, SM, EA, and JV contributed to the critical revisions of the manuscript. All the authors approved the final version of the manuscript and agree to be accountable for all aspects of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by grants from NIH [R01AG044829 (JV and NB), P01AG021654 (NB), R01AG046949 (NB), and RO1AG042188 (GA)]; the Nathan Shock Center of Excellence for the Biology of Aging [P30AG038072 (NB), K23AG051148 (SM); and Glenn Center for the Biology of Human Aging Paul Glenn Foundation Grant (NB)]. The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fmed.2018.00105/full#supplementary-material.
Footnotes
References
1. Fried LP, Darer J, Walston J. Frailty. In: Cassel CK, Leipzig R, Cohen HJ, Larson EB, Meier DE, editors. Geriatric Medicine. New York: Springer (2003). p. 1067–76.
2. Fried LP, Ferrucci L, Darer J, Williamson JD, Anderson G. Untangling the concepts of disability, frailty, and comorbidity: implications for improved targeting and care. J Gerontol A Biol Sci Med Sci (2004) 59(3):M255–63. doi:10.1093/gerona/59.3.M255
3. Lally F, Crome P. Understanding frailty. Postgrad Med J (2007) 83(975):16–20. doi:10.1136/pgmj.2006.048587
4. Clegg A, Young J, Iliffe S, Rikkert MO, Rockwood K. Frailty in elderly people. Lancet (2013) 381(9868):752–62. doi:10.1016/S0140-6736(12)62167-9
5. Fried LP, Tangen CM, Walston J, Newman AB, Hirsch C, Gottdiener J, et al. Frailty in older adults evidence for a phenotype. J Gerontol A Biol Sci Med Sci (2001) 56(3):M146–57. doi:10.1093/gerona/56.3.M146
6. Levers MJ, Estabrooks CA, Ross Kerr JC. Factors contributing to frailty: literature review. J Adv Nurs (2006) 56(3):282–91. doi:10.1111/j.1365-2648.2006.04021.x
7. Gobbens RJ, van Assen MA, Luijkx KG, Schols JM. Testing an integral conceptual model of frailty. J Adv Nurs (2012) 68(9):2047–60. doi:10.1111/j.1365-2648.2011.05896.x
8. Walston J. Frailty – the search for underlying causes. Sci Aging Knowledge Environ (2004) 2004(4):4. doi:10.1126/sageke.2004.4.pe4
9. Walston JD. Biological markers and the molecular biology of frailty. In: Carey JR, Robine JM, Pierre Michel J, Christen Y, editors. Longevity and Frailty. Research and Perspectives in Longevity Berlin, Heidelberg: Springer (2005). p. 83–90.
10. Walston J, Arking D, Fallin D, Li T, Beamer B, Xue Q, et al. IL-6 gene variation is not associated with increased serum levels of IL-6, muscle, weakness, or frailty in older women. Exp Gerontol (2005) 40(4):344–52. doi:10.1016/j.exger.2005.01.012
11. Bortz WM. A conceptual framework of frailty: a review. J Gerontol A Biol Sci Med Sci (2002) 57(5):M283–8. doi:10.1093/gerona/57.5.M283
12. Bellizzi D, D’Aquila P, Montesanto A, Corsonello A, Mari V, Mazzei B, et al. Global DNA methylation in old subjects is correlated with frailty. Age (2012) 34(1):169–79. doi:10.1007/s11357-011-9216-6
13. Fulop T, Larbi A, Witkowski JM, McElhaney J, Loeb M, Mitnitski A, et al. Aging, frailty and age-related diseases. Biogerontology (2010) 11(5):547–63. doi:10.1007/s10522-010-9287-2
14. Afilalo J, Karunananthan S, Eisenberg MJ, Alexander KP, Bergman H. Role of frailty in patients with cardiovascular disease. Am J Cardiol (2009) 103(11):1616–21. doi:10.1016/j.amjcard.2009.01.375
15. Newman AB, Gottdiener JS, McBurnie MA, Hirsch CH, Kop WJ, Tracy R, et al. Associations of subclinical cardiovascular disease with frailty. J Gerontol A Biol Sci Med Sci (2001) 56(3):M158–66. doi:10.1093/gerona/56.3.M158
16. Perkisas S, Vandewoude M. Where frailty meets diabetes. Diabetes Metab Res Rev (2016) 32(S1):261–7. doi:10.1002/dmrr.2743
17. Veronese N, Stubbs B, Fontana L, Trevisan C, Bolzetta F, De Rui M, et al. Frailty is associated with an increased risk of incident type 2 diabetes in the elderly. J Am Med Dir Assoc (2016) 17(10):902–7. doi:10.1016/j.jamda.2016.04.021
18. Buchman AS, Boyle PA, Wilson RS, Tang Y, Bennett DA. Frailty is associated with incident Alzheimer’s disease and cognitive decline in the elderly. Psychosom Med (2007) 69(5):483–9. doi:10.1097/psy.0b013e318068de1d
19. Buchman AS, Schneider JA, Leurgans S, Bennett DA. Physical frailty in older persons is associated with Alzheimer disease pathology. Neurology (2008) 71(7):499–504. doi:10.1212/01.wnl.0000324864.81179.6a
20. Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet (2005) 6(2):95–108. doi:10.1038/nrg1521
21. Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet (2012) 90(1):7–24. doi:10.1016/j.ajhg.2011.11.029
22. Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science (2007) 316(5830):1491–3. doi:10.1126/science.1142842
23. Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med (2007) 357(5):443–53. doi:10.1056/NEJMoa072366
24. Helgadottir A, Thorleifsson G, Magnusson KP, Grétarsdottir S, Steinthorsdottir V, Manolescu A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet (2008) 40(2):217–24. doi:10.1038/ng.72
25. Björck H, Länne T, Alehagen U, Persson K, Rundkvist L, Hamsten A, et al. Association of genetic variation on chromosome 9p21. 3 and arterial stiffness. J Intern Med (2009) 265(3):373–81. doi:10.1111/j.1365-2796.2008.02020.x
26. Cluett C, McDermott MM, Guralnik J, Ferrucci L, Bandinelli S, Miljkovic I, et al. The 9p21 myocardial infarction risk allele increases risk of peripheral artery disease in older people. Circ Cardiovasc Genet (2009) 2:347–53. doi:10.1161/CIRCGENETICS.108.825935
27. Bilguvar K, Yasuno K, Niemelä M, Ruigrok YM, von und zu Fraunberg M, van Duijn CM, et al. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat Genet (2008) 40(12):1472–7. doi:10.1038/ng.240
28. Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet (2009) 41(8):920–5. doi:10.1038/ng.411
29. Li W-Q, Pfeiffer RM, Hyland PL, Shi J, Gu F, Wang Z, et al. Genetic polymorphisms in the 9p21 region associated with risk of multiple cancers. Carcinogenesis (2014) 35:2698–705. doi:10.1093/carcin/bgu203
30. Laaksovirta H, Peuralinna T, Schymick JC, Scholz SW, Lai S-L, Myllykangas L, et al. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol (2010) 9(10):978–85. doi:10.1016/S1474-4422(10)70184-8
31. Emanuele E, Lista S, Ghidoni R, Binetti G, Cereda C, Benussi L, et al. Chromosome 9p21. 3 genotype is associated with vascular dementia and Alzheimer’s disease. Neurobiol Aging (2011) 32(7):1231–5. doi:10.1016/j.neurobiolaging.2009.07.003
32. Schormair B, Kemlink D, Roeske D, Eckstein G, Xiong L, Lichtner P, et al. PTPRD (protein tyrosine phosphatase receptor type delta) is associated with restless legs syndrome. Nat Genet (2008) 40(8):946–8. doi:10.1038/ng.190
33. Mattheisen M, Samuels JF, Wang Y, Greenberg BD, Fyer AJ, McCracken JT, et al. Genome-wide association study in obsessive-compulsive disorder: results from the OCGAS. Mol Psychiatry (2015) 20(3):337–44. doi:10.1038/mp.2014.43
34. Silander K, Tang H, Myles S, Jakkula E, Timpson NJ, Cavalli-Sforza L, et al. Worldwide patterns of haplotype diversity at 9p21. 3, a locus associated with type 2 diabetes and coronary heart disease. Genome Med (2009) 1(5):51. doi:10.1186/gm51
35. Pasmant E, Sabbagh A, Vidaud M, Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J (2011) 25(2):444–8. doi:10.1096/fj.10-172452
36. Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron (2011) 72(2):257–68. doi:10.1016/j.neuron.2011.09.010
37. Veeriah S, Brennan C, Meng S, Singh B, Fagin JA, Solit DB, et al. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci U S A (2009) 106(23):9435–40. doi:10.1073/pnas.0900571106
38. Melzer D, Frayling TM, Murray A, Hurst AJ, Harries LW, Song H, et al. A common variant of the p16 INK4a genetic region is associated with physical function in older people. Mech Ageing Dev (2007) 128(5):370–7. doi:10.1016/j.mad.2007.03.005
39. Adams HH, Verlinden VJ, Callisaya ML, van Duijn CM, Hofman A, Thomson R, et al. Heritability and genome-wide association analyses of human gait suggest contribution of common variants. J Gerontol A Biol Sci Med Sci (2015) 71(6):740–6. doi:10.1093/gerona/glv081
40. Eny KM, Lutgers HL, Maynard J, Klein BE, Lee KE, Atzmon G, et al. GWAS identifies an NAT2 acetylator status tag single nucleotide polymorphism to be a major locus for skin fluorescence. Diabetologia (2014) 57(8):1623–34. doi:10.1007/s00125-014-3286-9
41. Roshandel D, Klein R, Klein BE, Wolffenbuttel BH, van der Klauw MM, van Vliet-Ostaptchouk JV, et al. A new locus for skin intrinsic fluorescence in type 1 diabetes also associated with blood and skin glycated proteins. Diabetes (2016) 65(7):2060–71. doi:10.2337/db15-1484
42. Bergman A, Atzmon G, Ye K, MacCarthy T, Barzilai N. Buffering mechanisms in aging: a systems approach toward uncovering the genetic component of aging. PLoS Comput Biol (2007) 3(8):e170. doi:10.1371/journal.pcbi.0030170
43. Lipton RB, Katz MJ, Kuslansky G, Sliwinski MJ, Stewart WF, Verghese J, et al. Screening for dementia by telephone using the memory impairment screen. J Am Geriatr Soc (2003) 51(10):1382–90. doi:10.1046/j.1532-5415.2003.51455.x
44. Mitnitski AB, Song X, Rockwood K. The estimation of relative fitness and frailty in community-dwelling older adults using self-report data. J Gerontol A Biol Sci Med Sci (2004) 59(6):M627–32. doi:10.1093/gerona/59.6.M627
45. Rockwood K, Mogilner A, Mitnitski A. Changes with age in the distribution of a frailty index. Mech Ageing Dev (2004) 125(7):517–9. doi:10.1016/j.mad.2004.05.003
46. Rockwood K, Mitnitski A. Frailty in relation to the accumulation of deficits. J Gerontol A Biol Sci Med Sci (2007) 62(7):722–7. doi:10.1093/gerona/62.7.722
47. Sternberg SA, Schwartz AW, Karunananthan S, Bergman H, Mark Clarfield A. The identification of frailty: a systematic literature review. J Am Geriatr Soc (2011) 59(11):2129–38. doi:10.1111/j.1532-5415.2011.03597.x
48. Fried TR, Bradley EH, Williams CS, Tinetti ME. Functional disability and health care expenditures for older persons. Arch Intern Med (2001) 161(21):2602–7. doi:10.1001/archinte.161.21.2602
49. Yesavage JA, Brink TL, Rose TL, Lum O, Huang V, Adey M, et al. Development and validation of a geriatric depression screening scale: a preliminary report. J Psychiatr Res (1983) 17(1):37–49. doi:10.1016/0022-3956(82)90033-4
50. Fugate Woods N, LaCroix AZ, Gray SL, Aragaki A, Cochrane BB, Brunner RL, et al. Frailty: emergence and consequences in women aged 65 and older in the Women’s Health Initiative Observational Study. J Am Geriatr Soc (2005) 53(8):1321–30. doi:10.1111/j.1532-5415.2005.53405.x
51. de Souto Barreto P, Greig C, Ferrandez A-M. Detecting and categorizing frailty status in older adults using a self-report screening instrument. Arch Gerontol Geriatr (2012) 54(3):e249–54. doi:10.1016/j.archger.2011.08.003
52. Johansen KL, Dalrymple LS, Delgado C, Kaysen GA, Kornak J, Grimes B, et al. Comparison of self-report − based and physical performance − based frailty definitions among patients receiving maintenance hemodialysis. Am J Kidney Dis (2014) 64(4):600–7. doi:10.1053/j.ajkd.2014.03.016
53. Ayers E, Shapiro M, Holtzer R, Barzilai N, Milman S, Verghese J. Symptoms of apathy independently predict incident frailty and disability in community-dwelling older adults. J Clin Psychiatry (2017) 78:e529–36. doi:10.4088/JCP.15m10113
54. Brach JS, Perera S, Studenski S, Newman AB. The reliability and validity of measures of gait variability in community-dwelling older adults. Arch Phys Med Rehabil (2008) 89(12):2293–6. doi:10.1016/j.apmr.2008.06.010
55. Holtzer R, Wang C, Verghese J. The relationship between attention and gait in aging: facts and fallacies. Motor Control (2012) 16(1):64. doi:10.1123/mcj.16.1.64
56. 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature (2010) 467(7319):1061–73. doi:10.1038/nature09534
57. Xue Q-L. The frailty syndrome: definition and natural history. Clin Geriatr Med (2011) 27(1):1–15. doi:10.1016/j.cger.2010.08.009
58. Verghese J, Wang C, Lipton RB, Holtzer R, Xue X. Quantitative gait dysfunction and risk of cognitive decline and dementia. J Neurol Neurosurg Psychiatry (2007) 78(9):929–35. doi:10.1136/jnnp.2006.106914
59. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (2005) 21(2):263–5. doi:10.1093/bioinformatics/bth457
60. Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science (2002) 296(5576):2225–9. doi:10.1126/science.1069424
61. Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics (2006) 22(15):1928–9. doi:10.1093/bioinformatics/btl268
62. GTEx Consortium. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (2015) 348(6235):648–60. doi:10.1126/science.1262110
63. Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res (2012) 22(9):1790–7. doi:10.1101/gr.137323.112
64. Guo L, Du Y, Qu S, Wang J. rVarBase: an updated database for regulatory features of human variants. Nucleic Acids Res (2015) 44(D1):D888–93. doi:10.1093/nar/gkv1107
65. Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res (2008) 36(Suppl 1):D820–4. doi:10.1093/nar/gkm904
66. Baynes C, Healey CS, Pooley KA, Scollen S, Luben RN, Thompson DJ, et al. Common variants in the ATM, BRCA1, BRCA2, CHEK2 and TP53 cancer susceptibility genes are unlikely to increase breast cancer risk. Breast Cancer Res (2007) 9(2):R27. doi:10.1186/bcr1669
67. Pharoah PD, Tyrer J, Dunning AM, Easton DF, Ponder BA, Investigators S. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet (2007) 3(3):e42. doi:10.1371/journal.pgen.0030042
68. Benjamin EJ, Rice KM, Arking DE, Pfeufer A, Van Noord C, Smith AV, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet (2009) 41(8):879–81. doi:10.1038/ng.416
69. Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene (2000) 249(1):31–45. doi:10.1016/S0378-1119(00)00140-2
70. Li B-H, Zhang L-L, Yin Y-W, Pi Y, Guo L, Yang Q-W, et al. Association between interleukin-1α C (−889) T polymorphism and Alzheimer’s disease: a meta-analysis including 12,817 subjects. J Neural Transm (2013) 120(3):497–506. doi:10.1007/s00702-012-0867-y
71. Harris L, Genovesi LA, Gronostajski RM, Wainwright BJ, Piper M. Nuclear factor one transcription factors: divergent functions in developmental versus adult stem cell populations. Dev Dyn (2015) 244(3):227–38. doi:10.1002/dvdy.24182
72. Roy S, Bantel H, Wandrer F, Schneider AT, Gautheron J, Vucur M, et al. miR-1224 inhibits cell proliferation in acute liver failure by targeting the antiapoptotic gene Nfib. J Hepatol (2017) 67:966–78. doi:10.1016/j.jhep.2017.06.007
73. Rolando C, Erni A, Grison A, Beattie R, Engler A, Gokhale PJ, et al. Multipotency of adult hippocampal NSCs in vivo is restricted by Drosha/NFIB. Cell Stem Cell (2016) 19(5):653–62. doi:10.1016/j.stem.2016.07.003
74. Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol (2005) 25(2):685–98. doi:10.1128/MCB.25.2.685-698.2005
75. Piper M, Barry G, Harvey TJ, McLeay R, Smith AG, Harris L, et al. NFIB-mediated repression of the epigenetic factor Ezh2 regulates cortical development. J Neurosci (2014) 34(8):2921–30. doi:10.1523/JNEUROSCI.2319-13.2014
76. Denny SK, Yang D, Chuang C-H, Brady JJ, Lim JS, Grüner BM, et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell (2016) 166(2):328–42. doi:10.1016/j.cell.2016.05.052
77. Waki H, Nakamura M, Yamauchi T, Wakabayashi K-I, Yu J, Hirose-Yotsuya L, et al. Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genet (2011) 7(10):e1002311. doi:10.1371/journal.pgen.1002311
78. Mirabello L, Koster R, Moriarity BS, Spector LG, Meltzer PS, Gary J, et al. A genome-wide scan identifies variants in NFIB associated with metastasis in patients with osteosarcoma. Cancer Discov (2015) 5(9):920–31. doi:10.1158/2159-8290.CD-15-0125
79. Lemmelä S, Solovieva S, Shiri R, Benner C, Heliövaara M, Kettunen J, et al. Genome-wide meta-analysis of sciatica in Finnish population. PLoS One (2016) 11(10):e0163877. doi:10.1371/journal.pone.0163877
80. Breitling LP, Saum K-U, Perna L, Schöttker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics (2016) 8(1):21. doi:10.1186/s13148-016-0186-5
81. Feser J, Tyler J. Chromatin structure as a mediator of aging. FEBS Lett (2011) 585(13):2041–8. doi:10.1016/j.febslet.2010.11.016
82. Lardenoije R, Iatrou A, Kenis G, Kompotis K, Steinbusch HW, Mastroeni D, et al. The epigenetics of aging and neurodegeneration. Prog Neurobiol (2015) 131:21–64. doi:10.1016/j.pneurobio.2015.05.002
83. Sato K, Nakagawa H, Tajima A, Yoshida K, Inoue I. ANRIL is implicated in the regulation of nucleus and potential transcriptional target of E2F1. Oncol Rep (2010) 24(3):701–7. doi:10.3892/or_00000910
84. Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, Lipton R, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA (2003) 290(15):2030–40. doi:10.1001/jama.290.15.2030
85. Atzmon G, Rincon M, Schechter CB, Shuldiner AR, Lipton RB, Bergman A, et al. Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLoS Biol (2006) 4(4):e113. doi:10.1371/journal.pbio.0040113
Keywords: frailty, fried index, 9p21-23 locus, aging, genetics
Citation: Sathyan S, Barzilai N, Atzmon G, Milman S, Ayers E and Verghese J (2018) Genetic Insights Into Frailty: Association of 9p21-23 Locus With Frailty. Front. Med. 5:105. doi: 10.3389/fmed.2018.00105
Received: 15 January 2018; Accepted: 29 March 2018;
Published: 01 May 2018
Edited by:
Claudio Franceschi, Università degli Studi di Bologna, ItalyReviewed by:
Wee Shiong Lim, Tan Tock Seng Hospital, SingaporeMario Ulises Pérez-Zepeda, Instituto Nacional de Geriatría, Mexico
Copyright: © 2018 Sathyan, Barzilai, Atzmon, Milman, Ayers and Verghese. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joe Verghese, joe.verghese@einstein.yu.edu
 Sanish Sathyan
Sanish Sathyan Nir Barzilai
Nir Barzilai Gil Atzmon
Gil Atzmon Sofiya Milman
Sofiya Milman Emmeline Ayers
Emmeline Ayers Joe Verghese
Joe Verghese