- 1 Gamaleya Research Institute, Moscow, Russia
- 2 Institute for Experimental and Clinical Pharmacology and Toxicology, Albert-Ludwigs University of Freiburg, Freiburg, Germany
Legionella causes severe pneumonia in humans. The pathogen produces an array of effectors, which interfere with host cell functions. Among them are the glucosyltransferases Lgt1, Lgt2 and Lgt3 from L. pneumophila. Lgt1 and Lgt2 are produced predominately in the post-exponential phase of bacterial growth, while synthesis of Lgt3 is induced mainly in the lag-phase before intracellular replication of bacteria starts. Lgt glucosyltransferases are structurally similar to clostridial glucosylating toxins. The enzymes use UDP–glucose as a donor substrate and modify eukaryotic elongation factor eEF1A at serine-53. This modification results in inhibition of protein synthesis and death of target cells.In addition to Lgts, Legionella genomes disclose several genes, coding for effector proteins likely to possess glycosyltransferase activities, including SetA (subversion of eukaryotic vesicle trafficking A), which influences vesicular trafficking in the yeast model system and displays tropism for late endosomal/lysosomal compartments of mammalian cells. This review mainly discusses recent results on the structure–function relationship of Lgt glucosyltransferases.
Multifaceted Mechanisms in Legionella – Host Cell Interaction
Legionella is a fastidious Gram-negative bacterium, causing severe pneumonia in humans named Legionnaires’ disease. Among known species of Legionella, the most important human pathogen is Legionella pneumophila, strains of which account for more than 90% of morbidity records due to legionellosis (Diederen, 2008). Despite the description of at least 15 serogroups in this species, L. pneumophila serogroup 1 is responsible for over 80% of cases of the disease (Yu et al., 2002). Legionella longbeachae and Legionella bozemanii are the next most common etiological agents of Legionnaires’ disease, accounting for up to 7% of Legionella infections in Europe and in the USA (Muder and Yu, 2002). Interestingly, in Australia and New Zealand L. longbeachae is responsible for ∼30% of Legionnaires’ disease cases (Yu et al., 2002). In contrast to well-studied L. pneumophila, virulence mechanisms utilized by L. longbeachae and L. bozemanii are largely unknown.
Legionella pneumophila is able to multiply inside eukaryotic cells – either in free-living unicellular organisms (amebae and ciliated protozoa) or in mammalian cells (macrophages, monocytes, epithelial cells; Jules and Buchrieser, 2007). After uptake by host cells, the Legionella-containing phagosome is subjected to specialized biogenesis steps, leading to transformation of this organelle into a niche that supports multiplication of the bacteria (Isberg et al., 2009). A specialized type IV secretion system (T4SS), encoded by dot and icm gene clusters, translocates numerous bacterial effectors (>300 effectors; Hubber and Roy, 2010) into target cells, which participate in the change of the phagosome into a “replicative vacuole” (Ninio and Roy, 2007; Ensminger and Isberg, 2009). Several Legionella effectors target small GTP-binding proteins, participating in regulation of vesicular trafficking of the host cell. These eukaryotic proteins are molecular switches, which are regulated by a GTPase cycle. Legionella effectors specifically switch on or switch off these GTP-binding proteins. For example, the mammalian Arf1 protein, which is involved in vesicle formation in the Golgi, is activated by Legionella protein RalF, which acts as a GDP/GTP exchange factor for this small GTPase (Nagai et al., 2002). Another example is the Ras-superfamily protein Rab1, which regulates various steps of vesicle trafficking in eukaryotic cells. Rab1 is manipulated by several Legionella effectors, including DrrA/SidM, LidA, and LepB (Machner and Isberg, 2006; Murata et al., 2006). DrrA/SidM is a multifunctional protein, containing guanine nucleotide exchange activity and adenylyltransferase activity (Ingmundson et al., 2007; Machner and Isberg, 2007; Muller et al., 2010). Legionella effector LidA enhances Rab1 recruitment by DrrA/SidM (Machner and Isberg, 2006) and the effector LepB behaves as a Rab1 GTPase-activating protein, which inactivates the Rab protein (Ingmundson et al., 2007).
However, not only vesicular trafficking is targeted during biogenesis steps of the Legionella-containing vacuole. L. pneumophila is able to maintain a neutral pH inside its phagosome (Horwitz and Maxfield, 1984). It was shown recently that T4SS effector SidK targets vacuolar ATPase by interacting with VatA (also called VMA1), one of the key components of the vesicular proton pump, which is involved in ATP hydrolysis. Legionella effector SidK inhibits ATP hydrolysis, thereby blocking proton translocation and vacuole acidification (Xu et al., 2010). Similar to other pathogens L. pneumophila is able to exploit the eukaryotic ubiquitin-conjugating system for establishing successful intracellular infection (Ivanov and Roy, 2009). To achieve this, the bacterium produces several T4SS effector proteins that function in the eukaryotic ubiquitination pathway (Kubori et al., 2008; Price et al., 2009, 2010; Lomma et al., 2010). Legionella-induced modulation of target cell survival is also observed during intracellular proliferation of the bacterium. Several Legionella effector proteins have been shown to participate in apoptotic and anti-apoptotic processes either directly or indirectly (Laguna et al., 2006; Abu-Zant et al., 2007; Banga et al., 2007). Moreover, L. pneumophila modulates inflammatory responses through NF-κB (Ge et al., 2009; Losick et al., 2010), induces mitochondrial recruitment and microfilament rearrangements (Chong et al., 2009) or regulates MAP kinase response to bacteria (Li et al., 2009). Thus, all these findings indicate an extremely complex Legionella-host cell interaction.
LGTs as a New Family of Glucosyltransferases in L. pneumophila
Recently it was shown that glycosyltransferases (GTs) are highly effective virulence factors of Legionella. These enzymes target eukaryotic substrates by covalent attachment of glycosyl moieties to eukaryotic proteins thereby altering their functions (Belyi and Aktories, 2010).
The first glucosyltransferase purified from L. pneumophila Philadelphia-1 strain was Legionella glucosyltransferase 1 (Lgt1). Lgt1 has a molecular mass of 59.7 kDa and modifies a ∼50-kDa component in cytoplasmic fraction of eukaryotic cells (Belyi et al., 2003). The enzymatic activity is sugar-specific, i.e., only UDP–glucose, but not UDP–galactose, UDP–N-acetyl-galactosamine, UDP–N-acetyl-glucosamine, UDP–glucuronic acid, or GDP-mannose serves as donor substrate in the reaction (Belyi et al., 2006).
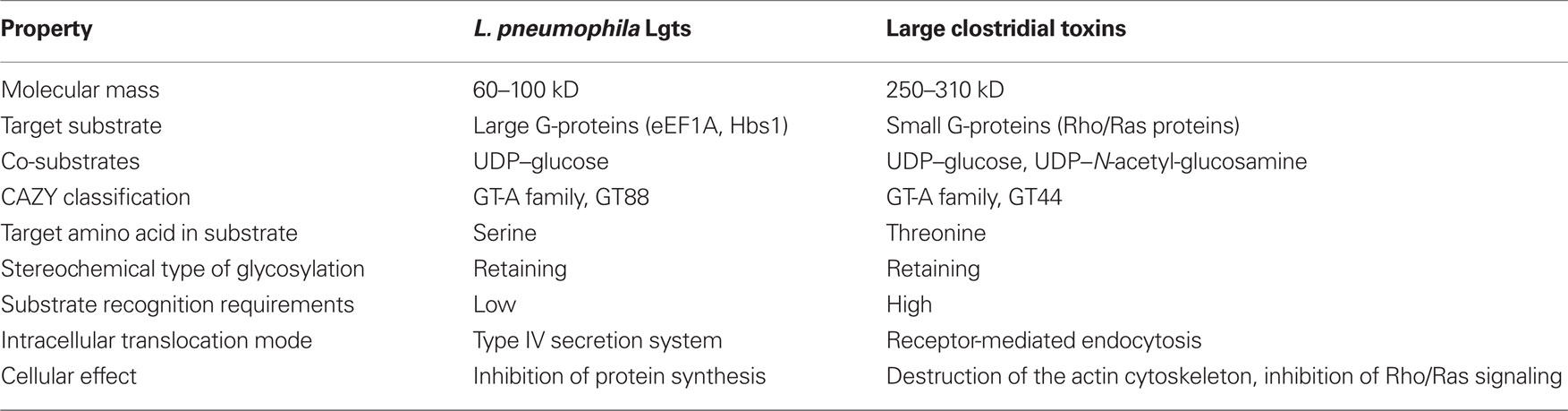
The primary amino acid sequence of Lgt1 shares little homology with known proteins. The only notable similarity is found between the central region of Lgt1 and the catalytic core of clostridial glucosylating toxins (CGT; Figure 1A; Table 1). In this region several groups of conserved amino acid residues could be identified, including the two aspartic amino acids D246 and D248, representing the DXD-motif – a known hallmark of GTs (Belyi et al., 2006).
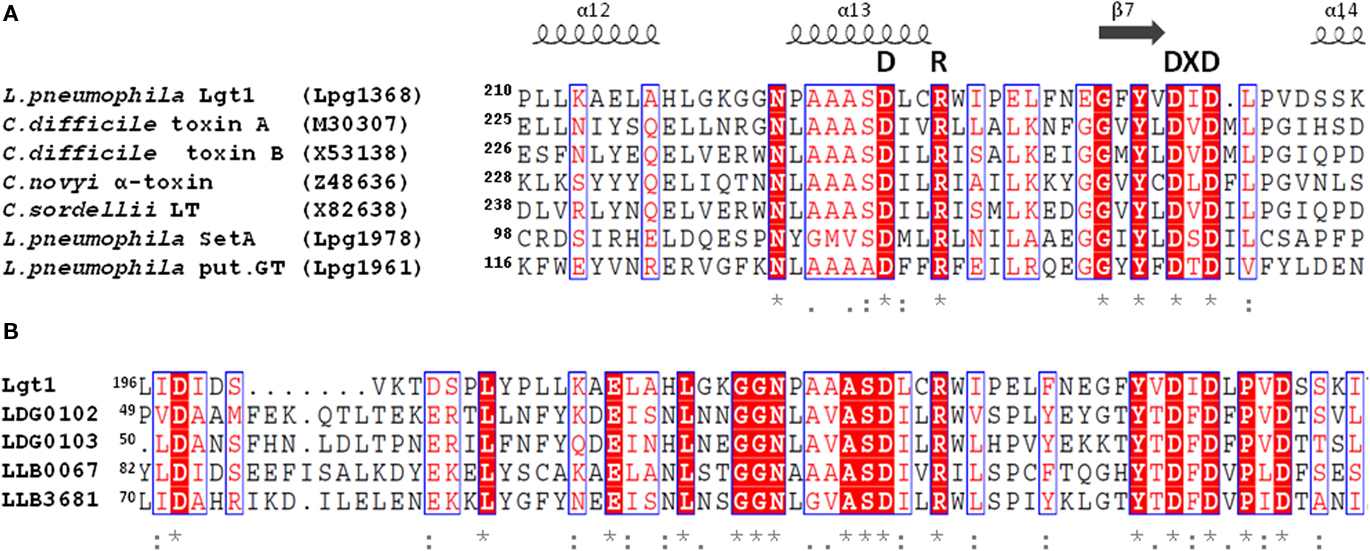
Figure 1. (A) Alignment of partial amino acid sequences of Lgt1, SetA, Lpg1961 from L. pneumophila Philadelphia-1 strain with that of proteins from a clostridial glucosylating toxin family: Toxins A and B from C. difficile, α-toxin from C. novyi, and lethal toxin from C. sordellii. Gene bank accession numbers of the corresponding coding sequences are shown in brackets. Essential amino acids mentioned in the text are highlighted (DXD-motif, GT-A triad). (B) Alignment of partial amino acid sequences of Lgt1 from L. pneumophila Philadelphia-1 with that of putative glycosyltransferases found in translated genomes of L. drancourtii LLAP12 and L. longbeachae D-4968. Identification codes for Lgt1 and putative glycosyltransferases in strain LLAP12 of L. drancourtii and strain D-4968 of L. longbeachae (two products in each strain) are Lpg1368, LDG0102/LDG0103, and LLB0067/LLB3681 respectively. Proteins LLO1578 and LLO1721 found in translated sequenced genome of L. longbeachae NSW150 were 100% identical to LLB0067 and LLB3681 from L. longbeachae D-4968 respectively and are not shown on the figure for simplicity reason. Identical amino acid residues are denoted by asterisks, highly conserved residues by double dots, and modestly conserved residues by dots. The secondary structural elements were deduced from the structure of Lgt1 (pdb 3JSZ). The alignment was prepared using ESPript 2.2 (http://espript.ibcp.fr).
Database searches in the sequenced genomes of six L. pneumophila strains (Philadelphia-1, Corby, Lens, Paris, 2300/99 Alcoy, and 130b) disclosed altogether 13 open reading frames with significant sequence homology with Lgt1 (Table 2). Based upon the level of identity, these gene products can be grouped into three families: Lgt1 through Lgt3 [in Philadelphia-1 strain the gene IDs (identification labels, used to distinguish coding sequences) are lpg1368, lpg2862, and lpg1488, coding for ∼60 kDa Lgt1, ∼70 kDa Lgt2, and ∼100 kDa Lgt3, respectively]. Only one copy of each gene family member is present in the corresponding genome. Philadelphia-1 strain contains the full set of the genes (i.e., lgt1, lgt2, and lgt3), whereas the other strains possess only lgt1 and lgt3. Representatives within each family are ∼90% identical in amino acid sequences whereas homology between the three groups’ members are in the range of 15–27%. Lgt1, Lgt2, and Lgt3 are serologically distinct and do not display antigenic cross-reactivity (Belyi et al., 2008).The enzymes are grouped into the glucosyltransferase family GT88 in the carbohydrate modifying enzymes database (http://www.cazy.org/GT88.html; Coutinho et al., 2003).
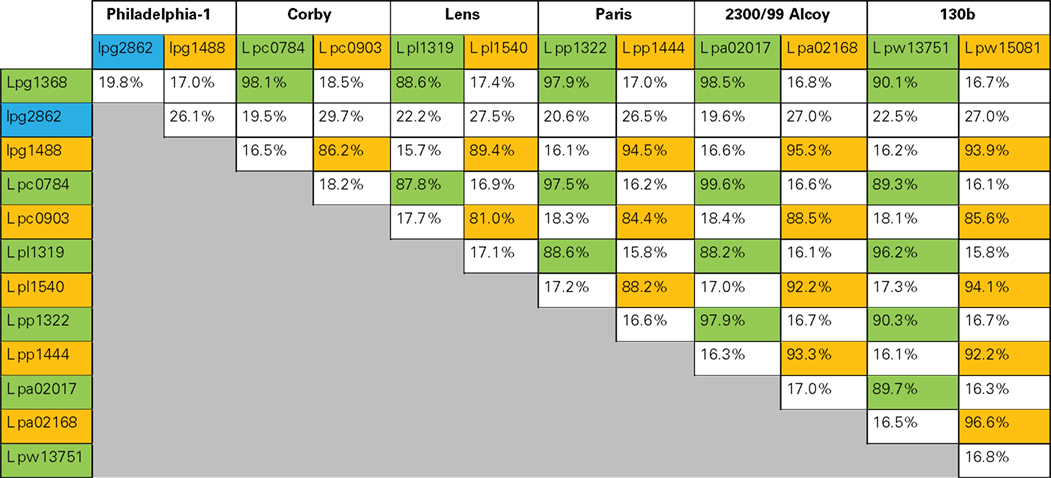
Table 2. Amino acid sequence identity of proteins of the Lgt-family of L. pneumophila (Philadelphia-1, Corby, Lens, Paris, 2300/99 Alcoy, and 130b). Proteins belonging to Lgt1, Lgt2, or Lgt3 groups were cross-aligned pair-wise to determine the degree of identical amino acid residues. The homology is shown as a percentage of identical amino acid residues. Lgt1-group glucosyltransferases are marked by green, Lgt2 – by blue, and Lgt3 – by yellow color.
To accomplish their functions bacterial virulence factors should be translocated into cytoplasm of a target cell. Legionella glucosyltransferases apparently miss a specific receptor-binding and translocation domain, which is typical for bacterial AB-type exotoxins. Accordingly, they do not produce toxic effects, when added into mammalian cell culture medium, indicating the requirement of a specialized secretion system. As shown in experiments using adenylate cyclase- or β-lactamase-chimeras, all Lgts are secreted via T4SS (de Felipe et al., 2005, 2008; Hurtado-Guerrero et al., 2010).
Often T4SS effectors are produced during the stationary phase of bacterial growth (Bruggemann et al., 2006; Zusman et al., 2007). At this stage bacterial cells become remarkably virulent and display a transmission phenotype (Byrne and Swanson, 1998). Also the production of Lgt1 and Lgt2 is strongly increased at the stationary phase of bacterial growth in broth; however, Lgt3 is detectable mainly in the pre-logarithmic phase of in vitro cultivation. Same results are obtained in in vivo experiments using the protozoan Acanthamoeba castellanii model as a host for L. pneumophila. Levels of mRNA coding for Lgt1 is maximal at late phase of co-infection, while lgt3 is expressed mainly at the initial stage of bacterium–ameba interaction (Belyi et al., 2008). These experiments suggest differential regulation of glucosyltransferase activity in L. pneumophila, which, in turn, indicates specific roles of each enzyme in bacterial virulence. One can speculate that Lgt3 is important for initiation of infection cycle, while Lgt1/Lgt2 is necessary for egress of Legionella from the host cell.
Targeting of eEF1A by Legionella Glucosyltransferases
Legionella glucosyltransferase 1, Lgt2, and Lgt3 glucosylate an ∼50-kDa component in mammalian cell extracts, which has been identified as elongation factor 1A (eEF1A). All these Legionella glucosyltransferases modify serine-53 of eEF1A (Belyi et al., 2006).
Elongation factor eEF1A, which is one of the most abundant proteins in eukaryotic cells, plays a key role in ribosome-dependent protein synthesis (Ramakrishnan, 2002). It possesses GTP-binding and GTPase activities and is required for the recruitment of aminoacylated tRNA to the A-site of mRNA-charged ribosomes. In addition, eEF1A was shown to be involved in several other cellular processes (Mateyak and Kinzy, 2010), including translational control, assembling/folding of newly synthesized proteins and proteosomal degradation of incorrectly folded peptides (Hotokezaka et al., 2002; Chuang et al., 2005), lipotoxic cell death (Borradaile et al., 2006), apoptosis (Ruest et al., 2002), nuclear export (Khacho et al., 2008), viral propagation (Matsuda et al., 2004), and regulation of actin cytoskeleton and cell morphology (Ejiri, 2002; Gross and Kinzy, 2005).
No structural data for mammalian eEF1A is available; however the very similar yeast elongation factor 1A from Saccharomyces cerevisiae has been crystallized and analyzed in detail (Andersen et al., 2000). The obtained structure shows that eEF1A is composed of three domains (Figure 2): domain 1 consists of ∼240 residues and is characterized by a Ras-like fold (Kjeldgaard et al., 1996). It contains consensus sequences of typical GTP-binding proteins and is termed therefore “G-domain.” Key features of this domain are binding and hydrolysis of GTP. Domains 2 and 3, consisting of 89 and 107 residues respectively, have a β-barrel structure and are involved in interaction with different targets like aminoacyl-tRNA and the elongation factor eEF1Bα, which is a GDP/GTP exchange factor of eEF1A (Andersen et al., 2000).
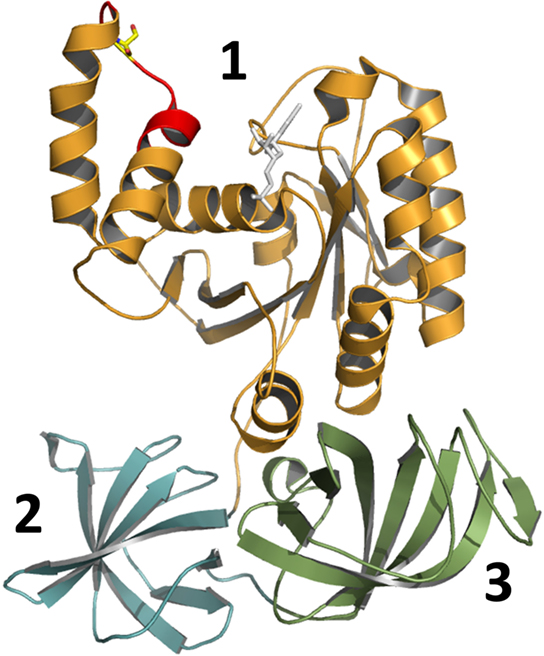
Figure 2. Structural view of yeast elongation factor eEF1A (adapted from pdb 1IJF). Elongation factor eEF1A consists of three main structural parts: domain 1 (G-domain), domain 2, and domain 3 (indicated by numbers). The decapeptide (GKGSFKYAWV), which is a sufficient substrate for glucosylation by Lgt, is shown in red. Serine-53, which is modified by glucosyltransferases Lgt, is shown in yellow. The complexed fragment of eEF1Bα molecule, which is present in the original structure, is omitted.
Serine-53 of eEF1A (Figure 2, shown in yellow), which is modified by Lgt, is located in the G-domain near the switch-1 region of the GTPase (Belyi et al., 2006, 2008). For the prokaryotic analog EF-Tu, it is known that the switch-1 region undergoes major conformational changes, depending on the nucleotide bound (GDP or GTP; Abel et al., 1996; Vetter and Wittinghofer, 2001). However in eEF1A the switch-1 region is not well defined, because two additional helices (A* and A′) are present, and no nucleotide-dependent structural changes in this region have been reported for eEF1A so far. Noteworthy, bacterial EF-Tu lacks Ser-53 excluding alteration of protein synthesis by glucosylation in Legionella.
Surprisingly, fragments of recombinant eEF1A are better substrates for glucosylation than full size eEF1A in vitro. Truncation analysis revealed that considerable portions of the elongation factor are dispensable for substrate recognition. Neither domains 2 nor 3 of eEF1A are necessary for glucosylation. Even the G-domain can be reduced to a decapeptide comprised of residues 50-GKGSFKYAWV-59. This peptide represents the loop of the helix–loop–helix region formed by helices A* and A′ of eEF1A and is part of the first turn of helix A′ (Figure 2, shown in red). Substitution of Ser-53, Phe-54, Tyr-56, or Trp-58 with alanine prevents or strongly decreases glucosylation. Even more surprising is the finding that modification of the decapeptide by Lgt1 is more efficient than the glucosylation of the isolated full length eEF1A. This suggests that the substrate properties of eEF1A depend on a specific conformation of the full length protein, which allows modification by the Legionella enzymes (Belyi et al., 2009).
Hbs1 Protein as a Novel Substrate of Legionella Glucosyltransferase Lgt
In silico screenings with the minimal peptide sequence, which is accepted as substrate for glucosylation by Lgts, retrieved the 70-kDa Hsp70 subfamily B suppressor 1 (Hbs1) as another possible target for Lgt1. Hbs1 shares significant sequence similarities with eEF1A (19% identity) and releasing factor eRF3 (24% identity) all over the protein. Moreover, yeast Hbs1 and human Hbs1-like proteins contain the decapeptides 210-GKSSFKFAWI-219 and 311-GKASFAYAWV-320, respectively, which are modified by Lgts. In vitro, all Lgt-family members are capable of glucosylating Hbs1. However, so far it is not known whether Hbs1 is a substrate of Legionella glucosyltransferases in intact cells.
The functional role of Hbs1 has been the topic of several investigations. First, it was shown that an increased copy number of Hbs1 suppresses the growth defect of the S. cerevisiae double mutant in ssb1 and ssb2 genes. Proteins Ssb1/2 are chaperones of the Hsp70 family that are associated with translating ribosomes and may aid in the passage of the nascent polypeptide through the ribosome channel into the cytosol (Nelson et al., 1992). Thus, these first experiments suggest a role of Hbs1 in the translational machinery although its precise function has not been established.
First direct indication toward the role of Hbs1 in eukaryotic cell physiology came from studies on the mechanism of RNA surveillance in yeast. Stalled translational complexes, which halt in elongation due to inhibitory structures or defects of translated mRNA (e.g., hairpin loops, rare codons, chemical damage), are subjected to specific degradation steps, termed “no-go-decay” (NGD). NGD starts with endonucleolytic cleavage of mRNAs near the site of the stall followed by degradation of produced 5′ and 3′ ribonucleic acid fragments (Doma and Parker, 2006). Such initial cleavage appears to depend on Hbs1 and another protein Dom34. Deletion of Dom34 avoid endonucleolytic cleavage, while deletion of Hbs1 strongly reduced but not prevented NGD. The latter observation suggests that Hbs1 although important is not absolutely required for this type of mRNA surveillance system. Recent studies by using in vitro reconstituted yeast translation system shed more light onto the function of Hbs1/Dom34 complex. According to these data, Hbs1/Dom34 directly destabilizes the mRNA:ribosome complex and promotes recycling of its functional components (Shoemaker et al., 2010). So far, however, it completely enigmatic how processes of NGD are related to the infection biology of Legionella.
Structural and Mechanistic Features of L. pneumophila Glucosyltransferase Lgt1
The crystal structure of Lgt1 was solved recently by two independent research groups almost simultaneously (Figure 3A; Hurtado-Guerrero et al., 2010; Lu et al., 2010). In principle the structures resemble two catalytic states. One structure represents the catalytic competent state with intact UDP–glucose and the divalent ion preformed for acceptor binding and modification (LplGT·UDP–glucose·Mg2+ pdb 2WZG, 3JSZ). The second structure most likely exhibits the product state with the donor substrate hydrolyzed (LppGT·UDP·glucose·Mn2+, pdb 2WZF).
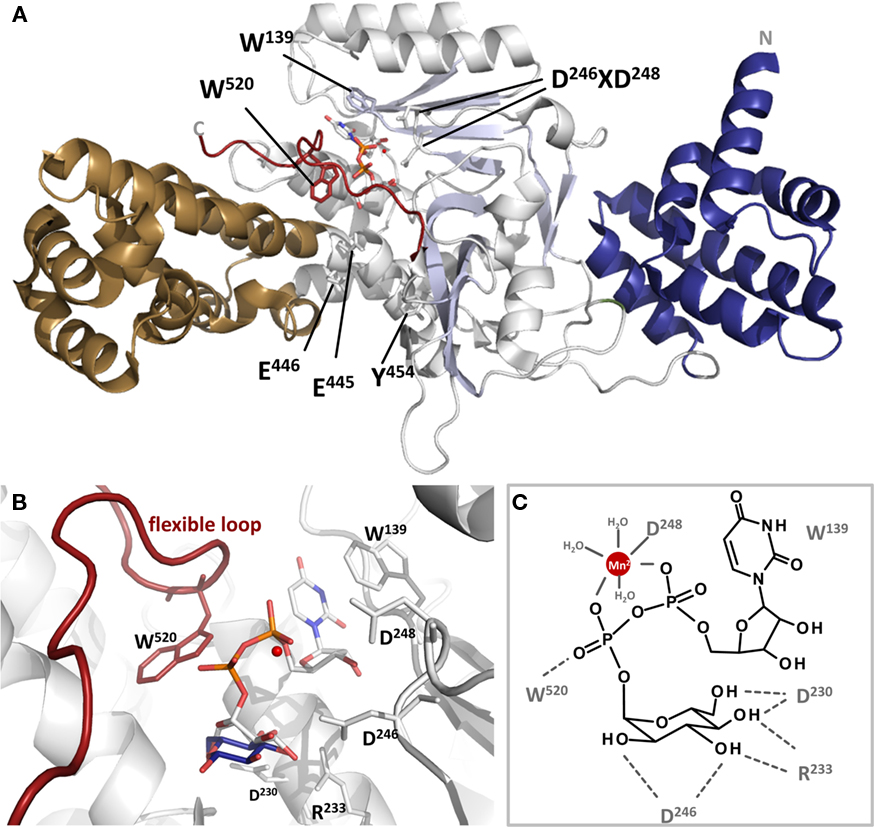
Figure 3. Cartoon presentation of Lgt1 crystal structure in complex with UDP–glucose and Mg2+ (pdb code 3JSZ). (A) The N-terminal domain is depicted in blue, the central domain in gray and the protrusion domain in brown. The central beta sheet is shaded in light blue. UDP–glucose is shown in sticks and Mg2+ as a red sphere. The flexible loop region is highlighted in dark red. Aspartic acid residues of the DXD-motif, Trp-520 of the flexible loop, Trp-139 stacking the base are shown in sticks. (B) Magnified view on the catalytic site of Lgt1 as in (A) with intact UDP–glucose (white) and glucose shifted about 1.6 Å after cleavage (dark blue; deduced from pdb 2WZF). Important amino acids are shown as sticks. Trp-139 is stacking the uracil ring of the donor, Asp-230, Arg-233, and Asp-246 are orientating the distal part of glucose as a triade. Asp-248 of the DXD-motif is coordinating the divalent ion (red) in conjunction with three additional water molecules. Trp-520 of the flexible loop is marked in red. (C) Schematic representation of the catalytic site as in (B) with important amino acids marked and highlighted.
In general, the structure of Lgt1 shows a mixed α/β-fold, which is grouped into the GT-A family of GTs. Lgt1 can be dissected into three different structural domains. Domain I consists of seven N-terminal α-helices (α1–α7) with yet unknown functions (Figure 3A). Domain II constitutes the typical GT GT-A core domain with a twisted continuous central β-sheet surrounded by α-helices presenting the double Rossmann fold-like signature (α8–α15/β1–β10). This nucleotide binding domain harbors the donor substrate-binding site and catalytic important residues. Domain III is a predominant α-helical “protrusion domain” (α16–α30/β11–β12) suggested to be involved in acceptor substrate-binding (Hurtado-Guerrero et al., 2010). As a common structural feature in GTs, Lgts possess a C-terminal flexible loop, which seems to be important for the proper arrangement of the acceptor binding site and the release of the reaction products (Figure 3A). Structural BLASTs with Lgt1 show highest similarity with the CGT toxin B from C. difficile, lethal toxin from C. sordellii, and α-toxin from C. novyi. Similarity is restricted to the catalytic core of the GTs, where interestingly several catalytically important residues are structurally very well conserved (Figure 1A). The N-terminal helical domains (domain I) of toxin B and Lgts are topologically unrelated and the function as a subcellular sorting signal, as found in CGTs, is not analyzed yet for Lgts (Mesmin et al., 2004; Kamitani et al., 2010). Sequence comparison of Lgt1 with the other family members Lgt2 and Lgt3 shows an overall identity of only 18–28%, due to several additional coiled-coil domains and C-terminal extensions. Nevertheless, remarkably high conservation is found in the nucleotide binding site, the suggested catalytic amino acids, and the acceptor binding groove. The unique function or structural importance of the C-terminal extension of Lgt3 remains elusive. It was shown that Lgt2 and Lgt3 and several other Legionella effectors have their type IV secretion signal in the C-terminus of the protein. In contrast Lgt1 seems to have its type IV secretion signal sequence in the N-terminal region (Hurtado-Guerrero et al., 2010). The exact position or motifs for this signal are not known yet. In Lgt1 it was speculated that this region is located within the first 10 amino acids, which were unfortunately disordered in the crystal structures.
UDP–Glucose Binding Pocket
As depicted in Figure 3B the binding of the sugar nucleotide proceeds via loops of the central β-sheet and the protrusion domain in a “curled under” conformation typical in GTs (Gibson et al., 2004; Qasba et al., 2005). In this tense conformation the glucose moiety is tucked underneath the pyrophosphate bridge positioning the anomeric carbon of glucose in such a manner to provide access for the incoming acceptor substrate. The nucleotide portion is bound by three loops (α12–α13, α4–α8, C-terminal loop) mainly via hydrogen bonding to the backbone. The uracil ring of UDP is sandwiched between Trp-139 and Pro-225 by hydrophobic stacking. The distal part of the glucosyl moiety of the donor substrate is bound by a typical triad binding geometry formed by Asp-230, Arg-233, and Asp-246 (Figure 3B; Negishi et al., 2003; Jank et al., 2007). This specific hydrogen bonding network might determine the sugar selectivity at the 4′-OH position, thus using glucose instead of galactose. Comparison of UDP–glucose bound to Lgt1 in the intact and cleaved form showed that in both states the nucleotides are bound in the same manner and adopt the same conformation. The main structural divergence is seen in a positional shift of the anomeric carbon of about 1.6 Å. Interestingly the same shift is recognized in the structure of C. difficile toxin B. Structural analysis of carbohydrate metabolizing enzymes as glycosidases reveal a similar movement of the anomeric atom of the sugar after hydrolysis, here called “electrophilic migration” (Vocadlo et al., 2001). This conserved movement substantiates the mechanistic importance of global structural rearrangements of the GT leading to a significant distortion of the donor substrate during transition state and hydrolysis.
DXD-motif
The DXD-motif (Asp-246 and Asp-248) upstream of a short hydrophobic patch is the remarkable motif for GTs of the GT-A type and crucial for divalent cation binding (Figures 1 and 3). In Lgt1 the cation is coordinated in an octahedral complex where two valences are occupied by the α- and β-phosphates of UDP. As in several other GTs only the second aspartic acid of the DXD-motif is involved in direct cation coordination, the first residue coordinates Mn2+ through a water molecule and hydrogen bonds a distal glucose hydroxyl. The remaining two valences are occupied by water molecules. Only mutation of the first aspartic acid lead to dramatic reduction in enzyme activity showing its fundamental importance (Hurtado-Guerrero et al., 2010). The role of the divalent metal ion in Lgts as in other GTs seems to be severalfold. Binding of the metal ion in conjunction with the donor substrate is a prerequisite for the induction of a conformational change in the C-terminal flexible loop region (Ziegler et al., 2008). Furthermore, the ion is necessary for the stabilization of the transition state during catalysis by compensating the negative charge of the β-phosphate of the nucleotide and facilitating the departure of the leaving group (Charnock and Davies, 1999; Qasba et al., 2005; Ramakrishnan et al., 2006; Hurtado-Guerrero et al., 2010).
Glycosyltransfer Mechanism
In general it is assumed that the reaction catalyzed by GTs follows a sequential ordered mechanism. Here, the metal ion and sugar nucleotide bind first followed by the acceptor (Qasba et al., 2005). After glycosyltransfer the product is ejected followed by the nucleotide and the metal ion. For Lgts there are some hints that the metal ion remains bound to the enzyme very tightly and is not ejected (unpublished data). The release of the products is accompanied by changes in the flexible loop region during which UDP is ejected. In Lgt1 there is one C-terminally located mobile loop (amino acid 513–525). This loop most likely rearranges upon binding to the donor substrate (Figure 3A; Hurtado-Guerrero et al., 2010). This conformational change is a general feature observed in GTs with GT-A and GT-B fold (Boix et al., 2001; Flint et al., 2005; Qasba et al., 2005; Gordon et al., 2006; Kubota et al., 2006; Ramakrishnan et al., 2006; Ziegler et al., 2008). In the structure of Lgt1 the loop shows only sufficient electron density in the UDP–glucose bound form (closed conformation) due to high mobility of the loop without intact donor substrate. For the closest homologous GTs, the CGT, the conformational changes of the flexible loops were crystallographically proven (Ziegler et al., 2008). In its apo-form the clostridial enzymes resemble an open conformation where the UDP–sugar has access to the donor substrate-binding pocket. Upon UDP–sugar-binding the loop closes and renders its C-terminal random coil structure to a rigid α-helix. Thereby rearrangement of a structurally conserved tryptophan residue (tryptophan-520 in toxin B) is induced resulting in an extensive movement of about 15 Å to bind the β-phosphate of the nucleotide. Mutation of this residue in C. difficile toxin B reduces the enzymatic and hydrolytic activity (transfer reaction to water instead of protein acceptor) of the GTs drastically implicating its decisive role in catalysis. Lgt1 harbors tryptophan-520 at the exact same position and suggests the same mechanistic function. Extended mutational analysis of Lgt1 reveals that tryptophan-520 is not directly involved in catalysis but has rather the function to bind the acceptor substrate (Lu et al., 2010).
Stereochemistry
Apart from the structural fold GTs are categorized and distinguished by its stereochemistry in glycosyl transfer mechanism, whether the anomeric configuration of the glycosyl moiety is retained or inversed (Coutinho et al., 2003). Lgt1 was shown to be a retaining GT. NMR structural analysis of glucosylated peptides revealed that the sugar is transferred to the acceptor with net retention of the α-anomeric configuration (Belyi et al., 2009). The mechanism for inverting GTs is well understood and follows a single nucleophilic substitution and thereby inversion of the sugar C1 configuration (Lairson et al., 2008). For a retaining mechanism two possible reaction schemes are highly discussed, the double displacement mechanism or a single SNi-like mechanism (Davies et al., 1997). In the double displacement strategy two subsequent SN2-reactions occur each with inversion of the anomeric bond. For GTs the double displacement theory seems to be rejected although there are several reports of trapped glycosyl-enzymes (Mosi et al., 1997; Uitdehaag et al., 1999; Gastinel et al., 2001; Lairson et al., 2004; Ramakrishnan et al., 2006; Soya et al., 2011). Mainly the lack of an appropriate positioned conserved nucleophilic amino acid on the β-face of the sugar argues against this theory (Lairson et al., 2008). In Lgts the conserved amino acid Asn-293 is located at the N-terminus of a structurally conserved central α-helix in the obvious access pathway of the substrate eEF1A. There it is moderately positioned to act as a nucleophil on the β-face of the sugar. Although mutations of this residue lead to severely reduced enzyme activity, Asp-293 seems not to be involved in the transfer mechanism itself. It rather functions in the guidance and/or binding of the acceptor substrate (Lu et al., 2010).
In contrast to the double displacement mechanism the SNi-like mechanism proceeds through a short-lived oxocarbenium intermediate. This intermediate is stabilized by the enzyme and shielded on the β-face of the sugar thereby preventing a nucleophilic attack from the opposite side of the reaction center (Lairson et al., 2008). An idea for Lgt1 reacting in this manner comes from the related crystal structure of lethal toxin from C. sordellii. The Ca2+-ligated structure presented a glucosyl half-chair conformation assuming an oxocarbenium intermediate state, the prerequisite for the SNi-like transfer mechanism (Ramakrishnan et al., 2006; Ziegler et al., 2008). Furthermore, studies with inhibitory glucomimetics and iminosugars, mimicking the oxocarbenium ion intermediate state conformation during catalysis, confirmed the SNi-like mechanism for the CGT biochemically as well as structurally (Jank et al., 2008). Lgts might react in the same manner as the related toxins but more studies are necessary.
In conclusion, the retaining reaction mechanism of the Lgts seems to follow a SNi-like mechanism. The glycosyltransfer reaction starts with the binding of the divalent cation to Asp-248 of the DXD-motif and the binding of the donor substrate UDP–glucose into the open cleft of the enzyme. Hereby, Trp-139 is stacking the base and Asp-230, Arg-233, and Asp-246 coordinate the distal part of the glucosyl moiety. Subsequently, the long C-terminal flexible loop rearranges to the closed conformation where Trp-520 flips into the catalytic pocket. In this state the catalytic competent conformation and the substrate-binding site is arranged. The Mn2+ and/or the pyrophosphoryl group abstract the proton of the incoming acceptor amino acid Ser-53 of eEF1A, leading to the positive charged oxocarbenium glucosyl-intermediate, which is then attacked by Ser-53 leading to the products UDP, Mn2+, and glucosyl–eEF1A. After dissociation of gluc–eEF1A the flexible loop opens and releases UDP and the cation to start a new reaction cycle.
Lgt1–eEF1A Interaction
The putative acceptor binding site of Lgt1 includes two acidic residues (Glu-445 and Glu-446) located at the funnel-like entrance to the active site representing an overall negative charge. Mutation of each of these residues leads to a slightly reduced glucosyltransferase activity without reducing the affinity of UDP–glucose to the catalytic site, implicating a role in acceptor substrate-binding (Hurtado-Guerrero et al., 2010). In the substrate eEF1A the acceptor amino acid Ser-53 resides on a loop between two helices (helix A*–loop–helix A′) of the GTPase domain. Serine-53 is flanked by two conserved lysine residues protruding as a positive charge from the GTPase domain. It was assumed that this opposite electrostatic surface potential is crucial for the Lgt1–eEF1A interaction and provides the necessary affinity for the enzyme–substrate complex (Hurtado-Guerrero et al., 2010; Lu et al., 2010). Contradictory results, however, came from comprehensive biochemical analysis, bordering the minimal substrate determinants of eEF1A. Here the authors showed that these lysine residues seem not to be of fundamental importance for an efficient transfer reaction (Belyi et al., 2009). This is in line with putative substrate Hbs1, which contains only one lysine residue adjacent to Ser-53 and is remarkably modified. Using the location of the acceptor site serine-53 and the overall shape of Lgt1 and eEF1A, docking simulations have been performed, revealing an interaction model of an enzyme–substrate complex. The flexible loop of the glucosyltransferase has to adopt a slightly different conformation upon substrate-binding (Lu et al., 2010). Interface mutagenesis of the key exposed amino acid tyrosine-454 and biochemical analysis support this interaction model (Hurtado-Guerrero et al., 2010). Considering that eEF1A by itself is a rather poor substrate and structural substrate determinants can be reduced to a decapeptide, it is assumed that a particular conformation of EF1A is the preferred substrate or the non-ambiguous substrate consists of additional host factors, which are not considered in the suggested interaction model (Belyi and Aktories, 2010).
Putative Glycosyltransferases in Legionella
Sequence analysis of the L. pneumophila genome allowed identification of other putative GTs chromosomally located in T4SS effector regions (Franco et al., 2009).
A screen of 127 confirmed and putative Dot/Icm substrates for their ability to generate lethal yeast phenotypes allowed identification of a ∼72-kDa protein termed subversion of eukaryotic vesicle trafficking A (SetA; Heidtman et al., 2009). Its coding sequence has ID lpg1978 in the genome of L. pneumophila Philadelphia-1 strain but is not present in the genome of L. longbeachae (Cazalet et al., 2010; Kozak et al., 2010). Expression of this gene in S. cerevisiae resulted in secretory defects detected by altered trafficking/processing of yeast markers carboxypeptidase Y and alkaline phosphatase. SetA seemed to be localized to late endosomal/lysosomal compartments, co-localizing with eukaryotic marker proteins LAMP-1 and Rab7. In L. pneumophila-infected cells SetA was secreted into eukaryotic cytosol in an Icm/DotA-dependent manner and demonstrated tropism for host cell membranes. One interesting structural feature of this protein is the occurrence of the GT-characteristic DXD-motif (D134SD136). Mutation of both aspartic acid residues in SetA to alanines alleviated the toxic phenotype, suggesting a link between possible GT activity of the protein and its lethal effect in yeast. Bearing in mind its subcellular distribution and altered trafficking phenotypes in yeast, transformed with the gene setA, these mutation experiments raise the possibility that SetA glycosylates and inactivates a factor of the endosomal protein sorting machinery.
BLAST search for proteins similar to SetA in the L. pneumophila Philadelphia-1 genome reveals a ∼59-kDa protein, representing a product of lpg1961 gene (Figure 1A). Product of this gene was also toxic for S. cerevisiae and caused selective defects on alkaline phosphatase processing in yeast (Heidtman et al., 2009). In accordance to several GTs this protein also possesses a DXD-motif with the canonical tyrosine, aspartic acid and arginine residues of the GT-A triad upstream the DXD-motif (Figure 1A). These features pointed toward a possible GT activity in this L. pneumophila product as well.
Recently, genome sequences of the two non-pneumophila species became available – Legionella drancourtii strain LLAP12 (Moliner et al., 2010) and L. longbeachae strains D-4968 and NSW150 (Cazalet et al., 2010; Kozak et al., 2010). BLAST searches within these strains disclose two proteins in L. drancourtii (LDG0102 and LDG0103) and two proteins in each L. longbeachae genomes (LLB0067/LLO1578 and LLB3681/LLO1721), showing identity of around 15% with Lgt1. Identical amino acid residues could be found predominantly in the first third of the proteins (Figure 1B), while homology outside this region was insignificant. Although all these four proteins possess a DXD-containing region resembling that of typical GTs, the nature of their enzymatic activity (if any), their targets and donor substrates remain to be determined. Furthermore, it is questionable whether these putative GTs are secreted Legionella effectors. It might be that their function lies not in virulence but rather in basic carbohydrate metabolism of the bacterium.
Functional Consequences of Glycosylation and Open Questions
Major targets of Lgt-catalyzed glucosylation are crucial components of translational machinery of eukaryotic cells, e.g., eEF1A and Hbs1. Addition of Lgt1, Lgt2, or Lgt3 to in vitro reticulocyte or yeast translational extracts resulted in a dose-dependent inhibition of protein synthesis. Furthermore, introduction of Lgts into mammalian cells by electroporation results in eEF1A modification, protein synthesis inhibition, and death (Belyi et al., 2006, 2008). Similarly, expression of genes coding for Lgt1, Lgt2, or Lgt3 in S. cerevisiae resulted in yeast cell death (Heidtman et al., 2009).
Up to date, the precise mechanism of protein inhibition by Lgt-induced glucosylation of Ser53 of eEF1A is still not clear. Moreover, the role of Lgt-induced protein synthesis inhibition in the infection biology of Legionella is not known. One speculation is that the action of Lgt strongly decreases general metabolism and thereby antibacterial activity and, thus, makes host cells “defenseless” against proliferation of invading bacteria. On the other hand, at final stages of the intracellular life cycle, Legionella has to kill and escape the eukaryotic cell and eEF1A-targeting glucosyltransferases may participate in such a task as strong lethal toxins.
An alternative hypothesis about the smart roles of bacterial effectors of Legionella has been proposed recently from investigations of the T4SS effector SidI (Shen et al., 2009). The ∼110-kDa protein SidI (Lpg2504) exhibits a toxic phenotype in yeast. It was demonstrated that SidI interacts with eEF1A and eEF1Bγ and inhibits protein synthesis both in vitro and in vivo. Another type of activity, associated with SidI is its participation in a stress response of eukaryotic cell.
It is known that stress response in mammalian cells is controlled by heat shock transcription factor 1 (HSF1), which is able to bind specific promoters (heat shock elements, HSE) and thus induces production of a panel of heat shock proteins, necessary to rescue eukaryotic cells, suffering under unfavorable conditions (Sarge et al., 1991). Activation of HSF1 is dependent upon formation of a multi-component complex, consisting of HSF1, eEF1A, and ∼0.6 kb non-coding RNA molecule, termed heat shock RNA 1 (HSR1; Shamovsky et al., 2006). Infection of macrophage-like cells U937 with virulent L. pneumophila, but not with a sidI-negative mutant, resulted in eukaryotic stress response detected by elevated level of HFS1/eEF1A complex, increased binding of HSF1 to HSE and stimulation of hsp70 expression. Similar phenomena were observed by transfection of target cells with SidI-coding plasmid (Shen et al., 2009). These results indicates that HSF1 is activated during L. pneumophila infection and SidI, shown initially to suppress protein synthesis, contributes to such an activation.
Thus, bearing in mind the multitude of its cellular functions, targeting elongation factor 1A by Lgt1/2/3 may lead to pleiotropic outcomes and the observed cytotoxicity may be a side effect of some other pro-bacterial consequence of eEF1A glucosylation (Ensminger and Isberg, 2009). The proposed modification of Hbs1 by the Legionella effectors further adds complexity to the list of events, which might be caused by the enzymatic activity of the glucosyltransferase Lgt.
Recent findings indicate that apart from Clostridia and Legionella, other bacteria can also possess GT activities as important virulence strategies. List of such putative glycosylating molecules includes several proteins found in Chlamydia trachomatis (Belland et al., 2001). One such protein, termed CT166, was shown to induce Rac-dependent actin re-organization and mammalian cell rounding, resembling action of glucosylating toxin B of C. difficile (Thalmann et al., 2010). Other toxins with possible glycosylation type of activity include LifA and toxin B of enteric pathogens (Stevens et al., 2004). These findings suggest that glucosylation is more often used by pathogens to prevail in a hostile environment as suggested before.
On the other hand, control of translational processes of host cells is a well-known mechanism used by various pathogenic bacteria and accomplished by different enzymatic activities, e.g., Pseudomonas aeruginosa exotoxin A and diphtheria toxin inhibit protein synthesis by ADP-ribosylation of eEF2, while Shiga- and Shiga-like toxins from Shigella and Escherichia coli, respectively, block host translation by N-glycosidase activity (Popoff, 1998). Biological purpose of inhibition of protein synthesis accomplished by the latter group of toxins is still not completely clear. But the fact that unrelated enzymatic activities (i.e., glucosylation, ADP-ribosylation, and N-glycosidation) result in termination of eukaryotic protein synthesis suggests critical importance of this targeting in host–pathogen interaction.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abel, K., Yoder, M. D., Hilgenfeld, R., and Jurnak, F. (1996). An alpha to beta conformational switch in EF-Tu. Structure 4, 1153–1159.
Abu-Zant, A., Jones, S., Asare, R., Suttles, J., Price, C., Graham, J., and Kwaik, Y. A. (2007). Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell. Microbiol. 9, 246–264.
Andersen, G. R., Pedersen, L., Valente, L., Chatterjee, I., Kinzy, T. G., Kjeldgaard, M., and Nyborg, J. (2000). Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha. Mol. Cell 6, 1261–1266.
Banga, S., Gao, P., Shen, X., Fiscus, V., Zong, W. X., Chen, L., and Luo, Z. Q. (2007). Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. U.S.A. 104, 5121–5126.
Belland, R. J., Scidmore, M. A., Crane, D. D., Hogan, D. M., Whitmire, W., McClarty, G., and Caldwell, H. D. (2001). Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. U.S.A. 98, 13984–13989.
Belyi, I., Popoff, M. R., and Cianciotto, N. P. (2003). Purification and characterization of a UDP-glucosyltransferase produced by Legionella pneumophila. Infect. Immun. 71, 181–186.
Belyi, Y., and Aktories, K. (2010). Bacterial toxin and effector glycosyltransferases. Biochim. Biophys. Acta 1800, 134–143.
Belyi, Y., Niggeweg, R., Opitz, B., Vogelsgesang, M., Hippenstiel, S., Wilm, M., and Aktories, K. (2006). Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proc. Natl. Acad. Sci. U.S.A. 103, 16953–16958.
Belyi, Y., Stahl, M., Sovkova, I., Kaden, P., Luy, B., and Aktories, K. (2009). Region of elongation factor 1A1 involved in substrate recognition by Legionella pneumophila glucosyltransferase Lgt1: identification of Lgt1 as a retaining glucosyltransferase. J. Biol. Chem. 284, 20167–20174.
Belyi, Y., Tabakova, I., Stahl, M., and Aktories, K. (2008). Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J. Bacteriol. 190, 3026–3035.
Boix, E., Swaminathan, G. J., Zhang, Y., Natesh, R., Brew, K., and Acharya, K. R. (2001). Structure of UDP complex of UDP-galactose: beta-galactoside-alpha-1,3-galactosyltransferase at 1.53-A resolution reveals a conformational change in the catalytically important C terminus. J. Biol. Chem. 276, 48608–48614.
Borradaile, N. M., Buhman, K. K., Listenberger, L. L., Magee, C. J., Morimoto, E. T., Ory, D. S., and Schaffer, J. E. (2006). A critical role for eukaryotic elongation factor 1A-1 in lipotoxic cell death. Mol. Biol. Cell 17, 770–778.
Bruggemann, H., Hagman, A., Jules, M., Sismeiro, O., Dillies, M. A., Gouyette, C., Kunst, F., Steinert, M., Heuner, K., Coppée, J. Y., and Buchrieser, C. (2006). Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell. Microbiol. 8, 1228–1240.
Byrne, B., and Swanson, M. S. (1998). Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66, 3029–3034.
Cazalet, C., Gomez-Valero, L., Rusniok, C., Lomma, M., Dervins-Ravault, D., Newton, H. J., Sansom, F. M., Jarraud, S., Zidane, N., Ma, L., Bouchier, C., Etienne, J., Hartland, E. L., and Buchrieser, C. (2010). Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 6, e1000851. doi: 10.1371/journal.pgen.1000851
Charnock, S. J., and Davies, G. J. (1999). Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38, 6380–6385.
Chong, A., Lima, C. A., Allan, D. S., Nasrallah, G. K., and Garduno, R. A. (2009). The purified and recombinant Legionella pneumophila chaperonin alters mitochondrial trafficking and microfilament organization. Infect. Immun. 77, 4724–4739.
Chuang, S. M., Chen, L., Lambertson, D., Anand, M., Kinzy, T. G., and Madura, K. (2005). Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol. Cell. Biol. 25, 403–413.
Coutinho, P. M., Deleury, E., Davies, G. J., and Henrissat, B. (2003). An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317.
Davies, G. J., Sinnott, M. L., and Withers, S. G. (1997). “Glycosyl transfer,” in Comprehensive Biological Catalysis, ed. M. L. Sinnott (London: Academic Press), 119–209.
de Felipe, K. S., Glover, R. T., Charpentier, X., Anderson, O. R., Reyes, M., Pericone, C. D., and Shuman, H. A. (2008). Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4, e1000117. doi: 10.1371/journal.ppat.1000117
de Felipe, K. S., Pampou, S., Jovanovic, O. S., Pericone, C. D., Ye, S. F., Kalachikov, S., and Shuman, H. A. (2005). Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187, 7716–7726.
Doma, M. K., and Parker, R. (2006). Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564.
Ejiri, S. (2002). Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Biosci. Biotechnol. Biochem. 66, 1–21.
Ensminger, A. W., and Isberg, R. R. (2009). Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12, 67–73.
Flint, J., Taylor, E., Yang, M., Bolam, D. N., Tailford, L. E., Martinez-Fleites, C., Dodson, E. J., Davis, B. G., Gilbert, H. J., and Davies, G. J. (2005). Structural dissection and high-throughput screening of mannosylglycerate synthase. Nat. Struct. Mol. Biol. 12, 608–614.
Franco, I. S., Shuman, H. A., and Charpentier, X. (2009). The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell. Microbiol. 11, 1435–1443.
Gastinel, L. N., Bignon, C., Misra, A. K., Hindsgaul, O., Shaper, J. H., and Joziasse, D. H. (2001). Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases. EMBO J. 20, 638–649.
Ge, J., Xu, H., Li, T., Zhou, Y., Zhang, Z., Li, S., Liu, L., and Shao, F. (2009). A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc. Natl. Acad. Sci. U.S.A. 106, 13725–13730.
Gibson, R. P., Tarling, C. A., Roberts, S., Withers, S. G., and Davies, G. J. (2004). The donor subsite of trehalose-6-phosphate synthase: binary complexes with UDP-glucose and UDP-2-deoxy-2-fluoro-glucose at 2 A resolution. J. Biol. Chem. 279, 1950–1955.
Gordon, R. D., Sivarajah, P., Satkunarajah, M., Ma, D., Tarling, C. A., Vizitiu, D., Withers, S. G., and Rini, J. M. (2006). X-ray crystal structures of rabbit N-acetylglucosaminyltransferase I (GnT I) in complex with donor substrate analogues. J. Mol. Biol. 360, 67–79.
Gross, S. R., and Kinzy, T. G. (2005). Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 12, 772–778.
Heidtman, M., Chen, E. J., Moy, M. Y., and Isberg, R. R. (2009). Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell. Microbiol. 11, 230–248.
Horwitz, M. A., and Maxfield, F. R. (1984). Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99, 1936–1943.
Hotokezaka, Y., Tobben, U., Hotokezaka, H., Van, L. K., Beatrix, B., Smith, D. H., Nakamura, T., and Wiedmann, M. (2002). Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J. Biol. Chem. 277, 18545–18551.
Hubber, A., and Roy, C. R. (2010). Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283.
Hurtado-Guerrero, R., Zusman, T., Pathak, S., Ibrahim, A. F., Shepherd, S., Prescott, A., Segal, G., and van Aalten, D. M. F. (2010). Molecular mechanism of elongation factor 1A inhibition by a Legionella pneumophila glycosyltransferase. Biochem. J. 426, 281–292.
Ingmundson, A., Delprato, A., Lambright, D. G., and Roy, C. R. (2007). Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450, 365–369.
Isberg, R. R., O’Connor, T. J., and Heidtman, M. (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24.
Ivanov, S. S., and Roy, C. R. (2009). Modulation of ubiquitin dynamics and suppression of DALIS formation by the Legionella pneumophila Dot/Icm system. Cell. Microbiol. 11, 261–278.
Jank, T., Giesemann, T., and Aktories, K. (2007). Clostridium difficile glucosyltransferase toxin B – essential amino acids for substrate-binding. J. Biol. Chem. 282, 35222–35231.
Jank, T., Ziegler, M. O., Schulz, G. E., and Aktories, K. (2008). Inhibition of the glucosyltransferase activity of clostridial Rho/Ras-glucosylating toxins by castanospermine. FEBS Lett. 582, 2277–2282.
Jules, M., and Buchrieser, C. (2007). Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics. FEBS Lett. 581, 2829–2838.
Kamitani, S., Kitadokoro, K., Miyazawa, M., Toshima, H., Fukui, A., Abe, H., Miyake, M., and Horiguchi, Y. (2010). Characterization of the membrane-targeting C1 domain in Pasteurella multocida toxin. J. Biol. Chem. 285, 25467–25475.
Khacho, M., Mekhail, K., Pilon-Larose, K., Pause, A., Cote, J., and Lee, S. (2008). eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol. Biol. Cell 19, 5296–5308.
Kjeldgaard, M., Nyborg, J., and Clark, B. F. C. (1996). The GTP binding motif: variations on a theme. FASEB J. 10, 1347–1368.
Kozak, N. A., Buss, M., Lucas, C. E., Frace, M., Govil, D., Travis, T., Olsen-Rasmussen, M., Benson, R. F., and Fields, B. S. (2010). Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J. Bacteriol. 192, 1030–1044.
Kubori, T., Hyakutake, A., and Nagai, H. (2008). Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 67, 1307–1319.
Kubota, T., Shiba, T., Sugioka, S., Furukawa, S., Sawaki, H., Kato, R., Wakatsuki, S., and Narimatsu, H. (2006). Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10). J. Mol. Biol. 359, 708–727.
Laguna, R. K., Creasey, E. A., Li, Z., Valtz, N., and Isberg, R. R. (2006). A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. U.S.A. 103, 18745–18750.
Lairson, L. L., Chiu, C. P., Ly, H. D., He, S., Wakarchuk, W. W., Strynadka, N. C., and Withers, S. G. (2004). Intermediate trapping on a mutant retaining alpha-galactosyltransferase identifies an unexpected aspartate residue. J. Biol. Chem. 279, 28339–28344.
Lairson, L. L., Henrissat, B., Davies, G. J., and Withers, S. G. (2008). Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555.
Li, Z., Dugan, A. S., Bloomfield, G., Skelton, J., Ivens, A., Losick, V., and Isberg, R. R. (2009). The amoebal MAP kinase response to Legionella pneumophila is regulated by DupA. Cell Host Microbe 6, 253–267.
Lomma, M., Dervins-Ravault, D., Rolando, M., Nora, T., Newton, H. J., Sansom, F. M., Sahr, T., Gomez-Valero, L., Jules, M., Hartland, E. L., and Buchrieser, C. (2010). The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell. Microbiol. 12, 1272–1291.
Losick, V. P., Haenssler, E., Moy, M. Y., and Isberg, R. R. (2010). LnaB: a Legionella pneumophila activator of NF-kappaB. Cell. Microbiol. 12, 1083–1097.
Lu, W., Du, J., Stahl, M., Tzivelekidis, T., Belyi, Y., Gerhardt, S., Aktories, K., and Einsle, O. (2010). Structural basis of the action of glucosyltransferase Lgt1 from Legionella pneumophila. J. Mol. Biol. 396, 321–331.
Machner, M. P., and Isberg, R. R. (2006). Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11, 47–56.
Machner, M. P., and Isberg, R. R. (2007). A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318, 974–977.
Mateyak, M. K., and Kinzy, T. G. (2010). eEF1A: thinking outside the ribosome. J. Biol. Chem. 285, 21209–21213.
Matsuda, D., Yoshinari, S., and Dreher, T. W. (2004). eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology 321, 47–56.
Mesmin, B., Robbe, K., Geny, B., Luton, F., Brandolin, G., Popoff, M. R., and Antonny, B. (2004). A phosphatidylserine-binding site in the cytosolic fragment of Clostridium sordellii lethal toxin facilitates glucosylation of membrane-bound Rac and is required for cytotoxicity. J. Biol. Chem. 279, 49876–49882.
Moliner, C., Fournier, P. E., and Raoult, D. (2010). Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev. 34, 281–294.
Mosi, R., He, S., Uitdehaag, J., Dijkstra, B. W., and Withers, S. G. (1997). Trapping and characterization of the reaction intermediate in cyclodextrin glycosyltransferase by use of activated substrates and a mutant enzyme. Biochemistry 36, 9927–9934.
Muder, R. R., and Yu, V. L. (2002). Infection due to Legionella species other than L. pneumophila. Clin. Infect. Dis. 35, 990–998.
Muller, M. P., Peters, H., Blumer, J., Blankenfeldt, W., Goody, R. S., and Itzen, A. (2010). The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949.
Murata, T., Delprato, A., Ingmundson, A., Toomre, D. K., Lambright, D. G., and Roy, C. R. (2006). The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8, 971–977.
Nagai, H., Kagan, J. C., Zhu, X., Kahn, R. A., and Roy, C. R. (2002). A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679–682.
Negishi, M., Dong, J., Darden, T. A., Pedersen, L. G., and Pedersen, L. C. (2003). Glucosaminylglycan biosynthesis: what we can learn from the X-ray crystal structures of glycosyltransferases GlcAT1 and EXTL2. Biochem. Biophys. Res. Commun. 303, 393–398.
Nelson, R. J., Ziegelhoffer, T., Nicolet, C., Werner-Washburne, M., and Craig, E. A. (1992). The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71, 97–105.
Ninio, S., and Roy, C. R. (2007). Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15, 372–380.
Popoff, M. R. (1998). Interactions between bacterial toxins and intestinal cells. Toxicon 36, 665–685.
Price, C. T., Al-Khodor, S., Al-Quadan, T., and Abu, K. Y. (2010). Indispensable role for the eukaryotic-like ankyrin domains of the ankyrin B effector of Legionella pneumophila within macrophages and amoebae. Infect. Immun. 78, 2079–2088.
Price, C. T., Al-Khodor, S., Al-Quadan, T., Santic, M., Habyarimana, F., Kalia, A., and Kwaik, Y. A. (2009). Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 5, e1000704. doi: 10.1371/journal.ppat.1000704
Qasba, P. K., Ramakrishnan, B., and Boeggeman, E. (2005). Substrate-induced conformational changes in glycosyltransferases. Trends Biochem. Sci. 30, 53–62.
Ramakrishnan, B., Ramasamy, V., and Qasba, P. K. (2006). Structural snapshots of beta-1,4-galactosyltransferase-I along the kinetic pathway. J. Mol. Biol. 357, 1619–1633.
Ruest, L. B., Marcotte, R., and Wang, E. (2002). Peptide elongation factor eEF1A-2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J. Biol. Chem. 277, 5418–5425.
Sarge, K. D., Zimarino, V., Holm, K., Wu, C., and Morimoto, R. I. (1991). Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 5, 1902–1911.
Shamovsky, I., Ivannikov, M., Kandel, E. S., Gershon, D., and Nudler, E. (2006). RNA-mediated response to heat shock in mammalian cells. Nature 440, 556–560.
Shen, X., Banga, S., Liu, Y., Xu, L., Gao, P., Shamovsky, I., Nudler, E., and Luo, Z. Q. (2009). Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell. Microbiol. 11, 911–926.
Shoemaker, C. J., Eyler, D. E., and Green, R. (2010). Dom34: Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330, 369–372.
Soya, N., Fang, Y., Palcic, M. M., and Klassen, J. S. (2011). Trapping and characterization of covalent intermediates of mutant retaining glycosyltransferases. Glycobiology 21, 547–552.
Stevens, M. P., Roe, A. J., Vlisidou, I., van Diemen, P. M., La Ragione, R. M., Best, A., Woodward, M. J., Gally, D. L., and Wallis, T. S. (2004). Mutation of toxB and a truncated version of the efa-1 gene in Escherichia coli O157: H7 influences the expression and secretion of locus of enterocyte effacement-encoded proteins but not intestinal colonization in calves or sheep. Infect. Immun. 72, 5402–5411.
Thalmann, J., Janik, K., May, M., Sommer, K., Ebeling, J., Hofmann, F., Genth, H., and Klos, A. (2010). Actin re-organization induced by Chlamydia trachomatis serovar D-evidence for a critical role of the effector protein CT166 targeting Rac. PLoS ONE 5, e9887. doi: 10.1371/journal.pone.0009887
Uitdehaag, J. C., Mosi, R., Kalk, K. H., van, D. V., Dijkhuizen, L, Withers, S. G., and Dijkstra, B. W. (1999). X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the alpha-amylase family. Nat. Struct. Biol. 6, 432–436.
Vetter, I. R., and Wittinghofer, A. (2001). The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304.
Vocadlo, D. J., Davies, G. J., Laine, R., and Withers, S. G. (2001). Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature 412, 835–838.
Xu, L., Shen, X., Bryan, A., Banga, S., Swanson, M. S., and Luo, Z. Q. (2010). Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 6, e1000822. doi: 10.1371/journal.ppat.1000822
Yu, V. L., Plouffe, J. F., Pastoris, M. C., Stout, J. E., Schousboe, M., Widmer, A., Summersgill, J., File, T., Heath, C. M., Paterson, D. L., and Chereshsky, A. (2002). Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186, 127–128.
Ziegler, M. O., Jank, T., Aktories, K., and Schulz, G. E. (2008). Conformational changes and reaction of clostridial glycosylating toxins. J. Mol. Biol. 377, 1346–1356.
Keywords: Legionella, virulence factor, glycosyltransferase, eEF1A, protein synthesis
Citation: Belyi Y, Jank T and Aktories K (2011) Effector glycosyltransferases in Legionella. Front. Microbio. 2:76. doi: 10.3389/fmicb.2011.00076
Received: 12 January 2011; Paper pending published: 14 February 2011;
Accepted: 31 March 2011; Published online: 12 April 2011.
Edited by:
Carmen Buchrieser, Pasteur Institute, FranceReviewed by:
José A. Bengoechea, Fundación Caubet-CIMERA Illes Balears, SpainShaynoor Dramsi, Institut Pasteur, France
Michel Popoff, Institut Pasteur, France
Copyright: © 2011 Belyi, Jank and Aktories. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Klaus Aktories, Institute for Experimental and Clinical Pharmacology and Toxicology, Albert-Ludwigs University of Freiburg, Albertstrasse 25, D-79104 Freiburg, Germany. e-mail: klaus.aktories@pharmakol.uni-freiburg.de