- 1 Instituto de Bioquímica Médica, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
- 2 Instituto Nacional de Metrologia Normalização e Qualidade Industrial, Rio de Janeiro, Brazil
- 3 Instituto de Biologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
- 4 Instituto Nacional de Controle da Qualidade em Saúde, Rio de Janeiro, Brazil
- 5 Departamento de Bioquímica, Universidade do Estado do Rio de Janeiro, Rio de Janeiro, Brazil
In this Opinion Article, we argue that tropical regions are, in fact, hot spots of extreme microbial diversity. The scope of the topic is vast and while this paper cannot be seen as exhaustive, it will hopefully serve to stimulate research interest in the reader to explore this fascinating area of microbiology and promote knowledge sharing and scientific collaboration. The article discusses the following points: (i) what extremophiles are and how useful they are to biotechnology; (ii) the importance of molecular techniques for extremophile identification due to their unculturability; and (iii) extremophile research performed in areas of different environmental conditions in Rio de Janeiro, Brazil.
Extremophiles are microorganisms able to grow optimally in extreme environments of temperature, pH, pressure, and salinity, often in places deemed inhospitable to life. This feature requires the stabilization of all cellular components, so that the functionality is maintained under conditions that would be harmful for most non- extremophile molecules (Hough and Danson, 1999). The elucidation of strategies to stabilize biomolecules, especially enzymes, represents a challenge for biology and industrial processes, including bio-engineering. Although the molecular strategies employed for survival in such environments are still not fully clarified, it is known that these microorganisms have adapted molecules, as well as different and peculiar biochemical pathways, which are of great interest for biochemical and biotechnological purposes (Alqueres et al., 2007). The extent and potential biotechnological applications of extremophile molecules have been reviewed in several articles. Their stability and activity at extreme conditions make them useful alternatives to labile mesophilic microorganisms. This is particularly true for their enzymes, which remain catalytically active under extremes of temperature, salinity, pH, and solvent conditions, and are seen as one potential answer to the biotechnological conflict between industrial conditions and the fragility of biological components.
Extremophiles often require specific growth conditions that are either difficult to achieve in a laboratory or even unknown. Furthermore, it is estimated that less than 1% of all microorganisms are able to grow in culture, reinforcing that metagenomic analysis provide the only way to obtain gene sequences and explore these organisms. In fact, the recent advent and application of high throughput next generation sequencing methods and computational analysis have enabled the discovery of novel molecules with biotechnological interest in these otherwise hidden organisms. As metagenomic tools become more accessible to the research community, allowing the investigation of microorganisms that cannot be cultured in the laboratory, exciting aspects of extremophilic biology are now being revealed (Cardoso et al., 2010). Metagenomic approaches provide an opportunity to describe the taxonomic components and metabolic potential of all microbes within the environment, and present a solid basis for understanding ecology, evolution, and also to reveal novel genes, new taxa, and metabolic pathways. Indeed, the discovery and production of sequence data from the enormous global microbial diversity may enable us to identify novel organisms and molecules with a natural ability to function under extreme conditions, improving industrial processes.
Although the extremophiles exist within the three domains of life, they are mainly of bacterial and archaeal origin, with the majority from Archaea (Cary et al., 1998). Extremophilic Archaea may give hints about molecular resistance mechanisms against high temperature, low and high pH, and high salt concentrations. This could allow for protein improvement by the addition of the desired features through genetic manipulation.
Brazil is well known for its great biodiversity of flora and fauna, however, only few studies have been published focusing on the Brazilian microbial diversity. Why then should people be inspired to come to this part of the world to study extremophiles while there are many other interesting environments around the globe in which the archaeal diversity is well known, such as the Yellowstone National Park in the United States? Perhaps the combination of physico-chemical conditions that we do not easily find in non-tropical areas makes the surrounding areas of Rio de Janeiro coastline a microbial diversity hotspot. Rio de Janeiro is the third largest metropolitan area in South America, its surrounding areas are overpopulated with limited sewage treatment and with a heavily industrialized park with an expected pollution impact. But it also has the largest urban forest in the world, peculiar environmental features and it is famous for its natural settings and beaches.
Rio de Janeiro exhibits complex and impacted ecosystems (Figure 1) such as Araruama lagoon (hypersaline), Sepetiba bay (impacted by heavy metal pollution), and Guanabara bay (an urban polluted estuary). These environments are home to intriguing habitats with salt lakes, pristine, and polluted estuaries, mangroves and anaerobic sediments containing a myriad of microorganisms waiting for biotechnological exploration and have the potential to become an important source of novel and unique microorganisms for bioremediation and the biotechnological industry.
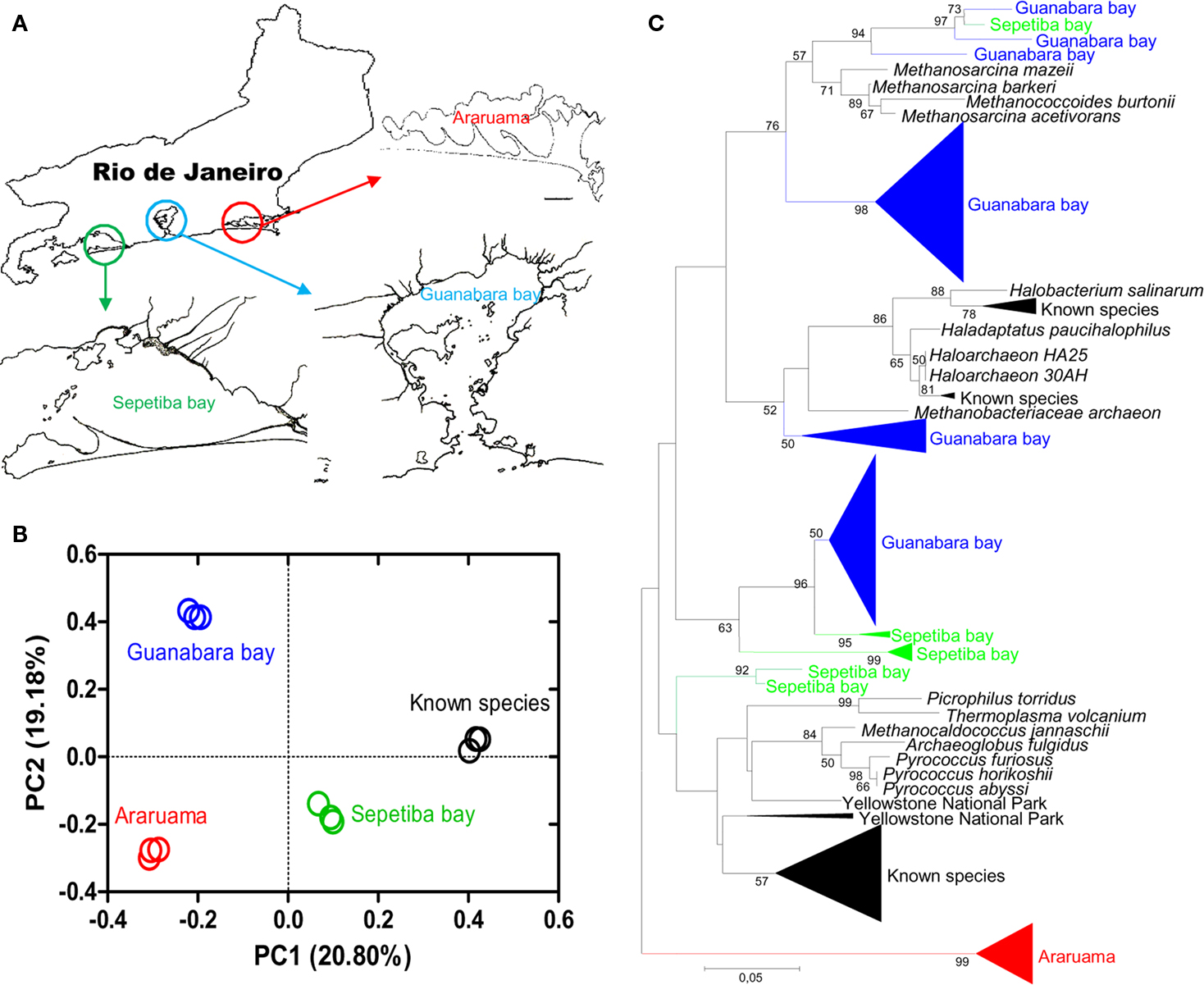
Figure 1. (A) Map of the described areas, (B) distribution patterns of archaeal communities (described archaea in relation to known species) using UniFrac metric analysis, and (C) summary tree of ribosomal sequences retrieved from the Rio area. The analyses were made as described in Silveira et al. (2011). The archaeal “known species” were obtained from RDP (Ribosomal Database Project).
This tropical region has scientific expertise with many universities, biotechnological companies, and good infrastructure. Extremophiles in tropical regions are highly unreported in the scientific literature; however the tropics hold numerous extreme and interesting ecological niches. Unfortunately, in most cases the path from an extremophilic molecule to a successful commercial application is not documented in peer reviewed scientific publications and can be only partially followed through patents and company websites where little scientific information is available. Knowledge should be shared widely.
Therefore, the archaeal diversity of this tropical region is an object of great interest for biotechnology industries and to science as a whole. Indeed, the detection of a high number of phylotypes related to uncultivated organisms points to the existence of new archaeal and bacterial lineages with great biotechnological potential (Clementino et al., 2007, 2008; Vieira et al., 2007; Almeida et al., 2009; Cardoso et al., 2010).
As mentioned above, hypersaline environments are widespread around Rio de Janeiro. The diversity of haloarchaea from different hypersaline habitats has been analyzed and compared by molecular phylogenetic techniques and cultivation approaches, revealing novel haloarchaeal lineages. Recently, we have shown (Clementino et al., 2008) that the retrieved archaeal clones from one of the largest hypersaline lagoons in the world, Araruama lagoon, were exclusively related to uncultured environmental Euryarchaeota. A remarkable characteristic of this lagoon system is the high salt content (an average of 5.2% total salts), nearly twice the concentration of seawater. The climate in this region is typically hot and dry. The low rainfall and high evaporation rates in this region contribute to the development of several salterns around the lagoon. Interestingly, in lagoon waters we found some OTUs related to Methanomicrobia and Methanothermococcus groups whereas, in saltern waters with salt saturating concentrations novel members related to the genus Halobacterium and Haloarcula were also detected.
Perhaps one of the most interesting aspects of hypersaline environments is how the salt affects adaptive strategies of halophilic Archaea (Clementino et al., 2008). Several different mechanisms allow protein adaptation to high salt concentrations which can be different for each protein. Recently, it was shown that it is possible to decrease the salt dependence of a typical halophilic protein to the level of a mesophilic form and engineer a protein from a mesophilic organism into an obligate halophilic form (Tadeo et al., 2009). Their extensive protein surface mutational studies also showed that the abundance of aspartic and glutamic acid residues, a characteristic of halophilic proteins, improves not only salt tolerance but also solubility, another valuable feature in typical high-salt environmental conditions (Hoff, 2009; Tadeo et al., 2009).
Another relevant study was performed to reveal the composition and structure of microbial communities in acid mine drainage waters associated with zinc mine tailings in Sepetiba bay (Almeida et al., 2009). This region is a semi-enclosed 520 km2 water body separated from the Atlantic Ocean by a natural sand barrier. Acidic and highly metal contaminated waters generated by dissolution of sulfidic minerals are the main pollution source in this area. Emission rate estimates suggest that the main Zn and Cd sources to Sepetiba Bay are effluents from the Inga zinc refinery processing plant, which added 3,660 t year−1 of Zn and 24 t year−1 of Cd into the bay’s basin until 1998, when the plant closed. Highly polluted environments with Zn, at 1 g/l, and Cd–Pb, at 1 μg/l, were analyzed by molecular methods revealing novel and fascinating microorganisms, contributing to a better understanding of the prokaryotic diversity, biogeography, bioremediation, and the role of microorganisms in zinc smelting acid mine drainage generation.
Several bacterial OTUs retrieved from acidic metal contaminated waters were related to uncultured environmental microorganisms found in similar habitats and to acidophilic genera such as Acidocella, Acidosphaera, and Acidiphilium. Most archaeal OTUs present in heavy metal acidic water were related to uncultivated Crenarchaeota found in a subsurface radioactive thermal spring, in subsurface geothermal water from a gold mine and in sulfidic marsh water. Thus, the occurrence of several OTUs related to uncultivated prokaryotes in this impacted Brazilian environment implies the possibility of isolating new species which could, for example, be used in remediation processes.
It is interesting to note that estuaries are extremely dynamic ecosystems, being at the interface between continental freshwater and adjacent coastal seawaters. Spatial and temporal variability of salinity, nutrients and pollution gradients found in these environments are due to patterns of several factors such as rain, currents, winds and tides, as well as anthropogenic inputs. We have characterized Archaea and Bacteria diversity in distinct environments within and around the tropical urban bay to learn about their biogeographic distribution, ecology, and niche adaptation.
Guanabara Bay is a tropical estuarine system located in Rio de Janeiro, which harbors the second largest urban and industrialized city in Brazil. The bay is under considerable degradation processes and is considered one of the most eutrophicated areas in the world (Mayr et al., 1989). The main human impacts recognized for the area are due to sewage discharges, oil pollution, garbage disposal, landfills, deforestation, and mangroves destruction. Since the impacts are not uniformly distributed, bay water quality differs spatially depending on pollution focuses, tidal influence, and water circulation patterns. Using molecular approaches we observed novel environmental Archaea (Vieira et al., 2007) and Bacteria communities (Vieira et al., 2008) with biotechnological potential in distinct habitats within and around Guanabara Bay estuarine ecosystems. Most bacterial OTUs were affiliated with the Proteobacteria and Bacteroidetes and several organisms related to hydrocarbon degradation were registered, in accordance with environmental status. Most archaeal OTUs were related to uncultured environmental Archaea and, in inner bay waters we observed the occurrence of OTUs related to Methanoplanus petrolearius isolated from petroleum wells. For both taxa (Archaea and Bacteria) we could observe that pollution and physico-chemical variables were very influential of population structure and composition. Thus Guanabara bay can be an interesting research model to allow a better comprehension of how bacterial and archaeal communities are distributed along pollution gradients in coastal metropolitan areas and, furthermore, it can also be viewed as a reservoir of novel microbes with great biotechnological potential.
Acknowledgments
This work was partially funded by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
References
Almeida, W. I., Vieira, R. P., Cardoso, A. M., Silveira, C. B., Costa, R. G., Gonzalez, A. M., Paranhos, R., Medeiros, J. A., Freitas, F. A., Albano, R. M., and Martins, O. B. (2009). Archaeal and bacterial communities of heavy metal contaminated acidic waters from zinc mine residues in Sepetiba Bay. Extremophiles 13, 263–271.
Alqueres, S. M. C., Almeida, R. V., Clementino, M. M., Vieira, R. P., Almeida, W. I., Cardoso, A. M., and Martins, O. B. (2007). Exploring the biotechnological applications in the archaeal domain. Braz. J. Microbiol. 38, 398–405.
Cardoso, A. M., Clementino, M. M., Vieira, R. P., Cavalcanti, J. J. V., Albano, R. M., and Martins, O. B. (2010). “Archaeal metagenomics: bioprospecting novel genes and exploring new concepts” in Metagenomics: Theory, Methods, and Applications, ed. D. Marco (Wymondham: Caister Academic Press), 159–169.
Cary, S. C., Shank, T., and Stein, J. (1998). Worms bask in extreme temperatures. Nature 391, 545–546.
Clementino, M. M., Fernandes, C. A., Vieira, R. P., Cardoso, A. M., Polycarpo, C. R., and Martins, O. B. (2007). Archaeal diversity in naturally occurring and impacted environments from a tropical region. J. Appl. Microbiol. 103, 141–151.
Clementino, M. M., Vieira, R. P., Cardoso, A. M., Nascimento, A. P. A., Silveira, C. B., Riva, T. C., Gonzalez, A. S. M., Paranhos, R., Albano, R. M., Ventosa, A., and Martins, O. B. (2008). Prokaryotic diversity in one of the largest hypersaline coastal lagoons in the world. Extremophiles 12, 595–604.
Hoff, M. (2009). Surviving salt: how do extremophiles do it? PLoS Biol. 7, e1000258. doi: 10.1371/journal.pbio.1000258
Mayr, L. M., Tenenbaun, D. R., Villac, M. C., Paranhos, R., Nogueira, C. R., Bonecker, S. L. C., and Bonecker, A. C. (1989). “Hydrobiological characterization of Guanabara Bay,” In Coastlines of Brazil, eds O. T. Magoon, and C. Neves (New York: American Society of Civil Engineers), 124–139.
Silveira, C. B., Vieira, R. P., Cardoso, A. M., Paranhos, R., Albano, R. M., and Martins, O. B. (2011). Influence of salinity on bacterioplankton communities from the Brazilian rain forest to the coastal atlantic ocean. PLoS One 6, e17789. doi: 10.1371/journal.pone.0017789
Tadeo, X., López-Méndez, B., Trigueros, T., Lain, A., Castano, D., and Millet, O. (2009). Structural basis for the aminoacid composition of proteins from halophilic archaea. PLoS Biol. 7, e1000257. doi: 10.1371/journal.pbio.1000257
Vieira, R. P., Clementino, M. M., Cardoso, A. M., Oliveira, D. N., Albano, R. M., Gonzalez, A. M., Paranhos, R., and Martins, O. B. (2007). Archaeal communities in a tropical estuarine ecosystem: Guanabara Bay, Brazil. Microb. Ecol. 54, 460–468.
Citation: Cardoso AM, Vieira RP, Paranhos R, Clementino MM, Albano RM and Martins OB (2011) Hunting for extremophiles in Rio de Janeiro. Front. Microbio. 2:100. doi: 10.3389/fmicb.2011.00100
Received: 25 February 2011; Accepted: 21 April 2011;
Published online: 02 May 2011.
Copyright: © 2011 Cardoso, Vieira, Paranhos, Clementino, Albano and Martins. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: amcardoso@bioqmed.ufrj.br
