New insights into the evolutionary history of biological nitrogen fixation
- Department of Chemistry and Biochemistry and the Astrobiology Biogeocatalysis Research Center, Montana State University, Bozeman, MT, USA
Nitrogenase catalyzed nitrogen fixation is the process by which life converts dinitrogen gas into fixed nitrogen in the form of bioavailable ammonia. The most common form of nitrogenase today requires a complex metal cluster containing molybdenum (Mo), although alternative forms exist which contain vanadium (V) or only iron (Fe). It has been suggested that Mo-independent forms of nitrogenase (V and Fe) were responsible for N2 fixation on early Earth because oceans were Mo-depleted and Fe-rich. Phylogenetic- and structure-based examinations of multiple nitrogenase proteins suggest that such an evolutionary path is unlikely. Rather, our results indicate an evolutionary path whereby Mo-dependent nitrogenase emerged within the methanogenic archaea and then gave rise to the alternative forms suggesting that they arose later, perhaps in response to local Mo limitation. Structural inferences of nitrogenase proteins and related paralogs suggest that the ancestor of all nitrogenases had an open cavity capable of binding metal clusters which conferred reactivity. The evolution of the nitrogenase ancestor and its associated bound metal cluster was controlled by the availability of fixed nitrogen in combination with local environmental factors that influenced metal availability until a point in Earth’s geologic history where the most desirable metal, Mo, became sufficiently bioavailable to bring about and refine the solution (Mo-nitrogenase) we see perpetuated in extant biology.
Introduction
Biological nitrogen fixation, the reduction of dinitrogen (N2) to ammonia, accounts for roughly two-thirds of the fixed nitrogen (N) produced on Earth today (Rubio and Ludden, 2008). The emergence of biological N2 fixation therefore enabled life to access the vast reserves of N present as N2 gas in our atmosphere (Rees, 1993), a feature that would have profoundly impacted the history of life on earth and the biogeochemical cycles that it modulates (Falkowski et al., 2008; Canfield et al., 2010). Today, biological N2 fixation is catalyzed by at least three genetically distinct but evolutionarily related nitrogenases. The majority of present-day biological nitrogen fixation is catalyzed by the molybdenum-nitrogenase (encoded by nifHDK), an oxygen-sensitive, metalloenzyme complex composed of the Fe protein (product of nifH) and the MoFe heterotetramer (products of nifDK; Rubio and Ludden, 2008). The Fe protein is a homodimer bridged by an intersubunit [4Fe–4S] cluster that serves as the obligate electron donor to the MoFe protein (Georgiadis et al., 1992). The MoFe protein is a α2β2 heterotetramer that houses the P-cluster, an [8Fe–7S] cluster that shuttles electrons to the FeMo-cofactor, a [Mo–7Fe–9S-homocitrate] cluster that provides the substrate reduction site (Kim and Rees, 1992). Two “alternative” forms of nitrogenase have also been identified in the genomes of organisms that encode for Nif (Joerger and Bishop, 1988; Raymond et al., 2004; Soboh et al., 2010; Boyd et al., 2011). The nitrogenase encoded by the vnfHDK genes is believed to contain vanadium in place of molybdenum in the active site cofactor, whereas the nitrogenase encoded by the anfHDK genes appears to contain only Fe as the metal constituent of its active site cofactor (Hales et al., 1986; Chisnell et al., 1988). When fixed nitrogen is limiting, the expression and activity of the alternative forms is regulated by the availability of Mo or V (Joerger and Bishop, 1988; Kessler et al., 1997; Hamilton et al., 2011).
Chemical stratigraphic measurements indicate that ancient oceans were limited in soluble Mo prior to the rise of oxygen ∼2.5 Ga (Anbar et al., 2007) due to the insolubility of Mo-sulfides under anoxic conditions (Helz et al., 1996). This prompted the proposal that Anf and Vnf represent primitive forms of nitrogenase that predate Nif (Anbar and Knoll, 2002; Raymond et al., 2004). Past phylogenetic analyses of the nitrogenase structural gene products have failed to provide convincing evidence for the trajectory of specific metal incorporation into the active site cofactor of nitrogenase during its evolution (Raymond et al., 2004; Soboh et al., 2010; Boyd et al., 2011). Here, we examine concatenations of protein homologs of the structural components shared between all known nitrogenases (H, D, and K; Raymond et al., 2004; Mehta and Baross, 2006; Dekas et al., 2009; Boyd et al., 2011). The results of our phylogenetic- and structure-based examination indicate an evolutionary path whereby Mo-dependent nitrogenase gave rise to the alternative forms suggesting that they arose later, perhaps in response to local Mo limitation. These results, when coupled with considerations of the physiology and the biochemistry of nitrogen fixation, lead to a new model for the stepwise evolution of nitrogenase and other related complex metalloproteins.
Materials and Methods
Phylogenetic Analysis
Representative homologs of Anf/Vnf/NifHDK and uncharacterized HDK (Table A1 in Appendix) were compiled as previously described (Soboh et al., 2010; Boyd et al., 2011). Individual H, D, and K homologs were aligned using CLUSTALX (version 2.0.8) specifying the Gonnet 250 protein substitution matrix and default gap extension and opening penalties (Larkin et al., 2007) as previously described (Boyd et al., 2011) with ChlLNB/BchLNB from Anabaena variabilis ATCC 29413 and Chlorobium limicola DSM 245 serving as outgroups. The individual alignment blocks were concatenated, subjected to evolutionary model prediction, and the phylogeny of each concatenated protein sequence evaluated using MrBayes (version 3.1.2; Huelsenbeck and Ronquist, 2001) and PhyML (version 3.0; Guindon and Gascuel, 2003) employing the WAG + I + G evolutionary model (Appendix) as identified by ProtTest (version 2.0; Abascal et al., 2005). In phylogenetic reconstructions using MrBayes, tree topologies were sampled every 500 generations over 450,000 generations (after a burnin of 50,000) at likelihood stationarity and after convergence of two separate Markov chain Monte Carlo runs (average SD of split frequencies <0.05). A consensus phylogenetic tree was projected from 1,800 trees using FigTree1 (version 1.2.2). One hundred bootstrap replicates were performed in phylogenetic reconstructions using PhyML. Matrices describing the Rao phylogenetic dissimilarity of concatenated HDK homologs, inferred using both MrBayes and PhyML, were generated using Phylocom (version 4.0.1; Webb et al., 2008).
Structural Analyses
The structures of the representative H, D, and K homologs selected for phylogenetic analysis were inferred using the CPH homology server for protein homology modeling (Nielsen et al., 2010) using the NifH (Georgiadis et al., 1992), NifD (Peters et al., 1997; Chiu et al., 2001; Mayer et al., 2002), and NifK (Peters et al., 1997; Chiu et al., 2001; Mayer et al., 2002) crystal structures from Azotobacter vinelandii AvOP. The inferred structures for each H, D, and K homolog were imported into PyMol2 (version 1.4). The root-mean-square-deviations (RMSD) in the Cαi positions were calculated for each individual inferred H, D, and K homolog structure in relation to the other inferred H, D, or K homolog structures resulting in a pairwise matrix describing the structural RMSD (e.g., structural dissimilarity) for H, D, and K homologs. RMSDs generated for H, D, and K homologs, were normalized to compensate for differing HDK protein lengths, and the normalized H, D, and K matrices were then averaged to produce an HDK RMSD matrix for use in statistical analyses. PyMol was also used to generate images of sequence conservation in the active site cavity of Anf/Vnf/NifDK homologs.
Statistical Analyses
Mantel regressions of dissimilarity matrices were performed using XL Stat (version 2009.5.01). Ten thousand permutations employing two-tailed t-tests were used to determine the strength and significance of the relationships between dissimilarity matrices, respectively.
Results and Discussion
Bayesian and maximum-likelihood phylogenetic analyses of concatenated protein homologs of the required structural components (H, D, and K) encoded by characterized and putative anf, vnf, and nif regulons (Raymond et al., 2004; Mehta and Baross, 2006; Dekas et al., 2009; Boyd et al., 2011) yielded congruent topologies (Figure A1 in Appendix) with well-supported lineages that correspond to the nitrogenase active site metal content (Figures 1 and A2 in Appendix). nif-Encoded HDK protein homologs formed two distinct lineages, one of which was comprised of proteins derived solely from hydrogenotrophic methanogens that branched at the base of the tree. The second Nif lineage was comprised of more recently evolved NifHDK homologs from both bacterial and methanogen genomes. These findings are consistent with the results of a recent phylogenetic analysis which indicate that Nif emerged within the hydrogenotrophic methanogen lineage ∼2.2 Ga (Boyd et al., 2011), at a time when Mo concentrations in oceans are thought to have begun to increase (Anbar et al., 2007).
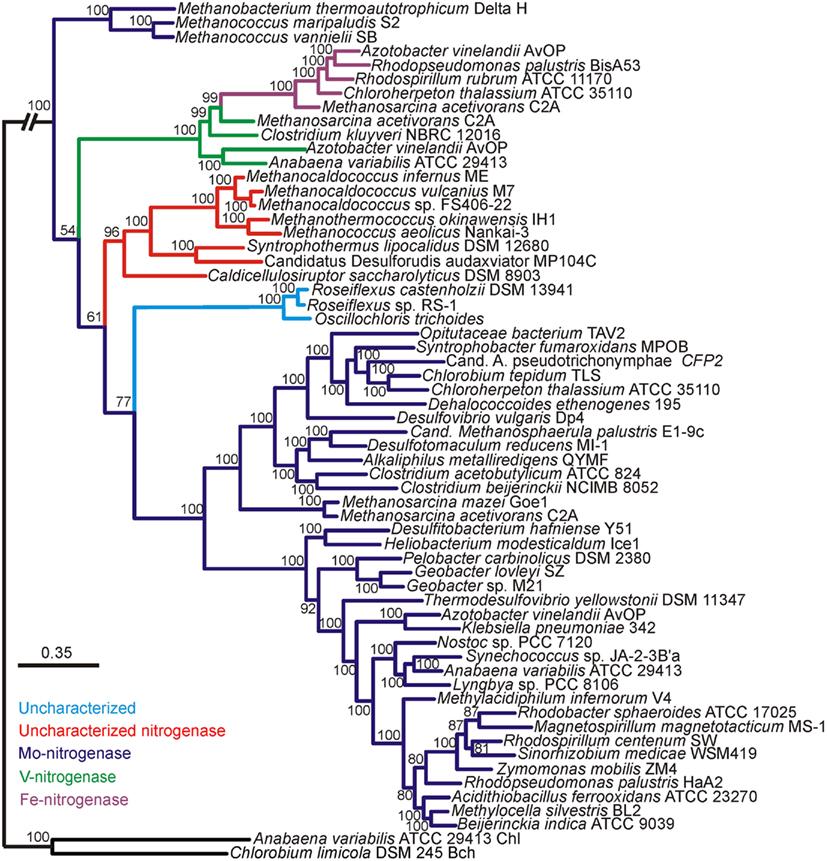
Figure 1. Bayesian inferred phylogenetic tree of concatenated HDK homologs (see Figure A1 in Appendix for maximum-likelihood inferred tree). Posterior probabilities are indicated above or below nodes. Branches are colored dark blue (Mo-nitrogenase, Nif), green (V-nitrogenase, Vnf), purple (Fe-nitrogenase, Anf), red (uncharacterized nitrogenase), and light blue (uncharacterized homolog). The hash at the root was introduced to conserve space.
Nested within the two Nif sublineages is a monophyletic lineage comprised of Vnf and Anf nitrogenase, indicating that Vnf and Anf are derived from Nif. It is likely that the ancestor of the Anf/Vnf lineage resulted from gene duplication within the hydrogenotrophic methanogen lineage and was not singly laterally transferred, a finding that is consistent with the observation that anf and vnf have yet to be identified in a genome that does not also encode for nif (Raymond et al., 2004; Boyd et al., 2011). VnfHDK homologs nest AnfHDK homologs with strong statistical support, providing evidence that Anf is derived from Vnf, both of which are derived from Nif. The latter conclusion appears to be supported by the results of a recent transcriptomic profiling of A. vinelandii, which indicated that a number of nif-encoded genes are up-regulated under conditions favoring the expression of Vnf and Anf (Hamilton et al., 2011). The observation that VnfHDK and AnfHDK from Methanosarcinales branch closely, coupled with the fact that these operons are located proximal in the genomes of these organisms, may suggest that anf is the result of a recent duplication of the vnf operon within the Methanosarcinales lineage. The acquisition of vnf within the Methanosarcinales lineage may have been the result of a lateral gene transfer (LGT) event with a firmicute, a finding that is consistent with the close spatial proximity noted between members of the Methanosarcinales and Firmicutes in a variety of anoxic environments (Stams, 1994) and with previous reports of LGT of individual genes and metabolic pathways between these two anaerobic lineages (Beiko et al., 2005; Fournier and Gogarten, 2008; Boyd et al., 2011). Importantly, evidence presented here and elsewhere (Raymond et al., 2004; Boyd et al., 2011) indicates that nif may have been acquired in the Methanosarcinales via LGT; however, it is unclear based on this dataset if that event predates the acquisition of vnf in this lineage and the subsequent duplication of vnf that resulted in anf. Nevertheless, considering that the biosynthesis of Anf and Vnf examined to date require nif-encoded gene products (Joerger et al., 1986; Kennedy and Dean, 1992; Hamilton et al., 2011), the acquisition of vnf and the duplication of vnf that led to anf are most likely to postdate the acquisition of nif within this lineage. Collectively, the evidence suggests that both Nif and Anf evolved in the methanogenic archaea, a guild of organisms which typically inhabit anoxic environments where Mo is in limited supply (Helz et al., 1996). Together with the fact that the expression of Anf and Vnf is tightly regulated by the availability of Mo and V (Joerger and Bishop, 1988; Hamilton et al., 2011), this set of observations suggests that the transient fluctuations in metal availability in anoxic environments may have been the impetus to incorporate new metals into the active site cluster of nitrogenase.
We also examined the evolutionary history of HDK homologs from the genomes of organisms that have been shown to fix N2 (Mehta and Baross, 2006; Dekas et al., 2009), but for which detailed biochemical analysis of the active site cofactor has yet to be performed (denoted as “uncharacterized nitrogenase” in Figures 1 and A2 in Appendix). These proteins formed a monophyletic lineage that branched after Nif derived from hydrogenotrophic methanogens and the Anf/Vnf lineages (Figure 1) indicating they emerged after Nif and Vnf, and possibly Anf. The “uncharacterized nitrogenase” lineage is comprised of proteins derived from strictly anaerobic taxa within the Firmicutes, as well as the methanogenic and methanotrophic archaea (Figure 1). A separate lineage comprised of uncharacterized HDK homologs that have only been identified in the genomes of filamentous anoxygenic phototrophic bacteria, branches after Nif, Vnf, and the uncharacterized nitrogenases indicating that they are the most recently evolved lineage of putative nitrogenase. A physiological or biochemical role for these proteins has yet to be conclusively demonstrated.
We inferred the protein structures of HDK homologs from anf, vnf, nif, and uncharacterized operons using homology modeling based on the structures of NifHDK from A. vinelandii (Georgiadis et al., 1992; Kim and Rees, 1992). Pairwise comparisons of the inferred protein structures enabled the generation of a matrix that describes their structural dissimilarity. A regression of this matrix and a matrix describing the phylogenetic dissimilarity of the concatenated HDK proteins revealed a significant and positive relationship (Mantel R2 = 0.23, p < 0.01; Figure A3 in Appendix). This indicates that the structure of nitrogenase has evolved significantly through time. Conserved residues that line the active site pocket in Anf/Vnf/NifD inferred through homology modeling (Figure A4 in Appendix) suggest that once the active site cavity evolved, the majority of the residues in the cavity and the cluster-coordinating ligands were maintained despite differences in the metal composition of the cofactor (Figure A5 in Appendix).
We examined phylogenetic and structural relationships among proteins that are evolutionarily related to nitrogenase, including those required to biosynthesize bacteriochlorophyll (BchN; Hearst et al., 1985; Burke et al., 1993) and those that have been proposed to catalyze an analogous reaction in Ni porphyrin F430 biosynthesis (NflD; Staples et al., 2007). Phylogenetic reconstruction of Anf/Vnf/NifD, BchN, and NflD revealed three lineages, with NflD proteins forming a lineage that bisects a lineage comprising Anf/Vnf/NifD and a lineage comprising ChlN/BchN (Figure 2). These findings are consistent with a previous phylogenetic analysis of concatenated Anf/Vnf/NifHD, BchLN, and NflHD proteins (Raymond et al., 2004), which together suggest that Nfl is ancestral to Anf/Vnf/Nif and Bch (Staples et al., 2007). Intriguingly, NflD proteins share little sequence conservation with the active site cavity of Anf/Vnf/NifD and BchN. Likewise, the cofactor coordinating ligands in Anf/Vnf/NifD are not conserved in BchN sequences, although the crystal structure of BchN reveals an open cavity for the binding of protochlorophyllide instead of the bound cofactor observed in nitrogenase (Muraki et al., 2010). Homology modeling of NflD from Methanocaldococcus jannaschii DSM 2661 threaded on the structure of BchN (Muraki et al., 2010) revealed an open cavity that is similar to that of BchN that may serve as the substrate binding site. Thus, the two derived states (Anf/Vnf/NifD and BchN) have maintained similar structural architecture to that of the inferred ancestral state (e.g., NflD) but appear to have fine-tuned cavity residues to bind target substrates as the paralogs diversified. This leads to a model for the emergence of nitrogenase (Figure 3) whereby a gene encoding for an ancestral protein complex with a cavity similar to that observed in the inferred structure of NflD duplicated, leading to the evolutionary precursor of BchN and Anf/Vnf/NifD. Serendipitously, metals (e.g., Fe) or metal clusters (e.g., 4Fe–4S) were bound in the cavity of the ancestor in a non-specific manner, resulting in an enzyme complex with altered reactivity, perhaps toward N2 reduction. In response to selective pressure of limited fixed nitrogen on early Earth, genes and associated gene products were presumably recruited to improve the enzyme stepwise through the modification of the metal cofactor (Rubio and Ludden, 2008; Hu and Ribbe, 2011). In parallel, the active site was refined to yield a cavity that binds the active site cofactor FeMo-co thereby fine-tuning the structural determinants for nitrogenase catalysis. In this mechanism, it is not inconceivable that the size and dimension of the nitrogenase cofactor were constrained somewhat by the structure of the ancestor. This might be supported by the observation that the end-to-end dimensions of the FeMo-cofactor of Mo-nitrogenase are not that different from those of bacteriochlorophyll or F430. Given that the as isolated FeMo-cofactor is not reactive toward N2 on its own, the aforementioned stepwise evolution of nitrogenase may be the only mechanism by which the biochemical pathway for cofactor biosynthesis could have evolved in response to the selective pressure of fixed nitrogen limitation.
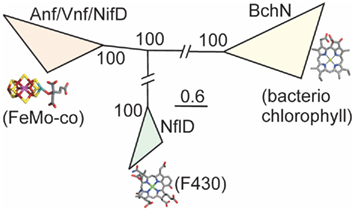
Figure 2. Bayesian inferred phylogenetic reconstruction of Anf/Vnf/NifD, BchN, and NflD proteins. The putative substrates and cofactors for each protein lineage are indicated below each respective clade. Posterior probabilities for each collapsed node are indicated. Nodes have been collapsed and hashes introduced to conserve space.
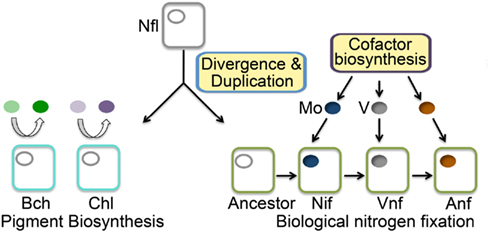
Figure 3. Model depicting the divergence of nitrogenase (NifD) and protochlorophyllide reductase (ChlN/BchN) from a NflD ancestor. The stepwise evolution of cofactor biosynthesis leading to the acquisition of metal specificity in the covalently bound active site metallocluster, where Mo acquisition and Mo-nitrogenase predates V acquisition and V-nitrogenase, and V acquisition predates Fe-only nitrogenase. ChlN/BchN bind substrates in their active site cavities non-covalently and release these substrates following reduction (Muraki et al., 2010). Abbreviations: Mo, molybdenum; V, vanadium; Nif, Mo-dependent nitrogenase; Vnf, V-dependent nitrogenase; Anf, Fe-only nitrogenase; Bch, BchN protein involved in bacteriochlorophyll biosynthesis; Chl, ChlN protein involved in chlorophyll biosynthesis.
In summary, the Mo-nitrogenase we see today in extant biology is not likely to be the first nitrogenase associated with early life on Earth, a finding that is in line with the dogma supported by geochemistry (Anbar and Knoll, 2002; Anbar, 2008). However, in contrast with what has been proposed previously (Anbar and Knoll, 2002; Raymond et al., 2004; Anbar, 2008), the results indicate that alternative nitrogenases (V- and Fe-only forms) are not ancestors of the Mo-nitrogenase but rather are derived from Mo-nitrogenase. The common ancestor of Nif/Vnf/Anf, Bch/Chl, and Nfl had a cavity capable of binding certain porphyrins and/or metal cluster fragments. The nature of the ancestral nitrogenase enzyme and its associated bound metal cluster was likely controlled by the selective pressure imposed by fixed nitrogen limitation in combination with local environmental metal availability until a point in Earth history (e.g., the “Great Oxidation Event”) when Mo became sufficiently bioavailable (Anbar and Knoll, 2002; Anbar, 2008) and the most favorable solution for biological nitrogen fixation (Mo-nitrogenase) emerged that is reflective of today’s extant enzyme. These results reveal a new paradigm for the evolution of biological nitrogen fixation and provide key insights into the manner in which early life forms might have exploited the reactivity of their mineral environment prior to evolving the refined complex metalloenzymes observed today.
Author Contributions
Eric S. Boyd designed the study and performed phylogenetic and statistical analyses. Trinity L. Hamilton generated the inferred protein structures. John W. Peters supervised the work. All authors contributed to the writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the NASA Astrobiology Institute (NAI) grant NNA08C-N85A to John W. Peters and Eric S. Boyd. Trinity L. Hamilton was supported by an NSF-Integrated Graduate Educational Research and Training fellowship grant and Eric S. Boyd was supported by a fellowship from the NAI Postdoctoral Program.
Footnotes
References
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 2, 2104–2105.
Anbar, A. D., Duan, Y., Lyons, T. W., Arnold, G. L., Kendall, B., Creaser, R. A., Kaufman, A. J., Gordon, G. W., Scott, C., Garvin, J., and Buick, R. (2007). A whiff of oxygen before the great oxidation event? Science 317, 1903–1906.
Anbar, A. D., and Knoll, A. H. (2002). Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science 297, 1137–1142.
Beiko, R. G., Harlow, T. J., and Ragan, M. A. (2005). Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 14332–14337.
Boyd, E. S., Anbar, A. D., Miller, S., Hamilton, T. L., Lavin, M., and Peters, J. W. (2011). A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology 9, 221–232.
Burke, D. H., Hearst, J. E., and Sidow, A. (1993). Early evolution of photosynthesis: clues from nitrogenase and chlorophyll iron proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 7134–7138.
Canfield, D. E., Glazer, A. N., and Falkowski, P. G. (2010). The evolution and future of Earth’s nitrogen cycle. Science 330, 192–196.
Chisnell, J. R., Premakumar, R., and Bishop, P. E. (1988). Purification of a second alternative nitrogenase from a nifHDK deletion strain of Azotobacter vinelandii. J. Bacteriol. 170, 27–33.
Chiu, H., Peters, J. W., Lanzilotta, W. N., Ryle, M. J., Seefeldt, L. C., Howard, J. B., and Rees, D. C. (2001). MgATP-Bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127 Delta-Fe-protein and the MoFe-protein. Biochemistry 40, 641–650.
Dekas, A. E., Poretsky, R. S., and Orphan, V. J. (2009). Deep-sea Archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326, 422–426.
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth’s biogeochemical cycles. Science 1034–1039.
Fournier, G. P., and Gogarten, J. P. (2008). Evolution of acetoclastic methanogenesis in Methanosarcina via horizontal gene transfer from cellulolytic Clostridia. J. Bacteriol. 190, 1124–1127.
Georgiadis, M. M., Komiya, H., Chakrabarti, P., Woo, D., Kornuc, J. J., and Rees, D. C. (1992). Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257, 1653–1659.
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704.
Hales, B. J., Case, E. E., Morningstar, J. E., Dzeda, M. F., and Mauterer, L. A. (1986). Isolation of a new vanadium-containing nitrogenase from Azotobacter vinelandii. Biochemistry 25, 7251–7255.
Hamilton, T. L., Ludwig, M., Dixon, R., Boyd, E. S., Dos Santos, P. C., Setubal, J. C., Bryant, D. A., Dean, D. R., and Peters, J. W. (2011). Transcriptional profiling of nitrogen fixation in Azotobacter vinelandii. J. Bacteriol. 193, 4477–4486.
Hearst, J. E., Alberti, M., and Doolittle, R. F. (1985). A putative nitrogenase reductase gene found in the nucleotide sequences from the photosynthetic gene cluster of R. capsulata. Cell 40, 219–220.
Helz, G. R., Miller, C. V., Charnock, J. M., Mosselmans, J. F. W., Pattrick, R. A. D., Garner, C. D., and Vaughan, D. J. (1996). Mechanism of molybdenum removal from the sea and its concentration in black shales: EXAFS evidence. Geochim. Cosmochim. Acta 60, 3631–3642.
Hu, Y., and Ribbe, M. W. (2011). Biosynthesis of nitrogenase FeMoco. Coord. Chem. Rev. 255, 1218–1224.
Huelsenbeck, J. P., and Ronquist, R. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754–755.
Joerger, R. D., and Bishop, P. E. (1988). Bacterial alternative nitrogen fixation systems. Crit. Rev. Microbiol. 16, 1–14.
Joerger, R. D., Premakumar, R., and Bishop, P. E. (1986). Tn5-induced mutants of Azotobacter vinelandii affected in nitrogen fixation under Mo-deficient and Mo-sufficient conditions. J. Bacteriol. 168, 673–682.
Kennedy, C., and Dean, D. (1992). The nifu, nifs and nifv gene products are required for activity of all three nitrogenases of Azotobacter Vinelandii. Mol. Gen. Genet. 231, 494–498.
Kessler, P. S., Mclarnan, J., and Leigh, J. A. (1997). Nitrogenase phylogeny and the molybdenum dependence of nitrogen fixation in Methanococcus maripaludis. J. Bacteriol. 179, 541–543.
Kim, J., and Rees, D. C. (1992). Crystallographic structure and functional implications of the nitrogenase molybdenum-iron protein from Azotobacter vinelandii. Nature 360, 553–560.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G.. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948.
Mayer, S. M., Gormal, C. A., Smith, B. E., and Lawson, D. M. (2002). Crystallographic analysis of the MoFe protein of nitrogenase from a nifV mutant of Klebsiella pneumoniae identifies citrate as a ligand to the molybdenum of iron molybdenum cofactor (FeMoco). J. Biol. Chem. 277, 35263–35266.
Mehta, M. P., and Baross, J. A. (2006). Nitrogen fixation at 92°C by a hydrothermal vent archaeon. Science 314, 1783–1786.
Muraki, N., Nomata, J., Ebata, K., Mizoguchi, T., Shiba, T., Tamiaki, H., Kurisu, G., and Fujita, Y. (2010). X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465, 110–114.
Nielsen, M., Lundegaard, C., Lund, O., and Petersen, T. N. (2010). CPHmodels-3.0-remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res. 38, W576–W581.
Peters, J. W., Stowell, M. H., Soltis, S. M., Finnegan, M. G., Johnson, M. K., and Rees, D. C. (1997). Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry 36, 1181–1187.
Raymond, J., Siefert, J. L., Staples, C. R., and Blankenship, R. E. (2004). The natural history of nitrogen fixation. Mol. Biol. Evol. 21, 541–554.
Rees, D. C. (1993). Dinitrogen reduction by nitrogenase: if N2 isn’t broken, it can’t be fixed. Curr. Opin. Struct. Biol. 3, 921–928.
Rubio, L. M., and Ludden, P. W. (2008). Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 62, 93–111.
Soboh, B., Boyd, E. S., Zhao, D., Peters, J. W., and Rubio, L. M. (2010). Substrate specificity and evolutionary implications of a NifDK enzyme carrying NifB-co at its active site. FEBS Lett. 584, 1487–1492.
Stams, A. (1994). Metabolic interactions between anaerobic bacteria in methanogenic environments. Antonie Van Leeuwenhoek 66, 271–294.
Staples, C. R., Lahiri, S., Raymond, J., Von Herbulis, L., Mukhophadhyay, B., and Blankenship, R. E. (2007). Expression and association of group IV nitrogenase NifD and NifH homologs in the non-nitrogen-fixing archaeon Methanocaldococcus jannaschii. J. Bacteriol. 189, 7392–7398.
Webb, C. O., Ackerly, D. D., and Kembel, S. W. (2008). Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100.
Appendix
Methods
Phylogenetic analyses
Representative homologs of Anf/Vnf/NifHDK (Table A1), as well as representative homologs of uncharacterized HDK, were compiled as previously described (Soboh et al., 2010; Boyd et al., 2011) and were aligned as previously described (Boyd et al., 2011). The H, D, and K alignment blocks were concatenated and the alignment block was subjected to evolutionary model prediction using ProtTest (version 2.0; Abascal et al., 2005). ProtTest identified the Whelan and Goldman (WAG) evolutionary model with gamma distributed rate variation with a proportion of invariable sites (I + G) as the best fit model for the data. In phylogenetic reconstructions using MrBayes, tree topologies were sampled every 500 generations over 450,000 generations (after a burnin of 50,000) at likelihood stationarity and after convergence of two separate Markov chain Monte Carlo runs (average SD of split frequencies <0.05). A consensus phylogenetic tree was projected from 1800 trees using FigTree (version 1.2.2; http://tree.bio.ed.ac.uk/UH). One hundred bootstrap replicates were performed in phylogenetic reconstructions using PhyML.
Results
The protein structures of HDK homologs from anf, vnf, nif, and uncharacterized operons were inferred using homology modeling based on the NifHDK from A. vinelandii. Pairwise comparisons of the inferred protein structures enabled the generation of a matrix that describes their structural dissimilarity. A regression of this matrix and a matrix describing the phylogenetic dissimilarity of the concatenated HDK proteins revealed a significant and positive relationship (Mantel R2 = 0.23, p < 0.01). This indicates that the structure of nitrogenase has evolved significantly through time. However, the slope of the linear regression indicated that a ∼2 unit increase in phylogenetic dissimilarity resulted in only a ∼1 unit increase in structural dissimilarity, suggesting that deviation in inferred tertiary structure is constrained to a greater extent than that observed in the primary sequence. Therefore, we examined the conservation of residues that line the active site pocket in Anf/Vnf/NifD to determine if the active site cavity has evolved to accommodate the varying cluster compositions associated with Anf, Vnf, Nif, and to potentially uncover evidence that could provide insight into the composition of the active site cofactor in uncharacterized nitrogenase homologs. This analysis revealed a number of residues that line the active site cavity that are conserved among Anf, Vnf, Nif, and uncharacterized nitrogenase including the two covalent ligands to the FeFe-, FeV-, and FeMo-cofactors Cys-275 and His-442 (numbered according to A. vinelandii NifD). In addition to ligands, a number of other residues in the binding cavity are conserved, including Val-70, Gln-191, His-195, and Phe-391. Together, these results suggest that once the active site cavity was evolved, it was maintained despite differences in the metal composition of the cofactor.
We next compared the conservation in these residues with NflD, a nitrogenase homolog that has been hypothesized to be involved in F430 biosynthesis (Staples et al., 2007), and BchN, a homolog involved in bacteriochlorophyll biosynthesis (Muraki et al., 2010). A previous phylogenetic reconstruction of Anf/Vnf/NifHD, BchLN, and NflHD revealed three lineages, with NflHD proteins forming a lineage that bisected the a lineage comprising Anf/Vnf/NifHD and a lineage comprising BchLN (Raymond et al., 2004), suggesting that that NflHD is ancestral to Anf/Vnf/NifHD and BchLB (Staples et al., 2007). Intriguingly, the active site cavity of NflD proteins shared little sequence conservation with that of Anf/Vnf/NifHD and BchLN. Likewise, the FeMo-co ligands in Anf/Vnf/NifD are not conserved in BchN sequences, although the crystal structure of BchNB reveals an open cavity for the binding of protochlorophyllide that is in the same position as to where FeMo-co is bound in nitrogenase. Thus, the two derived states (Anf/Vnf/NifD and BchN) have maintained similar structural architecture, but appear to have fine-tuned cavity residues to bind target substrates. To examine whether the open cavity architecture was a structural property of the ancestor of Anf/Vnf/NifD and BchN, we generated homology models of NflD from Methanocaldococcus jannaschii DSM 2661 (Accession no. NP_248427) threaded on A. vinelandii NifD and BchN from Rhodobacter capsulatus SB 1003 (Accession no.YP_003576837). The inferred structures reveal a cavity in NflD that is similar to that of BchN, the size of which would be capable of binding F430. These analyses suggest that nitrogenase evolved from an ancestral protein that exhibited an open cavity and adapted that cavity to covalently ligate the FeMo-co necessary for N2 reduction.
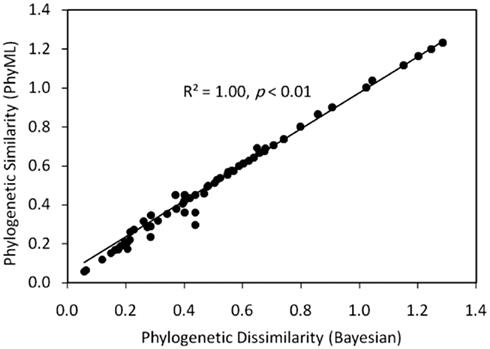
Figure A1. Plot of a Mantel regression of a matrix describing the Rao phylogenetic dissimilarity of concatenated HDK homologs inferred by PhyML as a function of the Rao phylogenetic dissimilarity of concatenated HDK homologs inferred by MrBayes. The strong positively trending correlation suggests that the topologies of the two trees are congruent.
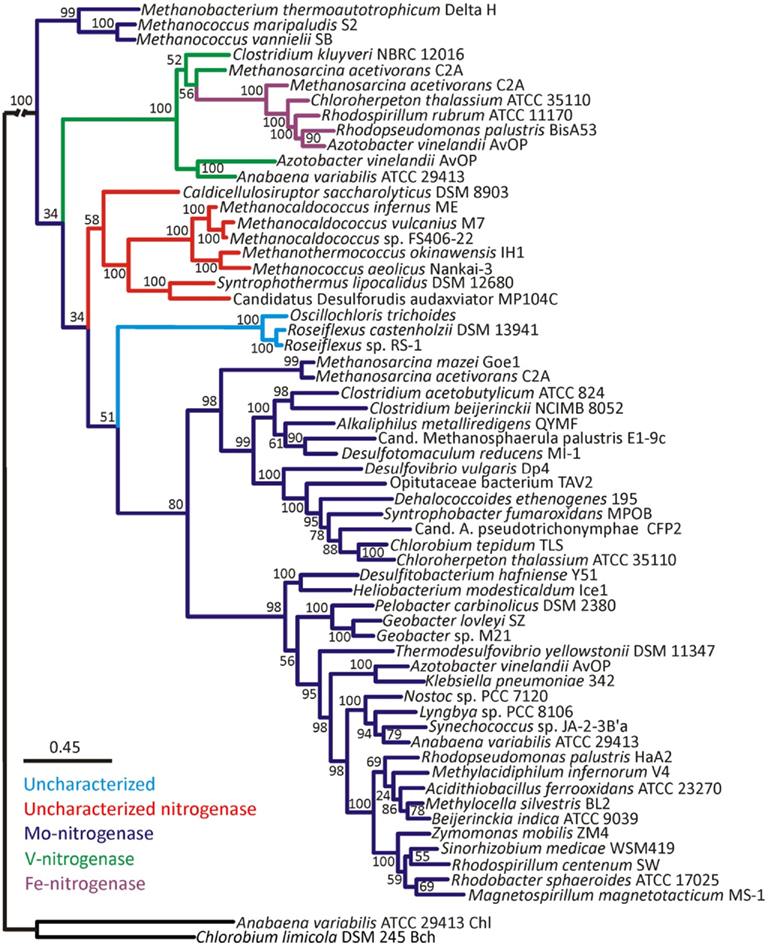
Figure A2. Maximum-likelihood inferred phylogenetic tree of concatenated HDK homologs. Bootstrap values based on 100 replicates are indicated at each node. Branches are colored dark blue (Mo-nitrogenase, Nif), green (V-nitrogenase, Vnf), purple (Fe-nitrogenase, Anf), red (uncharacterized nitrogenase), and light blue (uncharacterized homolog). The hash at the root was introduced to conserve space.
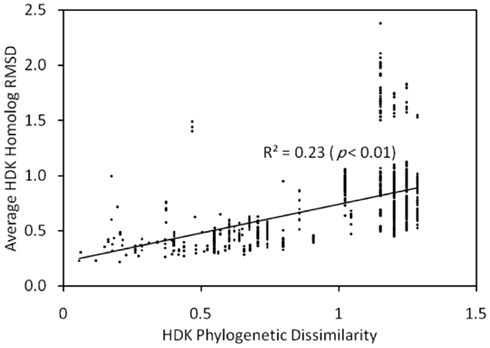
Figure A3. Plot of a Mantel regression of a matrix describing the average RMSD for H, D, and K protein structures inferred using homology modeling as a function of the Rao phylogenetic dissimilarity of concatenated HDK homologs inferred by MrBayes. The strong correlation suggests a relationship between the evolution of sequences and their inferred structures, implying that the HDK structure is evolving. The slope of the line linear regression (∼2) suggests that the evolution of protein structure is constrained to a greater extent than the evolution of the primary sequences.
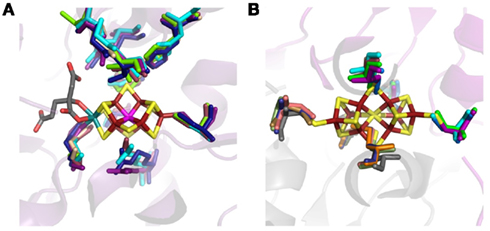
Figure A4. Structural alignment of the inferred structures of DK homologs indicating conservation in the active site (A) and P-cluster binding cavity (B). Ribbon diagram of the superimposition of NifDK from Azotobacter vinelandii AvOP (D, violet and K, gray), NifDK from Methanococcus maripaludis strain S2 (D, wheat and NifK, blue), UncDK from Methanocaldococcus sp. FS406-22 (D, cyan and K, orange), UncDK from Roseiflexus sp. RS-1 (D, marine, and K, sand), VnfDK from Methanosarcina acetivorans str. C2A (D, raspberry, and K, pale green), and AnfDK from Azotobacter vinelandii AvOP (D, green, and K, salmon), with the FeMo-co (A) and P-cluster (B) depicted as stick representations. Dark red, Fe; yellow, S; gray, C; red, O; teal, Mo; unknown, magenta. Protein Data Bank ID for Azotobacter vinelandii AvOP 1MIN.

Figure A5. Amino acid sequence conservation in selected residues that ligate FeMo-co (Cys-275, red box, and His-442, blue box) and that have been implicated as important in the FeMo-co binding pocket (indicated by a gray box). Representative NifD, VnfD, AnfD, and UncD (uncharacterized nitrogenase). Numbering is based on NifD from Azotobacter vinelandii AvOP. Abbreviations: A.v., Azotobacter vinelandii AvOP; M.a., Methanosarcina acetivorans str. C2A; M.m., Methanococcus maripaludis strain S2; R.c., Roseiflexus sp. RS-1; M.c. Methanocaldococcus sp. FS406-22. The conservation in the active site environment and active site custer ligands between classes of nitrogenase suggest that once the active site cavity evolved, it was maintained through time.
Keywords: nitrogenase, metalloenzyme, evolution, great oxidation event, oxygen molybdenum, vanadium, iron, sulfide
Citation: Boyd ES, Hamilton TL and Peters JW (2011) An alternative path for the evolution of biological nitrogen fixation. Front. Microbio. 2:205. doi: 10.3389/fmicb.2011.00205
Received: 09 August 2011;
Paper pending published: 28 August 2011;
Accepted: 10 September 2011;
Published online: 05 October 2011.
Edited by:
Stephen Wiley Ragsdale, University of Michigan, USAReviewed by:
Uwe Deppenmeier, University of Bonn, GermanyWilliam Lanzilotta, University of Georgia, USA
Copyright: © 2011 Boyd, Hamilton and Peters. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: John W. Peters, Department of Chemistry and Biochemistry, Montana State University, 103 Chemistry and Biochemistry Building, Bozeman, MT 59717, USA. e-mail: john.peters@chemistry.montana.edu