- 1 Biology Department, Occidental College, Los Angeles, CA, USA
- 2 Monterey Bay Aquarium Research Institute, Moss Landing, CA, USA
Tropical epiphytic plants within the family Bromeliaceae are unusual in that they possess foliage capable of retaining water and impounded material. This creates an acidic (pH 3.5–6.5) and anaerobic (<1 ppm O2) environment suspended in the canopy. Results from a Costa Rican rainforest show that most bromeliads (n = 75/86) greater than ~20 cm in plant height or ~4–5 cm tank depth, showed presence of methanogens within the lower anoxic horizon of the tank. Archaea were dominated by methanogens (77–90% of recovered ribotypes) and community structure, although variable, was generally comprised of a single type, closely related to either hydrogenotrophic Methanoregula or Methanocella, a specific clade of aceticlastic Methanosaeta, or Methanosarcina. Juvenile bromeliads, or those species, such as Guzmania, with shallow tanks, generally did not possess methanogens, as assayed by polymerase chain reaction specific for methanogen 16S rRNA genes, nor did artificial catchments (~100 ml volume), in place 6–12 months prior to sample collection. Methanogens were not detected in soil (n = 20), except in one case, in which the dominant ribotype was different from nearby bromeliads. Recovery of methyl coenzyme M reductase genes supported the occurrence of hydrogenotrophic and aceticlastic methanogens within bromeliad tanks, as well as the trend, via QPCR analysis of mcrA, of increased methanogenic capacity with increased plant height. Methane production rates of up to 300 nmol CH4 ml tank water−1 day−1 were measured in microcosm experiments. These results suggest that bromeliad-associated archaeal communities may play an important role in the cycling of carbon in neotropical forests.
Introduction
Because of the significant influence of methane on atmospheric chemistry and it’s prominent role as a greenhouse gas, it is becoming increasingly important to understand its sources and sinks on Earth (Crutzen, 1991; IPCC, 2001). Methanogenesis is the dominant terminal mineralization process in wetlands and freshwater sediments that experience prolonged flooding and are limited in more energetically favorable electron acceptors (e.g., sulfate, nitrate, and metal oxides; Whalen, 1993; Grosskopf et al., 1998; Galand et al., 2002; Cadillo-Quiroz et al., 2006). Methane emissions from these environments, both natural and man-made, are substantial (~200 Tg year−1) and are estimated to contribute up to 40% of total global CH4 emissions annually (Crutzen, 1991; Grosskopf et al., 1998; Watanabe et al., 1999; IPCC, 2001; Galand et al., 2002). Methane in these ecosystems is produced exclusively by the activity of certain archaea, during the final step in the anaerobic degradation of organic matter.
Methanogenic archaea can use a number of substrates as precursors of methane, with CO2 reduction and acetate fermentation being most important in wetland soils. Although the relative contributions of the two pathways can vary, aceticlastic methanogenesis generally contributes more to total terrestrial CH4 production than hydrogenotrophic CO2 reduction (Jetten et al., 1992; Conrad, 1999; Conrad et al., 2010). Specialist acetotrophic and more versatile methylotrophic (C1) methanogens belong predominantly to the Methanosarcinales, while H2/CO2-utilizing methanogens include members of the Methanocellales and Methanomicrobiales, to name a few. Methane production by these groups can be influenced by many factors, including availability of substrates and electron donors (e.g., H2), which are often formed catabolically by bacteria, quantity and quality of organic carbon, the presence of possible substrate competitors (e.g., bacteria), and environmental conditions such as pH and oxygen levels (Capone et al., 1983; Galand et al., 2002; Juottonen et al., 2005).
There is a general expectation that as the conditions for methanogenesis are discovered to be broader than predicted, so too will the number of habitats known to contribute methane to the atmosphere. Small-scale water catchments such as bromeliad tanks, Heliconia bracts, tree holes, etc., have the capacity to collect water, thereby hosting microbial communities, with the collective potential to possibly influence carbon cycling, when integrated over ecosystem scales. Epiphytic tank-forming bromeliads, in particular, are those that possess foliage arranged in a compact rosette capable of retaining water. They are predominant members of neotropical habitats, including rain and montane cloud forests, and by some estimates may trap as much as 50,000 l of suspended water in the canopy ha−1 (Sugden and Robins, 1979; Fish, 1983). In this way, bromeliad tanks provide a vast and unique freshwater niche that does not typically occur in other, especially above ground, locations in tropical forests. In fact, in a recent article by Yavitt (2010), the lentic ecosystems found in bromeliads were classified as “cryptic wetlands.” Accumulation of organic matter, such as leaves and carcasses of dead fauna, primarily arthropods, in bromeliad tanks under acidic, submerged conditions, provides the conditions necessary for the creation of an anaerobic microcosm capable of sustained decomposition and remineralization of terrestrial carbon and the potential for production and release of methane.
Martinson et al. (2010) first reported that tank bromeliads are important sources of atmospheric methane. Methane fluxes, measured from bromeliads in the Ecuadorian Andes, were directly correlated with tank diameter and the plant vascular system itself was determined to act as an important conduit for methane release (Martinson et al., 2010). By extrapolation, it was suggested that CH4 emissions from bromeliad-associated archaea (up to ~2 Tg CH4 year−1) might possibly explain the anomalously high methane levels observed previously over neotropical forests (Frankenberg et al., 2008; Martinson et al., 2010). Whether or not the contribution of methane by these ecosystems is globally significant, as suggested by Martinson et al. (2010), but downplayed by Yavitt (2010), it is clear that methanogens in bromeliad tank water play a key role in the cycling of carbon in neotropical environments.
Biomass degradation through methanogenesis remains largely unexplored with regard to suspended catchments, such as bromeliad tanks. Within bromeliad tanks, total organic carbon levels are elevated (~46% TOC), relative to nearby soil (4% TOC), the pH is neutral to acidic (6.5–3.5), and oxygen is low (<1 ppm; Bermudes and Benzing, 1991; Guimaraes-Souza et al., 2006; Goffredi et al., 2011). Additionally, bacteria related to Proteobacteria, Acidobacteria, Bacteroidetes, and Firmicutes, with varying capabilities of organic carbon breakdown, have been recovered from Costa Rican bromeliad tanks (Goffredi et al., 2011). These bacteria presumably contribute to organic matter decomposition, from recalcitrant chitin and cellulose breakdown to the production of volatile fatty acids and hydrogen, which facilitate the final production and evolution of methane gas by archaea. The objective of this study was to provide a molecular and functional assessment of the methanogen communities inhabiting bromeliad tanks in a lowland neotropical rainforest in Costa Rica, with emphasis on community structure and possible influences on methane production potential (e.g., environment and plant morphology). Bromeliads, as well as nearby soil and artificial catchments, were sampled over the course of 18 months, using both 16S rRNA genes, as well as methanogen-specific mcrA genes, to document variations in methanogen populations among tanks.
Materials and Methods
Sample Collection
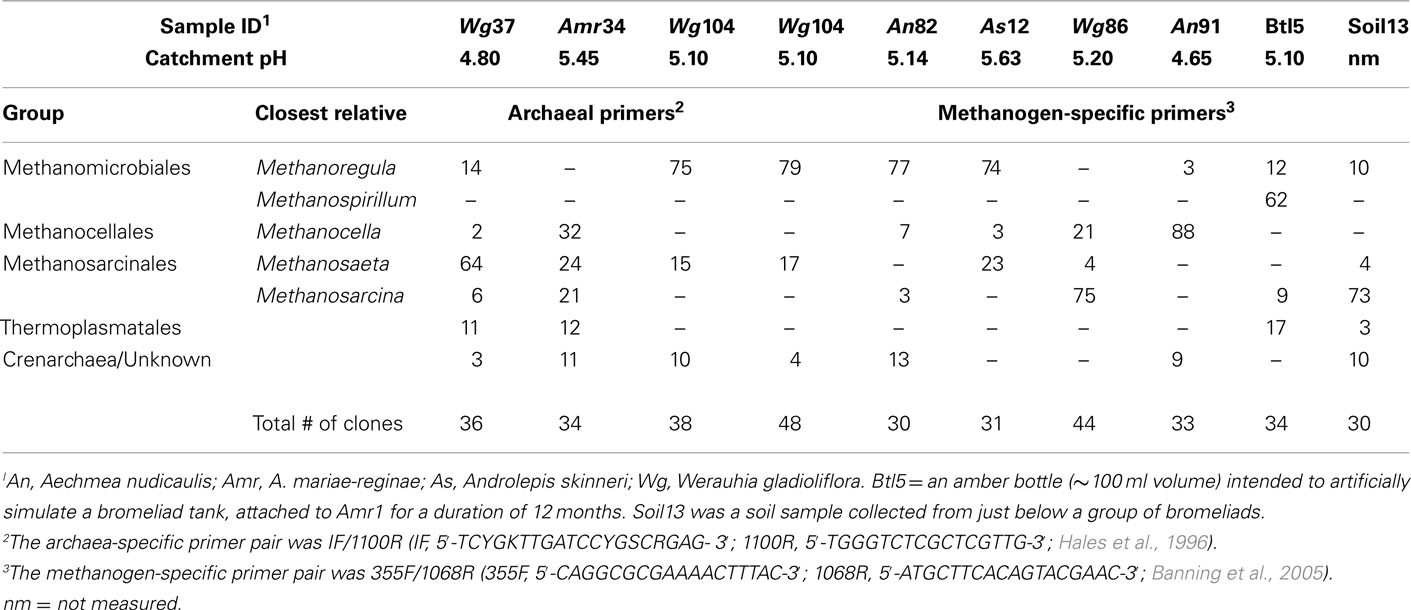
La Selva Biological Station, situated in a wet (4 m annual rainfall) lowland neotropical forest in northern Costa Rica, is located at the confluence of the Sarapiqui and Puerto Viejo rivers in the province of Heredia, Costa Rica (10°26′N, 83°59′W, 37–130 m elevation). The reserve, which covers approx 1600 ha, is home to dozens of bromeliads species, including those within the genera Werauhia (syn. Vriesea), Guzmania, Androlepis, and Aechmea, which were sampled in this study on three separate occasions over 18 months during 2009–2010 (Figure 1). This study included tank water from 106 adult and juvenile bromeliad specimens (Figure 1C), comprising six species; Aechmea mariae-reginae (“Amr”), Aechmea nudicaulis (“An”), Werauhia gladioliflora (“Wg”), Werauhia kupperiana (“Wk”), Androlepis skinneri (“As”), and Guzmania lingulata (“Gl”), which encompassed a range of tank morphologies (e.g., ~3–114 cm plant height) and pH conditions (3.6–6.5; Figure 1D). Additionally, 20 soil samples were collected from near and below sampled bromeliads, as well as eight amber bottles (~100 ml volume) intended to artificially simulate bromeliad tanks, attached to nearby bromeliads for a duration of 6–12 months (Figure 1B). For two bromeliads (Amr1 and Amr51), the pH was artificially depressed, from ~6.5 to ~4.5 for a total of 62 days, by the frequent addition of 1N hydrochloric acid to the tank (Goffredi et al., 2011). Several bromeliads (and paired soil) were sampled by climber from the canopy (~30 m), but most were within 2 m high on host trees, typically in either man-made or natural clearings. Tank water samples were collected via serological pipette from the bottommost horizon within the tank, with the exception of four bromeliads (with tank depths of ~10–15 cm), which were sampled stratigraphically every ~ 3–5 cm. Samples were transported to the lab in clean 15 ml plastic tubes and either processed or preserved immediately.
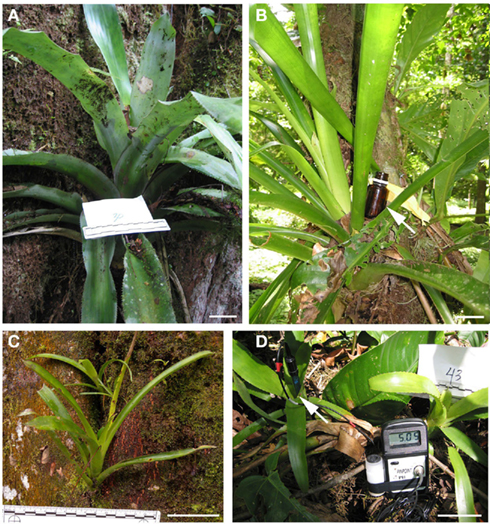
Figure 1. Costa Rican bromeliads used in this study. (A) Aechmea mariae-reginae with nearby note card and ruler for scale (B) Werauhia gladioliflora, with nearby amber bottle (arrow) intended to artificially simulate a bromeliad tank. (C) A juvenile specimen of W. gladioliflora. (D) Two specimens of W. gladioliflora, showing a pH electrode (arrow) and meter, reading 5.05 in the central catchment water of the left hand specimen. Scale bars, 5 cm.
Chemical Analyses
Tank pH was measured via hand-held pH electrode (Hanna Instruments HI-98103B) in the field, prior to sampling for DNA analysis. Oxygen was also measured in situ using a colorimetric dissolved oxygen test kit (CHEMetrics, Inc.).
Methane production by bromeliad tank communities was assayed via incubation of tank water (13–30 ml) in gas tight serum vials (total volume 73 ml). Bromeliad tank water for incubation experiments was collected by siphoning through a tube (tygon) inserted, at the other end, into a 100-ml serum vial. Minor amounts of partially decomposed leaf litter and invertebrate remains were entrained in the siphoned water. The serum vials were crimp sealed using a butyl rubber stopper and incubated, inverted, at a laboratory temperature of ~22°C in the dark. Over the course of 10–77 days, 10 ml headspace samples were taken via syringe after brief shaking. These samples were taken in the following manner to minimize pressure differences between the interior and exterior of the serum vial. A fully depressed 10-ml syringe with a 22-gauge needle was inserted into the headspace of the serum vial. A second 10-ml syringe with a 22-gauge needle filled with UHP nitrogen gas was inserted into the headspace. Pushing and pulling on the syringe plungers approximately 10–15 times mixed the gases. A 10-ml gas sample was pushed through a low volume magnesium perchlorate drying column prior to injection into the gas chromatograph via a 1-ml sample loop. The addition of nitrogen gas was accounted for when calculating methane concentrations. Headspace methane concentrations were determined using a Shimadzu mini-2 gas chromatograph equipped with a 1/8″ × 5-ft stainless steel column packed with Carbosieve G and a flame ionization detector (FID) and operated in isothermal mode (oven temperature 100°C; detector temperature 110°C) with a UHP nitrogen carrier gas. Methane gas standards (9.93 and 98.6 ppm CH4 in helium) were used for calibration. Compound specific isotopic values for methane from three selected samples were measured via gas chromatography-combustion-isotope ratio mass spectrometry using a system consisting of an Agilent 6890 GC, a combustion unit, and a mass spectrometer (Finnigan Delta V plus). Hydrocarbon components were separated by GC, and each individual component slated for isotopic analysis was combusted. The resultant CO2 was introduced directly into the mass spectrometer, and Finnigan’s Isodat software was utilized for peak detection and quantification (Isotech Laboratories Inc., Champaign, IL, USA).
Phylogenetic Analyses
Freshly collected bromeliad tank water (0.5 ml, including debris) was spun at 15,000 × g for 10 min and the resulting pellet was either extracted for total nucleic acids or preserved in 0.5–1.0 ml RNAlater (Ambion, Inc.). DNA was extracted using the Power Soil DNA extraction kit (MoBio Laboratories, Inc., Carlsbad, CA, USA), modified by two initial 5–10 min incubations at 65°C, one in the presence of solution S1, followed by 5–10 min vortexing. The remainder of the extraction procedure was carried out according to the manufacturer’s instructions, with the exception of a 4°C incubation in IRS solution (5 min) between solutions S2 and S3 to increase DNA yield and inhibitor removal. SSU (16S) rRNA was amplified by polymerase chain reaction (PCR) from extracted DNA, using the archaea-specific primer pair 1F/1100R (1F, 5′-TCYGKTTGATCCYGSCRGAG-3′; 1100R, 5′-TGGGTCTCGCTCGTTG-3′; Hales et al., 1996), the methanogen-specific primer pair 355F/1068R (355F, 5′-CAGGCGCGAAAACTTTAC-3′; 1068R, 5′-ATGCTTCACAGTACGAAC-3′; Banning et al., 2005), or a combination of 355F/1100R (Table 1). Thermal cycling conditions were as described, with an annealing temperature of 52°C, for 25 cycles (Hales et al., 1996; Banning et al., 2005). No difference in relative abundance and types of methanogens was detected between those recovered using the two main primer pairs (see Wg104; Table 1).
Presence/absence of methanogens was judged by electrophoretic separation and visualization of PCR products produced by the 355F/1068R primer set. For seven bromeliads, one artificial catchment, and one soil sample, PCR-amplified bacterial 16S rRNA genes were used for clone library construction. PCR products were pooled in triplicate prior to ligation. Transformants (30–48 clones analyzed for each bromeliad library; Table 1) were screened directly for the presence of inserts using M13F/R vector primers (8 min initial denaturation). M13 amplicons were cleaned prior to sequencing with MultiScreen HTS plates (Millipore Corporation, Bedford, MA, USA). Sequencing reactions were performed using the Genome Lab DTCS Quick Start Kit (Beckman Coulter, Fullerton, CA, USA), precipitated according to the manufacturer’s instructions, and run on a CEQ 8800 Genetic Analysis System (Beckman Coulter, Fullerton, CA, USA). Representative ribotypes, based on 97% sequence similarity, were selected for near full-length sequencing (Figure 4).
Sequences were assembled and edited using Sequencher v4.10.1 (Gene Codes Corporation). Initial sequence homology searches were performed using BLASTn (NCBI) and the Ribosomal Database Project classifier. Our 16S rRNA sequences along with additional sequences obtained from GenBank were compiled in ARB, after initial alignment using the SILVA Aligner function, with subsequent manual refinements (Ludwig et al., 2004; Preusse et al., 2007). For near full-length representatives and closest relatives, neighbor-joining (NJ) analysis was conducted with Felsenstein distance correction. In some cases, partial sequences recovered in our study were added to the NJ tree in ARB via parsimony insertion within a tree of longer sequences. NJ analysis was performed with 2000 bootstrap replicates to assign confidence levels to nodes, shown in Figure 4, if >70% confidence (PAUP*4.0b10; Swofford, 1998). Sequences obtained in this study have been deposited in the GenBank database under accession numbers JN810747–JN810784 (archaeal 16S rRNA ribotypes) and JN810785–JN810789 (mcrA genes).
Quantitative PCR
A SYBR green I assay was used to quantify total copy numbers of methyl coenzyme M reductase A (mcrA) genes present in bromeliad tank genomic DNA extracts. Methyl coenzyme M reductase is the key enzyme, unique among methanogenic archaea, which catalyzes the reduction of methyl coenzyme M, the final step in methanogenesis (Ermler et al., 1997; Lueders et al., 2001; Luton et al., 2002). The alpha subunit of the methyl coenzyme M reductase gene (mcrA) is highly conserved among all methanogens and is commonly used as an ecological and phylogenetic metric by which to investigate methanogens. mcrA primers, designed previously by Steinberg and Regan (2008) in a study of acidic peat, manure, and anaerobic wastewater, were used to amplify a 489-bp region of the mcrA subunit from methanogens (mlas; 5′-GGTGGTGTMGGDTTCACMCARTA-3′ and mcrA-rev; 5′-CGTTCATBGCGTAGTTVGGRTAGT-3′). pCR®4-TOPO-derived plasmids (Invitrogen, Eugene, OR, USA) containing known partial mcrA sequence copies were prepared according to the QIAPrep Spin Miniprep Kit (Qiagen, Valencia, CA, USA). During library construction for plasmid recovery, several mcrA clones were M13-amplified and sequenced, as described earlier (Table A1 in Appendix). Serial dilutions of the plasmids were used in the optimization of primer concentrations, as well as generation of a standard curve for cycle threshold (Ct) versus gene copy number. The Ct values from these plasmids were converted into copy numbers with consideration of the plasmid preparation nucleic acid concentration, length of the plasmid (3956 bp) and gene insert length (489 bp), average molecular weight of a double stranded DNA molecule (660 g mole−1), and Avogadro’s number, resulting in a DNA mass conversion of 1.096 × 10−21 g bp−1. The slope of the standard curve [regression line of Ct versus log(DNA)] was used to assess amplification efficiency and was empirically determined to be 3.3 for the mcrA primer set. Quantitative PCR amplifications (20 μl), performed in duplicate, contained 417 nmol of primer mcrA-mlas and 278 nmol of primer mcrA-rev, and 1× PCR buffer containing MgCl2, dNTPs, and Amplitaq Gold in the SYBR green master mix (Power SYBR® Green PCR Master Mix; Applied Biosystems, Foster City, CA, USA). Purified DNA from bromeliad tank water was added as template (~1–6 ng μl−1), in replicate dilution series. DNA concentrations for samples and standards were measured using the Quant-iT PicoGreen dsDNA assay kit (Invitrogen) and a DTX880 multimode detector and workstation (Beckman Coulter, Brea, CA, USA). Lambda-DNA was used as a standard, as per the manufacturer’s instructions. QPCR amplifications were carried out using an ABI Prism 7500 Sequence Detection System (Applied Biosystems). Cycling parameters included an initial denaturation of 95°C for 3 min, followed by 50 cycles of denaturation at 95°C (15 s) and 60 s of annealing/extension at 60°C. A dissociation curve from each reaction was examined to further ensure proper target sequence amplification (i.e., Tm > 80°C). Given the uncertainty of using plasmids as standards for absolute copy numbers (Hou et al., 2010), mcrA values are shown as relative in Figure 3.
Terminal Restriction Fragment Length Polymorphism and Diversity
Methanogen-specific terminal restriction fragment length polymorphism (T-RFLP) profiling was used to characterize the relative proportions, and corresponding diversity, of methanogen groups associated with bromeliad catchments. This approach has been used previously to quantify major methanogen phylotypes from acidic peatlands, a minerotrophic fen, and brackish lake sediment (Banning et al., 2005; Cadillo-Quiroz et al., 2006, 2008). A subset of 25 bromeliads, encompassing a range of morphological and physical–chemical parameters, was selected for T-RFLP fingerprinting. 16S rRNA genes from purified DNA samples, as described above, were PCR-amplified using methanogen-specific primers 355F/1100R [the latter was fluorescently labeled at position 1100 (E. coli numbering) with Well RED dye D4, Sigma-Proligo, St. Louis, MO, USA; Cadillo-Quiroz et al., 2006], using the conditions described above for unlabelled PCR amplifications. For each sample, duplicate PCR amplifications were performed and pooled (300–500 ng total) prior to digestion with HhaI/Sau961 (for 8 h at 37°C, with addition of bovine serum albumin; New England Biolabs, Beverly, MA, USA). Fluorescently labeled fragments were separated by capillary electrophoresis and analyzed on a CEQ 8800 Genetic Analysis System (Beckman Coulter, Fullerton, CA, USA). Fragment sizes were parsed by separation of >3 bp, and relative abundances were estimated using the CEQ 8800 Fragment Analysis software.
Statistical analyses were performed using the JMP statistical software (version 4.04; SAS Institute, Cary, NC, USA). Diversity estimates of T-RFLP data were performed using Primer v6 (Clarke, 1993; Clarke and Gorley, 2006). Hierarchical cluster analysis, using Bray–Curtis similarity resemblance, Euclidean distance, and no transformation of the data, was also performed using Primer v6.
Results
Methane Production in Bromeliad Tank Incubations
Methane production potential was assayed via incubation of bromeliad tank water in gas tight serum vials. Methane increased within a few days and reached a plateau, by day ~30, at ~900 nmol ml−1 CH4 in the headspace (= ~22000 ppm CH4; Figure 2). This plateau was likely due to constraints within the incubation vials as methane production resumed following a purge of the headspace, at day 76, with nitrogen (Figure 2). Methane production rates varied initially between 7 and 38 nmol CH4 ml tank water−1 day−1, and increased to 100–300 nmol CH4 ml−1 day−1 immediately following the purge. Additionally, δ13C stable isotopic signatures of the generated methane, taken at day 76, were −56.5 and −58.4‰, indicative of a combination of both hydrogenotrophic and aceticlastic methanogenesis. The δ13CH4 value of an additional sample (Wg104), not monitored for methane production, was −62.5‰.
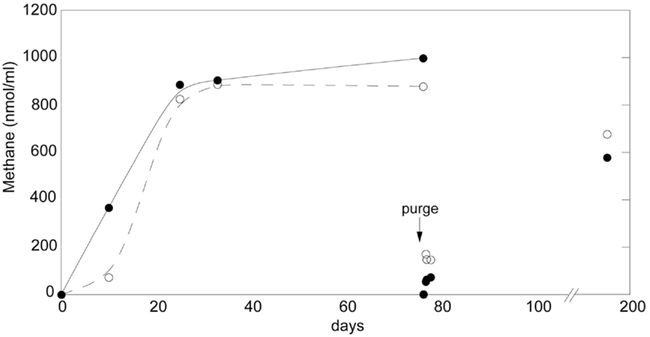
Figure 2. Methane production by bromeliad catchment water. Methane increased within a few days and reached a plateau, by day ~30, at ~900 nmol ml−1 CH4 in the headspace (= ~22000 ppm CH4). Following a purge of the headspace with nitrogen, at day 76, methane production resumed, and was measured once more at day 192. Methane production rates varied initially between 7 and 38 nmol CH4 ml tank water−1 day1, and increased to 100–300 nmol CH4 ml−1 day−1 immediately following the purge (days 77–78). Additionally, the δ13C isotopic signatures of the generated methane, taken at day 76 prior to the purge, were −56.5 and −58.4‰, for Wg37 and As12, closed and open circles, respectively.
Measures of, and Influences on, McrA Gene Copy Number
The methanogenic potential of the tank community was examined by QPCR assay of the methyl coenzyme M reductase A (mcrA) gene. Relative mcrA copy number ranged from 1.6 × 106 to 7.0 × 107 ng−1 DNA extract, however, actual copy numbers (and corresponding methanogen abundance) may have been overestimated due to use of plasmids as a standard curve for QPCR analysis (Hou et al., 2010). Nevertheless, there was a positive correlation between mcrA copy number and plant height, suggesting a greater capacity for methanogenesis in larger tanks (Figure 3, R2 = 0.61, P = 0.0009). For seven clonal specimens of A. nudicaulis (ranging from 2 to 7 cm tank depth and 4–20 cm estimated plant height), quantitative assessment of mcrA revealed the same trend of increased copy number in larger plants, and no detection of mcrA genes in the smallest individuals (Figure 3, inset). It should be noted that this relationship did not hold for plants with heights of >54 cm. Although they did possess mcrA genes (data not shown), greater stratification, and a sample volume of ~10–15 ml, likely resulted in an inability to quantitatively sample larger tanks. Additionally, QPCR analysis on four of the artificial catchments did not render detectable levels of the mcrA gene. Two artificial catchments, however, did possess methanogens, albeit at lower relative levels (40% of signal, as compared to the bromeliads with which they were paired).
Figure 3. Methanogenic potential of the tank community as examined by QPCR assay of the methyl coenzyme M reductase gene (mcrA). A direct relationship existed between relative mcrA copy number and plant height (R2 = 0.61, P = 0.0009), suggesting a greater potential for methanogenesis in larger tanks. This relationship did not exist in plants larger than 54 cm in height, likely due to an inability to quantitatively sample larger tanks. For seven clonal specimens of A. nudicaulis (connected via a rhizome and ranging from 2 to 7 cm tank depth; 4–20 cm estimated plant height), mcrA copy number revealed the same trend of increased relative copy # in larger plants, and no detection of mcrA genes in the smallest four individuals (inset). Note that actual mcrA copy numbers are not the focus and may have been affected (overestimated) by using plasmids as a QPCR standard.
Phylogenetic Characterization of Methanogenic Assemblages in Bromeliad Tanks
Archaeal community composition in bromeliad catchments was analyzed by cloning and sequencing of 16S rRNA genes. Archaea were dominated by methanogens (77–90% of recovered ribotypes) and community structure, although variable, revealed a predominance of various hydrogenotrophic methanogenic orders (e.g., Methanomicrobiales and Methanocellales), as well as both general and specific acetotrophs (Methanosarcinales; Table 1; Figure 4). Very few non-methanogen taxa were detected, using archaeal domain primers, but additional minor archaeal groups included possible members of Thermoplasmatales/Candidate Division II, and uncultured crenarchaea. Notably, the “methanogen-specific” primer set not only amplified the 16S rRNA genes from other Euryarchaeota, perhaps not surprisingly, but also Crenarchaeota in proportions not much different from the archaeal primers, which themselves, are somewhat biased toward Euryarchaeota. Many euryarchaeal ribotypes, using both archaeal- and methanogen-specific primers, were closely related (based on 97–100% similarity in partial 16S rRNA) to cultured representatives, including Methanoregula boonei, isolated from an acidic peat bog (Bräuer et al., 2006, 2011), Methanocella paludicola, isolated from rice paddy soil (Sakai et al., 2008), Methanosarcina barkeri, isolated from rice paddies and an underground gas storage facility (Joulian et al., 1998; Tarasov et al., 2011), and Methanosaeta concilii isolated from an anaerobic sewage sludge fermenter (Patel, 1984). Others were related to ribotypes associated with uncultured groups previously recovered from a range of environments, including estuaries, landfills, deep aquifers, fens, and bromeliads. In fact, several ribotypes recovered in the present study were most closely related to those recovered from Ecuadorian bromeliads (Figure 4; Martinson et al., 2010).
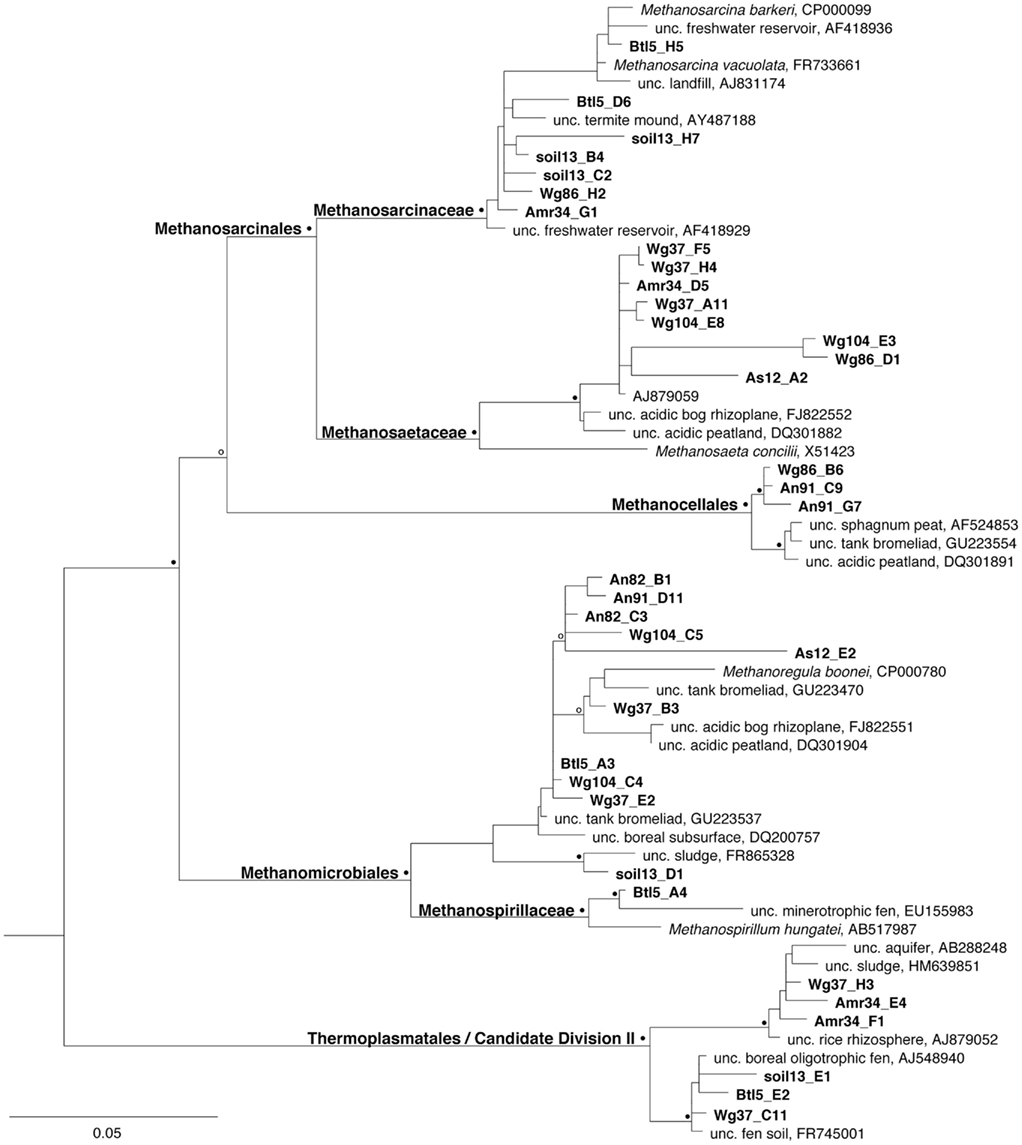
Figure 4. Phylogenetic relationships among archaea associated with Costa Rican bromeliads, a nearby soil sample, and an artificial catchment, relative to selected cultured and environmental sequences in public databases. Relationships are based on sequence divergence within the 16S rRNA gene, with emphasis on methanogenic archaea, as well as those related to the Thermoplasmatales/Candidate Division II, and an uncultured member of the Korarchaeota (DQ465910) as an out group (not shown). The symbols at the nodes represent bootstrap values from a Kimura-2 parameter neighbor-joining analysis obtained from 2000 replicate samplings (open symbol = 70–80%, closed symbol = 80+% bootstrap support). Sequences obtained in this study have been deposited in the GenBank database under accession numbers JN810747–JN810784.
The predominant group of methanogens recovered in clone libraries varied between bromeliads. For example, recovered ribotypes associated with Methanocella were numerically dominant (46 and 97% of methanogen 16S rRNA ribotypes recovered) in the tanks of two bromeliads (Amr34 and An91, respectively), Methanoregula-associated ribotypes were dominant (75 and 82%) in two other bromeliads (Wg104 and An82, respectively), while Methanosaeta- and Methanosarcina-associated ribotypes each dominated one bromeliad, as assayed via clone library construction (72 and 75% for Wg37 and Wg86, respectively; Table 1; Figure 4). Both the Methanosaeta- and Methanocella-associated clusters, in particular, appeared to be bromeliad-specific, in that ribotypes were most closely related to each other than to any others sequences deposited in the databases (Figure 4).
Out of 20 nearby soil samples (both ground level and canopy height, ~33 m), only one sample (soil13) produced a positive signal for methanogens. Methanogen ribotypes recovered from this soil sample were generally distinct from those recovered from nearby bromeliads, and were, instead, closely related to Methanosarcina vacuolata from sludge and Methanosarcina mazei from rice paddy soil (97–99% similarity in 16S rRNA; Table 1; Figure 4; Deppenmeier et al., 2002; Liu et al., 2009). Conversely, ribotypes associated with Methanoregula were recovered predominantly from a nearby bromeliad (data not shown). Additional methanogenic ribotypes in soil included those found in meadow soil and aquifers (Figure 4).
Artificial catchments (amber bottles), intended to simulate bromeliad tanks, were also analyzed for methanogen presence. Methanogens were generally not detected within the contents of these artificial catchments, suspended in the canopy near bromeliads, even though they had been in place, collecting debris, for 6–12 months. Despite holding similar volumes of water (~100 ml), oxygen levels in bottles were generally elevated (6–8 ppm O2), compared to <1 ppm O2 in the bottommost depths of paired bromeliad tanks. Two artificial catchments did show evidence of methanogens. Visual examination of these two bottles revealed that they contained the largest amount of organic debris. Moreover, they had the lowest pH values of all bottles measured (5.1 versus 6.6 average pH, comparatively). Unfortunately, oxygen levels were not measured in these specific bottles. Time in the field did not appear to matter, as both bottles that assayed positive for methanogens were in place for only 6 months, compared to others in place for up 12 months. The community composition within one bottle (“Btl5,” Table 1) was investigated further and determined to be distinct from nearby bromeliads, with ribotypes most closely related (97% similarity) to Methanospirillum hungatei, isolated from sewage sludge (Ferry et al., 1974), as the dominant methanogen (75% of known methanogen 16S rRNA ribotypes; Figure 4). Additional close relatives (99% similarity) included uncultured representatives isolated from minerotrophic fens and sludge, including a few Methanoregula-associated ribotypes (Figure 4; Cadillo-Quiroz et al., 2008; Rivière et al., 2009).
Ribotypes tentatively characterized as Thermoplasmatales-associated were recovered in significant numbers from two bromeliads (~12% of the recovered ribotypes; Amr34 and Wg37) and one artificial catchment (at least 17%; Btl5; Table 1). Thermoplasmatales do occur in low temperature soils (Horn et al., 2003; Juottonen et al., 2005), however, nearest neighbors in other studies (based on 16S rRNA) have been classified as distinct from Thermoplasmatales and may, in fact, represent new members within the Candidate Division II (Watanabe et al., 2002; Liu et al., 2009). Whether or not they play a role in methane metabolism is currently unknown.
The alpha subunit of the methyl coenzyme M reductase gene (mcrA) was also cloned and sequenced, using the QPCR primers described above, from the tank water of three bromeliads (Table A1 in Appendix). Based on recovered mcrA gene sequences, the dominant methanogens were closely related (86–96% amino acid similarity) to members of both the Methanosarcinales and Methanomicrobiales, many of which were previously recovered from similar habitats (e.g., rice paddy soil, acidic peat bogs, and wastewater sludge; Table A1 in Appendix).
Presence and Absence of Methanogens in Tank-Forming Bromeliads
Tank water samples from 106 specimens, comprising 6 bromeliad species (including A. mariae-reginae, A. nudicaulis, W. gladioliflora, and A. skinneri, as noted in the above section, and W. kupperiana and G. lingulata), were extracted for examination of methanogen presence by PCR. Methanogen-specific 16S rRNA genes were recovered for 75 of the 106 bromeliad tanks sampled and included all species, which ranged in plant height from 20 to 114 cm and tank pH of 3.6–6.5. Of the 31 tanks that were negative, or below the detection limit for methanogens, 10 were juvenile plants (less than 5 cm height, with small tank volumes of ~ 0.5–2 ml), 10 were small adult plants (<20 cm plant height and <5–7 cm tank depth), 3 were Guzmania species, which typically have very shallow tanks, leaving only eight that were within all specifications of those that did possess methanogens. Seven clonal, rhizomatous specimens of A. nudicaulis, ranging from 2 to 7 cm tank depth (= 4–20 cm estimated plant height for this species in general) were compared and found to possess methanogens only when tank depth exceeded ~4 cm (see also mcrA results; Figure 3).
Four bromeliads (with tank depths of ~10–15 cm) were sampled stratigraphically with three tank horizons sampled every ~3–5 cm depth. In all cases, only the deepest, bottommost layer tested positive for methanogens (data not shown). Strong pH and oxygen gradients were measured in the deeper horizons as well, with pH depressed by 0.2–0.6 pH units and oxygen dramatically reduced (<1 ppm O2 at depths of 6–7 cm, compared to 2–8 ppm O2 within 1 cm of the surface of the tank, n = 3; Walter T. Woodside, personal observation).
T-RFLP Analysis of Methanogenic Assemblage Structure and Diversity
For a subset of bromeliads (n = 25) that encompassed a range of morphologies and environmental conditions, T-RFLP fingerprinting was used to compare the diversity and relative ratios of associated methanogen-specific groups (Figure 5; Table A2 in Appendix). These bromeliads spanned a range of plant heights from 21 to 114 cm, photosynthetically active radiation (PAR) of 33–1282 μmol m−2 s−1, position off ground of 0.5–34 m, 2–51 mg ml−1 debris within the catchment fluid, and catchment water pH of 3.5–6.5 (including two bromeliads that underwent a manual pH adjustment from 4.5 to 6.5). Fragments (T-RFs) were assigned to particular phylogenetic groups, based on in silico analysis and direct comparison of T-RFs from methanogen clones recovered from bromeliad tank 16S rRNA libraries, as well as previous data from Cadillo-Quiroz et al., 2006, including: Methanosarcinaceae – 87, 142, 271 bp; Methanosaetaceae – 90, 119, 399 bp; Methanomicrobiales/Fen Cluster (group E2) or Methanocellales (Rice Cluster I) – 232 bp; Methanomicrobiales/Fen Cluster (group E1) – 291 bp; Methanocellales (RCI) – 363 bp. Although Cadillo-Quiroz et al. (2006) distinguished T-RF 230 bp (RCI) from 233 bp (Fen Cluster group E2), we have conservatively combined them, as did Martinson et al. (2010), as a representation of H2-consuming methanogens. Certain T-RF’s could not be phylogenetically resolved (e.g., 76–79, 135, and 143–150 bp). These unknown fragments typically represented <20% of the total (average 9.8 ± 1.6%), with the exception of five samples (cluster B, described below, Figure 5), for which the unknowns represented 30–49% of the community. In general, T-RFLP analysis resolved the dominant methanogen-affiliated taxa and supported the proportions of methanogens recovered (within ~10%) in the clone libraries. For Wg104 for example, the Methanomicrobiales/Fen Cluster represented ~78% of the clone library (Methanoregula spp. specifically) versus ~69% via T-RFLP analysis (T-RF 233 specifically). Likewise, the clone library for Wg104 recovered 17% Methanosaeta-associated relatives, whereas the T-RFLP analysis showed ~26% Methanosaeta (Table 1; Figure 5).
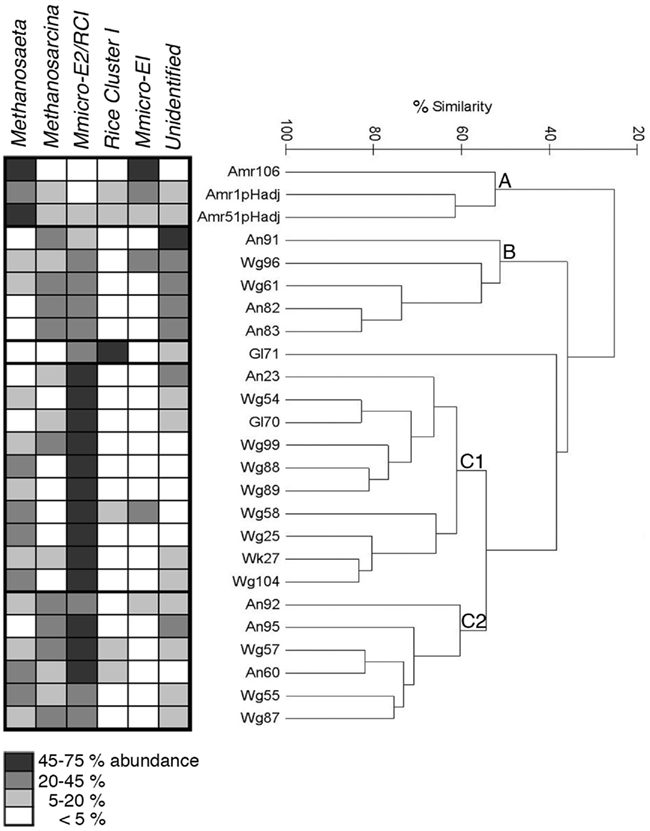
Figure 5. Hierarchical cluster analysis of methanogen community composition, as analyzed by T-RFLP fingerprinting, for the catchment water of 25 bromeliad individuals, comprising 6 species. Fragments (T-RFs) were assigned to particular phylogenetic groups, including: Methanosarcinaceae, Methanosaetaceae, Methanomicrobiales/Fen Cluster (group E2) or Methanocellales (Rice Cluster I), Methanomicrobiales/Fen Cluster (group E1), and Methanocellales (RCI) specifically. Hierarchical analysis revealed three main clusters of samples (A–C), with regard to diversity and relative ratios of associated methanogens, shown at left, shaded in relation to % abundance (performed using Primer v6, Bray-Curtis similarity resemblance, Euclidean distance, and no transformation of the data). “M-micro-E2” or “E1” = Methanomicrobiales/Fen Cluster group E2 or E1, respectively.
Hierarchical analysis revealed three main clusters of samples, with regard to diversity and relative ratios of associated methanogens, based on T-RF abundances, in bromeliad catchment water (>50% community similarity, Figure 5). Group A, which consisted of only specimens of A. mariae-reginae, was primarily distinguished by a high abundance of Methanosaetaceae (38.1 ± 8.8% average) and very few Methanomicrobiales (3.1 ± 0.8%, n = 3). Group B showed the highest abundance (40.6 ± 3.1%, n = 5) of the putative unknown methanogen ribotypes T-RF 143 bp. Group C, comprised of subgroups C1 and C2, had a high abundance of Methanomicrobiales (group E2)/Methanocellales (55.6 ± 3.4%, n = 16; Figure 5). Gl71 was distinct from the main groups, likely due to the very low diversity and dominance by Methanomicrobiales/Methanocellales (92% of T-RFs; Figure 5). Groups B and C1 also had moderately high abundances (~26 ± 3% average) of Methanosarcinaceae. Diversity indices ranged from 1.36 ± 0.08 (H′, Shannon index), 0.66 ± 0.03 (1λ′, Simpson index), 0.77 ± 0.02 (J′, Pielou’s evenness) for all bromeliads measured (n = 25; Table A2 in Appendix; Figure 5). Bromeliads that underwent manual perturbation of tank pH (manual adjustment from pH 4.5 to 6.5 for bromeliads Amr1 and Amr51) had the highest measures of methanogen diversity (2.01, 0.83, and 0.87, for H′, 1λ′, and J′, respectively; Table A2 in Appendix). The heterogeneous nature of methanogen community structure was similar to that observed in Martinson et al. (2010), and did not appear to correlate with any of the in situ parameters measured, including PAR, amount of solid debris within the tank, position off ground, specific location within the forest, or host plant species. Two notable exceptions included Group A, which were all A. mariae-reginae with high tank pH [~6.5, either naturally (Amr106) or experimentally induced (Amr1 and Amr51)] and Group B, which only included bromeliads collected from sites high in the canopy (11–34 m estimated height off ground).
Discussion
Plants within the family Bromeliaceae are known for their capacity for extreme epiphytism, sometimes growing on bare rock or suspended from vines. Tank-forming bromeliads, in particular, possess foliage arranged in a compact rosette capable of retaining water. In lieu of uptake via root systems, these plants are thought to rely on the catchments and products of decomposition of impounded material (litter and animals) for water and nutrients, respectively (Benzing, 1970; Winkler and Zotz, 2009). As a consequence, the suspended water within bromeliad tanks provides a unique niche in that water-saturated, acidic, anaerobic conditions do not typically occur in other locations in the canopy of tropical forests. These catchments are an ideal site for microbial decomposition, and possibly important habitats with regard to global biogeochemical cycling, including CO2 and CH4 efflux and organic carbon storage.
Methane Production Potential in Bromeliad Tanks
In vitro CH4 production potential measured for bromeliad tank water was comparable to methane production rates in ecosystems known to emit large quantities of methane. Methane production by bromeliad tanks varied between ~12 and 72 nmol CH4 ml−1 day−1, as compared to rice paddies (43–212 nmol CH4 g−1 day−1; Watanabe et al., 2009) and certain peatland ecosystems (13–47 nmol CH4 g−1 day−1; Basiliko et al., 2003). Methane dynamics in water-saturated ecosystems are complex and additional parameters must be known, including diffusion, conductance by plants, and oxidation by methanotrophs in overlying aerobic layers, in order to accurately extrapolate bromeliad tank water production rates to field emission rates. We have not indentified aerobic methanotrophs in bromeliad tank water, nor have we successfully amplified the key gene involved in aerobic methane oxidation (pmoA; Goffredi unpublished observation; Goffredi et al., 2011). To provide an estimate of net emission rate by tank bromeliads, we assume that aerobic methane oxidation is insignificant relative to methane production rate, and that methane transfer across the tank water (or plant tissue)-atmosphere interface is by diffusion and is in steady-state. Therefore, assuming an average bromeliad tank volume of 100 ml and a bromeliad density of 25,000–175,000 ha−1 (Sugden and Robins, 1979; Martinson et al., 2010), potential emission rates are of the order ~10–500 moles CH4 ha−1 year−1. These estimated emission rates are comparable to those measured for montane bromeliads (82 moles CH4 ha−1 year−1; Martinson et al., 2010) and plant-based pasture production (~190 moles CH4 ha−1 year−1; Parsons et al., 2006), but still fall short of the high, yet variable, values noted for acidic peatlands in general (72–210,000 moles CH4 ha−1 year−1 which depend primarily on moisture regime; Svensson and Rosswall, 1984; Galand et al., 2002; Horn et al., 2003).
The tank rosette itself may be important for the maintenance of the methanogenic microcosm. Stable, strong gradients, with regard to micro-limnological parameters (O2, pH, Corg) may, in part, be related to tank size and shape. Methane release from Ecuadorian bromeliads correlated, exponentially, with tank diameter (Martinson et al., 2010). Similarly, we observed a relationship between mcrA copy number and plant height, suggesting an influential role of plant (and consequently tank) height on methane production capacity, and possibly methanogen community structure. Taller, deeper tanks have time to collect more debris, assuming tank size is positively correlated with plant lifespan. Larger tanks may also experience less mixing and decreased susceptibility to desiccation (Benzing et al., 1972), thereby sustaining methanogen-favorable conditions. Similarly, oxygen levels appear to have some effect on methanogens, which were not observed in either the uppermost horizons within bromeliad tanks or throughout the water column in artificial catchments (i.e., amber bottles), both of which had elevated O2 levels (2–8 ppm O2), compared to the bottommost methanogen-dominated horizons within bromeliad tanks (<1 ppm O2). This implies that either tank shape, or other plant-related parameter, is required for establishing methanogen-favorable conditions (i.e., low oxygen, production of fermentation products).
Methanogenic Communities Associated with Bromeliad Tanks
Molecular analysis of the archaeal communities within bromeliad tanks revealed a dominance of methanogens (77–90% of the recovered ribotypes) and demonstrated the presence of members of both the Methanomicrobiales and Methanocellales, known to use H2/CO2, as well as the Methanosarcinales, which are versatile, but known commonly to be aceticlastic. Similar substrate heterogeneity, in terms of methane production, has been observed for methanogenic communities found in other freshwater environments, including fen soil, acidic peat bogs, and the Florida Everglades (Kotsyurbenko et al., 2004; Smith et al., 2007; Watanabe et al., 2009; Wüst et al., 2009; Martinson et al., 2010; Borrel et al., 2011; Kanokratana et al., 2011). For Ecuadorian bromeliads specifically, there was a similarly strong presence of ribotypes related to Methanosaeta, Methanocella, and what was called the Fen Cluster (or Methanomicrobiales), presumably Methanoregula (Martinson et al., 2010). Interestingly, the Methanobacteriales, which represented ~30% of the ribotypes recovered from montane bromeliad tanks (Martinson et al., 2010), were not observed in the lowland forest bromeliads of the present study. Although many parameters are required to determine more certainly the relative contributions of hydrogenotrophic versus aceticlastic methanogenesis in bromeliad catchments (Conrad, 1999, 2005; Whiticar, 1999; Kotsyurbenko et al., 2004), isotopic measures of methane in two tank microcosms reflected an environment in which methane was generally produced from both processes (δ13CH4 = approx −57‰ for Wg37 and As12), and one for which perhaps a slight shift in balance toward H2/CO2-based CH4 production was suggested (−62.5‰ δ13CH4 for Wg104). This assumption is consistent with clone library results from Wg37 and As12, which both showed ~ 30:70 ratios of the two major methanogenic guilds, whereas Methanoregula was the dominant methanogen in Wg104 (>80%; Table 1; Figure 2).
The relative contribution of acetate and H2/CO2 to CH4 production can vary within ecosystems (Conrad, 1999, 2005) and, among many influences on community composition, it appears that pH may play a role. In this study, individual bromeliads with lower tank pH values (<6.1) were overwhelmingly dominated by H2/CO2-utilizing methanogens, including those most closely related to Methanoregula (Fen Cluster group E2), and Methanocella (Rice Cluster I), both found previously to be abundant in similarly organic-rich, acidic habitats (Ramakrishnan et al., 2001; Galand et al., 2003; Sizova et al., 2003; Cadillo-Quiroz et al., 2006, 2008; Wüst et al., 2009). These two groups are tolerant of low pH and are known to use H2 as their primary electron donor (Sakai et al., 2008; Bräuer et al., 2011), and, in the case of Rice Cluster I, possess a genome specifically suggestive of a H2/CO2-dependent lifestyle (Erkel et al., 2006). Indeed, for methanogenic communities found in rice paddies, acidic peat bogs, and fens, hydrogenotrophic methanogenesis is commonly reported as the predominant process (Horn et al., 2003; Kotsyurbenko et al., 2004; Smith et al., 2007; Conrad et al., 2008; Watanabe et al., 2009).
There are several possible explanations for a higher contribution of hydrogenotrophic methanogenesis in bromeliad catchments. Goffredi et al. (2011) demonstrated an abundance of both hydrolytic and fermentative bacteria (e.g., Acidobacteria and Bacteroidetes), which may influence the ecological structure of methanogenic archaea by their production of volatile fatty acids and hydrogen during fermentation of organic matter. Both of these bacterial groups have been found to be essential for the cultivation of certain methanogens in the laboratory, providing further evidence for their supporting role in methane production (Sizova et al., 2003). Additionally, hydrogenotrophic methanogenesis can also dominate if there is incomplete degradation of organic matter (Miyajima et al., 1997; Conrad, 1999; Conrad et al., 2009). This is likely the case for bromeliad catchments, which have elevated total organic carbon levels (~46%) compared to nearby soil (4%; Goffredi et al., 2011), much of which is dead crustaceans (i.e., chitin) and lignified plant-derived cellulose. It is, thus, likely that part of the organic matter is refractory and incompletely oxidized even over very long time frames. In support of this idea, preliminary experiments in the field suggest that, although discoloration and leaching of leaf material can be observed, actual decrease in biomass occurs very slowly (i.e., precut dead leaves retained sharp, un-eroded edges after 3 months of submergence in bromeliad tanks in situ; Walter T. Woodside, unpublished observation).
Acetate-driven methanogenesis, which is estimated to account for a substantial amount of methane released in terrestrial and freshwater environments, appears to also be a common metabolic process in bromeliad tanks. Aceticlastic members of the Methanosaetaceae were recovered from certain bromeliad catchments, although often in lower abundance than their hydrogenotrophic counterparts (similarly observed in Martinson et al., 2010). Notably, Methanosaeta, which appeared to comprise a bromeliad-specific clade, were generally most prevalent in tanks with pH ~6.5 (Figure 5). Methanosaeta-associated strains, like many methanogens, have a pH optimum near neutrality and often predominate at higher pH values (Jones et al., 1987; Horn et al., 2003). In these same tanks, there was a striking decrease in Methanomicrobiales- and Methanocellales-associated ribotypes (<3%, as compared to ~40–80% in tanks with lower pH), perhaps suggesting a slight influence of tank pH on methanogen community structure. Similarly, the dominance of Methanosaeta-associated ribotypes in certain tanks strongly suggests that acetate concentrations were low, and illustrates the important need for future analysis of fatty acid concentrations maintained by bromeliad catchments.
The more versatile Methanosarcina, on the other hand, were only dominant in one bromeliad, by library construction, and occurred in moderate abundances (~26% average) in nine other bromeliad tanks, based on T-RFLP analysis. In the only other previous study of methanogenic communities within bromeliad catchments (from the high elevation Ecuadorian Andes), Methanosarcina were not observed (Martinson et al., 2010). Importantly, Methanosarcina are common in many soil habitats, including flooded rice paddies (Kudo et al., 1997; Ramakrishnan et al., 2001; Yuan et al., 2011), and we found them to also be the dominant methanogen in the only soil sample (out of 20) that showed positive presence of methanogens.
Conclusion
Epiphytic, tank-forming bromeliads are predominant members of neotropical habitats, including cloud and rainforests, and collectively suspend large amounts of water in the canopy, thus representing a large environmental resource. Our results indicate that in stratified bromeliad catchments, methane production occurs and that members of the Methanomicrobiales, Methanocellales, and Methanosarcinales (Methanosaetaceae in particular) are common. This, plus laboratory incubation of tank waters, provides compelling evidence of bromeliads as an important source of methane, produced by the activities of both hydrogenotrophic and aceticlastic methanogens during the final steps in the decomposition of organic matter. Although at this point linkages cannot be confidently drawn between environmental parameters, bromeliad methanogen community structure, and CH4 production dynamics, it is likely that a combination of factors, including substrate quality and availability, degree of anoxia and desiccation, tank pH, and tank morphology are most influential in regulating production and release of methane to the atmosphere above neotropical rainforests.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Drs. Gretchen North and Beth Braker (Occidental College) for advice about tropical plant biology; Bernal Matarrita, among others, for his laboratory support at La Selva Biological station; Dr. Deedra McClearn and members of the Organization for Tropical Studies for their support of this project; Dr. Victoria Orphan for use of the CEQ 8800 Genetic Analysis System at the California Institute of Technology as well as advice about methanogenic archaea; Josh Steele for running rarefaction curves on the clone library data; Rebecca Tribelhorn for laboratory assistance and data collection; and, finally, Dr. Catherine Cardelús (Colgate University) for canopy samples. Funding for this project included, in part, a NSF grant to B. Braker (Occidental College, OISE-0526551), a HHMI grant to Occidental College, as well as the Undergraduate Research Center (Academic Student and Summer Research Projects), and a Faculty Enrichment grant from Occidental College. Equipment used for this project was funded, in part, by a National Science Foundation MRI grant to S. Goffredi, G. North, and J. Schulz (Occidental College, MRI-0960254).
References
Banning, N., Brock, F., Fry, J. C., Parkes, R. J., Hornibrook, E. R., and Weightman, A. J. (2005). Investigation of the methanogen population structure and activity in a brackish lake sediment. Environ. Microbiol. 7, 947–960.
Basiliko, N., Yavitt, J. B., Dees, P. M., and Merkel, S. M. (2003). Methane biogeochemistry and methanogen communities in two northern peatland ecosystems, New York state. Geomicrobiol. J. 20, 563–577.
Benzing, D. H. (1970). Foliar permeability and the absorption of minerals and organic nitrogen by certain tank bromeliads. Bot. Gaz. 131, 23–31.
Benzing, D. H., Derr, J. A., and Titus, J. E. (1972). The water chemistry of microcosms associated with the bromeliad Aechmea bracteata. Am. Midl. Nat. 87, 60–70.
Bermudes, D., and Benzing, D. H. (1991). Nitrogen fixation in association with Ecuadorian bromeliads. J. Trop. Ecol. 7, 531–536.
Borrel, G., Jézéquel, D., Biderre-Petit, C., Morel-Desrosiers, N., Morel, J. P., Peyret, P., Fonty, G., and Lehours, A. C. (2011). Production and consumption of methane in freshwater lake ecosystems. Res. Microbiol. 162, 832–847.
Bräuer, S. L., Cadillo-Quiroz, H., Ward, R. J., Yavitt, J. B., and Zinder, S. H. (2011). Methanoregula boonei gen. nov., sp. nov., an acidiphilic methanogen isolated from an acidic peat bog. Int. J. Syst. Evol. Microbiol. 61, 45–52.
Bräuer, S. L., Yashiro, E., Ueno, N. G., Yavitt, J. B., and Zinder, S. H. (2006). Characterization of acid-tolerant H2/CO2-utilizing methanogenic enrichment cultures from an acidic peat bog in New York State. FEMS Microbiol. Ecol. 57, 206–216.
Cadillo-Quiroz, H., Bräuer, S., Yashiro, E., Sun, C., Yavitt, J., and Zinder, S. (2006). Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York State, USA. Environ. Microbiol. 8, 1428–1440.
Cadillo-Quiroz, H., Yashiro, E., Yavitt, J. B., and Zinder, S. H. (2008). Characterization of the archaeal community in a minerotrophic fen and terminal restriction fragment length polymorphism-directed isolation of a novel hydrogenotrophic methanogen. Appl. Environ. Microbiol. 74, 2059–2068.
Capone, D. G., Reese, D. D., and Kiene, R. P. (1983). Effects of metals on methanogenesis, sulfate reduction, carbon dioxide evolution, and microbial biomass in anoxic salt marsh sediments. Appl. Environ. Microbiol. 45, 1586–1591.
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143.
Conrad, R. (1999). Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 28, 193–202.
Conrad, R. (2005). Quantification of methanogenic pathways using stable carbon isotopic signatures: a review and a proposal. Org. Geochem. 36, 739–752.
Conrad, R., Claus, P., and Casper, P. (2009). Characterization of stable isotope fractionation during methane production in the sediment of a eutrophic lake, Lake Dagow, Germany. Limnol. Oceanogr. 54, 457–471.
Conrad, R., Klose, M., Claus, P., and Enrich-Prast, A. (2010). Methanogenic pathway, 13C isotope fractionation, and archaeal community composition in the sediment of two clear-water lakes of Amazonia. Limnol. Oceanogr. 55, 689–702.
Conrad, R., Klose, M., Noll, M., Kemnitz, D., and Bodelier, P. L. E. (2008). Soil type links microbial colonization of rice roots to methane emission. Glob. Chang Biol. 14, 657–669.
Deppenmeier, U., Johann, A., Hartsch, T., Merkl, R., Schmitz, R. A., Martinez-Arias, R., Henne, A., Wiezer, A., Bäumer, S., Jacobi, C., Brüggemann, H., Lienard, T., Christmann, A., Bömeke, M., Steckel, S., Bhattacharyya, A., Lykidis, A., Overbeek, R., Klenk, H. P., Gunsalus, R. P., Fritz, H. J., and Gottschalk, G. (2002). The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4, 453–461.
Erkel, C., Kube, M., Reinhardt, R., and Liesack, W. (2006). Genome of rice cluster I archaea – the key methane producers in the rice rhizosphere. Science 313, 370–372.
Ermler, U., Grabarse, W., Shima, S., Goubeaud, M., and Thauer, R. K. (1997). Crystal structure of methyl coenzyme M reductase: the key enzyme of biological methane formation. Science 278, 1457–1462.
Ferry, J. G., Smith, P. H., and Wolfe, R. S. (1974). Methanospirillum, a new genus of methanogenic bacteria, and characterization of Methanospirillum hungatii sp. nov. Int. J. Syst. Bacteriol. 24, 465–469.
Fish, D. (1983). “Phytotelmata: terrestrial plants as hosts for aquatic insect communities,” in Phytotelmata: Flora and Fauna, eds J. H. Frank and L. P. Lounibos (New Jersey: Plexus), 1–28.
Frankenberg, C., Bergamaschi, P., Butz, A., Houweling, S., Meirink, J. F., Notholt, J., Petersen, A. K., Schrijver, H., Warneke, T., and Aben, I. (2008). Tropical methane emissions: a revised view from SCIAMACHY onboard ENVISAT. Geophys. Res. Lett. 35, L15811.
Galand, P. E., Fritze, H., and Yrjälä, K. (2003). Microsite-dependent changes in methanogenic populations in a boreal oligotrophic fen. Environ. Microbiol. 5, 1133–1143.
Galand, P. E., Saarnio, S., Fritze, H., and Yrjälä, K. (2002). Depth related diversity of methanogen archaea in Finnish oligotrophic fen. FEMS Microbiol. Ecol. 42, 441–449.
Goffredi, S. K., Kantor, A. H., and Woodside, W. T. (2011). Aquatic microbial habitats within a neotropical rainforest: bromeliads and pH-associated trends in bacterial diversity and composition. Microb. Ecol. 61, 529–542.
Grosskopf, R., Janssen, P. H., and Liesack, W. (1998). Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64, 960–969.
Guimaraes-Souza, B. A., Mendes, G. B., Bento, L., Marotta, H., Santoro, A. L., Esteves, F. A., Pinho, L., Farjalla, V. F., and Enrich-Prast, A. (2006). Limnological parameters in the water accumulated in tropical bromeliads. Acta Limnologica Brasiliensia 18, 47–53.
Hales, B. A., Edwards, C., Ritchie, D. A., Hall, G., Pickup, R. W., and Saunders, J. R. (1996). Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62, 668–675.
Horn, M. A., Matthies, C., Kusel, K., Schramm, A., and Drake, H. L. (2003). Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69, 74–83.
Hou, Y., Zhang, H., Miranda, L., and Lin, S. (2010). Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: microalgal pcna as the model gene. PLoS ONE 5, e9545. doi:10.1371/journal.pone.0009545
IPCC. (2001). “Climate change 2001: the scientific basis,” in Contribution of Working Group I to the Third Assessment Report of the Intergovernmental Panel on Climate Change (IPCC), eds J. T. Houghton, Y. Ding, D. J. Griggs, M. Noguer, P. J. van der Linden, and D. Xiaosu (Cambridge: Cambridge University Press), 241–280.
Jetten, M. S. M., Stams, A. J. M., and Zehnder, A. J. B. (1992). Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Lett. 88, 181–197.
Jones, W. J., Nagle, D. P., and Whitman, W. B. (1987). Methanogens and the diversity of archaebacteria. Microbiol. Mol. Biol. Rev. 51, 135–177.
Joulian, C., Ollivier, B., Patel, B. K. C., and Roger, P. A. (1998). Phenotypic and phylogenetic characterization of dominant culturable methanogens isolated from rice field soils. FEMS Microbiol. Ecol. 25, 135–145.
Juottonen, H., Galand, P. E., Tuittila, E. S., Laine, J., Fritze, H., and Yrjälä, K. (2005). Methanogen communities and bacteria along an ecohydrological gradient in a northern raised bog complex. Environ. Microbiol. 7, 1547–1557.
Kanokratana, P., Uengwetwanit, T., Rattanachomsri, U., Bunterngsook, B., Nimchua, T., Tangphatsornruang, S., Plengvidhya, V., Champreda, V., and Eurwilaichitr, L. (2011). Insights into the phylogeny and metabolic potential of a primary tropical peat swamp forest microbial community by metagenomic analysis. Microb. Ecol. 61, 518–528.
Kotsyurbenko, O. R., Chin, K. J., Glagolev, M. V., Stubner, S., Simankova, M. V., Nozhevnikova, A. N., and Conrad, R. (2004). Acetoclastic and hydrogenotrophic methane production and methanogenic populations in an acidic West-Siberian peat bog. Environ. Microbiol. 6, 1159–1173.
Kudo, Y., Nakajima, T., Miyaki, T., and Oyaizu, H. (1997). Methanogen flora of paddy soils in Japan. FEMS Microbiol. Ecol. 22, 39–48.
Liu, R., Zhang, Y., Ding, R., Li, D., Gao, Y., and Yang, M. (2009). Comparison of archaeal and bacterial community structures in heavily oil-contaminated and pristine soils. J. Biosci. Bioeng. 108, 400–407.
Ludwig, W., Strunk, R., Westram, L., Richter, H., and Meier, H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Lueders, T., Chin, K. J., Conrad, R., and Friedrich, M. (2001). Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3, 194–204.
Luton, P. E., Wayne, J. M., Sharp, R. J., and Riley, P. W. (2002). The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148, 3521–3530.
Martinson, G. O., Werner, F. A., Scherber, C., Conrad, R., Corre, M. D., Flessa, H., Wolf, K., Klose, M., Robbert Gradstein, S., and Veldkamp, E. (2010). Methane emissions from tank bromeliads in neotropical forests. Nat. Geosci. 3, 766–769.
Miyajima, T., Wada, E., Hanba, Y. T., and Vijarnsorn, P. (1997). Anaerobic mineralization of indigenous organic matters and methanogenesis in tropical wetland soils. Geochim. Cosmochim. Acta 61, 3739–3751.
Parsons, A. J., Newton, P. C., Clark, H., and Kelliher, F. M. (2006). Scaling methane emissions from vegetation. Trends Ecol. Evol. (Amst.) 21, 423–424.
Patel, G. B. (1984). Characterization and nutritional properties of Methanothrix concilii sp. nov., a mesophilic, aceticlastic methanogen. Can. J. Microbiol. 30, 1383–1396.
Preusse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W. G., Peplies, J., and Glockner, F. O. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196.
Ramakrishnan, B. T., Lueders, T., Dunfield, P. F., Conrad, R., and Friedrich, M. W. (2001). Archaeal community structures in rice soils from different geographical regions before and after initiation of methane production. FEMS Microbiol. Ecol. 37, 175–186.
Rivière, D., Desvignes, V., Pelletier, E., Chaussonnerie, S., Guermazi, S., Weissenbach, J., Li, T., Camacho, P., and Sghir, A. (2009). Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 3, 700–714.
Sakai, S., Imachi, H., Hanada, S., Ohashi, A., Harada, H., and Kamagata, Y. (2008). Methanocella paludicola gen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage “rice cluster I,” and proposal of the new archaeal order Methanocellales ord. nov. Int. J. Syst. Evol. Microbiol. 58, 929–936.
Sizova, M. V., Panikov, N. S., Tourova, T. P., and Flanagan, P. W. (2003). Isolation and characterization of oligotrophic acido-tolerant methanogenic consortia from a Sphagnum peat bog. FEMS Microbiol. Ecol. 45, 301–315.
Smith, J. M., Castro, H., and Ogram, A. (2007). Structure and function of methanogens along a short-term restoration chronosequence in the Florida Everglades. Appl. Environ. Microbiol. 73, 4135–4141.
Steinberg, L. M., and Regan, J. M. (2008). Phylogenetic comparison of the methanogenic communities from an acidic, oligotrophic fen and an anaerobic digester treating municipal wastewater sludge. Appl. Environ. Microbiol. 74, 6663–6671.
Sugden, A. M., and Robins, R. J. (1979). Aspects of the ecology of vascular epiphytes in Colombian cloud forests, I. The distribution of the epiphytic flora. Biotropica 11, 173–188.
Svensson, B. H., and Rosswall, T. (1984). In situ methane production from acid peat in plant communities with different moisture regimes in a subarctic mire. Oikos 43, 341–350.
Swofford, D. L. (1998). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. (Sunderland: Sinauer Associates).
Tarasov, A. L., Borzenkov, I. A., Chernykh, N. A., and Belyayev, S. S. (2011). Isolation and investigation of anaerobic microorganisms involved in methanol transformation in an underground gas storage facility. Microbiology 80, 172–179.
Watanabe, A., Takeda, T., and Kimura, M. (1999). Evaluation of origins of CH4 carbon emitted from rice paddies. J. Geophys. Res. 104, 23623–23629.
Watanabe, K., Kodama, Y., Hamamura, N., and Kaku, N. (2002). Diversity, abundance, and activity of archaeal populations in oil-contaminated groundwater accumulated at the bottom of an underground crude oil storage cavity. Appl. Environ. Microbiol. 68, 3899–3907.
Watanabe, T., Kimura, M., and Asakawa, S. (2009). Distinct members of a stable methanogenic archaeal community transcribe mcrA genes under flooded and drained conditions in Japanese paddy field soil. Soil Biol. Biochem. 41, 276–285.
Whiticar, M. J. (1999). Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem. Geol. 161, 291–314.
Winkler, U., and Zotz, G. (2009). Highly efficient uptake of phosphorus in epiphytic bromeliads. Ann. Bot. 103, 477–484.
Wüst, P. K., Horn, M. A., and Drake, H. L. (2009). Trophic links between fermenters and methanogens in a moderately acidic fen soil. Environ. Microbiol. 11, 1395–1409.
Yuan, Y., Conrad, R., and Lu, Y. (2011). Transcriptional response of methanogen mcrA genes to oxygen exposure of rice field soil. Environ. Microbiol. Rep. 3, 320–328.
Appendix
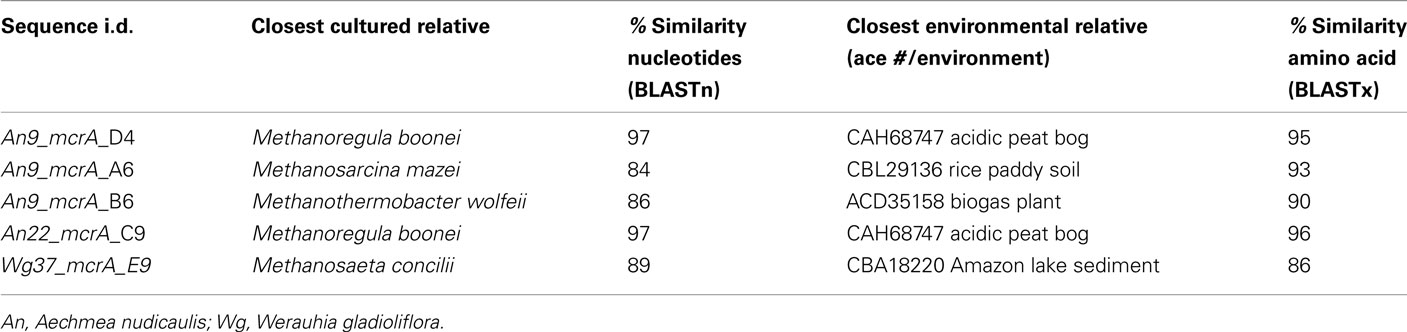
Table A1. Summary of archaeal methyl coenzyme M reductase (mcrA) clone library results from the tank water of three bromeliads specimens.
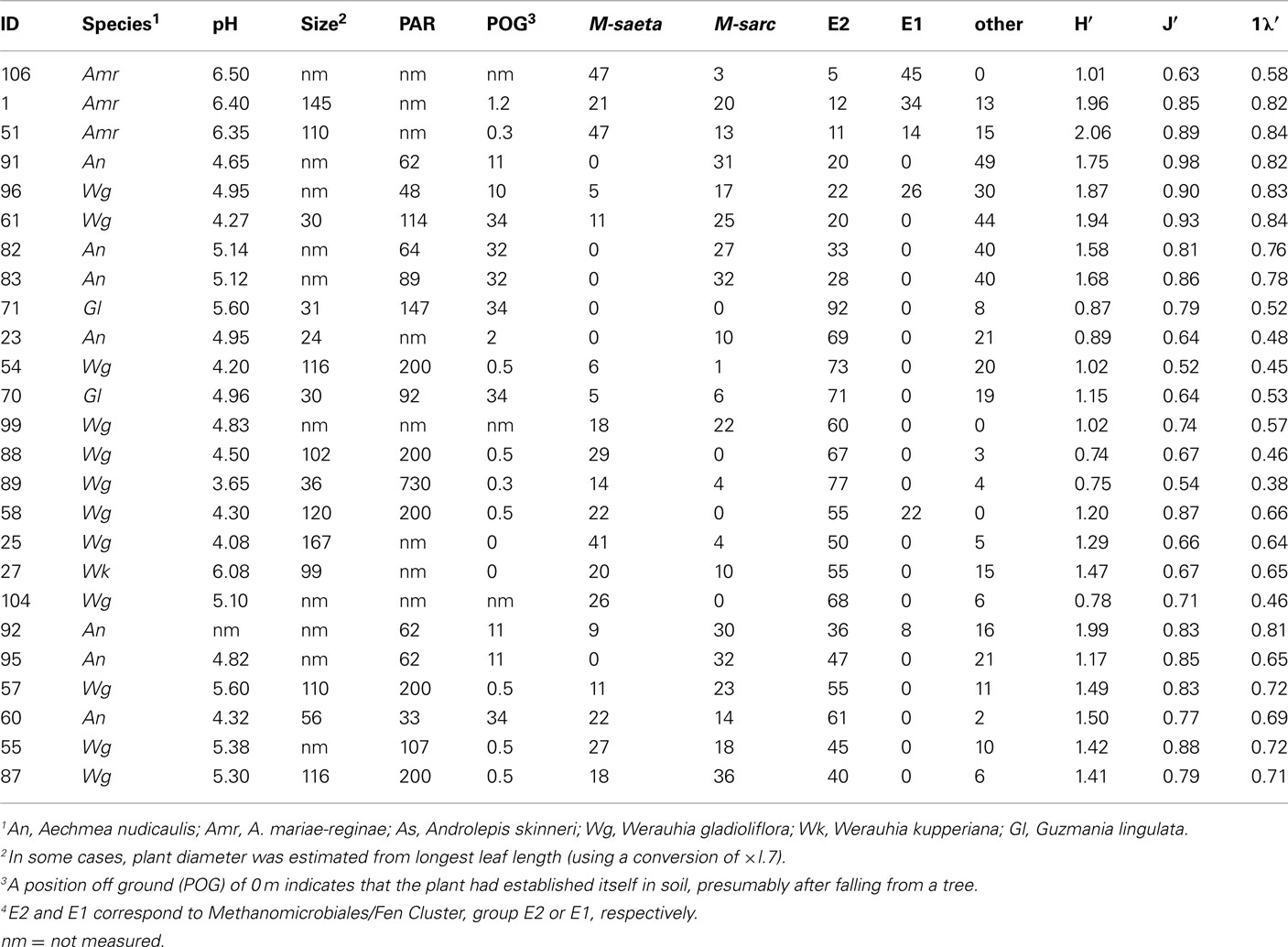
Table A2. Bromeliads used in T-RFLP analysis, including catchment pH, plant diameter (cm), photosynthetically active radiation (μmol m−2 s−1) exposure at the time of sample collection, position off ground (m), group-specific abundances (%) based on T-RFLP analysis, and corresponding diversity indices (Shannon index H′, Pielou’s evenness J′, and Simpson’s index 1λ′).
Keywords: bromeliad, methane, methanogen, Methanoregula, Methanosaeta, rainforest, hydrogenotrophic
Citation: Goffredi SK, Jang GE, Woodside WT and Ussler W III (2011) Bromeliad catchments as habitats for methanogenesis in tropical rainforest canopies. Front. Microbio. 2:256. doi: 10.3389/fmicb.2011.00256
Received: 23 August 2011;
Accepted: 02 December 2011;
Published online: 27 December 2011.
Edited by:
Josh David Neufeld, University of Waterloo, CanadaReviewed by:
Nathan Basiliko, University of Toronto, CanadaMarcus A. Horn, University of Bayreuth, Germany
Stephen Zinder, Cornell University, USA
Copyright: © 2011 Goffredi, Jang, Woodside and Ussler. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Shana K. Goffredi, Biology Department, Occidental College, 1600 Campus Road, Los Angeles, CA 90041, USA. e-mail: sgoffredi@oxy.edu