- Department of Chemistry, University of California, Davis, CA, USA
The growing need to address current energy and environmental problems has sparked an interest in developing improved biological methods to produce liquid fuels from renewable sources. While microbial ethanol production is well established, higher-chain alcohols possess chemical properties that are more similar to gasoline. Unfortunately, these alcohols (except 1-butanol) are not produced efficiently in natural microorganisms, and thus economical production in industrial volumes remains a challenge. Synthetic biology, however, offers additional tools to engineer synthetic pathways in user-friendly hosts to help increase titers and productivity of these advanced biofuels. This review concentrates on recent developments in synthetic biology to produce higher-chain alcohols as viable renewable replacements for traditional fuel.
Introduction
Depletion of finite liquid fuels is on the horizon, and the growing concern with current energy and environmental problems has ignited an interest in the production of liquid biofuels (Herrera, 2006; Zinoviev et al., 2010; Fairley, 2011). In 2010, petroleum consumption in the United States was about 36 quadrillion BTU, in which 70% consumed was imported (Fichman, 2011). To help increase independence from petroleum-derived fuels as well as its foreign importation, endeavors to develop and improve biological methods to produce renewable liquid fuels are in play (Stephanopoulos, 2007; Fortman et al., 2008; Lee et al., 2008; Sheehan, 2009). Ethanol ranks as the most prominent biofuel to date, reaching a production high of 13.2 billion gallons in 2010 (more than eight times produced a decade ago; Fichman, 2011). Despite its well-developed production and wide use, notably as a gasoline additive and as a high-percentage E85 blend, it falls short of being the ideal “drop-in” fuel to replace gasoline. Ethanol is hygroscopic, making it incompatible with current gasoline infrastructure. Moreover, ethanol is only 65% as energy dense as gasoline (20.8 versus 32.0 MJ/L, respectively; Li et al., 2010b). Higher-chain alcohols, on the other hand, are not hygroscopic, and provide energy densities that are more comparable to gasoline, and thus help to mitigate the problems that ethanol arises.
Unlike ethanol, higher-chain alcohols (except 1-butanol produced naturally by the Clostridium species; Jones and Woods, 1986) are neither produced efficiently in nature nor in efficient quantities, and so increased efforts are being made to use microorganisms to cost-effectively produce them from biomass as well as in industrial volumes. Much energy has been put into using metabolic engineering to improve natural producers of these alcohols in the past, but without success in reaching desired industrial-relevant titers and productivity (Lutke-Eversloh and Bahl, 2011). Part of this difficulty stems from a lack of tools and techniques for genetic modification, slow growth, and complex physiology of the natural producer (Ezeji et al., 2007). User-friendly hosts such as Escherichia coli and Saccharomyces cerevisiae, however, come with a wide range of tools and techniques, are faster growing, and possess a more simple physiology. With these helping hands, many efforts have been put into developing higher-chain alcohol production in non-native hosts.
Outside the advantageous inherent properties of using a well-characterized host and the ease in which metabolic engineering can help optimize pathways and maintain cell fitness, synthetic biology is a relatively nascent practice being used to aid in the development of higher-chain alcohol producers (Agapakis and Silver, 2009; Khalil and Collins, 2010). Synthetic biology provides the ability to piece together biological components from several different origins in order to redesign a natural or construct a novel pathway that the host uses to synthesize a valuable chemical (Keasling, 2008; Connor and Atsumi, 2010). In conjunction with metabolic engineering, development of a more efficient production platform in a non-native but user-friendly host for a desired chemical is made possible (Alper and Stephanopoulos, 2009). This review will concentrate on recent progress in using synthetic biology to engineer user-friendly microbes that produce C3–C10 alcohols.
CoA-Dependent Pathways
Natural production of 1-butanol occurs in the acetone–butanol–ethanol (ABE) pathway of the Clostridium species (Gheshlaghi et al., 2009), where starch and simple sugars are fermented into a mixture of solvents, including acetone, lactate, and acetoin. In this pathway, two acetyl-CoA molecules are converted to butyryl-CoA in four enzymatic steps using acetyl-CoA acetyltransferase, 3-hydroxybutyryl-CoA dehydrogenase, crotonase, and butyryl-CoA dehydrogenase (Bennett and Rudolph, 1995). From here, 1-butanol can be produced using a dual-function butyraldehyde/butanol dehydrogenase (Atsumi et al., 2008a).
Since natural alcohol producers like the Clostridium species have been difficult to engineer for 1-butanol production, great strides have been made to transfer and improve this traditional fermentation pathway in non-natural producers like E. coli and S. cerevisiae. For example, E. coli, a Gram-negative bacterium, has a long history of being studied and exploited as a model host, due to its rapid growth rate, relatively simple physiology and nutritional needs, as well as ease of cultivation and manipulation. Furthermore, elucidation and characterization of its genetics, physiology, and biochemistry has led to the development of a wide selection of genetic tools for manipulating cell behavior and function.
In general, overexpression of either native or heterologous genes involves construction of plasmids that fuse multiple genes transcriptionally. Overexpression of genes may also be integrated into the genomic DNA (gDNA) of a microorganism using homologous recombination via a plasmid containing the gene of interest and a selectable marker. This approach is used to engineer cyanobacteria such as Synechococcus elongatus (Golden et al., 1987). Furthermore, in order to gain control of expression levels, there are various promoters, both native and engineered (Gronenborn, 1976; Brosius et al., 1985; Studier and Moffat, 1986; Elvin et al., 1990; Mumberg et al., 1994; Lutz, 1997; Haldimann et al., 1998; Pátek et al., 2003), available to choose from. While heterologous gene expression – due to various factors like protein misfolding and aggregation (Thomas et al., 1997; Vabulas et al., 2010) – is not always successful, progress has been made to mechanistically understand why these processes can occur (Georgiou and Valax, 1996; Baneyx and Mujacic, 2004; Ami et al., 2009). As a result, several approaches have been developed to circumvent these types of pitfalls and increase the likelihood that heterologous expression is successful (Baneyx and Mujacic, 2004).
To minimize costs for industrial scale production, it is ideal that production can be done in minimum growth media rather than rich media. More often than not, however, rich media are used in order to maximize cell density and optimize cell condition for production in laboratory scale production (Tartoff and Hobbs, 1987; Amberg et al., 2005). Measurement of alcohols and other metabolites can be monitored using various chromatographic methods, typically gas chromatography–mass spectrometry (GC–MS), gas chromatography–flame ionization detector (GC–FID), and high-performance liquid chromatography (HPLC).
The following describes research to increase the titers of 1-butanol, isopropanol, and 1-hexanol in user-friendly hosts, as well as the utilization and engineering of the CoA-dependent pathway (Figure 1) to shunt production toward non-natural higher-chain alcohols.
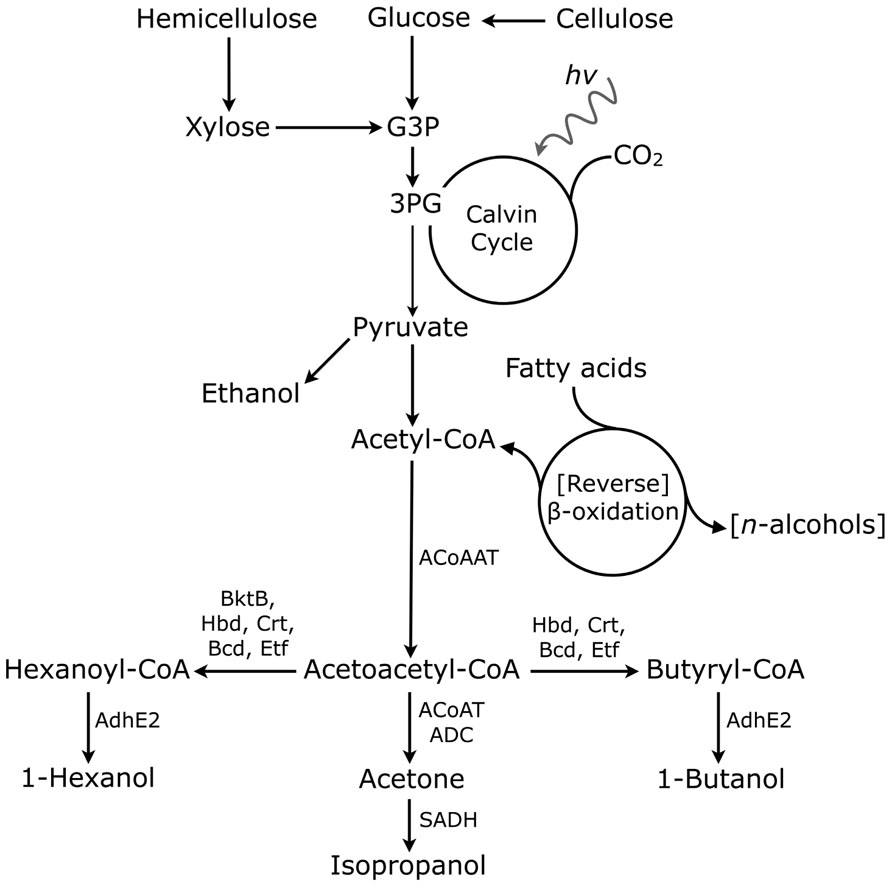
FIGURE 1. Metabolic representation of CoA-dependent pathways for biofuel production. G3P, glyceraldehyde-3-phosphate; 3PG, 3-phosphoglycerate; ACoAAT, acetyl-CoA acetyltransferase; BktB, 2-ketothiolase; Hbd, 3-hydroxybutyryl-CoA; Crt, crotonase; Bcd, butyryl-CoA dehydrogenase; Etf, electron transfer flavoprotein; ACoAT, acetoacetyl-CoA transferase; ADC, acetoacetate decarboxylase; AdhE2, aldehyde/alcohol dehydrogenase; SADH, primary–secondary alcohol dehydrogenase. Reversal of β-oxidation to make -alcohols (in the presence of a non-fatty acid carbon source) is indicated in brackets.
1-Butanol Production
In order to demonstrate the feasibility of this strategy, Atsumi et al. (2008a) transferred the 1-butanol pathway of Clostridium acetobutylicum into E. coli. Use of 1-butanol as an additive or replacement for gasoline has been growing in popularity due to its hygroscopicity and comparable energy density (29.2 MJ/L) to gasoline. For production through the Clostridium pathway, one 1-butanol molecule is produced per molecule of glucose and four NADH. The strain containing essential genes for 1-butanol production (thl, hbd, crt, bcd, etfAB, and adhE2) were cloned and expressed using a two-plasmid system, where thl and adhE2 were overexpressed under control of the IPTG-inducible PLlacO1 promoter (Lutz, 1997) in a colE (Kahn et al., 1979) origin plasmid, while crt, bcd, etfAB, and hbd were overexpressed under control of the PLlacO1 promoter in a p15A (Chang and Cohen, 1978) replication origin plasmid. This two-plasmid expression strain produced 13.9 mg/L 1-butanol in 40 h under anaerobic conditions. Elevating oxygen levels increased production in this strain, although these enzymes are from a strict anaerobe. To further improve production, genes responsible for production of metabolic byproducts in the E. coli genome were deleted. Namely, ΔldhA, ΔfrdBC, and ΔadhE knockouts were introduced in order to reduce lactate, ethanol, and succinate. Furthermore, pta (encoding phosphotransacetylase) was deleted to decrease amount of acetate produced. The fnr gene, encoding the regulator Fnr that deactivates the pyruvate dehydrogenase complex under anaerobic conditions, was deleted as well. The resulting strain boosted 1-butanol production to 373 mg/L. Finally, this strain, when grown in Terrific Broth (TB)-enriched, glycerol-supplemented media (Tartoff and Hobbs, 1987), increased 1-butanol titer to 552 mg/L.
A promising strategy to tweak alcohol production further in non-native hosts is to consider the various driving forces that are modified as a consequence to manipulating a host, and making efforts to alleviate negative affects in a resulting strain. When altering a native pathway, many factors such as growth and gene expression have the potential to be compromised, which can diminish cell fitness and create bottlenecks in the engineered system that prevent high titers of a desired product (Fischer et al., 2008). A recurring problem that must be tackled is the ubiquitous occurrence of reversible reactions in metabolic pathways that slow the optimal production of important intermediates essential for propelling a series of steps.
Bond-Watts et al. (2011) explored the chemical nature of enzymes to drive equilibria forward in the design of a 1-butanol platform in E. coli. First, in an E. coli strain containing phaA, phaB, and crt in a p15A origin (Chang and Cohen, 1978) plasmid, and ccr and adhE2 in a colE origin (Kahn et al., 1979) plasmid (both under control of the IPTG-inducible Ptrc promoter; Brosius et al., 1985), only 95 mg/L 1-butanol was produced after 6 days in TB supplemented with glucose. To push the equilibrium between crotonyl-CoA and butyryl-CoA forward, ter, encoding an NADH-dependent crotonyl-CoA reductase from Treponema denticola, replaced ccr. This gene replacement led to a 3.5-fold increase in titer (340 mg/L) after 3 days. Upon further investigation for additional bottlenecks, enzymes that generate stereo-specific products were found and tested. After replacing phaB with hbd, encoding NADH-dependent (S)-3-hydroxybutyryl-CoA dehydrogenase from C. acetobutylicum, and crt with phaJ, encoding an R-specific enoyl-CoA hydratase from Aeromonas caviae, production rose to 2.95 g/L. Finally, balance of reducing equivalents was taken into consideration. While four NADH molecules needed to make one molecule 1-butanol, glycolysis only produces two NADH per glucose molecule. To increase availability and direct consumption of acetyl-CoA and NADH toward alcohol production and away from lactate and acetate formation, aceEF-lpd, encoding the pyruvate dehydrogenase complex, was overexpressed. This enhancement provided the additional two NADH molecules needed to synthesize 1-butanol, and improved production levels to a final titer of 4.65 g/L at 28% the theoretical max from glucose.
Using a similar premise to improve the CoA-dependent pathway in E. coli, Shen et al. (2011) utilized a number of driving forces to generate 1-butanol in industrial-relevant quantities. In this study, atoB (E. coli), adhE2 (C. acetobutylicum), crt (C. acetobutylicum), hbd (C. acetobutylicum), and ter (T. denticola; used to replace the oxygen-sensitive, NADPH-dependent Bcd–EtfAB complex) genes were overexpressed in an E. coli ΔadhE ΔldhA Δfrd strain (JC166), which cannot grow without an additional NADH-consuming pathway. In this two-plasmid system, atoB, adhE2, crt, and hbd were overexpressed in a plasmid under control of the PLlacO1 promoter (Lutz, 1997) with a colE (Kahn et al., 1979) origin, while ter was overexpressed in a plasmid under PLlacO1 control with a colA origin (Zverev and Khmel, 1985). This strain anaerobically produced 1.8 g/L after 24 h in TB supplemented with glucose. Catalysis by Ter to convert crotonyl-CoA to butyryl-CoA was found irreversible in the presence of excess butyryl-CoA and NAD+ by in vitro enzyme assays. Furthermore, Bcd–EtfAB replacement by NADH-dependent Ter successfully restored anaerobic growth of JC166. Next, fdh, encoding a formate dehydrogenase from Candida boidinii, was overexpressed to reduce excess pyruvate. Without overexpression of fdh in the 1-butanol pathway, pyruvate serves as a sink for excess carbon flux and piles up in the cell. On the contrary, when fdh is overexpressed, formate is oxidized into CO2 and NADH to promote accumulation of the additional two NADH equivalents needed for 1-butanol production. Moreover, pta, which would have encoded a phosphate acetyltransferase, was deleted in order to build up acetyl-CoA and decrease acetate formation. As a result, 1-butanol production rose to 15 g/L after 3 days at 88% theoretical maximum yield. Lastly, anaerobic, pH-controlled fermentation was performed, in conjunction with continuous gas stripping, using a 1 L stirred-tank bioreactor. Final measurements indicated that 1-butanol levels shot up to 30 g/L after 7 days (~70% theoretical maximum).
Escherichia coli is not the only user-friendly host available to for 1-butanol production. The robust yeast, S. cerevisiae, has been a prominent host for industrial ethanol production as well as the non-native production of a variety of valuable chemicals. It is well-characterized, has a wide range of genetic tools for manipulation, its fermentation is relatively cheap, and its mass production and downstream processing is well-developed. Especially, S. cerevisiae has a vast history of being used for the production of ethanol for beer and wine from sugars. Like for E. coli, synthetic biology has contributed to the improvement of engineered processes in S. cerevisiae (Krivoruchko et al., 2011).
Steen et al. (2008) chose S. cerevisiae as the host for 1-butanol production, in which clostridial enzymes were substituted with isozymes from various other species. From an initial group of strains to elucidate a thiolase with the best activity [phaA (Ralstonia eutropha), atoB (E. coli), or ERG10 (S cerevisiae)], the highest producer (BY4747) containing phaA and phaB (R. eutropha), ccr (Streptomyces collinus), and crt and adhE2 (C. beijerinckii), under control of either the GAL1 or GAL10 galactose-dependent promoters in 2 μ origin (Huberman et al., 1987) plasmids, generated 1 mg/L 1-butanol. To improve upon this initial strain, an NADH-dependent hydroxybutyryl-CoA dehydrogenase, encoded by Hbd, replaced an NADPH-dependent enzyme, encoded by PhaB. The strain including ERG10 (encoding a native thiolase) and hbd resulted in 2.5 mg/L production in synthetic defined (SD) media (Amberg et al., 2005) supplemented with galactose.
In an interest to compare production in various hosts, Nielsen et al. (2009) reconstructed and engineered the 1-butanol pathway of C. acetobutylicum into Gram-negative Pseudomonas putida and Gram-positive Bacillus subtilis. P. putida and B. subtilis were chosen as hosts for this biosynthetic pathway due to higher 1-butanol tolerance relative to E coli (de Bont, 1998). P. putida has been exploited for bioremediation of soil, and possesses a versatile metabolism, an ability to thrive in extreme environmental conditions, simple nutritional requirements, and rapid growth rate (Dos Santos et al., 2004). The P. putida strain was cultured in TB under control of the IPTG-inducible T7lac promoter (Studier and Moffat, 1986) and produced 50 mg/L with glucose, and 122 mg/L with glycerol. Results also indicated that the Bcd–EtfAB complex of C. acetobutylicum was active in the obligate aerobe. B. subtilis, on the other hand, has been extensively studied as a model Gram-positive bacterium, and can live in several different environments, including soil, animal gastrointestinal tracts, and plant roots. Moreover, it possesses the distinct ability to become competent for transformation naturally (Earl et al., 2008). B. subtilis genes amyE (encoding an α-amylase), thrC (encoding a threonine synthase), and pyrD (encoding a dihydroorotate dehydrogenase) were deleted from the genome using double-crossover homologous recombination. The B. subtilis ΔamyE ΔthrC ΔpyrD strain was cultured in TB under control of the IPTG-inducible promoter (Studier and Moffat, 1986), and produced 23 mg/L with glucose, and 24 mg/L in glycerol under anaerobic conditions.
Production of higher-chain alcohols using bacteria that directly utilize CO2 and sunlight is another approach being explored. In addition, if an efficient production platform is successfully designed, then production of higher-chain alcohols directly from sunlight would give a higher-value advantage. One photosynthetic organism with a growing genetic toolbox, elucidated genome sequences, and relatively fast growth rate is known as cyanobacteria (Ruffing, 2011), and its capabilities have already been tapped to produce a range of different fuels and chemicals (Machado and Atsumi, 2012). In cyanobacterial systems, sunlight is utilized to fix carbon in the Calvin–Benson–Bassham cycle and produce intermediates that enter and are consumed in glycolysis. Thus, the need to grow and convert biomass into sugars is bypassed.
Lan and Liao (2011) introduced a 1-butanol pathway into S. elongatus PCC 7924 by integrating atoB (E. coli) and adhE2 (C. acetobutylicum), under Ptrc control (Brosius et al., 1985), into neutral site I (NSI; Bustos and Golden, 1992), and then integrating hbd, crt (C. acetobutylicum), and ter (T. denticola), under PLlacO1 control (Lutz, 1997), into neutral site II (NSII; Andersson et al., 2000) on the genome (Lan and Liao, 2011). In this strain, 3.04 mg/L 1-butanol accumulated after 7 days. In an improved strain containing gene rearrangement where a His-tagged ter was integrated, under Ptrc control, into NSI, and atoB, adhE2, hbd, and crt genes were integrated, under PLlacO1 control, into NSII, 13.16 mg/L 1-butanol was produced. Cultures in screw-capped flasks and air-bubbling cultures, however, did not produce 1-butanol. To determine whether light or oxygen halted 1-butanol production, various culturing conditions (i.e., oxic/anoxic, light/dark) in BG-11 media supplemented with NaHCO3 was tested. Delving further into oxygen effects, photosystem II was inhibited with 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU; Komenda et al., 2000), in order to prevent generation of oxygen under light. Between 2.5 and 5.5 mg/L 1-butanol was produced under continuous light and addition of varying concentrations of DCMU, indicating that elimination of oxygen is key for 1-butanol production in S. elongatus. Furthermore, experiments to test AdhE2 activity in oxic and anoxic conditions showed similar activities in both conditions, suggesting that low AdhE2 activity may not be related to the anoxic requirement for 1-butanol production. The best strain produced 14.5 mg/L 1-butanol in 7 days while decreasing in cell density (OD730 from 3.9 to 2.4) when incubated in dark anoxic conditions.
Isopropanol Production
Isopropanol production using the CoA-dependent pathway has also been explored. Even though isopropanol has an energy density of only 23.9 MJ/L, it has a lower hygroscopicity relative to ethanol. With these properties, isopropanol may be used as a gasoline or diesel additive.
Hanai et al. (2007) constructed the isopropanol pathway in E. coli, modeling it after the one in C. beijerinckii that converts acetyl-CoA via acetone into isopropanol. First, an acetyl-CoA acetyltransferase condenses two acetyl-CoA molecules into acetoacetyl-CoA. Next, an acetoacetyl-CoA transferase is used to form acetoacetate. Then, an acetoacetate decarboxylase is used to produce acetone and CO2. Finally, a secondary alcohol dehydrogenase (ADH) reduces acetone into isopropanol. Five combinations of the following genes were overexpressed for production under control of the PLlacO1 promoter (Lutz, 1997): atoB (E. coli) or thl (C. acetobultylicum), ctfAB (C. acetobutylicum) or atoAD (E. coli), adc (C. acetobutylicum), and adh (C. beijerinckii or Thermoanaerobacter brockii). All strains [except one where overexpression of an adh (T. brockii), encoding a primary–secondary alcohol (SADH), produced 1.1 g/L acetone] produced comparable isopropanol titers, ranging between 2.1 and 2.7 g/L, with minute quantities of acetone and ethanol. The strain that produced 2.7 g/L isopropanol overexpressed adh in a p15A origin (Chang and Cohen, 1978) plasmid, while thl, atoAD, and adc were overexpressed in a colE origin (Kahn et al., 1979) plasmid. Crude extract assays measured an SADH (C. beijerinckii) activity nearly 13 times higher than that of T. brockii, thus explaining the stark difference in isopropanol to acetone ratio. Further experiments in SD-8 media (Luli and Strohl, 1990), with varied starting amounts of glucose concentration resulted in isopropanol production peaking at 4.9 g/L after 30.5 h. Furthermore, maximum productivity was reported to be 0.41 g/L/h between 3 and 9.5 h at 43.5% theoretical yield.
Inokuma et al. (2010) further improved isopropanol production in E. coli by optimizing culture and media conditions, as well as using gas stripping for product recovery. First, pH-maintained, intermittent-fed fermentation in SD-8 media (Luli and Strohl, 1990) supplemented with glucose resulted in 40.1 g/L isopropanol after 60 h at 73.2% theoretical yield, and a production rate of 0.74 g/L/h between 6 and 60 h. To further increase titer, isopropanol was recovered using a gas-stripping system, where an optimized amount of air was bubbled through the culture in order to remove, transfer, and collect isopropanol and other volatile compounds into two collection bottles. By fusing fed-batch fermentation to the gas-stripping setup, isopropanol concentration increased to 79.6 g/L after 144 h, with a concentration of 19.8 g/L in the culture flask. However, cell density and production rate decreased over time. To alleviate this decrease, concentrated media was intermittently added in order to keep a fresh stock of nutrients available for consumption. Using this approach, titers reached 143 g/L after 240 h at 67.4% maximum theoretical yield, and demonstrated the potential for industrial fermentative production of isopropanol.
1-Hexanol Production
Dekishima et al. (2011) expanded the 1-butanol pathway in E. coli in order to produce 1-hexanol. The pathway constructed by Shen et al. (2011) was used as the starting point, where atoB (E. coli), adhE2 (C. acetobutylicum), hbd (C. acetobutylicum), and crt (C. acetobutylicum) were overexpressed in a plasmid under control of the PLlacO1 promoter (Lutz, 1997) with a colE origin (Kahn et al., 1979) of replication, and ter [Eg-ter (Euglena gracilis) or Td-ter (T. denticola)] was overexpressed in a plasmid under control of the PLlacO1 promoter and colA origin (Zverev and Khmel, 1985) to convert two molecules of acetyl-CoA into butyryl-CoA. Finally, butyryl-CoA is subsequently reduced by AdhE2 (C. acetobutylicum) into 1-butanol. In order to extend the potential alcohol by two carbons, bktB (R. eutropha; encodes a 2-ketothiolase) was overexpressed in a plasmid under control of the PLlacO1 promoter and colA origin to add an acetyl group to butyryl-CoA to generate 3-keto-hexanoyl-CoA. Similar to synthesis of 1-butanol, 3-keto-hexanoyl-CoA can then be converted to 1-hexanol using the same enzymes used to convert acetoacetyl-CoA. First, Hbd converts 3-keto-hexanoyl-CoA into 3-hydroxyhexanoyl-CoA, allowing Crt to catalyze the generation of hexenoyl-CoA. Next, Ter is used to make hexanoyl-CoA. Finally, alcohol/aldehyde dehydrogenase AdhE2 cleaves the CoA group from hexanoyl-CoA and reduces it into 1-hexanol in two steps. An E. coli ΔadhE ΔldhA ΔfrdBC strain was used to increase the NADH available to drive the pathway forward. In this strain where only one of the trans-enoyl-CoA reductases, Td-ter, was overexpressed, there was no 1-hexanol detected, but when both Td-ter and Eg-ter were co-expressed, 23 mg/L 1-hexanol after 68 h was produced in TB supplemented with glucose. To further attempt to increase titer, a strain containing ΔadhE ΔldhA ΔfrdBC Δpta knockouts was first constructed. 1-Hexanol titer using this strain was 27 mg/L after 68 h. Then, fdh was overexpressed in a plasmid under control of the PLlacO1 promoter and pSC101⋆ origin (Stoker et al., 1982) in this knockout strain in order to increase the amount of NADH. In a pH-maintained, anaerobic production where the culture was replenished with fresh TB media every 24 h, production of 1-hexanol at 48 h increased to 47 mg/L. Although 1-hexanol concentrations did not reach those of 1-butanol in this system, further improvements to increase enzymatic activity toward C6 relative to C4 intermediates can be done.
C5–C10 Alcohol Production Via β-Oxidation Pathway
In contrast to constructing a 1-butanol pathway by overexpressing genes from C. acetobutylicum, Gulevich et al. (2012) proposed the use of the native aerobic fatty acid β-oxidation cycle commonly found in bacteria and yeast. Normally, these species use this cycle to break down (Cn+2)-acyl-CoA molecules into acetyl-CoA and (Cn)-acetyl CoA molecules using four genes (Fujita et al., 2007). Since metabolic pathways are essentially reversible, however, 1-butanol could be made after one turn of this cycle (with chain elongation occurring by addition of acetyl-CoA to another thioester). That is, an acetyl-CoA C-acetyltransferase would first condense two acetyl-CoA molecules into acetoacetyl-CoA. Next, a 3-hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase would reduce the β-keto acid into 3-hydroxybutyryl-CoA, and subsequently to crotonyl-CoA. Then, crotonyl-CoA would be hydrogenated to a butyryl-CoA, where a native CoA-dependent alcohol/aldehyde dehydrogenase would then convert it into 1-butyraldehyde then 1-butanol in two steps. In an E. coli strain containing one chromosomal copy of each gene needed for 1-butanol synthesis (atoB, fadA, fadB, fadE, and adhE), an artificial genetic element, Ptrc-ideal-4-SDφ10 (containing a strong LacI-dependent promoter as well as a strong ribosome binding site from phage T7), replaced the native regulatory regions of each of the genes in the proposed pathway. When production was tested in semi-aerobic conditions in M9 media (Miller, 1992) containing glucose and glycerol or LB-containing glucose, 334 and 897 mg/L of 1-butanol was produced, respectively. In addition, 1-butanol in LB was higher, but amount of ethanol produced was the same, and the amount of acetic acid synthesized was lower. On the other hand, in comparison to semi-aerobic conditions, titer was lower in LB (674 mg/L), but higher in M9 media (615 mg/L) under anaerobic conditions.
Dellomonaco et al. (2011) also demonstrated reversal of the β-oxidation cycle to generate higher-chain alcohols and other chemicals. In the presence of a non-fatty acid carbon source, constitutive expression of the fad–ato regulon reversed the beta oxidation cycle (Dellomonaco et al., 2010). To increase efficiency, repressive regulation of the beta oxidation cycle in the presence of glucose was avoided by using a cAMP-independent mutant (crp⋆) to replace the native crp gene, and by deleting the arcA gene in order to eliminate any β-oxidation-encoding operon repression by ArcA (aerobic respiratory control A). In a strain containing these initial genomic modifications in pSC101⋆ origin (Stoker et al., 1982) plasmids under the control of the PLtetO1 promoter, production of 1-butanol was achieved by overexpression of native fucO, encoding a L-1,2-propanediol oxidoreductase, and yqeF, encoding an acyl transferase, as well as deletion of the following genes involved in competing pathways: adhE (lactate, ethanol, succinate), pta (acetate), and frdA (succinate), eutE and yqhD (ethanol). Production in M9 media supplemented with glucose synthesized 2.2 g/L 1-butanol after 24 h at a 28% theoretical yield. When production moved to a bioreactor, however, titer rose to 14 g/L after 36 h at 33% theoretical maximum and a rate of 2 g 1-butanol/g cell dry weight/h. In this pathway, many CoA-thioester intermediates produced could also be converted to the corresponding alcohols and carboxylic acids. By simultaneously overexpressing tesA or tesB encoding a thioesterase and yqeF encoding an acyltransferase, and deleting of both fadB encoding an acetyl-CoA C-acetyltransferase and ydiO encoding an acyl-CoA dehydrogenase, 150–500 mg/L amounts of various carboxylic acids were produced. Production of longer even-chain n-alcohols (C6, C8, and C10) was achieved by utilizing native ADHs (EutG, YiaY, and BetA). Lastly, generation of longer odd-chain n-alcohols (C5, C7, and C9) was achieved by overexpressing yiaY, encoding a Fe-containing ADH, and supplementing the media with propionate.
Bokinsky et al. (2011) used consolidated bioprocessing (CBP) (Lynd et al., 2002) to produce three different biofuels, including 1-butanol, using E. coli able to grow on and degrade ionic liquid-pretreated switchgrass (Li et al., 2010a), without the addition of exogenous glycoside hydrolases (Bokinsky et al., 2011). To degrade cellulose, they found a cellulase, when fused with OsmY, capable of being transported out of E. coli. Out of the two OsmY-cellulase fusions, the one that had the highest endocellulase activity was from Bacillus sp. D04. To degrade hemicellulose, a previously found endoxylanase from C. stercorarium, Xyn10B (Steen et al., 2010), was also fused to OsmY. Genes encoding this fusion were under control of a native PcspD promoter in a pSC101⋆ origin (Stoker et al., 1982) plasmid. To further break down the cello- and xylodextrins into glucose, a β-glucosidase from Cellvibrio japonicus, encoded by cel3A, was overexpressed under the control of the native PwrbA promoter to degrade cellobiose, while a xylobiosidase from C. japonicus, encoded by gly43F, was overexpressed under the control of the native PcstA promoter in a p15A origin (Chang and Cohen, 1978) plasmid to degrade xylodextrins. Resulting plasmids expressing cellulose and hemicellulose breakdown were named pCellulose and pXylan, respectively. By overexpressing a CoA-dependent 1-butanol pathway in an E. coli DH1 ΔadhE strain, 28 mg/L 1-butanol was produced in EZ-Rich media (Teknova) and 3.3% w/v ionic liquid-pretreated switchgrass as the only carbon source.
2-Keto Acid Pathways for C3–C8 Alcohols
Amino acid pathways are universal, and have also been harnessed for the ease in which amino acid precursors can be rerouted for production C3–C6 alcohols (Figure 2). The following describes research that emanates from utilization of the last two steps of the Ehrlich pathway (Sentheshanmuganathan and Elsden, 1958) to divert 2-keto acid intermediates to higher-chain alcohols in various hosts.
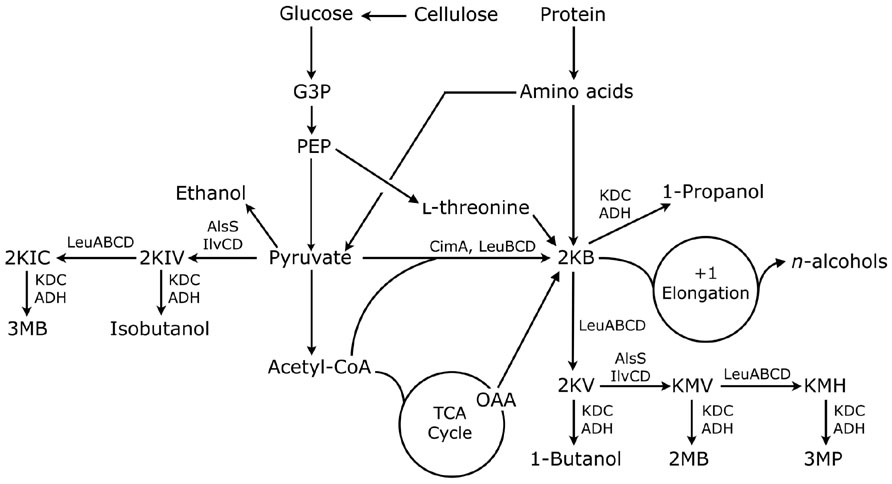
FIGURE 2. Metabolic representation of 2-keto acid pathways for biofuel production. G3P, glyceraldehyde-3-phosphate; PEP, phosphoenolpyruvate; AlsS, acetolactate synthase; IlvC, acetohydroxy acid isomeroreductase; IlvD, dihydroxy-acid dehydratase; LeuA, isopropylmalate synthase; LeuB, isopropylmalate dehydrogenase; LeuCD, isopropylmalate dehydratase; CimA, citramalate synthase; 2KIC, 2-ketoisocaproate; 3MB, 3-methyl-1-butanol; 2KIV, 2-ketoisovalerate; 2KB, 2-ketobutyrate; 2KV, 2-ketovalerate; KMV, 2-keto-3-methylvalerate; 2MB, 2-methyl-1-butanol; KMH, 2-keto-4-methylhexanoate; 3MP, 3-methyl-1-pentanol; OAA, oxaloacetate.
Atsumi et al. (2008b) reported production of several higher-chain alcohols, including isobutanol, 1-butanol, 2-methyl-1-butanol (2MB), 3-methyl-1-butanol (3MB), and 2-phenylethanol, from glucose by engineering the amino acid biosynthetic pathway in E. coli. In the pathway, 2-keto acids are converted to the corresponding aldehyde with a 2-keto acid decarboxylase (KDC) and then to the alcohol using an ADH. First, ADH2 (S. cerevisiae) was overexpressed with genes encoding five different KDCs [Pdc6, Aro10, and Thi3 (S. cerevisiae), Kivd (Lactococcus lactis), and Pdc (C. acetobutylicum)] in colE origin plasmids under control of the PLlacO1 promoter to determine which combination gave the highest longer-alcohol titer. Results indicated that Kivd was the most active and had the largest substrate range, and produced all of the expected alcohols. About 22 g/L isobutanol (86% theoretical max) was produced under microaerobic conditions in 112 h by overexpressing alsS (B. subtilis), ilvC, ilvD, kivd, and ADH2, by introducing ΔadhE, ΔldhA, ΔfrdAB, Δfnr, Δpta, and ΔpflB knockouts in order minimize production from competing pathways. Isobutanol has an energy density of 29.0 MJ/L. In addition, about 541 mg/L 1-propanol and 667 mg/L 1-butanol was produced in M9 media (supplemented with glucose) by the non-native alcohol producing pathway. In this pathway, 2-ketobutyrate (2KB), a precursor to isoleucine, is formed via the threonine pathway, and can be converted to 1-propanol using Kivd and Adh2. On the other hand, 2KB can be extended by one carbon using LeuABCD operon to produce the unnatural amino acid norvaline precursor 2-ketovalerate, which can be converted to 1-butanol using the same KDC and ADH.
Isobutanol Production Via the 2-Keto Acid Pathway
To improve upon the amino acid-based production of isobutanol, Baez et al. (2011) compared aerobic isobutanol production in a 1 L bioreactor connected to a gas-stripping system. Using the engineered isobutanol strain (Atsumi et al., 2008b), 50 g/L isobutanol was produced in the bioreactor after 72 h (68% maximum theoretical yield) under aerobic conditions and 30°C, which is over twice as high as the 22 g/L produced in a shake flask. By-product production (except acetate) was not detectable. Efforts to knockout several acetate-producing pathways (Δack Δpta ΔpoxB), however, did not prevent accumulation of acetate.
Microaerobic conditions have been used to produce isobutanol. For a large-scale production of isobutanol, however, anaerobic production is preferred. A major barrier that must be overcome is NADPH dependency. Since two of the enzymes in the pathway are NADPH-dependent, the pentose phosphate pathway and tricarboxylic acid cycles can regenerate this cofactor only in the presence of oxygen. Under anaerobic conditions, however, NADH is produced as a result of glycolysis. To overcome this cofactor imbalance issue, Bastian et al. (2011) constructed an NADH-dependent pathway by engineering a ketol-acid reductoisomerase IlvC (E. coli) and ADH AdhA (L. lactis). Directed evolution approaches (Romero and Arnold, 2009) were applied to IlvC and AdhA to switch cofactor dependence from NADPH to NADH. In an anaerobic, fully NADH-dependent strain containing AdhARE1 and IlvC6E6-his6, 13.4 g/L isobutanol at 100% theoretical maximum yield was produced in M9 media supplemented with glucose, yeast extract, and trace met als (hereafter modified M9). Overexpression of pntAB, which enabled reversible transfer of a hydride ion between NADH and NADPH, with ilvC and yqhD overcame the cofactor imbalance, but produced at a titer and theoretical yield slightly lower than the strain overexpressing NADH-utilizing engineered IlvC6E6-his6 and AdhARE1.
Another host used for isobutanol production is the cyanobacterium S. elongatus PCC7942, a photosynthetic organism that Atsumi et al. (2009) engineered to convert CO2 and light energy directly into isobutyraldehyde, a precursor to isobutanol. To install the isobutyraldehyde production pathway in S. elongatus, kivd (L. lactis), under Ptrc control, was integrated into NSI (Bustos and Golden, 1992), and alsS (B. subtilis), ilvC, and ilvD (E. coli), under PLlacO1 control, was integrated into NSII (Andersson et al., 2000) of the genome by homologous recombination (Golden et al., 1987). Production in a Roux culture bottle at 30°C and subsequent gas-stripping of isobutyraldehyde resulted in 723 mg/L after 12 days and a production rate of 2,500 μg/L·h. No isobutanol was detected, most likely because the endogenous ADH had no detectable activity toward isobutyraldehyde. One known bottleneck to the pathway is a low activity of ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) in fixing CO2 within the Calvin–Benson–Bassham cycle. To improve isobutyraldehyde production, the rbcLS genes from a related S. elongatus, PCC6301, was inserted downstream from the endogenous rbcLS genes. With an increased Rubisco activity, the resulting strain produced 1.1 g/L isobutyraldehyde after 8 days and a production rate of 6,230 μg/L·h. Isobutanol production was also measured by integrating NADPH-dependent ADH, encoded by yqhD, in addition to the isobutyraldehyde pathway. This strain produced 450 mg/L isobutanol after 9 days.
Isobutanol production in Gram-positive Corynebacterium glutamicum, an industrial amino acid producer (Kinoshita, 1972; Sahm et al., 1996; Wendisch, 2007) has also been achieved. Smith et al. (2010) explored isobutanol production using C. glutamicum because C. glutamicum was chosen for its rapid growth rate and vast range of tools for genetic manipulation. Furthermore, C. glutamicum has been modified to produce L-valine in industrial quantities, and the isobutanol pathway utilizes its precursors. In this system, alsS (B. subtilis), ilvC, ilvD, adhA (C. glutamicum), and kivd (L. lactis), in a Peftu-controlled, NG2 origin (Messerotti et al., 1990) plasmid, were overexpressed in C. glutamicum and resulted in 2.6 g/L isobutanol, 50 mg/L isobutyraldehyde, 0.4 g/L 3MB, as well as other various higher-chain alcohols, after 48 h in CGIII media supplemented with glucose (Menkel et al., 1989). To prevent direct formation of oxaloacetate from pyruvate, the pyc gene (encoding pyruvate carboxylase) was inactivated. In order to inactivate a desired gene in Gram-positive organisms like C. glutamicum, a pK-derived mobilizable cloning vector containing a fragment of the gene is first constructed (Schafer et al., 1994). Then, RP4-mediated conjugation occurs (Haase et al., 1995), where the pK-derived plasmid is first incorporated into the genome, then subsequently disrupted. Essentially, a double cross-over event is carried out. As a result of the Δpyc knockout, isobutanol titer increased to 4.3 g/L. Despite this increase, 2.6 g/L of lactate formed. To inhibit formation of lactate from pyruvate and NADH, an additional knockout, Δldh, which would have encoded lactate dehydrogenase, was introduced. In the highest-producing C. glutamicum Δpyc Δldh knockout strain (containing NG2 origin Peftu::alsS-ilvCD-adhA-kivd), 4.9 g/L isobutanol was produced after 120 h (23% theoretical maximum) using a fermenter.
Li et al. (2011) engineered B. subtilis for production of isobutanol. B. subtilis was a host of interest because it was found to have a higher isobutanol tolerance (2%) than both C. glutamicum and E. coli (1.9- and 3.8-fold, respectively). In a strain containing ΔamyE and a two-plasmid system (P43::ilvD-ilvC-alsS and P43::kivd-adh2), 2.02 g/L isobutanol was produced after 40 h under 37°C and microaerobic conditions. In addition, lower titers of ethanol, 1-phenylethanol, 2MB, and 3MB were produced. Acetate and lactate were also detected in the production culture. To improve titer, batch and fed-batch fermentations in 1 L shake flasks were performed under microaerobic conditions. After 48 h, the fed-batch fermentation in LB media supplemented with glucose, phosphate buffer, and trace metals produced 2.62 g/L isobutanol at a 0.086 g/L/h production rate.
Higashide et al. (2011) used the cellulolytic mesophile C. cellulolyticum to convert crystalline cellulose into isobutanol. C. cellulolyticum uses an extracellular, multi-enzymatic cellulosome to breakdown and consume cellulose and other complex polysaccharides efficiently (Desvaux, 2005). In a strain overexpressing kivd, yqhD, alsS, ilvC, and ilvD under the control of a constitutive ferredoxin C. pasteurianum promoter, 364 mg/L isobutanol was produced in 90 h from cellobiose, while 660 mg/L isobutanol in 9 days was produced from cellulose. Two major setbacks to the successful production of isobutanol in C. cellulolyticum using a CBP platform were (1) lack of an available inducible expression system and the resulting toxicity of gene products, and (2) lack of detectable activity of IlvC and IlvD (due to differences in GC content and codon usage frequencies between E. coli and C. cellulolyticum).
Chen et al. (2011) engineered S. cerevisiae to produce isobutanol. To construct the pathway, an acetolactate synthase, encoded by ILV2 (catalytic subunit) and ILV6 (regulatory subunit), converts two pyruvate molecules into 2-acetolactate, which is subsequently reduced to 2,3-dihydroxyisovalerate by an acetohydroxy acid reductoisomerase, encoded by ILV5. Then, 2-ketoisovalerate (2KIV) is produced using a dihydroxyacid dehydratase, encoded by ILV3. Next, two branched-chain amino acid aminotransferases, encoded by mitochondrial BAT1 and cytosolic BAT2, reversibly converts 2KIV into L-valine. Afterward, isobutanol is formed using pyruvate decarboxylases (Pdc6, 5, 1) and ADHs. In a strain overexpressing ILV2, ILV3, and ILV5 (in plasmids under control of the PGK1 promoter) led to 4.12 mg isobutanol/g glucose when cultivated under aerobic conditions in pH-maintained minimal media supplemented with yeast extract, peptone, dextrose, and 40 g/L glucose. Under anaerobic conditions and minimal media supplemented with 40 g/L glucose, isobutanol yield was 2.13 mg/g glucose in a strain that overexpressed BAT2 in addition to ILV2, ILV3, and ILV5.
To improve upon the isobutanol titer in S. cerevisiae, Kondo et al. (2012) searched for and overexpressed KDC and ADH genes from different sources. The following three KDC enzymes were tested: endogenous phenylpyruvate decarboxylase (Aro10), endogenous 2-ketoisocaproate decarboxylase (Thi3), and 2KIV decarboxylase (Kivd) from L. lactis. In addition, six ADH genes from S. cerevisiae, encoding for Adh1, Adh2, Adh5, Adh6, Adh7, and Sfa1, were overexpressed in plasmids under control of the PGK1 promoter. Different KDC–ADH combinations were used to measure isobutanol levels under additive additions of 8 g/L 2KIV, the precursor to isobutanol. A kivd-ADH6 combination gave the highest isobutanol level at 488 mg/L, and was used for isobutanol production. To reduce ethanol production, ILV2 encoding an acetolactate synthase was overexpressed (catalyzes the first committed step in valine biosynthesis from pyruvate). Furthermore, to prevent acetaldehyde production, pdc1, which encodes an isozyme of the pyruvate dehydrogenase complex, was deleted. This strain was cultured in synthetic dextrose media supplemented with yeast nitrogen base and glucose, and produced 75 mg/L after 72 h. Under microaerobic conditions, titer rose to 143 mg/L after 120 h at a yield of 6.6 mg/g glucose.
1-Propanol and 1-Butanol Production Via the 2-Keto Acid Pathway
Shen and Liao (2008) improved a keto acid-based pathway in E. coli to produce 1-propanol (contains an energy density of 27 MJ/L) and 1-butanol from glucose (Atsumi et al., 2008b; Shen and Liao, 2008). To deregulate threonine synthesis and increase 2-keto acid intermediates for alcohol production, a feedback-resistant ThrAfbr was used. In addition, several competing pathways were deleted to increase flux toward 2-keto acid production. The final strain containing ΔmetA Δtdh ΔilvB ΔilvI ΔadhE and overexpression of thrAfbrBC, ilvA, leuABCD (E. coli), kivd (L. lactis), and ADH2 (S. cerevisiae), in p15A (Chang and Cohen, 1978) or colE (Kahn et al., 1979) origin plasmids and all under the control of the PLlacO1 promoter, produced 2 g/L 1:1 coproduction of 1-butanol and 1-propanol after 72 h in modified M9 media under anaerobic conditions.
Atsumi and Liao (2008) improved production of 1-propanol and 1-butanol by evolving a citramalate synthase (CimA) from the extreme thermophile Methanococcus jannaschii. CimA is a homolog of LeuA that implements a more direct route than the threonine pathway toward 2KB. In addition, bypassing the threonine pathway to generate 2KB (ΔilvA and ΔtdcB) eliminates the liberation of NH3 due to deamination of threonine. CimA catalyzes the addition of an acetyl group from acetyl-CoA onto pyruvate to synthesize citramalate, which then undergoes transformations catalyzed by LeuBCD to produce 2KB. 1-Propanol and 1-butanol can then be produced using the same enzymes expressed by kivd (L. lactis) and ADH2 (S. cerevisiae; Atsumi et al., 2008b). To improve activity of CimA toward pyruvate, several rounds of random mutagenesis and DNA shuffling were used to mutate the cimA gene, and improved variants were screened for using a growth-based selection. That is, in a strain containing ΔilvA and ΔtdcB, isoleucine cannot be produced, and thus the strain cannot grow, unless CimA is active in the cell. The strain containing the evolved cimA, overexpressed in a PLlacO1-controlled, p15A origin (Chang and Cohen, 1978) plasmid, anaerobically produced 2.8 g/L 1-propanol and 393 mg/L 1-butanol after 40 h at 30°C in modified M9 media.
3-Methyl-1-Butanol Production Via the 2-Keto Acid Pathway
Connor and Liao (2008) took advantage of native amino acid biosynthetic pathways to produce 3MB, another higher-chain alcohol with an energy density of 30.5 MJ/L, in E. coli from glucose. To produce 3MB, the ilvIHCD genes were overexpressed to convert pyruvate into 2KIV, a precursor to the amino acid valine, which can be converted to 2-ketoisocaproate, a precursor to the amino acid leucine, by LeuABCD enzymes. Finally, 2-ketoisocaproate can then be redirected by Kivd and Adh2 to produce 3MB. The initial strain produced 56 mg/L 3MB after 18 h in modified M9 media under anaerobic conditions at 30°C. To increase 3MB titer, several modifications were introduced. First, alsS (B. subtilis), previously shown to increase isobutanol production via valine synthesis, was overexpressed in place of ilvIH in a pSC101 origin (Stoker et al., 1982) plasmid under PLlacO1 control (Lutz, 1997). To increase production of 2KIV, the leuABCD genes were overexpressed in a colE origin (Kahn et al., 1979), PLlacO1-controlled plasmid. Furthermore, genes expressing enzymes involved in competing pathways were deleted (ΔadhE ΔfrdBC ΔldhA Δpta Δfnr ΔpflB ΔilvE ΔtyrB) from the genome. Lastly, a feedback-insensitive LeuAfbr (Gusyatiner et al., 2002) was used to deregulate leucine synthesis and increase flux toward 2-ketoisocaproate. The final strain produced 1.28 g/L 3MB (0.11 g/g yield) and 0.2 g/L isobutanol after 28 h under anaerobic conditions.
Expanding upon the 3MB pathway (Connor and Liao, 2008), Connor et al. (2010) performed several rounds of random whole-cell mutagenesis with N-methyl-N′-nitro-N-nitrosoguanidine and selection with amino acid analog 4-aza-d,L-leucine (AZL) to produce a 3MB hyper-producer. If the cell can outcompete production of this L-leucine analog by producing much more natural L-leucine – in turn increasing production of 3MB precursor 2-ketoisocaproate – then the cell will circumvent the occurrence of fatal incorporation of AZL into polypeptides. Therefore, selecting for mutants able to adapt to the addition of toxic levels of AZL by elevating production of natural amino acids should increase 3MB titer. Using a mutant able to outcompete production of AZL, 4.4 g/L 3MB was produced with a theoretical yield of 30% after 36 h in under anaerobic conditions. In addition to mutagenesis and selection for higher 3MB producers, a two-phase fermentation strategy with oleyl alcohol was implemented in order to separate 3MB from the production phase and minimize toxic concentrations of 3MB, thus extending production lifetime of the engineered 3MB hyper-producer. Using this approach with a mutated 3MB strain resulted in a titer of 9.6 g/L at 33% theoretical maximum after 60 h. Further analysis of the two-phase culture revealed that 90% of 3MB and 80% of isobutanol produced was present in the oleyl alcohol phase.
2-Methyl-1-Butanol Production Via the 2-Keto Acid Pathway
Cann and Liao (2008) demonstrated the production of 2MB, a branched-chain alcohol with an energy density of 30.5 MJ/L, via the precursor for isoleucine, 2-keto-3-methylvalerate (KMV), in E. coli from glucose. In this study, different isozymes of key enzymes along the constructed 2MB pathway were tested and selected based on 2MB versus 1-propanol titer. Both 2MB and 1-propanol are derivatives of 2-keto acids, and involve the use of a broad-substrate range KDC to convert the corresponding 2-keto acid into an aldehyde, then reduce it into the alcohol using an ADH. Further, an acetohydroxy acid synthase (AHAS) catalyzes the first step in isoleucine synthesis, where 2KB and pyruvate are condensed to make 2-aceto-2-hydroxybutyrate, a precursor to 2MB. Therefore, an ideal AHAS for 2MB production was chosen based on how well it outcompeted KDC for 2KB. If the AHAS outcompetes KDC, then 2MB production would be favored over 1-propanol production. First, several AHAS isozymes were tested for rate of conversion of 2KB to 2MB versus 1-propanol. It was found that AHAS II (encoded by ilvGM) from Salmonella typhimurium produced the highest amount of 2MB and minimal 1-propanol in modified M9 media supplemented with 8 g/L 2KB. Next, various threonine deaminase isozymes were overexpressed in p15A origin (Chang and Cohen, 1978) plasmids under PLlacO1 control to analyze how well supplied L-threonine is consumed and converted into 2MB and 1-propanol. The strain containing the best threonine deaminase coded by ilvA (C. glutamicum) converted about 88% of supplied L-threonine into 2MB and 1-propanol. Then, the threonine pathway was overexpressed (thrABC). Lastly, several combinations of ΔmetA Δtdh ΔilvB ΔilvI ΔleuA and ΔilvE deletions were tested for 2MB production. The best combination was determined to be ΔmetA and Δtdh, which eliminated carbon flux toward methionine and consumption of threonine, respectively. The resulting E. coli strain produced, from glucose, 1.25 g/L 2MB (44% theoretical maximum) in 24 h at 30°C under anaerobic conditions.
C6–C8 Alcohol Production Via the 2-Keto Acid Pathway
Zhang et al. (2008) expanded the 2-keto acid-based pathway for the production of higher-chain alcohols (C > 5) by using structure-based protein engineering to expand the substrate range of 2-isopropylmalate synthase (LeuA) and 2KIV decarboxylase (Kivd) from L. lactis, which are both already known to be promiscuous enzymes. First, Kivd was engineered to be more selective toward 2-keto-4-methylhexanoate, a non-natural precursor of 3-methyl-1-pentanol. To do this, a sequence alignment was done on indolepyruvate decarboxylase (IPDC) of Enterobacter cloacae and pyruvate decarboxylase (PDC) from Zymomonas mobilis, each having a 40 and 31% homology to Kivd, respectively. Investigation of residues within the keto acid-binding pocket of each enzyme led to an F381L/V461A Kivd variant. To engineer LeuA, sequence alignment of LeuA from S. typhimurium and Mycobacterium tuberculosis showed 92 and 21% homology to that of E. coli, respectively. In addition to the G462D point mutation to deregulate valine synthesis, S139G mutation was inserted to accommodate the extra methyl group (S)-2-keto-3-methylvalerate contains. In an E. coli strain containing both Kivd and LeuA mutants, 793.5 mg/L 3-methyl-1-pentanol was produced in modified M9 media. Additional point mutations into smaller amino acids in Kivd were included to accommodate larger keto acid intermediates. Specifically, the addition of an N167A point mutation to the double mutant allowed for the production of 51.9 mg/L 4-methyl-1-hexanol. Moreover, incorporation of H97A to the triple mutant resulted in production of 57.3 mg/L 4-methyl-1-hexanol, as well as 22.0 mg/L 5-methyl-1-heptanol.
Marcheschi et al. (2012) modified LeuA further with structure-based protein engineering to increase its promiscuity toward larger 2-keto acids. These larger 2-keto acids, in turn, can be directed toward higher-chain alcohol production (C > 5). Furthermore, quantum mechanical modeling and protein–substrate complex modeling were used to simulate and analyze the different transition states of the carbon–carbon bond forming steps that are catalyzed by candidate LeuA variants (LeuA⋆) in order to determine the best conformers. Using this approach, mutants that cater to a 2-keto acid of interest can be predicted, whether it contains an aromatic, linear-chained, or branched-chain group. In a threonine hyperproducer strain containing a ΔrhtA (encoding threonine transporter gene), and three plasmid [with pSC101 (Stoker et al., 1982), p15A (Chang and Cohen, 1978), or colE (Kahn et al., 1979) replication origin], PLlacO1-controlled (Lutz, 1997) overexpression of thrABC (threonine pathway genes), ilvA (B. subtilis) leuA⋆BCD, kivd (F381L/V461A, L. lactis), and ADH6 (S. cerevisiae), production of C4–C8 alcohols in modified M9 media was measured. There were four notable LeuA⋆ variants. LeuA⋆ containing S139G/G462D produced 993 mg/L and 2.22 g/L 1-butanol and 1-pentanol, respectively. LeuA⋆ H97A/S139G/G462D produced 302 mg/L 1-hexanol. Lastly, LeuA⋆ H97A/S139G/P169A/G462D produced 80 and 2 mg/L 1-heptanol and 1-octanol, respectively.
Protein Conversion to C4–C5 Alcohols
Apart from the use of glucose as a feedstock for biofuel production, Huo et al. (2011) explored the use of protein biomass as a potential feedstock. E. coli was engineered to generate protein hydrolysates in order to breakdown proteins in three exogenous transamination and deamination cycles from bacteria, yeast, and microalgae into amino acids, which can then be converted to the corresponding 2-keto acid intermediates, and finally into C2–C6 alcohols by mimicking the Ehrlich pathway. Any reduced nitrogen, i.e., NH3, produced is excreted irreversibly from the cell, and can be collected and recycled as fertilizer. In addition, leftover amino acids can be used as raw material for various chemicals and pharmaceutical intermediates, or as a supplement for animal feedstocks. The final strain [YH19ΔglnAΔgdhAΔlsrA overexpressing alsS, ilvC, ilvD, and avtA in a p15A origin (Chang and Cohen, 1978), PLlacO1-controlled (Lutz, 1997) plasmid, kivD, yqhD, leuDH, and ilvE, in a colE origin (Kahn et al., 1979), PLlacO1-controlled plasmid, and sdaB and ilvA in a pSC101 origin (Stoker et al., 1982), PLlacO1-controlled plasmid] produced 4 g/L C4 and C5 alcohols at 56% theoretical yield from amino acids.
Isopentenol Production Via the Mevalonate Isoprenoid Pathway
Isoprenoids have a vast range of uses, including for nutraceuticals, therapeutics, polymers, and fragrances. They are hydrocarbons that stem from isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) precursors, which are produced from acetyl-CoA or pyruvate and glyceraldehyde-3-phosphate using the mevalonate (Goldstein and Brown, 1990) or methylerythritol (Lichtenthaler, 2000) pathways, respectively. The mevalonate isoprenoid pathway has been engineered and exploited for production of isopentenol from acetyl-CoA.
To produce isopentenol from isoprenoid precursors IPP and DMAP (Figure 3), Withers et al. (2007) first used a growth-based phenotype to screen a library of isoprene synthases from B. subtilis 6051 based on ability to minimize toxic levels of prenyl diphosphates by converting them into prenyl alcohols. One of two genes isolated from a pool of 19,000 clones, nudF, was overexpressed in a colE origin (Kahn et al., 1979) plasmid under control of a native Bacillus promoter in conjunction with the mevalonate pathway plasmids pMevT (p15 origin) and pMBI (Bbr origin) in the previously optimized E. coli DH1 strain (Martin et al., 2003), and produced 110 mg/L isopentenol after 43 h at 30°C under aerobic conditions in 250 mL shake flasks containing M9 media supplemented with 3-(N-morpholino)propanesulfonic acid, glucose, and yeast extract.
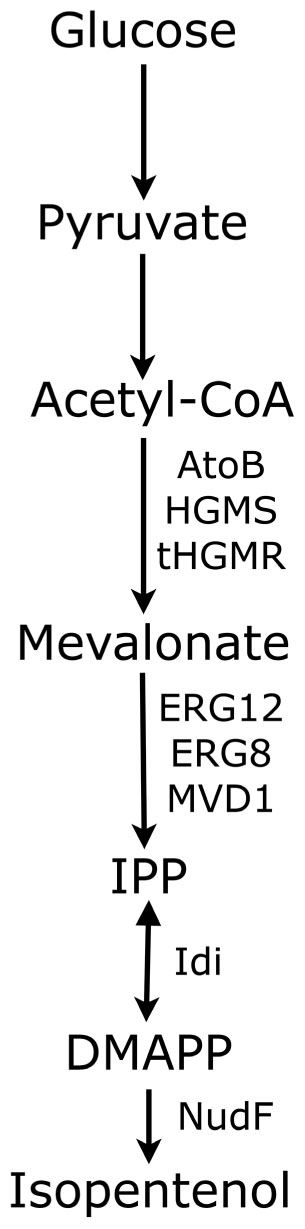
FIGURE 3. Metabolic representation of mevalonate isoprenoid pathway for biofuel production. AtoB, acetyl-CoA thiolase; HGMS, HMG-CoA synthase; tHGMR, truncated HMG-CoA reductase; ERG12, mevalonate kinase; ERG8, phosphomevalonate kinase; IPP, isopentenyl diphosphate; Idi, IPP isomerase; DMAP, dimethylallyl diphosphate; NudF, ADP-ribose pyrophosphatase.
Conclusion
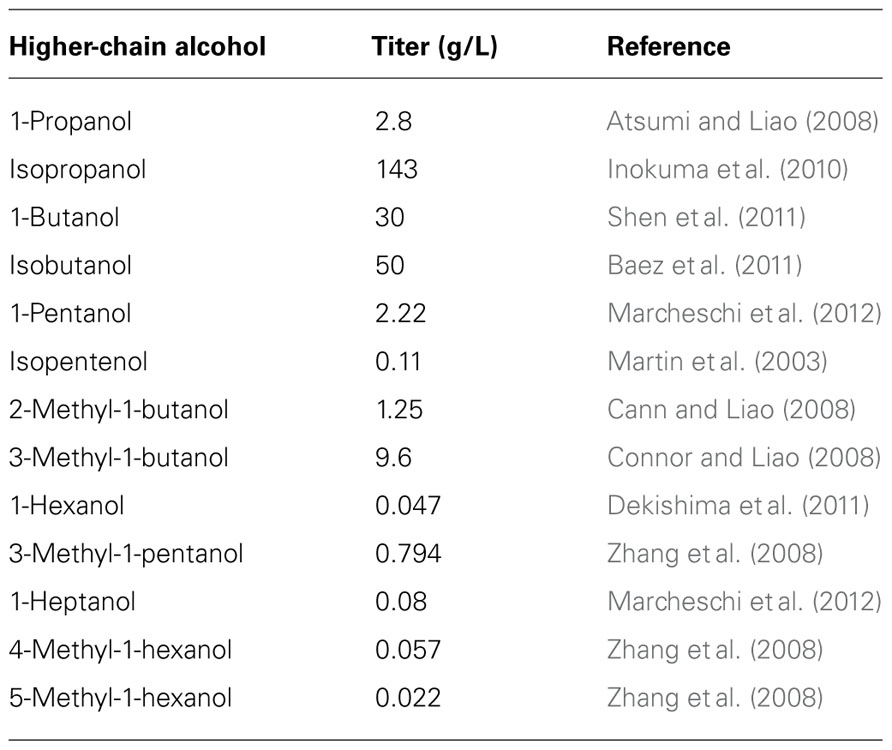
The evolving field of synthetic biology has helped widen the range of modular possibilities in which cell system and behavior can be modified, and will continue to be an influence in the development and improvement of biological methods for biofuel production. In recent years, there has been a growing number of biotech companies that are applying these methods in order to produce higher-chain alcohols commercially. Namely, both Gevo and Butamax have industrialized isobutanol production platforms. But as titers in an engineered host continue to elevate (Table 1), and experimental yields start to match up with theoretical maximums, the resulting increase in productivity comes with a price.
One prominent barrier is the toxic accumulation of the target molecule that stunts cell fitness during production, which, in turn, can hinder a newly engineered strain from producing at industrial-relevant titers. Thus, increasing the product tolerance of, as well as simultaneous product removal from, a host is desirable. Fortunately, there are several approaches that have been developed to help mitigate toxicity levels during the production phase (Zheng et al., 2009; Atsumi et al., 2010; Ezeji et al., 2010; Dunlop, 2011). There is, however, much to be elucidated about the convoluted physiological effects that occur in a host when exposed to elevated levels of a final product.
Development of a process to breakdown biomass effectively and efficiently is also vital to decreasing production time and costs. Until then, CBP is yet another approach worth considering in increasing titer, yield, and productivity. A unified production platform in which biomass can be broken down efficiently and subsequently converted into fuel can be practical, especially if paired with product removal methods. Meanwhile, development of a production platform that uses sunlight and CO2 directly, such as cyanobacteria and microalgae is well worth the effort, and is yet another approach that should be considered in biofuel research. Especially, this approach circumvents the need for a biomass source, and can be cultivated in non-arable areas, i.e., oceans and deserts.
The race to reach a renewable fuel production level of 36 billion gallons by 2022 continues as the production of advanced biofuels gains momentum. For 2012, the Renewable Fuel Standard (RFS2) has established a goal to produce 15.2 billion gallons of biofuels, which already approaches half the mandate for 2022. Although the process to become independent from petroleum-derived fuels is still in its initial stages, efforts to increase synergy between all aspects of strain design, development, optimization, and execution for renewable higher-chain alcohols and other liquid fuel production in non-native hosts will be a great aid in leading the way toward a permanent turn of tables.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported partly by NSF grant 1066182.
References
Agapakis, C. M., and Silver, P. A. (2009). Synthetic biology: exploring and exploiting genetic modularity through the design of novel biological networks. Mol. Biosyst. 5, 704–713.
Alper, H., and Stephanopoulos, G. (2009). Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat. Rev. Microbiol. 7, 715–723.
Amberg, D. C., Burke, D. J., and Strathern, J. N. (2005). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. New York: Cold Spring Harbor Laboratory Press.
Ami, D., Natalello, A., Schultz, T., Gatti-Lafranconi, P., Lotti, M., Doglia, S. M., and De Marco, A. (2009). Effects of recombinant protein misfolding and aggregation on bacterial membranes. Biochim. Biophys. Acta 1794, 263–269.
Andersson, C. R., Tsinoremas, N. F., Shelton, J., Lebedeva, N. V., Yarrow, J., Min, H., and Golden, S. S. (2000). Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 305, 527–542.
Atsumi, S., Cann, A. F., Connor, M. R., Shen, C. R., Smith, K. M., Brynildsen, M. P., Chou, K. J., Hanai, T., and Liao, J. C. (2008a). Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10, 305–311.
Atsumi, S., Hanai, T., and Liao, J. C. (2008b). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89.
Atsumi, S., Higashide, W., and Liao, J. C. (2009). Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 27, 1177–1180.
Atsumi, S., and Liao, J. C. (2008). Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Appl. Environ. Microbiol. 74, 7802–7808.
Atsumi, S., Wu, T. Y., Machado, I. M., Huang, W. C., Chen, P. Y., Pellegrini, M., and Liao, J. C. (2010). Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 6, 449.
Baez, A., Cho, K. M., and Liao, J. C. (2011). High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl. Microbiol. Biotechnol. 90, 1681–1690.
Baneyx, F., and Mujacic, M. (2004). Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 22, 1399–1408.
Bastian, S., Liu, X., Meyerowitz, J. T., Snow, C. D., Chen, M. M., and Arnold, F. H. (2011). Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab. Eng. 13, 345–352.
Bennett, G. N., and Rudolph, F. B. (1995). The central metabolic pathway from acetyl-CoA to butyryl-coA in Clostridium acetobutylicum. FEMS Microbiol. Rev. 17, 241–249.
Bokinsky, G., Peralta-Yahya, P. P., George, A., Holmes, B. M., Steen, E. J., Dietrich, J., Soon Lee, T., Tullman-Ercek, D., Voigt, C. A., Simmons, B. A., and Keasling, J. D. (2011). Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 19949–19954.
Bond-Watts, B. B., Bellerose, R. J., and Chang, M. C. (2011). Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat. Chem. Biol. 7, 222–227.
Brosius, J., Erfle, M., and Storella, J. (1985). Spacing of the -10 and -35 regions in the tac promoter. Effect on its in vivo activity. J. Biol. Chem. 260, 3539–3541.
Bustos, S. A., and Golden, S. S. (1992). Light-regulated expression of the psbD gene family in Synechococcus sp. strain PCC 7942: evidence for the role of duplicated psbD genes in cyanobacteria. Mol. Gen. Genet. 232, 221–230.
Cann, A. F., and Liao, J. C. (2008). Production of 2-methyl-1-butanol in engineered Escherichia coli. Appl. Microbiol. Biotechnol. 81, 89–98.
Chang, A. C., and Cohen, S. N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134, 1141–1156.
Chen, X., Nielsen, K. F., Borodina, I., Kielland-Brandt, M. C., and Karhumaa, K. (2011). Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol. Biofuels 4, 21.
Connor, M. R., and Atsumi, S. (2010). Synthetic biology guides biofuel production. J. Biomed. Biotechnol. 2010, 541698.
Connor, M. R., Cann, A. F., and Liao, J. C. (2010). 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation. Appl. Microbiol. Biotechnol. 86, 1155–1164.
Connor, M. R., and Liao, J. C. (2008). Engineering of an Escherichia coli strain for the production of 3-methyl-1-butanol. Appl. Environ. Microbiol. 74, 5769–5775.
Dekishima, Y., Lan, E. I., Shen, C. R., Cho, K. M., and Liao, J. C. (2011). Extending carbon chain length of 1-butanol pathway for 1-hexanol synthesis from glucose by engineered Escherichia coli. J. Am. Chem. Soc. 133, 11399–11401.
Dellomonaco, C., Clomburg, J. M., Miller, E. N., and Gonzalez, R. (2011). Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals. Nature 476, 355–359.
Dellomonaco, C., Rivera, C., Campbell, P., and Gonzalez, R. (2010). Engineered respiro-fermentative metabolism for the production of biofuels and biochemicals from fatty acid-rich feedstocks. Appl. Environ. Microbiol. 76, 5067–5078.
Desvaux, M. (2005). Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia. FEMS Microbiol. Rev. 29, 741–764.
Dos Santos, V. A., Heim, S., Moore, E. R., Stratz, M., and Timmis, K. N. (2004). Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6, 1264–1286.
Dunlop, M. J. (2011). Engineering microbes for tolerance to next-generation biofuels. Biotechnol. Biofuels 4, 32.
Earl, A. M., Losick, R., and Kolter, R. (2008). Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16, 269–275.
Elvin, C. M., Thompson, P. R., Argall, M. E., Hendry, P., Stamford, N. P., Lilley, P. E., and Dixon, N. E. (1990). Modified bacteriophage lambda promoter vectors for overproduction of proteins in Escherichia coli. Gene 87, 123–126.
Ezeji, T., Milne, C., Price, N. D., and Blaschek, H. P. (2010). Achievements and perspectives to overcome the poor solvent resistance in acetone and butanol-producing microorganisms. Appl. Microbiol. Biotechnol. 85, 1697–1712.
Ezeji, T. C., Qureshi, N., and Blaschek, H. P. (2007). Bioproduction of butanol from biomass: from genes to bioreactors. Curr. Opin. Biotechnol. 18, 220–227.
Fischer, C. R., Klein-Marcuschamer, D., and Stephanopoulos, G. (2008). Selection and optimization of microbial hosts for biofuels production. Metab. Eng. 10, 295–304.
Fortman, J. L., Chhabra, S., Mukhopadhyay, A., Chou, H., Lee, T. S., Steen, E., and Keasling, J. D. (2008). Biofuel alternatives to ethanol: pumping the microbial well. Trends Biotechnol. 26, 375–381.
Fujita, Y., Matsuoka, H., and Hirooka, K. (2007). Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66, 829–839.
Georgiou, G., and Valax, P. (1996). Expression of correctly folded proteins in Escherichia coli. Curr. Opin. Biotechnol. 7, 190–197.
Gheshlaghi, R., Scharer, J. M., Moo-Young, M., and Chou, C. P. (2009). Metabolic pathways of clostridia for producing butanol. Biotechnol. Adv. 27, 764–781.
Golden, S. S., Brusslan, J., and Haselkorn, R. (1987). Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 153, 215–231.
Goldstein, J. L., and Brown, M. S. (1990). Regulation of the mevalonate pathway. Nature 343, 425–430.
Gronenborn, B. (1976). Overproduction of phage lambda repressor under control of the lac promotor of Escherichia coli. Mol. Gen. Genet. 148, 243–250.
Gulevich, A. Y., Skorokhodova, A. Y., Sukhozhenko, A. V., Shakulov, R. S., and Debabov, V. G. (2012). Metabolic engineering of Escherichia coli for 1-butanol biosynthesis through the inverted aerobic fatty acid beta-oxidation pathway. Biotechnol. Lett. 34, 463–469.
Gusyatiner, M. M., Lunts, M. G., Kozlov, Y. I., Ivanovskaya, L. V., and Voroshilova, E. B. (2002). DNA coding for mutant isopropylmalate synthase L-leucine producing microorganism and method for producing L-leucine. U.S. Patent 6, 403, 342.
Haase, J., Lurz, R., Grahn, A. M., Bamford, D. H., and Lanka, E. (1995). Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J. Bacteriol. 177, 4779–4791.
Haldimann, A., Daniels, L. L., and Wanner, B. L. (1998). Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J. Bacteriol. 180, 1277–1286.
Hanai, T., Atsumi, S., and Liao, J. C. (2007). Engineered synthetic pathway for isopropanol production in Escherichia coli. Appl. Environ. Microbiol. 73, 7814–7818.
Higashide, W., Li, Y., Yang, Y., and Liao, J. C. (2011). Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl. Environ. Microbiol. 77, 2727–2733.
Huberman, J. A., Spotila, L. D., Nawotka, K. A., El-Assouli, S. M., and Davis, L. R. (1987). The in vivo replication origin of the yeast 2 microns plasmid. Cell 51, 473–481.
Huo, Y. X., Cho, K. M., Rivera, J. G., Monte, E., Shen, C. R., Yan, Y., and Liao, J. C. (2011). Conversion of proteins into biofuels by engineering nitrogen flux. Nat. Biotechnol. 29, 346–351.
Inokuma, K., Liao, J. C., Okamoto, M., and Hanai, T. (2010). Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. J. Biosci. Bioeng. 110, 696–701.
Jones, D. T., and Woods, D. R. (1986). Acetone-butanol fermentation revisited. Microbiol. Rev. 50, 484–524.
Kahn, M., Kolter, R., Thomas, C., Figurski, D., Meyer, R., Remaut, E., and Helinski, D. R. (1979). Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol. 68, 268–280.
Khalil, A. S., and Collins, J. J. (2010). Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367–379.
Komenda, J., Koblizek, M., and Prasil, O. (2000). Characterization of processes responsible for the distinct effect of herbicides DCMU and BNT on photosystem II photoinactivation in cells of the cyanobacterium Synechococcus sp. PCC 7942. Photosynth. Res. 63, 135–144.
Kondo, T., Tezuka, H., Ishii, J., Matsuda, F., Ogino, C., and Kondo, A. (2012). Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae. J. Biotechnol. 159, 32–37.
Krivoruchko, A., Siewers, V., and Nielsen, J. (2011). Opportunities for yeast metabolic engineering: lessons from synthetic biology. Biotechnol. J. 6, 262–276.
Lan, E. I., and Liao, J. C. (2011). Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 13, 353–363.
Lee, S. K., Chou, H., Ham, T. S., Lee, T. S., and Keasling, J. D. (2008). Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19, 556–563.
Li, C., Knierim, B., Manisseri, C., Arora, R., Scheller, H. V., Auer, M., Vogel, K. P., Simmons, B. A., and Singh, S. (2010a). Comparison of dilute acid and ionic liquid pretreatment of switchgrass: biomass recalcitrance, delignification and enzymatic saccharification. Bioresour. Technol. 101, 4900–4906.
Li, H., Cann, A. F., and Liao, J. C. (2010b). Biofuels: biomolecular engineering fundamentals and advances. Annu. Rev. Chem. Biomol. Eng. 1, 19–36.
Li, S., Wen, J., and Jia, X. (2011). Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression. Appl. Microbiol. Biotechnol. 91, 577–589.
Lichtenthaler, H. K. (2000). Non-mevalonate isoprenoid biosynthesis: enzymes, genes and inhibitors. Biochem. Soc. Trans. 28, 785–789.
Luli, G. W., and Strohl, W. R. (1990). Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 56, 1004–1011.
Lutke-Eversloh, T., and Bahl, H. (2011). Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Curr. Opin. Biotechnol. 22, 634–647.
Lutz, R. (1997). Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210.
Lynd, L. R., Weimer, P. J., Van Zyl, W. H., and Pretorius, I. S. (2002). Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577.
Machado, I. M. P., and Atsumi, S. (2012). Cyanobacterial biofuel production. J. Biotechnol. doi: 10.1016/j.jbiotec.2012.03.005 [Epub ahead of print].
Marcheschi, R. J., Li, H., Zhang, K., Noey, E. L., Kim, S., Chaubey, A., Houk, K. N., and Liao, J. C. (2012). A synthetic recursive “+1” pathway for carbon chain elongation. ACS Chem. Biol. 7, 689–697.
Martin, V. J., Pitera, D. J., Withers, S. T., Newman, J. D., and Keasling, J. D. (2003). Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21, 796–802.
Menkel, E., Thierbach, G., Eggeling, L., and Sahm, H. (1989). Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl. Environ. Microbiol. 55, 684–688.
Messerotti, L. J., Radford, A. J., and Hodgson, A. L. (1990). Nucleotide sequence of the replication region from the Mycobacterium–Escherichia coli shuttle vector pEP2. Gene 96, 147–148.
Miller, J. H. (1992). A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Mumberg, D., Muller, R., and Funk, M. (1994). Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22, 5767–5768.
Nielsen, D. R., Leonard, E., Yoon, S. H., Tseng, H. C., Yuan, C., and Prather, K. L. (2009). Engineering alternative butanol production platforms in heterologous bacteria. Metab. Eng. 11, 262–273.
Pátek, M., Nešvera, J., Guyonvarch, A., Reyes, O., and Leblon, G. (2003). Promoters of Corynebacterium glutamicum. J. Biotechnol. 104, 311–323.
Romero, P. A., and Arnold, F. H. (2009). Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10, 866–876.
Ruffing, A. M. (2011). Engineered cyanobacteria: teaching an old bug new tricks. Bioeng. Bugs 2, 136–149.
Sahm, H., Eggeling, L., Eikmanns, B., and Krämer, R. (1996). Construction of L-lysine-, L-threonine-, or L-isoleucine-overproducing strains of Corynebacterium glutamicum. Ann. N. Y. Acad. Sci. 782, 25–39.
Schafer, A., Tauch, A., Jager, W., Kalinowski, J., Thierbach, G., and Puhler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73.
Sentheshanmuganathan, S., and Elsden, S. R. (1958). The mechanism of the formation of tyrosol by Saccharomyces cerevisiae. Biochem. J. 69, 210–218.
Sheehan, J. J. (2009). Biofuels and the conundrum of sustainability. Curr. Opin. Biotechnol. 20, 318–324.
Shen, C. R., Lan, E. I., Dekishima, Y., Baez, A., Cho, K. M., and Liao, J. C. (2011). Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 77, 2905–2915.
Shen, C. R., and Liao, J. C. (2008). Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab. Eng. 10, 312–320.
Smith, K. M., Cho, K. M., and Liao, J. C. (2010). Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 87, 1045–1055.
Steen, E. J., Chan, R., Prasad, N., Myers, S., Petzold, C. J., Redding, A., Ouellet, M., and Keasling, J. D. (2008). Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb. Cell Fact. 7, 36.
Steen, E. J., Kang, Y., Bokinsky, G., Hu, Z., Schirmer, A., Mcclure, A., Del Cardayre, S. B., and Keasling, J. D. (2010). Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562.
Stephanopoulos, G. (2007). Challenges in engineering microbes for biofuels production. Science 315, 801–804.
Stoker, N. G., Fairweather, N. F., and Spratt, B. G. (1982). Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene 18, 335–341.
Studier, F., and Moffat, B. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130.
Tartoff, K. D., and Hobbs, C. A. (1987). Improved media for growing plasmid and cosmid clones. Bethesda Res. Lab. Focus 9, 12.
Thomas, J. G., Ayling, A., and Baneyx, F. (1997). Molecular chaperones, folding catalysts, and the recovery of active recombinant proteins from E. coli. To fold or to refold. Appl. Biochem. Biotechnol. 66, 197–238.
Vabulas, R. M., Raychaudhuri, S., Hayer-Hartl, M., and Hartl, F. U. (2010). Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol. 2, a004390.
Wendisch, V. F. (2007). Amino Acid Biosynthesis – Pathways, Regulation, and Metabolic Engineering (Microbiology Monographs). Berlin: Springer.
Withers, S. T., Gottlieb, S. S., Lieu, B., Newman, J. D., and Keasling, J. D. (2007). Identification of isopentenol biosynthetic genes from Bacillus subtilis by a screening method based on isoprenoid precursor toxicity. Appl. Environ. Microbiol. 73, 6277–6283.
Zhang, K., Sawaya, M. R., Eisenberg, D. S., and Liao, J. C. (2008). Expanding metabolism for biosynthesis of nonnatural alcohols. Proc. Natl. Acad. Sci. U.S.A. 105, 20653–20658.
Zheng, Y. N., Li, L. Z., Xian, M., Ma, Y. J., Yang, J. M., Xu, X., and He, D. Z. (2009). Problems with the microbial production of butanol. J. Ind. Microbiol. Biotechnol. 36, 1127–1138.
Zinoviev, S., Muller-Langer, F., Das, P., Bertero, N., Fornasiero, P., Kaltschmitt, M., Centi, G., and Miertus, S. (2010). Next-generation biofuels: survey of emerging technologies and sustainability issues. ChemSusChem 3, 1106–1133.
Keywords: biofuel, butanol, higher-chain alcohol, isobutanol, metabolic engineering, synthetic biology
Citation: Lamsen EN and Atsumi S (2012) Recent progress in synthetic biology for microbial production of C3–C10 alcohols. Front. Microbio. 3:196. doi:10.3389/fmicb.2012.00196
Received: 29 March 2012; Accepted: 14 May 2012;
Published online: 08 June 2012.
Edited by:
Weiwen Zhang, Tianjin University, ChinaCopyright: © 2012 Lamsen and Atsumi. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Shota Atsumi, Department of Chemistry, University of California, One Shields Avenue, Davis, CA, USA. e-mail: atsumi@chem.ucdavis.edu
 Edna N. Lamsen
Edna N. Lamsen