- 1Department of Environmental Microbiology, Eawag - Swiss Federal Institute of Aquatic Science and Technology, Dübendorf, Switzerland
- 2Department of Environmental Systems Sciences, Eidgenössische Technische Hochschule, Zurich, Switzerland
- 3Department of Process Engineering, Eawag - Swiss Federal Institute of Aquatic Science and Technology, Dübendorf, Switzerland
- 4Department of Civil, Environmental and Geomatic Engineering, Eidgenössische Technische Hochschule, Zurich, Switzerland
Nitrous oxide (N2O) is an environmentally important atmospheric trace gas because it is an effective greenhouse gas and it leads to ozone depletion through photo-chemical nitric oxide (NO) production in the stratosphere. Mitigating its steady increase in atmospheric concentration requires an understanding of the mechanisms that lead to its formation in natural and engineered microbial communities. N2O is formed biologically from the oxidation of hydroxylamine (NH2OH) or the reduction of nitrite (NO−2) to NO and further to N2O. Our review of the biological pathways for N2O production shows that apparently all organisms and pathways known to be involved in the catabolic branch of microbial N-cycle have the potential to catalyze the reduction of NO−2 to NO and the further reduction of NO to N2O, while N2O formation from NH2OH is only performed by ammonia oxidizing bacteria (AOB). In addition to biological pathways, we review important chemical reactions that can lead to NO and N2O formation due to the reactivity of NO−2, NH2OH, and nitroxyl (HNO). Moreover, biological N2O formation is highly dynamic in response to N-imbalance imposed on a system. Thus, understanding NO formation and capturing the dynamics of NO and N2O build-up are key to understand mechanisms of N2O release. Here, we discuss novel technologies that allow experiments on NO and N2O formation at high temporal resolution, namely NO and N2O microelectrodes and the dynamic analysis of the isotopic signature of N2O with quantum cascade laser absorption spectroscopy (QCLAS). In addition, we introduce other techniques that use the isotopic composition of N2O to distinguish production pathways and findings that were made with emerging molecular techniques in complex environments. Finally, we discuss how a combination of the presented tools might help to address important open questions on pathways and controls of nitrogen flow through complex microbial communities that eventually lead to N2O build-up.
Introduction
Nitric oxide (NO) and nitrous oxide (N2O) are atmospheric trace gases that influence atmospheric chemistry and the greenhouse effect. Biological and chemical processes produce N2O on the earth surface (Crutzen, 1979). Entering the stratosphere, N2O is converted to NO by photo-oxidation. NO together with nitrogen dioxide (NO2) participate in a set of reactions that transfer ozone (O3) to molecular oxygen (O2), thereby leading to O3 layer depletion. In fact, N2O is and will remain the dominant O3-depleting substance in the twenty-first century (Ravishankara et al., 2009), since the use of chlorofluorocarbons has been restricted by the Montreal Protocol. In addition, N2O is a potent greenhouse gas. The infrared radiative forcing of one N2O molecule is 206 times that of one carbon dioxide (CO2) molecule (Stein and Yung, 2003). Together with the long atmospheric lifetime of N2O (~120 years) this results in a ~300 times higher global warming potential of N2O than that of CO2 on a per molecule basis. Overall, N2O contributes 6–8% to the anthropogenic greenhouse effect, despite its relatively low atmospheric concentration (~322 ppbv) (Montzka et al., 2011).
Over the last 100 years atmospheric N2O concentrations have been steadily increasing due to the massive introduction of fixed nitrogen into the environment by humans (IPCC, 2001). Counteracting the further increase of N2O in the atmosphere will rely on (1) decreasing the introduction of fixed nitrogen into the environment by humans, (2) exactly quantifying the important environmental sources of N2O, and (3) implementing effective strategies to mitigate its formation in nitrogen-transforming, man-made ecosystems such as agriculture and wastewater treatment. Thus, there is an urgent need to understand the mechanisms that underpin the formation of N2O in natural and engineered microbial communities.
In this review, we will outline the current state-of-the-art on biological and chemical processes that can produce and consume N2O and NO—an important precursor of N2O in many biological pathways. We will discuss pathways that produce NO and N2O in natural and engineered microbial communities and experimental approaches that can be used to distinguish between different pathways in these systems. Importantly, NO and N2O formation can be highly dynamic and occur at small spatial scales. Thus, we will further introduce two novel technologies that provide such data and how they can lead to mechanistic insight: (1) NO and N2O microelectrodes and (2) the analysis of the site preference (SP) in N2O measured with quantum cascade laser absorption spectroscopy (QCLAS). In addition, we discuss the challenges of incorporating molecular biological techniques in this scheme.
Biological Pathways for NO and N2O Production
The study of laboratory cultures for pathways and controls of NO and N2O production in different organisms has generated considerable knowledge, which was partly reviewed recently (Stein, 2011; Chandran et al., 2011). Figure 1 shows that the sequential reduction of nitrite (NO−2) to NO and further to N2O can be performed by all organisms involved in the catabolic branch of the N-cycle. While all N-cycle organisms can perform these reactions it is currently believed that denitrifiers and ammonia oxidizing bacteria (AOB) and ammonia oxidizing archaea (AOA) are the most important environmental sources of N2O. However, in the following section we additionally review the evidence for NO and N2O production by nitrite oxidizing bacteria (NOB), anaerobic methane (N-AOM) and AOB (anammox), and bacteria that perform dissimilatory nitrate reduction to ammonia (DNRA). Even though it is clear that these bacteria can produce NO and N2O there is only few information on the controls, conditions and magnitude for NO and N2O production by these bacteria in the laboratory and in the environment. This should be an important aspect of future research as e.g., DNRA and anammox are the major N-conversion pathways in some important environments.
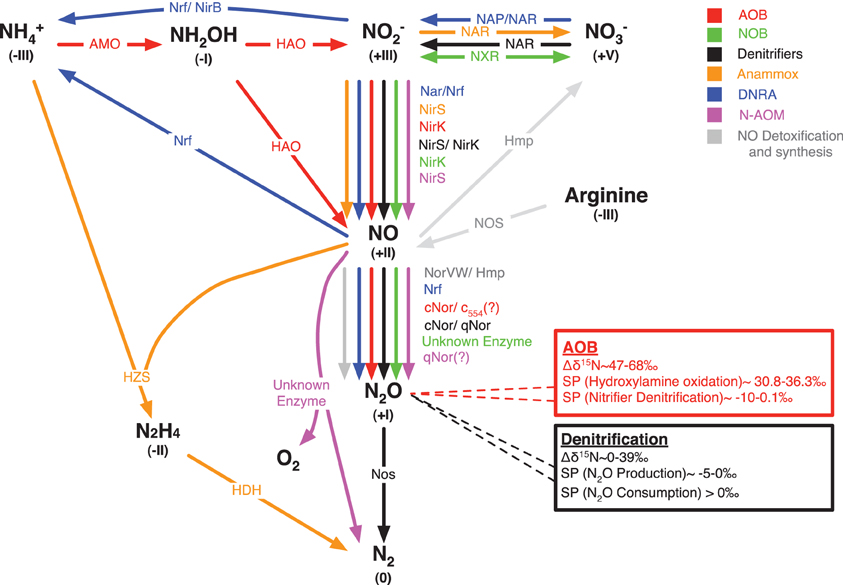
Figure 1. Biological pathways for NO and N2O turnover in the catabolic branch of the N-cycle plus NO synthesis and detoxification. Different colors are allocated to different microbial guilds or turnover pathways: AOB (red), ammonia oxidizing bacteria; NOB (green), nitrite oxidizing bacteria; anammox (orange), anaerobic oxidation of ammonia; DNRA (blue), dissimilatory nitrate/nitrite reduction to ammonia; N-AOM (purple), oxygenic nitrite-dependent anaerobic oxidation of methane. Key enzymes of each microbial guild are depicted that are known to mediate the conversion from one chemical N-species into another: AMO, ammonia monooxygenase; HAO, hydroxylamine oxidoreductase; NXR, nitrite oxidoreductase; Nar, membrane-bound nitrate reductase; Nap, periplasmic nitrate reductase; NirK, copper-containing nitrite reductase; NirS, cytochrome cd1 nitrite reductase; Nrf, cytochrome c nitrite reductase; NirB, cytoplasmic nitrite reductase; cNor, nitric oxide reductase that accepts electrons from c-type cytochromes; qNor, nitric oxide reductase that accepts electrons from quinols; c554, cytochrome c554; NorVW, flavorubredoxin, Hmp, flavohemoglobins; HZS, hydrazine synthase; HDH, hydrazine dehydrogenase; Nos, nitrous oxide reductase; NOS, nitric oxide synthase; unknown enzymes, nitric oxide dismutation to N2 and O2 during N-AOM and nitrous oxide producing enzyme in NOB. Roman numbers in brackets denote the oxidation state of the chemical N-species. The red and the black box denote the isotopic composition (δ15N) and the site preference (SP) in isotopomers of N2O produced by AOB and denitrifiers, respectively.
Denitrification
The key enzyme for NO formation during denitrification is nitrite reductase (Nir). Purification and characterization of Nir from several bacteria revealed two entirely different periplasmic enzymes: a heme-containing cytochrome cd1 Nir (NirS) and a copper-containing Nir (NirK) as reviewed by Cutruzzolà (1999). Reduction of NO to N2O is mediated by respiratory nitric oxide reductases (Nor). Respiratory Nor proteins are integral membrane proteins that fall into two groups: one is a cytochrome bc complex that can use c-type cytochromes as electron donors (cNor), whereas the other one lacks a cytochrome c component and accepts electrons from quinols (qNor; sometimes termed NorZ) (Hendriks et al., 2000; Zumft, 2005). Few bacteria use qNor for classical denitrification. Rather, qNor is mainly encoded by pathogenic bacteria that use it for NO detoxification and the survival of anoxic periods when expressed in concert with Nir, as shown for Neisseria spp. (Anjum et al., 2002; Rock et al., 2007). The final step in denitrification is mediated by nitrous oxide reductase (Nos), a multi-copper enzyme that reduces N2O to dinitrogen (N2) (Zumft and Kroneck, 2007).
N2O reduction by Nos is the only known N2O consuming process that can counteract release of N2O from ecosystems (Richardson et al., 2009). Accumulation of N2O is often observed in pure cultures (Baumann et al., 1996; Otte et al., 1996; Kester et al., 1997; Bergaust et al., 2010) and mixed microbial communities (Firestone and Tiedje, 1979; Firestone et al., 1980; Morley et al., 2008; Kampschreur et al., 2008b; Schreiber et al., 2009; Elberling et al., 2010; Pellicer-Nàcher et al., 2010; Liengaard et al., 2011) during transitions from anoxic to oxic conditions or vice versa (Table 1). Even in pure cultures the physiological basis for this is not well understood because it probably has multiple, strain-specific reasons. It has been hypothesized that Nos is—unlike Nir and Nor—inhibited by O2 (Morley et al., 2008), but in pure cultures evidence for O2-insensitive (Berks et al., 1993) and O2-sensitive (Otte et al., 1996) Nos have been reported. Likewise, it has been argued that expression of Nos is slower than that of the preceding denitrification enzymes (Firestone et al., 1980; Stief et al., 2009), but in Paracoccus denitrificans Nos synthesis is faster (Baumann et al., 1996; Bergaust et al., 2010) and in Pseudomonas stutzeri Nos is even constitutively expressed at low levels (Körner and Zumft, 1989). More studies on Nos expression in relation to N2O production pathways and on Nos inhibition by O2 are needed with environmentally relevant isolates and mixed microbial communities. Additional factors that lead N2O accumulation are the slower turnover of Nos at low pH as compared to nitrate reductase (Nar), Nir, and Nor (Richardson et al., 2009; Bergaust et al., 2010), low pH during Nos assembly (Bergaust et al., 2010), inhibition of Nos by nitrous acid formed from NO−2 at low pH (Zhou et al., 2008), inhibition of Nos by exogenously produced NO (Frunzke and Zumft, 1986; Schreiber et al., unpublished results) or hydrogen sulfide (H2S) (Sørensen et al., 1980) and copper limitation (Granger and Ward, 2012).
Ammonia Oxidizing Bacteria (AOB)
High levels of NO and N2O can be produced by pure cultures of aerobic AOB (Lipschultz et al., 1981; Kester et al., 1997; Shaw et al., 2006), but the mechanism is not completely understood. Generally, two different pathways are inferred. First, the activity of nitrifier-encoded NirK and cNor reduces NO−2 to NO and N2O in a pathway termed nitrifier denitrification (Poth and Focht, 1985; Wrage et al., 2001; Schmidt et al., 2004b). A few reports exist on N2 formation by AOB during nitrifier denitrification, but a nosZ gene or functional Nos in AOB was not demonstrated (Poth, 1986; Schmidt et al., 2004b; Schmidt, 2009). The term nitrifier denitrification is somewhat misleading as it has until now not been shown that it is a true dissimilatory process for energy conservation and growth, but rather may be a detoxification mechanism to counteract the accumulation of NO−2 to toxic concentrations (Beaumont et al., 2002, 2004a,b).
In the second pathway, N2O is formed by hydroxylamine (NH2OH) oxidation. The current model is that hydroxylamine oxidoreductase (HAO) oxidizes NH2OH to NO (Hooper, 1968; Hooper and Terry, 1979). NO is then reduced to N2O by a yet unidentified Nor; a potential candidate is cytochrome c554 (Upadhyay et al., 2006). However, the catalytic cycle of HAO, including its intermediates and its catalytic potential are a subject of ongoing debate (Hendrich et al., 2002; Cabail and Pacheco, 2003; Cabail et al., 2005; Fernández et al., 2008; Kostera et al., 2008) and as of yet direct formation of N2O from HAO or other reactions can not be excluded. Indeed, the difference in the SP of N2O produced by NH2OH oxidation and nitrifier denitrification indicates that N2O might be produced by HAO by a mechanism that (1) either does not involve NO reduction by canonical Nor used for nitrifier denitrification or (2) does proceed via a completely different mechanism without free NO as intermediate (discussed in section “site preference” and “HNO as intermediate of enzymatic hydroxylamine oxidation”). Both nitrifier denitrification and NH2OH oxidation require O2 to activate ammonia (NH3) with ammonia monooxygenase (AMO) to NH2OH, which serves as a substrate for HAO or as electron donor to nitrifier denitrification. A pathway in which AOB perform denitrification with organic substrates instead of NH3 as electron donor (Schmidt, 2009) should be considered heterotrophic denitrification performed by AOB. AOA have also been demonstrated to produce N2O probably by pathways akin to AOB (Santoro et al., 2011).
The relative importance of NH2OH oxidation and nitrifier denitrification for NO and N2O production is still debated. Based on pure culture investigations Yu et al. (2010) hypothesized that a high NH3 oxidation activity favors N2O production via NH2OH oxidation. Similarly, Wunderlin et al. (2012) found that NH2OH oxidation is favored by high NH3 and low NO−2 concentrations, and a high nitrification rate in a mixed culture for treating municipal wastewater. Moreover, stable nitrogen isotopes work with AOB pure cultures showed that NH2OH oxidation contributes to N2O production mainly at high O2 whereas nitrifier denitrification is more active at low O2 concentrations (Sutka et al., 2006).
Nitrite Oxidizing Bacteria (NOB)
NOB form NO and N2O during denitrification of nitrate (NO−3) or NO−2 with pyruvate or glycerol as electron donor under anoxic conditions (Freitag et al., 1987; Ahlers et al., 1990), but a known NO reductase could not be identified in the genomes of different Nitrobacter species and “Candidatus Nitrospira defluvii” (Starkenburg et al., 2006, 2008b; Lücker et al., 2010). Under anoxic conditions nitrite oxidoreductase (NXR) mediates NO−3 reduction to NO−2, while it mediates the reverse reaction under oxic conditions (Freitag et al., 1987). NOB actively express NirK, which co-purifies with NXR, in the presence of NO−2 and if O2 concentrations are low (Ahlers et al., 1990; Starkenburg et al., 2008a). NO generated by NOB-NirK is thought to direct cellular electron flux either toward O2 respiration at high O2 concentrations or toward NADH synthesis by reversibly inhibiting cytochrome oxidase at low O2 concentrations. An interesting question to explore in natural communities would be whether NO produced by AOB or denitrifying bacteria can influence the activity of NOB.
Dissimilatory Nitrate Reduction to Ammonia (DNRA)
NO and N2O turnover by bacteria that perform DNRA has been mainly investigated in Escherichia coli and Salmonella typhimurium. In E. coli, NO formation is mediated by cytochrome c nitrite reductase (Nrf) under anoxic conditions in the presence of NO−3 and NO−2 (Corker and Poole, 2003). NO detoxifying enzymes, such as flavorubredoxin, may further reduce NO to N2O. On the other hand, E. coli Nrf reduces NO to N2O or NH3 if electrons are donated to the enzyme at high or low potential, respectively (Costa et al., 1990), contributing to detoxification of exogenously generated NO (van Wonderen et al., 2008). Aerobic and anaerobic NO formation from NO−2 in S. typhimurium is mediated by membrane-bound nitrate reductase (Nar). Under aerobic conditions, activity of NO detoxifying Hmp (see below) oxidizes NO to NO−3 resulting in non-detectable NO concentrations in culture suspensions (Gilberthorpe and Poole, 2008).
Anaerobic Methane and Ammonia Oxidizing Bacteria
Bacteria that mediate the oxygenic nitrite-dependent oxidation of methane (N-AOM) and anaerobic ammonia oxidation (anammox) have been shown to use NO as an intracellular intermediate produced by NO−2 reduction via NirS while they consume exogenous NO without concurrent N2O formation (Ettwig et al., 2010; Kartal et al., 2010, 2011). Rather, N-AOM dismutates NO to form N2 and O2, while anammox couples the reduction of NO to a condensation with NH3 to produce hydrazine (N2H4). Both have the genetic potential to reduce NO to N2O; anammox bacteria encode for flavorubredoxin (Strous et al., 2006) and N-AOM encodes for qNor (Ettwig et al., 2010). However, physiological data for both indicates that they withstand rather high NO levels (N-AOM 20 μmol L−1, anammox 7 μmol L−1) without activating anaerobic NO detoxification mechanisms.
NO−2 → NO → N2O: Central Steps in the N-Cycle
Generally, the reduction of NO−2 to NO is a central step in the catabolic branch of the N-cycle, because it can be carried out by all involved organisms (Figure 1). The reduction of NO−2 to NO is central for energy conservation in denitrification, anammox and N-AOM. In contrast, during NO−2 oxidation and nitrifier denitrification the reduction of NO−2 to NO is involved in regulating metabolic homeostasis or the removal of toxic NO−2 (Beaumont et al., 2002, 2004a; Starkenburg et al., 2008a).
The reduction of NO to N2O is, besides a potential direct formation of N2O from NH2OH in AOB, the only known biochemical reaction that produces N2O. NO reduction to N2O is central for energy conservation only in denitrification (Zumft, 1997). The function of cNor in AOB is unclear. cNor is expressed and metabolically active during aerobic growth (Beaumont et al., 2004b). Knock-out mutants of cNor have lower growth rate and yield in chemostats (Schmidt et al., 2004b), but not in batch culture (Beaumont et al., 2004b). In chemostats, cNor regulates the free NO concentration to an optimal, non-toxic level and contributes to recovery of AOB from anaerobic conditions (Schmidt et al., 2004b). On the other hand, stripping NO from AOB cultures leads to the inhibition of growth, arguing for NO being an obligate intermediate of AOB (Zart et al., 2000).
NO Detoxification and NO Synthesis
Most bacteria encode for enzymes involved in NO detoxification. This is true for bacteria inside and outside the catabolic N-cycle. Flavohemoglobins (Hmp) mediate the O2-dependent detoxification of NO to NO−3 with NO dioxygenase activity (Gardner et al., 1998). In contrast, the anaerobic detoxification of NO is mediated by Flavodiiron NO reductase (flavorubredoxin [NorVW]) and Hmp by reducing NO to N2O (Kim et al., 1999; Gardner et al., 2002; Gomes et al., 2002).
An alternative, less explored route to N2O formation is via the synthesis of NO from arginine by NO synthases (NOS) and subsequent reduction of NO to N2O by cNor, qNor, Hmp or NorVW. Because NOS was discovered in the medical field it shares a similar abbreviation with N2O reductases (Nos). Until now, NOS has only been detected in a few bacterial –mostly gram-positive – species (Sudhamsu and Crane, 2009) and synthesized NO seems to remain intracellular (Shatalin et al., 2008; Schreiber et al., 2011). However, NOS activity has also been reported in blooming, pelagic diatoms (Vardi et al., 2006). More research is needed to elucidate if NOS-derived NO is a significant source for N2O emitted from phytoplankton blooms in oceans and freshwater.
Chemical Reactions in NO and N2O Turnover
Chemical production of NO and N2O from inorganic nitrogen compounds at ambient temperatures are well known phenomena in soil science (van Cleemput and Samater, 1996) and atmospheric chemistry (Lammel and Cape, 1996). In soil science, the chemical processes leading to NO and N2O are often summarized as chemo-denitrification (Chalk and Smith, 1983). NH2OH and NO−2 (or its acid HNO2) are the main precursors for chemical production of NO and N2O in wastewater or natural waters. In the following, we discuss chemical reactions involving HNO, NH2OH, and NO−2 that can be responsible for the release of NO and N2O. We will also discuss the possible significance of chemical N2O production during biological NH2OH oxidation.
Significance of HNO
In many studies on chemical N2O production, HNO is postulated as the direct precursor of N2O (see below): HNO dimerizes via hyponitrous acid (H2N2O2), to N2O and H2O (Bonner and Hughes, 1988).
It can be assumed that formation of HNO in natural and wastewater follows the same mechanisms that are used to synthesize HNO (DuMond and King, 2011) in the laboratory: (1) disproportionation of NH2OH derivatives containing good leaving groups attached to the nitrogen atom, and (2) decomposition of nitroso compounds (X–N=O, where X represents a good leaving group). Chemical HNO production are likely to occur during wastewater treatment, since nitrification can produce considerable amounts of both, HNO2, which is a precursor for nitrosation agents (e.g., dinitrogen trioxide N2O3, Bonner and Stedman, 1996), and NH2OH.
Recently, medical researchers have started to reevaluate the relevance of HNO for physiologically and biologically systems (Fehling and Friedrichs, 2011). The increased interest in HNO is due to the fact that HNO lifetime in aqueous solutions is much longer than previously assumed: the HNO dimerization rate constant has been reassessed to be on the order of 8 × 105 M−1 · s−1 instead of the previously reported value of 2 × 109 M−1 · s−1, and the pKa value of HNO has been redetermined to be 11.4 instead of the old value of 4.2 (Shafirovich and Lymar, 2002). It is likely that the importance of HNO has also been underestimated in the research on N2O emissions. Analytical determination of HNO is very challenging (Miranda, 2005), because HNO is short-lived. However, computer simulations could be a helpful tool to assess the importance of HNO in N2O formation (Law et al., 2012).
HNO2 Disproportionation
A well understood process for NO production is the disproportionation of HNO2 (Udert et al., 2005). Since the pKa value of the NO−2/HNO2 couple (pKa = 3.29; Schwartz and White, 1981) is far below 7, this process releases relevant amounts of NO only under acidic conditions. The disproportionation of HNO2 can be described with Equation 2. The products—NO and NO2—are in equilibrium with N2O3 (Equation 5) which is an important agent for nitrosation (Bonner and Stedman, 1996). Under aerobic conditions, NO will be further oxidized to NO2. Since NO2 reacts with H2O to form HNO2 and NO−3, the reaction scheme (Equations 2–4) is ultimately a chemical pathway for the oxidation of NO−2 to NO−3.
Since the kinetic and equilibrium constants for Equations 2–5 are known, the production of NO can be calculated (Udert et al., 2005). Depending on the aeration intensity, substantial losses of nitrogen oxides can occur during chemical HNO2 oxidation. The stripped nitrogen oxides are mainly HNO2, but also NO is lost.
Iron-Mediated Reduction of NO−2
Ferrous iron [Fe(II)] can reduce NO−2 to NO and, in the second reaction step, NO to N2O (Kampschreur et al., 2011).
The first reaction is thermodynamically not possible under standard conditions, but in natural waters ferric iron [Fe(III)] will precipitate and thereby draw the Gibbs free energy to negative values. Iron-mediated reduction of NO−2 was described as one of the sources of N2O in soils (van Cleemput, 1998). Recently, Kampschreur et al. (2011) postulated that this process can contribute significantly to N2O production in wastewater treatment, if NO−2 and Fe(II) are present concomitantly. One example for such a system is nitrogen removal from anaerobic digester effluents via nitritation/denitrification or nitritation/anammox. Digester supernatants can contain high amounts of Fe(II), because iron salts are used to precipitate phosphate and Fe(II) will be released in the anaerobic digester due to the reducing conditions. Hu et al. (2001) reported an additional reaction of NO−2 with iron: under acidic conditions NO−2 is reduced in the presence of metallic iron to N2 and NH3. They propose a mechanism, in which metallic iron is oxidized at low pH releasing Fe2+ ions and molecular hydrogen (H2). NO−2 is then reduced by H2 to N2 and NH3.
Oxidation of NH2OH by FE(III)
Iron not only mediates NO and N2O production from NO−2. As Fe(III), it also oxidizes NH2OH to N2O. This process can be used for the analytical determination of trace amounts of NH2OH (Butler and Gordon, 1986a). The general equation for the reaction is
In this reaction, N2O formation strongly depends on the pH value. In experiments with distilled water and natural seawater, Butler and Gordon (1986b) found that at pH 3, N2O recovery was 80%, while at a pH value of 9.5, N2O production was negligibly low. The authors hypothesized that at high pH values, HNO, reacts with O2 to produce NO−2 and H2O. However, it is also known that HNO can react with NH2OH to N2 (Bonner et al., 1978, Equation 10). Chemical production of N2O via NH2OH oxidation by Fe(III) is a likely process during nitrification, because Fe(III) compounds are ubiquitous in natural waters and wastewater treatment systems.
Reaction of NH2OH with HNO2 and HNO
Döring and Gehlen (1961) investigated the reaction of NH2OH and HNO2. They described the process as nitrosation of NH2OH. The overall reaction can be written as
In their reaction scheme, Döring and Gehlen (1961) included H2N2O2 (the dimer of HNO) as a direct precursor for N2O. At neutral pH values, N2O3 is the relevant nitrosation agent. There are several reaction pathways for N2O3 formation from HNO2. Formation of N2O3 from HNO2 is given by Equations 2 and 5. A kinetic constant for nitrosation of NH2OH is given by Döring and Gehlen (1961) and together with the kinetic constants for Equations 1 and 4 (Udert et al., 2005) the N2O production from NH2OH and HNO2 can be estimated. Some of the NH2OH can also react with the intermediate HNO to form N2 (Bonner et al., 1978)
Disproportionation of NH2OH
The disproportionation of NH2OH can be described with the following equation (Bonner et al., 1978):
In pure water, this process is very slow with slightly higher degradation rates at elevated pH values. At pH 3 and 25 ± 3°C, Bonner et al. (1978) observed no NH2OH disproportionation over 2 months, while 12–18% of the NH2OH was degraded over 2 months at pH 13.5. Complexes of transition metals can accelerate NH2OH disproportionation considerably (Alluisetti et al., 2004). Jenni et al. (2012) also observed N2O formation within minutes, although the experiment was conducted in a phosphate buffer solution without transition metals. The disproportionation might have been catalyzed by the steel surface of an electrode immersed in the reactor, but this hypothesis still has to be proven.
Autoxidation of NH2OH
Oxidation of NH2OH with O2 (autoxidation, Equation 12) is a slow process, although faster than NH2OH disproportionation.
Again, trace concentrations of metals can strongly accelerate the process. Anderson (1964) reported that in an aerated solution with 1 mmol·L−1 NH2OH and 1 μmol·L−1 cupric sulfate 30% of the NH2OH was oxidized within 1 h, while only 2.5% were degraded without cupric sulfate addition (pH between 7.8 and 7.9, 30°C). Cu is by far the most potent catalyzer for the autooxidation of NH2OH followed by Co(II), Fe(II), Mn(II), and Zn(II) (Moews and Audrieth, 1959). Since most wastewaters and natural waters contain some traces of metals, autoxidation of hydroxylamine cannot a priori be excluded as a source of N2O.
HNO as Intermediate of Enzymatic NH2OH Oxidation
Several authors postulated that HNO was a likely intermediate of HAO due to the observed N2O production (Anderson, 1964; Ritchie and Nicholas, 1972). Igarashi et al. (1997) could show that the crystal structure of HAO in Nitrosomonas europaea is in agreement with the following two step reaction
Based on this scheme, an imbalance of the two reaction steps could lead to an accumulation of HNO and subsequently to chemical N2O production (Equation 1). Law et al. (2012) developed four different metabolic computer models to elucidate the mechanisms of aerobic N2O production in a nitritation reactor. The best fit of the measurement data was achieved with a model based on chemical HNO production. The other models, which represented three different metabolic pathways for the enzymatic reduction of nitrite and NO to N2O, could not reproduce the measurement data satisfactorily. Indeed, we think that the positive SP of N2O produced during NH2OH oxidation can be explained by a kinetic isotope effect acting during the chemical cleavage of a symmetric intermediate such as H2N2O2 formed by dimerization of two HNO molecules (Equation 1; Toyoda et al., 2005). In addition, the studies of Law et al. (2012) and of Udert et al. (2005) exemplify that computer models are powerful tools to elucidate the mechanisms of N2O and NO production, especially when the processes contain microbial as well as chemical reaction steps.
Relevant Environments for Chemical Reactions
In the last years, nitrogen treatment of high-strength wastewaters such as digester supernatant, manure and urine have received considerable attention. Based on our literature review, these systems are particularly prone to chemical production of NO and N2O because of high NH3 oxidation rates and high concentrations of the intermediate NH2OH. Furthermore, some treatment schemes include NO−2 accumulation as a process step, for example SHARON®. Ubiquitous iron compounds, e.g., from phosphate precipitation or as sensors and reactor walls, are another factor that can support the production of NO and N2O. At the current stage of knowledge, it is hard to estimate the contribution of chemical processes to the overall NO and N2O production. Many chemical processes have been described, but with the exception of HNO2 disproportionation and the reaction of HNO2 with NH2OH, the kinetic data are insufficient for a reliable prediction of the production rates. Chemical production of NO and N2O can also occur in natural environments, where high ammonia inputs meet low pH values such as strongly fertilized soils (van Cleemput and Samater, 1996) or poorly buffered lakes (Schuurkes and Mosello, 1988). Furthermore, chemical oxidation of NO and N2O is an important process in the atmosphere (Lammel and Cape, 1996).
NO and N2O Formation in Natural Environments
Nitric Oxide
NO production and consumption has been studied in soils. The studies used inhibition of nitrification with low concentrations of acetylene (~10 Pa) to distinguish between NO turnover by nitrification and denitrification, assuming that acetylene does not inhibit N2O reductase at these concentrations. O2 availability, as regulated by soil moisture content, is the main factor controlling the mechanisms of NO release (Bollmann and Conrad, 1998). While denitrification is the only process that releases NO under anoxic conditions, nitrification dominates NO release under oxic conditions with highest rates at low O2 concentrations. In addition, soil pH, NH+4, NO−3, NO−2, and respiration are important soil variables that affect NO turnover (Gödde and Conrad, 2000).
Measurements of NO in seawater are rare, because concentrations are low and turnover is fast due to its reactivity. However, Zafiriou et al. (1980) found that surface water of the central equatorial Pacific is a NO source to the atmosphere. Here, NO is formed by photolysis of NO−2 during daytime and reaches concentrations in the picomolar range (Zafiriou and True, 1979). Moreover, NO is formed by microbial processes in the O2 minimum zone of the eastern tropical North Pacific (Ward and Zafiriou, 1988). Here, maximum NO turnover and concentration coincide with low O2 concentrations (10–100 μmol L−1) and some nitrification activity overlying the O2 minimum zone. In contrast, NO turnover and concentrations are low in the core of the O2 minimum zone. The exact source of NO remained unidentified, but it was hypothesized that nitrifiers produce NO under reduced O2 concentrations and that denitrifiers establish rather low NO concentrations in the core of the O2 minimum zone. NO formation has been measured in marine sediments (Schreiber et al., 2008) and a more detailed study of NO turnover has been performed in freshwater sediments (Schreiber et al., unpublished results). Both studies will be discussed in the section focusing on microelectrodes.
Nitrous Oxide
Generally, N2O formation has been investigated to greater detail and in a wider variety of habitats as compared to NO, because it is an environmental impact is considered to be stronger than that of NO and its turnover is easier to measure due to its chemical stability. At present, anthropogenic N2O emissions account for ~40% of the global N2O emissions (Montzka et al., 2011). Current estimates state that ~50% of the anthropogenic N2O is emitted from soils (Stein and Yung, 2003), 10% from estuaries and freshwater habitats (Beaulieu et al., 2011) and 3.2% are emitted from wastewater treatment plants (WWTP) (Kampschreur et al., 2009). We caution that future adjustments to these estimates are likely, and that these averages do not capture the high variability in emissions from selected environments. Recent work has suggested that emissions from WWTPs in particular are highly variable and may in some cases be up to an order of magnitude greater than previous estimates (Ahn et al., 2010; Lotito et al., 2012). Soils and aquatic habitats exposed to intense agricultural activities are the largest sources due to high N-input through fertilization. Since mixed microbial communities in soils are the largest anthropogenic source for N2O, its formation has been intensively studied and was recently reviewed (Baggs, 2011). N2O formation in WWTP has been reviewed by Kampschreur et al. (2009).
The ocean is an important source of N2O accounting for ~30% of the natural N2O emission (Stein and Yung, 2003). Large areas of the ocean are thought to be in equilibrium with the atmosphere, but regions of O2 depletion are significant sources of N2O (Elkins et al., 1978). In O2 minimum zones, N2O is generally produced to concentrations in the nanomolar range as O2 reaches low concentrations (Yoshida et al., 1989; Naqvi et al., 2000; Farias et al., 2007; Nicholls et al., 2007). High N2O accumulation was observed in surface water of the Arabian Sea and explained with frequent, turbulence-induced aeration of suboxic surface water (Naqvi et al., 2000). Likewise, O2 fluctuations, induced by the El Nino-Southern oscillation, have been proposed to affect N2O emission from the O2 minimum zone of the eastern South Pacific (Farias et al., 2007). Furthermore, marine and freshwater sediments emit N2O (Meyer et al., 2008; Nielsen et al., 2009). NO and N2O formation in sediments will be discussed in more detail in the section focusing on microelectrodes.
The occurrence of animals such as earthworms (Horn et al., 2003) in soils and macrofauna in fresh -or seawater habitats (Stief et al., 2009; Heisterkamp et al., 2010) enhances the emission of N2O in response to anthropogenic N-input. These animals ingest denitrifying bacteria and stimulate their activity probably with delayed expression of N2O reduction leading to enhanced N2O emissions.
Experimental Approaches
In most investigated habitats NO and N2O formation has been attributed to the NH2OH pathway by AOB, nitrifier denitrification and heterotrophic denitrification. There are three approaches to determine the contribution of the different pathways:
(1) Indirect inference of pathways by excluding the activity of all other possible pathways, which can be achieved by using inhibitors or by removing the substrate (Kampschreur et al., 2008b; Schreiber et al., 2009; Stief et al., 2009; Wunderlin et al., 2012).
(2) Measuring the isotopic signature of N2O (15N natural abundance or SP) and comparing the data to values of pure cultures (Yoshida, 1988; Yoshida et al., 1989; Sutka et al., 2006; Well et al., 2006; Charpentier et al., 2007; Wunderlin et al., unpublished results).
(3) Application of 15N isotopically-enriched substrates and mass spectrometric measurements of N2O (Bateman and Baggs, 2005; Baggs, 2008).
In complex systems all of these approaches suffer from the coupled nature of nitrification and denitrification. This especially applies to studies where bulk measurements have been done even though micro-environmental heterogeneities are expected; e.g., in aggregates in wastewater treatment systems or in soil particles. In addition, it has become clear that NO and N2O are dynamically produced in response to changing environmental conditions (Kampschreur et al., 2008b; Schreiber et al., 2009). Transient NO and N2O concentrations can be orders of magnitude higher than under steady state. Conventional mass spectrometric measurements do not allow measurements with high temporal and spatial resolution, making approach 2 and 3 inaccessible to microscale and dynamic analysis of NO and N2O.
Novel Analytical Methods
In the following sections, we will discuss different analytical methods (microelectrodes, mass spectrometry, and QCLAS) that can be used to allocate NO and N2O production to certain pathways by using one of the three approaches outlined above. Combining these methods and thus the different approaches will lead to a more firm pathway allocation. Microelectrodes can measure with high temporal and spatial resolution and in combination with other microelectrodes (NH+4, NO−3, NO−2, and O2) approach 1 can be used to allocate source pathways. Further, QCLAS can measure the SP in N2O dynamically and can be used to allocate N2O production pathways with approach 2. In addition, we will discuss the potential for other techniques that measure the isotopic composition of N2O and molecular methods to aid the understanding of NO and N2O formation in complex environments.
Microelectrodes to Capture Micro-Environmental Distribution and Temporal Dynamics of NO and N2O
NO and N2O Microelectrodes
Microelectrodes belong to the tool box of microbial ecologists since Revsbech et al. introduced an O2 microelectrode in the early 1980s (Revsbech et al., 1980). The first N2O microelectrode for microbial ecology (Revsbech et al., 1988) was a combined O2/N2O sensor where an O2-reducing gold cathode was placed in front of an N2O-reducing silver cathode (both polarized at −800 mV) to avoid the interference of O2 with N2O detection. These sensors where difficult to manufacture and had a short life-time. Thus, Andersen et al. (2001) introduced an improved O2-insensitive N2O microelectrode. Insensitivity to O2 is achieved by placing a reservoir filled with alkaline ascorbate solution for the chemical reduction of O2 in front of the N2O-reducing cathode, which is separated from the ascorbate reservoir with a gas permeable silicone membrane. These N2O microelectrodes have a sensitivity of ~0.5 μmol L−1 and a spatial resolution of ~60 μm.
Electrochemical NO sensors for the detection of NO in biological systems are available since the early 1990s (Shibuki, 1990). Amperometric sensing of NO is commonly achieved by the oxidation of NO at a working electrode polarized with 0.7–0.9 V vs. a reference electrode (Ag/AgCl or Calomel) leading to the following anodic reaction:
The resulting current is proportional to the NO concentration and can be detected as the analytical signal. Electrodes are reported as single anode-type electrodes or as combined sensors (Figure 2). In combined sensors, the reference electrode and the sensing electrode are placed together in an internal electrolyte compartment that is separated from the sample by a gas permeable, non-conductive membrane (Clark-type, Figure 2B), whereas single anode-type electrodes use the aqueous sample as an electrolyte and complete the measuring circuit by submerging an external reference electrode into it (Figure 2A). Charged interferences like NO−2 and ascorbate are typically repelled by constructing combined sensors with hydrophobic membranes like chloroprene (Shibuki, 1990), PTFE (Teflon™) (Lee et al., 2004), sol-gels (Shin et al., 2005), polystyrene (Kitamura et al., 2000) or silicone (Schreiber et al., 2008), or by depositing conductive Nafion™ on single anode-type electrodes (Malinski and Taha, 1992; Friedemann et al., 1996; Bedioui and Villeneuve, 2003).
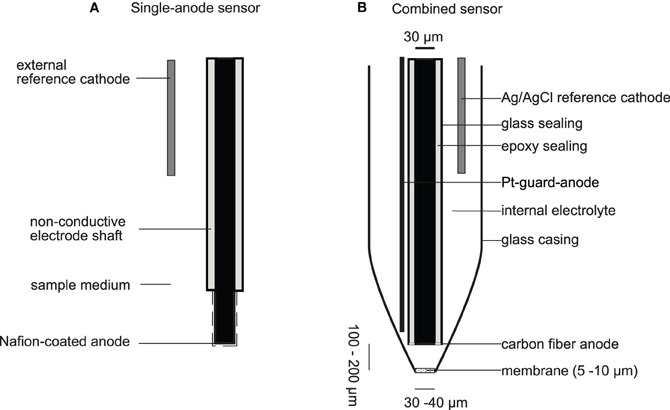
Figure 2. NO microelectrodes. (A) Depicts a typical single-anode type NO sensor with a long sensing anode, which is coated with Nafion to confer selectivity against charged interferences. The anode and reference cathode are directly emerged into the sample medium. Some sensor designs integrate the cathode into the electrode shaft. (B) Depicts the NO microelectrode for measurements in biofilms and sediments as reported by Schreiber et al. (2008). This sensor is also an example for a combined NO sensor (Clark-type) where sensing anode and reference cathode are separated from the sample medium by a gas permeable membrane. Drawing is not to scale.
Most of the previously described NO electrodes have been optimized to detect NO at low nanomolar or even picomolar concentration. This has been achieved by increasing the sensing surface with a subsequent loss of spatial resolution. Single-anode-type sensors commonly rely on carbon-fibers that have a length of up to several millimeters and combined sensors have openings in the high micrometer to millimeter range. Microelectrodes with long, exposed sensing surfaces are not applicable for profiling in stratified microbial systems because the concentration of the analyte might change along the sensing surface. The obtained signal is then an integrated measure of the concentrations along the electrode. Similarly, combined electrodes with wide openings are also problematic for profiling applications, since the step size of different measurement points in a depth profile should not be smaller than two times the outer diameter of the electrode (Gieseke and de Beer, 2004). In addition, single-anode sensors are not robust enough to be inserted in a sturdy sediment or soil sample since the particles will damage the Nafion™ membrane that confers selectivity against NO−2. Consequently, applications of NO electrodes—commercially supplied, e.g., by World Precision Instruments (Sarasota, Florida, USA)—in microbiology were restricted to detection of NO in pure culture suspensions (e.g., Corker and Poole, 2003).
Recently, an NO microelectrode was introduced that is applicable to study complex, stratified microbial communities in sediments and biofilms (Schreiber et al., 2008). The NO microelectrode is a combined (Clark-type) sensor with a carbon-fiber anode (+750 mV) placed behind a gas permeable silicon membrane (Figure 2B). The sensor has a detection limit of 0.030 μmol L−1 and a spatial resolution of ~60 μm. Thus, the sensor is optimized to provide sufficient sensitivity for NO concentrations produced in complex, N-cycling microbial communities and sufficient spatial resolution to measure in microbial biofilms, sediments and soils. The robust Clark-type design allows measurements in sturdy soil and sediment samples. It has been made commercially available through Unisense A/S (Arhus, Denmark), who also supplies N2O microelectrodes.
Interferences
H2S interferes with NO measurement as it passes the silicone membrane and is readily oxidized at the sensing anode. A sensitive H2S microsensor (Jeroschewski et al., 1996) should thus be used to rule out any interference of H2S in the measurements or –if possible- experiments must be designed to avoid active sulfate reduction in the sample by excluding sulfate from the medium. Jenni et al. (2012) investigated the interferences of CO2, O2, and various nitrogen compounds commonly found in wastewater treatment on NO and N2O sensors. They found that NO interfered with the N2O measurement, while the NO sensors were sensitive on NH3, NH2OH, HNO2, and N2H4. If high concentrations of these compounds are expected, it is recommended to check the concentrations of interfering compounds. No significant interferences were found by CO2 and O2. The cross-sensitivities can be corrected with calibration curves that are determined before the experiments. Jenni et al. (2012) also reported a significant temperature dependency. The NO signal increased by about 3.5% per 1°C and the N2O signal by 3.9% per 1°C. The temperature dependencies can be corrected with exponential functions.
Application of NO Microelectrodes
The novel NO microelectrode has been applied to study NO formation in permeable marine (Schreiber et al., 2008) and river (Schreiber et al., unpublished results) sediments. The results showed that in steady-state NO is produced in oxic/micro-oxic sediment strata reaching concentrations of 0.13 μmol L−1 in river and 0.5 μmol L−1 in marine sediments. In both sediments, NO produced in the oxic zone was consumed in the anoxic zone. It was hypothesized that NO was produced by AOB in the oxic zone. Labeling experiments with a 15N-labeled NO donor in the river sediment suggested that denitrification actively consumes exogenously produced NO.
Furthermore, the NO microelectrodes have been applied together with N2O microelectrodes in two N-cycling microbial biofilms; namely a complex NH+4-fed biofilm with nitrifying and denitrifying activity (Schreiber et al., 2009) and human dental plaque that was naturally exposed to high NO−3 and NO−2 in saliva (Schreiber et al., 2010). The study in dental plaque showed that plaque denitrified under aerobic conditions, that NO and N2O was produced by denitrification and that NO and N2O concentrations increased with decreasing pH. Aerobic denitrification has also been reported from permeable marine sediments (Gao et al., 2010) and from isolated (Patureau et al., 2000) or extracted soil bacteria (Morley et al., 2008). Until now, it is not known in which environments aerobic denitrification plays an important role, and if it is an environmentally significant NO and N2O emission pathway. NO, N2O, NO−2, NO−3, and O2 microelectrodes will be crucial to determine the importance of aerobic denitrification for NO and N2O release for complex ecosystems, because these sensors allow the simultaneous detection of NO, N2O, NO−2, NO−3, and O2 concentrations at high spatial resolution and their relation to denitrification activity.
Studying a complex N-cycling biofilm revealed the dynamics of NO and N2O formation upon perturbations in a system where nitrification and denitrification co-exist (Schreiber et al., 2009). The concomitant use of an O2 microelectrode and a set of control experiments enabled assignment of NO and N2O formation under oxic conditions to AOB and under anoxic conditions to denitrifiers. It also showed that AOB produce NO and N2O under fully oxic conditions if NO−2 concentrations are high. This is in agreement with other observations (Beaumont et al., 2004a,b; Shaw et al., 2006) and contradicts the assumption that that AOB require low O2 to release NO and N2O (Lipschultz et al., 1981; Poth and Focht, 1985; Kester et al., 1997; Beaumont et al., 2004a; Kampschreur et al., 2008b). The high temporal resolution of the microelectrodes allow to detect transient bursts (seconds to minutes) of NO and N2O. The bursts occurred by AOB upon O2 removal and upon NO−2 addition by both AOB and denitrifiers. The bursts only occurred if the perturbations were exerted upon metabolically active AOB and denitrifiers. In both scenarios NO and N2O are formed in parallel confirming that NO is the preceding intermediate of N2O in the N2O production pathways in this biofilm. An important contribution by Yu et al. (2010) showed that an AOB pure culture accumulated only NO, not N2O, upon transition from oxic to anoxic conditions. In mixed microbial communities were AOB and heterotrophic denitrifiers co-exist this could lead to NO release by AOB and immediate reduction to N2O by heterotrophic denitrifiers or anaerobic detoxification via NorVW and Hmp. This mixed source of N2O during transient oxic to anoxic conditions has to be taken into account when determining the pathways with isotopic techniques. It has been argued that N2O transiently accumulates during transition from anoxic to oxic conditions because O2 inhibits Nos while denitrification still proceeds, but direct evidence for this hypothesis is weak. Using both NO and N2O microelectrodes would allow to test this because N2O accumulation should not be accompanied by NO accumulation if the denitrification sequence is inhibited at the level of Nos.
Application of N2O Microelectrodes
In many habitats steady-state N2O concentrations are below or at the detection limit of the N2O microelectrode. Thus, the N2O microelectrode has commonly been used to estimate the denitrification potentials in stratified microbial communities such as sediments, biofilms, and aggregates in combination with the acetylene inhibition technique (Revsbech et al., 1988). Acetylene (~10 kPa) inhibits N2O reductase and leads to the accumulation of high amounts of N2O.
More recently, N2O microelectrodes have been used to study N2O production without acetylene inhibition in natural samples. These studies revealed that N2O concentrations in the micromolar range are expected when the system is exposed to a perturbation (Table 1). Transient accumulation of high N2O concentrations were achieved by any perturbation that affects the ambient O2 concentration: flooding of soils with water (Liengaard et al., 2011; Markfoged et al., 2011), creating an organic hotspot around a soil aggregate (Hojberg et al., 1994), thawing of permafrost soils (Elberling et al., 2010), and decreasing the O2 supply to wastewater-grown biofilms (Kampschreur et al., 2008a,b; Schreiber et al., 2009; Pellicer-Nàcher et al., 2010). In addition, increased input of NO−3, NO−2 or NH+4 to sediments, soils and biofilms (Hojberg et al., 1994; Meyer et al., 2008; Nielsen et al., 2009; Schreiber et al., 2009), organic inputs, salinity fluctuations in sediments (Nielsen et al., 2009) and changes of pH due to microbial activity in a denitrifying, dental biofilm (Schreiber et al., 2010) lead to increased micro-environmental N2O levels. Importantly, in many of these studies N2O accumulated in a transient manner making time-course measurements necessary to capture the N2O peak and the accumulation time span. The high spatial resolution of the N2O microelectrode allowed allocating processes that mitigate the emission of N2O to the atmosphere in soils, sediments and wastewater treatment biofilms. N2O that is produced by denitrification in deeper layers and is consumed during its diffusion toward the sediment-water interface in nutrient-enriched mangrove sediments (Meyer et al., 2008), toward the soil-atmosphere interface in a thawed permafrost soil (Elberling et al., 2010) or in a soil aggregate exposed to an organic hotspot (Hojberg et al., 1994). Likewise, N2O release from a membrane-aerated biofilm reactor was minimized by N2O-reducing microbes placed above AOB that produced N2O due to perturbations induced by an intermittent aeration regime (Pellicer-Nàcher et al., 2010).
Outlook
From the investigations of transient NO and N2O accumulation it emerges that two scenarios with distinct dynamics are important. First, N2O accumulates over hours to days, because it mirrors the onset of denitrification activity. Depending on the system it decreases because N2O reduction pathways are turned on with a delay or denitrification activity decreases due to substrate limitation. Ahn et al. (2011) even observed that peak NO and N2O emissions after a shift to O2-limitation in a nitrifying reactor were lasting for several month before adaptation on the metabolic or community level decreased the emissions. Second, perturbation of active AOB or denitrifiers leads to burst-like (within seconds to minutes) release of NO and N2O. The exact biochemical mechanisms for this require further research directly on the involved enzymes. Moreover, future research must show the contributions of the two types of transitions to the N2O budget and could use this as a framework to mitigate peak N2O releases to the atmosphere. Mitigation strategies could aid at avoiding perturbations or confining the N2O-releasing processes into a diffusion-limited environment that is overlaid with N2O-consuming microbial communities.
N2O Source Partitioning Based on the Nitrogen and Oxygen Isotopic Signature
In recent years, the isotopic signature of N2O has been used as a powerful tool to assign N2O production pathways to AOB and heterotrophic denitrifiers in different ecosystems such as soils, rivers, sea, wastewater treatment (Yoshida et al., 1989; Yamagishi et al., 2007; Baggs, 2008; Koba et al., 2009; Baulch et al., 2011; Park et al., 2011; Toyoda et al., 2011). N2O is a linear molecule (Nβ-Nα-O) with one nitrogen atom at the center position (Nα) bound to oxygen, and one at the end position (Nβ) bound to Nα. The three most abundant N2O isotopic species in the atmosphere are 14N15N16O (0.37%), 15N14N16O (0.37%) and 14N14N16O (>99%). Isotope abundances are usually reported in the δ-notation (in per-mil; ‰), δ15N = [(Rsample/Rreference)−1] × 1000, where R is the ratio of 15N/14N of a sample (Rsample) with respect to atmospheric N2 as the reference (Rreference) (Mariotti et al., 1981).
The intramolecular distribution of the nitrogen isotopes (14N15NO vs. 15N14NO) is termed SP and is expressed as the relative difference in δ15N between α and β position (SP = δ15Nα–δ15Nβ) (Toyoda and Yoshida, 1999). In analogy to the δ-notation, the isotopomer analysis denotes the relative difference of the 15N/14N isotope ratio for a given position (δ15Nα, δ15Nβ) with respect to the standard {e.g., δ15Nα = [(15Rα/15Rαreference)-1] × 1000, whereas 15Rα = (14N15N16O)/(14N14N16O) and 15Rαreference is the isotope ratio of the standard material (N2O) (see below)} (Toyoda and Yoshida, 1999). The SP has the advantage of being independent of the isotopic signature of the respective substrates (e.g., NH+4 or NO−3) but of being specific for pathways (enzymes) involved in N2O formation (Toyoda et al., 2005; Sutka et al., 2006).
Microbial (enzymatic) processes usually lead to an isotopic fractionation due to different transformation rates of 14N and 15N, resulting in isotopically lighter end-products than molecules in prior steps (Stein and Yung, 2003). Thus, the average 15N/14N ratio of N2O, termed as δ15NbulkN2O, can be used to distinguish different production pathways in complex samples if the isotopic signature of the pure bacterial culture is known. However, the meaning of δ15NbulkN2O can be limited since it is strongly-dependent on the isotopic signature of the substrate, which usually is unknown, as well as on the physiological activity (Mariotti et al., 1981). Additionally, the isotopic composition of an intermediate (e.g., N2O during heterotrophic denitrification) is affected by production (NO−3 reduction) as well as consumption (N2O reduction) processes.
In addition to nitrogen isotopes, oxygen isotope ratios are also increasingly used in order to better distinguish between the N2O formation pathways (Yoshinari and Wahlen, 1985; Kool et al., 2007; Baggs, 2008; Frame and Casciotti, 2010). In this case δ18O denotes the relative difference in the 18O/16O ratio of N2O (Rsample) with respect to the reference (Rreference), in per-mil (‰), usually being the Vienna Standard Mean Ocean Water (VSMOW) {δ18O = [(Rsample/Rreference)−1] × 1000} (Wahlen and Yoshinari, 1985).
Analysis of the Isotopic Signature of N2O
There are basically two different analytical techniques available to analyze N2O nitrogen isotopic signatures at natural abundance levels (Table 2): (1) the isotope-ratio mass spectrometry (IRMS) (Brenninkmeijer and Röckmann, 1999; Toyoda and Yoshida, 1999), and (2) the recently developed QCLAS (Waechter et al., 2008).
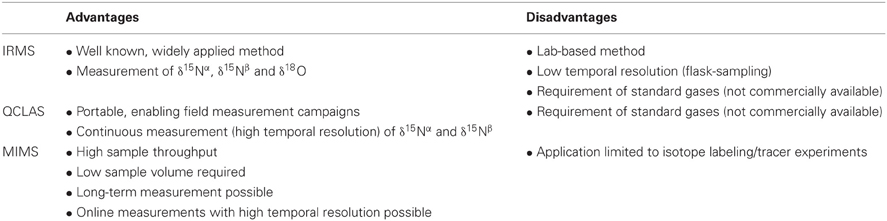
Table 2. Advantages and disadvantages of isotope-ratio mass spectrometry (IRMS), quantum cascade laser absorption spectroscopy (QCLAS) and membrane-inlet mass spectrometry (MIMS) adapted from Baggs (2008).
IRMS
IRMS-based method is widely applied with an excellent precision and accuracy (Mohn et al., 2010). Nevertheless, the calibration procedure of the intramolecular nitrogen isotope distribution in N2O is still under debate. Originally, two alternative approaches have been proposed, one by Toyoda and Yoshida (1999) and one by Brenninkmeijer and Röckmann (1999), which resulted in a difference in SP of about 30‰ for tropospheric N2O. The analysis of the SP by IRMS techniques relies on the N2O+ and NO+ fragment ions at the mass-to-charge ratio (m/z) 44, 45, 46 (for N2O) and m/z 30, 31 (for NO). However, both calibration approaches do not take into account the isotope effects associated with the formation of NO+ in the ion source of the mass spectrometer. Recently, Westley et al. (2007) investigated these discrepancies in more detail and found that these isotope effects have much smaller impact on the calibration procedure proposed by Toyoda and Yoshida (1999) (see below), and supported therefore this procedure as the most accurate basis for a community standard.
Furthermore, IRMS is a lab-based technique. Thus, the time resolution of N2O isotopic analysis during field measurement campaigns is therefore limited (Waechter et al., 2008). Nevertheless, in addition to nitrogen isotopes, the oxygen isotopic signature can also be analyzed routinely by IRMS.
QCLAS
QCLAS is a novel approach for site-specific analysis of nitrogen isotopes, with the advantage of a high sensitivity, time resolution, and portability, the latter of which enables field measurement campaigns (Waechter et al., 2008). This was demonstrated by Mohn et al. (2012), who recently presented first data of a high precision real-time analysis of site-specific isotopic signatures of atmospheric N2O above a grassland plot. The measurement campaign was run over 3 weeks with almost 550 analyzed gas samples. It was demonstrated that a continuous measurement of the N2O isotopic signature allowed improved detection of the dynamics of N2O production (before and after fertilizer application to the grassland plot), and thus opens a completely new field of applications. In another study, isotopic signature of N2O, produced during batch-scale experiments with activated sludge, were analyzed in real time, which permitted to trace short-term fluctuations in SP and δ15NbulkN2O, allowing to identify N2O production pathways in biological wastewater treatment (Wunderlin et al., unpublished results).
The QCLAS is based on direct absorption laser spectroscopy in the mid-infrared range for simultaneous measurement of the most abundant N2O isotopic species, such as 14N15N16O, 15N14N16O, and 14N14N16O (Waechter et al., 2008; Mohn et al., 2010). In order to enable high precision analysis (e.g., a precision of <0.1‰ for δ15Nα and δ15Nβ) (Waechter et al., 2008) a combination with a pre-concentration unit is essential at ambient or sub-ambient mixing ratios (Mohn et al., 2010, 2012). For example, with the liquid nitrogen-free, fully-automated pre-concentration unit built by Mohn et al. (2010), N2O can be concentrated by a factor of 200 (e.g., from ambient concentrations to around 60 ppm) from 10 L gas samples within 20 min.
Calibration
For both techniques, IRMS as well as QCLAS, an adequate calibration procedure needs to be applied, since instrumental nonlinearity and drifts impact the accuracy of the isotope ratio measurement (e.g., δ15NbulkN2O values depend on the N2O gas concentration) (Waechter et al., 2008). However, international standards are not commercially available so far. Therefore, they need to be prepared and analyzed from other laboratories (intercalibration) for δ15NbulkN2O, δ15Nα, and δ15Nβ, to ensure that measurements are performed on a common scale and that results are comparable between laboratories (Westley et al., 2007). So far, the calibration procedure proposed by Toyoda and Yoshida (1999), as mentioned above, is accepted as the provisional basis for a community standard: N2O is synthesized via thermal decomposition of isotopically characterized NH4NO3, since it is known that the nitrogen atom at the center (α) position of N2O originates from NO−3, while the end (β) nitrogen comes from NH+4. Using this calibration procedure a SP of tropospheric N2O of 18.7 ± 2.2‰ is measured (Westley et al., 2007).
Membrane-Inlet Mass Spectrometry (MIMS)
Membrane-inlet mass spectrometry (MIMS) was proposed as another promising tool to study the dynamics of N2O production in 15N labeling experiments. MIMS has a high sample throughput (within minutes), allows direct analysis of liquid or gas samples and requires only low sample amounts (Bauer, 1995; Baggs, 2008) (Table 2). Recently, it was coupled with an automated sampling and calibration unit (ASCU), and was tested in a long-term 15N-NO−3 tracer experiment over 7 days. It was confirmed that 15N measurements of N2 and N2O, detected as N2 at m/z 28, 29, and 30 (N2O was reduced to N2 in an elemental copper furnace prior to analysis), are in good agreement with IRMS-based analysis (Eschenbach and Well, 2011).
The membrane-inlet part can also be combined with a quadrupole mass spectrometer for simultaneous online measurement of different m/z ratios (e.g., 15,15N2O at m/z = 46, 14,15N2O at m/z = 45, 15,15N2 at m/z = 30, 14,15N2 at m/z = 29) with a time resolution of 1–2 min (Ettwig et al., 2010; Gao et al., 2010). Nevertheless, the interpretation of spectra corresponding to a certain gas mixture might be difficult since one peak can correspond to different atomic compositions (e.g., 14,14N+2 and CO+ at m/z = 28). This problem is reduced by applying 15N labeled substrates, where the only important remaining correction needed is for m/z = 30, which consist of the signal from the 15,15N+2 fragment of 15,15N2O, the 14NO+ fragment of 14,14N2O and 15,15N2) (Thomsen et al., 1994).
Isotopic Signature of N2O: Site Preference, δ15N and δ18O
Site Preference
The SP is a promising tool for N2O source partitioning since it is specific to pathways involved and independent of the respective substrates (Sutka et al., 2006) (Table 3). For N2O production via NH2OH oxidation by typical AOB pure cultures values in the range of 30.8 ± 5.9‰ to 35.6 ± 1.4‰ were measured (Sutka et al., 2003, 2004, 2006) which is in agreement with recently reported SP values of marine AOA (30.8 ± 4.4‰) (Santoro et al., 2011). In contrast, Frame and Casciotti (2010) estimated 36.3 ± 2.4‰ for a marine AOB. For nitrifier denitrification by AOB, the following SP values were reported: 0.1 ± 1.7‰ (Sutka et al., 2006), −0.8 ± 5.8‰ (Sutka et al., 2003, 2004) and −10.7 ± 2.9‰ (Frame and Casciotti, 2010). For N2O production via heterotrophic denitrification SP values in the range of −5.1‰ to 0‰ were reported (Toyoda et al., 2005; Sutka et al., 2006). Nitric oxide reductases (Nor) likely determine the SP of N2O during nitrifier denitrification as well as heterotrophic denitrification. The SP for both pathways is in the same range indicating that the involved Nor in AOB (cNor) and heterotrophic denitrifiers (cNor or qNor) (Stein and Yung, 2003; Stein, 2011) share a similar enzymatic mechanism. In case free NO is formed during NH2OH oxidation, any NO molecule that is funneled into nitrifier or heterotrophic denitrification (either directly or via initial oxidation to NO−2) would result in N2O with an SP of ~0‰ masking its initial NH2OH source.
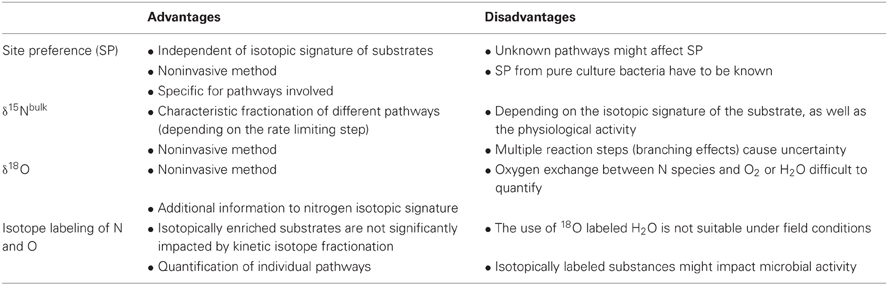
Table 3. Advantages and disadvantages of SP, δ15Nbulk, and δ18O, on a natural abundance or labeled level [adapted from Baggs (2008)].
The most probable explanation for a positive SP during NH2OH oxidation is a preferable 14N-16O bond cleavage of a symmetric intermediate such as hyponitrite (−16O14N15N16O−), leading to an enrichment of 14N-15N-16O (Schmidt et al., 2004a; Toyoda et al., 2005). In the current model of N2O formation from NH2OH oxidation, NH2OH is reduced to NO, which is further reduced to N2O by an unidentified Nor. However, the positive SP of N2O formed from NH2OH oxidation can only be explained, (1) if the involved Nor has a different mechanism than Nor's mediating nitrifier and heterotrophic denitrification or (2) if N2O is formed by a different mechanism, which does not involve free NO. We suggest mechanisms involving HNO: either by formation of free H2N2O2 with further chemical decomposition to N2O (discussed in section “HNO as intermediate of enzymatic NH2OH oxidation”) or a site specific enzymatic cleavage of −ONNO− as discussed above (Schmidt et al., 2004a; Toyoda et al., 2005). Further insights in the enzymatic mechanism of HAO and potentially HAO-associated Nor with careful chemical control experiments are needed to elucidate the biochemical mechanism of N2O formation during NH2OH oxidation.
Furthermore, a positive SP is, in addition to NH2OH oxidation, also an indicator for increasing importance of N2O reductase activity relative to N2O production (substantially greater activity than 10% compared to production) (Yamagishi et al., 2007; Jinuntuya-Nortman et al., 2008; Koba et al., 2009). As a consequence, N2O reduction to N2 might lead to an overestimation of N2O production by NH2OH oxidation, or vice versa. Nevertheless, further investigations are necessary in order to determine the individual signatures under conditions more representative for ecosystems with mixed culture populations (Wunderlin et al., unpublished results).
Under nitrifying conditions, N2O can theoretically be produced simultaneously via NH2OH oxidation as well as nitrifier denitrification. Thus, based on SP literature data, the individual contribution (FNN: NH2OH oxidation; FND: nitrifier denitrification) can be calculated from the following isotopomer mixing model:
where SPND and SPNN are the end-member SP signatures of the NH2OH oxidation and nitrifier denitrification pathway, respectively, as reviewed above, and SPtot the measured signature of the individual produced N2O (Frame and Casciotti, 2010).
δ15N
Wide ranges for δ15NbulkN2O were reported so far, mainly due to limited information about the isotopic signature of the substrates or to both a huge complexity determined by multiple transformation processes involving different enzymes, as well as variable reaction rates or mechanisms affecting isotopic fractionation (Perez et al., 2006) (Table 3). For example, it was shown that isotopic fractionation during NH3 oxidation is variable, depending mainly on the amino acid sequences for the α-subunit of AMO of the different investigated pure culture AOB (Casciotti et al., 2003). However, N2O produced by AOB during nitrifier denitrification or NH2OH oxidation is basically more strongly depleted in 15N (Δδ15N = δ15Nsubstrate− δ15 NbulkN2O; in the range of between 40‰ and 68‰) compared to heterotrophic denitrification, where N2O is an obligate intermediate and the fractionation therefore depends on both production and consumption processes (Δδ15N of 0–39‰) (Yoshida, 1988; Yoshida et al., 1989; Stein and Yung, 2003; Perez et al., 2006; Koba et al., 2009; Park et al., 2011).
δ18O
The oxygen isotopic signature of N2O (δ18O) is also used as a tool for N2O source partitioning, even though this approach faces a couple of difficulties: for example, N2O production via NH2OH oxidation as well heterotrophic N2O reduction result in a positive correlation between the δ18O in N2O and SP (Frame and Casciotti, 2010) (Table 3). Furthermore, δ18O enrichment factors are scarce and highly variable (Park et al., 2011), and are reported to be strongly influenced by oxygen exchange or incorporation, such as (1) oxygen incorporation (from dissolved O2) into NH2OH during the oxidation of NH+4 to NH2OH, (2) oxygen incorporation (from H2O) into NO−2 during the oxidation of NH2OH to NO−2, and (3) oxygen exchange between NO−2/NO−3 and H2O (Kool et al., 2007). For example, it was shown that 64–94% of the oxygen atoms in the precursors of N2O were exchanged with oxygen atoms in H2O (Snider et al., 2009; Park et al., 2011), which underscores the fact that the understanding and quantification of the effect of oxygen exchange between H2O and dissolved nitrogen species is and will remain challenging. Isotopic labeling is a promising approach to overcome such difficulties (see below), but up to now the natural abundance oxygen isotopic signature should be used with caution in N2O source partitioning studies (Kool et al., 2007, 2010).
N and O Labeling
Beside natural abundances, nitrogen and oxygen isotope labeling techniques have been applied to study and quantify N2O production pathways (Table 3). For example, Poth and Focht (1985) investigated the relative importance of the NH2OH oxidation and nitrifier denitrification pathway in Nitrosomonas europaea pure culture by applying 14N-NH+4 in combination with 15N-NO−2. Based on the large amounts of double-labeled 15,15N2O (m/z = 46), it was concluded that nitrifier denitrification is the dominant pathway. Baggs and Blum (2004) determined the relative contribution of nitrification and denitrification to 15N-N2O production by the application of 14NH154NO3 and 15NH154NO3. However, such conventional 15N labeling techniques do not allow to distinguish between NH2OH oxidation and nitrifier denitrification in mixed population systems (Kool et al., 2010). As a consequence, a dual isotope approach was applied, based on 18O-labeling of H2O as well as 15N-labeling of NH+4 or NO−3 (Wrage et al., 2005). The basic concept behind is, that AOB use oxygen from O2 for the oxidation of NH+4 to NH2OH, but oxygen from H2O for the oxidation of NH2OH to NO−2 (see above). As such, the 18O signature of N2O produced via nitrifier denitrification reflect to 50% the signature of O2 and to the other 50% the signature of H2O, which is in this study artificially enriched in 18O (Kool et al., 2007), under the assumption that no further oxygen is exchanged between NO−2 and H2O. In contrast, the 18O signature of N2O derived from NH2OH oxidation reflects to 100% the signature of O2 (Wrage et al., 2005; Kool et al., 2010). Nevertheless, the effect of oxygen exchange has to be taken into account.
Natural Samples
The analysis of the natural abundance isotopic signature of N2O emitted from ecosystems such as soils, rivers or biological wastewater treatment indicate that N2O from terrestrial and aquatic sources is depleted in 15N compared to tropospheric N2O (δ15N = 7‰ and δ18O = 20.7‰) (Stein and Yung, 2003), but also show a huge variability and complexity, making process identification ambiguous at large scale. For example, in biological wastewater treatment an average δ15NbulkN2O of −9.6‰, SP of 16‰ and δ18O of 22–44.3‰ were estimated (Yoshinari and Wahlen, 1985; Toyoda et al., 2011), indicating that nitrification as well as denitrification contributed to N2O production. N2O emitted from agricultural soils is reported to be strongly depleted in δ15NbulkN2O (e.g., −34‰) (Park et al., 2011), referring to nitrification dominated N2O production. Isotopic signatures of N2O emitted from rivers and streams are in the range of −18‰ to 2.4‰ (δ15Nbulk), −6‰ to 31‰ (SP) and 17‰ to 53‰ (δ18O) being in line with values reported above, which indicates to be highly influenced by sources such as agriculture or municipal wastewater treatment (Toyoda et al., 2009; Baulch et al., 2011). This is underscored by a recent study that investigates the oxygen and intramolecular nitrogen isotopic composition of N2O, confirming that nitrogen-based fertilizer application was largely responsible for the rise in N2O atmospheric concentration during the last 65 years (Park et al., 2012).
Outlook
In this section, the isotopic signature of N2O, especially the SP, is discussed to be a powerful tool to distinguish N2O production pathways. Recent technological advances, e.g., the development and application of the QCLAS, now allow a high temporal resolution in the analysis of the isotopic changes of N2O. Nevertheless, an adequate calibration procedure still needs to be applied, since instrumental nonlinearity and drifts impact the accuracy of the isotope ratio measurement, and calibration standards are not commercially available so far. It is a pressing issue to further investigate the characteristic isotopic signatures of the individual N2O production pathways in mixed microbial communities under controlled conditions, in order to more accurately interpret isotopic signatures from complex environmental systems. Further, it is important to study N2O isotopic signatures with respect to involved microbial communities, enzymatic reaction mechanisms and enzymatic transformation rates. The use of the oxygen isotopic signature of N2O as a reliable tool for pathway identification requires the elucidation of mechanisms and rates of oxygen exchange in the future.
Molecular Approaches to Understanding Microbial no and N2O Formation
While abiotic variables such as dissolved O2, pH, NO−2, and other nitrogen compounds have long been recognized to exert a strong influence on rates of microbial NO and N2O emissions, the importance of microbial community composition and dynamics to such emissions is still little understood (Wallenstein et al., 2006). As such, researchers have recently begun supplementing process-level NO and N2O emission measurements in a variety of environments with molecular techniques aimed at characterizing abundance, diversity, community structure, and activity of microbial guilds involved in nitrogen cycling. Here, we briefly introduce emerging molecular approaches to the delineation of key pathways, communities, and controls of NO and N2O production, and we summarize recent applications of these tools.
Quantifying the Genetic Potential for N2O Consumption
An appealing focus for application of molecular tools in environmental samples is direct quantification via the quantitative polymerase chain reaction (qPCR) of relevant functional genes (Smith and Osborn, 2008). Such an approach most commonly targets DNA, not RNA, and is thus a measure of genetic potential in the environment and not the activity.
Owing to the relative independence of each catabolic step, denitrification has been described as having a modular organization (Zumft, 1997). Indeed, Jones et al. (2008) concluded based on an analysis of 68 sequenced genomes of heterotrophic denitrifiers that approximately 1/3 lacked the nosZ gene encoding for N2O reductase and thus lack the genetic capacity for N2O reduction. Based on this assessment, researchers have hypothesized that the ratio of nosZ to the sum of nirK and nirS encoding for copper and cytochrome cd1-type nitrite reductases, respectively, is representative of the fraction of denitrifiers in a given environment that generate N2O as a catabolic end product. Environments with high nosZ/(nirK + nirS) ratios are likely associated with a high capacity for N2O consumption, and thus for low N2O emissions. Commonly used primers and qPCR conditions for genes relevant for NO and N2O turnover during N-cycling are available in the literature and are listed in Table 4, and thus the measurement of such ratios are feasible with little method development. Application of such tools has commonly shown a lower abundance of nosZ compared to other denitrifying reductases, particularly in soil environments (Henry et al., 2006; Hallin et al., 2009; Bru et al., 2011).
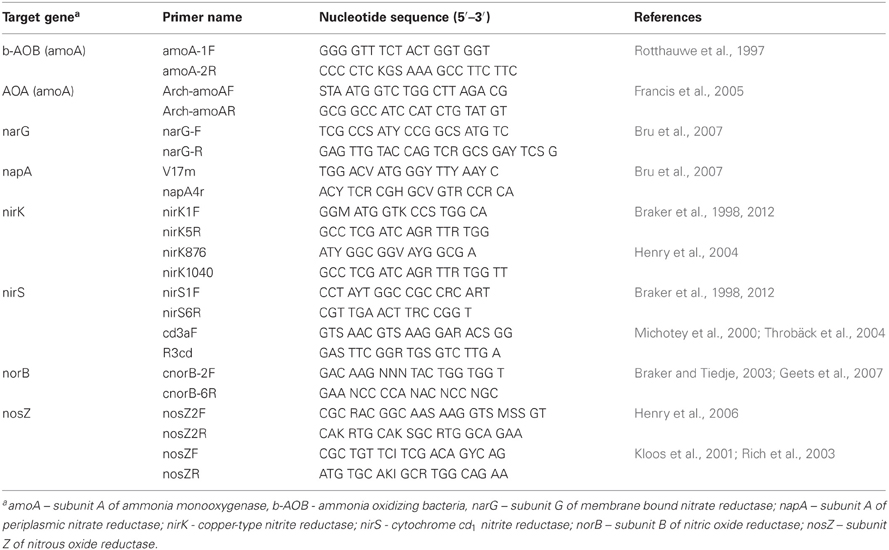
Table 4. Reported primers and literature references relevant for NO and N2O turnover during N-cycling.
First assessments of this hypothesis are somewhat conflicting. In favor for the hypothesis, Philippot et al. (2009) demonstrated a negative correlation between nosZ proportional abundance and N2O/(N2 + N2O) ratio in grassland pasture soil. In a follow-up study, Philippot et al. (2011) dosed three soils with several dilutions of a denitrifying bacterial isolate known to lack the nosZ gene, and measured the response at the DNA level of nirK, nirS, and nosZ genes via qPCR. N2O emissions increased in all soils upon dosing of the nosZ-deficient isolate. However, in two of the three soils, the increase in denitrification potential (relative to non-inoculated controls) was higher than the measured increase in N2O emissions, suggesting that the original denitrifier community was capable of acting as a sink for N2O production. Moreover, ratios of N2O emissions to total denitrifying end products (N2O + N2) in non-inoculated soils were not correlated to nosZ/(nirK + nirS). While the authors acknowledge that abundance of nosZ deficient denitrifiers may not be as important in soils with a high N2O uptake capacity, their results clearly demonstrate that abundance of denitrifiers incapable of N2O reduction can influence denitrification end products in natural environments. Similarly, Morales et al. (2010) document a strong positive correlation between the difference in nirS and nosZ gene abundance (nirS-nosZ; nirK was not quantified) and N2O emissions in 10 soils. Garcia-Lledo et al. (2011) suggested that a significant decrease in nosZ gene abundance during periods of high NO−3 content in a constructed wetland might be indicative of increased genetic capacity for (unmeasured) N2O emissions.
In contrast, Čuhel et al. (2010) detail a significant but, puzzlingly, positive correlation in grassland soil between nosZ/(nirS + nirK) ratios and N2O/(N2+N2O), but caution that the relative importance of denitrifier community composition and enzyme regulation relative to proportion of nosZ deficient community members remains uncertain. In line with this result, Braker and Conrad (2011) found similar ratios of nosZ/(nirS + nirK) via Most Probable Number (MPN-) PCR in three soils with profoundly different N2O/(N2+N2O) ratios, and concluded that the hypothesis that a higher abundance of denitrifiers lacking nosZ is linked to increased N2O emissions may be an oversimplification.
The genetic potential for N2O production via nitrifier denitrification in AOB (and possibly AOA) could theoretically be measured via qPCR of the nirK and norB genes. Design of such analyses is hampered due to the fact that AOB nirK and norB genes are not phylogenetically distinct from that of heterotrophic denitrifying organisms (Cantera and Stein, 2007; Garbeva et al., 2007). In addition, NorB is not the only NO reductase in AOB (Stein, 2011).
Community Structure and Diversity Impacts on NO and N2O Production
In addition to monitoring abundance of nosZ deficient denitrifiers, PCR-based tools are now being applied to the investigation of links between community structure and N2O emissions for both nitrifiers and denitrifiers. For this purpose, community structure is commonly profiled via cultivation-independent molecular fingerprinting methods, such as terminal restriction fragment length polymorphism (T-RFLP) or denaturing gradient gel electrophoresis (DGGE), targeting either 16S rRNA fragments specific to the functional guild of interest or functional genes (for example, nirK or amoA) directly. In addition, traditional cloning and Sanger sequencing and, increasingly, barcoded amplicon-based pyrosequencing of functional genes are often employed for robust phylogenetic comparisons. Readers are referred to Prosser et al. (2010) for a detailed methodological description of these and other nucleic-acid based methods. Multivariate statistical analyses such as canonical correspondence analysis (CCA), redundancy analysis (RDA) (Ramette, 2007; Wells et al., 2009), or path analysis (Avrahami and Bohannan, 2009) can then be used to explore the interplay between abiotic variables, community composition, and extant process rates.
It should be emphasized that the molecular and statistical tools highlighted above are most commonly used in microbial ecology to explore correlations, rather than causal associations, between community structure and function in complex microbial communities. As discussed in detail by Reed and Martiny (2007) directly testing causal relationships between microbial community composition or diversity and ecosystem processes is significantly more difficult, but experimental approaches often drawn from classical ecology are now being adapted to this challenge. We anticipate that future studies testing the functional significance of microbial community structure to NO or N2O production will benefit greatly from these approaches.
Studies targeting the relationship between nitrifier community composition and greenhouse gas production are sparse at present, despite the fact that ample molecular tools are available for this purpose. Avrahami and Bohannan (2009) employed a combination of qPCR and T-RFLP to explore the response of N2O emission rates and betaproteobacterial AOB abundance and composition in a California meadow to manipulations in temperature, soil moisture, and fertilizer concentration. While a complex interaction between factors was determined to directly and indirectly contribute to N2O emission rates, path analysis suggested that the major path by which NH+4 influenced emission rates in the high N fertilization treatment was indirectly via two specific AOB clusters. This observation suggested a significant relationship between AOB community structure and N2O emission rates. It is important to note that this study did not attempt to discriminate between the nitrifier denitrification and NH2OH oxidation pathways for AOB-linked N2O production, nor was the relative importance of heterotrophic denitrification vs. nitrification for overall N2O emissions directly compared.
Assessment of the importance of DNRA as a process, and diversity therein, to NO and N2O production is in its infancy. It has been suggested that our understanding of this little understood phenomena would benefit from the future investigations employing molecular techniques to quantify abundance and diversity of the nrf gene in conjunction with either modeling or stable isotope-based methods (Baggs, 2011). To our knowledge, such an assessment has yet to be conducted.
The relationship between denitrifier community composition and N2O emissions, while still ambiguous, has been studied in more detail. Palmer et al. (2010) investigated narG (encoding for membrane-bound nitrate reductase, Nar) and nosZ phylogenetic diversity in a low-pH fen via gene clone libraries and T-RFLP. They documented novel narG and nosZ genotypes and a phylogenetically diverse low-pH adapted denitrifier community, and suggested that the novel community structure may be responsible for complete denitrification and low N2O emissions under in situ conditions. In a more recent study, Palmer et al. (2012) investigated denitrifier gene diversity in peat circles in the arctic tundra via barcoded amplicon pyrosequencing of narG, nirK/nirS, and nosZ, and found evidence that high and low N2O emission patterns were associated with contrasting denitrifier community composition. Braker et al. (2012) found that, of three soils profiled, the soil with the most robust denitrification (lowest N2O/N2 ratio) harbored the most diverse denitrifier community, as measured via nosZ and nirK sequence diversity, suggesting that differences in community composition (higher diversity) are associated with ecosystem-level functional differences. In denitrifying bioreactors, population dynamics tracked via 16S rRNA-based T-RFLP were strongly correlated to NO−2 appearance and emissions of N2O (Gentile et al., 2007). In contrast, Rich and Myrold (2004) found little relationship between nosZ phylogenetic diversity as measured via T-RFLP in wet soils and creek sediments in an agrosystem, and suggested that activity and community composition were uncoupled in this ecosystem.
Taken together, the body of literature reviewed here suggests that, in at least some cases, community structure and diversity can play a functionally significant role in microbial N2O emissions. The importance of community composition relative to environmental parameters and metabolic adaptation in response to transient conditions (for example, shifts in patterns of gene expression or regulation) in determining N2O production, however, remains poorly understood. A worthwhile, but challenging future research direction would be to tease apart the influence of whole community metabolic adaptation versus community shifts on NO/N2O emissions in mixed microbial communities.
A Role for Variation in Regulatory Response
Differences in transcriptional and translational regulation as well as enzyme activity have also been highlighted as potentially critical modulators of microbial NO or N2O production (Richardson et al., 2009; Bergaust et al., 2011; Braker and Conrad, 2011). Such differences likely contribute to observed associations between community structure and greenhouse gas production discussed above. Strong regulation at the transcriptional, translational, and enzyme level is likely occurring in both nitrifier and denitrifier communities, and such regulation complicates attempts to directly relate abundance or diversity of functional guilds to process rates (Braker and Conrad, 2011). Similarly, transient near-instantaneous NO and N2O accumulation in active nitrifying and denitrifying biofilms in response to O2 or NO−2 perturbations, as measured with high temporal resolution via microelectrodes, strongly suggests that dynamics are controlled in some cases at the enzyme level (Schreiber et al., 2009). Indeed, culture-based assays targeting denitrifier isolates from two soils demonstrated substantial diversity in sensitivity of Nos enzymes to O2 and provided a physiological underpinning for a previously observed link between denitrifier community composition and rate of N2O production (Cavigelli and Robertson, 2000).
Gene expression can be readily quantified with reverse transcriptase quantitative PCR (RT-qPCR), and researchers are now beginning to explore the relationship between gene expression patterns for critical functional genes (amoA, hao, nirK, nirS, norB, and nosZ) and NO/N2O emissions. Yu et al. (2010) used such an approach to quantify expression of amoA, hao, nirK, and norB in chemostats of Nitrosomonas europaea during initiation and recovery from transient anoxic conditions. Surprisingly, expression profiles of nirK and norB were not strongly linked; strong overexpression of nirK concomitant with NO accumulation was observed upon initiation of anoxia, and at the same time norB, amoA, and hao gene transcripts declined in abundance. N2O emissions peaked during recovery to aerated conditions, but did not correlate strongly to gene expression. The methods of Yu et al. (2010) provide a robust road map for examining relationships between nitrifier gene expression and NO/N2O emissions in mixed communities in environmental settings, though it should be noted that such an analysis is complicated by the polyphyletic nature of the AOB nirK and norB genes.
RT-qPCR has also been used to assess the relationship between gene expression and NO/N2O production in systems dominated by denitrifiers. Liu et al. (2010) quantified the relationship between nirS, nirK, and nosZ gene pools, their transcription products, and gas kinetics (NO, N2O, and N2) as a function of pH in soils. Interestingly, neither gene pool abundance, nor transcription rates could explain a profound increase in N2O emissions at low pH. The authors attribute the observed N2O:N2 product ratio to post-transcriptional phenomenon, although it is also plausible that enhanced chemo-denitrification may play a role.
A worthy future contribution could be made via direct environmental metatranscriptomic assessment of patterns in microbial gene expression in environments with different or varying rates of NO or N2O production. Metatranscriptomics is the direct sequencing of cDNA generated via reverse transcription of environmental RNA transcripts, and therefore provides a picture of currently transcribed genes in a given environment (Morales and Holben, 2011). In line with the results of Liu et al. (2010), it is important to recognize that measurement of the size or diversity of the gene transcript pool neglects post-transcriptional regulation governing, for example, the assembly of N2O reductase and enzyme activity (Braker and Conrad, 2011). As of yet, variations in post-transcriptional regulation at the community level and its effect on NO/N2O production has been little explored in nitrifying and denitrifying pure cultures and communities. Critical insights in this regard may be possible in the future from an approach coupling metatranscriptomics and metaproteomics—that is, direct measurement of the composition of the proteome in an environment.
A Need for an Integrated Approach to NO and N2O Turnover in Complex Microbial Communities
NO and N2O can be produced by many different biological and chemical reactions. Considerable progress has been made to allocate NO and N2O production to certain biological pathways, but commonly some uncertainty remains, because many processes share the same reaction sequence for N2O production via NO and NO−2. We delineated basically three-independent approaches to allocate pathways (indirect inference; isotopic signature of N2O, and isotopic labeling). Parallel use of these approaches will increase confidence in the interpretation. The possibility for various chemical reaction that produce and consume NO and N2O additionally complicate the picture. Chemical reactions can be important in engineered systems that employ waters with concentrated N-contents and in natural systems, where low pH values coincide with high ammonia inputs. However, in most natural systems and in municipal wastewater treatment, chemical reactions will probably not be the main contributors of NO and N2O emissions. Nevertheless, the possibility of chemical NO and N2O production has to be considered when interpreting measurements results. Experiments with inactivated biomass could help to give a first estimation of the chemical production rates. However, care has to be taken since the chemical conditions that facilitate chemical NO and N2O production such as pH and trace metal availability are in turn shaped by microbial activity.
Molecular methods have largely been applied independently from the stable isotope and microelectrode approaches. Ample opportunities exist for integration of these techniques. Indeed, it is clear that such an integrated approach is critical to assessing the importance of microscale heterogeneity in environmental parameters, microbial community structure and stability, and genetic regulation to observed process-level N2O emission rates.
Joint use of stable isotope methods in conjunction with molecular techniques appears particularly important, given reported difference in isotope effects depending on the community structure of nitrifiers (Casciotti et al., 2003) or denitrifiers (Toyoda et al., 2005) present. In addition, linking source-partitioned N2O as measured via stable isotope techniques to the underlying microbial communities via molecular approaches may allow a more significant measure of the strength of coupling between microbial diversity and measured emissions (Baggs, 2008, 2011). One promising way forward is to assess environmental conditions that favor a shift of dominant N2O production pathway (for example, from denitrification to nitrification, or vice versa) as measured via stable isotope methods, and to simultaneously link such a shift to diversity and abundance of functional gene pools and transcripts via PCR-based molecular approaches. Such an approach has the potential to yield insights into the relative importance of dominant functional guilds, community composition, and activity in determining microbial NO/N2O production rates. A fruitful first application would be to combine stable isotope-based methods with the molecular approach pioneered by Yu et al. (2010) for delineating the relationship between transcriptional response of the model AOB Nitrosomonas europaea and NO/N2O production. This coupled approach would allow conclusive verification of conditions proposed by Chandran et al. (2011) to favor a switch between nitrifier denitrification and NH2OH oxidation as dominant sources NO and N2O production.
Similarly, it is clear that molecular tools and microelectrodes are complementary to study NO and N2O turnover. An excellent example of such integration is provided by Okabe et al. (2011), who profiled microscale gradients in N2O emissions in anammox granules and compared these profiles to spatial location of AOB, as measured via fluorescence in situ hybridization (FISH). Based on their results, the authors concluded that putative heterotrophic denitrifiers in the inner part of the granule, not AOB, were likely responsible for the majority of the extant N2O process emissions. A similar approach is likely applicable in a wide variety of environments, including flocs, sediments, soils, and microbial mats. In addition, use of either FISH probes with higher phylogenetic resolution or depth stratified DNA/RNA extraction coupled to PCR-based measurements may allow a direct microscale assessment of links between microbial diversity and activity and NO/N2O production profiles. Such a microscale assessment is important because stratified environments likely contain both regions of N2O production and consumption that are masked during bulk NO/N2O concentration measurements or DNA/RNA extractions. In addition, microelectrode measurements with high temporal resolution should be combined with qPCR to better understand the regulation of NO and N2O peak emissions from different environments.
The conditions for NO and N2O formation in pure cultures and by chemical reactions begin to be better understood. Furthermore, several recent technological advancements allow researcher to investigate the regulation of NO and N2O formation in complex environments at high spatial and temporal resolution. These advancements provide a cornerstone to understand and mitigate the release of NO and N2O from natural and engineered environments.
Conflict of Interest Statement
Frank Schreiber has a license agreement with Unisense A/S for the constrution and distribution of an NO microelectrode. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Joachim Mohn (Swiss Federal Laboratories for Materials Testing and Research, Empa) for helpful discussions during the preparation of the manuscript.
References
Ahlers, B., Konig, W., and Bock, E. (1990). Nitrite reductase activity in Nitrobacter vulgaris. FEMS Microbiol. Lett. 67, 121–126.
Ahn, J. H., Kim, S., Park, H., Rahm, B., Pagilla, K., and Chandran, K. (2010). N2O emissions from activated sludge processes, 2008–2009, results of a national monitoring survey in the United States. Environ. Sci. Technol. 44, 4505–4511.
Ahn, J. H., Kwan, T., and Chandran, K. (2011). Comparison of partial and full nitrification processes applied for treating high-strength nitrogen wastewaters: microbial ecology through nitrous oxide production. Environ. Sci. Technol. 45, 2734–2740.
Alluisetti, G. E., Almaraz, A. E., Amorebieta, V. T., Doctorovich, F., and Olabe, J. A. (2004). Metal-catalyzed anaerobic disproportionation of hydroxylamine. Role of diazene and nitroxyl intermediates in the formation of N2, N2O, NO+, and NH3. J. Am. Chem. Soc. 126, 13432–13442.
Andersen, K., Kjaer, T., and Revsbech, N. P. (2001). An oxygen insensitive microsensor for nitrous oxide. Sensor. Actuat. B Chem. 81, 42–48.
Anjum, M. F., Stevanin, T. M., Read, R. C., and Moir, J. W. B. (2002). Nitric oxide metabolism in Neisseria meningitidis. J. Bacteriol. 184, 2987–2993.
Avrahami, S., and Bohannan, B. J. M. (2009). N2O emission rates in a California meadow soil are influenced by fertilizer level, soil moisture and the community structure of ammonia-oxidizing bacteria. Glob. Change Biol. 15, 643–655.
Baggs, E. M. (2008). A review of stable isotope techniques for N2O source partitioning in soils: recent progress, remaining challenges and future considerations. Rapid Commun. Mass Spectrom. 22, 1664–1672.
Baggs, E. M. (2011). Soil microbial sources of nitrous oxide: recent advances in knowledge, emerging challenges and future direction. Curr. Opin. Environ. Sust. 3, 321–327.
Baggs, E. M., and Blum, H. (2004). CH4 oxidation and emissions of CH4 and N2O from Lolium perenne swards under elevated atmospheric CO2. Soil Biol. Biochem. 36, 713–723.
Bateman, E. J., and Baggs, E. M. (2005). Contributions of nitrification and denitrification to N2O emissions from soils at different water-filled pore space. Biol. Fertil. Soils 41, 379–388.
Bauer, S. (1995). Membrane introduction mass spectrometry; an old method that is gaining new interest through recent technological advances. Trends Anal. Chem. 14, 202–213.
Baulch, H. M., Schiff, S. L., Thuss, S. J., and Dillon, P. J. (2011). Isotopic character of nitrous oxide emitted from streams. Environ. Sci. Technol. 45, 4682–4688.
Baumann, B., Snozzi, M., Zehnder, A. J., and van der Meer, J. R. (1996). Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J. Bacteriol. 178, 4367–4374.
Beaulieu, J. J., Tank, J. L., Hamilton, S. K., Wollheim, W. M., Hall, R. O., Mulholland, P. J., et al. (2011). Nitrous oxide emission from denitrification in stream and river networks. Proc. Natl. Acad. Sci. U.S.A. 108, 214–219.
Beaumont, H. J. E., Hommes, N. G., Sayavedra-Soto, L. A., Arp, D. J., Arciero, D. M., Hooper, A. B., et al. (2002). Nitrite reductase of Nitrosomonas europaea is not essential for production of gaseous nitrogen oxides and confers tolerance to nitrite. J. Bacteriol. 184, 2557–2560.
Beaumont, H. J. E., Lens, S. I., Reijnders, W. N. M., Westerhoff, H. V., and van Spanning, R. J. M. (2004a). Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol. Microbiol. 54, 148–158.
Beaumont, H. J. E., van Schooten, B., Lens, S. I., Westerhoff, H. V., and van Spanning, R. J. M. (2004b). Nitrosomonas europaea expresses a nitric oxide reductase during nitrification. J. Bacteriol. 186, 4417–4421.
Bedioui, F., and Villeneuve, N. (2003). Electrochemical nitric oxide sensors for biological samples – principle, selected examples and applications. Electroanalysis 15, 5–18.
Bergaust, L., Bakken, L. R., and Frostegård, A. (2011). Denitrification regulatory phenotype, a new term for the characterization of denitrifying bacteria. Biochem. Soc. Trans. 39, 207–212.
Bergaust, L., Mao, Y., Bakken, L. R., and Frostegård, A. (2010). Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrous oxide reductase in Paracoccus denitrificans. Appl. Environ. Microbiol. 76, 6387–6396.
Berks, B. C., Baratta, D., Richardson, J., and Ferguson, S. J. (1993). Purification and characterization of a nitrous oxide reductase from Thiosphaera pantotropha. Implications for the mechanism of aerobic nitrous oxide reduction. Eur. J. Biochem. 212, 467–476.
Bollmann, A., and Conrad, R. (1998). Influence of O2 availability on NO and N2O release by nitrification and denitrification in soils. Glob. Change Biol. 4, 387–396.
Bonner, F. T., Dzelzkalns, L. S., and Bonucci, J. A. (1978). Properties of nitroxyl as intermediate in the nitric oxide-hydroxylamine reaction and in trioxodinitrate decomposition. Inorg. Chem. 17, 2487–2494.
Bonner, F. T., and Hughes, M. N. (1988). The aqueous solution chemistry of nitrogen in low positive oxidation states. Comments Inorg. Chem. 7, 215–234.
Bonner, F. T., and Stedman, G. (1996). “The chemistry of nitric oxide and redox-related species,” in Methods in Nitric Oxide Research, eds M. Feelisch and J. S. Stamler (New York, NY: John Wiley and Sons), 3–18.
Braker, G., and Conrad, R. (2011). Diversity, structure, and size of N2O producing microbial communities in soil – what matters for their functioning? Adv. Appl. Microbiol. 75, 33–69.
Braker, G., Dörsch, P., and Bakken, L. R. (2012). Genetic characterization of denitrifier communities with contrasting intrinsic functional traits. FEMS Microbiol. Ecol. 79, 542–554.
Braker, G., Fesefeldt, A., and Witzel, K.-P. (1998). Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64, 3769–3775.
Braker, G., and Tiedje, J. M. (2003). Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl. Environ. Microbiol. 69, 3476–3483.
Brenninkmeijer, C. A. M., and Röckmann, T. (1999). Mass spectrometry of the intramolecular nitrogen isotope distribution of environmental nitrous oxide using fragment-ion analysis. Rapid Commun. Mass Spectrom. 13, 2028–2033.
Bru, D., Ramette, A., Saby, N. P. A., Dequiedt, S., Ranjard, L., Jolivet, C., et al. (2011). Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 5, 532–542.
Bru, D., Sarr, A., and Philippot, L. (2007). Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl. Environ. Microbiol. 73, 5971–5974.
Butler, J. H., and Gordon, L. I. (1986a). An improved gas chromatographic method for the measurement of hydroxylamine in marine and fresh waters. Mar. Chem. 19, 229–243.
Butler, J. H., and Gordon, L. I. (1986b). Rates of nitrous oxide production in the oxidation of hydroxylamine by iron (III). Inorg. Chem. 25, 4573–4577.
Cabail, M. Z., Kostera, J., and Pacheco, A. A. (2005). Laser photoinitiated nitrosylation of 3-electron reduced Nm europaea hydroxylamine oxidoreductase: kinetic and thermodynamic properties of the nitrosylated enzyme. Inorg. Chem. 44, 225–231.
Cabail, M. Z., and Pacheco, A. A. (2003). Selective one-electron reduction of Nitrosomonas europaea hydroxylamine oxidoreductase with nitric oxide. Inorg. Chem. 42, 270–272.
Cantera, J. J. L., and Stein, L. Y. (2007). Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ. Microbiol. 9, 765–776.
Casciotti, K. L., Sigman, D. M., and Ward, B. B. (2003). Linking diversity and stable isotope fractionation in ammonia-oxidizing bacteria. Geomicrobiol. J. 20, 335–353.
Cavigelli, M. A., and Robertson, G. P. (2000). The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology 81, 1402–1414.
Chalk, P. M., and Smith, C. J. (1983). “Chemodenitrification,” in Gaseous Loss of Nitrogen from Plant-Soil Systems, eds J. R. Freney and J. R. Simpson (Dordrecht, The Netherlands: Kluwer), 65–89.
Chandran, K., Stein, L. Y., Klotz, M. G., and van Loosdrecht, M. C. M. (2011). Nitrous oxide production by lithotrophic ammonia-oxidizing bacteria and implications for engineered nitrogen-removal systems. Biochem. Soc. Trans. 39, 1832–1837.
Charpentier, J., Farias, L., Yoshida, N., Boontanon, N., and Raimbault, P. (2007). Nitrous oxide distribution and its origin in the central and eastern South Pacific Subtropical Gyre. Biogeosciences 4, 729–741.
Corker, H., and Poole, R. K. (2003). Nitric oxide formation by Escherichia coli-dependence on nitrite reductase, the NO-sensing regulator Fnr, and flavohemoglobin Hmp. J. Biol. Chem. 278, 31584–31592.
Costa, C., Macedo, A., Moura, I., Moura, J. G. G., Le Gall, J., Berlier, Y., et al. (1990). Regulation of the hexaheme nitrite/nitric oxide reductase of Desulfovibrio desulfuricans, Wolinella succinogenes and Escherichia coli: a mass spectrometric study. FEBS Lett. 1, 67–70.
Crutzen, P. J. (1979). The role of NO and NO2 in the chemistry of the troposphere and stratosphere. Ann. Rev. Earth Planet. Sci. 7, 443–472.
Čuhel, J., Šimek, M., Laughlin, R. J., Bru, D., Chèneby, D., Watson, C. J., et al. (2010). Insights into the effect of soil pH on N2O and N2 emissions and denitrifier community size and activity. Appl. Environ. Microbiol. 76, 1870–1878.
Döring, C., and Gehlen, H. (1961). Über die Kinetic der Reaktion zwischen Hydroxylamin und Salpetriger Säure. Zeitschrift für anorganische und allgemeine Chemie 312, 32–44.
DuMond, J. F., and King, S. B. (2011). The chemistry of nitroxyl-releasing compounds. Antioxid. Redox Signal. 14, 1637–1648.
Elberling, B., Christiansen, H. H., and Hansen, B. U. (2010). High nitrous oxide production from thawing permafrost. Nat. Geosci. 3, 332–335.
Elkins, J. W., Wofsy, S. C., McElroy, M. B., Kolb, C. E., and Kaplan, W. A. (1978). Aquatic sources and sinks for nitrous oxide. Nature 275, 602–606.
Eschenbach, W., and Well, R. (2011). Online measurement of denitrification rates in aquifer samples by an approach coupling an automated sampling and calibration unit to a membrane inlet mass spectrometry system. Rapid Commun. Mass Spectrom. 25, 1993–2006.
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M. M., et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548.
Farias, L., Paulmier, A., and Gallegos, M. (2007). Nitrous oxide and N-nutrient cycling in the oxygen minimum zone off northern Chile. Deep Sea Res. 54, 164–180.
Fehling, C., and Friedrichs, G. (2011). Dimerization of HNO in aqueous solution: an interplay of solvation effects, fast acid-base equilibria, and intramolecular hydrogen bonding? J. Am. Chem. Soc. 133, 17912–17922.
Fernández, M. L., Estrin, D. A., and Bari, S. E. (2008). Theoretical insight into the hydroxylamine oxidoreductase mechanism. J. Inorg. Biochem. 102, 1523–1530.
Firestone, M. K., Firestone, R. B., and Tiedje, J. M. (1980). Nitrous oxide from soil denitrification: factors controlling its biological production. Science 208, 749–751.
Firestone, M. K., and Tiedje, J. M. (1979). Temporal change in nitrous oxide and dinitrogen from denitrification following onset of anaerobiosis. Appl. Environ. Microbiol. 38, 673–679.
Frame, C. H., and Casciotti, K. L. (2010). Biogeochemical controls and isotopic signatures of nitrous oxide production by a marine ammonia-oxidizing bacterium. Biogeosciences 7, 2695–2709.
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A. 102, 14683–14688.
Freitag, A., Rudert, M., and Bock, E. (1987). Growth of nitrobacter by dissimilatoric nitrate reduction. FEMS Microbiol. Lett. 48, 105–109.
Friedemann, M. N., Robinson, S. W., and Gerhardt, G. A. (1996). o-Phenylenediamine-modified carbon fiber electrodes for the detection of nitric oxide. Anal. Chem. 68, 2621–2628.
Frunzke, K., and Zumft, W. (1986). Inhibition of nitrous-oxide respiration by nitric oxide in the denitrifying bacterium Pseudomonas perfectomarina. Biochim. Biophys. Acta 852, 119–125.
Gao, H., Schreiber, F., Collins, G., Jensen, M. M., Svitlica, O., Kostka, J. E., et al. (2010). Aerobic denitrification in permeable Wadden Sea sediments. ISME J. 4, 417–426.
Garbeva, P., Baggs, E. M., and Prosser, J. I. (2007). Phylogeny of nitrite reductase (nirK) and nitric oxide reductase (norB) genes from Nitrosospira species isolated from soil. FEMS Microbiol. Lett. 266, 83–89.
Garcia-Lledo, A., Vilar-Sanz, A., Trias, R., Hallin, S., and Baneras, L. (2011). Genetic potential for N2O emissions from the sediment of a free water surface constructed wetland. Water Res. 45, 5621–5632.
Gardner, A. M., Helmick, R. A., and Gardner, P. R. (2002). Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 277, 8172–8177.
Gardner, P. R., Gardner, A. M., Martin, L. A., and Salzman, A. L. (1998). Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. U.S.A. 95, 10378–10383.
Geets, J., de Cooman, M., Wittebolle, L., Heylen, K., Vanparys, B., De Vos, P., et al. (2007). Real-time PCR assay for the simultaneous quantification of nitrifying and denitrifying bacteria in activated sludge. Appl. Microbiol. Biotechnol. 75, 211–221.
Gentile, M. E., Lynn Nyman, J., and Criddle, C. S. (2007). Correlation of patterns of denitrification instability in replicated bioreactor communities with shifts in the relative abundance and the denitrification patterns of specific populations. ISME J. 1, 714–728.
Gieseke, A., and de Beer, D. (2004). “Use of microelectrodes to measure in situ microbial activities in biofilms, sediments, and microbial mats,” in Molecular Microbial Ecology Manual, eds G. Kowalchuk, F. de Bruijn, I. Head, A. Akkermans, and J. van Elsas (Heidelberg: Springer), 1581–1612.
Gilberthorpe, N. J., and Poole, R. K. (2008). Nitric oxide homeostasis in Salmonella typhimurium: roles of respiratory nitrate reductase and flavohemoglobin. J. Biol. Chem. 283, 11146–11154.
Gödde, M., and Conrad, R. (2000). Influence of soil properties on the turnover of nitric oxide and nitrous oxide by nitrification and denitrification at constant temperature and moisture. Biol. Fertil. Soils 32, 120–128.
Gomes, C. M., Giuffrè, A., Forte, E., Vicente, J. B., Saraiva, L. M., Brunori, M., et al. (2002). A novel type of nitric-oxide reductase. Escherichia coli flavorubredoxin. J. Biol. Chem. 277, 25273–25276.
Granger, J., and Ward, B. B. (2012). Accumulation of nitrogen oxides in copper-limited cultures of denitrifying bacteria. Limnol. Oceanogr. 48, 313–318.
Hallin, S., Jones, C. M., Schloter, M., and Philippot, L. (2009). Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 3, 597–605.
Heisterkamp, I., Schramm, A., de Beer, D., and Stief, P. (2010). Nitrous oxide production associated with coastal marine invertebrates. Mar. Ecol. Prog. Ser. 415, 1–9.
Hendrich, M. P., Upadhyay, A. K., Riga, J., Arciero, D. M., and Hooper, A. B. (2002). Spectroscopic characterization of the NO adduct of hydroxylamine oxidoreductase. Biochemistry 41, 4603–4611.
Hendriks, J., Oubrie, A., Castresana, J., Urbani, A., Gemeinhardt, S., and Saraste, M. (2000). Nitric oxide reductases in bacteria. Biochim. Biophys. Acta 1459, 266–273.
Henry, S., Baudoin, E., Lopezgutierrez, J., Martinlaurent, F., Brauman, A., and Philippot, L. (2004). Quantification of denitrifying bacteria in soils by gene targeted real-time PCR. J. Microbiol. Methods 59, 327–335.
Henry, S., Bru, D., Stres, B., Hallet, S., and Philippot, L. (2006). Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72, 5181–5189.
Hojberg, O., Revsbech, N. P., and Tiedje, J. M. (1994). Denitrification in soil aggregates analyzed with microsensors for nitrous oxide and oxygen. Soil Sci. Soc. Am. J. 58, 1691–1698.
Hooper, A. B. (1968). A nitrite-reducing enzyme from Nitrosomonas europaea: preliminary characterization with hydroxylamine as electron donor. Biochim. Biophys. Acta 162, 49–65.
Hooper, A. B., and Terry, K. R. (1979). Hydroxylamine oxidoreductase of Nitrosomonas. Production of nitric oxide from hydroxylamine. Biochim. Biophys. Acta 571, 12–20.
Horn, M. A., Schramm, A., and Drake, H. L. (2003). The earthworm gut: an ideal habitat for ingested N2O-producing microorganisms. Appl. Environ. Microbiol. 69, 1662–1669.
Hu, H. Y., Goto, N., and Fujie, K. (2001). Effect of pH on the reduction of nitrite in water by metallic iron. Water Res. 35, 2789–2793.
Igarashi, N., Moriyama, H., Fujiwara, T., Fukumori, Y., and Tanaka, N. (1997). The 2.8 Å structure of hydroxylamine oxidoreductase from a nitrifying chemoautotrophic bacterium, Nitrosomonas europaea. Nat. Struct. Biol. 4, 276–284.
Jenni, S., Mohn, J., Emmenegger, L., and Udert, K. M. (2012). Temperature dependence and interferences of NO and N2O microelectrodes used in wastewater treatment. Environ. Sci. Technol. 46, 2257–2266.
Jeroschewski, P., Steuckart, C., and Kühl, M. (1996). An amperometric microsensor for the determination of H2S in aquatic environments. Anal. Chem. 68, 4351–4357.
Jinuntuya-Nortman, M., Sutka, R. L., Ostrom, P. H., Gandhi, H., and Ostrom, N. E. (2008). Isotopologue fractionation during microbial reduction of N2O within soil mesocosms as a function of water-filled pore space. Soil Biol. Biochem. 40, 2273–2280.
Jones, C. M., Stres, B., Rosenquist, M., and Hallin, S. (2008). Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25, 1955–1966.
Kampschreur, M. J., Kleerebezem, R., de Vet, W. W. J. M., and Van Loosdrecht, M. C. M. (2011). Reduced iron induced nitric oxide and nitrous oxide emission. Water Res. 45, 5945–5952.
Kampschreur, M. J., van der Star, W. R. L., Wielders, H. A., Mulder, J. W., Jetten, M. S. M., and van Loosdrecht, M. C. M. (2008a). Dynamics of nitric oxide and nitrous oxide emission during full-scale reject water treatment. Water Res. 42, 812–826.
Kampschreur, M. J., Tan, N. C. G., Kleerebezem, R., Picioreanu, C., Jetten, M. S. M., and van Loosdrecht, M. C. M. (2008b). Effect of dynamic process conditions on nitrogen oxides emission from a nitrifying culture. Environ. Sci. Technol. 42, 429–435.
Kampschreur, M. J., Temmink, H., Kleerebezem, R., Jetten, M. S. M., and van Loosdrecht, M. C. M. (2009). Nitrous oxide emission during wastewater treatment. Water Res. 43, 4093–4103.
Kartal, B., Maalcke, W. J., de Almeida, N. M., Cirpus, I., Gloerich, J., Geerts, W., et al. (2011). Molecular mechanism of anaerobic ammonium oxidation. Nature 479, 127–130.
Kartal, B., Tan, N. C. G., Van de Biezen, E., Kampschreur, M. J., Van Loosdrecht, M. C. M., and Jetten, M. S. M. (2010). Effect of nitric oxide on anammox bacteria. Appl. Environ. Microbiol. 76, 6304–6306.
Kester, R. A., de Boer, W., and Laanbroek, H. J. (1997). Production of NO and N2O by pure cultures of nitrifying and denitrifying bacteria during changes in aeration. Appl. Environ. Microbiol. 63, 3872–3877.
Kim, S. O., Orii, Y., Lloyd, D., Hughes, M. N., and Poole, R. K. (1999). Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 445, 389–394.
Kitamura, Y., Uzawa, T., Oka, K., Komai, Y., Ogawa, H., Takizawa, N., et al. (2000). Microcoaxial electrode for in vivo nitric oxide measurement. Anal. Chem. 72, 2957–2962.
Kloos, K., Mergel, A., Sch, C., and Bothe, H. (2001). Denitrification within the genus Azospirillum and other associative bacteria. Funct. Plant Biol. 28, 991–998.
Koba, K., Osaka, K., Tobari, Y., Toyoda, S., Ohte, N., Katsuyama, M., et al. (2009). Biogeochemistry of nitrous oxide in groundwater in a forested ecosystem elucidated by nitrous oxide isotopomer measurements. Geochim. Cosmochim. Acta 73, 3115–3133.
Kool, D. M., Wrage, N., Oenema, O., Dolfing, J., and van Groenigen, J. W. (2007). Oxygen exchange between (de)nitrification intermediates and H2O and its implications for source determination of NO−3 and N2O: a review. Rapid Commun. Mass Spectrom. 21, 3569–3578.
Kool, D. M., Wrage, N., Zechmeister-Boltenstern, S., Pfeffer, M., Brus, D., Oenema, O., et al. (2010). Nitrifier denitrification can be a source of N2O from soil: a revised approach to the dual-isotope labelling method. Eur. J. Soil Sci. 61, 759–772.
Körner, H., and Zumft, W. G. (1989). Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl. Environ. Microbiol. 55, 1670–1676.
Kostera, J., Youngblut, M. D., Slosarczyk, J. M., and Pacheco, A. A. (2008). Kinetic and product distribution analysis of NO* reductase activity in Nitrosomonas europaea hydroxylamine oxidoreductase. J. Biol. Inorg. Chem. 13, 1073–1083.
Lammel, G., and Cape, J. N. (1996). Nitrous acid and nitrite in the atmosphere. Chem. Soc. Rev. 25, 361–369.
Law, Y., Ni, B.-J., Lant, P., and Yuan, Z. (2012). N2O production rate of an enriched ammonia-oxidising bacteria culture exponentially correlates to its ammonia oxidation rate. Water Res. 46, 3409–3419.
Lee, Y., Oh, B. K., and Meyerhoff, M. E. (2004). Improved planar amperometric nitric oxide sensor based on platinized platinum anode. 1. Experimental results and theory when applied for monitoring NO release from diazeniumdiolate-doped polymeric films. Anal. Chem. 76, 536–544.
Liengaard, L., Nielsen, L. P., Revsbech, N. P., Elberling, B., Priemé, A., Prast, A. E., et al. (2011). Effects of flooding cycles in the Pantanal on the turnover of soil nitrogen pools and emission of N2O. Biogeosci. Discuss. 8, 5991–6030.
Lipschultz, F., Zafiriou, O. C., Wofsy, S. C., McElroy, M. B., Valois, F. W., and Watson, S. W. (1981). Production of NO and N2O by soil nitrifying bacteria. Nature 294, 641–643.
Liu, B., Mørkved, P. T., Frostegård, Å., and Bakken, L. R. (2010). Denitrification gene pools, transcription and kinetics of NO, N2O and N2 production as affected by soil pH. FEMS Microbiol. Ecol. 72, 407–417.
Lotito, A. M., Wunderlin, P., Joss, A., Kipf, M., and Siegrist, H. (2012). Nitrous oxide emissions from the oxidation tank of a pilot activated sludge plant. Water Res. 46, 3563–3573.
Lücker, S., Wagner, M., Maixner, F., Pelletier, E., Koch, H., and Vacherie, B. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 13479–13484.
Malinski, T., and Taha, Z. (1992). Nitric oxide release from a single cell measured in situ by a porphyrinic-based microsensor. Nature 358, 676–678.
Mariotti, A., Germon, J. C., Hubert, P., Kaiser, P., Letolle, R., Tardieux, A., et al. (1981). Experimental determination of nitrogen kinetic isotope fractionation: some principles; illustration for the denitrification and nitrification processes. Plant Soil 62, 413–430.
Markfoged, R., Nielsen, L. P., Nyord, T., Ottosen, L. D. M., and Revsbech, N. P. (2011). Transient N2O accumulation and emission caused by O2 depletion in soil after liquid manure injection. Eur. J. Soil Sci. 62, 541–550.
Meyer, R. L., Allen, D. E., and Schmidt, S. (2008). Nitrification and denitrification as sources of sediment nitrous oxide production: a microsensor approach. Mar. Chem. 110, 68–76.
Michotey, V., Mejean, V., and Bonin, P. (2000). Comparison of methods for quantification of cytochrome cd 1-denitrifying bacteria in environmental marine samples. Appl. Environ. Microbiol. 66, 1564–1571.
Miranda, K. M. (2005). The chemistry of nitroxyl (HNO) and implications in biology. Coord. Chem. Rev. 249, 433–455.
Moews, P. C. Jr., and Audrieth, L. F. (1959). The autoxidation of hydroxylamine. J. Inorg. Nucl. Chem. 11, 242–246.
Mohn, J., Guggenheim, C., Tuzson, B., Vollmer, M., Toyoda, S., Yoshida, N., et al. (2010). A liquid-free preconcentration unit for measurements of ambient N2O isotopomers by QCLAS. Atmos. Meas. Tech. 3, 609–618.
Mohn, J., Tuzson, B., Manninen, A., Yoshida, N., Toyoda, S., Brand, W. A., et al. (2012). Site selective real-time measurements of ambient N2O isotopomers by laser spectroscopy. Atmos. Meas. Tech. Discuss. 5, 813–838.
Montzka, S. A., Dlugokencky, E. J., and Butler, J. H. (2011). Non-CO2 greenhouse gases and climate change. Nature 476, 43–50.
Morales, S. E., Cosart, T., and Holben, W. E. (2010). Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J. 4, 799–808.
Morales, S. E., and Holben, W. E. (2011). Linking bacterial identities and ecosystem processes: can “omic” analyses be more than the sum of their parts? FEMS Microbiol. Ecol. 75, 1–16.
Morley, N., Baggs, E. M., Dörsch, P., and Bakken, L. (2008). Production of NO, N2O and N2 by extracted soil bacteria, regulation by NO2(-) and O2 concentrations. FEMS Microbiol. Ecol. 65, 102–112.
Naqvi, S. W., Jayakumar, D. A., Narvekar, P. V., Naik, H., Sarma, V. V., et al. (2000). Increased marine production of N2O due to intensifying anoxia on the Indian continental shelf. Nature 408, 346–349.
Nicholls, J. C., Davies, C. A., and Trimmer, M. (2007). High-resolution profiles and nitrogen isotope tracing reveal a dominant source of nitrous oxide and multiple pathways of nitrogen gas formation in the central Arabian Sea. Limnol. Oceanogr. 52, 156–168.
Nielsen, M., Gieseke, A., de Beer, D., and Revsbech, N. (2009). Nitrate, nitrite, and nitrous oxide transformations in sediments along a salinity gradient in the Weser Estuary. Aquat. Microb. Ecol. 55, 39–52.
Okabe, S., Oshiki, M., Takahashi, Y., and Satoh, H. (2011). N2O emission from a partial nitrification-anammox process and identification of a key biological process of N2O emission from anammox granules. Water Res. 45, 6461–6470.
Otte, S., Grobben, N. G., Robertson, L. A., Jetten, M. S., and Kuenen, J. G. (1996). Nitrous oxide production by Alcaligenes faecalis under transient and dynamic aerobic and anaerobic conditions. Appl. Environ. Microbiol. 62, 2421–2426.
Palmer, K., Biasi, C., and Horn, M. A. (2012). Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. ISME J. 6, 1058–1077.
Palmer, K., Drake, H. L., and Horn, M. A. (2010). Association of novel and highly diverse acid-tolerant denitrifiers with N2O fluxes of an acidic fen. Appl. Environ. Microbiol. 76, 1125–1134.
Park, S., Croteau, P., Boering, K. A., Etheridge, D. M., Ferretti, D., Fraser, P. J., et al. (2012). Trends and seasonal cycles in the isotopic composition of nitrous oxide since 1940. Nat. Geosci. 5, 261–265.
Park, S., Pérez, T., Boering, K. A., Trumbore, S. E., Gil, J., Marquina, S., et al. (2011). Can N2O stable isotopes and isotopomers be useful tools to characterize sources and microbial pathways of N2O production and consumption in tropical soils? Glob. Biogeochem. Cycles 25, GB1001.
Patureau, D., Zumstein, E., Delgenes, J. P., and Moletta, R. (2000). Aerobic denitrifiers isolated from diverse natural and managed ecosystems. Microb. Ecol. 39, 145–152.
Pellicer-Nàcher, C., Sun, S., Lackner, S., Terada, A., Schreiber, F., Zhou, Q., et al. (2010). Sequential aeration of membrane-aerated biofilm reactors for high-rate autotrophic nitrogen removal: experimental demonstration. Environ. Sci. Technol. 44, 7628–7634.
Perez, T., Garcia-Montiel, D., Trumbore, S., Tyler, S., de Camargo, P., et al. (2006). Nitrous oxide nitrification and denitrification 15N enrichment factors from amazon forest soils. Ecol. Appl. 16, 2153–2167.
Philippot, L., Andert, J., Jones, C. M., Bru, D., and Hallin, S. (2011). Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Glob. Change Biol. 17, 1497–1504.
Philippot, L., Čuhel, J., Saby, N. P. A., Chèneby, D., Chroňáková, A., Bru, D., et al. (2009). Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ. Microbiol. 11, 1518–1526.
Poth, M. (1986). Dinitrogen production from nitrite by a Nitrosomonas isolate. Appl. Environ. Microbiol. 52, 957–959.
Poth, M., and Focht, D. D. (1985). 15N kinetic analysis of N2O production by Nitrosomonas: an examination of nitrifier denitrification. Appl. Environ. Microbiol. 49, 1134–1141.
Prosser, J. I., Jansson, J. K., and Liu, W.-T. (2010). “Nucleic-acid-based characterization of community structure and function,” in Environmental Molecular Microbiology, eds W.-T. Liu and J. K. Jansson (Norfolk, UK: Caister Academic Press), 63–86.
Ravishankara, A. R., Daniel, J. S., and Portmann, R. W. (2009). Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125.
Reed, H. E., and Martiny, J. B. H. (2007). Testing the functional significance of microbial composition in natural communities. FEMS Microbiol. Ecol. 62, 161–170.
Revsbech, N. P., Jørgensen, B. B., and Blackburn, T. H. (1980). Oxygen in the sea bottom measured with a microelectrode. Science 207, 1355–1356.
Revsbech, N. P., Nielsen, L. P., Christensen, P. B., and Sørensen, J. (1988). Combined oxygen and nitrous oxide microsensor for denitrification studies. Appl. Environ. Microbiol. 54, 2245–2249.
Richardson, D., Felgate, H., Watmough, N., Thomson, A., and Baggs, E. M. (2009). Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle – could enzymic regulation hold the key? Trends Biotechnol. 27, 388–397.
Rich, J. J., Heichen, R. S., Bottomley, P. J., Cromack, K., and Myrold, D. D. (2003). Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Appl. Environ. Microbiol. 69, 5974–5982.
Rich, J. J., and Myrold, D. D. (2004). Community composition and activities of denitrifying bacteria from adjacent agricultural soil, riparian soil, and creek sediment in Oregon, USA. Soil Biol. Biochem. 36, 1431–1441.
Ritchie, G. A., and Nicholas, D. J. (1972). Identification of the sources of nitrous oxide produced by oxidative and reductive processes in Nitrosomonas europaea. Biochem. J. 126, 1181–1191.
Rock, J. D., Thomson, M. J., Read, R. C., and Moir, J. W. B. (2007). Regulation of denitrification genes in Neisseria meningitidis by nitric oxide and the repressor NsrR. J. Bacteriol. 189, 1138–1144.
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712.
Santoro, A. E., Buchwald, C., McIlvin, M. R., and Casciotti, K. L. (2011). Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science 333, 7–10.
Schmidt, H. L., Werner, R. A., Yoshida, N., and Well, R. (2004a). Is the isotopic composition of nitrous oxide an indicator for its origin from nitrification or denitrification? A theoretical approach from referred data and microbiological and enzyme kinetic aspects. Rapid Commun. Mass Spectrom. 18, 2036–2040.
Schmidt, I., van Spanning, R. J. M., and Jetten, M. S. M. (2004b). Denitrification and ammonia oxidation by Nitrosomonas europaea wild-type, and NirK- and NorB-deficient mutants. Microbiology 150, 4107–4114.
Schmidt, I. (2009). Chemoorganoheterotrophic growth of Nitrosomonas europaea and Nitrosomonas eutropha. Curr. Microbiol. 59, 130–138.
Schreiber, F., Beutler, M., Enning, D., Lamprecht-Grandio, M., Zafra, O., González-Pastor, J. E., et al. (2011). The role of nitric-oxide-synthase-derived nitric oxide in multicellular traits of Bacillus subtilis 3610, biofilm formation, swarming, and dispersal. BMC Microbiol. 11:111. doi: 10.1186/1471-2180-11-111
Schreiber, F., Loeffler, B., Polerecky, L., Kuypers, M. M. M., and de Beer, D. (2009). Mechanisms of transent nitric oxide and nitrous oxide production in a complex biofilm. ISME J. 3, 1301–1313.
Schreiber, F., Polerecky, L., and de Beer, D. (2008). Nitric oxide microsensor for high spatial resolution measurements in biofilms and sediments. Anal. Chem. 80, 1152–1158.
Schreiber, F., Stief, P., Gieseke, A., Heisterkamp, I. M., Verstraete, W., de Beer, D., et al. (2010). Denitrification in human dental plaque. BMC Biol. 8:24. doi: 10.1186/1741-7007-8-24
Schuurkes, J. A. A. R., and Mosello, R. (1988). The role of external ammonium inputs in freshwater acidification. Swiss J. Hydrol. 50, 71–87.
Schwartz, S. E., and White, W. H. (1981). Solubility equilibria of the nitrogen oxides and oxyacids in dilute aqueous solution. Adv. Environ. Sci. Eng. 4, 1–45.
Shafirovich, V., and Lymar, S. V. (2002). Nitroxyl and its anion in aqueous solutions: spin states, protic equilibria, and reactivities toward oxygen and nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 99, 7340–7345.
Shatalin, K., Gusarov, I., Avetissova, E., Shatalina, Y., McQuade, L. E., Lippard, S. J., et al. (2008). Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 105, 1009–1013.
Shaw, L. J., Nicol, G. W., Smith, Z., Fear, J., Prosser, J. I., and Baggs, E. M. (2006). Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ. Microbiol. 8, 214–222.
Shibuki, K. (1990). An electrochemical microprobe for detecting nitric-oxide release in brain-tissue. Neurosci. Res. 9, 69–76.
Shin, J. H., Weinman, S. W., and Schoenfisch, M. H. (2005). Sol-gel derived amperometric nitric oxide microsensor. Anal. Chem. 77, 3494–3501.
Smith, C. J., and Osborn, A. M. (2008). Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol. Ecol. 67, 6–20.
Snider, D. M., Schiff, S. L., and Spoelstra, J. (2009). 15N/14N and 18O/16O stable isotope ratios of nitrous oxide produced during denitrification in temperate forest soils. Geochim. Cosmochim. Acta 73, 877–888.
Sørensen, J., Tiedje, J. M., and Firestone, R. B. (1980). Inhibition by sulfide of nitric and nitrous oxide reduction by denitrifying Pseudomonas fluorescens. Appl. Environ. Microbiol. 39, 105–108.
Starkenburg, S. R., Arp, D. J., and Bottomley, P. J. (2008a). Expression of a putative nitrite reductase and the reversible inhibition of nitrite-dependent respiration by nitric oxide in Nitrobacter winogradskyi Nb-255. Environ. Microbiol. 10, 3036–3042.
Starkenburg, S. R., Chain, P. S. G., Sayavedra-Soto, L. A., Hauser, L., Land, M. L., Larimer, F. W., et al. (2006). Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl. Environ. Microbiol. 72, 2050–2063.
Starkenburg, S. R., Larimer, F. W., Stein, L. Y., Klotz, M. G., Chain, P. S. G., Luis, A., et al. (2008b). Complete genome sequence of nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus nitrobacter. Appl. Environ. Microbiol. 74, 2852–2863.
Stein, L. Y., and Yung, Y. L. (2003). Production, isotopic composition, and a atmospheric fate of biologically produced nitrous oxide. Annu. Rev. Earth Planet. Sci. 31, 329–356.
Stief, P., Poulsen, M., Nielsen, L. P., Brix, H., and Schramm, A. (2009). Nitrous oxide emission by aquatic macrofauna. Proc. Natl. Acad. Sci. U.S.A. 106, 4296–4300.
Strous, M., Pelletier, E., Mangenot, S., Rattei, T., Lehner, A., Taylor, M. W., et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440, 790–794.
Sudhamsu, J., and Crane, B. R. (2009). Bacterial nitric oxide synthases: what are they good for? Trends Microbiol. 17, 212–218.
Sutka, R. L., Ostrom, N. E., Ostrom, P. H., Breznak, J. A., Gandhi, H., Pitt, A. J., et al. (2006). Distinguishing nitrous oxide production from nitrification and denitrification on the basis of isotopomer abundances. Appl. Environ. Microbiol. 72, 638–644.
Sutka, R. L., Ostrom, N. E., Ostrom, P. H., Gandhi, H., and Breznak, J. A. (2003). Nitrogen isotopomer site preference of N2O produced by Nitrosomonas europaea and Methylococcus capsulatus bath. Rapid Commun. Mass Spectrom. 17, 738–745.
Sutka, R. L., Ostrom, N. E., Ostrom, P. H., Gandhi, H., and Breznak, J. A. (2004). Nitrogen isotopomer site preference of N2O produced by Nitrosomonas europaea and Methylococcus capsulatus bath. Rapid Commun. Mass Spectrom. 18, 1411–1412.
Thomsen, J. K., Geest, T., and Cox, R. P. (1994). Mass spectrometric studies of the effect of pH on the accumulation of intermediates in denitrification by Paracoccus denitrificans. Appl. Environ. Microbiol. 60, 536–541.
Throbäck, I. N., Enwall, K., Jarvis, Å., and Hallin, S. (2004). Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49, 401–417.
Toyoda, S., Iwai, H., Koba, K., and Yoshida, N. (2009). Isotopomeric analysis of N2O dissolved in a river in the Tokyo metropolitan area. Rapid Commun. Mass Spectrom. 23, 809–821.
Toyoda, S., Mutobe, H., Yamagishi, H., Yoshida, N., and Tanji, Y. (2005). Fractionation of N2O isotopomers during production by denitrifier. Soil Biol. Biochem. 37, 1535–1545.
Toyoda, S., Suzuki, Y., Hattori, S., Yamada, K., Fujii, A., Yoshida, N., et al. (2011). Isotopomer analysis of production and consumption mechanisms of N2O and CH4 in an advanced wastewater treatment system. Environ. Sci. Technol. 45, 917–922.
Toyoda, S., and Yoshida, N. (1999). Determination of nitrogen isotopomers of nitrous oxide on a modified isotope ratio mass spectrometer. Anal. Chem. 71, 4711–4718.
Udert, K. M., Larsen, T. A., and Gujer, W. (2005). Chemical nitrite oxidation in acid solutions as a consequence of microbial ammonium oxidation. Environ. Sci. Technol. 39, 4066–4075.
Upadhyay, A. K., Hooper, A. B., and Hendrich, M. P. (2006). NO reductase activity of the tetraheme cytochrome C554 of Nitrosomonas europaea. J. Am. Chem. Soc. 128, 4330–4337.
van Cleemput, O. (1998). Subsoils: chemo- and biological denitrification, N2O and N2 emissions. Nutr. Cycl. Agroecosyst. 52, 187–194.
van Cleemput, O., and Samater, A. H. (1996). Nitrite in soils: accumulation and role in the formation of gaseous N compounds. Fertil. Res. 45, 81–89.
van Wonderen, J. H., Burlat, B., Richardson, D. J., Cheesman, M. R., and Butt, J. N. (2008). The nitric oxide reductase activity of cytochrome c nitrite reductase from Escherichia coli. J. Biol. Chem. 283, 9587–9594.
Vardi, A., Formiggini, F., Casotti, R., De Martino, A., Ribalet, F., Miralto, A., et al. (2006). A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 4:e60. doi: 10.1371/journal.pbio.0040060
Waechter, H., Mohn, J., Tuzson, B., Emmenegger, L., and Sigrist, M. W. (2008). Determination of N2O isotopomers with quantum cascade laser based absorption spectroscopy. Opt. Express 16, 9239–9244.
Wahlen, M., and Yoshinari, T. (1985). Oxygen isotope ratios in N2O from different environments. Nature 313, 780–782.
Wallenstein, M. D., Myrold, D. D., Firestone, M., and Voytek, M. (2006). Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol. Appl. 16, 2143–2152.
Ward, B. B., and Zafiriou, O. C. (1988). Nitrification and nitric-oxide in the oxygen minimum of the eastern tropical North Pacific. Deep Sea Res. 35, 1127–1142.
Well, R., Kurganova, I., Lopesdegerenyu, V., and Flessa, H. (2006). Isotopomer signatures of soil-emitted N2O under different moisture conditions—a microcosm study with arable loess soil. Soil Biol. Biochem. 38, 2923–2933.
Wells, G. F., Park, H.-D., Yeung, C.-H., Eggleston, B., Francis, C. A., and Criddle, C. S. (2009). Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ. Microbiol. 11, 2310–2328.
Westley, M. B., Popp, B. N., and Rust, T. M. (2007). The calibration of the intramolecular nitrogen isotope distribution in nitrous oxide measured by isotope ratio mass spectrometry. Rapid Commun. Mass Spectrom. 21, 391–405.
Wrage, N., Van Groenigen, J. W., Oenema, O., and Baggs, E. M. (2005). A novel dual-isotope labelling method for distinguishing between soil sources of N2O. Rapid Commun. Mass Spectrom. 19, 3298–3306.
Wrage, N., Velthof, G. L., Beusichem, M. L. V., and Oenema, O. (2001). Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 33, 1723–1732.
Wunderlin, P., Mohn, J., Joss, A., Emmenegger, L., and Siegrist, H. (2012). Mechanisms of N2O production in biological wastewater treatment under nitrifying and denitrifying conditions. Water Res. 46, 1027–1037.
Yamagishi, H., Westley, M. B., Popp, B. N., Toyoda, S., Yoshida, N., Watanabe, S., et al. (2007). Role of nitrification and denitrification on the nitrous oxide cycle in the eastern tropical North Pacific and Gulf of California. J. Geophys. Res. Biogeosci. 112, G02015.
Yoshida, N., Morimoto, H., Hirano, M., Koike, I., Matsuo, S., Wada, E., et al. (1989). Nitrification rates and N-15 abundances of N2O and NO−3 in the Western North Pacific. Nature 342, 895–897.
Yoshinari, T., and Wahlen, M. (1985). Oxygen isotope ratios in N2O from nitrification at a wastewater treatment facility. Nature 317, 349–350.
Yu, R., Kampschreur, M. J., van Loosdrecht, M. C. M., and Chandran, K. (2010). Mechanisms and specific directionality of autotrophic nitrous oxide and nitric oxide generation during transient anoxia. Environ. Sci. Technol. 44, 1313–1319.
Zafiriou, O. C., Mcfarland, M., and Bromund, R. H. (1980). Nitric-oxide in seawater. Science 207, 637–639.
Zafiriou, O. C., and True, M. B. (1979). Nitrite photolysis in seawater by sunlight. Mar. Chem. 8, 9–32.
Zart, D., Schmidt, I., and Bock, E. (2000). Significance of gaseous NO for ammonia oxidation by Nitrosomonas eutropha. Antonie van Leeuwenhoek 77, 49–55.
Zhou, Y., Pijuan, M., Zeng, R. J., and Yuan, Z. (2008). Free nitrous acid inhibition on nitrous oxide reduction by a denitrifying-enhanced biological phosphorus removal sludge. Environ. Sci. Technol. 42, 8260–8265.
Zumft, W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616.
Zumft, W. G. (2005). Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme-copper oxidase type. J. Inorg. Biochem. 99, 194–215.
Keywords: isotopic signature, microsensors, molecular tools, dinitrogen oxide, nitrogen monoxide, pathway identification, quantum cascade laser absorption spectroscopy (QCLAS), site preference
Citation: Schreiber F, Wunderlin P, Udert KM and Wells GF (2012) Nitric oxide and nitrous oxide turnover in natural and engineered microbial communities: biological pathways, chemical reactions, and novel technologies. Front. Microbio. 3:372. doi: 10.3389/fmicb.2012.00372
Received: 30 April 2012; Paper pending published: 11 June 2012;
Accepted: 28 September 2012; Published online: 23 October 2012.
Edited by:
Boran Kartal, Radboud University, NetherlandsReviewed by:
Robbert Kleerebezem, Delft University of Technology, NetherlandsKartik Chandran, Columbia University, USA
Copyright © 2012 Schreiber, Wunderlin, Udert and Wells. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Frank Schreiber, Department of Environmental Microbiology, Eawag-Swiss Federal Institute of Aquatic Science and Technology, Überlandstrasse 133, P.O. Box 611, 8600 Dübendorf, Switzerland. e-mail: frank.schreiber@eawag.ch