- Department of Food Science, Canadian Research Institute for Food Safety, University of Guelph, Guelph, ON, Canada
Lactobacillus helveticus is one of the species of lactic acid bacteria (LAB) most commonly used in the production of fermented milk beverages and some types of hard cheese. The versatile nature of this bacterium is based on its highly efficient proteolytic system consisting of cell-envelope proteinases (CEPs), transport system and intracellular peptidases. Besides use of L. helveticus in cheese processing, the production of fermented milk preparations with health promoting properties has become an important industrial application. Studies have shown that fermented dairy products are able to decrease blood pressure, stimulate the immune system, promote calcium absorption, and exert an anti-virulent effect against pathogens. These beneficial effects are produced by a variety of peptides released during the hydrolysis of milk proteins by the proteolytic system of L. helveticus, which provides the bacterium with its nutritional requirements for growth. In recent years, studies have focused on understanding the factors that affect the kinetics of milk protein hydrolysis by specific strains and have concentrated on the effect of pH, temperature, growth phase, and matrix composition on the bacterial enzymatic system. This review focuses on the role of the proteolytic system of L. helveticus in the production of bioactive compounds formed during fermentation of dairy products. Taking advantage of the powerful proteolytic system of this bacterium opens up future opportunities to search for novel food-derived compounds with potential health promoting properties.
Introduction
Lactobacillus helveticus is a homofermentative, thermophilic lactic acid bacterium widely used in the manufacture of Swiss type and, Italian aged cheeses, and fermented milk drinks (Gatti et al., 2004; Vinderola et al., 2007a). L. helveticus has the ability to reduce bitterness and give characteristic flavor to cheese, which makes this bacterium an important component of starter cultures for the dairy industry. Consumer interest in functional food has motivated researchers to focus on the bioactive compounds formed during fermentation of dairy products by lactic acid bacteria (LAB) such as L. helveticus (Matar et al., 2003; Vinderola et al., 2007b; Rattanachaikunsopon and Phumkhachorn, 2010; Wakai and Yamamoto, 2012). Milk proteins are considered to be the most important source of bioactive peptides that can be released through hydrolysis by proteolytic microorganisms such as L. helvetius (Meisel and Bockelmann, 1999; Matar et al., 2001; LeBlanc et al., 2002; Fitzgerald and Meisel, 2003; Kenny et al., 2003; de Moreno de LeBlanc et al., 2005). During milk fermentation only 1–2% of milk proteins undergo proteolysis and the principal substrate is casein but limited degradation of whey proteins may also occur (Khalid et al., 1991; Szwajkowska et al., 2011). The effect of this proteolysis is that fermented milks have a higher content of peptides and free amino acids, especially valine, histidine, serine and proline, than non-fermented milk (Matar et al., 2003). LAB rely on their complex proteolytic system to obtain all the necessary free amino acids required for growth in milk. L. helveticus strains are among the most nutritionally fastidious LAB as they present multiple amino acid auxothrophies (Hebert et al., 2002). In order to assure its nutritional requirements when grown in milk, L. helveticus counts on a potent proteolytic system capable of producing short peptides and liberating amino acids from the casein matrix. This explains why it has higher proteolytic activity than most other lactobacilli and why it hydrolyses more casein in culture media than other species (Savijoki et al., 2006). The proteolytic system of L. helveticus and the majority of LAB consists of proteinases which initially cleave caseins to large peptides, peptidases (intracellular), which further degrade these peptides to small peptides and amino acids, and specific transport proteins which transport amino acids and peptides across the cytoplasmic membrane (Kenny et al., 2003).
Proteolytic System
Proteinases
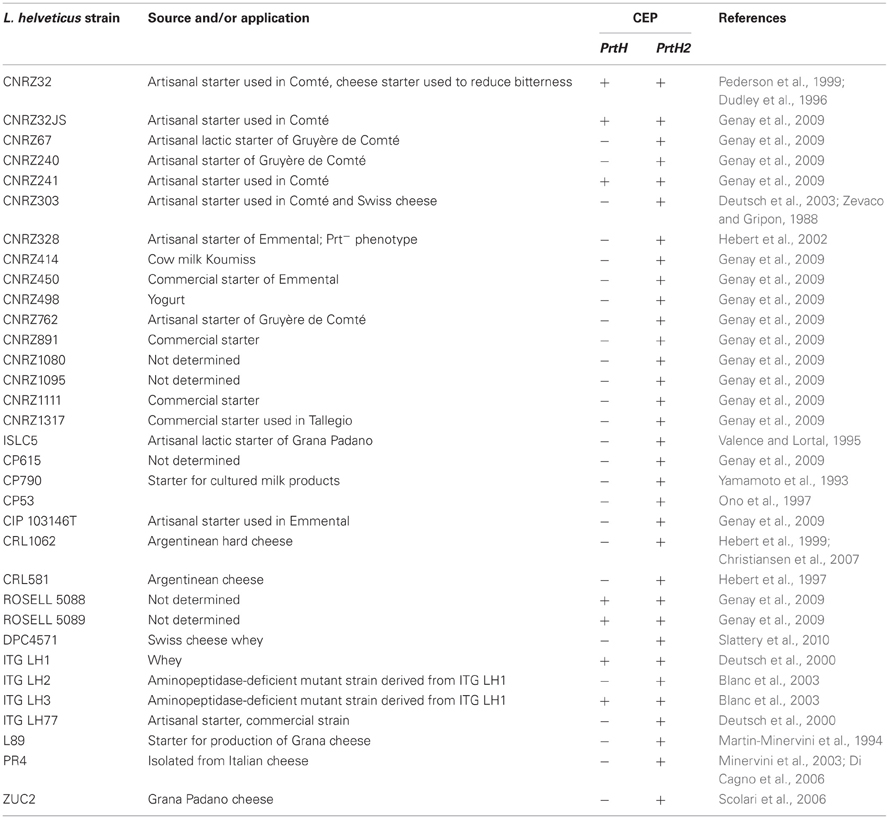
The main source of amino acids and peptides produced by LAB are derived from caseins, the most abundant protein in milk (Meisel and Bockelmann, 1999). The hydrolysis of caseins by LAB is initiated by a cell-envelope proteinase (CEP) that degrades the protein into oligopeptides. Moreover, gene deletion studies have shown that LAB are not able to grow in milk in the absence of a functional CEP (Mayo et al., 2010). This enzyme belongs to the family of subtilisin and it is a serine protease. CEPs are anchored to the cell wall via a mechanism involving the typical sortase A (SrtA) (Dandoy et al., 2011). Most of the LAB possess only one CEP but L. helveticus has been reported to have at least two: PrtH and PrtH2 (Gilbert et al., 1997). The CEPs of LAB are synthesized as preproteins of ~2000 residues and are composed of seven functional domains: S, PP, PR, A, B, H, and W (Fernandez-Espla et al., 2000). PrtH1 and PrtH2 present in L. helveticus differ from the CEPs of other LAB because they lack the anchor domain (AN), which is located at the C-terminus of the CEP sequence. Some authors consider that there are at least two other types of CEP in L. helveticus, namely PrtH3 and PrtH4 (Yamamoto et al., 1998; Savijoki et al., 2006; Broadbent et al., 2011; Sadat-Mekmene et al., 2011a). In L. helveticus CNRZ32, two genes encoding the CEPs, prtH, and prtH2 have been reported (Sadat-Mekmene et al., 2011b). Two additional putative genes encoding CEPs have been recently detected in L. helveticus CNRZ32, prtH3, and prtH4 (Jensen et al., 2009). In another study, an additional gene, prtH5, was detected (Smeianov et al., 2007). The presence of several genes encoding the proteolytic system of L. helveticus could explain the high efficiency of its proteolytic system. From the four identified genes, only prtH2 is common to all the characterized strains of L. helveticus (Genay et al., 2009). A single gene (prtH3) was found in strain DPC4571 (Broadbent et al., 2008), the prtH2 gene is present in strains CNRZ2 and MI 2359, while LHC2 seems to have just the prtH4 gene (Jensen et al., 2009). L. helveticus CM4, which has a very strong hydrolytic system, has been reported to have three CEP encoded genes: prtY, prtH2, and prtM2 (Wakai and Yamamoto, 2012). A comprehensive study of the proteolytic system of several strains of L. helveticus strains using comparative genome hybridization microarray based on L. helveticus CNZR 32 was published recently (Broadbent et al., 2011). This study showed that there is a high intraspecific diversity of genes encoding CEPs in the 51 strains of L. helveticus analyzed. Twelve percent of the strains tested contained all four CEP paralogs, 8% contained 3 paralogs, whereas 42% had genes for just two CEPs. Among the strains tested the most abundant paralog was prtH3, followed by prtH and prtH4 paralogs. L. helveticus BGRA43 isolated from human feces has strong proteolytic activity and is able to hydrolyze αs1-, β-, and κ-caseins completely (Lozo et al., 2011). Analysis of the proteinase from BGRA43 showed the only active gene was prtH. In L. helveticus as in other LAB species, CEP activation requires a maturation proteinase termed PrtM. In L. helveticus CNRZ32 two PrtMs (PrtM1 and PrtM2) have been identified (Savijoki et al., 2006). Further studies (Genay et al., 2009; Broadbent et al., 2011; Zhao et al., 2011) have reported that PrtM is required for activation of PrtH and PrtM2 plays a role in activation of other CEP paralogs in L. helveticus. The presence of the various CEPs in L. helveticus strains that have been characterized is presented in Table 1.
Peptide and Amino Acid Transport System
The second stage of protein degradation is the transport of di-, tri-, and oligo-peptides into the cell by different peptide transport systems. One of the first publications on the peptide transport system of L. helveticus concluded that Leu, Ile, Val, Thr, and Lys were transported by a secondary transport system, whereas the amino acids Asp, Glu, His, Arg, and Tyr seemed to be transported by an ATP-driven system (Nakajima et al., 1998). After L. helveticus strains DPC4571 (Callanan et al., 2008) and H10 (Zhao et al., 2011) were sequenced and information regarding their transport system became available, it was determined that strain DPC4571 possesses three peptide transport systems, the oligopeptide Opp transport system and the di- and tri-peptide transport system, Dpp and DtpT, respectively. Conversely, H10 has two peptide transport systems, the Opp and dtpT systems. These results indicate that the proteolytic system may differ between different strains in L. helveticus.
Peptidases
Peptidases are a very important part of the proteolytic system in LAB since they are involved in the hydrolysis of peptides to release essential amino acids (Christensen et al., 1999). Up until 2003 just 11 peptidases were known to be part of the proteolytic system of L. helveticus: two proline-specific endopepeptidases, PepE and PepO; a tripeptidase PepT; four aminopeptidases, PepX, PepI, PepQ, and PepR; and four dipeptidases, PepD, PepV, PepC, and PepN (Christensen et al., 1999; Savijoki and Palva, 2000; Kenny et al., 2003; Pan and Tanokur, 2004). Recently it was reported that strain CNRZ32 has seven oligoendopeptidases, three general aminopeptidases, five proline-specific peptidases, eight di- or tri-peptidases, and at least six other peptidases (Broadbent et al., 2011).
The proteolytic system of L. helveticus CNRZ32 has been widely studied (Khalid and Marth, 1990; Fernández et al., 1994; Shao et al., 1997; Chen et al., 2003; Christensen and Steele, 2003; Kilpi et al., 2007) due to its ability to reduce cheese bitterness (Fernández et al., 1994) and to release peptides during milk fermentation with angiotensin converting enzyme (ACE) inhibitory activity (Kilpi et al., 2007). It is well known that peptides derived from β-casein (f193–202) and α−s1 (f1–9) have been implicated in the development of bitterness in cheese (Christensen et al., 2003). For this reason, research has focused on studying the L. helveticus endopeptidases that are responsible for the hydrolysis of these caseins (Chen et al., 2003; Christensen et al., 2003). The endopeptidase PepO2 plays a pivotal role in the ability of L. helveticus CNRZ32 to reduce bitterness because of its specificity for bonds containing proline. The peptidase PepDA was not necessary for the liberation of essential amino acids since the deletion of the gene pepDA had no effect on the growth of strain CNRZ32 (Dudley et al., 1996). Similarly, the dipeptidase with proline activity (PepR) has been found not to be a limiting enzyme for growth of CNRZ32 (Shao et al., 1997). A recent study on the amino peptidase activity of six strains of L. helveticus grown in milk or MRS (de Man, Rogosa, and Sharpe) medium reported that the mechanism of regulation depended on the specific peptidase and possibly on the specific strain. Higher aminopeptidase activity against almost all substrates used for the study was found after growth in milk (Jensen and Ardo, 2010). The effect of amino-peptidases PepN and PepX present in L. helveticus CNRZ32 on the production of peptides with antihypertensive activity has been also a subject of study. Milk fermented with peptidases-negative mutants, induced higher production of ACE-inhibitory activity compared with milk fermented with wild-type strains (Kilpi et al., 2007). The importance of peptidases PepC, PepN, and PepX for the growth of L. helveticus CNRZ32 was evident when a multi-peptidase deletion mutant (pepC−, pepN−, and pepX−) was studied. The growth rate inhibition in milk exhibited by the mutants was overcome by supplementation with amino acids associated with the peptidase that was absent in each of the mutants (Christensen et al., 2003).
The essential role that L. helveticus peptidases have in the manufacture of Swiss cheese was confirmed when the proteolytic systems of selected LAB were compared (Deutsch et al., 2000). Peptidases of the L. helveticus strains tested showed higher proteolytic activity as reported previously (Valence et al., 1998, 2000) and the presence of one carboxypeptidase in all the strains tested was confirmed. An endopeptidase with the ability to hydrolyze peptides Val-Pro-Pro-Phe-Leu and Ile-Pro-Pro-Leu-Thr to produce Val-Pro-Pro and Ile-Pro-Pro, respectively, was isolated and characterized from L. helveticus CM4 (Ueno et al., 2004). Based on the results, it was suggested that the enzyme is a key player in the hydrolysis of the C-terminal during the production of Val-Pro-Pro and Ile-Pro-Pro in fermented milk. The highest aminopeptidase activity for all the strains was observed for the substrates Arg-, and Lys-pNA.
Bioactive Peptides Produced from Hydrolysis of Milk Proteins
Today, milk proteins are considered the most important source of bioactive peptides and an increasing number of bioactive peptides have been identified in milk protein hydrolysates and fermented dairy products (Clare et al., 2003; Silva and Malcata, 2005; Korhonen and Pihlanto, 2006). Milk can be the source of peptides with different bioactivities with potential interaction and synergism among them as well as with other non-peptide components. Upon oral administration, bioactive peptides may affect the major body systems. For this reason, the potential of distinct dietary peptide sequences to promote human health by reducing the risk of chronic diseases or boosting natural immune protection has aroused a great deal of scientific interest over the past few years (Schanbacher et al., 1998). Bovine milk contains 32 g/L of proteins, of which 80% are caseins and 20% are whey proteins. Casein, in addition to being a very efficient calcium and phosphate transport for the newborn, hides a series of bioactive molecules in its sequences. The caseins are a family of phosphoproteins and they have been designated αs1-, αs2-, β-, and κ-caseins, with αs1 being the major protein found in bovine milk. So far, no specific physiological property has been proposed for the whole casein system, whereas various peptides that are inactive in the amino-acid sequence have been proven to possess bioactivities (Pihlanto-Leppala et al., 1999; Florisa et al., 2003; Kilara and Panyam, 2003). CEPs of L. helveticus are highly specific since the sites of cleavage on the caseins differ from one strain to another depending on the type of casein (α- or β-casein) (Zevaco and Gripon, 1988; Jensen et al., 2009; Sadat-Mekmene et al., 2011a). A study conducted on the proteolytic system of L. helveticus Zuc2, reported the potential presence of two CEPs (Scolari et al., 2006). The most loosely bound CEP was partially characterized, which was able to hydrolyze primarily β-casein and to a lesser extent κ- and αs1-casein. The CEP cleaved αs1-CN (f1–23) in contrast with other CEPs from various L. helveticus strains. The proteolytic system of fifteen strains of L. helveticus more efficiently hydrolyzed β-casein than αs1-casein (Sadat-Mekmene et al., 2011b). Moreover, among all the tested strains there were no differences in the extent and time of hydrolysis of β-casein in strains with one CEP or more. Different studies have agreed that CEPs of L. helveticus can cleave αs1- or β-casein at different sites not specifically related to one type of amino acid (Zevaco and Gripon, 1988; Broadbent et al., 2011; Sadat-Mekmene et al., 2011b). For instance, strains L. helveticus CNRZ 303 and CP790 hydrolyzed several bonds at the C-terminal of β-casein, while strains CNRZ 32, CNRZ 303 and LHC2 hydrolyzed β-casein at the N-terminal (Jensen et al., 2009). Some strains of L. helveticus such as CP790 (Yamamoto et al., 1994), CP53 (Ono et al., 1997), and PR4 (Di Cagno et al., 2006) are able to hydrolyze both αs1- and β-casein. Two comprehensive studies have concluded that the hydrolysis of αs1-casein by L. helveticus strains predominantly occurs at the N-terminal (Jensen et al., 2009; Sadat-Mekmene et al., 2011a). However, it is clear that most of the characterized strains of L. helveticus are able to extensively hydrolyze β-casein. The CEPs of L. helveticus are also able to hydrolyze κ-casein and αs2-casein; although to a lesser extent than for the other caseins (Ezzat et al., 1985; Yamamoto et al., 1998; Scolari et al., 2006; Jensen et al., 2009; Sadat-Mekmene et al., 2011a).
Whey proteins have a globular structure and are more water-soluble than caseins. The principle fractions are β-lactoglobulin, α-lactalbumin, bovine serum albumin (BSA), and immunoglobulins (Ig). In the past, whey was considered a waste product or a raw material for relatively low-value commodities. However, in the last decade with the emergence of new separation technologies, the potential of whey proteins as bioactive compounds has been realized (Chatterton et al., 2006).
Production of bioactive peptides from enzymatic hydrolysis of whey proteins has been reviewed (Pihlanto-Leppala, 2001; Madureira et al., 2010). The opioid peptides, α-lactorphin and β-lactorphin were released during in vitro proteolysis of whey proteins and whey hydrolysates showed ACE-inhibitory activity after proteolysis with digestive enzymes, and several active peptides (Pihlanto-Leppala, 2001). To date, there are few publications that have focused on the production of bioactive peptides released during fermentation of whey with LAB. L. helveticus LH-2 was able to hydrolyze α-lactalbumin and release novel bioactive peptides with immunomodulatory properties (Tellez et al., 2010). Whey protein-based media have been used as a fermentation substrate for L. acidophilus La-5 and bioactive molecules produced during its growth are able to down-regulate genes involved in encoding the Type 3 Secretion System of E. coli O157:H7 (Medellin-Pena et al., 2007; Medellin-Pena and Griffiths, 2009; Zeinhom et al., 2012) and Salmonella (Bayoumi and Griffiths, 2012).
Physiological Role of the Bioactive Compounds Generated by L. helveticus
Traditionally milk proteins have been used as a raw material to obtain bioactive peptides because it is considered to be safe and relatively inexpensive. The production of bioactives in fermented milk and cheese has been widely studied (Leclerc et al., 2002; Matar et al., 2003; Gómez-Ruiz et al., 2004a,b; Fitzgerald and Murray, 2006; Jensen et al., 2009; López-Expósito et al., 2012). Upon oral administration, these bioactive peptides may have an impact on various functions in the human body depending on their amino acid sequence (Korhonen and Pihlanto, 2006). Various peptides with physiological functions, such as immunostimulating peptides, antimicrobial peptides, opioid peptides, mineral binding peptides and antihypertensive peptides have been isolated from products fermented with L. helveticus.
Antihypertensive
Antihypertensive peptides have been extensively reviewed (Ono et al., 1997; Yamamoto et al., 1998; Wakai and Yamamoto, 2012).
The ability of L. helveticus to produce antihypertensive peptides has been attributed to the CEP (Ono et al., 1997; Wakai and Yamamoto, 2012). To date, no relationship has been established between the antihypertensive effect and proteinase specificity. A number of researchers have concluded that the functional properties of L. helveticus depend on the specific strain (Slattery et al., 2010). Thus, strains CM4, CNRZ32, and DPC4571 that are able to hydrolyze milk proteins to produce antihypertensive peptides differ slightly in terms of their CEP genes when compared with strains that are incapable of producing them (Sadat-Mekmene et al., 2011b; Wakai and Yamamoto, 2012).
Immunomodulatory Activity
Milk fermented with L. helveticus can enhance specific and non-specific immunity (Laffineur et al., 1996; LeBlanc et al., 2004; Vinderola et al., 2007a,b; Tellez et al., 2010, 2011). Milk fermented with L. helveticus R389 was able to induce immune response and exert a protective effect against Salmonella Typhimurium and E. coli O157 (LeBlanc et al., 2004; Vinderola et al., 2007b). The positive effect on the mucosal and tumoral immunity was directly related to the proteolytic system of L. helveticus R389, since milk containing the non-proteolytic variant was not able to instigate an immune response (Matar et al., 2001). Although L. helveticus R389 is able to hydrolyze milk proteins to produce bioactive peptides that have a beneficial effect on the immune system, not much work has been done to characterize the components of the proteolytic system that is responsible for generation of these peptides, nor on the identification of the peptides responsible for the immunomodulatory effect. In vitro experiments have shown that milk fermented with L. helveticus LH-2 can enhance cellular immune responses (Ng and Griffiths, 2002; Tellez et al., 2010) and have a protective effect against Salmonella infection in vivo (Tellez et al., 2011). Four novel bioactive peptides derived from β-casein and α-lactalbumin following fermentation of milk with L. helveticus LH-2 were identified (Tellez et al., 2010). Three of the peptides share a common amino acid sequence with β-casein with variation at N- and C-terminals (f 148–154, f 145–160, and f 143–154). Peptides identified have in their structure a high percentage of proline and also contain residues of lysine and histidine.
The S-layer protein extract of L. helveticus R0052 has been shown to ameliorate the pathogenesis of E coli O157:H7 (Sherman et al., 2005; Johnson-Henry et al., 2007). Epithelial cells treated with S-layer protein extract prior to infection with E. coli O157 maintained their cellular integrity and barrier function. Beganovic et al. (2011) isolated and characterized the S-layer from L. helveticus M92 and tested its functionality in vivo. The slpA gene encoding the S-layer protein was sequenced and compared with other probiotic bacteria. The purified S-layer protein enhanced the immunomodulatory response in Salmonella-infected mice to a greater extent than L. helveticus M92 cells after the S-layer protein was removed.
The antimutagenic effect of milk fermented with L. helveticus L89 was attributed to the proteolytic system of this strain, since the non-proteolytic variant (Prt−) did not have the same effect (Matar et al., 1997).
Anti-Cancer
An in vivo study using a model of breast cancer in mice concluded that milk fermented with L. helveticus R389 or L. helveticus L89 delayed tumor development (de Moreno de LeBlanc et al., 2005). Modulation of cytokine production and apoptosis of tumor cells were observed in mice fed with fermented milk, which were related to the potent proteolytic ability of strains used in this study especially R389. In the same study, milk fermented with a proteolytic-deficient variant of L helveticus L89 did not exert the same regulatory response observed in mice fed with milk fermented with R389 or L89.
Calcium Binding
Recently, several strains of L. helveticus were tested to evaluate their calcium binding ability. Supernatant from milk fermented with L. helveticus LA, a highly proteolytic strain, exerted a high Ca-binding activity (Dimitrov, 2009). A peptide produced from the hydrolysis of alpha casein with A1 responsible for this effect was purified and sequenced.
L. helveticus 16H and LBK-16H have been reported to have a positive effect on calcium metabolism, bone volume, bone formation and bone mineral density (Narva et al., 2004a,b,c, 2007). An in vitro study (Narva et al., 2004b) on the effect of milk fermented with L. helveticus LBK-16H and its isolated peptides [isoleucyl-prolylproline (IPP) and valyl-prolyl-proline (VPP)] exerted an anabolic effect on bone. Milk fermented with L. helveticus 16H was also shown to increase bone mineral density and bone mineral content in growing rats (Narva et al., 2004a). The same strain has been reported to produce some bioactive peptides in milk, which exert a positive effect on calcium metabolism (Narva et al., 2004c). To test the effect of milk fermented with L. helveticus LBK-16H and synthetic peptide VPP, a study on ovariectomized rats was conducted (Narva et al., 2007). Fermented milk was able to prevent bone loss while VPP dissolved in water did not prevent ovariectomy-induced bone loss. Perhaps the synergies between different components produced during fermentation could be responsible for the functionality of fermented milk.
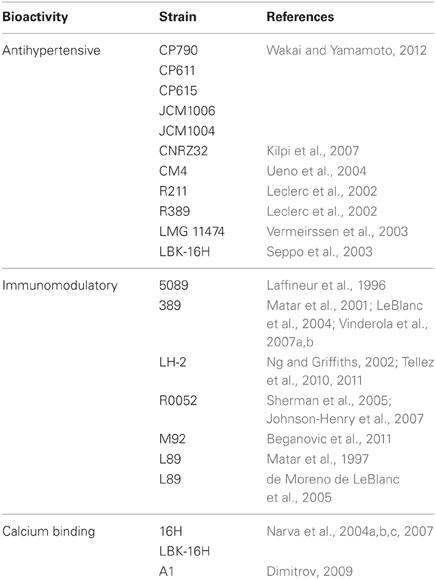
Table 2 presents a summary of the L. helveticus strains that have been reported to produce bioactive peptides from milk proteins.
Effect of the Growth Medium on the L. helveticus Proteolytic System
Even though there are a number of studies that have investigated the proteolytic ability of LAB, little is known about the effect of the growth media on their proteolytic system. It is well known that when milk is used as a growth media for LAB, their proteolytic system is activated (Wakai and Yamamoto, 2012). This is caused by the limited amount of amino acids present in milk and the nutritional needs of LAB. One of the few studies done on the modulation of CEPs by peptide supply (Hebert et al., 2002), concluded that the proteinase activity of the LAB strains used in this study was reduced in cells grown in a peptide-rich medium, such as MRS. Inparticular, the activity of L. helveticus CRL 1177 showed a 100-fold reduction in enzyme activity when cells were grown in MRS. Conversely, the peptidase activity of the L. helveticus strains tested were not affected when grown in a peptide-rich medium. These results agreed with previous findings of this research group (Hebert et al., 2000). Another study conducted on the effect of L. helveticus R0052 to inhibit E coli O157:H7 attachment (Johnson-Henry et al., 2007), tested the influence of different media on the S-layer protein. The authors concluded that, even though there was no differences in the structural formation of the S-layer, the physiological differences between bacteria grow under different conditions are unknown. Smeianov et al. (2007) showed that L. helveticus CNRZ 32 genes encoding CEPs, endopeptidases and oligopeptide transporters were upregulated when the strain was grown in milk compared with the levels of expression in MRS medium. In another study (Jensen et al., 2009), CEP activity of six L. helveticus strains on αs1- and β-casein in MRS and milk was evaluated. The CEPs of the different strains hydrolyzed intact αs1- and β-casein after growth in milk, but not in MRS.
The cell extract of six L. helveticus strains were analyzed for aminopeptidase activity (Jensen and Ardo, 2010). It was concluded that the peptidase activities were generally higher in the L. helveticus strains after growth in skim milk than in MRS, with the exception of Pro-pNA, which was affected by the growth medium in a strain dependent manner. Other studies on the peptidase activity of DPC 4571 and M10 in different growth media showed similar results (Kenny et al., 2003; Simova and Beshkova, 2007). The regulation of genes encoding peptidases of L. helveticus CNRZ 32 have been studied by Smeianov et al. (2007), who observed an up-regulation of the pepI gene during growth in MRS, up-regulation of the genes pepN, pepX, and pepR during growth in skim milk, and the same level of expression of pepC was seen in skim milk and MRS. The impact of the growth medium on proteinase activity was evident in a study conducted on L. helveticus CRL581 (Hebert et al., 1997). Proteinase activity decreased in the presence of an available nitrogen source and showed an increment of activity when the growth medium was milk.
Conclusions
Traditionally L. helveticus has been considered to be a very important bacterium for cheese manufacture because its ability to improve cheese quality. In the last 20 years it has became even more relevant due to its ability to produce bioactive compounds during milk fermentation. The industrial importance of L. helveticus depends on its effective proteolytic system, which seems to be the most efficient between LAB, as well as the specificity of different strains. We have gained some knowledge of the proteolytic system of this bacterium but there are still aspects that require further study. For instance, little is known regarding the specificity of the CEP on caseins, the transport system of the peptides, and the factors that can affect both the individual components of the system and the proteolytic system as a whole. Variations in the proteolytic system of different strains might not be a pivotal factor for growth of the L. helveticus but might have a big impact on the production of bioactive peptides. With the genetic and biochemical tools available it is possible to manipulate the proteolytic system, which can lead to production of bioactive peptides of particular interest or alternatively enzymes isolated from suitable microorganisms can be used alone. Technological challenges in cheese production can be overcome by gaining a deeper understanding of the kinetics of milk hydrolysis by L. helveticus. More research is necessary to shed light on the different components of the proteolytic system of L. helveticus and other LAB and their role in the production of health promoting compounds from milk fermentation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bayoumi, M., and Griffiths, M. W. (2012). In vitro inhibition of expression of virulence genes responsible for colonization and systemic spread of enteric pathogens using Bifidobacterium bifidum secreted molecules. Int. J. Food Microbiol. 156, 255–263.
Beganovic, J., Frece, J., Kos, B., Pavunc, A. L., Habjanic, K., and Suskovic, J. (2011). Functionality of the S-layer protein from the probiotic strain Lactobacillus helveticus M92. Antonie Van Leeuwenhoek 100, 43–53.
Blanc, B., Laloi, P., Atlan, D., Gilbert, C., and Portalier, R. (2003). Two cell-wall-associated aminopeptidases from Lactobacillus helveticus and the purification and characterization of APII from strain ITGLl. J. Gen. Microbiol. 139, 1441–1448.
Broadbent, J. R., Cai, H., Larsen, R. L., Hughes, J. E., Welker, D. L., De Carvalho, V. G., et al. (2011). Genetic diversity in proteolytic enzymes and amino acid metabolism among Lactobacillus helveticus strains. J. Dairy Sci. 94, 4313–4328.
Broadbent, J. R., Thompson, R. L., Hughes, J. E., Welker, D. L., Steele, J. L., Cai, H., et al. (2008). “Comparative genome analysis of the obligately homofermentative lactic acid bacterium Lactobacillus helveticus,” in 9th Symposium on Lactic Acid Bacteria (Netherlands), Poster A 063.
Callanan, M., Kaleta, P., O'Callaghan, J., O'Sullivan, O., Jordan, K., McAuliffe, O., et al. (2008). Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J. Bacteriol. 190, 727–735.
Chatterton, D., Smithers, G., Roupas, P., and Brodkorb, A. (2006). Bioactivity of b-lactoglobulin and a-lactalbumin-Technological implications for processing. Int. Dairy J. 16, 1229–1240.
Chen, Y. S., Christensen, J. E., Broadbent, J. R., and Steele, J. L. (2003). Identification and characterization of Lactobacillus helveticus PepO2, an endopeptidase with post-proline specificity. Appl. Environ. Microbiol. 69, 1276–1282.
Christensen, J. E., Broadbent, J. R., and Steele, J. L. (2003). Hydrolysis of casein-derived peptides alpha(S1)-casein(f1-9) and beta-casein(f193-209) by Lactobacillus helveticus peptidase deletion mutants indicates the presence of a previously undetected endopeptidase. Appl. Environ. Microbiol. 69, 1283–1286.
Christensen, J. E., Dudley, E. G., Pederson, J. A., and Steele, J. L. (1999). Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Van Leeuwenhoek 76, 217–246.
Christensen, J. E., and Steele, J. L. (2003). Impaired growth rates in milk of Lactobacillus helveticus peptidase mutants can be overcome by use of amino acid supplements. J. Bacteriol. 185, 3297–3306.
Christiansen, J. K., Hughes, J. E., Welker, D. L., Rodriguez, B. T., Steele, J. L., and Broadbent, J. R. (2007). Phenotypic and genotypic analysis of amino acid auxotrophy in Lactobacillus helveticus CNRZ 32. Appl. Environ. Microbiol. 74, 416–423.
Clare, D. A., Catignani, G. L., and Swaisgood, H. E. (2003). Biodefence properties of milk: the role of antimicrobial proteins and peptides. Curr. Pharm. Des. 9, 1239–1255.
Dandoy, D., Fremaux, C., Henry de Frahan, M., Horvath, P., Boyaval, P., Hols, P., et al. (2011). The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS. Microb. Cell Fact. 10, S1–S21.
de Moreno de LeBlanc, A., Matar, C., LeBlanc, N., and Perdigon, G. (2005). Effects of milk fermented by Lactobacillus helveticus R389 on a murine breast cancer model. Breast Cancer Res. 7, R477–R486.
Deutsch, S. M., Mollé, D., Gagnaire, V., Piot, M., Atlan, D., and Lortal, S. (2000). Hydrolysis of sequenced β-casein peptides provides new insight into peptidase activity from thermophilic lactic acid bacteria and highlights intrinsic resistance of phosphopeptides. Appl. Environ. Microbiol. 66, 5360–5367.
Deutsch, S. M., Neveu, A., Guezenec, S., Ritzenthaler, P., and Lortal, S. (2003). Early lysis of Lactobacillus helveticus CNRZ 303 in Swiss cheese is not prophage-related. Int. J. Food Microbiol. 81, 147–157.
Di Cagno, R., De Angelis, M., Limitone, A., Fox, P. F., and Gobbetti, M. (2006). Response of Lactobacillus helveticus PR4 to heat stress during propagation in cheese whey with a gradient of decreasing temperatures. Appl. Environ. Microbiol. 72, 4503–4514.
Dimitrov, Z. (2009). Characterization of bioactive peptides with calcium-binding activity released by specially designed cheese starter. Biotechnol. Biotechnol. Equip. 23, 927–930.
Dudley, E. G., Husgen, A. C., He, W., and Steele, J. L. (1996). Sequencing, distribution, and inactivation of the dipeptidase A gene (pepDA) from Lactobacillus helveticus CNRZ32. J. Bacteriol. 178, 701–704.
Ezzat, N., El-Soda, M., Bouillance, C., Zevaco, C., and Blanchard, P. (1985). Cell wall associated proteinases in Lactobacillus helveticus, Lactobacillus bulgaricus and Lactobacillus lactis. Milchwissenschaft 40, 140–143.
Fernandez-Espla, M. D., Garault, P., Monnet, V., and Rul, F. (2000). Streptococcus thermophilus cell wall-anchored proteinase: release, purification, and biochemical and genetic characterization. Appl. Environ. Microbiol. 66, 4772–4778.
Fernández, L., Bhowmik, T., and Steele, J. (1994). Characterization of the Lactobacillus helveticus CNRZ32 pepC gene. Appl. Environ. Microbiol. 60, 333–336.
Fitzgerald, R. J., and Meisel, H. (2003). “Milk protein hydrolysates and bioactive peptides,” in Advanced Dairy Chemistry, Vol. 1. Proteins. 3rd Edn., eds P. F. Fox and P. L. h. McSweeney (New York, NY: Kluwer Academic), 675–698.
Fitzgerald, R. J., and Murray, B. A. (2006). Bioactive peptides and lactic fermentations. Int. J. Dairy Technol. 59, 118–125.
Florisa, R., Recio, I., Berkhout, B., and Visser, S. (2003). Antibacterial and antiviral effects of milk proteins and derivatives thereof. Curr. Pharm. Des. 9, 1257–1275.
Gatti, M., Trivisano, C., Fabrizi, E., Neviani, E., and Gardini, F. (2004). Biodiversity among Lactobacillus helveticus strains isolated from different natural whey starter cultures as revealed by classification trees. Appl. Environ. Microbiol. 70, 182–190.
Genay, M., Sadat, L., Gagnaire, V., and Lortal, S. (2009). prtH2, not prtH, is the ubiquitous cell wall proteinase gene in Lactobacillus helveticus. Appl. Environ. Microbiol. 75, 3238–3249.
Gilbert, C., Blanc, B., Frot-Coutaz, J., Portalier, R., and Atlan, D. (1997). Comparison of cell surface proteinase activities within the Lactobacillus genus. J. Dairy Res. 64, 561–571.
Gómez-Ruiz, J. A., Ramos, M., and Recio, I. (2004a). Angiotensin converting enzyme-inhibitory activity of peptides isolated from Manchego cheese: stability under simulated gastrointestinal digestion. Int. Dairy J. 14, 1075–1080.
Gómez-Ruiz, J. A., Ramos, M., and Recio, I. (2004b). Identification and formation of angiotensin-converting enzyme-inhibitory peptides in Manchego cheese by high-performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 1054, 269–277.
Hebert, E. M., Raya, R. R., and de Giori, G. S. (1997). Characterization of a cell membrane-associated proteinase from Lactobacillus helveticus CRL 581. Curr. Microbiol. 35, 161–164.
Hebert, E. M., Raya, R. R., and Giori, G. S. (1999). Characterisation of a cell-envelope proteinase from Lactobacillus helveticus. Biotechnol. Lett. 21, 831–834.
Hebert, E. M., Raya, R. R., and de Giori, G. S. (2000). Nutritional requirements and nitrogen-dependent regulation of proteinase activity of Lactobacillus helveticus CRL (1062). Appl. Environ. Microbiol. 66, 5316–5321.
Hebert, E. M., Raya, R. R., and de Giori, G. S. (2002). Modulation of the cell-surface proteinase activity of thermophilic lactobacilli by the peptide supply. Curr. Microbiol. 45, 385–389.
Jensen, M. P., and Ardo, Y. (2010). Variation in aminopeptidase and aminotransferase activities of six cheese related Lactobacillus helveticus strains. Int. Dairy J. 20, 149–155.
Jensen, M. P., Vogensen, F. K., and Ardo, Y. (2009). Variation in caseinolytic properties of six cheese related Lactobacillus helveticus strains. Int. Dairy J. 19, 661–668.
Johnson-Henry, K. C., Hagen, K. E., Gordonpour, M., Tompkins, T. A., and Sherman, P. M. (2007). Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell. Microbiol. 9, 356–367.
Kenny, O., FitzGerald, R. J., O'Cuinn, G., Beresford, T., and Jordan, K. (2003). Growth phase and growth medium effects on the peptidase activities of Lactobacillus helveticus. Int. Dairy J. 13, 509–516.
Khalid, N. M., El-Soda, M., and Marth, E. H. (1991). Peptidase hydrolases of Lactobacillus helveticus and Lactobacillus delbrueckii ssp. bulgaricus. J. Dairy Sci. 74, 29–45.
Khalid, N. M., and Marth, E. H. (1990). Purification and partial characterization of a prolyl-dipeptidyl aminopeptidase from Lactobacillus helveticus CNRZ 32. Appl. Environ. Microbiol. 56, 381–388.
Kilara, A., and Panyam, D. (2003). Peptides from milk proteins and their properties. Crit. Rev. Food Sci. 43, 607–633.
Kilpi, E. R., Kahalaa, M. M., Steeleb, J. L., Pihlantoa, A. M., and Joutsjokia, V. V. (2007). Angiotensin I-converting enzyme inhibitory activity in milk fermented by wild-type and peptidase-deletion derivatives of Lactobacillus helveticus CNRZ32E. Int. Dairy J. 17, 976–984.
Korhonen, H., and Pihlanto, A. (2006). Bioactive peptides: production and functionality. Int. Dairy J. 16, 945–960.
Laffineur, E., Genetet, N., and Leonil, J. (1996). Immunodulatory activity of b-casein permeate medium fermented by lactic acid bacteria. J. Dairy Sci. 79, 2112–2120.
LeBlanc, A., Matar, C., LeBlanc, N., and Perdigón, G. (2005). Effects of milk fermented by Lactobacillus helveticus R389 on a murine breast cancer model. Breast Cancer Res. 7, R477–R486.
LeBlanc, J., Fliss, I., and Matar, C. (2004). Induction of a humoral immune response following an Escherichia coli O157:H7 infection with an immunomodulatory peptidic fraction derived from Lactobacillus helveticus-fermented milk. Clin. Diagn. Lab. Immunol. 11, 1171–1181.
LeBlanc, J. G., Matar, C., Valdez, J. C., LeBlanc, J., and Perdigon, G. (2002). Immunomodulating effects of peptidic fractions issued from milk fermented with Lactobacillus helveticus. J. Dairy Sci. 85, 2733–2742.
Leclerc, P., Gauthier, S. F., Bachelard, H., Santure, M., and Roy, D. (2002). Antihypertensive activity of casein-enriched milk fermented by Lactobacillus helveticus. Int. Dairy J. 12, 995–1004.
López-Expósito, I., Amigo, L., and Recio, I. (2012). A mini-review on health and nutritional aspects of cheese with a focus on bioactive peptides. Dairy Sci. Technol. 92, 419–438.
Lozo, J., Strahinic, I., Dalgalarrondo, M., Chobert, J., Haertlé, T., and Topisirovic, L. (2011). Comparative analysis of b-casein proteolysis by PrtP proteinase from Lactobacillus paracasei subsp. paracasei BGHN14, PrtR proteinase from Lactobacillus rhamnosus BGT10 and PrtH proteinase from Lactobacillus helveticus BGRA43. Int. Dairy J. 21, 863–868.
Madureira, A. R., Tavares, T., Gomes, A. M. P., Pintado, M. E., and Malcata, F. X. (2010). Physiological properties of bioactive peptides obtained from whey proteins. J. Dairy Sci. 93, 437–455.
Martin-Minervini, M. C., Alting, A. C., and Exterkate, F. A. (1994). Purification and characterization of the mature, membrane-associated cell-envelope proteinase of Lactobacillus helveticus L89. Appl. Microbiol. Biotechnol. 40, 828–834.
Matar, C., LeBlanc, J. G., Martin, L., and Perdigon, G. (2003). “Biologically active peptides released from fermented milk: role and functions,” in Handbook of Fermented Functional Foods. 1st Edn., ed E. R. Farnworth (Boca Raton, FL: CRC Press), 177–201.
Matar, C., Nadathur, S. S., Bakalinsky, A. T., and Goulet, J. (1997). Antimutagenic effects of milk fermented by Lactobacillus helveticus L89 and a protease-deficient derivative. J. Dairy Sci. 80, 1965–1970.
Matar, C., Valdez, J. C., Medinas, M., Rachid, M., and Perdigon, G. (2001). Immunomodulating effects of milks fermented by Lactobacillus helveticus and its non-proteolytic variant. J. Dairy Res. 68, 601–609.
Mayo, B., Aleksandrzak-Piekarczk, T., Fernandez, M., Kowalczyk, M., Alvarez-Martin, P., and Bardowski, J. (2010). “Updates in the metabolism of lactic acid bacteria,” in Biotechnology of Lactic Acid Bacteria-Novel Applications, eds F. Mozzi, R. R. Raya, and G. M. Vignolo (Iowa: Wiley-Blackwell), 3–33.
Medellin-Pena, M. J., and Griffiths, M. W. (2009). Effect of molecules secreted by Lactobacillus acidophilus strain La-5 on Escherichia coli O157:H7 colonization. Appl. Environ. Microbiol. 75, 1165–1172.
Medellin-Pena, M. J., Wang, H., Johnson, R., Anand, S., and Griffiths, M. W. (2007). Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl. Environ. Microbiol. 73, 4259–4267.
Meisel, H., and Bockelmann, W. (1999). Bioactive peptides encrypted in milk proteins: proteolytic activation and thropho-functional properties. Antoine Van Leeuwenhoek 76, 207–315.
Minervini, F., Algaron, F., Rizzello, C. G., Fox, P. F., Monnet, V., and Gobbetti, M. (2003). Angiotensin I-converting-enzyme-inhibitory and antibacterial peptides from Lactobacillus helveticus PR4 proteinase-hydrolyzed caseins of milk from six species. Appl. Environ. Microbiol. 69, 5297–5305.
Nakajima, H., Kunji, E. R. S., Poolman, B., and Konings, W. N. (1998). Amino acid transport in Lactobacillus helveticus. FEMS Microbiol. Lett. 158, 249–253.
Narva, M., Collin, M., Lamberg-Allardt, C., Kärkkäinen, M., Poussa, T., Vapaatalo, H., et al. (2004a). Effects of long-yerm intervention with Lactobacillus helveticus-Fermented milk on bone mineral density and bone mineral content in growing rats. Ann. Nutr. Metab. 48, 228–234.
Narva, M., Halleen, J., Väänänen, K., and Korpela, R. (2004b). Effects of Lactobacillus helveticus fermented milk on bone cells in vitro. Life Sci. 75, 1727–1734.
Narva, M., Nevala, R., Poussa, T., and Korpela, R. (2004c). The effect of Lactobacillus helveticus fermented milk on acute changes in calcium metabolism in postmenopausal women. Eur. J. Nutr. 43, 61–68.
Narva, M., Rissanen, J., Halleen, J., Vapaatalo, H., Väänänen, K., and Korpela, R. (2007). Effects of bioactive peptide, valyl-prolyl-proline (VPP), and Lactobacillus helveticus fermented milk containing VPP on bone loss in ovariectomized rats. Ann. Nutr. Metab. 51, 65–74.
Ng, K. Y., and Griffiths, M. W. (2002). Enhancement of macrophage cytokine release by cell-free fractions of fermented milk. Milchwissenschaft 57, 66–70.
Ono, H., Yamamoto, N., Maeno, M., and Takano, T. (1997). Purification and characterization of a cell-wall associated proteinase from Lactobacillus helveticus CP53. Milchwissenschaft 52, 373–377.
Pan, D., and Tanokur, M. (2004). Purification and characterization of an aminopeptidase from Lactobacillus helveticus JCM (1004). Food Chem. 88, 511–516.
Pederson, J. A., Mileski, G. J., Weimer, B. C., and Steele, J. L. (1999). Genetic characterization of a cell envelope-associated proteinase from Lactobacillus helveticus CNRZ32. J. Bacteriol. 181, 4592–4597.
Pihlanto-Leppala, A. (2001). Bioactive peptides derived from bovine whey proteins: opioid and ace- inhibitory peptides. Trends Food Sci. Technol. 11, 347–356.
Pihlanto-Leppala, A., Marnila, P., Hubert, L., Rokka, T., Korhonene, H. J. T., and Karp, M. (1999). The effect of α-lactalbumin and β-lactoglobulin hydrolysates on metabolic activity of Escherichia coli JM103. J. Appl. Microbiol. 87, 540–545.
Rattanachaikunsopon, P., and Phumkhachorn, P. (2010). Lactic acid bacteria: their antimicrobial compounds and their uses in food production. Ann. Biol. Res. 1, 218–228.
Sadat-Mekmene, L., Jardin, J., Corre, C., Mollé, D., Richoux, R., Delage, M. M., et al. (2011a). Simultaneous presence of PrtH and PrtH2 proteinases in Lactobacillus helveticus strains improves breakdown of the Pure s1-Casein. Appl. Environ. Microbiol. 77, 179–186.
Sadat-Mekmene, L., Genay, M., Atlan, D., Lortal, S., and Gagnaire, V. (2011b). Original features of cell-envelope proteinases of Lactobacillus helveticus. Int. J. Food Microbiol. 146, 1–13.
Savijoki, K., Ingmer, H., and Varmanen, P. (2006). Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 71, 394–406.
Savijoki, K., and Palva, A. (2000). Purification and molecular characterization of a tripeptidase (PepT) from Lactobacillus helveticus. Appl. Environ. Microbiol. 66, 794–800.
Schanbacher, F. L., Talhouk, R. S., Murray, F. A., Gherman, L. I., and Willett, L. B. (1998). Milk-born bioactive peptides. Int. Dairy J. 8, 393–403.
Scolari, G., Vescovo, M., Zacconi, C., and Vescovi, F. (2006). Extraction and partial characterization of proteolytic activities from the cell surface of Lactobacillus helveticus Zuc2. J. Dairy Sci. 89, 3800–3809.
Seppo, L., Jauhiainen, T., Poussa, T., and Korpela, R. (2003). A fermented milk high in bioactive peptides has a blood pressure-lowering effect in hypertensive subjects. Am. J. Clin. Nutr. 77, 326–330.
Shao, W., Yuksel, G. U., Dudley, E. G., Parkin, K. L., and Steele, J. L. (1997). Biochemical and molecular characterization of PepR, a dipeptidase, from Lactobacillus helveticus CNRZ32. Appl. Environ. Microbiol. 63, 3438–3443.
Sherman, P. M., Johnson-Henry, K. C., Yeung, H. P., Ngo, P. S. C., Goulet, J., and Tompkins, T. A. (2005). Probiotics reduce enterohemorrhagic Escherichia coli O157H7: H7-induced changes in polarized T84 epithelial cell monolayers by reducing bacterial adhesion and cytoskeletal rearrangements. Infect. Immun. 73, 5183–5188.
Simova, E., and Beshkova, D. (2007). Effect of growth phase and growth medium on peptidase activities of starter lactic acid bacteria. Lait 87, 555–573.
Slattery, L., O'Callaghan, J., Fitzgerald, G. F., Beresford, T., and Ross, R. P. (2010). Lactobacillus helveticus—A thermophilic dairy starter related to gut bacteria. J. Dairy Sci. 93, 4435–4454.
Smeianov, V. V., Wechter, P., Broadbent, J. R., Hughes, J. E., Rodriguez, B. T., Christensen, T. K., et al. (2007). Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl. Environ. Microbiol. 73, 2661–2672.
Szwajkowska, M., Wolanciuk, A., Barłowska, J., Król, J., and Litwiñczuk, Z. (2011). Bovine milk proteins as the source of bioactive peptides influencing the consumers' immune system – a review. Anim. Sci. Pap. Rep. 29, 269–280.
Tellez, A., Corredig, M., Brovko, L. Y., and Griffiths, M. W. (2010). Characterization of immune-active peptides obtained from milk fermented by Lactobacillus helveticus. J. Dairy Res. 77, 129–136.
Tellez, A., Corredig, M., Turner, P., Morales, R., and Griffiths, M. W. (2011). A peptidic fraction from milk fermented with L. helveticus protects mice against Salmonella infection. Int. Dairy J. 21, 607–614.
Ueno, K., Mizuno, S., and Yamamoto, N. (2004). Purification and characterization of an endopeptidase that has an important role in the carboxyl terminal processing of antihypertensive peptides in Lactobacillus helveticus CM4. Lett. Appl. Microbiol. 39, 313–318.
Valence, F., Deutsch, S. M., Richoux, R., Gagnaire, V., and Lortal, S. (2000). Autolysis and related proteolysis in Swiss cheese for two Lactobacillus helveticus strains. J. Dairy Res. 67, 261–271.
Valence, F., and Lortal, S. (1995). Zymogram and preliminary characterization of Lactobacillus helveticus autolysins. Appl. Environ. Microbiol. 61, 3391–3399.
Valence, F., Richoux, R., Thierry, A., Palva, A., and Lortal, S. (1998). Autolysis of Lactobacillus helveticus and Propionibacterium freundenreichii in Swiss cheese: first evidence by using species specific lysis markers. J. Dairy Res. 65, 609–620.
Vermeirssen, V., Van Camp, J., Decroos, K., Wijmelbeke, L. V., and Verstraete, W. (2003). The impact of fermentation and in vitro digestion on the formation of angiotesin-I-converting enzyme inhibitory acticity from Pea and Whey protein. J. Dairy Sci. 86, 429–438.
Vinderola, G., Matar, C., Palacios, J., and Perdigon, G. (2007a). Mucosal immunomodulation by the non-bacterial fraction of milk fermented by L. helveticus R389. Int. J. Food Microbiol. 115, 180–186.
Vinderola, G., Matar, C., and Perdigon, G. (2007b). Milk fermented by Lactobacillus helveticus R389 and its non-bacterial fraction confer enhanced protection against Salmonella enteritidis serovar Typhimurium infection in mice. Immunobiology 212, 107–118.
Wakai, T., and Yamamoto, N. (2012). “Antihypertensive peptides specific to Lactobacillus helveticus fermented milk,” in Biotechnology – Molecular Studies and Novel Applications for Improved Quality of Human Life, Biochemistry, Genetics and Molecular Biology, ed R. H. Sammour (InTech), ISBN 978-953-51-0151-2.
Yamamoto, N., Akino, A., and Takano, T. (1993). Purification and specificity of a cell-wall-associated proteinase from Lactobacillus helveticus CP790. J. Biochem. 114, 740–746.
Yamamoto, N., Akino, A., and Takano, T. (1994). Antihypertensive effect of the peptides derived from casein by an extracellular proteinase from Lactobacillus helveticus CP790. J. Dairy Sci. 77, 917–922.
Yamamoto, N., Ono, H., Maeno, M., Ueda, Y., Takano, T., and Momose, H. (1998). Classification of Lactobacillus helveticus strains by immunological differences in extracellular proteinases. Biosci. Biotechnol. Biochem. 62, 1228–1230.
Zeinhom, M., Tellez, A., Delcenserie, V., El-Kholy, A. M., El-Shinawy, S. H., and Griffiths, M. W. (2012). Yoghurt containing bioactive molecules produced by Lactobacilus acidophillus La-5 exerts a protective effect against Enterohaemorrhagic Escherichia coli (EHEC) in mice. J. Food Prot. 75, 1796–1805.
Zevaco, C., and Gripon, J. (1988). Properties and specificity of a ce II-wall proteinase from Lactobacillus helveticus. Le Lait 68, 393–408.
Keywords: proteolytic, Lactobacillus, helveticus, milk, proteins
Citation: Griffiths MW and Tellez AM (2013) Lactobacillus helveticus: the proteolytic system. Front. Microbiol. 4:30. doi: 10.3389/fmicb.2013.00030
Received: 15 November 2012; Paper pending published: 19 December 2012;
Accepted: 04 February 2013; Published online: 05 March 2013.
Edited by:
Giorgio Giraffa, Agriculture Research Council, ItalyReviewed by:
Luca Settanni, Università degli Studi di Palermo, ItalyPanagiotis Skandamis, Agricultural University of Athens, Greece
Copyright © 2013 Griffiths and Tellez. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: M. W. Griffiths, Department of Food Science, Canadian Research Institute for Food Safety, University of Guelph, 43 McGilvray Street, Guelph, ON N1G 2W1, Canada. e-mail: mgriffit@uoguelph.ca