- Department of Plant and Environmental Sciences, The Alexander Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem, Israel
Extremely halophilic microorganisms that accumulate KCl for osmotic balance (the Halobacteriaceae, Salinibacter) have a large excess of acidic amino acids in their proteins. This minireview explores the occurrence of acidic proteomes in halophiles of different physiology and phylogenetic affiliation. For fermentative bacteria of the order Halanaerobiales, known to accumulate KCl, an acidic proteome was predicted. However, this is not confirmed by genome analysis. The reported excess of acidic amino acids is due to a high content of Gln and Asn, which yield Glu and Asp upon acid hydrolysis. The closely related Halorhodospira halophila and Halorhodospira halochloris use different strategies to cope with high salt. The first has an acidic proteome and accumulates high KCl concentrations at high salt concentrations; the second does not accumulate KCl and lacks an acidic proteome. Acidic proteomes can be predicted from the genomes of some moderately halophilic aerobes that accumulate organic osmotic solutes (Halomonas elongata, Chromohalobacter salexigens) and some marine bacteria. Based on the information on cultured species it is possible to understand the pI profiles predicted from metagenomic data from hypersaline environments.
Introduction
In a study of the proteins of Halobacterium and Halococcus, Reistad (1970) noted an unusual amino acids composition of the cells’ bulk protein: a great excess of the acidic amino acids glutamate and aspartate compared to the basic amino acids lysine and arginine. Analysis of the genome of Halobacterium NRC-1 (Ng et al., 2000) and related organisms (Oren, 2013a) has confirmed the special properties of the proteins of this group of Archaea. The acidic proteins of the Halobacteriaceae typically require high salt concentrations for structural stability and activity, and the presence of such an acidic proteome was considered to be correlated with the accumulation of molar concentrations of KCl to provide osmotic balance to the cells (Lanyi, 1974; Mevarech et al., 2000; Oren, 2013b).
A different strategy of osmotic adaptation in halophilic and halotolerant microorganisms is the accumulation of organic osmotic solutes (glycine betaine, ectoine, glycerol, simple sugars, etc., often termed “compatible solutes”). Such molecules are generally uncharged or zwitterionic, and their presence in high intracellular concentrations does not require far-going adaptation of the proteins. The intracellular solute concentrations can be rapidly adjusted according to the outside salinity, so that microorganisms using this “low-salt-in” strategy can often adapt to life at a wide range of salinities (Galinski, 1995; Oren, 2002a,b, 2011, 2013b).
Early attempts to correlate the mode of osmotic adaptation used by halophilic microorganisms with their phylogenetic position yielded a relatively simple picture: the “high-salt-in” strategy was thought to be limited to the Halobacteriaceae within the Archaea domain. Among the Bacteria a single group was known that had high intracellular KCl concentrations and allegedly had a highly acidic proteome: the anaerobic fermentative Halanaerobiales (Firmicutes). Other groups of Bacteria use organic osmotic solutes, generally in a pattern correlated with their phylogenetic affiliation; organic solutes are also found in halophilic methanogens (Archaea) and in salt-adapted eukaryotic microorganisms (Oren, 2008; Trüper et al., 1991). This represented the state of our knowledge up to turn of the century. Since then the relatively simple picture got complicated by new information and insights. Some of these new data are discussed below.
Salinibacter ruber, its Mode of Osmotic Adaptation and the Properties of its Proteins
Salinibacter ruber (Bacteroidetes) is a red-pigmented aerobic heterotrophic extremely halophile, first isolated from Spanish saltern crystallizer ponds (Antón et al., 2002), but now known to be distributed worldwide in neutral-pH water bodies at or near salt saturation. This interesting organism shares many key properties with the Halobacteriaceae with which it shares its habitat (Oren, 2013c). These include the accumulation of molar concentrations of KCl intracellularly, insignificant concentrations of organic osmotic solutes (Oren et al., 2002), a highly acidic nature of the bulk protein, and a strict salt requirement of key enzymes (Oren and Mana, 2002). High intracellular KCl concentrations were also measured in the phylogenetically related Salisaeta longa (Vaisman and Oren, 2009; Vaisman and Oren, unpublished results).
Analysis of the Salinibacter genome (Mongodin et al., 2005) confirmed the highly acidic nature of most of its proteins. The median pI value of 5.92 for the proteins encoded by the S. ruber genome is slightly higher than that for Halobacterium NRC-1 (5.03; Figure 1). Salinibacter can be considered as an example of convergent evolution mediated by extensive gene exchange with archaeal halophiles found in the same habitat. The combination of the “salt-in” strategy and the possession of salt-dependent, highly acidic proteins is thus not necessarily limited to the Halobacteriaceae lineage of aerobic halophilic Archaea.
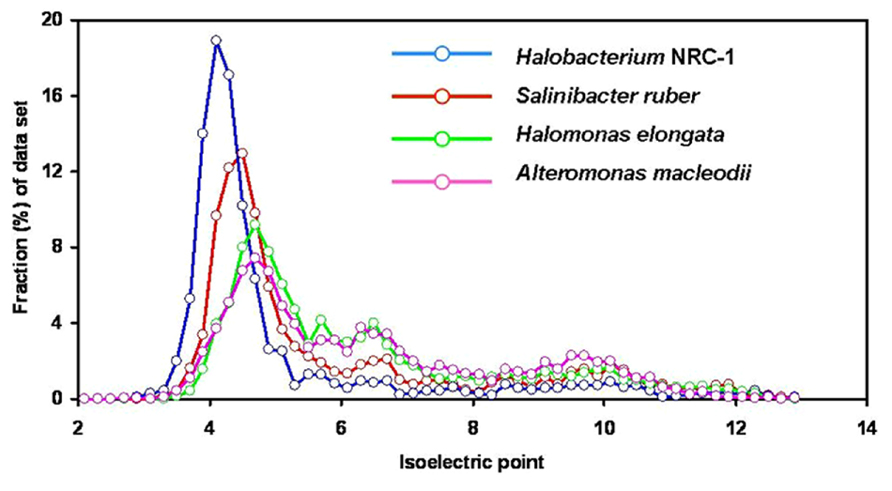
FIGURE 1. Isoelectric point profiles, calculated at 0.2 intervals, of predicted proteins encoded by the genomes of Halobacterium NRC-1 (2,675 proteins; median pI 5.03), the extreme halophilic bacterium Salinibacter ruber (2,845 proteins; median pI 5.92), the aerobic moderately halophilic bacterium Halomonas elongata (3,474 proteins; median pI 6.32) and the aerobic marine bacterium Alteromonas macleodii (4,396 proteins; median pI 6.46). From the genome annotations (Ng et al., 2000; Mongodin et al., 2005; Schwibbert et al., 2011) sequences encoding proteins or putative proteins were extracted, and for each protein sequence the predicted the pI value was calculated using the programs in the Galaxy platform (http://main.g2.bx.psu.edu; Giardine et al., 2005; Blankenberg et al., 2010; Goecks et al., 2010). With kind permission from Springer Science+Business Media: Extremophiles, Elevi Bardavid and Oren (2012b), compiled from data presented in Figures 1–3.
The Nature of the Proteome of the Halanaerobiales and other Halophilic Anaerobes within the Bacterial Domain
The order Halanaerobiales, families Halanaerobiaceae and Halobacteroidaceae (Rainey et al., 1995) forms a phylogenetically coherent group of anaerobic bacteria affiliated with the low G+C Firmicutes (Kivistö and Karp, 2011; Oren, 2013d). Members of the group have been found in sediments of Great Salt Lake, Utah, the Dead Sea, salterns, oil wells, and in alkaline hypersaline lakes (Lake Magadi, Kenya, Big Soda Lake, Nevada). Most species grow optimally at 10–15% NaCl, and some tolerate salt up to saturation. Most ferment sugars to acetate, ethanol, H2, CO2, and other products. One of these (Halothermothrix orenii, isolated from a warm saline lake in Tunisia), is a true thermophilic (up to 68°C; optimum at 60°C) halophile (growth up to 20% NaCl; Cayol et al., 1994). Some genera (Acetohalobium, Natroniella) have a homoacetogenic metabolism. Selenihalanaerobacter shriftii grows by anaerobic respiration with selenate or nitrate as electron acceptor.
Examination of the cytoplasm of members of the Halanaerobiales did not show significant concentrations of organic osmotic solutes (Oren, 1986; Rengpipat et al., 1988), with the possible exception of the finding of glycine betaine in Orenia salinaria grown in media containing yeast extract (Mouné et al., 2000). However, high ionic concentrations (K+, Na+, Cl-), sufficient to provide osmotic balance, were measured in Halanaerobium praevalens (Oren, 1986; Oren et al., 1997), Halanaerobium acetethylicum (Rengpipat et al., 1988), Halobacteroides halobius (Oren, 1986), and Natroniella acetigena (Detkova and Pusheva, 2006). Studies performed on glyceraldehyde-3-phosphate dehydrogenase, NAD-linked alcohol dehydrogenase, pyruvate dehydrogenase, and methyl viologen-linked hydrogenase from H. acetethylicum (Rengpipat et al., 1988), carbon monoxide dehydrogenase of N. acetigena (Detkova and Boltyanskaya, 2006; Detkova and Pusheva, 2006), the fatty acid synthetase complex of H. praevalens (Oren and Gurevich, 1993), and hydrogenase and carbon monoxide dehydrogenase of Acetohalobium arabaticum (Zavarzin et al., 1994) showed that all these enzymes function well in the presence of molar salt concentrations, and many need high salt for optimal activity. Therefore the “high-salt-in” strategy was assumed to be the mode of osmotic adaptation in this group (Kivistö and Karp, 2011; Oren, 2013d).
Based on these observations the proteome of the members of the Halanaerobiales was predicted to have a strongly acidic nature. Indeed, analysis of acid hydrolysates of Halanaerobium praevalens, Halanaerobium saccharolyticum, Natroniella acetigena, Halobacteroides halobius, and Sporohalobacter lortetii suggested that the bulk protein of all these species may have a strongly acidic nature (Oren, 1986; Detkova and Boltyanskaya, 2006). However, it must be remembered that during acid hydrolysis, the neutral asparagine and glutamine are deaminated to form aspartate and glutamate.
The first evidence against a highly acidic proteome in members of the Halanaerobiales was published in 1987 when it was shown that the H. praevalens ribosomal A-protein is not particularly rich in acidic amino acids (Matheson et al., 1987). Today genome sequences of three members of the group are available: H. praevalens GSLT (Ivanova et al., 2011), a haloalkaliphilic strain from Soap Lake, WA, USA, described as “Halanaerobium hydrogeniformans” (Brown et al., 2011), and Halothermothrix orenii H168T (Mavromatis et al., 2009). Analysis of these three genomes did not show preferential use of acidic amino acids and no low content of basic amino acids (Elevi Bardavid and Oren, 2012a; Figure 2). It was earlier suggested that the proteins of H. orenii may lack a pronounced acidic nature as a special adaptation toward growth at high temperatures (Mijts and Patel, 2001; Mavromatis et al., 2009). The properties of the other two genomes show that also the mesophilic species lack an acidic proteome. The bimodal distribution of the pI values with peaks around 4.6–4.8 and 9.8–10.2 is similar to that of the non-halophiles Bacteroides fragilis and Chlorobaculum tepidum (Mongodin et al., 2005). The main reason for the apparent discrepancy between the bulk protein analyses, showing a pronounced acid nature, and the analysis of the proteins encoded by the genomes, is the high content of glutamine and asparagine, which lose their amide group during the acid hydrolysis procedure involved in sample preparation for amino acid analysis.
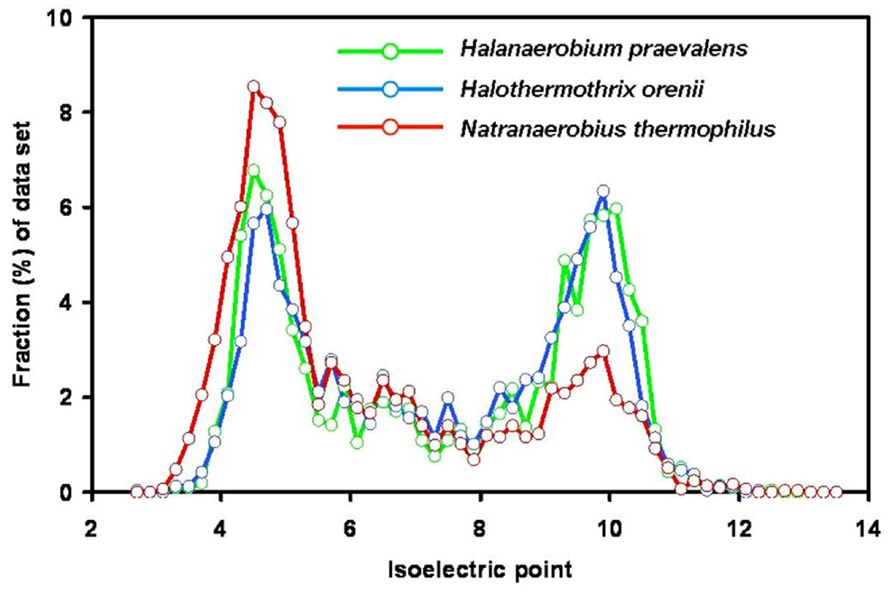
FIGURE 2. Isoelectric point profiles, calculated at 0.2 intervals, of predicted proteins encoded by the genomes of the three fermentative halophilic anaerobes: Halanaerobium praevalens GSLT (2,110 proteins; median pI 7.42), the thermophilic Halothermothrix orenii H168T (2,365 proteins, median pI 7.41) and the alkaliphilic and thermophilic Natranaerobius thermophilus JW.NM-WN-LFT (2,906 proteins; median pI 6.27). From the genome annotations (Mavromatis et al., 2009; Ivanova et al., 2011; Zhao et al., 2011) sequences encoding proteins or putative proteins were extracted, and pI profiles were calculated as for Figure 1. With kind permission from Springer Science+Business Media: Extremophiles, compiled from data presented in Elevi Bardavid and Oren (2012a) – Figure 1, and Elevi Bardavid and Oren (2012b) – Figure 4.
Still there is no reason to doubt the presence of high ionic concentrations within the cytoplasm to balance the osmotic pressure of the medium. Analysis of the three Halanaerobiales genomes did not show clear evidence for pathways leading to the synthesis of organic osmotic solutes. A gene for sucrose phosphate synthase was identified in H. orenii, which may point to the possibility of sucrose biosynthesis (Mavromatis et al., 2009). Whether indeed sucrose is present in the cells at high concentrations, remains to be ascertained. The possibility must be taken into account that the anaerobic halophilic of the Halanaerobiales group use a “high-salt-in” strategy of osmotic adaptation but have not adopted the pattern of acidic, low-pI proteins commonly associated with haloadaptation in the aerobic halophiles (Halobacteriaceae, Salinibacter). A renewed study of the special properties of the Halanaerobiales may therefore provide new insights into the strategies available to the prokaryote world to thrive at high salt concentrations (Elevi Bardavid and Oren, 2012a).
The genomes of two anaerobic fermentative halophiles belonging to other phylogenetic lineages were recently sequenced. One is Flexistipes sinusarabici MAS 10T, isolated from a deep-sea brine pool on the bottom of the Red Sea (Fiala et al., 1990; Lapidus et al., 2011). It was classified as a member of the Deferribacteres, a deep branch within the Bacteria; it grows between 3 and 10% salt and possibly higher. Its mode of osmotic adaptation is yet unknown. Its pI profile (bimodal, with a median pI of 7.47) resembles that of Halanaerobium. The second is Natranaerobius thermophilus JW.NM-WN-LFT an anaerobic halothermoalkaliphile isolated from the Wadi An Natrun lakes in Egypt. It was classified in the newly established order Natranaerobiales (Clostridia), requires 3.1–4.9 M Na+, and is markedly thermophilic (optimum growth at 53°C) and alkaliphilic (optimum pH 9.5; Mesbah et al., 2007). Analysis of its genome (Zhao et al., 2011) yielded a markedly acidic proteome (median pI 6.27; Elevi Bardavid and Oren, 2012b; Figure 2). Comparison of the proteins of five anaerobic halophiles of different phylogenetic lineages and with different temperature and pH optima thus shows great variations in the acidic nature of the proteome.
Disparate Osmotic Adaptation Strategies within the Genus Halorhodospira
The genus Halorhodospira currently contains four species: the type species H. halophila, H. neutriphila, H. halochloris and H. abdelmalekii. With respect to salt requirement and tolerance they are quite similar, and all tolerate NaCl at concentrations up to 25% or higher. They can be divided into two groups, phylogenetically separated on the basis of 16S rRNA gene sequences: H. halophila and H. neutriphila contain bacteriochlorophyll a and carotenoids of the spirilloxanthin group, while H. halochloris and H. abdelmalekii contain bacteriochlorophyll b and rhopdopin carotenoids (Imhoff and Süling, 1996; Hirschler-Réa et al., 2003; Oren, 2013e). With respect to their mode of osmotic adaptation they were always considered to be a prime example of organisms that use organic compatible solutes. H. halophila, H. halochloris and H. abdelmalekii were all shown to produce glycine betaine as osmotic solute, with minor amounts of ectoine and trehalose (Trüper et al., 1991). Ectoine, now known to be the most widespread osmotic solute in the prokaryote world, was first discovered in H. halochloris (Galinski et al., 1985).
In view of their common phylogeny and documented content of organic osmotic solutes, the finding of an acidic proteome and of high intracellular KCl concentrations in H. halophila but not in H. halochloris (Deole et al., 2013) came as a big surprise. While the latter does not accumulate KCl, the first contains high KCl when grown at high salt (35%) but not at low salt (5%). The genus Halorhodospira thus presents a thus far unique case in which different combinations of KCl concentrations, production of organic osmotic solutes, and presence of acidic vs. non-acidic proteomes are used for osmotic adaptation in phylogenetically closely related species. The authors concluded that “proteome acidity is not driven by stabilizing interactions between K+ ions and acidic side chains but by the need for maintaining sufficient solvation and hydration of the protein surface at high salinity …,” and they proposed that “obligate protein halophilicity is a non-adaptive property resulting from genetic drift in which constructive neutral evolution progressively incorporates weak K+-binding sites on an increasingly acidic protein surface” (Deole et al., 2013).
Acidic Proteomes in Moderately Halophilic Gammaproteobacteria
There is no a priori reason to assume that moderately halophilic aerobic heterotrophic bacteria that synthesize and/or accumulate organic compatible solutes should have a high acidic proteome adapted to function in the presence of high intracellular salt concentrations. A first survey of the proteins of the gammaproteobacterium Chromohalobacter salexigens DSM 3043T, based on 238 out of the 3,319 proteins encoded by its genome, indeed showed that most selected proteins were no more acidic than comparable proteins from non-halophilic counterparts. A notable exception was found for periplasmic proteins exposed to the high medium salinity (Oren et al., 2005).
Analysis of the entire C. salexigens genome, together with that of the phylogenetically related moderate halophile Halomonas elongata 1H9T (Schwibbert et al., 2011) showed large peaks of acidic proteins (maximum at pI 4.4–5.0 and 4.5–5.1, respectively) in the pI profiles of the predicted proteins. The median pI values for the proteins encoded by these genomes are 6.60 and 6.32, respectively (Elevi Bardavid and Oren, 2012b). These values are still in the low pI range, albeit somewhat higher than those reported for “high-salt-in” organisms such as Halobacterium and Salinibacter (Figure 2). Both organisms synthesize ectoine as compatible solute and accumulate glycine betaine when available in the medium.
Such acidic proteomes are found not only in halophilic and highly halotolerant members of the Gammaproteobacteria, but also in typically marine members of the group. Analysis of the pI distribution of the proteins predicted from the genomes of Alteromonas macleodii ATCC 27126T (Ivars-Martínez et al., 2008), a representative of a genus ubiquitous in the world’s oceans, and of the luminescent Aliivibrio fischeri strain MJ11 (Mandel et al., 2009) showed a pronounced peak in the acidic range (maximum at pI values of 4.6–4.8), with median pI values of 6.46 and 6.52, respectively (Elevi Bardavid and Oren, 2012b).
Acidic Metaproteomes in Hypersaline Environments
Metagenomic data from saline and hypersaline environments can be subjected to analyses similar to those shown above for microbial isolates. As shown by Rhodes et al. (2010), there is a general trend of increased average protein acidity (as expressed by the ratio of acidic to basic amino acids) with increased salinity. The highest salinity environments (the Dead Sea, saltern crystallizer ponds) have the greatest excess of acidic amino acids in the proteins encoded by the recovered DNA ([Glu + Asp]/[Lys + Arg + His] = 1.42–1.26). This could be expected as high-salt-in strategists (species of Halobacteriaceae, Salinibacter) with highly acidic proteomes dominate their biota. Metagenomes from different samples from the marine environment gave values in the range 0.86–0.95, and the benthic microbial mats in the 9% salt lagoons of Guerrero Negro, Mexico (Kunin et al., 2008), yielded an intermediate value of 1.01 on the average.
The finding that the pI distribution of the proteins encoded by the metagenome of the Guerrero Negro microbial mats showed an acid-shifted proteome (major peak at pI 4.5–4.9, median pI 6.8) as compared to non-halophilic or marine environments was at first puzzling, as at that salinity microorganisms are expected to use organic osmotic solutes, without the need to adapt their proteins to high salt. Kunin et al. (2008) concluded that the enhanced acidic nature of the proteins is linked to the increased salinity, explained as an example of species-independent molecular convergence in a microbial community. However, as documented above, many moderate halophiles, also those that do not accumulate organic osmotic solutes, show a broad peak at pI values 4.5–4.9 (Elevi Bardavid and Oren, 2012b). In comparison to the proteins encoded by the genomes of moderately halophilic aerobes and even certain marine bacteria (Figure 2), the metaproteome encoded by the metagenome of the different layers within the 9% salt Guerrero Negro microbial mats is not conspicuously acid-shifted.
The finding that marine metagenomes do not encode for metaproteomes enriched in acidic proteins (Rhodes et al., 2010) needs to be evaluated in view of the above-mentioned observation that some typically marine Gammaproteobacteria (Alteromonas and Vibrio spp.) are rich in low-pI proteins. A possible explanation is that other, possibly very abundant marine bacteria show the opposite trend. The small genome of “Pelagibacter ubique” (Alphaproteobacteria; the “SAR-11” phylotype; Giovannoni et al., 2005) encodes for 1,393 proteins with an overall excess of 2% (Lys + Arg) – (Glu + Asp). For comparison, Halobacterium, Salinibacter and H. elongata and Halomonas all show an excess of acidic amino acids (7.5, 4.1, and 2.8 mol %, respectively; Elevi Bardavid and Oren, 2012b). Most “Pelagibacter” proteins have pI values between 9.4 and 10.8, with a median value of 8.42.
Final Comments
The genomic and metagenomic data discussed above show that dominance of acidic proteins in halophilic microorganisms is by no means restricted to the Halobacteriaceae and to Salinibacter which resembles the Halobacteriaceae in many properties. Somewhat less acidic proteomes are found in many moderately halophilic and even in some marine bacteria, organisms that exclude salt from their cytoplasm to a large extent (Elevi Bardavid and Oren, 2012b). On the other hand, the analysis of the genomes of different anaerobic halophiles (Halanaerobiales and others) unexpectedly failed to show a highly acidic proteome (Elevi Bardavid and Oren, 2012a). The case of the two Halorhodospira species demonstrates that phylogenetically very closely related organisms may use completely different strategies for osmotic adaptation, and accordingly have highly different amino acid signatures of their proteins. The more or less coherent picture of a clear correlation between phylogenetic affiliation and modes of salt adaptation that was apparent in the past (Trüper et al., 1991; Oren, 2008) needs therefore drastic revision. We must rethink our concepts about the correlation between acidic proteomes, salt requirement and tolerance, accumulation of KCl, use of organic osmotic solutes, and microbial phylogeny and taxonomy.
A recently published analysis of the structure of primitive proteins that may have been formed from “prebiotic” amino acids expected to have been available at the time life originated on Earth showed that the predicted foldable proteins have a substantial acidification of pI and possessed halophilic properties (Longo et al., 2013). The question whether the environment for primordial life may have been hypersaline has been addressed earlier (Dundas, 1998). Therefore the issues discussed above may even have direct implications for our ideas on the origin of life and the properties of the earliest organisms that inhabited our planet.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Rahel Elevi Bardavid and Omri Finkel for their contributions to the data evaluation. This study was supported by grants no. 1103/10 and 343/13 from the Israel Science Foundation.
References
Antón, J., Oren, A., Benlloch, S., Rodríguez-Valera, F., Amann, R., and Rosselló-Móra, R. (2002). Salinibacter ruber gen. nov., sp. nov., a novel extreme halophilic member of the Bacteria from saltern crystallizer ponds. Int. J. Syst. Evol. Microbiol. 52, 485–491. doi: 10.1128/AEM.66.7.3052-3057.2000
Blankenberg, D., Von Kuster, G., Coraor, N., Ananda, G., Lazarus, R., Mangan, M., et al. (2010). Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19, Unit 19.10.1–21. doi: 10.1002/0471142727.mb1910s89
Brown, S. D., Begemann, M. B., Mormile, M. R., Wall, J. D., Han, C. S., Goodwin, L. A., et al. (2011). Complete genome sequence of the haloalkaliphilic, hydrogen-producing bacterium Halanaerobium hydrogeniformans. J. Bacteriol. 193, 3682–3683. doi: 10.1128/JB.05209-11
Cayol, J.-L., Ollivier, B., Patel, B. K. C., Prensier, G., Guezennec, J., and Garcia, J.-L. (1994). Isolation and characterization of Halothermothrix orenii gen. nov., sp. nov., a halophilic, thermophilic, fermentative, strictly anaerobic bacterium. Int. J. Syst. Bacteriol. 44, 534–540. doi: 10.1099/00207713-44-3-534
Detkova, E. N., and Boltyanskaya, Y. V. (2006). Relationships between the osmoadaptation strategy, amino acid composition of bulk protein, and properties of certain enzymes of haloalkaliphilic bacteria. Mikrobiologiya 75, 259–265.
Detkova, E. N., and Pusheva, M. A. (2006). Energy metabolism in halophilic and alkaliphilic acetogenic bacteria. Mikrobiologiya 75, 5–17. doi.org/10.1134/S0026261706010012
Deole, R., Challacombe, J., Raiford, D. W., and Hoff, W. D. (2013). An extremely halophilic proteobacterium combines a highly acidic proteome with a low cytoplasmic potassium content. J. Biol. Chem. 288, 581–588. doi: 10.1074/jbc.M112.420505
Dundas, I. (1998). Was the environment for primordial life hypersaline? Extremophiles 2, 375–377. doi: 10.1007/s007920050081
Elevi Bardavid, R., and Oren, A. (2012a). The amino acid composition of proteins from anaerobic halophilic bacteria of the order Halanaerobiales. Extremophiles 16, 567–572. doi: 10.1007/s00792-012-0455-y.
Elevi Bardavid, R., and Oren, A. (2012b). Acid-shifted isoelectric point profiles of the proteins in a hypersaline microbial mat – an adaptation to life at high salt concentrations? Extremophiles 16, 787–792. doi: 10.1007/s00792-012-0476-6
Fiala, G., Woese, C. R., Langworthy, T. A., and Stetter, K. O. (1990). Flexistipes sinusarabici, a novel genus and species of eubacteria occurring in the Atlantis II Deep brines of the Red Sea. Arch. Microbiol. 154, 120–126. doi: 10.1007/BF00423320
Galinski, E. A., Pfeiffer, H.-P., and Trüper, H. G. (1985). 1,4,5,6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid. A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur. J. Biochem. 149, 135–139. doi: 10.1111/j.1432-1033.1985.tb08903.x
Giardine, B., Riemer, C., Hardison, R. C., Burhans, R., Elnitski, L., Shah, P., et al. (2005). Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455. doi: 10.1101/gr.4086505
Giovannoni, S. J., Tripp, H. J., Givan, S., Podar, M., Vergin, K. L., Baptista, D., et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245. doi: 10.1126/science.1114057
Goecks, J., Nekrutenko, A., Taylor, J., and The Galaxy Team. (2010). Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86. doi: 10.1186/gb-2010-11-8-r86
Hirschler-Réa, A., Matheron, R., Riffaud, C., Mouné, S., Eatock, C., Herbert, R. A., et al. (2003). Isolation and characterization of spirilloid purple phototrophic bacteria forming red layers in microbial mats of Mediterranean salterns: description of Halorhodospira neutriphila sp. nov. and emendation of the genus Halorhodospira. Int. J. Syst. Evol. Microbiol. 53, 153–163. doi: 10.1099/ijs.0.02226-0
Imhoff, J. F., and Süling, J. (1996). The phylogenetic relationship among Ectothiorhodospiraceae: a reevaluation of their taxonomy on the basis of 16S rDNA analyses. Arch. Microbiol. 165, 106–113. doi: 10.1007/s002030050304
Ivanova, N., Sikorski, J., Chertkov, O., Nolan, M., Lucas, S., Hammon, N., et al. (2011). Complete genome sequence of the extremely halophilic Halanaerobium praevalens type strain (GSLT). Stand. Genomic Sci. 4, 312–321. doi:10.4056/sigs.1824509
Ivars-Martínez, E., D’Auria, G., Rodríguez-Valera, F., Sánchez-Porro, C., Ventosa, A., Ioint, I., et al. (2008). Biogeography of the ubiquitous marine bacterium Alteromonas macleodii determined by multilocus sequence analysis. Mol. Ecol. 17, 4092–4106. doi: 10.1111/j.1365-294X.2008.03883.x
Kivistö, A. T., and Karp, M. (2011). Halophilic anaerobic fermentative bacteria. J. Biotechnol. 152, 114–124. doi: 10.1016/j.jbiotec.2010.08.014
Kunin, V., Raes, J., Harris, J. K., Spear, J. R., Walker, J. J., Ivanova, N., et al. (2008). Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol. Syst. Biol. 4, 198. doi: 10.1038/msb.2008.35
Lanyi, J. K. (1974). Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol. Rev. 38, 272–290.
Lapidus, A., Chertkov, O., Nolan, M., Lucas, S., Hammon, N., Deshpande, S., et al. (2011). Genome sequence of the moderately thermophilic halophile Flexistipes sinusarabici strain (MAS10T). Stand. Genomic Sci. 5, 1. doi: 10.4056/sigs.2235024
Longo, L. M., Lee, J., and Blaber, M. (2013). Simplified protein design biased for prebiotic acids yields a foldable, halophilic protein. Proc. Natl. Acad. Sci. U.S.A. 110, 2135–2139. doi: 10.1073/pnas.1219530110
Mandel, M. J., Wollenberg, M. S., Stabb, E. V., Visick, K. L., and Ruby, E. G. (2009). A single regulatory gene is sufficient to alter bacterial host range. Nature 458, 215–218. doi: 10.1038/nature07660
Matheson, A. T., Louie, K. A., Tak, B. D., and Zuker, M. (1987). The primary structure of the ribosomal A-protein (L12) from the halophilic eubacterium Haloanaerobium praevalens. Biochimie 69, 1013–1020. doi: 10.1016/0300-9084(87)90001-0
Mavromatis, K., Ivanova, N., Anderson, I., Lykidis, A., Hooper, S. D., Sun, H., et al. (2009). Genome analysis of the anaerobic thermohalophilic bacterium Halothermothrix orenii. PLoS ONE 4:e4192. doi: 10.1371/journal.pone.0004192
Mesbah, N. M., Hedrick, D. B., Peacock, A. D., Rohde, M., and Wiegel, J. (2007). Natranaerobius thermophilus gen. nov., sp. nov., a halophilic, alkalithermophilic bacterium from soda lakes of the Wadi An Natrun, Egypt, and proposal of Natranaerobiaceae fam. nov. and Natranaerobiales ord. nov. Int. J. Syst. Evol. Microbiol. 57, 2507–2512. doi: 10.1099/ijs.0.65068-0
Mevarech, M., Frolow, F., and Gloss, L. M. (2000). Halophilic enzymes: proteins with a grain of salt. Biophys. Chem. 86, 155–164. doi: 10.1016/S0301-4622(00)00126-5
Mijts, B. M., and Patel, B. K. C. (2001). Random sequence analysis of genomic DNA of an anaerobic, thermophilic, halophilic bacterium, Halothermothrix orenii. Extremophiles 5, 61–69. doi: 10.1007/s007920000174
Mongodin, E. F., Nelson, K. E., Daugherty, S., DeBoy, R. T., Wister, J., Khouri, H., et al. (2005). The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. U.S.A. 102, 18147–18152. doi: 10.1073/pnas.0509073102
Mouné, S., Eatock, C., Matheron, R., Willison, J. C., Hirschler, A., Herbert, R., et al. (2000). Orenia salinaria sp. nov., a fermentative bacterium isolated from anaerobic sediments of Mediterranean salterns. Int. J. Syst. Evol. Microbiol. 50, 721–729. doi: 10.1099/00207713-50-2-721
Ng, W. V., Kennedy, S. P., Mahairas, G. G., Berquist, B., Pan, M., Shukla, H. D., et al. (2000). Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. U.S.A. 97, 12176–12181. doi: 10.1073/pnas.190337797
Oren, A. (1986). Intracellular salt concentrations of the anaerobic halophilic eubacteria Haloanaerobium praevalens and Halobacteroides halobius. Can. J. Microbiol. 32, 4–9. doi: 10.1139/m86-002
Oren, A. (2002a). Halophilic Microorganisms and their Environments. Dordrecht: Kluwer Scientific Publishers.
Oren, A. (2002b). Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 28, 56–63.
Oren, A. (2008). Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 4, 2. doi: 10.1186/1746-1448-4-2
Oren, A. (2011). “Diversity of halophiles,” in Extremophiles Handbook, ed. K. Horikoshi (Tokyo: Springer), 309–325.
Oren, A. (2013a). “The family Halobacteriaceae”, in The Prokaryotes. A Handbook on the Biology of Bacteria: Ecophysiology and Biochemistry, 4th Edn, eds E. Rosenberg, E. F. DeLong, F. Thompson, S. Lory, and E. Stackebrandt (New York, NY: Springer), in press.
Oren, A. (2013b). “Life at high salt concentrations,” in The Prokaryotes. A Handbook on the Biology of Bacteria: Ecophysiology and Biochemistry, 4th Edn, eds E. Rosenberg, E. F. DeLong, F. Thompson, S. Lory, and E. Stackebrandt (New York, NY: Springer), in press.
Oren, A. (2013c). Salinibacter, an extremely halophilic bacterium with archaeal properties. FEMS Microbiol. Lett. 342, 1–9.
Oren, A. (2013d). “The order Halanaerobiales, families Halanaerobiaceae and Halobacteroidaceae,” in The Prokaryotes. A Handbook on the Biology of Bacteria: Ecophysiology and Biochemistry, 4th Edn, eds E. Rosenberg, E. F. DeLong, F. Thompson, S. Lory, and E. Stackebrandt (New York, NY: Springer), in press.
Oren, A. (2013e). “Family Ectothiorhodospiraceae,” in The Prokaryotes. A Handbook on the Biology of Bacteria: Ecophysiology and Biochemistry, 4th Edn, eds E. Rosenberg, E. F. DeLong, F. Thompson, S. Lory, and E. Stackebrandt (New York, NY: Springer), in press.
Oren, A., and Gurevich, P. (1993). The fatty acid synthetase complex of Haloanaerobium praevalens is not inhibited by salt. FEMS Microbiol. Lett. 108, 287–290. doi: 10.1111/j.1574-6968.1993.tb06117.x
Oren, A., Heldal, M., and Norland, S. (1997). X-ray microanalysis of intracellular ions in the anaerobic halophilic eubacterium Haloanaerobium praevalens. Can. J. Microbiol. 43, 588–592. doi: 10.1139/m97-083
Oren, A., Heldal, M., Norland, S., and Galinski, E. A. (2002). Intracellular ion and organic solute concentrations of the extremely halophilic bacterium Salinibacter ruber. Extremophiles 6, 491–498. doi: 10.1007/s00792-002-0286-3
Oren, A., Larimer, F., Richardson, P., Lapidus, A., and Csonka, L. N. (2005). How to be moderately halophilic with a broad salt tolerance: clues from the genome of Chromohalobacter salexigens. Extremophiles 9, 275–279. doi: 10.1007/s00792-005-0442-7
Oren, A., and Mana, L. (2002). Amino acid composition of bulk protein and salt relationships of selected enzymes of Salinibacter ruber, an extremely halophilic bacterium. Extremophiles 6, 217–223. doi: 10.1007/s007920100241
Rainey, F. A., Zhilina, T. N., Boulygina, E. S., Stackebrandt, E., Tourova, T. P., and Zavarzin, G. A. (1995). The taxonomic status of the fermentative halophilic anaerobic bacteria: description of Halobacteriales ord. nov., Halobacteroidaceae fam. nov., Orenia gen. nov. and further taxonomic rearrangements at the genus and species level. Anaerobe 1, 185–199. doi: 10.1006/anae.1995.1018
Reistad, R. (1970). On the composition and nature of the bulk protein of extremely halophilic bacteria. Arch. Mikrobiol. 71, 353–360. doi: 10.1007/BF00417131
Rengpipat, S., Lowe, S. E., and Zeikus, J. G. (1988). Effect of extreme salt concentrations on the physiology and biochemistry of Halobacteroides acetoethylicus. J. Bacteriol. 170, 3065–3071.
Rhodes, M. E., Fitz-Gibbon, S., Oren, A., and House, C. H. (2010). Amino acid signatures of salinity on an environmental scale with a focus on the Dead Sea. Environ. Microbiol. 12, 2613–2623. doi: 10.1111/j.1462-2920.2010.02232.x
Schwibbert, K., Marin-Sanguino, A., Bagyan, I., Heidrich, G., Lentzen, G., Seitz, H., et al. (2011). A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas DSM 2581. Environ. Microbiol. 13, 1973–1994. doi: 10.1111/j.1462-2920.2010.02336.x
Trüper, H. G., Severin, J., Wolhfarth, A., Müller, E., and Galinski, E. A. (1991). “Halophily, taxonomy, phylogeny and nomenclature,” in General and Applied Aspects of Halophilism, ed. F. Rodriguez-Valera (New York, NY: Plenum Press), 3–7.
Vaisman, N., and Oren, A. (2009). Salisaeta longa gen. nov., sp. nov., a red halophilic member of the Bacteroidetes. Int. J. Syst. Evol. Microbiol. 59, 2571–2574. doi: 10.1099/ijs.0.010892-0
Zavarzin, G. A., Zhilina, T. A., and Pusheva, M. A. (1994). “Halophilic acetogenic bacteria,” in Acetogenesis, ed. H. L. Drake (New York: Chapman & Hall), 432–444.
Keywords: acidic proteins, osmotic adaptation, halophilic, marine bacteria, anaerobic, Halanaerobiaceae
Citation: Oren A (2013) Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front. Microbiol. 4:315. doi: 10.3389/fmicb.2013.00315
Received: 28 June 2013; Paper pending published: 05 October 2013;
Accepted: 06 October 2013; Published online: 05 November 2013.
Edited by:
Antonio Ventosa, University of Sevilla, SpainReviewed by:
Mohammad Ali Amoozegar, University of Tehran, IranMelanie R. Mormile, Missouri University of Science and Technology, USA
Copyright © 2013 Oren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aharon Oren, Department of Plant and Environmental Sciences, The Alexander Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Edmond Safra Campus, Jerusalem 91904, Israel e-mail: aharon.oren@mail.huji.ac.il
 Aharon Oren
Aharon Oren