- Department of Molecular and Cell Biology, University of Connecticut, Storrs, CT, USA
Dihydroxyacetone (DHA) is a ketose sugar that can be produced by oxidizing glycerol. DHA in the environment is taken up and phosphorylated to DHA-phosphate by glycerol kinase or DHA kinase. In hypersaline environments, it is hypothesized that DHA is produced as an overflow product from glycerol utilization by organisms such as Salinibacter ruber. Previous research has demonstrated that the halobacterial species Haloquadratum walsbyi can use DHA as a carbon source, and putative DHA kinase genes were hypothesized to be involved in this process. However, DHA metabolism has not been demonstrated in other halobacterial species, and the role of the DHA kinase genes was not confirmed. In this study, we examined the metabolism of DHA in Haloferax volcanii because putative DHA kinase genes were annotated in its genome, and it has an established genetic system to assay growth of mutant knockouts. Experiments in which Hfx. volcanii was grown on DHA as the sole carbon source demonstrated growth, and that it is concentration dependent. Three annotated DHA kinase genes (HVO_1544, HVO_1545, and HVO_1546), which are homologous to the putative DHA kinase genes present in Hqm. walsbyi, as well as the glycerol kinase gene (HVO_1541), were deleted to examine the effect of these genes on the growth of Hfx. volcanii on DHA. Experiments demonstrated that the DHA kinase deletion mutant exhibited diminished, but not absence of growth on DHA compared to the parent strain. Deletion of the glycerol kinase gene also reduced growth on DHA, and did so more than deletion of the DHA kinase. The results indicate that Hfx. volcanii can metabolize DHA and that DHA kinase plays a role in this metabolism. However, the glycerol kinase appears to be the primary enzyme involved in this process. BLASTp analyses demonstrate that the DHA kinase genes are patchily distributed among the Halobacteria, whereas the glycerol kinase gene is widely distributed, suggesting a widespread capability for DHA metabolism.
Introduction
Dihydroxyacetone (DHA) is a simple ketose sugar commonly used in sunless tanning lotions and sprays (Faurschou et al., 2004). DHA can be used as a carbon source by many different bacteria, yeast, and protists, and there are a number of different pathways in which it can be produced. In bacteria such as Klebsiella pneumoniae, DHA is produced anaerobically via glycerol oxidation by an NAD-dependent glycerol dehydrogenase (Forage and Lin, 1982). Gluconobacter oxydans and related bacteria also use glycerol oxidation to produce DHA, but they utilize a glycerol dehydrogenase that is pyrroloquinoline quinone (PQQ)-dependent and attached to the outer membrane. This pathway releases the DHA directly into the surrounding environment, which makes the Gluconobacter bacteria useful for industrial production of DHA (Deppenmeier et al., 2002). DHA can also be produced by methylotrophic yeast such as Candida boidinii by first oxidizing methanol to formaldehyde, after which a pyrophosphate-dependent transketolase transfers a two-carbon hydroxyethyl group to the formaldehyde to form DHA (Waites and Quayle, 1981).
Once DHA is obtained by a cell either via glycerol oxidation or uptake from the surrounding environment, it can then be phosphorylated and subsequently metabolized. Two types of kinases phosphorylate DHA: glycerol kinase and DHA kinase. Glycerol kinase is considered less specific, and it is capable of phosphorylating both glycerol and DHA using ATP (Hayashi and Lin, 1967; Weinhouse and Benziman, 1976; Jin et al., 1982). DHA kinase is more specific, and it is only able to phosphorylate DHA and its isomer, D-glyceraldehyde (Erni et al., 2006). There are two major families of DHA kinases. The first consists of two subunits (DhaK and DhaL) and which are ATP-dependent. The DhaK subunit binds to the DHA substrate, and the DhaL subunit binds to ATP and transfers a phosphate group from ATP to DhaK-DHA (Daniel et al., 1995; Siebold et al., 2003). In the second family, the DHA kinases are made up of three subunits (DhaK, DhaL, and DhaM) and are phosphoenolpyruvate (PEP)-dependent. This family of DHA kinases uses the PEP:sugar phosphotransferase system (PTS) to transfer a phosphate group from PEP to the DhaM subunit, a multidomain protein with one domain predicted to be a member of the mannose (EIIAMan) family of the PTS (Gutknecht et al., 2001; Zurbriggen et al., 2008). The DhaM then transfers the phosphate group to DhaL, which picks up the phosphate using an ADP cofactor bound to the subunit (Bachler et al., 2005). The phosphate is then transferred from DhaL to the DhaK subunit, which phosphorylates the bound DHA substrate to DHA phosphate. The ATP-dependent family of DHA kinases is present in eukaryotes and some bacteria, whereas the PEP-dependent family of DHA kinases is present only in bacteria and archaea (Erni et al., 2006).
DHA has been hypothesized as a potential carbon source in hypersaline environments for heterotrophic halobacterial species (Elevi Bardavid et al., 2008). This hypothesis is supported by previous studies on glycerol oxidation in Salinibacter ruber, a halophilic bacterium common in hypersaline environments. In a study by Sher et al. (2004), which examined the oxidation of radio-labeled glycerol by S. ruber, an unknown soluble product consisting of 20% of the radioactivity from the added glycerol was observed to be excreted by the cells. This soluble product was later analyzed in a study by Elevi Bardavid and Oren (2008) using a colorimetric assay, and was identified as DHA; indicating that S. ruber could produce DHA in hypersaline environments as an overflow product via glycerol oxidation.
The ability of Haloquadratum walsbyi, a common halobacterial species, to metabolize DHA further supports the hypothesis that DHA is a carbon source in hypersaline environments. Hqm. walsbyi was first hypothesized to metabolize DHA after examination of the sequenced genome in a study Bolhuis et al. (2006) identified an uptake system for DHA involving three genes (HQ2672A, HQ2673A, and HQ2674A) encoding the subunits of a putative PEP-dependent DHA kinase. The DHA kinase encoded by these genes was hypothesized to use a phosphate group from the PTS system to phosphorylate DHA to DHA phosphate, which could then be incorporated into the metabolism of the cell. Elevi Bardavid and Oren (2008) tested DHA metabolism in Hqm. walsbyi by adding DHA to a cell culture of Hqm. walsbyi and measuring the change in DHA concentration over time. A decrease in DHA concentration was observed, indicating that the DHA was being taken up and metabolized by the Hqm. walsbyi cultures.
Overall, the current evidence supports a model where halobacterial species Hqm. walsbyi metabolizes DHA in hypersaline environments produced by S. ruber; however, there is still little known about DHA metabolism in Halobacteria. While DHA metabolism has been observed to occur in Hqm. walsbyi, no other halobacterial species has been shown to be able to metabolize DHA. Additionally, the putative DHA kinase genes in Hqm. walsbyi were never confirmed to be involved in DHA phosphorylation and metabolism. In this study, we sought to elucidate our understanding of halobacterial metabolism of DHA by examining DHA utilization in Haloferax volcanii, a halobacterial species isolated from Dead Sea sediment (Mullakhanbhai and Larsen, 1975). We used Hfx. volcanii because it has three putative PEP-dependent DHA kinase genes that are homologous to Hqm. walsbyi (Anderson et al., 2011), and it has an established genetic system that can be used to delete genes and test their function (Bitan-Banin et al., 2003; Allers et al., 2004; Blaby et al., 2010). We also used DHA metabolism genes in Hfx. volcanii to search the other sequenced halobacterial genomes to better understand the distribution of these genes among the Halobacteria. Our data provide important new insights into the metabolism of DHA in halobacterial organisms.
Materials and Methods
Strains and Growth Conditions
Strains and plasmids used in this study are listed in Table 1. All Hfx. volcanii strains were grown in either Hv-YPC or Hv-CA medium at 42°C while shaking at 200 rpm. Hv-YPC and Hv-CA media were produced using the formulas outlined in The Halohandbook (Dyall-Smith, 2009). Hv-min medium used in growth experiments was modified from the formula in The Halohandbook to exclude a carbon source (Hv-min -C). Media were supplemented with uracil (50 μg/mL) and 5-fluoroorotic acid (50 μg/mL) as needed. For growth on Petri plates, 2% agar (w/v) was added to the media.
All Escherichia coli strains were grown in either S.O.C. media or LB-media at 37°C while shaking at 200 rpm. S.O.C. media was provided by Clontech (Cat. # 636763) and New England BioLabs (Cat. # B9020S). LB medium was produced by adding 5 g NaCl, 5 g tryptone, and 2.5 g of yeast extract to deionized water to a final volume of 500 mL and pH set to 7.0. LB was supplemented with ampicillin (100 μg/mL) as needed. When LB cell culture plates were produced, 1.5% agar (w/v) was added. LB plates were supplemented with 40 μL of X-gal (20 mg/mL) as needed.
PCR and DNA Isolation
All primers used in this study are listed in Table 2. DNA used for plasmid construction and screening was amplified via PCR. Reactions for PCR were assembled as 10 μL volumes and contained the following reagents: 5.9 μL of deionized water, 2 μL of 5x GC Phusion buffer (Thermo Scientific, Cat. # F-519), 1 μL of 100% DMSO (Thermo Scientific, Cat. # TS-20684), 0.4 μL of 10 mM dNTP (Promega, Cat. # U1511), 0.2 μL of 10 μM forward primer, 0.2 μL of 10 μM reverse primer, 0.2 μL of template DNA, and 0.1 μL of Phusion High-Fidelity DNA Polymerase (Thermo Scientific, Cat. # F-530S). When needed, water was substituted with 20% acetamide. The reactions were performed in a Mastercycler EP Gradient (Eppendorf) with the following cycle: a DNA melting step at 94°C for 22 s, an annealing step at 58.1°C for 35 s, and an extension step at 72°C for 90 s. This cycle was repeated 40 times, after which a final annealing step at 72°C for 5 min was performed. Template DNA included Hfx. volcanii DS2 genomic DNA (20 ng/μL), plasmid DNA listed in Table 1, and DNA from E. coli and Hfx. volcanii colonies.
Gel electrophoresis was performed to separate and analyze the PCR products using 0.8% (w/v) agarose in 1 × TAE buffer (40 mM Tris acetate, 2 mM EDTA). After gel electrophoresis, PCR products were excised from the gel and purified using the Wizard SV Gel and PCR Clean-Up System (Promega). Plasmids from E. coli strains were extracted and purified using the PureYield Plasmid Miniprep System (Promega). Plasmids linearized via digestion with restriction enzymes (BamHI, HindIII, XhoI, or XbaI) were also purified using the Wizard SV Gel and PCR Clean-Up System.
Gene Deletion in Hfx. volanii
Three Hfx. volcanii genes (dhaKLM; HVO_1544, HVO_1545, and HVO_1546), which encode homologs to the putative DHA kinase genes in Hqm. walsbyi, and a glycerol kinase gene (glpK; HVO_1541), were targeted for deletion in Hfx. volcanii strain H26 using the In-Fusion HD Cloning Kit (Clontech). The strategy for gene deletion was based on the methodology outlined in a study by Blaby et al. (2010) with a few modifications. Flanking regions of the targeted genes were developed to be between 800 and 1000 bp in length. The 15-bp linker used to combine the flanking regions was altered to so that EcoRI and BstOI sites were included for the dhaKLM deletion linker and BglI and BstOI sites were included for the glpK deletion linker. The pTA131 was linearized with HindIII and BamHI for the dhaKLM deletion and XhoI and XbaI for the glpK deletion. Constructed plasmids were transformed into Stellar Competent Cells (Clontech, Cat. # 636763), according to the directions of the provider, and were plated on LB-amp plates with X-gal. White colonies were screened via colony PCR using the external primers of the target gene flanking regions. Confirmed deletion plasmids (listed in Table 1) were subcloned in dam−/dcm− Competent E. coli (New England BioLabs, Cat. # C2925H) to produce demethylated plasmids for transformation of Hfx. volcanii. Hfx. volcanii H26 colonies were screened for deleted genes via PCR using the external primers of the target gene flanking regions. The size of PCR products of screened cells were compared to those produced with wild-type DNA (Figure 1). Smaller product size indicated that the gene had been deleted. The Hfx. volcanii H26 deletion strains produced by this process are listed in Table 1.
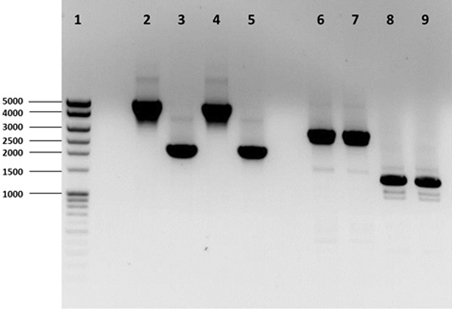
Figure 1. PCR analysis of H26, ΔdhaKLM, ΔglpK, and ΔdhaKLM ΔglpK. Analysis examined presence or absence of the dhaKLM operon (lanes 2–5) and glpK (lanes 6–9). Lane 1 contained exACTGene Mid Range Plus DNA Ladder (Fisher Scientific). Lanes 2 and 6 contained amplicons from H26. Lanes 3 and 7 contained amplicons from ΔdhaKLM. Lanes 4 and 8 contained amplicons from ΔglpK. Lanes 5 and 9 contained amplicons from ΔdhaKLM ΔglpK.
Complementation of Deleted Genes
The dhaKLM and glpK genes deleted in Hfx. volcanii H26 were resuscitated by constructing complementation plasmids. Primers were designed which amplified the upstream native promoter and the coding region of the targeted genes in Hfx. volcanii. The primers were also designed to have 15 bp of homology with pTA409. Restriction digestion of pTA409 was performed using BamHI and XhoI to linearize the plasmid. After the linearized pTA409 and gene fragments were gel-purified, the DNA fragments were combined together using the In-Fusion HD Cloning Kit according to the instructions of the provider. The constructed plasmids were cloned, screened, and demethylated as described in the above gene deletion protocol. Purified constructed plasmids (listed in Table 1) were then transformed into the Hfx. volcanii H26 deletion strains using the PEG mediated transformation of Haloarchaea protocol from The Halohandbook. PCR was used to confirm transformation success. The Hfx. volcanii complementation strains produced by this process are listed in Table 1.
DHA Growth Experiments
Hfx. volcanii strains listed in Table 1 were grown to late-exponential phase (OD600 = ~ 0.6−0.8) in Hv-YPC medium. The cell cultures were then centrifuged at 3220 RCF for 15 min and resuspended in Hv-min -C media supplemented with uracil. Centrifugation was repeated a total of three times to wash the cells of residual Hv-YPC media. During the final resuspension of the cells in Hv-min -C media, the cell cultures were diluted to OD600 ~0.01. Each cell culture was then distributed into the wells of a 96-well plate, with each well receiving 190 μL of cell culture. Also, 200 μL of Hv-min -C was added to the plate to be used as a blank. Three wells of each culture were treated with 10 μL of either 0.1 M DHA (final concentration of 5 mM DHA), 0.05 M DHA (final concentration of 2.5 mM DHA), 0.02 M DHA (final concentration of 1 mM DHA), or deionized water (negative control). The 96-well plate was then placed into a Multiscan FC plate reader (Fisher Scientific), which incubated the plate at 42°C while shaking it at low speed. The plate reader measured the OD620 of each well every hour for 72 h.
Bioinformatics
The amino acid sequences of the Hfx. volcanii putative DHA kinase gene dhaK (HVO_1546) and glycerol kinase gene glpK were used to perform BLASTp (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches of the NCBI database to determine other halobacterial species with DHA kinase and glycerol kinase genes. The amino acid sequences were retrieved from the NCBI database (dhaK GI number 292655696; glpK GI number 292655691). The search was restricted to the Halobacteriales (taxid 2235) with an E-value cut-off of 1e-20. Reciprocal BLASTp was performed to analyze only orthologous genes. The halobacterial genomes queried in this BLASTp search are listed in Table 3.
Results
DHA Kinase is Patchily Distributed Among the Halobacteria
Three DHA kinase genes (HQ2672A, HQ2673A, and HQ2674A) have been annotated in the genome of Hqm. walsbyi (Bolhuis et al., 2006), a halobacterial species which is able to metabolize external DHA (Elevi Bardavid and Oren, 2008). Homologs of these three genes are also annotated in Hfx. volcanii (HVO_1544, HVO_1545, and HVO_1546). In order to determine the prevalence of DHA kinase genes among the Halobacteria, the Hfx. volcanii dhaK gene (HVO_1546) was used to perform a BLASTp search against the database of Halobacteria genomes available on NCBI. The search yielded significant hits among 31 different halobacterial species (Table 4). Except for Haloferax larsenii and Haloferax elongans, all queried Haloferax species yielded significant hits in the BLASTp search. Species from the Halobiforma, Halococcus, Halorubrum, and Natronococcus genera also yielded significant hits, but not all queried species from these genera produced results. All representatives from the genera Haladaptatus, Halalkalicoccus, Halarchaeum, Haloquadratum, Halosarcina, and Salinarchaeum yielded significant hits. Halobacteria genera that did not yield significant hits in the BLASTp search (E-value cut-off of 1e-20) include Haloarcula, Halobacterium, Halobaculum, Halogeometricum, Halogranum, Halomicrobium, Halopiger, Haloplanus, Halorhabdus, Halosimplex, Halostagnicola, Haloterrigena, Halovivax, Natrialba, Natrinema, Natronobacterium, Natronolimnobius, Natronomonas, and Natronorubrum.
Growth on DHA in Hfx. volcanii is Concentration Dependent
Although putative DHA kinase genes are present in Hfx. volcanii, no previous research has demonstrated that Hfx. volcanii is able to grow on DHA as a carbon source. Therefore, experiments were performed to test the growth of Hfx. volcanii strain H26 on 5 mM, 2.5 mM, and 1 mM DHA. The results indicated that H26 was capable of growth on DHA as the sole carbon source. The cell density at which H26 reached stationary phase was also dependent on the initial concentration of DHA provided to the cells (Figure 2). H26 cells grown in medium supplemented with 1 mM DHA reached stationary phase at the lowest cell density, whereas cells grown with the highest tested concentration of 5 mM DHA reached stationary phase at the highest cell density. These data indicate that growth of Hfx. volcanii on DHA as a carbon source is concentration dependent.
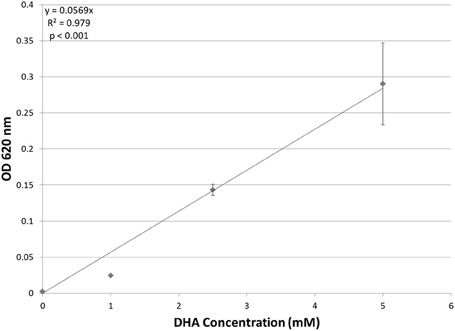
Figure 2. Cell density of H. volcanii H26 at stationary phase vs. concentration of DHA. Cell density is represented by the average optical density (OD620) reading of three cell culture replicates. Error bars depict the standard deviation of the averages. The depicted line represents the line of best fit for the data. ANOVA single factor, p < 0.001.
DHA Kinase is Used in DHA Metabolism in Hfx. volcanii
Evidence indicates that Hfx. volcanii, like Hqm. walsbyi, can use DHA as a carbon source. Although both organisms have DHA kinase genes, no previous studies demonstrated these putative DHA kinase genes have a role in DHA metabolism. In order to determine that DHA metabolism in Hfx. volcanii utilizes the annotated DHA kinase, the operon dhaKLM (HVO_1544—HVO_1546) was deleted in Hfx. volcanii strain H26. The growth of this deletion strain (ΔdhaKLM) on 5 mM DHA was then tested in comparison to the parent strain H26 as well as a complementation strain (ΔdhaKLM + pdhaKLM). The results indicate that the deletion of dhaKLM causes a reduction in growth on DHA, and that complementation of the deleted genes negates this growth deficiency (Figure 3). However, the ΔdhaKLM was still capable of growth on DHA, exhibiting a 33% decrease in growth compared to H26. These results indicate that the dhaKLM genes are used by Hfx. volcanii in DHA metabolism, most likely for the phosphorylation of DHA to DHA phosphate, and that the genes are apparently not essential. Since it is still capable of growth on DHA there must be additional genes involved in the phosphorylation step.
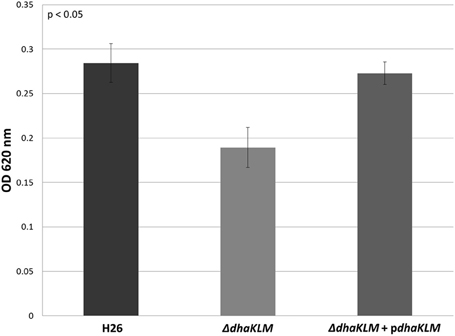
Figure 3. Cell density of H26, ΔdhaKLM, and ΔdhaKLM + pdhaKLM at stationary phase when grown on 5 mM DHA. Cell density is represented by the average optical density (OD620) reading of three cell culture replicates. Error bars depict the standard deviation of the averages. ANOVA single factor, p < 0.05.
Glycerol Kinase is More Important than DHA Kinase
In other organisms, glycerol kinase is also capable of phosphorylating DHA (Hayashi and Lin, 1967; Weinhouse and Benziman, 1976; Jin et al., 1982). Therefore, the other gene involved DHA metabolism in Hfx. volcanii was hypothesized to be the glycerol kinase gene glpK (HVO_1542). In order to test this hypothesis, the glpK gene was deleted in H26. The deletion strain (ΔglpK), and its complementation strain (ΔglpK + pglpK), were both grown on 5 mM DHA along with the parent strain H26. The results indicate that the deletion of glpK caused a reduction in growth on DHA even greater than deletion of dhaKLM, and that complementation of the glpK gene restores growth to normal levels (Figure 4). In comparison to the parent strain H26, ΔglpK strain demonstrated an 83% decrease in growth. This decrease is far greater than the 33% decrease exhibited by the ΔdhaKLM deletion mutant. These results indicate that the glpK gene is used by Hfx. volcanii in DHA metabolism, and that its role is potentially greater than that of the dhaKLM operon.
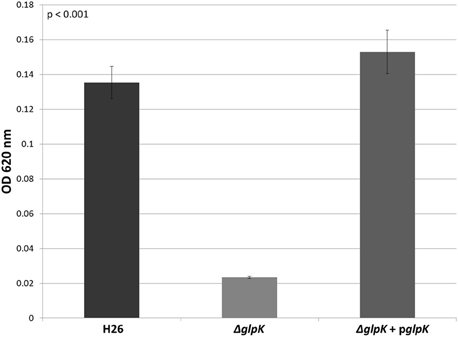
Figure 4. Cell density of H26, ΔglpK, and ΔglpK + pglpK at stationary phase when grown on 5 mM DHA. Cell density is represented by the average optical density (OD620) reading of three cell culture replicates. Error bars depict the standard deviation of the averages. ANOVA single factor, p < 0.001.
In order to further test the roles of the DHA kinase and glycerol kinase in DHA metabolism in Hfx. volcanii, the dhaKLM operon and glpK gene were both deleted in H26. This double deletion mutant (ΔdhaKLM ΔglpK), along with a DHA kinase complementation strain (ΔdhaKLM ΔglpK + pdhaKLM), a glycerol kinase complementation strain (ΔdhaKLM ΔglpK + pglpK), and the parent strain H26, were then grown on 5 mM DHA. The results indicate that the deletion of both kinases abolishes growth on DHA, and that complementation with glycerol kinase restores growth to a greater degree than complementation with DHA kinase (Figure 5). The ΔdhaKLM ΔglpK strain did not exhibit any growth, remaining at the initial OD620 of 0.0035. The ΔdhaKLM ΔglpK + pdhaKLM strain was able to grow on DHA, but demonstrated an 84% decrease compared to the H26 parent strain. The ΔdhaKLM ΔglpK + pglpK was also capable of limited growth on DHA, but demonstrated a 39% growth decrease from H26 and a 390% growth increase compared with ΔdhaKLM ΔglpK + pdhaKLM. Overall, these data confirm that glycerol kinase is more important for DHA metabolism in Hfx. volcanii than DHA kinase.
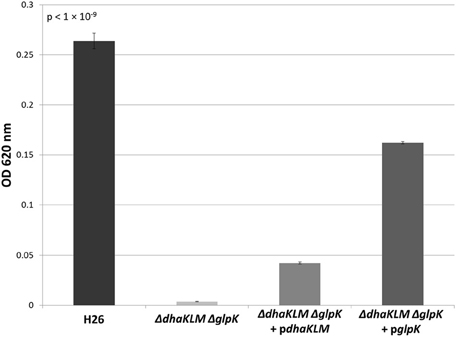
Figure 5. Cell density of H26, ΔdhaKLM ΔglpK, ΔdhaKLM ΔglpK + pdhaKLM, and ΔdhaKLM ΔglpK + pglpK at stationary phase when grown on 5 mM DHA. Cell density is represented by the average optical density (OD620) reading of three cell culture replicates. Error bars depict the standard deviation of the averages. ANOVA single factor, p < 1 × 10−9.
Glycerol Kinase is Widely Distributed among the Halobacteria
Since growth experiments indicated that glycerol kinase has a significant role in DHA metabolism, the presence of this gene in halobacterial species could potentially be a determinant of DHA metabolism in those species. Although the distribution of glpK homologs has been examined in previous studies (Sherwood et al., 2009; Anderson et al., 2011), a greater number of halobacterial genomes have become available since those studies. Therefore, the glpK gene in Hfx. volcanii was used to perform a BLASTp search against the halobacterial genomes available on NCBI. The search yielded 90 significant hits among 82 different species of Halobacteria (Table 5), indicating a much wider distribution of glycerol kinase compared to DHA kinase among the Halobacteria. Six species yielded more than one significant hit: Halogeometricum borinquense (3 hits), Haladaptatus paucihalophilus (3 hits), Haloferax prahovense (2 hits), Haloferax mucosum (2 hits), Haloferax gibbonsii (2 hits), and Natronomonas moolapensis (2 hits). The multiple hits indicate the presence of glpK paralogs in these species. Only 18 of the 100 queried halobacterial species did not yield significant hits: Haloarcula sp. AS7094, Halobacterium sp. DL1, Halobacterium sp. GN101, Halobaculum gomorrense, Halococcus sp. 197A, Halopiger sp. IIH2, Halopiger sp. IIH3, Haloplanus natans, Halorubrum ezzemoulense, Halosarcina pallida, Halostagnicola larsenii, Halovivax asiaticus, Halovivax ruber, Natrinema sp. CX2021, Natrinema sp. J7-1, Natronobacterium gregoryi, Natronobacterium sp. AS-7091, and Natronomonas pharaonis. It should be noted, however, that only the genomes of Halovivax ruber, Natronobacterium gregoryi, and Natronomonas pharaonis are completely sequenced, whereas the other genomes without significant hits are incomplete, leaving open the possibility that these species might have glpK homologs. With the exception of Halosarcina pallida, which has an incompletely sequenced genome, all halobacterial species that yielded significant hits in the dhaK BLASTp search also yielded significant hits in the glpK BLASTp search.
Discussion
Previously, Hqm. walsbyi was the only halobacterial species known to be able to utilize DHA as a carbon source (Elevi Bardavid and Oren, 2008). In this study, we have identified Hfx. volcanii as the second halobacterial species known to be capable of metabolizing DHA. When DHA was added to growth medium as the sole carbon source, Hfx. volcanii was capable of growth. This growth was variable based on the concentration of DHA present in the growth medium. The ability of Hfx. volcanii to metabolize DHA suggests that the substrate could be an important carbon source in the Dead Sea environment where Hfx. volcanii naturally lives. Elevi Bardavid and Oren (2008) have suggested that Salinibacter might be a source of DHA in hypersaline environments, since it can produce DHA as an overflow product. However, Salinibacter has not been identified in the Dead Sea, making it an unlikely candidate for DHA producer. The DHA could potentially be produced as an overflow product from Dunaliella parva, a halophilic alga that is the most prominent photosynthetic organism in the Dead Sea and is able to produce DHA (Ben-Amotz and Avron, 1974; Oren and Shilo, 1982). Elevi Bardavid and Oren (2008) hypothesized that the Dunaliella cell membrane could be permeable to DHA, allowing excess DHA produced by the cells to leak into the external environment. If D. parva produces a significant amount of DHA overflow, the substrate would be readily available for Hfx. volcanii to utilize as a source of carbon.
When Elevi Bardavid and Oren (2008) demonstrated that Hqm. walsbyi could utilize DHA as a carbon source, they hypothesized that the organism used a system involving a PEP-dependent DHA kinase to phosphorylate DHA to DHA kinase, based on genomic analysis from Bolhuis et al. (2006). However, their study did not demonstrate a direct connection between the putative DHA kinase and DHA metabolism. In our model halobacterial organism, Hfx. volcanii, we have demonstrated that DHA kinase is involved in metabolism of DHA. When the DHA kinase operon dhaKLM is deleted, growth of Hfx. volcanii on DHA is impeded, and complementation of the deleted genes with the dhaKLM operon restores growth. The growth of Hfx. volcanii is not completely abolished, however, and further analysis using a strain wherein the glycerol kinase gene glpK has been deleted indicates that Hfx. volcanii also uses glycerol kinase for DHA metabolism. Deletion of the glpK gene reduces growth on DHA more dramatically than the dhaKLM deletion, indicating that the role of glycerol kinase is more pronounced in DHA metabolism than that of DHA kinase for Hfx. volcanii. This enzyme primacy is further supported by the observation that, in the double deletion mutant ΔdhaKLM ΔglpK, complementation with glpK restores growth better than complementation with dhaKLM.
The primacy of the glycerol kinase in DHA metabolism is unexpected, since DHA kinase is usually the primary enzyme involved in DHA phosphorylation in other organisms due to the lower affinity of glycerol kinase for DHA. In Klebsiella pneumoniae, the glycerol kinase has a Km of 1 × 10−3 M for DHA, whereas the DHA kinase has a Km of 1 × 10−5 M (Jin et al., 1982). The glycerol kinase in E. coli has a Km of 5 × 10−4 M for DHA (Hayashi and Lin, 1967), but the DHA kinase has a Km of 4.5 × 10−7 M (Gutknecht et al., 2001). One possible explanation for the primacy of the glycerol kinase in Hfx. volcanii DHA metabolism is the glycerol kinase might have a higher affinity than DHA kinase for DHA. Another possible explanation might be differences in expression of the kinases. DHA kinase might be expressed at lower levels than glycerol kinase early in the Hfx. volcanii growth cycle, which would cause the glycerol kinase to be the primary DHA phosphorylating enzyme despite a possible lower affinity for DHA. Later in the growth cycle, however, Hfx. volcanii may increase expression of DHA kinase, leading to the higher affinity enzyme becoming the new primary enzyme for DHA phosphorylation. Growth experiments of ΔdhaKLM ΔglpK + pdhaKLM, in which the strain was grown beyond 72 h on 5 mM DHA, support this hypothesis, since growth of the strain on DHA increased significantly after 80 h, and actually surpassed ΔdhaKLM ΔglpK + pglpK after 96 h (data not shown). In-depth analysis into the enzymatic activity and kinetic constants of these enzymes toward DHA, as well as their expression levels, would enhance understanding on glycerol kinase primacy in Hfx. volcanii DHA metabolism.
DHA metabolism among the Halobacteria may extend beyond Hfx. volcanii and Hqm. walsbyi. Our BLASTp results for dhaK indicate that 29 other halobacterial species have a DHA kinase gene homologous to dhaK in Hfx. volcanii and Hqm. walsbyi. Since our data indicate that the dhaKLM genes in Hfx. volcanii are involved in DHA metabolism, the homologs of these genes in other halobacterial species likely also have this function, allowing those species to utilize DHA. Halobacterial species without DHA kinase might also be capable of utilizing DHA if they possess a glpK gene, since our results indicate that glycerol kinase also plays a role in DHA metabolism. BLASTp results for glpK indicate that 82 halobacterial species have homologs, and 51 of these species do not have dhaKLM homologs. We suspect that these species are also able to metabolize DHA. Eighteen halobacterial species are missing DHA and glycerol kinase genes, suggesting that they cannot metabolize DHA. However, only three of those genomes, Halovivax ruber, Natronobacterium gregoryi, and Natronomonas pharaonis, are not in draft form, leaving open the possibility for a near universal distribution of DHA metabolism in Halobacteria.
The broad taxonomic distribution of DHA and glycerol kinase genes among the Halobacteria suggests two interwoven hypotheses: (i) DHA is a common carbon source in hypersaline environments and (ii) DHA metabolism is widespread among the Halobacteria. A study by Elevi Bardavid and Oren (2008) detailed the conversion by the halophilic bacterium S. ruber of glycerol to DHA, which was then used as a growth substrate by Hqm. walsbyi. They speculated that DHA could be a common carbon source due to incomplete oxidation of glycerol, and from it being an intermediate of glycerol synthesis in Dunaliella. Our data demonstrating the extensive incidence of DHA and glycerol kinase genes provides support for their hypothesis that DHA is a common carbon source, and extends it to include that many if not most Halobacteria are capable of metabolizing it. However, future research on DHA production and turnover rates, and analysis on strains we predict to have DHA metabolism is necessary to elucidate the significance of this substrate to hypersaline ecosystems and Halobacteria.
Authors Contributions
R. Thane Papke, Andrea M. Makkay, and Matthew Ouellette conceived the researched and wrote the manuscript. Andrea M. Makkay and Matthew Ouellette performed the research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Thorsten Allers from the University of Nottingham for supplying us with Haloferax volcanii strains and plasmids, and Dr. Aharon Oren from the Hebrew University of Jerusalem for supplying us with dihydroxyacetone. This research was supported by the National Science Foundation (award numbers, DEB0919290 and DEB0830024) and NASA Astrobiology: Exobiology and Evolutionary Biology Program Element (Grant Number NNX12AD70G).
References
Allers, T., Ngo, H. P., Mevarech, M., and Lloyd, R. G. (2004). Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953. doi: 10.1128/AEM.70.2.943-953.2004
Anderson, I., Scheuner, C., Goker, M., Mavromatis, K., Hooper, S. D., Porat, I., et al. (2011). Novel insights into the diversity of catabolic metabolism from ten haloarchaeal genomes. PLoS ONE 6:e20237. doi: 10.1371/journal.pone.0020237
Bachler, C., Flukiger-Bruhwiler, K., Schneider, P., Bahler, P., and Erni, B. (2005). From ATP as substrate to ADP as coenzyme: functional evolution of the nucleotide binding subunit of dihydroxyacetone kinases. J. Biol. Chem. 280, 18321–18325. doi: 10.1074/jbc.M500279200
Ben-Amotz, A., and Avron, M. (1974). Isolation, characterization, and partial purification of a reduced nicotinamide adenine dinucleotide phosphate-dependent dihydroxyacetone reductase from the halophilic alga Dunaliella parva. Plant Physiol. 53, 628–631. doi: 10.1104/pp.53.4.628
Bitan-Banin, G., Ortenberg, R., and Mevarech, M. (2003). Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185, 772–778. doi: 10.1128/JB.185.3.772-778.2003
Blaby, I. K., Phillips, G., Blaby-Haas, C. E., Gulig, K. S., El Yacoubi, B., and de Crecy-Lagard, V. (2010). Towards a systems approach in the genetic analysis of archaea: accelerating mutant construction and phenotypic analysis in Haloferax volcanii. Archaea 2010, 426239. doi: 10.1155/2010/426239
Bolhuis, H., Palm, P., Wende, A., Falb, M., Rampp, M., Rodriguez-Valera, F., et al. (2006). The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 7:169. doi: 10.1186/1471-2164-7-169
Daniel, R., Stuertz, K., and Gottschalk, G. (1995). Biochemical and molecular characterization of the oxidative branch of glycerol utilization by Citrobacter freundii. J. Bacteriol. 177, 4392–4401.
Deppenmeier, U., Hoffmeister, M., and Prust, C. (2002). Biochemistry and biotechnological applications of Gluconobacter strains. Appl. Microbiol. Biotechnol. 60, 233–242. doi: 10.1007/s00253-002-1114-5
Dyall-Smith, M. (2009). Protocols for Haloarchaeal Genetics © Ver. 7.1. Available online at: http://www.haloarchaea.com/resources/halohandbook
Elevi Bardavid, R., Khristo, P., and Oren, A. (2008). Interrelationships between Dunaliella and halophilic prokaryotes in saltern crystallizer ponds. Extremophiles 12, 5–14. doi: 10.1007/s00792-006-0053-y
Elevi Bardavid, R., and Oren, A. (2008). Dihydroxyacetone metabolism in Salinibacter ruber and in Haloquadratum walsbyi. Extremophiles 12, 125–131. doi: 10.1007/s00792-007-0114-x
Erni, B., Siebold, C., Christen, S., Srinivas, A., Oberholzer, A., and Baumann, U. (2006). Small substrate, big surprise: fold, function and phylogeny of dihydroxyacetone kinases. Cell. Mol. Life Sci. 63, 890–900. doi: 10.1007/s00018-005-5518-0
Faurschou, A., Janjua, N. R., and Wulf, H. C. (2004). Sun protection effect of dihydroxyacetone. Arch. Dermatol. 140, 886–887. doi: 10.1001/archderm.140.7.886
Forage, R. G., and Lin, E. C. (1982). DHA system mediating aerobic and anaerobic dissimilation of glycerol in Klebsiella pneumoniae NCIB 418. J. Bacteriol. 151, 591–599.
Gutknecht, R., Beutler, R., Garcia-Alles, L. F., Baumann, U., and Erni, B. (2001). The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J. 20, 2480–2486. doi: 10.1093/emboj/20.10.2480
Hayashi, S. I., and Lin, E. C. (1967). Purification and properties of glycerol kinase from Escherichia coli. J. Biol. Chem. 242, 1030–1035.
Holzle, A., Fischer, S., Heyer, R., Schutz, S., Zacharias, M., Walther, P., et al. (2008). Maturation of the 5S rRNA 5' end is catalyzed in vitro by the endonuclease tRNase Z in the archaeon H. volcanii. RNA 14, 928–937. doi: 10.1261/rna.933208
Jin, R. Z., Forage, R. G., and Lin, E. C. (1982). Glycerol kinase as a substitute for dihydroxyacetone kinase in a mutant of Klebsiella pneumoniae. J. Bacteriol. 152, 1303–1307.
Mullakhanbhai, M. F., and Larsen, H. (1975). Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch. Microbiol. 104, 207–214. doi: 10.1007/BF00447326
Oren, A., and Shilo, M. (1982). Population dynamics of Dunaliella parva in the Dead Sea. Limnol. Oceanogr. 27, 201–211. doi: 10.4319/lo.1982.27.2.0201
Sher, J., Elevi, R., Mana, L., and Oren, A. (2004). Glycerol metabolism in the extremely halophilic bacterium Salinibacter ruber. FEMS Microbiol. Lett. 232, 211–215. doi: 10.1016/S0378-1097(04)00077-1
Sherwood, K. E., Cano, D. J., and Maupin-Furlow, J. A. (2009). Glycerol-mediated repression of glucose metabolism and glycerol kinase as the sole route of glycerol catabolism in the haloarchaeon Haloferax volcanii. J. Bacteriol. 191, 4307–4315. doi: 10.1128/JB.00131-09
Siebold, C., Arnold, I., Garcia-Alles, L. F., Baumann, U., and Erni, B. (2003). Crystal structure of the Citrobacter freundii dihydroxyacetone kinase reveals an eight-stranded alpha-helical barrel ATP-binding domain. J. Biol. Chem. 278, 48236–48244. doi: 10.1074/jbc.M305942200
Waites, M. J., and Quayle, J. R. (1981). The interrelation transketolase and dihydroxyacetone synthase activities in the methylotrophic yeast Candida boidinii. J. Gen. Microbiol. 124, 309–316. doi: 10.1099/00221287-124-2-309
Weinhouse, H., and Benziman, M. (1976). Phosphorylation of glycerol and dihydroxyacetone in Acetobacter xylinum and its possible regulatory role. J. Bacteriol. 127, 747–754.
Keywords: dihydroxyacetone metabolism, dihydroxyacetone kinase, glycerol kinase, archaea, Halobacteria, Haloarchaea
Citation: Ouellette M, Makkay AM and Papke RT (2013) Dihydroxyacetone metabolism in Haloferax volcanii. Front. Microbiol. 4:376. doi: 10.3389/fmicb.2013.00376
Received: 16 October 2013; Paper pending published: 01 November 2013;
Accepted: 21 November 2013; Published online: 16 December 2013.
Edited by:
Aharon Oren, The Hebrew University of Jerusalem, IsraelReviewed by:
Jerry Eichler, Ben Gurion University of the Negev, IsraelMaria-Jose Bonete, University of Alicante, Spain
Copyright © 2013 Ouellette, Makkay and Papke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: R. Thane Papke, Department Molecular and Cell Biology, University of Connecticut, 91 N. Eagleville Rd. Unit 3125, Storrs, CT 06268, USA e-mail: thane@uconn.edu
 Matthew Ouellette
Matthew Ouellette Andrea M. Makkay
Andrea M. Makkay Thane Papke
Thane Papke