- Ocean Sciences Department, University of California at Santa Cruz, Santa Cruz, CA, USA
The Vibrionaceae (Vibrio) are a ubiquitous group of metabolically flexible marine bacteria that play important roles in biogeochemical cycling in the ocean. Despite this versatility, little is known about Vibrio diversity and abundances in upwelling regions. The seasonal dynamics of Vibrio populations was examined by analysis of 16S rRNA genes in Monterey Bay (MB), California from April 2006–April 2008 at two long term monitoring stations, C1 and M2. Vibrio phylotypes within MB were diverse, with subpopulations clustering with several different cultured representatives including Allivibrio spp., Vibrio penaecida, and Vibrio splendidus as well as with many unidentified marine environmental bacterial 16S rRNA gene sequences. Total Vibrio population abundances, as well as abundances of a Vibrio sp. subpopulation (MBAY Vib7) and an Allivibrio sp. subpopulation (MBAY Vib4) were examined in the context of environmental parameters from mooring station and CTD cast data. Total Vibrio populations showed some seasonal variability but greater variability was observed within the two subpopulations. MBAY Vib4 was negatively associated with MB upwelling indices and positively correlated with oceanic season conditions, when upwelling winds relax and warmer surface waters are present in MB. MBAY Vib7 was also negatively associated with upwelling indices and represented a deeper Vibrio sp. population. Correlation patterns suggest that larger oceanographic conditions affect the dynamics of the populations in MB, rather than specific environmental factors. This study is the first to target and describe the diversity and dynamics of these natural populations in MB and demonstrates that these populations shift seasonally within the region.
Introduction
The Vibrionaceae (Vibrio) are a group of physiologically-flexible marine bacteria that are ubiquitous in ocean waters and have been identified in most marine ecosystems (Wietz et al., 2010). Vibrio have the distinctive ability to break down and utilize many carbon, nitrogen, and phosphorus substrates (McDougald and Kjelleberg, 2006; Thompson and Polz, 2006; Dryselius et al., 2008; Lai et al., 2009; Salter et al., 2009) and their production of the external enzymes chitinase and laminarinase provide access to abundant nutrients that are unavailable to other organisms (Svitil et al., 1997; Riemann et al., 2000; Ansede et al., 2001; Alderkamp et al., 2007; Murray et al., 2007). In addition to their diverse metabolic capabilities, Vibrio species have developed adaptive responses to starvation and environmental stress which include conversion to an ultramicrobacterial morphology (<0.4 μm diameter) (Denner et al., 2002) and retention of a high concentration of rRNA as a “stimulation ready” response that may contribute to this group's rapid growth potential after nutrient influxes (Eilers et al., 2000). Despite these capabilities, there have been few studies in oceanic upwelling regions, where metabolic flexibility, rapid nutrient response, and highly developed stress protection mechanisms should prove to be ecologically advantageous during changing environmental regimes.
Monterey Bay (MB) is an open embayment along the California coast that is distinguished by a near-shore deep-water canyon where periodic upwelling events sustain diverse sea life. Circulation within MB is variable and influenced by recently upwelled waters as well as offshore waters from the California and Davidson Currents that enter MB during relaxation events (Rosenfeld et al., 1994). Three hydrographic seasons have been defined within MB—an upwelling or “cold water phase” that usually occurs from mid-February through August, an oceanic period or “warm water phase” from mid-August through mid-October and a Davidson Current or “low thermal gradient” phase between mid-November and mid-February (Skogsberg and Phelps, 1946; Breaker and Broenkow, 1994). The periodic influx of nutrient-enriched waters into MB via upwelling results in a highly productive ecosystem presenting a unique environment for the study of Vibrio population dynamics.
Vibrio species are common isolates from MB waters but most studies have concentrated on pathogenic representatives (Kenyon et al., 1984; Kaysner et al., 1987; Miller et al., 2006). One phylogenetic screening study found that Vibrio-specific sequences made up 3.1% of the genetic information within a FOSMID library developed from a 100 m deep sample (Suzuki et al., 2004). Vibrio populations are estimated to average only about 1% of marine bacterial populations worldwide (Thompson and Polz, 2006) but it has also been suggested that differences in vertical distributions may be more significant than differences in geographic distribution for Vibrio populations, and thus, may make up a more significant portion of subsurface bacterial populations (Simidu and Tsukamoto, 1985).
This study was designed to determine the seasonal variability of Vibrio populations in MB by examining total Vibrio population dynamics as well as two subpopulations, a Vibrio sp. subpopulation (MBAY Vib7) and an Allivibrio sp. subpopulation (MBAY Vib4), and to analyze how population diversity changes with upwelling, season, and environmental factors. MB has a system of long term monitoring stations (Pennington and Chavez, 2000) and two of the stations within this network, C1 and M2 (Figure 1), were chosen as sample sites for this study. C1 is a coastal site that is influenced by aged upwelled waters and summer relaxation events. M2 is an outer Bay site that is influenced by upwelled waters that mix with California Current and Davidson Current waters. The examination of Vibrio population dynamics at these two different locations can provide insight into the effect of upwelling characteristics on Vibrio abundances.
Figure 1. Map of sample sites C1 and M2 in Monterey Bay, California. Each individual blue dot represents a sampling event and the black circle is the identified coordinate of the mooring station at M2.
Materials and Methods
Sample Collection
Seawater samples were collected from MB at stations C1 (36.797 N; 121.847 W; Figure 1) and M2 (36.697 N; 122.378 W; Figure 1) during cruises of the MB Time Series project undertaken by the Biological Oceanography Group (BOG) at the Monterey Bay Aquarium Research Institute (MBARI). Samples were collected from April 2006 through April 2008 on a periodic basis spanning 19 sampling dates. Samples were collected using a SeaBird 911 CTD rosette equipped with physical sensors described by Pennington and Chavez (2000). A total of 82 samples were collected at station C1 at 5 m (15 samples), 10 m (17 samples), 20 m (18 samples), 30 m (17 samples), and 200 m (15 samples). At station M2 a total of 86 samples were collected at 5 m (12 samples), 10 m (14 samples), 20 m (14 samples), 40 m (16 samples), 100 m (16 samples), and 200 m (16 samples). For each sample 1–2 liters of seawater were filtered using gentle peristaltic pumping through sequential in-line 25 mm diameter 10 μm pore-size PE filters (GE Osmonics, Minnetonka, MN, USA) and 25 mm diameter 0.2 μm pore-size Supor 200 membrane filters (Pall Corporation, Port Washington, NY, USA). The filters were transferred to 1.5 mL polypropylene microcentrifuge tubes containing 0.2 g of 0.1 mm and 0.5 mm diameter autoclaved glass beads (BioSpec Products, Bartlesville, OK, USA). Samples were flash frozen in liquid nitrogen before transfer to a −80°C freezer onshore for storage until nucleic acids were extracted.
Environmental Data
Upwelling indices (UI) of both monthly and daily averaged upwelling conditions were obtained for 36°N 122°W from the Pacific Fisheries Environmental Laboratory (PFEL, Pacific Grove) for determination of upwelling patterns (http://www.pfeg.noaa.gov/products/PFEL/modeled/indices/PFELindices.html). Units are given as m3 s−1 100 m of coastline-1 as the average amount of water upwelled through the bottom of the Ekman layer each second along each 100 m of coastline on a scale of about 200 miles. Surface data for nitrate, chlorophyll a, and temperature was obtained from the M2 mooring from MBARI LOBOviz (http://www.mbari.org/lobo/loboviz.htm). The BOG at MBARI provided environmental data from CTD measurements as well as surface (<10 m) phytoplankton concentrations, which were analyzed as described by Chavez et al. (1990). Specific phytoplankton groups (Synechococcus, total diatom, total dinoflagellate, and total phytoplankton) were chosen for analysis to assess the influence of different phytoplankton regimes on the variability of Vibrio groups in the water column.
DNA Extraction
DNA was extracted from the 0.2 μm pore-size filters using the Qiagen DNeasy Plant Kit (Hilden, Germany) as described by Moisander et al. (2008) with modifications to improve DNA recovery. Cells were lysed by a triplicate run of freeze-thaw steps in liquid nitrogen followed by a 65°C water bath. Cells were further disrupted by a 2-min agitation of bead beating (Mini bead beater 96, BioSpec Products, Bartlesville, OK, USA) and DNA yield was increased with the addition of 0.45 μ L of Proteinase K and incubation at 55°C for 1 h. AE buffer was used as the elution medium. Two duplicate elutions of 25 μ L each were combined for a final elution volume of 50 μ L. Extracts were stored at −20°C until use.
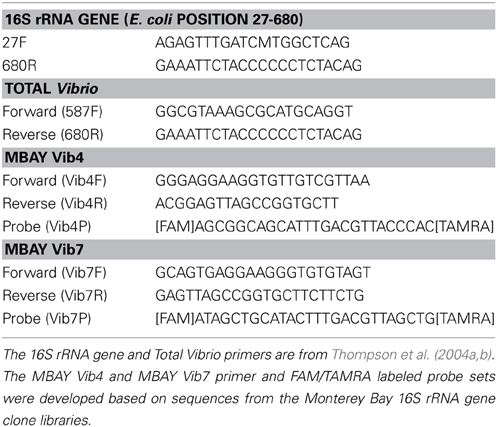
Amplification, Cloning and Sequencing of Vibrio-Specific 16S rRNA Gene
Samples were selected from 2007 to 2008 from 5 to 200 m depths at both station C1 and M2 to construct clone libraries of amplified 16S rRNA gene sequences from the MB. A total of 65 samples were processed for inclusion in the libraries. The PCR amplification utilized a universal forward primer (27F, Table 1) and a Vibrio specific reverse primer (680R, Table 1) and followed the two-phase PCR amplification technique outlined by Thompson et al. (2004b). Samples were amplified using Invitrogen Taq polymerase (Carlsbad, California), on a BioRad thermal cycler (Hercules, California). The PCR products were gel purified and cloned into pGEM-T vectors (Promega, Madison WI) using manufacturer's guidelines. Ten to fifteen clones were chosen from each sample, for a total of 911 clones, and prepared for sequencing with the Montage Plasmid Miniprep kit (Millipore, Billerica, MA) according to manufacturer's protocols. Cloned inserts were sequenced at the UC Berkeley DNA Sequencing Facility using T7 primers and analyzed on an Applied Biosystems 3730xl DNA Analyzer. Sequences were aligned and compared to published sequences using the Ribosomal Database Project (RDP) on-line interface (Cole et al., 2007, 2009) and were quality checked for chimeras using the RDP Chimera check program and Bellerophon (Huber et al., 2004). Phylogenetic analysis of 827 sequences was conducted in ARB (Ludwig et al., 2004) and neighbor-joining phylogenetic trees were constructed using the Jukes-Cantor correction. The distance matrix derived from the neighbor joining analysis was used in DOTUR for assignment of operational taxonomic units (OTUs) (Schloss and Handelsman, 2005). All sequences were submitted to the National Center for Biotechnology Information (NCBI) GenBank database as accession numbers KF941543–KF942369.
Quantitative PCR (qPCR)
Total Vibrio abundances were quantified using a SYBR Green qPCR method utilizing the Vibrio specific primers 567F and 680R (Table 1) from Thompson et al. (2004b) to ensure a broad spectrum of Vibrio species were included in the analysis. DNA template (2 μ l) was added to 12.5 μ l Applied Biosystems SYBR® Green PCR Master Mix, 8.25 μ l of 5 kD filtered water, 0.25 μ l of bovine serum albumin (BSA) and 1 μ l of each primer for a total volume of 25 μ l. The cycling conditions were 50°C for 2 min, 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 64°C for 1 min. Each run was followed by a dissociation step (95°C for 15 s and 60°C for 1 min 95°C for 15 s and 60°C for 15 s) to determine a melt curve for analysis of specificity.
Two qPCR assays utilizing TaqMan® chemistry were designed to target two Vibrio subpopulations within MB based on sequences of interest identified from the MB 16S rRNA clone library. The MBAY Vib4 group was closely associated with Alllivibrio sp. and the MBAY Vib7 group was associated with Vibrio penaecida. Primers and probes were designed with Primer Express (Table 1) (ABI, Foster City, CA) and synthesized by Sigma-Genosys (Woodlands, TX). The reactions for the sub-population assays contained 2 μ l of sample DNA, 12.5 μ l Applied Biosystems TaqMan® Universal PCR Master Mix, 8 μ l of 5 kD filtered water, 1 μ l each of the forward and reverse primer and 0.5 μ l of the probe. Samples were assessed using a TaqMan® qPCR method with cycling conditions of 50°C for 2 min, 95°C for 10 min followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. The qPCR primer sets were used to analyze the full range of samples at both stations.
Both qPCR assays compared CT values to standard curves (equivalent to 101–108 gene copies per reaction) derived from group specific environmental isolate clones from the MB clone library linearized with Nad1. An internal control at a concentration of 104 gene copies of linearized standard assessed the runs for inhibition. If detected, DNA samples were diluted 1:10 and amplification was repeated. All qPCR runs were performed on a 7500 Applied Biosystems Real-Time PCR instrument. Final concentrations are reported as gene copies l−1 of seawater sampled.
Statistical Analysis
Statistical analysis was performed in JMP version 9 (SAS Institute, North Carolina). Data was natural log transformed [ln (x + 1)] to scale variables for graphical interpretation and adjust for normalcy. A Principal Components Analysis (PCA) bi-plot was utilized to visualize relationships between Vibrio populations and physical and biological variables and Spearman's rank analysis (ρs) was used to define statistically significant relationships.
Results
Environmental Conditions in MB
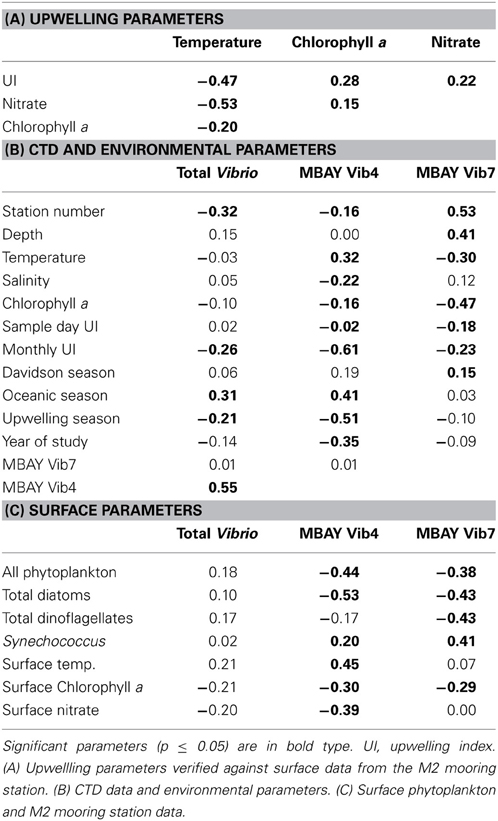
Water temperatures from the sample sites ranged from 7.7 to 15.4°C (Table 2). Salinity ranged from 32.7 to 34.4 ppt and chlorophyll a values ranged from 0 to 18.45 μ g liter−1 (Table 2). The Monthly Upwelling Index (MUI) ranged from 46 to 237 m3 s−1 100 m of coastline−1. The daily Upwelling Index was analyzed against surface conditions at M2 (Table 3A). Results were consistent with expected trends for upwelling events with a negative correlation to temperature (ρs = −0.47, p = 0.002) and a positive correlation to both nitrate (ρs = 0.22, p = 0.035) and chlorophyll a (ρs = 0.28, p = 0.009). Principal Component Analysis associated total diatoms, total dinoflagellates and all phytoplankton with the upwelling season and the monthly UI. Synechococcus was more closely associated with station M2 (Figure 2).
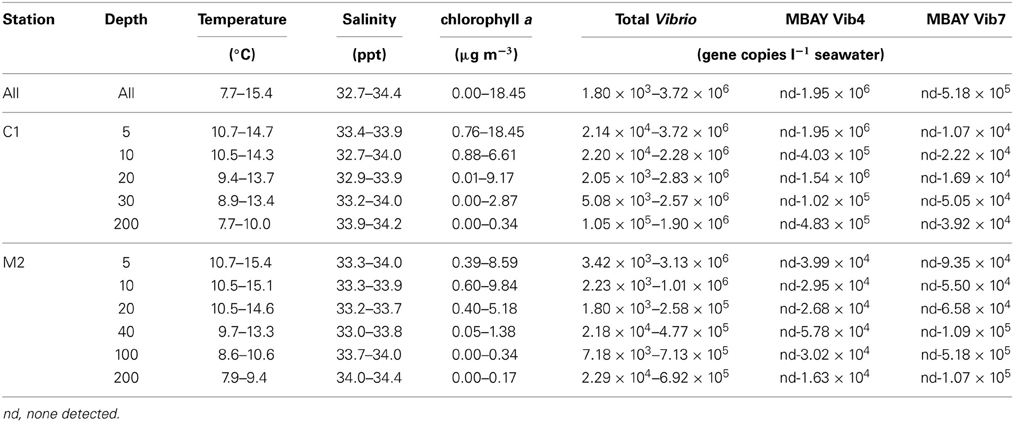
Table 2. CTD measurement and Vibrio concentration ranges over the study period for all stations and depths.
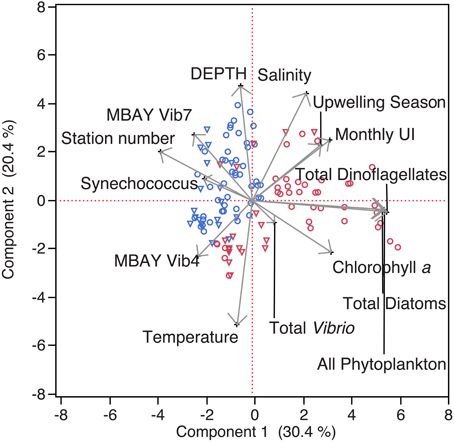
Figure 2. Principal Component Analysis of physical and biological parameters. MBAY Vib4 (O) MBAY Vib7 (Δ) from stations C1 (red) and M2 (blue).
16S rRNA Gene Phylogenetic Analysis
Multiple distinct 16S rRNA-based phylotypes were recovered from the MB clone libraries (Figure 3). The primer sets utilized to develop the MB clone libraries targeted Vibrio sp. strains but 67 sequences within the library clustered with the closely related Photobacterium sp. There were also a significant number of sequences that clustered with unidentified Vibrio strains and uncultured marine bacteria. Vibrio splendidus was defined in several distinct clades within the phlogenetic tree. Other MB sequences clustered with V. pomeroyi and V. lentus; V. pectenicida and V. rumoiensis; and V. aesturianus. One clade included sequences that clustered with V. agarivorans, V. hispanicus, V. haliticoli, V. ezurae, V. fortis, V. proteolyticus, V. sinaloensis, V. nigripulchritudo, V. harveyi, V. natriegens, and V. alginolyticus (Assorted Vibrio in Figure 3). Quantitative PCR (qPCR) assays were developed for two specific phylotypes recovered from the MB clone libraries. The MBAY Vib4 group (Figure 3B) clustered with Allivibrio sp., which includes A. salmonicida, A. fischeri, A. wodanis, and A. logei. MBAY Vib4 included sequences obtained from samples collected from both stations C1 and M2. Within the clone library the MBAY Vib7 group included sequences that were only obtained at station M2 and clustered with Vibrio penaecida (Figure 3C).
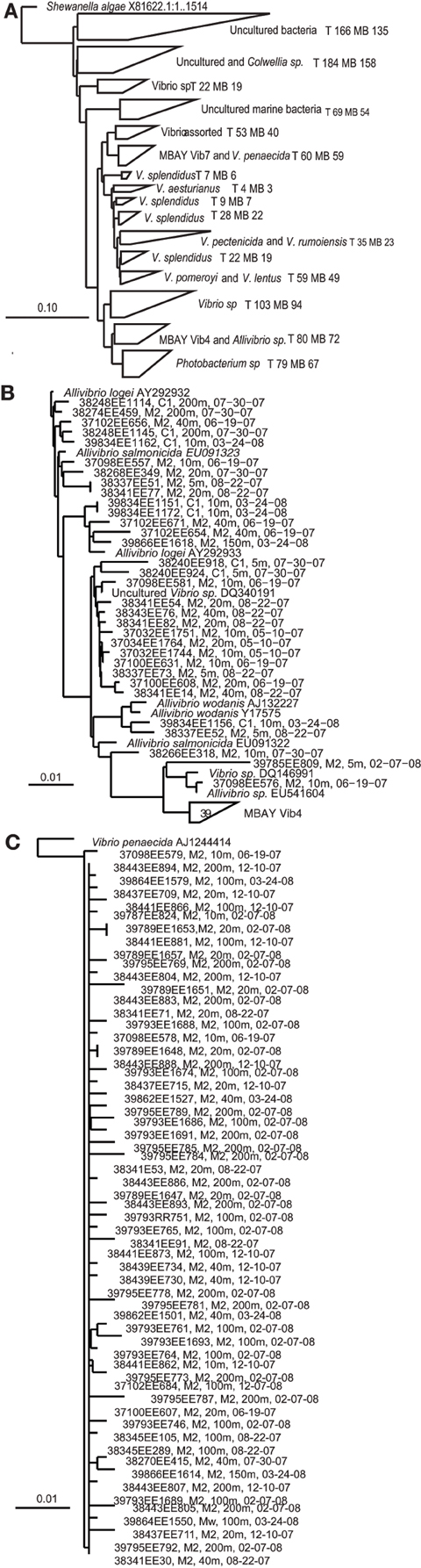
Figure 3. Neighbor-joining phylogenetic trees of Vibro sequences from Monterey Bay at stations C1 and M2. Tree constructed in ARB from partial 16S rRNA sequences (E. coli 27-680 bp) using ARB neighbor-joining distance matrix method with a Jukes-Cantor correction. T, total number of sequences in clade, MB, MB sequences in clade. Sample number, station, depth, and date are listed for Monterey Bay sequences. (A) Major groups of environmental Vibrio sequences recovered from the MB. Shewanella algae included as the tree root. (B) Allivibirio and MBAY Vib4 group sequences, and (C) Vibrio penaecida and MBAY Vib7 sequences. Scales represent 10% or 0.1% nucleotide substitutions.
Rarefaction curves were determined based on the neighbor-joining distance matrix of sequences within the MB clone libraries. Of the 827 sequences included in the total sequences analysis, 158 OTUs were defined at 97% identity, 103 OTUs at 95% identity, 61 OTUs at 93% identity, and 30 OTUs at 90% identity. Only at 93 and 90% identity did the rarefaction curves begin to reach an asymptote. Analysis revealed that there was no significant difference in species richness between station C1 and M2 but that both sampling efforts did not attain full coverage to estimate total population OTUs. Rarefaction analysis of the 327 sequences from station C1 defined 97 OTUs at 97% identity, 69 OTUs at 95% identity, 45 OTUs at 93% identity, and 23 OTUs at 90% identity. Rarefaction analysis of the 500 sequences from station M2 defined 98 OTUs at 97% identity, 66 OTUs at 95% identity, 41 OTUs at 93% identity, and 21 OTUs at 90% identity.
Quantification of Vibrio Population Abundances
There were observed differences in the Vibrio populations at both of the stations, with the MBAY Vib4 group having higher peak abundances at station C1 (Figure 4). The MBAY Vib4 group ranged in concentration from undetectable levels at all depths and stations up to 1.95 × 106 gene copies liter−1 seawater at 5 m at station C1 (Table 2). MBAY Vib4 was positively correlated to temperature (ρs = 0.32, p = 0.002), oceanic season (ρs = 0.41, p = 0.002), Synechococcus (ρs = 0.20, p = 0.02), and total Vibrio abundance (ρs = 0.55, p < 0.001) (Table 3). This group was negatively correlated to salinity (ρs = −0.22, p = 0.02), chlorophyll a (ρs = −0.16, p = 0.035), monthly UI (ρs = −0.61, p < 0.001), surface phytoplankton (ρs = −0.44, p < 0.001), and total diatoms (ρs = −0.55, p < 0.001) (Table 3).
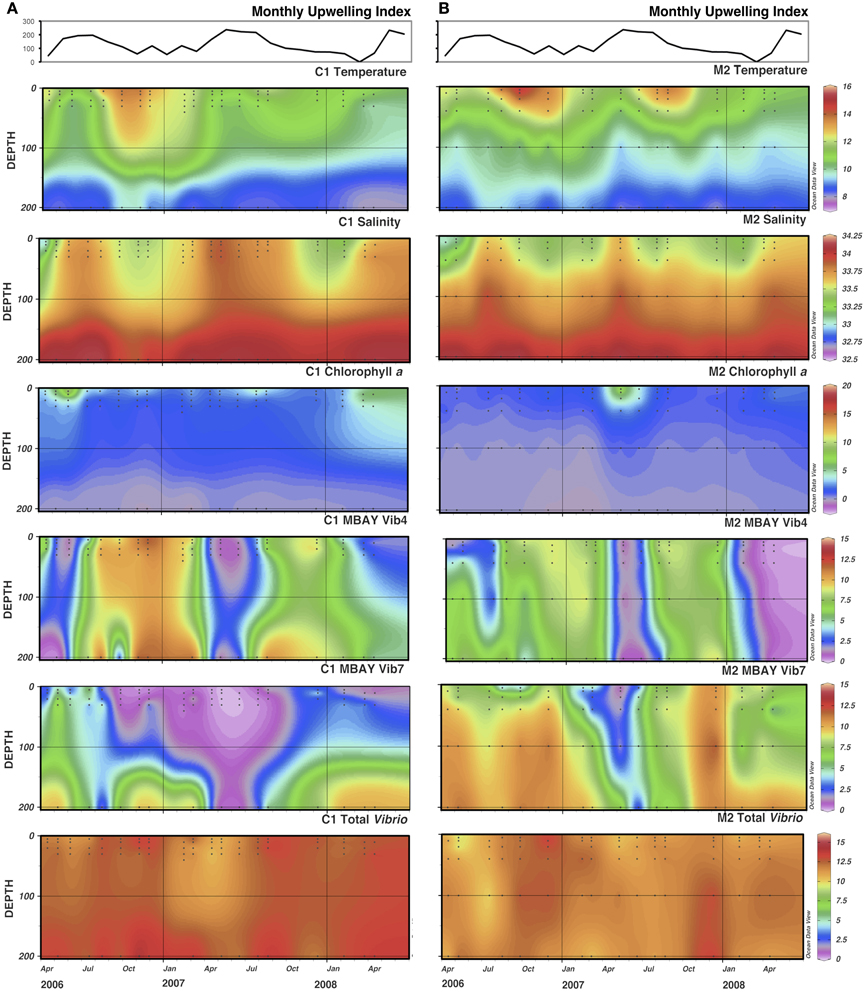
Figure 4. Monthly Upwelling Index (m3 s-1 100 m of coastline-1), temperature (°C), salinity (ppt), chlorophyll a (μg m−3) and total Vibrio, MBAY Vib4 and MBAY Vib7 concentrations (natural log of gene copies liter−1) for stations C1 (A) and M2 (B). Black dots represent discrete samples within the CTD cast.
The MBAY Vib7 population was not as abundant as MBAY Vib4 (Figure 4), with concentrations ranging from undetectable to 5.18 × 105 gene copies liter−1 seawater (Table 2). MBAY Vib7 was positively correlated with depth (ρs = 41, p < 0.001) (Table 3), with the highest detected concentration at 100 m at station M2 (Table 2). MBAY Vib7 populations were positively associated with station M2 (ρs = 0.53, p < 0.001), surface Synechococcus populations (ρs = 0.41, p < 0.001), and the Davidson season (ρs = 0.15, p = 0.0486) (Table 3). Temperature (ρs = −0.30, p = 0.0005), chlorophyll a (ρs = −0.47, p < 0.001) and monthly UI (ρs = −0.23, p = 0.012) were all negatively associated with MBAY Vib7 (Table 3). MBAY Vib7 was identified in samples from both station C1 and M2 but at lower concentrations and at deeper depths at station C1.
Total Vibrio populations showed some variability in abundance but were consistently present throughout the study at all depths and stations (Figure 4). Total Vibrio population abundances ranged from 1.80 × 103 to 3.72 × 106 gene copies liter−1 seawater (Table 2). The abundance of total Vibrio populations was positively correlated to MBAY Vib4 (ρs = 0.55, p < 0.001), and the oceanic season (ρs = 0.31, p < 0.001) and negatively correlated to station number (ρs = −0.32, p < 0.001) the monthly UI (ρs = −0.26, p = 0.0039) and upwelling season (ρs = −0.21, p = 0.0067) (Table 3). No other parameters examined in this study showed significant correlation with the abundance of the total Vibrio population.
Discussion
Genetic diversity within Vibrio populations can be up to 7% when examining the entire 16S rRNA gene (Dorsch et al., 1992; Kitatsukamoto et al., 1993). This variability is mostly located within 35 defined regions, 28 of which are located between the primers used to construct the clone library for this study (Wiik et al., 1995; Jensen et al., 2009). It is thus not surprising to have identified a significant number of OTUs and to observe deep branching within the phylogenetic tree of the MB clone library. Intraspecies sequence variability can also be high within Vibrio species (Jensen et al., 2009) and the identification of Vibrio splendidus within different branches of the tree is supported by observations of up to 2% difference within the 16S rRNA gene of this species (Jensen et al., 2009; Le Roux et al., 2009). Defining relationships within Vibrio populations is even more complex as their genomes contain multiple copies of the rRNA operon (Heidelberg et al., 2000). Thirteen copies have been identified in V. natriegens and 12 copies in both Allivibrio fischeri and A. salmonicida (Lee et al., 2009). Intragenomic heterogeneity within these copies may be high, as seen in V. parahemolyticus (Harth et al., 2007), or non-existent as is noted in other Vibrio species (Coenye and Vandamme, 2003). Since this study was not designed to differentiate between heterogeneous intragenomic copies and individuals, our results may overestimate diversity within the MB clone library. With that being said, sequences did cluster with over 20 known Vibrio species suggesting that MB does in fact contain diverse Vibrio populations.
Multiple copies of the rRNA operon may also affect the qPCR estimates of abundance presented in this study, but is not expected to significantly change the interpretations of our findings. Even an estimate of 20 gene copies per cell results in estimates of abundances that fall within the range of concentrations observed in other coastal waters (Thompson et al., 2004a), and are higher than observations from samples in similar temperature ranges (Randa et al., 2004; Eiler et al., 2006). Temperature is often cited as a significant driver in Vibrio population abundance (Heidelberg et al., 2002; Maeda et al., 2003; Eiler et al., 2006, 2007; Hsieh et al., 2008), and while temperature variability was correlated to the two subpopulations examined in this study, it was the upwelling index (MBAY Vib4) and depth and station (MBAY Vib7) that were more significant factors in defining subpopulation abundance.
MBAY Vib4 and MBAY Vib7 are separate subpopulations of the Vibrio community with distinct niches in the MB. The MBAY Vib4 is defined by close association with Allivibrio sp. and represents an oceanic season subpopulation. This season is characterized by a lack of upwelling, lower sea surface salinities, warmer sea surface temperatures and waters that are influenced by wind relaxation events and the slow flowing California Current (Rosenfeld et al., 1994). MBAY Vib7 represents a deeper water subpopulation of Vibrio sp. with a greater association with the offshore waters at station M2. This station is influenced by upwelled waters mixed with California Current waters, the California Undercurrent and the Davidson Current-a northward flowing current that develops in winter months (Tisch et al., 1992; Breaker and Broenkow, 1994; Rosenfeld et al., 1994). The California Undercurrent usually flows below 100 m along the California shelf but often nears the surface during the Davidson season (Tisch et al., 1992; Pierce et al., 2000; Tseng et al., 2005). The largest hydrographic changes around MB occur during transitions between upwelling regimes (i.e., during oceanic phases) and in winter when the horizontal and vertical thermal gradients are reduced due to the northward flow of the Davidson Current (Bac et al., 2003; Storlazzi et al., 2003; Warn-Varnas et al., 2007). Dynamics of the MB Vibrio populations examined in this study seem to reflect these hydrographic processes, and may be highly influenced by them-MBAY Vib4 during the oceanic phases and MBAY Vib7 during the winter phases.
In 2006, the MB had experienced 4 years of anomalous oceanographic conditions. Delayed and unusually shallow upwelling affected the food web in MB up through higher trophic levels (Goericke et al., 2007). Changes included a shift in dominant toxin-producing algal species from diatoms to dinoflagellates, poor recruitment of krill and low zooplankton biomass as well as seabird reproductive failure (Peterson et al., 2006; Goericke et al., 2007; Jester et al., 2009). MBAY Vib4 was positively correlated to this year possibly due to delayed upwelling, reduced chlorophyll a concentrations and warmer surface temperatures (Table 3). Increased abundances of Vibrio populations could also have been influenced by decreased competition from phytoplankton for resources or reduced top down control from zooplankton. Following the 2002–2006 lull events, unusually strong upwelling was observed in 2007 (Kaplan et al., 2009), and in 2008 a strong development of upwelling was observed at the beginning of the season (Figure 3). MBAY Vib4 populations showed significantly reduced abundance during those upwelling seasons. MBAY Vib7 and the total Vibrio populations did not seem to be strongly affected by these events.
Overall, the Vibrio populations examined in MB seemed to be influenced by larger scale upwelling events and shifts in currents and oceanographic seasons rather than individual environmental factors. In the future, more extensive analysis of nutrient availability and oceanographic parameters (i.e., flow velocities and wind patterns) may provide better insight into how upwelling characteristics and water mass flows play a role in Vibrio population dynamics. Since Vibrio populations might have significant influence over nutrient availability through recycling chitin and other bound nutrient resources, understanding the role of these bacterial populations in both surface and deeper waters can better our understanding of the productivity potential within upwelling regions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the Biological Oceanography Group at the Monterey Bay Aquarium Research Institute, especially Francisco Chavez and Tim Pennington for providing us with the phytoplankton data and ship time. The authors would also like to thank Kendra Turk-Kubo, Fitnat Yildiz and Shellie Bench for helpful discussions. Partial funding was provided by a Gordon and Betty Moore Foundation Marine Investigator award (Jonathan P. Zehr).
References
Alderkamp, A. C., Van Rijssel, M., and Bolhuis, H. (2007). Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol. Ecol. 59, 108–117. doi: 10.1111/j.1574-6941.2006.00219.x
Ansede, J., Friedman, R., and Yoch, D. (2001). Phylogenetic analysis of culturable dimethyl sulfide-producing bacteria from a Spartina-dominated salt marsh and estuarine water. Appl. Environ. Microbiol. 67, 1210–1217. doi: 10.1128/AEM.67.3.1210-1217.2001
Bac, M. G., Buck, K. R., Chavez, F. P., and Brassell, S. C. (2003). Seasonal variation in alkenones, bulk suspended POM, plankton and temperature in Monterey Bay, California: implications for carbon cycling and climate assessment. Org. Geochem. 34, 837–855. doi: 10.1016/S0146-6380(02)00248-6
Breaker, L. C., and Broenkow, W. W. (1994). “The circulation of Montery Bay and related processes,” in Oceanography and Marine Biology, Vol. 32, eds A. D. Ansell, R. N. Gibson, M. Barnes (Aberdeen: Aberdeen University Press), 1–64.
Chavez, F. P., Buck, K. R., and Barber, R. T. (1990). Phytoplankton taxa in relation to primary production in the equatorial Pacific. Deep Sea Res. Part A. Oceanographic Res. Papers 37, 1733–1752. doi: 10.1016/0198-0149(90)90074-6
Coenye, T., and Vandamme, P. (2003). Intragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomes. FEMS Microbiol. Lett. 228, 45–49. doi: 10.1016/S0378-1097(03)00717-1
Cole, J. R., Chai, B., Farris, R. J., Wang, Q., Kulam-Syed-Mohideen, A. S., Mcgarrell, D. M., et al. (2007). The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35, D169–D172. doi: 10.1093/nar/gkl889
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145. doi: 10.1093/nar/gkn879
Denner, E. B. M., Vybiral, D., Fischer, U. R., Velimirov, B., and Busse, H. J. (2002). Vibrio calviensis sp nov., a halophilic, facultatively oligotrophic 0 center dot 2 mu m-filterable marine bacterium. Int. J. Syst. Evol. Microbiol. 52, 549–553. doi: 10.1099/ijs.0.01859-0
Dorsch, M., Lane, D., and Stackebrandt, E. (1992). Towards a phylogeny of the genus Vibrio based on 16S robosomal-RNA sequences. Int. J. Syst. Bacteriol. 42, 58–63. doi: 10.1099/00207713-42-1-58
Dryselius, R., Izutsu, K., Honda, T., and Iida, T. (2008). Differential replication dynamics for large and small Vibrio chromosomes affect gene dosage, expression and location. BMC Genomics 9:559. doi: 10.1186/1471-2164-9-559
Eiler, A., Gonzalez-Rey, C., Allen, S., and Bertilsson, S. (2007). Growth response of Vibrio cholerae and other Vibrio spp. to cyanobacterial dissolved organic matter and temperature in brackish water. FEMS Microbiol. Ecol. 60, 411–418. doi: 10.1111/j.1574-6941.2007.00303.x
Eiler, A., Johansson, M., and Bertilsson, S. (2006). Environmental influences on Vibrio populations in northern temperate and boreal coastal waters (Baltic and Skagerrak Seas). Appl. Environ. Microbiol. 72, 6004–6011. doi: 10.1128/AEM.00917-06
Eilers, H., Pernthaler, J., and Amann, R. (2000). Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66, 4634–4640. doi: 10.1128/AEM.66.11.4634-4640.2000
Goericke, R., Venrick, E., Koslow, T., Sydeman, W. J., Schwing, F. B., Bograd, S. J., et al. (2007). The State of the California Current, 2006-2007: regional and local processes dominate. Cal. Coop. Ocean. Fish. Reports 48, 33. Available online at: http://calcofi.org/publications/ccreports/25-vol48-2007.html
Harth, E., Romero, J., Torres, R., and Espejo, R. T. (2007). Intragenomic heterogeneity and intergenomic recombination among Vibrio parahaemolyticus 16S rRNA genes. Microbiology 153, 2640–2647. doi: 10.1099/mic.0.2007/009175-0
Heidelberg, J., Heidelberg, K., and Colwell, R. (2002). Seasonality of Chesapeake Bay bacterioplankton species. Appl. Environ. Microbiol. 68, 5488–5497. doi: 10.1128/AEM.68.11.5488-5497.2002
Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., et al. (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483. doi: 10.1038/35020000
Hsieh, J. L., Fries, J. S., and Noble, R. T. (2008). Dynamics and predictive modelling of Vibrio spp. in the Neuse river estuary, north carolina, USA. Environ. Microbiol. 10, 57–64. doi: 10.1111/j.1462-2920.2007.01429.x
Huber, T., Faulkner, G., and Hugenholtz, P. (2004). Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319. doi: 10.1093/bioinformatics/bth226
Jensen, S., Frost, P., and Torsvik, V. L. (2009). The nonrandom microheterogeneity of 16S rRNA genes in Vibrio splendidus may reflect adaptation to versatile lifestyles. FEMS Microbiol. Lett. 294, 207–215. doi: 10.1111/j.1574-6968.2009.01567.x
Jester, R., Lefebvre, K., Langlois, G., Vigilant, V., Baugh, K., and Silver, M. W. (2009). A shift in the dominant toxin-producing algal species in central California alters phycotoxins in food webs. Harmful Algae 8, 291–298. doi: 10.1016/j.hal.2008.07.001
Kaplan, D. M., Halle, C., Paduan, J., and Largier, J. L. (2009). Surface currents during anomalous upwelling seasons off central California. J. Geophys. Res. Oceans 114, 17. doi: 10.1029/2009JC005382
Kaysner, C. A., Abeyta, C. Jr., Wekell, M., Depaola, A. Jr., Stott, R., and Leitch, J. (1987). Incidence of Vibrio cholerae from estuaries of the United States West Coast. Appl. Environ. Microbiol. 53, 1344–1348.
Kenyon, J. E., Piexoto, D. R., Austin, B., and Gillies, D. C. (1984). Seasonal variations of Vibrio cholerae (non-O1) isolated from California coastal waters. Appl. Environ. Microbiol. 47, 1243–1245.
Kitatsukamoto, K., Oyaizu, H., Nanba, K., and Simidu, U. (1993). Phylogenetic relationships of marine bacteria, mainly members of the family Vibrionaceae, determined on the basis of 16S ribosomal-RNA sequences. Int. J. Syst. Bacteriol. 43, 8–19. doi: 10.1099/00207713-43-1-8
Lai, C. J., Chen, S. Y., Lin, I. H., Chang, C. H., and Wong, H. C. (2009). Change of protein profiles in the induction of the viable but nonculturable state of Vibrio parahaemolyticus. Int. J. Food Microbiol. 135, 118–124. doi: 10.1016/j.ijfoodmicro.2009.08.023
Lee, Z. M. P., Bussema, C. 3rd., and Schmidt, T. M. (2009). rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 37, D489–D493. doi: 10.1093/nar/gkn689
Le Roux, F., Zouine, M., Chakroun, N., Binesse, J., Saulnier, D., Bouchier, C., et al. (2009). Genome sequence of Vibrio splendidus: an abundant planctonic marine species with a large genotypic diversity. Environ. Microbiol. 11, 1959–1970. doi: 10.1111/j.1462-2920.2009.01918.x
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar., et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Maeda, T., Matsuo, Y., Furushita, M., and Shiba, T. (2003). Seasonal dynamics in a coastal Vibrio community examined by a rapid clustering method based on 16S rDNA. Fisheries Sci. 69, 385–394. doi: 10.1046/j.1444-2906.2003.00633.x
McDougald, D., and Kjelleberg, S. (2006). “Adaptive responses of Vibrios,” in Biology of Vibrios, eds F. L. Thompson, B. Austin, and J. Swings (Washington, DC: ASM Press).
Miller, W., Miller, M., Gardner, I., Atwill, E., Byrne, B., Jang, S., et al. (2006). Salmonella spp., Vibrio spp., Clostridium perfringens, and Plesiomonas shigelloides in marine and freshwater invertebrates from coastal California ecosystems. Microb. Ecol. 52, 198–206. doi: 10.1007/s00248-006-9080-6
Moisander, P. H., Beinart, R. A., Voss, M., and Zehr, J. P. (2008). Diversity and abundance of diazotrophic microorganisms in the South China Sea during intermonsoon. ISME J. 2, 954–967. doi: 10.1038/ismej.2008.51
Murray, A. E., Arnosti, C., De La Rocha, C. L., Grossart, H. P., and Passow, U. (2007). Microbial dynamics in autotrophic and heterotrophic seawater mesocosms. II. Bacterioplankton community structure and hydrolytic enzyme activities. Aquat. Microb. Ecol. 49, 123–141. doi: 10.3354/ame01139
Pennington, J. T., and Chavez, F. P. (2000). Seasonal fluctuations of temperature, salinity, nitrate, chlorophyll and primary production at station H3/M1 over 1989-1996 in Monterey Bay, California. Deep Sea Res. Pt. II 47, 947–973. doi: 10.1016/S0967-0645(99)00132-0
Peterson, W., Emmett, R., Goericke, R., Venrick, E., Mantyla, A., Bograd, S. J., et al. (2006). The state of the California current, 2005-2006: warm in the north, cool in the south. Cal. Coop. Ocean. Fish. Reports 47, 30. Available online at: http://calcofi.org/publications/ccreports/14-vol47-2006.html
Pierce, S. D., Smith, R. L., Kosro, P. M., Barth, J. A., and Wilson, C. D. (2000). Continuity of the poleward undercurrent along the eastern boundary of the mid-latitude north Pacific. Deep. Sea Res. Pt. II 47, 811–829. doi: 10.1016/S0967-0645(99)00128-9
Randa, M. A., Polz, M. F., and Lim, E. (2004). Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl. Environ. Microbiol. 70, 5469–5476. doi: 10.1128/AEM.70.9.5469-5476.2004
Riemann, L., Steward, G. F., and Azam, F. (2000). Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66, 578–587. doi: 10.1128/AEM.66.2.578-587.2000
Rosenfeld, L. K., Schwing, F. B., Garfield, N., and Tracy, D. E. (1994). Bifrucated flow from an upwelling center- a cold water source for Monterey Bay. Cont. Shelf Res. 14, 931–964. doi: 10.1016/0278-4343(94)90058-2
Salter, I., Zubkov, M. V., Warwick, P. E., and Burkill, P. H. (2009). Marine bacterioplankton can increase evaporation and gas transfer by metabolizing insoluble surfactants from the air-seawater interface. FEMS Microbiol. Lett. 294, 225–231. doi: 10.1111/j.1574-6968.2009.01572.x
Schloss, P. D., and Handelsman, J. (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005
Simidu, U., and Tsukamoto, K. (1985). Habitat segregation and isochemical activites of marine members ot the family Vibrionaceae. Appl. Environ. Microbiol. 50, 781–790.
Skogsberg, T., and Phelps, A. (1946). Hydrography of Monterey Bay, California. Thermal conditions, Part II (1934–1937). Proc. Am. Philol. Soc. 90, 350–386.
Storlazzi, C. D., Mcmanus, M. A., and Figurski, J. D. (2003). Long-term, high-frequency current and temperature measurements along central California: insights into upwelling/relaxation and internal waves on the inner shelf. Cont. Shelf Res. 23, 901–918. doi: 10.1016/S0278-4343(03)00045-1
Suzuki, M. T., Preston, C. M., Beja, O., De La Torre, J. R., Steward, G. F., and Delong, E. F. (2004). Phylogenetic screening of ribosomal RNA gene-containing clones in bacterial artificial chromosome (BAC) libraries from different depths in Monterey Bay. Microb. Ecol. 48, 473–488. doi: 10.1007/s00248-004-0213-5
Svitil, A. L., Chadhain, S. M. N., Moore, J. A., and Kirchman, D. L. (1997). Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl. Environ. Microbiol. 63, 408–413.
Thompson, F. L., Iida, T., and Swings, J. (2004a). Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431. doi: 10.1128/MMBR.68.3.403-431.2004
Thompson, J., Randa, M., Marcelino, L., Tomita-Mitchell, A., Lim, E., and Polz, M. (2004b). Diversity and dynamics of a north Atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70, 4103–4110. doi: 10.1128/AEM.70.7.4103-4110.2004
Thompson, J. R., and Polz, M. F. (2006). “Dynamics of Vibrio populations and their role in environmental nutrient cycling,” in The Biology of Vibrios, eds F. Thompson, B. Austin, and J. Swings (Washington, DC: ASM Press), 190–203.
Tisch, T. D., Ramp, S. R., and Collins, C. A. (1992). Observations of the geostrophic current and water mass characteristics off Point Sur, California, from May 1988 through November 1989. J. Geophys. Res. Oceans 97, 12535–12555. doi: 10.1029/92JC01094
Tseng, Y. H., Dietrich, D. E., and Ferziger, J. H. (2005). Regional circulation of the Monterey Bay region: hydrostatic versus nonhydrostatic modeling. J. Geophys. Res. Oceans 110:C09015. doi: 10.1029/2003JC002153
Warn-Varnas, A., Gangopadhyay, A., and Hawkins, J. A. (2007). Water masses in the Monterey Bay during the summer of 2000. Cont. Shelf Res. 27, 1379–1398. doi: 10.1016/j.csr.2007.01.004
Wietz, M., Gram, L., Jorgensen, B., and Schramm, A. (2010). Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat. Microb. Ecol. 61, 179–189. doi: 10.3354/ame01443
Keywords: Vibrio, upwelling, Monterey Bay, seasonal variability, 16S rRNA gene diversity
Citation: Mansergh S and Zehr JP (2014) Vibrio diversity and dynamics in the Monterey Bay upwelling region. Front. Microbiol. 5:48. doi: 10.3389/fmicb.2014.00048
Received: 01 October 2013; Paper pending published: 26 October 2013;
Accepted: 22 January 2014; Published online: 12 February 2014.
Edited by:
Daniela Ceccarelli, University of Maryland, USAReviewed by:
Luigi Vezzulli, University of Genoa, ItalyRodrigo Costa, Centre of Marine Sciences, Portugal
Copyright © 2014 Mansergh and Zehr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan P. Zehr, Ocean Sciences Department, University of California Santa Cruz, 1156 High Street, Santa Cruz, CA 95064, USA e-mail: zehrj@ucsc.edu
 Sarah Mansergh
Sarah Mansergh Jonathan P. Zehr
Jonathan P. Zehr