- 1Departamento de Biología Celular, Genética y Fisiología, Facultad de Ciencias, Universidad de Malaga, Málaga, Spain
- 2Laboratoire de Microbiologie des Environnements Extrêmes, UMR 6197, Institut Français de Recherche pour l’Exploitation de la Mer, Université de Bretagne Occidentale, Plouzané, France
- 3CNRS, UMR 6197, Laboratoire de Microbiologie des Environnements Extrêmes, Plouzané, France
Replication slippage or slipped-strand mispairing involves the misalignment of DNA strands during the replication of repeated DNA sequences, and can lead to genetic rearrangements such as microsatellite instability. Here, we show that PolB and PolD replicative DNA polymerases from the archaeal model Pyrococcus abyssi (Pab) slip in vitro during replication of a single-stranded DNA template carrying a hairpin structure and short direct repeats. We find that this occurs in both their wild-type (exo+) and exonuclease deficient (exo-) forms. The slippage behavior of PabPolB and PabPolD, probably due to limited strand displacement activity, resembles that observed for the high fidelity P. furiosus (Pfu) DNA polymerase. The presence of PabPCNA inhibited PabPolB and PabPolD slippage. We propose a model whereby PabPCNA stimulates strand displacement activity and polymerase progression through the hairpin, thus permitting the error-free replication of repetitive sequences.
Introduction
Low complexity DNA sequences such as microsatellites (1–9 nt repeat length), including mono, di, and trinucleotide repeats, and minisatellites (unit ≥10 nt) are frequently associated with mutagenesis “hot-spots” in both eukaryotic and prokaryotic genomes (Bierne et al., 1991; Michel, 2000; Aguilera and Gomez-Gonzalez, 2008). These types of sequences are characterized by high instability, consisting of the addition or deletion of repeated units, leading to variations in repeat copy number. Such genetic variations have been termed “dynamic mutations” (Richards and Sutherland, 1992; Pearson et al., 2005). Arrest of the replication machinery within a repeated region is associated with such instability, where primer and template become misaligned (reviewed in Michel, 2000). This process, known as replication slippage, is involved in the generation of deletions or insertions within repeat regions (Viguera et al., 2001a; Lovett, 2004).
Replication slippage has been proposed to occur within homopolymeric runs (Kroutil et al., 1996) as well as in short and long tandem repeat sequences (Trinh and Sinden, 1993; Madsen et al., 1993; Tran et al., 1995; Bierne et al., 1996; Feschenko and Lovett, 1998). Repeated DNA sequences are generally characterized by the formation of non-B DNA structures, the majority of which can form intra-strand hairpin loops (Samadashwily et al., 1997; McMurray, 1999; Mirkin, 2007; Sinden et al., 2007). A direct role for replication slippage in the deletion of repeated sequences within hairpin structures has been demonstrated in vitro and in vivo (d’Alencon et al., 1994; Canceill and Ehrlich, 1996). Slippage-mediated deletions are believed to occur via a three step mechanism as illustrated in Figure 1 (Viguera et al., 2001a). In this model, the polymerase pauses as it reaches the base of the hairpin after copying the first direct repeat (DR), followed by polymerase dissociation. The 3′ end of the nascent strand then unpairs from the template before reannealing to the second DR. This new primer/template complex is recognized by the polymerase, allowing replication to continue but also generating a deletion.
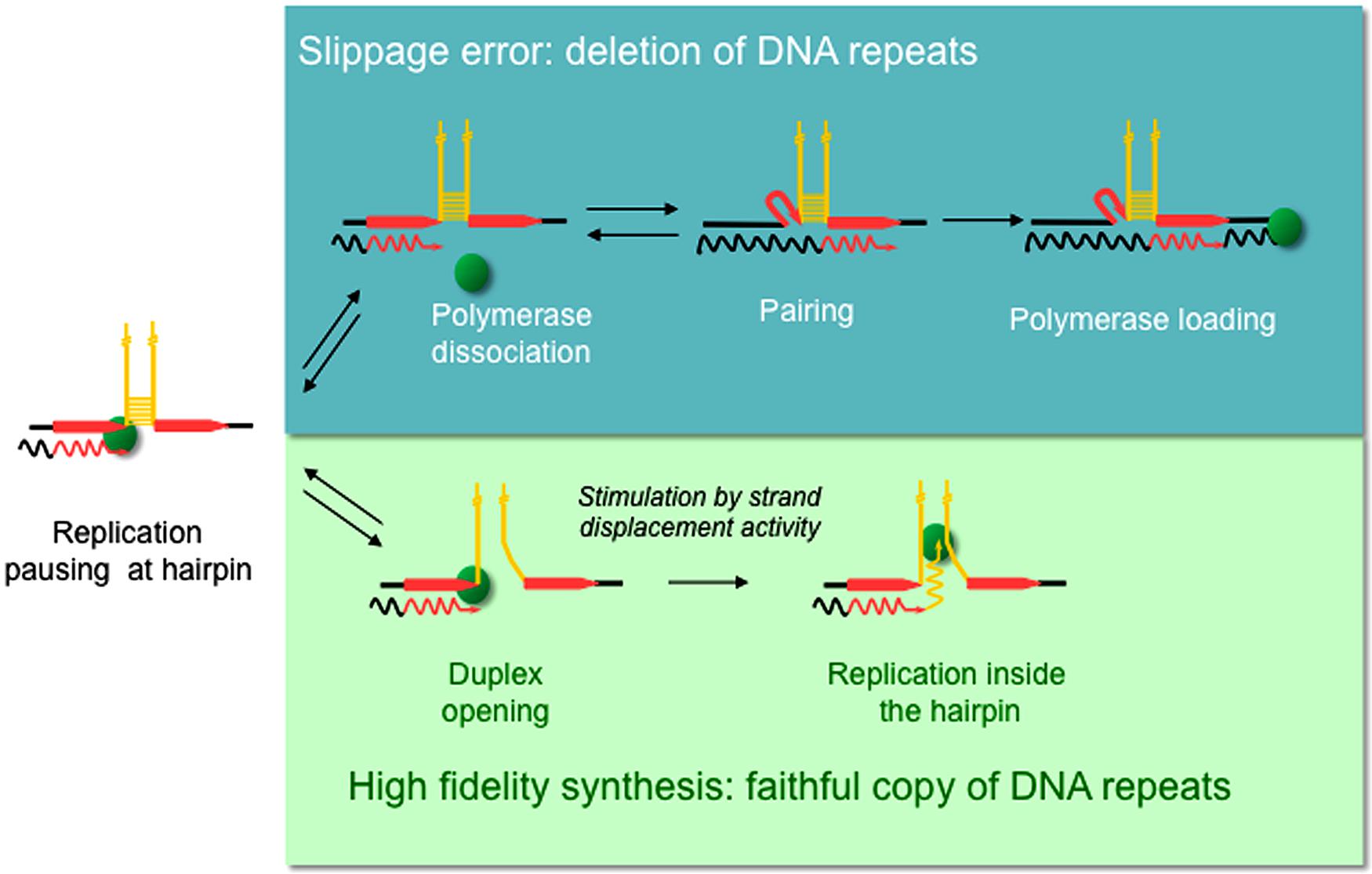
FIGURE 1. Model proposed for replication slippage between direct repeats promoted by hairpin-containing templates. Diagram shows part of a single stranded template (straight lines) with newly synthesized DNA (wavy lines) and the DNA polymerase (green sphere). Direct repeats (red arrows) flank inverted repeats (yellow) that anneal to form a hairpin structure. Slippage-mediated deletion is proposed to involve three sieps (Top): (1) the polymerase pauses within the first direct repeat at the base of the hairpin, (2) polymerase dissociation, and (3) strand misalignment, where the 3’ end of the nascent strand unpairs from the complementary strand and reanneals with the second direct repeat, thus generating a deletion (Viguera et al., 2001a). However, a polymerase with high strand displacement activity is able to open the hairpin duplex (bottom) and replicate the repeat-containing template faithfully.
Several DNA polymerases have been tested for their propensity to slip in vitro when replicating hairpin-containing templates. Surprisingly, the replicative DNA polymerase Pol III holoenzyme (HE) from Escherichia coli can slip in vitro (Canceill and Ehrlich, 1996; Canceill et al., 1999). This is of utmost importance because high fidelity replication is required to maintain genome integrity. Studies on DNA polymerases involved in DNA repair such as E. coli Pol I, E. coli Pol II, and the T4, T7, and ϕ29 phage DNA polymerases revealed that the strand displacement activity of a DNA polymerase is inversely related to their propensity to slip. DNA polymerases with high strand displacement activity such as ϕ29 or T7 pol exo- (SequenaseTM) can progress through template hairpin structures and consequently do not slip. On the other hand, DNA polymerases devoid of strand displacement activity such as E. coli DNA Pol II or T4 are blocked at the base of the hairpin, promoting DNA repeat misalignment and subsequent loss of repeat sequences. Depending on the template and the strand displacement activity of a DNA polymerase it is possible for the deletion error rate to exceed the base substitution rate (Kunkel and Bebenek, 2000). In the context of the model proposed for slippage-mediated deletions (Figure 1), a polymerase with high strand displacement activity would be able to open the hairpin duplex, avoiding the polymerase dissociation and nascent strand reannealing steps, and thus replicate the repeat-containing template faithfully.
Several thermostable DNA polymerases utilized for PCR can also slip even under the high temperatures used during PCR amplification (Viguera et al., 2001b); Table 1. DNA polymerases with high fidelity in terms of base substitution rate, such as P. furiosus (PfuPol), consistently undergo slippage and introduce deletions while replicating hairpin-containing templates (Viguera et al., 2001b). In contrast, a low fidelity DNA polymerase such as Thermus aquaticus (TaqPol) can replicate the same hairpin sequence without introducing deletions, although this is dependent on the magnesium concentration used. Other thermostable DNA polymerases endowed with a high strand displacement activity such as Thermococcus fumicolans (TfuPol) or Bacillus stearothermophilus (BstPol) also do not slip when replicating hairpin-containing sequences (Viguera et al., 2001b).
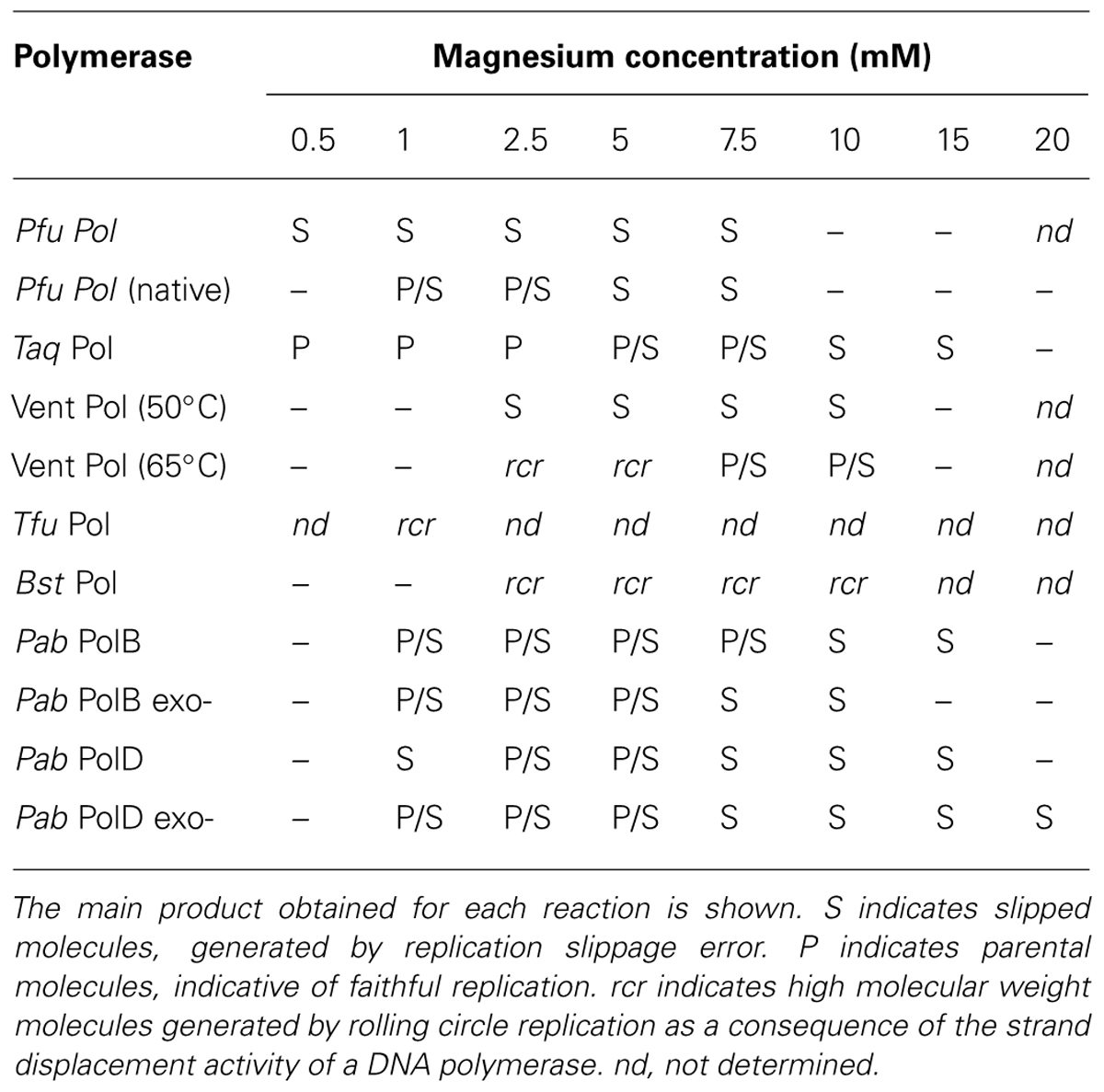
TABLE 1. Effect of magnesium concentration on the slippage of Pfu, Taq, Vent (50°C), Vent (65°C), Tfu and Bst polymerases determined previously (Viguera et al., 2001b) and Pab PolB, Pab PolB exo-, Pab PolD, and Pab PolD exo- determined in this work.
We have studied here the biochemical properties of DNA polymerases PolB (PabPolB) and PolD (PabPolD) from the hyperthermophilic euryarchaeon P. abyssi in terms of slippage during in vitro primer extension reactions. Archaeal replication proteins are more closely related to their eukaryotic than their bacterial equivalents. Euryarchaeal members contain DNA polymerases that belong to both the ubiquitous B family as well as the D family, which is unique to archaea (Ishino et al., 1998; Barry and Bell, 2006; Raymann et al., 2014). Both PolB and PolD have associated 3′-5′ exonuclease activity and moderate strand displacement activity, although PolB cannot displace a RNA-DNA hybrid (Henneke, 2012). However, in the presence of Pab Proliferating cell nuclear antigen (PCNA), both Pab polymerases show strand displacement activity (Henneke et al., 2005; Rouillon et al., 2007). Moreover, PabPCNA can be loaded onto DNA in the absence of the clamp-loader replication factor C (RF-C), although the presence of this factor does enhance its loading (Rouillon et al., 2007).
In this work, we report that both P. abyssi DNA polymerases slip in vitro on a template that consists of single-stranded DNA (ssDNA) with a hairpin structure flanked by short direct repeats. In addition, we find that PabPCNA increases replication fidelity of this template by triggering the strand displacement activity of PabpolB. Furthermore, we describe the effect of magnesium concentration on the replication slippage of both Pab DNA polymerases. These results help toward understanding the dynamics of replication through common non-B DNA structures and identifying the key DNA polymerases involved in replication slippage; a crucial step for understanding genome stability in these organisms.
Materials and Methods
Proteins
PabPCNA, Pabpol D, and exonuclease-deficient Pabpol D were obtained from G. Henneke (Ifremer, Brest, France). They were cloned, expressed and purified as described (Gueguen et al., 2001; Henneke et al., 2002, 2005; Palud et al., 2008). PabpolB (IsisTM) and PabpolB exonuclease-deficient (PyraTM exo-) were purchased from MP Biomedicals. One unit of Pab pols corresponds to the incorporation of 1 nmol of total dTMP into acid precipitable material per minute at 65°C in a standard assay containing 0.5 μg (nucleotides) of poly(dA)/oligo(dT)10:1 M13 gene protein II (gp II) was purified to homogeneity as described (Greenstein and Horiuchi, 1987). Thermus thermophilus SSB was a kind gift from Drs. C. Perales and J. Berenguer (CBM-SO, Madrid). Native Pfu Pol was from Stratagene. Taq Pol was from Roche Molecular Biochemicals.
ssDNA Template
Construction of the pHP727FXc plasmid has been described previously (Canceill and Ehrlich, 1996). Preparation of ssDNA templates was carried out essentially as described (Canceill and Ehrlich, 1996) with the following modifications: plasmid DNA was extracted using a Maxi Plasmid Kit (Qiagen). Briefly, a specific nick was introduced into the f1 replication origin (+) strand of purified FXc plasmid DNA using the M13 gpII protein. The reaction was stopped with 20 mM EDTA and the products treated with 200 μg/ml proteinase K for 10 min at 55°C, phenol extracted and dialyzed against TE buffer. Nicked strands were removed by exonuclease III digestion (10–40 units per μg of DNA for 1 h at 37°C). Finally, Exo III, nucleotides and oligonucleotides were removed using QIAquick® PCR (Qiagen) purification kits.
Primer Extension Reactions
Pyrococcus abyssi pols were tested in a primer extension reaction performed as described (Canceill and Ehrlich, 1996; Canceill et al., 1999). Briefly, 24.3 fmol of a 5′-end fluorescein labeled primer (Applied Biosystems) designated 1233 (5′AGC GGA TAA CAA TTT CAC ACA GGA 3′), were annealed 1235 bases upstream of the palindrome. All 10 μl primer extension reactions contained 25 ng (12.2 fmol) of primed ssDNA, and Pab pols that were added to the reaction mixture as indicated in the figure legends. Additionally, reactions contained unless otherwise mentioned 50 mM Tris-HCl (pH 8.8), 50 mM KCl, 10 mM DTT, 2 mM MgCl2, and 200 μM dNTPs. Reactions were performed at 60°C for 30 min, synthesis was arrested by the addition of 25 mM EDTA and 500 μg/ml proteinase K, and the mixture was further incubated for 15 min at 55°C. Reaction products were analyzed by electrophoresis using 0.8% agarose gels under native conditions, run in TBE buffer (89 mM Tris-borate, 2 mM EDTA, pH 8.3) at 2 V/cm for 16 h and visualized with a Typhoon 9400 Variable Mode Imager (Amersham Biosciences, GE Healthcare). Analysis of the results was performed using Image Quant 5.2 software. Quantification analysis was performed with Visilog 6.3 (Visualization Sciences Group. Noesis). A common fixed area was selected at the center of the bands corresponding to parental and heteroduplex molecules. The average gray value of all pixels of each area was obtained and the proportion of parental/heteroduplex was calculated.
Pfu Pol and Taq Pol were tested as above except that 200 μM dGTP, dATP, and dTTP (each), 40 μM dCTP and 50 μM (2.5 μCi) (α-32P)dCTP was used. The reaction buffers were prepared magnesium free as those furnished by the suppliers and contained, in addition to 30 mM NaCl brought by the primed ssDNA, the following ingredients: (i) for Taq Pol: 10 mM Tris-HCl pH 8.3, 50 mM KCl; (ii) for Native Pfu Pol: 20 mM Tris-HCl pH 8.0, 10 mM KCl, 6 mM (NH4)2SO4, 0.1% Triton® X-100, 10 μg/ml BSA. After gel electrophoresis, DNA was visualized by direct exposure of the dried gels to Imaging Plates (IP BAS-MP 2040S) and analyzed on a Fujifilm-BAS 1500.
Results
Experimental System
To study whether P. abyssi thermostable DNA polymerases (Pab pols) promote replication slippage, we performed primer-extension assays using the circular ssDNA template, FXc (Canceill and Ehrlich, 1996; Canceill et al., 1999; Viguera et al., 2001a,b). This template contains two 27 bp direct repeats (DRs) that flank a pair of 300 bp inverted repeats (IR) separated by a 1.3 kb insert as shown in Figure 2. The IRs anneal to form a stem-loop with the DRs at its base.
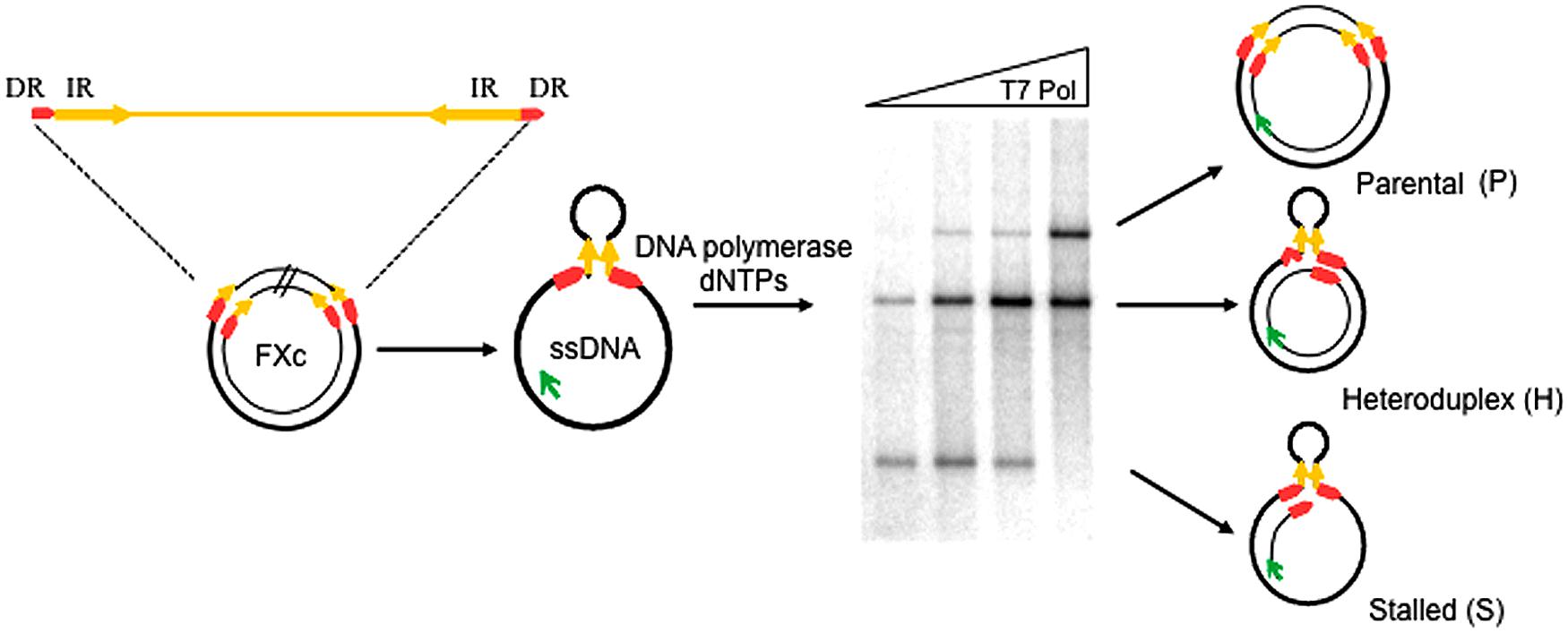
FIGURE 2. Experimental assay for the detection of replication slippage. Schematic representation of the ssDNA template and the different replication products expected after a primer extension reaction, FXc represents the double-stranded pHP727FXc plasmid containing a central 1370 bp region (insert) flanked by 300-bp inverted repeats (IR: yellow arrows) and 27-bp direct repeats (DR; red arrows). ssDNA FXc template is prepared in vitro and primer extension reactions performed at 60°C in the presence of a fluorescein-labeled primer (green arrow) and DNA polymerase as described in the “Materials and Methods.” Reaction products are then separated by agarose gel electrophoresis. An assay testing different T7 DNA Pol concentrations is shown in the example. P indicates fully replicated parental molecules. H represents heteroduplex molecules generated after slippage, with one strand lacking one direct repeat unit and the hairpin. S indicates stalled molecules generated by arrest of the polymerase at the base of the hairpin. Bands migrating between S and H correspond to DNA polymerase arrest inside the hairpin. Bands migrating above P corresponds to high molecular weight molecules generated by displacement of the extended primer (Viguera et al., 2001b).
DNA synthesis was carried out with a fluorescently labeled primer and the reaction products analyzed by agarose gel electrophoresis. Faithful replication of the FXc template generates complete double-stranded parental (P) molecules, which migrate in a retarded position on the gel. A slippage event generates a heteroduplex molecule (H), composed of a parental strand annealed to a recombinant strand lacking one of the DRs and the 1370 bp region between them. Heteroduplex molecules migrate ahead of parental molecules. Stalled (S) replication as the polymerase reaches the base of the hairpin results in a truncated molecule that migrates further than either parental or heteroduplex molecules.
Slippage of P. abyssi PolB and PolD Replicative Polymerases
It has been proposed that PolB and PolD have different roles in the cell, both participating at the replication fork in a manner analogous to Bacillus subtilis and the eukaryotic replisome. The current model for P. abyssi DNA replication proposes that PabPolD performs RNA-primed DNA synthesis and is later displaced by PabPolB to carry out processive DNA synthesis, at least on the leading strand (Henneke et al., 2005; Rouillon et al., 2007). Because of the capacity of PabPolD to displace RNA primers in a PCNA-dependent manner, it has been suggested that PabPolD is involved in lagging strand replication. However, definitive confirmation using genetic approaches such as those employed with eukaryotic polymerases has yet to be performed.
In order to test whether their putatively separate roles in leading and lagging strand replication also imply different slippage properties, we examined the slippage efficiency of wildtype PabPolB and PabPolD enzymes using the FXc template (Figure 2). PabPolB generated both parental and heteroduplex molecules, which indicate a mixture of normal FXc replication and slippage events (Figure 3, lanes 1–3). Similar proportions of parental and heteroduplex molecules were produced by PabPolB, with a slightly higher ratio of parental molecules as the polymerase concentration was increased. PabPolD behaved in a similar way although the proportion of heteroduplex molecules was higher and overall synthesis was improved at higher polymerase concentrations (Figure 3, lanes 7–9). These results indicate that PabPolB and PabPolD can slip under our assay conditions. The behavior of PabPolB and PabPolD is similar to that observed for Pol III HE, T7 Pol, or Taq Pol that also produce both parental and heteroduplex molecules (Canceill and Ehrlich, 1996; Canceill et al., 1999; Viguera et al., 2001b).
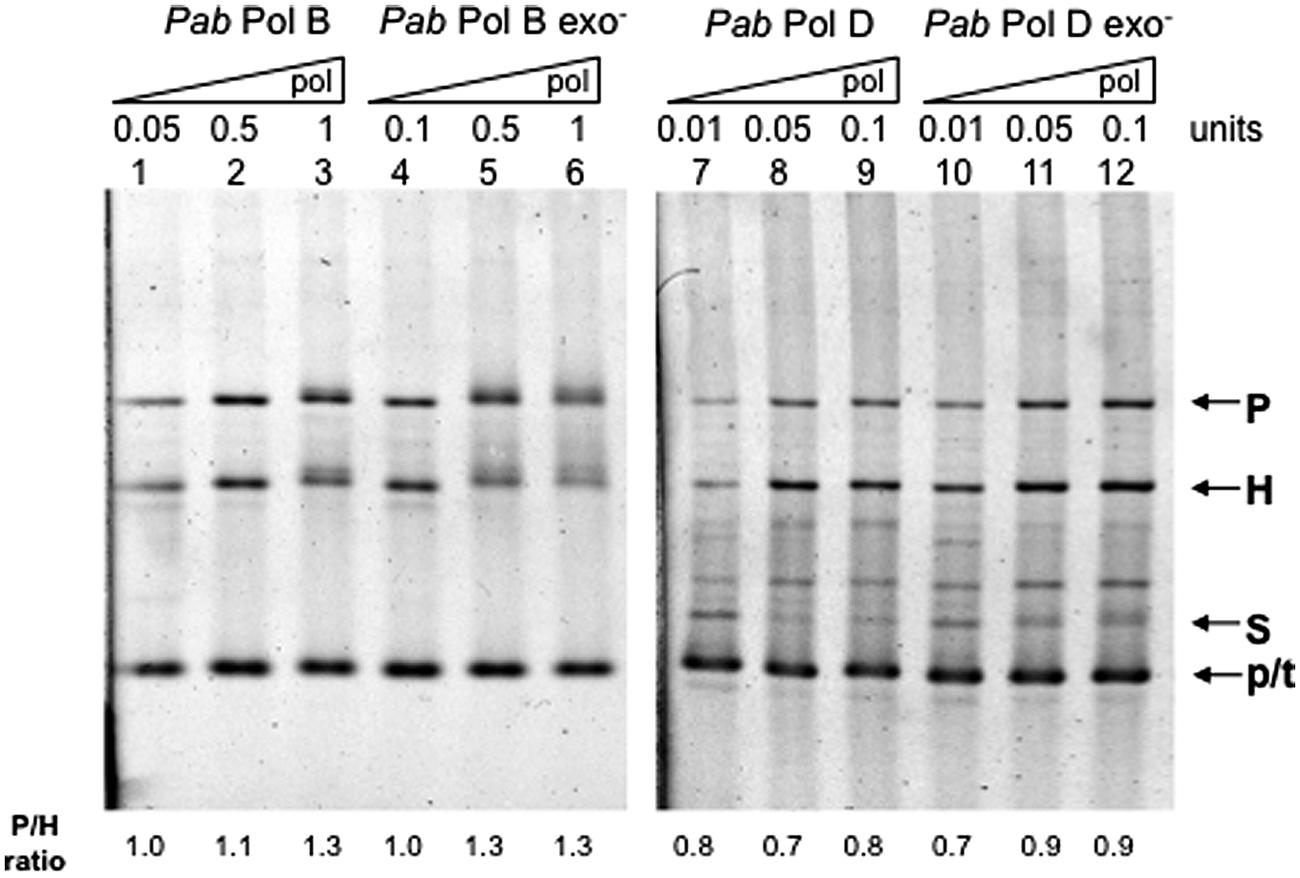
FIGURE 3. Effect of DNA polymerase concentration on Pab DNA polymerase slippage. Primer extension reactions were carried out as described in the “Materials and Methods” using increasing amounts of the appropriate Pab DNA polymerase. One polymerase unit represents 27 pmol PabPolB: 4 pmol PabPolB exo-; 96 pmol PabPolD; and 104 pmol of PabPolD exo-. P, H, S, and p/t refer to parental, heteroduplex, stalled molecules, and primer-template, respectively. The ratio P/H is indicated below the figure.
To generate parental molecules, a DNA polymerase must open the hairpin formed by the annealed inverted repeats of the single-stranded template (Figure 2), which is largely dependent on a DNA polymerase’s strand displacement activity. As a consequence, polymerases with high strand displacement activity (e.g., ϕ29 DNA polymerase) do not slip while DNA polymerases devoid of strand displacement activity (e.g., E. coli Pol II or T4 DNA pol) generate heteroduplex molecules as the sole product of the reaction (Canceill et al., 1999).
Strand displacement activity is modified in some DNA polymerase exo- mutants. For example the T7 DNA polymerase has relatively low strand displacement activity (Canceill et al., 1999). However, the T7 pol exo- (SequenaseTM), carrying a 28 amino acid deletion that inactivates its proofreading activity (Engler et al., 1983; Lechner et al., 1983), has increased strand displacement activity that prevents T7 pol exo- slippage (Canceill et al., 1999). Similarly, the E. coli Pol II exo- mutant gains a degree of strand displacement activity and the ability to synthesize parental molecules (Canceill et al., 1999). However, not all exo- forms exhibit increased strand displacement activity. For example, an exo- form of F 29 caused by a point mutation shows a 90% reduction in strand displacement activity compared to the native enzyme (Soengas et al., 1992).
To test whether Pab pol exo- variants have modified slippage properties, we performed FXc template primer extension assays using exo- mutant forms of PabPolB and PabPolD carrying single point mutations (D215A and H451A, respectively; see Palud et al., 2008). Our results show that the exo- forms of PabPolB and PabPolD both generated heteroduplex and parental molecules. However, unlike their native forms, increasing polymerase concentration inhibited slippage and resulted in a higher proportion of parental molecules (Figure 3, lanes 4–6 and 10–12, respectively). Both the native PabPolD enzyme and its exo- form produced some molecules that migrate between the heteroduplex and stalled molecules, possibly the result of inefficient polymerase progression through the hairpin (Viguera et al., 2001a).
Magnesium Concentration Affects the Slippage of P. abyssi Polymerases
The concentration of divalent cations needs to be precisely controlled during DNA synthesis as it affects enzyme activity, enzyme fidelity, primer/template annealing, and the stability of secondary structures, such as the stem-loop used in our assay. The fidelity of Taq and Pfu DNA polymerases in terms of base substitution and frameshift errors is dependent on magnesium concentration (Eckert and Kunkel, 1990; Cline et al., 1996). Moreover, trinucleotide repeat expansions are produced in vitro by Taq, E. coli Pol I, and the Pol I Klenow fragment at certain magnesium concentrations (Lyons-Darden and Topal, 1999). With respect to slippage, magnesium concentration differentially affects the slippage errors produced by thermostable DNA polymerases (Viguera et al., 2001b). In vitro experiments showed that slippage error-derived heteroduplex molecules account for almost all the product generated by Pfu DNA polymerase over the magnesium concentration range that permits efficient DNA synthesis (0.5–7.5 mM MgSO4). In contrast, from the same template Taq DNA polymerase faithfully generates parental molecules at a low magnesium concentration (0.5 mM MgCl2), heteroduplex molecules at high magnesium concentrations (10–20 mM MgCl2) and a mixture of parental and heteroduplex molecules at intermediate magnesium concentrations (1–7.5 mM MgCl2; Viguera et al., 2001b).
These observations prompted us to analyze the effect of magnesium concentration on the slippage errors produced by the wildtype and exo- forms of PabPolB and PabPolD. We found that varying magnesium concentration affected both slippage and overall DNA synthesis (Figure 4). There was almost no synthesis by PabPolB, PabPolD or their exo- forms at low magnesium concentrations (0.1–0.5 mM; Figure 4A, lanes 2–3 and 12–13; Figure 4B, lanes 2–3 and 12–13). Synthesis was also inhibited at the highest concentrations tested (15–20 mM; Figure 4A, lanes 9–10 and 19–20; Figure 4B, lanes 9–10 and 19–20). Parental molecules were readily detectable together with heteroduplex molecules at low to medium magnesium concentrations (1–5 mM; Figure 4A, lanes 4–6). Increasing magnesium concentration up to 15 mM decreased the proportion of parental molecules and resulted in heteroduplex molecules as the main reaction product (Figure 4A, lanes 7–9). This latter result could be due to stabilization of the hairpin structure by high magnesium concentrations making polymerase progression more difficult inside the hairpin (Canceill and Ehrlich, 1996). The effect of magnesium concentration on PabPolB was similar at the 40 and 100 μM nucleotide concentrations tested (data not shown). PabPolB exo- behaved in a similar way to the wildtype enzyme (Figure 4A, lanes 14–18).
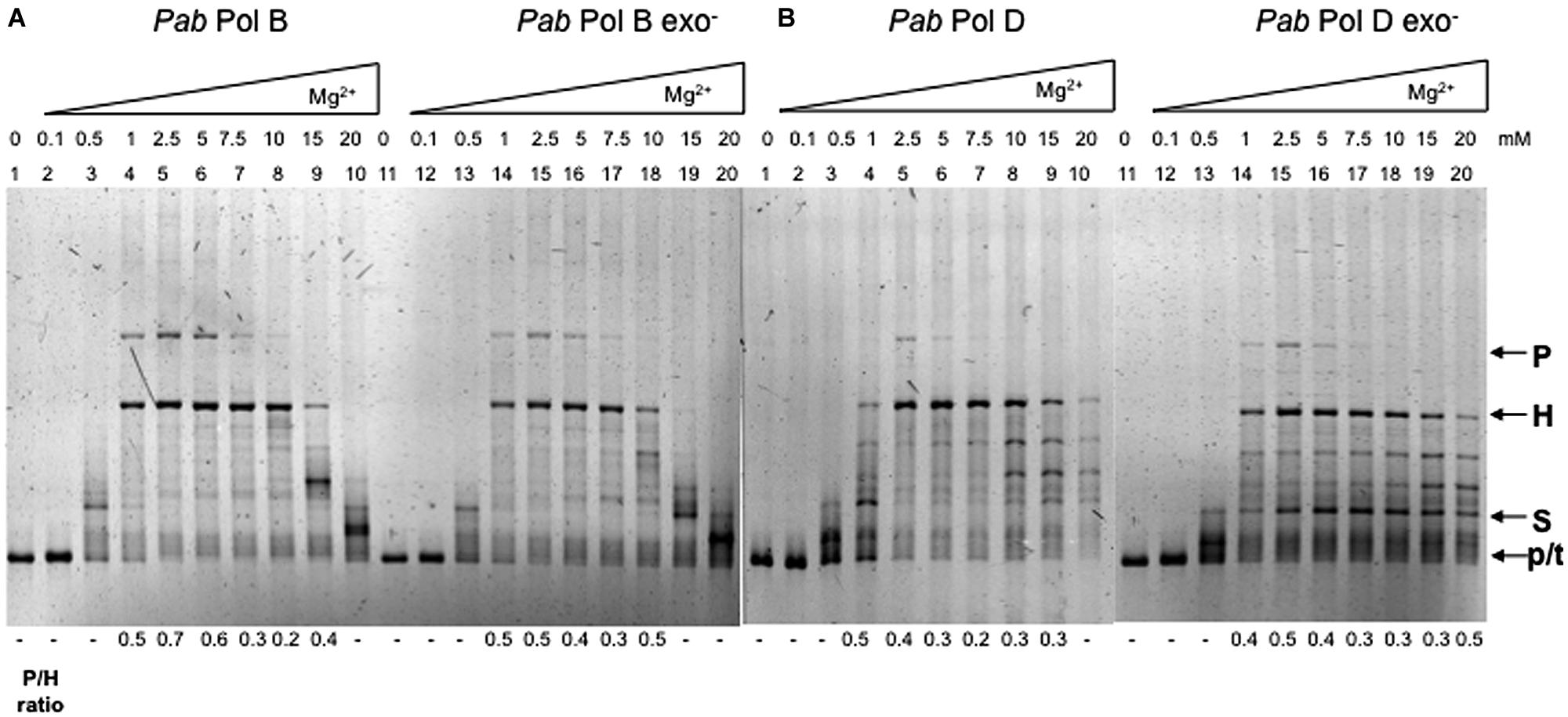
FIGURE 4. Effect of magnesium concentration on Pab DNA polymerase slippage. Primer extension reactions were carried out as described in the “Materials and Methods.” Reactions contained 0.5 units of PabPolB, (1.35 pmol), PabPolB exo- (2 pmol), PabPolD (4.8 pmol), and PabPolD exo- (5.2 pmol) with increasing concentrations of MgCI2. P, H, S, and p/t refer to parental, heteroduplex, stalled molecules, and primer-template, respectively.
Although both PabPolD and PabPolD exo- generated parental molecules, the main reaction products were heteroduplex molecules whenever synthesis was efficient (Figure 4B, lanes 4–10 and lanes 14–20). Additionally, some of the molecules generated by PabPolD and PabPolD exo- migrated between heteroduplex and stalled molecules (Figure 4B, lanes 8–9 and 15–20) that probably represent partially replicated DNA molecules due to inefficient polymerase progression within the hairpin.
In order to confirm that the magnesium concentrations used in the previous experiment are compatible with efficient PabPolB and PabPolD DNA synthesis, we performed primer extension experiments using a 5′ fluorescently labelled primer (33 mer) and a short single-stranded linear DNA template (87 mer) that has the potential to form a 28 bp secondary structure but lacks DRs (Henneke, 2012). This assay should only assess the replication efficiency of a template with a small hairpin without the possibility of slippage between DRs at its base. Fully replicated molecules were generated for both PabPolB and PabPolD using the same range of magnesium concentrations used for the primer extension assays (Figure S1), which indicates that the reaction conditions used were optimal for DNA synthesis.
We conclude that in spite of their high fidelity in terms of base substitution, PabPolB and PabPolD are highly prone to slip on ssDNA templates upon encountering secondary structures flanked by DRs, generating parental and heteroduplex molecules in a magnesium concentration-dependent manner. This is in agreement with previous results describing similar behavior for Taq DNA polymerase (Viguera et al., 2001b); Figure S2. In contrast, native Pfu DNA polymerase generates mostly heteroduplex molecules regardless of magnesium concentration (Viguera et al., 2001b); Figure S2.
Pyrococcus abyssi PCNA can Modulate the Slippage of PabPolB and PabPolD
The sliding clamp of Archaea, Eukarya, and Bacteria forms a ring around dsDNA that prevents the dissociation of DNA polymerases from their template, thus enhancing processivity (O’Donnell et al., 2013). Moreover, it acts as a platform that regulates polymerase switching, coupling DNA replication and DNA repair (López de Saro, 2009). The E. coli sliding clamp β homodimer subunit requires the clamp loader (or γ complex) to load it onto the template. The addition of β to primer extension reactions on a hairpin-containing template favors Pol III HE slippage as the synthesis of heteroduplex molecules is stimulated (Canceill and Ehrlich, 1996).
Pyrococcus abyssi, possesses a single processivity clamp, PCNA, that forms a homotrimer (Castrec et al., 2009). In contrast to Bacteria and Eukarya, the archaeal PCNA can be loaded onto DNA without a clamp-loader. PabRF-C and PabPolB, but not PabPolD, enhance PCNA loading. PabRF-C and PabPolB associate with PabPCNA, forming a stable complex on primed DNA (Rouillon et al., 2007).
We therefore investigated the role of PCNA on PabPolB and PabPolD slippage (Figure 5). The addition of equimolar amounts of PabPCNA to the FXc replication assay reduced PabPolB slippage. Formation of parental molecules was stimulated and the proportion of heteroduplex molecules diminished (Figure 5, lanes 1–3) with respect to reactions performed in the absence of PCNA (compare with Figure 3, lanes 1–3). PabPCNA addition also reduced PabPolB exo- slippage (Figure 5, lanes 4–6, compare with Figure 3, lanes 4–6). Furthermore, the addition of PCNA resulted in the appearance of slowly migrating high molecular weight molecules (Figure 5, lanes 5–6). These molecules could be the result of rolling circle replication (rcr), which implies that after completion of one round of replication, the newly synthesized strand becomes displaced allowing synthesis to continue (Canceill et al., 1999).
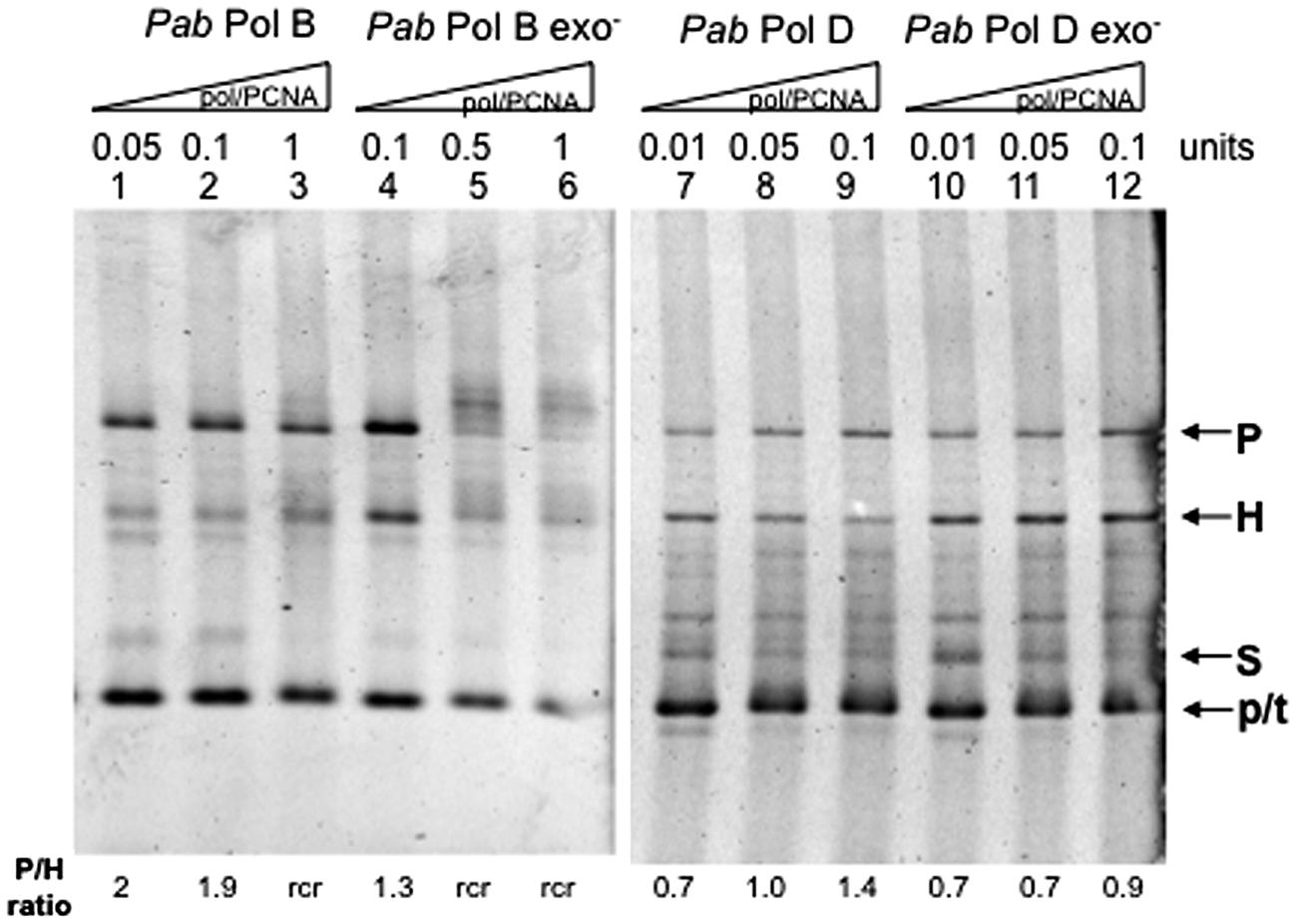
FIGURE 5. Effect of PabPCNA on Pab DNA polymerase slippage. Primer extension reactions were carried out as described in the “Materials and Methods” on 25 ng (12.2 fmol)of FXc template using increasing amounts of Pab DNA polymerase and equimolar amounts of PabPCNA. One polymerase unit represents 27 pmol PabPolB: 4 pmol PabPolB exo-: 96 pmol PabPolD and 104 pmol of PabPolD exo-, P, H, S, and p/t refer to parental heteroduplex, stalled molecules, and primer-template, respectively. The ratio P/H Is Indicated below the figure. Rolling circle replication (rcr) is indicative of slippage inhibition because of the strand displacement activity of the polymerase.
The presence of PabPCNA also increases the proportion of parental versus heteroduplex molecules generated by PabPolD (Figure 5, lanes 7–9), indicating that it also represses slippage by this DNA polymerase. However, PabPCNA had only a slight effect on PabPolD exo- slippage (Figure 5, lanes 10–12), as the proportion of parental molecules was only slightly higher.
We conclude from these experiments that PabPCNA stimulates the ability of PabPolB and PabPolD to replicate through a hairpin structure by inhibiting slippage, with the strongest effect in terms of slippage inhibition observed on PabPolD exo-. These results agree with those obtained by Henneke et al. (2005) in which PabPCNA stimulated the strand displacement activity of PabPolB and PabPolD (Henneke, 2012).
The Thermus thermophilus Single-Stranded DNA Binding (SSB) Protein Does not Affect Slippage Errors Produced by Pab DNA Pols
The amount of slippage exhibited by different DNA polymerases has been shown to be modulated by SSB proteins (Canceill and Ehrlich, 1996; Canceill et al., 1999). E. coli SSB stimulates the slippage of Pol III HE, inhibits the slippage of E. coli polymerase I and T7 DNA polymerase, and has no effect on E. coli pol II or T4 DNA polymerase. On the other hand, T4 SSB protein (gp32) inhibits T4 DNA pol slippage but does not affect the slippage properties of the Pol I Klenow fragment. These contrasting effects of SSB proteins on the same DNA template cannot be understood solely in terms of interaction with DNA, but rather as an interaction between SSB proteins and the different polymerases that alters their strand displacement activity (Canceill et al., 1999).
We therefore investigated whether a SSB protein could modify the slippage properties of Pab pols. Thermus thermophilus (Tth) SSB stimulates DNA synthesis of Tth DNA polymerase and the heterologous DNA polymerase from the Archaea P. furiosus (Perales et al., 2003). Furthermore, TthSSB increases the fidelity of proofreading deficient Thermus thermophilus DNA polymerase (Perales et al., 2003). We tested the slippage properties of PabPolB, PabPolD, and their exo- forms in the presence of increasing amounts of TthSSB. Results obtained for PabPolB and its exo- form are shown in Figure 6A, lanes 1–8. Heteroduplex products were detected in both the presence and absence of TthSSB. Similar results were obtained for PabPolD and its exo- form (Figure 6B, lanes 1–6). We conclude that TthSSB neither stimulated overall synthesis efficiency nor the slippage of Pab polymerases under the reaction conditions assayed.
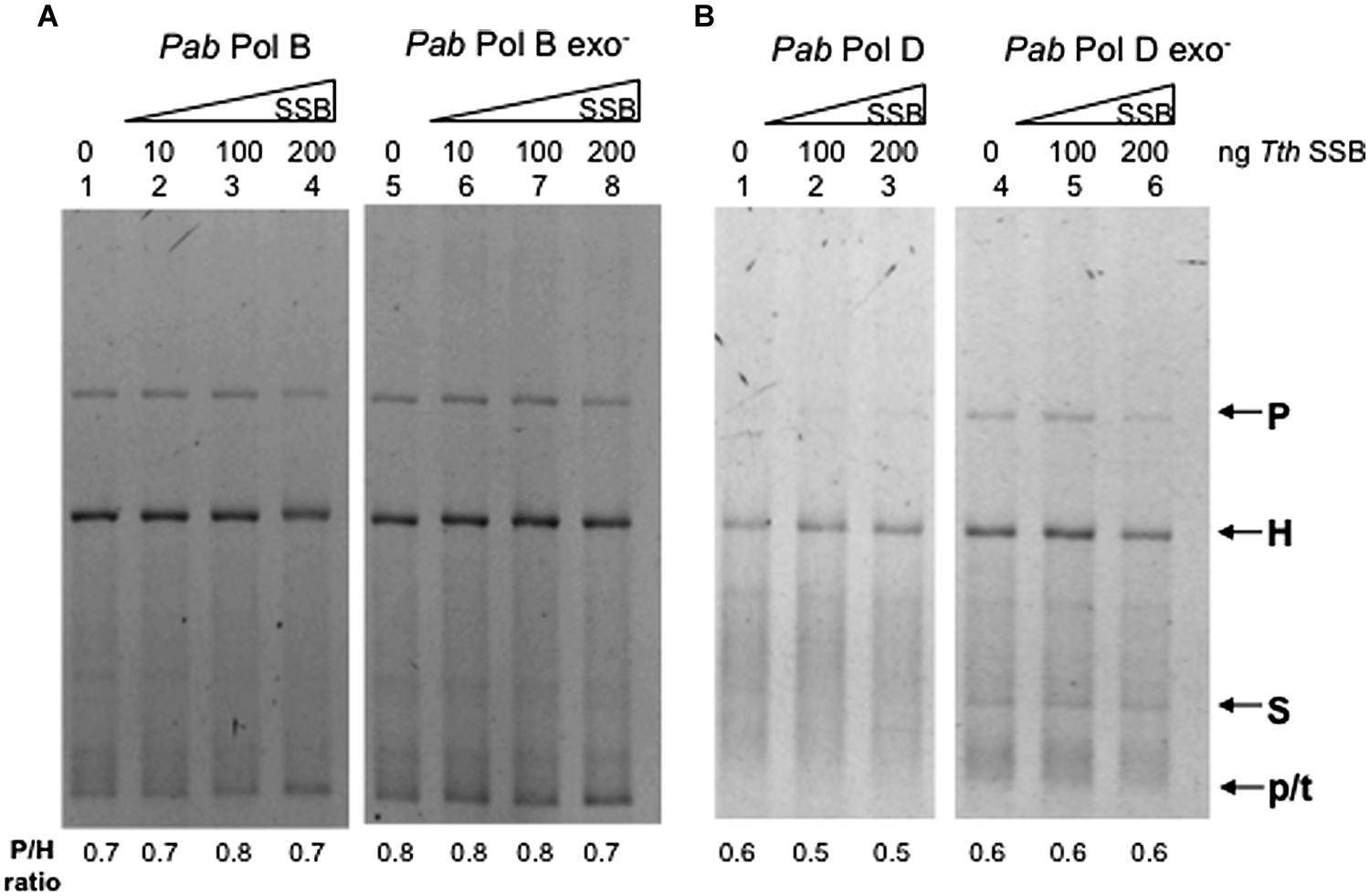
FIGURE 6. Effect of Thermus thermophilus SSB (T. thermophilus SSB) on Pab DNA polymerase slippage. Primer extension reactions were carried out as described in the “Materials and Methods” on 25 ng (12.2 fmol) of FXc template. Reactions contained 1 unit of PabPol B and PabPol B exo- (27 and 4 pmol, respectively, A), 0.05 units of PabPolD and PabPolD exo- (4.3 and 5.2 pmol respectively, B), and were pre-incubated at 60°C with increasing concentrations of T. thermophilus SSB (1 ng of Tth SSB corresponds to 16.78 fmol). P, H, S, and p/t refer to parental heteroduplex. stalled molecules and primer-template, respectively.
Discussion
Interest in DNA repeat instability has increased dramatically since links were established between expansions of trinucleotide repeats and neurodegenerative diseases (Bacolla and Wells, 2009; reviewed in Kim and Mirkin, 2013), microsatellite instability and certain types of cancer (Shah et al., 2010) and the identification of frameshift-mediated regulation of gene expression at simple sequence contingency loci of pathogenic bacteria such as Neisseria or Haemophilus (Moxon et al., 1994; Bayliss et al., 2001; reviewed in Gemayel et al., 2010).
Because of the association between DNA repeat instability and DNA replication, DNA polymerases have been analyzed in vitro to establish their ability to replicate repeated DNA sequences. We have shown previously that the main replicative DNA polymerase of the model bacteria E. coli, DNA Pol III HE, is able to slip in vitro on hairpin-containing templates despite the high fidelity required for genome replication (Canceill et al., 1999). Moreover, slippage is stimulated by factors that affect Pol III processivity such as the presence of β-clamp or SSB proteins (Canceill and Ehrlich, 1996).
Thermostable DNA polymerases are widely used for a number of applications, mostly involving PCR amplification. We have previously shown that replication slippage occurs efficiently even during the first PCR amplification cycle of Taq Pol, Pfu Pol, PyraTM Pol (Pab PolB exo-), or the ExpandTM mixture (Taq Pol and Pwo Pol; Viguera et al., 2001b). However, no slippage was detected during PCR performed by Tfu Pol or Vent® Pol. Since, high fidelity DNA polymerases can undergo slippage in terms of base substitution, slippage is only inhibited in those polymerases endowed with high strand displacement activity. Thus, the use of DNA polymerases with high strand displacement activity is advisable when amplifying DNA templates with potential strong secondary structures.
In this study, we have shown that replicative P. abyssi DNA polymerases are able to slip in vitro on hairpin containing templates as heteroduplex molecules, indicative of slippage error, as well as parental molecules were obtained at every enzyme concentration assayed. This result is quite different to those obtained for the thermostable polymerase B from P. furiosus (Pfu) or the mesophilic DNA polymerases E. coli Pol II and T4, where heteroduplex molecules are the only reaction product (Viguera et al., 2001b). Although native Pfu Pol generated some parental molecules, the main reaction products were heteroduplex structures (Figure S2B). In contrast, Taq DNA pol, Pol III HE, Pol I, and T7 DNA pol generate heteroduplex and/or parental molecules depending on the polymerase concentration used in the assay (Canceill et al., 1999; Viguera et al., 2001b; Figure S2A). High polymerase concentrations are believed to promote step-by-step progression inside the hairpin via multiple association/dissociation events (Canceill et al., 1999), thus slippage assay sensitivity to polymerase concentration is consistent with a polymerase possessing some degree of strand displacement activity.
Our interpretation is that the different slippage properties of the closely related PabPolB and Pfu Pol are most likely due to PabPolB having higher strand displacement activity, which allows it to generate a higher proportion of parental molecules.
Both PabPolB and PabPolD generated heteroduplex molecules alone or a combination of parental and heteroduplex products at the different magnesium concentration tested whenever synthesis was efficient. This result was somewhat similar to that obtained for Taq DNA pol, where either parental or heteroduplex molecules were obtained depending on the magnesium concentration (Viguera et al., 2001b), although we did not find any reaction condition where parental molecules were the sole product of either of the Pab polymerases. The strand displacement activity of PabPolB and PabPolD is probably insufficient to reliably open and progress through the hairpin structure even at the lowest magnesium concentration tested; a condition that should reduce DNA duplex stability. Consequently, even if PabPolB and PabPolD have different roles at the replication fork, they do not differ in terms of their slippage properties. This result prompted us to study other cellular factors that could inhibit the slippage errors detected by our in vitro assay.
We have shown that the PabPCNA sliding clamp promotes the synthesis of parental molecules by PabPolB. This effect is even more prominent for the exonuclease-deficient PabPolB (Figure 5). In comparison to PabPolB, inhibition of slippage by PCNA was weaker for PabPolD (Figure 5). PabPolB has been identified as the leading strand DNA polymerase (Henneke et al., 2005; Rouillon et al., 2007). PabPCNA interacts with PabPolB in a DNA-dependent way and stimulates its processivity, clamping PabPolB to DNA (Henneke et al., 2005; Henneke, 2012). The higher processivity of the PabPCNA-PolB complex and stimulation of strand displacement activity (Henneke et al., 2005) would facilitate opening of the DNA duplex leading to the synthesis of parental molecules. The effect of PabPCNA is further increased for PabPolB exo-. One possibility is that strand displacement activity is increased to some degree in this mutant and that this facilitates parental formation as has been observed for T7 pol exo- (SequenaseTM) and Pol II exo- (Canceill et al., 1999).
In our opinion, this result confirms PabPCNA-PolB as a competent and stable complex, capable of continuously synthesizing the leading strand. Upon encountering secondary structures (such as hairpin loops), DNA synthesis is unperturbed and the PabPCNA-PolB complex is capable of continuing strand elongation.
The reason that PabPCNA inhibited PabPolD replication slippage to a lesser extent than PabPolB is probably due to insufficient stimulation of strand displacement activity (Henneke et al., 2005) under the conditions tested. PabPCNA binds PolB and PolD in different ways (Castrec et al., 2009). Two PCNA-interacting protein (PIP) boxes are needed for PabPolD binding to PabPCNA whereas only one PIP motif is essential for PabPolB binding. This suggests that the mechanism involved in the PabPCNA-mediated stimulation of PabPolD may be different from that involved in PabPolB.
Previous work (Canceill and Ehrlich, 1996) has shown that in E. coli, the β-clamp does not stimulate the generation of parental molecules by Pol III HE but instead increases the formation of heteroduplex ones. Our finding that the addition of PabPCNA promotes parental formation by PabPolB and PabPolD, indicates that interaction between DNA polymerases and PCNA in the archaeal P. abyssi promotes faithful replication of DNA secondary structures. The functional homology between archaeal and eukaryal proteins, i.e., human PCNA can be loaded onto DNA by the P. abyssi RF-C complex (Henneke et al., 2002), suggests that slippage by replicative eukaryal DNA polymerases may also be inhibited by presence of the sliding clamp.
Conflict of Interest Statement
The Review Editor Bernard Connolly declares that, despite having collaborated and published with Ghislaine Henneke, the review process was handled objectively. All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. C. Perales and Dr. J. Berenguer (CBM-SO, Madrid) for their generous gift of TthSSB and Dr. J. R. Pearson for scientific English editing of the manuscript. This work was supported by grants BFU2007-64153 from the Ministerio de Educación y Ciencia and P09-CVI-5428 from the Junta de Andalucía to Enrique Viguera. Ghislaine Henneke was financially supported by grant ANR-10-JCJC-1501 01 from the National Research Agency. Melissa Castillo-Lizardo acknowledges the short-term fellowship from EMBO and the FPI predoctoral Fellowship BES-2005-10150 from Ministerio de Educación y Ciencia, Spain.
Suppplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00403/abstract
References
Aguilera, A., and Gomez-Gonzalez, B. (2008). Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9, 204–217. doi: 10.1038/nrg2268
Bacolla, A., and Wells, R. D. (2009). Non-B DNA conformations as determinants of mutagenesis and human disease. Mol. Carcinog. 48, 273–285. doi: 10.1002/mc.20507
Barry, E. R., and Bell, S. D. (2006). DNA replication in the archaea. Microbiol. Mol. Biol. Rev. 70, 876–887. doi: 10.1128/MMBR.00029-06
Bayliss, C. D., Field, D., and Moxon, E. R. (2001). The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Invest. 107, 657–662. doi: 10.1172/JCI12557
Bierne, H., Ehrlich, S. D., and Michel, B. (1991). The replication termination signal terB of the Escherichia coli chromosome is a deletion hot spot. EMBO J. 10, 2699–2705.
Bierne, P., Ehrlich, S. D., and Michel, B. (1996). Deletions at stalled replication forks occur by two different pathways. EMBO J. 16, 3332–3340. doi: 10.1093/emboj/16.11.3332
Canceill, D., and Ehrlich, S. D. (1996). Copy-choice recombination mediated by DNA polymerase III holoenzyme from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 93, 6647–6652. doi: 10.1073/pnas.93.13.6647
Canceill, D., Viguera, E., and Ehrlich, S. D. (1999). Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J. Biol. Chem. 274, 27481–27490. doi: 10.1074/jbc.274.39.27481
Castrec, B., Rouillon, C., Henneke, G., Flament, D., Querellou, J., and Raffin, J. P. (2009). Binding to PCNA in Euryarchaeal DNA Replication requires two PIP motifs for DNA polymerase D and one PIP motif for DNA polymerase B. J. Mol. Biol. 394, 209–218. doi: 10.1016/j.jmb.2009.09.044
Cline, J., Braman, J., and Hogrefe, H. (1996). PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucl. Acids Res. 24, 3546–3551. doi: 10.1093/nar/24.18.3546
d’Alencon, E., Petranovic, M., Michel, B., Noirot, P., Aucouturier, A., Uzest, M.,et al. (1994). Copy-choice illegitimate DNA recombination revisited. EMBO J. 13, 2725–2734.
Eckert, K. A., and Kunkel, T. A. (1990). High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucl. Acids Res. 18, 3739–3744. doi: 10.1093/nar/18.13.3739
Engler, M. J., Lechner, R. L., and Richardson, C. C. (1983). Two forms of the DNA polymerase of bacteriophage T7. J. Biol. Chem. 258, 11165–11173.
Feschenko, V. V., and Lovett, S. T. (1998). Slipped misalignment mechanisms of deletion formation: analysis of deletion endpoints. J. Mol. Biol. 276, 559–569. doi: 10.1006/jmbi.1997.1566
Gemayel, R., Vinces, M. D., Legendre, M., and Verstrepen, K. J. (2010). Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44, 445–477. doi: 10.1146/annurev-genet-072610-155046
Greenstein, D., and Horiuchi, K. (1987). Interaction between the replication origin and the initiator protein of the filamentous phage f1. Binding occurs in two steps. J. Mol. Biol. 197, 157–174. doi: 10.1016/0022-2836(87)90115-X
Gueguen, Y., Rolland, J. L., Lecompte, O., Azam, P., Le Romancer, G., Flament, D.,et al. (2001). Characterization of two DNA polymerases from the hyperthermophilic euryarchaeon Pyrococcus abyssi. Eur. J. Biochem. 268, 5961–5969. doi: 10.1046/j.0014-2956.2001.02550.x
Henneke, G. (2012). In vitro reconstitution of RNA primer removal in Archaea reveals the existence of two pathways. Biochem. J. 447, 271–280. doi: 10.1042/BJ20120959
Henneke, G., Flament, D., Hubscher, U., Querellou, J., and Raffin, J. P. (2005). The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 350, 53–64. doi: 10.1016/j.jmb.2005.04.042
Henneke, G., Gueguen, Y., Flament, D., Azam, P., Querellou, J., Dietrich, J.,et al. (2002). Replication factor C from the hyperthermophilic archaeon Pyrococcus abyssi does not need ATP hydrolysis for clamp-loading and contains a functionally conserved RFC PCNA-binding domain. J. Mol. Biol. 323, 795–810. doi: 10.1016/S0022-2836(02)01028-8
Ishino, Y., Komori, K., Cann, I. K., and Koga, Y. (1998). A novel DNA polymerase family found in Archaea. J. Bacteriol. 180, 2232–2236.
Kim, J. C., and Mirkin, S. M. (2013). The balancing act of DNA repeat expansions. Curr. Opin. Genet. Dev. 23, 280–288. doi: 10.1016/j.gde.2013.04.009
Kroutil, L. C., Register, K., Bebenek, K., and Kunkel, T. A. (1996). Exonucleolytic proofreading during replication of repetitive DNA. Biochemistry 35, 1046–1053. doi: 10.1021/bi952178h
Kunkel, T. A., and Bebenek, K. (2000). DNA replication fidelity. Annu. Rev. Biochem. 69, 497–529. doi: 10.1146/annurev.biochem.69.1.497
Lechner, R. L., Engler, M. J., and Richardson, C. C. (1983). Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J. Biol. Chem. 258, 11174–11184.
López de Saro, F. J. (2009). Regulation of interactions with sliding clamps during DNA replication and repair. Curr. Genomics 10, 206–215. doi: 10.2174/138920209788185234
Lovett, S. T. (2004). Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 52, 1243–1253. doi: 10.1111/j.1365-2958.2004.04076.x
Lyons-Darden, T., and Topal, M. (1999). Effects of temperature, Mg2+ concentration and mismatches on triplet- repeat expansion during DNA replication in vitro. Nucl. Acids Res. 27, 2235–2240. doi: 10.1093/nar/27.11.2235
Madsen, C. S., Ghivizzani, S. C., and Hauswirth, W. W. (1993) In vivo and in vitro evidence for slipped mispairing in mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 90, 7671–7675. doi: 10.1073/pnas.90.16.7671
McMurray, C. T. (1999). DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl. Acad. Sci. U.S.A. 96, 1823–1825. doi: 10.1073/pnas.96.5.1823
Michel, B. (2000). Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25, 173–178. doi: 10.1016/S0968-0004(00)01560-7
Mirkin, S. M. (2007). Expandable DNA repeats and human disease. Nature 447, 932–940. doi: 10.1038/nature05977
Moxon, E. R., Rainey, P. B., Nowak, M. A., and Lenski, R. E. (1994). Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4, 24–33. doi: 10.1016/S0960-9822(00)00005-1
O’Donnell, M., Langston, L., and Stillman, B. (2013). Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 5, pii: a010108. doi: 10.1101/cshperspect.a010108
Palud, A., Villani, G., L’Haridon, S., Querellou, J., Raffin, J. P., and Henneke, G. (2008). Intrinsic properties of the two replicative DNA polymerases of Pyrococcus abyssi in replicating abasic sites: possible role in DNA damage tolerance? Mol. Microbiol. 70, 746–761. doi: 10.1111/j.1365-2958.2008.06446.x
Pearson, C. E., Nichol Edamura, K., and Cleary, J. D. (2005). Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6, 729–742. doi: 10.1038/nrg1689
Perales, C., Cava, F., Meijer, W. J., and Berenguer, J. (2003). Enhancement of DNA, cDNA synthesis and fidelity at high temperatures by a dimeric single-stranded DNA-binding protein. Nucl. Acids Res. 31, 6473–6480. doi: 10.1093/nar/gkg865
Raymann, K., Forterre, P., Brochier-Armanet, C., and Gribaldo, S. (2014). Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in Archaea. Genome Biol. Evol. 6, 192–212. doi: 10.1093/gbe/evu004
Richards, R. I., and Sutherland, G. R. (1992). Dynamic mutations: a new class of mutations causing human disease. Cell 70, 709–712. doi: 10.1016/0092-8674(92)90302-S
Rouillon, C., Henneke, G., Flament, D., Querellou, J., and Raffin, J. P. (2007). DNA Polymerase Switching on Homotrimeric PCNA at the Replication Fork of the Euryarchaea Pyrococcus abyssi. J. Mol. Biol. 369, 343–355. doi: 10.1016/j.jmb.2007.03.054
Samadashwily, G. M., Gordana, R., and Mirkin, S. M. (1997). Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 17, 298–304. doi: 10.1038/ng1197-298
Shah, S. N., Hile, S. E., and Eckert, K. A. (2010). Defective mismatch repair, microsatellite mutation bias, and variability in clinical cancer phenotypes. Cancer Res. 70, 431–435. doi: 10.1158/0008-5472.CAN-09-3049
Sinden, R. R., Pytlos-Sinden, M. J., and Potaman, V. N. (2007). Slipped strand DNA structures. Front. Biosci. 12:4788–4799. doi: 10.2741/2427
Soengas, M. S., Esteban, J. A., Lazaro, J. M., Bernard, A., Blasco, M. A., Salas, M.,et al. (1992). Site-directed mutagenesis at the Exo III motif of phi 29 DNA polymerase; overlapping structural domains for the 3’-5’ exonuclease and strand-displacement activities. EMBO J. 11, 4227–4237.
Tran, H. T., Degtyareva, N. P., Koloteva, N. N., Sugino, A., Masumoto, H., Gordenin, D. A.,et al. (1995). Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol. Cell. Biol. 15, 5607–5617.
Trinh, T. Q., and Sinden, R. R. (1993). The influence of primary and secondary DNA structure in deletion and duplication between direct repeats in Escherichia coli. Genetics 134, 409–422.
Viguera, E., Canceill, D., and Ehrlich, S. D. (2001a). Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 20, 2587–2595. doi: 10.1093/emboj/20.10.2587
Keywords: slippage, primer-template misalignment, DNA polymerases, strand displacement activity, Archaea
Citation: Castillo-Lizardo M, Henneke G and Viguera E (2014) Replication slippage of the thermophilic DNA polymerases B and D from the Euryarchaeota Pyrococcus abyssi. Front. Microbiol. 5:403. doi: 10.3389/fmicb.2014.00403
Received: 30 May 2014; Accepted: 17 July 2014;
Published online: 07 August 2014.
Edited by:
Zvi Kelman, University of Maryland, USAReviewed by:
Juergen Reichardt, James Cook Univerrsity, AustraliaBernard Connolly, University of Newcastle, UK
Copyright © 2014 Castillo-Lizardo, Henneke and Viguera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Enrique Viguera, Departamento de Biología Celular, Genética y Fisiología, Facultad de Ciencias, Universidad de Málaga, Campus Universitario de Teatinos, 29071, Málaga, Spain e-mail: eviguera@uma.es
 Melissa Castillo-Lizardo
Melissa Castillo-Lizardo Ghislaine Henneke
Ghislaine Henneke Enrique Viguera
Enrique Viguera