- 1School of Freshwater Sciences, University of Wisconsin-Milwaukee, Milwaukee, WI, USA
- 2Laboratorio de Patología de Organismos Acuáticos y Biotecnología Acuícola, Facultad de Ciencias, Universidad Andrés Bello, Viña del Mar, Chile
- 3Interdisciplinary Center for Aquaculture Research (INCAR), Concepción, Chile
- 4Unit of Microbiology, Department of Basic Health Sciences, School of Medicine and Health Sciences, Institut d'Investigació Sanitaria Pere Virgili, University Rovira i Virgili, Reus, Spain
Arcobacter species are highly abundant in sewage where they often comprise approximately 5–11% of the bacterial community. Oligotyping of sequences amplified from the V4V5 region of the 16S rRNA gene revealed Arcobacter populations from different cities were similar and dominated by 1–3 members, with extremely high microdiversity in the minor members. Overall, nine subgroups within the Arcobacter genus accounted for >80% of the total Arcobacter sequences in all samples analyzed. The distribution of oligotypes varied by both sample site and temperature, with samples from the same site generally being more similar to each other than other sites. Seven oligotypes matched with 100% identity to characterized Arcobacter species, but the remaining 19 abundant oligotypes appear to be unknown species. Sequences representing the two most abundant oligotypes matched exactly to the reference strains for A. cryaerophilus group 1B (CCUG 17802) and group 1A (CCUG 17801T), respectively. Oligotype 1 showed generally lower relative abundance in colder samples and higher relative abundance in warmer samples; the converse was true for Oligotype 2. Ten other oligotypes had significant positive or negative correlations between temperature and proportion in samples as well. The oligotype that corresponded to A. butzleri, the Arcobacter species most commonly isolated by culturing in sewage studies, was only the eleventh most abundant oligotype. This work suggests that Arcobacter populations within sewer infrastructure are modulated by temperature. Furthermore, current culturing methods used for identification of Arcobacter fail to identify some abundant members of the community and may underestimate the presence of species with affinities for growth at lower temperatures. Understanding the ecological factors that affect the survival and growth of Arcobacter spp. in sewer infrastructure may better inform the risks associated with these emerging pathogens.
Introduction
The genus Arcobacter, described by Vandamme et al. (1992), belongs to the family Campylobacteraceae within the epsilon-Proteobacteria. Arcobacter spp. were originally grouped within genus Campylobacter, but differ from campylobacters in their ability to grow under aerobic conditions and lower temperatures. The genus Arcobacter currently contains 18 species (Levican et al., 2013a; Sasi Jyothsna et al., 2013) isolated from diverse environments (water, plant roots, food) and hosts (humans, poultry, pigs, shellfish) (Collado and Figueras, 2011). Many Arcobacter species have been isolated from multiple locations, suggesting that these organisms are metabolically flexible and can survive under an array of environmental conditions.
Three Arcobacter species, A. butzleri, A. cryaerophilus, and A. skirrowii, have emerged in recent years as potential human pathogens (Collado and Figueras, 2011). Strains of A. butzleri and A. cryaerophilus in particular have been isolated from human stool and blood samples, and pathogenicity can range from diarrhea to bacteremia (Figueras et al., 2014). Some A. butzleri isolates contain a suite of virulence genes (cadF, ciaB, cj1349, hecA, hecB, irgA, mviN, pldA, and tlyA) (Douidah et al., 2012; Levican et al., 2013a) and can adhere to and invade Caco-2 cells (a gut epithelial cell line) in vitro (Levican et al., 2013a). The development of new DNA-based screening methods for clinical samples shows that arcobacters can often be mistaken for Campylobacter spp., and therefore, the potential human pathogenicity of these microbes is likely underestimated as is their role in water- and food-borne disease (Collado and Figueras, 2011; Figueras et al., 2014).
Studies of sewage and sewage-contaminated environmental waters reveal that Arcobacter spp. are often found in association with raw (untreated) sewage and even treated effluent water (Stampi et al., 1993; Collado et al., 2008, 2010; Cai et al., 2014). The species A. butzleri and A. cryaerophilus are the most commonly found in isolation studies, and appear to have high genetic diversity within species (Collado et al., 2008, 2010). The species A. defluvii and A. cloacae have been recently discovered in sewage samples (Collado et al., 2011; Levican et al., 2013b) as well. A culture-independent analysis of sewage using 454 pyrosequencing showed that Arcobacter populations accounted for approximately 4% of sewage bacterial communities, but had low diversity based on V6 pyrotag amplification (Vandewalle et al., 2012). The dominant V6 pyrotag also could not be mapped to a specific species, as this region has relatively low diversity among eight named arcobacters. So while Arcobacter appears to be an important component of sewage communities, relatively little is known about the diversity of these organisms or the ecological niche they may occupy in sewer infrastructure.
Here we provide an in-depth, DNA-based analysis of the Arcobacter community from 37 sewage samples collected in the US and Spain. The oligotyping approach sorted over 400,000 sequences into ecologically meaningful subgroups and allowed us to track changes in the Arcobacter populations across seasons and geography. Our findings reveal potential new species yet to be cultivated and temperature-based trends in the dominant organisms found in sewage.
Materials and Methods
Sample Collection and Processing
We selected a subset of sewage samples from a larger study that contained a complete set of metadata in order to better assess the ecological factors that contribute to the distribution of total Arcobacter and also individual species within and among sewage samples (Tables S1, S2). All sewage samples represent a single replicate taken from municipal wastewater treatment facilities: 36 primary influent (untreated) samples collected from 12 facilities in the US on three occasions (August 2012, January 2013, and April 2013) and one sample collected from Reus, Spain in September 2012 (Table S1). These samples represent a range of geographic location, regional climate, and seasonal variation (Table S2).
Technicians at the US sewage treatment plants shipped samples on ice within 24 h of collection to our laboratory in Milwaukee, WI, USA, for processing. A volume of 25 mL of sewage was filtered (0.22 μm, 47 mm S-Pak® Millipore® filters) for each sample and filters were stored at −80°C. The sample from Reus (Spain) was a composite sample collected overnight from 8:00 p.m. to 8:00 a.m. from the inflow of the WWTP of this city. This sample was immediately taken to the laboratory at the Medical School in Reus where it was filtered. DNA was extracted following the protocol described below provided by the Milwaukee laboratory. The DNA was shipped on ice the same day to Milwaukee.
DNA Extraction, Amplicon Sequencing, and Bioinformatic Processing
We extracted DNA as previously described (Newton et al., 2013). Briefly, the FastSpin Soil DNA kit (MP Biomedicals, Santa Ana, CA) was employed according to the manufacturer's instructions using the material contained in the crushed filters. The DNA purity and concentration was assessed using the NanoDrop® spectrophotometer (Thermo Scientific, Waltham, MA) and by performing an electrophoresis in 1% TAE agarose gel.
The Josephine Bay Paul Center at the Marine Biological Laboratories in Woods Hole, MA, provided Illumina amplicon sequencing. Primers amplified the V4V5 region of the bacterial 16S rRNA gene, and the Illumina MiSeq platform produced the sequence reads. Primers, sequencing protocols, quality control measures, and bioinformatic trimming procedures for Illumina MiSeq are described in detail elsewhere (Morrison et al., 2013). The Global Alignment for Sequence Taxonomy (GAST) software (Huse et al., 2008) assigned taxonomy to our high-quality reads; this study uses only the sequences that mapped to the genus Arcobacter. The sequences obtained in this study are available in the National Center for Biotechnology Information (NCBI) Short Read Archive under accession number SRP047513.
Oligotyping
GAST taxonomic classification of sequence reads (Huse et al., 2008) to the genus Arcobacter resulted in 408,878 sequences for oligotyping. We implemented the oligotyping pipeline (Eren et al., 2013) to determine ecologically relevant sequence groupings. Gap characters added to the ends of shorter sequences produced sequences of equal length that are required by the analysis pipeline. The “entropy-analysis” script in the oligotyping pipeline calculated the Shannon entropy at each nucleotide along the length of the sequences. The Shannon entropy provides a measure of nucleotide variation at a given position; sites that have A, G, C, and T present in approximately equal proportions among sequences have the highest entropy values, whereas highly conserved sites have a minimum entropy value near zero. Starting with the highest entropy positions along the length of the sequence, we selected 31 positions (4, 41, 54, 55, 56, 57, 58, 65, 70, 78, 85, 105, 112, 114, 115, 118, 120, 128, 130, 133, 159, 203, 212, 226, 250, 287, 301, 308, 332, 335, 343) over a read length of 375 nucleotides until entropy peaks were eliminated in individual oligotypes. We required each oligotype to have a minimum substantive abundance (−M, the abundance of the dominant sequence representing the oligotype) of 408 in order to reduce noise in the dataset and to focus our analysis on the more abundant oligotypes. Oligotyping with no noise filtering produced over 3800 oligotypes; elimination of oligotypes with a minimum substantive abundance of less than 408 reads (equivalent to 0.1% of the total abundance of Arcobacter sequence reads in the dataset) resulted in 26 oligotypes. Over 90% of sequences (372,028) were retained in the final analysis; discarded sequences were distributed evenly across samples.
Statistical Analyses
We used the vegan (Oksanen et al., 2013) and stats packages in R (R Development Core Team, 2012) for statistical analyses. Hierarchical clustering and non-metric multi-dimensional scaling (NMDS) analyses were based on Bray-Curtis dissimilarities, using oligotype matrix-count data as input. We determined the influence of environmental parameters on clusters produced by NMDS using permutation analysis of variance (ADONIS in the vegan package) with 999 permutations. The non-parametric Spearman rho correlation coefficients and corresponding p values (cor.test in the stats package) were used to determine the relationships between temperature and oligotype proportion.
Phylogenetic Analyses
ClustalW in the MEGA5 package (Tamura et al., 2011) aligned DNA sequences. The 16S rRNA sequences of Arcobacter reference strains represented cultivated species (Levican et al., 2013b; Sasi Jyothsna et al., 2013). We aligned the oligotype representative sequences (375 bp in length) to nearly full-length reference sequences, then trimmed to 375 bases for phylogenetic analysis. The V4V5 amplicons overlap the reference sequences from nucleotides 544–928 based on E. coli numbering. The Jukes-Cantor method estimated evolutionary distances, and we generated 1000 replicate trees using the Neighbor-Joining algorithm. Campylobacter jejuni served as an outgroup.
Results
Sample Environmental Factors and Distribution of Oligotypes Among Samples
The percentage of the total bacterial community that mapped to the genus Arcobacter ranged between 0.8 and 19.6% for most samples, and 27/37 samples had more than 5% Arcobacter. Two outlier samples contained 73 and 85% Arcobacter (Figure 1A). Over 40,000 unique sequences were present in the set of 408,878 total Arcobacter sequences. Alignments of the nearly complete (>1400 bp) 16S rRNA gene of Arcobacter reference strains (Figure S1) allowed the calculation of the Shannon entropy within the V4V5 region compared to the other variable regions; entropy within the V4V5 amplicon sequences is shown as well. The V4V5 region of Arcobacter reference sequences contains many high entropy nucleotide positions, although fewer than in the V2 region. Amplicons have similar high entropy nucleotide positions, but very low entropy also occurs uniformly across the length of the amplicon sequences as well. Resolution of high entropy positions produced 26 relatively abundant oligotypes (Figure 1B), while the low entropy nucleotide positions represent the high number of unique sequences that derive from microdiversity within the genus but also from sequencing noise. All 26 oligotypes in the noise-filtered analysis had a significant relative proportion (>0.9%) in at least one sample and appeared across different treatment plants and from different collection dates (Figure 1B). Oligotypes are numbered based on their total abundance rank within the dataset (i.e., Oligotype 1 had the highest overall abundance). The sewage Arcobacter communities from different sites were dominated by 1–3 oligotypes, with extremely high microdiversity in the minor members (represented by the noise-filtered sequences). Overall, the nine most abundant oligotypes within the Arcobacter genus accounted for >80% of the total Arcobacter sequences in all samples analyzed. The US samples, which were all untreated sewage, contained ~20 oligotypes (19.8 average, 19.5 median), but the Spain sample had only 11 oligotypes, all of which were also found in US samples. In Figure 1B, the oligotype distribution within samples is shown with samples grouped by average site temperature and by sample date within each site. Overall, oligotype distribution showed more similar patterns within sites (ADONIS r2 = 0.712, p < 0.001), but also appeared to have trends that corresponded to sample temperatures.
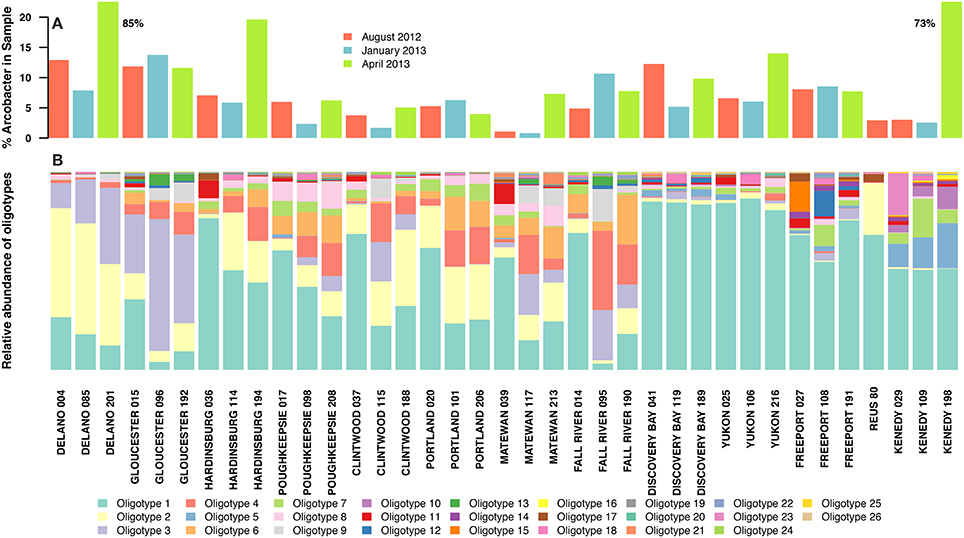
Figure 1. (A) Proportion of sequence reads in each sewage sample that mapped to the genus Arcobacter. Samples are color coded by the dates they were sampled: red = August 2012 (September 2012 for the Reus sample), blue = January 2013, green = April 2013. Two samples were outliers with significantly higher Arcobacter percentages than the rest of the samples; their values are shown as text next to the bars. (B) Proportions of 26 abundant oligotypes generated from sequence reads that mapped to the genus Arcobacter using the oligotyping pipeline. Samples are grouped by site, ordered from coldest to warmest average site temperature, then by sample collection date within sites. The legend shows the colors that represent each oligotype in (B); oligotypes are numbered based on the rank of their abundance summed over the whole dataset.
Temperature Dynamics of Arcobacter Oligotypes
Figure 2A shows a hierarchical clustering analysis based on Bray Curtis dissimilarities of Arcobacter population compositions as described by oligotypes. Arcobacter populations divided on the basis of the sample temperature, with the division occurring at temperatures higher or lower than 20°C. Samples of similar temperatures taken from the same site also tended to group together. Sample temperatures in the “warm” cluster ranged from 20 to 29.5°C, with two outliers that were 17°C (Figure 2B). The “cool” cluster sample temperatures ranged from 9.8 to 19.8°C, with one outlier at 21.2°C.
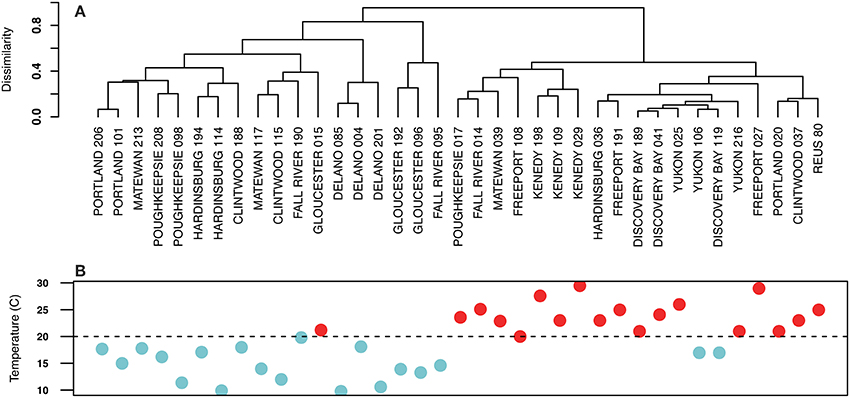
Figure 2. (A) Hierarchical clustering of sewage samples based on Bray Curtis dissimilarities of Arcobacter populations. Samples clustered into two distinct groups that were related to the temperature of the sample. The clade of samples on the left was composed of lower temperature samples than the clade on the right. (B) Temperatures of the sewage samples in (A) are shown below the sample name. Blue dots signify samples with temperatures below 20°C (indicated by the dashed line); red dots represent sample temperatures ≥20°C.
Oligotype proportion in samples was significantly correlated to temperature for 12 of the 26 oligotypes. Eight of these tended to have a higher proportion with higher temperature (positively correlated), and 4 negatively correlated with higher temperature. The Spearman correlation coefficients and p values for all oligotypes are shown in Table S3. No other metadata (total suspended solids, biochemical oxygen demand, total nitrogen, total phosphorus, population size, or average daily flow) had a significant correlation to the proportion of a given oligotype present in a sample (data not shown). The two most abundant oligotypes, which are denoted as “Oligotype 1” and “Oligotype 2” displayed opposing dynamics that coincided with the temperature of a sample; i.e., they correlated positively and negatively with temperature, respectively (Figure 3). While Oligotype 1 had by far the highest proportion overall and made up over 50% of almost all the high temperature samples, its relative abundance was notably lower in most lower temperature samples. Oligotype 1 accounted for >80% of the Arcobacter sequences represented by oligotypes in the two 17°C samples (Discovery Bay-119 and Yukon-106) that grouped with the “warm” cluster, while the 21.2°C sample from the “cool” cluster (Gloucester-015) had <50% Oligotype 1.
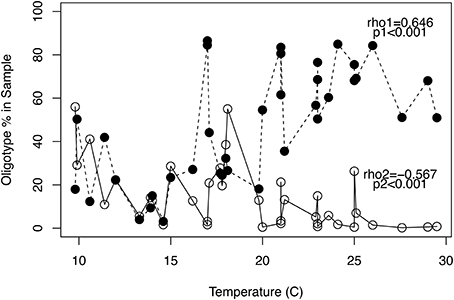
Figure 3. Changes in oligotype proportions with temperature. The two dominant oligotypes based on their relative abundance across all samples showed opposing dynamics with changes in temperature. The proportion of Oligotype (•) increased at temperatures >20°C, while Oligotype (°) proportions decreased above 20°C. The non-parametric correlation coefficient (Spearman's rho) and significance values for the relationships between oligotype proportion and temperature are shown by their respective oligotype. Temperature-proportion correlation coefficients and p values for all 26 oligotypes are given in Table S3.
Arcobacter Species Represented by Oligotypes
Seven of the sequences representing Arcobacter oligotypes shared 100% identity with previously characterized Arcobacter species based on BLAST comparison of sequences against the NCBI nucleotide database (Table S4). Oligotype 1 and Oligotype 2 matched exactly to A. cryaerophilus subgroups 1B and 1A, whose 16S rRNA genes differ in the V4V5 region by only a single nucleotide. Other oligotypes with 100% identity to the type strains of Arcobacter species included Oligotype 4 (A. suis), Oligotype 11 (A. butzleri), and Oligotype 14 (A. ellisii), and Oligotype 17 (A. cibarius). Oligotype 23 shared 100% identity with strains of two different species (A. cloacae and A. defluvii) that were identical in V4V5 region. The majority of oligotypes had no exact matches to the type strains or other strains within cultivated species, although all were at least 98% similar to a characterized Arcobacter species. Figure 4 shows the phylogenetic groupings of the sequences representing the 26 oligotypes in relation to known Arcobacter species. Oligotype 3 formed a distinct new clade along with Oligotypes 12 and 13, and Oligotypes 5, 10, 16, and 25 also formed a clade that might represent new species. However, as the phylogenetic analysis was limited to only 375 nucleotides, the groupings of the oligotypes are more illustrative than definitive; full-length sequences would be needed to confirm the true phylogenetic relationships.
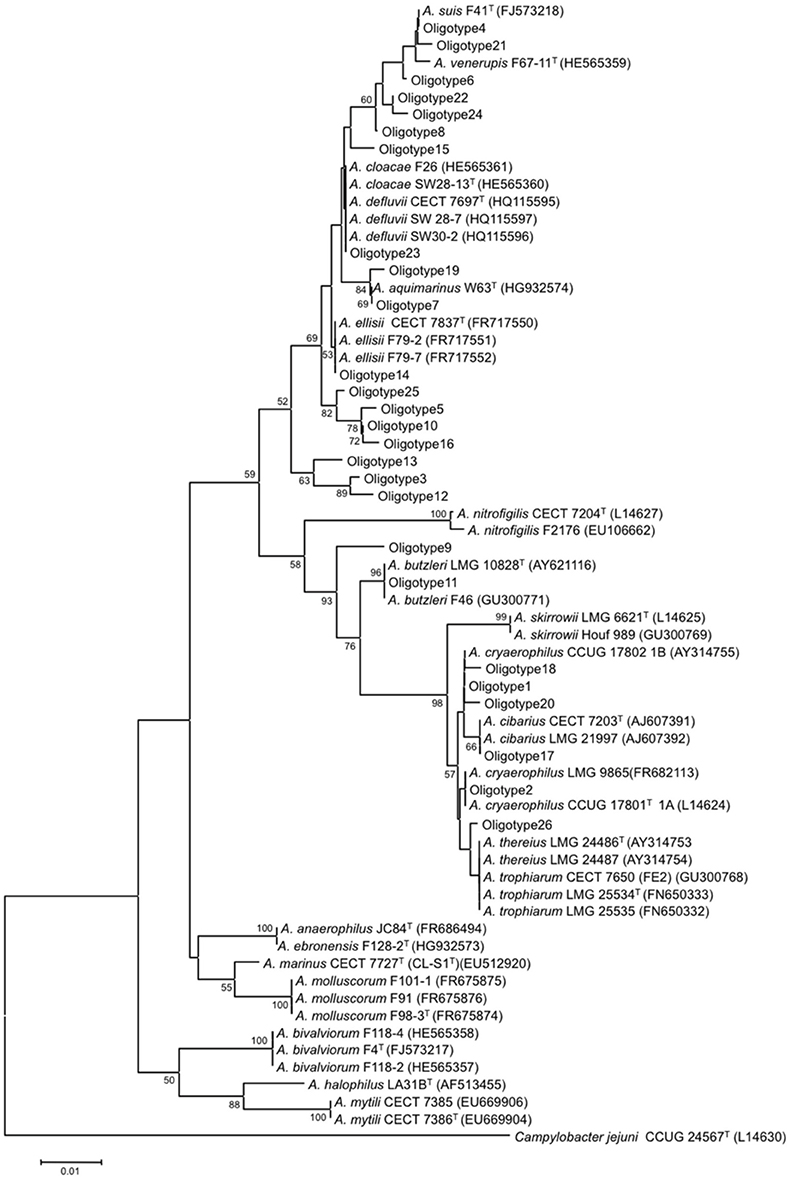
Figure 4. Neighbor-joining tree of Arcobacter reference strains and sewage oligotypes. Evolutionary distances were estimated using the Jukes-Cantor algorithm. This tree represents a composite of one thousand replicate trees; branches that occurred >50% of the time are noted at the nodes. The scale bar represents 1% nucleotide substitution. Campylobacter jejuni served as the outgroup.
Discussion
Oligotyping Discerns Ecologically Relevant Patterns within the Genus Arcobacter
In our study, different Arcobacter species were present in higher numbers depending on the sample temperature, similar to a previous study from estuarine water using conventional culturing methods and genetic identification of the isolates (Levican et al., 2014). For instance, Oligotype 11 (A. butzleri) was present in almost all samples collected during August but only in a few samples collected during January or April (Figure 1B) and correlated positively with higher environmental temperatures (Table S3). Along the same lines, Levican et al. (2014) observed a seasonal distribution of the species A. butzleri with a significantly higher recovery during summer. Moreover, in the later study the species A. cryaerophilus, A. skirrowii, and A. nitrofigilis were only isolated from environmental samples when water temperatures were lower (from 7.9°C to 18.2°C); however, the low number of strains recovered did not allow significant correlations to be made between species and either the water temperature or with the culturing approach (Levican et al., 2014). Conversely, the larger dataset used for oligotyping in the present study allowed us to infer a significant correlation between 12 and 26 oligotypes and the environmental temperature.
The fact that the proportions of the most abundant oligotypes varied by site and by temperature suggests that while a set group of organisms may be adapted to the ecological niche represented by a locale, changes in the environment within that system may favor different species at different times. Sewer systems appear to supply a unique niche where Arcobacter species thrive. Multiple samples taken from the different WWTPs demonstrate the consistency of community members at each site, but also the seasonal dynamics within populations that occur (Vandewalle et al., 2012). Dominant oligotypes were consistently present in samples collected from the same site, which may indicate a kind of ecological adaptation to general regional conditions such as climate, or more specifically, to the conditions found in particular sewerage systems.
Abundant Oligotypes Represent Both Characterized and Uncultivated Species
Oligotyping is often used to compare samples from sites that have obvious ecological differences and where one might expect differentiation of populations based on environmental influences (Eren et al., 2013; Reveillaud et al., 2014). We also used oligotyping to assess the relevant nucleotide signature positions that determine speciation (Eren et al., 2013). Operational Taxonomic Unit analysis can group sequences at a fine level (>97–99% similarity), but the 16S rRNA sequences of known Arcobacter species do not vary by a fixed percent. By using changes in the evolutionarily relevant nucleotide positions, we were able to identify significant groupings within the Arcobacter genus regardless of the overall degree of sequence similarity. This analysis showed that there are a limited number of dominant ecotypes, despite the high microdiversity within Arcobacter sequences.
In a previous study of sewage samples using 454 pyrosequencing of the 60 bp V6 region, a single dominant sequence comprised >80% of the sewage reads that mapped to Arcobacter (Vandewalle et al., 2012). The reduced diversity observed in these samples could either be due to a single dominant strain (as was observed for Discovery Bay samples), or because the V6 region is highly conserved among several Arcobacter species. The sequence of the dominant V6 pyrotag had a 100% match to A. cibarius, A. cloacae, A. cryaerophilus, A. defluvii, A. skirrowii, A. suis, and A. venerupis (data not shown). The genetic information contained within the V4V5 amplicons in this study vs. the V6 region in the previous study allowed better resolution of these reference strains from each other, but still failed to resolve some species (e.g., A. trophiarum and A. thereius). Amplicon sequencing of the V2 region (which has the highest diversity among named Arcobacter species) might therefore reveal even greater Arcobacter diversity in sewage, but more importantly, might better resolve the most abundant ecotypes and clarify their relation to known species.
Dominant oligotypes in the sewage samples examined here share 16S rRNA gene sequence identity with named species or ecotypes as well as yet to be characterized, possible new species. Several sewage oligotypes map to arcobacters cultivated from diverse sources: e.g., A. defluvii and A. cloacae were originally isolated from sewage (Collado et al., 2011; Levican et al., 2013b); A. venerupis, A. suis, and A. ellisii are associated with food products (Figueras et al., 2011; Levican et al., 2012; Hausdorf et al., 2013); and A. cryaerophilus and A. butzleri are found in diseased animals including humans (Collado and Figueras, 2011).
At least five different groups, each containing multiple oligotypes, appear to represent new uncultivated clades. Eight of the fifteen most abundant oligotype representative sequences had no exact match, and two had closest matches to uncharacterized environmental isolates. The third most abundant had only a 98% match to the closest named species and 100% shared identity with an environmental (non-sewage) isolate. Public sequence databases such as NCBI also contain many 16S rRNA gene sequences of uncultivated and not-yet-described Arcobacter species, some of which come from activated sludge and sewage (Collado and Figueras, 2011). Our results demonstrate that oligotyping can be used as a highly reproducible alternative to other sequence grouping methods to elucidate the population diversity of species within a given genus and may even enable recognition of new species.
The two most abundant oligotypes in this study, Oligotype 1 and Oligotype 2, corresponded with 100% match to two different subgroups of the species A. cryaerophilus (subgroups 1B and 1A, respectively). Isolates from subgroup 1B are the second most commonly isolated Arcobacter species after A. butzleri using traditional culturing and molecular identification methods, but subgroup 1A is rarely recovered in this manner (Collado and Figueras, 2011). Conversely, A. butzleri, the predominant species typically recovered from sewage by culturing in other studies (González et al., 2007; Collado et al., 2008, 2010) was only represented by the eleventh most abundant oligotype. The predominance of A. butzleri by culturing could be due to the fact that most of studies include an enrichment step that favors the growth of this species; however, this assertion still needs to be experimentally verified (Levican et al., 2014).
Implications for Culturing and Identification of New Arcobacter Species
Conventional media and isolation conditions (i.e., incubation temperature, atmosphere, etc.) for the recovery of Arcobacter are good for isolating certain species, particularly A. butzleri (Collado et al., 2010). However, many uncultured strains appear to be present in sewage as well that are missed by currently used methods (Collado and Figueras, 2011). Regarding the prevalence of A. cryaerophilus in different studies, subgroup 1B is much more prevalent than 1A, while both groups have so far been isolated simultaneously only from food products and from animal and human clinical samples (Collado and Figueras, 2011 and references therein). The A. cryaerophilus strains isolated from sewage thus far have all belonged to subgroup 1B. It is not clear whether this higher prevalence of subgroup 1B is a consequence of the isolation methods used, or due to specific adaptations of these species to different ecologic niches as observed in the present study.
Different culturing methods and incubation conditions can impact the prevalence and diversity of Arcobacter spp. recovered from different sources (food, water, sewage, blood) (Houf et al., 2002; Levican et al., 2014). In fact, the use of direct culturing in parallel to post enrichment cultivation allowed the discovery of the species A. defluvii and A. cloacae from sewage samples (Collado et al., 2011; Levican et al., 2013b). As previously noted, the large numbers of unclassified sequences indicate that many more potential new species reside in sewage that have yet to be isolated (Collado and Figueras, 2011). Knowing which species or ecotypes grow best at different temperatures may assist in cultivating underrepresented members of the sewage community. Future studies examining both the genetic potential (through genome sequencing) and phenotypic behavior of isolates will help to better determine how these organisms grow and thrive in the sewer systems and how they may impact human health. We lack a full understanding of how the Arcobacter spp. in sewage relate to the arcobacters known to be pathogenic to humans and animals. Comparison of sewage oligotypes to the 16S rRNA sequences of clinical isolates may be a first step in this direction, as identification of clinical isolates by sequencing becomes more routine practice (Prouzet-Mauléon et al., 2006; Collado and Figueras, 2011).
Oligotyping Tracks Arcobacter Population Dynamics
Almost all sewage samples grouped by hierarchical clustering based on Arcobacter community similarity separated at a breakpoint of 20°C, which (perhaps not coincidentally) is the temperature that delineates mesophilic bacteria from psychrophilic bacteria (Willey et al., 2008). Only three samples (Gloucester-August 2012, Yukon-January 2013, and Discovery Bay-January 2013) deviated from the group prescribed by their sample temperatures. In many of the sewage samples, the top two most abundant oligotypes had opposite temperature dynamics. Additionally, Oligotype 3, which may represent a new Arcobacter species and Oligotype 4 (corresponding to A. suis) were nearly absent from the warmest sites. Similar trends in seasonal/temperature-based variation for Acinetobacter populations were observed in sewage samples from Milwaukee, WI. The two most abundant Acinetobacter V6 pyrotags oscillated in abundance over the course of the year (Vandewalle et al., 2012), and relative proportions of other genera associated with sewage infrastructure (as opposed to the fecal component of sewage) also varied seasonally (Vandewalle et al., 2012).
It is difficult to ascertain how much variation in oligotype distribution is based strictly on sample temperature, as many other factors can contribute to population dynamics. However, observed trends based on both the temperature of the sample at the time of collection and the average site temperature (approximated as the mean of collected sample temperatures) suggest that temperature may contribute significantly to determining, at the very least, the relative proportions of Arcobacter species present (D'Sa and Harrison, 2005; Levican et al., 2014). Knowledge of Arcobacter population dynamics may lead to a better understanding of risks associated with environmental releases of these organisms in the case of combined/sanitary sewerage overflows (Ashbolt et al., 2010) and provide guidance for better management in food preparations (Van Driessche and Houf, 2008; Kjeldgaard et al., 2009).
Selective Growth of Arcobacter Species in Sewage
Arcobacters make up <0.001% of the human gut microbial community (Gevers et al., 2012; Koskey et al., 2014) but they make up a significant portion of sewage samples collected from geographically diverse locations (Shanks et al., 2013; Cai et al., 2014; Koskey et al., 2014). Since Arcobacter is found in the human gut, albeit in low proportions, humans may be the source of Arcobacter to sewerage systems; however, the sewer pipe environment appears to select for their survival and growth over more dominant gut bacteria, such as the Lachnospiraceae (McLellan et al., 2013). Three genera previously recognized to comprise a significant portion of sewage (Trichococcus, Acinetobacter, and Aeromonas) (Vandewalle et al., 2012) are also present, but in low relative abundance, in human feces (Gevers et al., 2012; Koskey et al., 2014). Thus, the specific ecological conditions present in sewage infrastructure, generally speaking, provide an ideal niche for certain organisms, not only to thrive, but also to maintain diversity within their own populations.
The factors contributing to the survival and growth of different arcobacters are of interest because some species have been identified as emerging human pathogens (Prouzet-Mauléon et al., 2006; Collado and Figueras, 2011; Figueras et al., 2014). Species sharing genetic similarity to pathogenic strains have also been isolated from environmental waters impacted by sewage inputs (Fong et al., 2007; Collado et al., 2010) or detected in impacted waters by molecular analyses (Collado et al., 2008; Lee et al., 2012), suggesting that at least some sewage-based Arcobacter species are viable after sewage releases. These organisms can survive or grow at temperatures found in sewerage systems, a range of environmental water temperatures (Levican et al., 2014), and laboratory isolation conditions, which may be ≥30°C (Stampi et al., 1993; Prouzet-Mauléon et al., 2006; Levican et al., 2014). Thus, although different Arcobacter species may have relatively high or low optimum growth temperatures, many seem to have a wide range of survival temperatures (D'Sa and Harrison, 2005; Van Driessche and Houf, 2008). If pathogenic Arcobacter strains also possess this trait, they may pose a threat even at very low abundance. These findings further stress the need to better understand the genetic crossovers between human and animal pathogenic strains, sewage ecotypes, and cultured isolates in order to ascertain risks associated with Arcobacter species.
The results of our study can be used as a general strategy for interpreting sequence data from populations of environmental bacteria, as oligotyping provides high resolution among species, even without full-length sequences. Here we show an approach that allows differentiation of known and unknown species, and also provides information on how environmental factors can modulate the presence and relative abundance of different ecotypes. Oligotyping may provide the means to establish the links between Arcobacter communities from food production, water sources, sewage, and diseased humans and animals, in order to better discern patterns of survival, growth, and infection.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by a grant to Sandra L. McLellan from the National Institutes of Health (1R01-AI091829) and was also supported in part by funding to María José Figueras from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement 311846 and by project AGL2011-30461-C02-02 by the Ministerio de Ciencia e Innovación (Spain). Arturo Levican was supported by a doctoral grant from the Universitat Rovira i Virgili (Spain), CONICYT (Chile) for the Postdoctoral research project FONDECYT 3140296, and by CONICYT/FONDAP grant number 15110027. The authors are solely responsible for the content of this publication. It does not represent the opinion of the European Commission. The European Commission is not responsible for any use that might be made of data appearing therein.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00525/abstract
References
Ashbolt, N. J., Schoen, M. E., Soller, J. A., and Roser, D. J. (2010). Predicting pathogen risks to aid beach management: the real value of quantitative microbial risk assessment (QMRA). Water Res. 44, 4692–4703. doi: 10.1016/j.watres.2010.06.048
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cai, L., Feng, J., and Zhang, T. (2014). Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl. Microbiol. Biotechnol. 98, 3317–3326. doi: 10.1007/s00253-013-5402-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Collado, L., and Figueras, M. J. (2011). Taxonomy, epidemiology and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 24, 174–192. doi: 10.1128/CMR.00034-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Collado, L., Inza, I., Guarro, J., and Figueras, M. J. (2008). Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environ. Microbiol. 10, 1635–1640. doi: 10.1111/j.1462-2920.2007.01555.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Collado, L., Kasimir, G., Perez, U., Bosch, A., Pinto, R., Saucedo, G., et al. (2010). Occurrence and diversity of Arcobacter spp. along the Llobregat River catchment, at sewage effluents and in a drinking water treatment plant. Water Res. 44, 3696–3702. doi: 10.1016/j.watres.2010.04.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Collado, L., Levican, A., Perez, J., and Figueras, M. J. (2011). Arcobacter defluvii sp. nov., isolated from sewage samples. Int. J. Syst. Evol. Microbiol. 61, 2155–2161. doi: 10.1099/ijs.0.025668-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Douidah, L., de Zutter, L., Baré, J., De Vos, P., Vandamme, P., Vandenberg, O., et al. (2012). Occurrence of putative virulence genes in Arcobacter species isolated from humans and animals. J. Clin. Microbiol. 50, 735–741. doi: 10.1128/JCM.05872-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
D'Sa, E., and Harrison, A. (2005). Effect of pH, NaCl content, and temperature on growth and survival of Arcobacter spp. J. Food Prot. 68, 18–25.
Eren, A. M., Maignien, L., Sul, W. J., Murphy, L. G., Grim, S. L., Morrison, H. G., et al. (2013). Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 4, 1111–1119. doi: 10.1111/2041-210X.12114
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Figueras, M. J., Levican, A., Collado, L., Isabel Inza, M., and Yustes, C. (2011). Arcobacter ellisii sp. nov., isolated from mussels. Syst. Appl. Microbiol. 34, 414–418. doi: 10.1016/j.syapm.2011.04.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Figueras, M. J., Levican, A., Pujol, I., Ballester, F., Rabada Quilez, M. J., and Gomez-Bertomeu, F. (2014). A severe case of persistent diarrhoea associated with Arcobacter cryaerophilus but attributed to Campylobacter sp. and a review of the clinical incidence of Arcobacter. New Microbes New Infect. 2, 31–37. doi: 10.1002/2052-2975.35
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fong, T. T., Mansfield, L. S., Wilson, D. L., Schwab, D. J., Molloy, S. L., and Rose, J. B. (2007). Massive microbiological groundwater contamination associated with a waterborne outbreak in Lake Erie, South Bass Island, Ohio. Environ. Health Perspect. 115, 856–864. doi: 10.1289/ehp.9430
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gevers, D., Knight, R., Petrosino, J. F., Huang, K., McGuire, A. L., Birren, B. W., et al. (2012). The human microbiome project: a community resource for the healthy human microbiome. PLoS Biol. 10:e1001377. doi: 10.1371/journal.pbio.1001377
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
González, A., Botella, S., Montes, R., Moreno, Y., and Ferrús, M. (2007). Direct detection and identification of Arcobacter species by multiplex PCR in chicken and wastewater samples from Spain. J. Food Prot. 70, 341–347.
Hausdorf, L., Neumann, M., Bergmann, I., Sobiella, K., Mundt, K., Froehling, A., et al. (2013). Occurrence and genetic diversity of Arcobacter spp. in a spinach-processing plant and evaluation of two Arcobacter-specific quantitative PCR assays. Syst. Appl. Microbiol. 36, 235–243. doi: 10.1016/j.syapm.2013.02.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Houf, K., De Zutter, L., Van Hoof, J., and Vandamme, P. (2002). Assessment of the genetic diversity among arcobacters isolated from poultry products by using two PCR-based typing methods. Appl. Environ. Microbiol. 68, 2172–2178. doi: 10.1128/AEM.68.5.2172-2178.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huse, S. M., Dethlefsen, L., Huber, J. A., Welch, D. M., Relman, D. A., and Sogin, M. L. (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. doi: 10.1371/journal.pgen.1000255
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kjeldgaard, J., Jørgensen, K., and Ingmer, H. (2009). Growth and survival at chiller temperatures of Arcobacter butzleri. Int. J. Food Microbiol. 131, 256–259. doi: 10.1016/j.ijfoodmicro.2009.02.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Koskey, A., Fisher, J., Eren, A., Ponce Terashima, R., Reis, M., Blanton, R., et al. (2014). Blautia and Prevotella sequences distinguish human and animal fecal pollution in Brazil surface waters. Environ. Microbiol. doi: 10.1111/1758-2229.12189. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, C., Agidi, S., Marion, J. W., and Lee, J. (2012). Arcobacter in Lake Erie beach waters: an emerging gastrointestinal pathogen linked with human-associated fecal contamination. Appl. Environ. Microbiol. 78, 5511–5519. doi: 10.1128/AEM.08009-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Levican, A., Alkeskas, A., Günter, C., Forsythe, S. J., and Figueras, M. J. (2013a). Adherence to and invasion of human intestinal cells by Arcobacter species and their virulence genotypes. Appl. Environ. Microbiol. 79, 4951–4957. doi: 10.1128/AEM.01073-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Levican, A., Collado, L., Aguilar, C., Yustes, C., Dieguez, A. L., Romalde, J. L., et al. (2012). Arcobacter bivalviorum sp. nov. and Arcobacter venerupis sp. nov., new species isolated from shellfish. Syst. Appl. Microbiol. 35, 133–138. doi: 10.1016/j.syapm.2012.01.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Levican, A., Collado, L., and Figueras, M. J. (2013b). Arcobacter cloacae sp. nov. and Arcobacter suis sp. nov., two new species isolated from food and sewage. Syst. Appl. Microbiol. 36, 22–27. doi: 10.1016/j.syapm.2012.11.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Levican, A., Collado, L., Yustes, C., Aguilar, C., and Figueras, M. J. (2014). Higher water temperature and incubation under aerobic and microaerobic conditions increase the recovery and diversity of Arcobacter spp. from shellfish. Appl. Environ. Microbiol. 80, 385–391. doi: 10.1128/AEM.03014-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McLellan, S. L., Newton, R. J., Vandewalle, J. L., Shanks, O. C., Huse, S. M., Eren, A. M., et al. (2013). Sewage reflects the distribution of human faecal Lachnospiraceae. Environ. Microbiol. 15, 2213–2227. doi: 10.1111/1462-2920.12092
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morrison, H., Grim, S., Vineis, J., and Sogin, M. L. (2013). 16S amplicon fusion primers and protocol for Illumina platform sequencing. figshare. doi: 10.6084/m9.figshare.833944. [Epub ahead of print].
Newton, R. J., Bootsma, M. J., Morrison, H. G., Sogin, M. L., and McLellan, S. L. (2013). A microbial signature approach to identify fecal pollution in the waters off an urbanized coast of Lake Michigan. Microb. Ecol. 65, 1011–1023. doi: 10.1007/s00248-013-0200-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., et al. (2013). Vegan: Community Ecology Package. R package version 2.0–8. Available online at: http://CRAN.R-project.org/package=vegan
Prouzet-Mauléon, V., Labadi, L., Bouges, N., Ménard, A., and Mégraud, F. (2006). Arcobacter butzleri: underestimated enteropathogen. Emerg. Infect. Dis. 12, 307–309. doi: 10.3201/eid1202.050570
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
R Development Core Team. (2012). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org
Reveillaud, J., Maignien, L., Eren, A. M., Huber, J. A., Apprill, A., Sogin, M. L., et al. (2014). Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 8, 1198–1209. doi: 10.1038/ismej.2013.227
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sasi Jyothsna, T. S., Rahul, K., Ramaprasad, E. V., Sasikala, C., and Ramana, C. V. (2013). Arcobacter anaerophilus sp. nov., isolated from an estuarine sediment and emended description of the genus Arcobacter. Int. J. Syst. Evol. Microbiol. 63, 4619–4625. doi: 10.1099/ijs.0.054155-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shanks, O. C., Newton, R. J., Kelty, C. A., Huse, S. M., Sogin, M. L., and McLellan, S. L. (2013). Comparison of the microbial community structures of untreated wastewaters from different geographic locales. Appl. Environ. Microbiol. 79, 2906–2913. doi: 10.1128/AEM.03448-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stampi, S., Varoli, O., Zanetti, F., and De Luca, G. (1993). Arcobacter cryaerophilus and thermophilic campylobacters in a sewage treatment plant in Italy: two secondary treatments compared. Epidemiol. Infect. 110, 633–639. doi: 10.1017/S0950268800051050
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vandamme, P., Vancanneyt, M., Pot, B., Mels, L., Hoste, B., Dewettinck, D., et al. (1992). Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skiwowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 42, 344–356.
Vandewalle, J. L., Goetz, G. W., Huse, S. M., Morrison, H. G., Sogin, M. L., Hoffmann, R. G., et al. (2012). Acinetobacter, Aeromonas and Trichococcus populations dominate the microbial community within urban sewer infrastructure. Environ. Microbiol. 14, 2538–2552. doi: 10.1111/j.1462-2920.2012.02757.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Driessche, E., and Houf, K. (2008). Survival capacity in water of Arcobacter species under different temperature conditions. J. Appl. Microbiol. 105, 443–451. doi: 10.1111/j.1365-2672.2008.03762.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: oligotyping, Arcobacter, sewage, population dynamics, V4V5, Illumina MiSeq
Citation: Fisher JC, Levican A, Figueras MJ and McLellan SL (2014) Population dynamics and ecology of Arcobacter in sewage. Front. Microbiol. 5:525. doi: 10.3389/fmicb.2014.00525
Received: 07 August 2014; Accepted: 21 September 2014;
Published online: 07 November 2014.
Edited by:
Lois Maignien, University of Western Brittany - Université Bretagne Occidentale, FranceReviewed by:
Raffaella Balestrini, Consiglio Nazionale delle Ricerche, ItalyBrett J. Baker, University of Texas Austin, USA
Copyright © 2014 Fisher, Levican, Figueras and McLellan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandra L. McLellan, School of Freshwater Sciences, University of Wisconsin-Milwaukee, 600 E. Greenfield Ave., Milwaukee, WI 53204, USA e-mail: mclellan@uwm.edu
 Jenny C. Fisher
Jenny C. Fisher Arturo Levican
Arturo Levican María J. Figueras
María J. Figueras Sandra L. McLellan
Sandra L. McLellan