- 1CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 2Departments of Medicine and Microbiology and Immunology, Albert Einstein College of Medicine, Bronx, NY, USA
- 3Beijing Key Laboratory of Microbial Drug Resistance and Resistome, Beijing, China
Hydrogen peroxide (H2O2) is one of a variety of reactive oxygen species (ROS) produced by aerobic organisms. Host production of toxic H2O2 in response to pathogen infection is an important classical innate defense mechanism against invading microbes. Understanding the mechanisms by which pathogens, in response to oxidative stress, mediate defense against toxic ROS, can reveal anti-microbial targets and shed light on pathogenic mechanisms. In this study, we provide evidence that a Mycobacterium smegmatis hemerythrin-like protein MSMEG_2415, designated MsmHr, is a H2O2-modulated repressor of the SigF-mediated response to H2O2. Circular dichroism and spectrophotometric analysis of MsmHr revealed properties characteristic of a typical hemerythrin-like protein. An msmHr knockout strain of M. smegmatis mc2155 (ΔmsmHr) was more resistant to H2O2 than its parental strain, and overexpression of MsmHr increased mycobacterial susceptibility to H2O2. Mutagenesis studies revealed that the hemerythrin domain of MsmHr is required for the regulation of the H2O2 response observed in the overexpression study. We show that MsmHr inhibits the expression of SigF (MSMEG_1804), an alternative sigma factor that plays an important role in bacterial oxidative stress responses, including those elicited by H2O2, thus providing a mechanistic link between ΔmsmHr and its enhanced resistance to H2O2. Together, these results strongly suggest that MsmHr is involved in the response of mycobacteria to H2O2 by negatively regulating a sigma factor, a function not previously described for hemerythrins.
Introduction
Hydrogen peroxide (H2O2) is a universal oxidative stress molecule produced by aerobic organisms from all three domains of life (Imlay, 2008). The production of H2O2 by the host in response to pathogen infection is also an important innate defense mechanism (Fang, 2004). This ROS can be damaging via direct toxic effects or disruption of redox balance, the latter being critical for metabolic homeostasis and hence survival. An important intracellular pathogen, Mycobacterium tuberculosis has evolved many strategies to detoxify H2O2, some of which are unique to mycobacteria (Kumar et al., 2011; Trivedi et al., 2012). For example, M. tuberculosis, which lacks the conventional redox buffer glutathione, uses mycothiol (MSH), a low-molecular-weight thiol that exists in millimolar quantities in the cytoplasm, to generate a reducing environment (Farhana et al., 2010); and MSH-deficient mycobacterial mutants are hyper-susceptible to H2O2(Rawat et al., 2002). The precise mechanisms by which MSH protects against oxidative stress and redox imbalance remain to be determined. M. tuberculosis also lacks classic sensors such as OxyR and SoxR for the detection of redox signals (Imlay, 2013). The tubercle bacillus, however, expresses DosS and DosT (Kumar et al., 2007), two sensor histidine kinases whose heme iron plays a critical role in their response to the levels of O2, nitric oxide (NO) and carbon monoxide (CO). Interaction with these various gasses activates the kinase activity of DosS and DosT, relaying the signals to the response regulator DosR. WhiB3 is an Fe-S cluster transcription factor that controls the expression of the hypoxia regulon of M. tuberculosis (Bhat et al., 2012). Indeed, accumulating evidence suggests that the M. tuberculosis WhiB family of Fe-S cluster proteins plays important roles in regulating a wide spectrum of microbial functions including responses to oxidative stress and virulence (Burian et al., 2012; Saini et al., 2012a). Together, the DosS-DosT/WhiB3 paradigm underscores the importance of iron-containing proteins in the response of M. tuberculosis to redox signals, including those imposed by reactive oxygen intermediates.
The non-heme, di-iron, O2-binding hemerythrin-like proteins are present in all domains of life (Bailly et al., 2008; French et al., 2008). Bioinformatics analyses have revealed over 400 hemerythrin-like proteins in available prokaryotic genomes (Bailly et al., 2008; French et al., 2008). These proteins harbor the conserved hemerythrin domain either singly or jointly with another distinct functional domain (Bailly et al., 2008; French et al., 2008). Experimental evidence suggests that they can function as oxygen sensors and reserves, as well as mediate the delivery and transport of this diatomic gas (French et al., 2008). Dcr (Desulfovibrio chemoreceptor) H was the first bacterial hemerythrin-like protein to be identified. It has been proposed that the C-terminal hemerythrin domain of DcrH, a member of the Dcr family of putative methyl-accepting chemotaxis proteins of the anaerobic sulfate-reducing bacterium Desulfovibrio vulgaris, functions to sense O2 (Xiong et al., 2000); the signal thus generated is proposed to be transduced to the transmitting domains to mediate chemotaxis (Xiong et al., 2000). The first single-domain hemerythrin-like protein was identified in Methylococcus capsulatus. The expression of the M. capsulatus hemerythrin-like protein is enhanced significantly with increasing concentrations of copper, and is thought to be an oxygen carrier that supplies copper-containing methane monooxygenase with oxygen (Kao et al., 2004; Karlsen et al., 2005; Chen et al., 2012). Despite the wide distribution of hemerythrin-like proteins in a wide variety of bacterial species, including M. tuberculosis (Xiong et al., 2000; Karlsen et al., 2005; Isaza et al., 2006; Justino et al., 2007; Onoda et al., 2011; Schaller et al., 2012), functional characterization studies are scarce.
Transcriptional regulation is critical to bacterial survival in response to various stresses. Sigma factors are the primary transcriptional regulators of bacterial gene expression. M. tuberculosis has 13 sigma factors, and SigH, SigE, SigL, and SigF play important roles in ROS detoxification (Rodrigue et al., 2006). SigF, a highly conserved sigma factor in the genus Mycobacterium (Rodrigue et al., 2006), is highly induced by various environmental stresses and during stationary phase (Demaio et al., 1996). In M. smegmatis, sigF deletion also increases susceptibility to oxidative stress (Gebhard et al., 2008; Humpel et al., 2010). Recently, a genome-wide gene expression study (Humpel et al., 2010) and work from our laboratory (Wu et al., 2012) have provided evidence that SigF regulates the expression of oxidative stress defense genes such as katA, dps1, and sodA, but not katG and ahpC, genes that have been linked to mycobacterial resistance to isoniazid (Silva et al., 2003). These studies suggest that SigF-mediated resistance to H2O2 is independent of KatG and AhpC. This notion is also supported by the fact that SigF-deficient mutants do not display decreased susceptibility to isoniazid (Demaio et al., 1996; Humpel et al., 2010). Regulation of SigF in mycobacteria is generally thought to be predominantly at the post-transcriptional level via the action of anti-sigma and anti-anti-sigma factors (Michele et al., 1999; Beaucher et al., 2002; Singh and Singh, 2008). SigF is transcribed from two promoters, Pmsmeg_1802 and Prbsw (Gebhard et al., 2008), and transcriptional reporter fusion studies have shown that promoter Pmsmeg_1802 responds to entry into the stationary phase and promoter Prbsw is inducible upon treatment with D-cycloserine (Gebhard et al., 2008).
In this study, to investigate the role of hemorythrin-like proteins in mycobacteria, we cloned, expressed, and characterized the M. smegmatis mc2155 strain MsmHr protein (encoded by msmeg2415). Using a genetic and biochemical approach, we show that (i) MsmHr displays circular dichroism (CD) and UV-vis spectrophotometric features typical of a hemerythrin-like protein; (ii) relative to wild-type bacilli, the msmHr knockout strain (ΔmsmHr) is more resistant to H2O2 and mc2155 overexpressing MsmHr exhibits enhanced H2O2 susceptibility; (iii) the H2O2 response is dependent on the hemerythrin domain; (iv) MsmHr represses sigF transcription through the promoter Prbsw, and thus participates in regulating the SigF-mediated H2O2 response. Our results indicate that MsmHr, the first mycobacterial hemorhythrin-like protein to be characterized, is involved in the H2O2 response in mycobacteria and provide insight into its mechanism.
Materials and Methods
Culture Medium and Growth Conditions
M. smegmatis cultures were grown in Middlebrook 7H9 medium (Becton Dickinson, Sparks, MD) supplemented with ADS enrichment (Albumin-Dextrose Saline containing 5% (w/v) Bovine serum albumin fraction V, 2% (w/v) D-Dextrose and 8.1% (w/v) NaCl) (Jacobs et al., 1991), 0.05% (v/v) Tween 80, and 0.5% (v/v) glycerol (Beijing Modern Eastern Finechemical Co. Ltd, Beijing). Middlebrook 7H10 medium supplemented with ADS enrichment and 0.5% (v/v) glycerol was used as the solid medium for examination of growth status. Growth was also examined in minimal Sauton's medium (4 g asparagine, 2 g sodium citrate, 0.5 g K2HPO4·3H2O, 0.5 g MgSO4·7H2O, 0.05 g ferric ammonium citrate, 60 g glycerol in 1 L of H2O supplemented with 0.05% (v/v) Tween 80) supplemented with antibiotics as indicated. Hygromycin (75 mg/L for M. smegmatis, 150 mg/L for Escherichia coli; Roche) and kanamycin (25 mg/L for M. smegmatis, 50 mg/L for Escherichia coli; Amresco) were added to the medium as needed. All bacterial strains used in this study are listed in Table S1.
Generation of Knockout Mutant Strains, Complementation Strains and Overexpression Strains
The msmHr (msmeg_2415) and sigF (msmg_1804) deletion mutants were generated via a specialized transducing phage delivery system as previously described (Bardarov et al., 2002). The 5'-flanking region of msmHr was amplified by polymerase chain reaction (PCR) with the 2415LL/2415LR primer pair and the 3'-flanking region of msmHr was amplified with the 2415RL/2415RR primer pair (all primers are listed in Supplemental Table S2. The flanking regions of sigF were generated by amplifying the upstream and downstream regions of sigF using the 1804LL/1804LR and 1804RL/1804RR primer pairs, respectively. Amplified fragments were ligated with plasmid p0004S, digested with PflMI (msmHr) or AlwnI (sigF), and allelic-exchange plasmids thus constructed were digested with PacI, and then ligated with PacI-digested phAE159. Phage packaging was performed using a MaxPlax packaging extract (Epicenter Biotechnologies, USA) to yield the knockout phages for msmHr (phAE-msmHr) and sigF (phAE-sigF). Specialized transduction was carried out as described previously (Bardarov et al., 2002). The knockout clones were screened by PCR using the primer pairs 2415InL/2415InR, 2415LLL/IL(R) and IR(F)/2415RRR for msmHr and 1804InL/1804InR, 1804LLL/IL(R), and IR(F)/1804RRR for sigF. Primer positions with respect to the appropriate genes are shown in Figure 2A and Figure S1. No msmHr or sigF mRNA was detected in the corresponding deletion strains by qRT-PCR using the appropriate primer pairs (Figure 2A and Figure S1). Complementation strains were constructed as described previously (Stover et al., 1991). Briefly, the full-length sequence of msmHr or sigF amplified from M. smegmatis genomic DNA was cloned into the integrating vector pMV361 (Stover et al., 1991) and the resultant plasmids were electroporated into the corresponding knockout strains to yield C-ΔmsmHr (ΔmsmHr::Phsp60-msmHr) and C-ΔsigF (ΔsigF::Phsp60-sigF). To over-express msmHr, the msmHr fragment was subcloned into pMV261 (Stover et al., 1991) to yield pMV261-msmHr for transformation into M. smegmatis (O-msmHr).
Determination of Survival Phenotypes Under Stress
Early phase cultures (OD600 = 0.3) of all tested strains were serially diluted (1:10) and spotted (3 μl) onto solid 7H10 medium supplemented with ADS enrichment and stress-inducing chemical agents (20 μM streptonigrin, 250 μM NaNO2) or subjected to low-pH stress (7H9 supplemented with ADS enrichment and 0.05% Tween 80, pH 5.5). For peroxide stress, early-phase cultures were treated with 5 mM H2O2 for 3 h, serially diluted (1:10) and spotted (3 μl) onto solid 7H10 medium supplemented with 10% ADS. The optical density at 600 nm (OD600) was measured at the indicated times in the presence of various stresses. Survival under heat shock stress at 50°C was determined by the number of colony forming units during the time indicated. Statistical analyses were performed using unpaired two-tailed t-tests. P-values are only shown where significant differences were found. *P < 0.05 and **P < 0.01.
Cloning, Expression and Purification of MsmHr in Escherichia coli
The coding sequence of msmHr was amplified from M. smegmatis mc2155 genomic DNA and cloned into the expression vector pET23b (+) (Novagen, USA), in-frame fused with a C-terminal 6xHis-tag sequence to generate the plasmid pET23b-msmHr, which was transformed into E. coli BL21 (AI) (Invitrogen, USA) for expression. Recombinant MsmHr was induced by incubation with 0.1% arabinose at 28°C for 3 h. Cells were harvested by centrifugation at 10,000 g for 5 min, resuspended in lysis buffer (20 mM Tris-HCl pH 8.0; 1 M NaCl, 10% (v/v) glycerol, 20 mM imidazole, 0.1% (v/v) Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mg/ml lysozyme) and lysed by sonication. Lysates were centrifuged at 12,000 g for 30 min at 4°C to remove debris before purification. The supernatants were incubated with Ni-NTA agarose (Qiagen, USA) with rotation (15 g) for 4 h at 4°C. Beads were then washed three times with washing buffer (20 mM Tris-HCl pH 8.0, 0.5 M NaCl, 10% (v/v) glycerol, 50 mM imidazole, 0.1% (v/v) Triton X-100, 1 mM PMSF, 25 mM MgCl2). The proteins were eluted with elution buffer (50 mM Tris-HCl pH 7.5, 0.5 M NaCl, 25 mM MgCl2, 10% (v/v) glycerol, 400 mM imidazole) and protein concentration was measured using the bicinchoninic acid protein assay reagent and a bovine serum albumin standard. Purified protein was examined using 12% sodium dodecyl sulfate polyacrylaminde gel electrophoresis to verify molecular weight and purity (Figure S2).
Spectrophotometric and Circular Dichroic (CD) Analysis of MsmHr
Purified MsmHr was diluted in 20 mM Tris-Cl buffer (pH 7.5). Deoxy samples were obtained by adding a 10-fold molar excess of Na2S2O4 to MsmHr. UV-Vis spectrophotometric spectra were obtained in 1 mm path length quartz cuvettes on a UV-2802H UV-Vis spectrophotometer (Unico Shanghai Instruments Co., Ltd., China). Spectra of deoxy-MsmHr were collected in an anaerobic incubator (Shanghai Yuejin Medicial Instruments Co., Ltd, China). CD measurements were performed using a Chirascan Circular Dichroism Spectrometer (Applied Photophysics Ltd. UK). The analysis software provided with the instrument was used for analysis of the results.
Determination of the Minimum Inhibitory Concentration of Isoniazid in M. smegmatis
The susceptibility of M. smegmatis to isoniazid (INH) was determined using the broth microdilution method (Wallace et al., 1986). After two-fold dilutions of INH in 7H9 supplemented with ADS enrichment and 0.5% (v/v) glycerol, 40-μl aliquots were mixed with 40 μl of M. smegmatis suspension (105 cells/ml) and deposited into wells of 96-well microtiter plates. The highest concentration of INH was 100 μg/ml. Plates were incubated at 37°C for 2 days and OD600 values of cultures were then measured using a microplate reader (FLUOstar OPTIMA, BMG Labtech). The minimum inhibitory concentration (MIC) was defined as the lowest concentration of drug that inhibited the visible bacterial growth of M. smegmatis after a 2-day incubation (OD600 < 0.05). INH susceptibility tests were repeated at least 3 times.
RNA Isolation, RT-PCR and Quantitative PCR
Log phase cultures (OD600 = 0.8–1.0) of all tested strains were diluted 1:100 in 7H9 media supplemented with ADS enrichment, 0.5% (v/v) glycerol and 0.05% (v/v) Tween 80. Strains were cultured until the OD600 reached 0.3 and then divided into control and treatment groups. In the treatment group, the cells were treated with 5 mM H2O2 for 30 min, and then collected by centrifugation at 12,000 g. Bacterial pellets were resuspended in TRIzol (Invitrogen, USA) and RNA was purified according to the manufacturer's instructions. cDNA was synthesized using the SuperScriptTM III First-Strand Synthesis System (Invitrogen, USA). Quantitative real-time PCR (qRT-PCR) was performed in a Bio-Rad iCycler using 2x SYBR real-time PCR pre-mix (Takara Biotechnology Inc., Japan). The following cycling program was used: 95°C for 1.5 min followed by 40 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 15 s, followed for 72°C for 6 min. M. smemgatis rpoD encoding RNA polymerase sigma factor SigA was selected as a reference to normalize gene expression. The 2−ΔΔCT method was used (Livak and Schmittgen, 2001) to evaluate the relative gene expression in different strains and/or different treatments. Primers used are listed in Table S2.
Construction of Promoter-lacZ Fusion Expression Vectors
SigF is reported to be transcribed from two promoters, Pmsmeg_1802 and Prbsw (Gebhard et al., 2008). To construct the Pmsmeg_1802-lacZ and PrbsW-lacZ plasmids, we used the reported primer pairs PsigFF/PsigFR and P1802F/P1802R (Gebhard et al., 2008), and PmsmHr-lacZ using P2415F/P2415R (Supplemental Table S2). The promoter sequences of sigF, msmeg_1802 and msmHr were then cloned into the upstream region of the lacZ gene in pLACZint (Vasudeva-Rao and McDonough, 2008), generating the plasmids PrbsW-lacZ, Pmsmeg_1802-lacZ and PmsmHr−lacZ, respectively.
β-Galactosidase (lacZ) Activity Assay
The β-galactosidase activity of various strains were determined as described previously (Gebhard et al., 2008). Briefly, cultures were collected at early logarithmic phase (OD600 ≈ 0.3) and resuspended in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 and 50 mM β-mercaptoethanol). Cells were then lysed using a Fastprep bead-beater (Biospec). The enzyme reaction was initiated by adding ortho-nitrophenyl-β-D-galactopyranoside (2 mg/ml) and terminated by adding Na2CO3(0.8 M). β-galactosidase activity was assessed by measuring the OD420 value of the reactions mixtures. Assays were performed in triplicate, and the Miller units (MU) were calculated as follows: 1000 × OD420/(OD600 of assayed culture × assayed volume × time).
Construction of msmHr Point Mutants
Mutation of specific amino acids was incorporated into the hemerythrin-like domain of msmHr in pMV261-msmHr by mismatched PCR primers (Table S2). Site-directed mutagenesis was performed following using a QuikChange® Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. Mutant clones were confirmed by DNA sequencing (BGI-Shenzhen, Shenzhen China). pMV261-msmHr mutants were transformed into M. smegmatis mc2155. Mutated sites are as follows (altered amino acids are in underlined bold font):
H1M: 55AVHETAEEMV65 → 55AVLETAAAMV65
H2M: 86EEHKAKQQLS96 → 86EELKAKAALS96
H3M: 122AAHEEAEEFV132 → 122AALEEAAAFV132
Results
MsmHr is a Bacterial Hemerythrin-Like Protein
Detoxification strategies for scavenging host immune defense system-derived H2O2 are important for the intracellular survival of mycobacterial pathogens. Expression of Fe-related proteins, such as the Fe-S cluster transcription factor WhiB3, and heme-iron sensors DosS and DosT, is an important strategy for regulating redox balance (Bhat et al., 2012). Hemerythrin proteins are iron-binding proteins known to be involved in oxygen transport and storage, but their biological functions in mycobacteria have yet to be elucidated. Three proteins in M. smegmatis, MsmHr, Msmeg_3312 and Msmeg_6612, are predicted to be hemerythrin-like proteins, however, as preliminary experiments indicated that only MsmHr is related to the H2O2 response (data not shown), we focused our attention on MsmHr.
We first constructed a multiple alignment of MsmHr with other hemerythrin-like proteins from different bacterial species (Figure 1A). Residues H24, H57, E61, H66, H88, H121, and E126 (numbering based on the MsmHr sequence) matched the characteristic motifs H… HxxxE … HxxxH… HxxxxD/E of hemerythrin domains. The secondary structure predicted by SWISS-MODEL (http://swissmodel.expasy.org/) (Guex and Peitsch, 1997; Schwede et al., 2003; Arnold et al., 2006; Kiefer et al., 2009) suggested that MsmHr has a typical hemerythrin structure with four α-helices (residues 16–40, 41–68, 80–103, and 104–132) (Figure 1B).
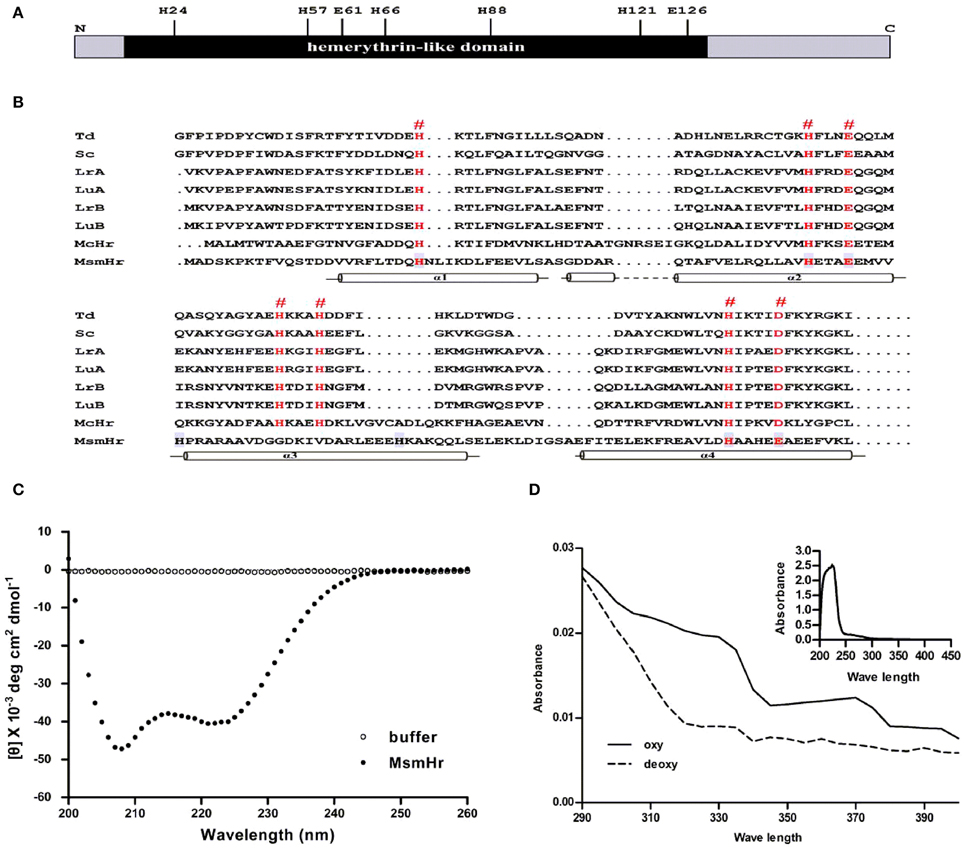
Figure 1. MsmHr is a hemerythrin-like protein. (A) Conserved residues of hemerythrin-like domain in MsmHr. H: histidine; E: glutamate. (B) MsmHr sequence analysis. Alignment of the hemerythrin-like domain of MsmHr with other hemerythrin family proteins. Hemerythrin sequences compared are from Themiste dyscrita (Td), Siphonosoma cumanense (Sc), Lingula reevii (α chain and β chain; LrA and LrB), and Lingula unguis (α chain and β chain; LuA and LuB), Methylococcus capsulatus (McHr). The predicted Fe2+ binding sites (#) are indicated in red font. Helices (α1, α2, α3, and α4) are indicated below the sequences. (C) CD spectral analysis of MsmHr. Blank control: 20 mM Tris-Cl buffer, pH 7.5 alone (clear circle); MsmHr: 100 mg/L MsmHr in 20 mM Tris-Cl buffer, pH 7.5 (black circle). Measurements were obtained at room temperature. Images are representative of 5 independent experiments. (D) UV/Vis spectral analysis of MsmHr. Reduced (deoxy) and oxidized (oxy) MsmHr. Inset: representative spectra for 100 mg/L MsmHr. Images shown are representative of 3 independent experiments.
To confirm the predicted helical structure of MsmHr, we purified MsmHr-His6protein from E. coli. Circular dichroism (CD) spectra of the E. coli-purified MsmHr showed two minima at 208 and 222 nm (Figure 1C). Such a pattern, which is characteristic of protein α-helical structures, is typical of the circular dichroism spectra of previously analyzed bacterial hemerythrins (Wirstam et al., 2003). We also performed a CD analysis of gelactin AAL (Agrocybe aegerita lectin, PDB 2ZGU), a protein which consists mainly of β-sheets. The CD spectra showed only one minima at 210 nm (Supplemental Figure S3). The UV-visible absorption spectra of MsmHr showed peaks at 327 and 376 nm, a pattern ascribable to the di-iron-center of hemerythrin-like proteins (Karlsen et al., 2005). Absorbance peaks were abrogated upon reduction with Na2S2O4 to generate the deoxy form by removing oxygen (Figure 1D). Since MsmHr has no Trp and only one Phe, UV/Vis spectrophotometric analysis of the purified protein did not detect absorbance at 280 nm (Figure 1D inset). Taken together, these bioinformatics, CD and spectrophotometric results strongly suggest that MsmHr is a hemerythrin-like protein.
MsmHr is Involved in the H2O2 Stress Response
To define the biological functions of MsmHr, a deletion mutant, ΔmsmHr, was generated by specialized transduction (Bardarov et al., 2002) (Figure 2A). The loss of msmHr was confirmed by PCR (Figure 2A) and no msmHr mRNA was detected in ΔmsmHr (Figure 2B). To determine if deletion of msmHr had polar effects on the msmeg_2414 and msmeg_2416 genes, their mRNA levels were compared in the wild type mc2155 and ΔmsmHr strains by RT-PCR. Statistically significant differences in msmeg_2414 and msmeg_2416 mRNA were not detected (Figure 2B), indicating that msmHr knockout did not affect the transcriptional levels of msmeg_2414 and msmeg_2416.
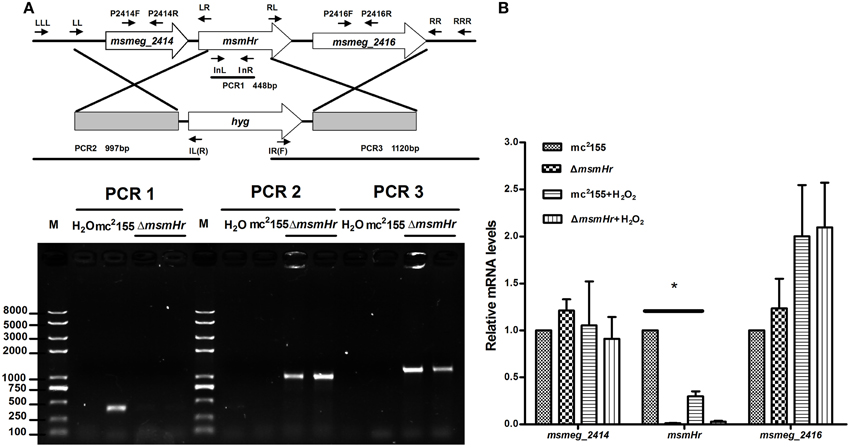
Figure 2. Generation of the ΔmsmHr M. smegmatis strain. (A) Genomic organization of the msmHr gene locus. (Upper panel) Genes are shown as large arrows in their native orientation. Small arrows represent the forward and reverse primers used for PCR, and sizes of the amplified products are indicated. Location and orientation of the hygromycin cassette are also indicated (Bottom panel). No PCR product was obtained using primers 2415InL and 2415InR to amplify the coding sequences of msmHr. PCR products for the upstream and downstream regions of msmHr were amplified using the primer pairs 2415LLL/IL(R) and 2415RRR/IR(F), respectively. (B) Effect of msmHr deletion on msmeg_2414 and msmeg_2416 expression. Quantitative real-time PCR (qRT-PCR) analysis of msmHr cluster transcription. The primer pairs 2414qF/2414qR and 2416qF/2416qR were used for qRT-PCR. Results are shown as the means ± standard deviations of three replicates (*P < 0.05).
We then compared the growth rates of the M. smegmatis parental strain mc2155 with the MsmHr-deficient mutant ΔmsmHr in both 7H9 rich medium and Sauton's minimal medium. No growth abnormalities were detected in ΔmsmHr relative to mc2155, demonstrating that MsmHr does not influence M. smegmatis growth in rich (7H9) or minimal medium (Sauton) (Figure 3A). To identify possible biological roles of MsmHr, the growth kinetics and survival of ΔmsmHr were examined under various stress conditions, including NO, hypoxia, H2O2, heat shock, and acidic pH. No growth defects in ΔmsmHr were detected under the conditions tested, with the exception of H2O2 stress (Figure 3B). ΔmsmHr exhibited mild H2O2 resistance compared to the wild type mc2155 strain after treatment with 5 mM H2O2 for 3 h, while no difference in the growth of the two strains was observed under non-H2O2 treatment conditions. In addition, the resistance phenotype was abrogated in the complemented strain, C-ΔmsmHr (Figure 3B). Moreover, overexpression of msmHr in mc2155 (O-msmHr) was associated with H2O2 susceptibility after treatment with 5 mM H2O2 for 3 h (Figure 3B). Differences in mRNA levels of msmeg_2414 and msmeg_2416 in wild type mc2155 and ΔmsmHr were not detected after treatment with H2O2(Figure 2B). A statistically significant reduction in msmHr mRNA levels was found between mc2155 in the presence and absence of H2O2 (Figure 2B). Taken together, our data suggest that msmHr plays an inhibitory role in the response to H2O2 stress.
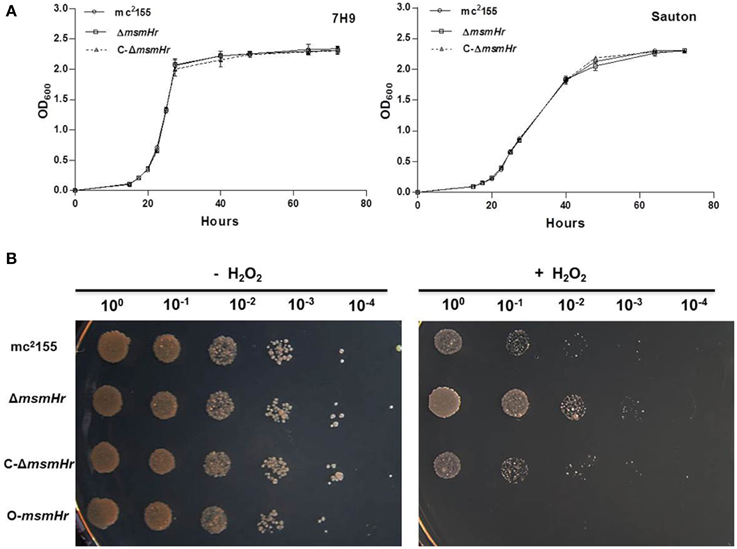
Figure 3. MsmHr is involved in the H2O2 stress response. (A) MsmHr is not necessary for normal growth. Growth curves of mc2155, ΔmsmHr and the complementary strain (C-ΔmsmHr) in 7H9 rich media (left) and Sauton minimal media (right). The data shown are representative of four independent experiments. (B) MsmHr plays a negative role in the bacterial H2O2 response. The left panel represents serial dilutions (1:10) of mc2155, ΔmsmHr, the complemented strain C-ΔmsmHr and the msmHr overexpression strain (O-msmHr: pMV261-msmHr/mc2155). Diluted log phase M. smegmatis cultures were spotted (3 μl) onto solid 7H10 medium supplemented with 10% ADS. Right panel, the corresponding strains were spotted on solid 7H10 medium supplemented with 10% ADS after treatment with 5 mM H2O2. Photographs were taken after 2–3 days incubation at 37°C. Images shown are representative of at least 3 experiments.
The Hemerythrin-Like Domains of MsmHr are Required for the Response to H2O2 Stress
To evaluate the potential roles of the hemerythrin-like domain of MsmHr in the H2O2 response, we constructed 3 mutants, H1M, H2M, and H3M, in which the respective conserved amino acid motifs HxxxEE, HxxxQQ, and HxxxEE were all mutated to LxxxAA (Figure 4A). As overexpression of msmHr showed higher susceptibility to H2O2, overexpression vector pMV261 containing msmHr alleles harboring the point mutations H1M, H2M or H3M was introduced into mc2155 which was then assessed for resistance to H2O2. All strains overexpressing the corresponding H1M, H2M or H3M mutant msmHr proteins behaved like mc2155. By contrast, strains containing intact msmHr proteins were susceptible to H2O2 (Figure 4B). This result indicates that the hemerythrin-like domain of MsmHr is required for H2O2 susceptibility.
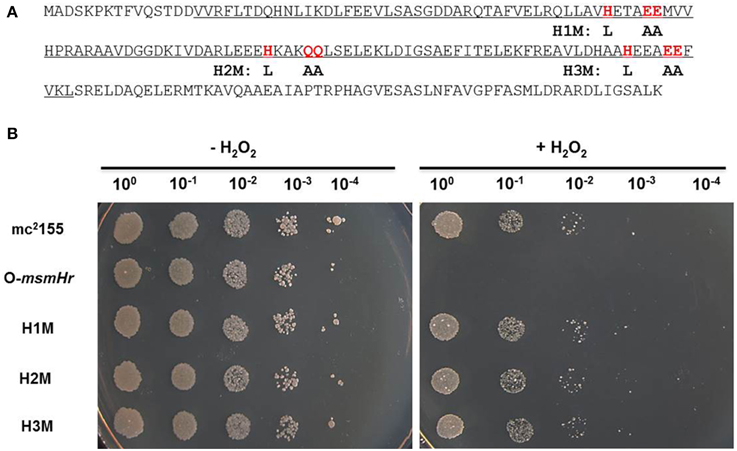
Figure 4. The hemerythrin domain is essential to MsmHr function. (A) The hemerythrin domain (underlined) of MsmHr and conserved amino acids (H… EE/QQ: red and boxed) in the hemerythrin domain are indicated. Mutations used in the following experiments are indicated under the sequence. (B) Serial dilutions (1:10) of mc2155, pMV261-msmHr/mc2155 (O-msmHr), pMV261-msmHrH1M/mc2155 (H1M), pMV261-msmHrH2M/mc2155 (H2M) and pMV261-msmHrH3M/mc2155 (H3M) treated with 0 (left) or 5 mM (right) H2O2 were spotted (3 μl) onto solid 7H10 agar. Photographs were taken after 2–3 days incubation at 37°C. Images shown are representative of at least 3 experiments.
MsmHr Represses sigF Expression Through the Promoter Prbsw
There are two independent oxidative stress pathways in mycobacteria: the KatG-, isoniazid (INH)-related pathway and the SigF-related, INH-unrelated related pathway (Gebhard et al., 2008; Wu et al., 2012). INH is an important first-line anti-mycobacterial pro-drug and is activated by the bacterial catalase-peroxide enzyme encoded by katG (msmeg_3461). Resistance to H2O2 has been shown to correlate with susceptibility to INH (Bulatovic et al., 2002). To determine which pathway MsmHr is involved in, we measured the minimum inhibitory concentration (MIC) of INH against ΔmsmHr and wild type mc2155. No difference in the MICs of these two strains was detected (3.125 mg/L in both ΔmsmHr and mc2155), suggesting that MsmHr is not involved in the INH-related H2O2 response. We then examined whether MsmHr is involved in the SigF-mediated H2O2 response. As previous studies have shown that sigF is transcribed from two promoters, we constructed two vectors with the two promoter regions, Pmsmeg_1802 and Prbsw, fused to lacZ (Pmsmeg_1802-lacZ and Prbsw-lacZ, respectively) (Humpel et al., 2010). We measured the indicated promoter activities at the early logarithmic phase: β –galactosidase activity associated with PmsmHr-lacZ was 6.9 ± 0.2 MU in mc2155 and 7.6 ± 0.1 MU in ΔmsmHr, indicating that MsmHr does not self-regulate at the transcriptional level. Wild type mc2155 harboring Pmsmeg_1802–lacZ had a β –galactosidase activity of 3.6 ± 0.3 MU, while the activity of ΔmsmHr harboring Pmsmeg_1802–lacZ was 3.7 ± 0.2 MU, indicating that MsmHr does not influence the promoter activity of Pmsmeg_1802 (Figure 5B left panel). In contrast, ΔmsmHr harboring Prbsw-lacZ had a significantly higher β –galactosidase activity (15.7 ± 1.9 MU) than that of wild type mc2155 (Figure 5B right panel). We also measured the mRNA levels of sigF in mc2155, ΔmsmHr and its complementary strain C-ΔmsmHr. Consistent with results for the promoter, the knockout msmHr led to a 1.9 ± 0.2 fold increase relative to the mRNA level of sigF to wild type mc2155, while levels of sigF mRNA were not significantly different (Figure 5C). Taken together, our results suggest that MsmHr affects the mRNA level of sigF via Prbsw.
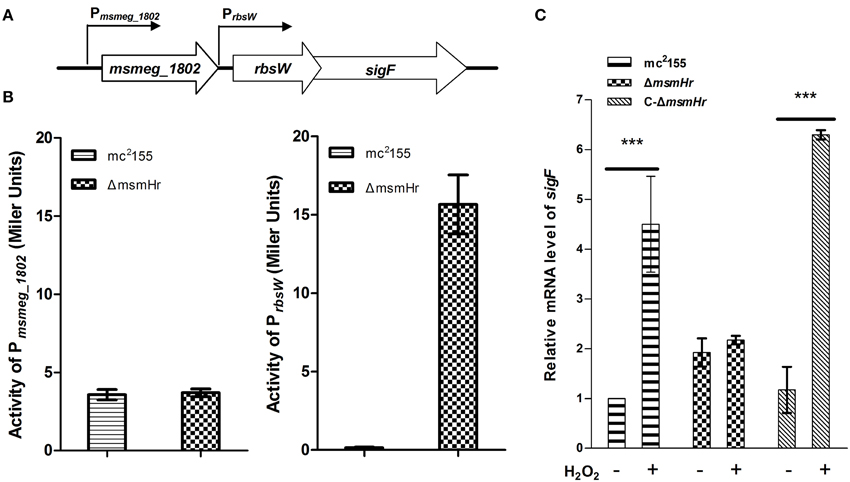
Figure 5. MsmHr regulates sigF expression through the promoter Prbsw. (A) The genomic organization of the sigF locus. Genes are shown as large arrows in their transcriptional orientation, and the locations of Pmsmeg_1802 and Prbsw are indicated. (B) MsmHr negatively regulates the activity of the sigF promoter Prbsw. Pmsmeg_1802-lacZ and Prbsw-lacZ were each transformed into mc2155 and the mutant strain ΔmsmHr. The β-galactosidase activity was measured when bacterial growth reached an OD600 of 0.3. Results representative of 5 independent experiments are shown. (C) The msmHr deletion is associated with an increase in sigF mRNA levels. Early-phase cultures of M. smegmatis mc2155, ΔmsmHr and the complemented strain C-ΔmsmHr were treated with 0 (−) or 5 (+) mM H2O2 for 30 min, and sigF expression levels were determined by qRT-PCR. Results are shown as the mean ± standard deviations of three replicates (***P < 0.01).
MsmHr is Necessary for the SigF Mediated H2O2 Response
The above results indicate that MsmHr suppresses sigF expression at the early logarithmic phase (Figure 5). We next examined the influence of MsmHr on the SigF-mediated H2O2 pathway. We compared the mRNA level of sigF between mc2155, ΔmsmHr and the ΔmsmHr complementary strain C-ΔmsmHr after 5 mM H2O2 treatment for 30 min. As shown in Figure 5C, sigF mRNA was induced by H2O2 in mc2155, while induction of sigF mRNA by H2O2 was not detected in ΔmsmHr. The increase in sigF mRNA induced by H2O2 was restored in C-ΔmsmHr. These results show that MsmHr is required for the SigF-mediated response to H2O2.
In addition, we used qRT-PCR to measure the mRNA levels of redox-related genes in both mc2155 and ΔmsmHr in response to H2O2 treatment, using rpoD mRNA as an internal invariant control (Table S3). We then chose the high H2O2-induced genes msmeg_4753 and msmeg_1782, which belong to the SigF regulon, to evaluate their mRNA level in response to H2O2 treatment in mc2155, ΔsigF, ΔmsmHr and C-ΔmsmHr. The level of msmeg_4753 mRNA increased 5.6 ± 0.9 fold in mc2155 after treatment with H2O2, but induction of msmeg_4753 was abrogated in ΔmsmHr and ΔsigF after treatment with H2O2 (Figure 6B). In C-ΔmsmHr, an increase in msmeg_4753 RNA was observed in response to H2O2 (Figure 6B). The level of Msmeg_1782 mRNA increased two-fold in both mc2155 and C-ΔmsmHr when treated with H2O2, while no changes in mRNA level were observed in response to H2O2 in ΔmsmHr and ΔsigF (Figure 6A). Taken together, this data indicates that MsmHr is required for the SigF-mediated H2O2 response.
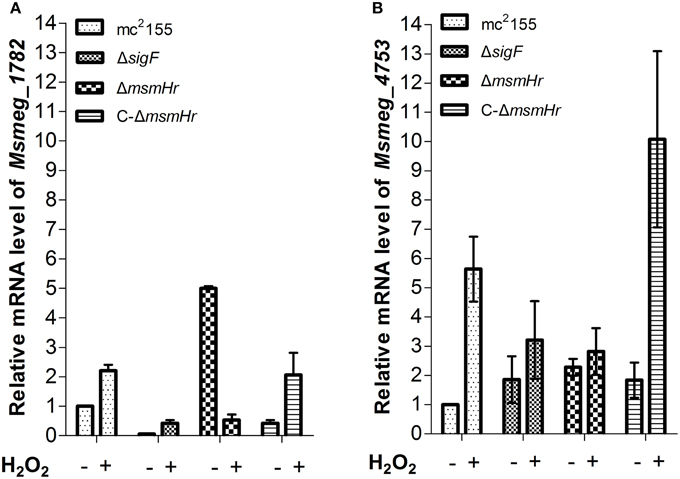
Figure 6. (A) Corresponding mRNA level of msmeg_1782 was determined by qRT-PCR. (B) Corresponding mRNA level of msmeg_4753 was determined by qRT-PCR. Bacteria were treated as described in Figure 5. Data shown are representative of three independent experiments.
Discussion
In this study, we have identified a mycobacterial hemerythrin-like protein MsmHr, which regulates sigF expression via promoter Prbsw and is necessary for the SigF-mediated H2O2 response. To our knowledge, MsmHr is the first hemerythrin-like protein to be characterized in mycobacteria.
Specific roles for hemerythrin-like proteins are just beginning to be characterized (Xiong et al., 2000; Justino et al., 2007; Schaller et al., 2012). On the basis of their sequences, hemerythrin-like proteins have been postulated to have diverse physiological functions related to oxygen and/or iron (Bailly et al., 2008; French et al., 2008). For example, the E. coli hemerythrin-like protein YtfE confers protection against both NO and H2O2 stresses (Justino et al., 2005, 2007). Here, however, we did not observe any growth differences between mc2155 and ΔmsmHr under NO stress, suggesting that the functions of hemerythrin-like proteins in mycobacteria might be distinct from those in E. coli. The variation in biological functions of hemerythrin-like proteins might be due to different selective evolutionary environments (Saini et al., 2012b; Martin-Duran et al., 2013).
No difference in sensitivity to INH was observed between the wild-type mc2155 and ΔmsmHr strains, suggesting that MsmHr is not involved in the INH-related oxidative stress response pathway but rather in an alternative SigF-related H2O2 pathway (Gebhard et al., 2008; Wu et al., 2012). It will be interesting to explore why mycobacteria use two-independent H2O2 scavenging pathways and which sensors trigger each of these signaling pathways. Recent reports show that a hemerythrin-like domain of FBXL5 can sense if endogenous iron is limiting and respond to iron stress in the mammalian system (Salahudeen et al., 2009; Vashisht et al., 2009). The correlation between iron- and oxygen- binding MsmHr and SigF-dependent H2O2 responses needs to be further explored.
The complexity of the transcription regulatory network allows for efficient and prompt change in levels of gene transcription in response to environmental changes. The protein encoded by mycobacterial sigF has closest homology to Streptomyces coelicolor SigF, Bacillus subtilis SigF and B. subtilis SigB (Demaio et al., 1996, 1997; Gebhard et al., 2008). In B. subtilis, sigB is activated upon entry into the stationary phase and by environmental stresses such as heat, oxidative stress and hyper osmosis. The transcription of sigB has been shown to be controlled by two promoters (Wise and Price, 1995). Similarly, SigF may be a potential general stress regulator; SigF is not only activated upon entry into the stationary phase, but is also induced by environmental stresses such as heat shock, acidic pH and oxidative stress (Wise and Price, 1995; Gebhard et al., 2008). Expression of sigF has been shown to be regulated by two promoters, Pmsmeg_1802 and Prbsw. While promoter Pmsmeg_1802 is known to respond to entry into the stationary phase (Gebhard et al., 2008), the role of Prbsw is less well understood. In this study, comparisons of the activity of the promoters of sigF and the mRNA level of sigF in the wild type mc2155 and mutant ΔmsmHr strains (Figure 5) showed that MsmHr regulates sigF expression via the Prbsw promoter.
We show here that MsmHr is essential for the SigF-mediated H2O2 response (Figures 5, 6). MsmHr hinders sigF promoter activation and inhibits sigF transcription during normal growth. When msmHr is deleted, inhibition of sigF is abrogated, and sigF maintains a higher transcript level. The transcription level of sigF did not vary in response to H2O2 treatment in mutant ΔmsmHr strains, possibly because the mRNA level of sigF was maintained at a higher level in ΔmsmHr (Figure 5). We measured mRNA level changes of members of the SigF-regulon, msmeg_4753 and msmeg_1782, before, between, and after treatment with H2O2. Our data show that in wild type mc2155, mRNA levels of both msmeg_4753 and msmeg_1782 increased in response to H2O2treatment, but no specific response to H2O2 was observed in ΔsigF and ΔmsmHr (Figure 6), suggesting that MsmHr is required for induction of msmeg_4753 and msmeg_1782 transcription in response to H2O2. The presence of inhibitory protein MsmHr suggests that the transcriptional regulation of SigF itself and the SigF regulon, or at least part of the SigF regulon, is based on the balance between the activation and inhibition of the H2O2 response. In ΔmsmHr, the inhibition of sigF is abrogated and sigF maintains high transcript levels. The expression of one of the SigF regulon genes, msmeg_1782, is high in ΔmsmHr during normal growth compared with mc2155, but decreases in response to H2O2 treatment. This is consistent with the finding that MsmHr is necessary for the SigF-dependent H2O2 pathway and that the response of sigF to H2O2 is also abrogated in the absence of msmHr.
In summary, we have identified a mycobacterial hemerythrin-like protein that negatively regulates SigF via the Prbsw promoter in response to oxidative stress.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Defeng Li for the gift of the protein gelactin AAL. This work was supported by the Ministry of Science and Technology of China (2014CB744402 and 2012CB518700).
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00800/abstract
References
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. doi: 10.1093/bioinformatics/bti770
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bailly, X., Vanin, S., Chabasse, C., Mizuguchi, K., and Vinogradov, S. N. (2008). A phylogenomic profile of hemerythrins, the nonheme diiron binding respiratory proteins. BMC Evol. Biol. 8:244. doi: 10.1186/1471-2148-8-244
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bardarov, S., Bardarov, S. Jr., Pavelka, M. S. Jr., Sambandamurthy, V., Larsen, M., Tufariello, J., et al. (2002). Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148, 3007–3017.
Beaucher, J., Rodrigue, S., Jacques, P. E., Smith, I., Brzezinski, R., and Gaudreau, L. (2002). Novel Mycobacterium tuberculosis anti-sigma factor antagonists control sigmaF activity by distinct mechanisms. Mol. Microbiol. 45, 1527–1540. doi: 10.1046/j.1365-2958.2002.03135.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhat, S. A., Singh, N., Trivedi, A., Kansal, P., Gupta, P., and Kumar, A. (2012). The mechanism of redox sensing in Mycobacterium tuberculosis. Free Radic. Biol. Med. 53, 1625–1641. doi: 10.1016/j.freeradbiomed.2012.08.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bulatovic, V. M., Wengenack, N. L., Uhl, J. R., Hall, L., Roberts, G. D., Cockerill, F. R. III. et al. (2002). Oxidative stress increases susceptibility of Mycobacterium tuberculosis to isoniazid. Antimicrob. Agents Chemother. 46, 2765–2771. doi: 10.1128/AAC.46.9.2765-2771.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Burian, J., Ramon-Garcia, S., Sweet, G., Gomez-Velasco, A., Av-Gay, Y., and Thompson, C. J. (2012). The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J. Biol. Chem. 287, 299–310. doi: 10.1074/jbc.M111.302588
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, K. H., Wu, H. H., Ke, S. F., Rao, Y. T., Tu, C. M., Chen, Y. P., et al. (2012). Bacteriohemerythrin bolsters the activity of the particulate methane monooxygenase (pMMO) in Methylococcus capsulatus (Bath). J. Inorg. Biochem. 111, 10–17. doi: 10.1016/j.jinorgbio.2012.02.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Demaio, J., Zhang, Y., Ko, C., and Bishai, W. R. (1997). Mycobacterium tuberculosis sigF is part of a gene cluster with similarities to the Bacillus subtilis sigF and sigB operons. Tuber. Lung Dis. 78, 3–12.
Demaio, J., Zhang, Y., Ko, C., Young, D. B., and Bishai, W. R. (1996). A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 93, 2790–2794.
Fang, F. C. (2004). Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2, 820–832. doi: 10.1038/nrmicro1004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Farhana, A., Guidry, L., Srivastava, A., Singh, A., Hondalus, M. K., and Steyn, A. J. (2010). Reductive stress in microbes: implications for understanding Mycobacterium tuberculosis disease and persistence. Adv. Microb. Physiol. 57, 43–117. doi: 10.1016/B978-0-12-381045-8.00002-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
French, C. E., Bell, J. M., and Ward, F. B. (2008). Diversity and distribution of hemerythrin-like proteins in prokaryotes. FEMS Microbiol. Lett. 279, 131–145. doi: 10.1111/j.1574-6968.2007.01011.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gebhard, S., Humpel, A., McLellan, A. D., and Cook, G. M. (2008). The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology 154, 2786–2795. doi: 10.1099/mic.0.2008/018044-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guex, N., and Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723. doi: 10.1002/elps.1150181505
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Humpel, A., Gebhard, S., Cook, G. M., and Berney, M. (2010). The SigF regulon in Mycobacterium smegmatis reveals roles in adaptation to stationary phase, heat, and oxidative stress. J. Bacteriol. 192, 2491–2502. doi: 10.1128/JB.00035-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Imlay, J. A. (2008). Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776. doi: 10.1146/annurev.biochem.77.061606.161055
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Imlay, J. A. (2013). The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454. doi: 10.1038/nrmicro3032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Isaza, C. E., Silaghi-Dumitrescu, R., Iyer, R. B., Kurtz, D. M. Jr., and Chan, M. K. (2006). Structural basis for O2 sensing by the hemerythrin-like domain of a bacterial chemotaxis protein: substrate tunnel and fluxional N terminus. Biochemistry 45, 9023–9031. doi: 10.1021/bi0607812
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jacobs, W. R. Jr., Kalpana, G. V., Cirillo, J. D., Pascopella, L., Snapper, S. B., Udani, R. A., et al. (1991). Genetic systems for mycobacteria. Meth. Enzymol. 204, 537–555.
Justino, M. C., Almeida, C. C., Teixeira, M., and Saraiva, L. M. (2007). Escherichia coli di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulfur clusters. J. Biol. Chem. 282, 10352–10359. doi: 10.1074/jbc.M610656200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Justino, M. C., Vicente, J. B., Teixeira, M., and Saraiva, L. M. (2005). New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 280, 2636–2643. doi: 10.1074/jbc.M411070200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kao, W. C., Chen, Y. R., Yi, E. C., Lee, H., Tian, Q., Wu, K. M., et al. (2004). Quantitative proteomic analysis of metabolic regulation by copper ions in Methylococcus capsulatus (Bath). J. Biol. Chem. 279, 51554–51560. doi: 10.1074/jbc.M408013200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Karlsen, O. A., Ramsevik, L., Bruseth, L. J., Larsen, O., Brenner, A., Berven, F. S., et al. (2005). Characterization of a prokaryotic haemerythrin from the methanotrophic bacterium Methylococcus capsulatus (Bath). FEBS J. 272, 2428–2440. doi: 10.1111/j.1742-4658.2005.04663.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kiefer, F., Arnold, K., Kunzli, M., Bordoli, L., and Schwede, T. (2009). The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392. doi: 10.1093/nar/gkn750
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, A., Farhana, A., Guidry, L., Saini, V., Hondalus, M., and Steyn, A. J. (2011). Redox homeostasis in mycobacteria: the key to tuberculosis control? Expert Rev. Mol. Med. 13, e39. doi: 10.1017/S1462399411002079
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, A., Toledo, J. C., Patel, R. P., Lancaster, J. R. Jr., and Steyn, A. J. (2007). Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U.S.A. 104, 11568–11573. doi: 10.1073/pnas.0705054104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martin-Duran, J. M., De Mendoza, A., Sebe-Pedros, A., Ruiz-Trillo, I., and Hejnol, A. (2013). A broad genomic survey reveals multiple origins and frequent losses in the evolution of respiratory hemerythrins and hemocyanins. Genome Biol. Evol. 5, 1435–1442. doi: 10.1093/gbe/evt102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Michele, T. M., Ko, C., and Bishai, W. R. (1999). Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigF gene: implications for chemotherapy against mycobacterial persistors. Antimicrob. Agents Chemother. 43, 218–225.
Onoda, A., Okamoto, Y., Sugimoto, H., Shiro, Y., and Hayashi, T. (2011). Crystal structure and spectroscopic studies of a stable mixed-valent state of the hemerythrin-like domain of a bacterial chemotaxis protein. Inorg. Chem. 50, 4892–4899. doi: 10.1021/ic2001267
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rawat, M., Newton, G. L., Ko, M., Martinez, G. J., Fahey, R. C., and Av-Gay, Y. (2002). Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob. Agents Chemother. 46, 3348–3355. doi: 10.1128/AAC.46.11.3348-3355.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rodrigue, S., Provvedi, R., Jacques, P. E., Gaudreau, L., and Manganelli, R. (2006). The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30, 926–941. doi: 10.1111/j.1574-6976.2006.00040.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saini, V., Farhana, A., Glasgow, J. N., and Steyn, A. J. (2012a). Iron sulfur cluster proteins and microbial regulation: implications for understanding tuberculosis. Curr. Opin. Chem. Biol. 16, 45–53. doi: 10.1016/j.cbpa.2012.03.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saini, V., Raghuvanshi, S., Khurana, J. P., Ahmed, N., Hasnain, S. E., Tyagi, A. K., et al. (2012b). Massive gene acquisitions in Mycobacterium indicus pranii provide a perspective on mycobacterial evolution. Nucleic Acids Res. 40, 10832–10850. doi: 10.1093/nar/gks793
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Salahudeen, A. A., Thompson, J. W., Ruiz, J. C., Ma, H. W., Kinch, L. N., Li, Q., et al. (2009). An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326, 722–726. doi: 10.1126/science.1176326
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schaller, R. A., Ali, S. K., Klose, K. E., and Kurtz, D. M. Jr. (2012). A bacterial hemerythrin domain regulates the activity of a Vibrio cholerae diguanylate cyclase. Biochemistry 51, 8563–8570. doi: 10.1021/bi3011797
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003). SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385. doi: 10.1093/nar/gkg520
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Silva, M. S., Senna, S. G., Ribeiro, M. O., Valim, A. R., Telles, M. A., Kritski, A., et al. (2003). Mutations in katG, inhA, and ahpC genes of Brazilian isoniazid-resistant isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 41, 4471–4474. doi: 10.1128/JCM.41.9.4471-4474.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singh, A. K., and Singh, B. N. (2008). Conservation of sigma F in mycobacteria and its expression in Mycobacterium smegmatis. Curr. Microbiol. 56, 574–580. doi: 10.1007/s00284-008-9126-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stover, C. K., De La Cruz, V. F., Fuerst, T. R., Burlein, J. E., Benson, L. A., Bennett, L. T., et al. (1991). New use of BCG for recombinant vaccines. Nature 351, 456–460. doi: 10.1038/351456a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Trivedi, A., Singh, N., Bhat, S. A., Gupta, P., and Kumar, A. (2012). Redox biology of tuberculosis pathogenesis. Adv. Microb. Physiol. 60, 263–324. doi: 10.1016/B978-0-12-398264-3.00004-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vashisht, A. A., Zumbrennen, K. B., Huang, X., Powers, D. N., Durazo, A., Sun, D., et al. (2009). Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721. doi: 10.1126/science.1176333
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vasudeva-Rao, H. M., and McDonough, K. A. (2008). Expression of the Mycobacterium tuberculosis acr-coregulated genes from the DevR (DosR) regulon is controlled by multiple levels of regulation. Infect. Immun. 76, 2478–2489. doi: 10.1128/IAI.01443-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wallace, R. J. Jr., Nash, D. R., Steele, L. C., and Steingrube, V. (1986). Susceptibility testing of slowly growing mycobacteria by a microdilution MIC method with 7H9 broth. J. Clin. Microbiol. 24, 976–981.
Wirstam, M., Lippard, S. J., and Friesner, R. A. (2003). Reversible dioxygen binding to hemerythrin. J. Am. Chem. Soc. 125, 3980–3987. doi: 10.1021/ja017692r
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wise, A. A., and Price, C. W. (1995). Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals. J. Bacteriol. 177, 123–133.
Wu, H., Hu, X., Xiao, J., Zhang, J., Tao, J., Huang, H., et al. (2012). Sigma factor F regulates Mycobacterium smegmatis hydrogen peroxide resistance. Wei Sheng Wu Xue Bao 52, 1352–1359.
Xiong, J., Kurtz, D. M. Jr., Ai, J., and Sanders-Loehr, J. (2000). A hemerythrin-like domain in a bacterial chemotaxis protein. Biochemistry 39, 5117–5125. doi: 10.1021/bi992796o
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: mycobacteria, hemerythrin-like protein, MsmHr, SigF, hydrogen peroxide
Citation: Li X, Tao J, Hu X, Chan J, Xiao J and Mi K (2015) A bacterial hemerythrin-like protein MsmHr inhibits the SigF-dependent hydrogen peroxide response in mycobacteria. Front. Microbiol. 5:800. doi: 10.3389/fmicb.2014.00800
Received: 19 September 2014; Accepted: 27 December 2014;
Published online: 15 January 2015.
Edited by:
Biswarup Mukhopadhyay, Virginia Tech, USAReviewed by:
C. Martin Lawrence, Montana State University, USAJames Moir, University of York, UK
Leif Kirsebom, Uppsala University, Sweden
Copyright © 2015 Li, Tao, Hu, Chan, Xiao and Mi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaixia Mi, Institute of Microbiology, NO. 1 Beichen West Road, Chaoyang District, Beijing 100101, China e-mail: mik@im.ac.cn
†These authors have contributed equally to this work.
 Xiaojing Li
Xiaojing Li Jun Tao
Jun Tao Xinling Hu1†
Xinling Hu1† Jing Xiao
Jing Xiao Kaixia Mi
Kaixia Mi