- 1UMR 7144, Université Pierre et Marie Curie Université Paris 06, Sorbonne Universités, Station Biologique de Roscoff, Roscoff, France
- 2Centre National de la Recherche Scientifique, UMR 7144, Station Biologique de Roscoff, Roscoff, France
- 3Centre National de la Recherche Scientifique and Université Pierre et Marie Curie, FR2424, ABiMS, Station Biologique, Roscoff, France
- 4Department of Genetics and Evolution, University of Geneva, Geneva, Switzerland
- 5Commissariat à l'Energie Atomique et aux Energies Alternatives, Genoscope, France
Rhizaria are an important component of oceanic plankton communities worldwide. A number of species harbor eukaryotic microalgal symbionts, which are horizontally acquired in the environment at each generation. Although these photosymbioses are determinant for Rhizaria ability to thrive in oceanic ecosystems, the mechanisms for symbiotic interactions are unclear. Using high-throughput sequencing technology (i.e., 454), we generated large Expressed Sequence Tag (EST) datasets from four uncultured Rhizaria, an acantharian (Amphilonche elongata), two polycystines (Collozoum sp. and Spongosphaera streptacantha), and one phaeodarian (Aulacantha scolymantha). We assessed the main genetic features of the host/symbionts consortium (i.e., the holobiont) transcriptomes and found rRNA sequences affiliated to a wide range of bacteria and protists in all samples, suggesting that diverse microbial communities are associated with the holobionts. A particular focus was then carried out to search for genes potentially involved in symbiotic processes such as the presence of c-type lectins-coding genes, which are proteins that play a role in cell recognition among eukaryotes. Unigenes coding putative c-type lectin domains (CTLD) were found in the species bearing photosynthetic symbionts (A. elongata, Collozoum sp., and S. streptacantha) but not in the non-symbiotic one (A. scolymantha). More particularly, phylogenetic analyses group CTLDs from A. elongata and Collozoum sp. on a distinct branch from S. streptacantha CTLDs, which contained carbohydrate-binding motifs typically observed in other marine photosymbiosis. Our data suggest that similarly to other well-known marine photosymbiosis involving metazoans, the interactions of glycans with c-type lectins is likely involved in modulation of the host/symbiont specific recognition in Radiolaria.
Introduction
Marine planktonic microorganisms are major constituents of oceanic ecosystems, being responsible for half of global primary production and participating massively to carbon sequestration to the deep ocean (Falkowski and Raven, 2013). Despite its recognized global impact, plankton represents one of the least explored compartments of the biosphere, particularly with regards to biotic interaction such as symbiosis. Radiolaria are abundant and widespread unicellular plankton, their large cells, typically ranging from 0.1 to 1 mm in size, exhibit mineral skeletons made up either of silica for Polycystinea or strontium sulfate for Acantharia. Radiolaria can account for a significant proportion of plankton communities in the water column (Dennett et al., 2002; Nimmergut and Abelmann, 2002) and are considered active predators (Swanberg and Caron, 1991). A number of radiolarian species exhibit also a mixotrophic metabolism, hosting eukaryotic photosymbiotic microalgae such as dinoflagellates (Gast and Caron, 1996), chlorophytes (Gast et al., 2000), and haptophytes (Decelle et al., 2012a). The intracellular algae provide photosynthetic products to the host which in turn creates a nutrient-rich micro-environment facilitating algal growth, the symbiotic interactions turning out to be critical for marine food webs and biogeochemical processes (Stoecker et al., 2009). Protists without a clear role as photosymbionts may also be present within radiolarian cells, as found for the acantharian Acanthochiasma sp. (Decelle et al., 2012b). Similarly marine invertebrates such as corals, anemones and sponges are known to harbor diverse microbial communities along with their photosymbionts (Radax et al., 2012; Sun et al., 2014).
Well-known photosymbiotic interactions in the marine environment include those occurring between Cnidaria and dinoflagellate from the genus Symbiodinium (La Jeunesse, 2001; Schwarz et al., 2008; Weis, 2008), and to a lesser extent those involving sponges (La Jeunesse, 2001) and benthic Foraminifera (Lee, 2006). Little is known about the interaction mechanisms between the host and the symbionts: lectin/glycan interactions were found to be involved in host/symbiont recognitions for different cnidarian taxa (Wood-Charlson et al., 2006; Vidal-Dupiol et al., 2009; Wood-Charlson and Weis, 2009). c-Type lectins are carbohydrate binding proteins involved in a range of different functions including cell adhesion, pathogen recognition and phagocytosis (Zelensky and Gready, 2005). Although a role of c-type lectins in symbiosis has been demonstrated for metazoa (Meyer and Weis, 2012) and to a lesser extent fungi (Singh and Walia, 2014), these proteins are present in a range of organisms and genome comparison based on hidden-Markov models suggest that putative c-type lectins are also present in several protists (http://supfam.org/SUPERFAMILY/cgi-bin/taxviz.cgi?sf=56436). Planktonic Rhizaria are essentially uncultured and tedious to collect, making investigations of symbiotic interactions difficult. Consequently, the mechanisms for host/symbiont recognition in Rhizaria are essentially unknown. In this context, our primary objective was to provide a general overview of the genes occurring in these species for which genomic data are not available and to identify genes potentially involved in symbiotic processes. To achieve our goal we produced and compared large Expressed Sequence Tag (EST) datasets from three photosymbiotic Radiolaria, an acantharian (Amphilonche elongata) and two polycystines (Collozoum sp. and Spongosphaera streptacantha), and one heterotrophic Rhizaria, the phaeodarian Aulacantha scolymantha (Cercozoa).
Materials and Methods
Sample Collection
The Phaeodaria (Cercozoa) A. scolymantha, and the Polycystinea (Radiolaria) Collozoum sp. and S. streptacantha were collected with a plankton net in the bay of Villefranche-sur-mer (43°41 N, 7°18 E, Mediterranean Sea) whereas the Acantharia A. elongata was collected in the Gulf of Eilat (29°32 N, 34°57 E, Red Sea).
The specimens were sorted manually from the plankton mix using a stereomicroscope by pipetting out of the samples, one by one, the cells of interest. The taxonomically homogeneous pool of sorted cells (one colony was isolated for Collozoum sp., about 150 cells for A. elongata, and 50 cells for both A. scolymanta and S. streptacantha) were maintained at least 2 h (up to 8 h) in a petri dish filled with 0.22 μm-filtered seawater. Since digestion of preys usually occurs within the time frame of minutes (Not personal observation) this procedure allowed minimizing sample contamination by marine microorganisms other than those closely associated with the sorted organisms. Cells were then collected individually from the petri dish and rapidly transferred (ca. 1 min) through two successive rinsing steps in clean, 0.22 μm-filtered seawater, to a sterile 1 mL tube. RNAlater was added to the tube which was stored overnight at 4°C and then transferred to −80°C until further analysis.
RNA Isolation, cDNA Library Preparation and Sequencing
Total RNA was isolated from the samples using the NucleoSpin XS (Macherey-Nagel, Germany) following the manufacturer instructions. Immediately after RNA isolation, cDNA synthesis and amplification was performed using the SMARTer RACE cDNA amplification kit (TaKaRa BIO/Clontech, Mountain View, CA) followed by the library preparation protocol available on Matzlab website (http://www.bio.utexas.edu/research/matz_lab/). Approximately 2.5 μg of the cDNA pool from prepared libraries were used for a titration run using one-quarter of a plate for each sample on the Roche 454 Genome Sequencer FLX using GS-FLX Titanium series reagents. Sequencing was carried out at Genoscope Institute (Evry, France). Data have been submitted to the GenBank under the Sequence Read Archive (SRA) numbers SRR1734678 (A. elongata), SRR1744093 (Collozoum sp.), SRR1734679 (S. streptacantha), and SRR1734688 (A. scolymantha).
Assembly, Data Cleaning, and Annotation
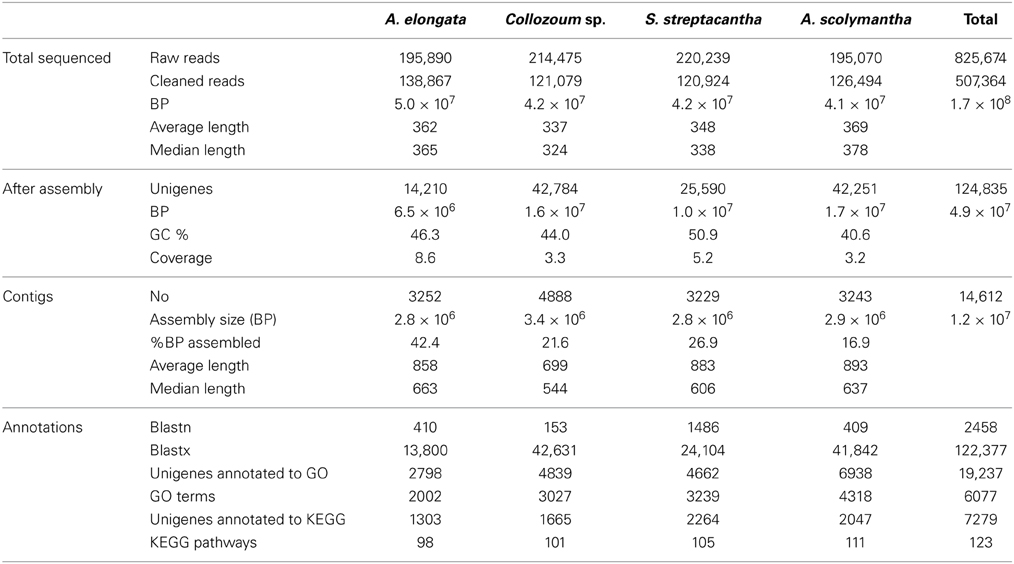
Raw 454 reads were cleaned and assembled using Newbler rd454_mapasm_08172010 (454.com/products/analysis-software). Sequencing adaptors were trimmed from the dataset using the –vt option and reads were finally assembled in contigs using the Newbler option -cDNA, which specifies a cDNA based assembly project. All sequences shorter than 200 bp were removed from the dataset. Seqclean (https://sourceforge.net/projects/seqclean) was then used to remove sequences of low quality. Overall the number of contigs recovered was 3252 for A. elongata, 4888 for Collozoum sp., 3229 for S. streptacantha, and 3243 for A. scolymantha (Table 1).
Contigs and singletons associated to rRNA were identified and extracted from the dataset by a blastn similarity search (Altschul et al., 1997) against the NCBI-nt database and the following keywords were searched within the description line of the blast results: 5.8S, 16S, 18S, 23S, 26S, 28S, ITS, rRNA, rDNA, ribosomal DNA, ribosomal RNA, LSU, and SSU. Reads matching with reference sequences containing one of those keywords, sharing >80% identity with them, and an alignment length >100 bp were considered as ribosomal sequences. The remaining sequences were further screened for the presence of rRNA reads as described in Radax et al. (2012). Unigenes were blasted against both LSU and SSU databases (releases 115, http://www.arb-silva.de/download/archive) (Quast et al., 2013) using the SILVA NGS platform (https://www.arb-silva.de/ngs/) and no additional sequences related to rRNA were found in the in the dataset. The remaining sequences were then considered as mRNA and annotated by homology using blastx similarity search against the NCBI –nr database (version 04/2012).
The taxonomic composition of both ribosomal and non-ribosomal sequences was inferred from the blast results using Megan (Huson and Mitra, 2012). The GC ratio distribution from the different sequences of our putative mRNA dataset was inferred using a homemade python script (github.com/frederic-mahe/bioinformatics-scripts/blob/master/GC_content.py).
Gene Ontology (GO) Term Distribution and Metabolic Pathways
Gene ontology terms were assigned to unigenes against the non-redundant protein database NR (April 2012) using the Blast2GO tool (Conesa et al., 2005). Overall our EST were assigned to 6264 different GO terms (Supplementary Table S1) which were then grouped into 117 categories (Supplementary Table S2) using the web-based tool CateGOrizer (Zhi-Liang et al., 2008) with the GO_slim classification method (http://geneontology.org/page/go-slim-and-subset-guide). To reconstruct the known metabolic pathways occurring in our samples, the assembled unigenes were annotated with corresponding enzyme commission (EC) numbers against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000) using the Blast2Go program (http://www.blast2go.com/b2ghome).
Clustering
Similarity between our different samples was evaluated by clustering the four mRNA libraries. Sequences were translated into the six possible reading frames and clustered using Cd-hit (Li and Godzik, 2006) with 50% sequence identity as described in Toseland et al. (2013). Clusters were examined and sorted using a Ruby script (https://github.com/georgeG/bioruby-cd-hit-report). A Venn diagram was constructed to compare the number of clusters shared by each specimen, using the VennDiagram package with the R version 2.15.0 (http://cran.r-project.org).
Symbiosis-Related Genes Search
To evaluate the occurrence of ESTs likely coding for symbiosis-related proteins, keyword searches were carried out on the description lines of both blastx and GO results. Enzymes and receptors involved in different functions to the interactions between cnidaria and dinoflagellates were searched. Specifically we searched for genes overexpressed in the coral Acropora palmata while in symbiosis with Symbiodinium spp., compared to non-symbiotic stages of the same species (Iversen and Seuthe, 2011). Similarly we analyzed the occurrence of candidate symbiosis genes which were previously found in different anemone and coral species (Meyer and Weis, 2012). In coral/dinoflagellate symbiosis these genes are involved in several functions such as pattern recognition, cell adhesion, vesicular trafficking, regulation of incoming light, apoptosis, nutrient and metabolite transport, lipid storage and transport, and response to oxygen reactive species (Meyer and Weis, 2012).
c-Type Lectin Extraction and Phylogenetic Analyses
The presence of genes putatively expressing c-type lectins was evaluated by a tblastn similarity search of all the six possible reading frames of the four samples mRNA against a selection of c-type lectins domains downloaded from Genbank using Geneious software (Drummond et al., 2011). The lectins used in this search were derived from the scleractinian corals Pocillopora damicornis (Vidal-Dupiol et al., 2009) and Acropora millepora (Kvennefors et al., 2008), the sea anemone Nematostella vectensis (Wood-Charlson and Weis, 2009), and the nematodes Laxus oneistus and Stilbonema magnum (Bulgheresi et al., 2011). Several unigenes from S. streptacantha, and two unigenes from both A. elongata and Collozoum sp. showed similarities with reference c-type lectin domains. All but one unigene extracted from our S. streptacantha library contained several different putative c-type lectin domains. According to previous studies (Zelensky and Gready, 2005) we define a c-type lectin domain (CTLD) as a sequence containing four conserved cysteine motifs and WIGL-like regions. Unigenes from S. streptacantha containing multiple CTLD were then split in the different domains and all the 28 domains obtained from S. streptacantha were then aligned to check similarity and remove redundant sequences. We finally obtained eight unique c-type lectin domains from S. streptacantha, which were aligned with putative c-type lectin domains from A. elongata, Collozoum sp. as well as reference c-type lectins from marine symbiosis described above and seven putative c-type lectins from other protists such as Apicomplexa (Cryptosporidium parvum, Cryptosporidium muris, Gregarina niphandrodes, Neospora canicum), Pelagophyceae (Aureococcus anophagefferens), Cryptophyta (Guillardia theta), and Foraminifera (Reticulomyxa filosa). Sequences were aligned using clustal omega (Sievers et al., 2011) and the alignment was cured using gblocks (Castresana, 2000). The final alignment contained 43 sequences with 246 unambiguously aligned positions, phylogenetic inferences were conducted using phyml (Guindon et al., 2010). The maximum likelihood results are shown as an unrooted tree.
Results
Assemblage Quality
A total of 825,674 raw reads were generated, and after removal of bad quality and short (<200 bp) sequences we obtained 507,364 reads for a sequencing size of 1.7 × 108 bp (Table 1). Both the number of reads and the sequencing size did not differ significantly (<20%) between the different holobionts. Reads assembled using Newbler led to a number of unigenes (contigs + singletons) from 14,210 (A. elongata) to 42,784 (Collozoum sp.). Collozoum sp. yielded the highest number of contigs (4888) and largest assembly size (3.4 × 106 bp). A. elongata ESTs were the best assembled in terms of coverage (8.1X) and proportion of bp assembled (42.4%, Table 1), whereas A. scolymantha and Collozoum sp. holobionts were the most poorly assembled with a coverage of 3.2X and 3.3X and 21.9 and 26.6% of bp assembled, respectively (Table 1). Most (93–97%) of our contigs were ≤2000 bp, and the median contig length ranged from 544 (Collozoum sp.) to 663 (A. elongata, Table 1) bp and the contig length peaked at 500 bp for all the samples (Figure 1).
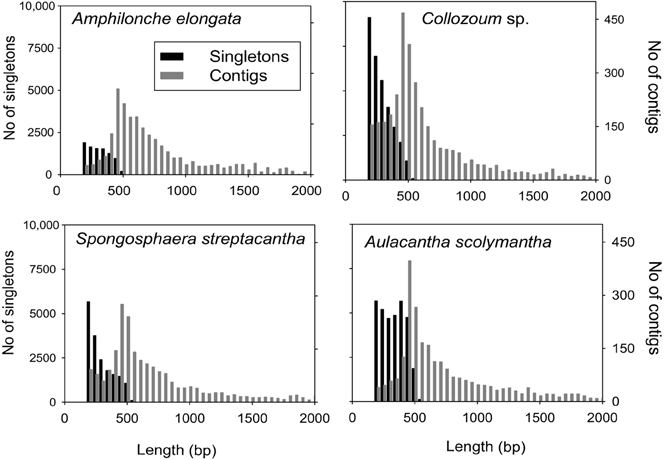
Figure 1. Distribution of contig and singleton lengths for A. elongata, Collozoum sp., S. streptacantha, and A. scolymantha. Contigs longer than 2000 bp (n = 139 for A. elongata, max length 2400 bp, n = 220 for A. scolymantha, max length 11,307, n = 110 for Collozoum sp., max length 4000, n = 75 for S. streptacantha, max length 30,120) are not shown.
Ribosomal rRNA
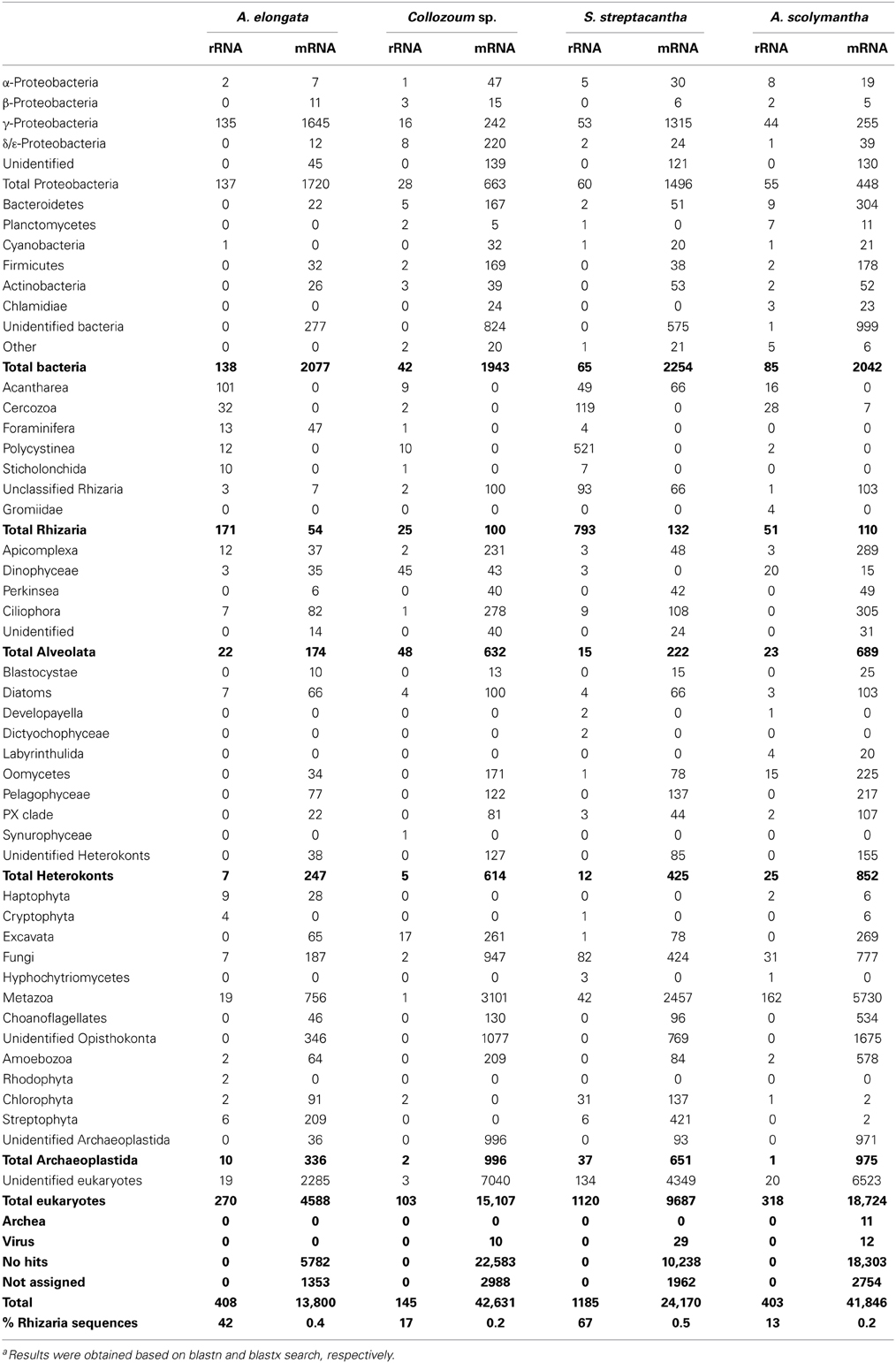
All the unigenes obtained were blasted against the GenBank nucleotide database (blastn) to discriminate between rRNA and putative mRNA sequences. Blastn hits from rRNA reads included both eukaryotic (18S, ITS, and 28S rRNA) and as prokaryotic (16S, ITS and 23S rRNA) genes (Table 2). The number of rRNA unigenes ranged from 153 (Collozoum sp.) to 1406 (S. streptacantha). Among rRNA unigenes 10% (S. streptacantha) to 33% (A. elongata) were affiliated to prokaryotes. Prokaryotic unigenes were dominated by γ-Proteobacteria and Bacteroidetes, with α-Proteobacteria and Planctomycetes also frequently occurring throughout our samples (Table 2).
Eukaryotic rRNA unigenes included from 13% (A. scolymantha) to 71% (S. streptacantha) of rhizarian sequences. Hundred and one rRNA unigenes from A. elongata were affiliated to several Acantharia genera, with only five unigenes associated with the genus Amphilonche (Supplementary Table S3). Polycystinea were poorly represented in the rRNA library of Collozoum sp. since only 10 out of 103 eukaryotic rRNA reads shared high similarities with Polycystinea. In contrast, about 25% of rRNA reads from S. streptacantha were affiliated to Polycystinea (Table 2), but none of them could be affiliated to the genus Spongosphaera, very likely because reference sequences from this genus are missing in GenBank. Rhizarian rRNA unigenes from A. scolymantha were mostly affiliated with Cercozoa (Table 2), with only one out of 28 cercozoan unigenes assigned to A. scolymantha (Supplementary Table S3).
Other eukaryotic sequences recovered within the rRNA from our holobionts were mainly affiliated to Alveolata (Table 2), specifically Apicomplexa for A. elongata, ciliates for S. streptacantha and dinoflagellates for Collozoum sp. and A. scolymantha. rRNA reads from known acantharian symbionts such as Phaeocystis sp. (Decelle et al., 2012a) were found for A. elongata holobionts along with reads from other photosynthetic protists such as Bacillariophyceae, Prasinophyceae, and Cryptophyceae (Supplementary Table S3). S. streptacantha holobiont also contained >100 microalgal rRNA reads associated with the Lotharella sp. (Chlorarachniophyceae) and some reads from Bacillariophyceae and Cryptophyceae.
Putative mRNA: Taxonomic Composition
Putative mRNA represented the majority of our dataset, corresponding to 97% of the total number of unigenes for A. elongata, 99.7% for Collozoum sp., 88% for S. streptacantha, and 98.3% for A. scolymantha. The putative mRNA unigenes correspond to transcripts occurring in our holobionts at the time of sampling and likely derive from the hosts, symbionts and/or other microorganisms present within and around the rhizarian cells. Overall we blasted 122,377 putative mRNA unigenes against the GenBank protein database (blastx) to obtain a general annotation of the expressed genes and to assign a putative metabolic function based on similarity. Most of the mRNA unigenes matched eukaryotic sequences or did not have significant hits with known reference sequences (no hits). The most represented matches were associated with land plants (Streptophyta), Metazoa and Fungi (Table 2), very likely reflecting the most important occurrence of genomes from these groups in current databases. The scarcity of genomic sequences from rhizarian groups other than Foraminifera and Cercozoa (i.e., no genomes of Polycystinea) implied that A. elongata, Collozoum sp. and S. streptacantha libraries included a number of transcripts blasting with Foraminifera. A few A. scolymantha unigenes were related to mRNA sequences from the cercozoan Bigelowiella sp. A minor proportion of the unigenes were assigned to Bacteria (Table 2).
GC Content
We analyzed the GC content of our samples to assess whether our holobionts showed a plurimodal distribution reflecting the occurrence of different microorganisms. The overall GC content of our samples ranged from 40.6% for A. scolymantha to 50.9% for S. streptacantha (Table 1). We then compared, within each of our holobionts, the GC-content distribution from the different domain level groups (Bacteria, Eukaryotes, no-hits, and not-assigned) and found that the GC content from our bacterial mRNA unigenes differed from that of the eukaryotic unigenes, especially for S. streptacantha (Supplementary Figure S1).
However the GC-content of the eukaryotic mRNA from our holobionts showed a unimodal distribution and we could not discriminate the contribution of the different microorganisms within our libraries. Significant differences occurred between the different samples, including the taxonomically closest ones, our two polycystinean species, with most unigenes from Collozoum sp. having a lower GC content than the majority of S. streptacantha reads (Figure 2).
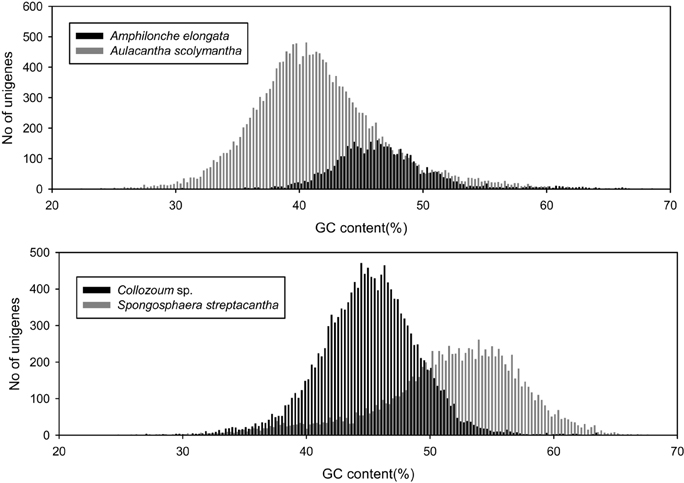
Figure 2. GC content of putative eukaryotic mRNA unigenes for the four specimens analyzed in the present study.
Gene Ontology and KEGG
Overall 19,237 unigenes from our mRNA dataset were annotated to a total of 6264 different GO terms which were then grouped into 117 GOslim categories (Supplementary Tables S1,S2). The most represented categories were related to catalytic activity and cell components (Figure 3). The proportion of unigenes expressed for each category was similar for the different samples, preventing identification of GO categories under-expressed in our non-photosymbiotic sample (A. scolymantha) and thus likely related to photosynthetic activities.
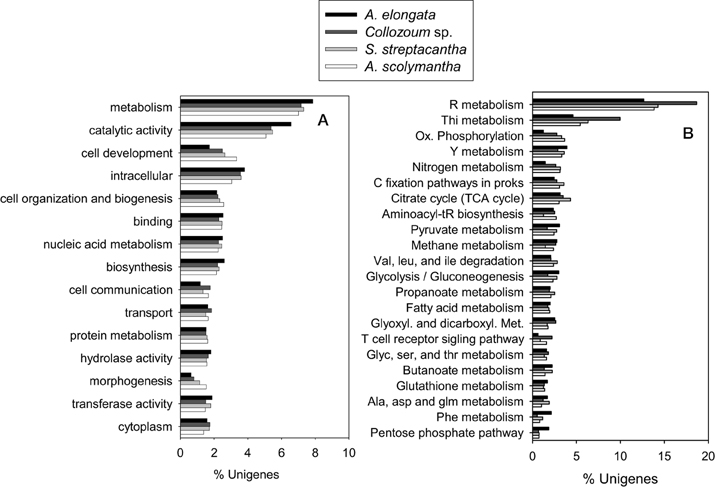
Figure 3. Comparison of the (A) Gene ontology terms summarized into Goslim categories and (B) KEGG assignments from the transcriptomes of the holobionts analyzed in the present study.
Moreover 1303–2264 unigenes from our different holobionts were also annotated to KEGG and a total of 123 KEGG pathways were identified within our dataset (Supplementary Table S4). The most represented pathways were related to nucleotides, thiamine and nitrogen metabolisms as well as citrate cycle and carbon fixation pathways in prokaryotes (Figure 3). A higher proportion of Collozoum sp. unigenes were related to purine and thiamine metabolisms compared to the other holobionts, whereas the proportion of ESTs associated with oxidative phosphorylation and nitrogen metabolisms was significantly lower for A. elongata. Consistent with GOslim data we could not find any KEGG pathway associated with non-symbiotic A. scolymantha over-represented or under-represented with respect to the other holobionts. Therefore, it was not possible to identify genes or pathways likely associated with photosymbiosis.
Clustering
The majority of the clusters identified here, based on a 50% similarity, only include sequences from one sample (Figure 4). For example 83% of the clusters containing sequences from A. elongata do not contain unigenes from other samples (Figure 4), and this percentage rises up to 94% for A. scolymantha. Very few clusters thus contain sequences from multiple samples. In spite of the taxonomic relatedness of our specimens only 130 clusters include unigenes from all our samples whereas higher proportions of the clusters are shared between A. elongata and Collozoum sp. (899), Collozoum sp. and S. streptacantha (1170), and A. elongata and S. streptacantha (951) compared to the clusters shared between A. scolymantha and any of the three other holobionts (Figure 4). These differences might be related to the lack of photosymbionts within A. scolymantha, in contrast with the other samples.
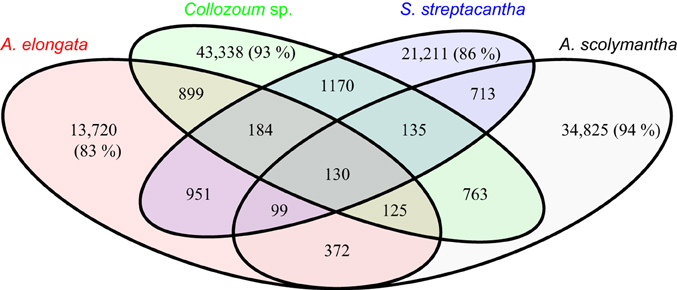
Figure 4. Venn diagram for pooled sequence clustering based on cd-hit. A total of 124,979 clusters were obtained using 50% similarity. Values indicate the number of clusters relative to each diagram section. Percentages show portions of clusters which only occur in one specimen.
Symbiosis-Related Genes and c-Type Lectin Domains
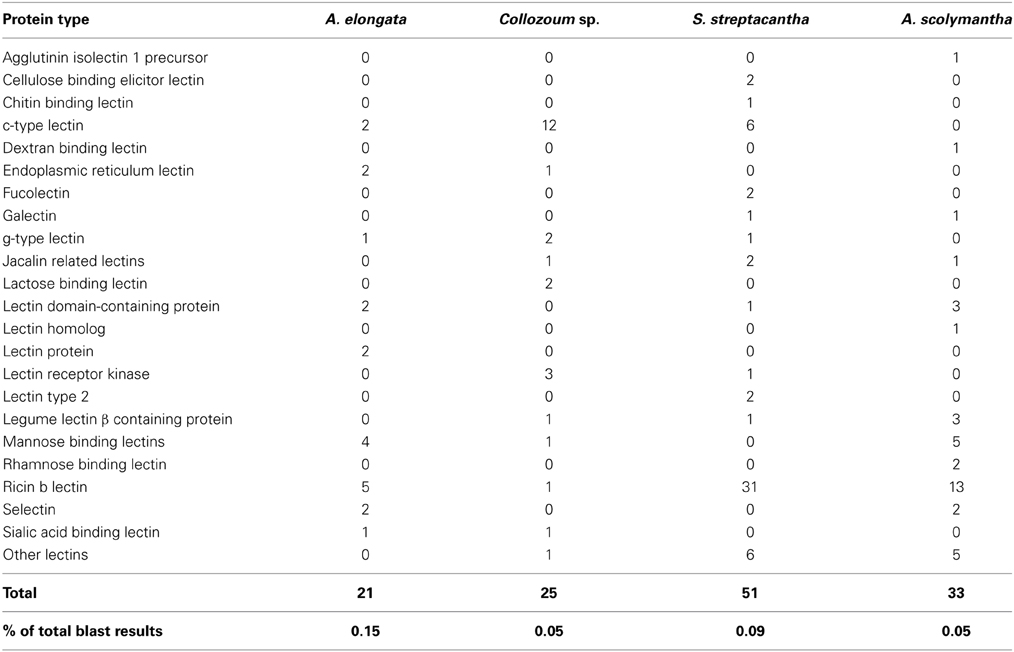
We searched for the presence of genes previously found overexpressed in the symbiotic stages of cnidaria/dinoflagellate holobionts (Kvennefors et al., 2008; Iversen and Seuthe, 2011; Meyer and Weis, 2012; Yuyama et al., 2012). A number of putative mRNA reads from our dataset are related to genes coding glutamate dehydrogenase, glutathione S-transferase, lectins, glutamine synthetase, lipase, phospholipase and ferritin (Supplementary Table S5). Within these genes we examined specifically the occurrence of lectins. Overall 142 unigenes from all our samples shared similarities with lectins (Table 3). Their number ranged from 19 (A. elongata) to 57 (S. streptacantha). We blasted reference sequences from CTLDs involved in marine symbiosis (Wood-Charlson et al., 2006; Vidal-Dupiol et al., 2009; Bulgheresi et al., 2011) against our dataset of mRNA translated into amino acid sequences in order to identify putative CTLD within our data. Overall eight sequences from the holobionts of A. elongata (two), Collozoum sp. (two), and S. streptacantha (four) showed similarities with CTLDs.
The alignment of putative CTLD from our samples with CTLD from other marine holobionts or putative CTLD from other protists consisted of four highly conserved cysteine residues as well as four motifs (Supplementary Figure S3). The WIGL-like motif which is crucial in the identification of CTLD (Zelensky and Gready, 2005) was found to vary for our holobionts, being WIGV for A. elongata, WLGF for Collozoum sp. and WIGL, WLGL, and WVGL for the different CTLD from S. streptacantha (Supplementary Figure S3). The glucose/mannose binding motif EPN (Zelensky and Gready, 2005) was found only in S. streptacantha whereas such motif was replaced by DSS in A. elongata and by WSD in Collozoum sp. Similarly the galactose binding motif LND (Zelensky and Gready, 2005) was only found in S. streptacantha CTLD. In A. elongata the LND motif was replaced by VND whereas we could not find an equivalent motif in Collozoum sp. (Supplementary Figure S3). The resulting phylogenetic tree reconstruction is shown as an unrooted tree (Figure 5), and includes putative CTLD extracted from the holobionts as well as a range of reference sequences from Metazoa and protists. Five well-supported c-lectin groups can be identified from the phylogenetic tree. Group A strictly includes coral CTLDs that could be related to several ungrouped sequences from alveolates. In group B we find the protist CTDLs of A. elongata and A. anophagefferens more closely related to other metazoans CTDLs than to other acantharians and apicomplexan protists, respectively. Group C is almost exclusively composed by S. streptacantha except for a sequence from N. vectensis. Group D is an assembly of various metazoan CTDLs with the presence of the protist G. theta. Finally, group E contains the CTDLs of Collozoum sp. and the lectins of the Salpingoeca rosetta and Cyprinus carpio.
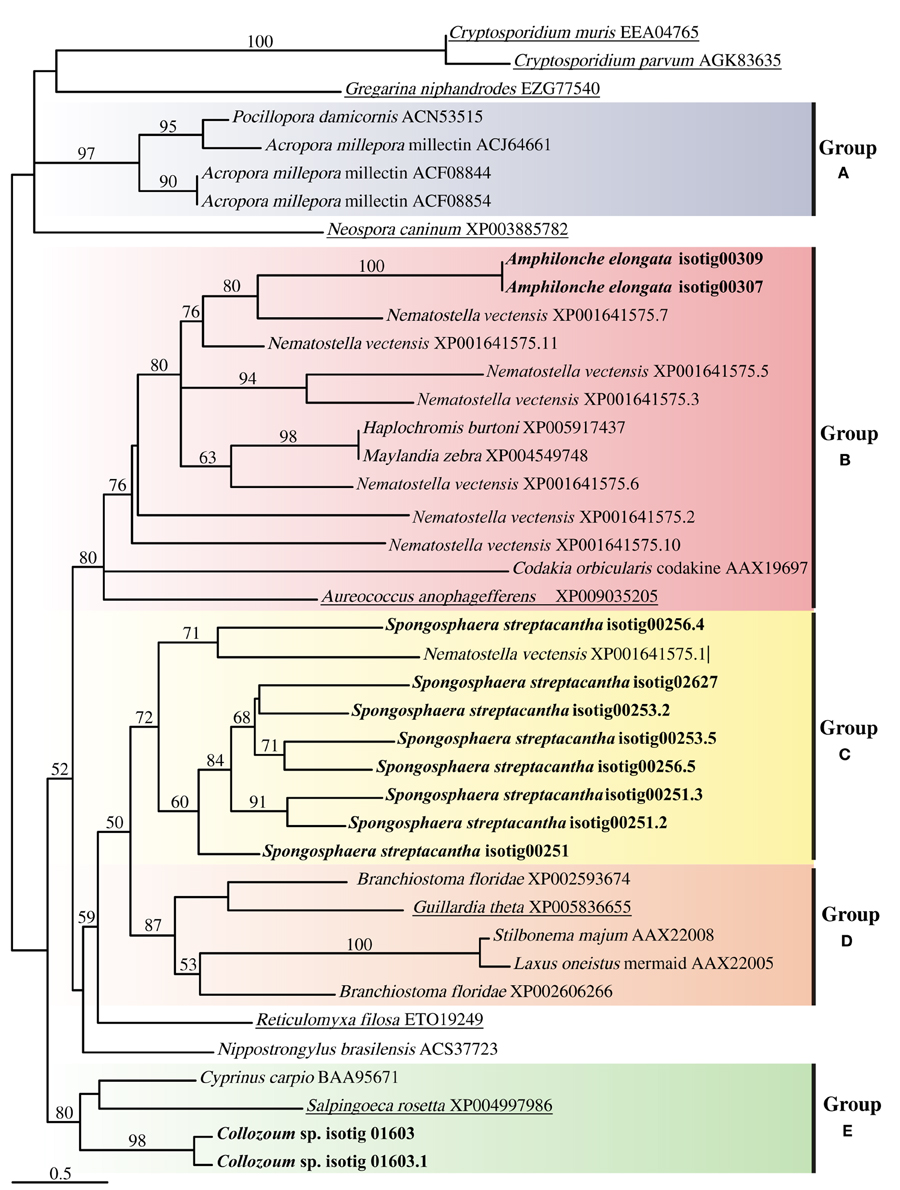
Figure 5. Unrooted tree describing the relatedness of c-type lectins of radiolarians in the present study, other protists and Metazoa harboring photosynthetic symbionts. The consensus tree resulting from phyml analyses of 246 unambiguously aligned positions and 43 protein sequences. Topological support of >50% obtained from aLRT approach are shown in each branch label. Sequences corresponding to this study are in bold whereas sequences from other protists are underlined.
The phylogeny found here does not reflect the taxonomic position of the species from which the sequences were extracted but depicts the relatedness of the lectin proteins present in the species shown (Figure 5). Several clusters, each including sequences from both protists and Metazoa were indeed found. Putative CTLDs within each holobionts group together forming well-supported clusters (100 support for A. elongata, 98 for Collozoum sp., and 72 for S. streptacantha CTLDs). Contrarily to what would be expected, the putative CTLDs extracted from the radiolarian holobionts showed to be more closely related to other protists or metazoan lectins than to radiolarians themselves (Figure 5).
Discussion
Holobiont Sequencing
Nowadays, high throughput sequencing approaches are largely applied to investigate the interactions of microbes with multicellular hosts, especially in the marine environment. Such approaches were used to elucidate symbiotic interactions in Cnidaria (Sabourault et al., 2009; Yuyama et al., 2011) and sponges (Radax et al., 2012). In contrast fewer studies investigated interactions when both partners are protists. Recently, EST analyses have been applied to understand the ability of Foraminifera to maintain chloroplasts deriving from previously ingested diatoms (Pillet and Pawlowski, 2013). In our investigation, because of the relatively low sequencing depth, our coverage is not sufficient to infer expression patterns but allows a qualitative assessment of the genes expressed within radiolarian holobionts. Still we observed different coverage between our datasets, with fewest unigenes recovered from the best assembled sample (A. elongata), suggesting some variability in transcript diversity between the holobionts analyzed, the lower coverage the higher diversity. Also, the high proportion of unassigned mRNA unigenes (Table 2) might be related to the fact that Rhizaria is the least sequenced eukaryotic super-group to date (Burki and Keeling, 2014; del Campo et al., 2014).
Only, one rhizarian genome, from Bigelowiella natans (Chloroarachniophyta), has been fully sequenced to date (Bhattacharya et al., 2013) and other data only include partially sequenced genomes and transcriptomes, mostly from Foraminifera (Burki et al., 2010; Bulman et al., 2011; Habura et al., 2011; Pillet and Pawlowski, 2013; Sierra et al., 2013). In the present study a number of A. elongata reads were assigned to Foraminifera likely because of the evolutionary relatedness of Acantharia and Foraminifera (Sierra et al., 2013). These two classes show higher genetic distances from Polycystinea implying that high proportions of reads from Collozoum sp. and S. streptacantha were not assigned, even at high taxonomic level.
The high proportion of mRNA sequences related to Streptophyta, unidentified Archaeplastida, Fungi, and Metazoa within our eukaryote-assigned transcripts could be explained by some contamination. Yet considering that proportions of rRNA reads affiliated to these taxa does not match with the mRNA assignments, it is also likely that it exists a bias related to the unevenness of taxonomic coverage within the public database (Keeling et al., 2014). As for a number of eukaryotes, a transcriptomic approach provides a valuable alternative to genome sequencing for uncultured protistan taxa. The use of transcriptomics is also particularly suited to study uncultured Radiolaria as their symbionts are mainly dinoflagellates (Gast et al., 2003; Probert et al., 2014), which have an enormous genome size (Lin, 2011) making sequencing very difficult. In the present study, investigations of the holobionts are important because they bring a holistic view of highly complex unicellular organisms living in close contact with diverse microbial communities.
Microbial Communities Associated with Rhizarian Hosts
Although the technique used for the cDNA library preparation targets specifically eukaryotic mRNA because of poly-T primers used which bind to the poly-A region of transcript, eukaryotic and prokaryotic rRNA as well as prokaryotic mRNA genes were recovered in our libraries. Blastn results of our putative ribosomal reads indicate a wide taxonomic diversity within our samples. Ten bacterial genera typically deriving from contamination while extracting nucleic acids (Salter et al., 2014) were present in very low abundances within the rRNA libraries, and mostly for A. scolymantha. In particular five to seven rRNA hits from the genera Mesorhizobium, Rhizobium and Flavobacterium were found in the A. scolymantha rRNA library. The impact of potential lab-contaminant to our data was thus minimal, even for A. scolymantha, which seems the specimen mostly affected by contamination.
The presence of symbiotic dinoflagellates in several Polycystinea including Collozoum (Gast and Caron, 2001; Probert et al., 2014) and that of symbiotic haptophytes in Acantharia (Decelle et al., 2012a,b) is also reflected here by the taxonomic composition of both rRNA and mRNA unigenes, where dinoflagellates and haptophytes accounted for an important portion of the libraries from Collozoum sp. and A. elongata, respectively (Table 2). In contrast to previous findings for other spumellarian species (Gast and Caron, 1996; Dolven et al., 2007), the S. streptacantha isolated here was unlikely to harbor dinoflagellate photobionts, since very few dinoflagellate rRNA sequences were recovered from this specimen. The high proportion of rRNA reads related to the chlorarachniophyte Lotharella sp. (Supplementary Table S6) found within our S. streptacantha sample suggests the occurrence of chlorarachniophyte photobionts, which have not been reported yet. However, one cannot exclude that Lotharella sp. cells might have been occurring in S. streptacantha as preys since the latter species has been recorded to predate on ciliates and small flagellates (Matsuoka, 2007). Accordingly a number of rRNA sequences from ciliates were found within our S. streptacantha sample (Table 2). Data from our A. scolymantha could illustrate an active predatory activity, since the rRNA reads from this specimens are dominated by marine Metazoa but also include reads from other protists (Table 2) which might be associated with preys engulfed by the host before sampling. As the basic ecology of Rhizaria is often elusive, EST analyses can thus be a useful tool to analyze the composition of microbial communities within our samples and also to address food preferences from rhizarian hosts.
EST analyses provided also useful information on the prokaryotic communities associated with our radiolarian hosts although the role of the different taxa found is unknown. The dominant bacterial classes found in the present study, γ-Proteobacteria and Bacteroidetes, are commonly abundant in surface seawater (Biers et al., 2009; Yooseph et al., 2010). Still, both γ-Proteobacteria and Bacteroidetes can dominate the intracellular environment of distinct mixotrophic protists (Martinez-Garcia et al., 2012) as well as particle-attached bacteria (Simon et al., 2002), which would explain their presence in our holobionts. Surprisingly cyanobacteria, mostly affiliated to the genus Synechococcus accounted for a very low proportion of our bacterial rRNA, although cyanobacterial endosymbionts were previously reported to occur within the cercozoan genus Paulinella (Bodyl et al., 2007; Bhattacharya et al., 2013), and some polycystinean such as Dictyocoryne spp. (Foster et al., 2006a,b; Yuasa et al., 2012) and Spongostaurus sp. (Foster et al., 2006b).
Overall, the rRNA sequence composition of our dataset suggests the occurrence of a diverse community in close contact or within our holobionts and confirms the photosymbiont identity for both A. elongata and Collozoum sp. (Decelle et al., 2012a; Probert et al., 2014) whereas the photosymbiotic status of S. streptacantha is unclear.
Specificities of the Holobionts Analysis
The differences in mRNA composition found here between our different holobionts (Table 2) are corroborated by our clustering data (Figure 4), since most (83–94%) of the clusters obtained from our mRNA unigenes based on 50% similarity include reads from one single species. These differences might be related to both the hosts and the symbionts. On the one hand clustering data suggest a significant genomic diversity between our hosts although they belong to the same phylum. On the other hand the microbial communities associated with our hosts (Table 2) likely expressed different, unrelated genes, contributing to the low similarities in mRNA found here.
The unimodal distribution of the GC content (Figure 2) within our eukaryotic putative mRNA unigenes does not allow discriminating the host-derived ESTs from those derived from other microorganisms. Such discrimination was complicated by the fact that our data likely include transcripts from a range of different eukaryotic microorganisms making the contribution from each taxon to the overall GC distribution complicate to disentangle. In previous studies, where genomic data from both host and symbionts were available, the contributions from each partner to the transcriptome were successfully discriminated based on the GC%. For example the contributions from the distinct partners were successfully discriminated for the symbiotic coral Acropora tenuis, because the holobiont transcriptome was compared with the genome of the symbiont-free larva (Kii et al., 2007). In contrast, Radiolaria are uncultivable, which makes more difficult to obtain symbionts-free radiolarian species. In the present study we used A. scolymantha as a photosymbiont free species, however the comparison of A. scolymantha mRNA with that obtained from the other species did not allow the discrimination between host- and symbiont-derived transcripts for two reasons. First although photobiont-free, A. scolymantha is still likely to harbor a diverse microbial assemblage, as suggested by the rRNA composition (Table 2), consisting in a combination of transcripts from the host and multiple symbionts from which the proportion of purely host transcripts is unknown. Second, since A. scolymantha is a phaeodarian branching within Cercozoa, it is phylogenetically distant from both Polycystinea and Acantharia (Krabberod et al., 2011; Sierra et al., 2013); therefore host derived transcripts from A. scolymantha might have a different GC profile from A. elongata, Collozoum sp. and S. streptacantha.
c-Type Lectins
The alignment of our sequences with CTLDs from marine organisms strongly suggests the occurrence of c-type lectins in our specimens of A. elongata, Collozoum sp. and S. streptacantha holobionts. c-type lectins have been described controlling the onset of photosymbiosis in several cnidarian (Wood-Charlson et al., 2006; Vidal-Dupiol et al., 2009), nematodes (Bulgheresi et al., 2006), and fungal (Singh and Walia, 2014) species. Although c-type lectins have been mostly described from Metazoa so far (Zelensky and Gready, 2003) we did not find putative CTLD in our A. scolymantha holobiont, which contained high proportions of rRNA and mRNA reads affiliated to Metazoa (Table 2). This suggests that the putative c-type lectins found here in A. elongata, Collozoum sp. and S. streptacantha are unlikely to derive from metazoan preys or debris present within the holobionts but are rather associated with Rhizaria or other microbial eukaryotes present within the holobionts.
The presence of unigenes related to c-type lectins among our symbiotic taxa only (i.e., A. elongata, Collozoum sp., and S. streptacantha) suggests that these proteins could play a role in the symbiotic process. Although a role in functions other than symbiosis cannot be excluded, since c-type lectins were demonstrated to trigger the onset of photosymbiosis in nematodes and corals, they might also play a role in the recognition of the symbionts in some Rhizaria.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Micro B3 project as well as the Swiss National Science Foundation grant 31003A-125372 (JP, RS). The Micro B3 project is funded from the European Union's Seventh Framework Programme (Joint Call OCEAN.2011-2: Marine microbial diversity – new insights into marine ecosystems functioning and its biotechnological potential) under the grant agreement No. 287589.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00098/abstract
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J. H., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bhattacharya, D., Price, D. C., Bicep, C., Bapteste, E., Sarwade, M., Rajah, V. D., et al. (2013). Identification of a marine cyanophage in a protist single-cell metagenome assembly. J. Phycol. 49, 207–212. doi: 10.1111/jpy.12028
Biers, E. J., Sun, S., and Howard, E. C. (2009). Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl. Environ. Microbiol. 75, 2221–2229. doi: 10.1128/AEM.02118-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bodyl, A., Mackiewicz, P., and Stiller, J. W. (2007). The intracellular cyanobacteria of Paulinelia chromatophora: endosymbionts or organelles? Trends Microbiol. 15, 295–296. doi: 10.1016/j.tim.2007.05.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bulgheresi, S., Gruber-Vodicka, H. R., Heindl, N. R., Dirks, U., Kostadinova, M., Breiteneder, H., et al. (2011). Sequence variability of the pattern recognition receptor Mermaid mediates specificity of marine nematode symbioses. ISME J. 5, 986–998. doi: 10.1038/ismej.2010.198
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bulgheresi, S., Schabussova, I., Chen, T., Mullin, N. P., Maizels, R. M., and Ott, J. A. (2006). A new C-type lectin similar to the human immunoreceptor DC-SIGN mediates symbiont acquisition by a marine nematode. Appl. Environ. Microbiol. 72, 2950–2956. doi: 10.1128/AEM.72.4.2950-2956.2006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bulman, S., Candy, J. M., Fiers, M., Lister, R., Conner, A. J., and Eady, C. C. (2011). Genomics of biotrophic, plant-infecting plasmodiophorids using in vitro dual cultures. Protist 162, 449–461. doi: 10.1016/j.protis.2010.09.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Burki, F., and Keeling, P. J. (2014). Rhizaria. Curr. Biol. 24, 103–107. doi: 10.1016/j.cub.2013.12.025
Burki, F., Kudryavtsev, A., Matz, M. V., Aglyamova, G. V., Bulman, S., Fiers, M., et al. (2010). Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol. Biol. 10:377. doi: 10.1186/1471-2148-10-377
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Decelle, J., Probert, I., Bittner, L., Desdevises, Y., Colin, S., de Vargas, C., et al. (2012a). An original mode of symbiosis in open ocean plankton. Proc. Natl. Acad. Sci. U.S.A. 109, 18000–18005. doi: 10.1073/pnas.1212303109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Decelle, J., Siano, R., Probert, I., Poirier, C., and Not, F. (2012b). Multiple microalgal partners in symbiosis with the Acantharia Acanthochiasma sp. (Radiolaria). Symbiosis 58, 233–244. doi: 10.1007/s13199-012-0195-x
del Campo, J., Sieracki, M. E., Molestina, R., Keeling, P., Massana, R., and Ruiz-Trillo, I. (2014). The others: our biased perspective of eukaryotic genomes. Trends Ecol. Evol. 29, 252–259. doi: 10.1016/j.tree.2014.03.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dennett, M. R., Caron, D. A., Michaels, A. F., Gallager, S. M., and Davis, C. S. (2002). Video plankton recorder reveals high abundances of colonial Radiolaria in surface waters of the central North Pacific. J. Plankton Res. 24, 797–805. doi: 10.1093/plankt/24.8.797
Dolven, J. K., Lindqvist, C., Albert, V. A., Bjorklund, K. R., Yuasa, T., Takahashi, O., et al. (2007). Molecular diversity of alveolates associated with neritic North Atlantic radiolarians. Protist 158, 65–76. doi: 10.1016/j.protis.2006.07.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Drummond, A. J., Ashton, B., Buxton, S., Cheung, M., Cooper, A., Duran, C., et al. (2011). Geneious Version 5.4 Created by Biomatters. Available online at: http://www.geneious.com
Falkowski, P. G., and Raven, J. A. (2013). Aquatic Photosynthesis. New York, NY: Princeton University Press. doi: 10.1515/9781400849727
Foster, R. A., Carpenter, E. J., and Bergman, B. (2006a). Unicellular cyanobionts in open ocean dinoflagellates, radiolarians, and tintinnids: ultrastructural characterization and immuno-localization of phycoerythrin and nitrogenase. J. Phycol. 42, 453–463. doi: 10.1111/j.1529-8817.2006.00206.x
Foster, R. A., Collier, J. L., and Carpenter, E. J. (2006b). Reverse transcription PCR amplification of cyanobacterial symbiont 16S rRNA sequences from single non-photosynthetic eukaryotic marine planktonic host cells. J. Phycol. 42, 243–250. doi: 10.1111/j.1529-8817.2006.00185.x
Gast, R. J., Beaudoin, D. J., and Caron, D. A. (2003). Isolation of symbiotically expressed genes from the dinoflagellate symbiont of the solitary radiolarian Thalassicolla nucleata. Biol. Bull. 204, 210–214. doi: 10.2307/1543561
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gast, R. J., and Caron, D. A. (1996). Molecular phylogeny of symbiotic dinoflagellates from plankton foraminifera and radiolaria. Mol. Biol. Evol. 13, 1192–1197. doi: 10.1093/oxfordjournals.molbev.a025684
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gast, R. J., and Caron, D. A. (2001). Photosymbiotic associations in planktonic foraminifera and radiolaria. Hydrobiologia 461, 1–7. doi: 10.1023/A:1012710909023
Gast, R. J., McDonnell, T. A., and Caron, D. A. (2000). srDNA-based taxonomic affinities of algal symbionts from a planktonic foraminifer and a solitary radiolarian. J. Phycol. 36, 172–177. doi: 10.1046/j.1529-8817.2000.99133.x
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Habura, A., Hou, Y. B., Reilly, A. A., and Bowser, S. S. (2011). High-throughput sequencing of Astrammina rara: sampling the giant genome of a giant foraminiferan protist. BMC Genomics 12:169. doi: 10.1186/1471-2164-12-169
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Huson, D. H., and Mitra, S. (2012). Introduction to the analysis of environmental sequences: metagenomics with MEGAN. Methods Mol. Biol. 856, 415–429. doi: 10.1007/978-1-61779-585-5_17
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Iversen, K. R., and Seuthe, L. (2011). Seasonal microbial processes in a high-latitude fjord (Kongsfjorden, Svalbard): I. Heterotrophic bacteria, picoplankton and nanoflagellates. Polar Biol. 34, 731–749. doi: 10.1007/s00300-010-0929-2
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keeling, P. J., Burki, F., Wilcox, H. M., Allam, B., Allen, E. E., Amaral-Zettler, L. A., et al. (2014). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889. doi: 10.1371/journal.pbio.1001889
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kii, S. I., Tanaka, J., and Watanabe, T. (2007). Guanine-cytosine contents of the host and symbiont cDNA in a symbiotic coral. Fish. Sci. 73, 1362–1372. doi: 10.1111/j.1444-2906.2007.01479.x
Krabberod, A. K., Brate, J., Dolven, J. K., Ose, R. F., Klaveness, D., Kristensen, T., et al. (2011). Radiolaria divided into polycystina and spasmaria in combined 18S and 28S rDNA phylogeny. PloS ONE 6:e23526. doi: 10.1371/journal.pone.0023526
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kvennefors, E. C. E., Leggat, W., Hoegh-Guldberg, O., Degnan, B. M., and Barnes, A. C. (2008). An ancient and variable mannose-binding lectin from the coral Acropora millepora binds both pathogens and symbionts. Dev. Comp. Immunol. 32, 1582–1592. doi: 10.1016/j.dci.2008.05.010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
La Jeunesse, T. C. (2001). Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the its region: in search of a “species” level marker. J. Phycol. 37, 866–880. doi: 10.1046/j.1529-8817.2001.01031.x
Li, W. Z., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lin, S. J. (2011). Genomic understanding of dinoflagellates. Res. Microbiol. 162, 551–569. doi: 10.1016/j.resmic.2011.04.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martinez-Garcia, M., Brazel, D., Poulton, N. J., Swan, B. K., Gomez, M. L., Masland, D., et al. (2012). Unveiling in situ interactions between marine protists and bacteria through single cell sequencing. ISME J. 6, 703–707. doi: 10.1038/ismej.2011.126
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Matsuoka, A. (2007). Living radiolarian feeding mechanisms: new light on past marine ecosystems. Swiss J. Geosci. 100, 273–279. doi: 10.1007/s00015-007-1228-y
Meyer, E., and Weis, V. M. (2012). Study of cnidarian-algal symbiosis in the “omics” age. Biol. Bull. 223, 44–65.
Nimmergut, A., and Abelmann, A. (2002). Spatial and seasonal changes of radiolarian standing stocks in the Sea of Okhotsk. Deep-Sea Res. Pt. I 49, 463–493. doi: 10.1016/S0967-0637(01)00074-7
Pillet, L., and Pawlowski, J. (2013). Transcriptome analysis of Foraminiferan Elphidium margaritaceum questions the role of gene transfer in kleptoplastidy. Mol. Biol. Evol. 30, 66–69. doi: 10.1093/molbev/mss226
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Probert, I., Siano, R., Poirier, C., Decelle, J., Biard, T., Tuji, A., et al. (2014). Brandtodinium gen. nov. and B.nutricula comb. Nov. (Dinophyceae), a dinoflagellate commonly found in symbiosis with polycystine radiolarians. J. Phycol. 50, 388–399. doi: 10.1111/jpy.12174
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Radax, R., Rattei, T., Lanzen, A., Bayer, C., Rapp, H. T., Urich, T., et al. (2012). Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ. Microbiol. 14, 1308–1324. doi: 10.1111/j.1462-2920.2012.02714.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sabourault, C., Ganot, P., Deleury, E., Allemand, D., and Furla, P. (2009). Comprehensive EST analysis of the symbiotic sea anemone, Anemonia viridis. BMC Genomics 10:333. doi: 10.1186/1471-2164-10-333
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Salter, S. J., Cox, M. J., Turek, E. M., Calus, S. T., Cookson, W. O., Moffatt, M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12:87. doi: 10.1186/s12915-014-0087-z
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schwarz, J. A., Brokstein, P. B., Voolstra, C., Terry, A. Y., Miller, D. J., Szmant, A. M., et al. (2008). Coral life history and symbiosis: functional genomic resources for two reef building Caribbean corals, Acropora palmata and Montastraea faveolata. BMC Genomics 9:97. doi: 10.1186/1471-2164-9-97
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sierra, R., Matz, M. V., Aglyamova, G., Pillet, L., Decelle, J., Not, F., et al. (2013). Deep relationships of Rhizaria revealed by phylogenomics: a farewell to Haeckel's Radiolaria. Mol. Phylogenet. Evol. 67, 53–59. doi: 10.1016/j.ympev.2012.12.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Simon, M., Grossart, H. P., Schweitzer, B., and Ploug, H. (2002). Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28, 175–211. doi: 10.3354/ame028175
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Singh, R. S., and Walia, A. K. (2014). Characteristics of lichen lectins and their role in symbiosis. Symbiosis 62, 123–134. doi: 10.1007/s13199-014-0278-y
Stoecker, D. K., Johnson, M. D., de Vargas, C., and Not, F. (2009). Acquired phototrophy in aquatic protists. Aquat. Microb. Ecol. 57, 279–310. doi: 10.3354/ame01340
Sun, W., Zhang, F. L., He, L. M., and Li, Z. Y. (2014). Pyrosequencing reveals diverse microbial community associated with the Zoanthid Palythoa australiae from the South China Sea. Microb. Ecol. 67, 942–950. doi: 10.1007/s00248-014-0395-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Swanberg, N. R., and Caron, D. A. (1991). Patterns of sarcodine feeding in epipelagic oceanic plankton. J. Plankton Res. 13, 287–312. doi: 10.1093/plankt/13.2.287
Toseland, A., Daines, S. J., Clark, J. R., Kirkham, A., Strauss, J., Uhlig, C., et al. (2013). The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Clim. Change 3, 979–984. doi: 10.1038/nclimate1989
Vidal-Dupiol, J., Adjeroud, M., Roger, E., Foure, L., Duval, D., Mone, Y., et al. (2009). Coral bleaching under thermal stress: putative involvement of host/symbiont recognition mechanisms. BMC Physiol. 9:14. doi: 10.1186/1472-6793-9-14
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Weis, V. M. (2008). Cellular mechanisms of Cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 211, 3059–3066. doi: 10.1242/jeb.009597
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wood-Charlson, E. M., Hollingsworth, L. L., Krupp, D. A., and Weis, V. M. (2006). Lectin/glycan interactions play a role in recognition in a coral/dinoflagellate symbiosis. Cell. Microbiol. 8, 1985–1993. doi: 10.1111/j.1462-5822.2006.00765.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wood-Charlson, E. M., and Weis, V. M. (2009). The diversity of C-type lectins in the genorne of a basal metazoan, Nematostella vectensis. Dev. Comp. Immunol. 33, 881–889. doi: 10.1016/j.dci.2009.01.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yooseph, S., Nealson, K. H., Rusch, D. B., McCrow, J. P., Dupont, C. L., Kim, M., et al. (2010). Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468, 60–66. doi: 10.1038/nature09530
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yuasa, T., Horiguchi, T., Mayama, S., Matsuoka, A., and Takahashi, O. (2012). Ultrastructural and molecular characterization of cyanobacterial symbionts in Dictyocoryne profunda (polycystine radiolaria). Symbiosis 57, 51–55. doi: 10.1007/s13199-012-0174-2
Yuyama, I., Harii, S., and Hidaka, M. (2012). Algal symbiont type affects gene expression in juveniles of the coral Acropora tenuis exposed to thermal stress. Mar. Environ. Res. 76, 41–47. doi: 10.1016/j.marenvres.2011.09.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yuyama, I., Watanabe, T., and Takei, Y. (2011). Profiling differential gene expression of symbiotic and aposymbiotic corals using a High Coverage Gene Expression Profiling (HiCEP) analysis. Mar. Biotechnol. 13, 32–40. doi: 10.1007/s10126-010-9265-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zelensky, A. N., and Gready, J. E. (2003). Comparative analysis of structural properties of the C-type-lectin-like domain (CTLD). Proteins 52, 466–477. doi: 10.1002/prot.10626
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zelensky, A. N., and Gready, J. E. (2005). The C-type lectin-like domain superfamily. FEBS J. 272, 6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhi-Liang, H., Bao, J., and Reecy, J. M. (2008). CateGOrizer: a web-based program to batch analyze gene ontology classification categories. Online J. Bioinform. 9, 108–112. Available online at: http://onljvetres.com/bioinfo.htm
Keywords: radiolarian, ESTs, c-type lectins, plankton, photosymbiosis, Rhizaria
Citation: Balzano S, Corre E, Decelle J, Sierra R, Wincker P, Da Silva C, Poulain J, Pawlowski J and Not F (2015) Transcriptome analyses to investigate symbiotic relationships between marine protists. Front. Microbiol. 6:98. doi: 10.3389/fmicb.2015.00098
Received: 18 November 2014; Accepted: 26 January 2015;
Published online: 17 March 2015.
Edited by:
Manuel Martinez Garcia, University of Alicante, SpainReviewed by:
Iñaki Ruiz-Trillo, Institut de Biologia Evolutiva (Commissariat à l'Energie Atomique et aux Energies Alternatives), SpainMichael Cunliffe, Marine Biological Association of the United Kingdom, UK
Copyright © 2015 Balzano, Corre, Decelle, Sierra, Wincker, Da Silva, Poulain, Pawlowski and Not. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sergio Balzano, Department of Marine Organic Biogeochemistry, Royal Netherlands Institute for Sea Research, PO Box 59, 1790 AB Den Burg, Netherlands e-mail: sergio.balzano@nioz.nl
†Present address: Sergio Balzano, Department of Marine Organic Biogeochemistry, Royal Netherlands Institute for Sea Research, Den Burg, Netherlands
 Sergio Balzano
Sergio Balzano Erwan Corre
Erwan Corre Johan Decelle
Johan Decelle Roberto Sierra
Roberto Sierra Patrick Wincker5
Patrick Wincker5 Corinne Da Silva
Corinne Da Silva Jan Pawlowski
Jan Pawlowski Fabrice Not
Fabrice Not