- 1Beijing Key Laboratory for Prevention and Control of Infectious Diseases in Livestock and Poultry, Institute of Animal Husbandry and Veterinary Medicine, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2Department of Animal Science, The University of Tennessee, Knoxville, TN, USA
Upon colonization in the host gastrointestinal tract, the enteric bacterial pathogen Campylobacter jejuni is exposed to a variety of signaling molecules including the catecholamine hormones epinephrine (Epi) and norepinephrine (NE). NE has been observed to stimulate the growth and potentially enhance the pathogenicity of C. jejuni. However, the underlying mechanisms are still largely unknown. In this study, both Epi and NE were also observed to promote C. jejuni growth in MEMα-based iron-restricted medium. Adhesion and invasion of Caco-2 cells by C. jejuni were also enhanced upon exposure to Epi or NE. To further examine the effect of Epi or NE on the pathobiology of C. jejuni, transcriptomic profiles were conducted for C. jejuni NCTC 11168 that was cultured in iron-restricted medium supplemented with Epi or NE. Compared to the genes expressed in the absence of the catecholamine hormones, 183 and 156 genes were differentially expressed in C. jejuni NCTC 11168 that was grown in the presence of Epi and NE, respectively. Of these differentially expressed genes, 102 genes were common for both Epi and NE treatments. The genes differentially expressed by Epi or NE are involved in diverse cellular functions including iron uptake, motility, virulence, oxidative stress response, nitrosative stress tolerance, enzyme metabolism, DNA repair and metabolism and ribosomal protein biosynthesis. The transcriptome analysis indicated that Epi and NE have similar effects on the gene expression of C. jejuni, and provided insights into the delicate interaction between C. jejuni and intestinal stress hormones in the host.
Introduction
Since the late 1970s, Campylobacter jejuni has emerged as the leading bacterial cause of foodborne human diseases in many industrialized countries (Olson et al., 2008). This pathogenic organism causes watery diarrhea and/or hemorrhagic colitis in humans and is also associated with Guillain-Barre syndrome, an acute flaccid paralysis that may lead to respiratory muscle compromise and death (Nachamkin et al., 1998). In contrast to its increased prevalence, limited progress has been made in the development of effective intervention strategies against Campylobacter infections in humans and animal reservoirs (Lin, 2009), which is primarily due to lack of good understanding of Campylobacter pathogenesis.
Through oral ingestion, C. jejuni enters the host intestine via the stomach acid barrier and colonizes the distal ileum and cecum. Once inside the intestine, C. jejuni is faced with various compounds and multiple levels of stresses, which may serve as cues to modulate C. jejuni gene expression for successful colonization in the intestine (Everest, 2007). Recently, gut catecholamines, a group of host stress hormones that could be released by the local enteric nervous system, have been reported to play an important role in host-bacteria communication in the intestine (Everest, 2007; Freestone, 2013). Specifically, catecholamines, such as epinephrine (Epi) and norepinephrine (NE), can modulate bacterial infectivity in different ways, such as promoting bacterial growth, stimulating virulence gene expression, and enhancing attachment to host tissue (Freestone et al., 2008; Lyte et al., 2011; Freestone, 2013). These unique findings have led to a new multidisciplinary research area called “Microbial Endocrinology” (Lyte, 1993, 2004; Freestone et al., 2008).
Recently, the catecholamine hormone NE was also observed to increase the virulence-associated properties of C. jejuni (Cogan et al., 2007) and stimulate the growth of C. jejuni under iron-limited condition (Cogan et al., 2007; Zeng et al., 2009; Xu et al., 2010). However, the underlying mechanisms of NE-mediated growth promotion and virulence enhancement in C. jejuni are still unknown. To address this issue, in this study, we used microarray to analyze transcriptomic profile of C. jejuni NCTC 11168 in response to catecholamine hormones Epi or NE under iron-restricted condition. The findings from this study provide a foundation for us to elucidate molecular basis of C. jejuni response to gut catecholamines in the future.
Materials and Methods
Bacterial Strain and Culture Conditions
C. jejuni NCTC 11168 was routinely grown in Mueller-Hinton (MH) broth (Difco, Sparks, MD) or on agar at 42°C in a microaerophilic incubator (5% O2, 10% CO2, 85% N2). To assess the effects of Epi or NE on the bacteria, C. jejuni NCTC 11168 was grown in the iron-restricted minimal essential medium alpha (MEMα) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Hyclone).
Bacterial Growth Assay
The effects of Epi and NE on the growth of C. jejuni NCTC 11168 were assessed using standard bacterial growth assay. Briefly, Campylobacter cells were grown in MH broth to log phase and were then pelleted, washed once and resuspended in MEMα medium. The bacterial suspension was inoculated into MEMα medium at a final concentration of 1 × 102 cfu/ml with or without 100 μM of (−)-epinephrine (+)-bitartrate salt (Epi) or L-(−)-norepinephrine (+)-bitartrate salt monohydrate (NE) (Sigma-Aldrich). Cultures were grown in 6-well plates with 5 ml of culture per well for 48 h. The optical density of each well at 600 nm was determined every 12 h of incubation using Eppendorf BioPhotometer. All assays were performed in three independent experiments with triplicate setting for each experiment. Results represent the mean ± the standard deviation (SD) of three independent experiments. The paired Student's t-test was used for statistical analysis at 5% level of significance.
Adhesion and Invasion Assay of Caco-2 Cells
The ability of Campylobacter to adhere and invade Caco-2 cells was assessed using assays as described in previous publications (Cogan et al., 2007; Ganan et al., 2010) with some modifications. Briefly, Caco-2 cells were grown and maintained in DMEM (Invitrogen) containing 1% non-essential amino acids and 10% fetal bovine serum. The Caco-2 cells were seeded into 24-well plates at 1 × 105 cells/well and incubated at 37°C in a 5% CO2 atmosphere until confluence. The C. jejuni NCTC 11168 was grown in MEMα medium in the presence or absence of Epi or NE for 36 h (mid-log phase) under microaerophilic condition as described above. The C. jejuni cells were pelleted, washed three times with MEMα medium, and finally resuspended in MEMα medium. The C. jejuni inoculum was added into triplicate wells of confluent monolayer of Caco-2 cells to give a MOI of 100:1. Following incubation at 37°C for 2 h, the non-adherent bacteria were removed by washing four times with PBS. To determine bacterial adherence, the cell monolayer were lysed with 1% Triton X-100 (Sigma-Aldrich) and total intracellular and extracellular bacteria were enumerated by serial dilutions. To determine bacterial invasion, the cell monolayer was incubated in fresh PBS containing 1% FBS and 150 μg/ml gentamicin for 2 h to kill remaining viable extracellular bacteria. Then the cell monolayer was washed three times with PBS and lysed with 1% Triton X-100 to determine the intracellular bacteria by serial dilutions. Results were expressed as percent of adhered or invasive bacteria relative to inoculum as described in previous study (Ganan et al., 2010). The experiments were repeated three times, and the results shown are the mean ± the standard deviation (SD). The paired Student's t-test was used for statistical analysis at 5% level of significance.
Microarray Protocol
The microarray slides for C. jejuni NCTC 11168 were designed using Agilent eArray program and manufactured by Agilent Technologies. Each customized microarray contained spots in 8 replicates with 1683 60-mer oligonucleotides specifically targeting the 1683 protein-coding genes in C. jejuni NCTC 11168 genome (Parkhill et al., 2000; Gundogdu et al., 2007).
As described in the bacterial growth assay, the C. jejuni NCTC 11168 cells harvested from MH broth were inoculated into MEMα medium at a final concentration of 1 × 102 cfu/ml with or without 100 μM Epi or NE. The Campylobacter cells were grown in 6-well plates with 5 ml per well at 42°C under microaerophilic condition. At the mid-log phase (approximately 36 h of growth), the Epi or NE treated and untreated C. jejuni NCTC 11168 cultures were collected and treated with RNAlater stabilization solution (Ambion, Life Technologies). Three (for Epi or NE treated cultures) or four (for untreated control cultures) independent biological replicates were performed. The harvested C. jejuni cells were pelleted to remove the RNAlater solution and the cell pellets were used for RNA extraction using RiboPure™-Bacteria Kit (Ambion, Life Technologies) according to the manufacturer's instruction. The quality and quantity of total RNA were determined by an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, US). The extracted RNA was further purified by RNeasy micro kit and RNase-Free DNase Set (QIAGEN, GmBH, Germany). Total RNA was amplified and labeled by Low Input Quick Amp Labeling Kit, One-Color (Agilent technologies, Santa Clara, CA, US) by following the manufacturer's instructions. The labeled cRNA were purified by RNeasy mini kit (QIAGEN, GmBH, Germany). Each slide was hybridized with 600 ng of Cy3-labeled cRNA using Gene Expression Hybridization Kit (Agilent technologies, Santa Clara, CA, US). Following 17 h of hybridization, the slides were washed in staining dishes with Gene Expression Wash Buffer Kit (Agilent technologies, Santa Clara, CA, US). Slides were scanned by Agilent Microarray Scanner (Agilent technologies, Santa Clara, CA, US) with default settings, dye channel: green, scan resolution = 5 μm, PMT 100%, 10%, 16 bit.
The microarray data were collected with Feature Extraction software 10.7 (Agilent technologies). Raw data were normalized by Quantile algorithm, Gene Spring Software 11.0 (Agilent technologies). All the data are MIAME compliant and the raw data has been deposited in the NCBI GEO database under the number GSE65187. Subsequently, the normalized data were statistically analyzed using a mixed-linear model with the SAS statistical package, accounting for fixed effects of treatment and random effects of replications, samples, and slides. P-values were adjusted by a False Discovery Rate of 5% and differential expression declared when P < 0.05. For this study, we chose P < 0.05 and fold change equal to or greater than 1.5 as the cutoff for significant differential expression between different growth conditions.
The differentially expressed genes were categorized by their Clusters of Orthologous Groups (COG) annotations (Tatusov et al., 2003). Genes with no COG annotation were included in the poorly characterized category. Moreover, the differentially expressed genes of C. jejuni following exposure to Epi or NE were analyzed and compared with two previously published datasets which obtained using microarray (Palyada et al., 2004) or RNA-Seq (Butcher and Stintzi, 2013) approaches under iron replete and iron limited conditions.
Real-Time Quantitative RT-PCR
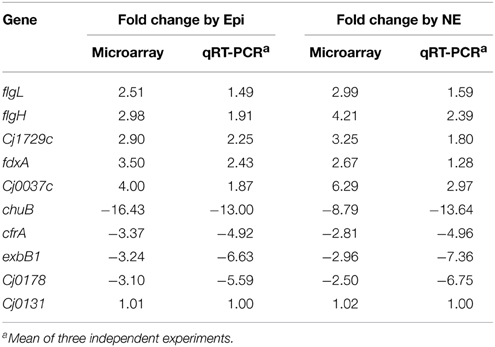
Total 10 genes were selected from microarray data analysis and validated by qRT-PCR analysis. The selected genes and the primers used for qRT-PCR are listed in Table S1. Briefly, RNA was extracted from the treatment and control cultures as described above. The qRT-PCR was performed using iQ SYBR green Supermix kit (Bio-Rad) with the IQ5 Real-time-PCR Detection System (Bio-Rad). The qRT-PCR parameters consisted of a cDNA synthesis at 50°C for 10 min and reverse transcriptase inactivation at 95°C for 5 min, followed by PCR cycling and detection of 95°C for 10 s and 60°C for 30 s (40 cycles). Similar to a previous study by which the relative expression level of each gene was normalized to the invariant expression genes slyD or ilvC (Palyada et al., 2004), the relative transcription level of each gene in this study was normalized to the Cj0131 gene which displayed no changes under different growth conditions. Quantitative values were obtained with the comparative threshold cycle (ΔΔCT) method. The transcript level from each RNA sample was assayed three times, and the mean CT value used for further analysis. The difference in the transcription of each specific gene was calculated by the 2−ΔΔCT method.
Results
Both Epi and NE Promote the Growth of C. jejuni
As shown in Figure 1, the growth of C. jejuni in the presence of Epi or NE was significantly enhanced compared with control culture at all tested time points (P < 0.01). By 24 h of growth, the addition of Epi or NE resulted in 3 and 2 folds increase in growth (reflected by OD600), respectively. The growth assay also showed that the growth rate of C. jejuni was higher in NE-supplemented culture than in Epi-supplemented culture (P < 0.01).
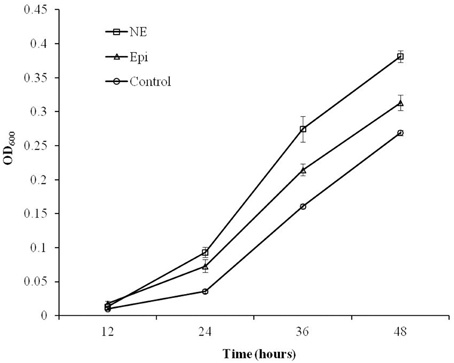
Figure 1. Growth response of C. jejuni NCTC 11168 to Epi or NE in iron-restricted MEMα medium containing 10% FBS. Results represent the mean plus the standard deviation (SD) of three independent experiments.
Enhancement of Adhesion and Invasion of Caco-2 Cells by Epi and NE
As showed in Figure 2, the adherence and invasion rates for the Campylobacter treated by Epi or NE were significantly higher than the untreated control group (P < 0.01). Specifically, the adherence rates are 0.012, 0.026, 0.051% for control, Epi, and NE group, respectively while invasion rates are 0.00032, 0.00098, 0.00056% for control, Epi, and NE group, respectively. Moreover, NE treated bacteria showed significantly higher adhesive (P < 0.01) and lower invasive (P < 0.05) capacities than bacteria treated by Epi.
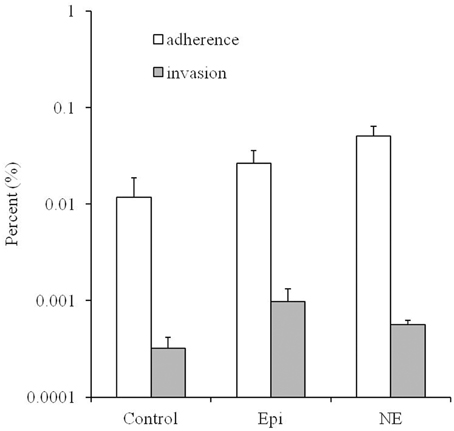
Figure 2. Effects of Epi and NE on adhesion and invasion of Caco-2 cells by C. jejuni NCTC 11168. Results were expressed as percent of adhered or invasive bacteria relative to inoculum. Results represent the mean plus the standard deviation (SD) of three independent experiments.
Effects of Epi or NE on the Transcriptome of C. jejuni
To investigate the transcriptional profiles of C. jejuni NCTC 11168 in response to Epi or NE, microarrays were used to compare the transcriptome of the C. jejuni grown in Epi or NE-supplemented MEMα medium to those grown in the absence of the catecholamines. The genes whose expression was altered by at least 1.5-fold (P < 0.05) were designated as differentially expressed as described in previous studies (Dowd, 2007; Li et al., 2012). The numbers of genes differentially expressed upon exposure to Epi or NE were calculated and analyzed (Table 1). A total of 183 (96 up-regulated genes and 87 down-regulated genes) and 156 genes (86 up-regulated genes and 70 down-regulated genes) were differentially expressed by Epi and NE, respectively (Tables S3, S4), with 102 differentially expressed genes (56 up-regulated genes and 46 down-regulated genes) common between the two treatments (Tables S3, S4).
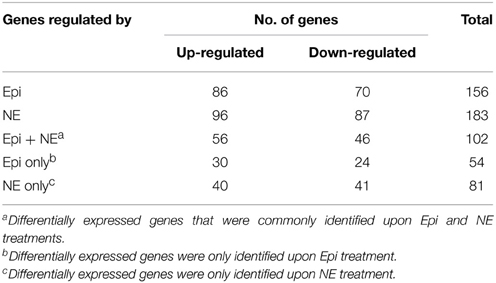
Table 1. Summary of differentially expressed genes in Campylobacter jejuni NCTC 11168 upon exposure to Epi or NE.
To validate the microarray data, the expression of 10 genes was selected and confirmed by qRT-PCR. As shown in Table 2, the qRT-PCR results are consistent with the data from microarray, demonstrating the reliability of the microarray data.
The individual genes that were found to be differentially expressed in response to Epi or NE were sorted by COG category. The differentially expressed genes for Epi or NE treatment were classified into 20 different functional categories respectively (Figure 3). Among differentially expressed genes by Epi, approximately 28% genes are poorly characterized (R+S); the main COG categories were J (20%), P (10.3%), E (5.8%), C (5.8%), N (5.1%), and U (4.5%). For differentially expressed genes by NE, in addition to the poorly characterized (R+S) genes (31%), the main COG categories include J (16%), M (8.7%), P (7.1%), E (6%), C (5.5%), and U (4.9%). There is only slight difference observed between Epi-regulated and NE-regulated genes. The wide diversity in functional categories suggests that Epi or NE treatment has a significant influence on metabolism, transcription and translation, cellular processes and signaling of C. jejuni NCTC 11168.
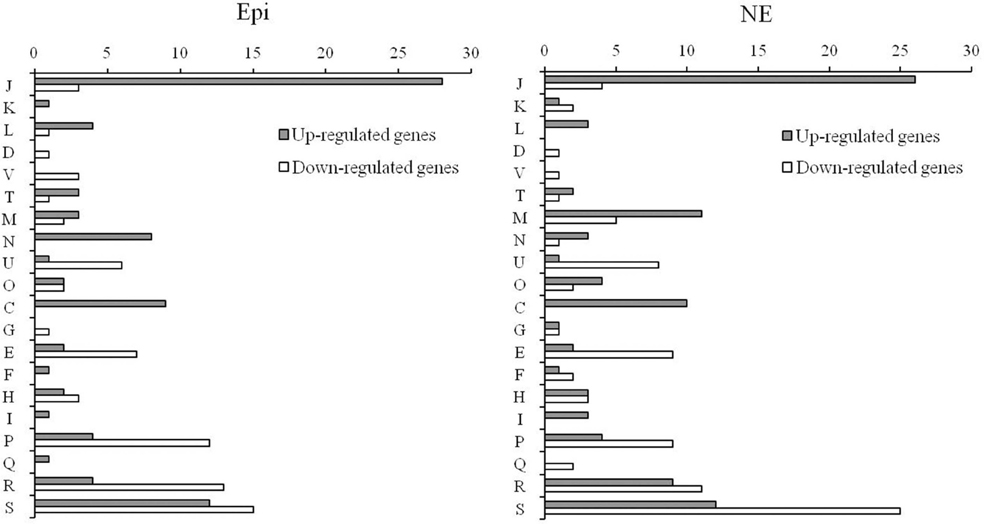
Figure 3. Functional classification of differentially expressed genes in C. jejuni NCTC 11168 upon exposure to Epi or NE. Gene functions are sorted according to COG categories: Information storage and processing (J: Translation, ribosomal structure, and biogenesis; K: Transcription; L: Replication, recombination and repair); Cellular processes and signaling (D: Cell cycle control, cell division, chromosome partitioning; V: Defense mechanisms; T: Signal transduction mechanisms; M: Cell wall/membrane/envelope biogenesis; N: Cell motility; U: Intracellular trafficking, secretion, and vesicular transport; O: Posttranslational modification, protein turnover, chaperones); Metabolism (C: Energy production and conversion; G: Carbohydrate transport and metabolism; E: Amino acid transport and metabolism; F: Nucleotide transport and metabolism; H: Coenzyme transport and metabolism; I: Lipid transport and metabolism; P: Inorganic ion transport and metabolism; Q: Secondary metabolites biosynthesis, transport and catabolism); Poorly characterized (R: General function prediction only; S: Function unknown or not in COG database).
Based on the COG category, the majority of differentially expressed genes in response to Epi or NE can be further classified into several main functional groups as described below.
Iron Uptake Systems
As shown in Table 3, the genes down-regulated by both Epi and NE include almost all the known iron uptake transporters in C. jejuni, including the ferric enterobactin receptor (cfrA), rhodotorulic acid (Cj1658, Cj1660-1662), hemin (chuABCD), and transferrin (cfbpABC, Cj0176c-0178) (Miller et al., 2009). Interestingly, no significant differential expression was observed for genes belonging to ABC transport system ceuBCDE and the rhodotorulic acid transporter p19. In addition, the genes belong to the part of energy transduction systems (exbB1-exbD1, exbB2-exbD2) were down-regulated by Epi or NE while the expression of tonB1, tonB2, tonB3, and exbB3-exbD3 was not affected by Epi or NE. Expression of the ferric uptake regulator Fur was not affected by Epi or NE either.
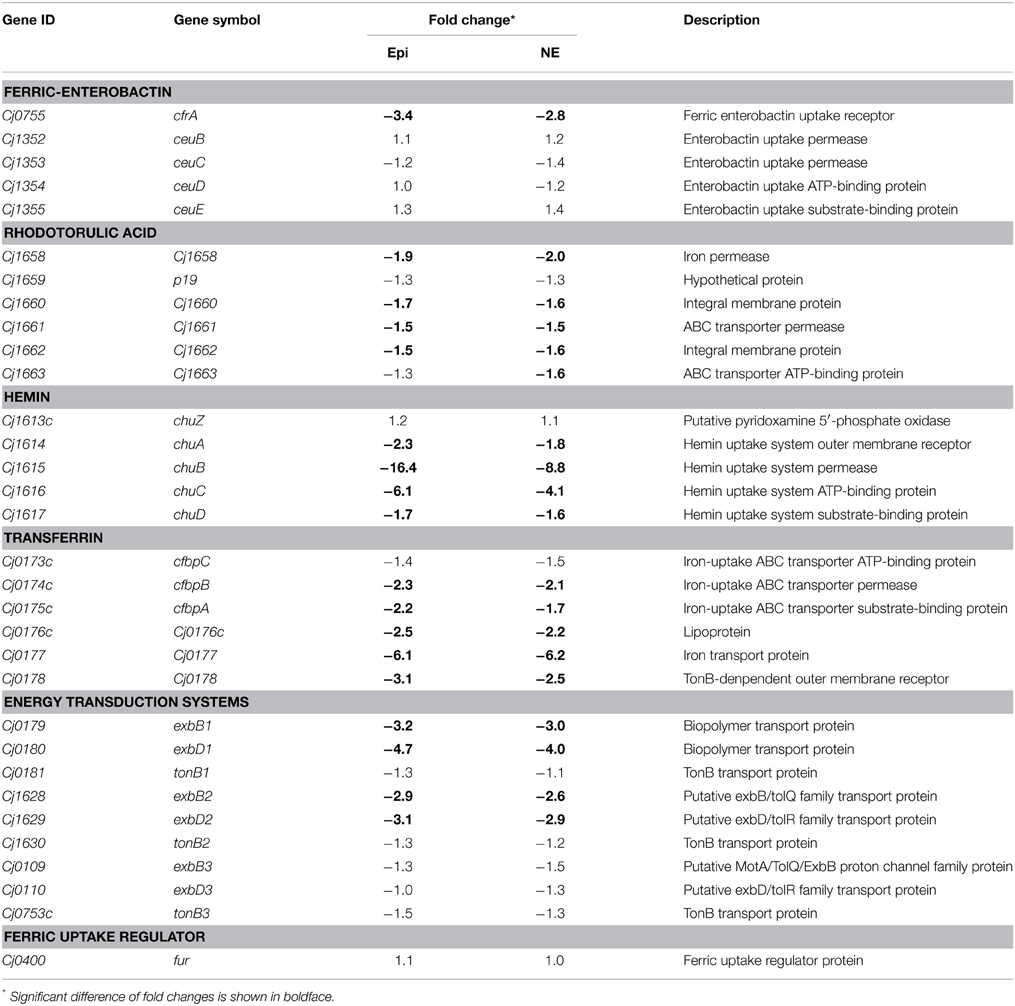
Table 3. Fold change of iron-uptake system components in Campylobacter jejuni upon exposure to Epi or NE.
Bacterial Motility (Flagellar Assembly Pathway)
Many of the up-regulated genes are involved in flagellar assembly pathway (Tables S3, S4). In the presence of Epi or NE, the flagellar genes involved into the flagellar biogenesis were up-regulated, which include the genes encoding hook-filament junction (flgL), hook (flgE and flgE2), FlgE chaperones (flgD and fliK), and rod (flgG2, flgH, and flgI) (Table 4). No difference was observed for the expression of the genes encoding other flagellar components including filament (flaA, flaG, and fliD), rod (fliE, fliF, fliG, and fliH) and regulatory proteins (fliS, flgM, flgR, and fliA) (Table 4). Chemotaxis also plays an essential role in flagella locomotion and bacterial motility in the presence of specific chemoeffectors (Zautner et al., 2012). The genes encoding chemotaxis histidine kinase CheA (Cj0284c) and the coupling scaffold protein CheW (Cj0283c) were both up-regulated with 2.1 and 1.8-fold by Epi, respectively. Expression of CheA and CheW genes was also increased about 2.4 and 2.1-fold by NE, respectively.
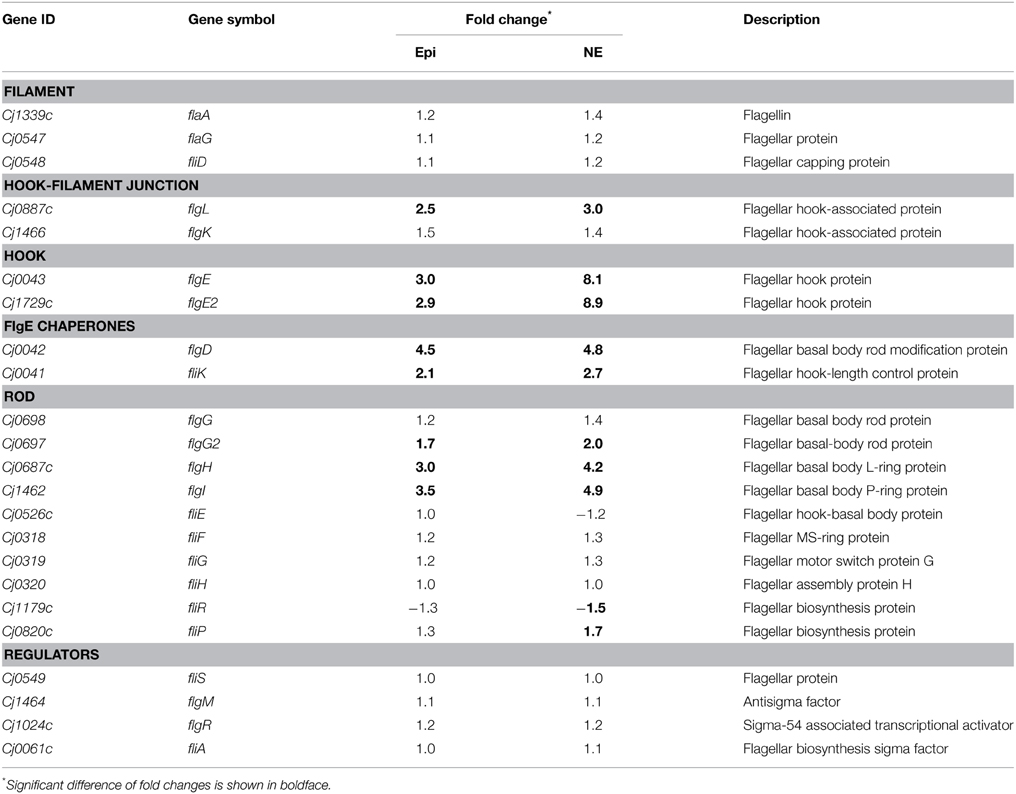
Table 4. Fold change of the flagella assembly components in Campylobacter jejuni upon exposure to Epi or NE.
Oxidative Stress Responses
Production of reactive oxygen species (ROS) during normal metabolic processes is detrimental to C. jejuni. Therefore, C. jejuni has evolved defense mechanisms to protect it under oxidative stress. In this study, genes of katA and Cj1386 were observed to be down-regulated by the catecholamines (2.8 and 1.7-fold by Epi; 2.9 and 1.8-fold by NE). The genes of a non-haem iron protein Rrc (Cj0012c) and ferredoxin FdxA (Cj0333c) were up-regulated by Epi or NE. The hemerythrin HerB (Cj1224) involved in protecting the activity of oxidoreductases from oxygen damage (Kendall et al., 2014) was also up-regulated by Epi.
Nitrosative Stress Tolerance
The anaerobic-respiration pathways are very important in C. jejuni growth and colonization (Weingarten et al., 2008). The periplasmic nitrite reductase NrfH-NrfA (Cj1358c-Cj1357c), the nitrate reductase NapA/B (Cj0780/Cj0783) and the cbb3-terminal oxidase ccoNOQ operon (Cj1490c-Cj1488c) were up-regulated by Epi or NE, which indicated that catecholamines enhance the ability of nitrosative stress tolerance in C. jejuni. In contrast, the yedYZ-like operon Cj0379c-Cj0378c which play a possible role in the reduction of reactive nitrogen species in the periplasm (Hitchcock et al., 2010) was down-regulated by Epi or NE. Notably, a nitrosative stress regulator NssR (Cj0466) was reduced 1.9-fold in expression by NE.
Virulence-Associated Genes
The Cj0125c, encoding a DksA-like protein which was reported to control expression of virulence genes in pathogenic bacteria (Yun et al., 2008), was up-regulated by Epi or NE. The Cj0040, Cj1025c, and Cj1242, which were identified as novel virulence factors co-regulated with flagella (Guerry, 2007), were also up-regulated by Epi or NE. The Cj1583c gene (a homolog of sapB) and the putative trigger factor Tig (Cj0193c), that were involved into antimicrobial peptides resistance (Hoang et al., 2012), were up-regulated by Epi or NE.
Enzyme Metabolism Genes
A bifunctional enzyme IspDF (Cj1607) involved into the isoprenoid biosynthesis in the nonmevalonate pathway (Gabrielsen et al., 2004) was up-regulated by Epi or NE. The genes downstream of Cj1607 (Cj1608 and Cj1609) were also up-regulated by Epi. The Pta gene (Cj0688) that encodes phosphotransacetylase for acetate production (Wright et al., 2009) was up-regulated by Epi or NE. Genes of HydB (Cj1266c) and NikAB (Cj1584c-Cj1583c) that were involved into a NiFe-type uptake system to maintain hydrogenase activity (Howlett et al., 2012) were also up-regulated by Epi or NE. In contrast, the Cj1161c gene, encoding a putative CopA homolog (Hall et al., 2008), was down-regulated by Epi or NE. Moreover, its downstream genes Cj1160c and Cj1158c were also down-regulated by Epi or NE.
DNA Repair and Metabolism
The UvrB (Cj0680c) and RecA (Cj1673c) genes involved into DNA repair mechanisms in C. jejuni (Gaasbeek et al., 2009) were up-regulated by Epi or NE. The gene encoding competence factor ComEA (Cj0011c), which function as a periplasmic DNA receptor contributing to the natural transformation of C. jejuni (Jeon and Zhang, 2007), was up-regulated by Epi. However, the competence factor Cj1211, which plays a critical role in natural transformation in Campylobacter (Jeon et al., 2008), was down-regulated by Epi or NE. The Mfd gene (Cj1085c), which encodes a transcription-repair coupling factor involved in strand-specific DNA repair and fluoroquinolone resistance (Han et al., 2008), was down-regulated by Epi.
Ribosomal Protein Biosynthesis
The 22 genes of rpsJ-rplR (Cj1708c-Cj1691c), rpsI-rplM (Cj1479c-Cj1480c), and rpsM-rpsK (Cj1592-Cj1593) involved into ribosomal protein biosynthesis were all up-regulated by Epi or NE. Moreover, genes of rpsU (Cj0370), rpmB (Cj0450c), rplO-rpsE (Cj1690c-Cj1689c) increased in expression by Epi and rpsD (Cj1594) increased in expression by NE.
Genes with opposite Expression Patterns between Catecholamines and Iron
Notably, the serum-MEMα medium used in this study was regarded as iron limited condition. Supplementation of such medium with Epi or NE may enable C. jejuni to utilize ferric-catecholamine complex as observed in our previous studies (Zeng et al., 2009; Xu et al., 2010). Thus, presence of Epi or NE in the iron-restricted MEMα medium could facilitate catecholamine-mediated iron uptake, consequently leading to increased iron level in cytoplasm. Therefore, the transcriptome profiling of C. jejuni NCTC 11168 upon exposure to Epi or NE in this study was also used to compare with two previous publications which analyzed the transcriptome of the bacteria under iron-replete and iron-limited growth conditions by microarray (Palyada et al., 2004) and RNA-seq (Butcher and Stintzi, 2013) respectively (Table S2). It's not surprising that a wide variety of genes involved in iron-uptake, motility, oxidative stress response, and other functions showed a similar expression profiling between this study (in response to catecholamine treatment) and the other two studies focused on the response to iron availability (Palyada et al., 2004; Butcher and Stintzi, 2013). However, it was notable that the transcriptional profiles of some genes in this study were not completely consistent with previous iron transcriptome studies. In particular, expression of some genes was totally in an opposite pattern. For example, this study demonstrated that the operon Cj1172c-Cj1169c encoding a hypothetical protein Cj1172c, a peptidyl-prolyl cis-trans isomerase Ppi (Cj1171c), a 50 kDa outer membrane protein precursor Omp50 (Cj1170c) and a putative periplasmic protein Cj1169c were all up-regulated by Epi or NE. In contrast, all the 4 genes were down-regulated in iron-replete medium (Palyada et al., 2004; Butcher and Stintzi, 2013). Other genes that were activated by Epi or NE in this study but were repressed by high iron level include typA (Cj0039c), thrS (Cj0206), panB (Cj0298c), fabG (Cj0435), aspS (Cj0640c), and fliP (Cj0820c). Moreover, genes that were down-regulated by Epi or NE in this study but were observed to be activated by iron restriction include birA (Cj0099), pyrE (Cj0233c), Cj0760-Cj0761, thiE (Cj1081c), mfd (Cj1085c), acpP3 (Cj1304), Cj1435c-Cj1436c, ctsD (Cj1474c), and Cj1684c. This finding strongly suggest that catecholamine-mediated impacts on Campylobacter do not solely rely on the iron chelator nature of catecholamines for iron acquisition; catecholamines may also serve as cues to modulate pathobiology of C. jejuni through other mechanisms.
Discussion
In the intestinal lumen, bacteria inevitably encounter the neurotransmitter, in particular, the NE released by noradrenergic neurons of the enteric nervous system (Freestone et al., 2008; Freestone, 2013). The medium chosen to investigate neurotransmitter responsiveness is crucially important in determining the mechanism by which the hormone modulates bacterial growth or virulence (Freestone et al., 2008). The iron-rich broth media such as Luria-Bertani (LB) and tryptic soy broth (TSB) have been used to investigate global effects of catecholamines on gene expression of Escherichia coli (Bansal et al., 2007), Salmonella enteric (Karavolos et al., 2008), and Actinobacillus pleuropneumoniae (Li et al., 2012). However, there have been some concerns for using such iron-replete media to examine the effect of catecholamines on systematic gene expression (Freestone et al., 2008). In particular, interactions of catecholamine with ferric iron in iron-rich medium can lead to the generation of oxygen-derived free radicals and subsequent oxidative stress influencing bacterial gene expression (Freestone et al., 2008). Therefore, it is important to choose appropriate growth medium to mimic in vivo environment for studying bacterial response to intestinal catecholamine hormones (Freestone, 2013). In other organisms, the serum-SAPI minimal medium has been used in microarray analysis of E. coli (Dowd, 2007) and S. enterica (Bearson et al., 2008) upon exposure to catecholamines. These iron-restricted, chemically defined media may minimize confounding factors during transcriptome analysis in response to catecholamine hormones. In this study, the MEMα medium containing 10% FBS was used to study C. jejuni growth and gene expression in response to Epi or NE. Significant growth promotion of C. jejuni by Epi or NE was also observed in this medium. Notably, the MEMα medium has been successfully used for examining C. jejuni transcriptome profiling in response to iron availability in previous C. jejuni studies (Palyada et al., 2004; Butcher and Stintzi, 2013).
The findings from this study indicate Epi and NE have similar effect on the transcriptome profile of C. jejuni grown in MEMα medium, likely due to their structural similarity for chelating iron (Table 1). However, presence of a panel of unique NE-modulated and Epi-modulated genes (Table 1) suggests that these two catecholamines may exert different effects on C. jejuni physiology. A recent study observed significant difference on gene expression of A. pleuropneumoniae between Epi and NE treatment (Li et al., 2012).
Epi and NE have been found to enhance the growth ability of C. jejuni under iron-restricted conditions in this study and previous publications (Cogan et al., 2007; Zeng et al., 2009; Xu et al., 2010). The effects of Epi or NE on bacterial growth promotion is likely due to catecholamine-mediated iron uptake because the catechol moiety in Epi and NE displays high affinity for ferric iron and can compete the ferric iron sequestered by host iron-binding proteins transferrin and lactoferrin (Freestone et al., 2000, 2008; Lyte, 2004). Subsequently, the ferric-catecholamine complex could be transported into intracellular via iron acquisition system for utilization. An early study suggested that growth response of E. coli to NE requires a functional Ent uptake system (Burton et al., 2002). Our previous studies indicated that FeEnt receptor CfrA (but not CfrB) is involved in NE utilization by C. jejuni (Zeng et al., 2009; Xu et al., 2010). As expected, most iron uptake players including CfrA, ChuA, and Cj0178 were down-regulated in the presence of Epi or NE. However, we cannot rule out the possibility that catecholamines may enhance bacterial growth and/or pathobiology features through other mechanisms.
To better understand the mechanisms by which bacteria sense catecholamines, the holistic approach microarray has been widely used. In recent studies, global transcriptome profiling of bacteria in response to catecholamines have been analyzed in E. coli (Bansal et al., 2007; Dowd, 2007; Kendall et al., 2007), S. enterica (Bearson et al., 2008; Karavolos et al., 2008), and A. pleuropneumoniae (Li et al., 2012). The catecholamine-modulated genes in other bacteria include those involved in the regulation of various physiological properties (Bansal et al., 2007; Dowd, 2007; Kendall et al., 2007; Bearson et al., 2008; Karavolos et al., 2008; Li et al., 2012). Similarly, in this study, we also observed that similar genes were modulated by Epi or NE, such as those involved in iron uptake, motility, stress response, and other functions.
In this study, we took advantage of previous iron transcriptome data (Palyada et al., 2004; Butcher and Stintzi, 2013) for comparative analysis (Table S2). We observed that almost all the genes involved in iron uptake and motility showed the similar expression patterns between catecholamines and iron though differentially expressed levels exist with some genes. For example, the differentially expressed genes activated by Epi or NE were observed in the flagella assembly components including hook-filament junction, hook, rod and chaperones (Table 4) while almost all the components of the flagellar assembly pathway were up-regulated under iron-replete condition (Butcher and Stintzi, 2013). Comparative analysis of the genes involved in stress response also showed the similar results. In previous studies, the genes involved in oxidative stress defense were repressed by iron, which include katA, Cj1386, perR, ahpC, trxB, and tpx (Van Vliet et al., 1999; Palyada et al., 2004, 2009; Holmes et al., 2005; Butcher and Stintzi, 2013). Similarly, the genes of katA, perR, and ahpC were also down-regulated by inactivation of the transcriptional regulator Cj1556, which plays a role in the oxidative and aerobic stress response in C. jejuni (Gundogdu et al., 2011). The similar expression profiles of katA and Cj1386 were observed in this study. Moreover, the three enzymes (NrfH-NrfA, NapA/B, and ccoNOQ) involved into nitrosative stress response in C. jejuni were increased in expression in response to Epi or NE, similar to transcriptional profiling of the three enzymes in C. jejuni during cecal colonization in vivo (Woodall et al., 2005) as well as the previous reports on induction of NrfH-NrfA and NapA/B expression by iron (Palyada et al., 2004; Holmes et al., 2005).
Notably, according to comparative analysis of transcriptome data (Table S2), we also observed a panel of genes were uniquely regulated by the presence of NE or Epi in C. jejuni, further supporting a hypothesis that catecholamine-mediated iron acquisition is not the only mechanism by which catecholamines influence on the bacterial pathobiology (Freestone et al., 2008). In E. coli (Sperandio et al., 2003) and S. Typhimurium (Bearson and Bearson, 2008), other regulatory factors have been observed to be involved in the interaction between bacteria and catecholamine hormones. The QseC and QseE sensor kinases have been identified as bacterial adrenergic receptors for catecholamines in E. coli (Clarke et al., 2006; Reading et al., 2009). Both the sensors belong to the two-component regulatory systems (QseBC and QseEF) and play a role in the catecholamine-mediated bacterial pathogenesis (Hughes et al., 2009; Reading et al., 2009). The two-component systems QseBC and QseEF also have been proposed to be catecholamine receptors in S. Typhimurium (Moreira and Sperandio, 2012; Karavolos et al., 2013). In this study, the C. jejuni two-component regulator gene Cj1608 was observed to be up-regulated 2.1 and 2.3-fold by Epi and NE, respectively. In addition, a putative two-component sensor kinase gene Cj1492c was down-regulated 1.6 and 2.1-fold by Epi and NE, respectively (Table S2). Specific role of these regulators in C. jejuni-catecholamine interaction needs to be examined in the future.
In this study, some genes showed opposite expression pattern when compared to previous iron transcriptome studies (Palyada et al., 2004; Butcher and Stintzi, 2013); examination of these genes was also highly warranted in the future. For example, among these specific genes, the Omp50 has been characterized as a species-specific outer membrane monomeric porin (Bolla et al., 2000). Recent C. jejuni work has demonstrated that Omp50 is a tyrosine phosphorylated kinase that controls capsule polysaccharide biosynthesis or export, and involves in bacterial motility, invasiveness and oxidative stress response (Corcionivoschi et al., 2012). Interestingly, treatment of C. jejuni with erythromycin increased expression of Omp50, which represents an adaptive mechanism by reducing cell permeability to Ery (Xia et al., 2013). The enhanced expression of Omp50 in response to NE and Epi observed in this study may have a significant implication in C. jejuni infection in the intestine.
Author Contributions
FX, and JL contributed with the conception and design of the study. FX, CW, FG, GC, XZ, BY, and JL were involved in the collection, assembly, analysis and interpretation of data; drafting and revision of the article; and final approval of the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Maojun Zhang (National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention) for support to this study. This study was supported by grant from the National Natural Science Foundation of China (31172344).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00452/abstract
References
Bansal, T., Englert, D., Lee, J., Hegde, M., Wood, T. K., and Jayaraman, A. (2007). Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect. Immun. 75, 4597–4607. doi: 10.1128/IAI.00630-07
Bearson, B. L., and Bearson, S. M. (2008). The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog. 44, 271–278. doi: 10.1016/j.micpath.2007.10.001
Bearson, B. L., Bearson, S. M., Uthe, J. J., Dowd, S. E., Houghton, J. O., Lee, I., et al. (2008). Iron regulated genes of Salmonella enterica serovar Typhimurium in response to norepinephrine and the requirement of fepDGC for norepinephrine-enhanced growth. Microbes Infect. 10, 807–816. doi: 10.1016/j.micinf.2008.04.011
Bolla, J. M., De, E., Dorez, A., and Pages, J. M. (2000). Purification, characterization and sequence analysis of Omp50,a new porin isolated from Campylobacter jejuni. Biochem. J. 352(Pt 3), 637–643. doi: 10.1042/0264-6021:3520637
Burton, C. L., Chhabra, S. R., Swift, S., Baldwin, T. J., Withers, H., Hill, S. J., et al. (2002). The growth response of Escherichia coli to neurotransmitters and related catecholamine drugs requires a functional enterobactin biosynthesis and uptake system. Infect. Immun. 70, 5913–5923. doi: 10.1128/IAI.70.11.5913-5923.2002
Butcher, J., and Stintzi, A. (2013). The transcriptional landscape of Campylobacter jejuni under iron replete and iron limited growth conditions. PLoS ONE 8:e79475. doi: 10.1371/journal.pone.0079475
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C., and Sperandio, V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10420–10425. doi: 10.1073/pnas.0604343103
Cogan, T. A., Thomas, A. O., Rees, L. E., Taylor, A. H., Jepson, M. A., Williams, P. H., et al. (2007). Norepinephrine increases the pathogenic potential of Campylobacter jejuni. Gut 56, 1060–1065. doi: 10.1136/gut.2006.114926
Corcionivoschi, N., Alvarez, L. A., Sharp, T. H., Strengert, M., Alemka, A., Mantell, J., et al. (2012). Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe 12, 47–59. doi: 10.1016/j.chom.2012.05.018
Dowd, S. E. (2007). Escherichia coli O157:H7 gene expression in the presence of catecholamine norepinephrine. FEMS Microbiol. Lett. 273, 214–223. doi: 10.1111/j.1574-6968.2007.00800.x
Everest, P. (2007). Stress and bacteria: microbial endocrinology. Gut 56, 1037–1038. doi: 10.1136/gut.2006.117150
Freestone, P. (2013). Communication between Bacteria and their hosts. Scientifica (Cairo) 2013:361073. doi: 10.1155/2013/361073
Freestone, P. P., Lyte, M., Neal, C. P., Maggs, A. F., Haigh, R. D., and Williams, P. H. (2000). The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J. Bacteriol. 182, 6091–6098. doi: 10.1128/JB.182.21.6091-6098.2000
Freestone, P. P., Sandrini, S. M., Haigh, R. D., and Lyte, M. (2008). Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol. 16, 55–64. doi: 10.1016/j.tim.2007.11.005
Gaasbeek, E. J., Van Der Wal, F. J., Van Putten, J. P., De Boer, P., Van Der Graaf-Van Bloois, L., De Boer, A. G., et al. (2009). Functional characterization of excision repair and RecA-dependent recombinational DNA repair in Campylobacter jejuni. J. Bacteriol. 191, 3785–3793. doi: 10.1128/JB.01817-08
Gabrielsen, M., Rohdich, F., Eisenreich, W., Grawert, T., Hecht, S., Bacher, A., et al. (2004). Biosynthesis of isoprenoids: a bifunctional IspDF enzyme from Campylobacter jejuni. Eur. J. Biochem. 271, 3028–3035. doi: 10.1111/j.1432-1033.2004.04234.x
Ganan, M., Campos, G., Munoz, R., Carrascosa, A. V., De Pascual-Teresa, S., and Martinez-Rodriguez, A. J. (2010). Effect of growth phase on the adherence to and invasion of Caco-2 epithelial cells by Campylobacter. Int. J. Food Microbiol. 140, 14–18. doi: 10.1016/j.ijfoodmicro.2010.02.021
Guerry, P. (2007). Campylobacter flagella: not just for motility. Trends Microbiol. 15, 456–461. doi: 10.1016/j.tim.2007.09.006
Gundogdu, O., Bentley, S. D., Holden, M. T., Parkhill, J., Dorrell, N., and Wren, B. W. (2007). Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics 8:162. doi: 10.1186/1471-2164-8-162
Gundogdu, O., Mills, D. C., Elmi, A., Martin, M. J., Wren, B. W., and Dorrell, N. (2011). The Campylobacter jejuni transcriptional regulator Cj1556 plays a role in the oxidative and aerobic stress response and is important for bacterial survival in vivo. J. Bacteriol. 193, 4238–4249. doi: 10.1128/JB.05189-11
Hall, S. J., Hitchcock, A., Butler, C. S., and Kelly, D. J. (2008). A Multicopper oxidase (Cj1516) and a CopA homologue (Cj1161) are major components of the copper homeostasis system of Campylobacter jejuni. J. Bacteriol. 190, 8075–8085. doi: 10.1128/JB.00821-08
Han, J., Sahin, O., Barton, Y. W., and Zhang, Q. (2008). Key role of Mfd in the development of fluoroquinolone resistance in Campylobacter jejuni. PLoS Pathog. 4:e1000083. doi: 10.1371/journal.ppat.1000083
Hitchcock, A., Hall, S. J., Myers, J. D., Mulholland, F., Jones, M. A., and Kelly, D. J. (2010). Roles of the twin-arginine translocase and associated chaperones in the biogenesis of the electron transport chains of the human pathogen Campylobacter jejuni. Microbiology 156, 2994–3010. doi: 10.1099/mic.0.042788-0
Hoang, K. V., Wang, Y., and Lin, J. (2012). Identification of genetic loci that contribute to Campylobacter resistance to fowlicidin-1, a chicken host defense peptide. Front. Cell. Infect. Microbiol. 2:32. doi: 10.3389/fcimb.2012.00032
Holmes, K., Mulholland, F., Pearson, B. M., Pin, C., McNicholl-Kennedy, J., Ketley, J. M., et al. (2005). Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151, 243–257. doi: 10.1099/mic.0.27412-0
Howlett, R. M., Hughes, B. M., Hitchcock, A., and Kelly, D. J. (2012). Hydrogenase activity in the foodborne pathogen Campylobacter jejuni depends upon a novel ABC-type nickel transporter (NikZYXWV) and is SlyD-independent. Microbiology 158, 1645–1655. doi: 10.1099/mic.0.054130-0
Hughes, D. T., Clarke, M. B., Yamamoto, K., Rasko, D. A., and Sperandio, V. (2009). The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC). PLoS Pathog. 5:e1000553. doi: 10.1371/journal.ppat.1000553
Jeon, B., Muraoka, W., Sahin, O., and Zhang, Q. (2008). Role of Cj1211 in natural transformation and transfer of antibiotic resistance determinants in Campylobacter jejuni. Antimicrob. Agents Chemother. 52, 2699–2708. doi: 10.1128/AAC.01607-07
Jeon, B., and Zhang, Q. (2007). Cj0011c, a periplasmic single- and double-stranded DNA-binding protein, contributes to natural transformation in Campylobacter jejuni. J. Bacteriol. 189, 7399–7407. doi: 10.1128/JB.01012-07
Karavolos, M. H., Spencer, H., Bulmer, D. M., Thompson, A., Winzer, K., Williams, P., et al. (2008). Adrenaline modulates the global transcriptional profile of Salmonella revealing a role in the antimicrobial peptide and oxidative stress resistance responses. BMC Genomics 9:458. doi: 10.1186/1471-2164-9-458
Karavolos, M. H., Winzer, K., Williams, P., and Khan, C. M. (2013). Pathogen espionage: multiple bacterial adrenergic sensors eavesdrop on host communication systems. Mol. Microbiol. 87, 455–465. doi: 10.1111/mmi.12110
Kendall, J. J., Barrero-Tobon, A. M., Hendrixson, D. R., and Kelly, D. J. (2014). Hemerythrins in the microaerophilic bacterium Campylobacter jejuni help protect key iron-sulphur cluster enzymes from oxidative damage. Environ. Microbiol. 16, 1105–1121. doi: 10.1111/1462-2920.12341
Kendall, M. M., Rasko, D. A., and Sperandio, V. (2007). Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 75, 4875–4884. doi: 10.1128/IAI.00550-07
Li, L., Xu, Z., Zhou, Y., Sun, L., Liu, Z., Chen, H., et al. (2012). Global effects of catecholamines on Actinobacillus pleuropneumoniae gene expression. PLoS ONE 7:e31121. doi: 10.1371/journal.pone.0031121
Lin, J. (2009). Novel approaches for Campylobacter control in poultry. Foodborne Pathog. Dis. 6, 755–765. doi: 10.1089/fpd.2008.0247
Lyte, M. (1993). The role of microbial endocrinology in infectious disease. J. Endocrinol. 137, 343–345. doi: 10.1677/joe.0.1370343
Lyte, M. (2004). Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol. 12, 14–20. doi: 10.1016/j.tim.2003.11.004
Lyte, M., Vulchanova, L., and Brown, D. R. (2011). Stress at the intestinal surface: catecholamines and mucosa-bacteria interactions. Cell Tissue Res. 343, 23–32. doi: 10.1007/s00441-010-1050-0
Miller, C. E., Williams, P. H., and Ketley, J. M. (2009). Pumping iron: mechanisms for iron uptake by Campylobacter. Microbiology 155, 3157–3165. doi: 10.1099/mic.0.032425-0
Moreira, C. G., and Sperandio, V. (2012). Interplay between the QseC and QseE bacterial adrenergic sensor kinases in Salmonella enterica serovar Typhimurium pathogenesis. Infect. Immun. 80, 4344–4353. doi: 10.1128/IAI.00803-12
Nachamkin, I., Allos, B. M., and Ho, T. (1998). Campylobacter species and Guillain-Barre syndrome. Clin. Microbiol. Rev. 11, 555–567.
Olson, C. K., Ethelberg, S., Pelt, W. V., and Tauxe, R. V. (2008). “Epidemiology of Campylobacter jejuni infections in industrialized nations,” in Campylobacter, 3rd Edn., eds I. Nachamkin, C. M. Szymanski, and M. J. Blaser (Washington, DC: ASM Press), 163–189.
Palyada, K., Sun, Y. Q., Flint, A., Butcher, J., Naikare, H., and Stintzi, A. (2009). Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481. doi: 10.1186/1471-2164-10-481
Palyada, K., Threadgill, D., and Stintzi, A. (2004). Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186, 4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Reading, N. C., Rasko, D. A., Torres, A. G., and Sperandio, V. (2009). The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 5889–5894. doi: 10.1073/pnas.0811409106
Sperandio, V., Torres, A. G., Jarvis, B., Nataro, J. P., and Kaper, J. B. (2003). Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U.S.A. 100, 8951–8956. doi: 10.1073/pnas.1537100100
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41
Van Vliet, A. H., Baillon, M. L., Penn, C. W., and Ketley, J. M. (1999). Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181, 6371–6376.
Weingarten, R. A., Grimes, J. L., and Olson, J. W. (2008). Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl. Environ. Microbiol. 74, 1367–1375. doi: 10.1128/AEM.02261-07
Woodall, C. A., Jones, M. A., Barrow, P. A., Hinds, J., Marsden, G. L., Kelly, D. J., et al. (2005). Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect. Immun. 73, 5278–5285. doi: 10.1128/IAI.73.8.5278-5285.2005
Wright, J. A., Grant, A. J., Hurd, D., Harrison, M., Guccione, E. J., Kelly, D. J., et al. (2009). Metabolite and transcriptome analysis of Campylobacter jejuni in vitro growth reveals a stationary-phase physiological switch. Microbiology 155, 80–94. doi: 10.1099/mic.0.021790-0
Xia, Q., Muraoka, W. T., Shen, Z., Sahin, O., Wang, H., Wu, Z., et al. (2013). Adaptive mechanisms of Campylobacter jejuni to erythromycin treatment. BMC Microbiol. 13:133. doi: 10.1186/1471-2180-13-133
Xu, F., Zeng, X., Haigh, R. D., Ketley, J. M., and Lin, J. (2010). Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter. J. Bacteriol. 192, 4425–4435. doi: 10.1128/JB.00478-10
Yun, J., Jeon, B., Barton, Y. W., Plummer, P., Zhang, Q., and Ryu, S. (2008). Role of the DksA-like protein in the pathogenesis and diverse metabolic activity of Campylobacter jejuni. J. Bacteriol. 190, 4512–4520. doi: 10.1128/JB.00105-08
Zautner, A. E., Tareen, A. M., Gross, U., and Lugert, R. (2012). Chemotaxis in Campylobacter jejuni. Eur. J. Microbiol. Immunol. (Bp) 2, 24–31. doi: 10.1556/EuJMI.2.2012.1.5
Keywords: Campylobacter jejuni, catecholamine hormones, microarray, growth promotion, virulence, adhesion and invasion
Citation: Xu F, Wu C, Guo F, Cui G, Zeng X, Yang B and Lin J (2015) Transcriptomic analysis of Campylobacter jejuni NCTC 11168 in response to epinephrine and norepinephrine. Front. Microbiol. 6:452. doi: 10.3389/fmicb.2015.00452
Received: 05 March 2015; Accepted: 27 April 2015;
Published: 18 May 2015.
Edited by:
Fiona Walsh, National University of Ireland Maynooth, IrelandCopyright © 2015 Xu, Wu, Guo, Cui, Zeng, Yang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuzhou Xu, Institute of Animal Husbandry and Veterinary Medicine, Beijing Academy of Agriculture and Forestry Sciences, No. 9, Shuguang Huayuan Middle Road, Beijing 100097, China, fuzhouxu@163.com
 Fuzhou Xu
Fuzhou Xu Cun Wu
Cun Wu Fangfang Guo
Fangfang Guo Guolin Cui
Guolin Cui Ximin Zeng
Ximin Zeng Bing Yang1
Bing Yang1 Jun Lin
Jun Lin