- 1Microbiology Department, University of Massachusetts, Amherst, MA, USA
- 2Graduate Program in Organismic and Evolutionary Biology, University of Massachusetts, Amherst, MA, USA
- 3Marine Biological Laboratories, Woods Hole, MA, USA
As Earth's climate warms, the massive stores of carbon found in soil are predicted to become depleted, and leave behind a smaller carbon pool that is less accessible to microbes. At a long-term forest soil-warming experiment in central Massachusetts, soil respiration and bacterial diversity have increased, while fungal biomass and microbially-accessible soil carbon have decreased. Here, we evaluate how warming has affected the microbial community's capability to degrade chemically-complex soil carbon using lignin-amended BioSep beads. We profiled the bacterial and fungal communities using PCR-based methods and completed extracellular enzyme assays as a proxy for potential community function. We found that lignin-amended beads selected for a distinct community containing bacterial taxa closely related to known lignin degraders, as well as members of many genera not previously noted as capable of degrading lignin. Warming tended to drive bacterial community structure more strongly in the lignin beads, while the effect on the fungal community was limited to unamended beads. Of those bacterial operational taxonomic units (OTUs) enriched by the warming treatment, many were enriched uniquely on lignin-amended beads. These taxa may be contributing to enhanced soil respiration under warming despite reduced readily available C availability. In aggregate, these results suggest that there is genetic potential for chemically complex soil carbon degradation that may lead to extended elevated soil respiration with long-term warming.
Introduction
The size of the soil carbon pool exceeds that of atmospheric and terrestrial vegetation carbon pools combined (Jobbágy and Jackson, 2000), making the fate of soil carbon a key variable in global climate models (McGuire et al., 2001; Wieder et al., 2013). Environmental perturbations that reduce the soil carbon pool or affect the microbes which process it may exacerbate soil carbon loss if this reduction in soil carbon stock feeds back to the initial stressor, as may occur under climate change (Davidson and Janssens, 2006). Indeed, experimental warming almost ubiquitously increases soil respiration (Rustad et al., 2001; Lu et al., 2013). Such increased decomposition is likely to be the consequence of some combination of both the direct effects of temperature on the organisms, and the indirect effects of altered community and/or soil organic matter composition (Bradford et al., 2008, 2010).
Accelerated microbial enzyme kinetics in response to warming can lead to increased soil respiration. In order to access polymeric carbon outside the cell, soil microorganisms produce extracellular enzymes that are affiliated with cell membrane surfaces or extracellular polysaccharides, or that are released into the environment (Wallenstein et al., 2011). These extracellular enzymes may have thermal optima greater than the temperatures they experience in the soil (Parham and Deng, 2000; Yan et al., 2010; Schipper et al., 2014), such that moderately elevated temperatures favor increased activity, as predicted by the Arrhenius equation (Stone et al., 2012; Baldrian et al., 2013). Physical explanations proposed for this phenomenon include increased flexibility of the active site and increased desorption of enzyme and substrate from mineral surfaces (Conant et al., 2011; Wallenstein et al., 2011). From a biological perspective, increased metabolic rates may enable greater enzyme production, since intracellular metabolism of soil microbiota is also responsive to temperature (Wallenstein et al., 2011). In addition, substrates that were previously degraded at relatively low rates due to the high activation energy of the decomposition reaction may now be more readily degraded, although it is unclear if this holds for soil enzymes (Davidson and Janssens, 2006; Baldrian et al., 2013; Erhagen et al., 2015).
In contrast to these direct kinetic effects of warming on microbial activity, this research examines how changes in microbial physiology or community structure may relate to elevated rates of soil carbon loss under warming. Warming may make microbial growth less efficient, likely through some combination of facilitating microbial access to lower-quality substrate pools (Devêvre and Horwáth, 2000) that require greater investment of resources to process, and by causing physiological changes in the organisms (Manzoni et al., 2012). Since only some soil bacteria have the capacity to rapidly respond to the presence of the most chemically complex compounds (Goldfarb et al., 2011), the reduction in the quality of soil carbon pools seen in some warming studies (Melillo et al., 2002; Bradford et al., 2008; Xu et al., 2012) may affect community structure. Indeed, although degradation of polymeric litter carbon is generally considered a fungal process (Boer et al., 2005; Moore-Kucera and Dick, 2008; Schneider et al., 2010, 2012; Baldrian et al., 2012), fungal:bacterial ratios have declined in a number of warming studies (Frey et al., 2008; Flury and Gessner, 2011; Zelikova et al., 2012; Sistla et al., 2013). Given the wide functional diversity and broad array of terminal electron acceptors they can use, bacteria are key decomposers of complex carbon under certain conditions (DeAngelis et al., 2011), but the extent to which these bacterial abilities are important for accelerated carbon cycling under extended warming is still unclear.
At our research site in a temperate deciduous forest in New England, 23 years of artificially warming soils has led to considerable changes in the stocks and flows of soil carbon. After 15 years, soil organic matter and carbon available to microbes had declined (Bradford et al., 2008; Frey et al., 2008). Soil respiration at this site has shown an initial increase in respiration with warming, accompanied by a decrease in fungal biomass (Frey et al., 2008), which disappeared after a decade (Melillo et al., 2002). Recently, we also observed substantial changes in microbial communities with warming, including a more than 80% increase in Alphaproteobacteria ribosomal RNA gene counts (DeAngelis et al., 2015). Using 13C-phenol and glucose, Frey and colleagues found evidence that warming has selected for a community specifically adapted to the more efficient utilization of structurally stable carbon at elevated temperatures (Frey et al., 2013). Research at an adjacent warming study indicates that while microbes are adapted to more rapid growth at higher temperatures, they are still limited overall by access to readily available carbon (Rousk et al., 2012). Together, these results indicate that increased utilization of chemically complex carbon may be driving the observed changes in carbon cycling at this site.
Here we evaluate whether the structure and enzymatic potential of the microbial community associated specifically with chemically complex carbon decomposition at this site has changed, using the heteropolymeric compound Kraft lignin as a proxy for chemically complex carbon. Lignin, which comprises 10–30% of leaf litter biomass (Aber et al., 1990), is degraded primarily by non-specific oxidative enzymes which may also break down soil organic matter (Fontaine et al., 2003; Creamer et al., 2015). By baiting soil microbes with lignin, we tested the hypothesis that experimental warming treatment has increased the diversity of the lignin-associated bacterial community.
Materials and Methods
Experimental Design
Our experiment was conducted within a long-term warming experiment in a mixed hardwood stand at the Harvard Forest LTER, Petersham, MA. At this site, soil temperatures have been artificially raised by 5°C using buried resistance cables since 1991 (Melillo et al., 2002). The dominant trees are Acer rubrum, Betula papyrifera, Quercus velutina, and Acer pensylvanicum, and the soils are coarse-loamy inceptisols (Peterjohn et al., 1994). The climate is temperate moist, with mean monthly temperatures ranging from −6°C in January to +20°C in July and a mean annual precipitation of 118 cm since the onset of the experiment (Boose and Gould, 1999; Boose, ongoing).
To each of four replicate heated and four replicate disturbance control plots, we horizontally deployed separate pouches containing lignin-amended (Sigma no. 471003) or unamended Bio-Sep bead (Microbial Insights, Knoxville TN) pouches (details in next section). Pouches were deployed as either “surface” bags, or as “subsurface” bags. Surface bags were placed on the surface of the soil under the leaf litter layer. Subsurface bead bags were buried in the soil between the organic horizon and the mineral soil. Where the depth of the organic horizon exceeded 3 cm, the beads were deployed to a depth of 3 cm. Bags were positioned in pairs, such that the lignin amended and unamended bead bags were side-by-side, but the surface and subsurface bead bags were at an approximate horizontal distance of at least 5 cm. We incubated a total of 32 bead pouches at our research site. Bags were deployed on August 5, 2013 and remained buried until October 23, 2013, a total of 11 weeks. This coincides with the seasonal period over which the greatest difference in soil respiration between heated and control plots can be seen (Melillo et al., 1999), and when the largest influx of complex plant litter to the soil occurs (Bowden et al., 2014).
Bio-Sep Bead Pouches
Bio-Sep beads are ~3–4 mm diameter porous spheres consisting of activated charcoal (25%) in a Nomex matrix (75%) (Williams et al., 2013). The beads are biochemically inert but are able to sorb nutrients. Selected substrates can be attached to the surfaces of the beads by covalent bonding via a proprietary method that enables microbial access to substrates while preventing leaching. Bio-Sep beads have been used in aquatic systems (Anderson et al., 2003; Peacock et al., 2004; Sublette et al., 2006; Baldwin et al., 2008; Williams et al., 2013) as well as terrestrial systems (DeAngelis et al., 2011; Omotayo et al., 2011) to monitor microbial activity. Bead pouches used in this study consisted of 5 g (~20 ml) of Kraft lignin amended or unamended beads in an 8 cm diameter circular window screen mesh pouch (Phifer silver gray fiberglass screen, product 4788811608, approximately 1 mm mesh size) that was heat-sealed. These pouches were then encased in a 9.5 × 9.5 cm square hardware cloth (YardGuard® 1/4 inch mesh (23 gage) with galvanized zinc coating, product 308231B), which kept the beads approximately two layers thick.
Enzyme Assays
Total oxidative enzyme assays were completed using 25 mM L-DOPA +0.3% hydrogen peroxide (Saiya-Cork et al., 2002). Slurry was prepared by vortexing 2 g of beads in 40 ml pH 4.7 Modified Universal Buffer (Östling and Virtama, 1946). An equal volume of substrate was added to each well. Plates were incubated at room temperature (23°C) in the dark for 48 h, and absorbance of a 50 μl aliquot was measured at 460 nm at five points during this time. Activity was calculated as the maximum change in absorbance over any three time points and standardized to total cell count assuming an extinction coefficient of 7.9 (Bach et al., 2013). Direct cell counts were completed on the initial bead slurry after staining with DAPI (4′,6-diamidino-2-phenylindole) (15–20 fields per sample); enzyme assays are reported as rates of cell normalized substrate converted per hour, and per gram of field-moist beads.
DNA Extraction
DNA was extracted between three and seven times from 0.3 g Bio-Sep beads (depending on yields) following a modified CTAB bead-beating procedure in tubes with three 5 mm glass beads (DeAngelis et al., 2010). To clear residual phenol, all extractions for a given sample were pooled and brought up to 200 μl with sterile Tris-Cl (pH 8.5), washed with 25:1 chloroform:isoamyl alcohol, and precipitated in 100% cold ethanol and sodium acetate (pH 5.2) at a final concentration of 0.3 M. Samples were desalted with 70% ethanol. DNA quality was verified using a NanoDrop 2000C (Thermo Scientific, Inc., Waltham MA), and quantified using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen).
16S rRNA Gene Sequencing
DNA was prepared for sequencing using a previously published method with a few modifications (Caporaso et al., 2011). Briefly, the V4 region of the 16S rRNA gene in the DNA template was amplified in triplicate using the primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′, Turner et al., 1999) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′, Caporaso et al., 2011) with sequencer adapters and sample-specific Golay barcodes on the forward primer. The 25 μl reaction mix contained 2.5 μl of each primer at a final concentration of 200 pM, 1.875 units of Takara ExTaq polymerase, 25 μg BSA, 200 μM each dNTPs, and 10 ng template. The amplification cycle consisted of an initial denaturation at 95°C for 5 min followed by 31 cycles of 30 s at 95°C, 25 s at 50°C, 120 s at 72°C and a final elongation of 10 min at 72°C. After verifying successful PCR amplification using agarose gel electrophoresis, technical triplicate reactions were pooled for cleanup using Qiagen MinElute kit. These were then quantified using PicoGreen® and assessed for quality using Nanodrop. Equimolar quantities of each of the samples were pooled into a single tube for paired-end 2 × 150 bp Illumina MiSeq sequencing at the Molecular Biology Core Facility at the Dana-Farber Cancer Institute.
FastQC was used to check for overall sequencing quality (Andrews, 2010). Paired end reads were subsequently merged using FLASH with default parameters except sequence length, which was limited to 253 ± 1 bp (Magoč and Salzberg, 2011). All subsequent stages of the sequence data processing were completed in QIIME v. 1.8.0 (Caporaso et al., 2010b). Demultiplexing and initial quality filtering were completed with a minimum Phred score of 20. We picked operational taxonomic units (OTUs) using QIIME's subsampled open reference picking protocol using UClust (Edgar, 2010) to bin sequences into OTUs at 99% identity using RDP (Wang et al., 2007) with Greengenes (V13.5, May 2013, McDonald et al., 2012; Werner et al., 2012) as the reference database. Sequences were aligned against Greengenes using PyNAST (Caporaso et al., 2010a) and an amplicon region specific lanemask. Chimeric sequences were identified using ChimeraSlayer (Haas et al., 2011). After removing doubletons from the dataset, we rarefied the community (Magurran and McGill, 2011; Unterseher et al., 2011). This process reduced the 8.5 million reads and 64,351 OTUs to 2.4 million reads and 56,835 OTUS in 32 bacterial (56,815 OTUs, 99.998% of reads) and 2 archaeal phyla (20 OTUs) (Table S1). All sequences are available under BioProject ID PRJNA242968.
TRFLP of Fungal Its Region
Terminal restriction fragment length polymorphism analysis was used to assess whether 23 years of warming has affected lignin-amended and unamended bead-associated fungal communities. The fungal-specific (Klamer et al., 2002) forward and reverse primers (ITS1F; 5′-CTTGGTCATTTAGAGGAAGTAA-3 /ITS4; 5′- TCCTCCGCTTATTGATATGC-3′, Gardes and Bruns, 1993) were both labeled with a fluorescent dye (ITS1F-FAM and ITS4-VIC, respectively). The 25 μl reaction mix contained 2.5 μl of each primer at a final concentration of 300 pM, 1.875 units of Takara ExTaq polymerase, 25 μg BSA, 200 μM each dNTPs, and 10 ng template DNA. Reaction conditions were: 1 min at 95°C, followed by 35 cycles of 95°C for 1 min, 51°C for 1 min, and 72°C for 3 min, with a final extension at 72°C for 8 min. PCR product was digested with 20U HaeIII restriction enzyme (Thermo Scientific). Approximately 70 ng digested PCR product was mixed with 0.5 μl of the GeneScan 1200 LIZ size standard (ABI) and 7.7 μl deionized formamide, and submitted for sequencing on an ABI 3130XL. TRFLP profiles were checked for quality and peaks heights >50 RFU were extracted using the Local Southern method in Peak Scanner Software v. 2.0 (Life Technologies) (Blackwood et al., 2003). Peak sizes were then rounded to the nearest integer and forward and reverse fragments separated to normalize peak heights by the sum of peak heights for a dye in a sample, then the two data sets were combined for analysis. This resulted in an average of 64.7 peaks per sample, with a total of 458 different TRFs across samples.
Identification of Potentially Lignolytic Bacteria
In order to identify bacterial genera capable of lignin degradation, we used a PubMed literature search with the keywords “lignin degradation” +bacteria on 13th March 2014, and then manually filtered the results to exclude irrelevant content (i.e., papers on fungi, or on other biopolymer degradation) and to include references cited within papers. We also included other papers that were not included in this search but were already familiar to us. The genera appearing in this search but not found in our dataset were Aeromonas (Gupta et al., 2001), Aneurinibacillus (Chandra et al., 2007), Azotobacter (Morii et al., 1995), Cladosporium (Ji et al., 2014), Desulfovibrio (Kim et al., 2009), Enterobacter (DeAngelis et al., 2013), Kocuria (Parshetti et al., 2011), Microbulbifer, Sagittula (González et al., 1996; Chen et al., 1997) Micrococcus (Taylor et al., 2012), Pantoea (Xiong et al., 2014), Thaurea (Kim et al., 2009), Thermobifida (Chen et al., 2013), and Xanthomonas (Odier and Monties, 1978).
Statistical Analysis
The experimental design included four biological replicates unless otherwise noted, with 16 technical replicates for enzyme assays. All statistical analyses were completed in R (R Development Core Team, 2008). Data were tested for normality and homogeneity of variances using the Shapiro-Wilk test (Royston, 1982) and Brown–Forsythe tests (Fox, 2008), respectively, with a p-value cutoff of 0.1. For data which did not meet parametric assumptions, the function boxcoxfit (geoR, Ribeiro and Diggle, 2001) was used to determine the optimal lambda for power transformation. Lignin-amended and unamended bead pouches for each plot and soil depth were subsequently treated as paired samples for analysis in three-way full factorial ANOVA with warming treatment, soil, and lignin amendment as main effects. Significant interaction effects were further assessed with Tukey's Honestly Significant Difference test (Tukey, 1949).
To determine whether lignin beads recruited known lignolytic organisms, relative abundance (percent reads) of genera with proposed lignin-degraders were pooled by soil effect and warming treatment. OTUs without genus assignment were removed before analysis, and samples were normalized by sum to account for different degrees of genus-level assignment before calculating p-values with a paired Wilcoxon rank-sum test. Reported p-values are corrected for multiple testing using Benjamini-Hochberg correction (Benjamini and Hochberg, 1995).
Parameters of alpha diversity were calculated using the vegan package (Oksanen et al., 2013) with criteria outlined in Magurran and McGill as a guide for selecting uncorrelated metrics (Magurran and McGill, 2011). Since some diversity metrics are non-linear (e.g., the Shannon-Weiner index), we converted these values to effective number of species using Neff = exp(H) prior to comparison (Jost, 2006). Faith's phylogenetic diversity (Faith, 1992) was calculated using picante (Kembel et al., 2010).
TRF profiles of the fungal community were compared using Hellinger distance (Legendre and Gallagher, 2001), and variables driving community structure were assessed using a permutational manova (Anderson, 2001). For the bacterial community, we completed principal coordinates analyses on weighted UniFrac distances (Lozupone and Knight, 2005) of the prokaryotic community using the phyloseq package (McMurdie and Holmes, 2013). Experimental factors driving relative abundance of major phyla and classes were assessed using step-down regression with Aikake's Information Criterion to select the best model while minimizing parameters (stepAIC, MASS, Venables and Ripley, 2002).
Dominant OTUs responsible for driving observed community trends in lignin-amendment and heating treatment were identified using paired t-tests or an indicator species test (Cáceres and Legendre, 2009; Roberts, 2013) with the Benjamini-Hochberg correction for multiple comparisons (Benjamini and Hochberg, 1995). Only OTUs shared by both lignin and unamended beads were considered in this analysis so as to separate ability to grown in the different bead types from a warming effect. Trees were drawn in iTOL (Letunic and Bork, 2007).
Results
Long-Term Warming Recruits More Microbes with Lignolytic Potential
To evaluate whether lignin-amended beads successfully recruited lignolytic organisms, we examined both oxidative enzymes assays as well as indicator species analysis of bead bacterial communities cross-referenced with a comparative literature search. We expected communities associated with lignin-amended beads to demonstrate greater potential to degrade model lignin compounds, and to contain a greater fraction of OTUs closely related to known lignin degraders compared to unamended beads.
In the surface beads, warmed communities showed 4.7x higher cell-normalized oxidative activity compared to controls, while in the subsurface beads there was no warming effect (Figure 1B; ANOVA warming*soil interaction p < 0.01, F = 8.69; three replicates used for surface lignin beads). Looking at the effect of lignin amendment, unamended beads showed approximately 4.5x greater bead weight normalized oxidative enzyme activity than lignin-amended beads (ANOVA, p < 0.001, F = 39.43, Figure 1A), and this was also true when accounting for differences in cell counts (ANOVA, p < 0.001, F = 17.65). DAPI-based cell counts of the enzyme slurry indicated that the number of cells per gram of beads was unaffected by the experimental factors (Three-Way full factorial ANOVA, p > 0.1 for all).
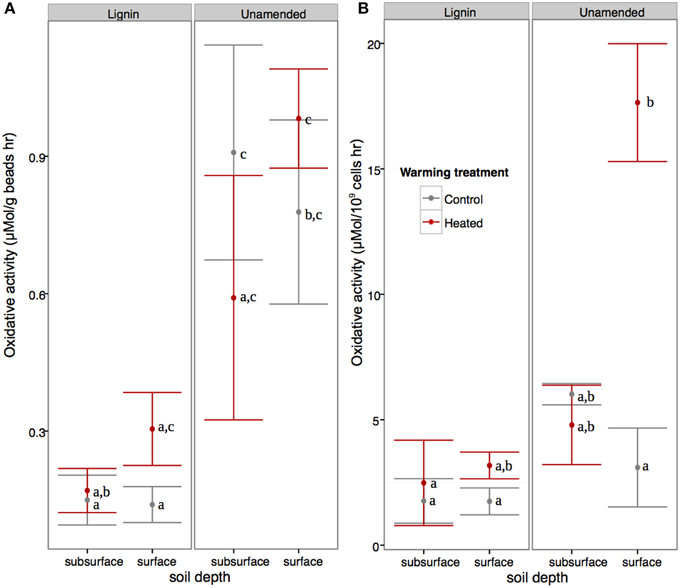
Figure 1. Effect of warming and soil on field-moist bead (A) and cell-normalized total oxidative enzyme activity (B). Shared letters indicates soil depth*warming treatment*lignin amendment effects are not significantly different at P = 0.05 using Tukey's HSD.
Sequencing of the 16S ribosomal RNA gene V4 region was used to identify the microbes associated with lignin-amended and unamended beads. Lignin-amended beads recruited many genera containing known lignin-degraders (Table 1). Overall, unamended beads had a greater fraction of reads assigned to genera with known lignin-degraders than lignin-amended beads did, but this was heavily skewed by Burkholderia, which accounted for almost 70% of the OTUs. Of the 27 genera we identified as containing known lignin-degrading taxa in a literature search, 15 had a significantly greater relative abundance in lignin than unamended beads, seven were unaffected by amendment, and five were enriched on unamended relative to lignin-amended beads (paired Wilcoxon, p < 0.05 for all). Those enriched for by lignin amendment included Sphingomonas, Acinetobacter, and Agrobacterium, respectively present at 6.4, 22, and 130x greater relative abundance in lignin than in unamended beads.
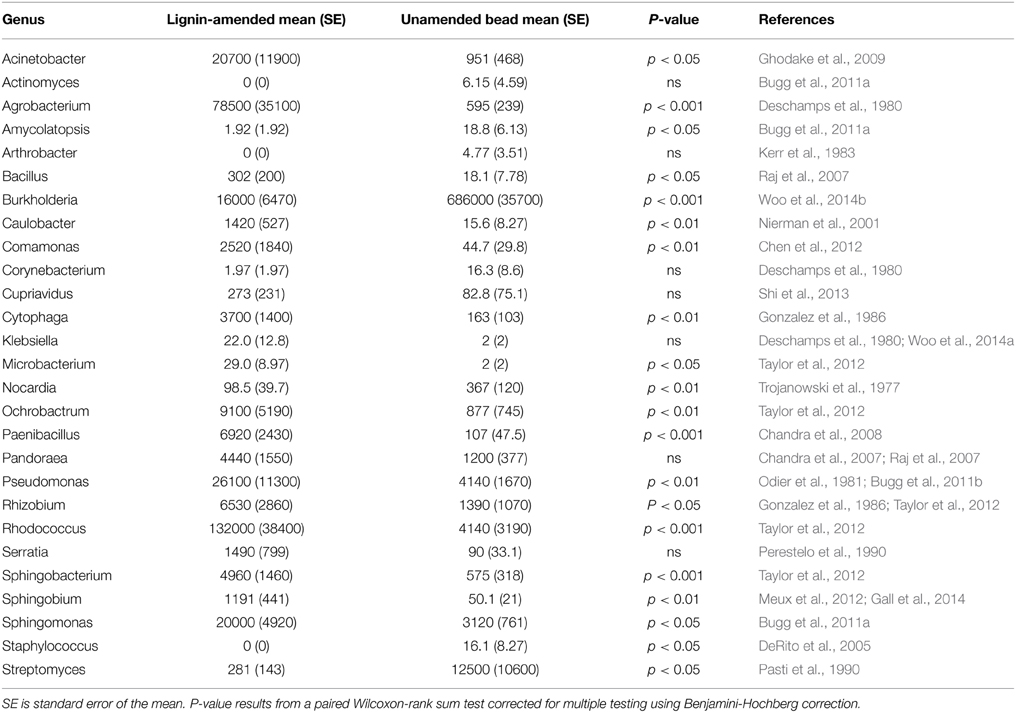
Table 1. Relative abundance (reads per million reads) of genera with proposed lignin-degraders in lignin-amended and unamended bead samples.
Warming Was Associated with Increased Diversity in Bacterial Communities
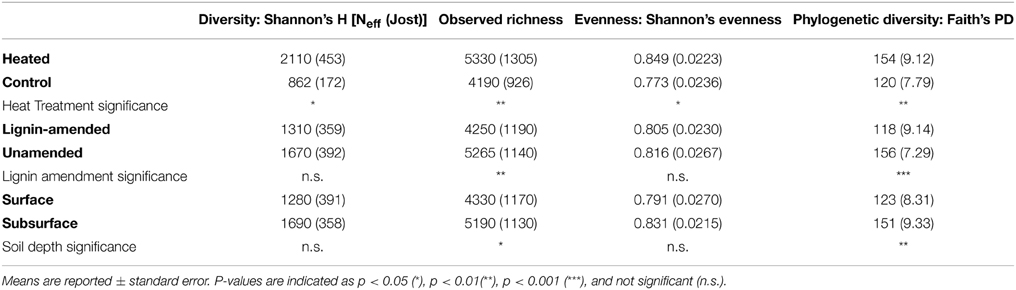
The bacterial communities in warmed plots were on average 32% richer (p < 0.05), 10% more even (p < 0.05), and had a phylogenetic diversity that was 28% greater (p < 0.01) than communities from control plots (Table 2). Examining communities associated with the lignin beads alone, warming increased just one metric of diversity, Faith's PD. There was no evidence for interactions between any of the experimental factors.
Bead Amendment Drives Beta Diversity among Bacteria and Fungi
Bead amendment was the primary force driving bacterial community structure, based on a permutational ANOVA of the UniFrac distances (F = 32.74; p = 0.001; R2 = 0.487; Figure 2A). Warming treatment (F = 3.93; p = 0.013; R2 = 0.059) played a secondary role in shaping overall bacterial beta diversity, with a greater fraction of the variation explained in the lignin-amended (R2 = 0.253) than the unamended beads (R2 = 0.141). We also analyzed factors driving relative abundance of each phylum individually (or subphyla, for the Proteobacteria) using step-down regression (Table S2). Consistent with whole community data, bead amendment was a primary driver of relative abundance of most phyla and subphyla considered, with Alphaproteobacteria at higher abundances on lignin-amended compared to unamended beads, and Acidobacteria, Actinobacteria, and Gammaproteobacteria at lower abundance on lignin-amended compared to unamended beads. Betaproteobacteria also tended to have reduced relative abundances in lignin-amended beads, but the fit for the model was poor (R2 adj = 0.19) compared to most of the other classes and phyla considered.
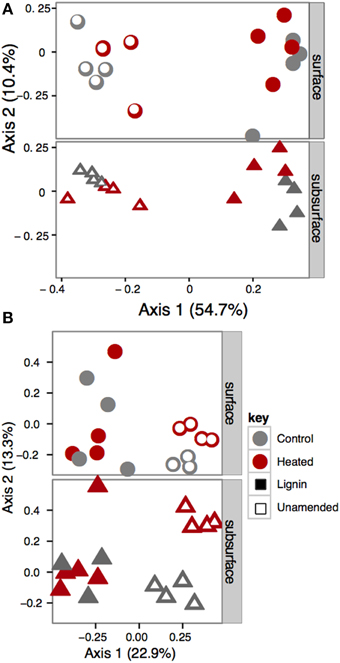
Figure 2. Principal coordinates plot of (A) UniFrac distances of bacterial communities and (B) Hellinger distance of Fungal communities. Lignin-amended and unamended beads support distinct communities, as demonstrated by the wide separation on the first axis.
Fungal community structure was also driven by bead amendment (F = 8.75, p = 0.001, R2 = 0.209), warming treatment (F = 2.33; p = 0.008, R2 = 0.056), and soil effect (F = 2.10; p = 0.028; R2 = 0.050; Figure 2B), as determined by permutational ANOVA using Hellinger distance of the fungal TRFLP profiles (Blackwood et al., 2003). Warming (adonis, F = 2.85; p = 0.002; R2 = 0.153) and soil effect (F = 2.537; p = 0.002; R2 = 0.136) also played a significant role in structuring the fungal community in unamended beads when examined alone, but not lignin-amended beads.
OTUs Enriched on Lignin-Amended Beads Are Phylogenetically Clustered
Since soil depth was not a significant driver of bacterial beta diversity, we pooled samples by soil depth and plot to identify OTUs whose abundance was affected by lignin amendment and/or warming. Lignin-amended and unamended beads showed distinct communities at the level of the OTU, sharing just 9924 (17.5%) of the 56,835 total OTUs present. Examining the 9924 OTUs shared between lignin-amended and unamended beads and re-rarefying the community, we were able to identify 213 OTUs significantly enriched on lignin-amended beads compared to unamended beads, and 536 significantly enriched on unamended beads compared to lignin-amended beads (based on a Wilcoxon rank-sum test with corrections for multiple testing, Table S3). OTUs in the phylum Acidobacteria were exclusively enriched on unamended beads, and many more Actinobacteria OTUs were enriched on unamended than lignin-amended beads. By contrast, an approximately equal number of Betaproteobacteria OTUs were enriched on unamended as on lignin-amended beads; unamended beads were enriched in a number of members of the family Burkholderiaceae, while lignin-amended beads were enriched in OTUs of the family Comamonadaceae. This taxonomic trend was representative of the whole dataset; overall, Burkholderiaceae were 49.9x more abundant in the unamended than the lignin-amended beads, and Comamonadaceae were 9.2x more abundant in the lignin-amended compared to the unamended beads. The only orders that included members enriched only on the lignin-amended beads were the Cytophagales and Flavobacteriales (phylum Bacteroidetes, two and four OTUs, respectively), Bacilliales (three OTUs in the phylum Firmicutes), Planctomycetales (one OTU in the phylum Planctomycetes), Methylophilales (one OTU in the class Betaproteobacteria), Pseudomonadales (six OTUs in the class Gammaproteobacteria) (Table S3).
OTUs Enriched by Warming on Lignin Beads Are Not Phylogenetically Clustered
For the subset of 9924 OTUs shared by both bead types, we used indicator species analysis (IndVal, Cáceres and Legendre, 2009) with a Benjamini-Hochberg correction for multiple testing to identify those OTUs differentially affected by warming on unamended and lignin-amended beads. Overall, 109 OTUs were negatively affected by warming on lignin-amended beads, while 256 increased in relative abundance, and the majority of these showed a warming response that was unique to the lignin beads (Figure 3; Table S4). While many taxonomic groups showed mixed responses to warming, individual members of the phyla Acidobacteria and Actinobacteria, and families Bradyhizobiaceae, Sphingomonadaceae, and Comamonadaceae showed clear increases in abundance with warming on lignin-amended beads, while members of the Caulobacteraceae, Oxalobacteraceae and Moraxellaceae decreased in relative abundance.
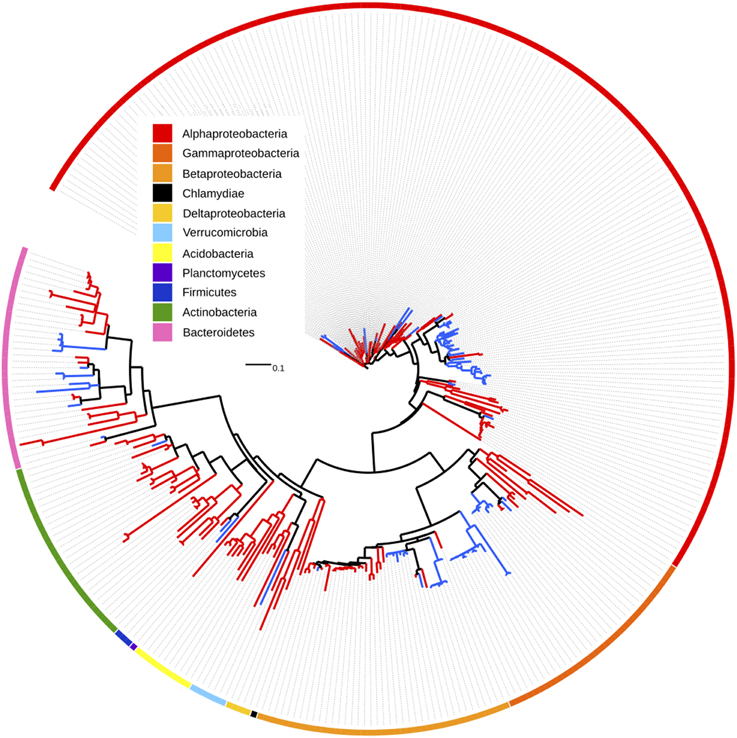
Figure 3. Tree depicting OTUs which are significantly enriched in warmed (red branches) or control (blue branches) plots on lignin-amended BioSep beads. OTUs also affected by warming treatment in the unamended beads were excluded from the tree. Each branch is a single OTU. Outer ring denotes phylum or class.
Discussion
In this study, we evaluated the hypothesis that two decades of experimental warming had selected for a microbial community better able to degrade chemically complex carbon. We based our expectation on observations that soil respiration at our experimental warming site has remained high despite considerable loss of soil organic matter (Melillo et al. in preparation) and of microbially available C (Bradford et al., 2008). To test this hypothesis, we used oxidative enzyme activity as a proxy for complex organic matter decomposition potential, and evaluated whether warming treatment affected the composition and diversity of the microbial community associated with lignin degradation.
The microbial communities subject to prolonged warming experienced reduced carbon availability (Bradford et al., 2008) and in this study showed increased total oxidative enzyme activity per billion cells, but the effect was only significant in the surface samples. Since there may be considerable overlap in pathways used to degrade lignin and soil organic matter (Fontaine et al., 2003; Carney et al., 2007), this result indicates the microbial community accessing complex carbon in the organic horizon was more strongly affected by warming treatment than the community accessing mineral-sorbed SOM. Microbial communities in the organic horizon are typically dominated by fungi (Baldrian et al., 2012), which are able to efficiently degrade lignin (Ahmad et al., 2010). However, fungal lipid biomass (Frey et al., 2008) and ITS region counts (DeAngelis et al., 2015) have been noted to be reduced by warming, so increased oxidative enzyme activity in surface soils with warming is unexpected. However, on a per gram basis, enzyme activity was unaffected by warming in surface bead samples, indicating that the smaller number of cells present in the heated samples may be more effective at degrading lignin-like compounds, a phenomenon that has been previously observed (Frey et al., 2013).
In our study, warming treatment preferentially affected microbes and oxidative activity in the surface compared to subsurface, where surface microbes are adapted to relatively “free” lignin compared to the mineral-stabilized SOM in the subsurface soils. However, since only half (9 of 16) of the subsurface bead bags were at the mineral-organic interface, it is difficult to directly assess this hypothesis. Factors such as a more stable temperature and moisture in the subsurface beads may have enabled microbes to invest more energy into producing extracellular enzymes, and less into maintenance than was possible for the surface beads. In addition to the showing greater enzymatic response to warming treatment, the surface beads were also home to a less phylogenetically-diverse bacterial community than the subsurface beads. This may in part be due to the increased moisture fluctuations faced by the surface beads, which may reduce diversity of the water-bound bacterial community (Lavelle, 1997).
Based on the fact that total oxidative enzyme activity was higher in unamended beads than lignin amended beads, it is unclear whether lignin amended Bio-Sep beads successfully recruited a lignolytic community. While it is possible that lignin was depleted on the lignin amended beads at the time of collection, we believe this is unlikely. In a similar experiment using these beads in tropical forest soils, DeAngelis et al. (2011) found that lignin amended beads had higher oxidative enzyme activity even after 30 weeks in the field, a time much longer than that used in the present study. Instead, higher oxidative enzyme activity in the unamended beads may be due to their better suitability for fungal colonization. DeAngelis et al. previously noted that fungi formed a smaller fraction of the eukaryotic community in lignin amended than unamended beads, and the small pores on the exterior of Bio-Sep beads have been reported to limit eukaryotic access (Williams et al., 2013). We observed that lignin amended beads are 44% denser than unamended beads, indicating that lignin amendment may have made it even more difficult for larger organisms to colonize the interior of the beads. Thus, the unexpected higher enzyme activity in unamended compared to lignin amended beads in our study may be indicative of the dominant lignin degraders being unable to access the substrate held within.
Lignin amended beads recruited a phylogenetically diverse collection of bacteria, including many in genera where known lignin degraders are found. These genera with significantly higher relative abundance in lignin amended than unamended beads included Rhodococcus and Pseudomonas but didn't include a number of the better-known lignin degrading genera such as Streptomyces, which are well-known for their biopolymer degrading capacity (Kampfer, 2012). Rhodococcus jostii RHA1 and Pseudomonas putida mt-2 are both able to degrade lignin without exogenous hydrogen peroxide and at a rate equivalent to some lignolytic fungi (Ahmad et al., 2010). We also found that Burkholderia, a cosmopolitan genus known for its diverse metabolism (Garrity et al., 2012), and Nocardia, one of the better-known lignin degraders (Bugg et al., 2011a), were present at significantly higher relative abundance in unamended than lignin amended beads. A mixture of OTUs in orders known to contain aromatic compound degraders, such as Sphingobacteriales (Taylor et al., 2012), and Xanthomonadales (Odier and Monties, 1978), and orders not previously noted to degrade lignin, such as Saprospirales, were found to be enriched in lignin compared to unamended beads. Likewise, warming treatment specifically enriched for OTUs in a combination of orders with and without known lignin degraders. For instance, eight members of Acidobacteria were enriched by warming on lignin beads, although to our knowledge direct evidence for their ability to degrade lignin has not yet been reported. It is also possible that their enrichment is due to ability to degrade lignin by-products produced by other lignin-degrading organisms. Dedysh and colleagues isolated Acidobacteria from phenolic-rich peats and mosses using humic acid (Dedysh et al., 2006; Pankratov and Dedysh, 2010), indicating these organisms may have lignolytic potential. However, it is probable that our lignin beads recruited organisms feeding on lignin decomposition products, rather than lignin, per se. A recent paper also noted the possibility that organisms in lignin-rich environments may gain the ability to degrade complex carbon sources through rapid horizontal gene transfer (Strachan et al., 2014).
In addition to specifically selecting for potential lignin degraders, warming may enhance the ability of the microbial community to degrade chemically complex carbon by increasing overall community diversity. Increased community diversity is associated with the acceleration of multiple ecosystem processes, including decomposition and respiration (Strickland et al., 2009; Cadotte et al., 2012; Pold and DeAngelis, 2013). However, across warming studies, the response of microbial community diversity and composition to experimental warming treatment has been variable (Pold and DeAngelis, 2013). At our study site, the community profile based on FAMEs had shifted after 12 years of warming in the mineral horizon (Frey et al., 2008). When first studied using sequencing after 20 years of warming, the bacterial community was noted to be more diverse overall and to have shifted in composition in the organic horizon (DeAngelis et al., 2015). Thus, the increase in diversity and altered community structure observed here for the bacterial and fungal communities was in line with previous results at our site. It remains unclear whether the diversity or identity of lignin-associated bacteria seen here may be driving observed changes in carbon cycling in our site.
Ultimately, the ability of the microbial community to efficiently convert soil carbon to biomass is the key first step in the release of stored soil carbon (Allison et al., 2010). Chemically complex compounds such as lignin are generally utilized by microbes at lower efficiencies than simpler ones (Manzoni et al., 2012), and microbes can differ in the efficiency with which they utilize litter carbon sources. Previous observations that the warmed plot microbial community at our site is adapted to more efficient use of phenolic compounds than that of the control plots (Frey et al., 2013) lead to the hypothesis that changes in microbial communities captured on our lignin beads may be contributing to the altered fate of carbon in soil. Though the lignin in our beads was not under the same physical and chemical constraints for degradation that soil organic matter may be, the potential overlap in degradation pathways for complex soil organic matter and lignin indicates that at least some of the organisms preferentially enriched for on lignin-amended beads are likely to also degrade soil organic matter. Yet we also recognize that the presence of an organism in a lignin-amended bead may be indicative of tolerance to oxidative stress as well as ability to degrade the lignin present (DeAngelis et al., 2011). Because of this, we refrain from drawing conclusions in the comparison of the lignin-amended compared to the unamended beads. However, the diversity of observed bacterial genera that were recruited to the lignin-amended beads, and the consistency of the taxonomy of many of these organisms with known lignin degraders, indicates that we have captured a functionally important subset of the community.
Conclusions
We found evidence for a change in the structure of the bacterial community adapted to degradation of a major litter component, lignin, as we have seen for the “whole-soil” bacterial community after two decades of warming at this site (DeAngelis et al., 2015). While we cannot conclude that the continued loss of soil carbon at our study site is due to the changes in the microbial community observed here, our research is consistent with previous research (Frey et al., 2013) in indicating that those microbes potentially capable of degrading complex carbon substrates are at least responsive to the direct or indirect effects of two decades of elevated temperature. Ultimately, the fate of soil organic matter in a warming world will depend upon a complex interplay between the efficiency with which the microbial community converts litter to biomass, and the ease with which this biomass is recycled into new biomass or physically stabilized by the soil matrix. Future approaches to partition enzymes to domains, as well as an assessment of the temperature sensitivity of oxidative enzyme activity, will help elucidate mechanisms driving observed increases in rates of soil organic matter loss at our site.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge Kerry Sublette for the donation of lignin-amended BioSep beads for this study; Will Werner for facilitating access to the field site; Matt Tuttle for assistance with TRFLP; Zach Herbert at the DFCI Molecular Biology Core Facility for providing technical assistance with sequencing; and Erin Nuccio at Lawrence Livermore National Laboratory for providing the Greengenes reference files truncated to the V4 region for OTU calling. This work was funded by the University of Massachusetts, Amherst and the National Science Foundation Long-Term Ecological Research (LTER) Program. This research was supported by the National Science Foundation LTER IV and LTER V grants to Harvard Forest (0620443 and 1237491) and the University of Massachusetts.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00480/abstract
References
Aber, J. D., Melillo, J. M., and McClaugherty, C. A. (1990). Predicting long-term patterns of mass loss, nitrogen dynamics, and soil organic matter formation from initial fine litter chemistry in temperate forest ecosystems. Botany 68, 2201–2208. doi: 10.1139/b90-287
Ahmad, M., Taylor, C. R., Pink, D., Burton, K., Eastwood, D., Bending, G. D., et al. (2010). Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders. Mol. Biosyst. 6, 815–821. doi: 10.1039/b908966g
Allison, S. D., Wallenstein, M., and Bradford, M. A. (2010). Soil-carbon response to warming dependent on microbial physiology. Nat. Geosci. 3, 336–340. doi: 10.1038/ngeo846
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Anderson, R. T., Vrionis, H. A., Ortiz-Bernad, I., Resch, C. T., Long, P. E., Dayvault, R., et al. (2003). Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69, 5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003
Andrews, S. (2010). FASTQC. A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Bach, C., Warnock, D., Van Horn, D., Weintraub, M. N., Sinsabaugh, R., Allison, S. D., et al. (2013). Measuring phenol oxidase and peroxidase activities with pyrogallol, l-DOPA, and ABTS: effect of assay conditions and soil type. Soil Biol. Biochem. 67, 183–191. doi: 10.1016/j.soilbio.2013.08.022
Baldrian, P., Kolařík, M., Štursová, M., Kopecký, J., Valášková, V., Větrovský, T., et al. (2012). Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 6, 248–258. doi: 10.1038/ismej.2011.95
Baldrian, P., Šnajdr, J., Merhautová, V., Dobiášová, P., Cajthaml, T., and Valášková, V. (2013). Responses of the extracellular enzyme activities in hardwood forest to soil temperature and seasonality and the potential effects of climate change. Soil Biol. Biochem. 56, 60–68. doi: 10.1016/j.soilbio.2012.01.020
Baldwin, B. R., Peacock, A. D., Park, M., Ogles, D. M., Istok, J. D., McKinley, J. P., et al. (2008). Multilevel samplers as microcosms to assess microbial response to biostimulation. Groundwater 46, 295–304. doi: 10.1111/j.1745-6584.2007.00411.x
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300.
Blackwood, C. B., Marsh, T., Kim, S.-H., and Paul, E. A. (2003). Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69, 926–932. doi: 10.1128/AEM.69.2.926-932.2003
Boer, W., de Folman, L. B., Summerbell, R. C., and Boddy, L. (2005). Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29, 795–811. doi: 10.1016/j.femsre.2004.11.005
Boose, E., and Gould, E. (1999). Shaler Meteorological Station at Harvard Forest 1964–2002. Available online at: http://harvardforest.fas.harvard.edu:8080/exist/apps/datasets/showData.html?id=hf000 (Accessed November 29, 2014).
Bowden, R. D., Deem, L., Plante, A. F., Peltre, C., Nadelhoffer, K. J., and Lajtha, K. (2014). Litter input controls on soil carbon in a temperate deciduous forest. Soil Sci. Soc. Am. J. 78, S66–S75. doi: 10.2136/sssaj2013.09.0413nafsc
Bradford, M. A., Davies, C. A., Frey, S. D., Maddox, T. R., Melillo, J. M., Mohan, J. E., et al. (2008). Thermal adaptation of soil microbial respiration to elevated temperature. Ecol. Lett. 11, 1316–1327. doi: 10.1111/j.1461-0248.2008.01251.x
Bradford, M. A., Watts, B. W., and Davies, C. A. (2010). Thermal adaptation of heterotrophic soil respiration in laboratory microcosms. Glob. Change Biol. 16, 1576–1588. doi: 10.1111/j.1365-2486.2009.02040.x
Bugg, T. D., Ahmad, M., Hardiman, E. M., and Singh, R. (2011a). The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22, 394–400. doi: 10.1016/j.copbio.2010.10.009
Bugg, T. D. H., Ahmad, M., Hardiman, E. M., and Rahmanpour, R. (2011b). Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 28, 1883–1896. doi: 10.1039/c1np00042j
Cáceres, M. D., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Cadotte, M. W., Dinnage, R., and Tilman, D. (2012). Phylogenetic diversity promotes ecosystem stability. Ecology 93, S223–S233. doi: 10.1890/11-0426.1
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Carney, K. M., Hungate, B. A., Drake, B. G., and Megonigal, J. P. (2007). Altered soil microbial community at elevated CO2 leads to loss of soil carbon. Proc. Natl. Acad. Sci. U.S.A. 104, 4990–4995. doi: 10.1073/pnas.0610045104
Chandra, R., Raj, A., Purohit, H. J., and Kapley, A. (2007). Characterisation and optimisation of three potential aerobic bacterial strains for kraft lignin degradation from pulp paper waste. Chemosphere 67, 839–846. doi: 10.1016/j.chemosphere.2006.10.011
Chandra, R., Singh, S., Krishna Reddy, M. M., Patel, D. K., Purohit, H. J., and Kapley, A. (2008). Isolation and characterization of bacterial strains Paenibacillus sp. and Bacillus sp. for kraft lignin decolorization from pulp paper mill waste. J. Gen. Appl. Microbiol. 54, 399–407. doi: 10.2323/jgam.54.399
Chen, C.-Y., Hsieh, Z.-S., Cheepudom, J., Yang, C.-H., and Meng, M. (2013). A 24.7-kDa copper-containing oxidase, secreted by Thermobifida fusca, significantly increasing the xylanase/cellulase-catalyzed hydrolysis of sugarcane bagasse. Appl. Microbiol. Biotechnol. 97, 8977–8986. doi: 10.1007/s00253-013-4727-y
Chen, F., Gonzalez, J. M., Dustman, W. A., Moran, M. A., and Hodson, R. E. (1997). In situ reverse transcription, an approach to characterize genetic diversity and activities of prokaryotes. Appl. Environ. Microbiol. 63, 4907–4913.
Chen, Y. H., Chai, L. Y., Zhu, Y. H., Yang, Z. H., Zheng, Y., and Zhang, H. (2012). Biodegradation of kraft lignin by a bacterial strain Comamonas sp. B-9 isolated from eroded bamboo slips. J. Appl. Microbiol. 112, 900–906. doi: 10.1111/j.1365-2672.2012.05275.x
Conant, R. T., Ryan, M. G., Ågren, G. I., Birge, H. E., Davidson, E. A., Eliasson, P. E., et al. (2011). Temperature and soil organic matter decomposition rates – synthesis of current knowledge and a way forward. Glob. Change Biol. 17, 3392–3404. doi: 10.1111/j.1365-2486.2011.02496.x
Creamer, C. A., de Menezes, A. B., Krull, E. S., Sanderman, J., Newton-Walters, R., and Farrell, M. (2015). Microbial community structure mediates response of soil C decomposition to litter addition and warming. Soil Biol. Biochem. 80, 175–188. doi: 10.1016/j.soilbio.2014.10.008
Davidson, E. A., and Janssens, I. A. (2006). Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440, 165–173. doi: 10.1038/nature04514
DeAngelis, K. M., Allgaier, M., Chavarria, Y., Fortney, J. L., Hugenholtz, P., Simmons, B., et al. (2011). Characterization of trapped lignin-degrading microbes in tropical forest soil. PLoS ONE 6:e19306. doi: 10.1371/journal.pone.0019306
DeAngelis, K. M., Pold, G., Topcuoglu, B. D., van Diepen, L. T. A., Varney, R., Blanchard, J., et al. (2015). Long-term forest soil warming alters microbial communities in temperate forest soils. Terr. Microbiol. 6:104. doi: 10.3389/fmicb.2015.00104
DeAngelis, K. M., Sharma, D., Varney, R., Simmons, B., Isern, N. G., Markilllie, L. M., et al. (2013). Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1. Front. Microbiol. 4:280. doi: 10.3389/fmicb.2013.00280
DeAngelis, K. M., Silver, W. L., Thompson, A. W., and Firestone, M. K. (2010). Microbial communities acclimate to recurring changes in soil redox potential status. Environ. Microbiol. 12, 3137–3149. doi: 10.1111/j.1462-2920.2010.02286.x
Dedysh, S. N., Pankratov, T. A., Belova, S. E., Kulichevskaya, I. S., and Liesack, W. (2006). Phylogenetic analysis and in situ identification of bacteria community composition in an acidic sphagnum peat bog. Appl. Environ. Microbiol. 72, 2110–2117. doi: 10.1128/AEM.72.3.2110-2117.2006
DeRito, C. M., Pumphrey, G. M., and Madsen, E. L. (2005). Use of field-based stable isotope probing to identify adapted populations and track carbon flow through a phenol-degrading soil microbial community. Appl. Environ. Microbiol. 71, 7858–7865. doi: 10.1128/AEM.71.12.7858-7865.2005
Deschamps, A. M., Mahoudeau, G., and Lebeault, J. M. (1980). Fast degradation of kraft lignin by bacteria. Eur. J. Appl. Microbiol. Biotechnol. 9, 45–51. doi: 10.1007/BF00500001
Devêvre, O. C., and Horwáth, W. R. (2000). Decomposition of rice straw and microbial carbon use efficiency under different soil temperatures and moistures. Soil Biol. Biochem. 32, 1773–1785. doi: 10.1016/S0038-0717(00)00096-1
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Erhagen, B., Ilstedt, U., and Nilsson, M. B. (2015). Temperature sensitivity of heterotrophic soil CO2 production increases with increasing carbon substrate uptake rate. Soil Biol. Biochem. 80, 45–52. doi: 10.1016/j.soilbio.2014.09.021
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Flury, S., and Gessner, M. O. (2011). Experimentally simulated global warming and nitrogen enrichment effects on microbial litter decomposers in a marsh. Appl. Environ. Microbiol. 77, 803–809. doi: 10.1128/AEM.01527-10
Fontaine, S., Mariotti, A., and Abbadie, L. (2003). The priming effect of organic matter: a question of microbial competition? Soil Biol. Biochem. 35, 837–843. doi: 10.1016/S0038-0717(03)00123-8
Fox, J. (2008). Applied Regression Analysis and Generalized Linear Models, 2nd Edn. Thousand Oaks, CA: SAGE.
Frey, S. D., Drijber, R., Smith, H., and Melillo, J. (2008). Microbial biomass, functional capacity, and community structure after 12 years of soil warming. Soil Biol. Biochem. 40, 2904–2907. doi: 10.1016/j.soilbio.2008.07.020
Frey, S. D., Lee, J., Melillo, J. M., and Six, J. (2013). Soil carbon cycling: the temperature response of microbial efficiency and its feedback to climate. Nat. Clim. Change 3, 395–398. doi: 10.1038/nclimate1796
Gall, D. L., Kim, H., Lu, F., Donohue, T. J., Noguera, D. R., and Ralph, J. (2014). Stereochemical features of glutathione-dependent enzymes in the Sphingobium sp. strain SYK-6 β-aryl etherase pathway. J. Biol. Chem. 289, 8656–8667. doi: 10.1074/jbc.M113.536250
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Garrity, G. M., Brenner, D. J., Krieg, N. R., and Staley, J. T. (2012). Bergey's Manual® of Systematic Bacteriology - Volume Two: The Proteobacteria (Part C). Springer. Available online at: http://www.springer.com/life+sciences/book/978-0-387-24145-6 (Accessed April 25, 2014).
Ghodake, G. S., Kalme, S. D., Jadhav, J. P., and Govindwar, S. P. (2009). Purification and partial characterization of lignin peroxidase from Acinetobacter calcoaceticus NCIM 2890 and its application in decolorization of textile dyes. Appl. Biochem. Biotechnol. 152, 6–14. doi: 10.1007/s12010-008-8258-4
Goldfarb, K. C., Karaoz, U., Hanson, C. A., Santee, C. A., Bradford, M. A., Treseder, K. K., et al. (2011). Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front. Microbiol. 2:94. doi: 10.3389/fmicb.2011.00094
Gonzalez, B., Merino, A., Almeida, M., and Vicna, R. (1986). Comparative growth of natural bacterial isolates on various lignin-related compounds. Appl. Environ. Microbiol. 52, 1428–1432.
González, J. M., Whitman, W. B., Hodson, R. E., and Moran, M. A. (1996). Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl. Environ. Microbiol. 62, 4433–4440.
Gupta, V. K., Minocha, A. K., and Jain, N. (2001). Batch and continuous studies on treatment of pulp mill wastewater by Aeromonas formicans. J. Chem. Technol. Biotechnol. 76, 547–552. doi: 10.1002/jctb.417
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Ji, L., Yang, J., Fan, H., Yang, Y., Li, B., Yu, X., et al. (2014). Synergy of crude enzyme cocktail from cold-adapted Cladosporium cladosporioides Ch2-2 with commercial xylanase achieving high sugars yield at low cost. Biotechnol. Biofuels 7, 130. doi: 10.1186/s13068-014-0130-x
Jobbágy, E. G., and Jackson, R. B. (2000). The vertical distribution of soil organic carbon and its relation to climate and vegetation. Ecol. Appl. 10, 423–436. doi: 10.1890/1051-0761(2000)010[0423:TVDOSO]2.0.CO;2
Kampfer, P. (2012). “Streptomyces,” in Bergey's Manual of Systematic Bacteriology: The Actinobacteria, Part A, eds W. B. Whitman, M. Goodfellow, P. Kämpfer, H.-J. Busse, M. E. Trujillo, W. Ludwig, K.-i. Suzuki, and A. Parte (New York, NY: Springer), 1455–1475.
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Kerr, T. J., Kerr, R. D., and Benner, R. (1983). Isolation of a bacterium capable of degrading peanut hull lignin. Appl. Environ. Microbiol. 46, 1201–1206.
Kim, J.-H., Kim, M., and Bae, W. (2009). Effect of oxidized leachate on degradation of lignin by sulfate-reducing bacteria. Waste Manag. Res. 27, 520–526. doi: 10.1177/0734242X08096899
Klamer, M., Roberts, M. S., Levine, L. H., Drake, B. G., and Garland, J. L. (2002). Influence of elevated CO(2) on the fungal community in a coastal scrub oak forest soil investigated with terminal-restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 68, 4370–4376. doi: 10.1128/AEM.68.9.4370-4376.2002
Lavelle, P. (1997). Faunal activities and soil processes: adaptive strategies that determine ecosystem function. Adv. Ecol. Res. 27, 93–132. doi: 10.1016/S0065-2504(08)60007-0
Legendre, P., and Gallagher, E. D. (2001). Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280. doi: 10.1007/s004420100716
Letunic, I., and Bork, P. (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Lu, M., Zhou, X., Yang, Q., Li, H., Luo, Y., Fang, C., et al. (2013). Responses of ecosystem carbon cycle to experimental warming: a meta-analysis. Ecology 94, 726–738. doi: 10.1890/12-0279.1
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Magurran, A. E., and McGill, B. J. (2011). Biological Diversity: Frotiers in Measurement and Assessment. Oxford: Oxford University Press.
Manzoni, S., Taylor, P., Richter, A., Porporato, A., and Ågren, G. I. (2012). Environmental and stoichiometric controls on microbial carbon-use efficiency in soils. New Phytol. 196, 79–91. doi: 10.1111/j.1469-8137.2012.04225.x
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
McGuire, A. D., Sitch, S., Clein, J. S., Dargaville, R., Esser, G., Foley, J., et al. (2001). Carbon balance of the terrestrial biosphere in the Twentieth Century: analyses of CO2, climate and land use effects with four process-based ecosystem models. Glob. Biogeochem. Cycles 15, 183–206. doi: 10.1029/2000GB001298
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. doi: 10.1371/journal.pone.0061217
Melillo, J. M., Steudler, P. A., Aber, J. D., Newkirk, K. M., Lux, H., Bowles, F. P., et al. (2002). Soil warming and carbon-cycle feedbacks to the climate system. Science 298, 2173–2176. doi: 10.1126/science.1074153
Melillo, J. M., Steudler, P. A., and Mohan, J. E. (1999). Prospect hill soil warming experiment at harvard forest since 1991. Harv. For. Data Arch. Available online at: http://harvardforest.fas.harvard.edu:8080/exist/apps/datasets/showData.html?id=hf005 (Accessed March 11, 2014).
Meux, E., Prosper, P., Masai, E., Mulliert, G., Dumarçay, S., Morel, M., et al. (2012). Sphingobium sp. SYK-6 LigG involved in lignin degradation is structurally and biochemically related to the glutathione transferase ω class. FEBS Lett. 586, 3944–3950. doi: 10.1016/j.febslet.2012.09.036
Moore-Kucera, J., and Dick, R. P. (2008). Application of 13C-labeled litter and root materials for in situ decomposition studies using phospholipid fatty acids. Soil Biol. Biochem. 40, 2485–2493. doi: 10.1016/j.soilbio.2008.06.002
Morii, H., Nakamiya, K., and Kinoshita, S. (1995). Isolation of a lignin-decolorizing bacterium. J. Ferment. Bioeng. 80, 296–299. doi: 10.1016/0922-338X(95)90835-N
Nierman, W. C., Feldblyum, T. V., Laub, M. T., Paulsen, I. T., Nelson, K. E., Eisen, J., et al. (2001). Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. U.S.A. 98, 4136–4141. doi: 10.1073/pnas.061029298
Odier, E., Janin, G., and Monties, B. (1981). Poplar lignin decomposition by gram-negative aerobic bacteria. Appl. Environ. Microbiol. 41, 337–341.
Odier, E., and Monties, B. (1978). Biodegradation de la lignine de ble par Xanthomonas 23. Ann. Microbiol. 129A, 361–377.
Oksanen, J., Blanchet, G., Kindt, R., Legendre, P., Minchin, P., O'Hara, R. B., et al. (2013). Vegan: Community Ecology Package. Available online at: http://CRAN.R-project.org/package=vegan
Omotayo, A. E., Ilori, M. O., Amund, O. O., Ghosh, D., Roy, K., and Radosevich, M. (2011). Establishment and characterization of atrazine degrading cultures from Nigerian agricultural soil using traditional and Bio-Sep bead enrichment techniques. Appl. Soil Ecol. 48, 63–70. doi: 10.1016/j.apsoil.2011.01.006
Östling, S., and Virtama, P. (1946). A modified preparation of the universal buffer described by Teorell and Stenhagen. Acta Physiol. Scand. 11, 289–293. doi: 10.1111/j.1748-1716.1946.tb00349.x
Pankratov, T. A., and Dedysh, S. N. (2010). Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 60, 2951–2959. doi: 10.1099/ijs.0.021824-0
Parham, J. A., and Deng, S. P. (2000). Detection, quantification and characterization of β-glucosaminidase activity in soil. Soil Biol. Biochem. 32, 1183–1190. doi: 10.1016/S0038-0717(00)00034-1
Parshetti, G. K., Parshetti, S., Kalyani, D. C., Doong, R., and Govindwar, S. P. (2011). Industrial dye decolorizing lignin peroxidase from Kocuria rosea MTCC 1532. Ann. Microbiol. 62, 217–223. doi: 10.1007/s13213-011-0249-y
Pasti, M. B., Pometto, A. L., Nuti, M. P., and Crawford, D. L. (1990). Lignin-solubilizing ability of actinomycetes isolated from termite (Termitidae) gut. Appl. Environ. Microbiol. 56, 2213–2218.
Peacock, A. D., Chang, Y.-J., Istok, J. D., Krumholz, L., Geyer, R., Kinsall, B., et al. (2004). Utilization of microbial biofilms as monitors of bioremediation. Microb. Ecol. 47, 284–292. doi: 10.1007/s00248-003-1024-9
Perestelo, F., Falcón, M. A., and De la Fuente, G. (1990). Biotransformation of kraft lignin fractions by Serratia marcescens. Lett. Appl. Microbiol. 10, 61–64. doi: 10.1111/j.1472-765X.1990.tb00265.x
Peterjohn, W. T., Melillo, J. M., Steudler, P. A., Newkirk, K. M., Bowles, F. P., and Aber, J. D. (1994). Responses of trace gas fluxes and N availability to experimentally elevated soil temperatures. Ecol. Appl. 4, 617–625. doi: 10.2307/1941962
Pold, G., and DeAngelis, K. (2013). Up against the wall: the effects of climate warming on soil microbial diversity and the potential for feedbacks to the carbon cycle. Diversity 5, 409–425. doi: 10.3390/d5020409
Raj, A., Reddy, M. M. K., Chandra, R., Purohit, H. J., and Kapley, A. (2007). Biodegradation of kraft-lignin by Bacillus sp. isolated from sludge of pulp and paper mill. Biodegradation 18, 783–792. doi: 10.1007/s10532-007-9107-9
R Development Core Team. (2008). R: a Language and Environment for Statistical Computing. Available online at: http://www.R-project.org
Ribeiro, P. J., and Diggle, P. (2001). geoR: a package for geostatistical analysis. R-NEWS 1, 15–18.
Roberts, D. W. (2013). labdsv: Ordination and Multivariate Analysis for Ecology. Available onlie at: http://cran.r-project.org/web/packages/labdsv/index.html (Accessed November 1, 2014).
Rousk, J., Frey, S., and Bååth, E. (2012). Temperature adaptation of bacterial communities in experimentally warmed forest soils. Glob. Change Biol. 18, 3252–3258. doi: 10.1111/j.1365-2486.2012.02764.x
Royston, J. P. (1982). An extension of shapiro and Wilk's W test for normality to large samples. J. R. Stat. Soc. Ser. C Appl. Stat. 31, 115–124. doi: 10.2307/2347973
Rustad, L. E., Campbell, J. L., Marion, G. M., Norby, R. J., Mitchell, M. J., Hartley, A. E., et al. (2001). A meta-analysis of the response of soil respiration, net nitrogen mineralization, and aboveground plant growth to experimental ecosystem warming. Oecologia 126, 543–562. doi: 10.1007/s004420000544
Saiya-Cork, K., Sinsabaugh, R., and Zak, D. (2002). The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 34, 1309–1315. doi: 10.1016/S0038-0717(02)00074-3
Schipper, L. A., Hobbs, J. K., Rutledge, S., and Arcus, V. L. (2014). Thermodynamic theory explains the temperature optima of soil microbial processes and high Q10 values at low temperatures. Glob. Change Biol. 20, 3578–3586. doi: 10.1111/gcb.12596
Schneider, T., Gerrits, B., Gassmann, R., Schmid, E., Gessner, M. O., Richter, A., et al. (2010). Proteome analysis of fungal and bacterial involvement in leaf litter decomposition. Proteomics 10, 1819–1830. doi: 10.1002/pmic.200900691
Schneider, T., Keiblinger, K. M., Schmid, E., Sterflinger-Gleixner, K., Ellersdorfer, G., Roschitzki, B., et al. (2012). Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions. ISME J. 6, 1749–1762. doi: 10.1038/ismej.2012.11
Shi, Y., Chai, L., Tang, C., Yang, Z., Zhang, H., Chen, R., et al. (2013). Characterization and genomic analysis of kraft lignin biodegradation by the beta-proteobacterium Cupriavidus basilensis B-8. Biotechnol. Biofuels 6:1. doi: 10.1186/1754-6834-6-1
Sistla, S. A., Moore, J. C., Simpson, R. T., Gough, L., Shaver, G. R., and Schimel, J. P. (2013). Long-term warming restructures Arctic tundra without changing net soil carbon storage. Nature 497, 615–618. doi: 10.1038/nature12129
Stone, M., Weiss, M., Goodale, C., Adams, M. B., Fernandez, I. J., German, D. P., et al. (2012). Temperature sensitivity of soil enzyme kinetics under N-fertilization in two temperate forests. Glob. Change Biol. 18, 1173–1184. doi: 10.1111/j.1365-2486.2011.02545.x
Strachan, C. R., Singh, R., VanInsberghe, D., Ievdokymenko, K., Budwill, K., Mohn, W. W., et al. (2014). Metagenomic scaffolds enable combinatorial lignin transformation. Proc. Natl. Acad. Sci. U.S.A. 111, 10143–10148. doi: 10.1073/pnas.1401631111
Strickland, M., Lauber, C. L., Fierer, N., and Bradford, M. A. (2009). Testing the functional significance of microbial community composition. Ecology 90, 441–451. doi: 10.1890/08-0296.1
Sublette, K., Peacock, A., White, D., Davis, G., Ogles, D., Cook, D., et al. (2006). Monitoring subsurface microbial ecology in a sulfate-amended, gasoline-contaminated aquifer. Groundw. Monit. Remediat. 26, 70–78. doi: 10.1111/j.1745-6592.2006.00072.x
Taylor, C. R., Hardiman, E. M., Ahmad, M., Sainsbury, P. D., Norris, P. R., and Bugg, T. D. (2012). Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J. Appl. Microbiol. 113, 521–530. doi: 10.1111/j.1365-2672.2012.05352.x
Trojanowski, J., Haider, K., and Sundman, V. (1977). Decomposition of 14C-labelled lignin and phenols by a Nocardia sp. Arch. Microbiol. 114, 149–153. doi: 10.1007/BF00410776
Tukey, J. W. (1949). Comparing individual means in the analysis of variance. Biometrics 5, 99–114. doi: 10.2307/3001913
Turner, S., Pryer, K. M., Miao, V. P., and Palmer, J. D. (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 46, 327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x
Unterseher, M., Jumpponen, A., Opik, M., Tedersoo, L., Moora, M., Dormann, C. F., et al. (2011). Species abundance distributions and richness estimations in fungal metagenomics–lessons learned from community ecology. Mol. Ecol. 20, 275–285. doi: 10.1111/j.1365-294X.2010.04948.x
Venables, W. N., and Ripley, B. D. (2002). Modern Applied Statistics with S. 4th Edn. New York, NY: Springer. doi: 10.1007/978-0-387-21706-2
Wallenstein, M., Allison, S. D., Ernakovich, J., Steinweg, J. M., and Sinsabaugh, R. (2011). “Controls on the temperature sensitivity of soil enzymes: a key driver of in situ enzyme activity rates,” in Soil Enzymology Soil Biology (Heidelberg: Springer-Verlag Berlin), 245–258. Available online at: http://allison.bio.uci.edu/publications/wallenstein2010b.pdf
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Werner, J. J., Koren, O., Hugenholtz, P., DeSantis, T. Z., Walters, W. A., Caporaso, J. G., et al. (2012). Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 6, 94–103. doi: 10.1038/ismej.2011.82
Wieder, W. R., Bonan, G. B., and Allison, S. D. (2013). Global soil carbon projections are improved by modelling microbial processes. Nat. Clim. Change 3, 909–912. doi: 10.1038/nclimate1951
Williams, N., Hyland, A., Mitchener, R., Sublette, K., Key, K. C., Davis, G., et al. (2013). Demonstrating the in situ biodegradation potential of phenol using bio-sep® bio-traps® and stable isotope probing. Remediat. J. 23, 7–22. doi: 10.1002/rem.21335
Woo, H. L., Ballor, N. R., Hazen, T. C., Fortney, J. L., Simmons, B., Davenport, K. W., et al. (2014a). Complete genome sequence of the lignin-degrading bacterium Klebsiella sp. strain BRL6-2. Stand. Genomic Sci. 9:19. doi: 10.1186/1944-3277-9-19
Woo, H. L., Utturkar, S., Klingeman, D., Simmons, B. A., DeAngelis, K. M., Brown, S. D., et al. (2014b). Draft genome sequence of the lignin-degrading Burkholderia sp. strain LIG30, isolated from wet tropical forest soil. Genome Announc. 2:e00637-14. doi: 10.1128/genomeA.00637-14
Xiong, X. Q., Liao, H. D., Ma, J. S., Liu, X. M., Zhang, L. Y., Shi, X. W., et al. (2014). Isolation of a rice endophytic bacterium, Pantoea sp. Sd-1, with ligninolytic activity and characterization of its rice straw degradation ability. Lett. Appl. Microbiol. 58, 123–129. doi: 10.1111/lam.12163
Xu, X., Sherry, R., Niu, S., Zhou, J., and Luo, Y. (2012). Long-term experimental warming decreased labile soil organic carbon in a tallgrass prairie. Plant Soil 361, 307–315. doi: 10.1007/s11104-012-1265-9
Yan, J., Pan, G., Ding, C., and Quan, G. (2010). Kinetic and thermodynamic parameters of B-glucosidase immobilized on various colloidal particles from a paddy soil. Colloids Surf. B Biointerfaces 79, 298–303. doi: 10.1016/j.colsurfb.2010.04.015
Keywords: chemically complex carbon, climate change, microbial ecology, soil organic matter, Bio-Sep beads, lignin degradation, in-situ enrichment
Citation: Pold G, Melillo JM and DeAngelis KM (2015) Two decades of warming increases diversity of a potentially lignolytic bacterial community. Front. Microbiol. 6:480. doi: 10.3389/fmicb.2015.00480
Received: 10 February 2015; Accepted: 30 April 2015;
Published: 20 May 2015.
Edited by:
Eoin L. Brodie, Lawrence Berkeley National Laboratory, USAReviewed by:
Christopher Blackwood, Kent State University, USALaurel Anne Kluber, Oak Ridge National Laboratory, USA
Copyright © 2015 Pold, Melillo and DeAngelis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kristen M. DeAngelis, Microbiology Department, University of Massachusetts, 639 North Pleasant Street, 203 Morrill IVN, Amherst, MA 01003, USA, deangelis@microbio.umass.edu
 Grace Pold
Grace Pold Jerry M. Melillo3
Jerry M. Melillo3 Kristen M. DeAngelis
Kristen M. DeAngelis