
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 June 2015
Sec. Extreme Microbiology
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00627
The SOS response addresses DNA lesions and is conserved in the bacterial domain. The response is governed by the DNA binding protein LexA, which has been characterized in model microorganisms such as Escherichia coli. However, our understanding of its roles in deep-sea bacteria is limited. Here, the influence of LexA on the phenotype and gene transcription of Shewanella piezotolerans WP3 (WP3) was investigated by constructing a lexA deletion strain (WP3ΔlexA), which was compared with the wild-type strain. No growth defect was observed for WP3ΔlexA. A total of 481 and 108 genes were differentially expressed at 20 and 4∘C, respectively, as demonstrated by comparative whole genome microarray analysis. Furthermore, the swarming motility and dimethylsulfoxide reduction assay demonstrated that the function of LexA was related to temperature. The transcription of the lexA gene was up-regulated during cold acclimatization and after cold shock, indicating that the higher expression level of LexA at low temperatures may be responsible for its temperature-dependent functions. The deep-sea microorganism S. piezotolerans WP3 is the only bacterial species whose SOS regulator has been demonstrated to be significantly influenced by environmental temperatures to date. Our data support the hypothesis that SOS is a formidable strategy used by bacteria against various environmental stresses.
Since it was first described by Radman (1975), the detailed pathway of the SOS response to DNA lesions had been thoroughly investigated in the model bacterium Escherichia coli. In addition to well-known classical DNA damaging agents such as UV-irradiation and antibiotics, several intracellular and extracellular processes and signals, including oxidative stress, acidic or alkaline stress, and high-pressure and acoustic cavitation, have also been revealed to be capable of triggering the SOS response (Erill et al., 2007). Stress-survival studies showed that an SOS-deficient mutant of Listeria monocytogenes was less resistant to heat (55°C), H2O2 and acid exposure (pH 3.4) compared to the wild-type cells (van der Veen et al., 2010). Additionally, the swarming motility was influenced by the SOS regulator RecA and conferred fitness to Salmonella enterica (Medina-Ruiz et al., 2010). Thus the SOS response has been recognized as an important strategy for bacterial survival and adaptation to changing environments. Furthermore, studies have demonstrated the crucial role of the SOS response in promoting the spread of mobile genetic elements (Beaber et al., 2003) and integron recombination, which are responsible for incorporating and expressing genes grouped as cassettes (Guerin et al., 2009). Thus, SOS also plays a considerable role in lateral gene transfer and evolution.
The LexA protein is a transcriptional regulator situated in the central position of the SOS pathway. The induction process of the SOS response has been well-described in several reviews (Michel, 2005; Kelley, 2006; Baharoglu and Mazel, 2014). Briefly, damage of the cellular DNA by external stressors produces single-stranded DNA (ssDNA); then, RecA bind to the ssDNA and becomes activated. The active RecA promotes the self-cleavage of the LexA dimer, releasing it from the promoter of the SOS regulon genes and derepressing their transcription. LexA homologues can be found in almost all sequenced bacterial genomes, suggesting both an ancient origin and widespread distribution of lexA and the SOS response (Erill et al., 2007). The LexA binding sites (SOS boxes) are rather conservative and have been used as a marker to indicate the evolutionary history of the SOS pathway in the bacterial domain (Abella et al., 2004; Mazón et al., 2004). Although genes of the LexA regulons are diverse in different bacteria and are not always equivalent to the SOS regulon (Kelley, 2006), there is no doubt that LexA is the key regulator in the SOS pathway (Butala et al., 2009). The regulatory function of LexA has been examined in various bacteria and was demonstrated to play important roles in growth, survival, hydrogenase expression, sporulation and antibiotic resistance (Tapias et al., 2000; Gutekunst et al., 2005; Rocha et al., 2008; Jochmann et al., 2009; Walter et al., 2014). However, the composition of the SOS regulon and the function of LexA in the deep-sea environment (which accounts for the majority of the ocean) is largely unknown. Considering the abyssal environment is characterized with permanent low temperatures (2–4°C), it would be interesting to testify whether bacteria which have evolved in a relatively constant habitat could be hyper-sensitive to temperature fluctuations, and thus systems associated with stress adaptation such as SOS pathway could manifest themselves differently depending on the thermal regime. Therefore, we focused on the investigation of the mechanism by which deficient SOS mutants of deep-sea bacterium respond to different temperatures in this study.
The Shewanella species are well-known for their versatile respiration ability and are widely distributed in aquatic environments, including the deep-sea. Shewanella piezotolerans WP3 (hereafter referred to as WP3) was previously isolated from a West Pacific sediment at a depth of 1914 m (Wang et al., 2004; Xiao et al., 2007). The temperature range of WP3 growth was characterized between 4 and 28°C at 0.1 MPa, with maximal growth at 20°C (Xiao et al., 2007). The cold adaptation mechanism has been investigated in WP3, and fatty acid biosynthesis and nitrate reductase were shown to respond to changes in temperature (Wang et al., 2009; Chen et al., 2011). In this study, the SOS regulon was characterized using whole genome microarray analysis of the lexA deletion mutant. The results indicated that the differentially expressed genes (DEGs) were significantly influenced by temperature. Further physiological testing confirmed the temperature-dependent function of LexA because the transcription of the lexA gene was up-regulated at 4°C and after cold-shock. To the best of our knowledge, this is the first report of the characterization of LexA in a deep-sea microorganism and the evaluation of the effect of temperature on LexA regulation. Taken together, our data implied an important role for LexA in the evolution of WP3 in response to the cold deep-sea environment.
All bacterial strains and plasmids used in this study are listed in Table 1. The Shewanella strains were cultured in 2216E marine medium (2216E; 5 g/l tryptone, 1 g/l yeast extract, 0.1 g/l FePO4, and 34 g/l NaCl) with shaking at 220 rpm at different temperatures under atmospheric pressure condition. E. coli strain WM3064 was incubated in lysogeny broth (LB) medium (10 g/l tryptone, 5 g/l yeast extract, and 10 g/l NaCl) at 37°C with the addition of 50 μg/ml of DL-α,𝜀-diaminopimelic acid (DAP). For solid medium, agar-A (Bio Basic Inc., Markham, ON, Canada) was added at a concentration of 1.5% (w/v). The antibiotics chloramphenicol (Cm) and kanamycin (Kan; Sigma, St. Louis, MO, USA) was added to the medium at concentrations of 12.5 and 50 μg/ml for the Shewanella strains, respectively. The anaerobic growth of the WP3 strain was determined using turbidity measurements at 600 nm with modified 2216E medium (Chen et al., 2011) supplemented with dimethylsulfoxide (DMSO; Sigma, St. Louis, MO, USA) as the sole electron acceptor.
A lexA deletion mutant was constructed using the previously described method (Wang et al., 2009). First, the upstream and downstream fragments flanking both sides of the lexA gene and a Kan resistance gene were amplified with the PCR primer pairs (Table 2). These three fragments were used as templates in a second fusion PCR, resulting in a fragment with a deletion in the lexA gene and insertion of the kanR gene. Then, the PCR product was cloned into pRE112 as a Sac I-Kpn I fragment, yielding pRE112-lexA. This plasmid was transformed into E. coli WM3064 and then into WP3 by two-parent conjugation. The transconjugant was selected by Cm and Kan resistance and verified by PCR. The WP3 strain with pRE112-lexA inserted into the chromosome was plated onto 2216E agar medium supplemented with Kan and 10% sucrose. A successful lexA deletion mutant was screened and confirmed by PCR. For complementation, the Shewanella- E. coli shuttle vector pSW2 was used as previously described (Chen et al., 2011). Briefly, the lexA gene with its native promoter region was amplified with the pfu DNA polymerase (Tiangen, Beijing, China). Both the PCR product and pSW2 were digested with Sac I and Sph I and ligated to generate pSW2-lexA. The recombinant plasmid was introduced into WM3064 and then into WP3ΔlexA by conjugation. The complementation strain was confirmed by PCR and enzyme digestion.
The cells used to assess the cold shock response were prepared as described previously (Jian et al., 2012). The samples were harvested by centrifugation and immediately placed in liquid nitrogen prior to RNA extraction. The WP3 strains were inoculated into 2216E medium at different temperatures as indicated in the text, and the culture was collected immediately when the cells reached the mid-exponential phase (OD600≈0.8). Total RNA extraction, reverse transcription and real-time quantitative real-time PCR (qPCR) were performed as described previously (Jian et al., 2013). The primer pairs (Table 2) used to amplify the selected genes in qPCR were designed using Primer Express software (Applied Biosystems, Foster City, CA, USA).
A microarray that contained 95% of the total predicted gene content of WP3 was designed and manufactured (CapitalBio, Beijing, China). The preparation of fluorescent dye-labeled DNA and hybridization image acquisition, data processing and clustering were performed as previously described (Jian et al., 2013). Briefly, the total RNA was reverse transcribed with SuperScript II (Invitrogen, Carlsbad, CA, USA), and the cDNAs were labeled with Cy3 and Cy5 using the Klenow enzyme (Takara Bio Inc., Japan) according to the manufacturers’ instructions. Labeled cDNA was purified with the PCR purification kit (Macherey-Nagel, Düren, Germany) and resuspended in elution buffer. The microarray slides were hybridized with cDNA prepared from 3 biological replicate samples. As a measure of technical replication, the dye-swap experiment was performed on each sample so that a total of six data points were available for every ORF on the microarray. A LuxScan 10K scanner and microarray scanner 2.3 software (CapitalBio, Beijing, China) were used for the array image acquisition. The linear normalization method was used for data analysis based on the expression levels of WP3 housekeeping genes in combination with the yeast external controls. The normalized data were log-transformed and loaded into MAANOVA under the R environment for multiple testing by fitting a mixed-effects ANOVA model (Wu et al., 2003). Microarray spots with P values <0.001 in the F-test were regarded as DEGs. All of the DEGs were confirmed with the Significance Analysis of Microarrays (SAM) software (Tusher et al., 2001).
For the motility assays, a 1 μl culture of each strain was placed on the swarming plates (2216E medium with 0.7% agar, Eiken Chemical, Tokyo, Japan). The plates were incubated at 20 and 4°C for 3 and 7 days, respectively. Motility was assessed by examining the migration distance of the bacteria from one side to another side of the colony edge (maximal swarming distance). For each strain, the assays were performed on at least three times with three independent cultures spotted onto three plates. The data were analyzed with Student’s two-tailed t-test using the Excel software (Microsoft Corporation, USA).
The SOS Box was identified by searching the 5′ regions (1–700 bp upstream of the start codon) of the genes in WP3 using the conservative sequence of SOS box in γ-proteobacterium. The information matrix for the generation of the LexA Logo was produced by aligning the WP3 LexA binding sequences predicted by the RegPredict web server, which is available at http://regpredict.lbl.gov (Novichkov et al., 2010b). A graphical representation of the matrix through a Logo graph was obtained with Weblogo software, which is available at http://weblogo.berkeley.edu (Crooks et al., 2004).
The intact lexA gene coding region was deleted from the WP3 genome, and the mutated strain was designated WP3ΔlexA. Initially, the putative impact of LexA on the growth of WP3 was investigated at different temperatures (Supplementary Figure S1). No growth deficiency of WP3ΔlexA was observed when the strain was cultivated either at 20°C (the optimal growth temperature of WP3) or 4°C (the in situ environmental temperature). Furthermore, although no significant filament phenotype was observed, the cell length of the lexA mutant was notably increased at 20°C, indicating that the lexA deletion had an effect on cell division and morphology (Supplementary Figure S2).
Whole-genome microarray analysis was performed to identify possible LexA regulation targets by comparing the gene transcription profiles of WP3 and WP3ΔlexA at 20 and 4°C. Overall, 481 genes were found to be differentially expressed between these two strains at 20°C, including 211 up-regulated and 270 down-regulated genes (Figure 1D, Supplementary Tables S1 and S2). However, only 108 DEGs (35 and 73 genes with increased and decreased transcription levels, respectively) were detected in WP3ΔlexA at 4°C, indicating a temperature-dependent regulatory function for LexA in WP3.
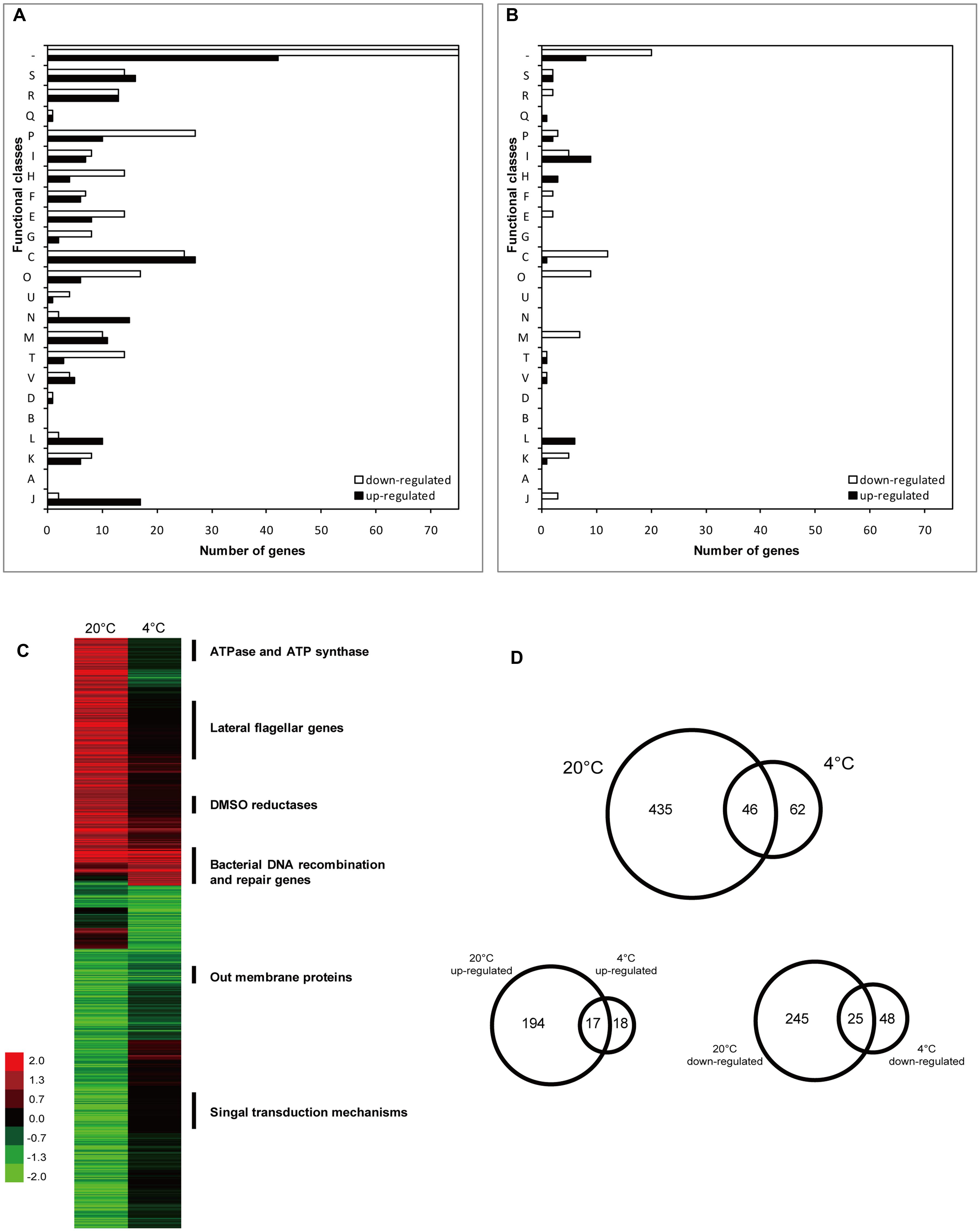
FIGURE 1. Number and functional annotation of the differentially expressed genes (DEGs) according to their COG categories. Bars indicate the number of genes in each group that showed significant changes in expression in WP3 after deletion of lexA gene at 20°C (A) and 4°C (B), respectively. Genes were divided into functional categories according to NCBI (http://www.ncbi.nlm.nih.gov/COG/). Functional categories are abbreviated as follows: J, translation, ribosomal structure and biogenesis; A, RNA processing and modification; K, transcription; L, replication, recombination, and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, and chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; U, intracellular trafficking, secretion and vesicular transport; O, posttranslational modification, protein turnover, and chaperones; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary-metabolite biosynthesis, transport, and catabolism; R, General function prediction only; S, Function unknown; and –, no COG identified. (C) Hierarchical clustering analysis of DEGs in WP3ΔlexA. The function of the enriched genes with different expression patterns at 20 and 4°C were indicated. Color legend is to the left, showing increased (green) and decreased (red) transcription levels. (D) Venn diagram displaying the numbers of DEGs at different temperatures.
To validate the microarray data, seven genes, including those that were up-regulated, down-regulated or unchanged, were selected for qPCR. It should be noted that the same samples were used for the microarray and qPCR. The relative mRNA levels for each gene were calculated and log-transformed. The correlation coefficients (R2) between the data obtained by microarray and qPCR were 0.948 and 0.899 at 20 and 4°C, respectively (Supplementary Figure S3), demonstrating that the microarray data were reliable and could be used for the follow-up analysis. The microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession numbers GSE60724 and GSE60727.
Functional classification of differentially transcribed genes was performed using the Clusters of Orthologous Groups (COGs) of Proteins database (Figures 1A,B). Approximately 30.6 and 29.6% of the genes with altered expression levels at 20 and 4°C, respectively, had unknown functions or no COG identified. The enriched clusters at 20°C were associated with energy production and conversion (52/239, number of DEGs/total number of genes in the specific COG category), protein modification and turn over (23/162), inorganic ion transport and metabolism (37/172) and cell motility (17/134). However, only a few of the functional groups with high numbers of DEGs were identified at 4°C in WP3ΔlexA, including energy production and conversion (13/239) and lipid transport and metabolism (14/120).
Hierarchical clustering analysis was performed to investigate the effect of temperature on the regulatory function of LexA. This analysis demonstrated a significant difference in the expression patterns at 20 and 4°C (Figure 1C). More than 90% of the DEGs at 20°C did not show differential expression at 4°C, suggesting that LexA regulated more genes at higher temperatures. Interestingly, most of the common DEGs at the different temperatures showed the same tendency, including 17 induced and 25 reduced genes (Figure 1D).
The common up-regulated genes were annotated as DNA recombination and repair. These genes consisted of the core LexA regulon as reported in other bacteria (Abella et al., 2004; Erill et al., 2007), such as recA (bacterial DNA recombination protein), recN (DNA repair protein), recG (ATP-dependent DNA helicase), dinP (DNA damage inducible protein P), topA (DNA topoisomerase III) and dnaE (DNA polymerase III α subunit), indicating that the regulation of the core regulon by LexA was not affected by temperature. The functions of the common down-regulated gene were shown to be associated with the ribosomal subunit interface protein (swp3757 and swp3408), outer membrane protein (swp0247, swp0248, and swp3209), Acyl-CoA dehydrogenase (swp2312 and swp2982), fumarate reductase flavoprotein (swp0430) and twin-arginine translocation pathway signal (swp5029), suggesting that a similar strategy could exist in WP3 for coping with DNA damage at different temperatures.
Because most of the DEGs exhibited different expression levels at 20°C, the characterization of these genes were of interest. Notably, a large portion of the lateral flagellar (laf) genes were up-regulated after lexA deletion at 20°C, but exhibited no significant difference in expression at 4°C (Figure 1C, Supplementary Tables S1 and S2). It is noteworthy that only one DEG was found in the polar flagellar gene cluster, indicating that LexA specifically regulated laf gene expression. To confirm this phenotypically, swarming motility assays were performed at different temperatures. The lexA mutant demonstrated a significantly higher (p < 0.01) swarming ability compared with the wild-type strain at 20°C, while no significant difference was detected at 4°C (Figure 2); this finding is consistent with the transcriptomic data.
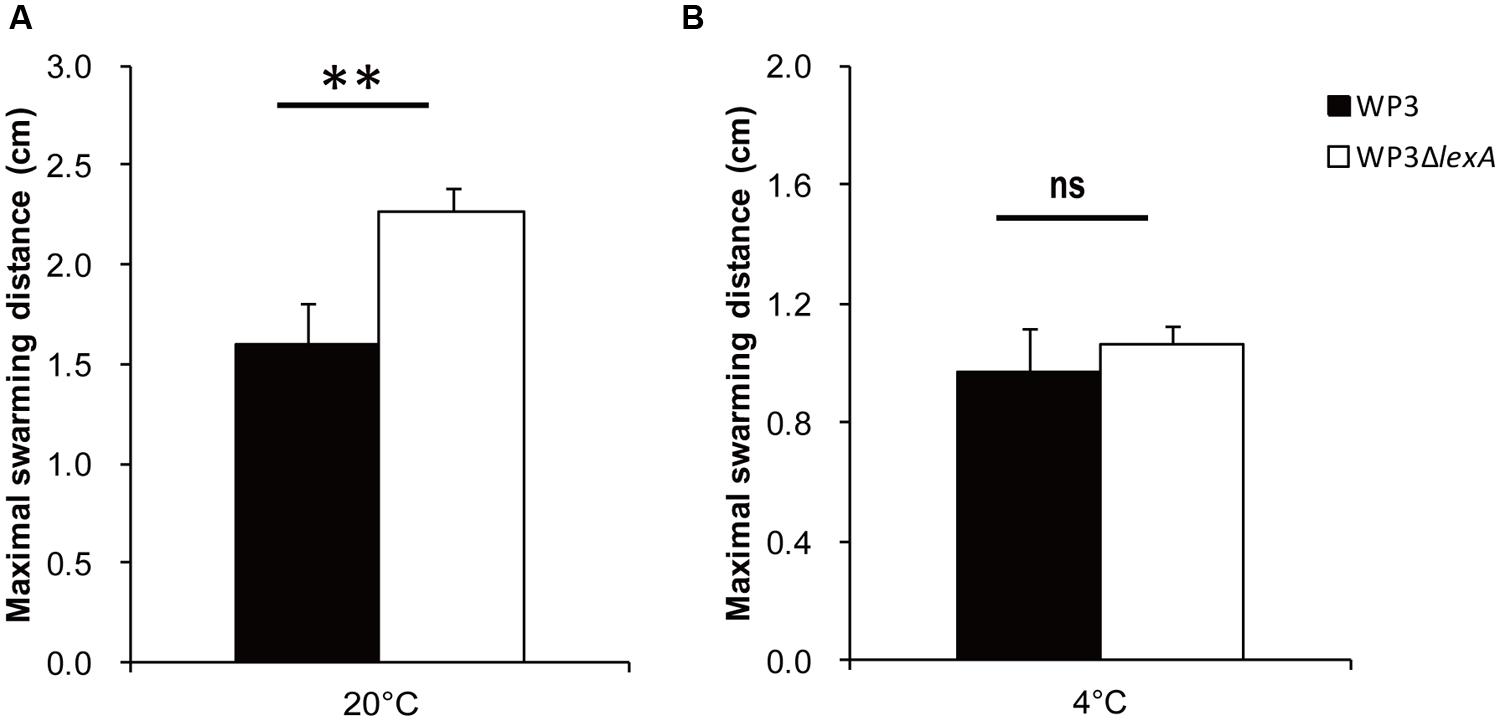
FIGURE 2. The swarming motility assays of the lexA gene mutant at 20°C (A) and 4°C (B). The data shown represent the results of two independent experiments, and the error bars indicate the SD. The data were analyzed by Student’s t-test, ∗∗P < 0.01; ns: not significantly different.
Another similar case was the two DMSO reduction gene clusters (swp0721–swp0725 and swp3457–3459), which were significantly up-regulated at 20°C but not at 4°C. Surprisingly, the growth assay using DMSO as the sole electron acceptor indicated a more profound effect of the lexA mutation at 4°C (Figure 3), suggesting that a post-transcriptional effect may be involved in the LexA-regulated DMSO reduction process in WP3. Regardless, our data showed that LexA was involved in the regulation of anaerobic respiration of WP3 using DMSO in response to changes in temperature.
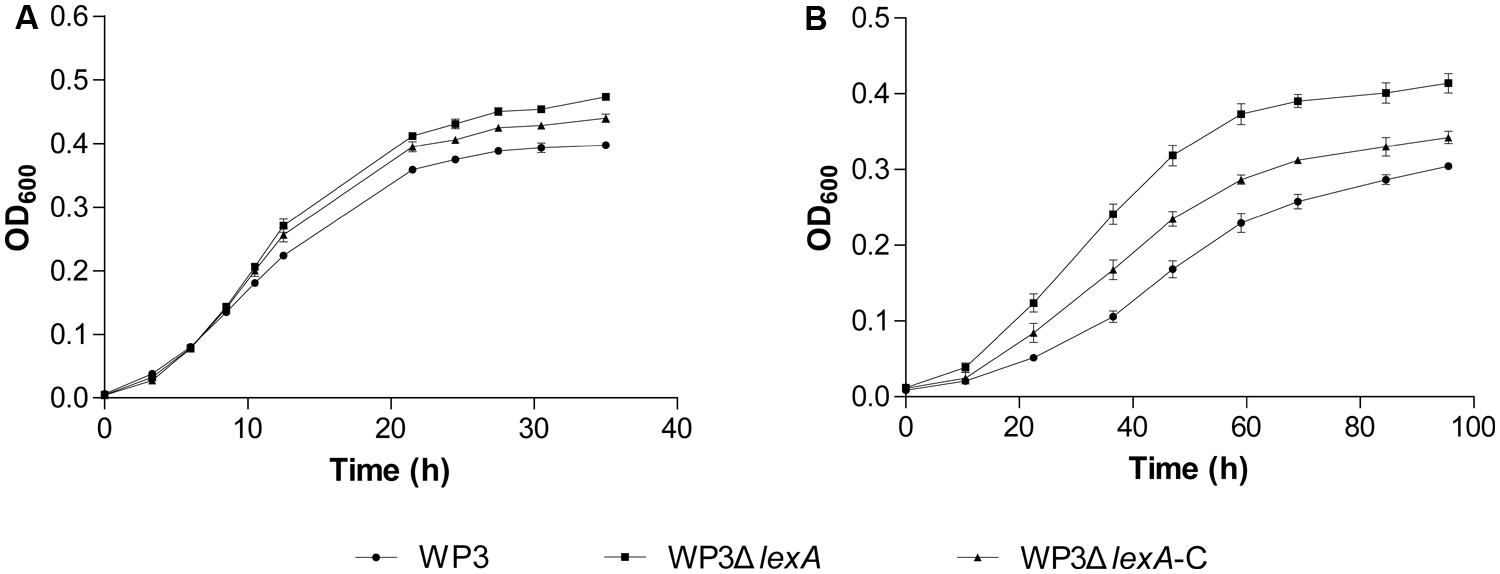
FIGURE 3. Growth curve of WP3ΔlexA at 20°C (A) and 4°C (B) with DMSO as the sole electron acceptor. All of the assays were performed in 2216E medium anaerobically supplemented with dimethylsulfoxide (DMSO). The average values and SD displayed by the error bars resulted from three replicates. All of the data shown represent at least two independent experiments.
As a transcriptional regulator, LexA plays a role as a DNA-binding protein. Thus, temperature may affect the LexA regulatory function in two ways: by influencing the quantity or the binding ability. First, the transcriptional level of the lexA gene was determined at different temperatures. A significantly higher mRNA (2.7-fold) level was observed at 4°C compared to 20°C (Figure 4A). We also investigated the transcription of the lexA gene in response to cold shock, and found a remarkable increase in the mRNA quantity of the lexA gene 2 h after the temperature shift from 20 to 4°C (Figure 4B).
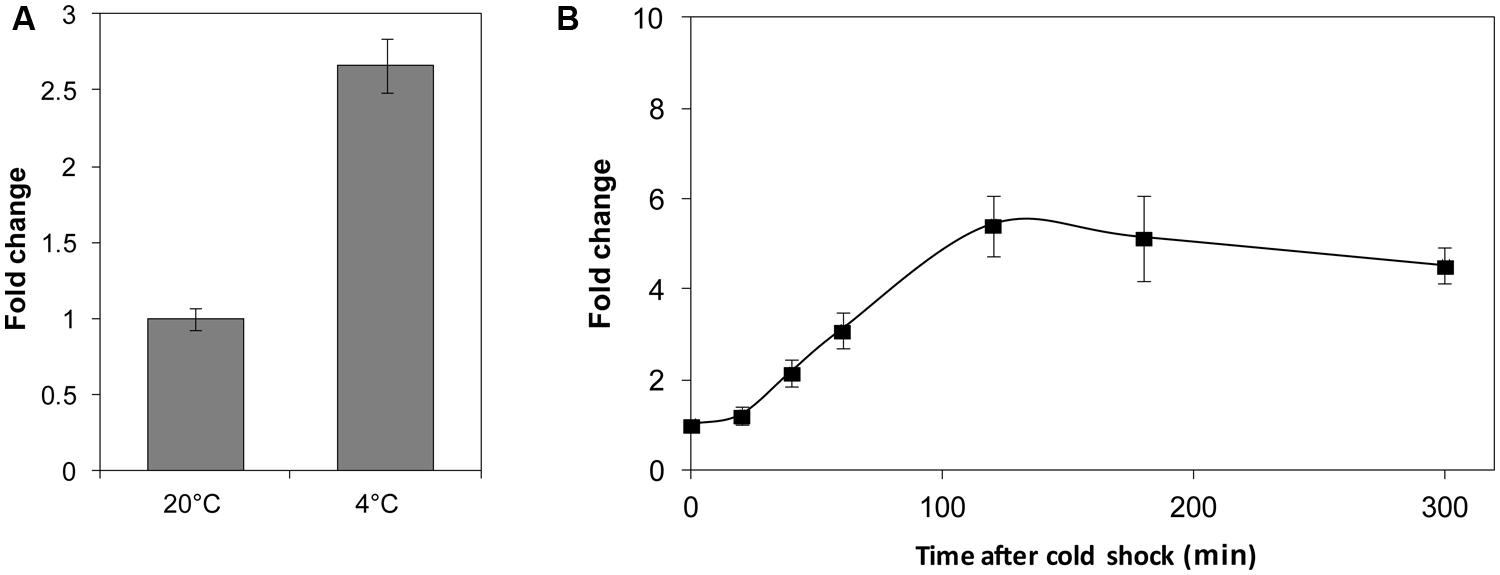
FIGURE 4. The relative transcription level assay of the lexA gene at different temperatures (A) and after cold shock (B). The transcription levels of WP3 at 20°C and before the 4°C cold shock were set as 1. The data shown represent two independent experiments, and the error bars indicate the SD.
Finally, we searched the WP3 genome for LexA binding sites using the conservative sequence of the SOS box in γ-proteobacterium (CTGTN8ACAG; Erill et al., 2003). The putative LexA binding sites located in the coding region were ruled out, and 20 SOS boxes were identified in the promoter region of 19 genes (Table 3). The highly conserved SOS box logo of WP3 is depicted (Figure 5); this sequence was found to be quite similar to other Shewanella stains (Erill et al., 2007; Novichkov et al., 2010a). Most of the genes with upstream LexA binding sites showed differential expression in WP3ΔlexA, but all showed similar changes at different temperatures. These results suggested that it was unlikely that temperature affected the binding of LexA to these target genes.
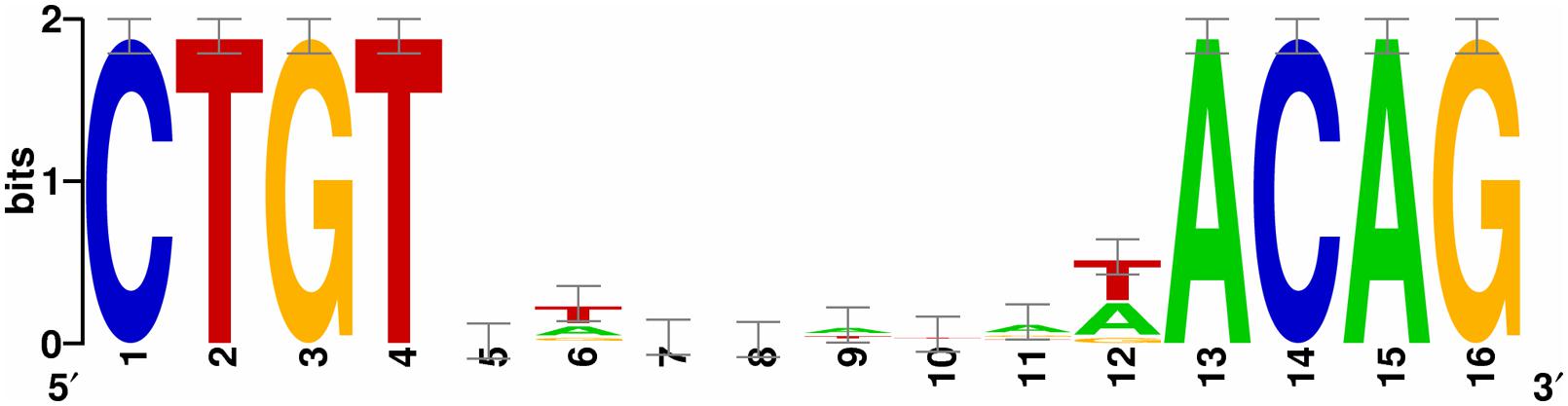
FIGURE 5. The identification of the LexA binding box in Shewanella piezotolerans WP3. A consensus LexA binding sequence logo was obtained from an unbiased search of the WP3 genome using the web-based tool RegPredict (http://regpredict.lbl.gov). The error bars indicate the SD of the sequence conservation.
Previous studies showed that the lexA gene deletion was lethal for E. coli and other γ-proteobacteria, such as Pseudomonas putida, P. aeruginosa, Aeromonas hydrophila, Erwinia carotovora, and Salmonella typhimurium (Walker, 1984; Calero et al., 1993; Riera and Barbe, 1995). Therefore, viable E. coli lexA-defective strains must be constructed based on the mutation of the sfiA gene (Walker, 1984). In this study, the lexA deletion mutant of deep-sea bacterium WP3 was constructed directly; no growth deficiency was observed in the growth assay (Supplementary Figure S1), which is different from the decreased growth rate of lexA mutants reported for other bacteria (Rocha et al., 2008; Jochmann et al., 2009). However, there is also a report indicating that the growth and viability of Rhodobacter sphaeroides lexA (Def) cultures do not show any change relative to the wild-type strain (Tapias et al., 2000). Therefore, the deletion of lexA leading to the defect of cell growth should not be considered as a rule within the bacterial domain. No SOS boxes were found in the promoter region of the sulA gene, which encodes the cell division suppressor (Adams and Errington, 2009). Moreover, no increased transcription of sulA was observed, indicating that this gene does not belong to the SOS regulon in WP3. Additionally, the cell length of WP3ΔlexA was increased significantly at 20°C but not at 4°C (Supplementary Figure S2), suggesting a profound influence of the lexA deletion at higher temperatures. In term of this phenotype, the posttranscriptional regulation of sulA gene expression or alternative cell division regulators could be responsible. The loss of control of cell division and reproduction by LexA in WP3 may be due to one of the adaptive strategies developed for survival in the deep-sea sediment, which is characterized by aphotic and relatively stable environmental factors. Thus the DNA lesions were speculated to be too low to necessitate LexA control of cell division inhibition in the bathypelagic microorganism.
The binding sites of LexA in WP3 were analyzed and the conservative sequence was demonstrated to share high similarity with other Shewanella strains, indicating an evolutionary history of LexA in this widespread genus. Interestingly, the genes harboring LexA boxes did not show differential expression at 20 and 4°C (Table 2), implying that the transcription of these genes regulated by LexA was not affected by temperature. Because our qPCR results indicated that the transcriptional level of the lexA gene increased during cold acclimation and after cold shock, the temperature-dependent regulation of LexA may be due to its different concentrations at various temperatures. Indeed, the intracellular level of LexA in E. coli varied markedly throughout the E. coli growth cycle (Dri and Moreau, 1993).
In this study, the LexA-regulated genes were demonstrated to respond to the change in temperature, with the number of DEGs at 4°C accounting for less than 1/4 of the DEGs at 20°C (Figure 1). The LexA regulon of E. coli was reported to be controlled by pH, and shifts of the bacterial cell to extreme pH increased the cellular concentration of the LexA repressor (Dri and Moreau, 1994). The growth of a LexA-depleted Synechococcus strain was found to be strongly dependent on the availability of inorganic carbon (Domain et al., 2004). Interestingly, a physiological test confirmed the temperature-dependent function of LexA in swarming motility and DMSO reduction (Figures 2 and 3). The analysis of gene expression profile in lexA gene mutant at 20 MPa and 4°C also demonstrated that the transcription of one DMSO reduction gene cluster (swp0721–swp0725) was significantly changed (Jian and Wang, 2015). Considering that the deep-sea is generally an oligotrophic environment with limited energy resources (Orcutt et al., 2011), LexA may be involved in the adjusting of the life-strategy and adaptation of WP3 to deep-sea sediments by regulation of anaerobic respiration such as DMSO reduction.
In addition to modulate the SOS response by preventing SulA inhibition of cell division in E. coli, it was revealed that the ATP-dependent protease LonS, regulates swarmer cell differentiation of Vibrio parahaemolyticus (Stewart et al., 1997). However, the SulA was demonstrated not responsible for laf gene expression (Gode-Potratz et al., 2011). As SOS boxes were identified both in the promoter region of polar (fliC) and laf gene (lafB), the LonS and LexA were speculated to play a role in the regulation of flagella synthesis and motility. Moreover, the thermo-regulation of swarming motility and laf gene has been reported in several bacteria (Ulitzur, 1975; Bresolin et al., 2008; Soo et al., 2008; Tambalo et al., 2010; Hockett et al., 2013). In this study, the increasing of swarming motility in WP3ΔlexA was only observed at 20°C. Furthermore, the transcriptional level of laf genes were up-regulated at 4°C according to our previous study (Wang et al., 2008). In conclusion, our data support the notion that laf system is an inducible system, and it is regulated by environmental factors including SOS response (Merino et al., 2006; Eloe et al., 2008; Lauro and Bartlett, 2008).
The comparative analysis of gene expression profile in lexA mutant at different temperature and pressures has been performed, and 19 DEGs were identified under all of the tested conditions (Supplementary Figure S4). These genes are mainly belonging to core SOS regulon (recA, recN, dinP, dinG, dnaE, and TopA). However, the majority of DEGs (206 genes) were shown to be specific at low-temperature and high-pressure, indicating the regulatory function of LexA is remarkablely influenced by environmental factors. Thus the flexible LexA regulon and conserved core genes and binding sites indicate the evolution of the SOS network during the adaptation process. Taken together, our data renewed the understanding of the role of LexA in sensing and adaptation to different environmental conditions. This finding will certainly stimulate the analysis of the SOS system to address global stress in bathypelagic ecosystems.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was financially supported by the National Natural Science Foundation of China (Grant No. 31290232), China Ocean Mineral Resources R & D Association (Grant No. DY125-15-T-04) and the National Natural Science Foundation of China (Grant No. 41306129).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00627
Abella, M., Erill, I., Jara, M., Mazón, G., Campoy, S., and Barbé, J. (2004). Widespread distribution of a lexA-regulated DNA damage-inducible multiple gene cassette in the Proteobacteria phylum. Mol. Microbiol. 54, 212–222. doi: 10.1111/j.1365-2958.2004.04260.x
Adams, D. W., and Errington, J. (2009). Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642–653. doi: 10.1038/nrmicro2198
Baharoglu, Z., and Mazel, D. (2014). SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 38, 1126–1145. doi: 10.1111/1574-6976.12077
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2003). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74. doi: 10.1038/nature02241
Bresolin, G., Trček, J., Scherer, S., and Fuchs, T. M. (2008). Presence of a functional flagellar cluster Flag-2 and low-temperature expression of flagellar genes in Yersinia enterocolitica W22703. Microbiology 154, 196–206. doi: 10.1099/mic.0.2007/008458-0
Butala, M., Žgur-Bertok, D., and Busby, S. J. (2009). The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 66, 82–93. doi: 10.1007/s00018-008-8378-6
Calero, S., Garriga, X., and Barbe, J. (1993). Analysis of the DNA damage mediated induction of Pseudomonas putida and Pseudomonas aeruginosa lexA genes. FEMS Microbiol. Lett. 110, 65–70. doi: 10.1111/j.1574-6968.1993.tb06296.x
Chen, Y., Wang, F., Xu, J., Mehmood, M. A., and Xiao, X. (2011). Physiological and evolutionary studies of NAP systems in Shewanella piezotolerans WP3. ISME J. 5, 843–855. doi: 10.1038/ismej.2010.182
Crooks, G. E., Hon, G., Chandonia, J.-M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Domain, F., Houot, L., Chauvat, F., and Cassier-Chauvat, C. (2004). Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 53, 65–80. doi: 10.1111/j.1365-2958.2004.04100.x
Dri, A.-M., and Moreau, P. L. (1993). Phosphate starvation and low temperature as well as ultraviolet irradiation transcriptionally induce the Escherichia coli LexA-controlled gene sfiA. Mol. Microbiol. 8, 697–706. doi: 10.1111/j.1365-2958.1993.tb01613.x
Dri, A.-M., and Moreau, P. L. (1994). Control of the LexA reguion by pH: evidence for a reversible inactivation of the LexA repressor during the growth cycle of Escherichia coli. Mol. Microbiol. 12, 621–629. doi: 10.1111/j.1365-2958.1994.tb01049.x
Edwards, R. A., Keller, L. H., and Schifferli, D. M. (1998). Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157. doi: 10.1016/S0378-1119(97)00619-7
Eloe, E. A., Lauro, F. M., Vogel, R. F., and Bartlett, D. H. (2008). The deep-sea bacterium Photobacterium profundum SS9 utilizes separate flagellar systems for swimming and swarming under high-pressure conditions. Appl. Environ. Microbiol. 74, 6298–6305. doi: 10.1128/AEM.01316-08
Erill, I., Campoy, S., and Barbé, J. (2007). Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31, 637–656. doi: 10.1111/j.1574-6976.2007.00082.x
Erill, I., Escribano, M., Campoy, S., and Barbé, J. (2003). In silico analysis reveals substantial variability in the gene contents of the gamma proteobacteria LexA-regulon. Bioinformatics 19, 2225–2236. doi: 10.1093/bioinformatics/btg303
Gao, H., Yang, Z. K., Wu, L., Thompson, D. K., and Zhou, J. (2006). Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 188, 4560–4569. doi: 10.1128/JB.01908-05
Gode-Potratz, C. J., Kustusch, R. J., Breheny, P. J., Weiss1, D. S., and Mccarter, L. L. (2011). Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol. Microbiol. 79, 240–263. doi: 10.1111/j.1365-2958.2010.07445.x
Guerin, E., Cambray, G., Sanchez-Alberola, N., Campoy, S., Erill, I., Re, S. D., et al. (2009). The SOS response controls integron recombination. Science 324, 1034. doi: 10.1126/science.1172914
Gutekunst, K., Phunpruch, S., Schwarz, C., Schuchardt, S., Schulz-Friedrich, R., and Appel, J. (2005). LexA regulates the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 as a transcription activator. Mol. Microbiol. 58, 810–823. doi: 10.1111/j.1365-2958.2005.04867.x
Hockett, K. L., Burch, A. Y., and Lindow, S. E. (2013). Thermo-regulation of genes mediating motility and plant interactions in Pseudomonas syringae. PLoS ONE 8:e59850. doi: 10.1371/journal.pone.0059850
Jian, H., and Wang, F. (2015). Microarray analysis of lexA gene deletion mutant of deep-sea bacterium Shewanella piezotolerans WP3 at low-temperature and high-pressure. Genomics Data 4, 130–132. doi: 10.1016/j.gdata.2015.03.013
Jian, H., Xiao, X., and Wang, F. (2013). Role of filamentous phage SW1 in regulating the lateral flagella of Shewanella piezotolerans strain WP3 at low temperatures. Appl. Environ. Microbiol. 79, 7101–7109. doi: 10.1128/AEM.01675-13
Jian, H., Xu, J., Xiao, X., and Wang, F. (2012). Dynamic modulation of DNA replication and gene transcription in deep-sea filamentous phage SW1 in response to changes of host growth and temperature. PLoS ONE 7:e41578. doi: 10.1371/journal.pone.0041578.g001
Jochmann, N., Kurze, A.-K., Czaja, L. F., Brinkrolf, K., Brune, I., Hüser, A. T., et al. (2009). Genetic makeup of the Corynebacterium glutamicum LexA regulon deduced from comparative transcriptomics and in vitro DNA band shift assays. Microbiology 155, 1459–1477. doi: 10.1099/mic.0.025841-0
Kelley, W. L. (2006). Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol. Microbiol. 62, 1228–1238. doi: 10.1111/j.1365-2958.2006.05444.x
Lauro, F. M., and Bartlett, D. H. (2008). Prokaryotic lifestyles in deep sea habitats. Extremophiles 12, 15–25. doi: 10.1007/s00792-006-0059-5
Mazón, G., Erill, I., Campoy, S., Cortés, P., Forano, E., and Barbé, J. (2004). Reconstruction of the evolutionary history of the LexA-binding sequence. Microbiology 150, 3783–3795. doi: 10.1099/mic.0.27315-0
Medina-Ruiz, L., Campoy, S., Latasa, C., Cardenas, P., Alonso, J. C., and Barbé, J. (2010). Overexpression of the recA gene decreases oral but not intraperitoneal fitness of Salmonella enterica. Infect. Immun. 78, 3217–3225. doi: 10.1128/IAI.01321-09
Merino, S., Shaw, J. G., and Tomás, J. M. (2006). Bacterial lateral flagella: an inducible flagella system. FEMS Microbiol. Lett. 263, 127–135. doi: 10.1111/j.1574-6968.2006.00403.x
Michel, B. (2005). After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 3:e255. doi: 10.1371/journal.pbio.0030255
Novichkov, P. S., Laikova, O. N., Novichkova, E. S., Gelfand, M. S., Arkin, A. P., Dubchak, I., et al. (2010a). RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38, D111–D118. doi: 10.1093/nar/gkp894
Novichkov, P. S., Rodionov, D. A., Stavrovskaya, E. D., Novichkova, E. S., Kazakov, A. E., Gelfand, M. S., et al. (2010b). RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 38, W299–W307. doi: 10.1093/nar/gkq531
Orcutt, B. N., Sylvan, J. B., Knab, N. J., and Edwards, K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422. doi: 10.1128/MMBR.00039-10
Radman, M. (1975). SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 5A, 355–367. doi: 10.1007/978-1-4684-2895-7_48
Riera, J., and Barbe, J. (1995). Cloning, sequence and regulation of expression of the lexA gene of Aeromonas hydrophila. Gene 154, 71–75. doi: 10.1016/0378-1119(94)00836-H
Rocha, R. P. D., Paquola, A. C. D. M., Marques, M. D. V., Menck, C. F. M., and Galhardo, R. S. (2008). Characterization of the SOS regulon of Caulobacter crescentus. J. Bacteriol. 190, 1209–1218. doi: 10.1128/JB.01419-07
Soo, P.-C., Horng, Y.-T., Wei, J.-R., Shu, J.-C., Lu, C.-C., and Lai, H.-C. (2008). Regulation of swarming motility and flhDCSm expression by RssAB signaling in Serratia marcescens. J. Bacteriol. 190, 2496–2504. doi: 10.1128/JB.01670-07
Stewart, B. J., Enos-Berlage, J. L., and Mccarter, L. L. (1997). The lonS gene regulates swarmer cell differentiation of Vibrio parahaemolyticus. J. Bacteriol. 179, 107–114.
Tambalo, D. D., Yost, C. K., and Hynes, M. F. (2010). Characterization of swarming motility in Rhizobium leguminosarum bv. viciae. FEMS Microbiol. Lett. 307, 165–174. doi: 10.1111/j.1574-6968.2010.01982.x
Tapias, A., Campoy, S., and Barbé, J. (2000). Analysis of the expression of the Rhodobacter sphaeroides lexA gene. Mol. Gen. Genet. 263, 957–965. doi: 10.1007/PL00008696
Tusher, V. G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121. doi: 10.1073/pnas.091062498
Ulitzur, S. (1975). Effect to temperature, salts, pH and other factors on the development of peritrichous flagella in Vibrio alginolyticus. Arch. Microbiol. 104, 285–288. doi: 10.1007/BF00447338
van der Veen, S., Van Schalkwijk, S., Molenaar, D., De Vos, W. M., Abee, T., and Wells-Bennik, M. H. (2010). The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology 156, 374–384. doi: 10.1099/mic.0.035196-0
Walker, G. C. (1984). Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev. 48, 60–93.
Walter, B. M., Rupnik, M., Hodnik, V., Anderluh, G., Dupuy, B., Paulič, N., et al. (2014). The LexA regulated genes of the Clostridium difficile. BMC Microbiol. 14:88. doi: 10.1186/1471-2180-14-88
Wang, F., Wang, J., Jian, H., Zhang, B., Li, S., Wang, F., et al. (2008). Environmental adaptation: genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS ONE 3:e1937. doi: 10.1371/journal.pone.0001937.g001
Wang, F., Wang, P., Chen, M., and Xiao, X. (2004). Isolation of extremophiles with the detection and retrieval of Shewanella strains in deep-sea sediments from the west Pacific. Extremophiles 8, 165–168. doi: 10.1007/s00792-003-0365-0
Wang, F., Xiao, X., Ou, H., Gai, Y., and Wang, F. (2009). Role and regulation of fatty acid biosynthesis in the response of Shewanella piezotolerans WP3 to different temperatures and pressures. J. Bacteriol. 191, 2574–2584. doi: 10.1128/JB.00498-08
Wu, H., Kerr, M., Cui, X., and Churchill, G. (2003). “MAANOVA: a software package for the analysis of spotted cDNA microarray experiments,” The Analysis of Gene Expression Data, eds G. Parmigiani, E. S. Garrett, R. A. Irizarry, and S. L. Zeger (New York, NY: Springer), 313–341. doi: 10.1007/0-387-21679-0_14
Keywords: SOS response LexA microarray temperature deep-sea
Citation: Jian H, Xiong L, He Y and Xiao X (2015) The regulatory function of LexA is temperature-dependent in the deep-sea bacterium Shewanella piezotolerans WP3. Front. Microbiol. 6:627. doi: 10.3389/fmicb.2015.00627
Received: 21 April 2015; Accepted: 08 June 2015;
Published: 18 June 2015
Edited by:
Philippe M. Oger, UMR CNRS 5276 Ecole Normale Supérieure de Lyon, FranceReviewed by:
Doug Bartlett, Scripps Institution of Oceanography, USACopyright © 2015 Jian, Xiong, He and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Xiao, State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, No. 800 Dongchuan Road, Shanghai 200240, China,xoxiang@sjtu.edu.cn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.