- 1Institute of Plant and Microbial Biology, Academia Sinica, Taipei, Taiwan
- 2Department of Life Sciences, Graduate School of Arts and Sciences, The University of Tokyo, Tokyo, Japan
- 3PRESTO, Japan Science and Technology Agency, Saitama, Japan
- 4CREST, Japan Science and Technology Agency, Saitama, Japan
Phosphatidylglycerol (PG) is an indispensable phospholipid class with photosynthetic function in plants and cyanobacteria. However, its biosynthesis in eukaryotic green microalgae is poorly studied. Here, we report the isolation and characterization of two homologs (CrPGP1 and CrPGP2) of phosphatidylglycerophosphate synthase (PGPS), the rate-limiting enzyme in PG biosynthesis, in Chlamydomonas reinhardtii. Heterologous complementation of Synechocystis sp. PCC 6803 pgsA mutant by CrPGP1 and CrPGP2 rescued the PG-dependent growth phenotype, but the PG level and its fatty acid composition were not fully rescued in the complemented strains. As well, oxygen evolution activity was not fully recovered, although electron transport activity of photosystem II was restored to the wild-type level. Gene expression study of CrPGP1 and CrPGP2 in nutrient-starved C. reinhardtii showed differential response to phosphorus and nitrogen deficiency. Taken together, these results highlight the distinct and overlapping function of PGPS in cyanobacteria and eukaryotic algae.
Introduction
Photosynthetic membranes are highly specialized biological membranes that contain distinct yet conserved classes of polar glycerolipids MGDG, DGDG, SQDG, and PG among cyanobacteria, eukaryotic microalgae, and land plants (Omata and Murata, 1983; Dorne et al., 1990; Sakurai et al., 2006). Gene knockout affecting biosynthesis of these lipids causes severe photosynthetic defects in a cyanobacterium, Synechocystis sp. PCC 6803 (Mendiola-Morgenthaler et al., 1985; Hagio et al., 2000; Sato et al., 2000; Aoki et al., 2004; Awai et al., 2007; Sakurai et al., 2007b), the eukaryotic green microalga Chlamydomonas reinhardtii (Dubertret et al., 1994; Sato et al., 1995) and the seed plant Arabidopsis thaliana (Dörmann et al., 1995; Hagio et al., 2002; Babiychuk et al., 2003; Kelly et al., 2003; Yu and Benning, 2003; Kobayashi et al., 2007, 2015). Biosynthesis of these lipids may be critical for photosynthesis.
Except for variation in fatty acid composition, the structural features of these four lipid classes are highly similar; however, the biosynthetic pathways may be diverse among cyanobacteria, eukaryotic algae and higher plants. For example, the most abundant lipid class, MGDG, is synthesized in Synechocystis sp. PCC 6803 by two steps (Sato and Murata, 1982): first DAG is glucosylated with UDP-glucose to form MGlcDG by MGlcDG synthase (Awai et al., 2006), which is then isomerized to MGDG by an epimerase (Awai et al., 2014). However, in A. thaliana and other seed plants, MGDG is synthesized by one-step galactosylation with UDP-galactose by MGDG synthases (Shimojima et al., 1997). In contrast, DGDG is produced by the further galactosylation of MGDG by DGDG synthases in both Synechocystis sp. PCC 6803 (Awai et al., 2007) and A. thaliana (Kelly and Dormann, 2002; Kelly et al., 2003).
Phosphatidylglycerol is the only major phospholipid class present in the photosynthetic membrane. It has a distinct contribution to photosynthesis. Gene knockout study revealed the crucial role of PG biosynthesis: disruption of pgsA, encoding PGPS in Synechocystis sp. PCC 6803, affects cell growth and photosynthetic activity unless PG is supplemented exogenously (Hagio et al., 2000). Arabidopsis possesses two PGPS, PGP1 and PGP2; knocking out PGP1 severely impairs chloroplast biogenesis but not mitochondrial function (Hagio et al., 2002; Babiychuk et al., 2003), and double knockout of PGP1 and PGP2 further reduces PG levels to a trace amount, and causing an embryonic-lethal phenotype (Tanoue et al., 2014).
Much less is known about the biosynthesis of photosynthetic membrane lipids in C. reinhardtii. However, a distinct feature of chloroplast lipid metabolism has been shown: an involvement of chloroplastic galactoglycerolipid lipase in triacylglycerol production in nitrogen-starved C. reinhardtii (Li et al., 2012). Because the possible contribution of chloroplastic glycerolipids in triacylglycerol production is uniquely observed in C. reinhardtii, dissecting the similarity and distinctiveness of chloroplastic lipid biosynthesis in C. reinhardtii with reference to Synechocystis sp. PCC 6803 and A. thaliana is important.
In this study, we isolated two PGPS genes in C. reinhardtii, CrPGP1 and CrPGP2, and assessed the molecular function by transforming them into a pgsA mutant of Synechocystis sp. PCC 6803. Moreover, gene expression profiles were examined in C. reinhardtii under phosphorus or nitrogen-starved conditions. The result showed distinct and overlapping functions of PGPS between cyanobacteria and eukaryotic algae.
Materials and Methods
Strains and Growth Conditions
The wild-type and pgsA cells of Synechocystis sp. PCC 6803 were grown photoautotrophically at 30°C in BG-11 medium supplemented with 20 μM PG as described previously (Sakurai et al., 2003). Growth of cultures was monitored by determining optical density at 730 nm (OD730). Light was provided by fluorescent lamps with approximately 50–60 μmol photons m-2 s-1. C. reinhardtii strain CC-4351 was obtained from the Chlamydomonas Resource Center and transformed with empty pChlamiRNA2 plasmid (Molnar et al., 2009). Cells were photoheterotrophically grown in TAP medium (Gorman and Levine, 1965) at 22°C. Nutrient deficiency was induced by collecting the cells by centrifugation (5 min at 3000 × g), washed twice with the respective media and subsequently resuspended in TAP medium, TAP medium without nitrogen (TAP-N), or phosphorus (TAP-P) by omitting NH4Cl, or replacing potassium phosphate with 1.5 mM KCl, respectively (Quisel et al., 1996).
Molecular Cloning
CrPGP1 (Cre03.g162601)
A 937-bp fragment was amplified using cDNA synthesized from the total RNA of C. reinhardtii strain CC-503 (cw92 mt+) as the template with the primers CH223 and CH224 and cloned into pENTR/D_TOPO to construct pCH067. Then, the cloned fragment was amplified with the primers CH772 and CH773 and inserted into NdeI and HpaI sites of pTCP2031V to construct pCH167.
CrPGP2 (Cre02.g095106)
A 790-bp fragment was amplified using cDNA synthesized from the total RNA of C. reinhardtii strain CC-503 (cw92 mt+) as the template with the primers CH225 and CH226 and cloned into pENTR/D_TOPO to construct pCH068. Then, the cloned fragment was amplified with the primers CH774 and CH775 and inserted into NdeI and HpaI sites of the pTCP2031V vector which was designed to incorporate a gene of interest at a neutral site (slr2031) with the psbA2 (slr1311) promoter and a chloramphenicol-resistance cassette (Satoh et al., 2001). The resulting plasmid pCH160 was used to transform the pgsA mutant of Synechocystis sp. PCC 6803 by homologous recombination. The strains, plasmids, and oligonucleotide primers used in this study are described in Supplementary Tables 1–3, respectively.
Complementation Assay of the Synechocystis sp. PCC 6803 pgsA Mutant by CrPGP1 and CrPGP2
For culture on BG-11 agar plates, 5 μl of liquid culture was used with serial 10-fold dilution for spotting from left to right starting at OD730 0.05 onto the plate with or without 20 μM PG. Plates were incubated photoautotrophically under 50∼60 μmol photons m-2 s-1 for 5 days at 30°C to observe the growth phenotype. For liquid culture, cells were photoautotrophically grown in BG-11 media. Initial growth was started at OD730 0.1 by the stirring culture at 180 rpm, 50∼60 μmol photons m-2 s-1 at 30°C.
Genotype Analysis
Genomic DNA was isolated from cells of the wild type, pgsA CrPGP1 and pgsA CrPGP2 of Synechocystis sp. PCC 6803. The primers CH784 and CH785 were used for PCR analysis of genetic background; a 1,141-bp fragment could be amplified from wild-type genomic DNA, whereas a 2,341-bp fragment could be amplified from genomic DNA of pgsA CrPGP1 or pgsA CrPGP2. PCR analysis of the insertion at slr2031 involved the primers for CH982 and CH1000, for an expected 575-bp fragment from the wild type, 2,938-bp fragment from pgsA CrPGP1 and 2,791-bp fragment from pgsA CrPGP2.
Lipid Analysis
Lipids were extracted from intact cells as described in (Bligh and Dyer, 1959) and analyzed previously (Nakamura et al., 2003).
RNA Extraction and qRT-PCR Analysis
Total RNA from Synechocystis sp. PCC 6803 cells grown in BG-11 media by stirring culture at 180 rpm, 50∼60 μmol photons m-2 s-1 at 30°C was extracted using TRI reagent (Ambion) including DNase treatment and reverse-transcribed with SuperScript III (Invitrogen, Carlsbad, CA, USA) for cDNA synthesis. Quantitative RT-PCR involved the ABI 7500 Real Time PCR System (Applied Biosystems) with the oligonucleotide primers for pgsA (slr 1522; CH1028 and CH1029), PGP1 (Cre03.g162601; CH919 and CH920), PGP2 (Cre02.g095106; CH953 and CH954) and RNase P subunit B (rnpB; CH947 and CH948). Gene expression was normalized to that of rnpB (Yuzawa et al., 2014). Data were averaged by three technical replicates in the same run and three biological replicates in separate runs.
Total RNA extraction and cDNA synthesis from C. reinhardtii cells grown by stirring culture at 200 rpm, 50∼60 μmol photons m-2 s-1 at 22°C, follow the method as described above. Oligonucleotide primers used are: Chlamydomonas G-protein beta subunit-like polypeptide (CBLP; Cre06.g278222; CH1076 and CH1077), PGP1 (Cre03.g162601; CH919 and CH920), PGP2 (Cre02.g095106; CH953 and CH954), monogalactosyldiacylglycerol synthase1 (MGD1; Cre13.g585301; CH890 and CH891), digalactosyldiacylglycerol synthase1 (DGD1; Cre13.g583600; CH1060 and CH1061), UDP-sulfoquinovose synthase (SQD1; Cre16.g656400; CH1062 and CH1063), sulfoquinovosyldiacylglycerol synthase (SQD2; Cre01.g038550; CH1064 and CH1065), nitrate reductase1 (NIT1; Cre09.g410950; CH1072 and CH1073) and phosphorus starvation response protein 1 (PSR1; Cre12.g495100; CH1066 and CH1067). Gene expression was normalized to that of CBLP (Schloss, 1990; Chang et al., 2005). Data were averaged by three technical replicates in the same run and two biological replicates in separate runs. Annotation of genes for the lipid metabolism is according to (Li-Beisson et al., 2015). The primers used are listed in Supplementary Table 3.
Assay of Photosynthetic Parameters
Photosynthetic oxygen-evolving activity from H2O to CO2 of intact cells was measured with a Clark-type oxygen electrode (Hansatech Instruments, Kings Lynn, UK) as described in (Gombos et al., 1991). Light from an incandescent lamp through a red optical filter was used for all oxygen evolution measurements at the light intensity 900 μmol photons m-2 s-1. The Chl concentration of cells, determined by the method of Porra et al. (1989) with 100% methanol extraction, was adjusted to 5 μg mL-1.
For measurements of 77 K fluorescence emission spectra, intact cells were suspended in BG-11 at 10 μg Chl mL-1. Fluorescence emission spectra of intact cells under 435 or 600 nm excitation were recorded at 77 K with a spectrofluorometer (RF-5300PC; Shimadzu) as described (Sakurai et al., 2007a).
Measurements of relaxation of flash-induced Chl fluorescence yield were performed in intact cells (5 μg Chl mL-1) by profiling the spectra following single flash excitation with or without 10 μM DCMU (Vass et al., 1999). Samples were incubated in darkness for 10 min before DCMU was added at a final concentration of 10 μM. The levels of Fo and Fm were normalized.
For measurements of absorption spectra, cells were grown by stirring culture at 180 rpm, 50∼60 μmol photons m-2 s-1 at 30°C. Absorption spectra of pigments were determined directly in intact cell suspension (OD730≈0.1) by spectrophotometry (Beckman Coulter DU 800 Spectrophotometer). Spectra were normalized at 625 nm, the maximum absorption of phycobiliproteins, as described (Gombos et al., 2002).
Results
Isolation of Genes Encoding Putative PGPS from C. reinhardtii
To isolate genes encoding functional PGPS for the biosynthesis of PG in C. reinhardtii, we performed a homology search with the amino acid sequence of AtPGP1 (At2g39290), which plays a major role in biosynthesis of PG and is essential for thylakoid membrane development in A. thaliana (Hagio et al., 2002; Xu et al., 2002; Babiychuk et al., 2003). Two genes were found as putative genes of PGPS in C. reinhardtii and named CrPGP1 (Cre03.g162601) and CrPGP2 (Cre02.g095106), encoding 32.1 and 28.7 kDa protein, respectively. We compared the amino acid sequence of CrPGP1 and CrPGP2 with characterized PGPs in A. thaliana (AtPGP1 and AtPGP2) (Hagio et al., 2002; Xu et al., 2002; Babiychuk et al., 2003) and Synechosystis sp. PCC 6803 (PgsA) (Hagio et al., 2000) by creating multiple amino acid sequence alignment (Figure 1A). As can be seen, the amino acid residues conserved in all sequences of proteins with the CDP-OH-P motif (PF01066.9) indicated by asterisks were conserved in CrPGP1 and CrPGP2, suggesting that these proteins are functional PGPs. Interestingly, CrPGP1 has longer N-terminal sequence than CrPGP2, as is found between AtPGP1 and AtPGP2. Next, we compared the identity and similarity of these PGPs (Figure 1B). The value ranges from 31.8 to 81.1% in identity and from 62.6 to 91.0% in similarity. The identity and similarity of PGPs were higher between C. reinhardtii and A. thaliana than CrPGP1 and CrPGP2, and those with Synechocystis PgsA were similar between C. reinhardtii and A. thaliana.
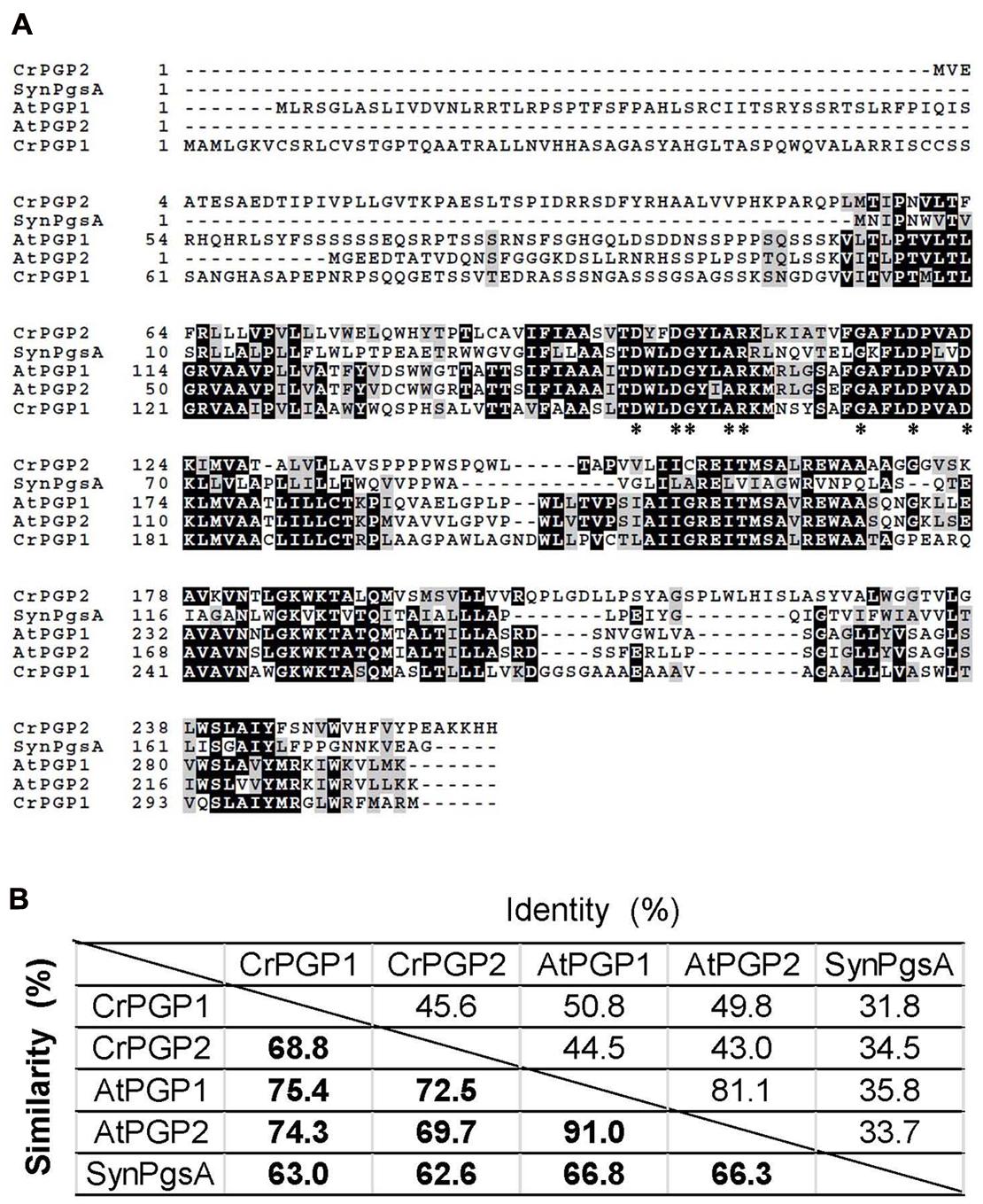
FIGURE 1. Comparison of amino acid sequences of PGPs. (A) Multiple amino acid sequence alignment of two PGPs of Chlamydomonas reinhardtii (CrPGP1 and CrPGP2) with reference to two PGPs of Arabidopsis thaliana (AtPGP1 and AtPGP2) and PgsA of Synechocystis sp. PCC 6803 (SynPgsA). Asterisks indicate the amino acid residues conserved in all sequences of proteins with the CDP-OH-P motif (PF01066.9). (B) The identity and similarity of amino acid sequences among PGPs aligned in (A).
To examine whether these two genes encode functional PGPS, we cloned CrPGP1 and CrPGP2 under the control of the psbA2 promoter and stably transformed them into a neutral genomic site (slr2031) (Satoh et al., 2001) of the Synechocystis sp. PCC 6803 pgsA mutant, which is defective in PGPS activity (Hagio et al., 2000) (Figure 2A). To confirm the successful homologous recombination of CrPGP1 and CrPGP2 into the neutral genomic site in the pgsA mutant, we performed PCR-based genotyping for both pgsA (slr1522) and slr2031 (Figure 2B). Compared to the wild type, both pgsA CrPGP1 and pgsA CrPGP2 mutants maintained the pgsA background and the introduced genes were fully segregated at the slr2031 locus by the homologous recombination. We further investigated whether CrPGP1 and CrPGP2 were expressed in pgsA by comparing mRNA levels of CrPGP1 in pgsA CrPGP1, CrPGP2 in pgsA CrPGP2, and pgsA in wild type. As shown in Figure 2C, both CrPGP1 and CrPGP2 were expressed, whose levels were fivefold and eightfold higher than that of pgsA. Thus, under the psbA2 promoter control, both CrPGP1 and CrPGP2 were expressed in pgsA CrPGP1 and pgsA CrPGP2.
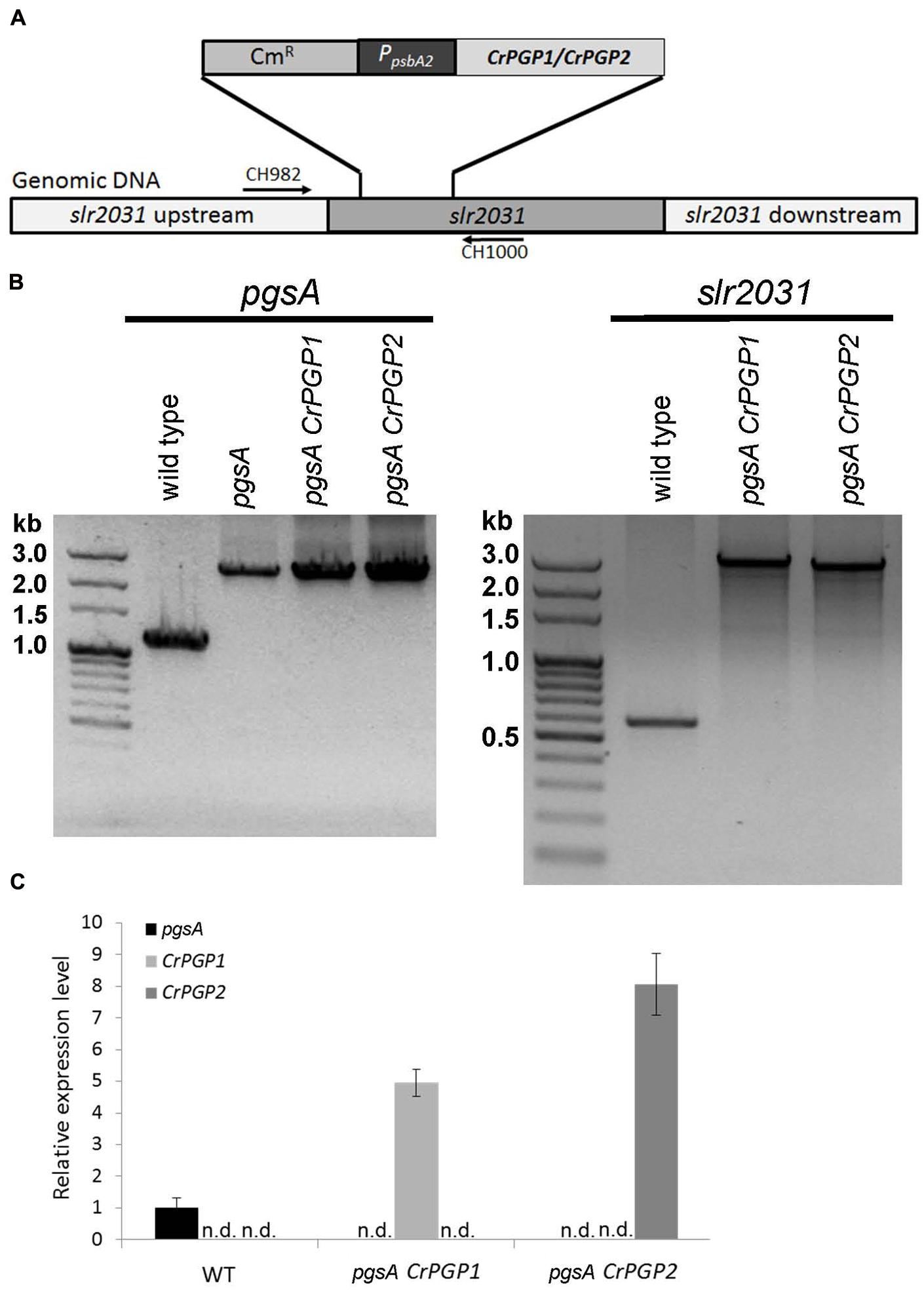
FIGURE 2. Functional complementation of Synechocystis sp. PCC 6803 pgsA mutant with PGP1 or PGP2 of Chlamydomonas reinhardtii. (A) Schematic illustration of homologous recombination of CrPGPs at slr2031 locus. (B) PCR-based genotyping analysis of the transformants for pgsA (left panel) and slr2031 (right panel). (C) Expression level of CrPGP1 and CrPGP2 in the wild type, pgsA CrPGP1 and pgsA CrPGP2 relative to that of pgsA in the wild type. Levels are normalized to that of rnpB. Data are mean ± SD from three biological replicates. n.d., not detected.
Complementation of Synechocystis sp. PCC 6803 pgsA Mutant with CrPGPs
Previous study showed that the pgsA mutant of Synechocystis sp. PCC 6803 requires exogenous supplementation of PG for growth (Hagio et al., 2000), so we compared the growth of the wild type, pgsA, pgsA CrPGP1, and pgsA CrPGP2 on solid BG-11 media with or without 20 μM PG. The wild-type cells grew similarly in the presence or absence of PG, whereas the pgsA mutant showed rescued growth only with PG (Figure 3A). The pgsA CrPGP1 and pgsA CrPGP2 showed rescued growth even in the absence of PG, which suggests that CrPGP1 and CrPGP2 are functional PGPS to rescue the growth defect of pgsA. To further investigate the growth profiles of pgsA CrPGP1 or pgsA CrPGP2, we monitored the growth in liquid BG-11 media without PG. Although the growth rate was significantly restored in the transgenic strains, it was inferior to that of the wild type, with the pgsA CrPGP1 showed slightly better growth than pgsA CrPGP2 (Figure 3B). Therefore, CrPGP1 and CrPGP2 could functionally complement the lethal phenotype of the pgsA mutant, although their growth remain slightly retarded as compared with the wild type.
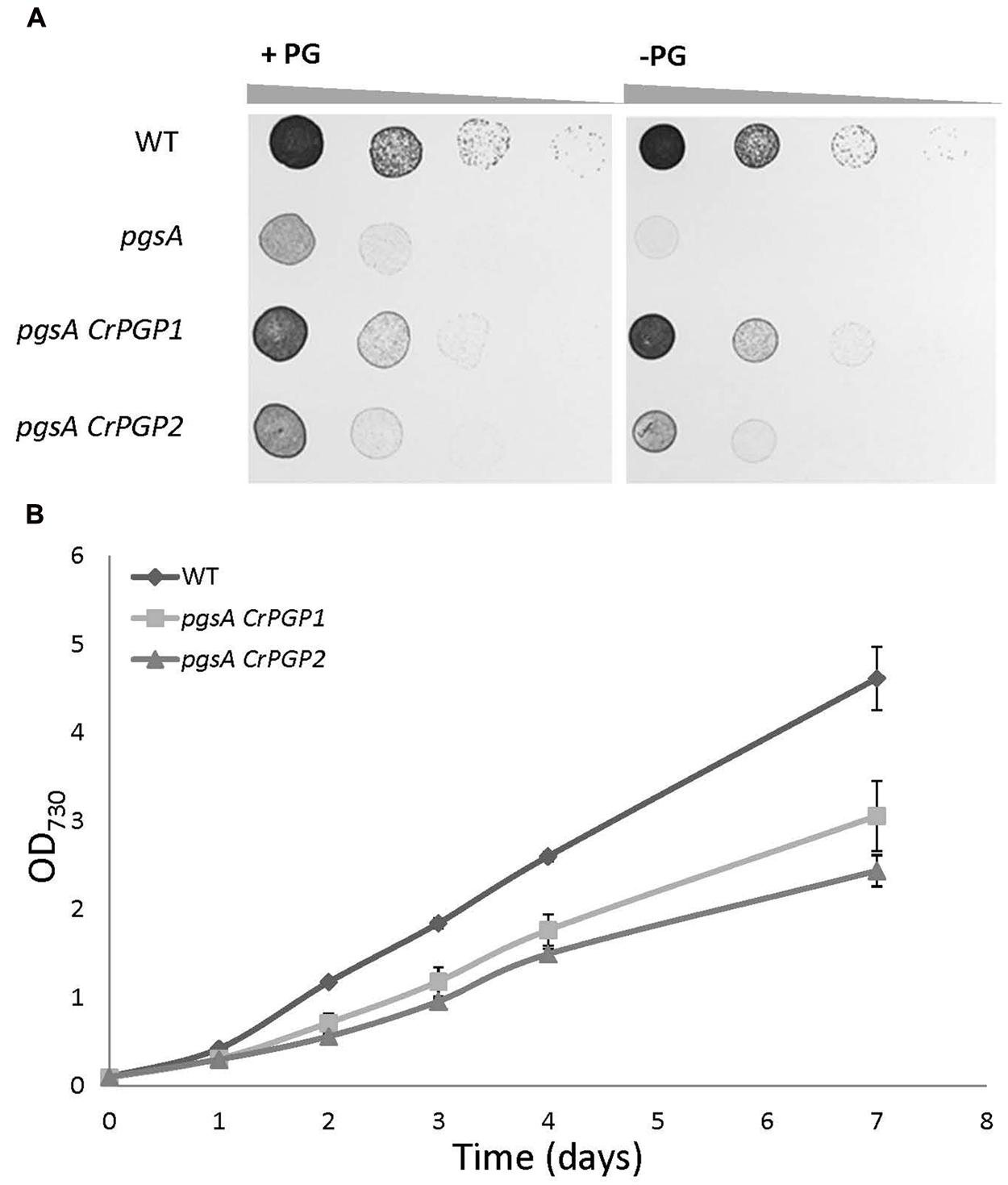
FIGURE 3. Growth of pgsA CrPGP1 or pgsA CrPGP2. (A) Growth of the wild type, pgsA, pgsA CrPGP1, or pgsA CrPGP2 on solid BG-11 media with or without PG supplementation. Spotting involved serial 10-fold dilution from left to right starting at OD730 0.05, with 5 μl each spotted onto a BG-11 agar plate with or without 20 μM PG and incubation under 50∼60 μmol photons m-2 s-1 for 5 days at 30°C. Images are representative of three biological replicates. (B) Growth profile of the wild type, pgsA CrPGP1 or pgsA CrPGP2 in liquid BG-11 media. Growth was initiated at OD730 0.1 by stirring culture at 180 rpm, 50∼60 μmol photons m-2 s-1 at 30°C. Data are mean ± SD from three biological replicates.
Lipid Composition of pgsA CrPGP1 and pgsA CrPGP2
To examine whether the levels of PG and other polar glycerolipids are restored in pgsA CrPGP1 or pgsA CrPGP2, we analyzed the composition of membrane lipid classes and their fatty acid composition. PG composition in the wild type was 11 mol% but was 8 and 5 mol% in pgsA CrPGP1 and pgsA CrPGP2, respectively (Figure 4A). Moreover, composition of DGDG was slightly higher in the transgenic strains than the wild type. Next, we analyzed the fatty acid composition of these lipid classes (Figures 4B–E). The fatty acid composition of MGDG, DGDG, and SQDG was fairly similar among the three strains, but that of PG was markedly decreased in 18:3 and 18:2 composition and increased in 16:0 in pgsA CrPGP1 and pgsA CrPGP2 (Figure 4E). Hence, CrPGP1 and CrPGP2 produced a significant level of PG in pgsA, but the level was slightly lower and fatty acid composition differed from that of the wild type.
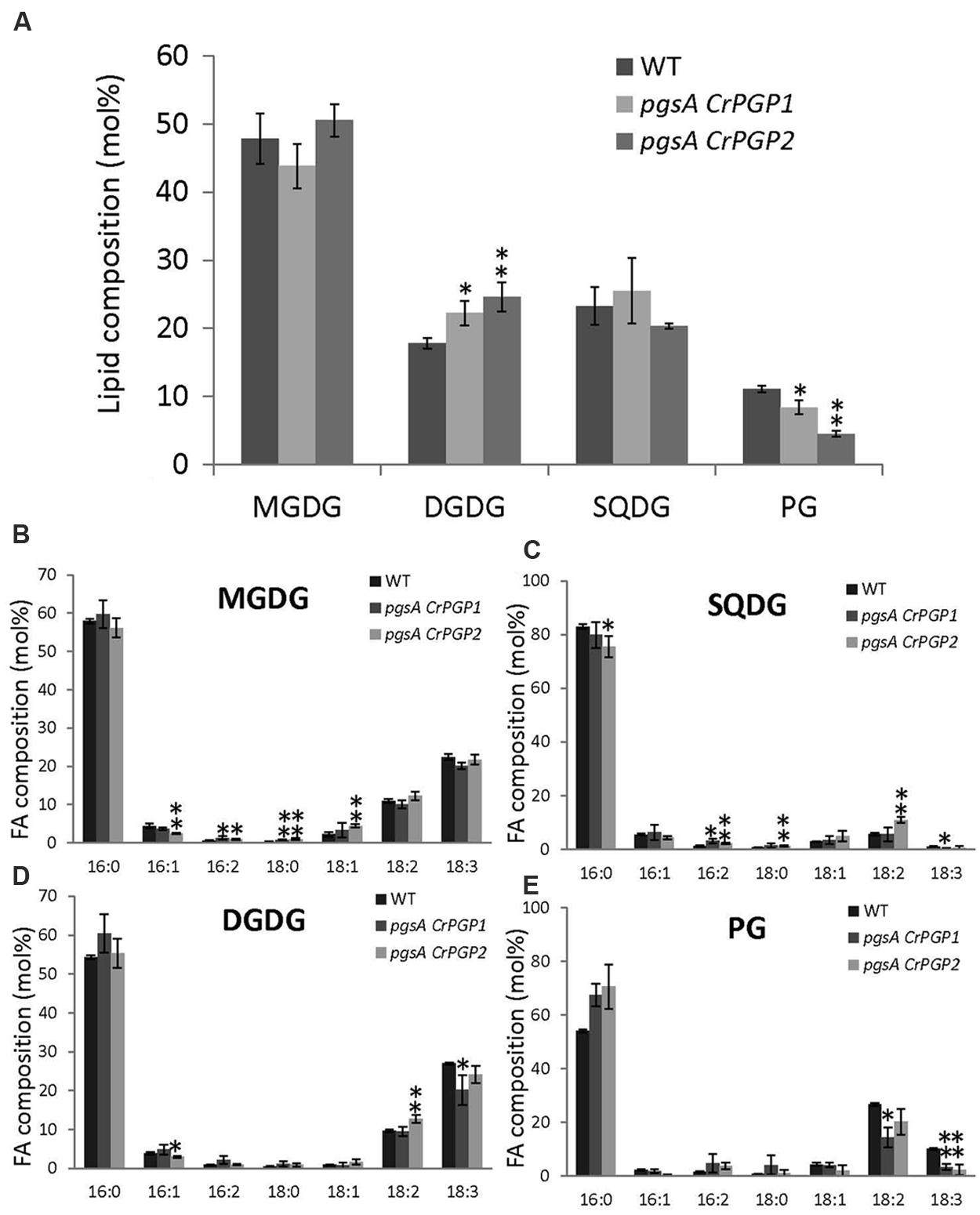
FIGURE 4. Polar glycerolipid composition (A) and fatty acid composition of (B) MGDG; (C) SQDG; (D) DGDG; and (E) PG of the wild type, pgsA CrPGP1 and pgsA CrPGP2. Data are mean ± SD from three biological replicates. Asterisks show significance (∗p < 0.05, ∗∗p < 0.01) from the wild type. Unsaturated fatty acids detected in all lipid classes contain only cis double bonds.
Pigment Content and Oxygen Evolution Activity of pgsA CrPGP1 and pgsA CrPGP2
Previous studies have shown that PG deficiency reduces Chl content in Synechocystis sp. PCC 6803 and other cyanobacteria (Gombos et al., 2002; Wu et al., 2006; Bogos et al., 2010). To assess whether the expression of CrPGPs can restore the Chl content in pgsA, we examined absorption spectra of pigments in the whole cells of the wild type, pgsA CrPGP1 and pgsA CrPGP2 (Figure 5). The spectra was normalized at 625 nm, the maximum absorption of phycobiliproteins. Absorptions at ∼683 and ∼441 nm by Chl were lower for both pgsA CrPGP1 and pgsA CrPGP2 than the wild type. Indeed, the cellular Chl content in both complemented strains was decreased to 63% and 68% of the wild-type level, respectively (Table 1). To elucidate whether CrPGPs can restore the photosynthetic activity in pgsA, we compared the net oxygen evolution rate in complemented strains and the wild type. On a Chl content basis, oxygen evolution activities in pgsA CrPGP1 and pgsA CrPGP2 cells were 57 and 61% less than wild-type activities, respectively, which corresponded to 73 and 73.5% less than the wild type on a cell density basis. Therefore, the introduction of CrPGP1 or CrPGP2 can partially but not fully complement the loss of pgsA in Synechocystis sp. PCC 6803 in terms of photosynthetic activity as well as Chl accumulation.
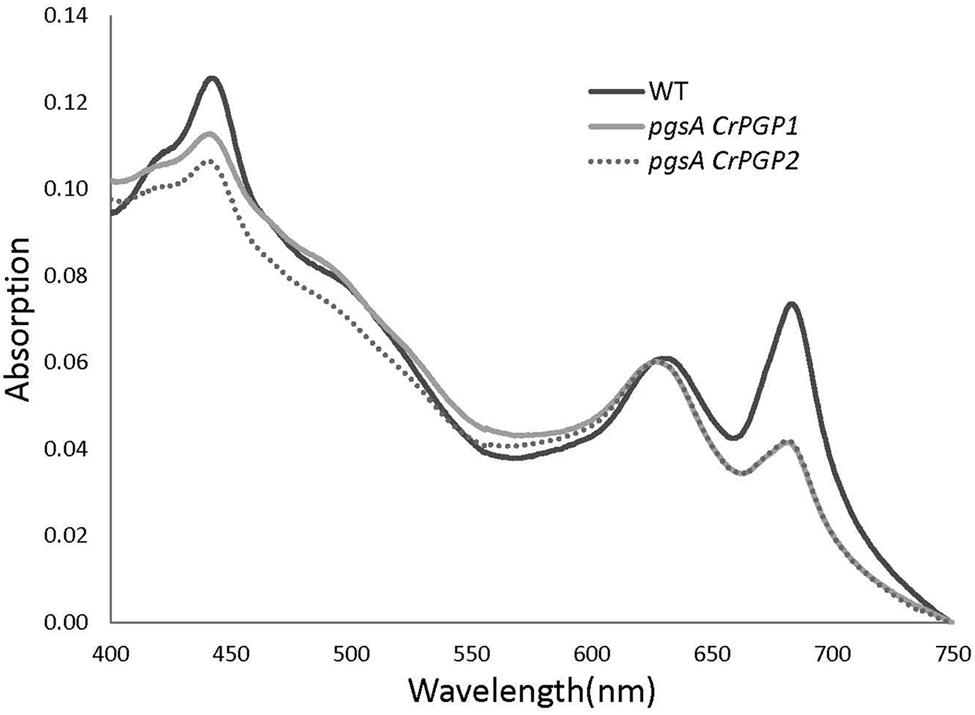
FIGURE 5. Absorption spectra of wild type, pgsA CrPGP1 and pgsA CrPGP2 in BG-11 medium. The spectra were measured with whole cells (OD730 ≈ 0.1) and normalized at 625 nm, the maximum absorption of phycobiliproteins.
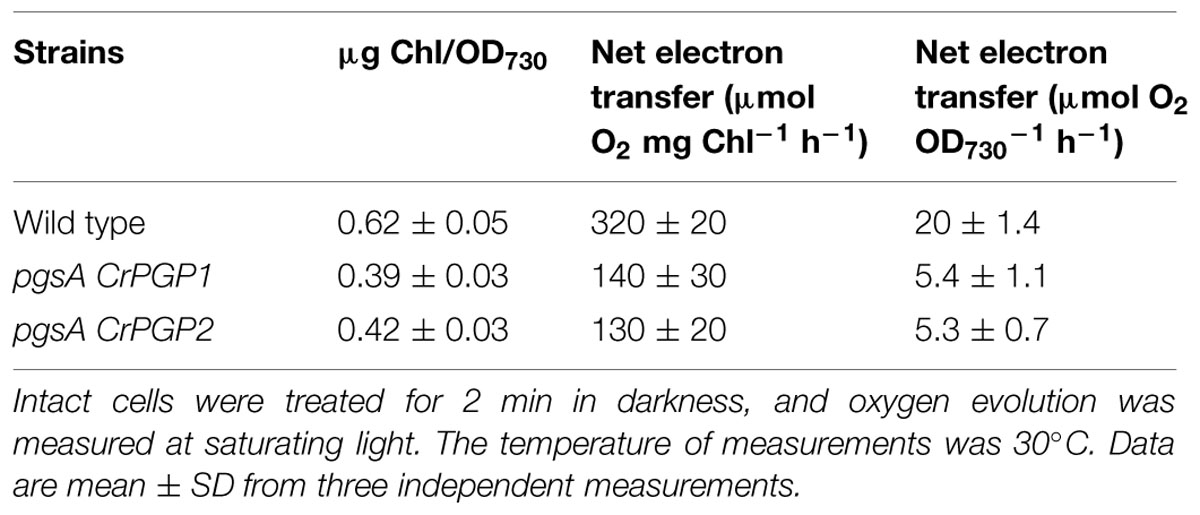
TABLE 1. Photosynthetic oxygen-evolving activities and Chl contents in intact cells of wild type, pgsA CrPGP1, and pgsA CrPGP2 of Synechocystis sp. PCC 6803.
Electron Transfer Kinetics in the Acceptor and Donor Side of PSII in pgsA CrPGP1 and pgsA CrPGP2
The low oxygen evolution activities in the pgsA CrPGP1 or pgsA CrPGP2 imply impaired photosynthetic electron transport in these strains. To characterize the functionality of electron transfer within PSII in the complemented strains, we evaluated the reoxidation kinetics of QA, the primary electron acceptor plastoquinone (PQ) of PSII, by analyzing the decay of Chl fluorescence after a single saturating flush. The kinetics of accepter-side electron transfer from QA- to the PQ pool was evaluated without DCMU (Figure 6A), and the kinetics from QA- to the donor-side components were evaluated with DCMU, which inhibits the electron transfer from QA to QB and causes charge recombination between QA- and the oxidizing-side components (Figure 6B). In both pgsA CrPGP1 and pgsA CrPGP2 cells, the fluorescence decay kinetics were almost identical to those in the wild type without or with DCMU. Thus, both complemented strains may assemble the functional PSII complex as in the wild type.
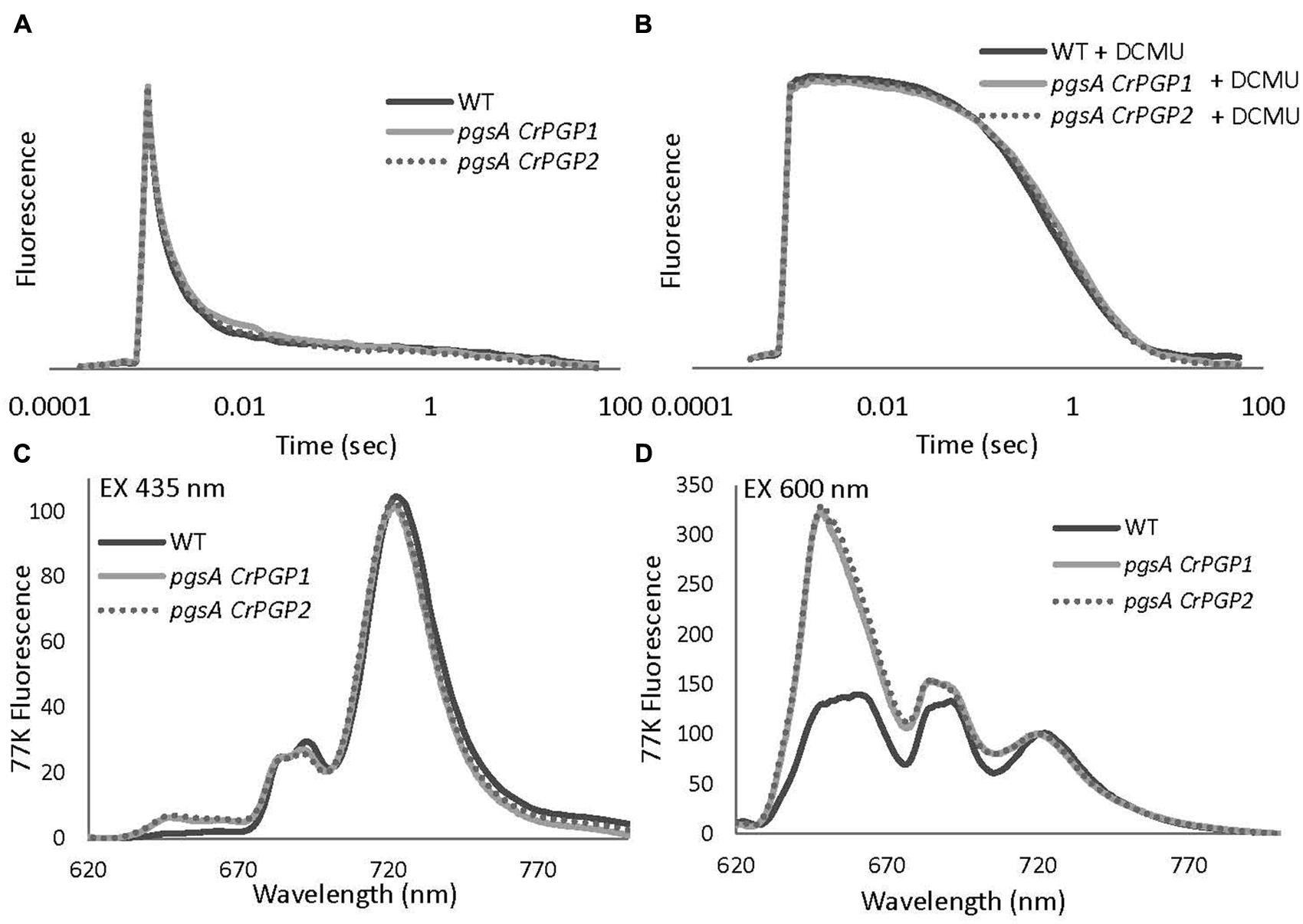
FIGURE 6. Relaxation of flash-induced Chl fluorescence yield (A,B) and 77 K fluorescence emission spectra (C,D) of wild type, pgsA CrPGP1 and pgsA CrPGP2 mutant cells in BG-11 medium. The spectra resulted from a single flash excitation without (A) or with (B) 10 μM DCMU. Data are the mean from three independent experiments. In each case, Chl concentrations were adjusted to 5 μg mL-1. Samples were incubated in darkness for 10 min before DCMU was added to the final concentration of 10 μM. The levels of Fo and Fm were normalized. (C,D) 77 K fluorescence emission spectra from the wild type, pgsA CrPGP1 and pgsA CrPGP2 mutant cells were measured under 435 nm (C) or 600 nm (D) excitation. Data are the mean from three independent experiments. Spectra were normalized at 720 nm to 100. In each case, Chl concentrations were adjusted to 10 μg mL-1.
Chl Fluorescence Emission Spectra at 77 K in pgsA CrPGP1 and pgsA CrPGP2
The core-antenna complexes of PSI and PSII are largely disordered in the PG-deficient pgsA mutant. To evaluate whether CrPGPs restore PS-antenna complexes in pgsA, we examined emission spectra of Chl fluorescence at 77 K. In both wild-type and mutant cells, the preferential excitation of Chl at 435 nm resulted in four emission peaks, at ∼647, ∼681, ∼691, and ∼720 nm (Figure 6C). The emission peak at ∼647 nm can be attributed to phycocyanin (Rakhimberdieva et al., 2007); peaks at ∼681 nm and ∼691 nm primarily originate from CP43 and CP47, which are functionally coupled to the PSII reaction center, respectively; and the peak at ∼720 nm originates from PSI. For pgsA CrPGP1 and pgsA CrPGP2 cells, we found only a slight decrease in emission peak at ∼691 nm as compared with the wild type, which suggests a small change in the PSII complex in these complemented strains. By contrast, the emission from phycocyanin at 647 nm was slightly increased in the complemented strains. To examine the energy coupling between phycobilisomes and PS cores in the complemented lines, phycobilins were preferentially excited at 600 nm at 77 K (Figure 6D). Both pgsA CrPGP1 and pgsA CrPGP2 cells showed prominent emissions at ∼647 and ∼681 nm as compared with wild-type cells. The emission at ∼647 nm originates from phycocyanin, whereas that at ∼681 nm is contributed by both PSII Chl and terminal phycobilin emitters. The strong enhancement of the emission peaks at ∼647 and ∼681 nm in the complemented lines suggests that the energy transfer from phycobilisomes to the PSII reaction center is uncoupled in these cells.
Gene Expression Profiles of CrPGP1 and CrPGP2 under Nutrient-Limited C. reinhardtii
To obtain physiological insight into the role of CrPGP1 and CrPGP2, we analyzed gene expression profiles of CrPGP1 and CrPGP2 together with related genes for glycerolipid metabolism during phosphorus or nitrogen starvation. During phosphorus starvation, significant portion of phospholipids is replaced by non-phosphorus galactolipid DGDG, termed membrane lipid remodeling (Nakamura, 2013). Moreover, SQDG is increased and compensates for the decreased PG and maintains the total amount of anionic lipid classes (Riekhof et al., 2003). However, gene expression profiles of enzymes related to this metabolic change were not studied previously. Here, we took two time points after switching C. reinhardtii to phosphorus starvation: 4 h for the early response and 5 days for the late response (Figure 7). CrPSR1 is a marker gene for phosphorus starvation response (Wykoff et al., 1999). Both CrPGP1 and CrPGP2 showed a transient decrease at 4 h but recovered at 5 days. In contrast, the expression of CrSQD1 and CrSQD2, which are required for SQDG biosynthesis, was induced at 4 h. Moreover, the gene expression of CrDGD1 was increased but that of CrMGD1 was not, which supports an increase in DGDG but not MGDG upon phosphorus starvation. These results showed that expression profiles of CrPGP1 and CrPGP2, as well as other related genes examined in Figure 7, are altered under phosphorus starvation. Next, we examined nitrogen starvation using CrNIT1 as a marker gene (Hipkin et al., 1980). This condition severely affects photosynthesis but induces accumulation of triacylglycerol (Wang et al., 2009). As shown in Figure 8, CrPGP1 but not CrPGP2 showed decreased gene expression under nitrogen starvation. CrSQD1 and CrSQD2 showed transient changes in gene expression level, and CrMGD1 and CrDGD1 both decreased the gene expression level at different time points. Thus, CrPGP1 and CrPGP2 showed differential gene expression profiles under phosphorus starvation and nitrogen starvation conditions.
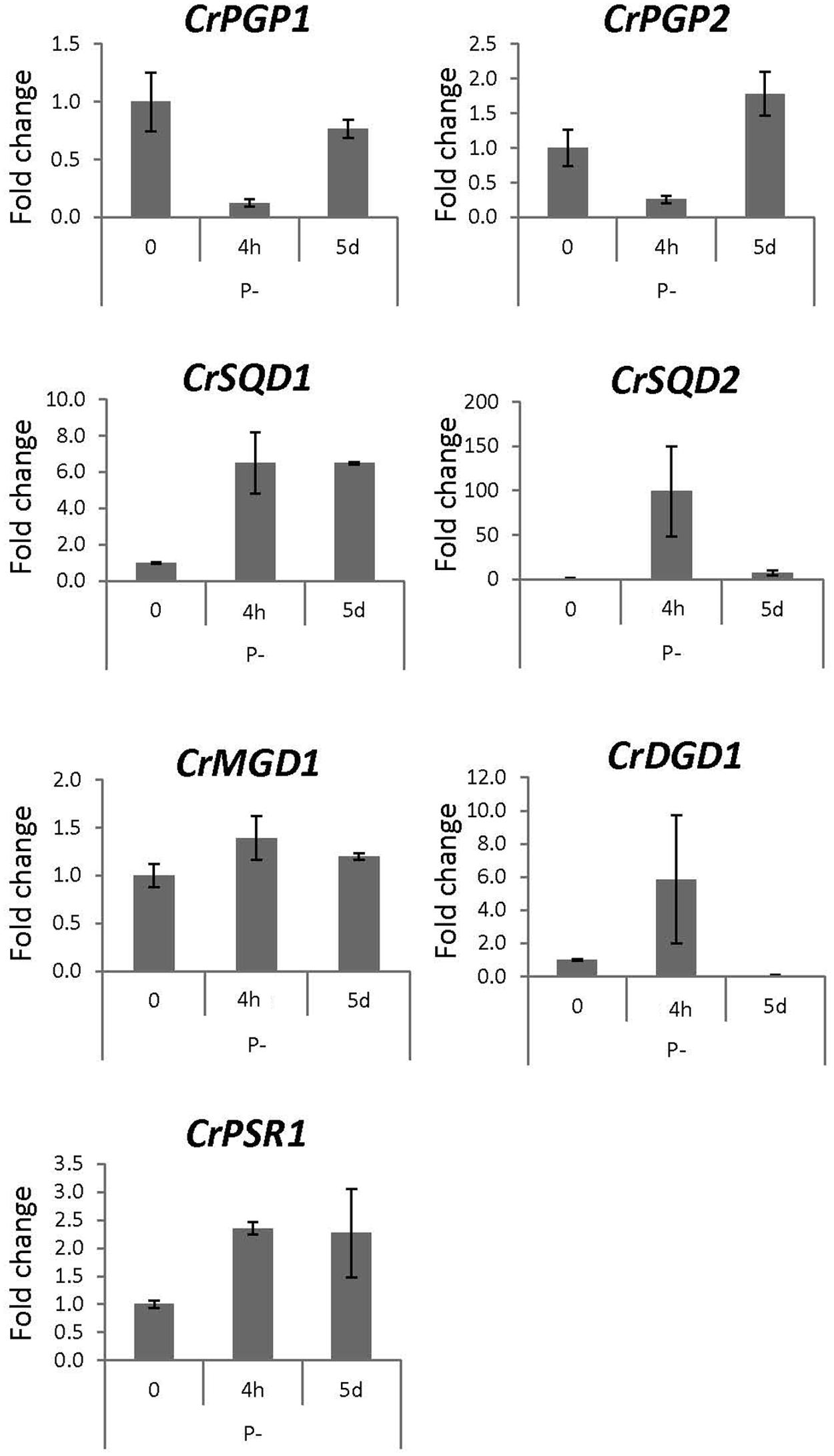
FIGURE 7. Gene expression profile of enzymes for plastidial polar glycerolipid biosynthesis under phosphorus starvation. Wild-type C. reinhardtii strain CC-4351 grown in TAP media was transferred to the media devoid of phosphate and harvested at 4 h and 5 days. Total RNA was extracted to synthesize cDNA for qRT-PCR analysis. Gene expression was normalized to that of CBLP. Data were averaged by three technical replicates in the same run and two biological replicates in separate runs. SQD1, UDP-sulfoquinovose synthase; SQD2, sulfoquinovosyldiacylglycerol synthase; MGD1, MGDG synthase1; DGD1, DGDG synthase1; PSR1, phosphorus starvation response1.
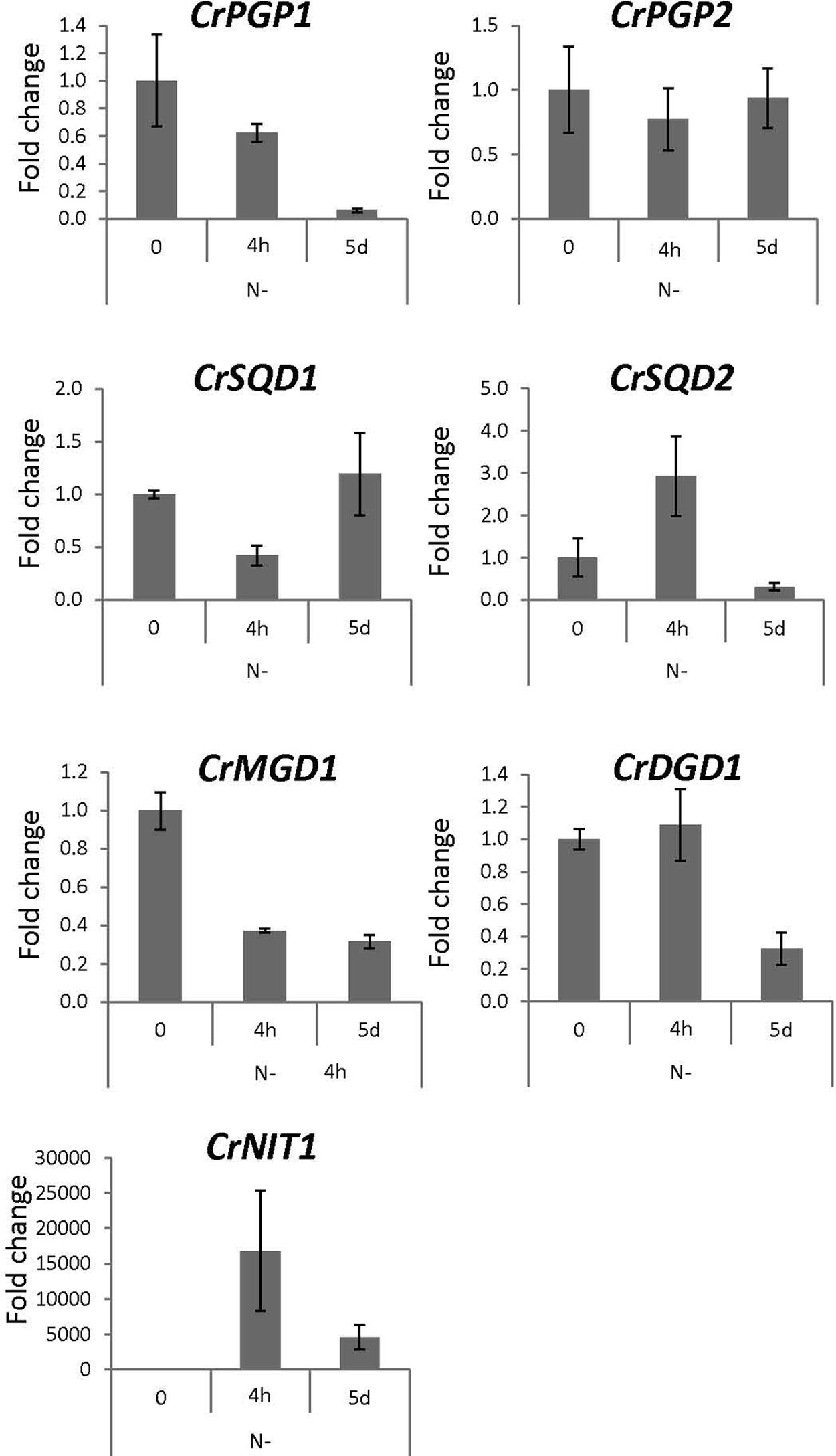
FIGURE 8. Gene expression profile of enzymes for plastidial polar glycerolipid biosynthesis under nitrogen starvation. NIT1, nitrate reductase 1. See legend of Figure 7 for sampling conditions, experimental methods, and abbreviations.
Discussion
Phosphatidylglycerol synthesis is critical to maintain the stability of membrane protein complexes in photosynthesis components and in the respiratory electron transfer chain (Hagio et al., 2000; Ostrander et al., 2001; Domonkos et al., 2004; Hasan et al., 2013). This study isolated a pair of PGPS genes, CrPGP1 and CrPGP2, in a model eukaryotic green microalga C. reinhardtii. The molecular function of these newly isolated PGPs was revealed by transforming them into the pgsA mutant of Synechocystis sp. PCC 6803, a representative cyanobacterium. Moreover, gene expression profiles of CrPGP1 and CrPGP2 were investigated in C. reinhardtii under phosphorus or nitrogen starvation. Strong responses of CrPGPs, particularly CrPGP1, to these conditions imply their involvements in membrane lipid remodeling in response to nutrient starvation. The results of cell growth, lipid analysis, and photosynthetic parameters in pgsA CrPGP1 and pgsA CrPGP2 showed distinct and overlapping function of PGPS between cyanobacteria and eukaryotic algae.
The heterologous complementation of the pgsA mutant phenotype by CrPGP1 or CrPGP2 supports that they encode a functional PGPS of C. reinhardtii (Figures 3A,B). However, the slower growth profile in pgsA CrPGP1 or pgsA CrPGP2 than the wild type under liquid culture suggests that CrPGPs cannot fully rescue the defect of pgsA mutant (Figure 3B). Our lipid analysis agrees with this idea: as compared with the wild type, pgsA CrPGP1 and pgsA CrPGP2 showed reduced PG contents by 73 and 45%, respectively. Although the expression level of CrPGP2 was higher than CrPGP1 (Figure 2C), expression of the CrPGP1 protein complemented the pgsA mutant phenotype more effectively than that of the CrPGP2. The protein sequence similarity with PgsA was slightly higher for CrPGP1 (63.0%) than CrPGP2 (62.6%). Moreover, in silico prediction of possible subcellular localization by ChloroP (Emanuelsson et al., 1999), TargetP (Emanuelsson et al., 2000), or PledAgro (Tardif et al., 2012) suggests that CrPGP1 may be localized at the chloroplasts or mitochondria whereas CrPGP2 may be localized somewhere other than the chloroplasts, the mitochondria or the secretory pathway. Indeed, longer N-terminal sequence was found in CrPGP1 (Figure 1A). Because the different subcellular localization was shown between AtPGP1 and AtPGP2 (Babiychuk et al., 2003; Tanoue et al., 2014), it is possible that CrPGP1 and CrPGP2 also have different subcellular localizations. It is possible that CrPGP1 is functionally more similar to PgsA because of the chloroplast localization. The fatty acid composition of PG showed reduced composition of 18:3 (Figure 4E). Because 18:3 in PG is produced by the fatty acid desaturase that desaturates acyl groups of PG, thus it is possible that PG produced by CrPGP1 and CrPGP2 may not be properly desaturated in pgsA, which highlights the different enzymatic features between PGP of Synechocystis sp. PCC 6803 and C. reinhardtii.
Many PG molecules are found in the PSII complex (Sakurai et al., 2006) and are present near the reaction center (Guskov et al., 2009; Umena et al., 2011). In fact, PG is indispensable for both donor- and acceptor-side activities of PSII (Hagio et al., 2000; Sato et al., 2000; Gombos et al., 2002; Sakurai et al., 2003, 2007a). Our data in Figure 6 reveals that the expression of CrPGPs in pgsA sufficiently restored electron transfer activities of PSII to the wild-type level, although the PG levels and fatty acid composition in the complemented strains differed somewhat from those in the wild type (Figure 4). Thus, CrPGP activities would be enough to supply PG molecules to the PSII complex in the pgsA background. Meanwhile, the oxygen evolution activity in pgsA was not fully recovered by introducing the CrPGPs and remained at low levels (Table 1). Because phycobilisomes, which are not present in green algae including Chlamydomonas, may be energetically uncoupled with the PSII reaction center in the pgsA mutants harboring CrPGPs (Figure 6D), loss of energy transfer from phycobilisomes to the PSII reaction center in part likely reduces net photosynthesis activities in the complemented strains. The PG-deficient pgsA mutant showed energetic uncoupling between phycobilisomes and the PSII reaction center and not fully restored by the supplementation of PG (Sakurai et al., 2007a). Thus, in situ PG synthesis by native PgsA may be important for assembly of phycobilisomes with the PSII complex in Synechocystis sp. PCC 6803.
The expression of CrPGP1 and CrPGP2, as well as the other genes showed differential profiles under two different nutrient starvation conditions (Figures 7 and 8). Most genes involved in glycoglycerolipid biosynthesis (CrSQD1, CrSQD2, and CrDGD1) showed rapid upregulation upon phosphorus starvation, whereas expression of CrPGP1 and CrPGP2 was decreased at 4 h after phosphorus starvation. This profile is in agreement with an increase in DGDG and SQDG levels and a decrease in PG levels under phosphorus starvation (Riekhof et al., 2003). However, the reduced expression of CrPGP1 and CrPGP2 was recovered at 5 days after phosphorus starvation. In addition to the membrane lipid remodeling, C. reinhardtii cells induce triacylglycerol biosynthesis by long-term phosphorus starvation with retaining thylakoid membrane networks (Iwai et al., 2014) and photosynthetic activity (Wykoff et al., 1998). Thus, the recovered expression of CrPGP1 and CrPGP2 may function to keep proper balance of lipid composition of thylakoid membranes and maintain photosynthetic activity even under long-term phosphorus starvation, at which triacylglycerol is produced possibly for an energy storage. Meanwhile, expression of most of the genes in Figure 8 was decreased in response to nitrogen starvation. These changes agree with the actual decrease in MGDG, DGDG, SQDG, and PG, degradation of thylakoid membranes, and loss of photosynthetic activity under nitrogen starvation (Siaut et al., 2011; Iwai et al., 2014; Sakurai et al., 2014). Notably, CrPGP1 but not CrPGP2 showed decreased expression, whose profile was similar to that of CrMGD1 upon nitrogen starvation. This suggests distinct roles between CrPGP1 and CrPGP2, which was also presumed by possible differences in subcellular localization and enzyme property of these isozymes. Differential expression profiles of some of lipid biosynthesis genes were previously observed. For example, the gene expression levels of CrSQD1 showed partial recovery at later time points, CrSQD2 expression showed fluctuation while decreasing, and CrMGD1 expression showed faster decrease than that of CrDGD1 upon nitrogen starvation (Boyle et al., 2012). These data are generally consistent with our results in Figure 8, although some differences were observed probably because of differences in growth conditions and/or time point analyzed. The distinct expression profiles among genes involved in membrane lipid biosynthesis including CrPGP1 and CrPGP2 may allow C. reinhardtii cells to fine-tune lipid metabolism in response to various nutrient conditions.
Conclusion
Our results reveal that both CrPGP1 and CrPGP2 encode functional PGPS, which can contribute to PG production and the formation of photosynthetic complexes in the PG-deficient Synechocystis pgsA mutant. Further analyses are required to elucidate the roles of CrPGP1 and CrPGP2 in lipid metabolism and photosynthesis in C. reinhardtii.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by PRESTO (to YN) and CREST (to HW), Japan Science and Technology Agency, and operation budget by Institute of Plant and Microbial Biology, Academia Sinica to YN.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00842
Abbreviations
Chl, chlorophyll; DAG, sn-1,2-diacylglycerol; DCMU, 3-(3,4-dichlorophenyl)-1,1-dimethylurea; DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; MGlcDG, monoglucosyldiacylglycerol; PG, phosphatidylglycerol; PGPS, phosphatidylglycerophosphate synthase; PS, photosystem; SQDG, sulfoquinovosyldiacylglycerol.
References
Aoki, M., Sato, N., Meguro, A., and Tsuzuki, M. (2004). Differing involvement of sulfoquinovosyl diacylglycerol in photosystem II in two species of unicellular cyanobacteria. Eur. J. Biochem. 271, 685–693. doi: 10.1111/j.1432-1033.2003.03970.x
Awai, K., Kakimoto, T., Awai, C., Kaneko, T., Nakamura, Y., Takamiya, K., et al. (2006). Comparative genomic analysis revealed a gene for monoglucosyldiacylglycerol synthase, an enzyme for photosynthetic membrane lipid synthesis in cyanobacteria. Plant Physiol. 141, 1120–1127. doi: 10.1104/pp.106.082859
Awai, K., Ohta, H., and Sato, N. (2014). Oxygenic photosynthesis without galactolipids. Proc. Natl. Acad. Sci. U.S.A. 111, 13571–13575. doi: 10.1073/pnas.1403708111
Awai, K., Watanabe, H., Benning, C., and Nishida, I. (2007). Digalactosyldiacylglycerol is required for better photosynthetic growth of Synechocystis sp. Plant Cell Physiol. 48, 1517–1523. doi: 10.1093/pcp/pcm134
Babiychuk, E., Muller, F., Eubel, H., Braun, H. P., Frentzen, M., and Kushnir, S. (2003). Arabidopsis phosphatidylglycerophosphate synthase 1 is essential for chloroplast differentiation, but is dispensable for mitochondrial function. Plant J. 33, 899–909. doi: 10.1046/j.1365-313X.2003.01680.x
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Bogos, B., Ughy, B., Domonkos, I., Laczko-Dobos, H., Komenda, J., Abasova, L., et al. (2010). Phosphatidylglycerol depletion affects photosystem II activity in Synechococcus sp. PCC 7942 cells. Photosynth. Res. 103, 19–30. doi: 10.1007/s11120-009-9497-0
Boyle, N. R., Page, M. D., Liu, B., Blaby, I. K., Casero, D., Kropat, J., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287, 15811–15825. doi: 10.1074/jbc.M111.334052
Chang, C. W., Moseley, J. L., Wykoff, D., and Grossman, A. R. (2005). The LPB1 gene is important for acclimation of Chlamydomonas reinhardtii to phosphorus and sulfur deprivation. Plant Physiol. 138, 319–329. doi: 10.1104/pp.105.059550
Domonkos, I., Malec, P., Sallai, A., Kovacs, L., Itoh, K., Shen, G., et al. (2004). Phosphatidylglycerol is essential for oligomerization of photosystem I reaction center. Plant Physiol. 134, 1471–1478. doi: 10.1104/pp.103.037754
Dörmann, P., Hoffmann-Benning, S., Balbo, I., and Benning, C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7, 1801–1810. doi: 10.1105/tpc.7.11.1801
Dorne, A. J., Joyard, J., and Douce, R. (1990). Do thylakoids really contain phosphatidylcholine? Proc. Natl. Acad. Sci. U.S.A. 87, 71–74. doi: 10.1073/pnas.87.1.71
Dubertret, G., Mirshahi, A., Mirshahi, M., Gerard-Hirne, C., and Tremolieres, A. (1994). Evidence from in vivo manipulations of lipid composition in mutants that the delta 3-trans-hexadecenoic acid-containing phosphatidylglycerol is involved in the biogenesis of the light-harvesting chlorophyll a/b-protein complex of Chlamydomonas reinhardtii. Eur. J. Biochem. 226, 473–482. doi: 10.1111/j.1432-1033.1994.tb20072.x
Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Protein Sci. 8, 978–984. doi: 10.1110/ps.8.5.978
Gombos, Z., Varkonyi, Z., Hagio, M., Iwaki, M., Kovacs, L., Masamoto, K., et al. (2002). Phosphatidylglycerol requirement for the function of electron acceptor plastoquinone Q(B) in the photosystem II reaction center. Biochemistry 41, 3796–3802. doi: 10.1021/bi011884h
Gombos, Z., Wada, H., and Murata, N. (1991). Direct evaluation of effects of fatty-acid unsaturation on the thermal-properties of photosynthetic activities, as studied by mutation and transformation of Synechocystis PCC6803. Plant Cell Physiol. 32, 205–211.
Gorman, D. S., and Levine, R. P. (1965). Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardii. Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669. doi: 10.1073/pnas.54.6.1665
Guskov, A., Kern, J., Gabdulkhakov, A., Broser, M., Zouni, A., and Saenger, W. (2009). Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16, 334–342. doi: 10.1038/nsmb.1559
Hagio, M., Gombos, Z., Varkonyi, Z., Masamoto, K., Sato, N., Tsuzuki, M., et al. (2000). Direct evidence for requirement of phosphatidylglycerol in photosystem II of photosynthesis. Plant Physiol. 124, 795–804. doi: 10.1104/pp.124.2.795
Hagio, M., Sakurai, I., Sato, S., Kato, T., Tabata, S., and Wada, H. (2002). Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana. Plant Cell Physiol. 43, 1456–1464. doi: 10.1093/pcp/pcf185
Hasan, S. S., Stofleth, J. T., Yamashita, E., and Cramer, W. A. (2013). Lipid-induced conformational changes within the cytochrome b6f complex of oxygenic photosynthesis. Biochemistry 52, 2649–2654. doi: 10.1021/bi301638h
Hipkin, C. R., Al-Bassam, B. A., and Syrett, P. J. (1980). The roles of nitrate and ammonium in the regulation of the development of nitrate reductase in Chlamydomonas reinhardii. Planta 150, 13–18. doi: 10.1007/BF00385608
Iwai, M., Ikeda, K., Shimojima, M., and Ohta, H. (2014). Enhancement of extraplastidic oil synthesis in Chlamydomonas reinhardtii using a type-2 diacylglycerol acyltransferase with a phosphorus starvation-inducible promoter. Plant Biotechnol. J. 12, 808–819. doi: 10.1111/pbi.12210
Kelly, A. A., and Dormann, P. (2002). DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J. Biol. Chem. 277, 1166–1173. doi: 10.1074/jbc.M110066200
Kelly, A. A., Froehlich, J. E., and Dormann, P. (2003). Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15, 2694–2706. doi: 10.1105/tpc016675
Kobayashi, K., Fujii, S., Sato, M., Toyooka, K., and Wada, H. (2015). Specific role of phosphatidylglycerol and functional overlaps with other thylakoid lipids in Arabidopsis chloroplast biogenesis. Plant Cell Rep. 34, 631–642. doi: 10.1007/s00299-014-1719-z
Kobayashi, K., Kondo, M., Fukuda, H., Nishimura, M., and Ohta, H. (2007). Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 17216–17221. doi: 10.1073/pnas.0704680104
Li, X., Moellering, E. R., Liu, B., Johnny, C., Fedewa, M., Sears, B. B., et al. (2012). A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24, 4670–4686. doi: 10.1105/tpc.112.105106
Li-Beisson, Y., Beisson, F., and Riekhof, W. (2015). Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 82, 504–522. doi: 10.1111/tpj.12787
Mendiola-Morgenthaler, L., Eichenberger, W., and Boschetti, A. (1985). Isolation of chloroplast envelopes from Chlamydomonas - lipid and polypeptide composition. Plant Sci. 41, 97–104. doi: 10.1016/0168-9452(85)90109-8
Molnar, A., Bassett, A., Thuenemann, E., Schwach, F., Karkare, S., Ossowski, S., et al. (2009). Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58, 165–174. doi: 10.1111/j.1365-313X.2008.03767.x
Nakamura, Y. (2013). Phosphate starvation and membrane lipid remodeling in seed plants. Prog. Lipid Res. 52, 43–50. doi: 10.1016/j.plipres.2012.07.002
Nakamura, Y., Arimitsu, H., Yamaryo, Y., Awai, K., Masuda, T., Shimada, H., et al. (2003). Digalactosyldiacylglycerol is a major glycolipid in floral organs of Petunia hybrida. Lipids 38, 1107–1112. doi: 10.1007/s11745-006-1166-x
Omata, T., and Murata, N. (1983). Isolation and characterization of the cytoplasmic membranes from the Blue-Green Alga (Cyanobacterium) Anacystis nidulans. Plant Cell Physiol. 24, 1101–1112.
Ostrander, D. B., Zhang, M., Mileykovskaya, E., Rho, M., and Dowhan, W. (2001). Lack of mitochondrial anionic phospholipids causes an inhibition of translation of protein components of the electron transport chain. A yeast genetic model system for the study of anionic phospholipid function in mitochondria. J. Biol. Chem. 276, 25262–25272. doi: 10.1074/jbc.M103689200
Porra, R. J., Thompson, W. A., and Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Quisel, J. D., Wykoff, D. D., and Grossman, A. R. (1996). Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 111, 839–848. doi: 10.1104/pp.111.3.839
Rakhimberdieva, M. G., Vavilin, D. V., Vermaas, W. F., Elanskaya, I. V., and Karapetyan, N. V. (2007). Phycobilin/chlorophyll excitation equilibration upon carotenoid-induced non-photochemical fluorescence quenching in phycobilisomes of the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1767, 757–765. doi: 10.1016/j.bbabio.2006.12.007
Riekhof, W. R., Ruckle, M. E., Lydic, T. A., Sears, B. B., and Benning, C. (2003). The sulfolipids 2’-O-acyl-sulfoquinovosyldiacylglycerol and sulfoquinovosyldiacylglycerol are absent from a Chlamydomonas reinhardtii mutant deleted in SQD1. Plant Physiol. 133, 864–874. doi: 10.1104/pp.103.029249
Sakurai, I., Hagio, M., Gombos, Z., Tyystjarvi, T., Paakkarinen, V., Aro, E. M., et al. (2003). Requirement of phosphatidylglycerol for maintenance of photosynthetic machinery. Plant Physiol. 133, 1376–1384. doi: 10.1104/pp.103.026955
Sakurai, I., Mizusawa, N., Ohashi, S., Kobayashi, M., and Wada, H. (2007a). Effects of the lack of phosphatidylglycerol on the donor side of photosystem II. Plant Physiol. 144, 1336–1346. doi: 10.1104/pp.107.098731
Sakurai, I., Mizusawa, N., Wada, H., and Sato, N. (2007b). Digalactosyldiacylglycerol is required for stabilization of the oxygen-evolving complex in photosystem II. Plant Physiol. 145, 1361–1370. doi: 10.1104/pp.107.106781
Sakurai, I., Shen, J. R., Leng, J., Ohashi, S., Kobayashi, M., and Wada, H. (2006). Lipids in oxygen-evolving photosystem II complexes of cyanobacteria and higher plants. J. Biochem. 140, 201–209. doi: 10.1093/jb/mvj141
Sakurai, K., Moriyama, T., and Sato, N. (2014). Detailed identification of fatty acid isomers sheds light on the probable precursors of triacylglycerol accumulation in photoautotrophically grown Chlamydomonas reinhardtii. Eukaryot. Cell 13, 256–266. doi: 10.1128/EC.00280-13
Sato, N., Hagio, M., Wada, H., and Tsuzuki, M. (2000). Requirement of phosphatidylglycerol for photosynthetic function in thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 97, 10655–10660. doi: 10.1073/pnas.97.19.10655
Sato, N., and Murata, N. (1982). Lipid biosynthesis in the blue-green alga, Anabaena variabilis I. Lipid classes. Biochim. Biophys. Acta 710, 271–278.
Sato, N., Tsuzuki, M., Matsuda, Y., Ehara, T., Osafune, T., and Kawaguchi, A. (1995). Isolation and characterization of mutants affected in lipid metabolism of Chlamydomonas reinhardtii. Eur. J. Biochem. 230, 987–993. doi: 10.1111/j.1432-1033.1995.0987g.x
Satoh, S., Ikeuchi, M., Mimuro, M., and Tanaka, A. (2001). Chlorophyll b expressed in Cyanobacteria functions as a light-harvesting antenna in photosystem I through flexibility of the proteins. J. Biol. Chem. 276, 4293–4297. doi: 10.1074/jbc.M008238200
Schloss, J. A. (1990). A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol. Gen. Genet. 221, 443–452. doi: 10.1007/BF00259410
Shimojima, M., Ohta, H., Iwamatsu, A., Masuda, T., Shioi, Y., and Takamiya, K. (1997). Cloning of the gene for monogalactosyldiacylglycerol synthase and its evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 94, 333–337. doi: 10.1073/pnas.94.1.333
Siaut, M., Cuine, S., Cagnon, C., Fessler, B., Nguyen, M., Carrier, P., et al. (2011). Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11:7. doi: 10.1186/1472-6750-11-7
Tanoue, R., Kobayashi, M., Katayama, K., Nagata, N., and Wada, H. (2014). Phosphatidylglycerol biosynthesis is required for the development of embryos and normal membrane structures of chloroplasts and mitochondria in Arabidopsis. FEBS Lett. 588, 1680–1685. doi: 10.1016/j.febslet.2014.03.010
Tardif, M., Atteia, A., Specht, M., Cogne, G., Rolland, N., Brugiere, S., et al. (2012). PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 29, 3625–3639. doi: 10.1093/molbev/mss178
Umena, Y., Kawakami, K., Shen, J. R., and Kamiya, N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A. Nature 473, 55–60. doi: 10.1038/nature09913
Vass, I., Kirilovsky, D., and Etienne, A. L. (1999). UV-B radiation-induced donor- and acceptor-side modifications of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry 38, 12786–12794.
Wang, Z. T., Ullrich, N., Joo, S., Waffenschmidt, S., and Goodenough, U. (2009). Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8, 1856–1868. doi: 10.1128/EC.00272-09
Wu, F., Yang, Z., and Kuang, T. (2006). Impaired photosynthesis in phosphatidylglycerol-deficient mutant of cyanobacterium Anabaena sp. PCC7120 with a disrupted gene encoding a putative phosphatidylglycerophosphatase. Plant Physiol. 141, 1274–1283. doi: 10.1104/pp.106.083451
Wykoff, D. D., Davies, J. P., Melis, A., and Grossman, A. R. (1998). The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117, 129–139. doi: 10.1104/pp.117.1.129
Wykoff, D. D., Grossman, A. R., Weeks, D. P., Usuda, H., and Shimogawara, K. (1999). Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 96, 15336–15341. doi: 10.1073/pnas.96.26.15336
Xu, C., Hartel, H., Wada, H., Hagio, M., Yu, B., Eakin, C., et al. (2002). The pgp1 mutant locus of Arabidopsis encodes a phosphatidylglycerolphosphate synthase with impaired activity. Plant Physiol. 129, 594–604. doi: 10.1104/pp.002725
Yu, B., and Benning, C. (2003). Anionic lipids are required for chloroplast structure and function in Arabidopsis. Plant J. 36, 762–770. doi: 10.1046/j.1365-313X.2003.01918.x
Yuzawa, Y., Shimojima, M., Sato, R., Mizusawa, N., Ikeda, K., Suzuki, M., et al. (2014). Cyanobacterial monogalactosyldiacylglycerol-synthesis pathway is involved in normal unsaturation of galactolipids and low-temperature adaptation of Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1841, 475–483. doi: 10.1016/j.bbalip.2013.12.007
Keywords: Chlamydomonas reinhardtii, chloroplast, glycerolipid, phosphatidylglycerol, photosynthesis
Citation: Hung C-H, Endo K, Kobayashi K, Nakamura Y and Wada H (2015) Characterization of Chlamydomonas reinhardtii phosphatidylglycerophosphate synthase in Synechocystis sp. PCC 6803. Front. Microbiol. 6:842. doi: 10.3389/fmicb.2015.00842
Received: 02 June 2015; Accepted: 31 July 2015;
Published: 24 August 2015.
Edited by:
Weiwen Zhang, Tianjin University, ChinaReviewed by:
Jiangxin Wang, Arizona State University, USAJunbiao Dai, Tsinghua University, China
Norihiro Sato, Tokyo University of Pharmacy and Life Sciences, Japan
Copyright © 2015 Hung, Endo, Kobayashi, Nakamura and Wada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuki Nakamura, Institute of Plant and Microbial Biology, Academia Sinica, 128 Section 2, Academia Road, Nankang, Taipei 11529, Taiwan, nakamura@gate.sinica.edu.tw
 Chun-Hsien Hung
Chun-Hsien Hung Kaichiro Endo
Kaichiro Endo Koichi Kobayashi
Koichi Kobayashi Yuki Nakamura
Yuki Nakamura Hajime Wada
Hajime Wada